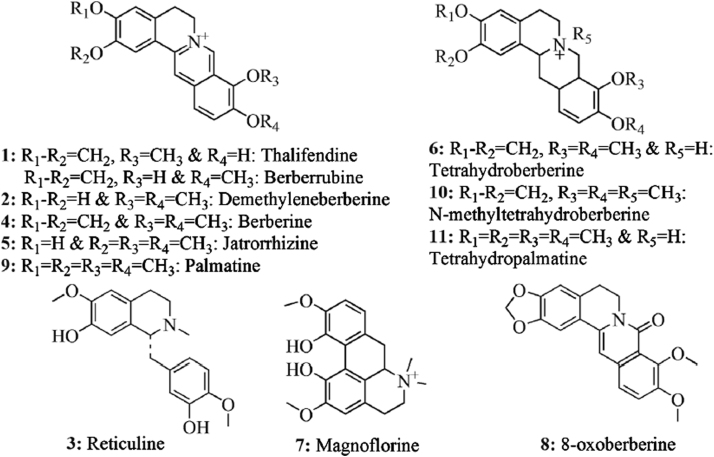
Abstract
Berberis petiolaris Wall. ex G. Don, an unexplored medicinal plant belonging to the family Berberidaceae, is a large deciduous shrub found in Western Himalaya between 1800–3000 m. Chemical profiling of fruit, leaf, root and stem was done by direct analysis in real time mass spectrometry followed by multivariate analysis for discrimination among the plant parts. The bioactive compounds, including magnoflorine, berberine, jatrorrhizine, thalifendine/berberrubine, demethyleneberberine, reticuline, 8-oxoberberine, N-methyltetrahydroberberine, tetrahydropalmatine, tetrahydroberberine and palmatine, were identified by their exact mass measurement and the corresponding molecular formula of each compound. A comparative study of distribution pattern for all these bioactive alkaloids showed qualitative and quantitative variations in different parts of B. petiolaris. Principal component analysis clearly discriminated each part of B. petiolaris plant.
Keywords: Berberis petiolaris, Alkaloids, Profiling, DART–TOF–MS, Statistical analysis
1. Introduction
Berberis petiolaris (B. petiolaris) is an unexplored deciduous shrub that belongs to the family Berberidaceae found in Western Himalaya between 1800–3000 m. This plant mainly contains different classes of alkaloids [1], [2], but only berberine, jatrorrhizine and palmatine were previously identified and isolated from B. petiolaris [3]. Decoction of root and stem of the plant is used for the treatment of malarial fever, diarrhea, conjunctivitis, and jaundice [4]. Recent developments in newer ionization methods have made it possible to obtain mass data directly from the plant parts without any sample preparation. Among the methods, direct analysis in real time (DART) is an ambient ionization technique requiring no sample preparation [5], [6]. It is, therefore, an appropriate technique for the rapid chemical profiling of plant species [7], [8]. The aim of present work was to develop a method for identification of biologically active compounds in B. petiolaris directly from intact plant parts. Different parts of the plant such as leaf, stem, root and fruit were screened to study comparative detection of the bioactive compounds in different parts of the plant. Hence, it was decided to profile the chemical constituents using DART time-of-flight mass spectrometry (DART–TOF–MS) and to study their variations in different plant parts of B. petiolaris.
2. Materials and methods
Plant samples of B. petiolaris were collected from Pandukholi forest, Almora Uttarakhand (India) and identified according to the flora of district Garhwal, northwest Himalaya and a forest flora of Kumaon. Herbarium specimen of B. petiolaris (KRA 24410) was housed in the departmental herbarium, Central Drug Research Institute (Lucknow, India). For DART–TOF–MS analysis, intact plant parts were used directly as the samples. The plant samples were thoroughly washed with tap water and distilled water in order to remove any foreign particles attached to their surface and kept in oven to dry at 40 °C.
The mass spectrometer was a Jeol the Accu TOF JMS-T100LC atmospheric pressure ionization time-of-flight mass spectrometer (Jeol, Tokyo, Japan) fitted with a DART ion source. The mass spectrometer was operated in positive-ion mode with a resolving power of 6000 (full-width at half-maximum). The orifice 1 potential was set to 28 V, resulting in minimal fragmentation. The ring lens and orifice 2 potential were set to 13 and 5 V, respectively. Orifice 1 was set to a temperature of 100 °C. The RF ion guide potential was 300 V. The DART ion source was operated with helium gas flowing at approximately 4.0 L/min. The gas heater was set to 300 °C. The potential on the discharge needle electrode of the DART source was set to 3000 V; electrode 1 was 100 V and the grid was at 250 V. Freshly cuted pieces of plant parts were positioned in the gap between the DART source and mass spectrometer for measurements. Data acquisition was from m/z 10 to 1050. Exact mass calibration was accomplished by including a mass spectrum of neat polyethylene (PEG) glycol (1:1 mixture of PEG 200 and PEG 600) in the data file. The mass calibration was accurate within ±0.002 u. Using the Mass Centre Main software (version 1.3.m; JEOL Japan), the elemental composition could be determined on selected peaks. Principal component analysis (PCA) analysis was carried out using Statistica windows version 7.0 (Stat Soft Inc., USA) statistical analysis software. This software normalizes observations with respect to mean and variance followed by PCA.
3. Results and discussion
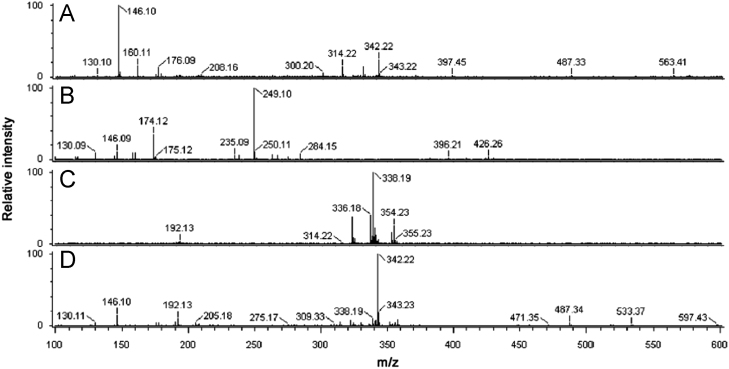
The identification from direct analysis of pharmaceutical products without any sample preparation is an ultimate achievement in pharmaceutical analysis for quality control and assurance of pharmaceutical products and herbal materials. Screening of bioactive constituents for identification of right selection of plant/parts for better efficacy is important for quality control of herbal products. DART–TOF–MS profiles or fingerprints of different plant parts such as fruit, leaf, root and stem of B. petiolaris were obtained after analysis in the present study. All these plant parts were subjected to DART–TOF–MS analysis under the same conditions. On the basis of literature report [9], [10] and our findings described below, some of the expected phytochemical components in B. petiolaris are shown in Fig. 1. These components are directly ionized from the plant part during analysis as molecular species in the resulting spectra. For instance, the DART mass spectra of the fruit, leaf, root and stem of B. petiolaris are given in Fig. 2. Peaks corresponding to the molecular species of thalifendine/berberrubine 1 (m/z 322), demethyleneberberine 2 (m/z 324), reticuline 3 (m/z 330), berberine 4 (m/z 336), jatrorrhizine 5 (m/z 338), tetrahydroberberine 6 (m/z 339), magnoflorine 7 (m/z 342), 8-oxoberberine 8 (m/z 352), palmatine 9 (m/z 352), N-methyltetrahydroberberine 10 (m/z 354) and tetrahydropalmatine 11 (m/z 355) were observed in the DART mass spectra. Thalifendine and berberrubine 1 have the same molecular formula and exact mass. Therefore, they cannot be distinguished on the basis of high resolution mass spectrometry (HRMS). Measured mass, calculated mass [2] and molecular formula [1] obtained from all the above constituents are reported in Table 1. Reticuline 3 (m/z 330) was not detected in B. petiolaris root or stem but detected in fruit and leaf. However, berberine 4 (m/z 336) was identified only in root and stem according to literature reports from other Berberis species [11]. Similarly, magnoflorine 7 was present in fruit, root and stem but absent in leaf. A peak at m/z 352 [M+H]+ corresponding to 8-oxoberberine 8 and a peak at m/z 352 [M]+ corresponding to palmatine 9 were observed in stem and root but not in leaf or fruit. N-methyltetrahydroberberine 10 (m/z 354) was also absent in the fruit, leaf and stem of B. petiolaris but showed significant presence in the root. DART–TOF–MS analysis of the fruit, leaf, root and stem of B. petiolaris showed differences in their spectra. Maximum abundance of compounds thalifendine/berberrubine 1 (m/z 322), berberine 4 (m/z 336), jatrorrhizine 5 (m/z 338) and N-methyltetrahydroberberine 10 (m/z 354) was observed in root, whereas magnoflorine 7 (m/z 342) showed its maximum abundance in fruit followed by stem. These observations confirmed that results obtained from DART–TOF–MS data were good, and it was the instrument of choice for the screening of natural products.
Fig. 1.
Chemical structure of indentified compounds.
Fig. 2.
DART mass spectra of (A) fruit, (B) leaf, (C) root and (D) stem.
Table 1.
Exact mass data from the DART mass spectra of B. petiolaris (R=root; S=stem; F=fruit; L=leaf).
| S.no. | Measured mass (m/z) | Calculated mass (m/z) | Error (mmu) | Molecular formula | Remarks | Peak type | Plant part |
|---|---|---|---|---|---|---|---|
| 1 | 322.10727 | 322.10793 | −0.66 | C19H16NO4 | Thalifendine/berberrubine | [M]+ | R |
| 2 | 324.12547 | 324.12358 | 1.89 | C19H18NO4 | Demethyleneberberine | [M]+ | R, S |
| 3 | 330.17039 | 330.17053 | −0.14 | C19H24NO4 | Reticuline | [M+H]+ | F, L |
| 4 | 336.12296 | 336.12358 | −0.62 | C20H18NO4 | Berberine | [M]+ | R, S |
| 5 | 338.13802 | 338.13923 | −1.21 | C20H20NO4 | Jatrorrhizine | [M]+ | R, S |
| 6 | 340.15262 | 340.15488 | −2.26 | C20H22NO4 | Tetrahydroberberine | [M+H]+ | R, S |
| 7 | 342.16916 | 342.17053 | −1.73 | C20H24NO4 | Magnoflorine | [M]+ | R, S, F |
| 8 | 352.12012 | 352.1185 | 1.62 | C20H18NO5 | 8-oxoberberine | [M+H]+ | R, S |
| 9 | 352.15538 | 352.15488 | 0.5 | C21H22NO4 | Palmatine | [M]+ | R, S |
| 10 | 354.17028 | 354.17053 | −0.25 | C21H24NO4 | N-methyltetrahydroberberine | [M]+ | R |
| 11 | 356.18700 | 356.18618 | 0.82 | C21H26NO4 | Tetrahydropalmatine | [M+H]+ | R, S |
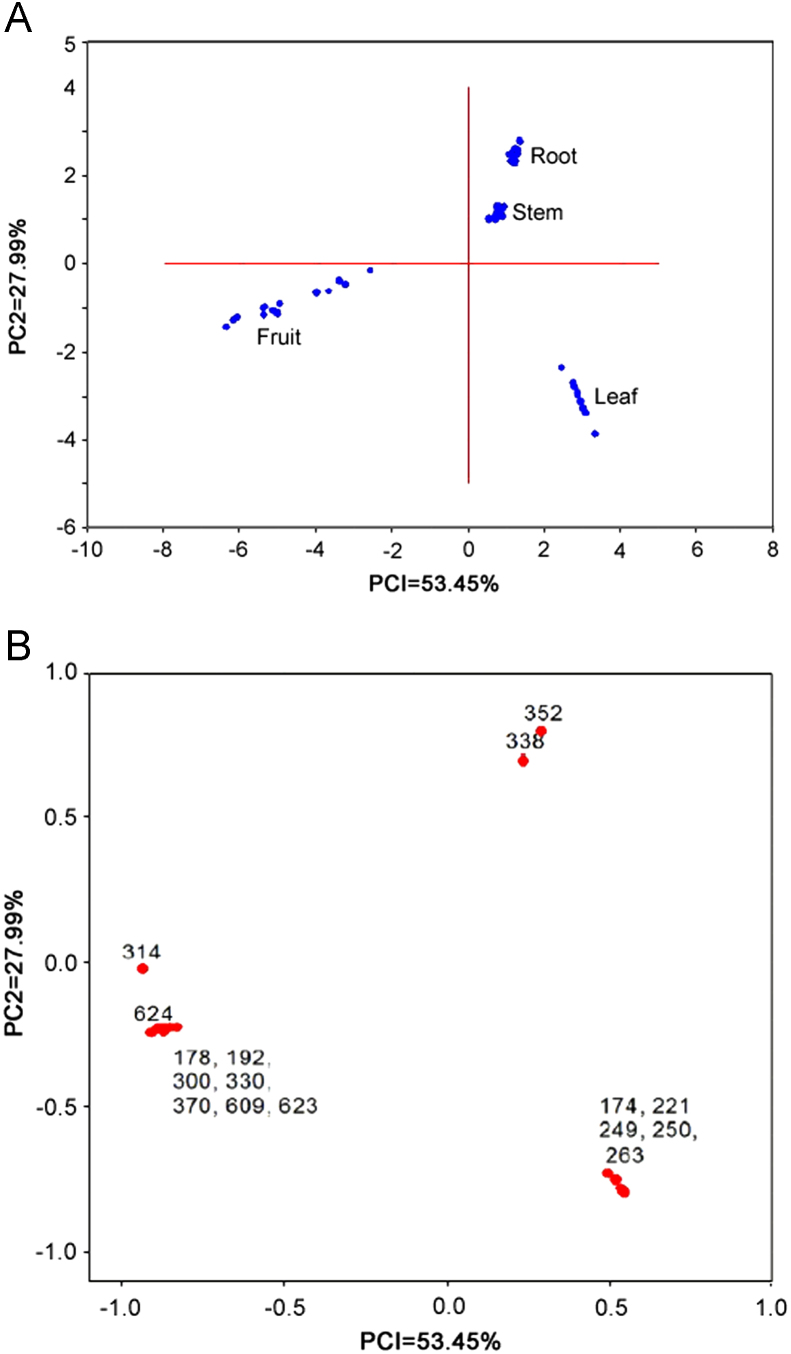
In metabolic profiling, identification of metabolite concentration changes by visual inspection of data is cumbersome and almost impractical for large sample sizes. Therefore, it is necessary to resort to multivariate techniques such as PCA, factor analysis, and partial least squares, which are important and proven techniques for complex data analysis [12]. We selected PCA for dimensionality reduction in an attempt to distinguish characteristic profiles from the DART–TOF–MS data. Accordingly, Fig. 3A shows the PCA plot which discriminates plant parts of B. petiolaris and their score plot (PC1 vs. PC2) obtained is given in Fig. 3B. It can be seen in Fig. 3A that the fruit, leaf, root and stem of B. petiolaris show clustering of the data according to the parts. Fig. 3A shows the distinct collection of PC scores in the biplot. The clustering of scores clearly shows the position of each plant part with a reasonable distance. This indicates that the first two PCs can easily discriminate the plant parts. Similar clustering and differentiation are clearly observed for the fruit, leaf, root and stem in Fig. 3B. Totally 16 peaks (m/z 178, 174, 192, 221, 249, 250, 263, 300, 314, 330, 338, 352, 370, 609, 623 and 624) were selected to study PCA for all the plant parts relying on percent (%) ionization of peaks (Table 2). PCA score plot clearly brings out relationship among all the plant parts. On the basis of eigen values, it was observed that the whole information of data matrix (81.44%) has been explained by two principal components. The contribution of PC1 was 53.45% and that of PC2 was 27.99%. Different parts of the same plant could be discriminated effectively by this technique and the presence and absence of peaks (as shown in Table 2) are complementary to PCA results. Peaks at m/z 338 and 352 were identified as markers for root and stem, while m/z 174, 221, 249, 250 and 263 for leaf and m/z 178, 192, 300, 314, 330, 370, 609, 623, 624 for fruit. It is, therefore, clear that DART–TOF–MS followed by PCA is an appropriate method for the clear differentiation of plant parts and rapid identification of compounds. The DART mass spectrometric technique has been applied for the first time for the profiling of alkaloids in B. petiolaris. Chemical profiling of fruit, leaf, root and stem was done successfully. It was observed that there were significant differences in mass spectra obtained from the fruit, leaf, root and stem of B. petiolaris. PCA analysis showed that DART–TOF–MS data could be used to differentiate these alkaloids containing plant species and to distinguish the plant parts. Peaks at m/z 338 [M]+ of jatrorrhizine, m/z 352 [M+H]+ of 8-oxoberberine and m/z 352 [M]+ of palmatine were identified as markers for root and stem. This will help with identification and characterization of genuine parts used for drug preparation and the effective quality control and efficacy of drug candidates in future.
Fig. 3.
(A) PCA plot discriminating plant parts of Berberis petiolaris. (B) Score plot discriminating plant parts of Berberis petiolaris.
Table 2.
Identified peaks which discriminated plant parts of B. petiolaris.
| Peaks | Remarks | Fruit | Leaf | Root | Stem |
|---|---|---|---|---|---|
| 174 | Unknown | − | + | − | − |
| 178 | Unknown | + | − | − | − |
| 192 | Unknown | + | − | − | − |
| 221 | Unknown | − | + | − | − |
| 249 | Unknown | − | + | − | − |
| 250 | Unknown | − | + | − | − |
| 263 | Unknown | − | + | − | − |
| 300 | Unknown | + | − | − | − |
| 314 | Unknown | + | − | + | + |
| 330 | Reticuline | + | + | − | − |
| 338 | Jatrorrhizine | − | − | + | + |
| 352 | 8-oxoberberine/palmatine | − | − | + | + |
| 370 | Unknown | + | − | − | − |
| 609 | Unknown | + | − | − | − |
| 623 | Unknown | + | − | − | − |
| 624 | Unknown | + | − | − | − |
Acknowledgments
Grateful acknowledgment is made to the Department of Science and Technology, India for Grant SB/EMEQ-095. Authors are also thankful to SAIF-CDRI, Lucknow, where all the mass spectral studies were carried out. Awantika Singh and Vikas Bajpai are also thankful to UGC and CSIR, respectively, for providing Senior Research Fellowship. CDRI communication number is 8994.
Footnotes
Peer review under responsibility of Xi׳an Jiaotong University.
References
- 1.Singh A., Bajpai V., Srivastava M. Rapid profiling and structural characterization of bioactive compounds and their distribution in different parts of Berberis petiolaris Wall. ex G. Don applying hyphenated mass spectrometric techniques. Rapid Commun. Mass Spectrom. 2014;28:2089–2100. doi: 10.1002/rcm.7001. [DOI] [PubMed] [Google Scholar]
- 2.Bajpai V., Singh A., Arya K.R. Rapid screening for the adulterants of Berberis aristata using direct analysis in real-time mass spectrometry and principal component analysis for discrimination. Food Addit. Contam. Part A. 2015;32:799–807. doi: 10.1080/19440049.2015.1022885. [DOI] [PubMed] [Google Scholar]
- 3.Bhardwaj D., Kaushik N. Phytochemical and pharmacological studies in genus Berberis. Phytochem. Rev. 2012;11:523–542. [Google Scholar]
- 4.Karimov A. Berberis alkaloids. Chem. Nat. Compd. 1993;29:415–438. [Google Scholar]
- 5.Bajpai V., Sharma D., Kumar B. Profiling of Piper betle linn cultivars by direct analysis in real time mass spectrometric technique. Biomed. Chromatogr. 2010;24:1283–1286. doi: 10.1002/bmc.1437. [DOI] [PubMed] [Google Scholar]
- 6.Cody R.B., Laramee J.A., Durst H.D. Versatile new ion source for the analysis of materials in open air under ambient conditions. Anal. Chem. 2005;77:2297–2302. doi: 10.1021/ac050162j. [DOI] [PubMed] [Google Scholar]
- 7.Kim S.W., Kim H.J., Kim J.H. A rapid, simple method for the genetic discrimination of intact Arabidopsis thaliana mutant seeds using metabolic profiling by direct analysis in real-time mass spectrometry. Plant Methods. 2011;7:14–24. doi: 10.1186/1746-4811-7-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim H.J., Jee E.H., Ahn K.S. Identification of marker compounds in herbal drugs on TLC with DART-MS. Arch. Pharm. Res. 2011;33:1355–1359. doi: 10.1007/s12272-010-0909-7. [DOI] [PubMed] [Google Scholar]
- 9.Grycova L., Dostal J., Marek R. Quaternary protoberberine alkaloids. Phytochemistry. 2007;68:150–175. doi: 10.1016/j.phytochem.2006.10.004. [DOI] [PubMed] [Google Scholar]
- 10.Arayne M.S., Sultana N., Bahadur S.S. The Berberis story-Berberis vulgaris in therapeutics. Pak. J. Pharm. Sci. 2007;20:83–92. [PubMed] [Google Scholar]
- 11.Cromwell B.T. Experiments on the origin and function of Berberine in Berberis darwini. Biochem. J. 1993;27:860–872. doi: 10.1042/bj0270860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li M., Zhou X., Zhao Y. Quality assessment of Curcuma longa L. by gas chromatography mass spectrometry fingerprint principle component analysis and hierarchical clustering analysis. Bull. Korean Chem. Soc. 2009;30:2287–2293. [Google Scholar]