Abstract
Background and Purpose:
This study explored the association between a single-nucleotide polymorphism of prostate stem cell antigen and prostate cancer in Chinese patients undergoing prostate biopsy.
Materials and Methods:
DNA from 416 patients undergoing prostate biopsy was typed for the prostate stem cell antigen rs1045531 single-nucleotide polymorphism. The frequency of the rs1045531 polymorphism in patients with prostate cancer and in patients with benign prostatic hyperplasia was compared. Associations between the polymorphism and the risk of prostate cancer, prostate special antigen, Gleason score, and clinical stage were analyzed.
Results:
Statistically significant differences in the distribution of the rs1045531 genotypes and alleles were found between prostate cancer and benign prostatic hyperplasia in patients undergoing prostate biopsy (P = .035 and .046, respectively). We found that the rs1045531 AC genotype was significantly associated with a high risk of prostate cancer in the heterozygote model (AC vs CC; odds ratio = 2.383, 95% confidence interval: 1.198-4.741, χ2 = 6.229, P = .013) and the dominant model (AA/AC vs CC; odds ratio = 2.169, 95% confidence interval: 1.112-4.229, χ2 = 5.228, P = .022). However, susceptibility of prostate cancer was decreased in the homozygote model (AA vs CC; odds ratio = 0.828, 95% confidence interval: 0.143-4.805, P = .601). When considering clinical factors, the rs1045531 showed an association with prostate special antigen of 10 ng/mL or greater, a Gleason score of 7 or greater, and a size of T2 or greater.
Conclusion:
Men with the rs1045531 AC genotype of prostate stem cell antigen were at higher risk of prostate cancer in Chinese patients undergoing prostate biopsy.
Keywords: single-nucleotide polymorphism, prostate stem cell antigen, prostate cancer, prostate biopsy, Chinese population
Introduction
Prostate cancer (PCa) is the most commonly diagnosed cancer and the second leading cause of cancer-related death in men in Western countries. According to the American Cancer Society, approximately 161 360 new cases of PCa will be diagnosed and 26 730 men will die from this disease during 2017.1 While the incidence of PCa is relatively low in China, it is increasing rapidly, especially in developed metropolitan areas.2 The etiology of PCa is largely unknown and is likely to be multifactorial. Genetic susceptibility is a major risk factor for PCa and is estimated to account for 42% variation of the disease.3
To date, genome-wide association studies (GWAS) have identified more than 50 potential PCa susceptibility genomic variants in European, African American, Japanese, and Chinese populations.4–7 Furthermore, with the development of gene sequencing technology, more PCa-related single-nucleotide polymorphisms (SNPs) will be discovered.8 The prostate stem cell antigen (PSCA) gene is located on chromosome 8q24.2, consists of 3 exons and 2 introns, and encodes a 123-amino acid glycoprotein, which belongs to the LY-6/Thy-1 family of cell surface antigens.9 Prostate stem cell antigen SNPs have been used in the study of bladder cancer, cervical cancer, gastric cancer, and duodenal ulcer disease.10–12 However, there have been few studies on the relationships between PSCA SNPs and PCa.
Joung et al found that men in South Korea with the PSCA rs1045531 AA genotype were at higher risk of PCa.13 However, a relationship between PSCA SNPs and PCa risk in China, especially in patients undergoing prostate biopsy, has not been reported. Therefore, the present study aimed to comprehensively examine the potential association of the PSCA rs1045531 SNP with the risk of PCa in a Chinese population undergoing prostate biopsy.
Materials and Methods
This study included 416 consecutive patients undergoing prostate biopsy for the detection of PCa based on digital rectal examination (DRE), prostate special antigen (PSA), and prostate nodules14 at a tertiary hospital in Suzhou, China, between January 2015 and September 2016. The participants in this study were representative of prostate biopsy patients in metropolitan areas of East China. Transrectal ultrasound-guided biopsy using an 18-gauge, 20-cm biopsy needle (Bard Peripheral Vascular Inc, Tempe, Arizona) was performed. All biopsy specimens were reviewed by a pathologist. Prior to performing a warranted prostate biopsy, a complete history was collected and a physical examination was performed on each patient, including PSA levels, magnetic resonance imaging, and emission computed tomography. Clinical tumor size (T) stages were determined at initial diagnosis via DRE, prostate biopsy, and imaging. Enlarged lymph nodes (N) and distant metastases (M) were detected via prostate magnetic resonance imaging with or without emission computed tomography, and the scans were interpreted by a uroradiologist.15 Biopsied tumors were graded according to the Gleason scoring system, and staging was performed according to the 2002 TNM classification . Written informed consent was obtained from each patient. All procedures performed in studies involving human participants were in accordance with the ethical standards of the First Affiliated Hospital of Suzhou University and with the 1964 Helsinki Declaration and its later amendments or comparable ethical standards.
Peripheral blood samples were collected in a standard tube and stored at −80°C. Using a standard protocol, genomic DNA was extracted from peripheral blood samples by QIAamp DNA Mini kit (Qiagen Co Ltd, Shanghai, China). One pair of primers was designed using Primer Premier 5.0 software to amplify fragments of the rs1045531. Primers sequences were 5′CAGCTTGAACTGCGTGGAT-3′and 5′TCAGACTTGCGTTAGGATGTG-3′. The PCR (Polymerase Chain Reaction) amplified products were sequenced by an ABI3730 sequencer (BioAsia Biotechnology Co Ltd, Shanghai, China; Figure 1), and the related data were managed using Sequenom Typer 4.0 software.16
Figure 1.
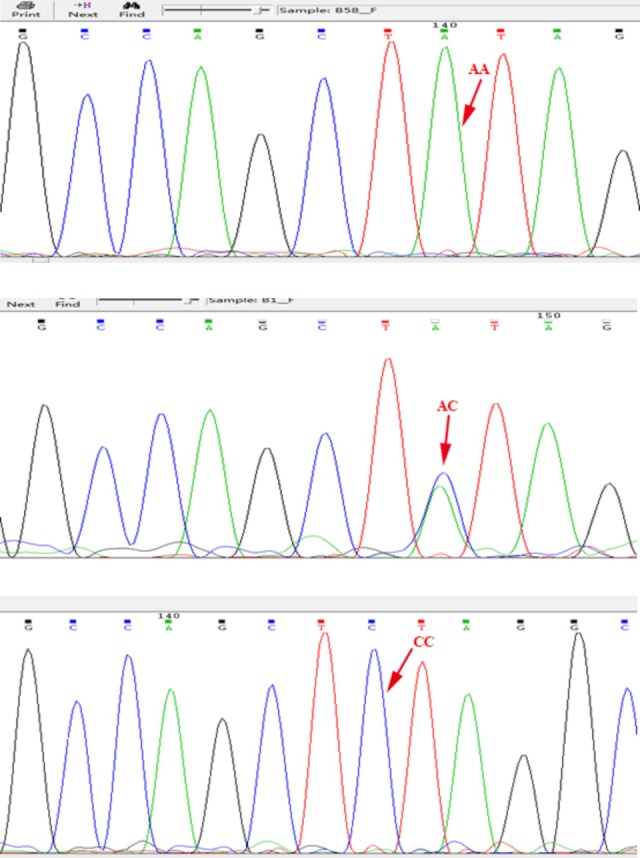
Prostate stem cell antigen (PSCA) rs1045531 genotype.
Microsoft Excel and SPSS software (version 21.0; SPSS Inc, Chicago, Illinois) were used for all analyses. Allele and genotype frequencies were calculated by counting. The χ2 test was utilized to evaluate the significant departure from the Hardy-Weinberg equilibrium. The exact test was used to examine the distribution of SNPs among groups, as well as their accordance with the Hardy-Weinberg equilibrium. Odds ratio (OR) and 95% confidence interval (CI) were adjusted for age, body mass index (BMI), and PSA and estimated by multiple logistic regression analysis to investigate the association between the rs1045531 polymorphism and susceptibility of PCa. All tests were 2 tailed; a P value of <.05 was considered statistically significant.
Results
Demographic and Clinical Characteristics of the Study Participants
The demographic and clinical characteristics of the patients with PCa and benign prostatic hyperplasia (BPH) are shown in Table 1. A total of 416 patients were recruited for this study, including 193 patients with PCa and 223 patients with BPH. There were no differences in the distribution of age and BMI between patients with PCa and BPH (P = .667 and .659, respectively). Prostate special antigen levels were higher in patients with PCa than in patients with BPH (P = .02).
Table 1.
The Characteristics of Biopsy Participants.
| Characteristics | PCa | BPH | P Value | |
|---|---|---|---|---|
| Number | 193 | 223 | ||
| Age, years | 72.00 ± 6.95 | 64.00 ± 7.96 | T = −0.43 | .667 |
| BMI, kg/m2 | 23.30 ± 3.38 | 23.53 ± 2.90 | T = −0.44 | .659 |
| PSA, ng/mL | 19.72 (8.25-64.47) | 13.98 (9.20-24.72) | U = −2.33 | .020 |
| Gleason score | ||||
| <7 | 67 | |||
| ≥7 | 126 |
Abbreviations: BMI, body mass index; BPH, benign prostatic hyperplasia; PCa, prostate cancer; PSA, prostate special antigen; T, t value from Student's t test; U, U value from Mann-Whitney U test.
Association Between PSCA rs1045531 SNP and the Risk of PCa
Detailed allele frequencies and genotype distributions of the rs1045531 are shown in Table 2. The allele frequencies and genotype distributions were in accordance with the Hardy-Weinberg equilibrium (P > .05). There were statistically significant differences in the distribution of the rs1045531 genotypes and alleles between patients with PCa and BPH (P = .035 and .046, respectively). When the CC genotype was used as the reference, a significantly increased susceptibility to PCa was found in the heterozygote comparison (AC vs CC; OR = 2.383, 95% CI: 1.198-4.741, χ2 = 6.229, P = .013) and dominant model (AA/AC vs CC; OR = 2.169, 95% CI: 1.112-4.229, χ2 = 5.228, P = .022). No significant associations were found in the homozygote model, recessive model, or allele model (P = .601, .686, or .101, respectively; Table 3).
Table 2.
Genotypes and Alleles Frequencies of PSCA Polymorphisms in Biopsy Participants.
| Genotype Frequencies | Allele Frequencies | ||||
|---|---|---|---|---|---|
| AA | AC | CC | A | C | |
| PCa (n = 193) | 6 (3.0%) | 104 (53.7%) | 83 (43.3%) | 116 (30.1%) | 270 (69.9%) |
| BPH (n = 223) | 12 (5.4%) | 72 (32.3%) | 139 (62.3%) | 96 (21.5%) | 350 (78.5%) |
| Total (n = 416) | 18 (4.3%) | 176 (42.3%) | 222 (53.4%) | 212 (25.5%) | 620 (74.5%) |
| χ2 = 2.13, P = .035 | χ2 = 2.69, P = .046 | ||||
Abbreviations: BPH, benign prostatic hyperplasia; PCa, prostate cancer; PSCA, prostate stem cell antigen.
Table 3.
Association Between the PSCA Polymorphisms and Susceptibility of PCa.
| Comparisons | OR (95% CI) | χ2 Value | P Value |
|---|---|---|---|
| Homozygote model (AA vs CC) | 0.828 (0.143-4.805) | – | .601a |
| Heterozygote model (AC vs CC) | 2.383 (1.198-4.741)) | 6.229 | .013b |
| Dominant model (AA/AC vs CC) | 2.169 (1.112-4.229) | 5.228 | .022b |
| Recessive model (AA vs AC/CC) | 0.562 (0.100-3.167) | – | .686a |
| Allele model (A vs C) | 1.560 (0.955-2.661) | 2.686 | .101b |
Abbreviations: CI, confidence interval; OR, odds ratio; PCa, prostate cancer; PSCA, prostate stem cell antigen.
aFisher exact test.
bχ2 test.
Associations Between the PSCA rs1045531 SNP and Clinical and Pathological Features of PCa
We evaluated the association of the rs1045531 SNP with various clinical and pathological features including PSA, Gleason score, T, N, and M, as shown in Tables 4 and 5. Stratification analyses for PSA, Gleason score, and T showed statistically significant differences in the distribution of the rs1045531 genotypes and alleles (P = .026, .031; 0, .002 and .03, .008). However, stratification analyses for N and M revealed no significant associations with the rs1045531 genotypes and alleles (P = .813, .792 and .835, 0.631, respectively).
Table 4.
Associations Between the PSCA Polymorphisms and Parameters of Patients With PCa.
| Genotype Frequencies | Allele Frequencies | ||||
|---|---|---|---|---|---|
| AA | AC | CC | A | C | |
| Gleason score | |||||
| <7 | 4 (66.7%) | 22 (21.2%) | 41 (49.4%) | 30 (25.9%) | 104 (38.5%) |
| ≥7 | 2 (33.3%) | 82 (78.8%) | 42 (50.6%) | 86 (74.1%) | 166 (61.5%) |
| P = 0a | χ2 = 5.74, P = .002b | ||||
| PSA | |||||
| <10 | 0 (0%) | 17 (16.3%) | 20 (24.1%) | 17 (14.7%) | 57 (21.1%) |
| 10-20 | 0 (0%) | 40 (38.5%) | 52 (62.7%) | 40 (34.5%) | 144 (53.3%) |
| >20 | 6 (100%) | 47 (45.2%) | 11 (13.2%) | 59 (50.8%) | 69 (25.6%) |
| P = .026a | χ2 = 6.918, P = .031b | ||||
Abbreviations: PCa, prostate cancer; PSA, prostate special antigen; PSCA, prostate stem cell antigen.
a Fisher exact test.
b χ2 test.
Table 5.
Associations Between the PSCA Polymorphisms and Clinical Characteristics of Patients With PCa.
| Genotype Frequencies | Allele Frequencies | ||||
|---|---|---|---|---|---|
| AA | AC | CC | A | C | |
| T stage | |||||
| T1 | 0 (0%) | 9 (8.7%) | 23 (27.7%) | 9 (7.8%) | 55 (20.4%) |
| T2 | 0 (0%) | 49 (47.1%) | 46 (55.4%) | 49 (42.2%) | 141 (52.2%) |
| T3 | 6 (100%) | 29 (27.9%) | 11 (13.3%) | 41 (35.3%) | 51 (18.9%) |
| T4 | 0 (0%) | 17 (16.3%) | 3 (3.6%) | 17 (14.7%) | 23 (8.5%) |
| P = .030a | P = .008b | ||||
| N stage | |||||
| N0 | 6 (100%) | 90 (86.5%) | 69 (83.1%) | 102 (87.9%) | 228 (84.4%) |
| N1 | 0 (0%) | 14 (13.5%) | 14 (16.9%) | 14 (12.1%) | 42 (15.6%) |
| P = .813a | P = .792b | ||||
| M stage | |||||
| M0 | 6 (100%) | 87 (83.7%) | 66 (79.5%) | 99 (85.3%) | 219 (81.1%) |
| M1 | 0 (0%) | 17 (16.3%) | 17 (20.5%) | 17 (14.7%) | 51 (18.9%) |
| P = .835a | P = .631b | ||||
Abbreviations: M, distant metastasis; N, lymph node metastasis; PCa, prostate cancer; PSCA, prostate stem cell antigen; T, tumor size.
aFisher exact test.
bχ2 test.
Discussion
Genetic studies have provided insight into various diseases, including cancers. Understanding the relationships between different genes and cancers may help with prevention, as well as the improvement in treatments and prognosis of certain diseases. Recently, numerous studies have revealed associations between SNP of PSCA and many malignant tumors, such as breast cancer and gastric cancer.17,18
Accumulating epidemiological and genetic evidence suggests that genetic variation plays an important role in PCa etiology. A GWAS showed that genetic polymorphisms of several genes at 17q12 and 17q24.3 are significantly associated with PCa. Another GWAS indicated significant relationship between genetic polymorphism at 8q24 and PCa risk in the Asian population.19–21 Prostate stem cell antigen is highly expressed in PCa tissue and is significantly associated with the grade and prognosis of PCa.22 The PSCA gene, located on chromosome 8q24.2, encodes a 123-amino acid protein. Currently, an association between SNP of PSCA and PCa in Chinese patients undergoing prostate biopsy has not been reported. We first found statistically significant differences in the distribution of the rs1045531 genotypes and alleles between Chinese patients with PCa and BPH undergoing prostate biopsy. Compared to the rs1045531 CC genotype, the susceptibility to PCa was significantly increased in the heterozygote model (AC vs CC; OR = 2.383) and dominant model (AA/AC vs CC; OR = 2.169). However, the susceptibility to PCa was decreased in the homozygote model (AA vs CC; OR = 0.828). A study based on Korean patients with PCa reported that the ORs of the rs1045531 AC, AA, and AC/AA genotypes were 1.31, 1.99, and 1.52, respectively.13 Given the heterogeneous nature of PCa, the discrepancies between our findings and those of Joung et al 13 may be explained by various factors, including region, lifestyle, ethnicity, and study design.
Important parameters of PCa, PSA, and Gleason score play an important role in choosing treatment and evaluating prognosis. We found statistically significant differences in the distribution of the rs1045531 genotypes in different PSA and Gleason score groups. Furthermore, the trend of increased rs1045531 AC genotype with increasing PSA and Gleason score was evident. This trend is consistent with the findings of Wang et al,23 who reported a significant association between the SNP of 8q24 and PCa risk in their study. However, Joung et al 13 found that the PSCA rs1045531 SNP was only associated with PSA, not with Gleason score. To further validate the association of this SNP with clinical stages of PCa, we demonstrated an association between the rs1045531 genotypes and alleles and T by comparing differences of this SNP in TNM groups. However, the rs1045531 genotypes and alleles were not associated with N and M.
Despite our novel finding of PCa-associated polymorphisms of the PSCA gene, our study has several limitations. First, the single-center design may preclude the extrapolation of our findings to other patient populations or ethnic groups. Second, patients undergoing prostate biopsy are not representative of all patients, and polymorphisms of PSCA were compared only between patients with PCa and BPH undergoing prostate biopsy, which may result in selection bias. Third, the sample size of this study is relatively small, which may affect the estimate of its association and the statistical power for subset analyses. Therefore, a multiple-center, well-designed prospective study with a larger sample size is needed to better validate our findings.
Conclusion
To our knowledge, this is the first study to show that men with the rs1045531 AC genotype of PSCA are at higher risk of PCa in Chinese patients undergoing prostate biopsy. We found that the rs1045531 polymorphism was significantly associated with PSA, Gleason score, and T. Therefore, this SNP may be used for evaluating the risk and prognosis of PCa in patients undergoing prostate biopsy.
Abbreviations
- BMI
body mass index
- BPH
benign prostatic hyperplasia
- CI
confidence interval
- DRE
digital rectal examination
- GWAS
genome-wide association studies
- M
distant metastasis
- N
lymph node metastasis
- OR
odds ratio
- PCa
prostate cancer
- PSCA
prostate stem cell antigen
- PSA
prostate special antigen
- SNP
single-nucleotide polymorphisms
- T
tumor size
Footnotes
Authors’ Note: Xuefeng Zhang, Qin Hu, and Ye Chen have contributed equally to this work.
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) received no financial support for the research, authorship, and/or publication of this article.
References
- 1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA Cancer J Clin. 2017;67(1):7–30. [DOI] [PubMed] [Google Scholar]
- 2. Na R, Liu F, Zhang P, et al. Evaluation of reported prostate cancer risk-associated SNPs from genome-wide association studies of various racial populations in Chinese men. Prostate. 2013;73(15):1623–1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Amundadottir LT, Sulem P, Gudmundsson J, et al. A common variant associated with prostate cancer in European and African populations. Nat Genet. 2006;38(6):652–658. [DOI] [PubMed] [Google Scholar]
- 4. Kote-Jarai Z, Olama AA, Giles GG, et al. Seven prostate cancer susceptibility loci identified by a multi-stage genome-wide association study. Nat Genet. 2011;43(8):785–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Haiman CA, Chen GK, Blot WJ, et al. Genome-wide association study of prostate cancer in men of African ancestry identifies a susceptibility locus at 17q21. Nat Genet. 2011;43(6):570–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Takata R, Akamatsu S, Kubo M, et al. Genome-wide association study identified five new susceptibility loci for prostate cancer in the Japanese population. Nat Genet. 2010;42(9):751–754. [DOI] [PubMed] [Google Scholar]
- 7. Xu J, Mo Z, Ye D, et al. Genome-wide association study in Chinese men identifies two new prostate cancer risk loci at 9q31.2 and 19q13.4. Nat Genet. 2012;44(11):1231–1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ishak MB, Giri VN. A systematic review of replication studies of prostate cancer susceptibility genetic variants in high-risk men originally identified from genome-wide association studies. Cancer Epidemiol Biomarkers Prev. 2011;20(8):1599–1610. [DOI] [PubMed] [Google Scholar]
- 9. Reiter RE, Gu Z, Watabe T, et al. Prostate stem cell antigen: a cell surface marker overexpressed in prostate cancer. Proc Natl Acad Sci U S A. 1998;95:1735–1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. García-González MA, Bujanda L, Quintero E, et al. Association of PSCA rs2294008 gene variants with poor prognosis and increased susceptibility to gastric cancer and decreased risk of duodenal ulcer disease. Int J Cancer. 2015;137(6):1362–1373. [DOI] [PubMed] [Google Scholar]
- 11. Wang S, Wu S, Zhu H, et al. PSCA rs2294008 polymorphism contributes to the decreased risk for cervical cancer in a Chinese population. Sci Rep. 2016;6:23465–23472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zheng KW, Chen ZG, Tian Y, Hao G. Association between PSCA mRNA expression levels and rs2294008 polymorphism in transitional cell cancer of bladder. Oncol Lett. 2015;9(2):557–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Joung JY, Lee YS, Park S, et al. Haplotype analysis of prostate stem cell antigen and association with prostate cancer risk. J Urol. 2011;185(6):2112–2118. [DOI] [PubMed] [Google Scholar]
- 14. Jiang H, Liu F, Wang Z, et al. Prediction of prostate cancer from prostate biopsy in Chinese men using a genetic score derived from 24 prostate cancer risk-associated SNPs. Prostate. 2013;73(15):1651–1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kim SH, Park WS, Kim SH, et al. Prostate stem cell antigen expression in radical prostatectomy specimens predicts early biochemical recurrence in patients with high risk prostate cancer receiving neoadjuvant hormonal therapy. PLoS One. 2016;11(3):1646–1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Thomas RK, Baker AC, Debiasi RM, et al. High-throughput oncogene mutation profiling in human cancer. Nat Genet. 2007;39(3):347–351. [DOI] [PubMed] [Google Scholar]
- 17. Wang M, Wang X, Fu SW, et al. Single-nucleotide polymorphisms in PSCA and the risk of breast cancer in a Chinese population. Oncotarget. 2016;7(19):27665–27675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Qiu LX, Cheng L, He J, et al. PSCA polymorphisms and gastric cancer susceptibility in an eastern Chinese population. Oncotarget. 2016;7(8):9420–9428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Thomas G, Jacobs KB, Yeager M, et al. Multiple loci identified in a genome-wide association study of prostate cancer. Nat Genet. 2008;40(3):310–315. [DOI] [PubMed] [Google Scholar]
- 20. Zheng SL, Sun J, Wiklund F, et al. Cumulative association of five genetic variants with prostate cancer. N Engl J Med. 2008;358(9):910–919. [DOI] [PubMed] [Google Scholar]
- 21. Gudmundsson J, Sulem P, Manolescu A, et al. Genome-wide association study identifies a second prostate cancer susceptibility variant at 8q24. Nat Genet. 2007;39(5):631–637. [DOI] [PubMed] [Google Scholar]
- 22. Gu Z, Thomas G, Yamashiro J, et al. Prostate stem cell antigen (PSCA) expression increases with high Gleason score, advanced stage and bone metastasis in prostate cancer. Oncogene. 2000;19(10):1288–1296. [DOI] [PubMed] [Google Scholar]
- 23. Wang L, McDonnell SK, Slusser JP, et al. Two common chromosome 8q24 variants are associated with increased risk for prostate cancer. Cancer Res. 2007;67(7):2944–2950. [DOI] [PubMed] [Google Scholar]


