Abstract
Tropomodulin family of proteins includes several isoforms of tropomodulins (Tmod) and leiomodins (Lmod). These proteins can sequester actin monomers or nucleate actin polymerization. Although it is known that their actin-binding properties are isoform-dependent, knowledge on how they vary in strengths of interactions with G-actin is missing. While it is confirmed in many studies that Tmods have two actin-binding sites, information on number and location of actin-binding sites in Lmod2 is controversial. We used atomic force microscopy to study interactions between G-actin and proteins of the tropomodulin family. Unbinding forces between G-actin and Tmod1, Tmod2, Tmod3, or Lmod2 were quantified. Our results indicated that Tmod1 and Tmod3 had unimodal force distributions, Tmod2 had a bimodal distribution and Lmod2 had a trimodal distribution. The number of force distributions correlate with the proteins abilities to sequester actin or nucleate actin polymerization. We assigned specific unbinding forces to the individual actin-binding sites of Tmod2 and Lmod2 using mutations that destroy actin-binding sites of Tmod2 and truncated Lmod2. Our results confirm the existence of the N-terminal actin-binding site in Lmod2. Altogether, our data demonstrate how the differences between the number and the strength of actin-binding sites of Tmod or Lmod translate to their functional abilities.
Keywords: actin, leiomodin, tropomodulin, atomic force microscopy (AFM), nucleation
INTRODUCTION
Actin filaments drive many cellular processes such as cell motility, membrane transport, chemotaxis, cellular morphogenesis and force generation [1, 2]. The formation and determination of the correct lengths of actin filaments is essential for many cellular processes to take place in an orderly manner. Actin monomers (G-actin) polymerize to form arrays of actin filaments (F-actin). F-actin has two structurally and biochemically distinct ends: a barbed end and a pointed end. Polymerization and depolymerization occur at both ends but polymerization is faster at the barbed end. In the absence of actin-binding proteins, F-actin is said to “treadmill” when it reaches steady state; G-actin continuously polymerizes at the barbed end and depolymerizes from the pointed end [3, 4].
Actin is capable of polymerizing spontaneously. However, this process is relatively slow and kinetically unfavorable in cells. Formation of actin dimers and trimers, which can easily disassemble due to their instability, is a rate-determining step in actin polymerization [5, 6]. The shape and the length of F-actin are regulated by actin-binding proteins, which assist actin polymerization and depolymerization [1, 2, 7–9]). Some actin-binding proteins can also sequester G-actin and prevent it from being added to the filament.
Proteins from the tropomodulin family, tropomodulin (Tmod) and leiomodin (Lmod), can bind both G-actin and F-actin (see reviews [10, 11]). By binding G-actin, they can sequester actin monomers or nucleate actin polymerization. They bind at the pointed end of F-actin in a tropomyosin (Tpm)-dependent fashion. Tmod caps the pointed ends of F-actin to inhibit polymerization, whereas Lmod binds at the same end but still allows filament elongation although at lower rates than in the absence of Lmod [12, 13].
Tmod and Lmod have several isoforms that are differentially expressed in various cell types [14]. Tmod1 is expressed in, but not limited to, erythrocytes, cardiac and skeletal muscle cells [15–17]. Tmod2 is expressed only in the brain [18, 19]. Tmod3 is expressed in many cell types [14, 20] and Tmod4 is expressed in adult skeletal muscle cells [21]. Of Tmod isoforms, Tmod3 is the best actin-sequestering isoform with weak nucleation ability, while Tmod2 is the best actin-nucleating isoform [22–24]. Of the three known Lmod isoforms, Lmod1 is expressed in smooth muscle cells, whereas Lmod2 and Lmod3 are expressed in cardiac and skeletal muscle cells [14, 25, 26]. Lmods were shown to be potent nucleators of actin polymerization [27, 28].
Both Tmod and Lmod are indispensable for cytoskeleton structure and function and vital for organisms. In mice, Tmod1 knockout leads to embryonic lethality due to cardiac defects [29–32], Tmod2 knockout causes reduced sensorimotor gating, impaired learning and memory [33], and Tmod3 knockout is lethal due to anemia [34]. The knockout of Lmod2 in mice causes dilated cardiomyopathy, resulting in juvenile death [35]. Mutations in the LMOD3 gene found in human patients [28] or the knockout of Lmod3 in mice [36] were shown to cause severe nemaline myopathy. In addition, the knockout of Lmod3 or Tmod4 in frog disrupted the sarcomeric assembly [37]. These findings highlight the necessity of Tmod and Lmod isoforms in maintaining normal cellular properties in various type of tissues. Understanding structure/function relationship for the members of the tropomodulin family is necessary to unravel their exact roles in cells.
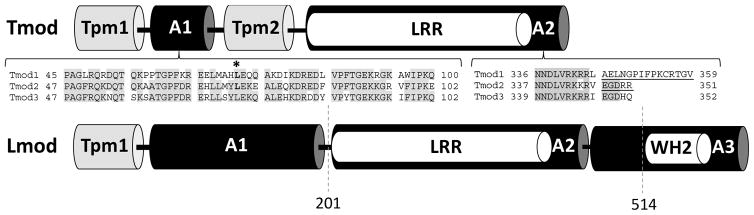
Tmod and Lmod have similar domain structure, however, there are essential differences in the number of actin- and Tpm-binding sites (Figure 1). Tmod has two Tpm-binding sites and two actin-binding sites [38–40]. Lmod, a bigger homolog of Tmod [14, 26], has the C-terminal extension that comprises a WH2 (Wiskott-Aldrich syndrome Homology 2) domain and a proline-rich region. Lmod2, the most studied Lmod isoform, contains a single Tpm-binding site [41, 42] and three actin-binding sites [12, 13, 27]. The presence of the first actin-binding site of Lmod2 is debatable. There are contradicting data obtained with Lmod2 fragments arguing the actin-binding ability of this site [12, 43, 44]. In the recent paper, Boczkowska et al. [43] stated that Lmods lost pointed-end capping elements present in Tmods. This statement was based on isothermal titration calorimetry (ITC) experiments where Tmod1’s N-terminal fragment was shown to interact with actin while Lmod2’s N-terminal fragment did not. The authors made the conclusion that Lmod2 lacks the N-terminal actin-binding site. On the other hand, using nuclear magnetic resonance (NMR), we showed that both Lmod2’s and Tmod1’s N-terminal fragments bind actin [12]. In the recent review, Fowler and Dominguez highlighted the need of additional experiments using full-length proteins to test the actin-binding function of the N-terminal region in Lmod2 [11] and to resolve existing contradiction.
Figure 1.
Domain structures of Tmod and Lmod. Tpm1, Tpm2: tropomyosin-binding sites; A1, A2, A3: actin-binding sites, and LRR: leucine-rich repeats. Insets show the sequence alignments between Tmod isoforms, where conserved residues between isoforms are highlighted. The location of the mutations and truncations that disrupt actin binding are shown with asterisk or underlined, respectively. Dashed lines show the locations, where Lmod2 was truncated.
Until now, the molecular interactions of Tmod or Lmod with actin had been assessed in pyrene-actin polymerization assays [13, 23, 27, 28, 44, 45] or directly measured using non-denaturing polyacrylamide gel-electrophoresis (for Tmod isoforms only [23]), ITC (for Tmod1 and Lmod2) [43, 46], and for Lmod2 only, bioLayer interferometry [44] and NMR spectroscopy [12]. Although the use of these techniques has given valuable insight, there may be challenges in their interpretation. The latter three methods needed high concentrations and used Lmod2 fragments. Crystal structures of complexes between Lmod2 or Tmod1 and actin also were obtained using fragments [43, 44, 46]. The use of fragments is convenient in understanding the function of multidomain proteins, as well as in estimating the binding kinetics between Tmod or Lmod and their binding partners. However, they may not reflect the binding expected between full-length proteins. Additionally, the tendency of actin to polymerize rapidly under physiological conditions complicates the measurement of its binding constants with Tmod or Lmod, as well as prevents crystallization [44]. In order to overcome this problem, previous efforts for measuring the interactions of Tmod or Lmod with actin involved the use of latrunculin B [43] or mutated actin [44] to stabilize the monomeric state of actin and prevent polymerization.
Atomic force microscopy (AFM) with its ability to measure interactions between single molecules allows quantification of protein-protein interactions with high accuracy under native conditions and using full-length proteins [47]. This technique is an exciting complement to previous studies of the structure/function relationship between Tmods and actin. In this study, we utilized AFM to characterize the unbinding forces between G-actin and proteins of the tropomodulin family. By creating mutants or fragments of Tmod2 and Lmod2, we assigned specific unbinding forces to their individual actin-binding sites. Our findings demonstrated that the N-terminal domain of Lmod2 interacts with actin and confirmed the existence of the N-terminal actin-binding site.
MATERIALS AND METHODS
Protein Sequences
Sequences of Tmod1 (NP_990358.1), Tmod2 (NP_001033799.1), Tmod3 (NP_058659.1) and Lmod2 (NP_001186644.2) were downloaded from NCBI.
Plasmid Construction
The plasmids for expression of Tmod1, Tmod11-344[L71D], Tmod2 and Tmod21-346 (pET(His)Tmod1, pET(His)Tmod11-344[L71D], pET(His)Tmod1 and pET(His)Tmod21-346, respectively) were generated previously [24, 39, 48]. pET(His)Tmod2[L73D] was generated by site-directed mutagenesis using pET(His)Tmod2 as a template by using Pfu Turbo DNA polymerase (Agilent Technologies, USA) and two complementary oligonucleotides, according to manufacturer’s instructions. The complementary set of oligonucleotides was synthesized by Integrated DNA Technologies (Coralville, IA) and the sequence of the sense primer was: 5′-CGA GAA CAC CTG CTC ATG TAC GAC GAG AAA GAG GCT TTG G-3′. The mutated triplet is underlined. Mutagenesis was confirmed by DNA sequencing at GENEWIZ, Inc. (South Plainfield, NJ).
Protein Expression and Purification
WT-Tmod1 and Tmod11-344[L71D] were expressed and purified as described in [45]. Tmod2, Tmod21-346 and Tmod2[L73D] were expressed and purified according to the method described in [49]. Tmod3 was expressed and purified as in [24]. Lmod2, Lmod21-514 and Lmod21-201 were a generous gift from Dr. Carol Gregorio (University of Arizona, Tucson, AZ). G-actin was purified from acetone powder using gel-filtration chromatography and pyrene-iodoacetamide labeled G-actin (P-actin) was prepared as described in [12].
Protein concentrations were determined by the difference method as described in [45] and/or using BCA Protein Assay Kit (Pierce) according to manufacturer’s instructions.
G-actin immobilization for AFM experiments
G-actin was adsorbed on mica as previously described [50]. Briefly, 50 μl of 3 μM G-actin in G-buffer (2 mM Tris-HCl pH 8.0, 0.2 mM CaCl2, 0.01% NaN3, 0.2 mM ATP and 0.5 mM DTT) was placed on a freshly cleaved mica disk and allowed to sit for 10 minutes at room temperature. The mica disk was then rinsed with G-buffer to remove the loosely bound G-actin. The mica disk was taped on a steel disk and was immediately used for AFM experiments without drying.
Cantilever modification with Tmod or Lmod
Silicon nitride (Si3N4) cantilevers (DNP-10, Bruker Inc., Camarillo, CA) were cleaned in 95% ethanol and pure methanol solutions (J.T. Baker) for 5 minutes each, respectively. Surfaces of cantilevers were oxidized by ultraviolet radiation for 10 minutes. Amine termination was achieved by immersion of cantilevers into 1.5% (v/v) 3-aminopropyl(diethoxy)methylsilane (APDMES) (Sigma-Aldrich, St. Louis, MO) in toluene for 2 hours. The excess APDMES was washed by immersing of the cantilevers into methanol and deionized water for 5 minutes each, respectively. A 3 μl of a 100 mM 1-3-dimethylaminopropyl-3-carbodimide (EDC) (Thermo Fisher Scientific, Waltham, MA) aqueous solution reacted with a 100 μl of 25 μg/ml Tmod or Lmod solution in cross-linking buffer (50 mM sodium phosphate pH 7.4, 150 mM NaCl and 0.5 mM DTT) for 30 minutes at room temperature. A 3 μl of 40 mM N-hydroxylsulfosuccinimide (NHS) (Thermo Fisher Scientific, Waltham, MA) aqueous solution was added to the mixture above and allowed to react for 30 minutes at room temperature. NH2-activated Si3N4 cantilevers were then incubated with the NHS/EDC-activated Tmod or Lmod for 2 hours at room temperature to allow for amine coupling. After that, cantilevers were rinsed with the cross-linking buffer and immediately used for AFM unbinding force measurements. The successful Tmod functionalization of cantilevers was confirmed via fluorescence imaging of Tmod labeled by primary rabbit antibodies against Tmod1 (custom made by Thermo Fisher Scientific, Waltham, MA) and goat anti-rabbit IgG (H+L) secondary antibody, Alexa Fluor® 488 conjugate (Thermo Fisher Scientific, Waltham, MA). The Tmod-modified cantilevers with attached primary and secondary antibodies showed green fluorescence, whereas APDMES modified cantilevers incubated with both primary and secondary antibodies showed no fluorescence (data not shown).
AFM imaging of G-actin
Topography images of immobilized G-actin on mica were acquired via a scanning probe microscope equipped with a J-scanner and a Nanoscope controller V in peakforce tapping mode™ (Bruker Inc., Santa Barbara, CA) under G-buffer. A Scanasyst-fluid+ probe (Bruker Inc., Santa Barbara, CA) with a 2 nm tip radius was used for imaging as recommended by the manufacturer. Scan rate was varied between 1 to 2 Hz to achieve high quality images. Scan sizes of 2×2 and 1×1 μm2 are reported to visualize the surface coverage with G-actin.
AFM unbinding force measurements
A PicoForce™ scanning probe microscope with a Nanoscope IIIa controller (Bruker Inc., Santa Barbara, CA) was utilized in the point and shoot mode for force measurements. Unbinding force measurements between Tmod or Lmod isoforms and G-actin were performed in G-buffer under ambient temperature and humidity conditions. The spring constant of Tmod or Lmod modified cantilevers was determined before each experiment using the spectral density of thermal noise fluctuations [51]. The mean of spring constants of cantilevers used was 0.07±0.008 N/m (N=4), which is close to the manufacturer’s reported value of 0.06 N/m. Prior to force-distance curve collection, deflection sensitivity of the cantilevers was determined on a hard mica disk and the cantilevers were moved to the area where G-actin was immobilized on. A 2×2 μm2 scan area of the sample with G-actin adsorbed on it was imaged in tapping mode. Once an image was obtained, the oscillation of the cantilever was stopped and force-distance curves were collected in an array of 16×16 using a relative trigger of 1.0 nN. A scan rate of 1.0 Hz with a z-ramp size of 500 nm, both which translate to 1.0 μm/sec pulling speed were used in all measurements. Only, the specific unbinding forces were selected for the analysis of binding strength between Tmod or Lmod and G-actin. The selection of specific unbinding forces followed the approach described elsewhere [52]. Briefly, if the peak was curved in shape as quantified by a changing slope of the force/distance curve as a function of distance, then the peak will represent a specific force. Whereas, for nonspecific forces, the slope of force/distance curve will be constant as a function of distance and the peak will have a triangular shape [52, 53]. Here, we only selected the unbinding forces where the slope of forces changed in the retraction curves. In addition to the shape and the slope of the peak, the magnitude of adhesion force of the peak was considered. In general, protein/ligand or protein/protein unbinding forces vary between 10 to 300 pN depending on the pulling speed [52, 54]. We chose a medium pulling speed (1.0 μm/sec) to achieve a medium range of unbinding forces to avoid the noise limitations. Here, our intention was not to study the kinetics of unbinding forces; therefore, only one pulling speed was used in unbinding force measurements. In addition to the shape of the curves, we only selected the unbinding forces below 250 pN as specific binding events to avoid incorporation of multiple binding events between Tmod modified cantilevers and G-actin. The absolute values of the unbinding forces are reported.
Analysis of retraction curves to determine the unbinding forces between Tmod or Lmod and G-actin
Over 500 retraction force curves collected in an array of 16×16 were screened for each isoform and only specific binding events were used for the quantification of the average of unbinding forces. Overall, about 10% to 20% of the curves included specific unbinding forces for a given isoform. Magnitudes of unbinding forces were determined using the Nanoscope Analysis 1.5 software (Bruker, Camarillo, CA). Each retraction curve was considered individually due to the heterogeneities expected among retraction curves measured.
Fluorescence measurements
Time-dependent actin polymerization was measured by the change in P-actin fluorescence (excitation, 366 nm, and emission, 387 nM) using a PTI fluorometer (Lawrenceville, NJ) with 2 nM slit widths. Briefly, 1 μM actin (10% P-actin) in G-buffer was mixed with 5, 10 or 25 nM of Lmod2-WT, Lmod21-514 or Lmod21-201 or 25 nM of Tmod2 and incubated for 1 minute at room temperature. Then, 20x polymerization buffer was added to 1x final concentration (25 mM Imidazole pH 7.0, 100 mM KCl, 2 mM MgCl2, 1 mM EGTA) immediately before the start of each experiment. Polymerization was measured for 50 minutes and initial rates (R) were calculated as the first derivatives of the polymerization curves at time zero using Exponential Rise to Maximum or Exponential Growth fit settings in SigmaPlot 12.0. The rate of nucleation of actin polymerization by Lmod2 or Tmod2 was calculated as Rexp/Rcontrol. Rcontrol is the rate of spontaneous actin polymerization (control) and Rexp is the rate of actin polymerization in the presence of Lmod2 or Tmod2.
Statistics
Statistical analysis was performed using Origin 9.0 (Northampton, MA). One way analysis of variance (ANOVA) followed by a Tukey test was used to determine whether significant differences existed between treatment groups. Statistical differences with a 95% confidence level were reported (p<0.05). Only most frequent unbinding forces as quantified via log-normal or Gaussian peak function were reported and compared to each other. Log-normal peak function was used for single peak distributions due to skewed data, whereas Gaussian peak function was used for multimodal distribution due to relative low skewed data compared to single peak distributions. For multimodal peak distribution, each histogram was broken down into two or three populations. Then each population was compared to each other to determine the statistical difference. In order to identify the end of a single population, the point where a log-normal or a Gaussian peak function reaches to the minimum probability value in a given data set was determined. Then this point was used to break down the bimodal or trimodal distribution into single populations for the further statistical comparison.
RESULTS AND DISCUSSION
Immobilization of G-actin on the mica surface shows uniform distribution
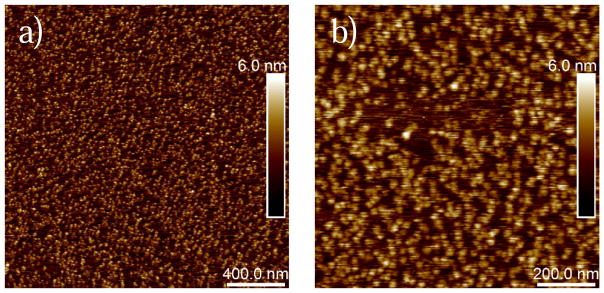
G-actin was immobilized on a mica disk prior to unbinding force measurements, and topography images were captured in order to verify the surface coverage with G-actin. Successful immobilization of G-actin on mica with almost a full coverage of both at 2×2 and 1×1 μm2 scan area are shown in Figure 2. Our images show globular particles on the mica surface and they correlate well in terms of shape with previous AFM images of G-actin [50].
Figure 2.
AFM topography images of G-actin immobilized on mica in G-buffer. a) A 2×2 μm2 topography image showing almost a full surface coverage with G-actin. b) A 1×1 μm2 topography image of G-actin showing surface details of globular actin. Note that the dense but uniform surface coverage provided chances for relatively frequent measurements of specific unbinding events in the force-distance curves.
Interactions of actin with Tmod2 have bimodal unbinding force distribution, while interactions with other Tmod isoforms have unimodal distribution
In order to perform the force measurements between G-actin immobilized on mica and its binding partners, the AFM cantilever was functionalized with Lmod (or Tmod) through its C-terminus. Cantilevers modified with Lmod or Tmod were approached to G-actin while interaction forces between the proteins were recorded simultaneously. After the modified cantilever contacted G-actin, the two proteins were retracted from each other. If a binding event occurred in the retraction portion of the force cycle between Tmod or Lmod and G-actin, this caused a bending of the cantilever. The bending resulted in a deflection in the laser beam reflected from the back of the cantilever and the deflection was measured and then converted to a distance and finally to a force [55]. These attractive rupture forces called unbinding forces [52] were measured between G-actin and Tmod isoforms or Lmod.
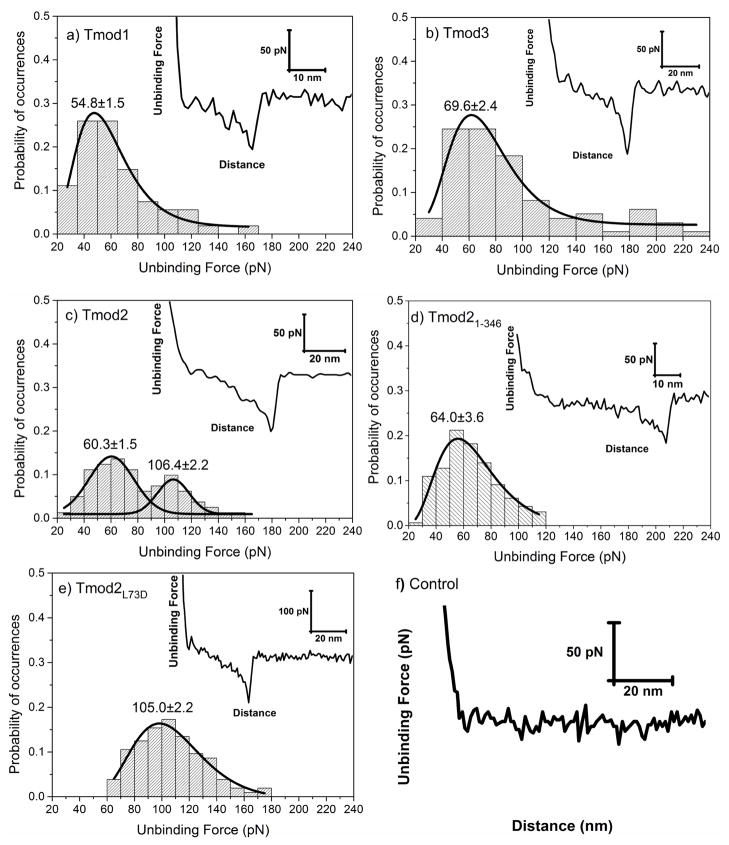
Figure 3 shows the distributions of the unbinding forces between Tmod isoforms and G-actin. Retraction curves (representative ones shown in the insets) were used for quantification of specific unbinding forces. Although the shapes of the peaks in the retraction curves shown in Figure 3 were similar for all isoforms, the distributions of the collective unbinding forces found for ~50 curves varied. For Tmod1 and Tmod3, we observed unimodal distributions with a single population of unbinding forces (Figure 3a, b). The most probable unbinding forces of Tmod1 and Tmod3 were 54.8±1.5 and 69.6±2.4 pN, respectively. The peak value of the unbinding forces of Tmod3, 69.6±2.4 pN, was significantly higher than that measured for Tmod1 (p<0.05).
Figure 3.
Distribution of unbinding forces measured between G-actin and a) Tmod1, b) Tmod3, c) Tmod2, d) Tmod21-346, and e) Tmod2[L73D], respectively. Insets show representative retraction force-curves with specific protein-protein unbinding force peaks. Solid lines show a dynamic peak function model fits to the data presented in the histograms (Origin 9.0, OriginLab Corp., Northampton, MA) (R2>0.95). Lognormal peak function was used for unimodal distribution whereas Gaussian peak function was used for bimodal distributions. Peak values estimated from dynamic peak function fitting representing the most probable values are given as insets. Tmod2 shows bimodal distribution suggesting that multiple actin binding sites are involved in the interactions to G-actin. Note that peak value of Tmod21-346 (64.0±3.6) is similar to the first peak value of Tmod2 (60.3±1.5) (p>0.05). f) Retraction force curve measured between Tmod11-344[L71D] and G-actin. The interactions between Tmod11-344[L71D] and G-actin were purely repulsive displaying no specific or nonspecific interactions.
The presence of two actin-binding sites in Tmods was studied and confirmed in experiments with F-actin [38, 40]. A unimodal distribution in binding to G-actin can be explained by two possible scenarios: (1) these Tmod isoforms bind one molecule of G-actin under conditions investigated or (2) there are two binding phenomena with indistinguishable unbinding forces. Since Tmod1 does not nucleate actin polymerization, while Tmod3 is able to weakly nucleate it [23, 24], the first scenario is true for Tmod1, while the second scenario may be true for Tmod3. However, in binding experiments using gel-chromatography, Yamashiro and co-authors demonstrated that Tmod3 binds G-actin in 1:1 ratio and the mutation L73D in the site A1 greatly reduced this binding [23]. Therefore, in our experiments with Tmod1 and Tmod3, most likely, we observe binding to the N-terminal actin-binding site A1 only. The strongest binding of G-actin to one site only explains why Tmod3 is the best actin-sequestering isoform [22].
Unlike these isoforms, Tmod2’s binding to G-actin had a bimodal distribution of the unbinding forces that are represented with well-separated populations (Figure 3c). The two peak values of the unbinding forces quantified via Gaussian peak function for Tmod2 were 60.3±1.5 and 106.4±2.2 pN. We assumed that each population represents binding of G-actin to one of the two binding sites in Tmod2.
Tmod1 with mutations in the actin-binding sites does not bind G-actin
To confirm that the forces we measure result from interactions and not unfolding, we carried out AFM measurements with Tmod11-344[L71D] as a negative control. This protein contained two mutations, L71D in the actin-binding site A1 and the C-terminal truncation in the actin-binding site A2 (Figure 1). These mutations drastically decrease Tmod1’s actin-binding ability in the corresponding sites [39, 40, 45]. Tmod11-344[L71D] was attached to AFM cantilevers and its interaction forces with G-actin were measured. Retraction force curves measured were purely repulsive displaying no specific or nonspecific interactions at all between Tmod11-344[L71D] and G-actin (Figure 3f). This result confirmed that Tmod11-344[L71D] does not bind actin and therefore confirmed that unbinding forces measured between wild-type Tmods and G-actin reflect specific interactions expected between them. The fact that the retraction curves collected for Tmod11-344[L71D] were purely repulsive points to that during the retraction process, unfolding of Tmod does not occur. The attractive forces measured between Tmods and G-actin are due to interaction between the two and not the unfolding of Tmod.
Assigning Tmod2 unbinding forces populations to actin-binding sites
We assumed that the two unbinding force populations shown in the distribution of Tmod2-actin interaction forces correspond to Tmod2’s two actin-binding sites. In order to assign each population of events to a corresponding actin-binding site, we performed experiments with two Tmod2 constructs: Tmod2[L73D] and Tmod21-346. The L73D mutation was designed based on the homologous mutation L71D that disrupted Tmod1’s actin-binding site A1 (Figure 1). In another construct, Tmod21-346, five C-terminal residues were removed (Figure 1). This truncation was shown to disrupt Tmod2’s interaction with actin through its actin-binding site A2 and affected actin nucleation ability of Tmod2 [24].
Both Tmod2[L73D] and Tmod21-346 had unimodal unbinding force distributions (Figure 3d, e). This indicated that, unlike wild-type Tmod2, each mutant bound only one molecule of actin. Furthermore, the peak value of unbinding force that was obtained for Tmod21-346 (64.0±3.6 pN) was statistically similar to that of the first population (60.3±1.5 pN) obtained for wild-type Tmod2 (Figure 3c, d) (p>0.05). The peak value of unbinding force that was obtained for Tmod2[L73D] (105.0±2.2 pN), was statistically similar to that of the second population (106.4±2.2) (Figure 3c, e) (p>0.05). These results confirm that the two unbinding force populations observed for Tmod2 in fact correspond to the two individual actin-binding sites of Tmod2.
While unbinding forces of Tmod2’s actin-binding site A1 are close to those of other Tmod isoforms, the Tmod2’s actin-binding site A2 demonstrated much higher forces. The actin-nucleating ability of Tmod2 is greater than any other Tmod isoform [23, 24]. Therefore, the ability to bind two molecules of G-actin and stronger binding in the site A2 explains Tmod2’s higher nucleation ability.
Lmod2-actin interactions have trimodal unbinding force distribution
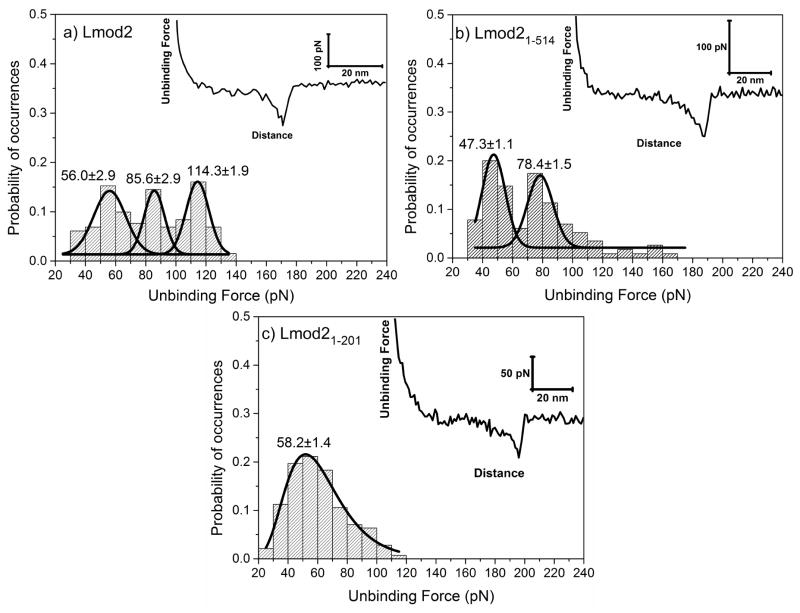
Binding sites of recently discovered Lmods are still under investigation. For instance, there are contradicting reports in the literature about whether Lmod2 contains the N-terminal actin-binding site A1 or not [12, 43, 44]. To find out if there is an actin-binding site A1 in Lmod2, first, we carried out AFM measurements with full-length Lmod2. Figure 4a shows a trimodal distribution of unbinding forces between Lmod2 and G-actin with peak values of 56.0±2.9, 85.6±2.9 and 114.3±21.9 pN. This result suggests the presence of three actin-binding sites in Lmod2.
Figure 4.
Distributions of specific unbinding forces measured between a) Lmod2, b) Lmod21-514, and c) Lmod21-201 isoforms and G-actin, respectively. Insets show representative retraction force-curves showing specific protein-protein unbinding force peaks. Solid lines show a dynamic peak function model fits to the data presented in the histograms (Origin 9.0, OriginLab Corp., Northampton, MA) (R2>0.95). Lognormal peak function was used for unimodal distribution whereas Gaussian peak function was used for bimodal or trimodal distributions. Peak values estimated from dynamic peak function fitting representing the most probable values are given as insets. Lmod2 shows a trimodal distribution whereas Lmod21-514 shows a bimodal distribution suggesting that multiple actin binding sites are involved in the interactions to G-actin. Note that the first peak values of Lmod2 (56.0±2.9) and Lmod21-514 (47.3±1.1) is similar to the peak value of Lmod21-201 (58.2±1.4). Similarly, second peak value of Lmod2 (85.6±2.9) is similar to the second peak value of Lmod21-514 (78.4±1.5). Slight shifts in the peak values could be due to synergistic effect of multiple sites on the unbinding forces.
To assign each peak to the actin-binding sites of Lmod2, we used two fragments of Lmod2, Lmod21-514 and Lmod21-201. The two fragments contained a different number of actin-binding sites. Specifically, Lmod21-514 should contain the sites A1 and A2, whereas Lmod21-201 should contain the site A1 (Figure 1). Lmod21-514 showed a bimodal distribution with peak values of 47.3±1.1 and 78.4±1.5 pN (Figure 4b) that were close to populations #1 (56.0±2.9 pN) and #2 (85.6±2.9 pN) of the full-length Lmod2, respectively. Yet, there was statistically significant difference (p<0.05) between the population #1 of Lmod21-514 and that of full-length Lmod2. This difference might be due to a synergistic effect of multiple binding sites on the unbinding forces. Removing the C-terminal binding site in Lmod2 might affect other actin-binding sites. Lmod21-201 showed a unimodal distribution with a peak value 58.2±1.4 pN (Figure 4c) that was close to population #1 (56.0±2.9 pN) of full-length Lmod2. Similarly, there was a statistically significant difference between Lmod21-201 and populations #1 of full-length Lmod2 (p<0.05).
The comparison of the unbinding forces of the individual binding sites of Lmod2 indicated that the interactions measured between actin and the WH2 domain were the strongest (114.3±1.9 pN) and the interactions measured between actin and the N-terminal actin-binding site (A1) were the weakest (56.0±2.9 pN) and close in strength to interactions measured for Tmods. This result is in contradiction with a previous study using bio-layer interferometry, where the three actin-binding sites were found to have similar dissociation constants with G-actin [44]. It is worth to note that Chen et al. used smaller fragments of Lmod2 and a nonpolymerizable Drosophila 5C actin mutant to prevent actin polymerization during measurements, while we used WT α-skeletal muscle actin from chicken in this study. This difference, in addition to different sensitivity levels of the utilized methods, may account for the inconsistency between the results of these two studies.
Correlation between actin-nucleating abilities and unbinding force distributions
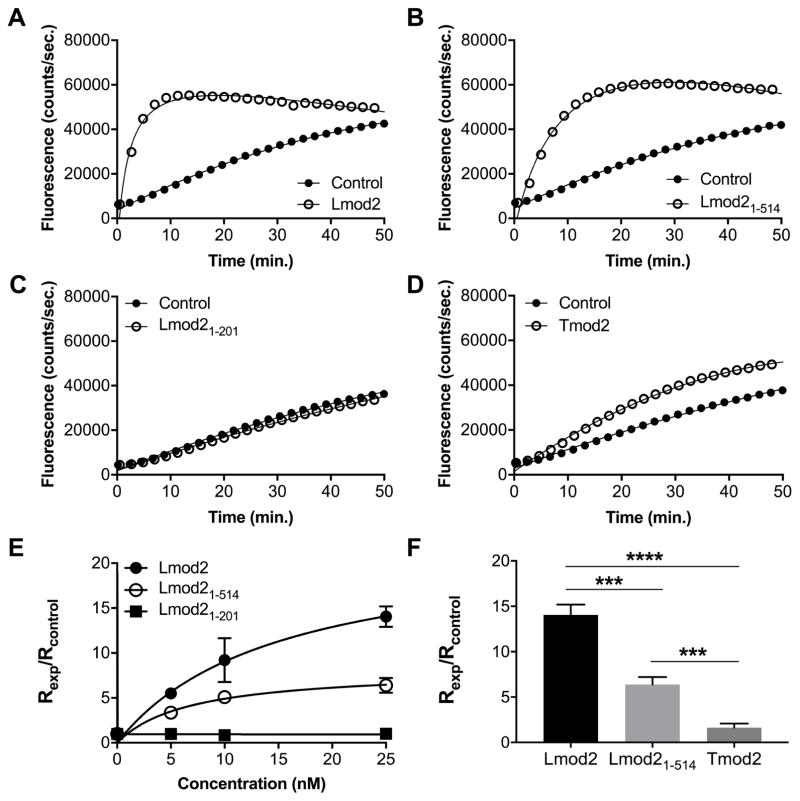
Pyrene-actin fluorescence experiments with Lmod2 and its fragments demonstrated that both full-length Lmod2 and Lmod21-514 nucleate actin polymerization in a concentration-dependent fashion (Figure 5a, b) and confirmed previous results [13, 27]. The polymerization rate of actin in the presence of Lmod21-514 was ~2-fold slower than that in the presence of full-length Lmod2 (Figure 5e, f). Lmod21-201 did not nucleate actin polymerization; the rate of actin polymerization in the presence of Lmod21-201, was essentially the same as the rate of spontaneous actin polymerization (control) at all tested concentrations (Figure 5c).
Figure 5.
Nucleation of actin polymerization by Lmod2. 1 μM actin was polymerized in the presence of Tmod2 or Lmod2, full-length or fragments. Representative actin polymerization curves in the presence of 25 nM Lmod2 (A–C) or Tmod2 (D) (control: actin only). E: Concentration dependence of the polymerization rates in the presence of Lmod2. F: Comparison of the polymerization rates in the presence of 25 nM Tmod2 or Lmod2. Rates of polymerization (Rexp) were calculated from fluorescence curves and normalized by dividing Rexp by the rate of spontaneous actin polymerization (Rcontrol). Data points are averages and error bars are standard deviations (n=3–4). Asterisks demonstrate statistically significant differences between groups that were determined by Student’s t-test (***p<0.001, ****p<0.0001).
Lmod21-514 exhibited two populations of unbinding forces, whereas WT-Lmod2 demonstrated three populations of unbinding forces. This finding indicates that the absence of the WH2 domain, which is a well-known actin-binding site [56], in Lmod21-514 resulted in the loss of its binding to at least one actin molecule. The weaker actin-nucleating ability of Lmod21-514 compared to that of full-length Lmod2 also supports this finding. Lmod21-201 demonstrated a single unbinding force population, which suggests that it interacts with one molecule of actin. Additionally, Lmod21-201 failed to nucleate actin polymerization entirely. This result was not surprising since the nucleation of actin polymerization requires at least two actin molecules to be recruited. The binding of Lmod21-201 to only one actin molecule fails to create a kinetically favorable seed for polymerization.
Our data support the models of Lmod2 binding three actin molecules [12, 44]. The magnitude of the unbinding forces obtained for Lmod2 and Tmod2 in the AFM experiments were very similar, although Lmod2 is a more potent nucleator (Figure 5). The presence of additional actin-binding sites in Lmod2 explains this phenomenon. However, Lmod21-514, which lacks the third actin-binding site, can still nucleate actin polymerization at least ~5-times better than Tmod2 (Figure 5d, f). This can be explained by suggestion that the LRR domain in Lmod2 alone can recruit two actin molecules [43].
Possible effects of non-conserved residues on divergent actin-binding abilities of Tmod isoforms
Although the LRR domains of proteins from the tropomodulin family are well conserved, there are striking differences in their abilities to bind G-actin and nucleate actin polymerization. The variances in amino acid sequences that form homologous domains provide specialized functions to Tmod and Lmod. To find out what residues in the LRR domain may cause these differences, we aligned sequences of the LRR domains from Tmod1, Tmod2, Tmod3 and Lmod2 (Figure S1A). We chose residues that were either different for all isoforms or similar in Tmod1 and Tmod3 only. The residues that have difference in charge of their side chains, such as polar vs non-polar, were considered. We checked positions and interactions of the chosen residues in known structures of Tmod1 (PDB # 4PKI) and Lmod2 (PDB # 4RWT and 5WFN) with actin.
Of twelve residues, ones in positions that correspond to N192, F242, S278, T280, and N297 in Tmod1 have exposed side chains not available for interactions with actin. Therefore, the residues in these positions should not be responsible for the differences. Residues in positions that correspond to S267 and Q321 in Tmod1 have exposed side chains available for interaction with actin. These residues are within eleven residues that were changed in Tmod1 to corresponding residues in Lmod2 sequence [43]. These mutations made the LRR domain of Tmod1 able to nucleate actin polymerization similar to the LRR domain of Lmod2. Corresponding residues in Tmod2 and Tmod3 are more similar to those in Tmod1 than in Lmod2. Therefore, while these residues are responsible for high nucleation ability of Lmod2, most likely they are not responsible for differences in G-actin binding between Tmod isoforms.
Residues in positions that correspond to Q191, E198, and A217 in Tmod1 are available for forming salt bridges with residues in an adjacent LRR helix. Formation or absence of the salt bridges may change the LRR’s curvature that consequently may affect interactions with actin monomers. A residue in the position that corresponds to N330 in Tmod1 may affect hydrophobic interactions with actin when a residue with a polar side chain is present. However, without knowing crystal structures of the LRR domains of Tmod2 and Tmod3 it is difficult to make an exact prediction.
Among all residues we examined, the most interesting is a residue in the position that corresponds to N348 of Tmod1. In the crystal structure of the complex of actin with the C-terminal domain of Lmod2 (PDB # 4RWT and 5WFN), Lmod2’s C-terminal helix in the LRR is two residues longer than in the Tmod1’s crystal structure and it includes K377 (R377 in our sequence) that corresponds to N348 of Tmod1. Tmod2 has Arg, and Tmod3 has His in this position. There is Glu in position 366 in an actin’s helix close to K377. Although side chains of E366 and K377 do not form a salt bridge in the crystal structure (Figure S1B), the distance between backbones of the helices is short enough for the salt bridge to form. If the side chains are more flexible in solution than in a crystal form, they may interact (Figure S1C). This salt bridge may also be formed in a complex of a Tmod2, but not Tmod1 or Tmod3, with actin. This residue may be at least one of reasons why Tmod2 is different from Tmod1 and Tmod3 in the binding G-actin via the LRR domain. Note, that when this residue was removed along with four other C-terminal residues (Figure 1), Tmod2 became unable to nucleate actin polymerization [24].
Solving the crystal structures of Tmod2 and Tmod3 alone and with actin monomers will be helpful for comparing the properties of tropomodulin isoforms. However, the most convenient tool to fully understand differences in function of proteins of tropomodulin family will be high-resolution electron microscopy (EM). Determining structures of Tmod or Lmod in a complex with actin filament by EM will shed light upon the fundamental differences between the actin-binding modes of Tmod and Lmod.
CONCLUSIONS
The unbinding forces between G-actin and proteins from the tropomodulin family were quantified and characterized using single molecule force spectroscopy. By creating mutants or fragments of Tmod2 and Lmod2, we assigned specific unbinding forces to their individual actin-binding sites. Our results demonstrate how the differences between the number and the strength of the actin-binding sites translate to the actin-sequestering and -nucleating abilities of Lmod and Tmod. Finally, our results confirm that Lmod2 binds at least three molecules of actin. In agreement with our recent report [12] and data published by Chen and co-authors [44], we conclude that the N-terminal domain of Lmod2 contains an actin-binding site.
AFM provides a valuable approach to quantify interactions between biomolecules [52]. AFM’s ability to quantify biomolecular forces with pico-newton resolution in liquid environments [47, 57] allowed researchers to investigate protein-ligand [58], protein-DNA [59], protein-protein [60], and protein-biomolecule [61] interactions at the nanoscale. The ability to work with full-length proteins, the use of relatively low protein concentrations (~0.5 to 3 μM in this study) and stability of actin in a monomeric form make AFM a promising technique for measuring forces of interaction of G-actin with other actin-nucleating and actin-sequestering proteins.
Supplementary Material
Highlights.
Unbinding forces between G-actin and Tmod1, Tmod2, Tmod3 or Lmod2 were quantified.
Tmod1 and Tmod3 have unimodal unbinding force distributions.
Tmod2 has a bimodal force distribution and Lmod2 has a trimodal distribution.
Specific unbinding forces were assigned to actin-binding sites of Tmod2 and Lmod2.
The existence of the N-terminal actin-binding site in Lmod2 was confirmed.
Acknowledgments
The authors thank Christopher Keller for assistance in Tmod2[L73D] preparation. This work was supported by the National Institutes of Health grants GM081688 and GM120137 to ASK. We thank Dr. Carol Gregorio for providing WT-Lmod2, Lmod21-514 and Lmod21-201.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Blanchoin L, Boujemaa-Paterski R, Sykes C, Plastino J. Actin dynamics, architecture, and mechanics in cell motility. Physiological reviews. 2014;94(1):235–63. doi: 10.1152/physrev.00018.2013. [DOI] [PubMed] [Google Scholar]
- 2.Pollard TD, Cooper JA. Actin, a central player in cell shape and movement. Science. 2009;326(5957):1208–12. doi: 10.1126/science.1175862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bugyi B, Carlier MF. Control of actin filament treadmilling in cell motility. Annu Rev Biophys. 2010;39:449–70. doi: 10.1146/annurev-biophys-051309-103849. [DOI] [PubMed] [Google Scholar]
- 4.Kudryashov DS, Reisler E. ATP and ADP actin states. Biopolymers. 2013;99(4):245–56. doi: 10.1002/bip.22155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cooper JA, Buhle EL, Jr, Walker SB, Tsong TY, Pollard TD. Kinetic evidence for a monomer activation step in actin polymerization. Biochemistry. 1983;22(9):2193–202. doi: 10.1021/bi00278a021. [DOI] [PubMed] [Google Scholar]
- 6.Sept D, McCammon JA. Thermodynamics and kinetics of actin filament nucleation. Biophysical journal. 2001;81(2):667–74. doi: 10.1016/S0006-3495(01)75731-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.dos Remedios CG, Chhabra D, Kekic M, Dedova IV, Tsubakihara M, Berry DA, Nosworthy NJ. Actin binding proteins: regulation of cytoskeletal microfilaments. Physiological reviews. 2003;83(2):433–73. doi: 10.1152/physrev.00026.2002. [DOI] [PubMed] [Google Scholar]
- 8.Fletcher DA, Mullins RD. Cell mechanics and the cytoskeleton. Nature. 2010;463(7280):485–92. doi: 10.1038/nature08908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Campellone KG, Welch MD. A nucleator arms race: cellular control of actin assembly. Nat Rev Mol Cell Biol. 2010;11(4):237–51. doi: 10.1038/nrm2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Colpan M, Moroz NA, Kostyukova AS. Tropomodulins and tropomyosins: working as a team. Journal of muscle research and cell motility. 2013;34(3–4):247–60. doi: 10.1007/s10974-013-9349-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fowler VM, Dominguez R. Tropomodulins and Leiomodins: Actin Pointed End Caps and Nucleators in Muscles. Biophysical journal. 2017;112(9):1742–1760. doi: 10.1016/j.bpj.2017.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ly T, Moroz N, Pappas CT, Novak SM, Tolkatchev D, Wooldridge D, Mayfield RM, Helms G, Gregorio CC, Kostyukova AS. The N-terminal tropomyosin- and actin-binding sites are important for leiomodin 2’s function. Molecular biology of the cell. 2016;27(16):2565–75. doi: 10.1091/mbc.E16-03-0200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tsukada T, Pappas CT, Moroz N, Antin PB, Kostyukova AS, Gregorio CC. Leiomodin-2 is an antagonist of tropomodulin-1 at the pointed end of the thin filaments in cardiac muscle. Journal of cell science. 2010;123(Pt 18):3136–45. doi: 10.1242/jcs.071837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Conley CA, Fritz-Six KL, Almenar-Queralt A, Fowler VM. Leiomodins: larger members of the tropomodulin (Tmod) gene family. Genomics. 2001;73(2):127–39. doi: 10.1006/geno.2000.6501. [DOI] [PubMed] [Google Scholar]
- 15.Fowler VM. Identification and purification of a novel Mr 43,000 tropomyosin-binding protein from human erythrocyte membranes. The Journal of biological chemistry. 1987;262(26):12792–800. [PubMed] [Google Scholar]
- 16.Fowler VM, Sussmann MA, Miller PG, Flucher BE, Daniels MP. Tropomodulin is associated with the free (pointed) ends of the thin filaments in rat skeletal muscle. The Journal of cell biology. 1993;120(2):411–20. doi: 10.1083/jcb.120.2.411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sussman MA, Sakhi S, Barrientos P, Ito M, Kedes L. Tropomodulin in rat cardiac muscle. Localization of protein is independent of messenger RNA distribution during myofibrillar development. Circulation research. 1994;75(2):221–32. doi: 10.1161/01.res.75.2.221. [DOI] [PubMed] [Google Scholar]
- 18.Watakabe A, Kobayashi R, Helfman DM. N-tropomodulin: a novel isoform of tropomodulin identified as the major binding protein to brain tropomyosin. Journal of cell science. 1996;109(Pt 9):2299–310. doi: 10.1242/jcs.109.9.2299. [DOI] [PubMed] [Google Scholar]
- 19.Sussman MA, Sakhi S, Tocco G, Najm I, Baudry M, Kedes L, Schreiber SS. Neural tropomodulin: developmental expression and effect of seizure activity. Brain research Developmental brain research. 1994;80(1–2):45–53. doi: 10.1016/0165-3806(94)90088-4. [DOI] [PubMed] [Google Scholar]
- 20.Cox PR, Zoghbi HY. Sequencing, expression analysis, and mapping of three unique human tropomodulin genes and their mouse orthologs. Genomics. 2000;63(1):97–107. doi: 10.1006/geno.1999.6061. [DOI] [PubMed] [Google Scholar]
- 21.Almenar-Queralt A, Lee A, Conley CA, Ribas de Pouplana L, Fowler VM. Identification of a novel tropomodulin isoform, skeletal tropomodulin, that caps actin filament pointed ends in fast skeletal muscle. The Journal of biological chemistry. 1999;274(40):28466–75. doi: 10.1074/jbc.274.40.28466. [DOI] [PubMed] [Google Scholar]
- 22.Fischer RS, Yarmola EG, Weber KL, Speicher KD, Speicher DW, Bubb MR, Fowler VM. Tropomodulin 3 binds to actin monomers. The Journal of biological chemistry. 2006;281(47):36454–65. doi: 10.1074/jbc.M606315200. [DOI] [PubMed] [Google Scholar]
- 23.Yamashiro S, Speicher KD, Speicher DW, Fowler VM. Mammalian tropomodulins nucleate actin polymerization via their actin monomer binding and filament pointed end-capping activities. The Journal of biological chemistry. 2010;285(43):33265–80. doi: 10.1074/jbc.M110.144873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Colpan M, Moroz NA, Gray KT, Cooper DA, Diaz CA, Kostyukova AS. Tropomyosin-binding properties modulate competition between tropomodulin isoforms. Arch Biochem Biophys. 2016;600:23–32. doi: 10.1016/j.abb.2016.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nanda V, Miano JM. Leiomodin 1, a new serum response factor-dependent target gene expressed preferentially in differentiated smooth muscle cells. The Journal of biological chemistry. 2012;287(4):2459–67. doi: 10.1074/jbc.M111.302224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Conley CA. Leiomodin and tropomodulin in smooth muscle. American journal of physiology Cell physiology. 2001;280(6):C1645–56. doi: 10.1152/ajpcell.2001.280.6.C1645. [DOI] [PubMed] [Google Scholar]
- 27.Chereau D, Boczkowska M, Skwarek-Maruszewska A, Fujiwara I, Hayes DB, Rebowski G, Lappalainen P, Pollard TD, Dominguez R. Leiomodin is an actin filament nucleator in muscle cells. Science. 2008;320(5873):239–43. doi: 10.1126/science.1155313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yuen M, Sandaradura SA, Dowling JJ, Kostyukova AS, Moroz N, Quinlan KG, Lehtokari VL, Ravenscroft G, Todd EJ, Ceyhan-Birsoy O, Gokhin DS, Maluenda J, Lek M, Nolent F, Pappas CT, Novak SM, D’Amico A, Malfatti E, Thomas BP, Gabriel SB, Gupta N, Daly MJ, Ilkovski B, Houweling PJ, Davidson AE, Swanson LC, Brownstein CA, Gupta VA, Medne L, Shannon P, Martin N, Bick DP, Flisberg A, Holmberg E, Van den Bergh P, Lapunzina P, Waddell LB, Sloboda DD, Bertini E, Chitayat D, Telfer WR, Laquerriere A, Gregorio CC, Ottenheijm CA, Bonnemann CG, Pelin K, Beggs AH, Hayashi YK, Romero NB, Laing NG, Nishino I, Wallgren-Pettersson C, Melki J, Fowler VM, MacArthur DG, North KN, Clarke NF. Leiomodin-3 dysfunction results in thin filament disorganization and nemaline myopathy. J Clin Invest. 2014;124(11):4693–708. doi: 10.1172/JCI75199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chu X, Chen J, Reedy MC, Vera C, Sung KL, Sung LA. E-Tmod capping of actin filaments at the slow-growing end is required to establish mouse embryonic circulation. American journal of physiology Heart and circulatory physiology. 2003;284(5):H1827–38. doi: 10.1152/ajpheart.00947.2002. [DOI] [PubMed] [Google Scholar]
- 30.Fritz-Six KL, Cox PR, Fischer RS, Xu B, Gregorio CC, Zoghbi HY, Fowler VM. Aberrant myofibril assembly in tropomodulin1 null mice leads to aborted heart development and embryonic lethality. The Journal of cell biology. 2003;163(5):1033–44. doi: 10.1083/jcb.200308164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McKeown CR, Nowak RB, Moyer J, Sussman MA, Fowler VM. Tropomodulin1 is required in the heart but not the yolk sac for mouse embryonic development. Circulation research. 2008;103(11):1241–8. doi: 10.1161/CIRCRESAHA.108.178749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ono Y, Schwach C, Antin PB, Gregorio CC. Disruption in the tropomodulin1 (Tmod1) gene compromises cardiomyocyte development in murine embryonic stem cells by arresting myofibril maturation. Dev Biol. 2005;282(2):336–48. doi: 10.1016/j.ydbio.2005.03.015. [DOI] [PubMed] [Google Scholar]
- 33.Cox PR, Fowler V, Xu B, Sweatt JD, Paylor R, Zoghbi HY. Mice lacking Tropomodulin-2 show enhanced long-term potentiation, hyperactivity, and deficits in learning and memory. Mol Cell Neurosci. 2003;23(1):1–12. doi: 10.1016/s1044-7431(03)00025-3. [DOI] [PubMed] [Google Scholar]
- 34.Sui Z, Nowak RB, Bacconi A, Kim NE, Liu H, Li J, Wickrema A, An XL, Fowler VM. Tropomodulin3-null mice are embryonic lethal with anemia due to impaired erythroid terminal differentiation in the fetal liver. Blood. 2014;123(5):758–67. doi: 10.1182/blood-2013-03-492710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pappas CT, Mayfield RM, Henderson C, Jamilpour N, Cover C, Hernandez Z, Hutchinson KR, Chu M, Nam KH, Valdez JM, Wong PK, Granzier HL, Gregorio CC. Knockout of Lmod2 results in shorter thin filaments followed by dilated cardiomyopathy and juvenile lethality. Proceedings of the National Academy of Sciences of the United States of America. 2015;112(44):13573–8. doi: 10.1073/pnas.1508273112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cenik BK, Garg A, McAnally JR, Shelton JM, Richardson JA, Bassel-Duby R, Olson EN, Liu N. Severe myopathy in mice lacking the MEF2/SRF-dependent gene leiomodin-3. J Clin Invest. 2015;125(4):1569–78. doi: 10.1172/JCI80115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nworu CU, Kraft R, Schnurr DC, Gregorio CC, Krieg PA. Leiomodin 3 and tropomodulin 4 have overlapping functions during skeletal myofibrillogenesis. Journal of cell science. 2015;128(2):239–50. doi: 10.1242/jcs.152702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fowler VM, Greenfield NJ, Moyer J. Tropomodulin contains two actin filament pointed end-capping domains. The Journal of biological chemistry. 2003;278(41):40000–9. doi: 10.1074/jbc.M306895200. [DOI] [PubMed] [Google Scholar]
- 39.Kostyukova AS, Rapp BA, Choy A, Greenfield NJ, Hitchcock-DeGregori SE. Structural requirements of tropomodulin for tropomyosin binding and actin filament capping. Biochemistry. 2005;44(12):4905–10. doi: 10.1021/bi047468p. [DOI] [PubMed] [Google Scholar]
- 40.Kostyukova AS, Choy A, Rapp BA. Tropomodulin binds two tropomyosins: a novel model for actin filament capping. Biochemistry. 2006;45(39):12068–75. doi: 10.1021/bi060899i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Colpan M, Tolkatchev D, Grover S, Helms GL, Cort JR, Moroz N, Kostyukova AS. Localization of the binding interface between leiomodin-2 and alpha-tropomyosin. Biochimica et biophysica acta. 2016;1864(5):523–30. doi: 10.1016/j.bbapap.2016.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kostyukova AS. Leiomodin/tropomyosin interactions are isoform specific. Arch Biochem Biophys. 2007;465(1):227–30. doi: 10.1016/j.abb.2007.05.012. [DOI] [PubMed] [Google Scholar]
- 43.Boczkowska M, Rebowski G, Kremneva E, Lappalainen P, Dominguez R. How Leiomodin and Tropomodulin use a common fold for different actin assembly functions. Nat Commun. 2015;6:8314. doi: 10.1038/ncomms9314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen X, Ni F, Kondrashkina E, Ma J, Wang Q. Mechanisms of leiomodin 2-mediated regulation of actin filament in muscle cells. Proceedings of the National Academy of Sciences of the United States of America. 2015;112(41):12687–92. doi: 10.1073/pnas.1512464112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kostyukova AS, Hitchcock-DeGregori SE. Effect of the structure of the N terminus of tropomyosin on tropomodulin function. The Journal of biological chemistry. 2004;279(7):5066–71. doi: 10.1074/jbc.M311186200. [DOI] [PubMed] [Google Scholar]
- 46.Rao JN, Madasu Y, Dominguez R. Mechanism of actin filament pointed-end capping by tropomodulin. Science. 2014;345(6195):463–7. doi: 10.1126/science.1256159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hinterdorfer P, Dufrene YF. Detection and localization of single molecular recognition events using atomic force microscopy. Nat Methods. 2006;3(5):347–355. doi: 10.1038/nmeth871. [DOI] [PubMed] [Google Scholar]
- 48.Kostyukova A, Maeda K, Yamauchi E, Krieger I, Maeda Y. Domain structure of tropomodulin: distinct properties of the N-terminal and C-terminal halves. European journal of biochemistry/FEBS. 2000;267(21):6470–5. doi: 10.1046/j.1432-1327.2000.01738.x. [DOI] [PubMed] [Google Scholar]
- 49.Moroz N, Guillaud L, Desai B, Kostyukova AS. Mutations changing tropomodulin affinity for tropomyosin alter neurite formation and extension. PeerJ. 2013;1:e7. doi: 10.7717/peerj.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fritz M, Radmacher M, Cleveland JP, Allersma MW, Stewart RJ, Gieselmann R, Janmey P, Schmidt CF, Hansma PK. Imaging Globular and Filamentous Proteins in Physiological Buffer Solutions with Tapping Mode Atomic Force Microscopy. Langmuir. 1995;11(9):3529–3535. [Google Scholar]
- 51.Hutter JL, Bechhoefer J. Calibration of Atomic-Force Microscope Tips. Review of Scientific Instruments. 1993;64(7):1868–1873. [Google Scholar]
- 52.Lee CK, Wang YM, Huang LS, Lin S. Atomic force microscopy: determination of unbinding force, off rate and energy barrier for protein-ligand interaction. Micron. 2007;38(5):446–61. doi: 10.1016/j.micron.2006.06.014. [DOI] [PubMed] [Google Scholar]
- 53.Willemsen OH, Snel MM, van der Werf KO, de Grooth BG, Greve J, Hinterdorfer P, Gruber HJ, Schindler H, van Kooyk Y, Figdor CG. Simultaneous height and adhesion imaging of antibody-antigen interactions by atomic force microscopy. Biophysical journal. 1998;75(5):2220–8. doi: 10.1016/S0006-3495(98)77666-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dammer U, Hegner M, Anselmetti D, Wagner P, Dreier M, Huber W, Güntherodt HJ. Specific antigen/antibody interactions measured by force microscopy. Biophysical journal. 1996;70(5):2437–2441. doi: 10.1016/S0006-3495(96)79814-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Binnig G, Quate CF, Gerber C. Atomic Force Microscope. Physical Review Letters. 1986;56(9):930–933. doi: 10.1103/PhysRevLett.56.930. [DOI] [PubMed] [Google Scholar]
- 56.Dominguez R. The WH2 Domain and Actin Nucleation: Necessary but Insufficient. Trends Biochem Sci. 2016;41(6):478–490. doi: 10.1016/j.tibs.2016.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lee GU, Kidwell DA, Colton RJ. Sensing Discrete Streptavidin-Biotin Interactions with Atomic Force Microscopy. Langmuir. 1994;10(2):354–357. [Google Scholar]
- 58.Hinterdorfer P, Baumgartner W, Gruber HJ, Schilcher K, Schindler H. Detection and localization of individual antibody-antigen recognition events by atomic force microscopy. Proceedings of the National Academy of Sciences of the United States of America. 1996;93(8):3477–3481. doi: 10.1073/pnas.93.8.3477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hamon L, Pastré D, Dupaigne P, Breton CL, Cam EL, Piétrement O. High-resolution AFM imaging of single-stranded DNA-binding (SSB) protein—DNA complexes. Nucleic Acids Research. 2007;35(8):e58–e58. doi: 10.1093/nar/gkm147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nakajima H, Kunioka Y, Nakano K, Shimizu K, Seto M, Ando T. Scanning force microscopy of the interaction events between a single molecule of heavy meromyosin and actin. Biochemical and biophysical research communications. 1997;234(1):178–82. doi: 10.1006/bbrc.1997.6612. [DOI] [PubMed] [Google Scholar]
- 61.Arslan B, Colpan M, Ju X, Zhang X, Kostyukova A, Abu-Lail NI. The Effects of Noncellulosic Compounds on the Nanoscale Interaction Forces Measured between Carbohydrate-Binding Module and Lignocellulosic Biomass. Biomacromolecules. 2016;17(5):1705–15. doi: 10.1021/acs.biomac.6b00129. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.