Abstract
Deciphering the human brain pathophysiology remains one of the greatest challenges of the 21st century. Neurological disorders represent a significant proportion of diseases burden; however, the complexity of the brain physiology makes it challenging to model its diseases. Simple in vitro models have been very useful for precise measurements in controled conditions. However, existing models are limited in their ability to replicate complex interactions between various cells in the brain. Studying human brain requires sophisticated models to reconstitute the tangled architecture and functions of brain cells. Recently, advances in the development of three-dimensional (3D) brain cell culture models have begun to recapitulate various aspects of the human brain physiology in vitro and replicate basic disease processes of Alzheimer’s disease, amyotrophic lateral sclerosis, and microcephaly. In this review, we discuss the progress, advantages, limitations, and future directions of 3D cell culture systems for modeling the human brain development and diseases.
Keywords: 3D culture, biomaterials, disease modeling, microfluidics, organoids
1. Introduction
Neurological disorders including Alzheimer’s disease (AD), Parkinson’s diseases (PD), schizophrenia, amyotrophic lateral sclerosis (ALS), stroke, and brain injuries represent a significant proportion of diseases burden, and affect up to one billion people globally irrespective of sex, age and education or income.[1] Despite significant advances in the past decades in the study of neurological diseases, no effective treatment exists for many of these diseases. Several factors contribute to this situation, including the limitations of current experimental tools. Much of our knowledge about the human cerebral development, function and diseases is based on a limited number of techniques that help explore the human brain, e.g., imaging the brain shape and activity and analysis of samples obtained from postmortem brain.[2] Studies using animal models have also contributed tremendously to our current knowledge about how the brain functions. However, these techniques and models face significant challenges that stem from the complexity of the brain cellular interactions.[3,4] The human brain holds an estimated 86 billion neurons and 85 billion non-neuronal cells,[5] which makes it one of the most difficult organs to diagnose and treat. Thus, dissecting these interactions, to understand which drive the pathological processes and which are essential for protection against disease, is extremely difficult. One approach that worked well in other areas of human diseases is to separate specific interactions and study them in controlled conditions in vitro.
One of the most commonly used systems for studying cell-cell interactions in vitro is the two-dimensional (2D) cell culture. It provides low cost and simplified approaches for studying the brain development and its diseases. Recently, pharmaceutical companies and research labs have enhances the 2D cell culture studies by utilizing human cells, including primary cells, embryonic stem cells, and induced pluripotent stem cells (iPSC). However, despite the progress on the availability of cells for 2D culture, it fails to mimic the complexity of the human brain and its unique features and functions. Animal models offer complexity but are often unable to recapitulate the human brain pathophysiology accurately.[3,4] Other drawbacks include high cost and uncertainties in the results involved with the animal models. This inability to model faithfully causes many drugs to fail in transition from animal to human clinical trials. New cell culture systems that enable us to reconstruct the elementary architectural components and the three-dimensional (3D) microenvironments of the human brain pathophysiology are needed.
Over the past decade, there has been a dramatic effort to develop 3D in vitro brain models thanks to the emergence of enabling technologies including stem cells, biomaterials and microfabrication techniques. They allow manipulation of cellular microenvironment and endow scientists with a new tool set that raises the complexity of tissues containing multiple cell types in the laboratory to new levels of sophistication in organs-on-chip constructs.[6,7] These 3D physiologically relevant cell culture systems aim to closely mimic the human tissues and provide high-throughput and reproducible studies. The 3D cell-cell interactions and physiological cues provided by the extracellular matrix (ECM) tend to offer an in vivo-like environment to the cells. Several of these ideas have been applied towards building 3D models of the central nervous system.[8] Two main approaches are inspired from organs-on-chip work: (i) bottom-up fabrication relies on designing the scaffolding architecture and then populating it with the cells of choice,[9] and (ii) top-down fabrication relies on self-organization of pluripotent stem cells or neural progenitor cells into structures within the natural or artificial ECM (e.g., cerebral organoids, and neurospheroids).[10–13] Several recent models of 3D cell culture systems have shown great potential to surpass the current 2D cell culture and to contribute to the study of the human brain development and pathology.
In this review, we will focus on the 3D cell culture models of the human brain. We will discuss the promises and limitations of such models in the context of neurological diseases and towards developing new cures (Figure 1). We will also discuss the recent advances of organs-on-chip technology and the emergence of synthetic biomaterials to mimic the in vivo micro-environments of the brain. We will underline the strengths and limitations of current techniques and comment on the next challenges that must be addressed in order to develop increasingly sophisticated and physiologically relevant in vitro 3D brain models.
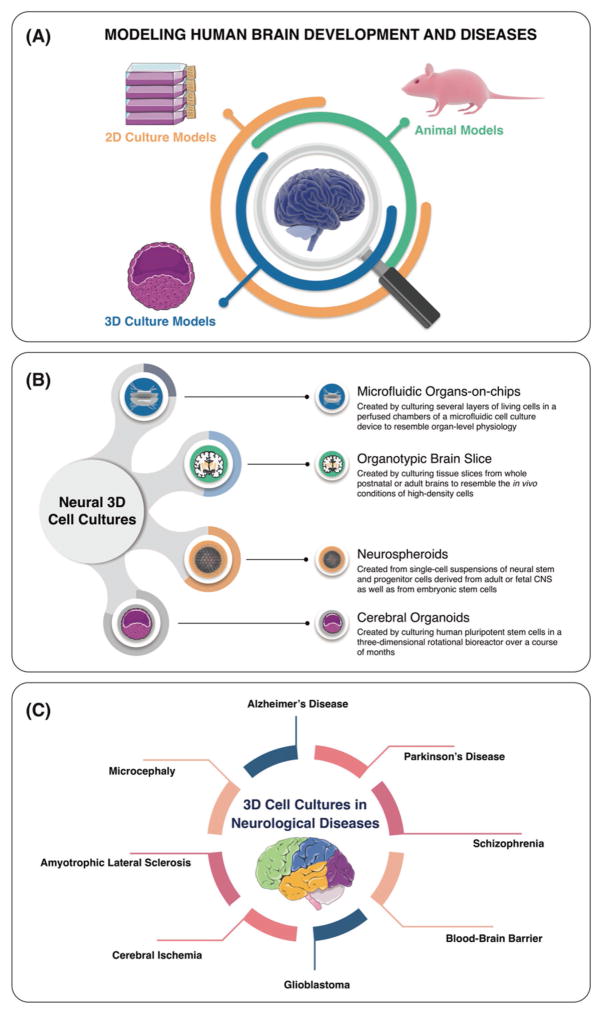
Figure 1.
Schematic representation of modeling the human brain development and its diseases (A), different neural 3D cell culture systems (B), and applications of 3D cellular models for neurological diseases (C).
2. Recapitulating the Human Brain Pathophysiology
2.1. Introduction on the Cell Biology of the Human Brain
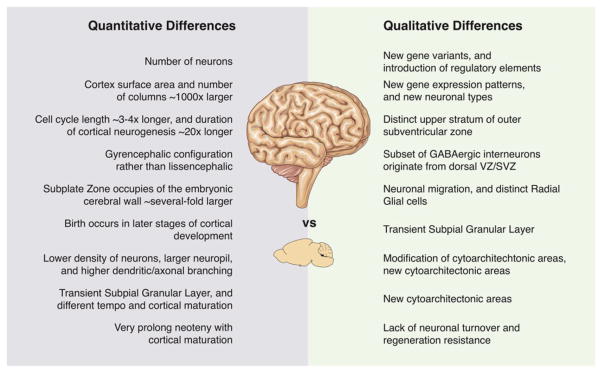
After 150 years of Darwin’s The Origin of Species, the majority of scientists concur that the human brain has extraordinary capacity correlated with human cognitive abilities and functions including thinking, memory, language, motor skills, and other cognitive functions.[14] Understanding the cell biology of the human brain development offers insights into the pathogenesis of the human brain diseases such as AD, PD, schizophrenia, autism spectrum disorder, and other human-specific neuropsychiatric disorders and possibly discover new therapeutics. Comparative anatomy studies revealed the evolution of human brain over time and the substantial expansion of cerebral cortex in comparison to other hominids. This created quantitative and qualitative differences between human and other mammals neocortex (Figure 2).[14]
Figure 2.
Quantitative and qualitative differences between developing human and mouse neocortex. Adapted with permission.[14] Copyright 2013, Elsevier.
During brain development, neural stem and progenitor cells generate all the neurons and glial cells (astrocytes, oligodendrocytes and Schwann cells) via a process called neurogenesis.[15,16] Neurogenesis process is followed by neuronal migration, differentiation, dendrite, axon formation, synaptogenesis, and neuronal network formation. These processes occur in parallel with other non-neuronal processes including origination of glial cells, myelination, angiogenesis, and generation of the blood-brain barrier (BBB). The neurogenesis process occurs in all regions of the neural tube either by symmetric or asymmetric cells divisions. In symmetric cell divisions, neural stem cells generate two cells with same fate, whereas the asymmetric cell divisions contribute to the generation of vast array of different cell types. Particularly, during the mammalian neocortex development, neuroepithelial cells (or stem cells) first undergo symmetric cell divisions followed by several asymmetric cell divisions.[15,16] In this process, neurons result from asymmetric divisions, whereas symmetric divisions initiate self-renewal of progenitor cells. Typically, the mammalian neocortex consists of six layers of neurons and glial cells. This process relys on spindle orientation, which has central implications in the human brain development and disease circumstances.[17] For example, microcephaly can begin by mutations in genes with certain roles in spindle orientation in the human cerebral cortex.[18]
2.2. The Emergence of Organs-on-Chip Models
The last decade has seen a growing trend towards engineering microphysiological systems (MPSs) called ‘organs-on-chip’ – that are, microfluidic-based microsystems capable of mimicking in vivo human physiology at small scale.[6] MPSs reconstitute the key physiological elements and functions of real organs including lung, liver, kidney, gut, heart, and brain in a miniaturized, and well-controlled microenvironment.[19–24] Aside from ethical considerations, MPSs overcome the high cost of animal care, the complexity of tissue isolation, the need for transgenic animals, as well as many of the uncertainities of translation of animal models to human physiology. These systems allow us to use cells isolated directly from patients, resulting in a more physiologically relevant system for mimicking the comprehensive disease pathology. Organs-on-chip also accelerate pharmaceutical testing by harnessing the potential of microfluidic high-throughput technologies to lower cost, increase reproducibility, and speed up drug screening for adsorption, distribution, metabolism, excretion and toxicity (ADME-Tox) compared to animal models that tend to be expensive and poor predictors.[25]
2.3. Microfluidics for 2D In Vitro Culture of Brain Cells; Simple Circuits
Microfluidic systems offer new opportunities for addressing unresolved challenges in neurobiology by precise control of the cellular microenvironment in both space and time. Over the past decade, the world has seen a significant increase in the development of microfluidic devices for both fundamental and applied neurobiology research, including neuronal culture and manipulation, neuropharmacology, neural stem cell differentiation, and neuro-electrophysiology. The use of microfluidic systems and tools for neurobiology applications has been recently reviewed.[26–30] In this section, we limit the discussion to representative examples of microfluidic systems in the content of neuropathology.
Microfluidic devices with micrometer-scale channels and nano-liter volumes are addressing the needs to control extra-cellular signals and intercellular responses by controlling spatial, chemical and temporal cellular microenvironment and allowing high-resolution measurements and imaging of brain cells and circuits in stable culture platform. Microfluidics offers substantial advantages for studying neuronal development and manipulation at subcellular, and cellular levels.[27] The main advantages of microfluidic technology over conventional cell culture models for studying neuronal development and manipulation include (i) high reproducibility, (ii) easy assembly, (iii) fluidic control, (iv) material versatility, (v) design flexibility, and (vi) experimental feasibility.
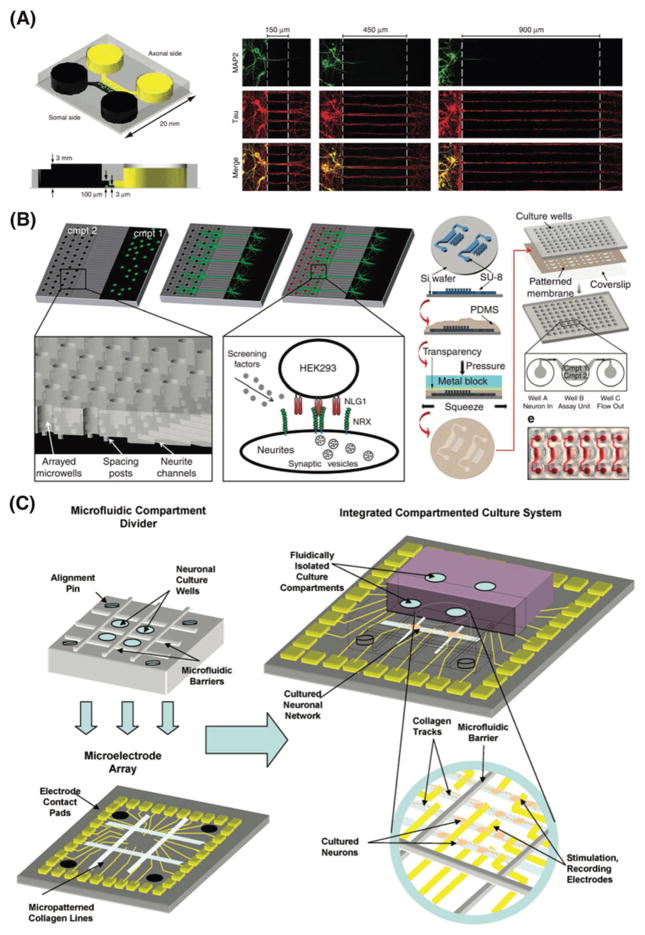
Microfluidic compartmentalized systems were established for neuronal development and manipulation.[31–33] These microfluidic-based multi-compartment platforms incorporate microgrooves to grow and direct axons in which presynaptic and postsynaptic neurons are isolated in well-controlled microenvironment (Figure 3A).[31–33] These microfluidic platforms provide excellent tools to separately analyze axons and cell bodies of neurons, which can be extremely useful to study axonal injury, to screen molecules that are secreted from different compartments, and to build co-culture models with other supporting cell types (e.g., oligodendrocytes). Overall, the ability to direct axonal growth and to precisely control the microenvironments of axons offers a new tool to overcome several limitations in answering multiple aspects of axonal biology that have been previously hindered by a lack of appropriate in vitro cell culture platform. Adaptations of the compartmentalized design have been utilized to investigate biochemical sensors in hippocampal neurons,[34] isolating axonal mRNA from cortical neurons,[35,36] dendrite-to-nucleus signaling,[37] presynaptic differentiation,[38] netrin-dependent axon guidance,[39] distal projections of human embryonic stem cells,[40] axonal diodes,[41] functional imaging of neuron-astrocyte interactions,[42] quantitative analysis of axonal transport,[43] co-culture of neurons and glia cells,[44,45] and neuronal activities on microelectrode arrays.[46–48]
Figure 3.
Mimicking the human brain physiology using microphysiological systems. A) Microfluidic compartmentalized cell culture system for directing axonal growth and isolating axons without soma or dendrites. Reproduced with permission.[32] Copyright 2005, Nature Publishing Group. B) Schematic representation of the high-throughput synapse microarray platform for quantitative screening of synaptogenesis in large-scale. Reproduced with permission.[50] Copyright 2011, Nature Publishing Group. C) Schematic representation of compartmented microfluidic culture system integrated with microelectrode array for neural pharmacology and electrophysiology. Reproduced with permission.[46] Copyright 2007, Elsevier.
Microfluidic technologies are exploited in the development of high-throughput and cost-effective assays for drug screening, including more reliable assessment of drug toxicity, efficacy, and pharmacokinetics of drug candidates.[30,49,50] For example, a high-throughput microarray utilizing the compartmentalized microfluidic platform was recently employed for quantitative screening of synaptogenesis in large-scale (Figure 3B).[50] By using this high-throughput platform, authors screened a chemical library that leads to discovery of class I histone deacetylase inhibitors as potential regulators of neuroligin-1-induced synaptogenesis pathway. This technology could also provides an excellent tool to study other neurological functions such as cell-cell interactions and neuronal development. Elegant combinations of integrated compartmented microfluidic culture systems with micro-electrode arrays enabled concurrent studies of neural pharmacology and electrophysiology (Figure 3C).[46] Aside from compartmentalized microfluidic devices, researchers have been engineering other novel designs of brain-on-a-chip models to recapitulate numerous aspects of the human brain physiology in vitro.[51–53]
Undoubtedly, microfluidic models advanced our understanding of the brain biology and have the power to answer fundamental questions in a sophisticated manner, although the era of microfluidic technology in neurobiology is still in its infancy. There are undeniable issues with the use of microfluidic devices including (i) shear stress, (ii) gas solubility, permeability and diffusibility, (iii) absorption, (iv) adsorption, (v) desorption, and (vi) evaporation that could potentially limit neuronal development in these systems. New designs and improvements in recent and upcoming years will minimize or eliminate unfavorable effects of these microfluidic devices for recapitulating human brain physiology in vitro. For more detailed discussion on the advantages and disadvantages of microfluidic devices, readers are referred to a recent review.[27]
2.4. Recent Advances in Organs-on-Chip for Neurological Diseases
Neurological diseases represent a significant proportion of diseases burden, however the development of functional and effective human brain models remains challenging due to the inherent complexity of the human brain physiology.[4] Although, great advances have been made in the last decades using animal models such as transgenic mice, many of these animal models fail to fully recapitulate the human brain pathophysiology. It is also difficult to study key cellular and molecular mechanisms underlying brain diseases pathology in whole-animal models. To tackle these challenges, engineers in collaboration with biologists started to leverage recent advances in stem cells, biomaterials, and microfabrication techniques with the ultimate goal to develop in vitro models of the human brain. Such in vitro models offer a very powerful platform for understanding the human brain as well as drug development and discovery in pharmaceutical industry for neurological diseases such as AD, PD, traumatic brain injury and related damage to the brain. Additionally, these experimental models could be invaluable test beds for assessing toxicology and the responses of the human brain cells to a variety of therapeutics. Overall, we have seen increasing interest in the last few years in the development of in vitro models of the brain, however; such models are still in their infancy. In the following sections, we provide some examples of recent experimental models and platforms and their impacts in neurobiology field.
2.4.1. Alzheimer’s Disease Models
Alzheimer’s disease (AD), a devastating and still incurable neurodegenerative disease, is characterized by two main pathological hallmarks: amyloid-β (Aβ) plaques, and neurofibrillary tangles.[54–58] Scientists could not understand well the mechanism of Aβ plaques at the subcellular level, partly due to the challenges associated with 2D cell cultures to isolate axons and/or dendrites from the cell body. A great deal of research into recapitulating Aβ pathology in vitro has focused on exploiting the compartmentalized microfluidic devices that enable us to study the mechanism locally and thereby advancing our understanding of the Aβ pathology.[59–63]
Our group studied microglia accumulation in the vicinity of Aβ plaques in AD pathology using microfluidic chemotaxis platform.[60] We found that soluble monomeric and oligomeric Aβ plays as a “recruiting signal”, whereas fibrillar and oligomeric Aβ serves as a “targeting signal” throughout microglia recruitment and localization. This reveals that soluble and insoluble Aβ species have synergistic role on microglia accumulation locally in close distance from Aβ plaques. Assessing the correlation between different forms of Aβ plaques (oligomeric versus fibril) and their neurotoxicity, it was found that although the number of Aβ fibrils increase over time, they do not control the neuronal cell toxicity.[62] This indicates the potential neurotoxicity of oligomeric Aβ compared to fibril Aβ in the AD pathology.
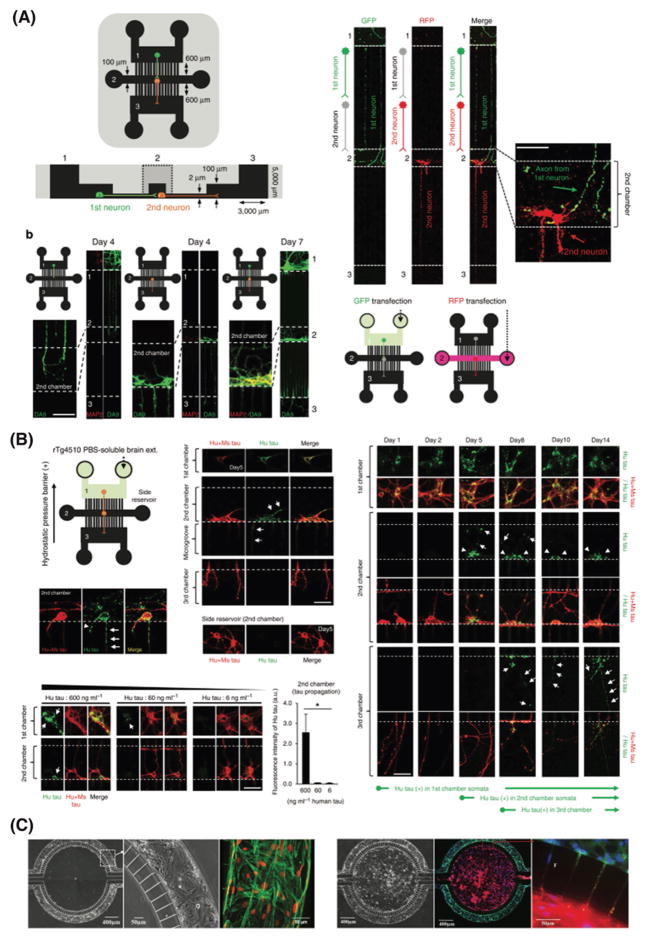
We also studied the neuron-to-neuron propagation of brain-derived tau species and the lifetime of internalized high-molecular-weight (HMW) tau using three-chamber compartmentalized microfluidic device (Figure 4A and B).[61] We found that the HMW becomes very stable once taken up by neurons, likely due to the hyperphosphorylation of tau species. Trans-synaptic propagation of tau species occurs rapidly and the released tau is taken up by the next neuron. This shows the central role of the release and uptake of the HMW tau species in propagation of tau across brain regions.[61]
Figure 4.
Mimicking neurological diseases using microfluidic systems. A) Three-chamber compartmentalized microfluidic devices used to study the neuron-to-neuron propagation of brain-derived tau species and (B) neuron-to-neuron transfer of mouse brain derived human tau. Reproduced with permission.[61] Copyright 2015, Nature Publishing Group. C) 3D in vitro BBB model composed of two-compartments chamber with a vascular channel cultured with rat brain endothelial cells (green) under shear flow and a tissue compartment seeded with astrocytes under static flow (red). Reproduced with permission.[96] Copyright 2015, PLoS ONE.
2.4.2. Parkinson’s Disease Models
Compartmentalized microfluidic devices have been used for modeling another neurodegenerative disease: Parkinson’s disease (PD). Histologically, PD, the second common neurodegenerative disease after AD, is primarily defined by the death of dopaminergic neurons in the substantia nigra pars compacta (SNpc) and intracellular aggregations of Lewy Bodies (LBs).[64] Volpicelli-Daley et al. studied the intracellular propagation of pathologic protein α-synuclein (α-syn) aggregates, a major constituent of Lewy bodies using compartmentalized microfluidic system.[65] They found that α-syn fibrils aggregations induce Lewy body pathology and leads to synaptic dysfunction and neuronal cell death. Lu et al. observed the transport of mitochondria along dopaminergic axons by developing another compartmentalized microfluidic platform.[66]
2.4.3. Cerebral Ischemia (Stroke) Models
Stroke is a devastating neurological disease caused by abrupt blockage of blood vessels supplying the brain, leading to hypoxia in the brain tissue and eventually death and dysfunction of brain cells.[67] We should understand the molecular mechanisms of neuronal death following stroke in order to foster drug discovery and screening.[68] In one recent example, Samson et al. replicated the spreading neurotoxicity following cerebral ischemia and traumatic brain injury using hippocampal neurons cultured in a microfluidic platform.[69] The neurons were environmentally isolated but synaptically connected – to assess neuroprotection and overcome the challenge in animal models of separating initial brain lesion from the entire brain response. Current in vitro models of stroke often use oxygen-glucose deprived (OGD) media over entire cell culture system. These models fail to physiologically mimic the pathology of stroke in particular focal ischemia.[70] Thus, it is of particular interest to develop experimental models that capable of applying hypoxia to specific regions of interests in the brain tissue. Several microfluidic devices have been developed over the last decade to generate oxygen gradients over cellular cultures by relying on the oxygen permeability of polydimethylsiloxane (PDMS).[71–76] Overall, researchers have investigated multiple approaches to model the ischemia stroke, however current microfluidic-based in vitro stroke models are still very challenging and further improvements are needed to faithfully recapitulate the disease pathology and understand the responses of brain cells following oxygen deprivation in stroke.
2.4.4. Amyotrophic Lateral Sclerosis
Amyotrophic lateral sclerosis (ALS), a progressive, late-onset neurodegenerative disease of motor neurons is characterized by two pathological hallmarks including progressive weakness and muscle atrophy, and spasticity, resulting in death of motor neurons and eventual fatal paralysis.[77] Despite enormous effort using in vivo models to understand the disease pathology and find a cure, limited therapeutics have been identified for slowing the disease course. Therefore, in vitro ALS models have emerged in recent years to tackle the challenges associated with large scale drug screening in animal models. For example, Kunze et al. used microfluidic systems to study non-cell autonomous conditions in ALS that initiate indirect interactions between neurons and astrocytes (extracellular metabolic communication) and to physically separate these from direct cell-cell contact.[78] They assessed neuronal cells activity in response to astrocytes by co-culturing neurons with genetically altered astrocytes that overexpress either a mutations in superoxide dismutase (SOD1) or a human wild-type (WT). This microfluidic platform allowed them to efficiently study the neuron-astrocytes metabolic interactions in vitro. Recent studies on mimicking neuromuscular junctions using micro-fluidic devices revealed that this technology could closely recapitulate ALS conditions in a dish.[79,80]
2.4.5. Mimicking the Blood-Brain Barrier in Neurological Diseases
Mimicking the physiochemical properties of the blood-brain barrier (BBB) has been a topic of interest in the last two decades due to the essential role of the neurovascular unit in multiple neurological diseases. 3D modeling of the neurovascular unit in vitro has been already reviewed in detail[81–83] and it is beyond the scope of this Progress Report to discuss the in vitro modeling of the BBB. Therefore, we will limit our discussion just to a few recent 3D models of the BBB using the micro-fluidic technology and their implications in understanding the human brain disease pathology. There is a growing evidence associating the neurovascular unit dysfunction, and leakages of the BBB with the pathogenesis of neurological diseases including AD, PD, ALS, multiple sclerosis, neuroinflammation, cerebral ischemia, and brain dementia.[84–92] Our group recently studied the BBB dysfunction in neuroinflammation and cerebral ischemia conditions using a microengineered 3D in vitro BBB model on a microfluidic chip developed by culturing endothelial cells monolayer in a tube-like platform.[93] In another study to investigate the responses of the BBB micro-fluidic model to neuroinflammatory stimulation with tumor necrosis factor-alpha (TNF-α), Herland et al. found that the integrity of endothelial monolayer strongly rely on the presence of other neurovascular cell types such as astrocytes and pericytes and concluded that these cells have distinct contributions to neuroinflammation.[94] In two subsequent studies, Prabhakarpandian and Deosarkar et al. reconstituted a BBB-on-chip constructed of two-compartments chamber with a microvascular channels cultured with rat brain endothelial cells under shear flow and a tissue compartment seeded with rat astrocytes under static flow as a 3D in vitro model to study neonatal neural pathology (Figure 4C).[95,96] Overall, current in vitro BBB models could not yet be fully applied to assess the mechanism pathways underlying human brain diseases. However, they are increasingly useful for assessing the toxicity of drugs for neurological diseases, together with in vitro 2D and in vivo models.
2.4.6. The Hope and the Hype of Organs-On-Chip in Brain Pathology
The inability of animal models to faithfully recapitulate human brain pathology, could be balanced by microfabrication technologies to reconstitute in vitro models using human brain cells. The rise of microfluidic technology allows manipulation of neuronal microenvironment and endows scientists with new tool set that raise the complexity of neurobiology to a new level of sophistication. In contrast to other models, organs-on-chip have emerged as a powerful platform for reverse engineering of the human physiology ex vivo, but in the meantime suffer from the unmatched simplicity of cellular components (i.e., a monolayer of cells), replicating the adult organ rather than the developing human brain compared to cellular models. It would be very interesting to harness the potential of the stem cell-derived brain organoids and neurospheroids in combination with the technologies borrowed from organs-on-chip to engineer the next generation of functional physiologically relevant brain-on-chip. This will provide new insights into the emerging developmental process of the human brain. In addition to this, microfluidics add the possibility to spatially and temporally refrain the biophysical and biochemical environment of the brain tissue ex vivo. Organs-on-chip has already captured the attention of many pharmaceutical and medical companies as well as government regulatory agencies including the United States Food and Drug Administration (FDA). We believe that combining stem cells with biomaterials and organs-on-chips discussed above could foster the development of in vivo-like cell culture systems that replicate brain pathophysiology and bring forward new opportunities of precise control over these systems.
2.5. Mimicking the Complexity of the Human Brain Diseases; 3D Cellular Models
The complexity of the nervous system makes treating and diagnosing brain diseases very challenging.[97] Lack of mechanistic understanding of the human brain pathology causes nonspecific and inefficient therapeutic approaches for brain disorders. As a result, pharmaceutical companies had a very low approval success rate of CNS drugs from the FDA.
Large-scale genetic studies helped us understand the etiology of brain diseases and discover the genes responsible for various neurological diseases. Although, many animal models have been utilized based on genetic risk conditions of specific diseases, unfortunately, these in vivo models fail to fully recapitulate the phenotypical characteristics of brain disorders. Moreover, animal models present critical challenges to generate polygenic models. Simple animal models with one or two genes cannot fully and accurately recapitulate brain diseases with many genetic variants. Induced pluripotent stem cells (iPSC) derived from patients with brain diseases, carrying their genetic background, contributed significantly to our knowledge of brain diseases. Although iPSC-based models generate polygenic models of brain disorders, they present some key challenges: (i) iPSC-derived neurons cannot yet mature as the adult human neurons, (ii) variability and reproducibility from batch-to-batch due to culture conditions and different genetic background, and (iii) lack of complex neuronal networks. Altogether, these challenges limit the abilities of current iPSC-based models to represent fully the brain diseases’ complexity. For example, the iPSC-derived neurons cannot yet fully reproduce the familial Alzheimer’s disease (FAD) in vitro, an example of inherited genetic neurodegenerative disease. The levels of Aβ species in the current iPSC-based cell culture models may not be high enough to recapitulate the pathological cascades of AD patients.[98–102]
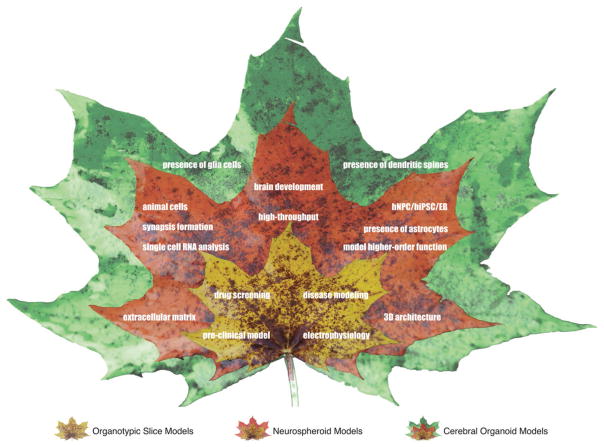
To establish a better model for FAD in vitro, our group created a unique human stem cells model grown in a Matrigel-based 3D environment.[12] We were able, for the first time, to show both hallmarks of AD in vitro; Aβ plaques and neurofibrillary tangles, by combining cell genetic manipulation and fluorescence-activated cell sorting (FACS) selection of cells overexpressing the mutated genes.[103] Other research groups have also showed 3D in vitro cell culture models able to partially reproduce AD pathology. In one study, Zhang et al. used human neuroepithelial-like stem cells (It-NES) in PuraMatrix hydrogel treated with synthetic Aβ,[104] and in another study, Choi et al. exploited 3D neurospheroids from rat cells in a microfluidic platform treated with synthetic Aβ.[105,106] Although, 3D platforms provided fundamental models for mimicking the brain microenvironment both to accelerate neuronal differentiation and to create complex neuronal network, these 3D models suffer from some limitations: (i) lack of physiologically relevant in vitro culture environments, (ii) inability to reconstitute specific brain regions, and (iii) lack of inflammatory cells.[39,107,108] Scientists recently begun to tackle these challenges by developing cerebral organoids containing cortical-layer like structure and multiple neuronal cell type, or by injecting human iPSC in the developing mouse/primate brains that provide the right environment for differentiation and maturation of the iPSC-derived neurons.[109–112] Throughout the following sections, we will review the primary approaches to develop the 3D cellular brain models (i.e., organotypic brain slices, neurospheroids and cerebral organoids) as physiologically relevant brain platforms, their similarities (Figure 5), differences, limitations, and recent progress sought to address one or a set of recapitulating the brain diseases’ challenges.
Figure 5.
Schematic representation of progress and similarities in 3D cellular brain models (organotypic brain slices, neurospheroids and cerebral organoids) to recapitulate the human brain development and diseases.
2.5.1. Brain Organotypic Models
Brain organotypic slices have been the first attempt to bridge in vitro and in vivo models by creating a platform that resemble the brain in vivo environment while keeping key in vitro characteristics.[113–116] These models now represent an established model for a variety of studies in brain molecular biology, electrophysiology, and immunohistochemistry. Brain organotypic slices models have several advantages such as easy preparation and low cost maintenance compared to animal models.[114,115] This also minimizes ethical issues associated with animal models and reduce the study timeframe, while providing the opportunity to study cell-cell interaction and molecular mechanisms. These models have been mainly used for assessing physiological and pharmacological properties of different tissues, however their applications lately has been extended to study neurodegenerative disorders, serving as ex vivo models for diseases such as AD, PD, Huntington’s, and cerebral ischemia.[113]
Organotypic brain slices models have been developed to study the corticostriatal pathway for PD modeling. For example, Humpel group used sagittal brain slices, and for the first time showed that dopamine neurons survive despite the non-functional striatonigral pathway.[117] Organotypic hippocampal slice cultures represent a good model for studying AD pathology, given that the hippocampal is a strategic region in AD patients that exhibits early neurodegeneration.[114,118] Several groups also used this model as widely accepted platform to study Aβ toxicity.[119–124] Besides neurodegenerative diseases, organotypic hippocampal slice cultures have been used as an ischemia model, in particular to study the effect of neuroinflammation on neurogenesis following oxygen/glucose deprivation.[114]
Brain organotypic slices represent a valuable platform for studying cell therapy by grafting cells into the slices and monitoring them. This allows researchers to assess cell-cell, and cell-cellular matrix interactions, cell migration, and stem cells phenotype changes.[125,126] Moreover, it is also feasible to investigate the electrophysiological properties as well as calcium and/or magnesium measurements using such brain organotypic slices models.[127,128] Despite all the advantages presented so far, organotypic brain slices show key disadvantages that halted people from using them widely as a standard model. These models can be maintained in cultures only for few weeks; they are very thin and fragile tissue (≈100–400 μm) and they can be distorted during the culture maintenance. Although organotypic slices derived from young animals (P3 to P10 – rats or mice) offer the most resilient slices, these slices do not represent a valuable model for adult neurodegenerative diseases. In a different approach, Moser et al. demonstrated that organotypic brain slices contain a strong network of laminin brain capillaries; laminin is a marker for cerebral vascular structures.[129,130] They showed that capillaries survive in absence of blood circulation and able to release molecules that influence other brain cells despite their functionality.[131] Daschil et al. used the brain organotypic slices as a model for screening pro- and anti-angiogenic drugs.[132] They tested calcium channel blockers in cortical organotypic slices derived from an AD mouse model and found that 60% of all Aβ plaques are associated with vessels. It was also found that the cells around the Aβ plaques have strong pro-angiogenic activity.[132]
Additionally, organotypic cultures of the neurogenic niches have been utilized to study neurogenesis in the CNS as well as implementing neurogenesis as a repair mechanism in brain disorders. In vivo neurogenesis is a multistep process that involves proliferation, migration, and differentiation of neural stem cells as well as integration into preexisting network and functionality.[133] Each of the mentioned steps can be assayed in an organotypic slice. Organotypic cultures are also used to study the mechanisms underlying integration of new cells into preexisting circuitries.[113] Brain organotypic slices became a valuable model in the past decades for testing drug effects on neurogenesis activation or improving cell fate specification. Taken together, organotypic brain slices could serve as a complementary platform for pre-clinical studies with the ultimate goal to test potential therapeutics with possibility to administer them in the brain by stereotaxy.[125,126]
2.5.2. Neurospheroids
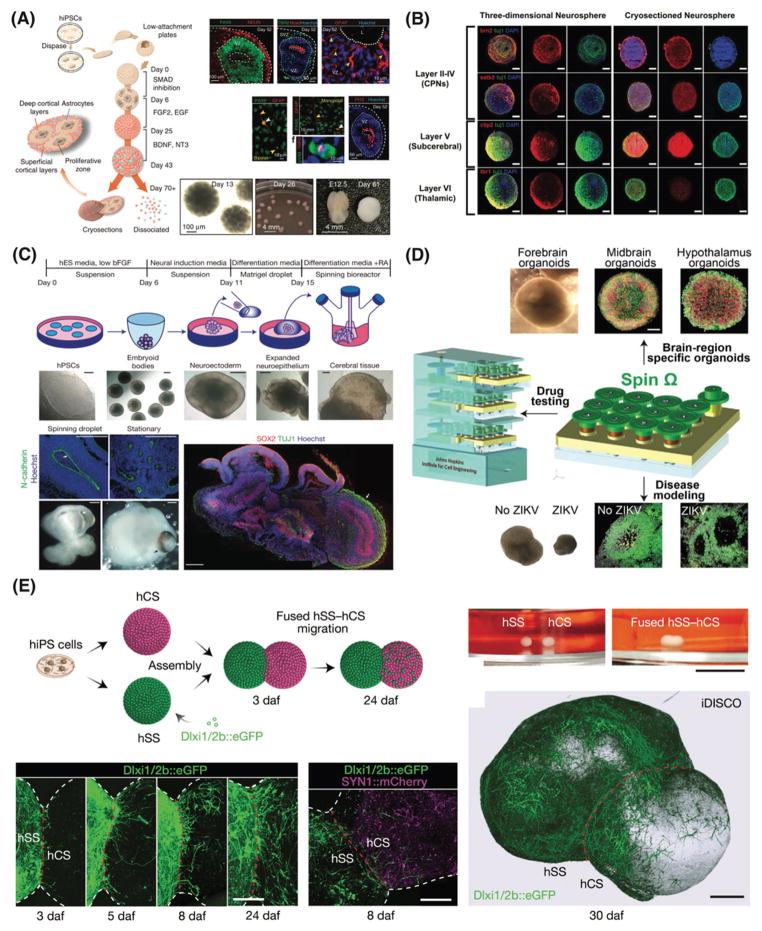
Neurospheroids models serve as a useful tool to analyze proliferation self-renewal capacity and multipotency of neural stem and progenitor cells.[134] Neurospheroids can originate from single-cell suspensions of neural stem and progenitor cells derived from the adult or fetal CNS, as well as from embryonic stem cells (ES).[135] Neurospheroids are mostly used as a respected brain region model for forebrain and/or cerebellar development. However, they still lack the ability to model other regions of the CNS such as ventral midbrain.[136–138] For example, Pasca et al. established a convenient and physiological system made of hiPSC-based cortical neurospheroids.[139] After culturing neurospheroids without scaffolds for 2.5 months, the system resembled a mid-fetal prenatal brain with cortical neurons and astrocytes and spontaneous synapses (Figure 6A). In this simple technique, the neurospheroids were grown in a serum-free neurobasal medium with b27 supplement up to 9 months, by floating in a low-attachment plate and avoiding external matrix. One could utilize this platform for all brain pathologies including synaptopathies and epilepsies.[139]
Figure 6.
3D cellular brain models. A) Schematic representation of the main stages for generating human cortical spheroids (hCS) and corticogenesis process in the hCS. Reproduced with permission.[139] Copyright 2015, Nature Publishing Group. B) 3D neurospheroids composed of cortical neuronal cells, which mimicking different horizontal layers of the brain. Reproduced with permission.[105] Copyright 2013, Elsevier. C) Schematic of cerebral organoid cell culture system and representative images of each stage as well as the neuroepithelial tissues generated using this technique. Reproduced with permission.[11] Copyright 2013, Nature Publishing Group. D) Modeling Zika virus using a miniaturized bioreactor for culturing brain-region-specific cerebral organoids generated from human iPSCs as a cost-effective platform. Reproduced with permission.[170] Copyright 2016, Elsevier. E) Generation of 3D neurospheroids from human pluripotent stem cells and assembly of the neurospheroids to recapitulate the saltatory migration of interneurons in the fused neurospheroids. Reproduced with permission.[176] Copyright 2017, Nature Publishing Group.
Disease Modeling Using Neurospheroids
Researchers used neurospheroids cell culture models for oncology studies.[140,141] For example, Joseph J. V. et al. showed that glioblastoma stem cells have similar characteristic of stem cells in terms of proliferation and migration.[140] They cultured floating neurospheroids in a serum-free medium with basic fibroblast growth factor (bFGF) and epidermal growth factor (EGF), allowing the neurospheroids to differentiate into different cell lineages and invasive into the brain parenchyma similar to stem cells. The authors found that differentiated cancer neurospheroids increase their migration and invasion properties compared to the non-differentiated ones.[141]
More recently, neurospheroid cell culture approach has been very useful for Zika virus studies. In one example, Garcez et al. showed that Zika virus induces cell death in human stem cells by caspace-2 activation and reduces organoids’ size.[142] The authors used iPSC-derived neurospheroids and brain organoids infected with the Zika virus for 24 hours. This model mimics a three-month old brain development environment, and these observations provide substantial value for understanding the mechanism by which the virus cause microcephaly in areas with risk of Zika virus.[142]
Drug Screening Using Neurospheroids
Different research groups have showed the application of the neurospheroids cell culture systems as a platform for drug screening. We showed that neurospheroids represent a good tool to study and test the effect of gamma-secretase inhibitors and modulators on neuronal differentiation, and cell adhesion.[143] This model could provide crucial data before the clinical phase of drug development. In a different approach, Lee et al. used a combination of differentiated iPSC and 3D neurospheroids to perform a drug screening in an AD model.[144] iPSC derived from AD patients were differentiated and treated with β-site amyloid precursor protein cleaving enzyme 1 (BACE1) inhibitors to assess their effect on Aβ levels. Interestingly, they found that higher concentrations of the inhibitor were needed in order to lower Aβ in neurospheroids model, likely due to the diminished surface exposure of neurospheroids, lower diffusion rate and more time required for the inhibitor to penetrate into the spheroids. This model represent a more physiologically relevant system to the human brain compared to other current cell culture systems and allows high-throughput quantification in addition to proteomics analyses.[144]
Generation of Neurospheroids Using Microwells Platforms
Kato-Negishi et al. proposed a method to create a robust neurospheroids network (NSN) with the possibility to transfer into the brain of PD patients to heal damaged tissue.[145] Neurospheroids were cultured on PDMS arrays, where they connected through their neurites, leading to the formation of centimeter-size NSN. They were able to transplant this network in specific areas of the brain with a simple method called ‘stamping’ that was possible thanks to the non-adhesive characteristic of PDMS. The NSN showed synapse formation and extended axons within the brain region in which was transplanted.[145] However, this strategy is limited mainly due to the small size of the graft that one can transplant into the brain; only a few damaged tissues could benefit from such potential strategy.
Choi et al. developed size-controlled networked neurospheroids using concave microwells made of PDMS to study Aβ toxicity in a cerebral cortex-like environment.[105] Neurospheroids derived from rat cortex were differentiated by plating cells in concave-shape microwells with four different diameters that determined the size of the ultimate neurospheroids (Figure 6B). The neurites processes outgrew and connected the neurospheroids between the different wells forming a neuronal network consisted of six horizontal layers. After Aβ treatment, they observed cell death and other pathological hallmarks of AD.[105] In another study, Jeong et al. reported an improved model of neurospheroids cultured in PDMS microwells.[146] They used deep hemi-cylindrical channels well networked to enhance the formation of neurospheroids that release laminin and induce higher differentiation of the neural progenitor cells into glia and neurons. They also showed electrical activity likely due to the networks formed between the neurospheroids.[146]
Neurospheroids Challenges
Neurospheroids are sensitive to any small variations such as cell density, procedure, number of passage, and medium composition. All those variables lead to higher heterogeneity of neurospheroids, resulting in neurospheroids with different properties, stage of differentiation, and cell types. This makes it hard to consolidate and reproduce the data among the same study or between different groups. Therefore, studies based on neurospheroids culture should be considered as a mixed population of neural progenitor cells.[147–154] To clearly demonstrate the potential of this exciting class of 3D neural cell culture model for recapitulating the brain pathophysiology, future studies need to create a platform capable of producing viable, same-sized 3D neurospheroids from human stem cells, and assess their potential as physiologically relevant model for brain disease pathology as well as high-throughput drug screening assay. This would not be possible without harnessing the potential of recent technologies such as micro-fluidic, and stem cell engineering with the ultimate goal to reconstitute a better in vitro models to mimic the human brain.
2.5.3. Cerebral Organoids
Cerebral organoids – 3D cell cultures of brain-like tissue derived from human or mouse stem cells - represent a bridge between preclinical drug development and human trials.[155] The first cerebral organoids was developed four decades ago by growing rat brain tissue as cell suspension culture,[156] however, substantial accomplishments occurred in recent years.[10,155,157–160] These systems have huge advantages in recapitulating the 3D architecture of the brain tissue than previous in vitro 3D models developed so far.[10,155,157–159,161,162]
Generation of Cerebral Organoids
Cerebral organoids are produced by harnessing the spontaneous self-organizing properties of the human pluripotent stem cells (hPSCs) derived either from mouse or human to create regions of the forebrain during development. This is exemplified by cerebral organoids from hPSCs formed in a rotating bioreactor (Figure 6C).[11] In this technique, the spinning movement enables and favorites the hPSCs to compartmentalize in multiple and different brain regions, leading to a specific brain model compared to the existed 3D neural cell culture systems. For example, a cortex-like model was formed, with a region that produces CSF and a zone that contains radial glia cells (oRGs). The finding of oRGs in a human in vitro model is highly significant, given that these types of human specific progenitor cells never reported in mouse or previous human models.[157,163] In a different model, the embryoid bodies formed spontaneously, and then embedded in Matrigel matrix in order to enhance the buds expansion with the formation of fluid filled cavities mimicking the brain ventricles. In addition to this, the rotating bioreactor helped a better oxygen and nutrients diffusion within the forming cerebral organoids, and thereby lessening the core apoptosis and necrotic area.[158] In this work, cerebral organoids also recapitulated microcephaly pahtology in vitro by harnesssing the iPCS cells derived from patients. The cerebral organoids carrying the microcephaly muation showed smaller size compared to controls without the mutation. The premature differentation of progenitor cells could be the reason since disease-derived cerebral organoids contain fewer progenitors cells and higher number of neurons.[10,11]
A serum-free floating culture of embryoid body-like aggregates showed quick re-aggregation starting from 3D culture of mouse and human embryonic stem cells. This platform could reproduce the optic cup in vitro, and mimic corticogenesis resembling the first three months of the fetal life.[160,164,165] Similar models showed presence of oRG in their cerebral organoids. The main difference between the two models rely on different techniques used to induce cell aggregation. In one model, cortex-selective culture conditions were realized by adding growth factors to the culture medium in order to stimulate differentiation into specific cell types and create a more reproducible model. A second model relied on the inherent properties of stem cells spontaneous cell aggregation which results in non-selective differentiation and a relatively more random model. One of the key initial limitations of the first developed cerebral organoids was their limited continuity and expansion of neuroephitelial tissue by adding Matrigel or laminin as a matrix, however those groups may have overcome this problem lately.[166]
Following on the first cerebral organoid model recapitulating microcephaly by Lancaster and colleagues, Jo et al. reported a human midbrain-like organoid derived from hPSCs mimicking dopaminergic neurons in substantia nigra, which is an impotent area to recapitulate PD pathology. We have previously discussed the importance and need from the scientific community to represent such specific brain region that is selectively vulnerable in each neurodegenerative disease. Previous in vitro models derived from differentiated hPSCs were unable to reproduce all the features of dopaminergic cells; for example, these cerebral organoids models suffer from lack of neuromelanin. The midbrain organoids developed by Jo et al. not only made possible to study this pigment in a physiologically relevant human model in vitro for the first time,[167] but the neutrons also expressed specific neurotransmitters such as dopamine along with electrophysiological characteristics similar to mature human functional midbrain neurons.[168,169] In another interesting work, Qian et al. utilized the forebrain organoids platform for Zika virus infection modeling and for testing compounds (Figure 6D).[170] People studied Zika virus infections and based on recent clinical data, the virus show the most sever and harmful effect during the first trimester of pregnancy.[171–173] One day of infection in vitro led to severe microcephaly-like events such as the reduction of neuronal layer thickness and overall cerebral organoid size.[170]
Single Cell Sequencing of Cerebral Organoids
Over the past decade, cutting-edge 3D cerebral organoids models have allowed sophisticated biology of the brain development and evolution as well as how the brain tissues are affected by neurological diseases. Despite these rapid pace advancements, it still remained unanswered which cell type arises in the developed cerebral organoids, and how these cerebral organoids are varying from batch-to-batch. To tackle these questions, very recently, studies integrating cerebral organoids and single cell sequencing technique described key steps towards understanding the brain organoids cell composition.[174,175] A step forward in cerebral organoid characterization was to compare the human cortical-like organoids with fetal neocortex using single-cell RNA sequencing technique.[174] Transcriptome analyses was used to determine the cell types within the cerebral organoids and their differentation status along with the types of genes involved in corticogenesis. Cell types are similar between the cerebral organoids and human fetus, as well as the genes involved during development and evolution (≈80% of genes). It seems that these genes were not specific to the physiological conditions of brain development since they were influenced by external stimuli such as the environment in which the cerebral organoids were created.[174]
In another recent study, Arlotta lab analyzed the gene profile of ~80,000 cells isolated from 31 human brain organoids (3- to 6-month old) – the most comprehensive single-cell study of cerebral organoids conducted so far.[175] They found similar genes present in the in vitro cerebral organoids of human brain and retina. They identified the cell types that constantly replicated and the ones that sporadically expressed due to the heterogeneity of the model. The matured neurons within the cerebral organoids showed dendritic spines and spontaneous networks, but more importantly, for the first time, they showed the ability to respond to light stimulation through optogenetics technique. This model presented the ability to reproduce higher-order function and opened new avenues to study brain functions associated with cell-cell interactions, neuronal circuits in physiological and/or pathological conditions.[175] It is of great importance to mention that Arlotta lab found substantial differences in cell compositions from batch-to-batch organoids. This highlights the unmet need one more time for engineering a better technique to create cerebral organoids that minimally vary from batch-to-batch and improve the reproducibility of this cutting-edge platform for reverse engineering the human brain pathophysiology.
Overall, both studies revealed the powerful potential of high-throughput, single-cell transcriptome analysis to study cell composition and gene-expression of cerebral organoids originated from different batches, as well as studying disease pathology. Nevertheless, despite these exciting valuable tools for future studies, it is still unclear how transcriptomes are varying from batch-to-batch, from different stem-cell lines-derived cerebral organoids, and from different patients with the same disease pathology. This should be addressed in the future harnessing the power of the new advancements in technology.
From Neurospheroids to Cerebral Organoids
Recently, neurospheroids resembling different regions of the human brain have been placed next to each other into multi-region neural 3D cell cultures, the cells fused over to form forebrain-like organoids (Figure 6E).[176] Two neurospheroids, resembling the cortex and the subpallium, were placed in close vicinity in a conical tube. After a few days, the neurospheroids started to fuse into each other and the cells start to migrate, with a saltatory pattern followed by pauses from the subpallium to the cortex – similar to the in vivo route of interneurons migration. This phenomenon was not observed when the neurospheroids were plated on coverslips. A pharmacologic block of migration led to a decreased movement in terms of frequency, length, speed and change of direction. This system was also used to model the neurodevelopmental disorder Timothy syndrome. This syndrome is associated with mutation in a L-type calcium channel that regulates the interneurons migration. They first created their neurospheroids from hPSCs derived from patients affected by Timothy syndrome. These neurospheroids were able to differentiate, however they showed an increase in saltation frequency and a decrease in interneurons migration rate as compared to the controls. This effect was rescued using pharmacological compounds that can block mutated L-type calcium channels, con-firming the key role of ion channel in the disease pathology. Finally, the researchers used a single-cell transcriptome analysis on assembled forebrain-like organoids (≈ 4-week old) to characterize the migrated cells versus the non-migrated ones. The migrated cells showed different genes related to interneuron migration, better electrophysiological properties in comparison to non-migrated neurons, and producing synapses within the migrated region. Patch clamp analyses showed both excitatory and inhibitory postsynaptic currents.[176]
Cerebral Organoids Challenges
Although cerebral organoids represent the future of human in vitro models, we should not underestimate several limitations that come with this platform. For example, devices needed to support the culture platform and implement these systems for drug screening could add extra variables such as drug adsorption to the interpreted data.[162] Most of the cerebral organoids developed so far rely on the spontaneous and casual self-aggregation of the stem cells, which might lead to inconsistency and lack of reproducibility, similar to the neurospheroids models as discussed in Section 2.5.2.[177] Current cerebral organoids fail to recapitulate many late brain development events such as gliagenesis and myelination mainly due to the longer time needed for maturation. Microglia cells play a significant role in the brain by inducing the formation of mature dendritic spine and synapses, however current organoids systems lack these cells into their culture. Lancaster et al. recently proposed a way to overcome the lack of vascularization in the cerebral organoids by growing them on microfluidic devices that will allow the transport of fluids into the cerebral organoids.[178] The use of fluorescent reporters would be a valuable addition to select specific cell type and monitor their migrations.[179]
From an ethical point of view, the use of these complex and complete 3D in vitro human models may help reduce the use of animal models, but at the same time it raises its own ethical issues associated with donation, storage, and further use of cerebral organoids. The identity of the material derived from leftover of human specimens after clinical care or collected specifically for research, and stored in ‘biobanks’ for further use is one of these issues. While the genetic background of the patient and his medical history are essential in order to use the cerebral organoid for the donors’ benefit, donors may decline the de-identification of the sample. Once the consent is obtained, other ethical challenges could also emerge related to ownership and further use of the stored samples.[180] For further discussion on the cerebral organoids biobanking and ethical issues involved with this technology, readers are referred to a recent report published elsewhere.[181] It has been suggested that these systems should be used as a complementary platforms to animal studies and/or cell-based in vitro systems rather than in competition with other standard methodologies.[180]
3. Designing Synthetic Matrices for Recapitulating the Extracellular Matrix
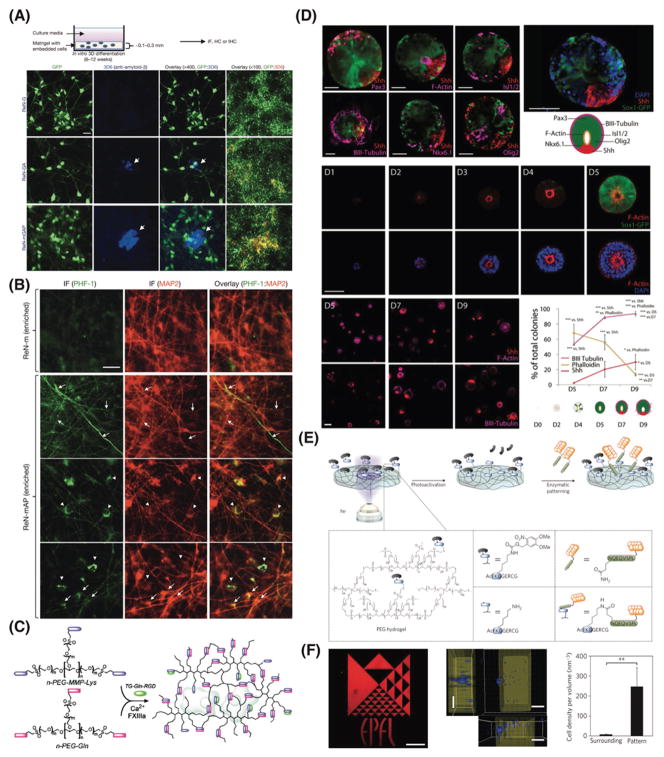
The necessity to work with cellular models that fully recapitulate the functions of living tissues encouraged scientists to move from 2D cell monolayers to 3D cell culture systems. Creating 3D cell culture models requires new cell lines and protocols for ordering cells in the relevant configuration, as well as sophisticated 3D matrices that mimic the native extracellular microenvironments’ architecture and function. Natural and synthetic matrices have been used to mimic the in vivo extracellular matrix (ECM) to direct cellular process, such as migration,[182–184] stem cell fate[185–189] and organogenesis[188,190] or as biomimetic 3D cultures for modeling human diseases pathology.[12] For example, Tang-Schomer et al. developed a 3D brain-like cortical tissue using silk-collagen scaffolds and primary cortical neurons, resembled the layered structure of the cerebral cortex with capability to study traumatic brain injury.[9,191] A 3D in vitro human neural cell culture model of AD using human neural progenitor cells embedded in Matrigel showed the two pathological hallmarks of AD: extracellular aggregation of Aβ and accumulation of intercellular hyperphosphorylated tau proteins.[12,192] Excessive accumulation of Aβ leads to the aggregation of hyperphosphorylated tau,[58,193] however, no animal model could verify this hypothesis prior to this study. Treatment with either β- or γ-secretase inhibitors dramatically decreased the Aβ pathology, but also attenuated tauopathy, most likely due to the inhibition of Aβ accumulation within the ECM (Figure 7A and B).[12] This 3D cell culture model has the potential to serve as a base for the development of 3D in vitro human models of other neurological disorders. Recently a unique chimeric human-mouse model was developed by transplanting stem-cell-derived human cortical neurons into mouse brain.[194] Remarkably, the new mice showed that Aβ species generated in the brain can induce AD hallmarks, including tau hyperphosphorylation, neurite dystrophy, and cell death in non-manipulated human neurons. This study highlights the unmet need for engineering more physiologically relevant matrices that could fully recapitulate the natural ECM architecture and function.
Figure 7.
Recapitulating the 3D native extracellular matrix using natural and synthetic matrices. A) 3D in vitro human neural cell culture model of AD using human neural progenitor cells embedded in Matrigel showing the two pathological hallmarks of AD, including extracellular aggregation of Aβ and (B) accumulation of intercellular hyperphosphorylated tau proteins. Reproduced with permission.[12] Copyright 2014, Nature Publishing Group. C) Enzymatically responsive multifunctional synthetic hydrogels using factor XIIIa for cross-linking two multi-arm PEG peptide conjugates in combination with coupling a cell adhesion peptide. Reproduced with permission.[198] Copyright 2007, American Chemical Society. D) Dorsal-ventral patterning and neural tube architecture in synthetic multi-arm PEG matrices to control early neural morphogenesis and to explore the role of ECM in the development of 3D neuroepithelial organoids. Cyst patterning shows key characteristics of neural tube architecture. Reproduced with permission.[201] Copyright 2016, National Academy of Sciences. E) Schematic representation of the concept of photo-responsive enzymatic peptide patterning of hydrogels, and (F) spatial patterning of hydrogels using ultraviolet light on a specific region within the hydrogel. Reproduced with permission.[204] Copyright 2013, Nature Publishing Group.
It is not clear which biophysical and biochemical factors of the ECM and its components are critical in the human brain function and development. In general, natural ECM such as collagen and Matrigel have an advantage of their inherent biological properties, receptor-binding ligands, and potential for cell-triggered degradation and natural remodeling. Despite these advantages, animal derived matrices suffer from poorly defined compositions and from batch-to-batch variations. We have yet to understand the biophysical and biochemical factors of the ECM and the role its components play in the human brain function and development. To tackle the challenges associated with natural ECM and to gain a better insight of the in vivo ECM, synthetic cell-compatible hydrogels composed of polymer backbones and enzymatically degradable peptides as cross-linkers have been developed. Hubbell and Lutolf pioneered the development of enzymatically degradable hydrogels formed through Michael-type reactions of multi-arm poly (ethylene glycol) (PEG) macromeres with di-functional oligopeptides linkers that mimic collagen of native ECM.[195–197] This chemical reaction generates a strong elastic and cell-compatible hydrogel in the presence of cells and/or tissues. Enzymatically responsive multifunctional synthetic hydrogels were also developed, by employing activated transglutaminase enzyme factor XIIIa for site-specific coupling of cell adhesions peptides and for cross-linking the multi-arm PEG hydrogel networks (Figure 7C).[198] This approach significantly enhanced previous material building blocks for 3D cell cultures based on the chemical cross-linking of PEG hydrogels with peptides, as it allowed to immobilize any biomolecules of choice in a very controlled fashion. Using this system, neuroepithelial cysts could be reconstituted from mouse embryonic stem cells directly embedded in 3D culture.[199] Three different 3D cell culture systems were compared: (i) Matrigel, (ii) laminin/entactin, and (iii) synthetic PEG hydrogel. ECM proteins improved cyst-forming ability – although they were not required to generate lumen-containing neuroepithelial cysts in the 3D culture system.
Recent developments in the field of biomaterials science and bioengineering helped engineer synthetic hydrogels as biomimetic 3D cell microenvironments with very well-defined biochemical and biophysical properties.[197,200,201] Modular synthetic matrices have been used to control early neural morphogenesis and to explore the role of ECM in the development of 3D neuroepithelial organoids.[200,201] A recent study used a 3D microarray platform based on cross-linked multi-arm PEG, to probe the effect of factors including matrix elasticity, degradability, and signaling proteins on mouse embryonic stem cells (Figure 7D).[201] Current approaches to study the neural tube morphogenesis also rely on commercial animal derived matrices, such as Matrigel. However, Matrigel leads to heterogeneities, whereas well defined synthetic matrices lead to more homogeneous and defined populations. New insights are emerging into the role of biophysical factors in neural morphogenesis and multifactorial cell-matrix interactions are key to control the growth and differentiation of stem cells in vitro.[201]
The native ECM of the brain has very permissive structure to cellular migrations and movements, resulting in a complex physical behavior, which impacts the biological processes such as morphogenesis. Only a few studies published in culturing multicellular epithelial. There has been a recent interest to replicate the native ECM properties such as porosity and fibrillar within synthetic matrices. Advances in the field of photochemistry have enabled the researchers to engineer 3D spatial and temporal patterning of mechanical and biochemical signals of the synthetic ECM.[186,190,202–207] This profoundly impacted the way we study the biology of cells. For example, PEG hydrogels tethered locally with a ‘caged’ synthetic peptide helped study cell migration.[204] In this work, a photo-responsive peptide substrate of activated transglutaminase factor XIII (FXIIIa) was incorporated within a 3D hydrogel and shining ultraviolet light on a specific region within the hydrogel lead to uncaging the FXIIIa domain and tethering of a biomolecule of interest at a desired time. This could control the migration of human mesenchymal stem cells within the ECM (Figure 7E and F). Compared with previous studies on directing stem cells migrations within the ECM using conventional peptide photo-patterning,[208,209] this approach utilized the site-specific nature of FXIIIa to pattern full length bioactive proteins. This light-responsive enzymatic patterning could help us to gain insights in the effects of dynamic ECM on the cells of choice within the 3D cell culture system. For further details on the engineering of synthetic matrices to recapitulate the native ECM, readers are referred to other excellent reviews.[190,197,202,210–214]
Overall, the last decade has marked a significant paradigm shift in designing criteria for cell-compatible synthetic biomaterials with greater control over materials properties. The integration of cells and molecular biology principles with dynamic biomaterials and molecular cues mimicking the in vivo ECM structure and function have led to considerable progress. Work remains to be done towards engineering biomaterials with precise control over the matrices properties. These advances will ultimately enable precision studies of the human brain development and diseases pathogenesis.
4. Conclusions and Future Perspectives
3D brain cell culture models including cerebral organoids and neurospheroids are a fundamental research tool in the human brain cell biology. The past decade has seen the rapid development of techniques to recreate aspects of morphogenesis ex vivo, which helped decipher the biology of the human brain and discover new drugs. Most recently, harnessing the potential of human embryonic and induced pluripotent stem cells for reconstructing the human brain (e.g., self-organizing) has been attracting a lot of interest. Organs-on-chip have emerged as a powerful platform for reverse engineering of the human pathophysiology ex vivo, and they provide increasingly insightful observations of the developmental processes of the human brain. Despite the importance and unquestionable role of stem-cell derived cell culture models such as cerebral organoids in furthering our understanding of the human brain, these systems have a number of serious drawbacks and several issues remain unresolved at present. For example, the existing cerebral organoids suffer from poor nutrient availability due to the lack of vascularization and fail to recreate the full-developed brain tissue in vitro. Further platforms should provide more nutrients and metabolic transport to these 3D cellular models by either imitating vascularization or sustaining a long-term growth of cells to allow full recapitulation of the human brain. Brain organoid models suffer from the absence of other neural cell types such as astrocytes and oligodendrocytes, which together compose the majority of cells in the adult human brain. Furthermore, we have limited control over the course of the cells morphogenesis that leads to the cerebral organoids. Despite their limitations and challenges, stem cell-derived brain organoids represent the most useful models at present to recapitulate the complexity and morphogenesis of the human brain. Today, no single model, device, method or platform can fully recapitulate the natural microenvironment of the brain development and its diseases in vitro. While remarkable insights have been gained from current innovative 3D cellular models, we anticipate that sophisticated microfluidics, advances in materials science, and stem cells engineering will develop in synergy and enable complex, multifunctional platforms for neurobiology research.
Acknowledgments
This work was supported by the National Institutes of Health (NS045776, AG048080, AG15379, and AG014713), Cure Alzheimer’s Fund, and BrightFocus Foundation.
Biographies

Mehdi Jorfi is a Research Fellow in the Center for Engineering in Medicine at Massachusetts General Hospital, and Harvard Medical School. He received his Ph.D. in Bioengineering from the University of Fribourg in 2014. Prior to joining Harvard, he was a postdoctoral fellow at the Department of Chemical Engineering at Massachusetts Institute of Technology (MIT) in Cambridge, USA. His research interests are at the interface of engineering and life sciences, particularly on using biomaterials and microtechnologies toward understanding and impacting neurological disorders. His current research focuses on developing three-dimensional human neural cell culture models of Alzheimer’s disease and blood-brain barrier.

Carla D’Avanzo is an Instructor in Neurology at Massachusetts General Hospital, and Harvard Medical School. She received her Ph.D. in Neuroscience at University of Naples Federico II (Italy), and then joined Dr. Kovacs lab at MGH as a Research Fellow. She worked on the characterization of novel Amyloid Precursor Protein interacting proteins and their potential effect of APP metabolism and Amyloid β production. She then moved to Dr. Kim lab and been part of a project aimed to design and characterize a novel three-dimensional human neural cell culture system that shows key events in the pathogenic cascade of Alzheimer’s disease.

Doo Yeon Kim is an Assistant Professor of Neurology at Massachusetts General Hospital (MGH), and Harvard Medical School. He received his Ph.D. in Biology at Korea Advanced Institute of Technology (South Korea), and then moved to MGH. Since then, he has been studying pathogenic mechanisms of Alzheimer’s disease for 16 years. Recently, his team, together with Dr. Rudolph E. Tanzi (MGH), developed a novel three-dimensional human neural cell culture of Alzheimer’s disease, which recapitulated pathological hallmarks of Alzheimer’s disease including robust β-amyloid plaques and β-amyloid-driven neurofibrillary tangles for the first time.

Daniel Irimia is a bioengineer, a medical doctor by training, and a leading researcher in the areas of microfluidics, inflammation, and sepsis. He is an Associate Professor in the Department of Surgery at Massachusetts General Hospital and Harvard Medical School. He is the organizer of the Dicty World Race, aimed at encouraging biologists to integrate microfluidic tools in cell chemotaxis research. He was recently awarded the “Pioneers of Miniaturization” prize from the Chemical and Biological Microsystems Society for his work on microfluidic devices for studying neutrophils and other leukocytes.
Footnotes
Conflict of Interest
The authors declare no conflict of interest.
Contributor Information
Dr. Mehdi Jorfi, Center for Engineering in Medicine, Massachusetts General Hospital, Harvard Medical School, Charlestown, Massachusetts 02129, USA
Dr. Carla D’Avanzo, Genetics and Aging Research Unit, MassGeneral Institute for Neurodegenerative Disease, Massachusetts General Hospital, Harvard Medical School, Charlestown, Massachusetts 02129, USA
Prof. Doo Yeon Kim, Genetics and Aging Research Unit, MassGeneral Institute for Neurodegenerative Disease, Massachusetts General Hospital, Harvard Medical School, Charlestown, Massachusetts 02129, USA
Prof. Daniel Irimia, Center for Engineering in Medicine, Massachusetts General Hospital, Harvard Medical School, Charlestown, Massachusetts 02129, USA
References
- 1.Neurological Disorders: Public Health Challenges. W. H. Organization; Geneva: 2006. [Google Scholar]
- 2.Bae BI, Walsh CA. Science. 2013;342:200. doi: 10.1126/science.1245812. [DOI] [PubMed] [Google Scholar]
- 3.Lui JH, Hansen DV, Kriegstein AR. Cell. 2011;146:18. doi: 10.1016/j.cell.2011.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shuler ML, Hickman JJ. Proc Natl Acad Sci. 2014;111:13682. doi: 10.1073/pnas.1414484111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Azevedo FAC, Carvalho LRB, Grinberg LT, Farfel JM, Ferretti REL, Leite REP, Filho WJ, Lent R, Herculano-Houzel S. The Journal of Comparative Neurology. 2009;513:532. doi: 10.1002/cne.21974. [DOI] [PubMed] [Google Scholar]
- 6.Bhatia SN, Ingber DE. Nat Biotech. 2014;32:760. doi: 10.1038/nbt.2989. [DOI] [PubMed] [Google Scholar]
- 7.Yi Y, Park J, Lim J, Lee CJ, Lee SH. Trends Biotechnol. 2015;33:762. doi: 10.1016/j.tibtech.2015.09.007. [DOI] [PubMed] [Google Scholar]
- 8.Hopkins AM, DeSimone E, Chwalek K, Kaplan DL. Prog Neurobiol. 2015;125:1. doi: 10.1016/j.pneurobio.2014.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tang-Schomer MD, White JD, Tien LW, Schmitt LI, Valentin TM, Graziano DJ, Hopkins AM, Omenetto FG, Haydon PG, Kaplan DL. Proc Natl Acad Sci. 2014;111:13811. doi: 10.1073/pnas.1324214111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lancaster MA, Knoblich JA. Science. 2014;345:1247125. doi: 10.1126/science.1247125. [DOI] [PubMed] [Google Scholar]
- 11.Lancaster MA, Renner M, Martin CA, Wenzel D, Bicknell LS, Hurles ME, Homfray T, Penninger JM, Jackson AP, Knoblich JA. Nature. 2013;501:373. doi: 10.1038/nature12517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Choi SH, Kim YH, Hebisch M, Sliwinski C, Lee S, D’Avanzo C, Chen H, Hooli B, Asselin C, Muffat J, Klee JB, Zhang C, Wainger BJ, Peitz M, Kovacs DM, Woolf CJ, Wagner SL, Tanzi RE, Kim DY. Nature. 2014;515:274. doi: 10.1038/nature13800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Urich E, Patsch C, Aigner S, Graf M, Iacone R, Freskgard PO. Sci Rep. 2013;3:1500. doi: 10.1038/srep01500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Geschwind DH, Rakic P. Neuron. 2013;80:633. doi: 10.1016/j.neuron.2013.10.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gotz M, Huttner WB. Nat Rev Mol Cell Biol. 2005;6:777. doi: 10.1038/nrm1739. [DOI] [PubMed] [Google Scholar]
- 16.Taverna E, Gotz M, Huttner WB. Annu Rev Cell Dev Biol. 2014;30:465. doi: 10.1146/annurev-cellbio-101011-155801. [DOI] [PubMed] [Google Scholar]
- 17.Lancaster MA, Knoblich JA. Curr Opin Neurobiol. 2012;22:737. doi: 10.1016/j.conb.2012.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Glotzer M. Nat Rev Mol Cell Biol. 2009;10:9. doi: 10.1038/nrm2609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huh D, Matthews BD, Mammoto A, Montoya-Zavala M, Hsin HY, Ingber DE. Science. 2010;328:1662. doi: 10.1126/science.1188302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nakao Y, Kimura H, Sakai Y, Fujii T. Biomicrofluidics. 2011;5:22212. doi: 10.1063/1.3580753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jang KJ, Suh KY. Lab Chip. 2010;10:36. doi: 10.1039/b907515a. [DOI] [PubMed] [Google Scholar]
- 22.Kim HJ, Huh D, Hamilton G, Ingber DE. Lab Chip. 2012;12:2165. doi: 10.1039/c2lc40074j. [DOI] [PubMed] [Google Scholar]
- 23.Lee J, Choi JH, Kim HJ. Expert Rev Gastroenterol Hepatol. 2016;10:883. doi: 10.1080/17474124.2016.1200466. [DOI] [PubMed] [Google Scholar]
- 24.Kim HJ, Li H, Collins JJ, Ingber DE. Proc Natl Acad Sci USA. 2016;113:E7. doi: 10.1073/pnas.1522193112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guan A, Hamilton P, Wang Y, Gorbet M, Li Z, Phillips KS. Nat Biomed Eng. 2017;1:0045. [Google Scholar]
- 26.Wang JY, Ren L, Li L, Liu WM, Zhou J, Yu WH, Tong DW, Chen SL. Lab Chip. 2009;9:644. doi: 10.1039/b813495b. [DOI] [PubMed] [Google Scholar]
- 27.Millet LJ, Gillette MU. Trends Neurosci. 2012;35:752. doi: 10.1016/j.tins.2012.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Park JW, Kim HJ, Kang MW, Jeon NL. Lab Chip. 2013;13:509. doi: 10.1039/c2lc41081h. [DOI] [PubMed] [Google Scholar]
- 29.Taylor AM, Jeon NL. Curr Opin Neurobiol. 2010;20:640. doi: 10.1016/j.conb.2010.07.011. [DOI] [PubMed] [Google Scholar]
- 30.Neto E, Leitao L, Sousa DM, Alves CJ, Alencastre IS, Aguiar P, Lamghari M. J Neurosci. 2016;36:11573. doi: 10.1523/JNEUROSCI.1748-16.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Taylor AM, Rhee SW, Tu CH, Cribbs DH, Cotman CW, Jeon NL. Langmuir. 2003;19:1551. doi: 10.1021/la026417v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Taylor AM, Blurton-Jones M, Rhee SW, Cribbs DH, Cotman CW, Jeon NL. Nat Meth. 2005;2:599. doi: 10.1038/nmeth777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Park JW, Vahidi B, Taylor AM, Rhee SW, Jeon NL. Nat Protoc. 2006;1:2128. doi: 10.1038/nprot.2006.316. [DOI] [PubMed] [Google Scholar]
- 34.Sutton MA, Taylor AM, Ito HT, Pham A, Schuman EM. Neuron. 2007;55:648. doi: 10.1016/j.neuron.2007.07.030. [DOI] [PubMed] [Google Scholar]
- 35.Taylor AM, Berchtold NC, Perreau VM, Tu CH, Jeon NL, Cotman CW. J Neurosci. 2009;29:4697. doi: 10.1523/JNEUROSCI.6130-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Taylor AM, Wu J, Tai HC, Schuman EM. J Neurosci. 2013;33:5584. doi: 10.1523/JNEUROSCI.2944-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cohen MS, Orth CB, Kim HJ, Jeon NL, Jaffrey SR. Proc Natl Acad Sci USA. 2011;108:11246. doi: 10.1073/pnas.1012401108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pinto MJ, Alves PL, Martins L, Pedro JR, Ryu HR, Jeon NL, Taylor AM, Almeida RD. J Cell Biol. 2016;212:789. doi: 10.1083/jcb.201509039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Menon S, Boyer NP, Winkle CC, McClain LM, Hanlin CC, Pandey D, Rothenfusser S, Taylor AM, Gupton SL. Dev Cell. 2015;35:698. doi: 10.1016/j.devcel.2015.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bigler RL, Kamande JW, Dumitru R, Niedringhaus M, Taylor AM. Sci Rep. 2017;7:611. doi: 10.1038/s41598-017-00676-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Peyrin JM, Deleglise B, Saias L. Lab Chip. 2011;11:3663. doi: 10.1039/c1lc20014c. [DOI] [PubMed] [Google Scholar]
- 42.Gao Y, Broussard J, Haque A, Revzin A, Lin T. Microsys Nanoeng. 2016;2:15045. doi: 10.1038/micronano.2015.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim HJ, Park JW, Byun JH, Poon WW, Cotman CW, Fowlkes CC, Jeon NL. Acs Chem Neurosci. 2012;3:433. doi: 10.1021/cn3000026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Majumdar D, Gao Y, Li D. J Neurosci Methods. 2011;196:38. doi: 10.1016/j.jneumeth.2010.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shi M, Majumdar D, Gao Y. Lab Chip. 2013;13:3008. doi: 10.1039/c3lc50249j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ravula SK, Wang MS, Asress SA, Glass JD, Frazier AB. J Neurosci Methods. 2007;159:78. doi: 10.1016/j.jneumeth.2006.06.022. [DOI] [PubMed] [Google Scholar]
- 47.Kanagasabapathi TT, Massobrio P, Barone RA, Tedesco M, Martinoia S, Wadman WJ, Decre MMJ. J Neural Eng. 2012;9:036010. doi: 10.1088/1741-2560/9/3/036010. [DOI] [PubMed] [Google Scholar]
- 48.Hofmann F, Bading H. J Physiology-Paris. 2006;99:125. doi: 10.1016/j.jphysparis.2005.12.005. [DOI] [PubMed] [Google Scholar]
- 49.Chokshi TV, Bazopoulou D, Chronis N. Lab Chip. 2010;10:2758. doi: 10.1039/c004658b. [DOI] [PubMed] [Google Scholar]
- 50.Shi P, Scott MA, Ghosh B, Wan D, Wissner-Gross Z, Mazitschek R, Haggarty SJ, Yanik MF. Nat Commun. 2011;2:510. doi: 10.1038/ncomms1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kilic O, Pamies D, Lavell E, Schiapparelli P, Feng Y, Hartung T, Bal-Price A, Hogberg HT, Quinones-Hinojosa A, Guerrero-Cazares H, Levchenko A. Lab Chip. 2016;16:4152. doi: 10.1039/c6lc00946h. [DOI] [PubMed] [Google Scholar]
- 52.Dauth S, Maoz BM, Sheehy SP, Hemphill MA, Murty T, Macedonia MK, Greer AM, Budnik B, Parker KK. J Neurophysiol. 2017;117:1320. doi: 10.1152/jn.00575.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fan Y, Nguyen DT, Akay Y, Xu F, Akay M. Sci Rep. 2016;6:25062. doi: 10.1038/srep25062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Frigerio CS, De Strooper B. Annu Rev Neurosci. 2016;39:57. doi: 10.1146/annurev-neuro-070815-014015. [DOI] [PubMed] [Google Scholar]
- 55.De Strooper B, Karran E. Cell. 2016;164:603. doi: 10.1016/j.cell.2015.12.056. [DOI] [PubMed] [Google Scholar]
- 56.Lee VMY, Goedert M, Trojanowski JQ. Annu Rev Neurosci. 2001;24:1121. doi: 10.1146/annurev.neuro.24.1.1121. [DOI] [PubMed] [Google Scholar]
- 57.Iqbal K, Liu F, Gong CX. Nat Rev Neurol. 2016;12:15. doi: 10.1038/nrneurol.2015.225. [DOI] [PubMed] [Google Scholar]
- 58.Hardy J, Selkoe DJ. Science. 2002;297:353. doi: 10.1126/science.1072994. [DOI] [PubMed] [Google Scholar]
- 59.Li W, Xu Z, Xu B, Chan CY, Lin X, Wang Y, Chen G, Wang Z, Yuan Q, Zhu G, Sun H, Wu W, Shi P. Adv Healthcare Mater. 2017:6. doi: 10.1002/adhm.201600895. [DOI] [PubMed] [Google Scholar]
- 60.Cho H, Hashimoto T, Wong E, Hori Y, Wood LB, Zhao L, Haigis KM, Hyman BT, Irimia D. Sci Rep. 2013;3:1823. doi: 10.1038/srep01823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Takeda S, Wegmann S, Cho H, DeVos SL, Commins C, Roe AD, Nicholls SB, Carlson GA, Pitstick R, Nobuhara CK, Costantino I, Frosch MP, Muller DJ, Irimia D, Hyman BT. Nat Commun. 2015;6:8490. doi: 10.1038/ncomms9490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Choi YJ, Chae S, Kim JH, Barald KF, Park JY, Lee SH. Sci Rep. 2013;3:1921. doi: 10.1038/srep01921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Poon WW, Blurton-Jones M, Tu CH, Feinberg LM, Chabrier MA, Harris JW, Jeon NL, Cotman CW. Neurobiol Aging. 2011;32:821. doi: 10.1016/j.neurobiolaging.2009.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dauer W, Przedborski S. Neuron. 2003;39:889. doi: 10.1016/s0896-6273(03)00568-3. [DOI] [PubMed] [Google Scholar]
- 65.Volpicelli-Daley LA, Luk KC, Patel TP, Tanik SA, Riddle DM, Stieber A, Meaney DF, Trojanowski JQ, Lee VMY. Neuron. 2011;72:57. doi: 10.1016/j.neuron.2011.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lu X, Kim-Han JS, O’Malley KL, Sakiyama-Elbert SE. J Neurosci Methods. 2012;209:35. doi: 10.1016/j.jneumeth.2012.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lo EH, Dalkara T, Moskowitz MA. Nat Rev Neurosci. 2003;4:399. doi: 10.1038/nrn1106. [DOI] [PubMed] [Google Scholar]
- 68.Camos S, Mallolas J. Molecules. 2010;15:9104. doi: 10.3390/molecules15129104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Samson AJ, Robertson G, Zagnoni M, Connolly CN. Sci Rep. 2016;6:33746. doi: 10.1038/srep33746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Richard MJP, Saleh TM, El Bahh B, Zidichouski JA. J Neurosci Methods. 2010;190:20. doi: 10.1016/j.jneumeth.2010.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Oppegard SC, Nam KH, Carr JR, Skaalure SC, Eddington DT. PLoS One. 2009;4:e6891. doi: 10.1371/journal.pone.0006891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Adler M, Polinkovsky M, Gutierrez E, Groisman A. Lab Chip. 2010;10:388. doi: 10.1039/b920401f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Polinkovsky M, Gutierrez E, Levchenko A, Groisman A. Lab Chip. 2009;9:1073. doi: 10.1039/b816191g. [DOI] [PubMed] [Google Scholar]
- 74.Lo JF, Sinkala E, Eddington DT. Lab Chip. 2010;10:2394. doi: 10.1039/c004660d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Skolimowski M, Nielsen MW, Emneus J, Molin S, Taboryski R, Sternberg C, Dufva M, Geschke O. Lab Chip. 2010;10:2162. doi: 10.1039/c003558k. [DOI] [PubMed] [Google Scholar]
- 76.Mauleon G, Fall CP, Eddington DT. PLoS One. 2012;7:e43309. doi: 10.1371/journal.pone.0043309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Boillee S, Vande Velde C, Cleveland DW. Neuron. 2006;52:39. doi: 10.1016/j.neuron.2006.09.018. [DOI] [PubMed] [Google Scholar]
- 78.Kunze A, Lengacher S, Dirren E, Aebischer P, Magistretti PJ, Renaud P. Integr Biol. 2013;5:964. doi: 10.1039/c3ib40022k. [DOI] [PubMed] [Google Scholar]
- 79.Uzel SG, Platt RJ, Subramanian V, Pearl TM, Rowlands CJ, Chan V, Boyer LA, So PT, Kamm RD. Sci Adv. 2016;2:e1501429. doi: 10.1126/sciadv.1501429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Park HS, Liu S, McDonald J, Thakor N, Yang IH. Conf Proc IEEE Eng Med Biol Soc. 2013;2013:2833. doi: 10.1109/EMBC.2013.6610130. [DOI] [PubMed] [Google Scholar]
- 81.Wolff A, Antfolk M, Brodin B, Tenje M. J Pharm Sci. 2015;104:2727. doi: 10.1002/jps.24329. [DOI] [PubMed] [Google Scholar]
- 82.Wilhelm I, Krizbai IA. Mol Pharm. 2014;11:1949. doi: 10.1021/mp500046f. [DOI] [PubMed] [Google Scholar]
- 83.Naik P, Cucullo L. J Pharm Sci. 2012;101:1337. doi: 10.1002/jps.23022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zhao Z, Nelson AR, Betsholtz C, Zlokovic BV. Cell. 2015;163:1064. doi: 10.1016/j.cell.2015.10.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Nelson AR, Sweeney MD, Sagare AP, Zlokovic BV. Biochim Biophys Acta. 2016;1862:887. doi: 10.1016/j.bbadis.2015.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Takeshita Y, Ransohoff RM. Clin Exp Neuroimmunol. 2015;6:351. [Google Scholar]
- 87.Zlokovic BV. Nat Rev Neurosci. 2011;12:723. doi: 10.1038/nrn3114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zlokovic BV. Neuron. 2008;57:178. doi: 10.1016/j.neuron.2008.01.003. [DOI] [PubMed] [Google Scholar]
- 89.Shlosberg D, Benifla M, Kaufer D, Friedman A. Nat Rev Neurol. 2010;6:393. doi: 10.1038/nrneurol.2010.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Latour LL, Kang DW, Ezzeddine MA, Chalela JA, Warach S. Ann Neurol. 2004;56:468. doi: 10.1002/ana.20199. [DOI] [PubMed] [Google Scholar]
- 91.An J, Zhang C, Polavarapu R, Zhang X, Zhang X, Yepes M. Blood. 2008;112:2787. doi: 10.1182/blood-2008-02-141630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Sandoval KE, Witt KA. Neurobiol Dis. 2008;32:200. doi: 10.1016/j.nbd.2008.08.005. [DOI] [PubMed] [Google Scholar]
- 93.Cho HS, Seo JH, Wong KHK, Terasaki Y, Park J, Bong K, Arai K, Lo EH, Irimia D. Sci Rep. 2015;5:15222. doi: 10.1038/srep15222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Herland A, van der Meer AD, FitzGerald EA, Park TE, Sleeboom JJF, Ingber DE. PLoS One. 2016;11:e0150360. doi: 10.1371/journal.pone.0150360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Prabhakarpandian B, Shen MC, Nichols JB, Mills IR, Sidoryk-Wegrzynowicz M, Aschner M, Pant K. Lab Chip. 2013;13:1093. doi: 10.1039/c2lc41208j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Deosarkar SP, Prabhakarpandian B, Wang B, Sheffield JB, Krynska B, Kiani MF. PLoS One. 2015;10:e0142725. doi: 10.1371/journal.pone.0142725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Glass RI. Nature. 2015;527:S150. doi: 10.1038/nature16027. [DOI] [PubMed] [Google Scholar]
- 98.Muratore CR, Rice HC, Srikanth P, Callahan DG, Shin T, Benjamin LN, Walsh DM, Selkoe DJ, Young-Pearse TL. Hum Mol Genet. 2014;23:3523. doi: 10.1093/hmg/ddu064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Kondo T, Asai M, Tsukita K, Kutoku Y, Ohsawa Y, Sunada Y, Imamura K, Egawa N, Yahata N, Okita K, Takahashi K, Asaka I, Aoi T, Watanabe A, Watanabe K, Kadoya C, Nakano R, Watanabe D, Maruyama K, Hori O, Hibino S, Choshi T, Nakahata T, Hioki H, Kaneko T, Naitoh M, Yoshikawa K, Yamawaki S, Suzuki S, Hata R, Ueno S, Seki T, Kobayashi K, Toda T, Murakami K, Irie K, Klein WL, Mori H, Asada T, Takahashi R, Iwata N, Yamanaka S, Inoue H. Cell Stem Cell. 2013;12:487. doi: 10.1016/j.stem.2013.01.009. [DOI] [PubMed] [Google Scholar]
- 100.Koch P, Tamboli IY, Mertens J, Wunderlich P, Ladewig J, Stuber K, Esselmann H, Wiltfang J, Brustle O, Walter J. Am J Pathol. 2012;180:2404. doi: 10.1016/j.ajpath.2012.02.012. [DOI] [PubMed] [Google Scholar]
- 101.Israel MA, Yuan SH, Bardy C, Reyna SM, Mu Y, Herrera C, Hefferan MP, Van Gorp S, Nazor KL, Boscolo FS, Carson CT, Laurent LC, Marsala M, Gage FH, Remes AM, Koo EH, Goldstein LS. Nature. 2012;482:216. doi: 10.1038/nature10821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Yagi T, Ito D, Okada Y, Akamatsu W, Nihei Y, Yoshizaki T, Yamanaka S, Okano H, Suzuki N. Hum Mol Genet. 2011;20:4530. doi: 10.1093/hmg/ddr394. [DOI] [PubMed] [Google Scholar]
- 103.D’Avanzo C, Aronson J, Kim YH, Choi SH, Tanzi RE, Kim DY. BioEssays. 2015;37:1139. doi: 10.1002/bies.201500063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Zhang D, Pekkanen-Mattila M, Shahsavani M, Falk A, Teixeira AI, Herland A. Biomaterials. 2014;35:1420. doi: 10.1016/j.biomaterials.2013.11.028. [DOI] [PubMed] [Google Scholar]
- 105.Choi YJ, Park J, Lee SH. Biomaterials. 2013;34:2938. doi: 10.1016/j.biomaterials.2013.01.038. [DOI] [PubMed] [Google Scholar]
- 106.Park J, Lee BK, Jeong GS, Hyun JK, Lee CJ, Lee SH. Lab Chip. 2015;15:141. doi: 10.1039/c4lc00962b. [DOI] [PubMed] [Google Scholar]
- 107.Choi SH, Kim YH, Quinti L, Tanzi RE, Kim DY. Mol Neurodegener. 2016;11:75. doi: 10.1186/s13024-016-0139-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Choi SH, Kim YH, D’Avanzo C, Aronson J, Tanzi RE, Kim DY. US Neurol. 2015;11:102. doi: 10.17925/USN.2015.11.02.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Sandoe J, Eggan K. Nat Neurosci. 2013;16:780. doi: 10.1038/nn.3425. [DOI] [PubMed] [Google Scholar]
- 110.Passier R, Orlova V, Mummery C. Cell Stem Cell. 2016;18:309. doi: 10.1016/j.stem.2016.02.011. [DOI] [PubMed] [Google Scholar]
- 111.Kaiser T, Feng G. Nat Med. 2015;21:979. doi: 10.1038/nm.3935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Imaizumi Y, Okano H. J Neurochem. 2014;129:388. doi: 10.1111/jnc.12625. [DOI] [PubMed] [Google Scholar]
- 113.Cavaliere F, Benito-Muñoz M, Matute C. Stem Cells Int. 2016;2016:6. doi: 10.1155/2016/3540568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Humpel C. Neuroscience. 2015;305:86. doi: 10.1016/j.neuroscience.2015.07.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Shamir ER, Ewald AJ. Nat Rev Mol Cell Biol. 2014;15:647. doi: 10.1038/nrm3873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Gähwiler BH. J Neurosci Methods. 1981;4:329. doi: 10.1016/0165-0270(81)90003-0. [DOI] [PubMed] [Google Scholar]
- 117.Ullrich C, Daschil N, Humpel C. J Neurosci Methods. 2011;201:131. doi: 10.1016/j.jneumeth.2011.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.De Simoni A, Yu LM. Nat Protoc. 2006;1:1439. doi: 10.1038/nprot.2006.228. [DOI] [PubMed] [Google Scholar]
- 119.Prasanthi JR, Larson T, Schommer J, Ghribi O. PLoS One. 2011;6:e26420. doi: 10.1371/journal.pone.0026420. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 120.Alberdi E, Sanchez-Gomez MV, Cavaliere F, Perez-Samartin A, Zugaza JL, Trullas R, Domercq M, Matute C. Cell Calcium. 2010;47:264. doi: 10.1016/j.ceca.2009.12.010. [DOI] [PubMed] [Google Scholar]
- 121.Kreutz F, Frozza RL, Breier AC, de Oliveira VA, Horn AP, Pettenuzzo LF, Netto CA, Salbego CG, Trindade VM. Neurochem Int. 2011;59:648. doi: 10.1016/j.neuint.2011.06.007. [DOI] [PubMed] [Google Scholar]
- 122.Cavaliere F, Vicente ES, Matute C. J Neurosci Methods. 2010;188:205. doi: 10.1016/j.jneumeth.2010.02.008. [DOI] [PubMed] [Google Scholar]
- 123.Healy S, McMahon J, Owens P, FitzGerald U. Sci Rep. 2016;6:36410. doi: 10.1038/srep36410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Hellwig S, Masuch A, Nestel S, Katzmarski N, Meyer-Luehmann M, Biber K. Sci Rep. 2015;5:14624. doi: 10.1038/srep14624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Yagi T, Ito D, Okada Y, Akamatsu W, Nihei Y, Yoshizaki T, Yamanaka S, Okano H, Suzuki N. Hum Mol Genet. 2011;20:4530. doi: 10.1093/hmg/ddr394. [DOI] [PubMed] [Google Scholar]
- 126.Daviaud N, Garbayo E, Schiller PC, Perez-Pinzon M, Montero-Menei CN. Exp Neurol. 2013;248:429. doi: 10.1016/j.expneurol.2013.07.012. [DOI] [PubMed] [Google Scholar]
- 127.Jaderstad LM, Jaderstad J, Herlenius E. Regen Med. 2010;5:901. doi: 10.2217/rme.10.80. [DOI] [PubMed] [Google Scholar]
- 128.Tonnesen J, Parish CL, Sorensen AT, Andersson A, Lundberg C, Deisseroth K, Arenas E, Lindvall O, Kokaia M. PLoS One. 2011;6:e17560. doi: 10.1371/journal.pone.0017560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Moser KV, Schmidt-Kastner R, Hinterhuber H, Humpel C. Eur J Neurosci. 2003;18:85. doi: 10.1046/j.1460-9568.2003.02728.x. [DOI] [PubMed] [Google Scholar]
- 130.Moser KV, Reindl M, Blasig I, Humpel C. Brain Res. 2004;1017:53. doi: 10.1016/j.brainres.2004.05.013. [DOI] [PubMed] [Google Scholar]
- 131.Hutter-Schmid B, Kniewallner K, Humpel C. Front Cell Dev Biol. 2015;3:52. doi: 10.3389/fcell.2015.00052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Daschil N, Kniewallner KM, Obermair GJ, Hutter-Paier B, Windisch M, Marksteiner J, Humpel C. Neurobiol Aging. 2015;36:1333. doi: 10.1016/j.neurobiolaging.2014.12.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Kempermann G, Wiskott L, Gage FH. Curr Opin Neurobiol. 2004;14:186. doi: 10.1016/j.conb.2004.03.001. [DOI] [PubMed] [Google Scholar]
- 134.Jennings CG, Landman R, Zhou Y, Sharma J, Hyman J, Movshon JA, Qiu Z, Roberts AC, Roe AW, Wang X, Zhou H, Wang L, Zhang F, Desimone R, Feng G. Nat Neurosci. 2016;19:1123. doi: 10.1038/nn.4362. [DOI] [PubMed] [Google Scholar]
- 135.Tropepe V, Hitoshi S, Sirard C, Mak TW, Rossant J, van der Kooy D. Neuron. 2001;30:65. doi: 10.1016/s0896-6273(01)00263-x. [DOI] [PubMed] [Google Scholar]
- 136.Jensen JB, Parmar M. Mol Neurobiol. 2006;34:153. doi: 10.1385/MN:34:3:153. [DOI] [PubMed] [Google Scholar]
- 137.Pastrana E, Silva-Vargas V, Doetsch F. Cell Stem Cell. 2011;8:486. doi: 10.1016/j.stem.2011.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Chung S, Shin BS, Hwang M, Lardaro T, Kang UJ, Isacson O, Kim KS. Stem Cells. 2006;24:1583. doi: 10.1634/stemcells.2005-0558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Pasca AM, Sloan SA, Clarke LE, Tian Y, Makinson CD, Huber N, Kim CH, Park J-Y, O’Rourke NA, Nguyen KD, Smith SJ, Huguenard JR, Geschwind DH, Barres BA, Pasca SP. Nat Meth. 2015;12:671. doi: 10.1038/nmeth.3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Guerrero-Cázares H, Chaichana KL, Quiñones-Hinojosa A. In: Cancer Stem Cells: Methods and Protocols. Yu JS, editor. Humana Press; Totowa, NJ: 2009. p. 73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Joseph JV, Mv Roosmalen IA, Busschers E, Tomar T, Conroy S, Eggens-Meijer E, Peñaranda Fajardo N, Pore MM, Balasubramanyian V, Wagemakers M, Copray S, Ad Dunnen WF, Kruyt FAE. PLoS One. 2015;10:e0145393. doi: 10.1371/journal.pone.0145393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Garcez PP, Loiola EC, Madeiro da Costa R, Higa LM, Trindade P, Delvecchio R, Nascimento JM, Brindeiro R, Tanuri A, Rehen SK. Science. 2016;352:816. doi: 10.1126/science.aaf6116. [DOI] [PubMed] [Google Scholar]
- 143.D’Avanzo C, Sliwinski C, Wagner SL, Tanzi RE, Kim DY, Kovacs DM. FASEB J. 2015;29:3335. doi: 10.1096/fj.15-271015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Lee H-K, Velazquez Sanchez C, Chen M, Morin PJ, Wells JM, Hanlon EB, Xia W. PLoS One. 2016;11:e0163072. doi: 10.1371/journal.pone.0163072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Kato-Negishi M, Tsuda Y, Onoe H, Takeuchi S. Biomaterials. 2010;31:8939. doi: 10.1016/j.biomaterials.2010.08.008. [DOI] [PubMed] [Google Scholar]
- 146.Jeong GS, Chang JY, Park JS, Lee SA, Park D, Woo J, An H, Lee CJ, Lee SH. Mol Brain. 2015;8:17. doi: 10.1186/s13041-015-0109-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Arsenijevic Y, Weiss S, Schneider B, Aebischer P. J Neurosci. 2001;21:7194. doi: 10.1523/JNEUROSCI.21-18-07194.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Caldwell MA, He X, Wilkie N, Pollack S, Marshall G, Wafford KA, Svendsen CN. Nat Biotechnol. 2001;19:475. doi: 10.1038/88158. [DOI] [PubMed] [Google Scholar]
- 149.Hack MA, Sugimori M, Lundberg C, Nakafuku M, Gotz M. Mol Cell Neurosci. 2004;25:664. doi: 10.1016/j.mcn.2003.12.012. [DOI] [PubMed] [Google Scholar]
- 150.Morshead CM, Benveniste P, Iscove NN, van der Kooy D. Nat Med. 2002;8:268. doi: 10.1038/nm0302-268. [DOI] [PubMed] [Google Scholar]
- 151.Parmar M, Skogh C, Bjorklund A, Campbell K. Mol Cell Neurosci. 2002;21:645. doi: 10.1006/mcne.2002.1204. [DOI] [PubMed] [Google Scholar]
- 152.Eriksson C, Bjorklund A, Wictorin K. Exp Neurol. 2003;184:615. doi: 10.1016/S0014-4886(03)00271-1. [DOI] [PubMed] [Google Scholar]
- 153.Parmar M, Sjoberg A, Bjorklund A, Kokaia Z. Mol Cell Neurosci. 2003;24:741. doi: 10.1016/s1044-7431(03)00239-2. [DOI] [PubMed] [Google Scholar]
- 154.Suslov ON, Kukekov VG, Ignatova TN, Steindler DA. Proc Natl Acad Sci USA. 2002;99:14506. doi: 10.1073/pnas.212525299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Mason JO, Price DJ. Neuroscience. 2016;334:105. doi: 10.1016/j.neuroscience.2016.07.048. [DOI] [PubMed] [Google Scholar]
- 156.Honegger P, Lenoir D, Favrod P. Nature. 1979;282:305. doi: 10.1038/282305a0. [DOI] [PubMed] [Google Scholar]
- 157.Shamir ER, Ewald AJ. Nat Rev Mol Cell Biol. 2014;15:647. doi: 10.1038/nrm3873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Lancaster MA, Knoblich JA. Nat Protocols. 2014;9:2329. doi: 10.1038/nprot.2014.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Chambers SM, Tchieu J, Studer L. Cell Stem Cell. 2013;13:377. doi: 10.1016/j.stem.2013.09.010. [DOI] [PubMed] [Google Scholar]
- 160.Eiraku M, Takata N, Ishibashi H, Kawada M, Sakakura E, Okuda S, Sekiguchi K, Adachi T, Sasai Y. Nature. 2011;472:51. doi: 10.1038/nature09941. [DOI] [PubMed] [Google Scholar]
- 161.Bershteyn M, Kriegstein AR. Cell. 2013;155:19. doi: 10.1016/j.cell.2013.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Skardal A, Shupe T, Atala A. Drug Discov Today. 2016 doi: 10.1016/j.drudis.2016.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Hansen DV, Lui JH, Parker PR, Kriegstein AR. Nature. 2010;464:554. doi: 10.1038/nature08845. [DOI] [PubMed] [Google Scholar]
- 164.Watanabe K, Kamiya D, Nishiyama A, Katayama T, Nozaki S, Kawasaki H, Watanabe Y, Mizuseki K, Sasai Y. Nat Neurosci. 2005;8:288. doi: 10.1038/nn1402. [DOI] [PubMed] [Google Scholar]
- 165.Nasu M, Takata N, Danjo T, Sakaguchi H, Kadoshima T, Futaki S, Sekiguchi K, Eiraku M, Sasai Y. PLoS One. 2012;7:e53024. doi: 10.1371/journal.pone.0053024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Kadoshima T, Sakaguchi H, Nakano T, Soen M, Ando S, Eiraku M, Sasai Y. Proc Natl Acad Sci USA. 2013;110:20284. doi: 10.1073/pnas.1315710110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Miller JD, Ganat YM, Kishinevsky S, Bowman RL, Liu B, Tu EY, Mandal PK, Vera E, Shim JW, Kriks S, Taldone T, Fusaki N, Tomishima MJ, Krainc D, Milner TA, Rossi DJ, Studer L. Cell Stem Cell. 2013;13:691. doi: 10.1016/j.stem.2013.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Marton RM, Paşca SP. Cell Stem Cell. 2016;19:145. doi: 10.1016/j.stem.2016.07.017. [DOI] [PubMed] [Google Scholar]
- 169.Jo J, Xiao Y, Sun AX, Cukuroglu E, Tran H-D, Göke J, Tan ZY, Saw TY, Tan C-P, Lokman H, Lee Y, Kim D, Ko HS, Kim S-O, Park JH, Cho N-J, Hyde TM, Kleinman JE, Shin JH, Weinberger DR, Tan EK, Je HS, Ng H-H. Cell Stem Cell. 2016;19:248. doi: 10.1016/j.stem.2016.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Qian X, Nguyen HN, Song MM, Hadiono C, Ogden SC, Hammack C, Yao B, Hamersky GR, Jacob F, Zhong C, Yoon KJ, Jeang W, Lin L, Li Y, Thakor J, Berg DA, Zhang C, Kang E, Chickering M, Nauen D, Ho CY, Wen Z, Christian KM, Shi PY, Maher BJ, Wu H, Jin P, Tang H, Song H, Ming GL. Cell. 2016;165:1238. doi: 10.1016/j.cell.2016.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Cauchemez S, Besnard M, Bompard P, Dub T, Guillemette-Artur P, Eyrolle-Guignot D, Salje H, Van Kerkhove MD, Abadie V, Garel C, Fontanet A, Mallet HP. Lancet. 2016;387:2125. doi: 10.1016/S0140-6736(16)00651-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Cauchemez S, Besnard M, Garel C, Fontanet A, Mallet HP. Lancet. 2016;388:338. doi: 10.1016/S0140-6736(16)31017-0. [DOI] [PubMed] [Google Scholar]
- 173.Faria NR, do Azevedo RS, Kraemer MU, Souza R, Cunha MS, Hill SC, Theze J, Bonsall MB, Bowden TA, Rissanen I, Rocco IM, Nogueira JS, Maeda AY, Vasami FG, Macedo FL, Suzuki A, Rodrigues SG, Cruz AC, Nunes BT, Medeiros DB, Rodrigues DS, Nunes Queiroz AL, da Silva EV, Henriques DF, Travassos da Rosa ES, de Oliveira CS, Martins LC, Vasconcelos HB, Casseb LM, de Simith DB, Messina JP, Abade L, Lourenco J, Carlos Junior Alcan-tara L, de Lima MM, Giovanetti M, Hay SI, de Oliveira RS, da Lemos PS, de Oliveira LF, de Lima CP, da Silva SP, de Vasconcelos JM, Franco L, Cardoso JF, Vianez-Junior JL, Mir D, Bello G, Delatorre E, Khan K, Creatore M, Coelho GE, de Oliveira WK, Tesh R, Pybus OG, Nunes MR, Vasconcelos PF. Science. 2016;352:345. doi: 10.1126/science.aaf5036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Camp JG, Badsha F, Florio M, Kanton S, Gerber T, Wilsch-Bräuninger M, Lewitus E, Sykes A, Hevers W, Lancaster M, Knoblich JA, Lachmann R, Pääbo S, Huttner WB, Treutlein B. Proc Natl Acad Sci USA. 2015;112:15672. doi: 10.1073/pnas.1520760112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Quadrato G, Nguyen T, Macosko EZ, Sherwood JL, Min Yang S, Berger DR, Maria N, Scholvin J, Goldman M, Kinney JP, Boyden ES, Lichtman JW, Williams ZM, McCarroll SA, Arlotta P. Nature. 2017;545:48. doi: 10.1038/nature22047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Birey F, Andersen J, Makinson CD, Islam S, Wei W, Huber N, Fan HC, Metzler KRC, Panagiotakos G, Thom N, O’Rourke NA, Steinmetz LM, Bernstein JA, Hallmayer J, Huguenard JR, Paşca SP. Nature. 2017;545:54. doi: 10.1038/nature22330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Giandomenico SL, Lancaster MA. Curr Opin Cell Biol. 2017;44:36. doi: 10.1016/j.ceb.2017.01.001. [DOI] [PubMed] [Google Scholar]
- 178.Kelava I, Lancaster MA. Dev Biol. 2016;420:199. doi: 10.1016/j.ydbio.2016.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Quadrato G, Brown J, Arlotta P. Nat Med. 2016;22:1220. doi: 10.1038/nm.4214. [DOI] [PubMed] [Google Scholar]
- 180.Bredenoord AL, Clevers H, Knoblich JA. Science. 2017;355:eaaf9412. doi: 10.1126/science.aaf9414. [DOI] [PubMed] [Google Scholar]
- 181.Boers SN, van Delden JJ, Clevers H, Bredenoord AL. EMBO Rep. 2016;17:938. doi: 10.15252/embr.201642613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Doyle AD, Carvajal N, Jin A, Matsumoto K, Yamada KM. Nat Commun. 2015;6:8720. doi: 10.1038/ncomms9720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Schultz KM, Kyburz KA, Anseth KS. Proc Natl Acad Sci USA. 2015;112:E3757. doi: 10.1073/pnas.1511304112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Vu LT, Jain G, Veres BD, Rajagopalan P. Tissue Engineering Part B: Reviews. 2014;21:67. doi: 10.1089/ten.teb.2013.0782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Lutolf MP, Gilbert PM, Blau HM. Nature. 2009;462:433. doi: 10.1038/nature08602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Yang C, DelRio FW, Ma H, Killaars AR, Basta LP, Kyburz KA, Anseth KS. Proc Natl Acad Sci USA. 2016;113:E4439. doi: 10.1073/pnas.1609731113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Yang C, Tibbitt MW, Basta L, Anseth KS. Nat Mater. 2014;13:645. doi: 10.1038/nmat3889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Gjorevski N, Sachs N, Manfrin A, Giger S, Bragina ME, Ordóñez-Morán P, Clevers H, Lutolf MP. Nature. 2016:539. doi: 10.1038/nature20168. [DOI] [PubMed] [Google Scholar]
- 189.Yao X, Peng R, Ding J. Adv Mater. 2013;25:5257. doi: 10.1002/adma.201301762. [DOI] [PubMed] [Google Scholar]
- 190.Gjorevski N, Ranga A, Lutolf MP. Development. 2014;141:1794. doi: 10.1242/dev.101048. [DOI] [PubMed] [Google Scholar]
- 191.Chwalek K, Tang-Schomer MD, Omenetto FG, Kaplan DL. Nat Protocols. 2015;10:1362. doi: 10.1038/nprot.2015.091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Kim YH, Choi SH, D’Avanzo C, Hebisch M, Sliwinski C, Bylykbashi E, Washicosky KJ, Klee JB, Brustle O, Tanzi RE, Kim DY. Nat Protoc. 2015;10:985. doi: 10.1038/nprot.2015.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Selkoe DJ. Science. 2002;298:789. doi: 10.1126/science.1074069. [DOI] [PubMed] [Google Scholar]
- 194.Espuny-Camacho I, Arranz AM, Fiers M, Snellinx A, Ando K, Munck S, Bonnefont J, Lambot L, Corthout N, Omodho L, Vanden Eynden E, Radaelli E, Tesseur I, Wray S, Ebneth A, Hardy J, Leroy K, Brion JP, Vanderhaeghen P, De Strooper B. Neuron. 2017;93:1066. doi: 10.1016/j.neuron.2017.02.001. [DOI] [PubMed] [Google Scholar]
- 195.Lutolf MP, Lauer-Fields JL, Schmoekel HG, Metters AT, Weber FE, Fields GB, Hubbell JA. Proc Natl Acad Sci USA. 2003;100:5413. doi: 10.1073/pnas.0737381100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Lutolf MP, Hubbell JA. Biomacromolecules. 2003;4:713. doi: 10.1021/bm025744e. [DOI] [PubMed] [Google Scholar]
- 197.Lutolf M, Hubbell J. Nat Biotechnol. 2005;23:47. doi: 10.1038/nbt1055. [DOI] [PubMed] [Google Scholar]
- 198.Ehrbar M, Rizzi SC, Schoenmakers RG, San Miguel B, Hubbell JA, Weber FE, Lutolf MP. Biomacromolecules. 2007;8:3000. doi: 10.1021/bm070228f. [DOI] [PubMed] [Google Scholar]
- 199.Meinhardt A, Eberle D, Tazaki A, Ranga A, Niesche M, Wilsch-Bräuninger M, Stec A, Schackert G, Lutolf M, Tanaka EM. Stem Cell Rep. 2014;3:987. doi: 10.1016/j.stemcr.2014.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Ranga A, Gobaa S, Okawa Y, Mosiewicz K, Negro A, Lutolf M. Nat Commun. 2014;5:4234. doi: 10.1038/ncomms5324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Ranga A, Girgin M, Meinhardt A, Eberle D, Caiazzo M, Tanaka EM, Lutolf MP. Proc Natl Acad Sci USA. 2016:03529. doi: 10.1073/pnas.1603529113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Rosales AM, Anseth KS. Nature Reviews Materials. 2016;1:15012. doi: 10.1038/natrevmats.2015.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Alge DL, Anseth KS. Nat Mater. 2013;12:950. doi: 10.1038/nmat3794. [DOI] [PubMed] [Google Scholar]
- 204.Mosiewicz KA, Kolb L, van der Vlies AJ, Martino MM, Lienemann PS, Hubbell JA, Ehrbar M, Lutolf MP. Nat Mater. 2013;12:1072. doi: 10.1038/nmat3766. [DOI] [PubMed] [Google Scholar]
- 205.Lutolf MP. Nature. 2012;482:477. doi: 10.1038/482477a. [DOI] [PubMed] [Google Scholar]
- 206.Kloxin AM, Tibbitt MW, Anseth KS. Nat Protoc. 2010;5:1867. doi: 10.1038/nprot.2010.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Kloxin AM, Kasko AM, Salinas CN, Anseth KS. Science. 2009;324:59. doi: 10.1126/science.1169494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Luo Y, Shoichet MS. Nat Mater. 2004;3:249. doi: 10.1038/nmat1092. [DOI] [PubMed] [Google Scholar]
- 209.DeForest CA, Polizzotti BD, Anseth KS. Nat Mater. 2009;8:659. doi: 10.1038/nmat2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Tibbitt MW, Anseth KS. Sci Transl Med. 2012;4:160ps24. doi: 10.1126/scitranslmed.3004804. [DOI] [PubMed] [Google Scholar]
- 211.Tibbitt MW, Rodell CB, Burdick JA, Anseth KS. Proc Natl Acad Sci USA. 2015;112:14444. doi: 10.1073/pnas.1516247112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Seliktar D. Science. 2012;336:1124. doi: 10.1126/science.1214804. [DOI] [PubMed] [Google Scholar]
- 213.Cushing MC, Anseth KS. Science. 2007;316:1133. doi: 10.1126/science.1140171. [DOI] [PubMed] [Google Scholar]
- 214.Pampaloni F, Reynaud EG, Stelzer EHK. Nat Rev Mol Cell Biol. 2007;8:839. doi: 10.1038/nrm2236. [DOI] [PubMed] [Google Scholar]