Abstract
Engineering biomolecular motors can provide direct tests of structure-function relationships and customized components for controlling molecular transport in artificial systems1 or in living cells2. Previously, synthetic nucleic acid motors3–5 and modified natural protein motors6–10 have been developed in separate complementary strategies for achieving tunable and controllable motor function. Integrating protein and nucleic acid components to form engineered nucleoprotein motors may enable additional sophisticated functionalities. However, this potential has only begun to be explored in pioneering work harnessing DNA scaffolds to dictate the spacing, number, and composition of tethered protein motors11–15. Here, we describe myosin motors that incorporate RNA lever arms, forming hybrid assemblies in which conformational changes in the protein motor domain are amplified and redirected by nucleic acid structures. The RNA lever arm geometry determines the speed and direction of motor transport, and can be dynamically controlled using programmed transitions in lever arm structure7,9. We have characterized the hybrid motors using in vitro motility assays, single-molecule tracking, cryo-electron microscopy, and structural probing16. Our designs include nucleoprotein motors that reversibly change direction in response to oligonucleotides that drive strand-displacement17 reactions. In multimeric assemblies, the controllable motors walk processively along actin filaments at speeds of 10–20 nm s−1. Finally, to illustrate the potential for multiplexed addressable control, we demonstrate sequence-specific responses of RNA variants to oligonucleotide signals.
Myosins generate directed motion on actin filaments using a swinging crossbridge18 mechanism in which conformational changes in the catalytic domain are amplified by an extended lever arm structure. Protein engineering has previously been used to replace the myosin lever arm with alternative structures that can reproduce or modify wild-type behavior6,7,9,19, including modules that function as gearshifts that respond to optical or chemical signals7,9. Since nucleic acid engineering offers a complementary approach for precise design of programmable molecular structures and devices20,21, we asked whether a functional myosin lever arm could also be constructed from RNA. In order to propagate an angular change from the myosin head to the RNA structure, we sought to create an oriented rigid connection between the myosin and the RNA, distinct from the flexible tethering strategies commonly employed in DNA-scaffolded protein assemblies11–15.
We designed an engineered myosin that incorporates an RNA binding domain to attach an RNA lever arm (Figure 1). M6-RB (for Myosin VI - RNA-Binding) was generated by fusing myosin VI to the L7Ae kink-turn binding domain22. Following a design principle established in previous work on engineered myosin lever arms6,7,9,19, we made use of a helix-sharing junction in which the C-terminal helix of the truncated myosin VI lever arm was fused to the N-terminal helix of the L7Ae domain. Guided by crystal structures of L7Ae-RNA22 and of myosin VI23,24, we aligned the terminal helices and optimized the phasing of the junction to orient a bound kink-turn motif as a structural foundation from which to build extended RNA lever arms. The interaction of L7Ae with kink-turn motifs has been previously exploited in nanotechnology and synthetic biology applications25–27, and yields stable complexes with reported Kd values of ~1 nM and dissociation rates of ~2–7 × 10−4 s−1 25,27,28.
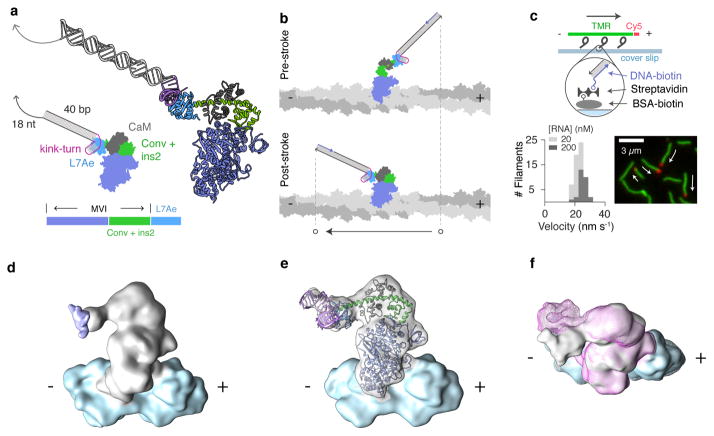
Figure 1. Design and characterization of an engineered myosin with an RNA lever arm.
a, Design of protein and RNA components. An annotated schematic is shown alongside a larger 3-dimensional ribbon diagram for M6-RB:ktL. Below: protein block diagram for M6-RB. The RNA-binding L7Ae domain is fused to myosin VI (MVI) after the converter domain and insert 2 (conv + ins2). b, Cartoon of predicted power stroke for M6-RB:ktL bound to actin. In the transition from the pre-stroke to the post-stroke state, the tip of the lever arm moves toward the minus end of the actin filament. c, Measuring directed motility using a gliding filament assay. Top: assay design. M6-RB:ktL is affixed to the surface by binding to a complementary biotinylated DNA strand immobilized via streptavidin and biotin-BSA. Propelled actin is fluorescently labeled with Cy5 at its plus end, and TMR along its body. Bottom: results. Image is taken from a movie of gliding filaments, with arrows showing direction of motion. A stacked histogram of filament velocities is shown for two surface [RNA] conditions (see also Supplementary Table 1 and Supplementary Movies 1a–b). d–f, Cryoelectron microscopy reconstructions of engineered myosin and RNA bound to actin, low-pass filtered at 13 Å. d, Reconstruction of M6-RB (apo, grey) bound to F-actin (blue). Segmented density is displayed corresponding to a single myosin motor bound to 2 actin subunits. A difference map of M6-RB:ktLshort (apo) minus M6-RB (apo) is displayed as a purple isosurface, unambiguously localizing the position of the RNA. e, Flexible fitting of myosin-RNA model (Supplementary Movies 3b,c) to the cryo-EM reconstruction of M6-RB:ktLshort (apo) bound to F-actin. The DireX flexible-fitting model of M6-RB:ktLshort (apo) is colored as in Fig. 1a. f, Nucleotide-dependent conformational change (Supplementary Movies 4a,b). Overlaid reconstructions were segmented as in Fig. 1e for the apo (grey surface) and ADP (pink mesh) nucleotide states of M6-RB:ktLshort bound to actin (blue surface) .
As an initial lever arm design, we fused a kink-turn with a 40 bp RNA duplex to create ktLinear (ktL, Fig. 1a, Supplementary Fig. 2a). A single-stranded overhang was added to the distal 3′ end of the RNA lever arm to enable surface attachment or assembly into multimeric complexes via binding to a complementary oligonucleotide. When the hybrid motor binds to actin and transitions from the pre-stroke to the post-stroke conformation, M6-RB:ktL is predicted to swing the tip of its lever arm from the plus-end toward the minus-end of actin (Fig. 1b), generating minus-end directed motility as seen for native myosin VI.
We tested the M6-RB:ktL complex for its designed function using dual-labeled gliding filament assays7,9 (Fig. 1c). Imaged actin filaments moved with their plus-ends leading (Fig. 1c, Supplementary Movie S1), corresponding to motors pulling toward the minus-end of actin filaments, as expected (Fig. 1c and Supplementary Tables 1,2). In control experiments omitting the RNA component, no filament gliding was seen (see Methods). We then used 3D cryo-electron microscopy to directly probe the structure of the hybrid motor, assess the orientation of the artificial lever arm, and test for transmission of nucleotide-dependent conformational changes from the catalytic head to reorient the RNA. Actin filaments were decorated with M6-RB, either alone or bound to a truncated variant of ktL (ktLshort, Supplementary Fig. 2b) with a reduced duplex extension (Supplementary Fig. 3 and Supplementary Movie 2). 3D reconstructions of the nucleotide-free motor, obtained in the presence and absence of RNA, show the expected overall motor conformation and RNA location (Fig. 1d and Supplementary Movie 3a). A model of M6-RB:ktLshort, obtained by flexible fitting to the experimental density (Fig. 1e and Supplementary Movie 3b), closely matches our design (Supplementary Fig. 3 and Supplementary Movie 3c). The lever arm orientation of myosin VI differs between the ADP and nucleotide-free states, swinging through a small angle that represents a substep of the larger power stroke29,30. We observed this reorientation when comparing reconstructions of the hybrid motor in the ADP and nucleotide-free states, as expected for a functional engineered lever arm (Fig. 1f, and Supplementary Movies 4a–b).
Changing the directionality of myosins by altering the structure of the lever arm has provided rigorous tests of the swinging crossbridge model and represents an additional demonstration that an attached structure functions as a lever6,7,9,19. Following a similar strategy used in protein engineering studies of myosins and kinesin-149,31, we aimed to reverse the direction of the motor by designing an RNA lever arm that inverted the projection of the power stroke, yielding plus-end directed motility. We designed ktReverseSwitch (ktRS1) by fusing the kink-turn to a previously described motif32 (the ‘reverse motif’, Fig. 2a, Supplementary Fig. 2c) that flips the orientation of the duplex at a single crossover point and stabilizes the reoriented duplex by a loop-receptor tertiary interaction (Fig. 2a), followed by a duplex extension. A model of ktRS1 bound to M6-RB shows the end of the lever arm oriented toward the plus-end of the actin filament in the post-stroke state (Fig. 2b).
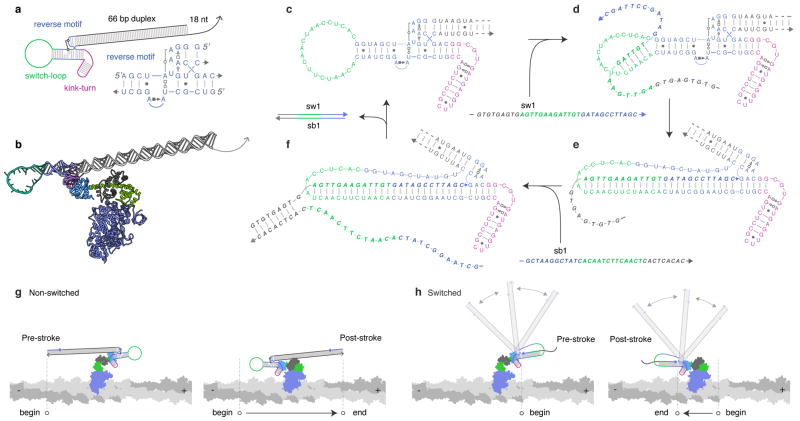
Figure 2. Design of an RNA lever arm for controllable bidirectional motion.
a, Secondary structure of ktRS1. The arm is composed of a kink-turn, reverse motif, switch-loop, and lever arm extension. Right and bottom: detailed diagram of the reverse motif32 using Leontis-Westhof (LW) notation36. A tetraloop (GAAA) binds the 11 nucleotide loop receptor32, stabilizing the orientation of the lever arm extension. b, 3-dimensional ribbon model of ktRS1 bound to M6-RB. c–f, Designed directional switching mechanism. c, In the non-switched state, the orientation of the lever arm extension is stabilized by the tertiary interaction. d, The switch strand (sw1) initially binds to a toehold in the switch-loop (green). e, Subsequent strand invasion by sw1 disrupts the tertiary interaction, and the distal portion of the lever arm is free to swivel. f, The arm converts back to the non-switched state when the switchback strand (sb1) binds sw1 at its overhang, removing it from ktRS1 to form the sw1:sb1 waste duplex. g, In the non-switched state, the end of the lever arm strokes from the minus-end to the plus-end of actin. f, In the switched state, the orientation of the distal portion of the lever arm is disordered (greyed out helices with motion arrows indicating an ensemble of conformations in this flexible state) and the effective end of the lever arm lies at the crossover point. The end of the arm thus strokes from the plus-end to the minus-end of actin.
We further designed ktRS1 to enable dynamic control over motor directionality. Rigid to flexible transitions have been used to dynamically control the speed and direction of engineered protein motors7,9, by introducing a point of flexibility that creates a new effective end of the lever arm. In DNA nanotechnology, rigid to flexible transitions triggered by strand displacement reactions have been used to control the state and geometry of DNA structures17. We designed a switching mechanism for ktRS1 that reversibly converts the RNA reverse motif from a rigid to flexible state. A stem-loop structure (Fig. 2a,c) acts as the nucleus for a reversible toehold-mediated strand displacement reaction17. The reaction is predicted to disrupt the receptor site of the tertiary interaction (Fig. 2e) and undock the reoriented arm. Similar to a previous light-activated design9, the reaction reversibly shifts the end-point of the effective lever arm. In the non-switched state, the end of the lever arm is predicted to stroke from the minus-end toward the plus-end (Fig. 2g). In the switched state, the new end of the effective lever arm (the crossover junction) is predicted to stroke from the plus-end toward the minus-end (Fig. 2h). The switched state can be generated by introducing a switch strand (sw), and the non-switched state can be recovered using a second strand displacement reaction driven by a switchback strand (sb1).
We tested M6-RB:ktRS1 for controllable bidirectional motion using gliding filament assays (Fig. 3a,b and Supplementary Movies 5–7). As expected, in the absence of control strands, M6-RB:ktRS1 drove plus-end directed motility, demonstrating successful redirection as a result of altering the RNA geometry, and strongly confirming lever arm function for the attached RNA structure. Gliding filament assays performed after introducing the switch strand showed minus-end directed motility, confirming strand-displacement control over directionality (Supplementary Tables 1,2). Finally, assays conducted after successively introducing both the switch and the switchback strands reverted to plus-end directed motility, confirming the reversibility of the transition. The switching behavior was further captured in a dynamic assay (Fig. 3b, Supplementary Movie 8) where the switch and switchback strand were cyclically introduced over two complete switching cycles to a flow cell containing M6-RB:ktRS1.
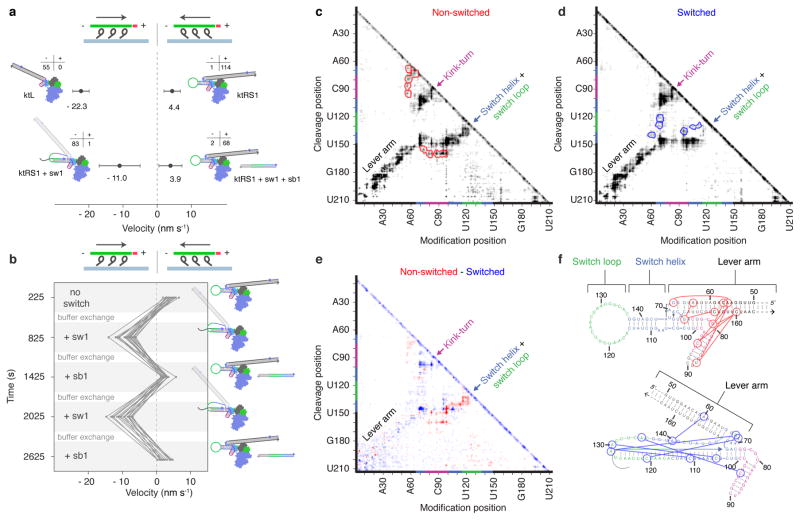
Figure 3. Functional and structural characterization of switching behavior.
a, Scatter plot of gliding velocities (including directionality statistics) for M6-RB:ktL and the three switching states of M6-RB:ktRS1 (see also Supplementary Movies 5–7 and Table 1); plotted data are for 20 nM RNA incubations. b, Plot of dynamic switching with M6-RB:ktRS1 (Supplementary Movie 8). Seventeen dual-labeled filaments were tracked while cycling conditions using buffer exchange. Velocities were measured within each observation phase (grey), and plotted in the middle of the phase. Buffer exchange (white) took place for 2.5 min between each phase. c–d, MOHCA-seq proximity maps16 of ktRS1 in non-switched (left) and switched (right) states. Cleavage position (y-axis) represents the position at which the RNA was cleaved by hydroxyl radicals, and modification position (x-axis) represents the position at which reverse transcription starting at the cleavage position was stopped. Sequence positions of designed structural elements are indicated by colored sections on axes. Helical elements appear as diagonal swaths of signal and are labeled by name on the maps. MOHCA-seq proximities unique to each state are indicated by red (non-switched) or blue (switched) contours on the corresponding map. e, Difference map of non-switched and switched ktRS1, with red indicating stronger MOHCA-seq signal in the non-switched state and blue indicating stronger signal in the switched state. f, Secondary structure diagrams of non-switched and switched states, with unique MOHCA-seq hits from c–d shown as lines connecting circled residues (see also Supplementary Fig. 4).
To directly probe the structure of the ktRS1 RNA lever arm and the conformational transition driven by strand displacement, we conducted multiplexed hydroxyl radical cleavage analysis with paired-end sequencing (MOHCA-seq)16 on ktRS1 in the presence and absence of the switch strand (Fig. 3c–f and Supplementary Fig. 4a). In MOHCA-seq, hydroxyl radicals are generated from point sources tethered to the RNA backbone, enabling a readout of spatially correlated oxidative damage events that correspond to positions in the RNA that are close together in three dimensions, usually 10–30Å apart. Correlated pairs of RNA positions are represented in a 2D proximity map (Fig. 3c–e). Helical elements appear as diagonal swaths of signal on the map, with additional peaks corresponding to tertiary proximities. MOHCA-seq proximities in the unswitched state (Fig. 3c) were similar to expectations based on our naïve model for ktRS1 (Supplementary Fig. 4b). In the switched state (Fig. 3d), MOHCA-seq directly detected disruption of the switch helix, as seen in a difference map (Fig. 3e). Additionally, MOHCA-seq revealed sets of distinct unique tertiary proximities in the switched and non-switched states (red and blue contours in Fig. 3c,d; colored lines in Fig. 3f), supporting a large conformational change between the states. The proximities in the flexible switched state are expected to arise from an ensemble of interconverting conformations rather than a single discrete structure.
As a further test of the modular construction of controllable ribonucleoprotein motor assemblies, we asked whether M6-RB:ktL and M6-RB:ktRS1 could act as components of processive complexes that walk on actin filaments (Fig, 4), a function that typically requires two or more motor heads working together8 to move long distances and transport cargo. Engineered myosin tetramers8, and varying numbers and arrangements of cytoskeletal motors scaffolded by DNA2,11–13, have previously exhibited long-range transport. We chose a tetrameric design in order to favor high processivity at saturating [ATP]; dimeric myosin VI motors with engineered lever arms have previously been found be only weakly processive, with tetramers yielding ~10X longer run lengths8. To assemble tetrameric constructs, we generated a cyclized ssDNA ring (tet) that binds four DNA strands, each of which provides a binding site for one RNA molecule. This design symmetrically organizes four M6-RB:RNA hybrids around tet (Fig. 4a, b), with flexible linkages between the monomer units. Assembly of tetrameric complexes was verified using native PAGE (Supplementary Fig. 5,6); in addition to the major tetramer product, we also detect a minority population of incompletely assembled complexes, which are expected to behave similarly to tetramers but with reduced processivity.
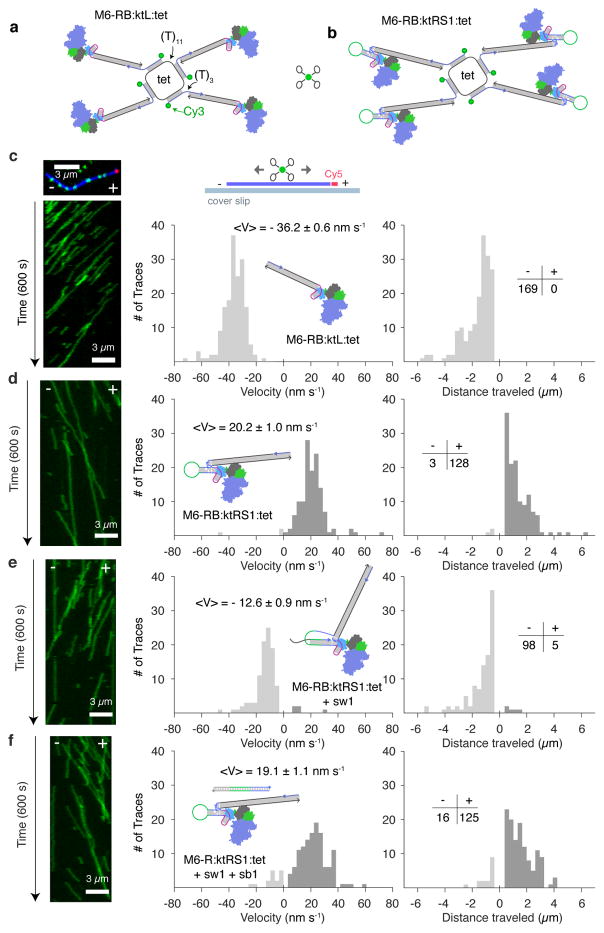
Figure 4. Design, characterization, and directional control of hybrid processive walkers.
a, Design of tetrameric assembly M6-RB:ktL:tet. Four M6-RB:ktL complexes are symmetrically bound around a cyclized DNA ring (tet). Bridging DNA strands are labeled with 5′-Cy3. b, M6-RB:ktRS1:tet. c–f, Processive directed motion of tetramers on polarity-labeled actin filaments. Actin was labeled with Cy5 at the plus-end, and Alexa-488 along the body, and immobilized on glass cover slips to record the movement and direction of Cy3 labeled motors (c, top left and top middle). Left: kymographs of Cy3-motors moving along the actin filaments, with time running from top to bottom. Right: Histograms of pooled single-molecule data; the signed velocity and total distance traveled was tabulated for each motor trace (see also Supplementary Table 3). Dark grey, plus-end directed; light grey, minus-end directed. c, M6-RB:ktL:tet. Kymograph is of actin filament shown above. d, M6-RB:ktRS1:tet in the unswitched state. e, M6-RB:ktRS1:tet in the switched state. f, M6-RB:ktRS1:tet switched-back state (see also Supplementary Table 3 and Supplementary Movies 10–13). Mini-table directionality statistics apply to both velocity and distance traveled histograms.
We imaged the multimers moving along immobilized polarity-labeled actin filaments, using three-color total internal reflection fluorescence (TIRF) microscopy (Fig. 4c–f, Supplementary Movies S9–12). As expected, M6-RB:ktL:tet complexes consistently moved processively toward the minus-end of actin filaments (Fig. 4c, Supplementary Movie 9), with an average velocity of −36.2 ± 0.6 nm s−1. In assays of multimeric reverse switch motors, M6-RB:ktRS1:tet also moved processively, with 98% (128/131) of the single-molecule traces moving towards the plus-end of actin as expected (Fig. 4d, Supplementary Movie 10) and an average velocity of 20.2 ± 1.0 nm s−1. When M6-RB:ktRS1:tet was incubated with the switch strand, 95 % (98/103) of traces moved toward the minus-end of actin, with an average velocity of −12.6 ± 0.9 nm s−1 (Fig. 4e, Supplementary Movie 11). Finally, when switched M6-RB:ktRS1:tet was incubated with the switchback strand (sb1), it reverted back to moving toward the plus-end of actin in 89% (125/141) of traces (Fig. 4f, Supplementary Movie 12). These statistics show robust but incomplete switching of individual multimeric complexes, in contrast to near-perfect switching of filament direction in gliding assays, where the individuality of motors may be masked by averaging over many myosins interacting with each filament.
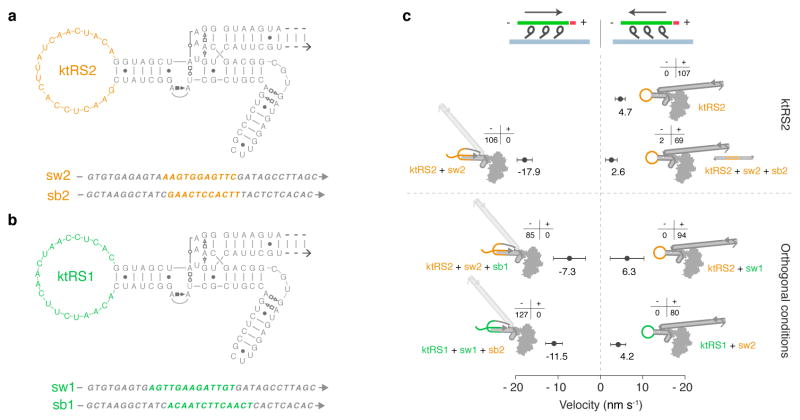
In comparison with previous designs controlled by metal ions7 or blue light9, a particular strength of oligononucleotide-controlled motors is the potential for straightforward multiplexing of orthogonal signals directed at distinct motor populations, taking advantage of the sequence addressability of strand displacement reactions17,21. To verify that our design strategy supports sequence-specific responses, we performed proof-of-concept gliding filament experiments in which hybrid motors incorporating two different RNA molecules were tested in the presence of cognate and non-cognate control strands (Fig. 5). We created a second reverse-switch RNA lever arm (ktRS2) in which only the switch-loop sequence differed from ktRS1, and also designed switch (sw2) and switchback (sb2) strands for controlling this construct (Fig 5a–b). As expected, M6-RB:ktRS2 shows (+) end directed gliding motility in the absence of control strands (Supplementary Movie 13). We conducted oligonucleotide-controlled gliding filament assays (as in Fig. 3a) for both M6-RB:ktRS1 and M6-RB:ktRS2, while varying the identities of the switch (sw1 or sw2) and switchback (sb1 or sb2) strands (Supplementary Movies 14a–b). As expected, directional switching was sequence-dependent: M6-RB:ktRS1 reversed direction in the presence of sw1 but not sw2, and could be switched back to (+) end directed motility by sb1 but not by sb2; similarly, M6-RB:ktRS2 responded specifically to sw2 and sb2 for directional switching (Fig. 5c). These experiments show that varying the switch loop alone is sufficient for achieving sequence-addressable control, without changing the sequence of structural elements such as the switch-helix — an example of simple modular design of receiver and actuator elements.
Figure 5. Orthogonal sequence control of directional switching.
a–b, Sequences of switchable RNA molecules and corresponding switch and switchback strands. a, ktRS2 was designed by replacing the sequence of the switch loop (colored residues) in b, ktRS1 (see Fig. 2). c, Scatter plot of gliding velocities (including directionality statistics) for M6-RB:ktRS2 and M6-RB:ktRS1 (see also Fig. 3c, Supplementary Movies 13–14, and Table 1). Top, ktRS2 changes direction in response to sw2 and switches back in response to sb2. Bottom, M6-RB:ktRS1 and M6-RB:ktRS2 do not change direction after incubation with orthogonal switch strands, and switched complexes do not change direction after incubation with orthogonal switchback strands.
Here we have demonstrated that engineered ribonucleoprotein machines can function as cytoskeletal motors with designable and controllable properties, including bidirectional processive motion reversibly controlled by sequence-specific oligonucleotide signals. Tetrameric complexes are capable of translocating for microns along the actin filament in both directions (Fig. 4, Supplementary Table 3). The controllable hybrid motors have velocities in the 10–20 nm s−1 range, much faster than typical programmable DNA-based motors, which have velocities in the nm min−1 range5, and also faster than previously designed bidirectional motors controlled by metal ions and light, which have velocities in the 1–3 nm s−1 range7,9. Replacing protein structural elements with nucleic acids allows for versatile design using easily controlled duplex lengths and a repertoire of modular RNA structural elements33, and hybrid motors may in future be integrated with RNA34 or DNA35 origami platforms to produce large functional assemblies with defined motor orientations, as seen in some biological motor assemblies such as myosin filaments15,18. Applications of hybrid motors may include artificial molecular transport systems in programmable devices1, where the designs described here provide the possibility of using DNA computational outputs21 to control the directions and speeds of sequence-addressable populations of biologically derived protein motors. Switch strands bind to the motor and thus function as both signals and cargos, which may be exploited in artificial transport systems; for example, a bidirectional shuttle that reverses direction after dropping off a cargo may be implemented by conjugating the switch strand to a cargo molecule and affixing the switchback strand to a dropoff site. In principle, these genetically encoded hybrid motors may also be expressed in living cells to enable new levels of control over mechanical functions2, although technical hurdles will likely need to be overcome related to export, processing, and stability of the engineered RNA structures33. With the rapid development of protein, DNA and RNA engineering, finding ways to integrate these three biomolecules into functional machines and motors will aid not only in our fundamental understanding of structure-function relationships, but also in the development of complex active systems that extend the functionality of nanotechnology and biology.
Methods
Information on Materials and Microscopy is provided below. The supplementary material for this paper includes additional information on Molecular Design, Cryo-EM, Gel Electrophoresis, MOHCA-seq, and RNA and DNA Sequences.
Materials
Protein expression and purification
A DNA construct for protein expression was assembled from fragments encoding porcine myosin VI (residues 1–817) and Archaeoglobus fulgidus L7Ae (residues 9–118), cloned into a pBiex-1 (Novagen) expression vector modified to include codons for a C-terminal eYFP, and FLAG tag (DYKDDDDK) with intervening GSG repeats (see Supplementary Fig. 1). Proteins were expressed by direct transfection of SF9 cells and affinity purified as previously described19,37.
Transcription template cloning
Sequences for ktL and ktRS were cloned into the pUC19 vector using one-step isothermal Gibson38 assembly. To prepare for the assembly reaction, Gibson cloning primers (see “RNA and DNA sequences”) were pre-annealed in two separate batches corresponding roughly to each half of the long hairpin-like structure of the RNA arm (ktL: half1[p1, p2] and half2[p3, p4]; ktRS: half1[t1, t1, t3, b1, b2, b3] and half2[t2_4, t2_5, t2_6, b2_4, b2_5, b2_6]). Primers were combined at 90 nM in 10 μl of TE buffer and quick-annealed (1 min at 90 C, 5 min at 65 C, 10 min at 50 C, 10 min at 37 C and finally 10 min at room temperature). The two halves were combined and annealed at RT for 30 min to form the final insert. For the Gibson reaction, 45 nM insert was combined with 1.25 ng/μl HindIII/PstI digested pUC19 vector and isothermal enzyme mix in a total volume of 20 μl.
Transcription and RNA purification
To prepare for run-off transcription, ktL and ktRS plasmids were linearized using PstI, phenol/chloroform extracted, ethanol precipitated, dried down on a vacuum concentrator, and reconstituted in water or 1X TE buffer. RNA was synthesized from these templates using the NEB HiScribe T7 High Yield RNA Synthesis Kit, following standard kit protocols, using .025 μg/μl of linearized template along with the high molecular weight mix provided with the kit, in a 100 μl reaction volume. Reactions were incubated for 3 to 4 hours at 37 C, ethanol precipitated, dried down on a vacuum concentrator, and gel purified on 8 % denaturing PAGE gels containing 8M Urea and 1X TBE. Bands were visualized by UV shadowing, cut, and eluted overnight at RT in buffer containing 20 mM Tris-HCl, 0.25 M sodium acetate, 1 mM EDTA, and 0.25 % (w/v) SDS. The samples were then ethanol precipitated, dried down, and reconstituted in water.
Synthetic RNA for cryoEM
The ktLshort RNA was synthesized and HPLC purified by IDT.
Preparation of circular tet DNA strand
The non-circularized tet strand was synthesized with a 5′ phosphate and column purified by PAN facilities, Stanford University. 700 pmol non-circularized tet was combined with 1.4 μmol tet-join in 1.4 mL T4 DNA ligase buffer (Invitrogen), and the solution was incubated at 90 C for 5 min, then left at RT for 10 min, to allow tet-join to form a splint connecting the 3′ and 5′ ends of tet. 50 U (Weiss units) of T4 DNA ligase (Invitrogen) was then added, and the reaction was incubated at RT for 3 hours. 200 U of ExoI (NEB) and 200 U of ExoIII (NEB) were then added, and the solution was incubated overnight at 37 C, phenol-chloroform extracted, ethanol precipitated, and resuspended in water. Samples were then gel purified using 8 % denaturing PAGE gels containing 8M Urea and 1X TBE. Bands were visualized by UV shadowing, cut, and eluted overnight at RT in buffer containing 500 mM ammonium acetate, 10 mM magnesium acetate, and 2 mM EDTA. The samples were then ethanol precipitated, dried down, and reconstituted in water.
Microscopy
Gliding filament assays
Monomers of the RNA-myosin chimeras were characterized by adapting a previously described dual-labeled gliding filament assay7 to enable immobilization of the RNA via a streptavidin-biotin interaction. RNA for gliding assays was prepared at 20 nM (or 200 nM as indicated) in tris assay buffer (TAB) containing 25 mM Tris-HCl (pH 7.5), 25 mM KCl, 10 mM dithiothreitol (DTT), 1 mM EGTA, and 2 mM MgCl2. Solutions were annealed for 2 minutes at 90 C and snap-cooled on ice, where they were kept (at least 1 hour, and up to 24 hours) until they were added to channels. The biotin modified DNA tether (t-btn) was prepared at 100 nM in TAB and stored on ice. Flow cells with ~ 2 mm wide channels were prepared by spin-coating glass coverslips with nitrocellulose and forming sandwich assemblies with glass slides using double-sided tape. For experiments without switch strands, channels were prepared by adding reagents in 12 steps (7 μl in step 1, 25 μl in steps 2–12): (1) 2 mg/ml biotin-BSA (Sigma), 2 min incubation; (2) TAB buffer with 2 mg/ml BSA (Sigma) (TAB_BSA), no incubation; (3) 0.5 mg/ml streptavidin (Life) in PBS buffer, 7 min incubation; (4) TAB_BSA, no incubation; (5) DNA tether, 7 min incubation; (6) TAB, no incubation; (7) RNA, 15 min incubation; (8) TAB, no incubation; (9) M6-RB at 9–12 nM in TAB with 1.8 mM trolox, and 5 μM calmodulin (TAB_CaM), 10 min incubation; (10) TAB_CaM, no incubation; (11) TMR/Cy5 labeled actin in TAB_CaM, 2 min incubation; and finally (12) GO buffer, containing 23 mM Tris-HCl (pH 7.5), 23 mM KCl, 9 mM dithiothreitol (DTT), 0.9 mM EGTA, 10 mM MgCl2, 2 mM ATP, 1.62 mM trolox, an ATP regeneration system containing 1.0 mM phosphocreatine and 0.95 μg/ml creatine phosphokinase, and an oxygen scavenging system containing 0.4% (wt/vol) glucose, 0.20 mg/ml glucose oxidase and 0.36 μg/ml catalase. When the switching reaction was performed, the GO buffer (step 12) contained 0.5 μM switch strand. When the switchback reaction was performed, the GO buffer with switch was incubated for 5 min, and an additional wash (step 13) was done with GO buffer containing 1 μM switchback strand. After channel preparation, the filaments were imaged by total internal reflection fluorescence through a Nikon 100X 1.49 NA objective using a 532 nm optically pumped semiconductor laser (Coherent) with TMR and Cy5 fluorescence channels collected simultaneously on an electron-multiplying charge-coupled device camera (Andor).
For the dynamic switching assay (Fig. 3b), flow cells were modified by using parafilm gaskets and input and output tubes to allow for buffer exchange during videomicroscopy39. Channel preparation was the same as for fixed condition gliding motility assays, except that in steps 2–12, 100 μl of solution was used for each step. The dynamic switching proceeded as follows: (1) ktRS1 in the non-switched state was imaged for 7.5 minutes; (2) channel buffer was exchanged with GO buffer containing switch strand at 0.5 μM, for 2.5 minutes; (3) ktRS1 was imaged with the switch buffer for 7.5 minutes; (4) channel buffer was exchanged with GO buffer containing switchback strand at 1 μM, for 2.5 minutes; (5) ktRS1 was imaged with the switchback buffer for 7.5 minutes; (6) channel buffer was exchanged with GO buffer containing switch strand at 0.5 μM, for 2.5 minutes; (7) ktRS1 was imaged with the switch buffer for 7.5 minutes; (8) channel buffer was exchanged with GO buffer containing switchback strand at 1 μM, for 2.5 minutes; (9) ktRS1 was imaged with the switchback buffer for 20 minutes.
Filament directions and velocities for both the single-state and dynamic switching assays were determined using only filaments whose polarity could be scored on the basis of a single Cy5-labeled seed. Velocities were determined in ImageJ by measuring displacements of the tips of reptating filaments that moved in a straight line over a fixed period of time. Control experiments were performed to verify that filament capture and motility depended on the RNA component. In side-by-side experiments conducted on three different days, surfaces prepared with ktL RNA (n=6 channels) all showed gliding actin filaments (92 ± 30 gliding filaments per field of view), whereas surfaces prepared identically except for the omission of RNA (n=5 channels) showed no gliding filaments and very few stuck filaments after searching many fields of view (in representative experiments we tabulated 1 stuck filament and 0 gliding filaments over 18 fields of view, n=2 channels).
Single-molecule tracking assays
Tetramers of the RNA-myosin chimeras were characterized by adapting a previously described single-molecule fluorescence tracking assay8,37 modified to use dual-labeled polarity-marked filaments. For three-color imaging, actin was body labeled with A8,37 lexa 488 phalloidin (Life) and plus-end labeled with gelsolin-capped Cy5-actin seeds, while motors were labeled with Cy3. Assay buffers were the same as in the motility assay, except TAB buffer was modified to contain 8 mM MgCl2, and TAB_BSA was modified to contain only 1 mg/ml BSA. RNA tetramers were prepared in three steps: (1) 180 nM ktRS1 was annealed in TAB for 2 min at 90 C, and then snap-cooled and stored on ice; (2) 100 nM tet and 600 nM of t-c3 (see RNA and DNA sequences) were annealed together in TAB for 5 min. at 90 C, 10 min. at 65 C, 15 min. at 45 C, 20 min. at 37 C, and then RT for at least 20 min.; and (3) 135 nM ktRS1 and 15 nM tet:t-c3 (‘tet:t-c3’ shortened to ‘tet’ when complexed with ktRS1) complex were annealed together at RT for 30 minutes, yielding a final ktRS1:tet concentration of 15 nM. Flow cells were prepared as in the gliding assays, except coverslips were not coated with nitrocellulose. Channel preparation was done in seven steps (all steps 25 μl): (1) N-ethyl maleimide-inactivated full-length skeletal muscle myosin (Cytoskeleton) was incubated for 2 min.; (2) TAB_BSA was incubated for 5 min.; (3) 488/Cy5 actin in TAB_CaM was incubated for 2 min.; (4) channel was washed with TAB, no incubation; and finally (5) M6-RB:ktRS1:tet (prepared by incubating 0.2 nM ktRS1:tet with 20–40 nM M6-RB for 3 minutes at RT) was added in GO buffer. For the switching reaction, GO with M6-RB:ktRS1:tet was supplemented with switch strand for a final concentration of 0.5 μM switch, and incubated for another 5 min before adding it to the channel. For the switchback reaction, GO with M6-RB:ktRS1:tet was incubated with switch at 0.5 μM for 5 minutes, and then the switchback strand was added at 1.5 μM, and the solution was added to the channel without additional incubation. Three fluorescence channels were imaged successively using total internal reflection microscopy: first Cy5 fluorescence was acquired to locate actin plus-ends, then movies of motors were obtained using a Cy3 filter set, and finally actin filament bodies were imaged using 488 nm laser excitation (Spectra Physics).
To generate histograms of distances traveled and velocities, motor traces were analyzed by first generating filament kymographs for polarity-labeled filaments having only a single Cy5-labeled seed. Kymographs were generated in ImageJ using the “reslice” command after tracing the filament with a piecewise linear approximation37. Trajectories were manually identified from the kymographs, and the total signed displacement and elapsed time for each trajectory was recorded. Velocities were determined after manually removing extended pauses using a piecewise linear approximation of each trajectory.
Data Availability Statement
The data that support the plots within this paper and other findings of this study are available from the corresponding author upon reasonable request.
Supplementary Material
Acknowledgments
We thank M. Nakamura, T. Schindler, H. Ennomani, and other members of the Bryant lab for discussions and assistance. This work was supported by NIH Fellowship F32GM09442 to T.O., the Division of Intramural Research of the National Heart, Lung, and Blood Institute (G.A.), a Women & Science Fellowship from the Rockefeller University to P.G., a Human Frontiers Science Program Long-Term Fellowship to P.V.R., NIH High-Risk Research Grants 1DP2 OD004690 (to Z.B.) and 7DP5OD17885 (to G.A.), NIH R01 Grants GM100953 and GM102519 (to R.D.), and a grant from the W.M. Keck Foundation to Manu Prakash and Z.B.
Footnotes
Author Contributions
T.O. and Z.B. conceived the project; T.O. designed molecules, performed research, and analyzed data; P.S.G. and L.Y.K. performed cryo-EM research; P.S.G. analyzed cryo-EM data; C.Y.C performed MOHCA-seq research and analyzed data; P.V.R. performed research and contributed methods; Z.B., G.M.A., and R.D. supervised research; T.O., Z.B., P.S.G., G.M.A., and C.Y.C. wrote the manuscript; all authors provided expertise, discussed the results, and commented on the manuscript.
Competing Financial Interests Statement
The authors declare no competing financial interests.
Additional Information
Supplementary information is available in the online version of the paper.
References
- 1.Korten T, Månsson A, Diez S. Towards the application of cytoskeletal motor proteins in molecular detection and diagnostic devices. Curr Opin Biotechnol. 2010;21:477–488. doi: 10.1016/j.copbio.2010.05.001. [DOI] [PubMed] [Google Scholar]
- 2.Goodman BS, Derr ND, Reck-Peterson SL. Engineered, harnessed, and hijacked: synthetic uses for cytoskeletal systems. Trends Cell Biol. 2012:1–9. doi: 10.1016/j.tcb.2012.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lund K, et al. Molecular robots guided by prescriptive landscapes. Nature. 2011;465:206–210. doi: 10.1038/nature09012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wickham SFJ, et al. A DNA-based molecular motor that can navigate a network of tracks. Nat Nanotechnol. 2012;7:169–173. doi: 10.1038/nnano.2011.253. [DOI] [PubMed] [Google Scholar]
- 5.Pan J, Li F, Cha TG, Chen H, Choi JH. Recent progress on DNA based walkers. Curr Opin Biotechnol. 2015;34:56–64. doi: 10.1016/j.copbio.2014.11.017. [DOI] [PubMed] [Google Scholar]
- 6.Manstein DJ. Molecular engineering of myosin. Philos Trans R Soc Lond B Biol Sci. 2004;359:1907–1912. doi: 10.1098/rstb.2004.1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen L, Nakamura M, Schindler TD, Parker D, Bryant Z. Engineering controllable bidirectional molecular motors based on myosin. Nat Nanotechnol. 2012;7:252–256. doi: 10.1038/nnano.2012.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schindler TD, Chen L, Lebel P, Nakamura M, Bryant Z. Engineering myosins for long-range transport on actin filaments. Nat Nanotechnol. 2014;9:33–38. doi: 10.1038/nnano.2013.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nakamura M, et al. Remote control of myosin and kinesin motors using light-activated gearshifting. Nat Nanotechnol. 2014;9:693–697. doi: 10.1038/nnano.2014.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Furuta A, et al. Creating biomolecular motors based on dynein and actin-binding proteins. Nat Nanotechnol. 2016 doi: 10.1038/nnano.2016.238. [DOI] [PubMed] [Google Scholar]
- 11.Miyazono Y, Hayashi M, Karagiannis P, Harada Y, Tadakuma H. Strain through the neck linker ensures processive runs: a DNA-kinesin hybrid nanomachine study. EMBO J. 2010;29:93–106. doi: 10.1038/emboj.2009.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Derr ND, et al. Tug-of-war in motor protein ensembles revealed with a programmable DNA origami scaffold. Science. 2012;338:662–665. doi: 10.1126/science.1226734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Furuta K, et al. Measuring collective transport by defined numbers of processive and nonprocessive kinesin motors. Proceedings of the National Academy of Sciences. 2013;110:501–506. doi: 10.1073/pnas.1201390110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wollman AJM, Sanchez-Cano C, Carstairs HMJ, Cross RA, Turberfield AJ. Transport and self-organization across different length scales powered by motor proteins and programmed by DNA. Nat Nanotechnol. 2013;9:44–47. doi: 10.1038/nnano.2013.230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hariadi RF. Mechanical coordination in motor ensembles revealed using engineered artificial myosin filaments. Nat Nanotechnol. 2015;10:696–700. doi: 10.1038/nnano.2015.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cheng CY, et al. Consistent global structures of complex RNA states through multidimensional chemical mapping. Elife. 2015;4:e07600. doi: 10.7554/eLife.07600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yurke B, Turberfield AJ, Mills AP, Simmel FC, Neumann JL. A DNA-fuelled molecular machine made of DNA. Nature. 2000;406:605–608. doi: 10.1038/35020524. [DOI] [PubMed] [Google Scholar]
- 18.Spudich JA. The myosin swinging cross-bridge model. Nat Rev Mol Cell Biol. 2001 doi: 10.1038/35073086. [DOI] [PubMed] [Google Scholar]
- 19.Liao JC, Elting MW, Delp SL, Spudich JA, Bryant Z. Engineered myosin VI motors reveal minimal structural determinants of directionality and processivity. J Mol Biol. 2009;392:862–867. doi: 10.1016/j.jmb.2009.07.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li H, et al. RNA as a stable polymer to build controllable and defined nanostructures for material and biomedical applications. Nano Today. 2015;10:631–655. doi: 10.1016/j.nantod.2015.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen YJ, Groves B, Muscat RA, Seelig G. DNA nanotechnology from the test tube to the cell. Nat Nanotechnol. 2015;10:748–760. doi: 10.1038/nnano.2015.195. [DOI] [PubMed] [Google Scholar]
- 22.Moore T, Zhang Y, Fenley MO, Li H. Molecular basis of box C/D RNA-protein interactions; cocrystal structure of archaeal L7Ae and a box C/D RNA. Structure. 2004;12:807–818. doi: 10.1016/j.str.2004.02.033. [DOI] [PubMed] [Google Scholar]
- 23.Ménétrey J, et al. The structure of the myosin VI motor reveals the mechanism of directionality reversal. Nature. 2005;435:779–785. doi: 10.1038/nature03592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ménétrey J, et al. Processive Steps in the Reverse Direction Require Uncoupling of the Lead Head Lever Arm of Myosin VI. Mol Cell. 2012;48:75–86. doi: 10.1016/j.molcel.2012.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Saito H, et al. Synthetic translational regulation by an L7Ae-kink-turn RNP switch. Nat Chem Biol. 2010;6:71–78. doi: 10.1038/nchembio.273. [DOI] [PubMed] [Google Scholar]
- 26.Ohno H, et al. Synthetic RNA-protein complex shaped like an equilateral triangle. Nat Nanotechnol. 2011;6:116–120. doi: 10.1038/nnano.2010.268. [DOI] [PubMed] [Google Scholar]
- 27.Stapleton JA, et al. Feedback control of protein expression in mammalian cells by tunable synthetic translational inhibition. ACS Synth Biol. 2012;1:83–88. doi: 10.1021/sb200005w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Turner B, Melcher SE, Wilson TJ, Norman DG, Lilley DMJ. Induced fit of RNA on binding the L7Ae protein to the kink-turn motif. RNA. 2005;11:1192–1200. doi: 10.1261/rna.2680605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wells AL, et al. Myosin VI is an actin-based motor that moves backwards. Nature. 1999;401:505–508. doi: 10.1038/46835. [DOI] [PubMed] [Google Scholar]
- 30.Lister I, et al. A monomeric myosin VI with a large working stroke. EMBO J. 2004;23:1729–1738. doi: 10.1038/sj.emboj.7600180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tsiavaliaris G, Fujita-Becker S, Manstein DJ. Molecular engineering of a backwards-moving myosin motor. Nature. 2004;427:558–561. doi: 10.1038/nature02303. [DOI] [PubMed] [Google Scholar]
- 32.Geary CC, Chworos AA, Jaeger LL. Promoting RNA helical stacking via A-minor junctions. Nucleic Acids Res. 2011;39:1066–1080. doi: 10.1093/nar/gkq748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ohno H, Saito H. RNA and RNP as Building Blocks for Nanotechnology and Synthetic Biology. Prog Mol Biol Transl Sci. 2016;139:165–185. doi: 10.1016/bs.pmbts.2015.12.004. [DOI] [PubMed] [Google Scholar]
- 34.Geary C, Rothemund PWK, Andersen ES. RNA nanostructures. A single-stranded architecture for cotranscriptional folding of RNA nanostructures. Science. 2014;345:799–804. doi: 10.1126/science.1253920. [DOI] [PubMed] [Google Scholar]
- 35.Yang YR, Liu Y, Yan H. DNA Nanostructures as Programmable Biomolecular Scaffolds. Bioconjug Chem. 2015;26:1381–1395. doi: 10.1021/acs.bioconjchem.5b00194. [DOI] [PubMed] [Google Scholar]
- 36.Leontis NBNB, Westhof EE. Geometric nomenclature and classification of RNA base pairs. RNA. 2001;7:499–512. doi: 10.1017/s1355838201002515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Elting MW, Bryant Z, Liao JC, Spudich JA. Detailed Tuning of Structure and Intramolecular Communication Are Dispensable for Processive Motion of Myosin VI. Biophys J. 2011;100:430–439. doi: 10.1016/j.bpj.2010.11.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gibson DG, Smith HO, Hutchison CA, Venter JC, Merryman C. Chemical synthesis of the mouse mitochondrial genome. Nat Methods. 2010;7:901–903. doi: 10.1038/nmeth.1515. [DOI] [PubMed] [Google Scholar]
- 39.Lebel P, Basu A, Oberstrass FC, Tretter EM, Bryant Z. Gold rotor bead tracking for high-speed measurements of DNA twist, torque and extension. Nat Methods. 2014;11:456–462. doi: 10.1038/nmeth.2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the plots within this paper and other findings of this study are available from the corresponding author upon reasonable request.