Abstract
Background
Cognitive impairment in humans with Alzheimer's disease (AD) and in animal models of Aβ-pathology can be ameliorated by treatments with the nuclear receptor peroxisome proliferator-activated receptor-gamma (PPARγ) agonists, such as rosiglitazone (RSG). Previously, we demonstrated that in the Tg2576 animal model of AD, RSG treatment rescued cognitive deficits and reduced aberrant activity of granule neurons in the dentate gyrus (DG), an area critical for memory formation.
Methods
We used a combination of mass spectrometry, confocal imaging, electrophysiology and split-luciferase assay and in vitro phosphorylation and Ingenuity Pathway Analysis.
Results
Using an unbiased, quantitative nano-LC-MS/MS screening, we searched for potential molecular targets of the RSG-dependent rescue of DG granule neurons. We found that S226 phosphorylation of fibroblast growth factor 14 (FGF14), an accessory protein of the voltage-gated Na+ (Nav) channels required for neuronal firing, was reduced in Tg2576 mice upon treatment with RSG. Using confocal microscopy, we confirmed that the Tg2576 condition decreased PanNav channels at the AIS of the DG, and that RSG treatment of Tg2576 mice reversed the reduction in PanNav channels. Analysis from previously published data sets identified correlative changes in action potential kinetics in RSG-treated T2576 compared to untreated and wildtype controls. In vitro phosphorylation and mass spectrometry confirmed that the multifunctional kinase GSK–3β, a downstream target of insulin signaling highly implicated in AD, phosphorylated FGF14 at S226. Assembly of the FGF14:Nav1.6 channel complex and functional regulation of Nav1.6-mediated currents by FGF14 was impaired by a phosphosilent S226A mutation. Bioinformatics pathway analysis of mass spectrometry and biochemistry data revealed a highly interconnected network encompassing PPARγ, FGF14, SCN8A (Nav 1.6), and the kinases GSK–3 β, casein kinase 2β, and ERK1/2.
Conclusions
These results identify FGF14 as a potential PPARγ-sensitive target controlling Aβ-induced dysfunctions of neuronal activity in the DG underlying memory loss in early AD.
Keywords: Alzheimer's disease, PPARgamma, Fibroblast growth factor 14, Mass spectrometry, Confocal microscopy
1. Background
Alzheimer's disease (AD) is the most common of debilitating dementias characterized by loss of memory and cognitive decline, and accounts for 50–60% of all overall cases of dementia in persons over 65 years of age (Francis et al., 1999). Epidemiological studies have demonstrated a link between insulin signaling, cognition, and risk for AD. Supporting this link, the class of insulin-sensitizing drug known as the thiazolidinediones, which includes rosiglitazone (RSG) and pioglitazone (PIO), and are potent and selective agonist of the peroxisome proliferator-activated receptor gamma (PPARγ), has shown efficacy in patients with MCI (Mild Cognitive Impairment) and concomitant insulin resistance (Risner et al., 2006; Harrington et al., 2007; Watson et al., 2005; Gad et al., 2015; Sato et al., 2011; Perez & Quintanilla, 2015). Likewise, RSG treatment rescues hippocampus-dependent cognitive function in the Tg2576 AD mouse model (Rodriguez-Rivera et al., 2011; Denner et al., 2012) by a mechanism that involves normalized intrinsic excitability and synaptic function of granule neurons in the dentate gyrus (DG), a critical location for new memory formation (Nenov et al., 2015). Dysregulation of the ERK-MAPK signaling pathway that subserves hippocampus-dependent cognition (Denner et al., 2012; Ahi et al., 2004; Atkins et al., 1998; Sweatt, 2004; McGaugh, 2000; Jahrling et al., 2014), the presynaptic VAMP-2 glutamate release machinery (Nenov et al., 2014), and voltage-gated ion channels (Nenov et al., 2015) may mediate the effect of RSG treatment on granule neuron function, but details of how PPARγ–agonism affects excitability in ion channel complexes are lacking. Because voltage-gated ion channels are the fundamental unit of excitability in the neuron, investigating how RSG treatment affects ion channel proteins may identify critical nodes in downstream pathways of Aβ pathology and provide useful information for therapeutic strategies (Rusinova et al., 2011; Imbrici et al., 2016; Catterall, 2012).
Quantitative phosphoproteomics can be combined with state-of-the-art mass spectrometry to identify potential signaling pathways that regulate post-translational modifications of cellular components (Baek et al., 2011; Berendt et al., 2010; Olsen et al., 2006). Recent studies have identified highly clustered areas containing phosphorylation sites on the voltage-gated Na + (Nav) Nav1.2 isoform, particularly at the I–II linker and the C-terminal intracellular regions (Baek et al., 2011; Berendt et al., 2010). These phosphorylation sites control channel gating, expression, and sub-cellular localization, which impact Nav channel function and consequently affect excitability (Berendt et al., 2010; Hsu et al., 2015, 2016; Wildburger et al., 2015). Previous studies have identified alterations in the phosphoproteome concomitant with neurologic disease: studies in humans with AD as well as an early onset mouse model of AD reveal significant alterations in global phosphorylation profiles with critical alterations in multiple cellular signaling pathways (Wang et al., 2013; Zahid et al., 2012), and phosphorylation as well as other post-translational modifications of Nav1.2 channels (James et al., 2015), such as methylation, are reciprocally modulated in epilepsy (Baek et al., 2014). These observations suggest that PTMs on Nav channel complexes may have important implications in the screening and diagnosis of brain disorders.
In addition to post-translational modifications, protein:protein interactions (PPIs) contribute to Nav channel function in native conditions (Wildburger et al., 2015). Nav channels are highly clustered at the axonal initial segment (AIS), where they exist in a large macromolecular complex with accessory modulatory proteins (Hsu et al., 2014). Among these, fibroblast growth factor 14 (FGF14) stands out for its specific modulatory roles on neuronal Nav channels. Recent studies highlight the importance of FGF14, a multivalent accessory protein of the Nav channel, in animal as well as human studies (Hsu et al., 2015, 2016; James et al., 2015; Ali et al., 2016; Alshammari et al., 2016a, b, c; Tempia et al., 2015; Ali et al., 2014; Shavkunov et al., 2013). Through direct monomeric binding to the Nav channel C-terminal tail, FGF14 forms a complex with the channel that is required for proper gating, expression and trafficking of the channel to the AIS as well as the control of neuronal excitability (Ali et al., 2014, 2016; Alshammari et al., 2016a; Laezza et al., 2007, 2009; Lou et al., 2005; Wang et al., 2000, 2002; Xiao et al., 2007; Goetz et al., 2009). Using the bioluminescence-based luciferase complementation assay (LCA) (Michnick et al., 2010; Remy & Michnick, 2006; Shavkunov et al., 2012; Villalobos et al., 2007, 2008), a PPI assay that quantitatively measures the complementation of reconstituted FGF14 and Nav1.6 channel complexes, we previously identified glycogen synthase kinase 3 (GSK–3), a multifunctional kinase dysregulated in Alzheimer's, depression and schizophrenia (Emamian, 2012; Jope et al., 2007; Jope & Roh, 2006; Liu et al., 2013; Budni et al., 2012; Scala et al., 2015; Maqbool et al., 2016; Morris & Berk, 2016; Provensi et al., 2016; Avila et al., 2010), as a key modulator of the FGF14:Nav1.6 complex. Inhibition of GSK–3 reduces the assembly of the FGF14:Nav channel complex, modifies FGF14-dependent regulation of Na+ currents, and induces dissociation and subcellular redistribution of the native FGF14:Nav channel complex (Shavkunov et al., 2013). Furthermore, CK2 is a known priming kinase of GSK–3, and recent studies have shown that inhibition of CK2 induces similar alterations in assembly and distribution of the FGF14:Nav complex (Hsu et al., 2016).
The significance of FGF14 extends beyond its regulation of Nav channels. In humans, a naturally occurring F145S mutation in the FGF14 gene is implicated in the development of spinocerebellar ataxia 27 (SCA27), a severe motor and cognitive neurodegenerative disorder (Laezza et al., 2009; Brusse et al., 2006; Wozniak et al., 2007; Coebergh et al., 2014), and SNPs in the Fgf14 gene have been associated with depression and schizophrenia (Hsu et al., 2014; Verbeek et al., 2012; Rodriguez-Murillo et al., 2014; Di Re et al., 2017). Recent transcriptomics studies from human post-mortem brains identify FGF14 as a new schizophrenia risk gene, a finding that is corroborated by fgf14−/− mice which exhibit phenotypes including changes in network activity, decreased gamma frequency, and cognitive impairments that mimic the human disease (Alshammari et al., 2016b). In addition, studies performed on the Alzheimer's Disease Neuroimaging Initiative (ADNI) WGS data have shown SNP rs17502999 in Fgf14 to be significantly associated with entorhinal cortex volume change in AD (Di Re et al., 2017; Yang et al., 2015). Given that these studies suggest that alterations in the sequence and/or expression of fgf14 are associated with neuropsychiatric and cognitive impairment symptoms seen in AD, investigating the signaling pathways that control the function of FGF14 is essential for discovering what role this protein might play in the pathogenesis of AD and other complex brain disorders associated with cognitive impairment.
Here we used a combined approach implementing large-scale phosphoproteomics, bioinformatics network analysis, mass spectrometry, and split-luciferase complementation assay (LCA) to identify relevant protein targets in an AD mouse model that is sensitive to the PPARγ agonist RSG. Among the hits, we identified Ser226 of FGF14 as a relevant target sensitive to RSG. Using confocal microscopy and image quantification, we show that the Tg2576 phenotype is characterized by a reduction in PanNav and the PanNav:FGF14 ratio at the AIS, which is reversed by treatment with RSG. Analysis of previously published electrophysiological data, which established a mechanistic link between cognitive improvement by thiazolidinediones and neuronal excitability in an AD mouse model (Nenov et al., 2015), provides correlative findings to the imaging studies. In addition, we show that phosphorylation of FGF14 at S226 by GSK–3 occurs in vitro, while the phosphosilent mutant FGF14S226A reduced complex formation with Nav1.6. These observations suggest a potential novel role for PPAR agonists in the modulation of an important signaling pathway that is dysregulated in an AD mouse model.
2. Methods
2.1. DNA constructs
All plasmids used in this study were previously described (Lou et al., 2005). The FGF14S226A-GFP construct was generated using QuikChange Lightning kits protocol (Agilent Technologies Santa Clara, CA) using the following primers FW: 5′–GGTGACGCCAGCTAAAAGCACAAGT RV: 5′–CCACTGCGGTCGATTTTCGTGTTCA, and confirmed by sequencing at the Molecular Genomics Core Facility at the University of Texas Medical Branch, Galveston, TX. U.S.A.
2.2. Chemicals
Triciribine (EMD Chemicals, San Diego, CA) was dissolved in 100% DMSO (Sigma-Aldrich, St. Louis, MO) to a working stock concentration of 20 mM, aliquoted, and stored at −20 °C. Further dilutions to a final concentration of 0.15% DMSO were made to be controls for experiments using 30 µM. Mass Spectrometry experiments were performed using LC-MS grade acetonitrile (ACN) and water (J.T. Baker, Philipsburg, NJ). Formic acid from Pierce (Rockford, IL) and iodoacetamide (IAA) and dithiotheritol (DTT) were all purchased from Sigma-Aldrich (St. Louis, MO). Sequencing grade endoproteinase Glu-C was purchased via Promega (Madison, WI). GSK–3 recombinant enzyme was purchased from New England Biolabs (Product P6040S, Ipswitch, MA).
2.3. Animal colonies
Hemizygous Tg2576 animals were bred and aged in the UTMB animal care facility by mating hemizygous Tg2576 males with B6SJL/F1J females (Jackson Laboratory, Cat#100012).Male and female 8 month old Tg2576 and WT mice were fed control or 30 mg/kg RSG or 50 mg/kg PIO diet (Bio-Serv) for 30 days ad libitum, as described previously (Rodriguez-Rivera et al., 2011; Miller et al., 2016). At 9.5 months of age, mice were sacrificed, transcardially perfused with 0.1 M phosphate buffer saline (PBS) with protease inhibitors and the brain removed for hippocampus dissection.
Fgf14−/− and fgf14+/+ male and female mice are maintained on an inbred C57/BL6J background with greater than ten generations of backcrossing to C57/BL6J. Animals were bred in the UTMB animal care facility: either heterozygous fgf14+/− males and females or, in few cases, homozygotes fgf14−/− males with fgf14+/− females; fgf14+/+ wild-type mice served as control. WT and fgf14−/− male and female mice were sacrificed at 4–6 months of age, unless otherwise stated.
The University of Texas Medical Branch operates in compliance with the United States Department of Agriculture Animal Welfare Act, the Guide for the Care and Use of Laboratory Animals, and IACUC approved protocols. Mice were housed, n ≤ 5 per cage, kept under a 12-h light/dark cycle with sterile food and water ad libitum. All genotypes described were confirmed by genotyping of the progeny using DNA extraction and PCR amplification following established protocols or conducted at Transnetyx Inc. (Cordova, TN).
2.4. Stable isotope labeling and phosphoproteomic mass spectrometry
Stable isotope labeling was used to quantify differential phosphorylation between Tg2576 and Tg2576 on an RSG diet as previously described (Nenov et al., 2014; Sadygov et al., 2010; Wu et al., 2013). Briefly, the DG region of the hippocampus was dissected under magnification with a dissecting microscope, homogenized in TRIzol (Life Technologies), and resuspended in guanidine. The sample was reduced, alkylated, digested with trypsin, and desalted with SepPak C18 cartridges. Lyophilized peptides were treated with immobilized trypsin (Applied Biosystems) in normal water (H2O16) for control-fed mice or heavy water (H2O18) for RSG-fed mice for trypsin-mediated exchange of oxygen atoms on the C-terminus. Desalted peptides were pooled, and phosphopeptides were enriched with TiO2 resin followed by strong cation exchange chromatography into 60 fractions. Each fraction was injected on a C18 peptide trap (Agilent), desalted, and separated on a reverse-phase nano-HPLC column with a 120 min linear gradient at 200 nL/min. Liquid chromatography-tandem mass spectrometry (nLC-MS/MS) experiments were performed with an LTQ linear ion trap MS (Thermo Finnigan) equipped with a nanospray source coupled to a ProteomX nano-HPLC system (Thermo Finnigan). The MS was operated in the data-dependent mode to trigger fragmentation of precursor ions with a neutral loss indicative of a phosphorylated peptide. The top three most intense ions in each MS survey scan were selected for zoom scans, followed by MS/MS. Each fraction was analyzed in triplicate. SEQUEST on the BioWorks 3.2 platform (Thermo Finnigan) was used to search for hits using filtering criteria of Sp ≥ 300, ΔCn ≥ 0.12, and Xcorr of 1.9, 2.0, and 3.0 for data from a singly, doubly, or triply charged precursor ions, respectively.
2.5. Peptide synthesis
The peptide FGF14 [KPGVTPSKSTSASAIMNGGK-NH2] was synthesized on a CEM Liberty Blue Discover automated microwave synthesizer (Matthews, NC). Solid phase coupling was completed on Wang resin using standard hydroxybenzotriazole/diisopropylcarbodiimide (HOBt/DIC) chemistries. Peptide resins were then cleaved in a solution of 95% trifluoroacetic acid (TFA)/2.5% H2O/2.5% triisopropylsilane (TIS) for 3 h, washed 3× in diethyl ether, and then allowed to dry overnight. Peptides were purified by reverse phase HPLC over a gradient of 20%–60% ACN:H2O over 60 min. Final product molecular weights were verified by MALDI-MS and purity was determined to be >95% by HPLC.
2.6. In vitro phosphorylation and sample preparation
In vitro phosphorylation of FGF14 was performed using recombinant GSK–3β expressed from a rabbit skeletal muscle cDNA library (Cat # P6040S) with methods from New England Biolabs. Briefly, 98 nmoles (25 µL of a 7.5 mg/mL stock solution) of FGF14 peptide [KPGVTPSKSTSASAIMNGGK-NH2] were incubated with 1× reaction buffer and 10,000 units (20 µL) of GSK–3 enzyme, and incubated for 2 h at 37 °C. Control studies were performed under identical conditions, except that experiments lacked the addition of the kinase to the reaction solution. Subsequent reduction of the peptide was performed using 10 mM DTT for 30 min at RT. The peptide was alkylated using 5 mM IAA (30 min at RT) and digested with sequencing grade Glu-C 1:00 (w/w) overnight at 37 °C. The digested samples were desalted using C8 Sep-Pak SPE columns (Waters, Milford, MA) and the eluent was lyophilized in vacuo.
2.7. Mass spectrometry
High resolution nanoLC-MS/MS analysis was performed using an Easy LC1000 (Proxeon Biosystems, Odense, Denmark) interfaced to a LTQ Orbitrap Elite (Thermo Fisher Scientific) as previously described (James et al., 2015). All LC gradient and MS data acquisition parameters were unchanged.
2.8. Data analysis for mass spectrometry
Mass spectrometry raw data files were imported into PEAKS database to search and identify phosphorylation sites. Parent ion mass tolerance was set to 10 ppm and fragment mass tolerance was set to 0.1 Da with a maximum two-miss cleavage allowance. Data was manually annotated using the MS-Product tool on the Protein Prospector website (prospector.ucsf.edu) to generate theoretical mass-to-charge ratio, m/z, values for all fragment ions as well as all phosphorylation sites identified by locating all present b and y ions in the sequence.
2.9. Immunohistochemistry
Performed as previously described (Shavkunov et al., 2013). Briefly, WT, Tg2576, and Tg2576 mice on a PIO diet were sacrificed, brain removed, hippocampus dissected and frozen in LN2, mounted with plastic 25 × 20 × 5 mm cryomolds with OCT embedding media (VWR, Atlanta, GA), and cut into 10 um sagittal slices and mounted on Superfrost® glass microscope slides (Fisher Scientific, Waltham, MA) and dried at room temperature. Sections were fixed by immersion in −20 °C EtOH for 5 min, followed by −20 °C acetone for 5 min and dried. After 30 min, sections were then washed with 1× PBS, blocked with 10% BSA/TBS for 1 h at RT, and incubated for 16 h with combinations of the following primary antibodies: mouse anti-FGF14 IgG1 (monoclonal, 1:400, UC Davis/NIH NeuroMab Facility, CA), mouse anti-PanNav IgG1 (monoclonal, 1:100, Sigma-Aldrich, MO), mouse anti-AnkyrinG IgG2b (monoclonal, 1:400, UC Davis/NIH NeuroMab Facility, CA). Samples were then washed (3× with PBS) and incubated with combinations of isotope-specific Alexa 488, 568, and 633 (1:400) antibodies for 1 h at RT, washed (6× with PBS), and mounted with Prolong Gold anti-fade reagent (Invitrogen, Carlsbad, CA). Confocal images were acquired with a Zeiss LSM-510 Meta confocal microscope with a Plan-Apochromat 63× (1.4 NA) oil-immersion objective. Multi-track acquisition was performed with excitation lines at 488 nm, 543 nm, and 633 nm for Alexa 488, Alexa 568, and Alexa 633 respectively. Respective emission filters were band-pass 505–530 nm, 560–615 nm, and low pass 650 nm. The optical slices were 0.8 µm; Z-stacks were collected at z-steps of 0.4 µm with a frame size of 1024 × 1024, pixel time of 2.5 µs, pixel size 0.1 × 0.1 µm (63×) and an 8 frame Kallman–averaging.
2.10. Image analysis
Confocal images were analyzed in ImageJ Fiji 1.48S. The gradient-based Robust Automatic Threshold Selection (RATS) algorithm (Wilkinson & Schut, 1998) was used on the FGF14 channel for segmentation with the following parameters: noise threshold = 200, lambda factor = 5, min leaf size = 205. After 1 pass of erode, particles were analyzed with the following parameters: size = 100-Infinity, circularity = 0.00–1.00. All particles outside the region of the DG were manually excluded from the selection. Statistics including particle area, mean intensity, and integrated density were exported to Excel for further analysis and binned by image.
2.11. Cell culture and transient transfections
HEK293 cells were maintained in DMEM (Invitrogen, Carlsbad, CA), supplemented with 10% fetal bovine serum, 100 U/mL penicillin, 100 µg/mL streptomycin, and 500 µg/mL G418 (Invitrogen), and incubated at 37 °C with 5% CO2. Cells were transfected at 90–100% confluency using Lipofectamine 2000 (Invitrogen), according to manufacturer's instructions. Prior to treatment with compounds, cells were maintained in serum-free, phenol-red free DMEM/F12 media.
2.12. Bioluminescence assays
Cells (~4.5 × 105 per 24-well plates-mm dish) were transiently cotransfected with pairs of plasmids or single plasmids as indicated by using Lipofectamine 2000 (Invitrogen, CA) according to the manufacturer's directions. For bioluminescence assays, transfected cells (200 µL of medium) were transferred to 96-well white-walled plates 24 h prior to luminescence readings, 48 h after transfection. Prior to treatment with compounds, cells were incubated 30 min in serum-free, phenol-red free DMEM/F12 media. Subsequently, growth media were replaced with media containing appropriate drugs or vehicle and cells incubated for the times indicated. Luminescence readings were performed with the Synergy® H4 Hybrid Multi-Mode Microplate Reader initiated by automated injection of 100 µL of substrate (in a 1:1 volume ratio) containing 1.5 mg/mL of d-luciferin (final concentration = 0.75 mg/mL), followed by 3 s of mild plate shaking, and measurements taken at 2 min intervals with 0.5 s integration time for a total duration of 30 min. Raw signal intensity was computed from the mean value of peak luminescence and two adjacent time points for the biological replicates for a particular dose-compound dataset. Normalized signal intensity was expressed as percentage of mean signal intensity relative to control treated with 0.5% DMSO. Independent untreated controls, controls treated with vehicle, and CLuc-FGF14 mono-transfected HEK293 cells were performed for each 96-well plate for technical verification and to obtain positive and negative controls.
2.13. In vitro electrophysiology experiments and data analysis
HEK-Nav1.6 cells transfected with FGF14-GFP or FGF14S226A-GFP were plated at low density on glass coverslips for 3–4 h, and then transferred to a recording chamber. Recordings were performed at room temperature using a MultiClamp 700B and Axopatch 200B amplifiers (Molecular Devices, Sunnyvale, CA). The extracellular recording solutions contained the following (mM): 140 NaCl, 3 KCl, 1MgCl2, 1 CaCl2, 10 HEPES, 10 glucose, pH 7.3; and the intracellular solution (mM): 130 CH3O3SCs, 1 EGTA, 10 NaCl, 10 HEPES, pH 7.3. Membrane capacitance and series resistance were estimated by the dial settings on the amplifier and electronically compensated by 70–80%. Data were acquired at 20 kHz and filtered at 2 kHz prior to digitization and storage. All experimental parameters were controlled by Clampex 9.2 software (Molecular Devices) and interfaced to the electrophysiological equipment using a Digidata 1322A analog-digital interface (Molecular Devices). Voltage-dependent inward currents for HEK-Nav1.6 cells were evoked by depolarizations to test potentials between−100 mV and + 60 mV from a holding potential of −70 mV followed by a voltage pre-step pulse of −120 mV (Nav1.6). Steady-state (fast) inactivation of Nav channels was measured with a paired-pulse protocol. From the holding potential, cells were stepped to varying test potentials between −120 mv (Nav1.6) and +20 mV (pre-pulse) prior to a test pulse to −20 mV. Current densities were obtained by dividing Na+ current (INa) amplitude by membrane capacitance. Current–voltage relationships were generated by plotting current density as a function of the holding potential. Conductance (GNa) was calculated by the following equation:
where INa is the current amplitude at voltage Vm, and Erev is the Na+ reversal potential. Steady-state activation curves were derived by plotting normalized GNa as a function of test potential and fitted using the Boltzmann equation:
where GNa,Max is the maximum conductance, Va is the membrane potential of half-maximal activation, Em is the membrane voltage and k is the slope factor. For steady-state inactivation, normalized current amplitude (INa/INa, Max) at the test potential was plotted as a function of prepulse potential (Vm) and fitted using the Boltzmann equation:
where Vh is the potential of half-maximal inactivation, Em is the membrane voltage, and k is the slope factor. Transient INa inactivation decay was estimated with standard exponential equation. Inactivation time constant (tau, τ) was fitted with the following equation:
where A1 and f1 are the amplitude and time constant, respectively. The variable C is a constant offset term along the Y axis. The goodness of fitting was determined by correlation coefficient (R), and the cutoff of R was set at 0.85. Data analysis was performed using Clampfit 9 software (Molecular Devices) and Origin 8.6 software (OriginLab Corporation, Northampton, MA). Results are expressed as mean ± SEM. Statistical significance was determined using Student's t-test.
2.14. Acute brain slice sectioning and whole-cell patch-clamp recording
This section refers to data that were previously collected (Nenov et al., 2015), then additional set of analyses were done seeking functional changes attributing to AIS reorganization and presented in this study. Slice preparation and whole-cell patch-clamp experiments were performed as described previously (Nenov et al., 2015). Briefly, acute hippocampal slices were sectioned from 9-mo-old wild-type littermate control and Tg2576 mice, treated with RSG. Brains were dissected and 250 µm horizontal hippocampal slices prepared with a vibratome VT1200 S (Leica, Buffalo Grove, IL) in iced, sucrose-based ACSF consisting of the following (mM): 56 NaCl, 100 sucrose, 2.5 KCl, 20 glucose, 5 MgCl2, 1 CaCl2, 30 NaHCO3, and 1.25 NaH2PO4; osmolarity 300–310, pH 7.4 was equilibrated by continuous bubbling with a mixture of 95% O2/5% CO2.
Standard ACSF consisting of the following (in mM): 130 NaCl, 3.5 KCl, 10 glucose, 1.5 MgCl2, 1.4 CaCl2, 23 NaHCO3, and 1.25 NaH2PO4; osmolarity 300–310, oxygenated and equilibrated to pH 7.4 with a mixture of 95% O2/5% CO2 at 31 °C was used as extracellular solution for whole-cell patch-clam recordings in slices. Borosilicate glass capillary electrodes (4–7 mΩ) were filled with intracellular solution of the following composition (in mM): 120 K-hydroxymethanesulfinate, 10 KCl, 10 HEPES, 10 glucose, 2 MgCl2, 0.5 EGTA, 2 MgATP, and 0.5 Na3GTP; osmolarity 280–290, pH 7.3, adjusted with KOH. Whole-cell somatic recordings were performed from visually identified dentate gyrus granule cells using an Axopatch 200A amplifier (Molecular Devices, Sunnyvale, CA), low-pass filtered at 5 kHz, and sampled at 10–20 kHz using a Digidata 1200 analog-to-digital interface and pCLAMP 7 acquisition software (Molecular Devices).
Evoked action potentials were induced by current steps (500 ms duration) of constant increments (10 pA per step) in granule cells recorded in current clamp mode. First action potential at the current threshold (minimal amount of current to induce action potential) was analyzed for time to rise and time to decay half-amplitude with ClampFit 9 (Molecular Devices). Observed differences between groups were quantified by Kruskal-Wallis with post hoc Dunn's multiple comparison tests. All data were analyzed with pCLAMP 9 (Molecular Devices) and GraphPad Prism 6 software (GraphPad Software, San Diego, CA).
2.15. Insulin responsiveness assay
Insulin responsiveness of synaptosomal preparations of frozen mouse brain were prepared as previously described (Franklin & Taglialatela, 2016). Briefly, 60 mg fresh frozen WT and fgf14−/− mouse brain were homogenized in SynPER buffer (Thermo Scientific) with protease and phosphatase inhibitors and centrifugated at 1230 ×g for 10 min. The supernatant was collected and centrifuged once more at 15000 ×g for 20 min. The pellet was resuspended in HEPES-buffered Krebs-like (HBK) buffer and divided into 2 tubes, unstimulated and insulin stimulated. All tubes received 8 mM ATP and insulin stimulation performed with 0.333 units/mL of U-100 insulin. All tubes were incubated at 37 °C for 15 min. Samples were pelleted and resuspended in 1× RIPA plus 1% protease and phosphatase inhibitors. Western blots were performed using P-IGF-IRβ (Cell Signaling) and IR (Cell Signaling) antibodies and imaged using the LI-COR Odyssey infrared imaging system.
2.16. Statistical analysis
Results were expressed as mean ± standard error (SEM). The statistical significance of observed differences among groups was determined by Student's t-test and one-way ANOVA, or corresponding non-parametric tests, Mann-Whitney rank sum and Kruskal-Wallis tests, respectively, based on the distribution of the samples underlying the populations. A p < 0.05 was regarded as statistically significant. Bonferroni or Dunnett's tests were used for ANOVA post-hoc analysis. Network analysis was performed using Ingenuity® Pathway Analysis by specifying target proteins and using the “Connect” function to discover mechanistic relationships between proteins in the target list.
3. Results
3.1. Rosiglitazone reduces FGF14 phosphorylation at S226 in Tg mice
In previous studies conducted in the Tg2576 animal model of AD, we provided evidence for the role of PPARγ agonist, rosiglitazone (RSG), in rescuing cognitive deficits associated with AD. At the cellular level, we identified relevant pathways sensitive to RSG in the Tg2576 mouse model and electrophysiological changes in the dentate gyrus (DG) circuitry that were restored to normal levels by the same RSG treatment (Nenov et al., 2015). In this study, we used unbiased quantitative nLC-MS/MS to identify differentially regulated phosphorylation sites that might be relevant for DG function. Tg2576 mice overexpressing human APP were fed control or RSG diet for one month starting at 8 months of age. After stable isotope labeling (16O control; 18O RSG diet) of tryptic peptides, samples were enriched for phosphopeptides and analyzed by nLC-MS/MS.
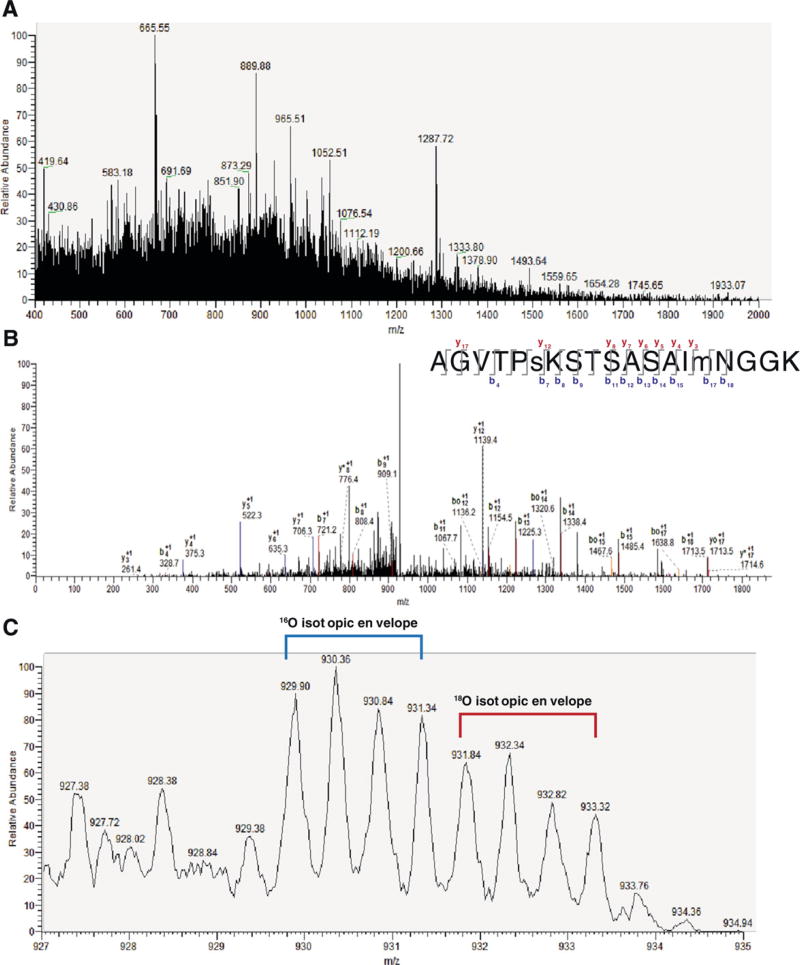
MS1 spectra identified peaks from m/z = 400–2000, which positively identified the doubly charged peptide AGVTPsKSTSASAImNGGK with high stringency criteria (XCorr = 2.935; ΔCn=0.0288), matching 19 of 54 b and y ions including the phosphorylated serine (Fig. 1A–B, Table 1). This peptide matched amino acids 221–239 of FGF14_MOUSE Fibroblast growth factor 14 (UniProt Acc. # P70379) phosphorylated on S226. Zoom scan of the precursor ion (Fig. 1C) gave the ratio of 18O/16O = 0.74, indicating that RSG decreased phosphorylation of this site compared to control diet. These results demonstrate that the phosphorylation level of S226 in FGF14 is up-regulated in Tg2576 and is decreased by RSG treatment of Tg2576. Thus, changes in the phosphorylation level of FGF14 might be part of the central PPARγ axis mediating cognitive dysfunction in the brain of Tg2576 mice.
Fig. 1.
Identification of peptide AGVTPsKSTSASAImNGGK and quantification of phosphorylation of S226 in FGF14 regulated by RSG diet. A) Complete MS spectrum at RT=143.46, m/z = 400–2000. B) MS2 of peptide AGVTPsKSTSASAImNGGK (s denotes phosphorylated serine; m denotes oxidized methionine), RT=143.57 with labeled b and y ion series. * denotes loss of NH3; o denotes loss of H2O. Inset, top right: labeled b/y ion series with 19 out of 54 potential ions identified. C) Zoom of A at RT=143.54 showing peptide AGVTPsKSTSASAImNGGK. The area under the 4 peaks of the 18O isotopic envelope cluster (red, RSG diet) was divided by the area of the 4 peaks of the 16O isotopic envelope cluster (blue, control diet). A ratio of 0.74 indicated that RSG decreased phosphorylation of S226 in Tg2576 mice. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Table 1.
Masses of ions in fragmentation spectrum for detected peptide AGVTPsKSTSASAImNGGK.
| Ion | Theoretical m/z (Da) | Experimental m/z (Da) | Error (Da) |
|---|---|---|---|
| y3 | 261.16 | 261.4 | −0.24 |
| b4 | 329.18 | 328.7 | 0.48 |
| y4 | 375.2 | 375.3 | −0.1 |
| y5 | 522.23 | 522.3 | −0.07 |
| y6 | 635.32 | 635.3 | 0.02 |
| y7 | 706.36 | 706.3 | 0.06 |
| b7 | 721.33 | 721.2 | 0.13 |
| y*8 | 776.36 | 776.4 | −0.04 |
| b8 | 808.36 | 808.4 | −0.04 |
| b9 | 909.41 | 909.1 | 0.31 |
| b11 | 1067.48 | 1067.7 | −0.22 |
| y12 | 1139.54 | 1139.4 | 0.14 |
| b12 | 1154.51 | 1154.5 | 0.01 |
| b13 | 1225.55 | 1225.3 | 0.25 |
| b14 | 1338.63 | 1338.4 | 0.23 |
| b15 | 1485.67 | 1485.4 | 0.27 |
| bo17 | 1638.72 | 1638.8 | −0.08 |
| b18 | 1713.75 | 1713.5 | 0.25 |
| y17 | 1714.77 | 1714.6 | 0.17 |
Values expressed as m/z to one significant figure for experimental masses.
denotes loss of NH3; o denotes loss of H2O.
3.2. Rosiglitazone reverses changes in Nav distribution in Tg2576 animals
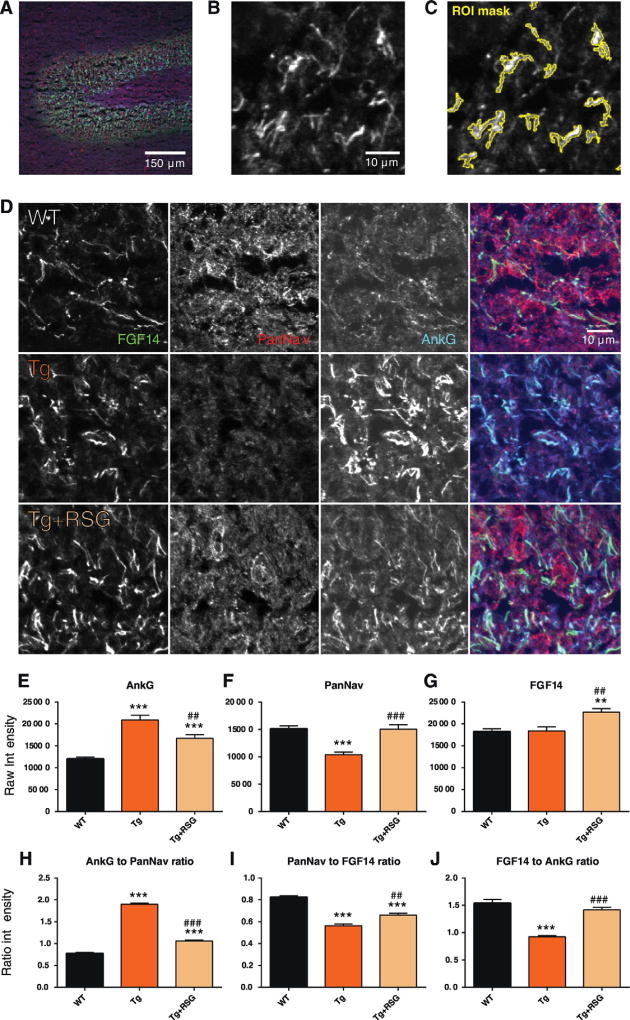
Confocal microscopy is an increasingly valuable tool to study distribution and localization of proteins in neuronal compartments. We examined the effects of diet supplemented with RSG on the distribution of FGF14, PanNav, and Ankyrin-G, a well-known marker of the AIS (Alshammari et al., 2016c; Grubb & Burrone, 2010; Bennett & Lambert, 1999), in the DG of Tg2576 animals, compared to untreated WT and Tg2576. After sacrifice of animals at 9.5 months of age, 10 µm sagittal slices of fresh frozen mouse hippocampus were sectioned for immunohistochemistry with confocal microscopy performed on the DG of the hippocampus (Fig. 2A–C).We observed that Tg2576 animals exhibited a substantial decrease in PanNav intensity in segmented regions corresponding to the AIS (−4731 ± 745 raw intensity mean difference, vs. WT, p < 0.001;WT N=1618 ROIs, 9 experiments, 2 animals; Tg N=574 ROIs, 6 experiments, 2 animals), which was reversed by treatment with RSG (+4619± 1101 raw intensity mean difference, vs. Tg, p < 0.001; Tg + RSG N = 1311 ROIs, 9 experiments, 1 animal) (Fig. 2D, F, Table 2). For Ankyrin-G, a corresponding increase in intensity was observed in Tg (+8891±1024 vs. WT, p < 0.001) and incompletely reversed by treatment with RSG (−4212 ± 1377 vs. Tg, p = 0.009) (Fig. 2E). Interestingly, while AIS in Tg2576 mice did not exhibit a significant change in FGF14 staining intensity compared to WT, treatment with RSG significantly increased FGF14 abundance compared to WT and Tg (+4309 ± 1265 vs. Tg, p = 0.005) (Fig. 2G). These phenotypes, which manifest as significant alterations in the ratios between FGF14, AnkG, and PanNav (Fig. 2H–J, Table 3), suggest that the hippocampi of Tg2576 animals are characterized by a shift in the PanNav and Ankyrin-G composition at the AIS of the DG, and that RSG partially reverses these alterations as well as increases the abundance of FGF14 at the DG. Further support for a role of PanNav, FGF14 and Ankyrin-G in the same AD animal model is provided by complementary confocal studies investigating the CA1 region of hippocampal neurons. In these studies, untreated Tg2576 animals and animals treated with another thiazolidinedione, pioglitazone, were compared to WT (Fig. S1). In this study, we also observed a similar suppression of PanNav in Tg and reversal by treatment with PIO with corresponding changes in the PanNav to FGF14 ratio (Fig. S1A, C, F); however, we observed that the CA1 region was in fact characterized by a relative suppression of FGF14 localization in treated Tg mice (Fig. S1D). These findings suggest that the role of PPARγ agonism in Tg2576 mice may involve a complex, region-dependent and agonist-specific mechanism.
Fig. 2.
Redistribution of Nav complex-associated proteins at the AIS in DG. Confocal images of the DG of the hippocampus in wild type (WT), Tg2576 (Tg), and Tg2576 + rosiglitazone (Tg +RSG) mice showing immunolabeling with mouse anti-FGF14, rabbit anti-PanNav, and mouse anti-AnkG antibodies. (A) 20× zoom of the WT DG treated with antibodies against AnkG (green), PanNav (red), FGF14 (blue) with scale bars shown. (B) 63× zoom, cropped, of the WT DG in A, showing the FGF14 channel (grayscale). (C) ROI generated by image segmentation and thresholding applied to B by ImageJ Fiji, shown in yellow outline (see methods). (D) 63× zoom, cropped, of WT, Tg, and Tg + RSG, treated with antibodies against FGF14 (green), PanNav (red), AnkG (blue). Far right: composite image of all three channels. (E–G) Raw intensities of AnkG, PanNav, and FGF14 channels for all groups. (H–J) Channel intensity ratios (AnkG/PanNav, PanNav/FGF14, FGF14/AnkG) for all groups. Statistical analysis: One-way ANOVA with Tukey's multiple comparisons test with single-pooled variance. Data mean ± SEM; versus WT: * p < 0.05, ** p < 0.01, *** p < 0.001; versus TG: # p < 0.05, ## p < 0.01, ### p < 0.001. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Table 2.
Channel intensity of Nav complex-associated proteins at the AIS for WT, Tg, and Tg + RSG dentate gyrus.
| Phenotype | Channel | Intensity | N (experiments) | p-Value, vs WT | p-Value, vs Tg |
|---|---|---|---|---|---|
| WT | PanNav | 15,130 ± 516 | 9 | ||
| WT | AnkG | 12,012 ± 422 | 9 | ||
| WT | FGF14 | 18,230 ± 680 | 9 | ||
| Tg | PanNav | 10,400 ± 477 | 6 | <0.0001 | |
| Tg | AnkG | 20,903 ± 1099 | 6 | <0.0001 | |
| Tg | FGF14 | 18,397 ± 927 | 6 | 0.8839 | |
| TG + RSG | PanNav | 15,019 ± 832 | 9 | 0.9111 | 0.0010 |
| TG + RSG | AnkG | 16,691 ± 853 | 9 | 0.0002 | 0.0091 |
| TG + RSG | FGF14 | 22,707 ± 825 | 9 | 0.0007 | 0.0047 |
Values expressed as mean ± SEM.
Table 3.
Channel pair ratios of Nav complex-associated proteins at the AIS forWT, Tg, and Tg+RSG dentate gyrus.
| Phenotype | Channel pair | Ratio | N | p-Value, vs WT | p-Value, vs Tg |
|---|---|---|---|---|---|
| WT | PanNav/FGF14 | 0.82 ± 0.01 | 9 | ||
| WT | AnkG/PanNav | 0.78 ± 0.02 | 9 | ||
| WT | FGF14/AnkG | 1.54 ± 0.07 | 9 | ||
| Tg | PanNav/FGF14 | 1.05 ± 0.04 | 6 | <0.0001 | |
| Tg | AnkG/PanNav | 1.90 ± 0.02 | 6 | <0.0001 | |
| Tg | FGF14/AnkG | 0.92 ± 0.02 | 6 | <0.0001 | |
| Tg + RSG | PanNav/FGF14 | 1.71 ± 0.05 | 9 | <0.0001 | 0.0017 |
| Tg + RSG | AnkG/PanNav | 1.06 ± 0.02 | 9 | <0.0001 | <0.0001 |
| Tg + RSG | FGF14/AnkG | 1.42 ± 0.04 | 9 | 0.1392 | <0.0001 |
Values expressed as mean ± SEM.
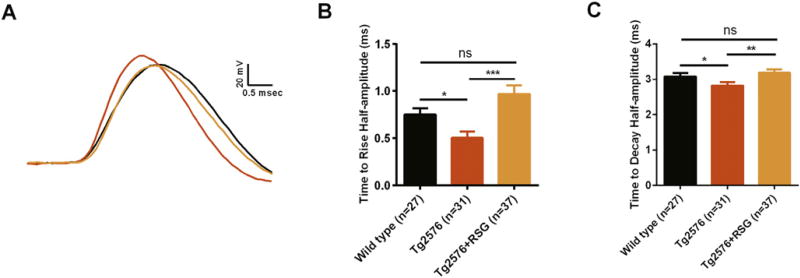
3.3. RSG treatment restores action potential time to rise and time to decay half-amplitude in Tg2576
Previously we showed that RSG diet ameliorates aberrant synaptic transmission and neuronal excitability in Tg2576 DG granule cells (Nenov et al., 2014, 2015). Treatment with RSG significantly restores repetitive firing in two types (type 1 and type 2) of electrophysiologically characterized DG granule cells via bi-directional modulation of spike after-depolarization (Nenov et al., 2014, 2015). Here, in order to characterize functional outcomes associated with changes in Tg2576 AIS expression profiles for PanNav channels, FGF14 and another AIS marker, Ankyrin-G, we assessed time to rise and time to decay half-amplitude of action potentials in two previously identified types of DG granule cells (Nenov et al., 2015; Tempia et al., 2015). Interestingly, we found reduction in time to rise and time to decay action potential half-amplitude in both type 1 and type 2 Tg2576 DG granule cells, and that treatment with RSG restores both parameters in each group to their wild type control values. Time to rise half-amplitude in Tg2576 DG granule cells (type 1 and type 2 combined) was significantly lower (0.5 ± 0.07 msec, n = 31) compared to wild type (0.75 ± 0.07 msec, n = 27; p < 0.05), and treatment with RSG resulted in significant increase of time to rise half-amplitude in Tg2576 (0.97± 0.09, n=36; p < 0.005) compared to untreated Tg2576 (Fig. 3A–B). Similarly, time to decay half-amplitude in Tg2576 DG granule cells was significantly lower (2.8 ± 0.1 msec, n = 31) compared to wild type (3.1 ± 0.09 msec, n = 27; p < 0.05), and treatment with RSG resulted in significant increase of time to decay half-amplitude in Tg2576 (3.2 ± 0.09, n = 36; p < 0.01) compared to untreated Tg2576 (Fig. 3A, C). Changes in the action potential rise and decay time have been attributed to shifts in the spike initiation site. Thus, the changes in action potential kinetics reported here might contribute to the aberrant excitability found in Tg2576 animals and reversed by RSG.
Fig. 3.
RSG affects time to rise and time to decay half-amplitude in Tg2576 dentate gyrus granule cells restoring them to the control level. (A) Representative traces of evoked action potential at current threshold for wild type, Tg2576 and RSG-treated Tg2576 dentate gyrus granule cells. (B) Bar graphs summarizing the effect of RSG diet on the time to rise half-amplitude of granule cells evoked action potential for wild type, Tg2576 and RSG-treated Tg2576 mice. (C) Bar graphs summarizing the effect of RSG diet on the time to decay half-amplitude of granule cells evoked action potential for wild type, Tg2576 and RSG-treated Tg2576 mice. “n” represents total number of neurons obtained for each group of animals: 27 neurons were recorded from 15 wild type, 31 from 16 Tg2576 and 37 from 21 Tg2576 on RSG diet mice, respectively. *p < 0.05, **p < 0.01, ***p < 0.005; Kruskal-Wallis with post hoc Dunn's multiple comparison tests.
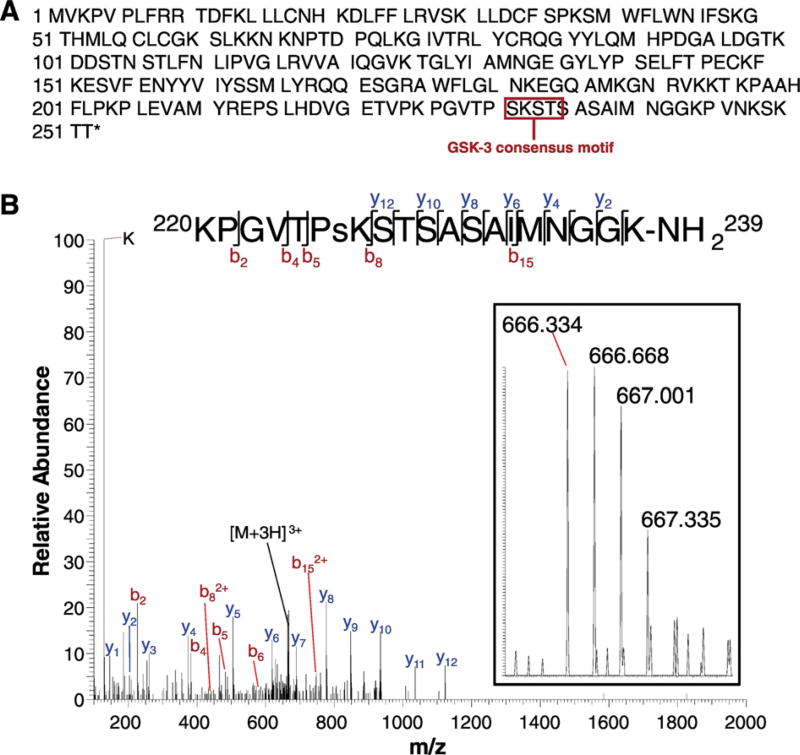
3.4. GSK–3 phosphorylates the FGF14 peptidomimetic KPGVTPSKSTSASAIMNGGK-NH2 at S226
Compelling evidence indicates that FGF14 is a relevant accessory protein of the Nav channel complex (Hsu et al., 2015; Ali et al., 2016; Alshammari et al., 2016a; Tempia et al., 2015; Shavkunov et al., 2013; Bosch et al., 2015), responsible for action potential initiation and propagation in neurons. In previous studies, we showed that FGF14 forms a complex with neuronal Nav1.6 channel, and this interaction is controlled by S/T kinases (Hsu et al., 2015, 2016; Shavkunov et al., 2013). The S226 residue of FGF14 is located in the C-terminal tail of the protein in a region enriched with other S/T sites (Fig. 4A). Although the C-tail is not part of the FGF14:Nav interface (Ali et al., 2016), its role in modulating this interaction has been previously postulated (Goetz et al., 2009). Thus, we hypothesized that changes in phosphorylation of residues at the FGF14 C-tail could affect the interaction with Nav channels. Our previous research has shown that GSK–3, a multifunctional kinase important for neuronal survival, cellular signaling, and stress response (Jope et al., 2007; Jope & Roh, 2006; Wu et al., 2013; Chen et al., 2011; Kim & Snider, 2011) and dysregulated in a number of AD and psychiatric disorders (Emamian, 2012; Jope et al., 2007; Jope & Roh, 2006; Liu et al., 2013; Budni et al., 2012; Scala et al., 2015; Maqbool et al., 2016; Morris & Berk, 2016; Provensi et al., 2016; Avila et al., 2010) affecting cognition, including depression and bipolar disorder (Gould et al., 2004; Koros & Dorner-Ciossek, 2007; Omata et al., 2011), critically modifies the interaction, assembly, localization, and activity of FGF14 and Nav channels (Hsu et al., 2015; James et al., 2015; Shavkunov et al., 2013). Furthermore, we observed that the C-tail of FGF14 contains a consensus GSK–3 phosphorylation motif (S/T)XXX(S/T), the first residue in this sequence corresponding to S226. (The phosphorylation motif for GSK–3 contains two serine/threonines: the first, at the P position, corresponds to the site phosphorylated by GSK–3, and the second, at the P + 4 position, corresponds to a priming phosphorylation site that increases the efficiency of GSK–3-dependent phosphorylation (Casaday et al., 2004; St-Denis et al., 2015; ter Haar et al., 2001)). Based on these observations, we conducted an in vitro phosphorylation assay using recombinant GSK–3β with a FGF14 peptide with the sequence KPGVTPSKSTSASAIMNGGK. After reduction, alkylation, and digestion with Glu-c, high-resolution MS detected the peak of m/z 666.334 (theoretical mass = 666.334; ppm < 1.5), corresponding to the [M+3H]3+ peak. The observed fragmentation pattern is consistent with KPGVTPsKSTSASAIMNGGK-NH2, corresponding to phosphorylation of S226 of full-length FGF14, with site-determining ions b5, , and y12 (Fig. 4B, Table 4). The absence of any phosphorylated y-ion species excludes the S/T in the post-motif sequence STSASAIMNGGK, and the flanking b ions (b5, ) excludes T224 as a possible phosphothreonine, leaving S226 as the only phosphorylated species. These observations confirm that GSK–3 phosphorylates FGF14 at S226 in vitro, supporting the evidence that changes in the phosphorylation state of this serine in FGF14 in native systems may be driven by endogenous signaling pathways.
Fig. 4.
Mass spectrometry identifying in vitro phosphorylation of FGF14 at S226. (A) Full sequence of FGF14-1b (H. sapiens), highlighting the GSK–3 consensus phosphorylation motif (SXXXS) beginning at S226. (B) Phosphorylation of peptide KPGVTPsKSTSASAIMNGGK-NH2 at S226 by GSK–3β. Main figure: MS/MS fragmentation spectra showing the b and y ion series and the dominant peak [M+3H]3+ (theoretical mass: 666.34; experimental mass: 666.34; ppm < 15). Top: The b and y ion series for KPGVTPsKSTSASAIMNGGK, with the site-determining ions b5, b82+, and y12. Inset: high resolution MS spectra showing the parent [M+3H]3+ peak (666.334).
Table 4.
Masses of ions in fragmentation spectrum for peptide KPGVTPsKSTSASAIMNGGK-NH2.
| Ion | Theoretical m/z (Da) | Experimental m/z (Da) | Error (Da) | |
|---|---|---|---|---|
| K | 129.10 | 129.10 | 0.00 | |
| y1 | 146.13 | 146.13 | 0.00 | |
| y2 | 203.15 | 203.15 | 0.00 | |
| b2 | 226.16 | 226.15 | −0.01 | |
| y3 | 260.17 | 260.17 | 0.00 | |
| y4 | 374.21 | 374.21 | 0.00 | |
| b4 | 382.24 | 382.25 | 0.01 | |
|
|
438.23 | 438.23 | 0.00 | |
| b5 | 483.29 | 483.29 | 0.00 | |
| y5 | 505.26 | 505.26 | 0.00 | |
| b6 | 580.35 | 580.36 | 0.01 | |
| y6 | 618.34 | 618.34 | 0.00 | |
| [M + 3H]3+ | 666.34 | 666.34 | 0.00 | |
| y7 | 689.38 | 689.38 | 0.00 | |
|
|
746.87 | 746.88 | 0.01 | |
| y8 | 776.41 | 776.41 | 0.00 | |
| y9 | 847.45 | 847.45 | 0.00 | |
| y10 | 934.48 | 934.48 | 0.00 | |
| y11 | 1035.53 | 1035.53 | 0.00 | |
| y12 | 1122.56 | 1122.56 | 0.00 |
Values expressed as m/z to two significant figures.
3.5. A phosphosilent mutant of FGF14 impairs Nav complementation
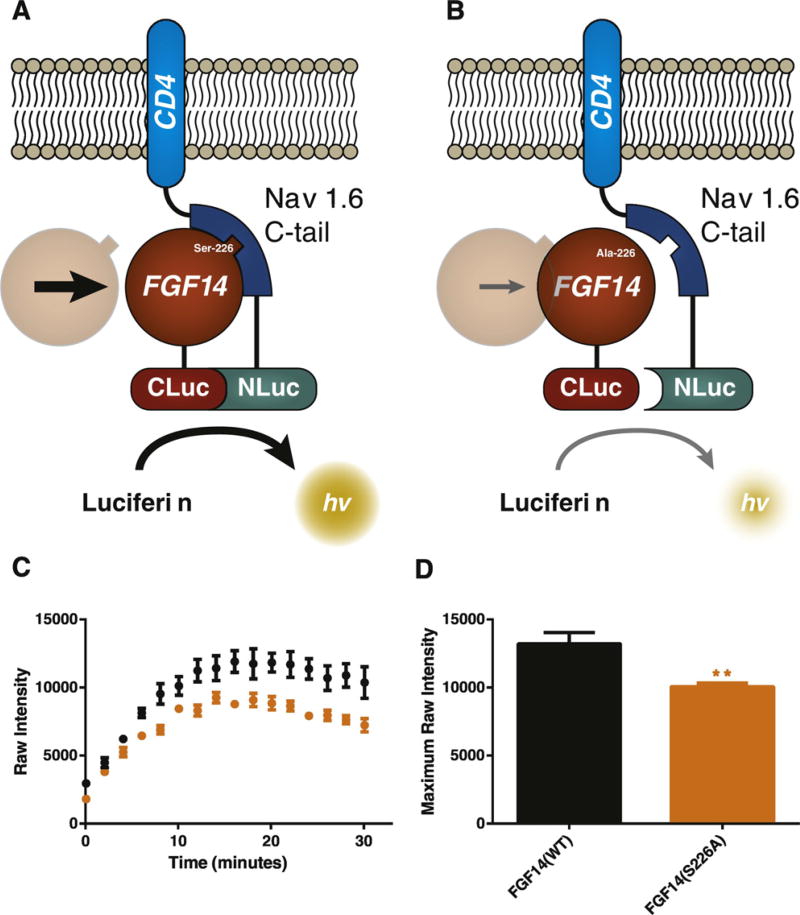
To assess whether a S226A phosphosilent mutation affects the interaction of FGF14 with Nav1.6, we reconstituted the FGF14:Nav1.6 C-tail complex using LCA (Fig. 5A–B) (Hsu et al., 2015, 2016; Ali et al., 2016; Ali et al., 2014; Shavkunov et al., 2013; Shavkunov et al., 2012; Shavkunov et al., 2015). HEK293 cells were transiently transfected with CLuc-FGF14 and CD4-Nav1.6-C-tail-NLuc plasmids expressing the coding sequence of FGF14 and the C-terminal tail of Nav1.6 fused to the single transmembrane domain protein CD4, respectively. While transient transfection with CD4-Nav1.6-C-tail-NLuc and CLuc-FGF14 gave rise to a robust luminescence signal, replacement of the construct with CLuc-FGF14S226A reduced the stability of the complex leading to a 24% (FGF14: 1.32 ± 0.08 × 104 RLU, n = 6 vs. 1.01 ± 0.03 × 104 RLU, n = 6 in FGF14S226A, p = 0.005) decrease in luminescence (Fig. 5C–D). These results indicate that the S226 residue influences the assembly of the FGF14:Nav1.6 complex.
Fig. 5.
Using the split-luciferase assay to assess the phenotype of a S226A phosphosilent mutant. (A) The split-luciferase assay produces a luminescence readout based on the complementation of N-terminal and C-terminal fragments of Photinus luciferase enzyme, which are attached to two interacting proteins (in this case, FGF14 and a CD4-Nav1.6-C-tail construct). Upon interaction of FGF14 and the C-tail of Nav1.6, the C-terminal and N-terminal fragments reconstitute into functional luciferase enzyme, producing a robust luminescence response when luciferin substrate is added. (B) Lack of a serine/threonine site, as in the FGF14S226A mutation, is projected to weaken the interaction between FGF14 and Nav1.6, resulting in a diminished luminescence response compared to wild type FGF14. (C) Real-time raw luminescence response of CLuc-FGF14/CD4-Nav1.6-C-tail-NLuc (black) vs CLuc-FGF14S226A/CD4-Nav1.6-C-tail-NLuc (orange). (D) Quantification of maximum observed intensity over the course of (C). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
3.6. FGF14S226A phosphosilent mutant reduces Nav1.6 mediated fast transient sodium currents
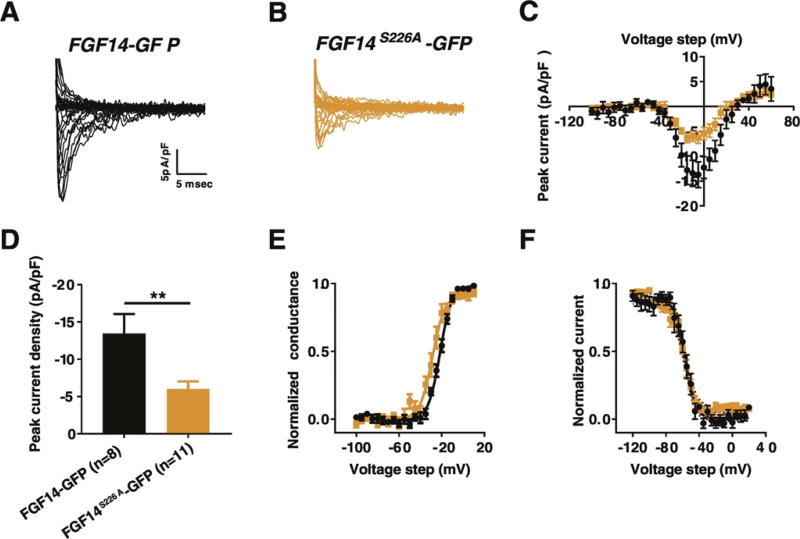
To test whether the S226A phosphosilent mutation alters FGF14-regulated Nav1.6 current, we used whole-cell patch-clamp electrophysiology recording from HEK293-Nav1.6 cells transiently transfected with FGF14-GFP or FGF14S226A-GFP (Fig. 6A–B). We show that the peak current densities were significantly lower in the FGF14S226A-GFP compared to FGF14-GFP transfected HEK293-Nav1.6 cells across multiple voltage steps (Fig. 6C). For instance, at –10 mV voltage step, the peak current density in HEK293-Nav1.6 cell expressing FGF14S226A-GFP was −6.02 ± 1.02pA/pF (n = 11) compared to −13.43 ± 2.63pA/pF (n = 8) in HEK293-Nav1.6 cell expressing FGF14-GFP (p < 0.01 with Student's t-test, Fig. 6D, Table 5). Subsequent analysis of Nav1.6 channel biophysical properties showed that the S226A mutation has no significant effect on both the voltage dependence of activation and steady-state inactivation (Fig. 6E and F, Table 5).
Fig. 6.
Comparison of the effects of FGF14-GFP and FGF14S226A-GFP on sodium currents in HEK293-Nav1.6 cells. (A, B) Representative traces of fast transient sodium currents recorded from HEK293-Nav1.6 cells transiently transfected with (A) FGF14-GFP or (B) FGF14S226A-GFP. (C) Current-voltage relationship of peak Na+ current densities recorded from HEK293-Nav1.6 transiently expressing either FGF14-GFP (black) or FGF14S226A-GFP (orange). (D) Bar graphs represents peak Na+ current density at voltage step of –10 mV recorded from HEK293-Nav1.6 transiently expressing either FGF14-GFP (black) or FGF14S226A-GFP (orange). (E) Voltage-dependence of Nav current activation and (F) steady-state inactivation plotted against membrane potential (mV) and fitted using the Boltzmann equation were obtained from HEK293-Nav1.6 transiently expressing either FGF14-GFP (black) or FGF14S226A-GFP (orange). **p < 0.01; Student t-test. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Table 5.
Comparison of peak current densities at voltage step of −10 mV, activation and steady-state inactivation for HEK 293-Nav1.6 cells transfected with FGF14-GFP or FGF14S226AGFP.
| Condition | Peak density (pA/pF) | Activation V1/2 (mV) | kact (mV) | Inactivation V1/2 (mV) | kinact (mV) |
|---|---|---|---|---|---|
| FGF14-GFP | −13.43 ± 2.63 (n = 8) | −21.5 ± 1 (n = 8) | 5 ± 0.25 (n = 8) | −57.2 ± 2.3 (n = 8) | 4.9 ± 0.57 (n = 8) |
| FGF14S226A-GFP | −6.02 ± 1.02** (n = 11) | −26.35 ± 2.8 (n = 11) | 5.5 ± 0.77 (n = 11) | −60.4 ± 2.3 (n = 11) | 5.8 ± 0.61 (n = 11) |
p < 0.01.
3.7. FGF14, Nav1.6, and GSK–3 form a highly-interconnected network that includes PPARγ
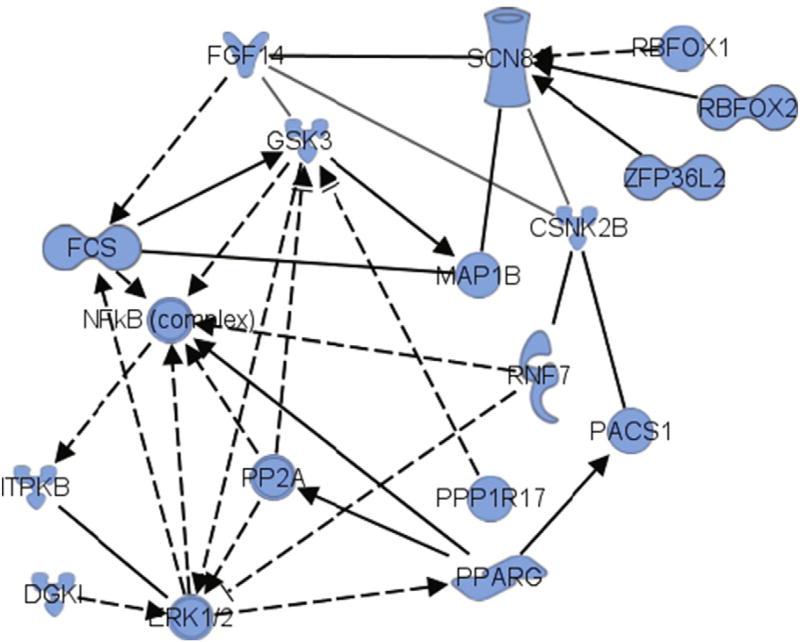
Based on the relationships described in this work including the biological agonism of PPARγ with RSG leading to in vivo phosphorylation of the S226 site on FGF14, the role of this site in association with Nav1.6 (SCN8A), and the ability of GSK–3 to phosphorylate this site, we used Ingenuity Pathway Analysis that included PPARγ as a mediator of RSG action. This analysis, based on prior mass spectrometry and biochemistry data on the effect of RSG in Tg2576 animals (Denner et al., 2012; Ahi et al., 2004; Atkins et al., 1998; Sweatt, 2004; McGaugh, 2000; Jahrling et al., 2014), revealed an extensive network including these known mediators in addition to the kinases casein kinase 2β (CSNK2B) and ERK1/2 (Fig. 7). This analysis further underscored putative mechanisms of these mediators in regulating signaling important for PPARγ agonism in cognitive enhancement in the Tg2576 mouse model of AD.
Fig. 7.
Bioinformatics using Ingenuity Pathways Analysis™ of proteins central to RSG regulation of a key network connecting FGF14, SCN8A, GSK, PPARG and ERK1/2. Key: FGF14, fibroblast growth factor 14; SCN8A, voltage-gated sodium channel Na1.6; GSK–3, glycogen synthase kinase 3; CSNK2B, casein kinase 2 beta; PPARG, peroxisome proliferator-activated receptor gamma; ERK1/2, p42/p44 mitogen activated protein kinase; NFκB, nuclear factor kappa B. (See www.ingenuity.com for a more detailed description of network statistical calculations, molecule naming and symbol descriptions).
3.8. WT and fgf14−/− demonstrate no significant difference in insulin sensitivity
There has been growing evidence that signaling through the PI3K-Akt-GSK3 pathway (Lee et al., 2011; Ong et al., 2010; Chaussade et al., 2007) may be one of the significant endpoints of insulin response; however, other studies have also demonstrated functional redundancy in some of these endpoints, and inhibition of one or more specific endpoints in the PI3K pathway was demonstrated to not be sufficient to suppress insulin signaling (Chaussade et al., 2007). Furthermore, thiazolidinedione drugs are insulin sensitizers and deficits in the insulin pathway are co-morbid with cognitive impairment in AD (Gad et al., 2015; Sato et al., 2011; Perez & Quintanilla, 2015; Rodriguez-Rivera et al., 2011; Jahrling et al., 2014; Xu et al., 2014; Cowley et al., 2012; Bosco et al., 2011; Escribano et al., 2010; Escribano et al., 2009). Based on this evidence, we posited that genetic deletion of FGF14 could affect the insulin pathway. To examine the role of FGF14 in insulin response in the brain, we examined the insulin sensitivity of WT and fgf14−/− mouse brain synaptosomal extract through detection of insulin receptor (IR) activation (Franklin & Taglialatela, 2016).We show that in the presence of insulin, the level of phospho-IR/IR, a measure of IR activation, is not significantly affected in fgf14−/− compared to WT (Fig. S2), demonstrating that the absence of fgf14 is not sufficient to suppress IR activation in mouse brain synaptosomal extract.
4. Discussion
In the current study, we used a combination of mass spectrometry, bioinformatics, confocal microscopy, patch-clamp electrophysiology, and protein binding studies to explore a novel mechanism of action of PPARγ agonists in an AD mouse model. Specifically, we show that in the DG of the Tg2576 mouse model, the level of phosphorylation of FGF14 at Ser226 is elevated, and is reduced with a one-month treatment regimen of the PPARγ agonist rosiglitazone (RSG) (Fig. 1). Confocal microscopy of the DG demonstrates alterations in Nav and AnkG localization in Tg2576 mice, with a large increase in the AnkG/PanNav ratio; this change was reversed in Tg2576 mice treated with RSG (Fig. 2). Interestingly, RSG further increased FGF14 localization in the AIS of the DG compared to untreated Tg2576 mice (Fig. 2G), which is associated with normalization of the Nav/FGF14 and FGF14/AnkG ratios (Fig. 2I–J). Furthermore, initial studies suggest that the effect of PPARγ agonists on the hippocampus is region-dependent; confocal microscopy of the CA1 region of the hippocampus demonstrates a relative suppression of FGF14 localization in the AIS of CA1 neurons in PIO-treated Tg2576 mice (Fig. S1). We note that the DG and CA1 differ substantially in terms of neurogenerative potential, cell type populations, and resistance to oxidative stress, all which may significantly affect downstream responses to PPAR agonism (Harrington et al., 2007; Jahrling et al., 2014; Nenov et al., 2014; Rusinova et al., 2011). Analysis of electrophysiological parameters from previously collected studies support an RSG-dependent effect on action potential time to rise and time to decay half-amplitude opposing Tg2576-dependent alterations in the action potential waveform (Fig. 3). These changes suggest that in DG granule cells of Tg2576 mice, rather than originating in the AIS, the action potential might originate at a subcellular location that is spatially closer to the soma (Foust et al., 2010; Popovic et al., 2011). Such changes could be the result of redistribution and loss of expression of Nav channel and functionally relevant accessory proteins at the AIS (Fig. 2). Treatment with RSG restores both the expression and distribution of Nav channels at the AIS and action potential properties, providing a tighter correlation between structural and functional phenotypes. These molecular changes likely contribute to deficits in memory formation associated with signal processing in the DG circuit in AD.
Mass spectrometry confirms that the putative GSK–3 phosphorylation site (S/TXXXS/T) at S226 of FGF14 is phosphorylated in vitro by GSK-3β, which supports a possible regulatory role for this site in the context of GSK–3 signaling (Fig. 4). Protein binding studies show that the phosphosilent mutant FGF14S226A reduces the formation of the FGF14:Nav1.6 complex (Fig. 5), and concomitantly reduced FGF14 function in mediating Nav1.6-mediated currents (Fig. 6). This suggests that phosphorylation sites on the FGF14 C-tail may act as an excitability-modulating switch sensitive to changes in GSK-3 activity. We posited that a potential upstream target of the GSK-3 pathway relevant for AD was insulin. However, the absence of FGF14 was not sufficient to suppress insulin sensitivity in mouse brain extract (Fig. S2), suggesting that GSK-3-dependent phosphorylation of FGF14 in the context of AD is not downstream of the insulin pathway or that compensatory mechanisms might correct for the deficit. Finally, bioinformatics pathway analysis revealed a highly-interconnected network encompassing PPARγ, FGF14, SCN8A (Nav 1.6), and the kinases GSK–3, casein kinase 2β, and ERK1/2. Overall, these findings support a highly inter-connected FGF14-Nav1.6-GSK–3 network that includes PPARγ, suggesting that signaling within this interactome may be crucial nodes in the mechanism of PPARγ agonists in AD mice (Fig. 7).
Previous research has shown that pharmacological treatment with PPARγ agonists, such as RSG or PIO, rescues cognitive impairment in a subset of AD patients (Risner et al., 2006; Harrington et al., 2007; Watson et al., 2005; Sato et al., 2011; Perez & Quintanilla, 2015). Analogously, a dietary regimen enriched in RSG in the well-studied Tg2576 AD mouse model, which exhibits early AD symptoms, DG synaptic alterations, and significant aberrations in neuronal excitability after 9 months of age, restores memory function and rescues synaptic deficits and intrinsic firing properties of granule cells in the DG (Nenov et al., 2014, 2015), the brain area required for memory formation and most affected in early phases of AD (Rodriguez-Rivera et al., 2011). An ERK-MAPK-dependent genetic program initiated by stimulation of PPARγ has been proposed as a plausible candidate pathway and signaling axis that globally restores neuronal activity by normalizing the expression of critical target molecules, ultimately affecting intracellular signaling (Harrington et al., 2007; Denner et al., 2012; Jahrling et al., 2014; Escribano et al., 2009; Mandrekar-Colucci et al., 2012). In addition, we and others have demonstrated additional beneficial effects of RSG, indicating links between insulin signaling, cognition and AD (Xu et al., 2014; Bosco et al., 2011; Hong et al., 2012; Mattos et al., 2012; Li et al., 2011). However, we currently lack mechanistic insights into how this network operates and affects key molecular targets. This study provides a potential new mechanistic link between PPARγ signaling and the target molecules that might mediate the changes in intrinsic excitability of granule cells in the DG circuit.
Together, these findings support a perspective that impaired hippocampus-dependent cognitive function in Tg2576 is centered on changes in ionic conductances, such as Nav channels, triggering early-stage deficits in memory loss (Wang et al., 2016) which prompted us to analyze candidate molecules that may complement axonal function such as FGF14. Previously, we and several other research groups have demonstrated the vital role that FGF14 has in controlling biophysical properties and trafficking/targeting of Nav channels in in vitro and in vivo mouse models (Hsu et al., 2016; Shavkunov et al., 2013; Laezza et al., 2007, 2009; Goetz et al., 2009; Shavkunov et al., 2012).
This PPI, established through direct binding of FGF14 with the intracellular C-tail of Nav channels, is required for neuronal excitability, synaptic transmission and plasticity. In mice, targeted disruption of FGF14 produces severe ataxia, paroxysmal dystonia, and cognitive impairment (Wang et al., 2002; Wozniak et al., 2007), with neurons that exhibit severe impairments in synaptic transmission and plasticity, and neuronal excitability (Alshammari et al., 2016a,b; Xiao et al., 2007; Wozniak et al., 2007). Significantly, the assembly and trafficking of FGF14 itself is controlled via a GSK–3 dependent signaling pathway that may critically regulate excitability through PPI at the level of the Nav complex (Hsu et al., 2015; Shavkunov et al., 2013). Additionally, the distribution of FGF14 is Nav channel-dependent; deletion of the Nav1.6 α subunit in mouse Purkinje neurons markedly increases FGF14 levels in the AIS, in parallel with increased expression of Nav1.1 and β-IV-spectrin (Xiao et al., 2013). As a result, through interactions with macromolecular complexes at the AIS, FGF14 plays an important role in the regulation of Nav channel activity. Correspondingly, deletion, mutations or genetic variants of fgf14 have been linked to brain disorders and diseases and the list is growing. First identified as the source of the hereditary spinocerebellar ataxia 27 (Brusse et al., 2006; van Swieten et al., 2003), a debilitating childhood-onset condition characterized by postural tremor, slowly progressive ataxia, and cognitive deficits, SNPs in fgf14 have been identified recently in a genome-wide study performed on a Dutchmajor depressive disorder cohort (Verbeek et al., 2012). Additionally, copy variants of fgf14 have been found in a pediatric population admitted for recurrent unprovoked seizures (Olson et al., 2014), IGF-1 induced neuroprotection against oxidative stresses is accompanied by downregulation of fgf14 (Genis et al., 2014), and correlative changes in FGF14 and markers of GABAergic inhibitory transmission have been found in transcriptomics from post-mortem schizophrenia patients (Alshammari et al., 2016b).
It is believed that the primary mode of action of FGF14 is to control channel gating and channel localization at the AIS, with direct influence on neuronal firing (Laezza et al., 2007, 2009; Lou et al., 2005). In recent studies we discovered through a HTS of small molecule inhibitors that the FGF14:Nav1.6 complex assembly is under the control of a network of kinases that exert modulatory actions on complex assembly, with some kinase inhibitors stabilizing the PPI complex and others instead promoting its dissociation (Hsu et al., 2015, 2016; Shavkunov et al., 2013). As a result, these studies revealed a network of kinases centered on GSK–3 that modulate the FGF14:Nav channel complex bidirectionally (Hsu et al., 2015). As shown previously, pharmacological inhibition and/or silencing of GSK–3 suppresses the interaction of FGF14 with Nav1.6 and Nav1.2 channels, modulates the functional activity of FGF14 on Na + currents, and control the sub-cellular distribution and co-localization of the two proteins in the axon and the somatodendritic compartment, phenotypes that might contribute to the role of GSK–3 in regulating neuronal excitability in the brain (James et al., 2015; Shavkunov et al., 2013; Laezza et al., 2007, 2009). These studies highlight the potential of using protein complementation assays to assess signaling pathways, a method that has been extended in the current study through the identification of a novel pathway that controls FGF14 function.
Primary sequence analysis of FGF14 reveals numerous predicted S/T phosphorylation sites, some throughout the core domain of FGF14 and others grouped within the C-terminal tail (FGF14T224 to FGF14T231) (Fig. 3A). Additionally, the S226 site corresponds to a consensus GSK–3 motif, (S/TXXXS/T), shown to be phosphorylated in vitro in this current study, which supports a possible regulatory role for this site in the context of GSK–3 signaling (Fig. 3B). Thus, by impacting the affinity of the Nav channels for its chaperone protein, FGF14, S/T phosphorylation of FGF14 is likely to be a critical nexus in neuronal excitability. Interestingly, S/T phosphorylation sites that match a GSK–3 motif are also found on Nav; the T1966 residue in the C-tail of Nav1.2 has been validated in vivo and also lies in a putative GSK–3 phosphorylation motif (Baek et al., 2011; Farghaian et al., 2011). This residue is conserved in Nav1.6 and might also be a candidate for the effect of GSK–3 phosphorylation that we observed in this study. Furthermore, we have recently shown that FGF14 is phosphorylated at S228 and S230 by casein kinase 2 (CK2), an important intracellular kinase that mediates the association of Nav and AnkG, and is known to be a significant priming kinase for GSK–3 phosphorylation (Hsu et al., 2016; Bréchet et al., 2008; Wildburger & Laezza, 2012). Given the role of CK2 and the GSK–3 consensus motif, we propose that S228 and S230 may be priming sites for GSK–3 dependent phosphorylation of S226, and that CK2 may act in concert with GSK–3 in fine-tuning regulation of FGF14 distribution and trafficking. In general, GSK–3 phosphorylation sites that could influence the assembly of the FGF14:Nav1.6 complex could be a direct link to extracellular signaling. This signaling could be relayed through tyrosine-kinase receptors, G-protein coupled receptors or other signal transduction pathways upstream of the PI3K/Akt/GSK–3 pathways, providing candidate targets for homeostatic regulation of firing and mechanistic links to brain circuitry hyperactivity found in brain disorders. Therefore, signaling through the FGF14/Nav1.6/GSK–3 axis may be a critical component of the mechanism of action of RSG on AD mouse models through a network of kinases related to GSK-3 (Fig. 7).
It has been shown that action potential waveform may significantly change due to availability and activity of Nav channels at the somato-axonal region in neurons (Khaliq & Raman, 2006). This prompted us to test whether changes in expression patterns for AnkG, Nav and FGF14 in the AIS of DG granule cells could correlate with modifications in action potential waveform in Tg2576. We measured time to half-amplitude of action potential, which reflects changes related to the action potential initiation site (Foust et al., 2010; Popovic et al., 2011). We found reductions in both rise time and decay time to half-amplitude in Tg2576, and that treatment with RSG can reverse the phenotypes to the level of wild type control (Fig. 5). Thus, aberrant expression of AnkG, Nav and FGF14 in Tg2576 granule cells could represent the molecular signature of change in the action potential waveform, which might associate with a shift in the action potential initiation site along the somato-axonal axis (Khaliq & Raman, 2006).
What could be the overarching model of changes in FGF14 phosphorylation downstream of PPARγ agonist treatment, and how is this model recapitulated in the framework of the AD mouse model? Under normal conditions, FGF14 might serve as a kinase scaffold or a phosphorylation relay that translates signaling information into changes in firing. Phosphorylation of FGF14 could result in changes in binding and/or recruitment of other proteins to the Nav channel complex with effects on channel targeting and biophysical properties. As such, FGF14 phosphorylation could act as a fast-acting, coincidence detector of signals that can be rapidly translated into neuronal firing. There is a growing interest in understanding how neuronal intrinsic excitability is changed in AD and whether restoring this function might have a therapeutic value. Recent studies indicate that neuronal firing undergoes early alterations in the pathogenesis of AD, preceding the deficits in synaptic transmission (Scala et al., 2015). Our discovery is in line with this new line of research. Future studies will focus on the in vivo effects of FGF14 site-specific mutants to probe the effects of modulating specific post-translational modifications on the assembly and function of the AIS, and whether fgf14−/− phenocopies RSG in the Tg2576 mouse model. The phosphorylation of FGF14 at S226 may regulate binding with Nav channels, but may also control PPI with other chaperone proteins and as well as convey signaling pathways that target Nav channels to the AIS. Because binding of FGF14 to Nav channels affects the localization and biophysical properties of the channels, we hypothesize that post-translational modification of FGF14 induced by signaling pathways may explain some of the deficits seen in the Tg2576 mouse model. Further research is needed to elucidate the exact mechanisms by which the Tg2576 mouse model produces the effects observed, and if PPARγ agonists such as RSG or related compounds can be beneficial in other animal models of dementia.
Recently, FGF14 SNPs have been associated with volumetric changes in the entorhinal cortex of patients with AD (Yang et al., 2015; Wu et al., 2016). Animal studies furthering investigating the role of FGF14 in the brain will be important translational models to elucidate this emerging role of FGF14 in humans afflicted with AD and other numerous brain disorders, including schizophrenia (Alshammari et al., 2016b; Gadelha et al., 2012; Brzustowicz et al., 1999; Need et al., 2009) and major depressive disorder (Verbeek et al., 2012), to which critical links with FGF14 have been found.
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.expneurol.2017.05.005.
Supplementary Material
Acknowledgments
Funding
This work was supported by a National Institutes of Health Grant R01MH095995 from NIMH (FL) R01MH111107-01A1, the pre-doctoral Kempner fellowship (WJH), the Wiktor W. Nowinski Endowment Fund travel award (WJH) and the Sealy Center for Molecular Medicine, UTMB (WJH), 1R01AG042890 (GT), R01 AG031859 (KTD and LD), and a kind gift from the Mohn Foundation (KTD and LD) and from Emmett and Miriam McCoy Foundation (LD), post-doctoral Kempner fellowship (OF).
Abbreviations
- ACN
acetonitrile
- AD
Alzheimer's disease
- AIS
axonal initial segment
- CK2
casein kinase 2
- DG
dentate gyrus
- DIC
diisopropylcarbodiimide
- DMEM
Dulbecco's modified Eagle medium
- DTT
dithiotheritol
- FGF14
fibroblast growth factor 14
- GSK–3
glycogen synthase kinase 3
- HEPES
(4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid) (buffer)
- HOBt
hydroxybenzotriazole
- HEK293
human embryonic kidney 293 (cells)
- IAA
iodoacetamide
- IACUC
Institutional Animal Care and Use Committee
- LCA
split-luciferase complementation assay
- MALDI
matrix-assisted laser desorption/ionization
- MIPS
monoisotopic precursor selection
- PIO
pioglitazone
- PPARγ
PPARgamma peroxisome proliferator-activated receptor-gamma
- PPI
protein:protein interaction
- PTM
post-translational modification
- RIPA
radioimmunoprecipitation assay (buffer)
- RSG
rosiglitazone
- SPE
solid phase extraction
- TFA
trifluoroacetic acid
- TIS
triisopropylsilane
Footnotes
Declarations
All animal procedures were performed in accordance to the University of Texas Medical Branch at Galveston, Institutional Animal Care and Use Committee (IACUC) approved protocols. The University of Texas Medical Branch at Galveston operates in compliance with the United States Department of Agriculture Animal Welfare Act, the Guide for the Care and Use of Laboratory Animals, and IACUC approved protocols.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
WJH performed confocal microscopy, protein interaction assays, statistical analysis, and prepared the manuscript. NW, CL performed mass spectrometry and data interpretation. SJH, RGS, LD performed and/or interpreted mass spectrometry analysis of Tg2576. MNN, OF performed and interpreted electrophysiological data. AS performed protein interaction assays. BCC, JSR provided synthesis support for the FGF14 peptide and contributed to mass spectrometry and in vitro phosphorylation data interpretation. WFF performed synaptosomal binding assays and data interpretation. IC provided animal care, sacrifice, and mouse brain sample preparation. KTD, GT, LD, FL provided laboratory space and funding and assisted in experimental design. WJH, FL wrote the manuscript. WJH, NW, FL, OF, MN edited the manuscript. FL supervised all data acquisition, interpretation and experimental design. RGS contributed software for quantifying relative protein ratios from unit resolution mass spectrometers.
References
- Ahi J, Radulovic J, Spiess J. The role of hippocampal signaling cascades in consolidation of fear memory. Behav. Brain Res. 2004;149:17–31. doi: 10.1016/s0166-4328(03)00207-9. [DOI] [PubMed] [Google Scholar]
- Ali S, Shavkunov A, Panova N, Stoilova-McPhie S, Laezza F. Modulation of the FGF14:FGF14 homodimer interaction through short peptide fragments. CNS Neurol. Disord. Drug Targets. 2014;13:1559–1570. doi: 10.2174/1871527313666141126103309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali SR, Singh AK, Laezza F. Identification of Amino Acid Residues in Fibroblast Growth Factor 14 (FGF14) Required for Structure-Function Interactions with Voltage-gated Sodium Channel Nav1.6. May 20. J Biol Chem. 2016;291(21):11268–11284. doi: 10.1074/jbc.M115.703868. http://dx.doi.org/10.1074/jbc.M115.703868 Epub 2016 Mar 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alshammari MA, Alshammari TK, Nenov MN, Scala F, Laezza F. Fibroblast Growth Factor 14 Modulates the Neurogenesis of Granule Neurons in the Adult Dentate Gyrus. Dec;53. Mol Neurobiol. 2016a;53(10):7254–7270. doi: 10.1007/s12035-015-9568-5. Epub 2015 Dec 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alshammari TK, Alshammari MA, Nenov MN, Hoxha E, Cambiaghi M, Marcinno A, James TF, Singh P, Labate D, Li J, et al. Genetic deletion of fibroblast growth factor 14 recapitulates phenotypic alterations underlying cognitive impairment associated with schizophrenia. Transl. Psychiatry. 2016b;6:e806. doi: 10.1038/tp.2016.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alshammari MA, Alshammari TK, Laezza F. Improved methods for fluorescence microscopy detection of macromolecules at the axon initial segment. Front. Cell. Neurosci. 2016c;10:5. doi: 10.3389/fncel.2016.00005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkins CM, Selcher JC, Petraitis JJ, Trzaskos JM, Sweatt JD. The MAPK cascade is required for mammalian associative learning. Nat. Neurosci. 1998;1:602–609. doi: 10.1038/2836. [DOI] [PubMed] [Google Scholar]
- Avila J, Wandosell F, Hernandez F. Role of glycogen synthase kinase-3 in Alzheimer's disease pathogenesis and glycogen synthase kinase-3 inhibitors. Expert. Rev. Neurother. 2010;10:703–710. doi: 10.1586/ern.10.40. [DOI] [PubMed] [Google Scholar]
- Baek JH, Cerda O, Trimmer JS. Mass spectrometry-based phosphoproteomics reveals multisite phosphorylation on mammalian brain voltage-gated sodium and potassium channels. Semin. Cell Dev. Biol. 2011;22:153–159. doi: 10.1016/j.semcdb.2010.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baek JH, Rubinstein M, Scheuer T, Trimmer JS. Reciprocal changes in phosphorylation and methylation of Mammalian brain sodium channels in response to seizures. J. Biol. Chem. 2014;289:15363–15373. doi: 10.1074/jbc.M114.562785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett V, Lambert S. Physiological roles of axonal ankyrins in survival of premyelinated axons and localization of voltage-gated sodium channels. J. Neurocytol. 1999;28:303–318. doi: 10.1023/a:1007005528505. [DOI] [PubMed] [Google Scholar]
- Berendt FJ, Park KS, Trimmer JS. Multisite phosphorylation of voltage-gated sodium channel alpha subunits from rat brain. J. Proteome Res. 2010;9:1976–1984. doi: 10.1021/pr901171q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosch MK, Carrasquillo Y, Ransdell JL, Kanakamedala A, Ornitz DM, Nerbonne JM. Intracellular FGF14 (iFGF14) is required for spontaneous and evoked firing in cerebellar Purkinje neurons and for motor coordination and balance. J. Neurosci. 2015;35:6752–6769. doi: 10.1523/JNEUROSCI.2663-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosco D, Fava A, Plastino M, Montalcini T, Pujia A. Possible implications of insulin resistance and glucose metabolism in Alzheimer's disease pathogenesis. J. Cell. Mol. Med. 2011;15:1807–1821. doi: 10.1111/j.1582-4934.2011.01318.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bréchet A, Fache MP, Brachet A, Ferracci G, Baude A, Irondelle M, Pereira S, Leterrier C, Dargent B. Protein kinase CK2 contributes to the organization of sodium channels in axonal membranes by regulating their interactions with ankyrin G. J. Cell Biol. 2008;183:1101–1114. doi: 10.1083/jcb.200805169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brusse E, de Koning I, Maat-Kievit A, Oostra BA, Heutink P, van Swieten JC. Spinocerebellar ataxia associated with a mutation in the fibroblast growth factor 14 gene (SCA27): a new phenotype. Mov. Disord. 2006;21:396–401. doi: 10.1002/mds.20708. [DOI] [PubMed] [Google Scholar]
- Brzustowicz LM, Honer WG, Chow EW, Little D, Hogan J, Hodgkinson K, Bassett AS. Linkage of familial schizophrenia to chromosome 13q32. Am. J. Hum. Genet. 1999;65:1096–1103. doi: 10.1086/302579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budni J, Lobato KR, Binfare RW, Freitas AE, Costa AP, Martin-de-Saavedra MD, Leal RB, Lopez MG, Rodrigues AL. Involvement of PI3K, GSK-3beta and PPARgamma in the antidepressant-like effect of folic acid in the forced swimming test in mice. J. Psychopharmacol. 2012;26:714–723. doi: 10.1177/0269881111424456. [DOI] [PubMed] [Google Scholar]
- Casaday RJ, Bailey JR, Kalb SR, Brignole EJ, Loveland AN, Cotter RJ, Gibson W. Assembly protein precursor (pUL80.5 homolog) of simian cytomegalovirus is phosphorylated at a glycogen synthase kinase 3 site and its downstream “priming” site: phosphorylation affects interactions of protein with itself and with major capsid protein. J. Virol. 2004;78:13501–13511. doi: 10.1128/JVI.78.24.13501-13511.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catterall WA. Voltage-gated sodium channels at 60: structure, function and pathophysiology. J. Physiol. 2012;590:2577–2589. doi: 10.1113/jphysiol.2011.224204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaussade C, Rewcastle GW, Kendall JD, Denny WA, Cho K, Gronning LM, Chong ML, Anagnostou SH, Jackson SP, Daniele N, Shepherd PR. Evidence for functional redundancy of class IA PI3K isoforms in insulin signalling. Biochem. J. 2007;404:449–458. doi: 10.1042/BJ20070003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Zhou W, Chen PC, Gaisina I, Yang S, Li X. Glycogen synthase kinase-3beta is a functional modulator of serotonin-1B receptors. Mol. Pharmacol. 2011;79:974–986. doi: 10.1124/mol.111.071092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coebergh JA, van de Putte DE, Fransen, Snoeck IN, Ruivenkamp C, van Haeringen A, Smit LM. A new variable phenotype in spinocerebellar ataxia 27 (SCA 27) caused by a deletion in the FGF14 gene. Eur. J. Paediatr. Neurol. 2014;18:413–415. doi: 10.1016/j.ejpn.2013.10.006. [DOI] [PubMed] [Google Scholar]
- Cowley TR, O'Sullivan J, Blau C, Deighan BF, Jones R, Kerskens C, Richardson JC, Virley D, Upton N, Lynch MA. Rosiglitazone attenuates the age-related changes in astrocytosis and the deficit in LTP. Neurobiol. Aging. 2012;33:162–175. doi: 10.1016/j.neurobiolaging.2010.02.002. [DOI] [PubMed] [Google Scholar]
- Denner LA, Rodriguez-Rivera J, Haidacher SJ, Jahrling JB, Carmical JR, Hernandez CM, Zhao Y, Sadygov RG, Starkey JM, Spratt H, et al. Cognitive enhancement with rosiglitazone links the hippocampal PPARgamma and ERK MAPK signaling pathways. J. Neurosci. 2012;32:16725–16735a. doi: 10.1523/JNEUROSCI.2153-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Re J, Wadsworth P, Laezza F. Intracellular fibroblast growth factor 14: emerging risk factor for brain disorders. Front. Cell. Neurosci. 2017;11:103. doi: 10.3389/fncel.2017.00103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emamian ES. AKT/GSK3 signaling pathway and schizophrenia. Front. Mol. Neurosci. 2012;5:33. doi: 10.3389/fnmol.2012.00033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escribano L, Simon AM, Perez-Mediavilla A, Salazar-Colocho P, Del Rio J, Frechilla D. Rosiglitazone reverses memory decline and hippocampal glucocorticoid receptor down-regulation in an Alzheimer's disease mouse model. Biochem. Biophys. Res. Commun. 2009;379:406–410. doi: 10.1016/j.bbrc.2008.12.071. [DOI] [PubMed] [Google Scholar]
- Escribano L, Simon AM, Gimeno E, Cuadrado-Tejedor M, Lopez de Maturana R, Garcia-Osta A, Ricobaraza A, Perez-Mediavilla A, Del Rio J, Frechilla D. Rosiglitazone rescues memory impairment in Alzheimer's transgenic mice: mechanisms involving a reduced amyloid and tau pathology. Neuropsychopharmacology. 2010;35:1593–1604. doi: 10.1038/npp.2010.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farghaian H, Chen Y, Fu AW, Fu AK, Ip JP, Ip NY, Turnley AM, Cole AR. Scapinin-induced inhibition of axon elongation is attenuated by phosphorylation and translocation to the cytoplasm. J. Biol. Chem. 2011;286:19724–19734. doi: 10.1074/jbc.M110.205781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foust A, Popovic M, Zecevic D, McCormick DA. Action potentials initiate in the axon initial segment and propagate through axon collaterals reliably in cerebellar Purkinje neurons. J. Neurosci. 2010;30:6891–6902. doi: 10.1523/JNEUROSCI.0552-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis PT, Palmer AM, Snape M, Wilcock GK. The cholinergic hypothesis of Alzheimer's disease: a review of progress. J. Neurol. Neurosurg. Psychiatry. 1999;66:137–147. doi: 10.1136/jnnp.66.2.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin W, Taglialatela G. A method to determine insulin responsiveness in synaptosomes isolated from frozen brain tissue. J. Neurosci. Methods. 2016;261:128–134. doi: 10.1016/j.jneumeth.2016.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gad ES, Zaitone SA, Moustafa YM. Pioglitazone and exenatide enhance cognition and downregulate hippocampal beta amyloid oligomer and microglia expression in insulin-resistant rats. Can. J. Physiol. Pharmacol. 2015:1–10. doi: 10.1139/cjpp-2015-0242. [DOI] [PubMed] [Google Scholar]
- Gadelha A, Ota VK, Cano JP, Melaragno MI, Smith MA, de Jesus, Mari J, Bressan RA, Belangero SI, Breen G. Linkage replication for chromosomal region 13q32 in schizophrenia: evidence from a Brazilian pilot study on early onset schizophrenia families. PLoS One. 2012;7:e52262. doi: 10.1371/journal.pone.0052262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genis L, Davila D, Fernandez S, Pozo-Rodrigalvarez A, Martinez-Murillo R, Torres-Aleman I. Astrocytes require insulin-like growth factor I to protect neurons against oxidative injury. F1000Res. 2014;3:28. doi: 10.12688/f1000research.3-28.v1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goetz R, Dover K, Laezza F, Shtraizent N, Huang X, Tchetchik D, Eliseenkova AV, Xu CF, Neubert TA, Ornitz DM, et al. Crystal structure of a fibroblast growth factor homologous factor (FHF) defines a conserved surface on FHFs for binding and modulation of voltage-gated sodium channels. J. Biol. Chem. 2009;284:17883–17896. doi: 10.1074/jbc.M109.001842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould TD, Einat H, Bhat R, Manji HK. AR-A014418, a selective GSK-3 inhibitor, produces antidepressant-like effects in the forced swim test. Int. J. Neuropsychopharmacol. 2004;7:387–390. doi: 10.1017/S1461145704004535. [DOI] [PubMed] [Google Scholar]
- Grubb MS, Burrone J. Building and maintaining the axon initial segment. Curr. Opin. Neurobiol. 2010;20:481–488. doi: 10.1016/j.conb.2010.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ter Haar E, Coll JT, Austen DA, Hsiao H-M, Swenson L, Jain J. Structure of GSK3[beta] reveals a primed phosphorylation mechanism. Nat. Struct. Mol. Biol. 2001;8:593–596. doi: 10.1038/89624. [DOI] [PubMed] [Google Scholar]
- Harrington WW, C SB, J GW, N OM, J GB, D CL, W RO, M CL, D MI. The effect of PPARalpha, PPARdelta, PPARgamma, and PPARpan agonists on body weight, body mass, and serum lipid profiles in diet-induced obese AKR/J mice. PPAR Res. 2007;2007:97125. doi: 10.1155/2007/97125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong S, Xin Y, HaiQin W, GuiLian Z, Ru Z, ShuQin Z, HuQing W, Li Y, Yun D. The PPARgamma agonist rosiglitazone prevents cognitive impairment by inhibiting astrocyte activation and oxidative stress following pilocarpine-induced status epilepticus. Neurol. Sci. 2012;33:559–566. doi: 10.1007/s10072-011-0774-2. [DOI] [PubMed] [Google Scholar]
- Hsu WC, Nilsson CL, Laezza F. Role of the axonal initial segment in psychiatric disorders: function, dysfunction, and intervention. Front. Psychiatry. 2014;5:109. doi: 10.3389/fpsyt.2014.00109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu WC, Nenov MN, Shavkunov A, Panova N, Zhan M, Laezza F. Identifying a kinase network regulating FGF14:Nav1.6 complex assembly using split-luciferase complementation. PLoS One. 2015 Feb 6;10(2):e0117246. doi: 10.1371/journal.pone.0117246. http://dx.doi.org/10.1371/journal.pone.0117246 eCollection 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu WC, Scala F, Nenov MN, Wildburger NC, Elferink H, Singh AK, Chesson CB, Buzhdygan T, Sohail M, Shavkunov AS, Panova NI, Nilsson CL, Rudra JS, Lichti CF, Laezza F. Free PMC ArticleCK2 activity is required for the interaction of FGF14 with voltage-gated sodium channels and neuronal excitability. FASEB J. 2016 Jun;30(6):2171–2186. doi: 10.1096/fj.201500161. http://dx.doi.org/10.1096/fj.201500161 Epub 2016 Feb 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imbrici P, Liantonio A, Camerino GM, De Bellis M, Camerino C, Mele A, Giustino A, Pierno S, De Luca A, Tricarico D, et al. Therapeutic approaches to genetic ion channelopathies and perspectives in drug discovery. Front. Pharmacol. 2016;7:121. doi: 10.3389/fphar.2016.00121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahrling JB, Hernandez CM, Denner L, Dineley KT. PPARgamma recruitment to active ERK during memory consolidation is required for Alzheimer's disease-related cognitive enhancement. J. Neurosci. 2014;34:4054–4063. doi: 10.1523/JNEUROSCI.4024-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James TF, Nenov MN, Wildburger NC, Lichti CF, Luisi J, Vergara F, Panova-Electronova NI, Nilsson CL, Rudra JS, Green TA, et al. The Nav1.2 channel is regulated by GSK3. Biochim. Biophys. Acta. 2015;1850:832–844. doi: 10.1016/j.bbagen.2015.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jope RS, Roh MS. Glycogen synthase kinase-3 (GSK3) in psychiatric diseases and therapeutic interventions. Curr. Drug Targets. 2006;7:1421–1434. doi: 10.2174/1389450110607011421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jope RS, Yuskaitis CJ, Beurel E. Glycogen synthase kinase-3 (GSK3): inflammation, diseases, and therapeutics. Neurochem. Res. 2007;32:577–595. doi: 10.1007/s11064-006-9128-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khaliq ZM, Raman IM. Relative contributions of axonal and somatic Na channels to action potential initiation in cerebellar Purkinje neurons. J. Neurosci. 2006;26:1935–1944. doi: 10.1523/JNEUROSCI.4664-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim WY, Snider WD. Functions of GSK-3 signaling in development of the nervous system. Front. Mol. Neurosci. 2011;4:44. doi: 10.3389/fnmol.2011.00044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koros E, Dorner-Ciossek C. The role of glycogen synthase kinase-3beta in schizophrenia. Drug News Perspect. 2007;20:437–445. doi: 10.1358/dnp.2007.20.7.1149632. [DOI] [PubMed] [Google Scholar]
- Laezza F, Gerber BR, Lou JY, Kozel MA, Hartman H, Craig AM, Ornitz DM, Nerbonne JM. The FGF14 (F145S) mutation disrupts the interaction of FGF14 with voltage-gated Na+ channels and impairs neuronal excitability. J. Neurosci. 2007;27:12033–12044. doi: 10.1523/JNEUROSCI.2282-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laezza F, Lampert A, Kozel MA, Gerber BR, Rush AM, Nerbonne JM, Waxman SG, Dib-Hajj SD, Ornitz DM. FGF14 N-terminal splice variants differentially modulate Nav1.2 and Nav1.6-encoded sodium channels. Mol. Cell. Neurosci. 2009;42:90–101. doi: 10.1016/j.mcn.2009.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CC, Huang CC, Hsu KS. Neuropharmacology. Vol. 61. Elsevier Ltd; England: 2011. Insulin promotes dendritic spine and synapse formation by the PI3K/Akt/mTOR and Rac1 signaling pathways; pp. 867–879. [DOI] [PubMed] [Google Scholar]
- Li YF, Cheng YF, Huang Y, Conti M, Wilson SP, O'Donnell JM, Zhang HT. Phosphodiesterase-4D knock-out and RNA interference-mediated knock-down enhance memory and increase hippocampal neurogenesis via increased cAMP signalling. J. Neurosci. 2011;31:172–183. doi: 10.1523/JNEUROSCI.5236-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu RJ, Fuchikami M, Dwyer JM, Lepack AE, Duman RS, Aghajanian GK. GSK-3 inhibition potentiates the synaptogenic and antidepressant-like effects of subthreshold doses of ketamine. Neuropsychopharmacology. 2013;38:2268–2277. doi: 10.1038/npp.2013.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lou JY, Laezza F, Gerber BR, Xiao M, Yamada KA, Hartmann H, Craig AM, Nerbonne JM, Ornitz DM. Fibroblast growth factor 14 is an intracellular modulator of voltage-gated sodium channels. J. Physiol. 2005;569:179–193. doi: 10.1113/jphysiol.2005.097220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandrekar-Colucci S, Karlo JC, Landreth GE. Mechanisms underlying the rapid peroxisome proliferator-activated receptor-gamma-mediated amyloid clearance and reversal of cognitive deficits in a murine model of Alzheimer's disease. J. Neurosci. 2012;32:10117–10128. doi: 10.1523/JNEUROSCI.5268-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maqbool M, Mobashir M, Hoda N. Pivotal role of glycogen synthase kinase-3: a therapeutic target for Alzheimer's disease. Eur. J. Med. Chem. 2016;107:63–81. doi: 10.1016/j.ejmech.2015.10.018. [DOI] [PubMed] [Google Scholar]
- Mattos RT, Bosco AA, Nogueira-Machado JA. Rosiglitazone, a PPAR-gamma agonist, inhibits VEGF secretion by peripheral blood mononuclear cells and ROS production by human leukocytes. Inflamm. Res. 2012;61:37–41. doi: 10.1007/s00011-011-0386-6. [DOI] [PubMed] [Google Scholar]
- McGaugh JL. Memory - a century of consolidation. Science. 2000;287:248–251. doi: 10.1126/science.287.5451.248. [DOI] [PubMed] [Google Scholar]
- Michnick SW, Ear PH, Landry C, Malleshaiah MK, Messier V. A toolkit of protein-fragment complementation assays for studying and dissecting large-scale and dynamic protein-protein interactions in living cells. Methods Enzymol. 2010;470:335–368. doi: 10.1016/S0076-6879(10)70014-8. [DOI] [PubMed] [Google Scholar]
- Miller WR, Fox RG, Stutz SJ, Lane SD, Denner L, Cunningham KA, Dineley KT. PPARγ agonism attenuates cocaine cue reactivity. Addict Biol. 2016 Nov 11; doi: 10.1111/adb.12471. http://dx.doi.org/10.1111/adb.12471 [Epub ahead of print] [DOI] [PMC free article] [PubMed]
- Morris G, Berk M. The putative use of lithium in Alzheimer's disease. Curr. Alzheimer Res. 2016;13:853–861. doi: 10.2174/1567205013666160219113112. [DOI] [PubMed] [Google Scholar]
- Need AC, Ge D, Weale ME, Maia J, Feng S, Heinzen EL, Shianna KV, Yoon W, Kasperaviciute D, Gennarelli M, et al. A genome-wide investigation of SNPs and CNVs in schizophrenia. PLoS Genet. 2009;5:e1000373. doi: 10.1371/journal.pgen.1000373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nenov MN, Laezza F, Haidacher SJ, Zhao Y, Sadygov RG, Starkey JM, Spratt H, Luxon BA, Dineley KT, Denner L. Cognitive enhancing treatment with a PPARgamma agonist normalizes dentate granule cell presynaptic function in Tg2576 APP mice. J. Neurosci. 2014;34:1028–1036. doi: 10.1523/JNEUROSCI.3413-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nenov MN, Tempia F, Denner L, Dineley KT, Laezza F. Impaired firing properties of dentate granule neurons in an Alzheimer's disease animal model are rescued by PPARgamma agonism. J. Neurophysiol. 2015;113:1712–1726. doi: 10.1152/jn.00419.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen JV, Blagoev B, Gnad F, Macek B, Kumar C, Mortensen P, Mann M. Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell. 2006;127:635–648. doi: 10.1016/j.cell.2006.09.026. [DOI] [PubMed] [Google Scholar]
- Olson H, Shen Y, Avallone J, Sheidley BR, Pinsky R, Bergin AM, Berry GT, Duffy FH, Eksioglu Y, Harris DJ, et al. Copy number variation plays an important role in clinical epilepsy. Ann. Neurol. 2014;75:943–958. doi: 10.1002/ana.24178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omata N, Chiu CT, Moya PR, Leng Y, Wang Z, Hunsberger JG, Leeds P, Chuang DM. Lentivirally mediated GSK-3beta silencing in the hippocampal dentate gyrus induces antidepressant-like effects in stressed mice. Int. J. Neuropsychopharmacol. 2011;14:711–717. doi: 10.1017/S1461145710000726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ong CS, Zhou J, Ong CN, Shen HM. Luteolin induces G1 arrest in human nasopharyngeal carcinoma cells via the Akt-GSK-3β-Cyclin D1 pathway. Cancer Lett. 2010;298:167–175. doi: 10.1016/j.canlet.2010.07.001. [DOI] [PubMed] [Google Scholar]
- Perez MJ, Quintanilla RA. Therapeutic actions of the Thiazolidinediones in Alzheimer's disease. PPAR Res. 2015;2015:957248. doi: 10.1155/2015/957248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popovic MA, Foust AJ, McCormick DA, Zecevic D. The spatio-temporal characteristics of action potential initiation in layer 5 pyramidal neurons: a voltage imaging study. J. Physiol. 2011;589:4167–4187. doi: 10.1113/jphysiol.2011.209015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Provensi G, Costa A, Passani MB, Blandina P. Donepezil, an acetylcholine esterase inhibitor, and ABT-239, a histamine H3 receptor antagonist/inverse agonist, require the integrity of brain histamine system to exert biochemical and procognitive effects in the mouse. Neuropharmacology. 2016;109:139–147. doi: 10.1016/j.neuropharm.2016.06.010. [DOI] [PubMed] [Google Scholar]
- Remy I, Michnick SW. A highly sensitive protein-protein interaction assay based on Gaussian luciferase. Nat. Methods. 2006;3:977–979. doi: 10.1038/nmeth979. [DOI] [PubMed] [Google Scholar]
- Risner ME, Saunders AM, Altman JF, Ormandy GC, Craft S, Foley IM, Zvartau-Hind ME, Hosford DA, Roses AD. Efficacy of rosiglitazone in a genetically defined population with mild-to-moderate Alzheimer's disease. Pharm. J. 2006;6:246–254. doi: 10.1038/sj.tpj.6500369. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Murillo L, Xu B, Roos JL, Abecasis GR, Gogos JA, Karayiorgou M. Fine mapping on chromosome 13q32-34 and brain expression analysis implicates MYO16 in schizophrenia. Neuropsychopharmacology. 2014;39:934–943. doi: 10.1038/npp.2013.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Rivera J, Denner L, Dineley KT. Rosiglitazone reversal of Tg2576 cognitive deficits is independent of peripheral gluco-regulatory status. Behav. Brain Res. 2011;216:255–261. doi: 10.1016/j.bbr.2010.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusinova R, Herold KF, Sanford RL, Greathouse DV, Hemmings HC, Jr, Andersen OS. Thiazolidinedione insulin sensitizers alter lipid bilayer properties and voltage-dependent sodium channel function: implications for drug discovery. J. Gen. Physiol. 2011;138:249–270. doi: 10.1085/jgp.201010529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadygov RG, Zhao Y, Haidacher SJ, Starkey JM, Tilton RG, Denner L. Using power spectrum analysis to evaluate (18)O-water labeling data acquired from low resolution mass spectrometers. J. Proteome Res. 2010;9:4306–4312. doi: 10.1021/pr100642q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato T, Hanyu H, Hirao K, Kanetaka H, Sakurai H, Iwamoto T. Efficacy of PPARgamma agonist pioglitazone in mild Alzheimer disease. Neurobiol. Aging. 2011;32:1626–1633. doi: 10.1016/j.neurobiolaging.2009.10.009. [DOI] [PubMed] [Google Scholar]
- Scala F, Fusco S, Ripoli C, Piacentini R, Li Puma DD, Spinelli M, Laezza F, Grassi C, D'Ascenzo M. Intraneuronal Abeta accumulation induces hippocampal neuron hyperexcitability through A-type K(+) current inhibition mediated by activation of caspases and GSK-3. Neurobiol. Aging. 2015;36:886–900. doi: 10.1016/j.neurobiolaging.2014.10.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shavkunov A, Panova N, Prasai A, Veselenak R, Bourne N, Stoilova-McPhie S, Laezza F. Bioluminescence methodology for the detection of protein-protein interactions within the voltage-gated sodium channel macromolecular complex. Assay Drug Dev. Technol. 2012;10:148–160. doi: 10.1089/adt.2011.413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shavkunov AS, Wildburger NC, Nenov MN, James TF, Buzhdygan TP, Panova-Elektronova NI, Green TA, Veselenak RL, Bourne N, Laezza F. The fibroblast growth factor 14/voltage-gated sodium channel complex is a new target of glycogen synthase kinase 3 (GSK3) J. Biol. Chem. 2013;288:19370–19385. doi: 10.1074/jbc.M112.445924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shavkunov AS, Ali SR, Panova-Elektronova NI, Laezza F. Split-luciferase complementation assay to detect channel-protein interactions in live cells. Methods Mol. Biol. 2015;1278:497–514. doi: 10.1007/978-1-4939-2425-7_33. [DOI] [PubMed] [Google Scholar]
- St-Denis N, Gabriel M, Turowec JP, Gloor GB, Li SS, Gingras AC, Litchfield DW. Systematic investigation of hierarchical phosphorylation by protein kinase CK2. J. Proteome. 2015;118:49–62. doi: 10.1016/j.jprot.2014.10.020. [DOI] [PubMed] [Google Scholar]
- Sweatt JD. Mitogen-activated protein kinases in synaptic plasticity and memory. Curr. Opin. Neurobiol. 2004;14:311–317. doi: 10.1016/j.conb.2004.04.001. [DOI] [PubMed] [Google Scholar]
- van Swieten JC, Brusse E, de Graaf BM, Krieger E, van de Graaf R, de Koning I, Maat-Kievit A, Leegwater P, Dooijes D, Oostra BA, Heutink P. A mutation in the fibroblast growth factor 14 gene is associated with autosomal dominant cerebellar ataxia [corrected] Am. J. Hum. Genet. 2003;72:191–199. doi: 10.1086/345488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tempia F, Hoxha E, Negro G, Alshammari MA, Alshammari TK, Panova-Elektronova N, Laezza F. Parallel fiber to Purkinje cell synaptic impairment in a mouse model of spinocerebellar ataxia type 27. Front. Cell. Neurosci. 2015;9:205. doi: 10.3389/fncel.2015.00205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verbeek EC, Bakker IM, Bevova MR, Bochdanovits Z, Rizzu P, Sondervan D, Willemsen G, de Geus EJ, Smit JH, Penninx BW, et al. A fine-mapping study of 7 top scoring genes from a GWAS for major depressive disorder. PLoS One. 2012;7:e37384. doi: 10.1371/journal.pone.0037384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villalobos V, Naik S, Piwnica-Worms D. Current state of imaging protein-protein interactions in vivo with genetically encoded reporters. Annu. Rev. Biomed. Eng. 2007;9:321–349. doi: 10.1146/annurev.bioeng.9.060906.152044. [DOI] [PubMed] [Google Scholar]
- Villalobos V, Naik S, Piwnica-Worms D. Detection of protein-protein interactions in live cells and animals with split firefly luciferase protein fragment complementation. Methods Mol. Biol. 2008;439:339–352. doi: 10.1007/978-1-59745-188-8_23. [DOI] [PubMed] [Google Scholar]
- Wang Q, McEwen DG, Ornitz DM. Subcellular and developmental expression of alternatively spliced forms of fibroblast growth factor 14. Mech. Dev. 2000;90:283–287. doi: 10.1016/s0925-4773(99)00241-5. [DOI] [PubMed] [Google Scholar]
- Wang Q, Bardgett ME, Wong M, Wozniak DF, Lou J, McNeil BD, Chen C, Nardi A, Reid DC, Yamada K, Ornitz DM. Ataxia and paroxysmal dyskinesia in mice lacking axonally transported FGF14. Neuron. 2002;35:25–38. doi: 10.1016/s0896-6273(02)00744-4. [DOI] [PubMed] [Google Scholar]
- Wang F, Blanchard AP, Elisma F, Granger M, Xu H, Bennett SA, Figeys D, Zou H. Phosphoproteome analysis of an early onset mouse model (TgCRND8) of Alzheimer's disease reveals temporal changes in neuronal and glia signaling pathways. Proteomics. 2013;13:1292–1305. doi: 10.1002/pmic.201200415. [DOI] [PubMed] [Google Scholar]
- Wang X, Zhang XG, Zhou TT, Li N, Jang CY, Xiao ZC, Ma QH, Li S. Elevated Neuronal Excitability Due to Modulation of the Voltage-Gated Sodium Channel Nav1.6 by Abeta1-42. Front. Neurosci. 2016;10:94. doi: 10.3389/fnins.2016.00094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson GS, Cholerton BA, Reger MA, Baker LD, Plymate SR, Asthana S, Fishel MA, Kulstad JJ, Green PS, Cook DG, et al. Preserved cognition in patients with early Alzheimer disease and amnestic mild cognitive impairment during treatment with rosiglitazone: a preliminary study. Am. J. Geriatr. Psychiatry. 2005;13:950–958. doi: 10.1176/appi.ajgp.13.11.950. [DOI] [PubMed] [Google Scholar]
- Wildburger NC, Laezza F. Control of neuronal ion channel function by glycogen synthase kinase-3: new prospective for an old kinase. Front. Mol. Neurosci. 2012;5:80. doi: 10.3389/fnmol.2012.00080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wildburger NC, Ali SR, Hsu WC, Shavkunov AS, Nenov MN, Lichti CF, LeDuc RD, Mostovenko E, Panova-Elektronova NI, Emmett MR, et al. Quantitative proteomics reveals protein-protein interactions with fibroblast growth factor 12 as a component of the voltage-gated sodium channel 1.2 (nav1.2) macromolecular complex in Mammalian brain. Mol. Cell. Proteomics. 2015;14:1288–1300. doi: 10.1074/mcp.M114.040055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson MHF, Schut F. Digital Image Analysis of Microbes: Imaging, Morphometry, Fluorometry and Motility Techniques and Applications. John Wiley & Sons; 1998. [Google Scholar]
- Wozniak DF, Xiao M, Xu L, Yamada KA, Ornitz DM. Impaired spatial learning and defective theta burst induced LTP in mice lacking fibroblast growth factor 14. Neurobiol. Dis. 2007;26:14–26. doi: 10.1016/j.nbd.2006.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu P, Zhao Y, Haidacher SJ, Wang E, Parsley MO, Gao J, Sadygov RG, Starkey JM, Luxon BA, Spratt H, et al. Detection of structural and metabolic changes in traumatically injured hippocampus by quantitative differential proteomics. J. Neurotrauma. 2013;30:775–788. doi: 10.1089/neu.2012.2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G, Shen D, Sabuncu M. Machine Learning and Medical Imaging. Elsevier Science; 2016. [Google Scholar]
- Xiao M, Xu L, Laezza F, Yamada K, Feng S, Ornitz DM. Impaired hippocampal synaptic transmission and plasticity in mice lacking fibroblast growth factor 14. Mol. Cell. Neurosci. 2007;34:366–377. doi: 10.1016/j.mcn.2006.11.020. [DOI] [PubMed] [Google Scholar]
- Xiao M, Bosch MK, Nerbonne JM, Ornitz DM. FGF14 localization and organization of the axon initial segment. Mol. Cell. Neurosci. 2013 doi: 10.1016/j.mcn.2013.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu S, Guan Q, Wang C, Wei X, Chen X, Zheng B, An P, Zhang J, Chang L, Zhou W, et al. Rosiglitazone prevents the memory deficits induced by amyloid-beta oligomers via inhibition of inflammatory responses. Neurosci. Lett. 2014;578c:7–11. doi: 10.1016/j.neulet.2014.06.010. [DOI] [PubMed] [Google Scholar]
- Yang T, Wang J, Sun Q, Hibar DP, Jahanshad N, Liu L, Wang Y, Zhan L, Thompson PM, Ye J. Detecting genetic risk factors for Alzheimer's disease in whole genome sequence data via lasso screening. Proc. IEEE Int. Symp. Biomed. Imaging. 2015;2015:985–989. doi: 10.1109/ISBI.2015.7164036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahid S, Oellerich M, Asif AR, Ahmed N. Phosphoproteome profiling of substantia nigra and cortex regions of Alzheimer's disease patients. J. Neurochem. 2012;121:954–963. doi: 10.1111/j.1471-4159.2012.07737.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.