Abstract
Metal ions are critical for RNA structure and enzymatic activity. We present the structure of an asymmetric RNA loop that binds metal ions and has an essential function in a bacteriophage packaging motor. Prohead RNA is a noncoding RNA that is required for genome packaging activity in phi29-like bacteriophage. The loops in GA1 and phi29 bacteriophage share a conserved adenine that forms a base triple, although the structural context for the base triple differs. NMR relaxation studies and femtosecond time-resolved fluorescence spectroscopy reveal the dynamic behavior of the loop in the metal ion bound and unbound forms. The mechanism of metal ion binding appears to be an induced conformational change between two dynamic ensembles rather than a conformational capture mechanism. These results provide experimental benchmarks for computational models of RNA–metal ion interactions.
Graphical abstract
Prohead RNA (pRNA) is a noncoding RNA in phi29-like bacteriophage packaging motors.1 Although the role of pRNA in the mechanism of the motor is poorly understood, mutations in the pRNA abolish packaging activity.2 The A-helix of pRNA has a role in the initiation phase of packaging,3 but the role of pRNA during packaging and the mechanism of the ATPase remain active areas of experiment and debate.4–7 High-resolution structural data is necessary to determine the packaging motor mechanisms. This research reports the NMR structure of a pRNA loop that interacts with the ATPase, compares the structural differences between two sequences of this loop, and identifies the common structural features of two RNA sequences that fulfill the same function in the packaging motor.
Interestingly, different species of bacteriophage have very different primary sequences for pRNA while the secondary structure and quaternary interactions are conserved.8,9 The helical stem sequences may vary,10 but the loop regions are critical for packaging motor function. The loop size, asymmetry, and sequences vary widely in different bacteriophage species.8,9,11 In this work, we compare the three-dimensional structures of the phi29 and GA1 bacteriophage loops. A conserved adenine forms a base triple in both loops, although the structural context differs significantly.
pRNA loop dynamics were probed using both NMR and femtosecond time-resolved fluorescence spectroscopy. Ultrafast fluorescence spectroscopy has previously provided insight into base stacking and the conformational capture mechanisms of RNA loops that bind proteins and antibiotics.12–22 Although RNA–metal ion interactions have been well-studied by NMR, fluorescence spectroscopy, and computational studies, for example,23–32 RNA base stacking changes during metal ion binding have not been previously studied at this time scale. Changes in hydration, metal ion residency, and the formation of encounter complexes occur at the nanosecond to femtosecond time scales.16,33–37 Unlike the conformational capture mechanisms observed for antibiotic and protein binding, metal ion binding to the GA1 and phi29 bacteriophage loops shows a conformational change with new dynamic components.
Metal ion binding in RNA is critical for RNA folding, catalysis, and ligand and antibiotic binding.38–41 Bound metal ions can stabilize RNA base stacking even though residency times can be as short as 5 ns.34 Metal ion departure can destabilize base stacking and thus initiate a transition from a trapped RNA conformation.34 Metal ion binding to RNA loops can change helix bending angles and thus the overall architecture of an RNA fold.42 In the preQ1 riboswitch, metal ion binding can shift ligand binding from an induced fit to a conformational selection mechanism.43 Different aspects of metal ion binding occur at time scales ranging from picosecond to millisecond time scales, and local metal ion binding events can affect large-scale RNA structure and biological function on the millisecond to second time scales. Here, we study the fast base stacking changes in pRNA loops upon metal ion binding with femtosecond time-resolved fluorescence spectroscopy. These results will be important for developing computational models of RNA–metal ion interactions.
NMR Structure and Dynamics of the GA1 pRNA Loop
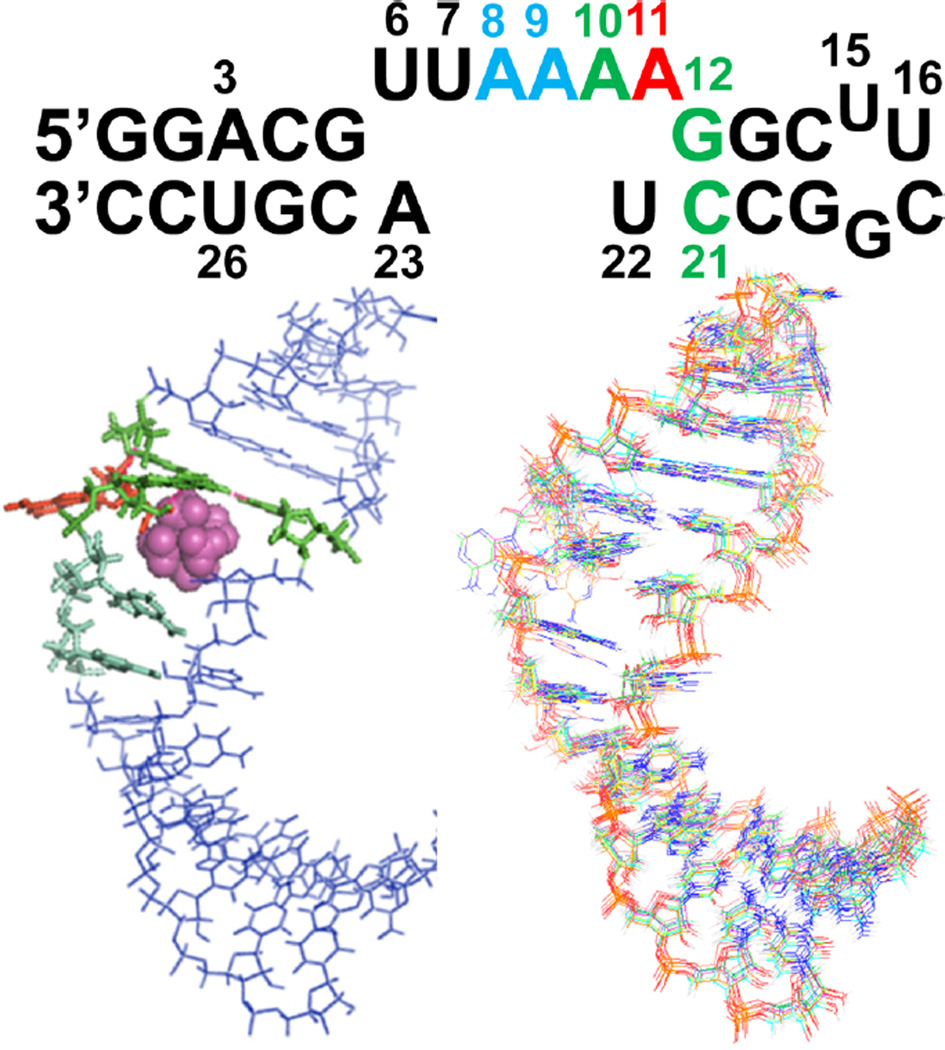
In the pRNA loop of GA1 packaging motors, the AU closing pairs do not form standard Watson–Crick pairs; therefore, the loop is actually an asymmetric loop of six nucleotides opposite two nucleotides (Figure 1). A total of 473 nuclear Overhauser effects (NOEs) (~17 per nucleotides), 255 dihedral angle restraints, and 29 residual dipolar couplings (RDCs) were used for structure calculations (Table 1). Ten structures with the lowest energy and no violations were chosen as a representative ensemble. Excluding nucleotide A11, which is flipped out of the loop (Figure 1), the ensemble structure yielded an RMSD of 1.10 Å for heavy atoms (Table 1).
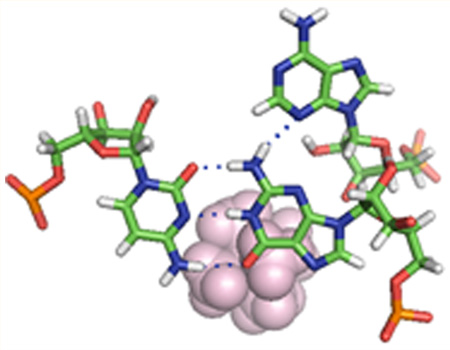
Figure 1.
Secondary structure of the 28 nucleotide hairpin used for NMR studies. On the left is the ensemble average structure with the base triple (A10-G12-C21) highlighted in green, A11 in red, and the stacking A8 and A9 adenines in light cyan. The Co(NH3)63+ ion is shown as magenta spheres. On the right is the overlay of 10 lowest-energy structures.
Table 1.
NMR Structural Statistics
| NOE-derived distance constraints | 473 |
| internucleotide NOE | 213 |
| intranucleotide NOE | 260 |
| dihedral constraints | 255 |
| hydrogen bond constraints | 52 |
| RDCs | 29 |
| Structure Statistics RMSD (Deviation) | |
| NOE constraints (Å) | 0.012 ± 0.001 |
| dihedral angle constraints (deg) | 0.875 ± 0.065 |
| Deviations from Idealized Geometry | |
| bond lengths (Å) | 0.005 ± 0.000 |
| bond angles (deg) | 0.797 ± 0.005 |
| impropers (deg) | 0.675 ± 0.017 |
| RMSD for all heavy atoms | 1.44 |
| RMSD for all heavy atoms (no A11) | 1.10 |
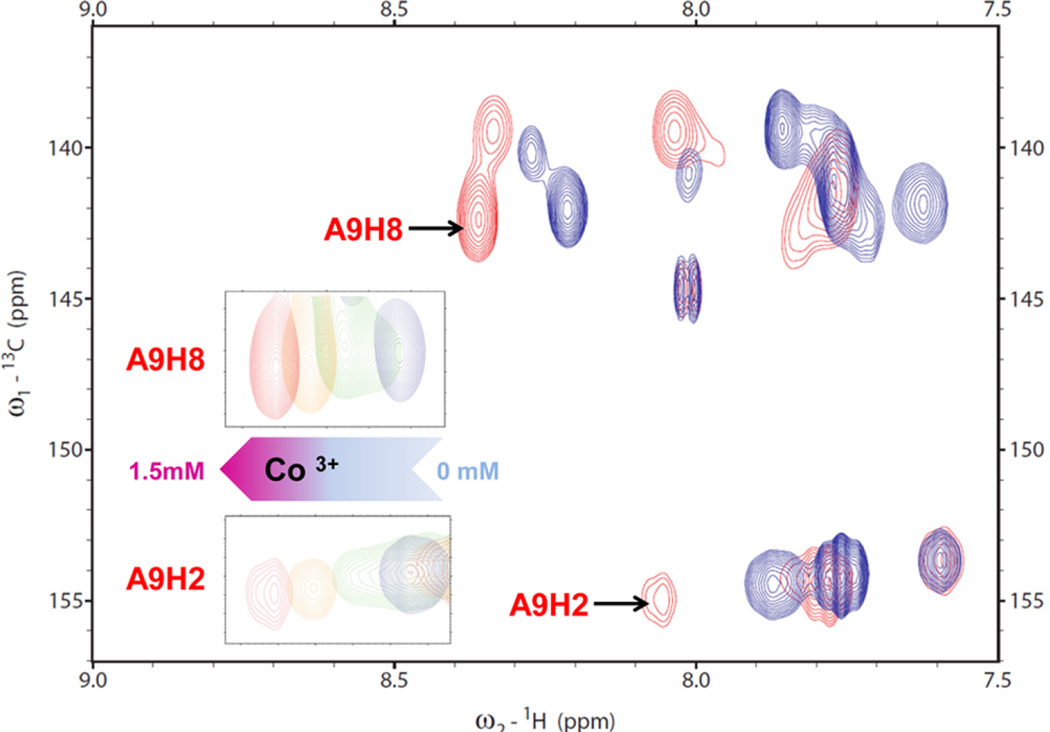
In the GA1 pRNA loop, U6 is stacked over G5 in the closing base pair of the loop (Figure 1). Rather than base pairing with U6, A23 is closer to A8 and A9. U22 is partly stacked over the C21. Instead of base pairing with U22, A11 is exposed to solvent, and thus, the position of A11 is less well-defined relative to the rest of the nucleotides. The nucleotides 6-UUAA-9 partly stack as in A-form RNA. H1′–H2′ cross peaks that indicate C2′-endo puckering occur in U16 and C17 in a UUCG tetra-loop but do not occur in the asymmetric loop. In 13C HSQC experiments, A9H2 and A9H8 show the largest chemical shift changes as the metal ion is titrated into the RNA NMR sample (Figures 2, S1, and S2). NOEs between the Co(NH3)63+ and A8, A9, A10, U22, and A23 define the location of the metal ion binding site (Figure S2). NOEs to the amine groups are unique to Co(NH3)63+ ions and enable future applications in NMR and computational studies of RNA–metal ion interactions. Co(NH3)63+ binds to the pRNA loop and increases its stability by 1.3 kcal/mol (Table S2). The excess of Co(NH3)63+ ensures that the structure is an ensemble of bound conformations and not a mixture of bound and unbound conformations. Experimental measurements of cobalt hexamine binding to RNA using phosphorus NMR and difference Raman spectroscopy show that cobalt hexamine binds approximately 5-to 50-fold tighter than hydrated magnesium.44,45 Cobalt hexamine does not form innersphere coordination with RNA but can outcompete innersphere-coordinated magnesium ions.44 The addition of cobalt hexamine chloride increases the thermodynamic stability and sharpens the line widths of the NMR resonances more than magnesium chloride (Table S2 and Figure S3) and thus for practical technical reasons, provides the best biophysical experimental conditions to study RNA dynamics.
Figure 2.
13C HSQC overlay spectrum for the 13C–15N–A- and –U-labeled NMR sample. Red and blue crosspeaks are from spectra collected in the presence and absence of Co(NH3)63+, respectively. Shown in the insets are the A9H2 and A9H8 crosspeaks moving at Co(NH3)63+ concentrations of 0 (blue), 0.5 (green), 1.0 (orange), and 1.5 mM (red), which suggests that Co(NH3)63+ induces a new conformation.
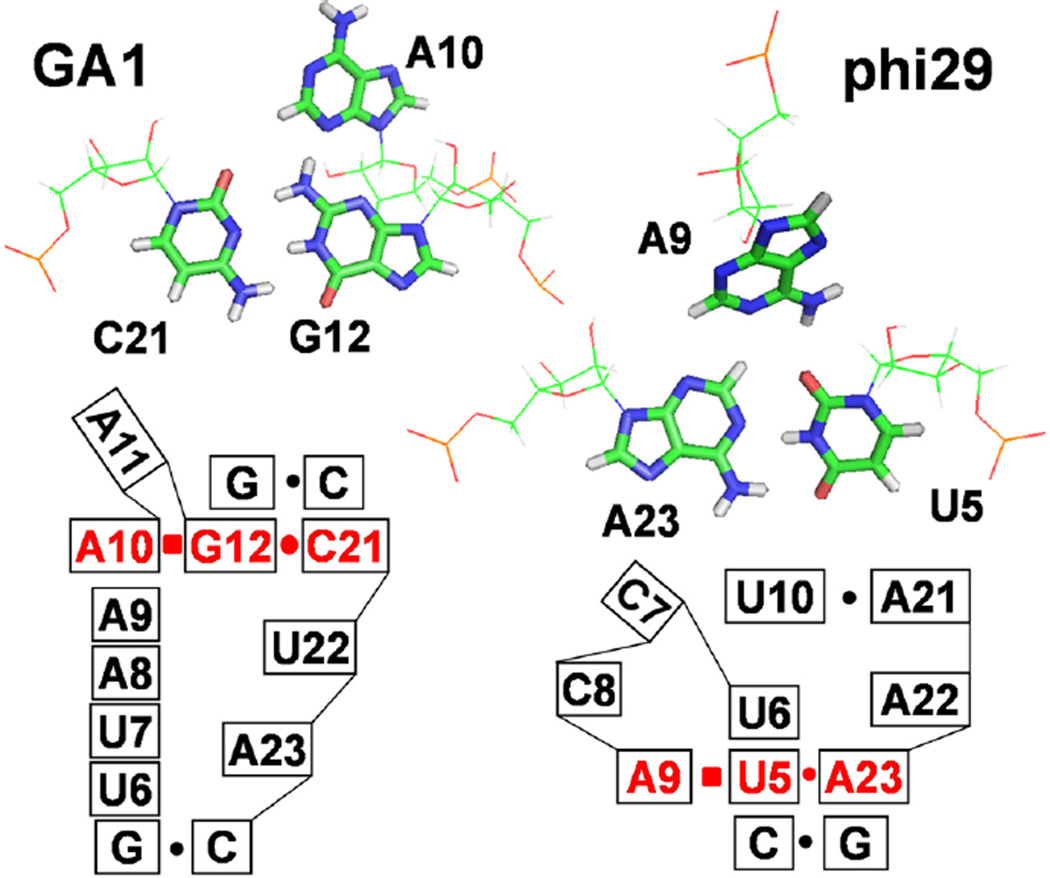
A10 N3 forms a hydrogen bond with G12 H2 and thus forms a base triple A10–G12–C21 (Figure 3). A10 is the only conserved nucleotide in the loop that binds the ATPase in pRNAs from GA1, phi29, M2, and SF5 bacteriophage. In the NMR structure of the phi29 pRNA loop,46 the conserved A at the same position forms a base triple with the AU base pair at the end of the helix (Figure 3). This base triple is critical for the backbone direction and the bending angle in the helical axis of the phi29 pRNA. The phi29 and GA1 pRNA have different sequences, loop sizes, loop asymmetry, and helical bending angles. Both the GA1 and phi29 loops have flexible dynamic nucleotides in the apo state. Thus, although the structural context of the base triple is different, both the GA1 and phi29 loops form a base triple with the conserved adenine in a dynamic asymmetric loop.
Figure 3.
Base triples in GA1 and phi29 pRNA loops.46 In the lower secondary structure models, lines represent the phosphodiester backbone between nucleotides. Stacked boxes indicate base stacking. The solid circles and squares represent cis-Watson–Crick and cis-Hoogsteen pairing.47 NOEs A10H2–C21H1′, A10H2–G13H1′, and A10H8–G12H1′ indicate base triple formation (Figure S1).
We used NMR to study the nucleotide dynamics in the GA1 loop with A and U nucleotides, specifically 13C–15N-labeled in the presence of Co(NH3)63+ (Figure 4). All relaxation decay curves were well fit by single exponentials, without any evidence for multiexponential relaxation.44 Some nuclei were excluded from the analysis of the relaxation data because the corresponding resonances were overlapped in the spectra. In the absence of Co(NH3)63+ ions, all of the resonances were too overlapped for analysis.
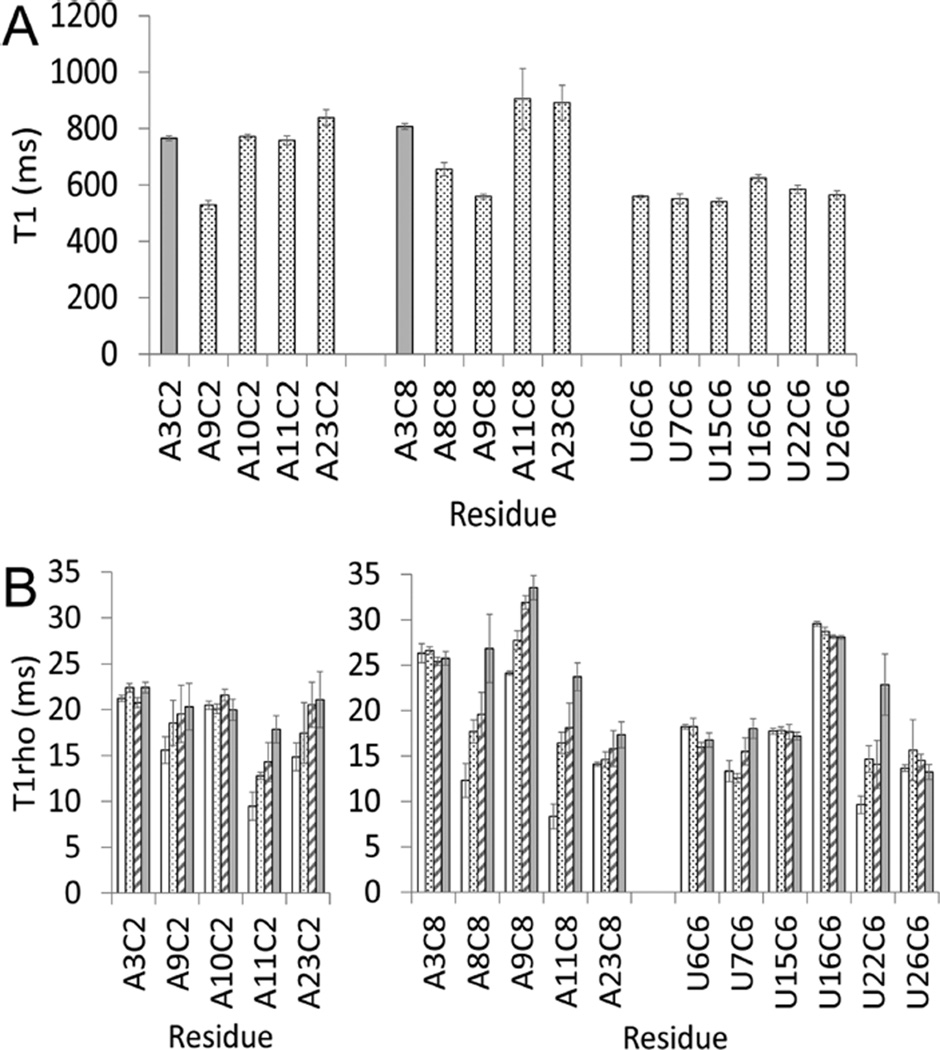
Figure 4.
Relaxation parameters T1 (A) and T1rho (B) for C2, C8, and C6 resonances plotted as a function of residue position. The four spin-lock powers for T1rho data were 1.0 (white), 1.4 (dot), 2.0 (stripe), and 2.8 kHz (gray). The data were collected at 800 MHz and 25 °C on a 1.5 mM RNA sample with a 1:1 ratio of RNA/Co(NH3)63+.
The values of T1 and T1rho for the Watson–Crick pair A3–U26 provide a reference point to assess the internal motion in other loop nucleotides on the nanosecond time scale, as well as the micro- to millisecond time scale. RNA dynamics occur on multiple time scales simultaneously. A10 forms a base triple with G12 and C21 (Figure 2), and the T1 and power-independent T1rho values for A10 exhibit motions similar to those of A3 in a Watson–Crick pair in a helix. This observation indicates that the base triple forms a stable structure in the presence of Co(NH3)63+ ions. A11 is flipped out of the loop, and the dependence of the T1rho values on power is consistent with its dynamic structure and the few NOEs observed to A11. Nucleotides that show NOEs between their base protons and the Co(NH3)63+ ion, that is, A8, A9, U22, and A23, show a strong power dependence in the T1rho values that may be the result of metal ion binding on the microsecond to millisecond time scale. The protons closest to the metal ion show different T1rho relaxation dynamics than the protons in a Watson–Crick base pair (i.e., A3), which suggests that interacting with the metal ion changes the nucleotide dynamics.
Ultrafast Fluorescence Spectroscopy Reveals an Induced Fit Mechanism for Metal Ion Binding
Femtosecond time-resolved fluorescence spectroscopy was used to probe the dynamic nature of the pRNA loops in the absence or presence of Co(NH3)63+ ions to understand the conformational distribution and dynamic motion of the metal ion binding motif. In these experiments, 2-aminopurine (2AP) (A in red, Table 2, Figure S4) replaces one of the purine bases in the loop, thus allowing measurement of the ultrafast quenching dynamics on a picosecond–nanosecond time scale and to define its stacking interactions with surrounding bases. 7-Deazaadenine (A in blue, Table 2) quenches the fluorescence of 2AP differently than the natural bases of adenine and thus facilitates identification of base stacking interactions. Changes in hydration, the formation of encounter complexes, and the residency time of metal ions bound to RNA occur on the femtosecond to nanosecond time scale.16,33,34,48,49 Femtosecond time-resolved spectroscopy provides new insight into the motions of RNA bases during these fast time scale events in the formation of RNA–metal ion interactions.
Table 2.
Parameters of Femtosecond Time-Resolved Fluorescence Decay Dynamicsa
| duplex | ± ion |
τ1 (ps) |
A1 (%) |
τ2 (ps) |
A2 (%) |
τ3 (ps) |
A3 (%) |
τ4 (ps) |
A4 (%) |
|
|---|---|---|---|---|---|---|---|---|---|---|
| A | SS | 1 | 46.0 | 55 | 30.0 | 13.0 | 24.0 | |||
| DS | 1 | 42.6 | 50 | 29.5 | 2.9 | 27.8 | ||||
| Co | 1 | 42.6 | 15 | 18.5 | 115 | 11.1 | 4.5 | 27.8 | ||
| B | DS | 3 | 39.5 | 70 | 28.9 | 4.1 | 31.6 | |||
| Co | 3 | 26.8 | 20 | 38.0 | 350 | 3.9 | 2.3 | 31.3 | ||
| C | DS | 3 | 36.2 | 60 | 29.8 | 2.1 | 34.0 | |||
| Co | 3 | 33.3 | 27 | 19.6 | 300 | 23.5 | 3.3 | 21.6 | ||
| D | DS | 3 | 38.8 | 70 | 38.8 | 2.2 | 22.4 | |||
| Co | 3 | 37.0 | 30 | 34.8 | 300 | 11.0 | 2.1 | 17.4 | ||
| E | DS | 3 | 31.1 | 70 | 28.9 | 2.0 | 40.0 | |||
| Co | 3 | 25.0 | 30 | 31.8 | 300 | 11.4 | 2.6 | 31.8 | ||
| F | DS | 3 | 38.6 | 40 | 29.6 | 2.5 | 31.8 | |||
| Co | 3 | 31.2 | 30 | 33.3 | 300 | 18.8 | 4.8 | 16.7 | ||
Fluorescence decay times (τ) are fit with the minimum number of components. Percent amplitudes have an error of 5%. Salt conditions are 100 mM NaCl with (Co) or without (DS) the addition of Co(NH3)63+ in a 2:1 ratio with RNA. Red adenines are 2AP. Blue adenines are 7-deazaadenine. SS indicates the single strand containing 2AP.
In the absence of the Co(NH3)63+ ion (Table 2), the fluorescence decay curves for each duplex were fit with a minimum of three unique decay terms. For example, in the GA1 duplex that has a 2AP replacing the conserved adenine (Table 2 row A, DS), the multiphasic decay profile suggests a heterogeneous stacking pattern for this base. The 2AP has a partial population (72.2%) that is stacked, reflected in the 1 (42.6%) and 50 ps (29.5%) decay components, and also a significant population that is completely unstacked from the loop (the long, 2.9 ns component, 27.8%). The phi29 loop with the same conserved A substituted with 2AP shows a similar dynamic profile (Table 2 row f) as the adenines in the GA1 loop (Table 2 rows A–E). In the presence of a Co(NH3)63+ ion, a new component with a lifetime of hundreds of picoseconds appears for each duplex, indicating that a new ensemble of dynamic conformations is induced by Co(NH3)63+ ion binding. All of the duplexes show a new τ3 component whether or not the nucleotide substituted with 2AP has direct interactions with Co(NH3)63+. This suggests that Co(NH3)63+ induces a global rearrangement of all of the adenines and creates a relaxation pathway that was not detectable in the previous conformational ensemble. The new dynamic ensemble is likely to be the formation of the base triple, where the high-energy backbone conformations for A10 and A11 are stabilized by favorable interactions with the metal ion. In a conformational capture model, one would expect a shift in the existing populations of the different components, but a new component on a new time scale, that is on the order of hundreds of picoseconds with a percentage above error estimates indicates a new set of conformations.
The conformational capture model for antibiotic and peptide binding to RNA was demonstrated previously by ultrafast fluorescence spectroscopy.13,14 In conformational capture mechanisms, a preferred conformation is selected from an ensemble of states when a ligand binds. In contrast, in induced fit mechanisms, a conformational change occurs when a ligand binds. In the case of metal ion binding to the pRNA loops, the appearance of a new component in the fit of the fluorescence decay curves suggests an induced fit mechanism. The data is most consistent with a conformational change between two dynamic ensembles upon metal ion binding. This highlights an important distinction in understanding the differences between these two mechanisms for ligand binding. The difference between induced fit and conformational capture is not simply the result of the observation time scale. The complex molecular events that comprise induced fit mechanisms occur on multiple time scales and include femtosecond to nanosecond time scales. An induced fit mechanism can occur between dynamic ensembles and not simply between two single static conformations. This is an important distinction for developing computational models of RNA metal ion binding, RNA structural dynamics, and ligand binding and improving metal ion force fields34,42,43,50
In GA1 and phi29 pRNA, a conserved adenine forms a base triple in a dynamic, asymmetric loop. For the GA1 loop, we used NMR spectroscopy to measure the dynamics of metal ion binding and found dynamics in the microsecond to millisecond time scale at which ligand binding occurs and in the nanosecond time scale at which water binding occurs. Metal ion binding to RNA has not been previously studied on the femtosecond to picosecond time scale, and thus, this research expands the range for the time scale profile of dynamic interactions between metal ions, water, and RNA to include very fast changes in base stacking that occur on the time scale of encounter complex formation and metal ion residency. Metal ion binding in the pRNA loops induces a conformational change into a new dynamic ensemble rather than capturing a particular conformation. These new insights will provide important benchmarks for computational studies of RNA–metal ion interactions.
Supplementary Material
Acknowledgments
M.T. and G.C. were supported by NIH Grant P41GM103399 (NIGMS). This study made use of the National Magnetic Resonance Facility at Madison, which is supported by NIH Grant P41GM103399 (NIGMS). Equipment was purchased with funds from the University of Wisconsin–Madison, the NIH (P41GM103399, S10RR02781, S10RR08438, S10RR023438, S10RR025062, S10RR029220), the NSF (DMB-8415048, OIA-9977486, BIR-9214394), and the USDA. Research was supported by grants from NSF 0844913 (S.J.S.), the Oklahoma Center for the Advancement of Science and Technology HR13-206 (S.J.S.), and NIH GM 095997 (D.Z.). The authors thank Gretchen Scheel for collecting some of the optical melting data in Table S2.
Footnotes
ASSOCIATED CONTENT
Supporting Information
- Detailed experimental methods, NMR spectra, and fluorescence spectra (PDF)
The authors declare no competing financial interest.
The NMR data and structure have been deposited to the PDB (2NCI) and the Biological Magnetic Resonance Database (26024).
REFERENCES
- 1.Guo P, Erickson S, Anderson D. A Small Viral RNA is Required for in Vitro Packaging of Bacteriophage phi29 DNA. Science. 1987;236:690–694. doi: 10.1126/science.3107124. [DOI] [PubMed] [Google Scholar]
- 2.Zhao W, Morais M, Anderson D, Jardine P, Grimes S. Roles of the CCA Bulge of Prohead RNA Bacteriophage phi29 in DNA Packaging. J. Mol. Biol. 2008;383:520–528. doi: 10.1016/j.jmb.2008.08.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhao W, Jardine PJ, Grimes S. An RNA Domain Imparts Specificity and Selectivity to a Viral DNA Packaging Motor. J. Virol. 2015;89:12457–12466. doi: 10.1128/JVI.01895-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hill AC, Bartley LE, Schroeder SJ. Prohead RNA: a Noncoding Viral RNA of Novel Structure and Function. WIREs RNA. 2016;7:428. doi: 10.1002/wrna.1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jardine PJ. Packaging Models versus Modeling Packaging. Biophys. J. 2016;110:287–288. doi: 10.1016/j.bpj.2015.10.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu S, Chistol G, Bustamante C. Mechanical Operation and Intersubunit Coordination of Ring-shaped Molecular Motors: Insights from Single-molecule Studies. Biophys. J. 2014;106:1844–1858. doi: 10.1016/j.bpj.2014.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schwartz C, Guo P. Ultrastable pRNA Hexameric Ring Gearing Hexameric phi29 DNA-packaging Motor by Revolving without Rotating and Coiling. Curr. Opin. Biotechnol. 2013;24:581–590. doi: 10.1016/j.copbio.2013.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gu X, Schroeder SJ. Different Sequences Show Similar Quaternary Interaction Stabilities in Prohead Viral RNA Self-Assembly. J. Biol. Chem. 2011;286:14419–14426. doi: 10.1074/jbc.M110.191064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bailey S, Wichitwechkarn J, Johnson D, Reilly BE, Anderson DL, Bodley JW. Phylogenetic Analysis and Secondary Structure of Bacillus subtilis Bacteriophage RNA Required for DNA Packaging. J. Biol. Chem. 1990;265:22365–22370. [PubMed] [Google Scholar]
- 10.Ding F, Lu C, Zhao W, Rajashankar K, Anderson D, Jardine P, Grimes S, Ke A. Structure and Assembly of the Essential RNA Ring Component of a Viral DNA Packaging Motor. Proc. Natl. Acad. Sci. U. S. A. 2011;108:7357–7362. doi: 10.1073/pnas.1016690108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hao Y, Kieft JS. Diverse Self-association Properties within a Family of Phage Packaging RNAs. RNA. 2014;20:1759–1774. doi: 10.1261/rna.045948.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xia T. Taking Femtosecond Snapshots of RNA Conformational Dynamics and Complexity. Curr. Opin. Chem. Biol. 2008;12:604–611. doi: 10.1016/j.cbpa.2008.08.033. [DOI] [PubMed] [Google Scholar]
- 13.Lu J, Nguyen L, Zhao L, Xia T, Qi X. A Cyclic Mimic of HIV Tat Differentiates Similar TAR RNAs on the Basis of Distinct Dynamic Behaviors. Biochemistry. 2015;54:3687–3693. doi: 10.1021/acs.biochem.5b00325. [DOI] [PubMed] [Google Scholar]
- 14.Lu J, Zhao L, Xia A, Xia T, Qi X. Dissect Conformational Distribution and Drug-induced Population Shift of Prokaryotic rRNA A-site. Biochemistry. 2013;52:1651–1653. doi: 10.1021/bi400053s. [DOI] [PubMed] [Google Scholar]
- 15.Xia T, Becker HC, Wan C, Frankel A, Roberts RW, Zewail AH. The RNA-Protein Complex: Direct Probing of the Interfacial Recognition Dynamics and its Correlation with Biological Functions. Proc. Natl. Acad. Sci. U. S. A. 2003;100:8119–8123. doi: 10.1073/pnas.1433099100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pal SK, Zhao L, Xia T, Zewail AH. Site- and Sequence-selective Ultrafast Hydration of DNA. Proc. Natl. Acad. Sci. U. S. A. 2003;100:13746–13751. doi: 10.1073/pnas.2336222100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lu J, Wong V, Zhang Y, Tran T, Zhao L, Xia A, Xia T, Qi X. Distinct Conformational Transition Patterns of Noncoding 7SK snRNA and HIV TAR RNAs upon Tat Binding. Biochemistry. 2014;53:675–681. doi: 10.1021/bi401131z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jain N, Zhao L, Liu JD, Xia T. Heterogeneity and Dynamics of the Ligand Recognition Mode in Purine-sensing Riboswitches. Biochemistry. 2010;49:3703–3714. doi: 10.1021/bi1000036. [DOI] [PubMed] [Google Scholar]
- 19.Kadakkuzha BM, Zhao L, Xia T. Conformational Distribution and Ultrafast Base Dynamics of Leadzyme. Biochemistry. 2009;48:3807–3809. doi: 10.1021/bi900256q. [DOI] [PubMed] [Google Scholar]
- 20.Lee SW, Zhao L, Pardi A, Xia T. Ultrafast Dynamics Show that the Theophylline and 3-Methylxanthine Aptamers Employ a Conformational Capture Mechanism for Binding their Ligands. Biochemistry. 2010;49:2943–2951. doi: 10.1021/bi100106c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lu J, Kadakkuzha BM, Zhao L, Fan M, Qi X, Xia T. Dynamic Ensemble View of the Conformational Landscape of the HIV-1 TAR RNA and Allosteric Recognition. Biochemistry. 2011;50:5042–5047. doi: 10.1021/bi200495d. [DOI] [PubMed] [Google Scholar]
- 22.Xia T, Wan C, Roberts RW, Zewail AH. RNA-protein Recognition: Single-Residue Ultrafast Dynamical Control of Structural Specificity and Function. Proc. Natl. Acad. Sci. U. S. A. 2005;102:13013–13018. doi: 10.1073/pnas.0506181102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rau MJ, Welty R, Tom Stump W, Hall KB. Formation of Tertiary Interactions during rRNA GTPase Center Folding. J. Mol. Biol. 2015;427:2799–2815. doi: 10.1016/j.jmb.2015.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bergonzo C, Hall KB, Cheatham TE., 3rd Stem-Loop V of Varkud Satellite RNA Exhibits Characteristics of the Mg(2+) Bound Structure in the Presence of Monovalent Ions. J. Phys. Chem. B. 2015;119:12355–12364. doi: 10.1021/acs.jpcb.5b05190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rau MJ, Hall KB. 2-Aminopurine Fluorescence as a Probe of Local RNA Structure and Dynamics and Global Folding. Methods Enzymol. 2015;558:99–124. doi: 10.1016/bs.mie.2015.01.006. [DOI] [PubMed] [Google Scholar]
- 26.Jean JM, Hall KB. 2-aminopurine Fluorescence Quenching and Lifetimes: Role of Base Stacking. Proc. Natl. Acad. Sci. U. S. A. 2001;98:37–41. doi: 10.1073/pnas.011442198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bonneau E, Girard N, Lemieux S, Legault P. The NMR Structure of the II-III-VI Three-way Junction from the Neurospora VS ribozyme Reveals a Critical Tertiary Interaction and Provides New Insights into the Global Ribozyme Structure. RNA. 2015;21:1621–1632. doi: 10.1261/rna.052076.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bonneau E, Legault P. NMR Localization of Divalent Cations at the Active Site of the Neurospora VS Ribozyme Provides Insights into RNA-Metal-ion Interactions. Biochemistry. 2014;53:579–590. doi: 10.1021/bi401484a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hayes RL, Noel JK, Whitford PC, Mohanty U, Sanbonmatsu KY, Onuchic JN. Reduced Model Captures Mg(2+)-RNA Interaction Free Energy of Riboswitches. Biophys. J. 2014;106:1508–1519. doi: 10.1016/j.bpj.2014.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McPhee SA, Huang L, Lilley DM. A Critical Base Pair in K-turns that Confers Folding Characteristics and Correlates with Biological Function. Nat. Commun. 2014;5:5127. doi: 10.1038/ncomms6127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Noeske J, Buck J, Furtig B, Nasiri HR, Schwalbe H, Wohnert J. Interplay of ‘Induced Fit’ and Preorganization in the Ligand Induced Folding of the Aptamer Domain of the Guanine Binding Riboswitch. Nucleic Acids Res. 2006;35:572–583. doi: 10.1093/nar/gkl1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Noeske J, Schwalbe H, Wohnert J. Metal-ion Binding and Metal-ion induced Folding of the Adenine-Sensing Riboswitch Aptamer Domain. Nucleic Acids Res. 2007;35:5262–5273. doi: 10.1093/nar/gkm565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Al-Hashimi HM, Walter NG. RNA dynamics: It is about Time. Curr. Opin. Struct. Biol. 2008;18:321–329. doi: 10.1016/j.sbi.2008.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xu X, Yu T, Chen SJ. Understanding the Kinetic Mechanism of RNA Single Base Pair Formation. Proc. Natl. Acad. Sci. U. S. A. 2016;113:116–121. doi: 10.1073/pnas.1517511113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tan C, Liu Z, Li J, Guo X, Wang L, Sancar A, Zhong D. The Molecular Origin of High DNA-repair Efficiency by Photolyase. Nat. Commun. 2015;6:7302. doi: 10.1038/ncomms8302. [DOI] [PubMed] [Google Scholar]
- 36.Chang CW, Guo L, Kao YT, Li J, Tan C, Li T, Saxena C, Liu Z, Wang L, Sancar A, Zhong D. Ultrafast Solvation Dynamics at Binding and Active Sites of Photolyases. Proc. Natl. Acad. Sci. U. S. A. 2010;107:2914–2919. doi: 10.1073/pnas.1000001107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Qin Y, Yang Y, Zhang L, Fowler JD, Qiu W, Wang L, Suo Z, Zhong D. Direct Probing of Solvent Accessibility and Mobility at the Binding Interface of Polymerase (Dpo4)-DNA Complex. J. Phys. Chem. A. 2013;117:13926–13934. doi: 10.1021/jp410051w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Piccirilli JA, Vyle JS, Caruthers MH, Cech TR. Metal Ion Catalysis in the Tetrahymena Ribozyme Reaction. Nature. 1993;361:85–88. doi: 10.1038/361085a0. [DOI] [PubMed] [Google Scholar]
- 39.Johnson-Buck AE, McDowell SE, Walter NG. Metal Ions: Supporting Actors in the Playbook of Small Ribozymes. Metal Ions in Life Sciences. 2011:175–196. doi: 10.1039/9781849732512-00175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang Y, Shen JK, Schroeder SJ. Nucleotide Dynamics at the A-site Cleft in the Peptidyltransferase Center of H. marismortui 50S Ribosomal Subunits. J. Phys. Chem. Lett. 2012;3:1007–1010. doi: 10.1021/jz3001882. [DOI] [PubMed] [Google Scholar]
- 41.Denesyuk NA, Thirumalai D. How do Metal Ions Direct Ribozyme Folding? Nat. Chem. 2015;7:793–801. doi: 10.1038/nchem.2330. [DOI] [PubMed] [Google Scholar]
- 42.Mouzakis KD, Dethoff EA, Tonelli M, Al-Hashimi H, Butcher SE. Dynamic Motions of the HIV-1 Frameshift Site RNA. Biophys. J. 2015;108:644–654. doi: 10.1016/j.bpj.2014.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Suddala KC, Wang J, Hou Q, Walter NG. Mg(2+) Shifts Ligand-Mediated Folding of a Riboswitch from Induced-fit to Conformational Selection. J. Am. Chem. Soc. 2015;137:14075–14083. doi: 10.1021/jacs.5b09740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gong B, Chen JH, Bevilacqua PC, Golden BL, Carey PR. Competition between Co(NH(3)(6)3+ and Inner Sphere Mg2+ Ions in the HDV Ribozyme. Biochemistry. 2009;48:11961–11970. doi: 10.1021/bi901091v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Maderia M, Horton TE, DeRose VJ. Metal Interactions with a GAAA RNA Tetraloop Characterized by (31)P NMR and Phosphorothioate Substitutions. Biochemistry. 2000;39:8193–8200. doi: 10.1021/bi000140l. [DOI] [PubMed] [Google Scholar]
- 46.Harjes E, Kitamura A, Zhao W, Morais MC, Jardine PJ, Grimes S, Matsuo H. Structure of the RNA Claw of the DNA Packaging Motor of Bacteriophage phi29. Nucleic Acids Res. 2012;40:9953–9963. doi: 10.1093/nar/gks724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Leontis N, Stombaugh J, Westof E. The Non-Watson-Crick Base Pairs and their Associated Isostericity Matrices. Nucleic Acids Res. 2002;30:3497–3531. doi: 10.1093/nar/gkf481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yang Y, Qin Y, Ding Q, Bakhtina M, Wang L, Tsai MD, Zhong D. Ultrafast Water Dynamics at the Interface of the Polymerase-DNA Binding Complex. Biochemistry. 2014;53:5405–5413. doi: 10.1021/bi500810a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lobsiger S, Blaser S, Sinha RK, Frey HM, Leutwyler S. Switching on the Fluorescence of 2-aminopurine by Site-selective Microhydration. Nat. Chem. 2014;6:989–993. doi: 10.1038/nchem.2086. [DOI] [PubMed] [Google Scholar]
- 50.Musiani F, Rossetti G, Capece L, Gerger TM, Micheletti C, Varani G, Carloni P. Molecular Dynamics Simulations Identify Time Scale of Conformational Changes Responsible for Conformational Selection in Molecular Recognition of HIV-1 Transactivation Responsive RNA. J. Am. Chem. Soc. 2014;136:15631–15637. doi: 10.1021/ja507812v. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.