Abstract
Human calprotectin (CP, S100A8/S100A9 oligomer) is a metal-sequestering host-defense protein that prevents bacterial acquisition of Mn(II). In this work, we investigate Mn(II) competition between CP and two solute-binding proteins that Staphylococcus aureus and Streptococcus pneumoniae, Gram-positive bacterial pathogens of significant clinical concern, use to obtain Mn(II) when infecting a host. Biochemical and electron paramagnetic resonance (EPR) spectroscopic analyses demonstrate that CP outcompetes staphylococcal MntC and streptococcal PsaA for Mn(II). This behavior requires the presence of excess Ca(II) ions, which enhance the Mn(II) affinity of CP. This report presents new spectroscopic evaluation of two Mn(II) proteins important for bacterial pathogenesis, direct observation of Mn(II) transfer from the bacterial Mn(II) acquisition proteins to CP, and molecular-level insight into the extracellular battle for metal nutrients that occurs during infection.
TOC graphic
Transition metal ions are essential nutrients for all organisms. In the context of the host/pathogen interaction, microbes must acquire nutrient metals from the host environment. In response to invading pathogens, the mammalian innate immune system launches a metal-withholding response, often termed “nutritional immunity,” to restrict the availability of essential metal nutrients.1 Human calprotectin (CP) is a metal-chelating protein that contributes to this innate immune response. It employs an unusual hexahistidine site for sequestering divalent first-row transition metals, including Mn(II).2 The ability of CP to affect Mn(II) availability was first recognized during studies of murine tissue abscesses infected with the Gram-positive opportunistic human pathogen Staphylococcus aureus3 and later examined for the Gram-negative gastrointestinal pathogen Salmonella enterica serovar Typhimurium.4 In the current work, performed to provide molecular understanding of the tug-of-war between CP and microbes for Mn(II), we employ biochemical and electron paramagnetic resonance (EPR) spectroscopic methods to evaluate metal competition between human CP and bacterial metal transport machinery. We focus on extracellular solute-binding proteins (SBPs) employed by two Gram-positive human pathogens for acquiring Mn(II) from the host, MntC of S. aureus5 and PsaA of Streptococcus pneumoniae.6 The results show that CP outcompetes both of these Mn(II)-acquisition proteins for Mn(II) under conditions of high Ca(II), as found in the extracellular environment.
S. aureus employs the ATP-binding cassette transport system MntABC when battling with CP for Mn(II).3,7 MntC is the SBP that scavenges Mn(II) from the extracellular space. Following Mn(II) capture, MntC delivers the Mn(II) ion to MntAB, which transports the divalent cation across the cell membrane to the cytoplasm. Although less is known about the competition between CP and S. pneumoniae for nutrient metals, a recent antibacterial activity study showed that CP inhibits the growth of S. pneumoniae.8 S. pneumoniae expresses the ATP-binding cassette transporter PsaABC for Mn(II) acquisition, which is important for virulence.9 PsaA is the SBP that scavenges Mn(II) from the extracellular space and brings the ion to PsaBC for cytoplasmic delivery.
Most reported studies that consider the interplay between CP and bacterial metal transport systems are at the cellular level and typically begin with a comparison of the growth inhibitory activity of CP against wild-type bacteria and knock-out strains that lack a component of the metal transport machinery under investigation.3,4,7 Biochemical and biophysical methods provide complementary methods for investigating metal competition at the host/pathogen interface. In particular, these approaches can afford direct assessment of the metal-ion speciation during competition between microbial and host proteins for metal ions.
In an effort to achieve a molecular picture of Mn(II) competition at the host/microbe interface, we overexpressed and purified staphylococcal MntC and streptococcal PsaA. Guided by reported studies of these proteins,10 we prepared MntC and PsaA that lack the N-terminal lipid anchors and obtained soluble proteins during overexpression. We overexpressed and purified each SBP without an affinity tag, which afforded the proteins in high yield and purity (Tables S1–S3, Figures S1,S2, Supporting Information). Because the SBPs accumulated bound metals during purification (Table S1), we removed contaminating metal ions after chromatographic purification of the proteins, and obtained MntC and PsaA with notably low metal content (Table S2). Moreover, Mn(II) competition titrations with the metal-ion sensor ZP1 indicated that both SBPs outcompete ZP1 (Kd,Mn(II) = 550 nM)11 for 1 equiv of Mn(II) (Figure S3), which confirmed that each SBP was obtained predominantly in the apo form and, as expected from prior studies,10b,d binds 1 equiv of Mn(II) with high affinity.
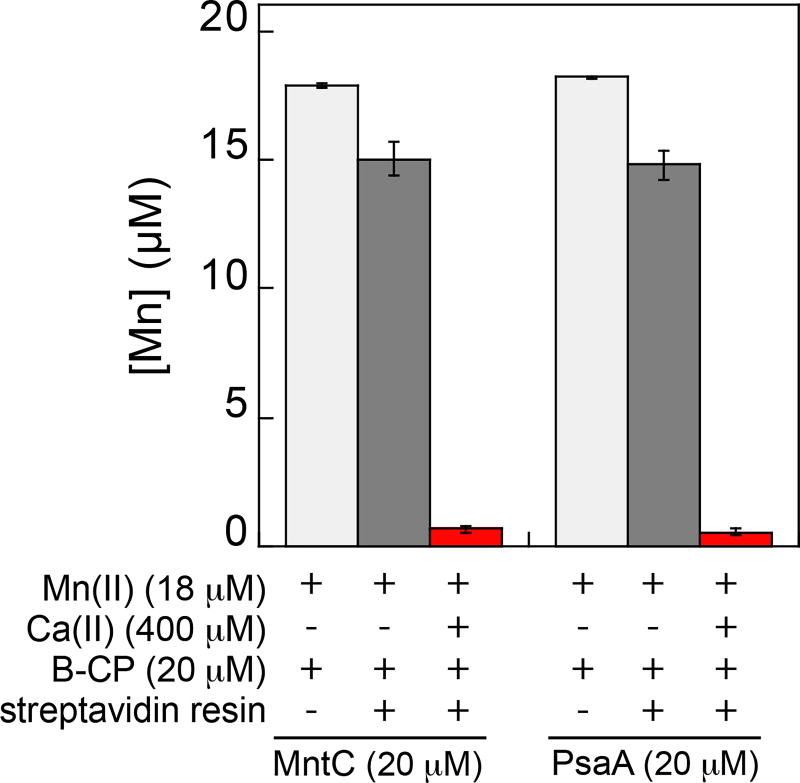
We designed a pull-down assay for evaluating Mn(II) speciation between the SBPs and CP. We prepared biotinylated CP (B-CP), which has a biotin moiety covalently attached to Cys3 of S100A9 and established that it displays Ca(II)-dependent heterotetramerization, antibacterial activity comparable to the unmodified protein, and coordinates Mn(II), Fe(II) and Zn(II) with the expected stoichiometries (Figures S4,S5). Next, we prepared solutions containing 18 µM Mn(II), 20 µM SBP, and 20 µM B-CP in the absence and presence of 400 µM Ca(II) at pH 7.5. We selected a long, 10-h time point for this preliminary experiment because we lacked information about timescale required for the mixture to reach equilibrium. Following removal of B-CP with streptavidin resin (Figure S6), we analyzed the Mn(II) content of the solution by inductively-coupled plasma mass spectrometry (Figure 1). In the absence of Ca(II), >80% of Mn(II) remained in solution, presumably bound by the SBP. In contrast, in the presence of Ca(II), <4% of Mn(II) remained in solution, indicating that B-CP outcompeted the SBPs for Mn(II) and the pull-down removed Mn(II) from solution. These data provide evidence that the Ca(II)-bound, high-affinity form of CP coordinates Mn(II) with greater affinity than MntC or PsaA. This Ca(II)-bound CP species is expected to exist in the extracellular space where Ca(II) concentrations are ≈2 mM.12,13
Figure 1.
Concentration of Mn(II) in solution following pull-down of mixtures containing B-CP, MntC or PsaA, and Mn(II) with or without Ca(II) (75 mM HEPES, 100 mM NaCl, pH 7.5). The mixtures were incubated for 10 h at room temperature before pull-down.
Because removal of B-CP from solution by streptavidin resin takes minutes, we limited application of this pull-down assay to the end-point analysis. Moreover, we sought to directly observe and discriminate Mn(II) bound to CP and the SBPs in the same assay mixture at varying time points. Low-temperature EPR spectroscopy is ideally suited for this purpose because previous structural studies demonstrate that the Mn(II)-binding sites of CP and the SBPs have notably distinct coordination environments and degrees of symmetry. The high-affinity Mn(II)-His6 site of CP shows a nearly idealized octahedral coordination geometry.2a–c In contrast, a crystal structure of MntC indicates that it coordinates Mn(II) at a 5-coordinate N2O3 motif composed of two His, one bidentate Glu, and one Asp residue.10b A crystal structure of Mn(II)-bound PsaA shows that it binds Mn(II) with the same residues as MntC, but the site has been described as tetrahedral where the Glu and Asp residue are reported to be monodendate ligands.10e,13 We reasoned that the different Mn(II) coordination environments in CP and the SBPs would result in readily distinguishable low-temperature Mn(II) EPR spectroscopic signatures,14 providing spectral discrimination between Mn(II)-CP and Mn(II)-SBP in a sample. Prior EPR spectroscopy of the Mn(II)-His6 site of CP revealed a six-line pattern centered at g = 2 (≈335 mT) with a low zero-field splitting (ZFS) of 485 MHz (E/D = 0.30) for the Mn(II) ion (S = 5/2), consistent with a highly symmetric site.2a,b On the basis of the crystallographic characterization of the Mn(II)-SBP sites, we expected that Mn(II) EPR spectra of these proteins would exhibit larger ZFS.14,15
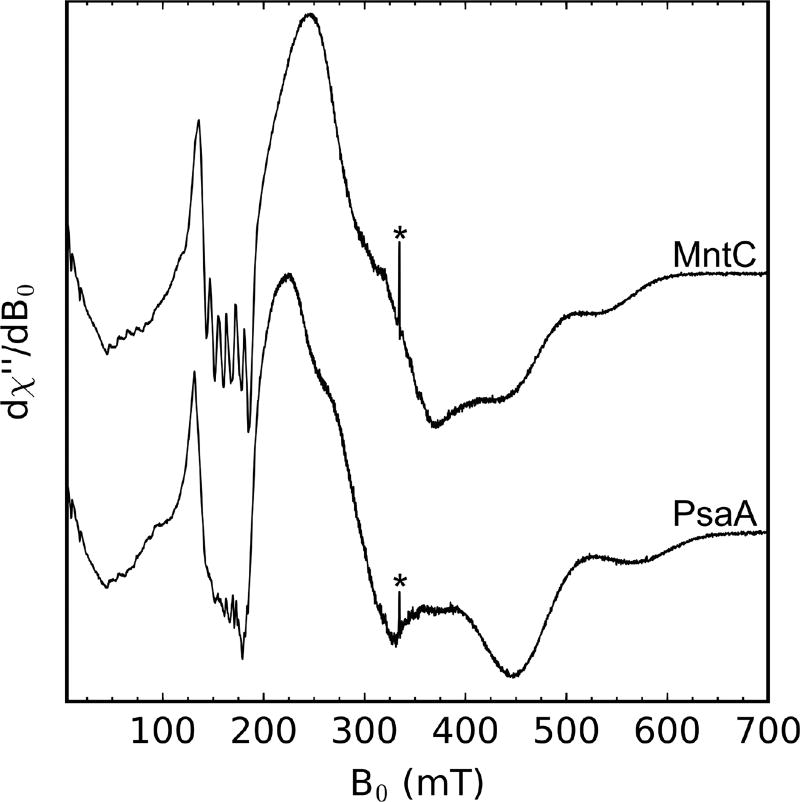
To evaluate the EPR spectra of Mn(II) bound to the SBPs, we prepared samples containing 1 mM SBP and 750 µM Mn(II) (75 mM HEPES, 100 mM NaCl, pH 7.5). Both of the Mn(II)-SBP spectra (Figure 2) indicate a high-spin Mn(II) ion (S = 5/2) with a large ZFS that is of comparable magnitude to the microwave quanta at X-band (≈9.4 GHz).14a,15b,16 The large ZFS (D>3 GHz) leads to these systems not being in the “high field” regime at X-band which is met when the electron Zeeman is the dominant term in the spin Hamiltonian and results in broad spectra that span >600 mT at X-band.15c Multi-frequency EPR studies are needed to further evaluate the electronic structure of the Mn(II) ions coordinated by MntC and PsaA; however, the current spectra provide suitable handles for monitoring Mn(II) competition between these proteins and CP. In contrast to the Mn(II)-PsaA EPR spectra in Figure 2, a previously reported Mn(II)-PsaA spectrum showed 6-line Mn(II) EPR features at g = 2 (≈335 mT),17 which likely arose from aqueous Mn(II) contamination.
Figure 2.
X-band CW EPR spectra of Mn(II) (750 µM) bound to MntC (1 mM) or PsaA (1 mM) (75 mM HEPES, 100 mM NaCl, pH 7.5). The asterisk denotes a quartz background radical. Spectrometer settings: vmw = 9.4 GHz, 0.5 mT modulation amplitude at 100 kHz, power = 200 µW, T = 10 K.
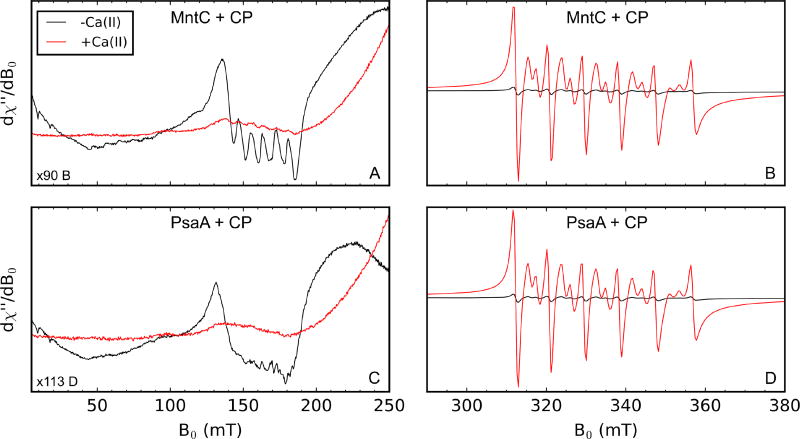
As anticipated, the EPR spectra of the Mn(II)-SBPs are markedly different than those of Mn(II)-CP or aqueous Mn(II), allowing simultaneous monitoring of Mn(II)-bound CP and MntC or PsaA (Figure S7). The sharp spectral features in the g = 4.5 region (≈150 mT) of the Mn(II)-SBP spectra do not overlap with the Mn(II)-CP signals in the g = 2 region (≈335 mT). Thus, to examine Mn(II) speciation between CP and the SBPs by EPR spectroscopy, we prepared samples containing a 1:1:1 ratio of Mn(II), CP, and SBP (500 µM each) in the absence and presence of 5 mM Ca(II) at pH 7.5 and incubated them for 10 h at room temperature. In the samples without added Ca(II), we observed a distinct low-field feature around g = 4.5 (≈150 mT, Figure 3A,C; black spectra) that corresponds to Mn(II)-MntC or Mn(II)-PsaA and a weak Mn(II)-CP-Ser signal centered at g = 2 (≈335 mT, Figure 3B,D; black spectra). In the presence of excess Ca(II), a strong Mn(II)-CP-Ser signal (Figure 3B,D; red spectra) and a negligible Mn(II)-SBP signal (Figure 3A,C; red spectra) occur, indicating that Ca(II)-bound CP outcompetes both SBPs for Mn(II). These results are consistent with the pull-down assays performed in the absence and presence of Ca(II), where negligible Mn(II) was found in the SBP-containing supernatant after pull-down of B-CP only when excess Ca(II) was present (Figure 1). Moreover, this work provides further information about the Mn(II) affinity of CP. Prior studies indicate that the His6 site coordinates Mn(II) with Kd ≤ 10 nM when Ca(II) ions are present.11 The current results indicate that this value is in the sub-nanomolar range based on the reported Kd,Mn(II) values of MntC (Kd ≈ 4 nM)10b and PsaA (Kd < 10 nM).10d
Figure 3.
X-band CW EPR of 1:1:1 mixtures of Mn(II):CP:SBP incubated for 10 h at room temperature in the absence (black) or presence (red) of 5 mM Ca(II) (75 mM HEPES, 100 mM NaCl, pH 7.5). Each sample contained 500 µM Mn(II), 500 µM SBP, and 500 µM CP after mixing. Panels A and C contain the same Mn(II) spectra of the samples containing CP and MntC, and panels B and D contain the same Mn(II) spectra of the samples containing CP and PsaA. Panels A and C are scaled differently from B and D along the y-axis to make the low-field, g = 4.5, features of each Mn(II)-SBP more apparent relative to the mid-field features, g = 2, of Mn(II)-CP. Spectrometer settings: vmw = 9.4 GHz, 0.5 mT modulation amplitude at 100 kHz, power = 200 µW, T = 10 K.
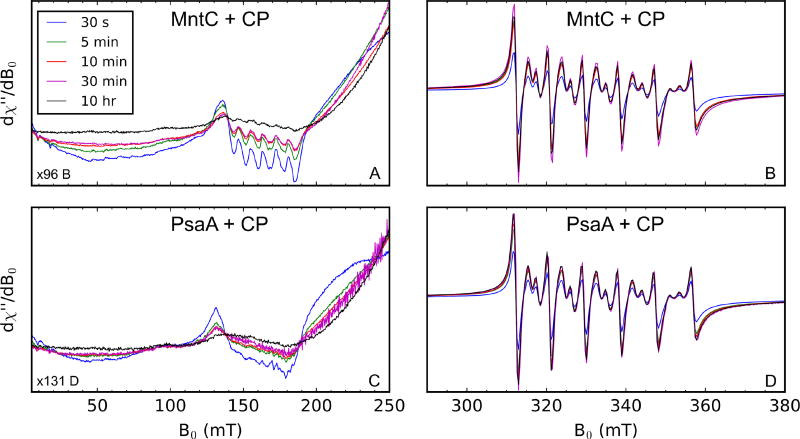
Next, to probe the timescale of Mn(II) transfer from each SBP to CP, we performed EPR experiments where a solution containing 1 mM CP and 10 mM Ca(II) was added to an equal volume of a solution containing 1 mM Mn(II)-SBP. Aliquots of this mixture were frozen at varying time points and analyzed by EPR spectroscopy (Figure 4). This assay revealed a time-dependent decrease in the Mn(II)-MntC or Mn(II)-PsaA signal (Figure 4A,C) and a corresponding increase in the Mn(II)-CP-Ser signal (Figure 4B,D). The greatest changes in signal intensity marking transfer of the Mn(II) ion from each SBP to CP occurred in the first 10 min of the time course. The short time-scale of Mn(II) transfer is striking and highlights the remarkable ability of CP to capture and retain labile divalent transition metals.
Figure 4.
Time course of Mn(II) transfer from Mn(II)-SBPs (500 µM) to CP (500 µM) in the presence of 5 mM Ca(II) monitored by X-band EPR spectroscopy (75 mM HEPES, 100 mM NaCl, pH 7.5). Panels A and C contain the same Mn(II) spectra of the samples containing CP and MntC, and panels B and D contain the same Mn(II) spectra of the samples containing CP and PsaA. Panels A and C are scaled differently from B and D along the y-axis to make the low-field, g = 4.5, features of each Mn(II)-SBP more apparent. Spectrometer settings: vmw = 9.4 GHz, 0.5 mT modulation amplitude at 100 kHz, power = 200 µW, T = 10 K.
In closing, to our knowledge, this work provides the first direct analysis of metal speciation between CP and a metal-capturing SBP. Together, these experiments demonstrate that the Ca(II)-bound, high-affinity form of CP outcompetes MntC and PsaA, two SBPs used by Gram-positive pathogens for Mn(II) acquisition. This conclusion informs the current model of how CP contributes to extracellular metal withholding at the host/pathogen interface, and provides an important benchmark for future studies of this system and other microbial metal transporters. In particular, we expect that investigations of metal competition between CP and fully reconstituted ABC transporter systems will be informative.
Supplementary Material
Acknowledgments
This work was supported by the NIH (R01GM118695 to EMN and R01GM104543 to RDB). R. C. Hadley is a recipient of the R. R. Schrock Graduate Fellowship, and T. G. Nakashige is a recipient of a NSF graduate fellowship. The MIT Biophysical Instrumentation Facility for the Study of Complex Macromolecular Systems is supported by NSF grant 0070319. The ICP-MS instrument is housed in the MIT CEHS Bioanalytical Core, which is supported by NIH grant P30-ES002109. We thank Prof. D. P. Giedroc for his comments on the manuscript, and Prof. Giedroc and Dr. John Lisher for contributing a S. pneumoniae D39 genomic DNA fragment containing the psaA sequence, and J. R. Stephan and E. M. Zygiel for assistance with protein purifications. S. aureus USA300 JE2 was obtained from the Network on Antimicrobial Resistance Against Staphylococcus aureus.
Footnotes
ASSOCIATED CONTENT
The Supporting Information is available free of charge on the ACS Publications website. Complete experimental methods, Tables S1–S3, Figures S1–S8.
References
- 1.(a) Weinberg ED. J. Am. Med. Assoc. 1975;231:39–41. [Google Scholar]; (b) Hood MI, Skaar EP. Nat. Rev. Microbiol. 2012;10:525–537. doi: 10.1038/nrmicro2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.(a) Hayden JA, Brophy MB, Cunden LS, Nolan EM. J. Am. Chem. Soc. 2013;135:775–787. doi: 10.1021/ja3096416. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Gagnon DM, Brophy MB, Bowman SEJ, Stich TA, Drennan CL, Britt RD, Nolan EM. J. Am. Chem. Soc. 2015;137:3004–3016. doi: 10.1021/ja512204s. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Damo SM, Kehl-Fie TE, Sugitani N, Holt ME, Rathi S, Murphy WJ, Zhang Y, Betz C, Hench L, Fritz G, Skaar EP, Chazin WJ. Proc. Natl. Acad. Sci. U. S. A. 2013;110:3841–3846. doi: 10.1073/pnas.1220341110. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Nakashige TG, Zhang B, Krebs C, Nolan EM. Nat. Chem. Biol. 2015;11:765–771. doi: 10.1038/nchembio.1891. [DOI] [PMC free article] [PubMed] [Google Scholar]; (e) Nakashige TG, Stephan JR, Cunden LS, Brophy MB, Wommack AJ, Keegan BC, Shearer JM, Nolan EM. J. Am. Chem. Soc. 2016;138:12243–12251. doi: 10.1021/jacs.6b06845. [DOI] [PMC free article] [PubMed] [Google Scholar]; (f) Nakashige TG, Zygiel EM, Drennan CL, Nolan EM. J. Am. Chem. Soc. 2017;139:8828–8836. doi: 10.1021/jacs.7b01212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Corbin BD, Seeley EH, Raab A, Feldmann J, Miller MR, Torres VJ, Anderson KL, Dattilo BM, Dunman PM, Gerads R, Caprioli RM, Nacken W, Chazin WJ, Skaar EP. Science. 2008;319:962–965. doi: 10.1126/science.1152449. [DOI] [PubMed] [Google Scholar]
- 4.Diaz-Ochoa VE, Lam D, Lee CS, Klaus S, Behnsen J, Liu JZ, Chim N, Nuccio S-P, Rathi SG, Mastroianni JR, Edwards RA, Jacobo CM, Cerasi M, Battistoni A, Ouellette AJ, Goulding CW, Chazin WJ, Skaar EP, Raffatellu M. Cell Host Microbe. 2016;19:814–825. doi: 10.1016/j.chom.2016.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gribenko AV, Liberator P, Anderson AS, Matsuka YV, Mosyak L. Encylopedia of Inorganic and Bioinorganic Chemistry. John Wiley & Sons, Ltd; 2015. [Google Scholar]
- 6.Eijkelkamp BA, McDevitt CA, Kitten T. Biometals. 2015;28:491–508. doi: 10.1007/s10534-015-9826-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kehl-Fie TE, Zhang Y, Moore JL, Farrand AJ, Hood MI, Rathi S, Chazin WJ, Caprioli RM, Skaar EP. Infect. Immun. 2013;81:3395–3405. doi: 10.1128/IAI.00420-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Makthal N, Nguyen K, Do H, Gavagan M, Chandrangsu P, Helmann JD, Olsen RJ, Kumaraswami M. EBioMedicine. 2017;21:131–141. doi: 10.1016/j.ebiom.2017.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.(a) Ogunniyi AD, Mahdi LK, Jennings MP, McEwan AG, McDevitt CA, Van der Hoek MB, Bagley CJ, Hoffmann P, Gould KA, Paton JC. J. Bacteriol. 2010;192:4489–4497. doi: 10.1128/JB.00064-10. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Rajam G, Anderton JM, Carlone GM, Sampson JS, Ades EW. Crit. Rev. Microbiol. 2008;34:131–142. doi: 10.1080/10408410802275352. [DOI] [PubMed] [Google Scholar]; (c) Eijkelkamp BA, Morey JR, Ween MP, Ong C-IY, McEwan AG, Paton JC, McDevitt CA. PLoS One. 2014;9:e89427. doi: 10.1371/journal.pone.0089427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.(a) Anderson AS, Scully IL, Timofeyeva Y, Murphy E, McNeil LK, Mininni T, Nuñez L, Carriere M, Singer C, Dilts DA, Jansen KU. J. Infect. Dis. 2012;205:1688–16896. doi: 10.1093/infdis/jis272. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Gribenko A, Mosyak L, Ghosh S, Parris K, Svenson K, Moran J, Chu L, Li S, Liu T, Woods VL, Jr, Jansen KU, Green BA, Anderson AS, Matsuka YV. J. Mol. Biol. 2013;425:3429–3445. doi: 10.1016/j.jmb.2013.06.033. [DOI] [PubMed] [Google Scholar]; (c) Salazar N, Castiblanco-Valencia MM, da Silva LB, Arantes de Castro Í, Monaris D, Masuda HP, Barbosa AS, Arêas APM. PLoS One. 2014;9:e112730. doi: 10.1371/journal.pone.0112730. 22. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) McDevitt CA, Ogunniyi AD, Valkov E, Lawrence MC, Kobe B, McEwan AG, Paton JC. PLoS Pathog. 2011;7:e1002357. doi: 10.1371/journal.ppat.1002357. [DOI] [PMC free article] [PubMed] [Google Scholar]; (e) Couñago RM, Ween MP, Begg SL, Bajaj M, Zuegg J, O'Mara ML, Cooper MA, McEwan AG, Paton JC, Kobe B, McDevitt CA. Nat. Chem. Biol. 2014;10:35–41. doi: 10.1038/nchembio.1382. [DOI] [PubMed] [Google Scholar]
- 11.You Y, Tomat E, Hwang K, Atanasijevic T, Nam W, Jasanoff AP, Lippard SJ. Chem. Commun. 2010;46:4139–4141. doi: 10.1039/c0cc00179a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brini M, Ottolini D, Cali T, Carafoli E. Met. Ions Life Sci. 2013;20:87–93. doi: 10.1007/978-94-007-7500-8_4. [DOI] [PubMed] [Google Scholar]
- 13.Brophy MB, Nolan EM. ACS Chem. Biol. 2015;10:641–651. doi: 10.1021/cb500792b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stich TA, Lahiri S, Yeagle G, Dicus M, Brynda M, Gunn A, Aznar C, DeRose VJ, Britt RD. Appl. Magn. Reson. 2007;31:321–341. doi: 10.1007/BF03166263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.(a) Walsby CJ, Telser J, Rigsby RE, Armstrong RN, Hoffman BM. J. Am. Chem. Soc. 2005;127:8310–8319. doi: 10.1021/ja044094e. [DOI] [PubMed] [Google Scholar]; (b) Whittaker JW, Whittaker MM. J. Am. Chem. Soc. 1991;113:5528–5540. [Google Scholar]; (c) Vance CK, Miller AF. Biochemistry. 2001;40:13079–13087. doi: 10.1021/bi0113317. [DOI] [PubMed] [Google Scholar]
- 16.(a) Un S, Tabares LC, Cortez N, Hiraoka BY, Yamakura F. J. Am. Chem. Soc. 2004;126:2720–2726. doi: 10.1021/ja036503x. [DOI] [PubMed] [Google Scholar]; (b) Tabares LC, Cortez N, Agalidis I, Un S. J. Am. Chem. Soc. 2005;127:6039–6047. doi: 10.1021/ja047007r. [DOI] [PubMed] [Google Scholar]
- 17.Li N, Yang XY, Guo Z, Zhang J, Cao K, Han J, Zhang G, Liu L, Sun X, He QY. J. Biol. Inorg. Chem. 2014;19:829–838. doi: 10.1007/s00775-014-1114-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.