Abstract
Molecularly imprinted polymers for dimethoate recognition were synthesized by the precipitation polymerization technique using methyl methacrylate (MMA) as the functional monomer and ethylene glycol dimethacrylate (EGDMA) as the cross-linker. The morphology, adsorption and recognition properties were investigated by scanning electron microscopy (SEM), static adsorption test, and competitive adsorption test. To obtain the best selectivity and binding performance, the synthesis and adsorption conditions of MIPs were optimized through single factor experiments. Under the optimized conditions, the resultant polymers exhibited uniform size, satisfactory binding capacity and significant selectivity. Furthermore, the imprinted polymers were successfully applied as a specific solid-phase extractants combined with high performance liquid chromatography (HPLC) for determination of dimethoate residues in the cucumber samples. The average recoveries of three spiked samples ranged from 78.5% to 87.9% with the relative standard deviations (RSDs) less than 4.4% and the limit of detection (LOD) obtained for dimethoate as low as 2.3 μg/mL.
Keywords: Molecularly imprinted polymer, Precipitation polymerization, Dimethoate, Cucumber, HPLC
1. Introduction
Dimethoate, as one of the major classes of organophosphorous pesticides, is widely used in the fruit, ornamentals, and field crops to promote the development of farming and satisfy the need for agricultural products [1] owing to its low persistence and biodegradation. However, excessive use of dimethoate could lead to superfluous residues of agricultural products accumulating in the human body through the food chain, which could disrupt cholinesterase enzyme and cause cholinergic dysfunction or even death [2]. Therefore, accurate and reliable analysis of dimethoate residual concentration in agricultural products is of importance to human health. Up to now, the most commonly used methods for detecting organophosphorous pesticide residues mainly include gas chromatography (GC), gas chromatography–mass spectrometry (GC–MS), gas chromatography–tandem mass spectrometry (GC–MS/MS), high performance liquid chromatography (HPLC), liquid chromatography–mass spectrometry (LC–MS), and liquid chromatography–tandem mass spectrometry (LC–MS/MS) [3], [4]. However, the sample pretreatment process is an extremely necessary step before instrumental analysis [5], [6] due to the complexity of the sample matrix. And the routine sample extraction methods, such as liquid–liquid extraction, require a large amount of solvent and multiple consequent steps, and solid-phase extraction usually results in co-extraction of interfering compounds on account of the nonspecific interaction between the analyte and the adsorbent. Thus, the development of new adsorbents with high affinity and specific recognition is greatly desired.
Molecularly imprinted polymers (MIPs), artificial ones, possess tailor-made binding sites complementary to the shapes, sizes and functional groups of the templates. MIPs have numerous merits, including low cost, easy synthesis, high stability to harsh chemical and physical conditions, and excellent reusability [7], which have led to their application in a variety of fields such as solid-phase extraction (SPE) [8], [9], [10], matrix solid-phase dispersion (MSPD) [11], [12], [13], membrane separation [14], [15], chromatographic analysis [16], [17], [18], sensors [19], [20], catalysis [21], [22], and drug controlled release [23], [24], [25]. Among these applications, MIPs used as efficient alternative adsorbents of SPE for extraction of environmental contaminants from the complex samples appear to be highly promising. To acquire data, a series of synthesis methods for MIPs have been developed, such as bulk polymerization [26], precipitation polymerization [27], suspension polymerization, surface polymerization [28], [29] and in-situ polymerization [30]. Among these, precipitation polymerization tends to decrease the guarant viscosity for great improvement of the operability of polymerization and produce a narrow range of particle sizes, which has gained a lot of attention.
In this paper, we prepared dimethoate molecularly imprinted polymers via a simple precipitation polymerization process for specific recognition of dimethoate in cucumber samples. The synthesis conditions were optimized in detail through single factor experiments and the morphology of the resulting products was characterized by SEM, the results exhibited spherical morphology and homogeneous distribution. The obtained imprinted polymers manifested satisfactory recognition and favorable selectivity towards the template molecule through binding experiments. Moreover, an effective analytical method combining the resultant imprinted materials which were used as absorbents for the pretreatment process with HPLC for determination of dimethoate in cucumber samples was established.
2. Experiment
2.1. Chemicals and apparatus
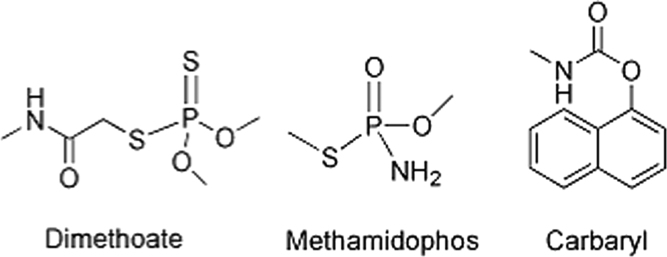
Dimethoate, carbaryl, methamidophos and ethylene glycol dimethacrylate (EGDMA) were purchased from Jingchun Scientifical Co., Ltd. (Shanghai, China). Methacrylic acid (MAA), methyl methacrylate (MMA), acrylamide (AM), azobisisobutyronitrile (AIBN), acetonitrile (ACN), toluene (phMe) and acetic acid were provided by Hongxin Chemical Company (Xi׳an, China). The structures of carbaral, dimethoate, and methamidophos are shown in Fig. 1. The highly purified water (18.0 MΩ/cm) was obtained from a WaterPro water system (Axlwater Corporation, TY10AXLC1805-2, China) and used throughout the experiments. All the reagents used were of at least analytical grade.
Fig. 1.
Molecular structures of dimethoate, methamidophos, and carbaryl.
The HPLC analyses were performed on a Hitachi L-2130 HPLC system equipped with an L-2130 pump, an L-2400 UV detector, and a Kromosil C18 column (150 mm×4.6 mm, 5 μm). D-2000 software was used to acquire and process the chromatographic data. The mobile phase was methanol-H2O (40/60, v/v) delivered at a flow rate of 1.0 mL/min. The injection volume was 20 µL, and the column effluent was monitored at 220 nm. Sample solutions were filtered through a 0.22 μm nylon filter before use. The morphology of MIPs was evaluated by a scanning electron microscope (SEM, VEGA TS5136XM).
2.2. Synthesis of MIPs
The dimethoate MIPs were prepared through a precipitation polymerization process. Briefly, template molecule dimethoate (10 mg) was dissolved in acetonitrile (20 mL) in a round-bottom flask, and then mixed with 20, 30 or 60 mg of functional monomer. The mixture was stirred until the solution became homogeneous, and put into the refrigerator for 6 h to gain a prepolymerization solution. After that, the cross-linker EGDMA (200, 250, 300 mg) and the initiator AIBN (100 mg) were added to the prepolymerization solution and the mixture was blended under ultrasonication for 10 min, and then purged with nitrogen for 5 min to degas oxygen. The solution was sealed with a parafilm and reacted on water bath oscillator at 20 oC for 24 h with a certain rotation speed. The resultant products were separated by centrifugation at 4000 rpm for 15 min, and washed with the mixture of methanol and acetic acid (8:2, v/v) until no template molecules were detected in the extraction solvent by HPLC. Then the polymer particles were washed several times with methanol and dried at 80 °C for 24 h under vacuum.
As a reference, non-imprinted polymers (NIPs) were prepared following the same procedure in the absence of the template.
2.3. Static adsorption experiment
To determine the isothermal binding capacity of the imprinted polymers, MIPs (20 mg) or NIPs (20 mg) were added to 4.0 mL of acetonitrile solution of dimethoate at varied initial concentration ranging from 0.1 to 2.0 mM. The mixture was shaken on a water bath reciprocating shaking-table for 4 h at room temperature. Then the supernatants and polymers were separated by centrifugation and the supernatants were filtered through a 0.22 μm nylon filter and the concentrations of dimethoate in the filtrate were measured by HPLC–UV.
2.4. Competitive adsorption experiment
To evaluate the selectivity of the as-prepared polymers, 20 mg of MIPs or NIPs was added into 4.0 mL of mixed acetonitrile solution of dimethoate, methamidophos and carbaryl at a concentration of 0.6 mM. After incubation for 4 h on a water bath oscillator at room temperature, the supernatants and polymers were separated by centrifugation and the supernatants were filtered through a 0.22 μm microporous filter and the concentrations of three kinds of pesticides in the filtrate were measured by HPLC–UV.
2.5. Real sample analysis
Cucumber samples used for this study were purchased from the local supermarket in Xi׳an. A total of 200 g of cucumber sample was homogenized by a food mixer, and then 50 g of the homogenized sample was immersed in 50 mL of acetonitrile solution with a certain amount of dimethoate standard substance in a covered conical flask. The mixture was incubated on a water bath oscillator for 5 h at 20 °C. Next, the blend was centrifuged at 4000 rpm for 5 min to collect the supernatant, and the sediment at the bottom of the centrifuge tube was washed with acetonitrile several times, then the supernatant and washings were concentrated with a rotary evaporator and the residue was redissolved with 20 mL of acetonitrile to obtain the spiked sample solution. The blank samples were prepared similarly to the spiked sample except that no dimethoate standard substances were added. Then, the blank and spiked samples were stored at 4 °C before analysis. 50 mg of MIPs was added to 10 mL of the spiked sample and after 2 h of incubation on an oscillator at room temperature, the MIPs were isolated by centrifugation and the supernatant solution was discarded. MIPs which adsorbed the target molecule were eluted with a mixture of methanol and acetic acid (8:2, v/v) solution, and then the elution was collected and evaporated to dryness under a stream of nitrogen. Next, the residue of the elution was dissolved in 0.5 mL of methanol. The blank, spiked, and eluted samples were filtered through a 0.22 μm microporous filter and measured by HPLC–UV.
3. Results and discussion
3.1. Single factor experiments
One of the important factors for successful molecular imprinting is the choice of appropriate functional monomer. The role of monomers is to assist in the creation of the specific binding capacity by exposing interacting chemical function groups after polymerization that are situated within the cavity in an optimal position for rebinding. According to the structure and features of the template, we selected MAA, MMA and AM as functional monomers to investigate the adsorption capacity of the corresponding polymers. The adsorption capacity (Q) of the imprinted polymers was calculated according to the following equation:
where Q (μmol/g) represents the adsorption capacity, Ci and Cf (mM) are the initial and final solution concentrations of the analyte, respectively, V (mL) is the volume of the solution, and M (mg) is the weight of the imprinted polymers.
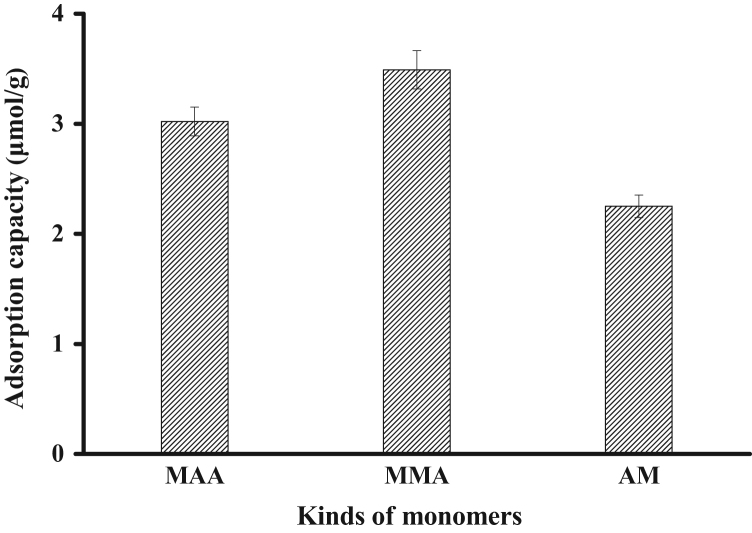
Compared with the adsorption capacities of MIPs using MAA, MMA, and AM as monomers, the results are shown in Fig. 2. We found that the imprinted polymers using MMA as a functional monomer had a higher capacity than that of MAA (3.10 μmol/g) and AM, which might result from the stronger interaction between functional monomer MMA and the template molecule dimethoate. Because of its superiority, MMA was chosen as the functional monomer in this work.
Fig. 2.
Adsorption capacities of MIP synthesized using MAA, MMA and AM as functional monomers, respectively. The initial concentration of dimethoate in acetonitrile was 0.30 mM.
Moreover, the mass ratio of the functional monomer-to-cross-linker-to-template has a very important influence on the specific affinity of MIPs and the number of recognition sites. As shown in Table 1, Q increased with the increased amount of functional monomer and cross-linker, due to the increase in the number of recognition cavities in MIPs. However, excessive amount of cross-linker used would lead to agglomeration of MIPs, which decreased the adsorption capacity of the imprinted polymers. Therefore, the optimum mass ratio of template molecule-to-functional monomer-to-cross-linker was 1:6:30.
Table 1.
The adsorption capacities of MIPs synthesized under different mass ratios of template, monomer and cross-linker. The initial concentration of dimethoate in acetonitrile was 0.30 mM.
| Template:monomer:cross-linker | 1:2:20 | 1:4:20 | 1:6:20 | 1:6:25 | 1:6:30 | 1:6:35 |
| Adsorption capacity (μmol/g) | 1.30 | 1.53 | 2.17 | 2.62 | 3.07 | 2.94 |
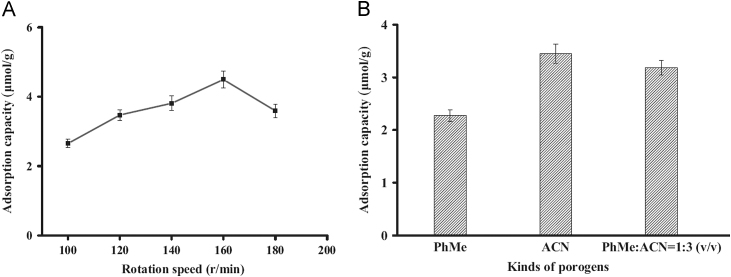
Furthermore, the porogen and rotation speed were also optimized, and the results are shown in Fig. 3. An increase in the adsorption of dimethoate with the increase of rotation speed ranging from 100 to 160 r/min can be seen, indicating that the reaction was conducted sufficiently under high rotation speed, while with further increase of the rotation speed, the adsorption capacity decreased, which would be ascribed to the fact that the reagent might stick to the wall of the container and have no chance to attain the reaction that led to the decrease in the number of recognition sites. Herein, the rotation speed of 160 r/min was adopted for the polymerization reaction. In the process of polymerization, three kinds of porogen solutions including toluene, acetonitrile and the mixture of toluene and acetonitrile (1:3, v/v) were selected. The results indicated that acetonitrile used as a porogen was much better than the others deduced from the higher adsorption capacity.
Fig. 3.
Effects of different rotation speeds (A) and porogens (B) on the imprinting of MIPs. The initial concentration of dimethoate in acetonitrile was 0.30 mM.
3.2. Characterization of MIPs
Under the optimized conditions, the morphology of resultant polymers was characterized by SEM, and the images in Fig. 4 exhibited that the imprinted polymers possessed a spherical shape and satisfactory dispersibility with an average diameter of about 3 µm.
Fig. 4.
SEM images of dimethoate molecularly imprinted polymers with different magnifications.
3.3. Equilibrium rebinding study
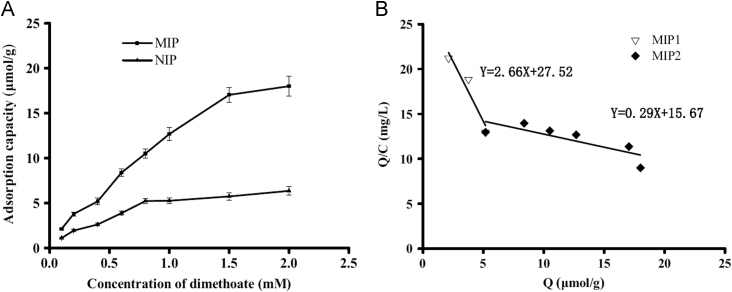
To evaluate the adsorption capability of MIPs and NIPs towards dimethoate, a static adsorption experiment was carried out with the initial concentration ranging from 0.1 to 2.0 mM. The adsorption isotherm curve is shown in Fig. 5A. It can be seen that the adsorption capacity increased rapidly along with the increasing initial concentration of dimethoate and reached adsorption saturation when the initial concentration was above 1.5 mM. Additionally, the adsorption capacity of MIPs was far greater than that of NIPs at the same initial concentration, revealing that MIPs had specific binding sites which could recognize dimethoate selectively.
Fig. 5.
Static adsorption of MIPs and NIPs for dimethoate (A) and Scatchard analysis to estimate the binding nature of MIPs.
To further estimate the binding properties of the imprinted polymers, the saturation binding data were further processed by Scatchard analysis, which was expressed as the following equation:
where C (mM) is the free dimethoate concentration at equilibrium, Q (µmol/g) is the amount of dimethoate bound to MIPs at equilibrium, Qmax (µmol/g) is the apparent maximum adsorption capacity and Kd is the dissociation constant. The values of Kd and Qmax can be calculated from the slope and intercept of the linear curve plotted as Q/C versus Q.
The Scatchard analysis of MIPs was performed. As shown in Fig. 5B, two straight lines were obtained in the plot region, which indicates that there exist two kinds of binding sites of high and low affinities. The linear regression equations for the left and right slopes of the biphasic curve are Q/C=27.52+2.66Q (r=0.9992) and Q/C=15.67+0.29Q (r=0.9993). From the slope and intercept of the biphasis curve obtained, the constant Kd1 and the apparent maximum amount Qmax1 for the higher affinity binding sites can be calculated to be 0.38 mM and 10.3 μmol/g, respectively, while Kd2 and Qmax2 for the lower affinity binding sites were calculated to be 3.4 mM and 53.3 μmol/g, respectively.
3.4. Selective adsorption experiment
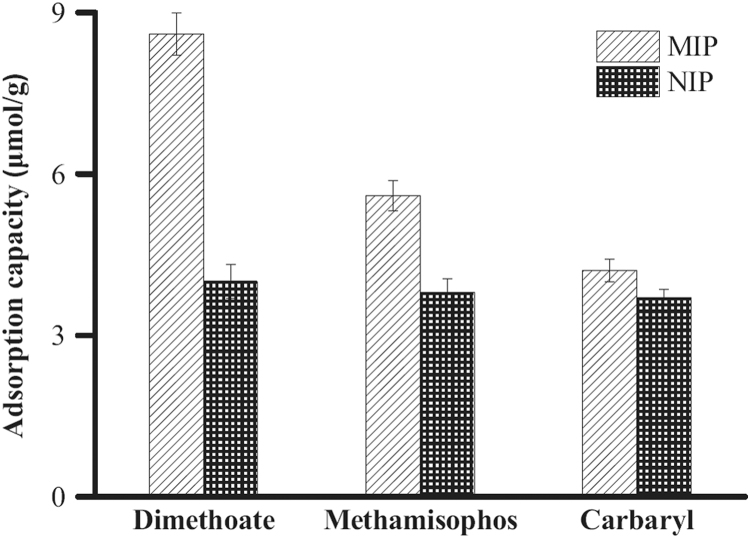
To assess the selectivity of MIPs and NIPs for dimethoate, two other pesticides were selected as analogs. The adsorption capacities of MIPs and NIPs to the mixture of dimethoate, methamisophos, and carbaryl with a concentration of 1.5 mM were investigated. The results are shown in Fig. 6, from which we can observe that the bound amount of dimethoate for MIPs was much higher than that of the other two pesticides, suggesting that the imprinted polymers have a relatively higher affinity for dimethoate than its analogs. The reason might be explained as that the orientation of functional groups and steric complementariness of imprinted cavities to the template molecule were formed during the preparation process. In contrast to MIPs, the adsorption capacities of NIPs for dimethoate, methamidophos and carbaryl were relatively lower, and nearly the same, which indicates that the NIPs have no specific selectivity for the three pesticides. These results further verified the satisfactory imprinting efficiency of the imprinted polymers for dimethoate in the present work.
Fig. 6.
The competitive adsorption capacity of MIPs and NIPs to dimethoate, methanisophos and carbaryl in the mixture solution. The initial concentration of every solution was 0.6 mM.
3.5. Validation of the method
The proposed method was validated by a series of experimental parameters including linear range, correlation coefficients (r2), limit of detection (LOD), and limit of quantification (LOQ). The results indicated that a good linearity was achieved in the range of 0.010 – 2.0 μg/mL with r2 of 0.9992 for dimethoate and the linear regression equation was y=31900x+57762, where y is peak area, x is the concentration of the analyte. The LOD and LOQ were 2.3 μg/mL and 7.7 μg/mL, which were calculated from three times and ten times of signal-to-noise ratio, respectively.
To evaluate the accuracy of the developed method, the cucumber samples spiked with three levels of dimethoate (0.1, 0.5, 1.0 μg/mL) were analyzed. At each concentration, three measurements were performed, and the results are shown in Table 2. The average recoveries of dimethoate of cucumber samples ranged from 78.5% to 87.9% and the relative standard deviation (RSD) was less than 4.4%. These results revealed that the proposed method was simple, reliable and sensitive.
Table 2.
Recoveries of MIPs binding dimethoate for the spiked cucumber samples (n=3).
| Sample | Spiked level (μg/mL) | Recovery (%) | RSD (%) |
|---|---|---|---|
| 1 | 0.1 | 78.5 | 3.5 |
| 2 | 0.5 | 80.8 | 4.4 |
| 3 | 1.0 | 87.9 | 3.7 |
3.6. Real sample analysis
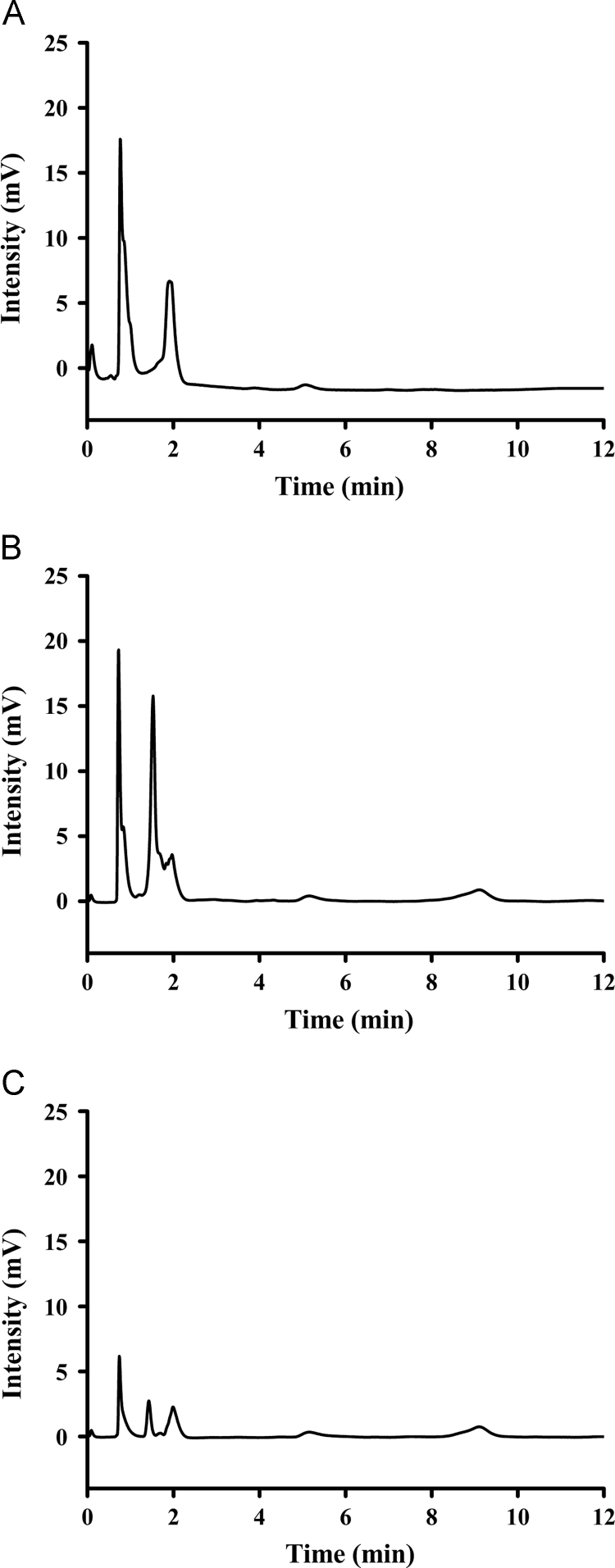
The application of MIPs to selective adsorption of dimethoate in a cucumber sample was investigated. After adsorption of the cucumber sample spiked with dimethoate at a concentration of 0.5 μg/mL, the adsorbed MIPs were washed with a mixture of methanol and acetic acid (8:2, v/v) solutions. The chromatograms of samples spiked with dimethoate at a concentration of 0.5 μg/mL, elution of adsorbed MIPs, and the cucumber sample without spiking are displayed in Fig. 7. The peak of dimethoate could not be seen from chromatograms of the cucumber sample without spiking (Fig.7A). After adsorption of the spiked sample with MIP (Fig.7B) and washing with an eluent (Fig.7C), the peak of dimethoate appeared distinctly at 8.52 min, which was consistent with the peak position of dimethoate in chromatograms of the spiked sample, but other peaks of irrelevant compounds in the cucumber sample were dramatically decreased compared with the spiked sample. These results revealed that MIPs could be directly applied for selective isolation and determination of dimethoate in the cucumber sample.
Fig. 7.
Chromatograms of dimethoate in the cucumber sample. Sample without spiking (A), samples spiked with dimethoate at a concentration of 0.5 μg/mL (B) and the elution of MIPs after adsorbing the cucumber spiked sample (C).
4. Conclusion
A type of dimethoate molecularly imprinted polymers with uniform shape and size was synthesized by precipitation polymerization, and the synthesis conditions were optimized by single factor experiments. The results revealed that when selecting MMA as the monomer, acetonitrile as the porogen, 160 r/min as rotation speed, and 1:6:30 as the mass ratio of template:monomer:cross-linker, the adsorption of MIP was significantly satisfactory. The optimized MIPs exhibited excellent recognition performance and favorable selectivity affinity towards dimethoate. Meanwhile, the resulting products were successfully used as solid-phase extractants coupled with the HPLC technique for specific isolation and detection of dimethoate from the cucumber sample, indicating a potential value in practical application.
Acknowledgments
The authors are grateful for financial support from the National Natural Science Foundation of China (No. 21305107) and the Fundamental Research Funds for the Central Universities (Nos. xjj2013041, 08142034).
Footnotes
Peer review under responsibility of Xi׳an Jiaotong University.
References
- 1.Lv Y.Q., Lin Z.X., Feng W. Selective recognition and large enrichment of dimethoate from tea leaves by molecularly imprinted polymers. Biochem. Eng. J. 2007;36:221–229. [Google Scholar]
- 2.Kanagaraj K., Affrose A., Sivakolunthu S. Highly selective fluorescent sensing of fenitrothion using per-6-amino-β-cyclodextrin: Eu(III) complex. Biosens. Bioelectron. 2012;35:452–455. doi: 10.1016/j.bios.2012.02.046. [DOI] [PubMed] [Google Scholar]
- 3.Sharma D., Nagpal A., Pakade Y.B. Analytical methods for estimation of organophosphorus pesticide residues in fruits and vegetables: a review. Talanta. 2010;82:1077–1089. doi: 10.1016/j.talanta.2010.06.043. [DOI] [PubMed] [Google Scholar]
- 4.Chung S.W.C., Chan B.T.P. Validation and use of a fast sample preparation method and liquid chromatography–tandem mass spectrometry in analysis of ultra-trace levels of 98 organophosphorus pesticide and carbamate residues in a total diet study involving diversified food types. J. Chromatogr. A. 2010;1217:4815–4824. doi: 10.1016/j.chroma.2010.05.043. [DOI] [PubMed] [Google Scholar]
- 5.Martín-Esteban A. Molecularly-imprinted polymers as a versatile, highly selective tool in sample preparation. Trends Anal. Chem. 2013;45:169–181. [Google Scholar]
- 6.Song X.L., Xu S.F., Chen L.X. Recent advances in molecularly imprinted polymers in food analysis. J. Appl. Polym. Sci. 2014;131:1–18. [Google Scholar]
- 7.Chen L.X., Xu S.F., Li J.H. Recent advances in molecular imprinting technology: current status, challenges and highlighted applications. Chem. Soc. Rev. 2011;40:2922–2942. doi: 10.1039/c0cs00084a. [DOI] [PubMed] [Google Scholar]
- 8.Guo Z.Y., Gai P.P., Hao T.T. Determination of malachite green residues in fish using a highly sensitive electrochemiluminescence method combined with molecularly imprinted solid phase extraction. J. Agric. Food Chem. 2011;59:5257–5262. doi: 10.1021/jf2008502. [DOI] [PubMed] [Google Scholar]
- 9.Vitor R.V., Martins M.C.G., Figueiredo E.C. Application of molecularly imprinted polymer solid-phase extraction for salivary cotinine. Anal. Bioanal. Chem. 2011;400:2109–2117. doi: 10.1007/s00216-011-4870-1. [DOI] [PubMed] [Google Scholar]
- 10.Zhang Z., Xu S.F., Li J.H. Selective solid-phase extraction of sudan I in chilli sauce by single-hole hollow molecularly imprinted polymers. J. Agric. Food Chem. 2012;60:180–187. doi: 10.1021/jf2041609. [DOI] [PubMed] [Google Scholar]
- 11.Crescenzi C., Bayoudh S., Cormack P.A.G. Determination of clenbuterol in bovine liver by combining matrix solid-phase dispersion and molecularly imprinted solid-phase extraction followed by liquid chromatography/electrospray ion trap multiple-stage mass spectrometry. Anal. Chem. 2001;73:2171–2177. doi: 10.1021/ac0014360. [DOI] [PubMed] [Google Scholar]
- 12.Wang T., Tong J., Sun M.L. Fast and selective extraction of chloramphenicol from soil by matrix solid-phase dispersion using molecularly imprinted polymer as dispersant. J. Sep. Sci. 2011;34:1886–1892. doi: 10.1002/jssc.201100046. [DOI] [PubMed] [Google Scholar]
- 13.Zhang H.Y., Wei Y.W., Zhou J.H. Preparation and application of a molecular imprinting matrix solid phase dispersion extraction for the determination of olaquindox in chicken by high performance liquid chromatography. Food Anal. Methods. 2012;6:915–921. [Google Scholar]
- 14.Faizal C.K.M., Kobayashi T. Molecularly imprinted membrane applied for selective separation. J. Appl. Sci. 2011;11:2411–2415. [Google Scholar]
- 15.Zang D.J., Yan M., Ge S.G. A disposable simultaneous electrochemical sensor array based on a molecularly imprinted film at a NH2-graphene modified screen-printed electrode for determination of psychotropic drugs. Analyst. 2013;138:2704–2711. doi: 10.1039/c3an00109a. [DOI] [PubMed] [Google Scholar]
- 16.Liu Z.S., Xu Y.L., Yan C. Preparation and characterization of molecularly imprinted monolithic column based on 4-hydroxybenzoic acid for the molecular recognition in capillary electrochromatography. Anal. Chim. Acta. 2004;523(2):243–250. [Google Scholar]
- 17.Javanbakht M., Moein M.M., Akbari-adergani B. On-line clean-up and determination of tramadol in human plasma and urine samples using molecularly imprinted monolithic column coupling with HPLC. J. Chromatogr. B. 2012;911:49–54. doi: 10.1016/j.jchromb.2012.10.019. [DOI] [PubMed] [Google Scholar]
- 18.Chen X.Q., Yang W.J., Zhou Y.M.H. In situ synthesis of monolithic molecularly imprinted stationary phases for liquid chromatographic enantioseparation of dibenzoyl tartaric acid enantiomers. J. Porous Mater. 2011;19:587–595. [Google Scholar]
- 19.Xie C.G., Gao S., Guo Q.B. Electrochemical sensor for 2,4-dichlorophenoxy acetic acid using molecularly imprinted polypyrrole membrane as recognition element. Microchim. Acta. 2010;169:145–152. [Google Scholar]
- 20.Zhang Z.H., Hu Y.F., Zhang H.B. Layer-by-layer assembly sensitive electrochemical sensor for selectively probing l-histidine based on molecular imprinting sol–gel at functionalized indium tin oxide electrode. Biosens. Bioelectron. 2010;26:696–702. doi: 10.1016/j.bios.2010.06.062. [DOI] [PubMed] [Google Scholar]
- 21.Resmini M. Molecularly imprinted polymers as biomimetic catalysts. Anal. Bioanal. Chem. 2012;402:3021–3026. doi: 10.1007/s00216-011-5671-2. [DOI] [PubMed] [Google Scholar]
- 22.Meng Z.H., Yamazaki T., Sode K. Enhancement of the catalytic activity of an artificial phosphotriesterase using a molecular imprinting technique. Biotechnol. Lett. 2003;25:1075–1080. doi: 10.1023/a:1024152229526. [DOI] [PubMed] [Google Scholar]
- 23.Say R., Erdem M., Ersöz A. Biomimetic catalysis of an organophosphate by molecularly surface imprinted polymers. Appl. Catal. A. 2005;286:221–225. [Google Scholar]
- 24.Sellergren B., Allender C.J. Molecularly imprinted polymers: a bridge to advanced drug delivery. Adv. Drug Deliv. Rev. 2005;57:1733–1741. doi: 10.1016/j.addr.2005.07.010. [DOI] [PubMed] [Google Scholar]
- 25.Javanbakht M., Mohammadi S., Esfandyari-Manesh M. Molecularly imprinted polymer microspheres with nanopore cavities prepared by precipitation polymerization as new carriers for the sustained release of dipyridamole. J. Appl. Polym. Sci. 2011;119:1586–1593. [Google Scholar]
- 26.Song X.L., Li J.H., Wang J.T. Quercetin molecularly imprinted polymers: preparation, recognition characteristics and properties as sorbent for solid-phase extraction. Talanta. 2009;80:694–702. doi: 10.1016/j.talanta.2009.07.051. [DOI] [PubMed] [Google Scholar]
- 27.Xu S.F, Li J.H., Chen L.X. Molecularly imprinted core–shell nanoparticles for determination of trace atrazine by reversible addition–fragmentation chain transfer surface imprinting. J. Mater. Chem. 2011;21:4346–4351. [Google Scholar]
- 28.Gao R.X., Su X.Q., He X.W. Preparation and characterisation of core–shell CNTs@MIPs nanocomposites and selective removal of estrone from water samples. Talanta. 2011;83:757–764. doi: 10.1016/j.talanta.2010.10.034. [DOI] [PubMed] [Google Scholar]
- 29.Xu S.F., Li J.H., Chen L.X. Molecularly imprinted polymers by reversible addition–fragmentation chain transfer precipitation polymerization for preconcentration of atrazine in food matrices. Talanta. 2011;85:282–289. doi: 10.1016/j.talanta.2011.03.060. [DOI] [PubMed] [Google Scholar]
- 30.Qiao F.X., Sun H.W., Yan H.Y. Molecularly imprinted polymers for solid phase extraction. Chromatographia. 2006;64:625–634. [Google Scholar]