Abstract
The interaction between fosfomycin (FOS) and bovine serum albumin (BSA) has been investigated effectively by multi-spectroscopic techniques under physiological pH 7.4. FOS quenched the intrinsic fluorescence of BSA via static quenching. The number of binding sites n and observed binding constant KA were measured by the fluorescence quenching method. The thermodynamic parameters ΔG0, ΔH0 and ΔS0 were calculated at different temperatures according to the van’t Hoff equation. The site of binding of FOS in the protein was proposed to be Sudlow’s site I based on displacement experiments using site markers viz. warfarin, ibuprofen and digitoxin. The distance r between the donor (BSA) and acceptor (FOS) molecules was obtained according to the Förster theory. The effect of FOS on the conformation of BSA was analyzed using synchronous fluorescence spectra (SFS), circular dichroism (CD) and 3D fluorescence spectra. A molecular modeling study further confirmed the binding mode obtained by the experimental studies.
Keywords: Fosfomycin, Serum albumin, Spectroscopic methods, Synchronous fluorescence, 3D spectra
1. Introduction
Serum albumins (52%–60% of the total amount of plasma proteins) are the major soluble protein constituents of the circulatory system, and they have many physiological functions, such as combination with many endogenous and exogenous compounds. They also play an important role in storage and transport of energy [1]. BSA solution is stable and homogeneous; the 3D structure of BSA is believed to be similar to that of human serum albumin (HSA). BSA has two tryptophan residues (Trp 134 and Trp 212) located in sub-domains IA and IIA, respectively [2].
Fosfomycin (FOS) is a naturally occurring antibiotic that was described originally under the name fosfonomycin in 1969 [3]. FOS is a clinically useful antibiotic for the treatment of limb-threatening diabetic foot infections and lower urinary tract infections [4]. It is reasonably soluble in water [5] and the drug substance is stable under normal storage conditions (2–3 years). Chemical structures of BSA and FOS are shown in Supplementary Fig. S1.
The literature survey revealed that attempts have not been made so far to investigate the binding mechanism of FOS with BSA by spectroscopic techniques like synchronous fluorescence spectra (SFS), circular dichroism (CD) and 3D spectra. We further investigated the effect of fluorescence resonance energy transfer (FRET) and the conformational change of BSA. In the present study of the binding mechanism between FOS and BSA regarding the binding parameters, the thermodynamic functions and the effect of FOS on the protein conformation were investigated in detail.
2. Experimental
2.1. Materials and methods
Fosfomycin disodium salt was obtained from HIMEDIA (India). Protease-free and essentially globulin-free BSA (Fraction V) was purchased from Sigma Chemical Co. (St. Louis, USA). The stock solution of BSA (65,000) and FOS was prepared in 0.1 M phosphate buffer of pH 7.4 containing 0.15 M NaCl. All other materials were of analytical reagent grade and millipore water (resistivity of millipore water is 20 MΩ) was used throughout the experiment.
2.2. Instruments
Fluorescence measurements were recorded using an RF-5301 PC Hitachi spectrofluorometer Model F-2000 (Hitachi, Tokyo, Japan) equipped with a 150 W Xenon lamp and a slit width of 5 nm. A 1.00 cm quartz cell and thermostatic cuvette holder were used for measurements. The absorption spectra were recorded on a double beam CARY 50-BIO UV–vis spectrophotometer (Varian, Australia) equipped with a 150 W Xenon lamp and a slit width of 5 nm. A 1.00 cm quartz cell was used for measurements. The pH of solution was measured with an Elico LI120 pH meter (Elico Ltd., India). Circular dichroism measurements were recorded on a Jasco J-715 spectropolarimeter (Tokyo, Japan) with a 0.2 cm quartz cell.
2.3. Methods
2.3.1. Fluorescence quenching study
Based on preliminary investigations, the concentration of BSA was kept constant at 5 μM [6], while that of the drug was varied from 5 to 45 μM. Fluorescence spectra were recorded at three temperatures in the range of 300–500 nm upon excitation at 296 nm.
2.3.2. UV measurements
The UV–vis spectra were obtained by scanning the solution on the spectrophotometer in the wavelength range of 240–320 nm. BSA concentration was fixed at 5 μM, while that of FOS was varied from 5 to 45 μM in the presence of phosphate buffer.
2.3.3. SFS
SFS were obtained by scanning the excitation and emission monochromator simultaneously. The spectrum behavior of tyrosine and tryptophan residues of BSA was observed (Δλ=15 nm and Δλ=60 nm). The spectra were recorded in the range of 250–320 nm.
2.3.4. 3D fluorescence spectra
The three-dimensional fluorescence spectra of BSA were recorded. 40 µL (5 µM) of protein solution was transferred to a quartz cell, diluted to 2.0 mL with phosphate buffer and mixed well. To this, 360 µL of 45 µM FOS was added and the 3D fluorescence spectra were recorded by scanning excitation wavelength in the range of 200–350 nm and emission wavelength from 200 to 600 nm at an interval of 10 nm. The scanning parameters were just the same as the fluorescence quenching experiments.
2.3.5. Competitive binding studies
The competitive binding studies were performed by using different competitors, viz., warfarin for site I, ibuprofen for site II and digitoxin for site III, by keeping the concentration of protein and the competitor constant (each of 5 μM). The fluorescence quenching titration was performed as before, to determine the binding constant of FOS–BSA in the presence of the above-said site probes.
2.3.6. Effect of common ions
The fluorescence spectra of FOS–BSA were recorded in the presence of various common ions, viz., Ca2+, Cu2+, Mg2+, Zn2+ and Ni2+, in the range of 300–500 nm upon excitation at 296 nm. The overall concentration of BSA and that of the common ions were fixed at 5 μM.
2.3.7. Circular dichroism
The CD measurements of protein (2.5 μM) in the presence and absence of FOS were made in the range of 200–260 nm. The ratio of BSA to drug concentration was varied (1:0, 1:1 and 1:3) and the CD spectra were recorded.
2.3.8. Molecular modeling
Molecular docking was performed with Surflex-Dock program that is interfaced with Sybyl-X 2.0. [7]. Crystal structure of BSA was composed from PDB under code 3V03 [8] and was extracted from the Brookhaven Protein Database (PDB http://www.rcsb.org/pdb). All the hydrogen atoms were added to define the correct configuration and tautomeric states. Then the modeled structure was energy-minimized using Tripos force field with distance dependent dielectric function and partial atomic charges were calculated by AMBER7F9902 method and finally water molecules were removed from the model. The geometry of the molecule FOS was subsequently optimized to minimal energy using the Powell energy minimization algorithm, Tripos force field with Gasteiger-Hückel charges. FOS was then separately docked into the binding pocket for docking–scoring analysis. To identify the ligand–protein interactions, the top pose and protein were loaded into work area and the Molecular Computer Aided Design (MOLCAD) program was employed to visualize the binding mode between the protein and ligand.
3. Results and discussion
3.1. Fluorescence studies of BSA quenched by FOS
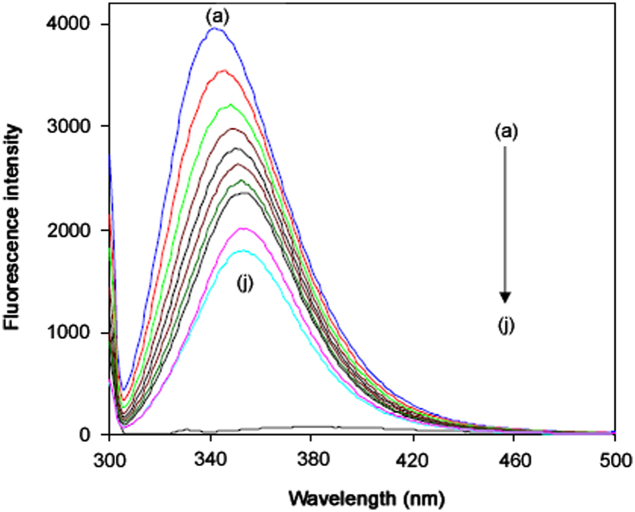
Fluorescence quenching refers to any process which decreases the fluorescence intensity of a sample [9]. The fluorescence emission spectra of BSA with increasing concentrations of FOS are shown in Fig. 1. We measured the competitive absorbance of protein and drug molecules at the excitation wavelength (296 nm) and observed that both of them did not contribute to the inner filter effect as evident from their very low absorbance values. Hence, there was no inner filter effect. The possible mechanisms of quenching include dynamic quenching, static quenching or both [9], [10]; dynamic and static quenching could be distinguished based on their differing dependence on temperature. For this, we carried out the quenching studies at different temperatures. For fluorescence quenching, the decrease in intensity is usually described by the Stern−Volmer equation [11] as shown below:
| (1) |
where F0 and F denote the steady-state fluorescence intensities in the absence and presence of quencher (FOS), respectively, KSV is the Stern−Volmer quenching constant, and [Q] is the concentration of the quencher. Hence, Eq. (1) was applied to determine KSV by linear regression of a plot F0/F against [Q] (Supplementary Fig. S2). The values of KSV at different temperatures are given in Table 1.
Fig. 1.
Fluorescence spectra of BSA (5 μM) in the presence of FOS: 0 (a) to 45 μM (j) at 298 K.
Table 1.
Stern–Volmer quenching constant KSV and Kq values.
| System | Temp (T) | KSV (×104) (L/mol) | Kq (×1012) (L/mol s) |
|---|---|---|---|
| BSA–FOS | 288 | 1.65±0.10 | 1.65±0.10 |
| 298 | 1.19±0.06 | 1.19±0.06 | |
| 308 | 1.03±0.05 | 1.03±0.05 |
These values were found to decrease with increase of temperature. This result confirmed that the quenching is mainly a static quenching process. Kq is the quenching rate constant of bio-molecule, τ0 is the average lifetime of bio-molecule without the quencher, and [Q] is the concentration of the quencher. Obviously,
| (2) |
Since the lifetime fluorescence of the biopolymer is 10−8 s [11], the values of Kq for FOS−BSA system were observed at different temperatures as shown in Table 1. The order of magnitude of the quenching rate constant Kq was observed to be 1012 in the present study. However, the maximum scatter collision quenching constant, Kq, of the various quenchers with the biopolymer [12] is 2×1010 (L/mol s). Thus, the rate constant calculated by protein quenching procedure was greater than Kq of scatter procedure. This indicated that a quenching mechanism was operative [13].
3.2. Binding parameters
3.2.1. Binding constant and binding sites
If it is assumed that there are similar and independent binding sites in the biomolecule, for the static quenching interaction, the binding constant (KA) and the number of sites (n) can be determined according to the method described by Wei et al. [14], using the equation:
| (3) |
where KA is the binding constant of FOS with BSA and n is the number of binding sites per albumin molecule, a plot of log(F0−F)/F versus log[Q] gives a straight line (Supplementary Fig. S3), whose slopes equal n and the intercept on Y-axis equals log KA. The corresponding values of KA and n at different temperature are given in Table 2.
Table 2.
Thermodynamic parameters of FOS–BSA system.
| System | Temp. (K) | Binding constant (L/mol×103) | No. of binding sites (n) | ∆H0 (kJ/mol) | ∆S0 (J/K mol) | ∆G0 (kJ/mol) |
|---|---|---|---|---|---|---|
| BSA–FOS | 288 | 7.05±0.35 | 0.916 | |||
| 298 | 4.71±0.23 | 0.903 | −34.93 | −47.36 | −20.81 | |
| 308 | 2.73±0.14 | 0.901 |
The values of n at the experimental temperatures were approximately equal to 1, which indicated that there was one class of binding site to FOS with BSA. In BSA, the tryptophan residues involved in binding can be either Trp134 or Trp212 of both tryptophans in BSA, and Trp134 is more exposed to a hydrophilic environment, whereas Trp212 is deeply buried in the hydrophobic loop [13]. So, from the value of n, it was proposed that FOS most likely binds to the hydrophobic pocket located in subdomain IIA. That is to say, Trp212 is near or within the binding site [15].
3.2.2. Binding mode
The forces acting between drug and biomolecule may include hydrogen bond, van der Waals force, electrostatic force and hydrophobic interaction force. The thermodynamic parameters, enthalpy change, entropy change and free energy change, are the main evidences to determine the binding mode [16], [17]. The binding studies were carried out at different temperatures and analyzed using the van’t Hoff equation [18] given below:
| (4) |
where KA is the binding constant at the corresponding temperature, and R is the gas constant and T is the temperature. The log KA versus 1/T plot enabled the determination of ∆H0 (kJ/mol) and ∆S0 (J/K mol) values for the binding process. The value of ∆G0 was calculated using the equation:
| (5) |
and the corresponding values are presented in Table 2. ∆H0 and ∆S0 for the interaction between FOS and BSA were found to be –34.93 kJ/mol and –47.36 J/K mol, respectively. Thus, the negative sign for ∆G0 meant that the binding process was spontaneous and the formation of complex was an exothermic reaction accompanied by negative ∆S0 value. Besides, the negative ∆H0 and ∆S0 values revealed that the interaction forces between FOS and BSA were owing to van der Waals force and hydrogen bonds. However, electrostatic interactions might play a role in the interaction [17].
3.2.3. Binding distance
Fluorescence resonance energy transfer (FRET) [19] occurs whenever the emission spectrum of the fluorophore (donor) overlaps with the absorption spectrum of the acceptor. Basically, efficiency of the FRET mainly depends on the following three parameters: (i) the distance between donor and acceptor (which must be within the specified Förster distance of 2–8 nm); (ii) appreciable overlap between the donor fluorescence and acceptor absorption band; and (iii) proper orientation of the transition dipole of the donor and acceptor. The distance between the donor and acceptor could be calculated according to Förster’s theory [20]. The efficiency of energy transfer, E, was calculated using
| (6) |
where F and F0 are the fluorescence intensities of donor in the presence and absence of the acceptor, respectively. r represents the acceptor–donor distance and R0 is the critical distance when the transfer efficiency is 50%. R0 can be expressed as
| (7) |
where k2 is the spatial orientation factor of the dipole, N is the refractive index of the medium, Φ is the fluorescence quantum yield of the donor and J is the overlap integral of the fluorescence emission spectrum of the donor and the absorption spectrum of the acceptor [21]. J is given by
| (8) |
where F(λ) is the fluorescence intensity of the fluorescent donor at wavelength λ and ε(λ) is the molar absorption coefficient of the acceptor at wavelength λ. In the present case, k2=2/3, n=1.36 and Ф=0.15 [22]. From Eqs. (6), (7), (8), J=1.865×10−14 (cm3 L/mol), R0=2.831 nm, E=0.074 and r=4.99 nm for FOS–BSA system can be calculated. The value of r indicates that the donor and acceptor are close to each other and hence they have strong binding between them [23]. Furthermore, the observed donor-to-acceptor distance r<8 nm revealed the presence of static quenching in the interaction [24].
3.2.4. Effect of metal ions on the interactions of FOS with BSA
Some common ions are widely distributed in humans and animals, which can affect the interactions of compounds with serum albumin. Therefore, we examined the effects of some cations (Ca2+, Cu2+, Mg2+, Ni2+ and Zn2+) on binding of FOS with protein at 298 K. The binding constant values (Table 3) of FOS–protein were noticed to decrease in the presence of the above metal ions. This indicated that the drug would be quickly cleared from the blood [25]. In such an event, it necessitated to readjust the dose limits of the drug in the presence of these metal ions.
Table 3.
Effect of common ions on binding constant of FOS–BSA.
| Systems (cations) | Binding constant (M×103) |
|---|---|
| BSA+FOS | 4.714 |
| BSA+FOS+Ca2+ | 3.526 |
| BSA+FOS+Cu2+ | 3.068 |
| BSA+FOS+Mg2+ | 2.288 |
| BSA+FOS+Ni2+ | 1.806 |
| BSA+FOS+Zn2+ | 1.530 |
3.2.5. Binding studies using UV–visible absorption spectra
UV–vis absorption measurement is a very simple method and applicable to explore the structural change and know the complex formation [26]. The UV–vis absorption spectra of FOS, BSA and the FOS–BSA system were also investigated to confirm the probable quenching mechanism. The UV–vis absorbance intensity decreased regularly with the increasing concentration of FOS [27], indicating that the BSA molecules were associated with FOS and formed a FOS–BSA complex. This confirmed again a static quenching mechanism.
3.2.6. Site probe studies
Sudlow et al. [28] suggested two main distinct binding sites (sites I and II) in BSA. Site I of BSA has affinity for warfarin and site II for ibuprofen. It was reported that the binding of digitoxin is independent of sites I and II [29] and binds to site III. In order to establish the binding site in the BSA for FOS, competitive binding studies were performed using site probes, warfarin, ibuprofen and digitoxin, in connection with Sudlow’s classification of the binding sites. Table 4 shows the binding constant of FOS–BSA in presence of different site markers. As evident from Table 4, FOS was not significantly displaced by ibuprofen or digitoxin. However, warfarin (site I) showed a displacement of FOS, suggesting that FOS bound to site I of BSA, which is located in the hydrophobic pocket of subdomain II A [6].
Table 4.
The comparison of binding constants of FOS–BSA before and after the addition of site probe.
| Systems | Binding constant (M) |
|---|---|
| BSA+FOS | 4.714×103 |
| BSA+FOS+Warfarin | 1.397×102 |
| BSA+FOS+Ibuprofen | 4.694×103 |
| BSA+FOS+Digitoxin | 4.348×103 |
3.3. Confirmation investigation
3.3.1. Synchronous fluorescence study
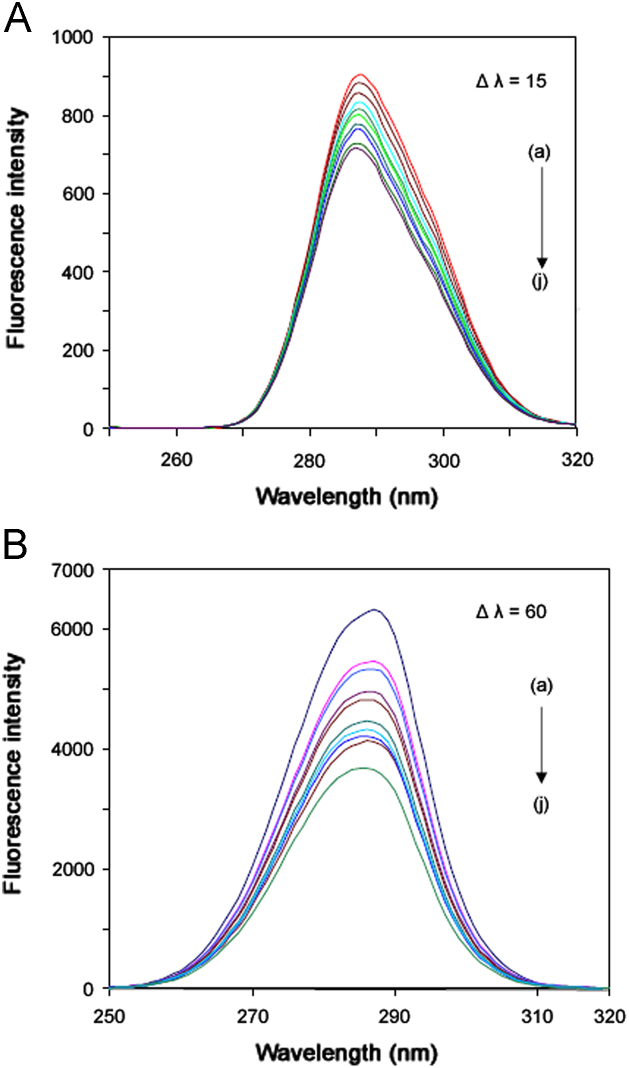
To explore the structural change of BSA by the addition of FOS, SFS (Fig. 2) of BSA were measured with various amounts of FOS. The SFS give information about the molecular environment in a vicinity of the chromosphere molecules and have several advantages, such as sensitivity, spectral simplification, spectral bandwidth reduction as well as avoiding different perturbing effects [30]. Yuan et al. [31] suggested a useful method to study the environment of amino acid residues by measuring the possible shift in wavelength emission maximum λmax, the shift in position of emission maximum corresponding to the changes of the polarity around the chromophore molecule. When the D-value (∆λ) between excitation wavelength and emission wavelength was stabilized at 15 or 60 nm, the SFS gave the characteristic information of tyrosine residues or tryptophan residues, respectively. The effect of FOS on protein SFS is shown in Fig. 2(A) and (B).
Fig. 2.
Synchronous fluorescence spectrum of BSA–FOS (T=298 K, pH 7.40): (A) ∆λ=15 nm and (B) ∆λ=60 nm. (a) [BSA]=5 μM; (b)–(j) [FOS]=5–45 μM.
In Fig. 2(A), it is observed that there is a gradual decrease of the fluorescence intensity of tyrosine residues and a blue shift at the maximum emission upon addition of FOS, which indicated that the binding between FOS and the protein is located in close proximity to the tyrosine residues. The conformation of BSA was changed, and it suggested a less polar (or more hydrophobic) environment of tyrosine residue [32]. While in Fig. 2(B), the maximum emission wavelength of tryptophan residues showed a blue shift, which indicates that the conformation of BSA was changed such that the polarity around the tryptophan residues decreased and the hydrophobicity was increased [33].
3.3.2. 3D fluorescence
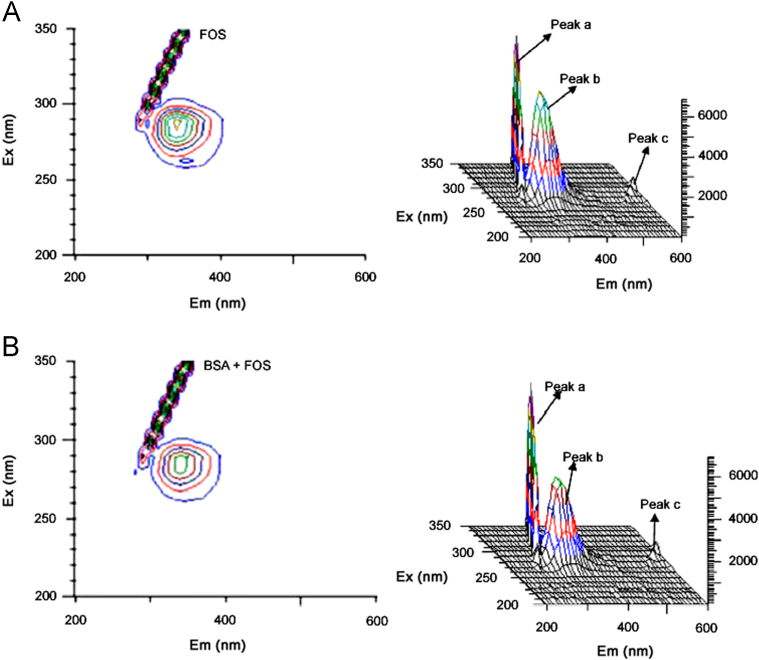
It is well known that 3D fluorescence spectra can provide more detailed information about the conformational changes of proteins. The maximum emission wavelength and the fluorescence intensity of the residues have a close relation to the polarity of their micro-environment [34], [31]. By comparing the 3D fluorescence spectral changes of BSA in the absence and presence of FOS, we can investigate the conformational and micro-environmental changes of BSA. The 3D fluorescence spectra of BSA and the BSA−FOS system are shown in Fig. 3. The changes of three dimension fluorescence spectra of BSA in the presence of different volumes of FOS are listed in Table 5.
Fig. 3.
Three dimensional fluorescence spectra of (A) BSA and (B) the FOS+BSA system.
Table 5.
Three-dimensional fluorescence spectral characteristics of HSA and the HSA–FOS system.
| System | Peak a | Peak b | |
|---|---|---|---|
| BSA | Peak position (Ex/Em) | 250/250–360/360 | 290/340 |
| Relative intensity (F) | 4.984–6617 | 5106 | |
| BSA–FOS | Peak position (Ex/Em) | 250/250–360/360 | 290/340 |
| Relative intensity (F) | 9.4–6933 | 3793 | |
In addition, the contour map provides bird’s eye view of 3D fluorescence spectra. Peak ‘a’ denotes the Raleigh scattering peak (λex=λem), whereas strong peak ‘b’ mainly reveals the spectral characteristics of Trp and Tyr residues on proteins, and another peak ‘c’ is the second-order scattering peak (λem=2λex).
In Fig. 3, the fluorescence intensity of peak ‘a’ increased with the addition of FOS. The possible reason was FOS–BSA complex formation after FOS was added. So the diameter of the macromolecule increased and resulted in the enhanced scattering effect [35]. Peak ‘b’ mainly reflected the spectral behavior of Trp and Tyr residues. The fluorescence intensity of peak ‘b’ decreased markedly and the maximum emission wavelength of the peak was changed following the addition of FOS. Peak ‘c’ was the second order scattering peak (λem=2λex), analyzing from the intensity changes of peak ‘a’ and peak ‘b’ revealed that the binding of FOS to BSA induced some conformational and micro-environmental changes in BSA [36].
3.3.3. Circular dichroism spectra
CD is a powerful tool in elucidating the modifications of the secondary structure of biopolymers as a result of interaction with small molecules. The far UV−CD spectra of BSA exhibit a typical shape of a α-helix-rich secondary structure (two minima at approximately 208 and 222 nm). BSA has a high percentage of α-helical structure, which shows characteristic strong double minimum signals at 222 and 208 nm, which represents the transition of π−π* and n–π* of α-helix structure [37]. The results of CD were expressed in terms of the mean residue ellipticity (MRE) in according to the following equation
| (9) |
where Cp is the molar concentration of the protein, n is the number of amino acid residues and l is the path length. The α-helical contents of free and combined BSA were calculated from MRE values at 208 nm using the following equation [38]
| (10) |
where MRE208 is the observed MRE value at 208 nm, 4000 is the MRE of the β-form and random coil conformation cross at 208 nm and 33,000 is the MRE value of a pure α-helix at 208 nm. The CD spectra of BSA in the absence and presence of different concentrations of FOS are shown in Fig. 4.
Fig. 4.
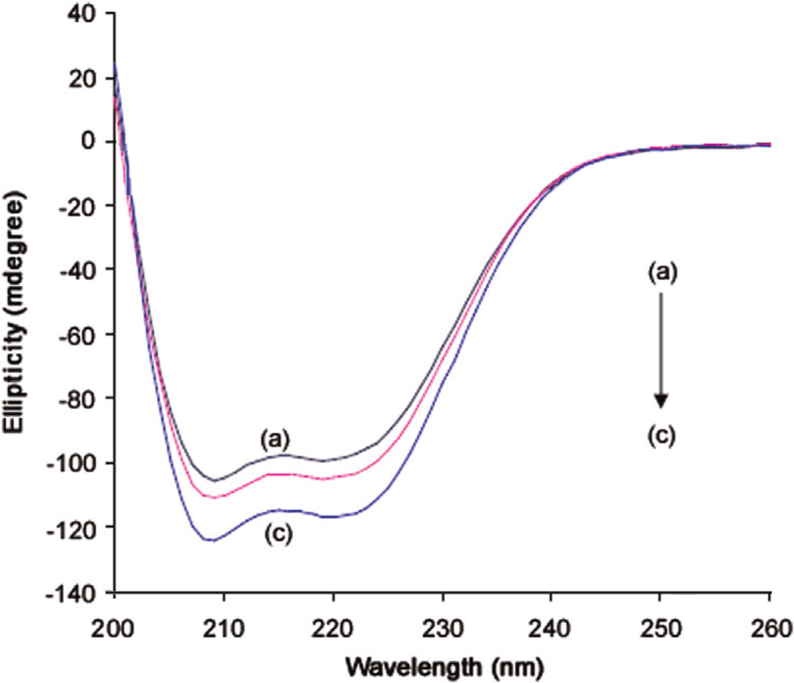
The CD spectra of the FOS–BSA system: (a) free BSA, (b) and (c) BSA+FOS obtained at 298 K and at pH=7.4.
From the above equation, the percentage of helicity of BSA is from 62% in free BSA (Fig. 4) to 67% and 79% in the presence of FOS–BSA, which showed that binding of FOS to BSA might induce some conformational changes. However, the shape of peaks and the peak maximum position remained almost the same. This indicated that BSA has predominantly α-helix nature even after binding to the drug. In the presence of FOS, no appreciable perturbation of secondary and tertiary structures in the protein is observed (Fig. 4). Thus, FOS, the aromatic and peptide regions do not show any appreciable change in the CD spectra.
3.3.4. Molecular modeling results
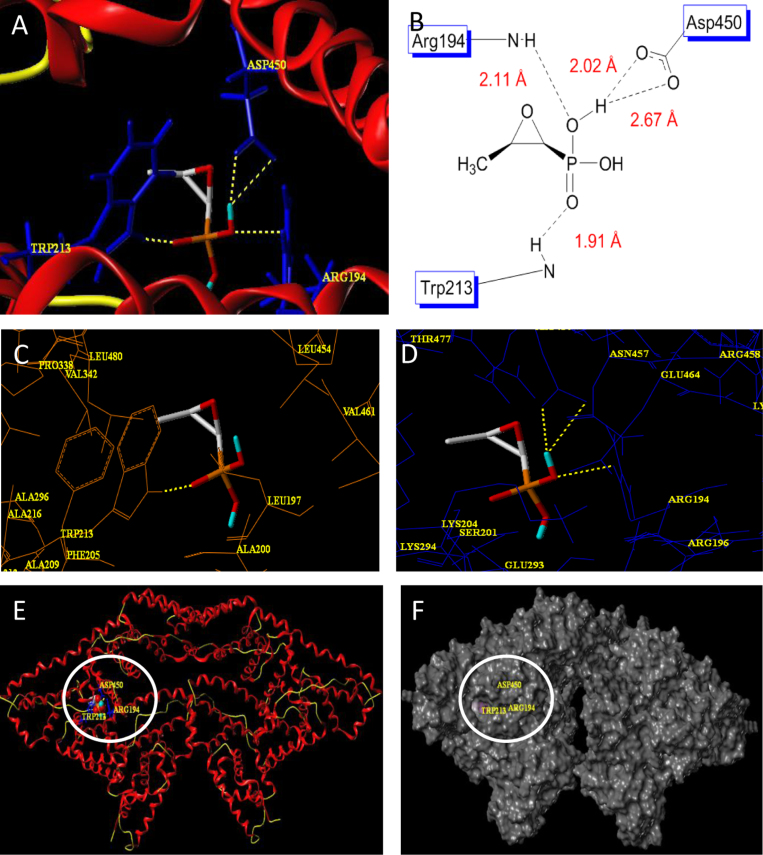
Surflex-docking was employed to understand the interaction between BSA and FOS and to ultimately elucidate the interaction mechanism. Results obtained by Surflex-docking tools presented 20 conformations of FOS. We selected the best conformation for further analysis, owing to its higher binding affinity (4.50 kcal/mol) and the lowest molecular energy. Fig. 5(A) and (B) with best confirmation shows four H-bonds, oxygen at phosphate making one H-bond with Trp213 (P=O––HN–Trp213, 1.91 Å), that of oxygen of OH making H-bond with Arg194 (HO––HN–Arg194, 2.11 Å), hydrogen of same OH making two H-bonds with Asp450 (OH––OC–Asp450, 2.02, 2.67 Å). FOS was surrounded with hydrophobic amino acids (Leu197, Ala200, Phe205, Trp213, Ala216, Ala296, Pro338, Val342, Leu454, Val461, Leu480) and hydrophilic amino acids (Arg194, Arg196, Ser201, Lys204, Glu293, Lys294, Asn457, Glu464) (Fig. 5(C) and (D)). Fig. 5(E) and (F) represents the binding domain for FOS at the active site of BSA chain A. The principal ligand binding site on BSA is located in the hydrophobic cavities in subdomains IIA and IIIA, which are consistent with sites I and II. It is important to note that Trp213 is in subdomain IIA, from the H-bonding interactions and surrounded amino acids, which indicated that the binding location was Sudlow’s site I in subdomain IIA.
Fig. 5.
(A) Details of the interactions between BSA and FOS in the docked model. (B) 2D schematic representation of hydrogen bond interactions. (C) Hydrophobic and (D) hydrophilic amino acids surrounded to FOS. (E) and (F) Binding domain for FOS at BSA chain A.
4. Conclusions
In this work, we used different approaches to explore the interactions between FOS and BSA under physiological conditions. The experimental results clearly showed that FOS quenches the fluorescence of BSA by a static quenching mechanism. This work provided a more comprehensive study on the distance between the donor (protein) and acceptor (FOS) based on fluorescence resonance energy transfer. Experimental results from the quantitative analysis of circular dichroism, three dimensional fluorescence studies and synchronous fluorescence spectrum demonstrated that the binding of FOS to BSA induced some micro-environmental and conformational change of serum albumin. Our study is expected to provide important insights into the interactions of the physiologically important protein with FOS, facilitating further investigation on the pharmacological behavior of FOS. This type of investigation on drug–protein interaction assumes importance in life sciences, chemistry and clinical medicine.
Acknowledgments
One of the authors Manjunath D. Meti expresses gratitude to DST New Delhi for providing Innovation in Science Pursuit for Inspired Research (INSPIRE) fellowship (IF110548).
Footnotes
Peer review under responsibility of Xi׳an Jiaotong University.
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.jpha.2015.01.004.
Appendix A. Supplementary materials
Supplementary data
References
- 1.Yua J., Lib B., Daia P. Molecular simulation of the interaction between novel type rhodanine derivative probe and bovine serum albumin. Spectrochim. Acta A: Mol. Biomol. Spectrosc. 2009;74:277–281. doi: 10.1016/j.saa.2009.06.013. [DOI] [PubMed] [Google Scholar]
- 2.Zhao H.W., Ge M., Zhang Z.X. Spectroscopic studies on the interaction between riboflavin and albumins. Spectrochim. Acta A: Mol. Biomol. Spectrosc. 2006;65:811–817. doi: 10.1016/j.saa.2005.12.038. [DOI] [PubMed] [Google Scholar]
- 3.Hendlin D., Stapley E.O., Jackson M. Phosphonomycin, a new antibiotic produced by strams of streptomyces. Science. 1969;166:122–123. doi: 10.1126/science.166.3901.122. [DOI] [PubMed] [Google Scholar]
- 4.Munos J.W., Moon S., Mansoorabadi S.O. Purification and characterization of the epoxidase catalyzing the formation of fosfomycin from pseudomonas syringae. Biochemistry. 2008;47:8726–8735. doi: 10.1021/bi800877v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bergogne-Berezin E. In: Fosfomycin and Derivatives in Antimicrobial Agents. Bryskier A., editor. ASM Press; Washington, DC: 2005. pp. 972–982. [Google Scholar]
- 6.Kandagal P.B., Seetharamappa J., Ashoka S. Study of the interaction between doxepin hydrochloride and bovine serum albumin by spectroscopic techniques. Int. J. Biol. Macromol. 2006;39:234–239. doi: 10.1016/j.ijbiomac.2006.03.027. [DOI] [PubMed] [Google Scholar]
- 7.Tripos International, Sybyl-X 2.0, Tripos International, St. Louis, MO, USA, 2012.
- 8.Majorek K.A., Porebski P.J., Dayal A. Structural and immunologic characterization of bovine, horse, and rabbit serum albumins. Mol. Immunol. 2012;52(3–4):174–182. doi: 10.1016/j.molimm.2012.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Silva D., Cortez C.M., Cunha-Bastos J. Methyl parathion interaction with human and bovine serum albumin. Toxicol. Lett. 2004;147:53–61. doi: 10.1016/j.toxlet.2003.10.014. [DOI] [PubMed] [Google Scholar]
- 10.Bi S., Yan L., Wang B. Spectroscopic and voltammetric characterizations of the interaction of two local anesthetics with bovine serum albumin. J. Lumin. 2011;131:866–873. [Google Scholar]
- 11.Lakowicz J.R. 3rd ed. Springer; New York: 2006. Principles of Fluorescence Spectroscopy. [Google Scholar]
- 12.Lakowicz J.R., Weber G. Quenching of fluorescence by oxygen. Probe for structural fluctuations in macromolecules. Biochemistry. 1973;12:1461. doi: 10.1021/bi00745a020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Deepa S., Mishra A.K. Fluorescence spectroscopic study of serum albumin-bromadiolone interaction: fluorimetric determination of bromadiolone. J. Pharm. Biomed. Anal. 2005;38:556–563. doi: 10.1016/j.jpba.2005.01.023. [DOI] [PubMed] [Google Scholar]
- 14.Wei Y.L., Li J.Q., Dong C. Investigation of the association behaviors between biliverdin and bovine serum albumin by fluorescence spectroscopy. Talanta. 2006;70:377–382. doi: 10.1016/j.talanta.2006.02.052. [DOI] [PubMed] [Google Scholar]
- 15.Cheng Z., Zhang Y. Spectroscopic investigation on the interaction of salidroside with bovine serum albumin. J. Mol. Struct. 2008;889:20–27. [Google Scholar]
- 16.Xiao J., Shi J., Cao H. Analysis of binding interaction puererin and bovine serum albumin by multi-spectroscopic method. Pharm. Biomed. Anal. 2007;45:609–615. doi: 10.1016/j.jpba.2007.08.032. [DOI] [PubMed] [Google Scholar]
- 17.Ju P., Fan H., Liu T. Probing the interaction of flower-like CdSe nanostructure particles targeted to bovine serum albumin using spectroscopic techniques. J. Lumin. 2011;131:1724–1730. [Google Scholar]
- 18.Zhang Y.Z., Xiang X., Dai P.M.J. Spectroscopic studies on the interaction of congo red with bovine serum albumin. Spectrochim. Acta A: Mol. Biomol. Spectrosc. 2009;72:907–914. doi: 10.1016/j.saa.2008.12.007. [DOI] [PubMed] [Google Scholar]
- 19.Lakowicz J.R. Plenum; New York: 2006. Principles of Fluorescence Spectroscopy. pp. 278, 281, 283. [Google Scholar]
- 20.Förster T., Sinanoglu O. Academic Press; NewYork, USA: 1996. Modern Quantum Chemistry. pp. 93–136. [Google Scholar]
- 21.Bi S., Song D., Tian Y. Molecular spectroscopic study on the interaction of tetracyclines with serum albumins. Spectrochim. Acta A: Mol. Biomol. Spectrosc. 2005;61:629–636. doi: 10.1016/j.saa.2004.05.028. [DOI] [PubMed] [Google Scholar]
- 22.Cui F.L., Fan J., Ma D.L. A study of the interaction between a new reagent and serum albumin by fluorescence spectroscopy. Anal. Lett. 2003;36:2151–2166. [Google Scholar]
- 23.Jiang C.Q., Gao M.X., Meng X.Z. Study of the interaction between daunorubicin and human serum albumin, and the determination of daunorubicin in blood serum samples. Spectrochim. Acta A: Mol. Biomol. Spectrosc. 2003;59:1605–1610. doi: 10.1016/s1386-1425(02)00362-1. [DOI] [PubMed] [Google Scholar]
- 24.Cui F.L., Fan J., Li J.P. Interactions between 1-benzoyl-4-p-chlorophenyl thiosemicarbazide and serum albumin: investigation by fluorescence spectroscopy. Bioorg. Med. Chem. 2004;12:151–157. doi: 10.1016/j.bmc.2003.10.018. [DOI] [PubMed] [Google Scholar]
- 25.Li Y., He W., Liu J. Binding of the bioactive component jatrorrhizine to human serum albumin. Biochim. Biophys. Acta. 2005;1722:15–21. doi: 10.1016/j.bbagen.2004.11.006. [DOI] [PubMed] [Google Scholar]
- 26.Guo X.J., Zhang L., Sun X.D. Spectroscopic studies on the interaction between sodium ozagrel and bovine serum albumin. J. Mol. Struct. 2009;928:114–120. [Google Scholar]
- 27.Zhang Y.Z., Zhang X.P., Hou H.N. Study on the interaction between Cu(phen)23+ and bovine serum albumin by spectroscopic methods. Biol. Trace Elem. Res. 2008;121:276–287. doi: 10.1007/s12011-007-8045-z. [DOI] [PubMed] [Google Scholar]
- 28.Sudlow G., Birkett D.J., Wade D.N. Further characterization of specific drug binding sites on human serum albumin. Mol. Pharmacol. 1976;12:1052–1061. [PubMed] [Google Scholar]
- 29.Sjoholm I., Ekman B., Kober A. Binding of drugs to human serum albumin:XI. The specificity of three binding sites as studied with albumin immobilized in microparticles. Mol. Pharmacol. 1979;16:767–777. [PubMed] [Google Scholar]
- 30.Hu Y.J., Liu Y., Pi Z.B. Interaction of cromolyn sodium with human serum albumin: a fluorescence quenching study. Bioorg. Med. Chem. 2005;13:6609–6614. doi: 10.1016/j.bmc.2005.07.039. [DOI] [PubMed] [Google Scholar]
- 31.Yuan T., Weljie A.M., Vogel H.J. Tryptophan fluorescence quenching by methionine and selenomethionine residues of calmodulin: orientation of peptide and protein binding. Biochemistry. 1998;37:3187–3195. doi: 10.1021/bi9716579. [DOI] [PubMed] [Google Scholar]
- 32.He L.L., Wang X., Liu B. Interaction between ranitidine hydrochloride and bovine serum albumin in aqueous solution. J. Solut. Chem. 2010;39:654–664. [Google Scholar]
- 33.Chi Z., Liu R., Teng Y. Binding of oxytetracycline to bovine serum albumin: spectroscopic and molecular modeling investigations. J. Agric. Food Chem. 2010;58:10262–10269. doi: 10.1021/jf101417w. [DOI] [PubMed] [Google Scholar]
- 34.Ding F., Li N., Han B. The binding of C.I. acid red 2 to human serum albumin: determination of binding mechanism and binding site using fluorescence spectroscopy. Dyes Pigments. 2009;83:249–257. [Google Scholar]
- 35.Tian F.F., Jiang F.L., Han X.L. Synthesis of a novel hydrazone derivative and biophysical studies of its interactions with bovine serum albumin by spectroscopic, electrochemical, and molecular docking methods. J. Phys. Chem. B. 2010;114:14842–14853. doi: 10.1021/jp105766n. [DOI] [PubMed] [Google Scholar]
- 36.Juarez J., Lopez S.G., Cambon A. Influence of electrostatic interactions on the fibrillation process of human serum albumin. J. Phys. Chem. B. 2009;113:10521–10529. doi: 10.1021/jp902224d. [DOI] [PubMed] [Google Scholar]
- 37.Zhang J., Yan Q., Liu J. Study of the interaction between 5-sulfosalicylic acid and bovine serum albumin by fluorescence spectroscopy. J. Lumin. 2013;134:747–753. [Google Scholar]
- 38.Guo X.J., Hao A.J., Han X.W. The Investigation of the interaction between ribavirin and bovine serum albumin by spectroscopic methods. Mol. Biol. Rep. 2011;38:4185–4192. doi: 10.1007/s11033-010-0539-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data