Abstract
The gut microbiota (GM) plays a critical role in human health and disease. Likewise, it is becoming increasingly evident that changes or disruptions to the GM can have significant effects on animal models and their expressed phenotypes, adding a complex and important variable into basic research and pre-clinical studies. In this article, we review some of the most common sources of GM variability in rodent models, and discuss measures to address this variability for improved reproducibility.
The past two decades have seen an explosion of research investigating the role of microbiota in health and disease. These complex microbial communities that include bacteria, viruses, archaea, protozoa, and fungi are found in virtually any niche of the human body. The intestinal tract has been the most studied as the vast majority of bacterial microbes reside there1. It is now well established that the gut microbiota (GM) plays a critical role in health with mechanisms ranging from aiding in digestion to promoting immune system development and colonization resistance against bacterial pathogens and pathobionts. Moreover, the advent of next-generation sequencing has allowed for robust surveys of these populations and many recent studies have shown that patients with a number of diseases, both enteric and systemic, often have GM that differs from their healthy counterparts2, 3. This occurs even in situations where the hosts are genetically identical4. As with many diseases, animal models are critical to establish cause-and-effect relationships between disease and microbiota.
While studies with rodent models can be performed using genetically identical animals in highly controlled environments, reproducibility of results using such models has also emerged as a topic of concern. Several reports have highlighted the lack of reproducibility in model-based biomedical research5, 6. Sources of variation that may explain this lack of reproducibility abound and include subtle differences in methodology, reagents, or the animals themselves. Interestingly, recent studies have shown that the GM of contemporary rodent colonies can differ markedly depending on the source of the animal and other factors associated with rodent husbandry. Thus it is logical to speculate that differences in GM may play a role in this lack of reproducibility and data are rapidly emerging to support this premise.
Factors affecting the composition of the gut microbiota
Multiple husbandry-related factors are likely capable of altering the GM of research animals. For example, the common practice of transfer of mice from one facility to another at the same institution can result in short term shift in GM that can normalize after acclimation to the new facility7. Even differing water decontamination methods can alter GM8, 9. Other factors, such as therapeutic use of antibiotics or rederivation to render animals free of unwanted pathogens, should also be considered as potential GM-modulating procedures.
Intuitively, diet has great potential to modulate GM and in humans, both short- and long-term changes in diet have been shown to alter the GM10. Rodent studies designed to uncover mechanisms of diet-induced microbiota changes—for example, studies assessing the link between diet, microbiota and obesity11—have confirmed that dramatic changes in diet can alter the rodent GM. Moreover, in a study comparing purified diets, customized diets, and chow diets, changes in GM were noted and these were associated with local and systemic immune response alterations12. However, most rodent studies are performed on standard rodent chows. While little is known about the effects on GM of changing chow sources or types (for example, maintenance vs. breeder chow), caution should be taken when doing so. For example, it is well-documented that the GM of most mammals, including humans and rodents, is dominated by two phyla, the Firmicutes and Bacteroidetes13. In general, increases in the Firmicutes to Bacteroidetes ratio are associated with increased food intake and obesity14. Such shifts enhance the ability of the GM to harvest energy from ingesta14, resulting in a positive feedback loop. Thus, any factor capable of modulating the ratio of these two phyla (for example, accidental consumption of breeder chow), could initiate a gradual shift toward increased energy harvest and adiposity. One must also remember that most commercial diets are closed formula. While they are milled to achieve targeted levels of specific nutrients, the source of grains and other ingredients can change from year to year15. Thus, studies performed months or years apart from each other may include data generated from animals consuming diets milled from different dietary ingredients. It is conceivable that a change to an ingredient that promotes increased energy harvest could in turn result in an unanticipated change in GM.
In addition to the composition of the diet, autoclaving and irradiation are also variables which influence the microbial exposure to research animals. For example, the label of one commercially available irradiated rodent chow lists the amount of thermophilic, mesophilic, aerobic and anaerobic spores recovered from “control” chow, suggesting that non-irradiated feed may be a variable source of bacteria and fungi. Anecdotally, segmented filamentous bacteria (SFB), a bacterium that can have significant impact on several mouse models of disease (see below), can also be acquired from food contaminated with spores.
Several groups have demonstrated substantial differences between the GM of mice purchased from different commercial vendors or repositories16–18, and our own studies19 have begun to characterize the nature and extent of those differences. One of the most recognized differences is the presence of the aforementioned SFB in most commercial colonies and its absence in the largest supplier of genetically engineered mice, The Jackson Laboratory19.
Because the primary source of GM to rodent pups is the birth dam, the latter should also be considered a source of variation. In established colonies, GM remains stable over generations20. However, if new dams are introduced into a colony, they may possess a GM that differs from that of the existing population (see vendor differences above). While the dam is the primary source of GM, the presence of a sire harboring a different GM may also present a confounding experimental factor due to horizontal transfer. Even when present for a limited time, such as during a timed mating, coprophagy of the sire’s feces by the dam could conceivably lead to downstream changes in the GM of the resulting pups.
A differing GM may also be introduced into a colony through rederivation, which is typically performed when populating new research facilities, when transferring animals from conventional housing to barrier settings, when resuscitating animals from cryopreserved germplasm, or when attempting to eliminate unwanted pathogens. The procedure usually entails surgical transfer of embryos into a commercially available surrogate dam known to be free of selected pathogens. The GM of the surrogate dam then seeds the newly rederived animals with resulting high concordance of dam and pup GM21. Less labor-intensive methods of eliminating unwanted bacteria (e.g., Helicobacter spp.), such as cross-fostering, will intuitively result in similar GM changes, although the amount of time between parturition and transfer to a surrogate will likely affect the degree to which the pup GM mimics that of the surrogate or birth dam.
Rederivation is also complicated by the fact that, like humans22, 23, the mode of delivery likely influences the GM composition of pups. Human neonates delivered vaginally are seeded primarily with microbes found in the maternal vagina or feces while Caesarian delivery results in increased proportions of microbes that inhabit the external body surfaces. These differences can have profound effects on host physiology. For example, exposure to lipopolysaccharides during vaginal delivery induces epithelial tolerance in the offspring, resulting in differential innate immune responses between mice delivered via the two modes24.
Experimental or therapeutic procedures also have potential to alter microbiota. For example, administration of antibiotics has well-documented and intuitive effects on the GM25–31. When purposefully attempting to alter GM, multiple, broad spectrum, bactericidal antibiotics with complementary spectra are administered orally25. Such administration can result in dramatic acute shifts in GM composition. Moreover, when animals are allowed to recover, the GM may either return to a composition and richness similar to that present before treatment or have incomplete restitution. This is highly dependent on the target spectrum of the drug or drug combinations, as well as the richness and diversity of the GM before antibiotic administration. In extreme situations, antibiotics administered chronically to adult rodents can even result in a near germ-free (GF) status32, 33. Little is known about the impact of therapeutic antibiotic regimens used routinely in rodent colonies (for example, the use of trimethoprim/sulfamethoxazole for the control of Pneumocystis spp.-associated morbidity in immunodeficient rodents). Based on the above studies, one can speculate that the broad spectrum and chronicity of use can modulate not just the agent of interest but also other resident microbiota. Additionally, the effects of transient administration of tetracycline derivatives in models with inducible transgenes (for example, Tet-On systems) is unclear.
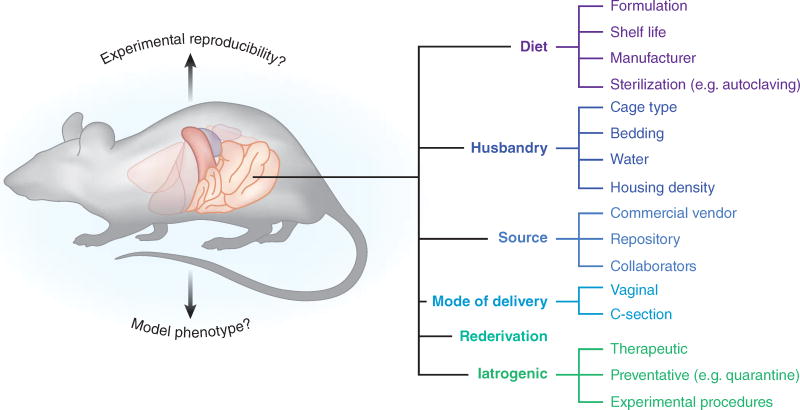
There are many other factors capable or suspected of modulating the microbiota of research animals including psychological stress34, 35, type of caging, type of bedding, and frequency of bedding change. In designing experiments that examine GM, changes in any of these factors should always be taken into consideration (see Fig. 1 for summary of factors discussed above).
Figure 1.
Illustration depicting several categories and sub-categories of factors that have been shown to significantly influence the rodent GM.
Impact of the gut microbiota on variability and reproducibility of disease models
It is now well accepted that changes in GM can have a dramatic impact on human health. Intuitively, the GM of research animals can similarly impact the phenotype of models of human disease. While this has been recognized anecdotally for many years, only recently have controlled studies been performed to support this hypothesis16, 18, 36–38. Frequently, a model will experience a loss or change in phenotype when an animal colony is relocated to a different institution, or undergoes a change in husbandry. Such scenarios raise the specter of a GM-mediated effect, especially when other potential contributory factors are optimally controlled. In addition to colony-wide changes in phenotype, it stands to reason that the inherent variability of the GM within a colony of mice may be reflected in the phenotypes of those models with GM-dependent mechanisms. For example, ApcMin mice, a model of colorectal cancer purportedly susceptible to GM-mediated effects on tumor load39, 40, develop a highly variable number of tumors. Could the difference between individual mice that develop 75 or 25 tumors be related to differences in the composition of the GM? This is an important consideration due to both the pathogenic mechanisms that may be revealed, but also due to the implications for animal welfare. If it is possible to reduce the variance of the GM within research animals via strict attention to, and control of, those variables capable of modulating the GM, it may be feasible to reduce the variance seen in the phenotype, potentially reducing the sample size needed to achieve statistical power.
The National Institutes of Health (NIH) has recently acknowledged issues of poor reproducibility in biomedical research, particularly in preclinical studies using animal models5, 6. Editorials41–43 have highlighted the costs associated with the inability to reproduce preclinical studies, and a wide range of contributing factors has been identified. Possible solutions to improve reproducibility include inclusion of both sexes in animal studies6, division of litters between treatment groups, and thorough documentation of all husbandry variables44. A less publicized solution suggests the need for reporting, and possibly standardizing, the GM of research animals45. Many consider it impractical to document the GM composition for all published animal-based research, but it should be noted that, in recent years, the cost of next-generation sequencing has been decreasing and its availability has increased dramatically. If these trends continue, such documentation could become financially viable in the near future. Lastly, producers and vendors of purpose-bred research animals are beginning to make efforts to account for these variables in their production processes and even document the GM composition of their production colonies.
Rodent helicobacters and reproducibility
As mentioned above, over the past three decades it has become increasingly apparent that several members of the GM can modulate model phenotypes, and their presence or absence can thus potentially impact reproducibility of these models. Perhaps the most studied are members of the genus Helicobacter that inhabit the large intestine of rodents. The first species identified was Helicobacter hepaticus in 1992; its impact on research reproducibility was immediately obvious. At that time, a previously unrecognized hepatitis and an increased incidence of hepatic neoplasia had been noted in untreated control mice on a long term carcinogenicity study at the National Cancer Institute Frederick Cancer Research and Development Center46, 47. Ultimately, these changes in disease phenotype were attributed to a H. hepaticus48, 49 and its presence invalidated this entire study as well as others50.
Concurrent with the recognition of H. hepaticus, several newly developed genetically engineered strains of mice found to have chronic inflammation of the intestinal tract were proposed as models of inflammatory bowel disease. Subsequently, many of these mouse strains were found to be colonized by H. hepaticus51. In most cases, inflammation of the intestinal tract was not evident in helicobacter-free mice51, 52. Moreover, experimental infection of IL-10 and TCR–αβ knockout mice with H. hepaticus caused more severe disease, further supporting the role of this bacterium in induction of intestinal inflammation and its potential to confound reproducibility of results from inflammatory bowel disease research52, 53. Many of these mutant mouse strains, most notably the IL-10 knockout mouse, are now used as models of inflammatory bowel disease and the presence of large intestinal helicobacters is critical to optimal development of their disease phenotype.
Similarly, the presence or absence of H. hepaticus can impact cancer studies using mice with mutations in transforming growth factor β1 (Tgfb1) and recombinase activating gene 2 (Rag2). These mice develop colonic inflammation and cancer; however, when rendered GF, these lesions do not develop54. To discern the role of H. hepaticus in inflammation and cancer, GF Tgfb1−/− Rag2−/− mice were reintroduced into either H. hepaticus-containing environments or H. hepaticus-free environments. Only when introduced into the H. hepaticus-containing environment did lesions reappear, suggesting that this bacterium was a key trigger of intestinal inflammation and subsequent cancer.
The study of Helicobacter spp.-induced models of inflammatory bowel disease has also demonstrated how the complexity of bacterial interactions impact reproducibility. To this end, IL-10 knockout mice raised in conventional settings and colonized by H. hepaticus readily develop intestinal inflammation52, 55. However, mono-association of germfree IL-10-deficient mice with H. hepaticus results in no intestinal inflammation56. This, as well as studies using mice harboring defined microbiota, has led to the belief that some bacteria, such as the rodent helicobacters, do not directly initiate inflammation, but rather serve as provocateurs for inflammation initiated against other microbial species57–60.
Because rodent helicobacters have been associated with such profound disease changes in certain strains of mice, many consider these bacteria to be pathobionts rather than commensal symbionts. However, it should be noted that in many strains, such as the common C57BL/6, Helicobacter spp. behave more like a symbiont with colonization resulting in no intestinal inflammation and only minimal changes in mucosal cytokine gene expression61, 62.
Segmented filamentous bacteria (SFB) and reproducibility
More recently, a bacterium historically considered part of the “normal” murine GM has been shown to have a dramatic impact on model reproducibility. In the late 2000s, Ivanov et al.18, 63 recognized differences in mucosal TH17 immune responses associated with mice from two different suppliers. Through an elegant series of experiments, they ultimately implicated SFB as a key modulator of this immune response. SFB had previously been considered a normal symbiont of the ileal microbiota and little attention was paid to its role in host physiology. Numerous subsequent studies have shown that this SFB-associated alteration of the mucosal TH17 and other immune effector mechanisms has dramatic effects on the phenotypes of models ranging from diabetes mellitus to multiple sclerosis to arthritis16, 63–65. Of note, many of these models represent diseases of non-gastrointestinal systems, which greatly broadens the impact of differing GM on model phenotype reproducibility. For a more detailed description of the physiological effects of SFB, readers are referred to a recent review on the topic66.
The literature is now rapidly expanding with examples of how other intestinal inhabitants previously considered to be part of the “normal” GM can alter host physiology and potentially impact model reproducibility. These now include protozoans such as Tritrichomonas spp67, 68. and a human isolate of Bifidobacterium adolescentis69. While single agents are being implicated, it is also highly likely that more complex communities of multiple agents will likely have equally important, if not more important, roles in model phenotypes70.
Microbiota considerations in optimizing reproducibility of animal models
Given the potential for differing individual microbes and complex microbiota to modulate phenotypes, what tools are available to perform studies to optimize reproducibility? The most controlled approaches use classical gnotobiotic technology to finely control the GM of mice on studies. Alternatively, researchers can carefully document and report the GM status of animals used in experiments and when lack of reproducibility arises, one can assess whether associated differences in GM exist. If differences are identified, subsequent studies can be performed to confirm the causal relationship of specific GM.
Classical gnotobiotics or gnotobiology (reviewed in71) is the study of animals that are free of all microorganisms or colonized only by known taxa. The term arises from the Greek gnotos, meaning “known” and bios, meaning “life”71. There are several categories of rodents that fall under the term gnotobiotic: 1) axenic (i.e., GF) rodents free of all microorganisms including bacteria, viruses (with the exception of endogenous retroviruses), fungi, protozoa and other parasites; 2) mono-associated animals generated by colonizing GF animals with a single agent; and 3) defined microbiota (flora) animals generated by reconstituting GF mice with “cocktails” or consortia of bacteria or other agents.
Germfree mice
In studies of microbiota, GF mice are ideal for asking whether or not microbiota, in general, plays any role in a model phenotype. If GF mice fail to develop the phenotype seen in their conventionally raised counterparts, microbiota can be implicated in the development of this phenotype. The use of these mice has been critical to our understanding of how intestinal microbes can influence health and disease, especially when coupled with mono-association, defined microbiota, or complex microbiota strategies4, 72–74. Data from GF mouse studies must also be interpreted with caution as several normal host physiologic parameters are altered in these mice. For example, GF mice have underdeveloped immune systems63, 75–79, slower intestinal epithelial turnover80, differences in epithelial gene expression81, 82, differing nutritional requirements, less body fat despite increased consumption83, and markedly enlarged ceca. The latter may lead to death from volvulus or may indirectly lower reproductive performance, presumably due to competition for space with the gravid uterus. GF mice are also markedly susceptible to infection by opportunistic agents, should a break in containment occur. Of note, the National Gnotobiotic Rodent Resource Center is an NIH-funded center that can aid investigators in studies using GF or gnotobiotic mice (http://www.med.unc.edu/ngrrc). Alternatively, many institutions have or are creating in-house gnotobiotic facilities. These are relatively easy to set up, but it is prudent to do a thorough cost analysis before doing so as there are added expenses of creating, maintaining, and monitoring GF mice as compared to conventional mice.
Mono-associated mice
GF mice are also important to microbiota research because they can be reconstituted with agents ranging from a single bacterium (mono-associated) to defined microbiota (for example, modified Schaedler’s flora) to complex microbiota to xenografted microbiota (for example, human microbiota). To create mono-associated mice, GF mice generated and maintained as described above are inoculated with a pure bacterial culture, usually by gastric gavage. Mono-associated mice allow for the study of responses to a single agent (most commonly bacterial) or identification of bacterial species responsible for specific bacterial products84. A notable example of the use of mono-associated mice in understanding host:microbe interactions comes from studies that identified the aforementioned segmented filamentous bacterium (SFB) as a key component of the intestinal microbiota that promotes the development of TH17 cells63, 76. Similarly, Bacteroides fragilis, through the production of polysaccharide A, is important in the induction of T regulatory cells85, 86. Other recent studies have taken this strategy beyond the study of resident bacteria. To this end, an enteric virus (murine norovirus) was shown to provide some beneficial functions similar to those of mutualistic bacteria87. While important data have been generated from studies using mono-associated mice, results should also be interpreted with caution because, like GF mice, physiologic and immunologic processes in these mice may be very different when compared to mice raised with a complex microbiota20. Moreover, as discussed above, in models of inflammatory bowel disease, the presence of Helicobacter spp. in isolation causes no disease but is critical to disease progression when other microbiota are present88. Collectively, these findings indicate that any beneficial or detrimental process ascribed to a microbe in mono-associated mice should be interpreted only after performing similar studies where the agent in question is in the presence of complex microbiota89.
Defined microbiota (flora) mice
To circumvent some of the physiologic disadvantages of GF and mono-associated mice while still maintaining a controlled microbiota, mice reconstituted with defined microbiota were established. Schaedler et al.90, 91 initiated these studies by defining key cultivable components of the intestinal microbiota and then experimentally inoculating GF mice with various “cocktails” of aerobes and anaerobes. The resulting “Schaedler defined flora” was refined and standardized by Orcutt et al.92 resulting in “altered Schaedler’s flora” (ASF) that is now commonly used in gnotobiotic research93–96. Mice reconstituted with ASF or similar defined microbiota offer several advantages for microbiota-based research. Like GF mice, they are very well-defined, but unlike GF mice, they develop a mucosal immune system, have cecal volumes approaching those of conventionally raised mice and have normal reproductive performance. Disadvantages of using defined microbiota mice center on the simplicity of this microbiota, which does not recapitulate the interactions that may occur in complex microbiota that typifies human and animal populations. Like GF mice, these mice must also be generated in isolators and monitored routinely for the presence of appropriate microbiota and the lack of contaminants. To reduce costs, defined microbiota mice can be generated in isolators, but then removed for study. This approach still requires strict biosecurity for study mice including housing in ventilated racks and appropriate barrier husbandry practices for cage changing, sanitation, animal handling and so forth. Advancements in defined microbiota continue, and recently criteria desirable in isobiotic mouse strains have been proposed97. The latter, if applied, will aid in optimizing reproducibility of these models.
Collectively, an overall advantage of classical gnotobiology is optimal control through use of strictly defined microbiota. Of note, GF mice are often reconstituted with microbiota at weaning or adulthood and then used as study subjects. Data from these studies must be interpreted with the caveat that they do not account for early life events that may occur if the mouse is immediately exposed at birth. Therefore, consideration should be given to use of progeny if early life events are of importance. An overall disadvantage is the cost associated with producing, maintaining, and monitoring these mice. Moreover, results from gnotobiotic mice must be interpreted with caution as the lack of microbiota in GF mice or lower microbial richness and diversity of defined microbiota mice do not fully recapitulate the complex interactions that occur within the microbiota of conventionally raised mice or the humans which they are intended to model98, 99. Studies using gnotobiotic animals should perhaps best be performed in multiple microbiota statuses. For example, GF mice may implicate microbiota, mono-association studies may aid in identification of putative causative agents of disease, and studies using defined microbiota and ultimately highly complex microbiota may confirm the role of these causative agents in a context that is more translatable to human health.
Reproducibility considerations in complex microbiota studies
While classical gnotobiology allows for optimal control of GM, it is now also possible to optimize reproducibility of studies that use animals with naturally occurring complex GM. To this end, the rapid and ongoing advances in next-generation sequencing allow for the characterization of complex GM to various taxonomic levels. Using this approach, one can readily correlate the presence of certain complex GM with optimal disease phenotypes and when phenotypes are lost or cannot be reproduced, one can assess whether this loss of reproducibility is associated with differing GMs. From here, investigators can use a variety of techniques to manipulate the GM (summarized below and reviewed in100) to begin to establish a cause and effect relationship. In many ways, this approach could be referred to as “next-generation gnotobiology” as we are now able to “know” what GM we are working with. This terminology should, however, be used with great caution for several reasons: 1) current databases are still growing and are insufficient to identify many intestinal bacteria to a species level; 2) many bacteria identified by sequencing tools have yet to be cultured and fully characterized; thus their phylogeny is minimally characterized and their functional attributes can only be assessed by comparison to others of similar taxonomy; 3) these factors are further confounded by the fact that many contemporary metagenomics survey studies rely on targeted sequencing strategies, such as for 16S rDNA gene, which can only provide predictive functionality analysis (for example, using Phylogenetic Investigation of Communities by Reconstruction of Unobserved States / PICRUSt analysis).
With these caveats, one can proceed with studies to assess the role of complex microbiota in model phenotypes and reproducibility. Possible scenarios that lead to such studies might include lack of reproducibility between two labs or loss of a phenotype associated with a change in husbandry practices or institution. In these cases, GM of mice of differing phenotypes can be defined with next-generation sequencing tools and compared. Lack-of-reproducibility studies require samples from both populations while loss-of-phenotype scenarios require before-and-after samples. Given the latter, if GM is known or expected to influence phenotype, one should always consider fecal banking when changes in husbandry or institution are looming. In either scenario, if correlative differences exist, one can proceed to proof-of-concept studies to assess whether restoration of a specific GM results in restoration of phenotype. This can be achieved in a number of ways, as described above.
To convincingly document changes in the GM, researchers must have the foresight to have collected fecal samples before the change in phenotype. While few investigators routinely bank fecal samples “just in case” their model phenotype is lost, the authors would recommend doing so before any foreseen changes in husbandry (for example, diet or bedding change) or relocation to a different institution.
Co-housing as a means to address lack of reproducibility
One of the simplest, least expensive, and perhaps most popular methods of assessing the influence of a complex GM on phenotype changes involves co-housing of animals with the desired phenotype and those that lack or have lost the phenotype. Co-housing relies on passive transfer of GM and results in hybridization of microbiota from the two cohoused animals. Because of this, successful phenotype transfer provides strong support for a GM influence, but a lack of phenotype transfer does not necessarily eliminate a contribution of the GM to the phenotype18, 101, 102.
Cross-fostering as a means to address lack of reproducibility
Cross-fostering of rodents to dams harboring a specific GM is also an affordable and simple strategy. Using cross-fostering, amidst a variety of other methods to manipulate the GM, Garrett et al. showed that mice lacking the transcription factor T-bet develop a colitogenic GM capable of inducing disease in T-bet-sufficient hosts103. The colitogenic104, 105 or protective106 capacity of the GM in several other models has been similarly demonstrated via cross-fostering. The caveat to cross-fostering is that it requires transfer of pups to a timed-mating foster dam as soon as possible following parturition, and there is typically a brief but unavoidable period during which pups are exposed to the GM of the biological dam. The effects of this transient exposure on the ontogeny of the pups’ GM is unknown, but it is likely that the biological dam’s vaginal and environmental microbiota contribute somewhat to the pups’ GM, resulting in a hybridized GM similar to that seen with co-housing. Thus, like co-housing, a negative transfer of phenotype in cross-fostered animals should be interpreted with caution and does not definitely rule out GM as a contributing factor. Given the caveats of these techniques, their use should be accompanied with next-generation sequencing analysis of microbiota of donors and recipient pups to optimally assess the efficacy of transfer.
Targeted strategies as means to address lack of reproducibility
More targeted strategies may also be considered when attempting to assess the role of GM when lack or loss of reproducibility is evident. These include reconstituting GF mice with specific GM that are associated with the phenotype, or using targeted rederivation procedures to establish specific GM. Reconstituting GF mice eliminates the possibility of GM hybridization, but comes with the disadvantages of GF and experimental reconstitution-based experiments, including: 1) expense, 2) the need to use second-generation animals to ensure that GM are present during early stages of development, and 3) the possibility of incomplete transfer of GM.
Rederivation via surgical embryo transfer (ET) can be also used to establish colonies of animals with disparate GMs. Pups generated via ET are exposed to the maternal GM beginning immediately at birth (avoiding the short period of exposure that occurs with cross-fostering) and acquire GM by natural means, which likely optimally transfers GM. The disadvantages of rederivation include the requirement for considerable expertise and specialized equipment and facilities. Moreover, in cases of loss or lack of reproducibility, this approach requires having surrogate mice that possess the desired GMs and have sufficient reproductive indices for ET (commonly used inbred strains may be inferior in these indices). Should the appropriate mice not be available for any of these above techniques, one can also survey mice for similar GM and use those as surrogates. To this end, the NIH-funded University of Missouri Mutant Mouse Resource and Research Center (http://www.mu-mmrrc.com/) created four colonies of outbred CD-1 mice (optimal reproductive indices) that stably possess GM from the four most commonly used vendors/producers of mice in the United States. These colonies thus likely possess GM similar to those that are present in most contemporary research mouse colonies. These mice can be used for any of the above mentioned techniques (co-housing, cross-fostering, GF transfer or rederivation), should optimal mice not be available. One important consideration when using such mice in the context of either cross-fostering or rederivation is the possibility of confounding maternal effects. Phenotypic differences detected in isogenic pups born to dams harboring distinct GM may be associated with differences in the GM, or they may be due to differing levels of maternal care or epigenetic factors such as methylation status107.
Many other techniques, including antibiotic administration, fecal microbiota transfer, and humanizing / xenografting microbiota have also been established. These are primarily used for prospective studies as opposed to trouble-shooting strategies. However, similar rigor, including use of next-generation sequencing to document and monitor GM, should be incorporated into these studies to optimize their reproducibility.
Future directions
In summary, with the exploding interest in microbiota, tools are now available to assess the impact of differing GM on reproducibility of rodent (and other species) models of disease. This review has only scratched the surface of considerations that need to be taken when evaluating the role of GM in model reproducibility and many challenges remain. For example, the vast majority of intestinal microbes remain uncultivable and novel culture methods or creative strategies to selectively eliminate these agents must be developed to fully investigate their role in the complex communities.
A clear distinction should also be made between the classical “one pathogen” model of infectious disease and investigations of the complex GM communities. While specific bacteria, such as Helicobacter hepaticus and SFB, have been implicated in model phenotype changes, it is highly likely that complex interactions of multiple agents have equally, if not more, important roles in model phenotype reproducibility. Study designs and associated statistical analyses, such as Random Forest, are being developed and refined to identify and implicate such complex interactions70.
Recently, several manuscripts have questioned whether the bacterial GM of contemporary laboratory rodents is of insufficient richness and species makeup to optimally model human conditions, and proposed to use GM from wild rodents or those obtained from the pet trade108–110. It is reasonable to conclude that the GM of laboratory mice has been simplified over the years as these rodents have been rendered free of pathogens. A return to the day when rodents were contaminated with these pathogens is unlikely, but can a happy medium be found where the richness of GM of laboratory mice can be increased without introducing unwanted pathogens? This scenario also raises questions about how other intestinal inhabitants, including viruses, protozoa, and fungi, influence the bacterial microbiota and model phenotypes.
Other questions that remain unanswered include: what common husbandry variables (for example, bedding, diet, and housing) affect the microbiota and how do we avoid institutional microbiota drift to optimize reproducibility? Are there strategies to decrease the expense of gnotobiotic housing, whole genome sequencing, transcriptomics, metabolomics, and bioinformatics, and are novel statistical methods required for these complex data sets? How can we take advantage of the inherent variability of the GM in our rodent populations and can this variability be exploited to model personalized medicine? All of these questions face the biomedical research and laboratory animal community but will surely be surmounted in these exciting times of this rapidly evolving field.
Footnotes
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Whitman WB, Coleman DC, Wiebe WJ. Prokaryotes: the unseen majority. Proc. Natl. Acad. Sci. USA. 1998;95:6578–6583. doi: 10.1073/pnas.95.12.6578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Clemente JC, Ursell LK, Parfrey LW, Knight R. The impact of the gut microbiota on human health: an integrative view. Cell. 2012;148:1258–1270. doi: 10.1016/j.cell.2012.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.de Vos WM, de Vos EA. Role of the intestinal microbiome in health and disease: from correlation to causation. Nutr. Rev. 2012;70(Suppl 1):S45–S56. doi: 10.1111/j.1753-4887.2012.00505.x. [DOI] [PubMed] [Google Scholar]
- 4.Ridaura VK, et al. Gut microbiota from twins discordant for obesity modulate metabolism in mice. Science. 2013;341:1241214. doi: 10.1126/science.1241214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Collins FS, Tabak LA. Policy: NIH plans to enhance reproducibility. Nature. 2014;505:612–613. doi: 10.1038/505612a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Perrin S. Preclinical research: Make mouse studies work. Nature. 2014;507:423–425. doi: 10.1038/507423a. [DOI] [PubMed] [Google Scholar]
- 7.Ma BW, et al. Routine habitat change: a source of unrecognized transient alteration of intestinal microbiota in laboratory mice. PLoS ONE. 2012;7:e47416. doi: 10.1371/journal.pone.0047416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sofi MH, et al. pH of drinking water influences the composition of gut microbiome and type 1 diabetes incidence. Diabetes. 2014;63:632–644. doi: 10.2337/db13-0981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sasada T, et al. Chlorinated Water Modulates the Development of Colorectal Tumors with Chromosomal Instability and Gut Microbiota in Apc-Deficient Mice. PLoS ONE. 2015;10:e0132435. doi: 10.1371/journal.pone.0132435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jin UH, et al. Microbiome-derived tryptophan metabolites and their aryl hydrocarbon receptor-dependent agonist and antagonist activities. Mol. Pharmacol. 2014;85:777–788. doi: 10.1124/mol.113.091165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Turnbaugh PJ, Backhed F, Fulton L, Gordon JI. Diet-induced obesity is linked to marked but reversible alterations in the mouse distal gut microbiome. Cell Host Microbe. 2008;3:213–223. doi: 10.1016/j.chom.2008.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ooi JH, et al. Dominant effects of the diet on the microbiome and the local and systemic immune response in mice. PLoS ONE. 2014;9:e86366. doi: 10.1371/journal.pone.0086366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schloss PD, Handelsman J. Status of the microbial census. Microbiol. Mol. Biol. Rev. 2004;68:686–691. doi: 10.1128/MMBR.68.4.686-691.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Turnbaugh PJ, et al. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027–1031. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- 15.Barnard DE, Lewis SM, Teter BB, Thigpen JE. Open- and closed-formula laboratory animal diets and their importance to research. J. Am. Assoc. Lab. Anim. Sci. 2009;48:709–713. [PMC free article] [PubMed] [Google Scholar]
- 16.Denning TL, et al. Functional specializations of intestinal dendritic cell and macrophage subsets that control Th17 and regulatory T cell responses are dependent on the T cell/APC ratio, source of mouse strain, and regional localization. J. Immunol. 2011;187:733–747. doi: 10.4049/jimmunol.1002701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hufeldt MR, Nielsen DS, Vogensen FK, Midtvedt T, Hansen AK. Variation in the gut microbiota of laboratory mice is related to both genetic and environmental factors. Comp. Med. 2010;60:336–347. [PMC free article] [PubMed] [Google Scholar]
- 18.Ivanov II, et al. Specific microbiota direct the differentiation of IL-17-producing T-helper cells in the mucosa of the small intestine. Cell Host Microbe. 2008;4:337–349. doi: 10.1016/j.chom.2008.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ericsson AC, et al. Effects of vendor and genetic background on the composition of the fecal microbiota of inbred mice. PLoS ONE. 2014 doi: 10.1371/journal.pone.0116704. (in the press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chung H, et al. Gut immune maturation depends on colonization with a host-specific microbiota. Cell. 2012;149:1578–1593. doi: 10.1016/j.cell.2012.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Friswell MK, et al. Site and strain-specific variation in gut microbiota profiles and metabolism in experimental mice. PLoS ONE. 2010;5:e8584. doi: 10.1371/journal.pone.0008584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Biasucci G, et al. Mode of delivery affects the bacterial community in the newborn gut. Early Hum. Dev. 2010;86(Suppl 1):13–15. doi: 10.1016/j.earlhumdev.2010.01.004. [DOI] [PubMed] [Google Scholar]
- 23.Dominguez-Bello MG, et al. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc. Natl. Acad. Sci. USA. 2010;107:11971–11975. doi: 10.1073/pnas.1002601107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lotz M, et al. Postnatal acquisition of endotoxin tolerance in intestinal epithelial cells. J. Exp. Med. 2006;203:973–984. doi: 10.1084/jem.20050625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Antonopoulos DA, et al. Reproducible community dynamics of the gastrointestinal microbiota following antibiotic perturbation. Infect. Immun. 2009;77:2367–2375. doi: 10.1128/IAI.01520-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bohnhoff M, Drake BL, Miller CP. Effect of streptomycin on susceptibility of intestinal tract to experimental Salmonella infection. Proc. Soc. Exp. Biol. Med. 1954;86:132–137. doi: 10.3181/00379727-86-21030. [DOI] [PubMed] [Google Scholar]
- 27.Hill DA, et al. Metagenomic analyses reveal antibiotic-induced temporal and spatial changes in intestinal microbiota with associated alterations in immune cell homeostasis. Mucosal Immunol. 2010;3:148–158. doi: 10.1038/mi.2009.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hoentjen F, et al. Antibiotics with a selective aerobic or anaerobic spectrum have different therapeutic activities in various regions of the colon in interleukin 10 gene deficient mice. Gut. 2003;52:1721–1727. doi: 10.1136/gut.52.12.1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Meynell GG. Some factors affecting the resistance of mice to oral infection by Salmonella typhimurium. Proc. R. Soc. Med. 1955;48:916–918. [Google Scholar]
- 30.Robinson CJ, Young VB. Antibiotic administration alters the community structure of the gastrointestinal micobiota. Gut Microbes. 2010;1:279–284. doi: 10.4161/gmic.1.4.12614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sekirov I, et al. Antibiotic-induced perturbations of the intestinal microbiota alter host susceptibility to enteric infection. Infect. Immun. 2008;76:4726–4736. doi: 10.1128/IAI.00319-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bongers G, et al. Interplay of host microbiota, genetic perturbations, and inflammation promotes local development of intestinal neoplasms in mice. J. Exp. Med. 2014;211:457–472. doi: 10.1084/jem.20131587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rakoff-Nahoum S, Paglino J, Eslami-Varzaneh F, Edberg S, Medzhitov R. Recognition of commensal microflora by toll-like receptors is required for intestinal homeostasis. Cell. 2004;118:229–241. doi: 10.1016/j.cell.2004.07.002. [DOI] [PubMed] [Google Scholar]
- 34.Cryan JF, Dinan TG. Mind-altering microorganisms: the impact of the gut microbiota on brain and behaviour. Nat. Rev. Neurosci. 2012;13:701–712. doi: 10.1038/nrn3346. [DOI] [PubMed] [Google Scholar]
- 35.O'Mahony SM, et al. Early life stress alters behavior, immunity, and microbiota in rats: implications for irritable bowel syndrome and psychiatric illnesses. Biol. Psychiatry. 2009;65:263–267. doi: 10.1016/j.biopsych.2008.06.026. [DOI] [PubMed] [Google Scholar]
- 36.Robosky LC, et al. Metabonomic identification of two distinct phenotypes in Sprague-Dawley (Crl:CD(SD)) rats. Toxicol. Sci. 2005;87:277–284. doi: 10.1093/toxsci/kfi214. [DOI] [PubMed] [Google Scholar]
- 37.Rohde CM, et al. Metabonomic evaluation of Schaedler altered microflora rats. Chem. Res. Toxicol. 2007;20:1388–1392. doi: 10.1021/tx700184u. [DOI] [PubMed] [Google Scholar]
- 38.Yang I, et al. Intestinal microbiota composition of interleukin-10 deficient C57BL/6J mice and susceptibility to Helicobacter hepaticus-induced colitis. PLoS ONE. 2013;8:e70783. doi: 10.1371/journal.pone.0070783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li Y, et al. Gut microbiota accelerate tumor growth via c-jun and STAT3 phosphorylation in APCMin/+ mice. Carcinogenesis. 2012;33:1231–1238. doi: 10.1093/carcin/bgs137. [DOI] [PubMed] [Google Scholar]
- 40.Mai V, Colbert LH, Perkins SN, Schatzkin A, Hursting SD. Intestinal microbiota: a potential diet-responsive prevention target in ApcMin mice. Mol. Carcinog. 2007;46:42–48. doi: 10.1002/mc.20233. [DOI] [PubMed] [Google Scholar]
- 41.Begley CG, Ellis LM. Drug development: Raise standards for preclinical cancer research. Nature. 2012;483:531–533. doi: 10.1038/483531a. [DOI] [PubMed] [Google Scholar]
- 42.Hutchinson L, Kirk R. High drug attrition rates-where are we going wrong? Nat. Rev. Clin. Oncol. 2011;8:189–190. doi: 10.1038/nrclinonc.2011.34. [DOI] [PubMed] [Google Scholar]
- 43.Prinz F, Schlange T, Asadullah K. Believe it or not: how much can we rely on published data on potential drug targets? Nat. Rev. Drug Discov. 2011;10:712. doi: 10.1038/nrd3439-c1. [DOI] [PubMed] [Google Scholar]
- 44.Kilkenny C, Browne WJ, Cuthill IC, Emerson M, Altman DG. Improving bioscience research reporting: the ARRIVE guidelines for reporting animal research. PLoS Biol. 2010;8:e1000412. doi: 10.1371/journal.pbio.1000412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bleich A, Hansen AK. Time to include the gut microbiota in the hygienic standardisation of laboratory rodents. Comp. Immunol. Microbiol. Infect. Dis. 2012;35:81–92. doi: 10.1016/j.cimid.2011.12.006. [DOI] [PubMed] [Google Scholar]
- 46.Ward JM, Anver MR, Haines DC, Benveniste RE. Chronic active hepatitis in mice caused by Helicobacter hepaticus. Am. J. Pathol. 1994;145:959–968. [PMC free article] [PubMed] [Google Scholar]
- 47.Ward JM, et al. Chronic active hepatitis and associated liver tumors in mice caused by a persistent bacterial infection with a novel Helicobacter species. J. Natl. Cancer Inst. 1994;86:1222–1227. doi: 10.1093/jnci/86.16.1222. [DOI] [PubMed] [Google Scholar]
- 48.Boutin SR, et al. Hepatic temporal gene expression profiling in Helicobacter hepaticus-infected A/JCr mice. Toxicol. Pathol. 2004;32:678–693. doi: 10.1080/01926230490524058. [DOI] [PubMed] [Google Scholar]
- 49.Fox JG, et al. Helicobacter hepaticus sp. nov., a microaerophilic bacterium isolated from livers and intestinal mucosal scrapings from mice. J. Clin. Microbiol. 1994;32:1238–1245. doi: 10.1128/jcm.32.5.1238-1245.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hailey JR, et al. Impact of Helicobacter hepaticus infection in B6C3F1 mice from twelve National Toxicology Program two-year carcinogenesis studies. Toxicol. Pathol. 1998;26:602–611. doi: 10.1177/019262339802600503. [DOI] [PubMed] [Google Scholar]
- 51.Foltz CJ, et al. Spontaneous inflammatory bowel disease in multiple mutant mouse lines: association with colonization by Helicobacter hepaticus. Helicobacter. 1998;3:69–78. doi: 10.1046/j.1523-5378.1998.08006.x. [DOI] [PubMed] [Google Scholar]
- 52.Kullberg MC, et al. Helicobacter hepaticus triggers colitis in specific-pathogen-free interleukin-10 (IL-10)-deficient mice through an IL-12- and gamma interferon-dependent mechanism. Infect. Immun. 1998;66:5157–5166. doi: 10.1128/iai.66.11.5157-5166.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chin EY, Dangler CA, Fox JG, Schauer DB. Helicobacter hepaticus infection triggers inflammatory bowel disease in T cell receptor alphabeta mutant mice. Comp. Med. 2000;50:586–594. [PubMed] [Google Scholar]
- 54.Engle SJ, et al. Elimination of colon cancer in germ-free transforming growth factor beta 1-deficient mice. Cancer Res. 2002;62:6362–6366. [PubMed] [Google Scholar]
- 55.Cahill RJ, et al. Inflammatory bowel disease: an immunity-mediated condition triggered by bacterial infection with Helicobacter hepaticus. Infect. Immun. 1997;65:3126–3131. doi: 10.1128/iai.65.8.3126-3131.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dieleman LA, et al. Helicobacter hepaticus does not induce or potentiate colitis in interleukin-10-deficient mice. Infect. Immun. 2000;68:5107–5113. doi: 10.1128/iai.68.9.5107-5113.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fox JG. Helicobacter bilis: bacterial provocateur orchestrates host immune responses to commensal flora in a model of inflammatory bowel disease. Gut. 2007;56:898–900. doi: 10.1136/gut.2006.115428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jergens AE, et al. Helicobacter bilis triggers persistent immune reactivity to antigens derived from the commensal bacteria in gnotobiotic C3H/HeN mice. Gut. 2007;56:934–940. doi: 10.1136/gut.2006.099242. doi:gut.2006.099242 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jiang HQ, Kushnir N, Thurnheer MC, Bos NA, Cebra JJ. Monoassociation of SCID mice with Helicobacter muridarum, but not four other enterics, provokes IBD upon receipt of T cells. Gastroenterology. 2002;122:1346–1354. doi: 10.1053/gast.2002.32959. [DOI] [PubMed] [Google Scholar]
- 60.Liu Z, et al. Helicobacter bilis colonization enhances susceptibility to Typhlocolitis following an inflammatory trigger. Dig. Dis. Sci. 2011;56:2838–2848. doi: 10.1007/s10620-011-1701-3. [DOI] [PubMed] [Google Scholar]
- 61.Myles MH, Dieckgraefe BK, Criley JM, Franklin CL. Characterization of cecal gene expression in a differentially susceptible mouse model of bacterial-induced inflammatory bowel disease. Inflamm. Bowel Dis. 2007;13:822–836. doi: 10.1002/ibd.20138. [DOI] [PubMed] [Google Scholar]
- 62.Myles MH, Livingston RS, Livingston BA, Criley JM, Franklin CL. Analysis of gene expression in ceca of Helicobacter hepaticus-infected A/JCr mice before and after development of typhlitis. Infect. Immun. 2003;71:3885–3893. doi: 10.1128/IAI.71.7.3885-3893.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ivanov II, et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. 2009;139:485–498. doi: 10.1016/j.cell.2009.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lee YK, Menezes JS, Umesaki Y, Mazmanian SK. Proinflammatory T-cell responses to gut microbiota promote experimental autoimmune encephalomyelitis. Proc. Natl. Acad. Sci. USA. 2011;108(Suppl 1):4615–4622. doi: 10.1073/pnas.1000082107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wu HJ, et al. Gut-residing segmented filamentous bacteria drive autoimmune arthritis via T helper 17 cells. Immunity. 2010;32:815–827. doi: 10.1016/j.immuni.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ericsson AC, Hagan CE, Davis DJ, Franklin CL. Segmented filamentous bacteria: commensal microbes with potential effects on research. Comp. Med. 2014;64:90–98. [PMC free article] [PubMed] [Google Scholar]
- 67.Chudnovskiy A, et al. Host-Protozoan Interactions Protect from Mucosal Infections through Activation of the Inflammasome. Cell. 2016;167:444–456. e414. doi: 10.1016/j.cell.2016.08.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Escalante NK, et al. The common mouse protozoa Tritrichomonas muris alters mucosal T cell homeostasis and colitis susceptibility. J. Exp. Med. 2016;213:2841–2850. doi: 10.1084/jem.20161776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tan TG, et al. Identifying species of symbiont bacteria from the human gut that, alone, can induce intestinal Th17 cells in mice. Proc. Natl. Acad. Sci. USA. 2016;113:E8141–E8150. doi: 10.1073/pnas.1617460113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zackular JP, Baxter NT, Chen GY, Schloss PD. Manipulation of the Gut Microbiota Reveals Role in Colon Tumorigenesis. mSphere. 2016;1 doi: 10.1128/mSphere.00001-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rahija RJ. In: The Mouse in Biomedical Research. Fox JG, et al., editors. Vol. 3. Academic Press; 2007. pp. 217–233. [Google Scholar]
- 72.Donohoe DR, et al. A gnotobiotic mouse model demonstrates that dietary fiber protects against colorectal tumorigenesis in a microbiota- and butyrate-dependent manner. Cancer Discov. 2014;4:1387–1397. doi: 10.1158/2159-8290.CD-14-0501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Goodman AL, et al. Extensive personal human gut microbiota culture collections characterized and manipulated in gnotobiotic mice. Proc. Natl. Acad. Sci. USA. 2011;108:6252–6257. doi: 10.1073/pnas.1102938108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Nguyen TL, Vieira-Silva S, Liston A, Raes J. How informative is the mouse for human gut microbiota research? Dis. Model. Mech. 2015;8:1–16. doi: 10.1242/dmm.017400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Atarashi K, et al. Induction of colonic regulatory T cells by indigenous Clostridium species. Science. 2011;331:337–341. doi: 10.1126/science.1198469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gaboriau-Routhiau V, et al. The key role of segmented filamentous bacteria in the coordinated maturation of gut helper T cell responses. Immunity. 2009;31:677–689. doi: 10.1016/j.immuni.2009.08.020. [DOI] [PubMed] [Google Scholar]
- 77.Helgeland L, Vaage JT, Rolstad B, Midtvedt T, Brandtzaeg P. Microbial colonization influences composition and T-cell receptor V beta repertoire of intraepithelial lymphocytes in rat intestine. Immunology. 1996;89:494–501. doi: 10.1046/j.1365-2567.1996.d01-783.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Macpherson AJ, Harris NL. Interactions between commensal intestinal bacteria and the immune system. Nat. Rev. Immunol. 2004;4:478–485. doi: 10.1038/nri1373. [DOI] [PubMed] [Google Scholar]
- 79.Umesaki Y, Setoyama H, Matsumoto S, Okada Y. Expansion of alpha beta T-cell receptor-bearing intestinal intraepithelial lymphocytes after microbial colonization in germ-free mice and its independence from thymus. Immunology. 1993;79:32–37. [PMC free article] [PubMed] [Google Scholar]
- 80.Savage DC, Siegel JE, Snellen JE, Whitt DD. Transit time of epithelial cells in the small intestines of germfree mice and ex-germfree mice associated with indigenous microorganisms. Appl. Environ. Microbiol. 1981;42:996–1001. doi: 10.1128/aem.42.6.996-1001.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Chowdhury SR, et al. Transcriptome profiling of the small intestinal epithelium in germfree versus conventional piglets. BMC Genomics. 2007;8:215. doi: 10.1186/1471-2164-8-215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hooper LV, et al. Molecular analysis of commensal host-microbial relationships in the intestine. Science. 2001;291:881–884. doi: 10.1126/science.291.5505.881. 291/5505/881 [pii] [DOI] [PubMed] [Google Scholar]
- 83.Backhed F, et al. The gut microbiota as an environmental factor that regulates fat storage. Proc. Natl. Acad. Sci. USA. 2004;101:15718–15723. doi: 10.1073/pnas.0407076101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wikoff WR, et al. Metabolomics analysis reveals large effects of gut microflora on mammalian blood metabolites. Proc. Natl. Acad. Sci. USA. 2009;106:3698–3703. doi: 10.1073/pnas.0812874106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Mazmanian SK, Liu CH, Tzianabos AO, Kasper DL. An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell. 2005;122:107–118. doi: 10.1016/j.cell.2005.05.007. [DOI] [PubMed] [Google Scholar]
- 86.Round JL, et al. The Toll-like receptor 2 pathway establishes colonization by a commensal of the human microbiota. Science. 2011;332:974–977. doi: 10.1126/science.1206095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kernbauer E, Ding Y, Cadwell K. An enteric virus can replace the beneficial function of commensal bacteria. Nature. 2014;516:94–98. doi: 10.1038/nature13960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Nell S, Suerbaum S, Josenhans C. The impact of the microbiota on the pathogenesis of IBD: lessons from mouse infection models. Nat. Rev. Microbiol. 2010;8:564–577. doi: 10.1038/nrmicro2403. [DOI] [PubMed] [Google Scholar]
- 89.Fritz JV, Desai MS, Shah P, Schneider JG, Wilmes P. From meta-omics to causality: experimental models for human microbiome research. Microbiome. 2013;1:14. doi: 10.1186/2049-2618-1-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Schaedler RW, Dubos R, Costello R. The Development of the Bacterial Flora in the Gastrointestinal Tract of Mice. J. Exp. Med. 1965;122:59–66. doi: 10.1084/jem.122.1.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Schaedler RW, Dubs R, Costello R. Association of Germfree Mice with Bacteria Isolated from Normal Mice. J. Exp. Med. 1965;122:77–82. doi: 10.1084/jem.122.1.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Orcutt RP, Gianni FJ, Judge RJ. Development of an “altered” Schaedler flora for NCI gnotobiotic rodents. Microecology and Therapy. 1987;17:59. [Google Scholar]
- 93.Dewhirst FE, et al. Phylogeny of the defined murine microbiota: altered Schaedler flora. Appl. Environ. Microbiol. 1999;65:3287–3292. doi: 10.1128/aem.65.8.3287-3292.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Robertson BR, et al. Mucispirillum schaedleri gen. nov., sp. nov., a spiral-shaped bacterium colonizing the mucus layer of the gastrointestinal tract of laboratory rodents. Int. J. Syst. Evol. Microbiol. 2005;55:1199–1204. doi: 10.1099/ijs.0.63472-0. [DOI] [PubMed] [Google Scholar]
- 95.Sarma-Rupavtarm RB, Ge Z, Schauer DB, Fox JG, Polz MF. Spatial distribution and stability of the eight microbial species of the altered schaedler flora in the mouse gastrointestinal tract. Appl. Environ. Microbiol. 2004;70:2791–2800. doi: 10.1128/AEM.70.5.2791-2800.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Wannemuehler MJ, Overstreet AM, Ward DV, Phillips GJ. Draft genome sequences of the altered schaedler flora, a defined bacterial community from gnotobiotic mice. Genome Announc. 2014;2:e00287. doi: 10.1128/genomeA.00287-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Macpherson AJ, McCoy KD. Standardised animal models of host microbial mutualism. Mucosal Immunol. 2015;8:476–486. doi: 10.1038/mi.2014.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Belenguer A, et al. Two routes of metabolic cross-feeding between Bifidobacterium adolescentis and butyrate-producing anaerobes from the human gut. Appl. Environ. Microbiol. 2006;72:3593–3599. doi: 10.1128/AEM.72.5.3593-3599.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Pfeiffer T, Bonhoeffer S. Evolution of cross-feeding in microbial populations. Am. Nat. 2004;163:E126–E135. doi: 10.1086/383593. [DOI] [PubMed] [Google Scholar]
- 100.Ericsson AC, Franklin CL. Manipulating the Gut Microbiota: Methods and Challenges. ILAR J. 2015;56:205–217. doi: 10.1093/ilar/ilv021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Bel S, et al. Reprogrammed and transmissible intestinal microbiota confer diminished susceptibility to induced colitis in TMF−/− mice. Proc. Natl. Acad. Sci. USA. 2014;111:4964–4969. doi: 10.1073/pnas.1319114111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Vijay-Kumar M, et al. Metabolic syndrome and altered gut microbiota in mice lacking Toll-like receptor 5. Science. 2010;328:228–231. doi: 10.1126/science.1179721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Garrett WS, et al. Communicable ulcerative colitis induced by T-bet deficiency in the innate immune system. Cell. 2007;131:33–45. doi: 10.1016/j.cell.2007.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Couturier-Maillard A, et al. NOD2-mediated dysbiosis predisposes mice to transmissible colitis and colorectal cancer. J. Clin. Invest. 2013;123:700–711. doi: 10.1172/JCI62236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Elinav E, et al. NLRP6 inflammasome regulates colonic microbial ecology and risk for colitis. Cell. 2011;145:745–757. doi: 10.1016/j.cell.2011.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Fuhrer A, et al. Milk sialyllactose influences colitis in mice through selective intestinal bacterial colonization. J. Exp. Med. 2010;207:2843–2854. doi: 10.1084/jem.20101098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Turecki G, Meaney MJ. Effects of the Social Environment and Stress on Glucocorticoid Receptor Gene Methylation: A Systematic Review. Biol. Psychiatry. 2016;79:87–96. doi: 10.1016/j.biopsych.2014.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Beura LK, et al. Normalizing the environment recapitulates adult human immune traits in laboratory mice. Nature. 2016;532:512–516. doi: 10.1038/nature17655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Reese TA, et al. Sequential Infection with Common Pathogens Promotes Human-like Immune Gene Expression and Altered Vaccine Response. Cell Host Microbe. 2016;19:713–719. doi: 10.1016/j.chom.2016.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Weldon L, et al. The Gut Microbiota of Wild Mice. PLoS ONE. 2015;10:e0134643. doi: 10.1371/journal.pone.0134643. [DOI] [PMC free article] [PubMed] [Google Scholar]