Abstract
Programmable nucleases, such as Cas9, are used for precise genome editing by homology-dependent repair (HDR)1–3. However, HDR efficiency is constrained by competition from other double-strand break (DSB) repair pathways, including non-homologous end-joining (NHEJ)4. We report the discovery of a genetically encoded inhibitor of 53BP1 that increases the efficiency of HDR-dependent genome editing in human and mouse cells. 53BP1 is a key regulator of DSB repair pathway choice in eukaryotic cells4, 5 and functions to favor NHEJ over HDR by suppressing end resection, which is the rate-limiting step in the initiation of HDR. We screened an existing combinatorial library of engineered ubiquitin variants6 for inhibitors of 53BP1. Expression of one variant, named i53 (inhibitor of 53BP1), in human and mouse cells blocked accumulation of 53BP1 at sites of DNA damage and improved gene targeting and chromosomal gene conversion with either double-stranded DNA or single-stranded oligonucleotide donors by up to 5.6-fold. Inhibition of 53BP1 is a robust method to increase efficiency of HDR-based precise genome editing.
In human cells, the dominant pathway that mends two-ended DSBs, such as those created by programmable nucleases is NHEJ. A key regulator of the choice between NHEJ and HDR is 53BP1 (encoded by TP53BP1 in human cells), a pro-NHEJ factor that limits HR in part by blocking DNA end resection but also by inhibiting BRCA1 recruitment to DSB sites7, 8. We therefore reasoned that 53BP1 might make a suitable target for increasing rates of precise gene editing by HDR.
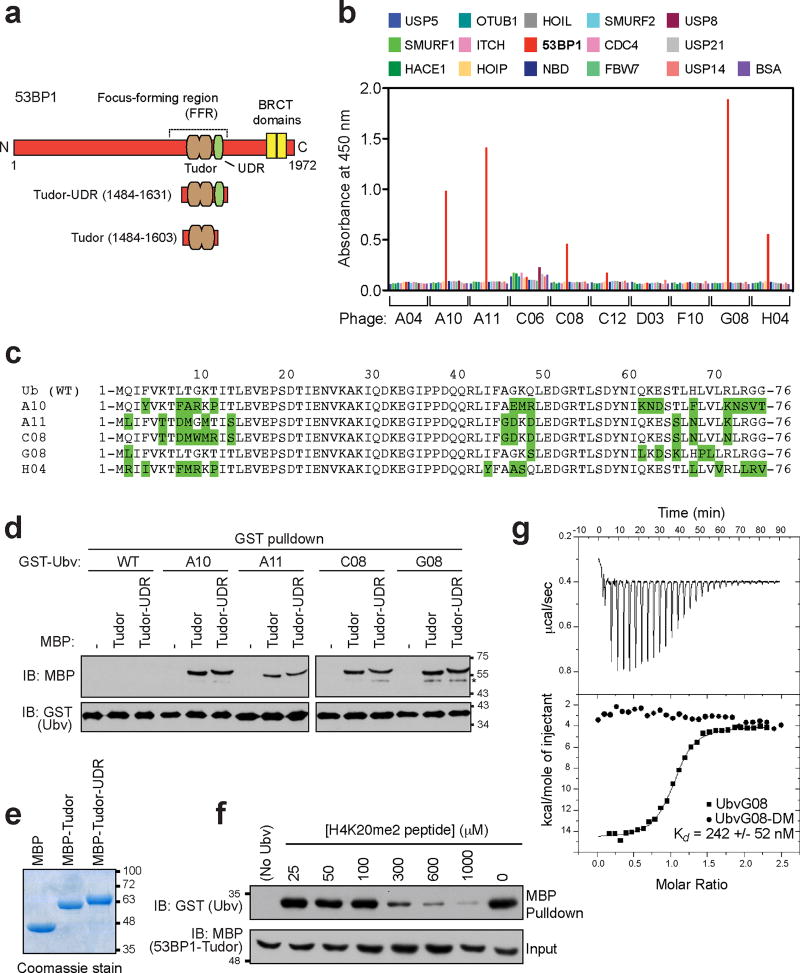
To identify inhibitors of 53BP1, we took advantage of a soft-randomized library of ubiquitin variants (Ubvs) that was initially developed to identify inhibitors of ubiquitin-binding proteins6. As 53BP1 recognizes histone H2A ubiquitylated on Lys15 (H2AK15ub) in order to accumulate at DSB sites9, we reasoned that it might be possible to identify Ubvs targeting the 53BP1 ubiquitin-dependent recruitment (UDR) domain involved in ubiquitylated histone recognition9. After 5 rounds of selection against a GST-53BP1 fragment containing the tandem Tudor domain and the UDR (residues 1484–1631; Fig. 1a), 10 unique phages were selected for re-testing in ELISA assays for binding to the 53BP1 Tudor-UDR region and to 14 other proteins, most of them known ubiquitin-binding proteins (Fig. 1b). We identified five distinct Ubvs that bound selectively to 53BP1 (A10, A11, C08, G08 and H04; Fig. 1bc). Using GST fusion proteins of 4 of these 5 Ubvs and testing them in GST pulldown assays against maltose-binding protein (MBP) fused to either the Tudor domain (residues 1484–1603) or the Tudor-UDR fragment of 53BP1, we found that each Ubv bound to the MBP fusion containing only the 53BP1 Tudor domain, in addition to those also containing the UDR (Fig. 1de). Because the UDR is apparently not required for binding to the Ubv, all further experiments were carried out with proteins containing solely the Tudor domain. We selected clone G08 for further analysis because the phage expressing it displayed strongest binding by ELISA (Fig. 1b) and contained only 7 mutations, the lowest number of amino acid substitutions among the selected Ubvs (Fig. 1c).
Figure 1. Identification of 53BP1-binding ubiquitin variants.
a, Schematic representation of 53BP1, highlighting the focus-forming region (FFR), which is necessary and sufficient for the recruitment of 53BP1 to DSB sites. b, Phage enzyme-linked immunosorbent assays (ELISAs) for binding to the following immobilized proteins (color coded as indicated in the panel): USP5, USP7, SMURF1, HACE, HOIP, HOIL, 53BP1 (Tudor-UDR region), NBD, SMURF2, CDC4, OTUB1, FBW7, USP8, ITCH, USP21, USP14 and BSA. Bound phages were detected spectrophotometrically (optical density at 450 nm), and background binding to neutravidin was subtracted from the signal. c, Sequence alignments of the 53BP1-binding Ubvs. d, Pulldown assays of the indicated GST-Ubv fusion with either MBP alone (−) or MBP fused to the Tudor or Tudor-UDR fragments of 53BP1. The asterisk (*) labels bands that we attribute as possible protein degradation products. e, the various MBP proteins used in the pulldown assays were separated by SDS-PAGE and stained with Coomassie brilliant blue. f, Competition assay in which the GST-UbvG08 was prebound to the MBP-Tudor fusion of 53BP1. Increasing amounts of a synthetic peptide derived from the region of H4K20me2 were added. After extensive washing, bound proteins were analyzed by immunoblotting against GST and MBP. g, Isothermal titration calorimetry profiles obtained by titration of UbvG08 (squares) or UbvG08-DM (circles) titrated into a solution of the 53BP1 Tudor protein. Curves were fitted with a one-set-of-sites model. The dissociation constant (Kd) for the UbvG08-53BP1 interaction is indicated (N=3). Please note that the gels and blots shown in panels d,e,f are cropped and uncropped version can be found as Supplementary Information.
Because the 53BP1 Tudor domain binds to dimethylated histone H4 Lys20 (H4K20me2)10, we next tested whether UbvG08- and H4K20me2-binding functions were mutually exclusive. We found that H4K20me2 peptides competed with UbvG08 for 53BP1 binding with a half-maximal competing concentration in the 100 µM to 300 µM range (Fig. 1f). Since the dissociation constant (Kd) of the H4K20me2 peptide-53BP1 Tudor interaction is 20 µM10, the result of the H4K20me2 peptide competition implied that 53BP1 bound to UbvG08 with higher affinity than methyl-lysine peptides. Indeed, as assessed by isothermal titration calorimetry (ITC), UbvG08 bound to the 53BP1 Tudor domain with Kd of 242 +/− 52 nM (N=3), two orders of magnitude tighter than the 53BP1-H4K20me2 interaction (Fig. 1g). In contrast, a version of UbvG08 that reverted the L69P and V70L mutations to wild type (mutant DM; see below for the rationale behind these mutations) did not display any detectable binding to the 53BP1 Tudor domain by ITC (Fig. 1g).
To gain insight into the mechanism by which UbvG08 binds to 53BP1, we solved the crystal structure of UbvG08 bound to the 53BP1 Tudor domain (see Supplementary Table 1 for Data collection and refinement statistics). Within the solved complex, the 53BP1 Tudor domain adopted a canonical mixed α-β fold identical to that reported in its apo state (1XNI; secondary structure RMSD of 1.0 Å) and in complex with a H4K20me2 derived peptide (2IG0; secondary structure RMSD of 1.1 Å) (Supplementary Fig. 1a). UbvG08 displayed the expected ubiquitin-like fold consisting of a five-strand β-sheet (β1–5) buttressed against a single α-helix (α1) and a short 310 helix. However, the register of strand β5 was shifted 4 positions from its expected position, resulting in an increase in the length of the loop preceding strand β5 by 4 residues and a shortening of the C-terminal tail of β5 by 4 residues (Supplementary Fig. 1b,c).
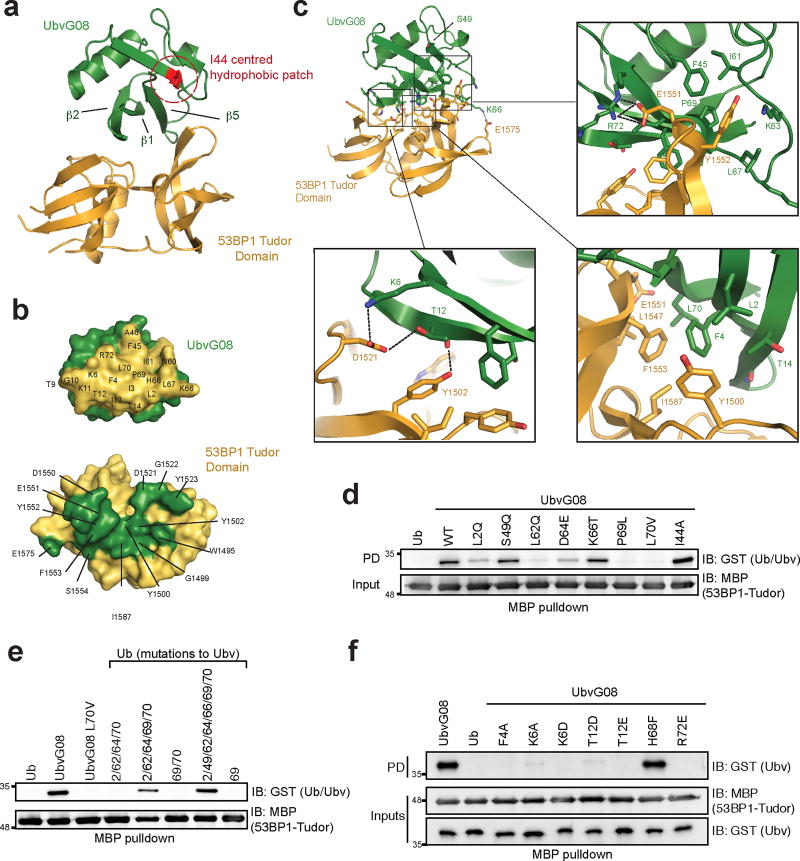
Complex formation was achieved by association of the β-sheet surface of UbvG08 centred on β1, β2 and β5, with the ligand-binding surface of the 53BP1 Tudor domain (Fig. 2a). This surface on the Ubv is adjacent to but distinct from the I44-centred hydrophobic patch that mediates the majority of Ub-protein interactions11. The contact surfaces were extensive (buried surface area=755.4 Å2), and comprised of a mixture of hydrophobic and hydrophilic residues (Fig. 2b, with notable interactions depicted in Fig. 2c).
Figure 2. Structure of the UbvG08 bound to the 53BP1 Tudor domain.
a, Ribbons representation of the UbvG08 (shown in green) – 53BP1 Tudor domain (shown in gold) complex. The hydrophobic patch centered on I44 of the UbvG08 structure is highlighted in red. b, Reciprocal interaction surfaces on UbvG08 (top) and 53BP1 Tudor domain (bottom). Contact residues are highlighted on their respective surfaces. c, Zoom-in of the UbvG08-53BP1 Tudor domain contact region. Hydrogen and salt interactions are denoted by black dotted lines. d, MBP pulldown assay of GST fused to ubiquitin (Ub) or to the indicated UbvG08 proteins, with the MBP-53BP1-Tudor protein. e, MBP-pulldown assay of GST fused to UbvG08, its L70V mutant or the indicated Ub proteins, with the MBP-53BP1-Tudor protein. f, MBP-pulldown assay of GST fused to ubiquitin (Ub) or to the indicated UbvG08 proteins, with the MBP-53BP1-Tudor protein. PD, pulldown. IB, immunoblot. Please note that the blots shown in panels d,e,f are cropped and uncropped version can be found as Supplementary Information.
The high-affinity binding between UbvG08 and the Tudor domain of 53BP1 can be rationalized as follows. Whereas the sequence of UbvG08 differs from ubiquitin by 7 residues, only 4 substitutions are well positioned on the contact surface to allow direct interaction of their side chains with 53BP1. Specifically, L70 (Val in Ub) forms favourable hydrophobic contacts with 53BP1 F1553 and L1547; L2 (Gln in Ub) forms favourable hydrophobic contacts with 53BP1 Y1500; and P69 (Leu in Ub) forms favourable hydrophobic contact with 53BP1 Y1552 (Fig. 2c). Additionally K66 (Thr in Ub) is well positioned to form an electrostatic interaction with 53BP1 E1575 (Fig. 2c).
Other substitutions in UbvG08 may contribute to enhanced binding by stabilizing a shift in the register of strand β5. The L62 mutation (Gln in Ub) appears most important, as it resides at the initiating position of the normally tight loop preceding β5 in Ub (Supplementary Fig. 1d). The L62 substitution causes a reorientation of the side chain from a solvent-exposed orientation (in Ub) to a buried position (in UbvG08) in the hydrophobic core, which would be disruptive to tight turn formation. Additionally, the substituted side chains of D64 (Glu in Ub) and K66 (Thr in Ub) occupy new positions in the enlarged solvent-exposed loop preceding β5, whereas in the absence of a register shift, they would occupy positions in strand β3 directly facing the Tudor domain where they might otherwise contribute suboptimal interactions with 53BP1 (Supplementary Fig. 1e). The register difference in strand β5 adds an additional layer of complexity due to the non-substituted R72 side-chain displaced by 17 Å from its expected position in Ub, allowing it to form a near ideal salt interaction with E1551 in the Tudor domain (Fig. 2c).
To validate the structural model, we interrogated the respective binding surfaces with site-directed mutagenesis. We first assessed the impact of reverting each of the 7 substitutions in UbvG08 to their Ub counterparts. The L2Q, L62Q, D64E, P69L and L70V reversions all reduced UbvG08 binding to 53BP1, with the P69L and L70V mutations having the strongest effect (Fig. 2d). Indeed, simultaneous reversions of P69 and L70 to their Ub counterparts (Ubv08-DM) completely abolished UbvG08 binding to the 53BP1 Tudor domain, as measured by ITC (Fig. 1g). In a converse set of experiments, simultaneous mutation of the equivalent residues in Ub into their UbvG08 counterparts were sufficient to convert Ub into a robust 53BP1-binding protein, as measured in pulldown assays (Fig. 2e). We also assessed the importance of the non-substituted (i.e. same as Ub) residues in UbvG08 (Fig. 2f) as well as the residues on the 53BP1 Tudor domain predicted by our model to be engaged in key interactions (Supplementary Fig. 2ab). These analyses strongly validated the structural model of the UbvG08-53BP1 interaction.
We next tested whether intracellular expression of UbvG08 inhibits 53BP1 in cells. We prepared Flag-tagged versions of UbvG08 and the DM mutant. The C-terminal di-glycine motif was removed to preclude its incorporation in the active ubiquitin pool and we also incorporated a I44A mutation, which disables the majority of ubiquitin-dependent interactions11 but does not impact the interaction of UbvG08 with 53BP1 (Fig. 2d). This version of Ubv-G08 is referred to hereafter as inhibitor of 53BP1 or i53 for reasons that will become apparent below.
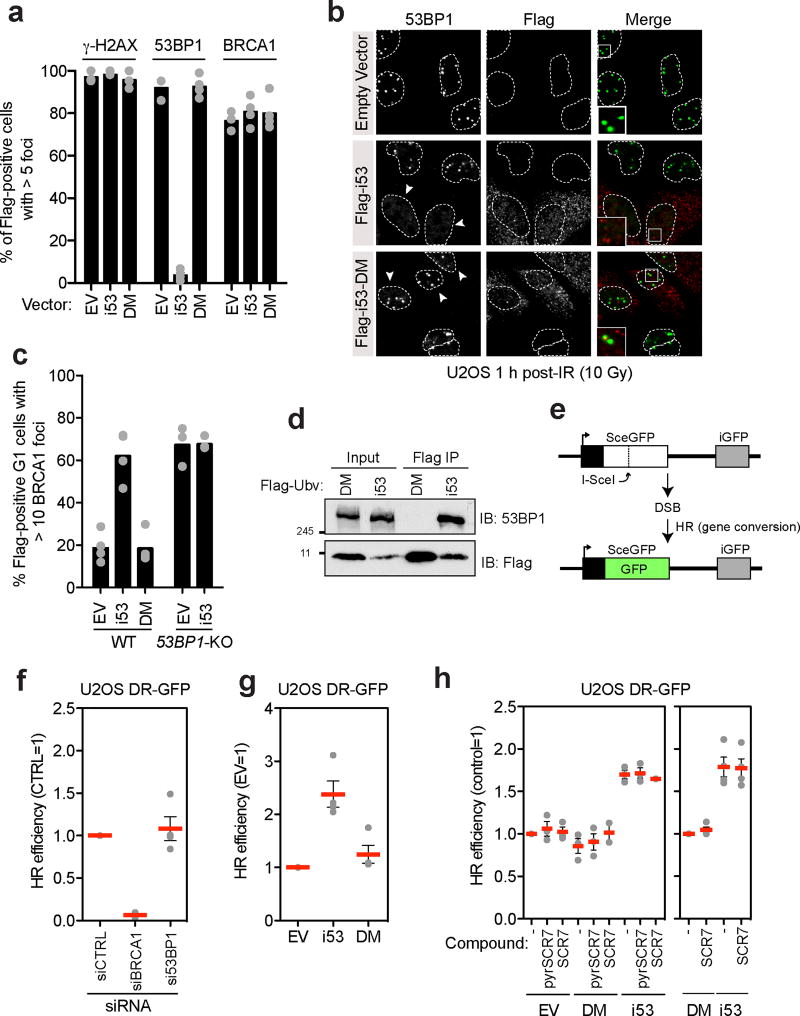
When U-2-OS (U2OS) cells transfected with vectors expressing i53 or its DM mutant were irradiated with a 10 Gy dose of X-rays, we observed that i53 but not the 53BP1-binding defective DM mutant strongly suppressed 53BP1 recruitment to DSB sites, as monitored by ionizing radiation focus formation (Fig. 3a,b). The inhibition of focus formation was specific to 53BP1, as i53 did not impact γ-H2AX and BRCA1 focus formation (Fig. 3a and Supplementary Fig. 3a). Transfection of i53 also induced BRCA1 accumulation at DSB sites in G1 cells7 to a similar extent as that caused by loss of 53BP17, 12, providing a first clue that i53 not only inhibits 53BP1 recruitment to damaged chromatin but also acts as an inhibitor of 53BP1 function (Fig. 3c and Supplementary Fig. 3b). i53, but not its DM mutant efficiently retrieved 53BP1 in co-immunoprecipitation experiments (Fig. 3d) suggesting that the inhibition of 53BP1 occurs through occlusion of the Tudor domain ligand binding site.
Figure 3. The i53 protein inhibits 53BP1 and activates gene conversion.
a–b, U2OS cells were transfected with vectors expressing i53, its 53BP1-binding deficient mutant (DM) or an empty vector (EV) control. Cells were then X-irradiated with a 10 Gy dose and processed for immunofluorescence with the indicated antibodies 1 h post-irradiation (IR). DAPI staining (not shown) was used to delineate the outline (dashed lines) of the cell nuclei. The region in the magnified inset is indicated with a square. Quantitation of the experiment is shown in panel (a) where each circle is a biological replicate and the bar is at the mean (N=3 for EV, N=4 for i53 and i53-DM), whereas in (b) representative micrographs are shown. Arrowheads indicate Flag-positive cells. Additional micrographs are shown in Supplementary Fig. 3a. c, Parental or 53BP1ΔU2OS cells transfected with vectors expressing i53, the DM mutant or an empty vector (EV) control were irradiated (2 Gy) 1 h before being processed for immunofluorescence. Cell cycle stage was assessed by Cyclin A staining. Each circle represents a biological replicate and the bar is at the mean; N=4 (U2OS) and N=3 (53BP1ΔU2OS). Micrographs are shown in Supplementary Fig. 3b. d, Immunoprecipitation (IP) of Flag-tagged proteins from extracts prepared from 293T cells transfected with vectors expressing Flag-i53 or the i53-DM mutant. Proteins were separated by SDS-PAGE and immunoblotted (IB) for Flag and 53BP1. e, Schematic of the DR-GFP assay. f. U2OS DR-GFP were first transfected with siRNAs targeting the 53BP1 or BRCA1 mRNAs along with a non-targeting siRNA (CTRL). 24 h post-transfection, cells were transfected with the I-SceI expression vector and the percentage of GFP-positive cells was determined 48 h post-I-SceI transfection for each condition. The values were normalized to the CTRL siRNA condition. Each point is a biological replicate and the bar is at the mean ± s.e.m; N=4. g, U2OS DR-GFP cells were transfected with the vectors expressing i53, the DM mutant or an empty vector control (EV) along with an I-SceI expression vector. The percentage of GFP-positive cells was determined 48 h post- transfection for each condition and was normalized to the empty vector condition. Each point is a biological replicate and the bar is at the mean ± s.e.m; N=4. h, U2OS DR-GFP cells were transfected with either an empty vector (EV) or vectors expressing Flag-tagged i53 or the DM mutant along with an I-SceI expression vector. Cells were treated either with DMSO (−) 1 µM SCR7 or 1 µM of the SCR7 pyrazine analog (pyrSCR7). The percentage of GFP-positive cells was determined 48 h post-transfection for each condition and was normalized either to the EV (left) or DM (right) conditions. Each point is a biological replicate and the bar is at the mean ± s.e.m (N=3 for all experiments on the left graph, except for SCR7+DM and SCR7+i53 where N=2; N=4 for all experiments on the right graph). Please note that the blots shown in panel d are cropped and uncropped version can be found as Supplementary Information.
Although UbvG08, the parent molecule of i53, shows a high degree of selectivity towards 53BP1 in ELISA assays (Fig. 1b), we determined the repertoire of cellular proteins bound by i53. We generated 293T Flp-In/T-Rex cell lines that expressed Flag-tagged i53 or i53-DM under the control of a tetracycline-inducible promoter as previously described13. Nine IP-MS experiments were analyzed (3 biological replicate IPs each for control, i53- and i53-DM expressing cell lines). The only protein found to interact with i53 in two or more experiments was 53BP1 (Supplementary Table 2). We conclude that i53 is a selective binder of 53BP1 in cells.
Loss of 53BP1 results in increased HDR levels14, making inhibitors of 53BP1 potential tools to manipulate DSB repair pathways during genome engineering reactions. However, the depletion of 53BP1 by siRNA, although near complete as determined by immunoblotting (Supplementary Fig. 4a), is often insufficient to induce HDR in the well-characterized direct-repeat (DR)-GFP assay15 (Fig. 3e,f). We therefore tested whether i53 impacted gene conversion frequency and observed that i53 led to a 2.4-fold (+/−0.25) increase in gene conversion when compared to the empty vector control, whereas the i53-DM mutant had virtually no impact on gene conversion (1.25-fold +/−0.17; Fig. 3g and Supplementary Fig. 4b). As a point of comparison, we compared i53 to SCR7, the reported inhibitor of the NHEJ factor DNA ligase IV16, which has been shown in some systems to increase homology-dependent repair17, 18. We also tested its related pyrazine analog, which has been proposed to be the active SCR7 analog (https://www.tocris.com/dispprod.php?ItemId=432017#.VvUhqt-rSRs). Under our experimental conditions, i53 was a more potent inducer of gene conversion, compared to both SCR7 and to SCR7 pyrazine, which had minimal impact in this assay (Fig. 3h and Supplementary Fig. 4c).
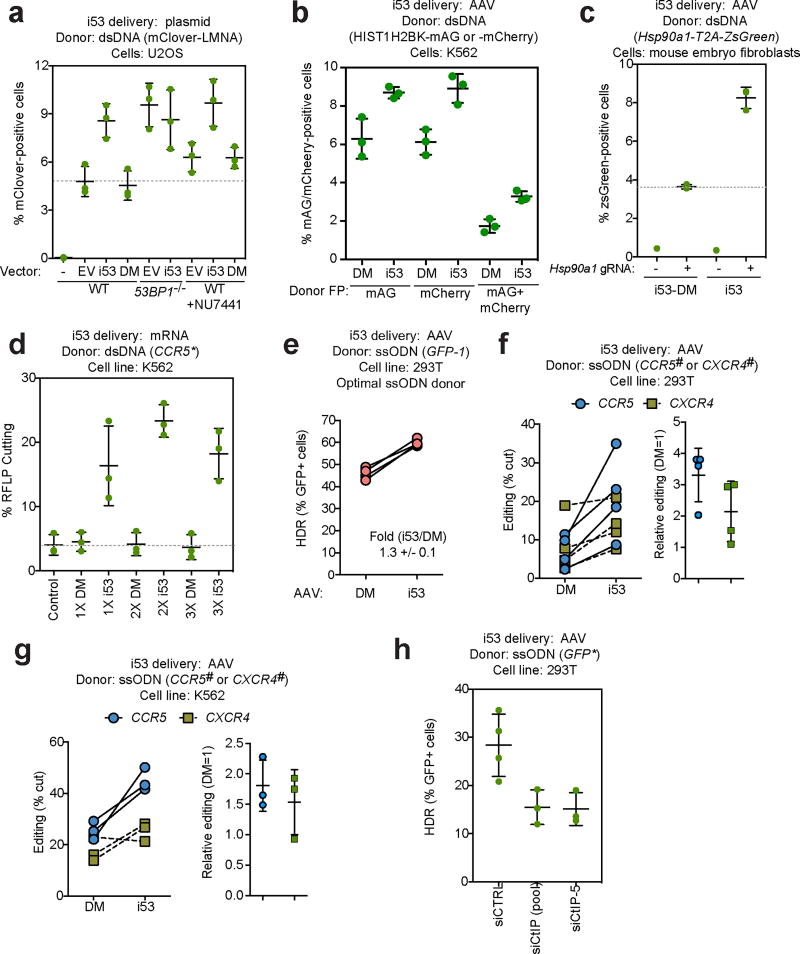
As an orthogonal approach, we next tested whether i53 expression increased the efficiency of gene targeting stimulated by CRISPR/Cas9, in assays that involve the introduction of the coding sequences of fluorescent proteins such as mClover, mAG or mCherry in frame with the coding regions of Lamin A (LMNA)12, 19 or histone H2B (HIST1H2BK)20. Precise HDR results in the expression of fluoresecent protein fusions that can be quantitated by flow cytometry. We first assessed gene targeting at the LMNA locus in U2OS cells, which are not responsive to SCR7 treatment19, suggesting that end-joining may not provide a strong barrier to HR at this locus. Similarly, inhibition of DNA-PK, a core NHEJ factor, with NU7441 only resulted in a 1.3-fold increase in gene targeting in this assay (Fig. 4a). However, we observed that i53, but not the DM mutant, increased gene-targeting nearly two-fold (from 4.8% +/− 0.5% for the empty vector control to 8.6% +/− 0.6% for the i53 condition). The gene-targeting efficiency in i53-expressing cells approached that of 53BP1-null cells (53BP1-KO)12, suggesting that the inhibition of 53BP1 was near complete. Introduction of i53 in 53BP1−/− cells did not result in a further increase in gene targeting, demonstrating that the effect of i53 on HR is via inhibition of 53BP1. Combining DNA-PK inhibition and i53 only modestly increased gene targeting, in line with the function of 53BP1 in promoting NHEJ.
Figure 4. Stimulation of HDR by i53 with dsDNA and ssODN donors.
a, Gene targeting efficiency at the LMNA locus12, 19 in parental or 53BP1Δ U2OS cells following transfection with vectors expressing Flag-tagged i53 or its DM mutant or an empty vector control (EV). The DNA-PK inhibitor NU7441 was also added where indicated. 24 h post-transfection, cells were analysed for mClover fluorescence. Individual experiments are presented along with the mean +/− s.d., (N=3). b, Mono- and bi-allelic gene targeting at the HIST1HB2K locus20 in K562 cells previously transduced with AAV coding for i53 or i53-DM (DM). The HIST1HB2K-mAG and HIST1HB2K-mCherry donors were introduced by nucleofection at the same time as a Cas9 RNP targeting HIST1HB2K. Control reactions where 72 h post-transfection, cells were analysed for mAG and mCherry fluorescence. Individual experiments are presented along with the mean +/− s.d., (N=3). c, Gene targeting efficiency at the Hsp90a1 locus in mouse embryo fibroblasts previously transduced with AAV coding for i53 or i53-DM (DM). The Hsp90a1-t2A-ZsGreen template vector was introduced by transfection at the same time as the vector coding for the sgRNA and Cas9. 8 days post-transfection, cells were analysed for ZsGreen fluorescence. Individual experiments are presented along with the mean +/− s.d., (N=3). The no sgRNA controls were only done once. d, HDR at the CCR5 locus22 in K562 cells nucleofected with ZFN mRNA and an XhoI inserting CCR5 plasmid donor (CCR5*), followed by nucleofection 24 h later with increasing concentrations of i53 mRNA or i53-DM (DM) mRNA, or left untreated (control). 24 h later, cells were collected and gene targeting was determined by RFLP analysis. Individual experiments are presented along with the mean +/− s.d., (N=3). e, BFP-to-GFP conversion by ssODN-mediated HDR in 293T cells previously transduced with AAV coding for i53 or i53-DM (DM). An optimal ssODN donor targeting GFP (GFP-1)26 and Cas9 RNP were then nucleofected and 96 h post-transfection, cells were analysed for GFP and BFP expression. Individual paired experiments are presented (N=4). f, g, HDR at the CCR5 and CXCR4 loci in 293T (f) and K562 (g) cells previously transduced with AAV coding for i53 or i53-DM (DM). ssODN donor stargeting CCR5 (CCR5#) or CXCR4 (CXCR4#)26 and Cas9 RNP were then nucleofected and 72 h post-transfection, editing was determined by RFLP analysis. Individual paired experiments are presented on the left graphs whereas the relative editing efficiency is presented in the right graphs where data is presented as the mean +/− s.d., (N=4 for f and N=3 for g). h, BFP-to-GFP conversion by ssODN-mediated HDR in 293T cells was carried out as panel e with the exception that cells were either transfected with a non-targeting siRNA (siCTRL), a pool of siRNAs targeting CtIP (siCtIP (pool)) or a different, custom-designed siRNA targeting CtIP (siCtIP-5). Individual experiments are presented along with the mean +/− s.d., (N=4 for siCTRL, N=3 for siCTIP conditions).
The experiments above were undertaken with i53 delivered using plasmid transfection. Since adeno-associated virus (AAV)-mediated gene delivery is a method of choice for the delivery of gene editing components in vivo, we tested whether AAV-mediated delivery of i53 also stimulates HDR. AAV-delivered i53 stimulated gene conversion by 2.5-fold over DM in U2OS cells using DR-GFP assays (Supplementary Fig. 4d,e), indicating that sufficient i53 levels can be achieved by AAV delivery. We next used AAV-mediated i53 delivery to assess its ability to stimulate homology-driven insertion of the coding sequence for the fluorescent protein mAG at the 3’ end of the HIST1H2BK open reading frame. In those experiments, Cas9-sgRNA ribonucleoprotein (RNP) complexes and dsDNA plasmid donors were nucleofected into 293T or K562 cells. i53 stimulated gene targeting at the HIST1H2BK locus in both 293T and K562 cells (1.3-fold and 1.8-fold, respectively), with the extent of stimulation being more pronounced in K562 cells (Supplementary Fig. 5a,b). The simultaneous nucleofection of donors inserting either mAG or mCherry at the HIST1H2BK locus next allowed us to estimate bi-allelic modification of this locus by monitoring cells that simultaneously express both fluorescent proteins20. We observed that i53 stimulated bi-allelic gene targeting 1.9-fold at the HIST1H2BK locus in K562 cells (Fig 4b and Supplementary Fig. 5c).
In parallel, we also assessed if AAV-delivered i53 could stimulate HDR in mouse cells. We employed a gene-targeting assay conceptually similar to those described above where the incorporation of a t2A-ZsGreen cassette at the Hsp90ab1 locus is monitored by flow cytometry. We found that i53 stimulated gene-targeting in mouse embryo fibroblasts 2.3-fold over DM (Fig 4c and Supplementary Fig. 6) indicating that AAV-mediated i53 stimulates HDR in both human and mouse cells.
In addition to AAV-mediated delivery, delivery of mRNA is another means for introducing gene-editing components in primary cells21, 22. We therefore tested whether electroporation of an i53-coding mRNA stimulated zinc finger nuclease (ZFN)-mediated gene targeting at the CCR5 locus in K562 using a dsDNA donor used previously22, 23. We found that mRNA delivery of i53 robustly increased gene targeting stimulated by a ZFN up to 5.6-fold over DM (Fig 4d) and resulted in a corresponding decrease of indels at the site of cleavage, as detected by the CEL1 nuclease (Supplementary Fig. 7a). These results indicate that i53-mediated stimulation of HDR is independent of nuclease or delivery methods, and they suggest that i53 acts by skewing DSB repair pathway choice towards HDR.
Canonical, RAD51-dependent HDR is the dominant pathway for gene conversion with large dsDNA donors24. In contrast, HDR using single-stranded oligonucleotide (ssODN) donors involves a poorly understood RAD51-independent annealing-type mechanism25. We therefore examined the impact of i53 on the ssODN-mediated conversion of GFP to BFP in 293T and MCF10a cells following a Cas9-induced DSB, as previously described26 (Fig 4e and Supplementary Fig. 7b,c). We observed, with both optimal and suboptimal ssODN donors, that i53 stimulated HDR in this assay by approximately 30%–40%, depending on the donor and cell line used. We next modified two endogenous loci, CXCR4 and CCR5, to introduce new restriction endonuclease sites by ssODN-mediated HDR in both 293T and K562 cells. We observed that i53 stimulated HDR with ssODN donors up to 3.3 (+/− 0.8) fold in 293T cells (Fig 4f and Supplementary Fig. 7d,e), with a concomitant decrease in indel formation at the same loci (Supplementary Fig. 7f). These data indicate that i53 stimulates HDR with ssODN donors at multiple loci and cell types. Notably, these data also suggested that DNA end resection plays an important role in ssODN-mediated HDR. In support of this idea, depletion of CtIP, a key end-resection factor4, reduced HDR by ssODN in 293T cells by nearly 50% in the GFP-to-BFP conversion assay (Fig 4h and Supplementary Fig. 8a,b).
DNA end resection inhibits NHEJ but can activate alternative end-joining pathways in addition to activating HR27. Resection can reveal regions of microhomology that may be rejoined in a process termed microhomology-mediated end joining (MMEJ)27. The impact of 53BP1 on this repair process is unclear due to conflicting results28, 29. To assess whether i53 increases MMEJ, we employed the EJ2-GFP reporter assay30, 31. We found that i53 expression increased MMEJ (1.4 +/− 0.2 fold over the empty vector; Supplementary Fig. 8c,d) but because the expression of the DM mutant also increased MMEJ to a similar extent (1.3 +/− 0.1 fold), it is unlikely that the increase was due to 53BP1 inhibition.
In summary, we report the development of a genetically encoded inhibitor of 53BP1 that robustly stimulates canonical and ssODN-mediated homology-directed repair of DSBs. In addition to gene targeting applications, i53 could be useful in additional gene editing reactions e.g. interparalog gene conversion reactions. Notably, i53 expression does not impact spontaneous or DNA damage-stimulated sister chromatid exchanges (SCEs), a potential threat to genome stability that occurs as a consequence of crossover reactions (Supplementary Fig. 8e–h).
The 53BP1 Tudor domain is highly conserved across a wide range of vertebrate species including agriculturally important animals such as pigs and cows; and we have shown that i53 can also stimulate HDR in mouse cells. Thus we expect that i53 could be used to stimulate HR in other species as well.
The observation that i53 stimulates HDR by ssODNs led us to ask whether CtIP, a key resection factor, also promotes this type of gene editing reaction. This is in line with the annealing-driven strand synthesis (ADSS) and single stranded template repair (SSTR) models of HDR by ssODNs25, 32, which both suggest that end resection creates annealing targets for oligonucleotides. Notably, CtIP depletion only led to a ~50% reduction in HDR by ssODN raising the possibility that multiple HDR pathway operate at DSBs or that Cas9-mediated cleavage might engage CtIP-independent resection. Deciphering how these pathways operate should suggest new opportunities for intervention and synergy with 53BP1 inhibition.
The versatility of the ubiquitin scaffold onto which i53 is built; along with the determination of the molecular basis of the i53-53BP1 interaction should enable us to improve 53BP1 inhibition either through protein engineering or through affinity maturation of the UbvG08 via additional rounds of mutagenesis and phage display selections. We therefore propose that 53BP1 inhibition could be a propitious alternative for boosting HDR rates, which can also be used in combination with other methods, such as the recently described marker-free co-selection strategy based on the modification of the ATP1A1 gene20.
ONLINE METHODS
Cell culture and treatments
U-2-OS (U2OS) and 293T cells were obtained from ATCC. 293T and HEK293 Flp-In/T-REx cells (Invitrogen) were propagated in DMEM medium supplemented with 10% fetal bovine serum (FBS, Gibco) and 2 mM L-alanyl-L-glutamine, and were maintained in a 37 °C and 5% CO2 atmosphere. U2OS cells were grown in McCoy’s medium supplemented with 10% FBS. U2OS DR-GFP and EJ2-GFP cells were a gift of Jeremy Stark. 53BP1Δ U2OS and U2OS cell lines stably expressing CtIP-T847E were previously described12. MEFs were cultured in Dulbecco’s Modified Eagle Medium (DMEM) supplemented with 10% fetal bovine serum (FBS) (Gibco), 2 mM L-glutamine (Sigma), 100 U/ml penicillin (Sigma), 0.1 g/ml streptomycin (Sigma), 0.1 mM non-essential amino acids (Invitrogen), and 1 mM sodium pyruvate (Sigma).
RPE1 hTERT cells were obtained from ATCC and maintained in DMEM + 10% FCS. A Flag-Cas9-2A-Blast expression cassette was integrated as described before33. Upon single clone selection, cells were maintained in the presence of 2 µg/mL blasticidin. The TP53 gene was knocked-out using transient transfection of the LentiGuide plasmid with Lipofectamine. 24 h post-transfection, cells were selected for 24 h with 15 µg/mL puromycin, followed by a 5-day recovery and 48 h selection with 10 µM of the MDM2 inhibitor Nutlin-3 (Cayman Chemical) after which single clones were isolated and verified for loss of p53 protein. Furthermore, CRISPR-generated indel mutations in the TP53 gene were verified by PCR amplification of the region surrounding the sgRNA target sequence, cloning of products into the pCR2.1 TOPO vector (TOPO TA Cloning kit, Thermo Fisher Scientific) and Sanger sequencing of individual bacterial clones (forward PCR-primer: GCATTGAAGTCTCATGGAAGC, reverse PCR-primer: TCACTGCCATGGAGGAGC). 53BP1 and/or BRCA1 gene knockouts were generated by electroporation of the respective LentiGuide vectors (Lonza Amaxa II Nucleofector, program T-023, 5 µg plasmid per 700,000 cells). 24 h post transfection, cells were selected for 24 hr with 15 µg/mL puromycin, followed by single clone isolation. The double 53BP1/BRCA1Δ cell line was created by deleting BRCA1 from the 53BP1 single knock-out cell line. Gene mutations were further confirmed by PCR amplification and sequencing as described above for TP53 (53BP1 forward PCR-primer: CCAGCACCAACAAGAGC, 53BP1 reverse PCR-primer: GGATGCCTGGTACTGTTTGG, BRCA1 forward PCR-primer: TCTCAAAGTATTTCATTTTCTTGGTGCC, BRCA1 reverse PCR-primer: TGAGCAAGGATCATAAAATGTTGG). Retrovirus of GFP (IRES-GFP), i53-IRES-GFP and DM-IRES-GFP was generated in 293T cells by transient transfection of the pMX-IRES-GFP vector together with the packaging vectors VSVG and Gag-Pol using LT1 transfection reagent (Mirus). Supernatants containing retrovirus were collected and filtered through 0.45 µm filters. RPE1 cells were transduced in two hits (24 h apart) to an MOI of ≈0.8 in the presence of 8 µg/mL polybrene and sorted for GFP 72 h after the second hit. All cells were >97% positive for GFP throughout the experiments, as based on FACS analysis. All cell lines tested negative for mycoplasma contamination and the identity of cell lines confirmed by STR analysis.
Plasmids
The phagemid (DDp2235) from the UbvG08 phage was obtained from the ubiquitin variant library described in6; see below for details. The UbvG08 open reading frame (ORF) lacking the C-terminal di-Gly residues was cloned into a pDONR vector using a product from PCR amplification of the phagemid template and Gateway recombination, yielding plasmid DDp2251 (UbvG08 ΔGG). The pETM-30-2-GST-UbvG08 (DDp2186) and pETM30-2-GST-ubiquitin (DDp2192) were cloned following PCR amplification from the UbvG08ΔGG or UbΔGG ORFs, respectively. The constructs encoding His6-GST-TEV and MBP fusions of 53BP1 Tudor-UDR (residues 1484–1631) and Tudor (residues 1484–1603) domains were described previously in9. The I44A mutation was introduced into DDp2186, which was then used as a template for amplification of the modified Ubv by PCR. The PCR product was cloned into the BamHI and NotI sites of a pcDNA3-Flag plasmid to yield pcDNA3-Flag-i53 (DDp2534). The BamHI-NotI fragment of DDp2534 was subsequently cloned into a pcDNA5-Flag-FRT/TO Flag vector to yield plasmid DDp2535. All other plasmids were generated by site-directed mutagenesis carried out by Quikchange (Agilent). The lentiviral vector coding for a siRNA-resistant Flag-tagged CtIP T847E construct was previously described12. The plasmids used for the LMNA assay were gifts of G. Dellaire19.
Single guide (sg)RNAs targeting TP53 (CAGAATGCAAGAAGCCCAGA), BRCA1 (AAGGGTAGCTGTTAGAAGGC) and 53BP1 (TCCAATCCTGAACAAACAGC) were cloned into lentiGuide-Puro (Addgene: #52963) as described34. The i53 and DM lentiviral expression vectors were prepared by PCR amplification that also introduced sequences coding for an N-terminal HA-tag and flanking PacI and NotI restriction sites. The PCR products were cloned in the PacI and NotI sites of pMX-IRES-GFP (a gift from A. Nussenzweig, National Institutes of Health). The Lenti-Cas9-2A-Blast construct was a gift from J. Moffat (University of Toronto). All constructs were sequence-verified.
To prepare AAV-i53 and i53-DM vectors, the region comprising Flag-i53 (or i53-DM) was PCR-amplified from pcDNA3-i53 and pcDNA3-i53-DM with the addition of a 5’ ClaI site and 3’ HindIII site. The GFP insert was excised from pAAV-GFP (Cell Biolabs) and replaced with the ClaI- and HindIII-flanked Flag-UbV PCR products to produce pAAV-Flag-i53-DM and pAAV-Flag-i53, which were verified by diagnostic digests and sequencing.
To facilitate monitoring of mono- versus bi-allelic editing, the mAG coding sequence in the HISTH2BK-mAG donor plasmid (gift of Y. Doyon) was exchanged for mCherry. The mCherry gene was PCR amplified from a pCDNA5-FRT/TO-mCherry plasmid, flanked by a 5’ KpnI site, and a 3’ BsrGI site. The reverse primer simultaneously served to silently mutate a naturally occurring BsrGI site at the 3’ end of the mCherry sequence. The PCR product and HISTH2BK-mAG1 plasmid were digested with KpnI and BsrGI, and gel-purified. The PCR fragment was then ligated into the HISTH2BK donor vector, and clones were screened by diagnostic digest, and confirmed by sequencing.
The vectors pcDNA3-Flag-i53 (#74939), pcDNA3-Flag-i53-DM (#74940), pAAV-Flag-i53 (#92170) and pAAV-Flag-i53-DM (#92171) have been deposited at Addgene.
Selection of and purification of the 53BP1-binding ubiquitin variants
The phage-displayed Ubv library used in this study was re-amplified from Library 2 as previously described6. Protein immobilization and subsequent phage selections were performed according to established protocols35. Briefly, purified 53BP1 protein fragments were coated on 96-well MaxiSorp plates (Thermo Scientific 12565135) by adding 100 µL of 1 µM proteins and incubating overnight at 4 °C. Afterwards, five rounds of selection using the phage-displayed Ubv library were performed against immobilized proteins. A total of 96 phage clones obtained from the fourth and the fifth round of binding selections (48 from each round) were subjected to clonal ELISA to identify individual phages that bound to 53BP1. The sequences of phage-displayed Ubvs were derived by sequencing of the phagemid DNA35. For phage ELISA, proteins in study (53BP1 and/or control proteins) were immobilized on 384-well MaxiSorp plates (Thermo Scientific 12665347) by adding 30 µL of 1 µM proteins for overnight incubation at 4 °C before adding amplified phages (1:3 dilution in PBS + 1% BSA + 0.05% Tween) and incubated overnight. Binding of phage was detected using anti-M13-HRP antibody (GE Healthcare 27942101).
Pulldowns
MBP and GST pulldowns were done essentially as described in ref9 with the modifications described below. We used the following buffer for the binding reactions: 50 mM Tris-Cl pH 7.5, 50 mM NaCl, 0.01% NP40 and 1% BSA. We also used 2.5 µg of the MBP- and GST-fusion proteins as baits. For peptide competition pulldowns 2.5 µg MBP-53BP1-Tudor was coupled to amylose resin (New England Biolabs) and 0.75 µg GST-UbvG08 was added simultaneously to a biotin-labeled peptide derived from histone H4K20me2 (Biotin-Mini-PEG-YGKGGAKRHRKme2VLRD; BioBasic Canada Inc.) for 2 h at 4°C. Peptide pulldowns were washed in binding buffer, eluted with SDS-PAGE sample buffer, and analyzed by immunoblotting. For all pulldowns, 1–2% of the total amount of the input proteins was separated by SDS-PAGE and probed for immunoblotting.
Protein expression, crystallization and structure determination
The 53BP1 Tudor domain (residues 1784–1603) and UbvG08 were individually expressed and purified from bacteria as GST-tagged fusion proteins. In brief, GST-tagged fusion proteins were purified from bacterial lysates on to glutathione-Sepharose (GE Healthcare), washed, and then eluted by TEV protease digestion to GST moieties, followed by purification by size exclusion chromatography (SEC). The 53BP1 Tudor-UbvG08 complex was formed by mixing purified proteins at equimolar concentration, incubating overnight at 4 °C, and purifying the complex by SEC in 10 mM Tris-Cl pH 7.5, 150 mM NaCl and 1 mM DTT column buffer. Crystals of the complex were grown at 20 °C using the hanging drop vapor diffusion method by mixing equal volumes (1 µL) of complex at 28.5 mg/ml with crystallization buffer consisting of 0.1 M MES pH 6.0, 0.2 M trimethylamine N-oxide and 25% (w/v) PEG MME 2000. Crystals were cryo-protected by a quick soak in crystallization buffer supplemented with 20% glycerol, prior to flash freezing. A single crystal dataset was collected at −180 °C on a home-source consisting of a Rigaku MicroMax-007 HF rotating anode generator, coupled to a R-axis 4++ detector (Rigaku) and VariMax multilayer optics. Data processing was performed using the XDS software suite. The structure of a single 53BP1 Tudor-UbvG08 complex in the asymmetric unit was solved by molecular replacement using the apo Tudor domain (PDB 2IG0) and ubiquitin (PDB 3NHE chain B) as search models in Phaser (Phenix suite). Structure refinement was performed using Refine (Phenix suite). See Supplementary Table 1 for data collection and refinement statistics.
Immunoprecipitation
293T cells were transfected with 10 µg of pcDNA3-Flag-i53-derived plasmids using polyethylenimine (PEI). 48 h post-transfection, cells were lysed in 1 mL high salt lysis buffer (50 mM Tris-HCl pH 7.6, 300 mM NaCl, 1 mM EDTA, 1% (v/v) Triton X-100, and 1× protease inhibitors (Complete, EDTA-free, Roche)) and cell lysates were clarified by centrifugation at 4 °C. 100 µL was removed as the input sample. The remaining lysate was incubated with ~15 µL anti-Flag (M2) affinity gel (Sigma) for 2 h at 4 °C. The immunoprecipitates were then washed twice with high salt lysis buffer, once with 50 mM Tris-HCl pH 8.0, 0.1 mM EDTA and eluted in 25 µL 2× Laemmli sample buffer for analysis by immunoblotting.
Antibodies
We employed the following antibodies: rabbit anti-53BP1 (A300-273A, Bethyl), mouse anti-γ-H2AX (clone JBW301, Millipore), mouse anti-53BP1 (#612523, BD Biosciences), rabbit anti-GST (sc-459, Santa Cruz), a mouse anti-HA (F-7, sc-7392, SantaCruz or clone 12CA5, gift from M. Tyers, University of Montréal), mouse anti-MBP (E8032S, NEB), mouse anti-Flag (clone M2, Sigma), rabbit anti-Flag (#2368, Cell Signaling), mouse anti-tubulin (clone DM1A, Calbiochem), mouse anti-p53 (sc-126, Santa Cruz), rabbit anti-ubiquitin (Z0458, DAKO), rabbit anti-BRCA1 (#07-434, Millipore or home-made antibody7). Goat anti-GFP (gift from L. Pelletier, Lunenfeld-Tanenbaum Research Institute), HRP-conjugated AffiniPure goat anti-rabbit IgG (Jackson ImmunoResearch), HRP-linked sheep anti-mouse IgG (NA931, GE Healthcare). Alexa Fluor 488 goat anti-mouse and anti-rabbit IgG, Alexa Fluor 555 goat anti-mouse and anti-rabbit (MolecularProbes).
RNA interference
All siRNAs employed in this study were single duplex siRNAs purchased from ThermoFisher. RNA interference (RNAi) transfections were performed using Lipofectamine RNAiMax (Invitrogen) in a forward transfection mode. The individual siRNA duplexes used were BRCA1 (D-003461-05), CtIP/RBBP8 (M-001376-00), 53BP1/T53BP1 (D-003549-01), KEAP1 (D-12453-02) or 53BP1/T53BP1 (D-003548-01), non-targeting control siRNA (D-001210-02). Except when stated otherwise, siRNAs were transfected 48 h before cell processing.
Inhibitors and fine chemicals
The following drugs and chemicals were used: DNA-PKcs inhibitor (NU7441; Genetex) at 10 µM, lovastatin (S2061; Selleck Chemicals) at 40 µM, doxycycline (#8634-1; Clontech), SCR7 (M60082-2; Xcessbio) at 1 µM. Olaparib was purchased from Selleck Chemicals.
AAV production
The viral supernatant of AAV293 cells (Agilent Technologies) was collected and used for infections 48 h after co-transfection with 3.5 µg each of expression plasmid, pDJ and pHelper (Cell BioLabs). For MEFs, the viral supernatant was first concentrated in Amicon Ultra-15 Centrifugal Filter Unit with Ultracel-100 membrane.
Preparation of RNPs
Purified SpCas9 was diluted to a concentration of 3.2 µg/µl in Cas9 buffer (20 mM HEPES (pH 7.5), 150 mM KCl, 1 mM MgCl2, 10% glycerol and 1 mM TCEP), and sgRNA was diluted to a concentration of 0.8 µg/µl in Cas9 buffer. 5 µl of diluted Cas9 was slowly mixed into 5 µl of diluted sgRNA, then incubated for 10–20 minutes at room temperature.
Immunofluorescence microscopy
Cells were grown on glass coverslips, fixed with 2% (w/v) paraformaldehyde in PBS for 20 min at room temperature, permeabilized with 0.3% (v/v) Triton X-100 for 20 min at room temperature and blocked with 5% BSA in PBS for 30 min at room temperature. Cells were then incubated with the primary antibody diluted in PBS-BSA for 2 h at room temperature. Cells were next washed with PBS and then incubated with secondary antibodies diluted in PBS-BSA supplemented with 0.8 µg ml−1 of DAPI (Sigma) to stain DNA for 1 h at room temperature. The coverslips were mounted onto glass slides with Prolong Gold mounting agent (Invitrogen). Confocal images were taken using a Zeiss LSM780 laser-scanning microscope.
Reporter-based DNA repair assays
The direct repeat (DR)-GFP assay to measure the frequency of HR and the strand annealing EJ2-GFP assay to measure the frequency of MMEJ were performed as previously described30. Briefly, U2OS DR-GFP or U2OS EJ2-GFP cells were transfected with 10 nM siRNA (Dharmacon) using Lipofectamine RNAiMAX (Invitrogen). 24 h later, the cells were transfected with the pCBASceI plasmid (Addgene #26477) and plasmids, using Lipofectamine 2000 (Invitrogen). 48 h post-plasmid transfection, the cells were trypsinized and the percentage of GFP-expressing cells was analyzed using the BD FACSCalibur flow cytometer.
The Lamin A (LMNA) assay to measure the frequency of introduction of the coding sequence for mClover at the 5’ end of LMNA using the CRISPR/Cas9 was performed as previously described12. Parental or 53BP1Δ U2OS cell lines were transfected with the indicated plasmids using Lipofectamine RNAiMAX (Invitrogen). 24 h later, the cells were electroporated with 2.5 µg of sgRNA plasmids and 2.5 µg of donor template using a Nucleofector (Lonza; protocol X-001). Under those condition, omission of the sgRNA gives negligible levels of mClover-positive cells12. Parental or 53BP1Δ U2OS cells stably expressing CtIP-T847E mutant were transfected with an siRNA against KEAP1 and the indicated plasmids and processed as previously described12.
A FACS-based gene targeting assay was developed to monitor the targeting efficiency of a Zsgreen reporter at the 3’end of Hsp90 locus. A homology repair template plasmid was generated with the coding sequence of Zsgreen and homology arms (600 base pairs) that correspond to sequence flanking the Hsp90 STOP codon. MEFs transduced with AAV i53 and control cells expressing DM were co-transfected with a repair template plasmid and a plasmid expressing Cas9 (pX330-U6-Chimeric_BB-CBh-hSpCas9) and a gRNA (5’-CGATGAGGATGCCTCGCGCA-3’). One million cells were transfected with 200ng of Cas9/gRNA plasmid and 800ng of template plasmid using Lipofectamine 3000 (Invitrogen) according to the manufacturer’s instruction. 16 hours later, cells were selected with puromycin (2 µg/ml) for 48 hours to enrich for Cas9-expressing cells. To determine the percentage of Zsgreen positive cells, cells were by analyzed by flow cytometry using a FACS Aria III cell sorter 8 days post-transfection.
The HISTH2BK (H2B) targeting assay was adapted from ref20 for use with Cas9 RNPs. For the targeting assay, 1.25 µg of plasmid donor was used per nucleofection.
BFP-to-GFP ssODN-based HDR assay
293T cells were transduced at a low MOI (<0.3) with a lentivirus expressing BFP under the control of an EF1α promoter (Addgene #71825) and sorted by flow cytometry to produce a pure population of BFP-expressing cells. One day post-AAV transduction, 2 × 105 cells were resuspended in 20 µl SF buffer (Lonza) and nucleofected with 10 µl sgBFP RNP and 100 pmol of ssODN donor, using program DS-150 on a Nucleofector 96-well Shuttle system (Lonza). Cells were then transferred to a 12-well plate containing pre-warmed medium, and grown for 4 days. BFP and GFP fluorescence were measured by flow cytometry on a BD Fortessa, and the results were analyzed using FlowJo v10 software.
ssODN-based RFLP HDR assay
The day post-AAV transduction 2 × 105 cells were resuspended in 20 µl SF buffer (Lonza) and nucleofected along with 10 µl sgCCR5 or sgCXCR4 RNPs and 100 pmol of ssODN donor, using program FF-120 (for K562 cells) or DS-150 (for 293T cells), respectively, on a Nucleofector 96-well Shuttle system (Lonza). Cells were collected 3d post-nucleofection and their genomic DNA isolated (Qiagen DNeasy kit). The CCR5 and CXCR4 loci were amplified by PCR from 400 ng of genomic DNA using Pfx Platinum Polymerase (Invitrogen). 200 ng of purified PCR product was digested overnight with PciI (NEB), then resolved on a 2% agarose gel and analyzed with ImageQuant software.
Mass spectrometry
Following immunoprecipitation of Flag-tagged UbvG08 and UbvG08DM from HEK293 Flp-In/T-REx cells, peptides were identified using LC-MS/MS. Proteins were digested in solution with trypsin (Sigma, T7575-1KT) and dried to completeness. For LC-MS/MS analysis, peptides were reconstituted in 5% formic acid and loaded onto a 12–15 cm fused silica column with pulled tip packed in-house with 3.5 µm Zorbax C18 (Agilent Technologies, CA, USA).
UbvG08 and UbvG08-DM were analyzed using an LTQ (Thermo Scientific) coupled to an Agilent 1100 Series HPLC (Agilent Technologies). Peptides were eluted from the column using a 90 min period cycle with a linear gradient from 0% to 40% ACN in 0.1% formic acid. Tandem MS spectra were acquired in a data-dependent mode for the top 5 most abundant ions using collision-induced dissociation. Acquired spectra were searched against the human Refseq_V53 database using Mascot (Matrix Science).
Isothermal titration calorimetry
Isothermal titration calorimetry was performed using a VP-ITC calorimeter (MicroCal). Untagged 53BP1 Tudor and UbvG08 (or the DM mutant) were dialyzed into PBS and degassed. 100 µM UbvG08 in the syringe was titrated into 10 µM 53BP1 Tudor protein in the sample cell using 30 consecutive 10 µl injections at 25 °C. Resultant binding isotherms were processed with Origin 5.0 software (Microcal). Curve fits were carried out using the one-set-of-sites model.
ZFN editing reagents
A ZFN pair targeting CCR5 has been previously described22, 36. The ZFN coding sequences were cloned into pVAX (Thermo Fisher Scientific, Waltham, MA), which was linearized by restriction enzyme digestion to generate templates for mRNA synthesis, using the mMESSAGE mMACHINE T7 ULTRA Kit (Thermo Fisher Scientific), according to the manufacturer’s instructions. A CCR5 homologous sequence22 containing an internal XhoI restriction site, was used in its plasmid form as a homology donor.
Analysis of gene disruption and editing by ZFNs
K562 cells were obtained from the American Type Culture Collection, and transfected by nucleofection as previously described23, using 16-well Nucleocuvette™ Strips in the 4D-Nucleofector™ System, and the SF Cell Line 4D-Nucleofector® X Kit S (Lonza, Cologne, Germany). Briefly, 2×105 cells were resuspended in 20 µL of SF solution, together with mRNA and/or plasmids, and nucleofected using the recommended program for K562 cells. Two sequential nucleofections were performed, 24 hours apart, to allow i53 or DM expression before delivery of ZFN mRNA and donor plasmids. Cells were recounted and normalized prior to the second nucleofection. A total of 2.1, 4.2, or 6.3 pmol of i53/DM mRNAs, 1.8 pmol of each ZFN mRNA, and/or 0.3 pmol of donor plasmids were used. K562 cell pellets were harvested 5 days after the last nucleofection and CCR5 gene disruption rates determined using GeneArt® Genomic Cleavage Detection Kit (Thermo Fisher Scientific) following the manufacturer’s instructions. Briefly, genomic DNA was extracted from cell pellets and subject to PCR amplification using human CCR5-specific primers22. The PCR product was then denatured and reannealed in conditions allowing small DNA mismatch reannealing, and a proprietary detection enzyme was added to cleave unmatched regions. Digested PCR products were subject to electrophoresis on a 10% polyacrylamide gel, and gene disruption rates determined based on densitometry analysis of cleaved and uncleaved products. Gene editing rates were determined using an RFLP assay, as described22, 23. Briefly, a pair of CCR5 primers located outside the region of homology contained within the CCR5 donor molecule were used for PCR amplification, followed by XhoI digestion, resolution on a 1% agarose gel, and analysis of products by densitometry.
sgRNA targeting sequences
| Target | Guide sequence |
Targeting Donor | Non-targeting donor |
|---|---|---|---|
| BFP-1 | ATGGCG TGCAGT GCTTCA GC |
GCCACCTACGGCAAGCTGACCCTGAA GTTCATCTGCACCACCGGCAAGCTGC CCGTGCCCTGGCCCACCCTCGTGACC ACCCTGACGTACGGCGTGCAGTGCTT CAGCCGCTACCCCGACCACATGA |
AGTGGCCAGAGTCCAGCTTGGGCCC ACGCAGGGGCCTGGCCAGCAGCAA GCAGCACTCTGCCCTCGTGGGTTTG TGGTTGCCCACACATGTCATTGGAG GTGACATCGATGTCCTCCCCATTGG CCT |
| CCR5# | TGACAT CAATTA TTATAC AT |
ACAAAACCAAAGATGAACACCAGTG AGTAGAGCGGAGGCAGGAGGCGGGC TGCGATTTGCTTCACATTGATTTTTTG GCAGGGCTCacATGTATAATAATTGAT GTCATAGATTGGACTTGACACTT |
ATGGATTGGTCATCCTGGTCATGGG TTACCAGAAGAAACTGAGAAGCAT GACGGACAAGTACAGGCTGCACCT GTCAGTGGCCGACATGTTCTTTGTC ATCACGCTTCCCTTCTGGGCAGTTG ATGC |
| CXCR4# | GAAGCG TGATGA CAAAGA GG |
ATGGATTGGTCATCCTGGTCATGGGT TACCAGAAGAAACTGAGAAGCATGA CGGACAAGTACAGGCTGCACCTGTCA GTGGCCGACATGTTCTTTGTCATCAC GCTTCCCTTCTGGGCAGTTGATGC |
ACAAAACCAAAGATGAACACCAGT GAGTAGAGCGGAGGCAGGAGGCGG GCTGCGATTTGCTTCACATTGATTTT TTGGCAGGGCTCacATGTATAATAAT TGATGTCATAGATTGGACTTGACAC TT |
| HISTH2BK | GGGGCT TTAAGA CGCTTA CT |
Agudelo et al. ref20 | N/A |
Data availability statement
Source data for figures 1–4 and supplementary figures 1–8 are provided with the paper as a supplementary file (Source Data). The structure data was deposited at the RCSB protein data bank (accession # PDB 5J26). Additional information can be found in the Life Sciences Reporting Summary.
Supplementary Material
Acknowledgments
We are grateful to R. Szilard for critical reading of the manuscript. We thank J. Stark for the DR- and EJ2-GFP U2OS cell lines, G. Dellaire for the LMNA assay plasmids and Y. Doyon for the HIST1H2BK-mAG targeting vector. AFT was a CIHR post-doctoral fellow and AO was a recipient of the Terry Fox Foundation Strategic Initiative for Excellence in Radiation Research for the 21st Century at CIHR fellowship. PAM-G received a Breast Cancer postdoctoral fellowship award from the US department of defense (BC134020). MDW held a long-term fellowship from the Human Frontier Science Program. SMN receives a postdoctoral fellowship from the Dutch Cancer Society (KWF). DD is the Thomas Kierans Chair in Mechanisms of Cancer Development and a Canada Research Chair (Tier 1) in the Molecular Mechanisms of Genome Integrity. Work was supported by NIH grant U19 HL129902 to PMC, CIHR grants MOP111149 and MOP136956 (to SSS), FDN143277 (to FS) and FDN143343 (to DD) and a Grant-in-Aid from the Krembil Foundation (to DD).
Footnotes
Author contributions
MDC generated the mutations for the analysis by pulldown and carried out most of the pulldown experiments, generated the protein crystals, conducted the ITC experiments, and carried out some of the initial assays demonstrating efficiacy of i53. NM carried out the DR- and EJ2-GFP assays, all assays with AAV-i53 in human cells along with all ssODN assays and the determination of 53BP1 foci in G1 cells. LW refined and analyzed the crystal structure and prepared the figures describing the structure. AFT produced the GST-53BP1 protein for Ubv selection and the GST-fusion Ubv proteins for the initial pulldown assays she carried out. YCJ supervised the crystallization and collected the diffraction data. AO carried out the LMNA gene targeting assays with the help of TN. WZ carried out the Ubv selections with AV and AE. DK and ES conducted HDR analyses in K562 cells. TC assessed the impact of i53 on p53 to answer a reviewer comment. PMC supervised DK and ES. PMG performed mouse gene targeting assay. SMN carried out the PARP inhibitor assays using cells generated by MZ. MDW helped with biochemical experiments. MM did the IP-MS and helped MDC, AFT and AO. SSS supervised WZ, AV and AE. FS supervised LW and YCJ. DD conceived and supervised the project and wrote the manuscript with LW, MDC and FS, with input from the other authors.
Competing financial interests
The authors declare competing financial interests as the use of 53BP1-blocking Ubvs for stimulating genome editing are the subject of a patent application and a license agreement.
References
- 1.Doudna JA, Charpentier E. Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science. 2014;346:1258096. doi: 10.1126/science.1258096. [DOI] [PubMed] [Google Scholar]
- 2.Cox DB, Platt RJ, Zhang F. Therapeutic genome editing: prospects and challenges. Nat Med. 2015;21:121–131. doi: 10.1038/nm.3793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chandrasegaran S, Carroll D. Origins of Programmable Nucleases for Genome Engineering. J Mol Biol. 2015 doi: 10.1016/j.jmb.2015.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hustedt N, Durocher D. The control of DNA repair by the cell cycle. Nat Cell Biol. 2016;19:1–9. doi: 10.1038/ncb3452. [DOI] [PubMed] [Google Scholar]
- 5.Panier S, Boulton SJ. Double-strand break repair: 53BP1 comes into focus. Nat Rev Mol Cell Biol. 2014;15:7–18. doi: 10.1038/nrm3719. [DOI] [PubMed] [Google Scholar]
- 6.Ernst A, et al. A strategy for modulation of enzymes in the ubiquitin system. Science. 2013;339:590–595. doi: 10.1126/science.1230161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Escribano-Diaz C, et al. A Cell Cycle-Dependent Regulatory Circuit Composed of 53BP1-RIF1 and BRCA1-CtIP Controls DNA Repair Pathway Choice. Molecular cell. 2013;49:872–883. doi: 10.1016/j.molcel.2013.01.001. [DOI] [PubMed] [Google Scholar]
- 8.Feng L, Fong KW, Wang J, Wang W, Chen J. RIF1 counteracts BRCA1-mediated end resection during DNA repair. The Journal of biological chemistry. 2013;288:11135–11143. doi: 10.1074/jbc.M113.457440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fradet-Turcotte A, et al. 53BP1 is a reader of the DNA-damage-induced H2A Lys 15 ubiquitin mark. Nature. 2013;499:50–54. doi: 10.1038/nature12318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Botuyan MV, et al. Structural basis for the methylation state-specific recognition of histone H4-K20 by 53BP1 and Crb2 in DNA repair. Cell. 2006;127:1361–1373. doi: 10.1016/j.cell.2006.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dikic I, Wakatsuki S, Walters KJ. Ubiquitin-binding domains - from structures to functions. Nature reviews. Molecular cell biology. 2009;10:659–671. doi: 10.1038/nrm2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Orthwein A, et al. A mechanism for the suppression of homologous recombination in G1 cells. Nature. 2015;528:422–426. doi: 10.1038/nature16142. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 13.O'Donnell L, et al. The MMS22L-TONSL complex mediates recovery from replication stress and homologous recombination. Molecular cell. 2010;40:619–631. doi: 10.1016/j.molcel.2010.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xie A, et al. Distinct roles of chromatin-associated proteins MDC1 and 53BP1 in mammalian double-strand break repair. Mol Cell. 2007;28:1045–1057. doi: 10.1016/j.molcel.2007.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moynahan ME, Chiu JW, Koller BH, Jasin M. Brca1 controls homology-directed DNA repair. Mol Cell. 1999;4:511–518. doi: 10.1016/s1097-2765(00)80202-6. [DOI] [PubMed] [Google Scholar]
- 16.Srivastava M, et al. An inhibitor of nonhomologous end-joining abrogates double-strand break repair and impedes cancer progression. Cell. 2012;151:1474–1487. doi: 10.1016/j.cell.2012.11.054. [DOI] [PubMed] [Google Scholar]
- 17.Chu VT, et al. Increasing the efficiency of homology-directed repair for CRISPR-Cas9-induced precise gene editing in mammalian cells. Nat Biotechnol. 2015;33:543–548. doi: 10.1038/nbt.3198. [DOI] [PubMed] [Google Scholar]
- 18.Maruyama T, et al. Increasing the efficiency of precise genome editing with CRISPR-Cas9 by inhibition of nonhomologous end joining. Nat Biotechnol. 2015;33:538–542. doi: 10.1038/nbt.3190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pinder J, Salsman J, Dellaire G. Nuclear domain 'knock-in' screen for the evaluation and identification of small molecule enhancers of CRISPR-based genome editing. Nucleic Acids Res. 2015 doi: 10.1093/nar/gkv993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Agudelo D, et al. Marker-free coselection for CRISPR-driven genome editing in human cells. Nat Methods. 2017 doi: 10.1038/nmeth.4265. [DOI] [PubMed] [Google Scholar]
- 21.Maggio I, Goncalves MA. Genome editing at the crossroads of delivery, specificity, and fidelity. Trends Biotechnol. 2015;33:280–291. doi: 10.1016/j.tibtech.2015.02.011. [DOI] [PubMed] [Google Scholar]
- 22.Wang J, et al. Homology-driven genome editing in hematopoietic stem and progenitor cells using ZFN mRNA and AAV6 donors. Nat Biotechnol. 2015;33:1256–1263. doi: 10.1038/nbt.3408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang J, et al. Targeted gene addition to a predetermined site in the human genome using a ZFN-based nicking enzyme. Genome Res. 2012;22:1316–1326. doi: 10.1101/gr.122879.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Verma P, Greenberg RA. Noncanonical views of homology-directed DNA repair. Genes Dev. 2016;30:1138–1154. doi: 10.1101/gad.280545.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Davis L, Maizels N. Two Distinct Pathways Support Gene Correction by Single-Stranded Donors at DNA Nicks. Cell Rep. 2016;17:1872–1881. doi: 10.1016/j.celrep.2016.10.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Richardson CD, Ray GJ, DeWitt MA, Curie GL, Corn JE. Enhancing homology-directed genome editing by catalytically active and inactive CRISPR-Cas9 using asymmetric donor DNA. Nat Biotechnol. 2016;34:339–344. doi: 10.1038/nbt.3481. [DOI] [PubMed] [Google Scholar]
- 27.Sfeir A, Symington LS. Microhomology-Mediated End Joining: A Back-up Survival Mechanism or Dedicated Pathway? Trends Biochem Sci. 2015;40:701–714. doi: 10.1016/j.tibs.2015.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Munoz MC, et al. Ring finger nuclear factor RNF168 is important for defects in homologous recombination caused by loss of the breast cancer susceptibility factor BRCA1. The Journal of biological chemistry. 2012 doi: 10.1074/jbc.M112.410951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xiong X, et al. 53BP1 promotes microhomology-mediated end-joining in G1-phase cells. Nucleic Acids Res. 2015;43:1659–1670. doi: 10.1093/nar/gku1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gunn A, Stark JM. I-SceI-Based Assays to Examine Distinct Repair Outcomes of Mammalian Chromosomal Double Strand Breaks. Methods Mol Biol. 2012;920:379–391. doi: 10.1007/978-1-61779-998-3_27. [DOI] [PubMed] [Google Scholar]
- 31.Bennardo N, Cheng A, Huang N, Stark JM. Alternative-NHEJ is a mechanistically distinct pathway of mammalian chromosome break repair. PLoS genetics. 2008;4:e1000110. doi: 10.1371/journal.pgen.1000110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Richardson CD, et al. CRISPR-Cas9 Genome Editing In Human Cells Works Via The Fanconi Anemia Pathway. bioRxiv. 2017 doi: 10.1038/s41588-018-0174-0. [DOI] [PubMed] [Google Scholar]
- 33.Hart T, et al. High-Resolution CRISPR Screens Reveal Fitness Genes and Genotype-Specific Cancer Liabilities. Cell. 2015;163:1515–1526. doi: 10.1016/j.cell.2015.11.015. [DOI] [PubMed] [Google Scholar]
- 34.Sanjana NE, Shalem O, Zhang F. Improved vectors and genome-wide libraries for CRISPR screening. Nat Methods. 2014;11:783–784. doi: 10.1038/nmeth.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tonikian R, Zhang Y, Boone C, Sidhu SS. Identifying specificity profiles for peptide recognition modules from phage-displayed peptide libraries. Nat Protoc. 2007;2:1368–1386. doi: 10.1038/nprot.2007.151. [DOI] [PubMed] [Google Scholar]
- 36.Perez EE, et al. Establishment of HIV-1 resistance in CD4+ T cells by genome editing using zinc-finger nucleases. Nat Biotechnol. 2008;26:808–816. doi: 10.1038/nbt1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Source data for figures 1–4 and supplementary figures 1–8 are provided with the paper as a supplementary file (Source Data). The structure data was deposited at the RCSB protein data bank (accession # PDB 5J26). Additional information can be found in the Life Sciences Reporting Summary.