Abstract
Pathogenic variants in genes which encode DNA repair and damage response proteins result in a number of genomic instability syndromes with features of accelerated aging. ERCC4 (XPF) encodes a protein that forms a complex with ERCC1 and is required for the 5′ incision during nucleotide excision repair. ERCC4 is also FANCQ, illustrating a critical role in interstrand crosslink repair. Pathogenic variants in this gene cause xeroderma pigmentosum, XFE progeroid syndrome, Cockayne syndrome, and Fanconi anemia. We performed massive parallel sequencing for 42 unsolved cases submitted to the International Registry of Werner Syndrome. Two cases, each carrying two novel heterozygous ERCC4 variants, were identified. The first case was a compound heterozygote for: c.2395C>T (p.Arg799Trp) and c.388+1164_792+795del (p.Gly130Aspfs*18). Further molecular and cellular studies indicated that the ERCC4 variants in this patient are responsible for a phenotype consistent with a variant of Cockayne syndrome. The second case was heterozygous for two variants in cis: c.[1488A>T; c.2579C>A] (p.[Gln496His; Ala860Asp]). While the second case also had several phenotypic features of accelerated aging, we were unable to provide biological evidence supporting the pathogenic roles of the associated ERCC4 variants. Precise genetic causes and disease mechanism of the second case remains to be determined.
Keywords: Cockayne syndrome, Xeroderma pigmentosum group F, ERCC4, Atypical Werner syndrome, Segmental progeroid syndromes, Molecular genetics, Mendelian disease
INTRODUCTION
Genomic instability is associated with many features of aging, including benign and malignant neoplasms. In the last decade or so, many proteins involved in sensing and responding to DNA damage have been characterized, thus enhancing our understanding of genotoxic stress responses. In most cases, genome instability is a consequence of errors made by the enzymes responsible for chromosome segregation, DNA replication, or DNA damage repair (Vijg and Suh, 2013). A wide range of variants and chromosomal aberrations can activate oncogenes or inactivate tumor suppressor genes and thus increase the risk of cancer (Marteijn, et al., 2014a).
Genomic instability syndromes can also be caused by variants at loci involved in DNA repair, including nucleotide excision repair (NER). NER has two major pathways; global genome NER (GG-NER) and transcription-coupled NER (TC-NER). These pathways are distinguished by their initial DNA damage recognition events (Evans, et al., 1997; Hanawalt, 2002; Hoeijmakers, 2009; Marteijn, et al., 2014). Pathogenic variants in these loci can lead to a wide spectrum of disorders that include Cockayne syndrome (CS; MIM# 216400 and 133540), xeroderma pigmentosum (XP; MIM# 278700, 610651, 278720, 278730, 278740, 278760, and 278780), and trichothiodystrophy (TTD; MIM# 616390, 616395, 234050, 616943, 601675, and 300953). In general, GG-NER causes XP and TC-NER leads to CS, but overlap in the syndromes can occur. CS is commonly caused by mutations in ERCC8 (CSA) or ERCC6 (CSB) and is characterized by microcephaly, neurologic dysfunction, postnatal growth failure, cataracts, enophthalmos, loss of body fat, and skin photosensitivity (Laugel, 2013; Wilson, et al., 2016). XP is characterized by photosensitivity, pigmentary changes, premature skin aging, and a 10,000-fold increased risk of skin cancer (English and Swerdlow, 1987) (Kraemer and DiGiovanna, 2014). The dermatological phenotypes in CS are milder than those of XP. Moreover, skin cancers are not a feature of classical CS.
An XPF endonuclease encoded by ERCC4, also called FANCQ, forms a complex with its regulatory protein encoded by ERCC1 (XPF-ERCC1 complex). XPF-ERCC1 plays an essential role during NER repair that is common to both GG-NER and TC-NER (Friedberg, et al., 2006; Schärer, 2013). In general, patients with constitutional ERCC4 mutations exhibit xeroderma pigmentosum, group F (XP-F; MIM# 278760) (Gregg, et al., 2011; Sijbers, et al., 1996). Homozygous or compound heterozygous ERCC4 mutations have also been reported in at least one case of CS, one case of XP-CS (Kashiyama, et al., 2013), two cases of Fanconi anemia, complementation group Q (FANCQ; MIM# 615272) (Bogliolo, et al., 2013) (Kashiyama, et al., 2013), as well as one case of XFE progeroid syndrome (XFEPS; MIM# 610965) (Niedernhofer, et al., 2006). There appears to be no consistent genotype-phenotype correlation with the ERCC4 variants (Kashiyama, et al., 2013).
Here, we report on two individuals, one compound heterozygote and the other with two in-cis ERCC4 variants, as revealed by massive parallel sequencing. They represent some of the initial results of sequencing efforts to discover novel genetic variants among a series of 42 cases referred to the International Registry of Werner Syndrome (www.wernersyndrome.org) because of some clinical features thought to overlap with those found in the Werner syndrome. After excluding causative variants in loci known to be associated with segmental progeroid phenotypes (including WRN, POLD and LMNA), these cases were operationally classified as examples of Atypical Werner syndrome (AWS) (Oshima and Hisama, 2014; Oshima, et al., 2016).
MATERIALS AND METHODS
Subject recruitment and ethical approval
Physicians refer patients they suspect of having a progeroid syndrome to the International Registry of Werner Syndrome Seattle at the University of Washington. After review of a questionnaire and clinical information, appropriate patients (and, when available, their nuclear family members) are enrolled for genetic research studies. All patients must provide written informed consent in order to be enrolled, including permission for the establishment of cell lines. All protocols have been approved by a University of Washington Institutional Review Board.
Candidate gene sequencing
For candidate gene analysis, we designed intronic primers to PCR amplify coding exons and the respective exon-intron boundaries by using genomic DNA from the affected individual. Primer pairs for the amplification of the WRN, LMNA, and POLD1 coding exons and approximately 50 bp of flanking intronic sequences (RefSeq accession NM_000553.4, NM_170707.3, and NM_002691.3) are available upon request. For Sanger sequencing of ERCC4, we utilized primer pairs for the amplification of exons 8 and 11 (ERCC4_EX8F [5′-GGCACAGGGAAACTAGGAGG-3′] and ERCC4_EX8R [5′-TGCAACTGGTATAGTTTCTGGG-3′]) (ERCC4_EX11F [5′-TTCAAGGGAAATTGCCTCTAC-3′] and ERCC4_EX11R [5′-AGGCAAGTTCAAGTGATGCC-3′] and for flanking regions of the deletion spanning intron 2 to 4 (ERCC4_del_F [5′-CTGATGTCCCCAAAGCTAATG-3′] and ERCC4_del-R [5′-GATACTGGGACTATCCTGAAC-3′]).
Targeted ERCC4 gene sequencing by next-generation sequencing (NGS) using long-acting (LA)-PCR
Long-acting PCR was performed to amplify approximately 32kbp spanning the ERCC4 genomic locus using 100 ng of the probands’ genomic DNA. Primer sequences were ERCC4_LA_F (5′-GAGTTCGGCCAACGCTTGCCTTCTC-3′) and ERCC4_LA_R (5′-GCCAGCTGGACTTAGGGATCTCTG-3′). PrimeSTAR GXL DNA Polymerase (TaKaRa) was used. Library preparation (Illumina Nextera XT DNA Library Preparation Kit) was conducted in order to obtain a shotgun-library according to the manufacturer’s instructions. Pooled, barcoded libraries were sequenced via paired-end 150-bp reads and an 8-bp barcode read on Illumina MiSeq sequencers. Following basic analysis steps and filtering, conditions for the identification of rare variants were equivalent to that of the exome sequencing as described below.
Exome sequencing in 18 AWS patients and filtering of variants
Exome sequencing was performed with Illumina HiSeq sequencers as previously described (Chong, et al., 2015). Demultiplexed read data (fastq files) were aligned to a human reference sequence (UCSC Genome Browser hg19) via the Burrows-Wheeler Aligner v.0.7.10 (Li and Durbin, 2009). All aligned read data were subjected to (1) removal of duplicate reads (Picard MarkDuplicates v.1.111) (https://broadinstitute.github.io/picard/), (2) indel realignment (Genome Analysis Toolkit [GATK] IndelRealigner v.3.2) (DePristo, et al., 2011), and (3) base-quality recalibration (GATK BaseRecalibrator v.3.2). Single-nucleotide variant (SNV) detection and genotyping were performed with the GATK HaplotypeCaller (v.3.2). Variants were annotated with the VariantEffectPredictor v.83 (McLaren, et al., 2010) after decomposition and normalization of variants in the raw VCF file with Vt v.0.1.0 (Tan, et al., 2015). Variant filtering was performed with GEMINI (v.0.19.1) (Paila, et al., 2013). We generated copy-number variant (CNV) calls from exome data by using CoNIFER (Copy Number Inference From Exome Reads)(Krumm, et al., 2012).
Filtering of candidate variants was done as previously described (Chong, et al., 2015). Briefly, we first excluded variants with alternate allele frequencies > 0.005 in any population within the Exome Aggregation Consortium (ExAC v.0.3) Browser (http://exac.broadinstitute.org), NHLBI Exome Sequencing Project Exome Variant Server dataset ESP6500 (http://evs.gs.washington.edu/EVS/), or 1000 Genomes (Abecasis, et al., 2010), as well as variants with allele frequencies > 0.05 in an internal database of more than 2,500 exomes. We excluded variants with “LOW” impact severities, as defined by the GEMINI software package (v0.19.1) (Paila, et al., 2013), which includes functional predictions such as: “synonymous_coding,” “intergenic,” “upstream,” “UTR,” “intron,” etc. Individual genotypes with a depth < 8 or genotype quality < 20 were treated as missing. The probands were also screened for larger CNVs via array comparative genomic hybridization on the Illumina Infinium Human-Core-24 BeadChip.
Cell lines and cell culture
Epstein–Barr virus transformed lymphoblastoid cell lines (LCLs) originating from MME1010 were established as previously described (Huang, et al., 2006). Primary human diploid fibroblast cell lines were established from biopsies of skin samples of CALIF1010 and were immortalized with the catalytic subunit of human telomerase (hTERT), as previously described (Saha, et al., 2013). Cellular assays utilized EBV transformed lymphoblastoid cells [Control1 (normal), GM07398 (normal), LB313 (XP-F), MME1010] and hTERT immortalized fibroblasts [Control2 (normal), C5RO (normal), XP51RO (XFE), CALIF1010]. All LCLs were grown in RPMI1640 + glutamax (Gibco) media with 10% fetal bovine serum and 1% penicillin streptomycin. Fibroblasts were grown in DMEM or F10 + glutamax (Gibco) media with 10% fetal bovine serum and 1% penicillin streptomycin. All cells were grown in an incubator at 37 °C, 5% CO2 at ambient oxygen concentration.
Cellular sensitivity to mitomycin C (MMC) was determined by adding final concentration of 200nM, 1μM or 5μM MMC into the culture media and counting the cell numbers after 24hrs and 4 days with a hemocytometer.
RT-PCR analysis
RT-PCR was done as described previously (Hisama, et al., 2011). To evaluate alternative transcription of ERCC4 isoforms caused by variant alleles, we performed RT-PCR reactions. Primer sequences used for amplification of the product spanning exons 1 through 7 in ERCC4 (RefSeq accession NM_005236.2) are XPF_CDS_F (5′-ATGGAGTCAGGGCAGCCGGCTCGAC-3′) and XPF_CDS_R (5′-GCACAAATCAGTACTTGACCTGGAC-3′) which are expected to give a wildtype product of 1,232 bp.
For Allele specific RT-PCR in MME1010, two forward primers with variations in their 3′ nucleotides were designed so that each primer could be specific for one of the two variants (ERCC4_F_wt [5′-CGGAAGTTGACCTTAACTCAA-3′] or ERCC4_F_mt [5′-CGGAAGTTGACCTTAACTCAT-3′]). Two reverse primers were prepared as well as forward primers (ERCC4_R_wt [5′-TCAAGGAGCGGCAGTTTTTGG-3′] or ERCC4_R_mt [5′-TCAAGGAGCGGCAGTTTTTGT-3′]). By using PCR in combination with each forward and reverse primer, the relative locations of the two variants could be investigated.
Quantitative PCR
Quantitative RT-PCR was used to measure total mRNA level for XPF and ERCC1. Human GAPDH (Fwd: 5′-TCATGGGTGTGAACCATGAGAA-3′; Rev: 5′-GGCATGGACTGTGGTCATGAG-3′), ERCC1 (Fwd: 5′-TTGTCCAGGTGGATGTGAAA-3′; Rev: 5′-GCTGGTTTCTGCTCATAGGC-3′), and XPF (Fwd: 5′-TTTGTGAGGAAACTGTATCTGTGG-3′; Rev: 5′-GTCTGTATAGCAAGCATGGTAGG-3′) were analyzed for expression. qPCR reactions were performed on duplicate samples of cDNAs from three independent collections of all cell lines.
Western blot analysis
Western blot analysis was done as described previously (Hisama, et al., 2011; Huang, et al., 2006) Isolated nuclei from the LCL cell lines or hTERT immortalized fibroblasts were lysed in 2xLaemmli buffer and were subjected to Western analysis. Commercial antibodies used for nuclear Western analysis were anti-XPF (dilution 1:500, Thermo Scientific, Clone 219), anti-lamin A/C (dilution 1:2,500, Santa Cruz, H-110), anti-β-actin (dilution 1:40,000, Sigma, clone AC-15) and corresponding biotinylated secondary antibodies against mouse or rabbit IgG (Vector Laboratories, BA-9200 and BA-1000, respectively). The reactions were visualized with western lightening chemiluminescence reagent (NEL100, Perkin Elmer) according to the manufacturer’s instructions.
Immunoblot detection was also used to compare expression of XPF in CALIF1010 to that of an established XFE progeroid patient XP51RO (Niedernhofer, 2006). Rabbit α-GAPDH antibody (1:10000)(Sigma Aldrich), ERCC1 mouse monoclonal antibody (D-10)(1:500) (Santa Cruz) and XPF mouse monoclonal antibody (SPM228)(1:1000) (Abcam) were used for detection.
Flow cytometric-based unscheduled DNA synthesis assay
This is a modification of a previously published unscheduled DNA synthesis assay (Ahmad, et al., 2010). 1–2×106 cells were aliquoted and resuspended in an equivalent volume of DPBS. For each cell line, one set of samples were irradiated using the cell culture hood’s UV-C lamp in an uncovered petri dish at 24 J/m2 while being mixed at 300 rpm using an orbital shaker. Samples were then centrifuged and resuspended in fresh media containing 10 μM EdU and then incubated at 37 °C for 2.5 hr. Following incubation, cells were centrifuged and washed with DPBS containing 1% BSA. Cells were resuspended in 1X Saponin diluted in PBS with 1% BSA and incubated for 30 min in darkness at room temperature to allow for cell membrane permeabilization. The cells were again centrifuged and washed with PBS containing 1% BSA. In order to conjugate the Alexa Fluor 647 fluorophore to the incorporated EdU in cells, samples were then incubated with Alexa-647 reaction cocktail for 30 min in the dark at 4 °C as per instructions from the Click-iT Alexa Fluor 647 flow cytometry assay kit (Thermo Fisher). The cells were centrifuged and washed once with DPBS containing 1% BSA and washed once with cold DPBS. The cells were resuspended in 200 μL DPBS and fixed by adding an equal volume of 4% formaldehyde dropwise while vortexing at a gentle speed and left to fix overnight at 4 °C. The following day, the samples were centrifuged and washed with DPBS before being resuspended in buffer containing PBS, 5% FBS, 0.5% Tween-20, and 10 μg/mL DAPI, and incubation at 4 °C for 2 hr. Following this, cells were centrifuged and washed with DPBS before being resuspended in ~300 μL DPBS for flow cytometric analysis. Cells were analyzed on Canto flow cytometer (BD) for median Alexa Fluor 647 fluorescence intensity in the G1 population determined by DAPI for cell cycle. Data analysis was performed using FlowJo 9.3.1 software.
Recovery of RNA synthesis (RRS)
RRS was measured as previously described (Velez-Cruz, et al., 2013). 1×105 cells were trypsinized, resuspended in 1 mL of DPBS, and deposited on a 60 mm cell culture dish lid for irradiation with 10 J/m2 UV-C. After irradiation, the cells were collected, spun down, and replated with complete media then collected at the indicated time points. Total RNA was isolated using Trizol extraction as specified by the manufacturer (Thermo-Fisher). cDNA was generated using the Transcriptor First Strand Kit (Roche) and qRT-PCR was performed on a StepOne thermocycler (Thermo-Fisher) using Universal SYBR with ROX (Roche). DHFR and GAPDH mRNA expression was normalized to 18s rRNA and then graphed as the ratio of expression in irradiated cells relative to sham-irradiated cells at each time point, as previously described (Velez-Cruz, et al., 2013). Reactions were performed in triplicate and each data point represents the mean of five independent experiments. Human primer sequences included: DHFR Fwd 5′CCACAACCTCTTCAGTAG-3″ DHFR Rev 5′-GAGGTTCCTTGAGTTCTCTG-3′; GAPDH Fwd 5′-TCATGGGTGTGAACCATGAGAA-3′, GAPDH Rev 5′-GGCATGGACTGTGGTCATGAG-3′; 18s Fwd 5′-CTACCACATCCAAGGAAGCA-3′, 18s Rev 5′-TTTTTCGTCACTACCTCCCCG-3′.
RESULTS
Case presentations
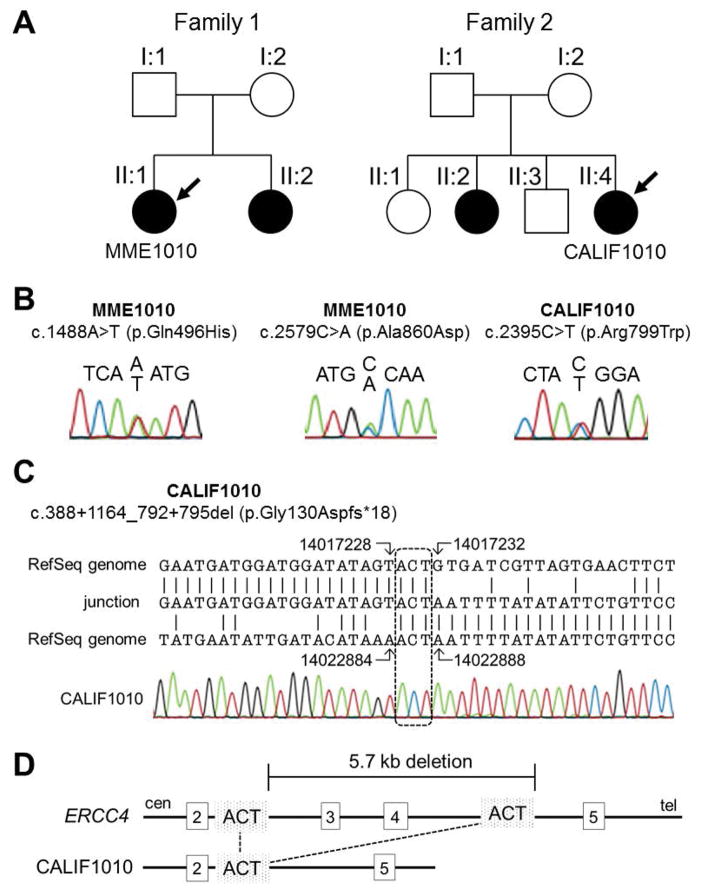
Registry# CALIF1010 is a woman who presented at the age of 35 years for genetic evaluation of microcephaly, enophthalmos and a prematurely aged appearance. She was born at term with a birth weight of 1,820g and was hospitalized for failure to thrive during infancy. Childhood photographs of her (at age 5 y and later childhood) showed normal skin, eyes, teeth and hair. She began to wear eye glasses in kindergarten and was intellectually disabled. She required special education and graduated from high school. She developed premature ovarian insufficiency and underwent menopause at age 29 years. All of her teeth were extracted in her 30’s. It is unknown whether she had enamel hypoplasia or severe dental caries. She developed bilateral orbital atrophy and enophthalmos. At 35 years of age, her height was normal at 158.5cm (25th percentile), body weight was 31.1kg (less than 3rd percentile), and her occipitofrontal circumference was 50.5cm (≪ 3rd percentile). She had an overall premature aged appearance exemplified by thin hair with gray streaks, loss of subcutaneous fat, senile lentigines on the face and hands, and thin, wrinkled, mottled skin. She had a prominent brow, sunken orbits, and was legally blind. Ophthalmological examination showed severe corneal scarring thought to be the cause of her blindness, no cataracts, and no salt-and-pepper retinopathy. Hearing loss was not reported, and audiometry was not performed. She was edentulous and she had poor breast development, genital exam was Tanner V. She had thin extremities and bilateral pes cavus. A brain MRI confirmed enophthalmos, brain abnormalities, including white matter lesions. Neither cerebral calcifications nor ventriculomegaly were noted. A cytogenetic analysis was normal (46, XX). The family history revealed a similarly affected sister with intellectual disability, enophthalmos, premature menopause, and complete dental extraction. Consanguinity was denied (Fig. 1A).
Figure 1.
ERCC4 variants in two unrelated segmental progeroid syndrome probands.
(A) Pedigrees of two progeroid syndrome families. Individuals, Registry# MME1010 and CALIF1010, are used in this study (see text). Symbols and colors are defined as follows: square, male; circle, female; white, unaffected; black, affected. (B) Sanger sequencing of ERCC4 single nucleotide variations (SNVs). Two SNVs were found in MME1010 and one SNV was found in CALIF1010. (C) Breakpoint sequence in the DNA of CALIF1010. The patient’s sequence (middle, described as “junction”) was aligned to intron 2 (top) and intron 4 (bottom) of the chr 16 RefSeq, NC_000016.9 using the genomic coordinates of the GRCh37/hg19 assembly. The dashed square shows the overlapping ACT sequences. (D) Diagram of the exon 2–5 region of the ERCC4 gene. The top line is a diagram of the wildtype allele with the locations of the deletion breakpoints. The bottom line is a diagram of the deleted allele. Overlapping ACT sequences involved in the deletions are shown.
Registry# MME1010 is a 68 year-old Mexican woman with bilateral cataracts (diagnosed at age 54), tight atrophic skin, graying and thinning of hair (starting at age 30), type II diabetes mellitus, osteoporosis and an overall aged appearance. Her height was 162 cm and her body weight was 40 kg. Her past history revealed a diagnosis of papillary thyroid cancer at age 56. There was no evidence of cognitive impairment or sun sensitivity. She has a sister who is possibly affected. Parental consanguinity was denied (Fig. 1A).
Identification of ERCC4 variants
From among the 42 individuals who had been referred to the International Registry of Werner Syndrome with the putative diagnosis of WS, and in whom we had previously excluded WRN, LMNA, and POLD1 mutations, we conducted exome sequencing in 18 subjects. We had to exclude subjects for whom consent for exome sequencing could not be obtained, typically because older versions of consent forms did not include explicit permission to perform exome sequencing. As a result of exome sequencing, two previously unreported heterozygous ERCC4 (RefSeq accession: NM_005236.2) variants, c.1488A>T (p.Gln496His) and c.2579C>A (p.Ala860Asp), were detected in a single proband, MME1010.
In an attempt to discover other probands with comparable variants in ERCC4, we screened ERCC4 from an additional 24 genetically undiagnosed Atypical WS patients for whom exome analysis could not be carried out using targeted amplification sequencing with an Illumina Miseq sequencer. This led to the identification of two rare heterozygous ERCC4 variants in an additional proband, CALIF1010.
NGS analysis of CALIF1010 revealed a heterozygous missense variant, c.2395C>T (p.Arg799Trp), which has been previously reported as pathogenic (Niedernhofer, et al., 2006; Sijbers, et al., 1998). This was confirmed by Sanger sequencing (Fig. 1B, Table 1). Sequencing analysis also revealed a novel heterozygous genomic deletion spanning from intron 2 through 4, which is expected to cause deletion of exons 3 and 4. Breakpoints of the genomic deletion were determined by genomic PCR using primers designed to amplify the flanking regions. Sequence alignment of the breakpoints confirmed a 5,656 bp deletion spanning introns 2 through 4, c.388+1164_792+795del, which would result in a premature termination, p.Gly130Aspfs*18 (Fig. 1C–D, Table 1). The presence of a short sequence, ACT, at both breakpoints suggested an intragenic homologous recombination as the primary mutational mechanism. Parental samples of CALIF1010 were unable to be obtained for additional study to confirm that these variants are in trans. The new pathogenic variants have been submitted to the Leiden Open Variation Database (http://www.lovd.nl/ERCC4).
Table 1.
Characteristics of the four ERCC4 variants identified in the proband
| MME1010 | CALIF1010 | |||
|---|---|---|---|---|
| Variant 1 | Variant 2 | Variant 1 | Variant 2 | |
| Genomic position (GRCh37/hg19) | chr16: 14029277 | chr16: 14042032 | chr16: 14017232_14022887del | chr16: 14041848 |
| DNA variant | c.1488A>T | c.2579C>A | c.388+1164_792+795del (5656bp del) | c.2395C>T |
| Protein alteration | p.Gln496His | p.Ala860Asp | p.Gly130Aspfs*18 | p.Arg799Trp |
| Predicted Effect of Variants | ||||
| CADD score | 16 | 23.4 | N/A | 27.8 |
| Polyphen score | 0.982 | 0.67 | N/A | 1 |
| SIFT score | 0.01 | 0.01 | N/A | 0 |
| Minor Allele Frequency | ||||
| ExAC | 0.0008154 | 0.0008072 | N/A | 0.0005107 |
| ESP6500 | 0.002924427 | 0.002924427 | N/A | 0.000769586 |
| 1000G | 0.00239617 | 0.00239617 | N/A | N/A |
Molecular analysis of ERCC4 variants
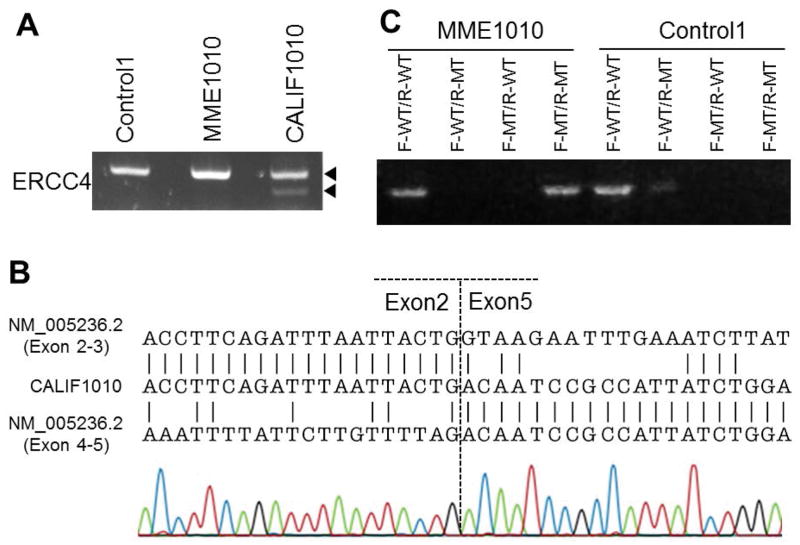
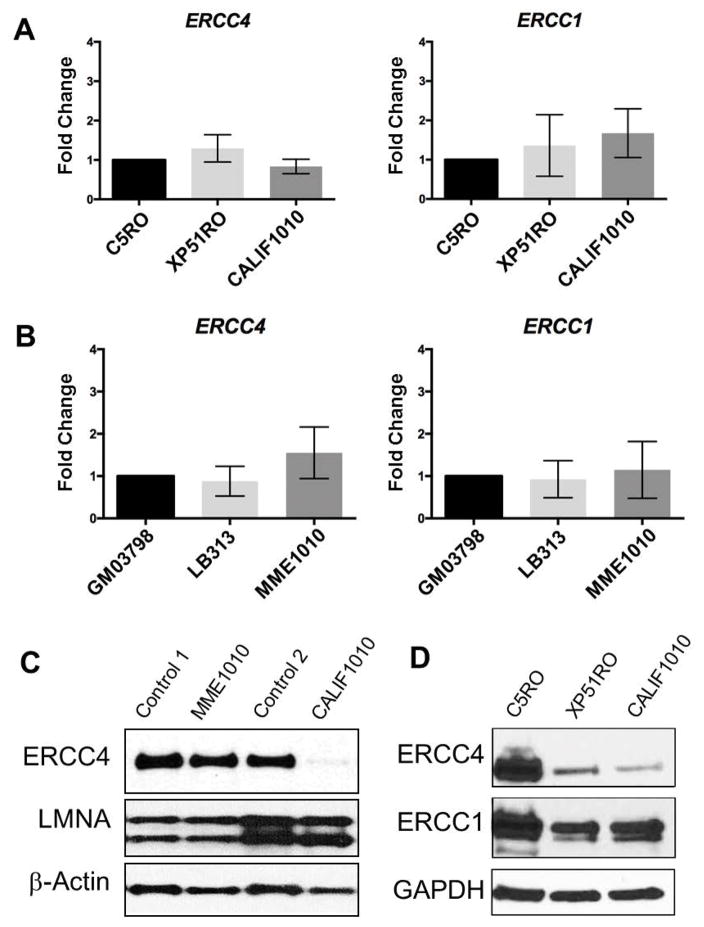
We next investigated mRNA expressions of ERCC4 using the LCL of patient MME1010 and the fibroblasts of patient CALIF1010. As shown in Fig. 2A, a faint extra band was detected in CALIF1010, suggesting the presence of a deletion at the mRNA level. Sanger sequencing of this shorter extra band revealed a deletion of exons 3 and 4 (Fig. 2B), further confirming the presence of a heterozygous deletion in CALIF1010. Levels of the full length mRNA for both ERCC4 and ERCC1 were compared to other patients with ERCC4 mutations and found to be within the normal range (Fig. 3A,B). There have been examples of cryptic splicing generated by nucleotide substitutions in coding regions. Therefore, the possibility of alternative transcripts was evaluated, and absent in MME1010, however, indicating that the novel variants found in MME1010 did not affect splicing.
Figure 2.
Mutation analyses of ERCC4 mRNA in the two probands
(A) Reverse transcription and PCR (RT-PCR) spanning exons 1 through 7 revealed a faint extra band located below the control band in CALIF1010. (B) Breakpoint sequence in the cDNA of CALIF1010. The patient sequence (middle) was aligned to exon 2 (top) and exon 5 (bottom) of the RefSeq accession, NM_005236.2. (C) Results of allele specific PCR analysis for cDNA of MME1010. The PCR product from the use a combination of forward wildtype specific primer (F-WT) and reverse wildtype specific primer (R-MT) serves as a control. Positive bands were obtained in combination with both F-WT/R-WT and F-MT/R-MT in proband cDNA.
Figure 3.
Expression analysis of XPF-ERCC1 complex. (A) Levels of full length mRNAs are shown for ERCC4/XPF and ERCC1 as measured by qRT-PCR in CALIF1010 fibroblasts. C5R0 is used as a normal control and set to 1. XP51RO is from a patient with XFE progeroid syndrome caused by a mutation in ERCC4. (B) For MME1010 LCL, GM03798 was used as a normal control and set to 1.0. LB313 cells are from an XP-F patient with low but detectable NER capacity. The bar graphs represent the mean values ± SD of the 6 reactions carried out for each gene. Statistical analysis (One-way ANOVA post hoc test) showed no significant differences in ERCC4 or ERCC1 mRNA levels among patients and control cell lines. (C) Western blot analysis of nuclear fractions of cells of patients’ LCL and fibroblast cells showing levels of ERCC4, LMNA and β-actin. LMNA used was used as a marker for nuclei and β-actin was utilized as an additional loading control. Control 1 and control 2 correspond to extracts from control LCLs and fibroblasts, respectively. (D) Immunodetection of ERCC4 and ERCC1 in healthy donor fibroblasts (C5RO) and cells from an ERCC4 progeroid patient (XP51RO) and the new CALIF1010 patient. GAPDH was used as the loading control. Both ERCC4 and ERCC1 protein levels are substantially lower in both progeroid patients compared to controls.
We then evaluated protein expression of ERCC4/XPF in the probands’ LCLs or fibroblasts (Fig. 3C). Immunoblot detection of ERCC4 protein in the nuclear fraction of cell lysates revealed no difference between MME1010 and normal controls. In contrast, CALIF1010 showed a dramatic reduction of ERCC4 to approximately 6% of the control. This is in agreement with the previous observation that c.2395C>T (p.Arg799Trp) causes protein instability and mislocalization of the mutant protein (Niedernhofer, et al., 2006; Sijbers, et al., 1998). Presence of truncated ERCC4 protein corresponding to p.Gly130fs was unable to be determined due to the location of antibody epitope. Comparison of ERCC4 levels in CALIF1010 to that of XP51RO, an XFE progeroid syndrome patient with significant impairment in DNA repair (Ahmad, et al., 2010; Niedernhofer, et al., 2006), revealed similar expression levels of ERCC4 in both cell lines (Fig. 3D). ERCC1 expression levels were also reduced in both progeroid patients demonstrating reduced levels of the DNA repair endonuclease.
The phases of the two variants, c.1488A>T and c.2579C>A, in MME1010 were determined by allele specific PCR of cDNA using combinations of primers specific for wild type or mutant because parental DNA samples were not available. In MME1010, RT-PCR products were obtained in the wildtype-wildtype and mutant-mutant primer combinations but not in wildtype-mutant primer combinations, while control RT-PCR gave rise to a RT-PCR product with wildtype-wildtype only (Fig. 2C). This indicated that the two variants in MME1010 are in cis (monoallelic), not in trans (biallelic), explaining why the variants did not affect the mRNA or protein levels of ERCC4.
Cellular consequences of ERCC4 variants
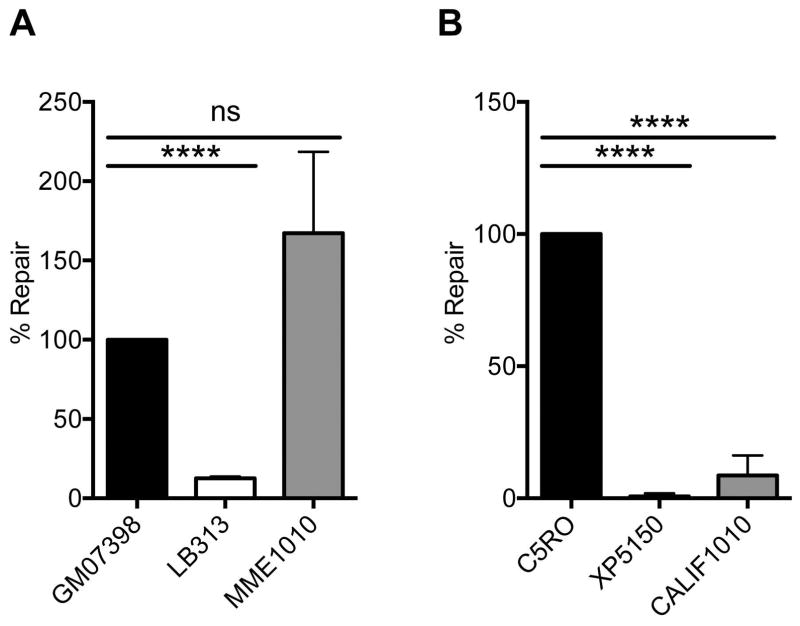
In order to evaluate the functional consequences of the ERCC4 variants found in these two subjects, we measured unscheduled DNA synthesis (UDS) in response to UV irradiation. UV-induced UDS (i.e. DNA synthesis in non-replicating cells) is pathognomonic for a defect in NER (Yamaguchi, et al., 1990), for which XPF is essential (Sijbers, et al., 1996). For MME1010 LCLs, GM07398 was used as a normal control and set to 100% repair and LB313 XP-F patient cells were used as a control with reduced UDS (Fig. 4A). MME1010 had normal levels of UDS following UV irradiation (Fig. 4A, Supp. Fig. S1A), indicating that NER is normal and therefore XPF function is intact. For CALIF1010, C5RO fibroblasts were used as a normal control and set to 100% NER (Fig. 4B). XP51RO cells from an XFE progeroid patient and very low UDS (Niedernhofer, et al., 2006) were used as a negative control and showed <5% normal UDS. CALIF1010 had <10% normal UDS (Fig. 4B, Supp. Fig. S1B), reflecting a severe defect in NER likely as a consequence of reduced XPF expression.
Figure 4.
Measuring UV-induced unscheduled DNA synthesis as an index of NER capacity. (A and B) A defect in NER is diagnostic of XP, and is measured by detection of DNA synthesis in non-replicating cells after UV irradiation. (A) In lymphoblastoid cells, GM07398 are from an unaffected individual and set to 100% of normal NER capacity. LB313 cells are from an XP-F patient with a reported UDS of 25%. Relative to these positive and negative controls, MME1010 has normal NER capacity. (B) In fibroblasts, C5RO represents normal NER of UV-induced DNA damage. XP51RO is a negative control from an XFE progeroid patient reported to have a UDS of <5% (Niedernhofer, et al., 2006). UDS levels were significantly reduced in the CALIF1010 cells compared to the positive control, consistent with substantial impairment in NER.
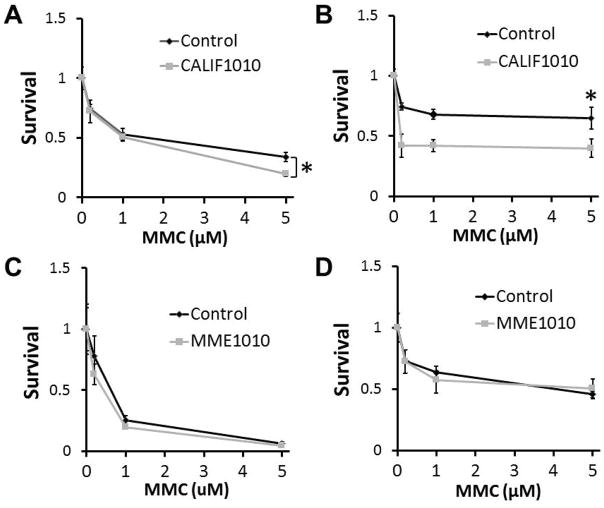
To test the impact of the ERCC4 mutations on interstrand crosslink (ICL) repair, we determined the cellular sensitivities to mitomycin C (MMC), to which ERCC4 or ERCC1 mutants are known to be hypersensitive (Niedernhofer, et al., 2004; Su, et al., 2012). After 24 hrs, dose-response survival data of CALIF1010 were 56%, 62% and 62% relative to a control hTERT line when challenged with 200nM, 1μM and 5μM of MMC, respectively. These were statistically significant results (Fig. 5A). After 4 days of treatment with 5μM MMC, the survival of CALIF1010 was 59% of control, a statistically significant result. Survivals of CALIF1010 cells at 200nM and 1μM, however, were not statistically different from that of the control cultures (Fig. 5B). For MEM1010, there was no difference in survivals compared with control LCL under the same conditions (Fig. 5C,D). Together, these results support a conclusion that the ERCC4 variants found in CALIF1010 are indeed responsible for progeroid features of the patient, but that the ERCC4 variants found in MME1010 are unlikely to be pathogenic.
Figure 5.
Cellular sensitivity to mitomycin C as an index of capacity for interstrand crosslink repair. Cells were treated with the indicated concentrations of MMC (μM) for 24 hrs (A and C) or 4 days (B and D). Cell counts were normalized to that of untreated cultures to obtain survival data. Standard deviations were calculated from the results of 3 experiments. * indicates the statistical significance of t-tests.
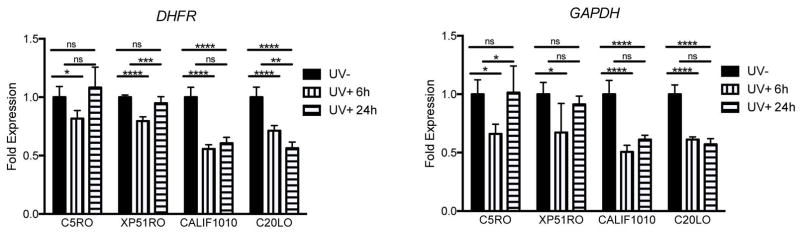
The recovery of RNA synthesis (RRS) post-UV irradiation is a measure of transcription-coupled NER, which is specifically impaired in cells from Cockayne Syndrome (CS) patients (Andrade-Lima, et al., 2015). Since mutations in ERCC4 have been linked to CS (Kashiyama, et al., 2013), RRS was measured in CALIF1010 at several time points following UV irradiation of the cells. Expression of the housekeeping genes DHFR and GAPDH were used in RRS as previously described (Velez-Cruz, et al., 2013). Normal fibroblasts (C5RO) were used as a negative control, while CS20LO cells from a patient with CS due to a mutation in ERCC1 (Kashiyama, et al., 2013) were used as a positive control. At 6 hrs post-irradiation, expression of DHFR and GAPDH were significantly reduced by approximately 25% in normal fibroblasts and cells from an XP-F patient without CS features (XP51RO) (Fig. 6). By 24 hrs post-irradiation, expression had returned to baseline. In contrast, in CS20LO and CALIF1010 cells, UV caused reduced expression of the housekeeping genes was by approximately 50% at 6 hrs post-irradiation and expression remained significantly reduced at 24 hrs. This is compatible with the clinical diagnosis of CS.
Figure 6.
Recovery of RNA synthesis after UV irradiation is impaired in CALIF1010 and CS patient cell lines. mRNA expression of DHFR and GAPDH was measured in the indicated cell lines after UV irradiation (10 J/m2). Expression was normalized to the amount of 18s rRNA and the results are plotted as the ratio of expression in irradiated vs. sham-irradiated cells. Each data point represents the mean of three qPCR reactions of five independent experiments. Error bars represent the SD. One-way ANOVA, *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001.
Structural analysis of ERCC4 variants
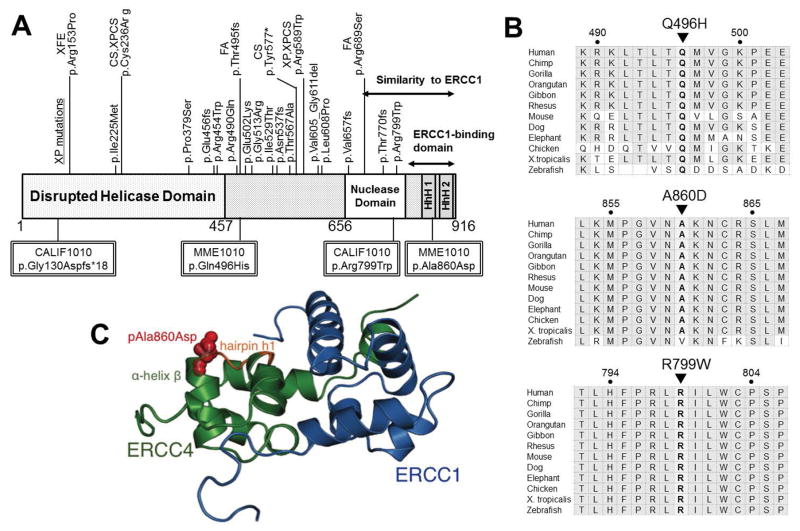
The ERCC4 endonuclease consists of an N-terminal domain with distant homology to DNA helicases of superfamily 2, a central domain containing the nuclease active site, and a C-terminal tandem helix-hairpin-helix (HhH)2 domain (Sgouros, et al., 1999). As shown in Fig. 7A, the previously reported ERCC4 mutations are widely distributed over the entire length of the protein. ERCC4 and ERCC1 have sequence similarities in their central and nuclease domains, and in their HhH domains (Gaillard and Wood, 2001). p.Arg799Trp, which is found in CALIF1010, has been reported to be a causal “hot spot” for XP-F (Ahmad, et al., 2010; Sijbers, et al., 1998). The other frame-shift variant of CALIF1010, p.Gly130Aspfs*18, is located near the N-terminus and resulted in the shortest ERCC4 transcript so far reported. The altered amino acids are highly conserved among various species (Fig. 7B).
Figure 7.
Distribution of the reported ERCC4 variants and conservation of the Gln496, Ala860, and Arg799 amino acids throughout evolution. (A) The structural domains in ERCC4 and the locations of amino acid alterations in previously reported patients. XP pathogenic variants are grouped above the functional domains. Pathogenic variants reported in other conditions are shown above the XP variants along with the corresponding diagnosis. ERCC4 includes an N-terminal disrupted helicase domain and a nuclease domain. The C-terminal domain, including the nuclease domain and helix-hairpin-helix (HhH) motifs, shows similarity to that of ERCC1. Double-lined squares show the variants found in MME1010 and CALIF1010. (B) Conservation of the altered amino acids. Amino acids colored in gray indicate conservation between groups of highly similar properties. (C) A proposed model for the impact of the MME1010 patient’s variant residing in the HhH domain of ERCC4. This C-terminal part of the protein is critical for interaction with ERCC1 and for heteroduplex stability. ERCC4/XPF in green. ERCC1 is represented in blue. The red spheres represent the mutant residue Asp860. The overlapping stick representation in orange represents the wildtype residue Ala860 superimposed, illustrating little structural deformity caused by the variant. The model was generated using Mutagenesis wizard of Pymol visualization software. The model was calculated on HADDOCK web server for electrostatics, desolvation and van der Waals energy refinement (van Dijk, et al., 2012; van Zundert, et al., 2016)
In MME1010, the p.Gln496His variant is located outside the disrupted helicase domain, where several XP and FA mutations have been reported (Fig. 7A). Structural analysis predicted that the p.Gln496His variant is likely benign and would only affect protein function if the Gln side chain were a component of an active site or a binding site for an interacting protein (Fig. 7B). In contrast, the pAla860Asp variant of MME1010 is within the (HhH)2 domain (Fig. 7A). The two functions of (HhH)2 domain are heteroduplex formation with ERCC1 and DNA binding, both of which are very important for structural and functional integrity of the XPF-ERCC1 endonuclease (Das, et al., 2012; Faridounnia, et al., 2015; Tripsianes, et al., 2005; Tsodikov, et al., 2005). pAla860Asp is located at the C-terminal of α-helix β in this interaction domain adjacent to a DNA binding site of ERCC4 (hairpin h1, and Lys861) (Das, et al., 2012), residue numbers adapted to the most recent residue numbering in ERCC4/XPF) (Fig. 7C). Based on helix propensity data, p.Ala860Asp substitution could alter the helix stability, leading to altered DNA binding properties, and/or a sidechain occupancy less likely to accommodate DNA substrates. Nevertheless, MME1010 cells had an NER capacity equivalent to normal cells (Fig. 4A) and normal sensitivity to the crosslinking agent MMC (Fig. 5C, D). Thus, either the second allele of ERCC4 is adequate to compensate for the p.Ala860Asp allele, or the p.Ala860Asp allele is benign and does not affect XPF function. We favor the former based on the observation that in mice, one copy of Xpf is adequate to support normal expression of the gene and to confer resistance to UV and MMC (Tian, et al., 2004).
DISCUSSION
Genomic instability is not only a cause of various cancers but is also suspected to be a major driver of human aging. NGS in 42 unsolved AWS patients revealed two cases, each of which had two heterozygous ERCC4 mutations. Both patients showed several features consistent with accelerated aging.
To the best of our knowledge, there has been no previous report of exon deletions within the ERCC4 gene. Our breakpoint analysis of the deletion mutation in CALIF1010 suggests that common ACT sequence-mediated intragenic recombination may be the mechanism underlying the ERCC4 deletion mutation. A similar mechanism has been reported for as reported for other loci which undergo non-allelic homologous recombination (NAHR) following ectopic cross-over of repeat sequences (Liu, et al., 2012; Oshima, et al., 2009).
The majority of pathogenic ERCC4 mutations present as XP-F and are characterized by relatively late onset of skin manifestation including increased sun sensitivity and skin cancer (Gregg, et al., 2011; Sijbers, et al., 1996). Cases of CS without any features of XP are quite rare among ERCC4 mutants (Kashiyama, et al., 2013). There are indeed rare cases which show combined features of CS and XP, referred to as combined xeroderma pigmentosum-Cockayne syndrome (XP-CS) (Kashiyama, et al., 2013). XP-CS individuals present with skin phenotypes common in XP patients, including severe photosensitivity, abnormal skin pigmentation, and skin cancer; additional features include developmental abnormalities common in CS patients. Although CS is associated with only TC-NER specific defects, XP-CS is caused by the defects within both of these pathways.
The recently published Cockayne Syndrome Natural History Study (Wilson, et al., 2016) highlighted the limitations of current criteria for the diagnosis of Cockayne syndrome, since only 20%–36% of their cohort would have been diagnosed as having CS. They suggest instead, that CS be suspected in a patient with postnatal growth failure, microcephaly, and at least two other features. Using these suggested criteria, CALIF1010 would be suspected of having CS based on weight (but not height, which was normal), microcephaly, loss of body fat, and enophthalmos.
Wilson et al (2016) also advocated for including patients with cerebro-oculofacioskeletal syndrome (MIM# 616570) caused by pathogenic variants of ERCC5 (XPG; MIM# 133530) in the spectrum of CS. ERCC5 (XPG) encodes an exonuclease involved in transcriptional coupled DNA repair, but unlike ERCC4, it generates a 3′ incision following UV-induced damage (Ito, et al., 2007).
CALIF1010 carries obligatory compound heterozygous mutations in ERCC4 and exhibited dramatically reduced ERCC4 protein expression. The frame-shift deletion of exon 3 and 4 (p.Gly130Aspfs*18) resulted in null expression of XPF. Sijbers et al (1998) reported that homozygosity for p.Arg799Trp is also associated with markedly reduced XPF expression (Sijbers, et al., 1998). The major clinical features of CALIF1010 consist of early failure to thrive, and. marked microcephaly, followed by development of sunken eyes, cognitive impairment, progressive loss of body fat, and premature aging of the skin. CALIF1010, therefore, appears to be the second case report of a patient with a Cockayne-like syndrome without XP and with progeroid features caused by biallelic ERCC4 variants (Kashiyama, et al., 2013).
MME1010 carries two novel variants in cis in ERCC4. CADD scores (Kircher, et al., 2014), which are utilized for pathogenicity predictions for p.Gln496His and p.Ala860Asp, are 16.0 and 23.4, respectively (Table 1), exceeding the recommended threshold of 15.0 for pathogenicity. The p.Ala860Asp variant is located in the C-terminal HhH domain, where XPF not only interacts with ERCC1 but binds to DNA (de Laat, et al., 1998). However, we were unable to obtain definitive biological evidence of NER or ICL defects in MME1010 cells. Taken together, we conclude that the phenotypes observed in our MME1010 patient is not due to the variants detected in the patient’s ERCC4 locus, and that the genetic cause of progeroid features remains unknown. Further clarification of the potential pathogenicity of p.Gln496His and p.Ala860Asp must await the identification of individuals homozygous for these variants.
As summarized in Fig. 7A, there appears to be no clear correlation between the clinical phenotypes and locations of ERCC4 variants. Moreover, cellular disease phenotypes are not always consistent. Bogliolo et al (2013) reported one FA case with biallelic ERCC4 variants, p.Thr495Asnfs*6 and p.Arg689Ser, without any skin and neurological phenotypes, concluding that FA-causing ERCC4 pathogenic variants disrupt the ERCC4 function in DNA ICL repair without severely compromising NER (Bogliolo, et al., 2013). These findings raise the possibility of modifying loci not only for clinical presentations but also for cellular disease phenotypes.
Supplementary Material
Acknowledgments
All authors declare no conflict of interest. This work was supported in part by NIH grants, R24AG42328 and R01CA210916 (GMM/JO). The University of Washington Center for Mendelian Genomics (UW-CMG) was funded by the NIH/NHGRI and NIH/NHLBI grant, U54HG006493 (DAN, MJB, and Suzanne Leal). LJN, MJY and MF were supported by the NIH grant, P01AG043376. The WeNMR project (European FP7 e-Infrastructure grant, contract no. 261572, www.wenmr.eu), supported by the European Grid Initiative (EGI) through the national GRID Initiatives of Belgium, France, Italy, Germany, the Netherlands, Poland, Portugal, Spain, UK, South Africa, Malaysia, Taiwan, the Latin America GRID infrastructure via the Gisela project and the US Open Science Grid (OSG) are acknowledged for the use of web portals, computing and storage facilities.
References
- Abecasis GR, Altshuler D, Auton A, Brooks LD, Durbin RM, Gibbs RA, Hurles ME, McVean GA Consortium GP. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–73. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmad A, Enzlin JH, Bhagwat NR, Wijgers N, Raams A, Appledoorn E, Theil AF, JHJH, Vermeulen W, NGJJ, Scharer OD, Niedernhofer LJ. Mislocalization of XPF-ERCC1 nuclease contributes to reduced DNA repair in XP-F patients. PLoS Genet. 2010;6:e1000871. doi: 10.1371/journal.pgen.1000871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrade-Lima LC, Veloso A, Paulsen MT, Menck CF, Ljungman M. DNA repair and recovery of RNA synthesis following exposure to ultraviolet light are delayed in long genes. Nucleic Acids Res. 2015;43:2744–56. doi: 10.1093/nar/gkv148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogliolo M, Schuster B, Stoepker C, Derkunt B, Su Y, Raams A, Trujillo JP, Minguillón J, Ramírez MJ, Pujol R, Casado JA, Baños R, Rio P, Knies K, Zúñiga S, Benítez J, Bueren JA, Jaspers NG, Schärer OD, de Winter JP, Schindler D, Surrallés J. Mutations in ERCC4, encoding the DNA-repair endonuclease XPF, cause Fanconi anemia. Am J Hum Genet. 2013;92:800–6. doi: 10.1016/j.ajhg.2013.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong JX, Burrage LC, Beck AE, Marvin CT, McMillin MJ, Shively KM, Harrell TM, Buckingham KJ, Bacino CA, Jain M, Alanay Y, Berry SA, Carey JC, Gibbs RA, Lee BH, Krakow D, Shendure J, Nickerson DA, Bamshad MJ. Autosomal-Dominant Multiple Pterygium Syndrome Is Caused by Mutations in MYH3. Am J Hum Genet. 2015;96:841–9. doi: 10.1016/j.ajhg.2015.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das D, Folkers GE, van Dijk M, Jaspers NG, Hoeijmakers JH, Kaptein R, Boelens R. The structure of the XPF-ssDNA complex underscores the distinct roles of the XPF and ERCC1 helix-hairpin-helix domains in ss/ds DNA recognition. Structure. 2012;20:667–75. doi: 10.1016/j.str.2012.02.009. [DOI] [PubMed] [Google Scholar]
- de Laat WL, Sijbers AM, Odijk H, Jaspers NG, Hoeijmakers JH. Mapping of interaction domains between human repair proteins ERCC1 and XPF. Nucleic Acids Res. 1998;26:4146–52. doi: 10.1093/nar/26.18.4146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, Philippakis AA, del Angel G, Rivas MA, Hanna M, McKenna A, Fennell TJ, Kernytsky AM, Sivachenko AY, Cibulskis K, Gabriel SB, Altshuler D, Daly MJ. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43:491–8. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- English JS, Swerdlow AJ. The risk of malignant melanoma, internal malignancy and mortality in xeroderma pigmentosum patients. Br J Dermatol. 1987;117:457–61. doi: 10.1111/j.1365-2133.1987.tb04925.x. [DOI] [PubMed] [Google Scholar]
- Evans E, Moggs JG, Hwang JR, Egly JM, Wood RD. Mechanism of open complex and dual incision formation by human nucleotide excision repair factors. EMBO J. 1997;16:6559–73. doi: 10.1093/emboj/16.21.6559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faridounnia M, Wienk H, Kovacic L, Folkers GE, Jaspers NG, Kaptein R, Hoeijmakers JH, Boelens R. The Cerebro-oculo-facio-skeletal Syndrome Point Mutation F231L in the ERCC1 DNA Repair Protein Causes Dissociation of the ERCC1-XPF Complex. J Biol Chem. 2015;290:20541–55. doi: 10.1074/jbc.M114.635169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedberg EC, Aguilera A, Gellert M, Hanawalt PC, Hays JB, Lehmann AR, Lindahl T, Lowndes N, Sarasin A, Wood RD. DNA repair: from molecular mechanism to human disease. DNA Repair (Amst) 2006;5:986–96. doi: 10.1016/j.dnarep.2006.05.005. [DOI] [PubMed] [Google Scholar]
- Gaillard PH, Wood RD. Activity of individual ERCC1 and XPF subunits in DNA nucleotide excision repair. Nucleic Acids Res. 2001;29:872–9. doi: 10.1093/nar/29.4.872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregg SQ, Robinson AR, Niedernhofer LJ. Physiological consequences of defects in ERCC1-XPF DNA repair endonuclease. DNA Repair (Amst) 2011;10:781–91. doi: 10.1016/j.dnarep.2011.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanawalt PC. Subpathways of nucleotide excision repair and their regulation. Oncogene. 2002;21:8949–56. doi: 10.1038/sj.onc.1206096. [DOI] [PubMed] [Google Scholar]
- Hisama FM, Lessel D, Leistritz D, Friedrich K, McBride KL, Pastore MT, Gottesman GS, Saha B, Martin GM, Kubisch C, Oshima J. Coronary artery disease in a Werner syndrome-like form of progeria characterized by low levels of progerin, a splice variant of lamin A. Am J Med Genet A. 2011;155A:3002–6. doi: 10.1002/ajmg.a.34336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoeijmakers JH. DNA damage, aging, and cancer. N Engl J Med. 2009;361:1475–85. doi: 10.1056/NEJMra0804615. [DOI] [PubMed] [Google Scholar]
- Huang S, Lee L, Hanson NB, Lenaerts C, Hoehn H, Poot M, Rubin CD, Chen DF, Yang CC, Juch H, Dorn T, Spiegel R, Oral EA, Abid M, Battisti C, Lucci-Cordisco E, Neri G, Steed EH, Kidd A, Isley W, Showalter D, Vittone JL, Konstantinow A, Ring J, Meyer P, Wenger SL, von Herbay A, Wollina U, Schuelke M, Huizenga CR, Leistritz DF, Martin GM, Mian IS, Oshima J. The spectrum of WRN mutations in Werner syndrome patients. Hum Mutat. 2006;27:558–67. doi: 10.1002/humu.20337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito S, Kuraoka I, Chymkowitch P, Compe E, Takedachi A, Ishigami C, Coin F, Egly JM, Tanaka K. XPG stabilizes TFIIH, allowing transactivation of nuclear receptors: implications for Cockayne syndrome in XP-G/CS patients. Mol Cell. 2007;26:231–43. doi: 10.1016/j.molcel.2007.03.013. [DOI] [PubMed] [Google Scholar]
- Kashiyama K, Nakazawa Y, Pilz DT, Guo C, Shimada M, Sasaki K, Fawcett H, Wing JF, Lewin SO, Carr L, Li TS, Yoshiura K, Utani A, Hirano A, Yamashita S, Greenblatt D, Nardo T, Stefanini M, McGibbon D, Sarkany R, Fassihi H, Takahashi Y, Nagayama Y, Mitsutake N, Lehmann AR, Ogi T. Malfunction of nuclease ERCC1-XPF results in diverse clinical manifestations and causes Cockayne syndrome, xeroderma pigmentosum, and Fanconi anemia. Am J Hum Genet. 2013;92:807–19. doi: 10.1016/j.ajhg.2013.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kircher M, Witten DM, Jain P, O’Roak BJ, Cooper GM, Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet. 2014;46:310–5. doi: 10.1038/ng.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraemer KH, DiGiovanna JJ. Global contributions to the understanding of DNA repair and skin cancer. J Invest Dermatol. 2014;134:E8–17. doi: 10.1038/skinbio.2014.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krumm N, Sudmant PH, Ko A, O’Roak BJ, Malig M, Coe BP, Quinlan AR, Nickerson DA, Eichler EE. Copy number variation detection and genotyping from exome sequence data. Genome Res. 2012;22:1525–32. doi: 10.1101/gr.138115.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laugel V. Cockayne syndrome: the expanding clinical and mutational spectrum. Mech Ageing Dev. 2013;134:161–70. doi: 10.1016/j.mad.2013.02.006. [DOI] [PubMed] [Google Scholar]
- Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–60. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P, Carvalho CM, Hastings PJ, Lupski JR. Mechanisms for recurrent and complex human genomic rearrangements. Curr Opin Genet Dev. 2012;22:211–20. doi: 10.1016/j.gde.2012.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marteijn JA, Lans H, Vermeulen W, Hoeijmakers JH. Understanding nucleotide excision repair and its roles in cancer and ageing. Nat Rev Mol Cell Biol. 2014;15:465–81. doi: 10.1038/nrm3822. [DOI] [PubMed] [Google Scholar]
- McLaren W, Pritchard B, Rios D, Chen Y, Flicek P, Cunningham F. Deriving the consequences of genomic variants with the Ensembl API and SNP Effect Predictor. Bioinformatics. 2010;26:2069–70. doi: 10.1093/bioinformatics/btq330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niedernhofer LJ, Garinis GA, Raams A, Lalai AS, Robinson AR, Appeldoorn E, Odijk H, Oostendorp R, Ahmad A, van Leeuwen W, Theil AF, Vermeulen W, van der Horst GT, Meinecke P, Kleijer WJ, Vijg J, Jaspers NG, Hoeijmakers JH. A new progeroid syndrome reveals that genotoxic stress suppresses the somatotroph axis. Nature. 2006;444:1038–43. doi: 10.1038/nature05456. [DOI] [PubMed] [Google Scholar]
- Niedernhofer LJ, Odijk H, Budzowska M, van Drunen E, Maas A, Theil AF, de Wit J, Jaspers NG, Beverloo HB, Hoeijmakers JH, Kanaar R. The structure-specific endonuclease Ercc1-Xpf is required to resolve DNA interstrand cross-link-induced double-strand breaks. Mol Cell Biol. 2004;24:5776–87. doi: 10.1128/MCB.24.13.5776-5787.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oshima J, Hisama FM. Search and insights into novel genetic alterations leading to classical and atypical Werner syndrome. Gerontology. 2014;60:239–46. doi: 10.1159/000356030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oshima J, Magner DB, Lee JA, Breman AM, Schmitt ES, White LD, Crowe CA, Merrill M, Jayakar P, Rajadhyaksha A, Eng CM, del Gaudio D. Regional genomic instability predisposes to complex dystrophin gene rearrangements. Hum Genet. 2009;126:411–23. doi: 10.1007/s00439-009-0679-9. [DOI] [PubMed] [Google Scholar]
- Oshima J, Sidorova JM, Monnat RJ. Werner syndrome: Clinical features, pathogenesis and potential therapeutic interventions. Ageing Res Rev. 2016 doi: 10.1016/j.arr.2016.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paila U, Chapman BA, Kirchner R, Quinlan AR. GEMINI: integrative exploration of genetic variation and genome annotations. PLoS Comput Biol. 2013;9:e1003153. doi: 10.1371/journal.pcbi.1003153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha B, Zitnik G, Johnson S, Nguyen Q, Risques RA, Martin GM, Oshima J. DNA damage accumulation and TRF2 degradation in atypical Werner syndrome fibroblasts with LMNA mutations. Front Genet. 2013;4:129. doi: 10.3389/fgene.2013.00129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schärer OD. Nucleotide excision repair in eukaryotes. Cold Spring Harb Perspect Biol. 2013;5:a012609. doi: 10.1101/cshperspect.a012609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sgouros J, Gaillard PH, Wood RD. A relationship betweena DNA-repair/recombination nuclease family and archaeal helicases. Trends Biochem Sci. 1999;24:95–7. doi: 10.1016/s0968-0004(99)01355-9. [DOI] [PubMed] [Google Scholar]
- Sijbers AM, de Laat WL, Ariza RR, Biggerstaff M, Wei YF, Moggs JG, Carter KC, Shell BK, Evans E, de Jong MC, Rademakers S, de Rooij J, Jaspers NG, Hoeijmakers JH, Wood RD. Xeroderma pigmentosum group F caused by a defect in a structure-specific DNA repair endonuclease. Cell. 1996;86:811–22. doi: 10.1016/s0092-8674(00)80155-5. [DOI] [PubMed] [Google Scholar]
- Sijbers AM, van Voorst Vader PC, Snoek JW, Raams A, Jaspers NG, Kleijer WJ. Homozygous R788W point mutation in the XPF gene of a patient with xeroderma pigmentosum and late-onset neurologic disease. J Invest Dermatol. 1998;110:832–6. doi: 10.1046/j.1523-1747.1998.00171.x. [DOI] [PubMed] [Google Scholar]
- Su Y, Orelli B, Madireddy A, Niedernhofer LJ, Scharer OD. Multiple DNA binding domains mediate the function of the ERCC1-XPF protein in nucleotide excision repair. J Biol Chem. 2012;287:21846–55. doi: 10.1074/jbc.M111.337899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan A, Abecasis GR, Kang HM. Unified representation of genetic variants. Bioinformatics. 2015;31:2202–4. doi: 10.1093/bioinformatics/btv112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian M, Shinkura R, Shinkura N, Alt FW. Growth retardation, early death, and DNA repair defects in mice deficient for the nucleotide excision repair enzyme XPF. Mol Cell Biol. 2004;24:1200–5. doi: 10.1128/MCB.24.3.1200-1205.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripsianes K, Folkers G, Ab E, Das D, Odijk H, Jaspers NG, Hoeijmakers JH, Kaptein R, Boelens R. The structure of the human ERCC1/XPF interaction domains reveals a complementary role for the two proteins in nucleotide excision repair. Structure. 2005;13:1849–58. doi: 10.1016/j.str.2005.08.014. [DOI] [PubMed] [Google Scholar]
- Tsodikov OV, Enzlin JH, Scharer OD, Ellenberger T. Crystal structure and DNA binding functions of ERCC1, a subunit of the DNA structure-specific endonuclease XPF-ERCC1. Proc Natl Acad Sci U S A. 2005;102:11236–41. doi: 10.1073/pnas.0504341102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dijk M, Wassenaar TA, Bonvin AM. A Flexible, Grid-Enabled Web Portal for GROMACS Molecular Dynamics Simulations. J Chem Theory Comput. 2012;8:3463–72. doi: 10.1021/ct300102d. [DOI] [PubMed] [Google Scholar]
- van Zundert GC, Rodrigues JP, Trellet M, Schmitz C, Kastritis PL, Karaca E, Melquiond AS, van Dijk M, de Vries SJ, Bonvin AM. The HADDOCK2.2 Web Server: User-Friendly Integrative Modeling of Biomolecular Complexes. J Mol Biol. 2016;428:720–5. doi: 10.1016/j.jmb.2015.09.014. [DOI] [PubMed] [Google Scholar]
- Velez-Cruz R, Zadorin AS, Coin F, Egly JM. Sirt1 suppresses RNA synthesis after UV irradiation in combined xeroderma pigmentosum group D/Cockayne syndrome (XP-D/CS) cells. Proc Natl Acad Sci U S A. 2013;110:E212–20. doi: 10.1073/pnas.1213076110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijg J, Suh Y. Genome instability and aging. Annu Rev Physiol. 2013;75:645–68. doi: 10.1146/annurev-physiol-030212-183715. [DOI] [PubMed] [Google Scholar]
- Wilson BT, Stark Z, Sutton RE, Danda S, Ekbote AV, Elsayed SM, Gibson L, Goodship JA, Jackson AP, Keng WT, King MD, McCann E, Motojima T, Murray JE, Omata T, Pilz D, Pope K, Sugita K, White SM, Wilson IJ. The Cockayne Syndrome Natural History (CoSyNH) study: clinical findings in 102 individuals and recommendations for care. Genet Med. 2016;18:483–93. doi: 10.1038/gim.2015.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi J, Mamada A, Kondo S, Satoh Y. Defective DNA repair in cultured melanocytes from xeroderma pigmentosum patients. J Dermatol. 1990;17:465–72. doi: 10.1111/j.1346-8138.1990.tb01677.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.