Abstract
HIV-1 requires a specialized nuclear export pathway to transport unspliced and partially spliced viral transcripts to the cytoplasm. Central to this pathway is the viral protein Rev, which binds to the Rev Response Element in stem IIB located on unspliced viral transcripts and subsequently oligomerizes in a cooperative manner. Previous work identified a number of cellular DEAD-box helicases as in vivo binding partners of Rev, and siRNA experiments indicated a functional role for many in the HIV replication cycle. Two DEAD-box proteins, DDX1 and DDX3, had previously been shown to play a role in HIV pathogenesis. In this study, another protein identified in that screen, DDX21, is tested for protein and RNA binding and subsequent enzymatic activities in the context of the Rev/RRE pathway. We found that DDX21 can bind to the RRE with high affinity, and this binding stimulates ATPase activity with an enzymatic efficiency similar to DDX1. Further, DDX21 is both an ATP-dependent and ATP-independent helicase, and both ATPase and ATP-dependent helicase activities are inhibited by Rev in a dose-dependent manner, though ATP-independent helicase activity is not. A conserved binding interaction between DDX protein’s DEAD-domain and Rev was identified, with Rev’s nuclear diffusion inhibitory signal motif playing a significant role in binding. Finally, DDX21 was shown to enhance Rev binding to the RRE in a manner similar to that previously described for DDX1, although DDX3 does not. These data indicate that DDX1 and DDX21 have similar biochemical activities with regard to the Rev/RRE system, while DDX3 differs.
Graphical abstract
Background
During HIV replication, transcription of proviral DNA is functionally linked to the splicing, export, and translation of HIV RNA. The early protein Regulator of Expression of Virion proteins, Rev, is transported into the nucleus and binds to the Rev-Responsive Element (RRE) RNA located in the env gene of unspliced or singly spliced viral RNA transcripts. This results in transport of those RNAs from the nucleus to the cytoplasm via the host cell CRM-1 export pathway[1].
Rev is a 116-amino acid protein, with a structured N-terminus responsible for RNA binding, Rev oligomerization and nuclear import, and an unstructured C-terminus responsible for nuclear export (Figure 1a). The shuttling of Rev between the cytoplasm and the nucleus occurs by temporally controlled engagement of either the Nuclear Localization Signal (NLS) or Nuclear Export Signal (NES). Rev monomers bind to the RRE RNA via a high affinity binding site located within stem II (Figure 1b) and subsequently oligomerize in a stepwise and cooperative manner[2–9]. This oligomerized RNP product is the substrate for binding to CRM-1, resulting in export of the viral transcript from the nucleus.
Figure 1.
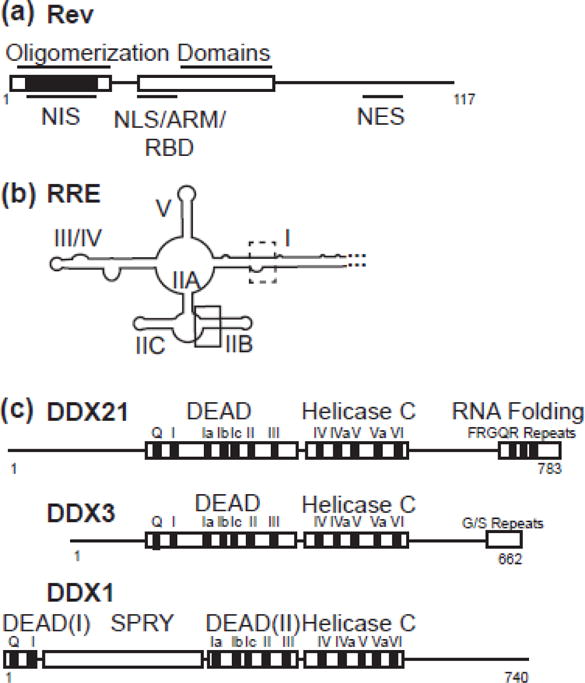
Schematic diagrams of proteins and RNA used in this study. a) Rev, with structured helix-turn helix domains boxed. Oligomerization domains, nuclear export inhibitory signal (nis), nuclear localization signal (NLS), arginine rich motif (ARM), RNA binding domain (RBD) and nuclear export signal (NES) labelled. NIS region has additionally been colored black. b) RRE RNA molecule, with stems I, IIA, IIB, IIC, III/IV, and V labelled. High affinity primary binding site boxed in black, and lower affinity binding site boxed in dotted line. c) DDX21, DDX3 and DDX1 diagrams aligned to DEAD domain and Helicase C domain interface. Accessory domains are labelled.
While many cellular components of the CRM-1 export machinery have been characterized, other factors that might be involved in Rev-related functions are still not well understood. Work to identify other Rev cofactors led to the identification of a number of DEAD-box helicases that are able to interact with Rev-containing complexes (RCCs) intracellularly[10]. Two of these DEAD-box helicases, DDX1 and DDX3, had been previously implicated in the HIV-1 Rev regulatory pathway[10–22]. DDX1 directly interacts with Rev and RCCs, playing a role in the correct localization of Rev within the nucleus in vivo, as well as affecting Rev’s association with and oligomerization on the RRE in vitro[10, 12, 14, 15, 23, 24]. DDX3 affects HIV production at the mRNA export, protein translation and budding steps of the viral lifecycle, though what role, if any, Rev may play in these processes remains unclear[10, 18, 19, 21]. The roles of the other helicases that were found to interact with Rev, including DDX21, have not been fully characterized, nor have their biophysical interactions with or biochemical implications to Rev and Rev-dependent processes.
DDX21, also known as nucleolar RNA helicase 2 and Gu, is highly expressed in various types of cancers[25, 26], and is implicated in multiple steps of ribosome biogenesis, namely rRNA processing[27], coordination between transcription and rRNA processing[28] and the recruitment of snoRNAs into pre-ribosome complexes[29]. DDX21 also inhibits Influenza A viral replication by interacting with viral PB1 protein and inhibiting the viral RNA polymerase [30], and has been implicated in the dsRNA response in dendritic cells[31]. Previous immunoprecipitation (IP) experiments followed by phenotypic characterization of protein knockdown indicated that HIV requires DDX21 for efficient viral production[10]. Further, this DDX21-dependent activity was shown to affect the distribution of unspliced RNA in the cell, similar to that seen with DDX1, and therefore may be a functional partner for Rev-related processes[10]. The DDX21 protein contains all the signature motifs (Q, I–VI) required for DEAD-helicase function (Figure 1c) (for a review of DEAD-box helicases, see [32]). In addition to the conserved helicase core domains seen in DDX1and DDX3, DDX21 contains several atypical FRGQR repeats located in its C-terminus that contribute to its RNA binding and RNA folding activities [33, 34].
In this study, the RNA-dependent ATPase, RNA binding, and protein binding activities of DDX21 are compared to the previously published results for DDX1 and DDX3 [13, 23, 35]. Further, these activities are discussed in the context of the Rev:RRE regulatory pathway, including the effect of each helicase on Rev oligomerization on the RRE.
Results
DDX21 is an RNA stimulated ATPase
Previous work established both DDX1 and DDX3 as RNA stimulated ATPases[13]. For DDX1, this activity was stimulated by various RNAs including tRNA, RRE RNA and stem IIB RRE RNA (the high affinity binding site of Rev). DDX21 ATP hydrolysis activity was measured using a steady-state assay where ATP hydrolysis is enzymatically coupled to the oxidation of NADH via pyruvate kinase and lactate dehydrogenase[36]. The consumption of NADH is optically monitored at 340nm as a read-out of ATPase activity. Steady state kinetic assays were carried out while titrating the ATP concentration in the presence of saturating amounts of stem IIB RNA (Figure 2a). The kcat for ATP hydrolysis was 0.67 min−1 with a KmATP of 90μM (Figure 2a). This leads to a kcat/Km value of 7.4 mM−1min−1, an enzymatic efficiency comparable with previously reported DDX1 data, although the underlying kcat and Km values are very different (Table 1). While both ATP hydrolysis efficiencies are within previously published ranges for other DEAD-box helicases, the values are 5–10 fold lower than published results for DDX3 (Table 1)[13, 35].
Figure 2.
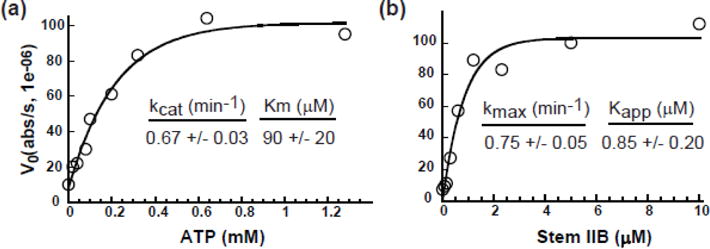
DDX21 ATPase activity is RNA dependent. a) Spectroscopic ATPase activity of DDX21 in the presence of 5 μM stem IIB RNA measured as a function of ATP concentration. Kcat and Km values noted. b) Increasing concentrations of stem IIB RNA were titrated into DDX21 in the presence of 2.5 mM ATP, with kmaxIIB and KappIIB values noted.
Table 1.
Steady-state ATPase kinetics of DDX21
| ATP titration (in presence of excess RNA) | ||||
|---|---|---|---|---|
| Construct | kcat (min−1) | Km (ATP) (mM) | kcat/Km (min−1mM−1) | |
| DDX21 | 0.67 +/− 0.03 | 0.09 +/− 0.2 | 7.4 | |
| DDX3* | 3.2 +/− 0.1 | .045 +/− .003 | 71.1 | |
| DDX1** | 15.2 +/− 0.6 | 1.0 +/− 0.1 | 15.2 | |
| RNA titration (in presence of excess ATP) | ||||
| Construct | RNA | kmax (min−1) | Kapp (RNA) (μM) | kmax/Kapp (min−1 μM−1) |
| DDX21 | IIB | 0.75 +/− 0.05 | 0.85 +/− 0.2 | 0.9 |
| Poly (U)20 | 1.08 +/− 0.05 | 0.20 +/− 0.04 | 5.4 | |
| DDX1** | IIB | 16.0 +/− 0.4 | 0.50 +/− 0.05 | 32.0 |
| RRE | 16.8 +/− 0.6 | 0.24 +/− 0.05 | 70.0 | |
| tRNA | 13.5 +/− 0.6 | 1.9 +/− 0.3 | 7.1 | |
| RRE + 1× Rev | 17 +/− 1 | 0.2 +/− 0.1 | 85.0 | |
| RRE + 2X Rev | 16 +/− 1 | 0.3 +/− 0.2 | 53.0 | |
| RRE + 4X Rev | 13.5 +/− 0.6 | 1.9 +/− 0.3 | 7.1 | |
Data from Garbelli et al, 2011
Data from Edgcomb et al, 2011
To evaluate the role of RNA in DDX21 ATPase activity, ATP hydrolysis was tested as a function of RRE stem IIB concentration in the presence of saturating ATP, resulting in a kmaxIIB of 0.75 min−1 and a KappIIB of 0.85 μM (Figure 2b, Table 1). DDX21’s relative enzymatic specificity (kmax/KappIIB) of 0.88 represents a ~36-fold decrease when compared to DDX1 in the presence of stem IIB (Table 1)[13]. Previous DDX3 and DDX21 ATP hydrolysis studies utilized a poly U RNA substrate with both high specificity and efficiency[35]. DDX21 ATPase assays were repeated using a similar poly (U)20 reagent and resulted in kmax and Kapp(U)20 values of 1.08 min−1 and 0.20 μM respectively (Table 1), or a ~ 7 fold increase in kmax/Kapp(U)20 substrate specificity when compared to stem IIB. This may be a result of either a preference of DDX21 for the single-stranded nature of the poly-U sequence over a highly base-paired RRE stem IIB, or a sequence preference for uridines. While there is a difference in substrate specificity, both RNA substrates efficiently stimulate ATPase activity at levels comparable to those reported for other DEAD-box helicases[13, 37, 38].
DDX21 helicase is stimulated by, but not dependent on, ATP
While RNA-stimulated ATPase activity is often described as a byproduct of DEAD-box helicase activity, it does not directly assess the ability of an enzyme to dissociate base-paired nucleotides. To determine the helicase activity of DDX21, a recently developed fluorescence-based unwinding assay was adapted [39]. Briefly, a fluorescein labeled 10mer RNA is hybridized to an RNA 26mer labelled with a dabcyl quencher, designed so that duplex formation results in quenching of the fluorescent signal. The unwinding activity is measured in the presence of an excess of unlabeled competitor 14mer RNA, where strand separation results in irreversible dissociation of the quencher modified RNA, and an increase in overall sample fluorescence (Figure 3a).
Figure 3.
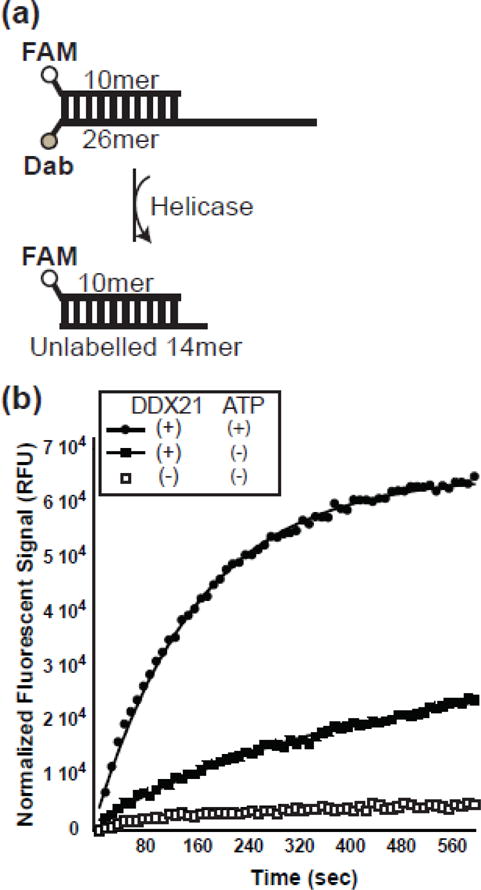
DDX21 helicase activity is stimulated by ATP. a) Schematic diagram of RNA molecules used in helicase assay. A fluorescently labelled RNA 10mer is annealed to a fluorescent quencher 26mer. Once the strands separate, the 10mer reanneals to the excess unlabeled 14mer, resulting in a fluorescent signal. b) Helicase assay results showing a graph of fluorescent intensity over time for RNA dimer alone (open boxes), DDX21 with RNA dimer (closed boxes) and DDX21 with ATP and RNA dimer (closed circles).
In the absence of DDX21 protein and ATP, there is little increase in fluorescence intensity over time, indicating a stable RNA hybrid (Figure 3b). In the presence of DDX21, and the absence of exogenous ATP, fluorescent signal increases over time, with an unwinding rate constant (kunw) of 0.11 +/− 0.01 min−1. It is possible that ATP was co-purified with the DDX21 protein, acting as the energy source for the observed “ATP-independent” helicase activity. This seems unlikely due to the fact that the amount of ATP that would be co-purified would be hydrolyzed quickly at the given RNA and protein concentrations. Another explanation would be contaminating RNase activity, but this also seems unlikely since the protein can be used in RNA binding assays (see below). Therefore, DDX21 exhibits a basal helicase activity that does not require the hydrolysis of excess ATP. When ATP is added to the DDX21 helicase assay, kunw increases threefold to 0.37 +/− 0.01 min−1. These data together indicate that while exogenous ATP may not be required, it does enhance RNA strand separation rates by DDX21.
Rev Inhibition of ATPase-related activities is due to RNA binding
DDX1 RNA-dependent ATPase activity is inhibited by the presence of Rev in a dose dependent manner (Table 1)[13]. Similarly, DDX21 shows a dose-dependent ATPase inhibition by Rev when using a stem IIB concentration of 3.3 μM and excess ATP (Figure 4). This decrease in activity could be due to Rev sequestering ATP from DDX21, sequestering RNA from DDX21, or directly inhibiting DDX21 activity through a protein-protein interaction. ATP hydrolysis assays were repeated in the presence of 1.65 μM stem IIB RNA to test the effects of RNA concentration on Rev-dependent inhibition, and indeed it inhibited DDX21 ATPase activity more efficiently at lower RNA concentrations (Figure 4a). Further, this inhibition was markedly relieved when an RNA-binding-defective Rev mutant, Rev N40A, was introduced instead of WT Rev (Figure 4a). This data together would indicate Rev-dependent inhibition of DDX21 activity is closely related to Rev’s ability to bind the RNA substrates, either titrating away the effective RNA reaction pools, or by directly inhibiting DDX21 activity in an RNA-bound form.
Figure 4.
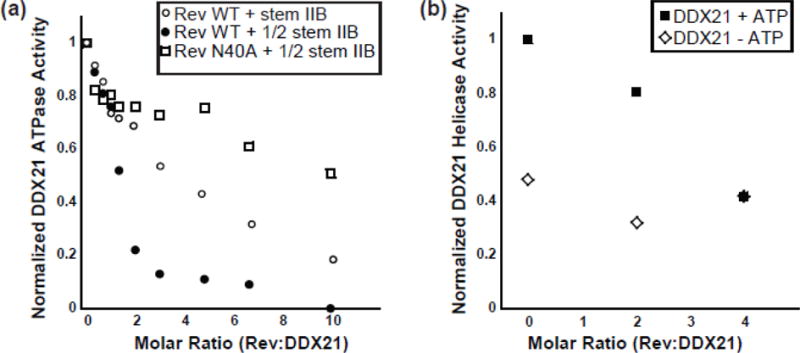
Rev inhibits DDX21 ATP hydrolysis and ATP-dependent helicase activities. a) Relative DDX21 ATP hydrolysis activity in the presence of 2 uM stem IIB and WT Rev (open circles), 1 uM stem IIB and WT Rev (closed circles) and 1 uM stem IIB and Rev N40A. b) Relative DDX21 RNA unwinding activity in the presence (closed boxes) and absence (open diamonds) of 2.5 mM ATP.
RNA helicase assays were performed using increasing concentrations of Rev to better understand the inhibition in DDX21-related enzymatic activities (Figure 4b, Table 2). In the presence of excess ATP, RNA helicase rates decreased when Rev was added in a dose-dependent manner. While this effect mirrors those observed with ATPase assays above, it is important to note that non-zero helicase activity remains at the endpoint of the titration. Indeed, DDX21 helicase activity levels out to rates seen for helicase assays performed in the absence of ATP (Figure 4b, Table 2). When the same experiment is performed in the absence of ATP, there is little change in the basal helicase activity, indicating Rev-dependent inhibition is somehow associated with ATP-dependent helicase activity only (Figure 4b, Table 2). This is somewhat surprising given that the same RNA substrate is used for both ATP dependent and independent activities. If Rev binding to the RNA substrate was inhibiting binding to DDX21, it would be expected that both ATP-dependent and ATP-independent activities are inhibited, which they clearly are not. This would indicate that the mechanism of inhibition is more complicated than titration of the RNA substrate from the active reaction pool by Rev, though it is unclear what that mechanism might be.
Table 2.
RNA helicase unwinding constants for DDX21
| DDX21 | ATP | kunw (min−1) |
|---|---|---|
| (−) | (−) | − |
| WT | (−) | 0.11 +/− 0.01 |
| WT | (+) | 0.37 +/− 0.01 |
| (183–620) | (−) | − |
| (183–620) | (+) | 0.29 +/− 0.01 |
| Rev Titration | ||
| WT + 0 Rev | (+) | 0.31 +/− 0.01 |
| WT + 2X Rev | (+) | 0.25 +/− 0.01 |
| WT + 4X Rev | (+) | 0.13 +/− 0.01 |
| WT + 0 Rev | (−) | 0.15 +/− 0.01 |
| WT + 2X Rev | (−) | 0.10 +/− 0.01 |
| WT + 4X Rev | (−) | 0.13 +/− 0.01 |
DDX21 interacts with Rev at low nM affinity directly through a conserved DEAD domain
Previous binding studies between DDX1 and Rev demonstrated a direct and high affinity interaction, with a dissociation constant of ~40 nM (Table 3)[13, 24]. Further, the binding site of DDX1 was mapped to the highly conserved DEAD domain of DDX1, and not the SPRY domain as previously suggested (Table 3)[24]. This led to the hypothesis that Rev interacts with DEAD-box proteins in a conserved manner. To assess this, previously established fluorescence binding assays were conducted between either DDX21 or DDX3 and fluorescently-labelled Rev[13]. DDX21 binds to Rev with a dissociation constant (Kd) of 21 +/− 7 nM, a value similar to DDX1, while DDX3 binds with a Kd of 122 +/− 15 nM (Figure 5a, Table 3). The DEAD domains of DDX3 and DDX21 were independently tested for binding to Rev, with near-WT binding dissociation constants of 60 +/− 9 nM and 200 +/− 83 nM respectively (Table 3). Therefore, the DEAD domain of each DEAD-box protein is sufficient for high affinity binding to Rev, again suggesting a conserved mechanism of binding between this family of proteins and Rev.
Table 3.
DDX Protein Equilibrium Dissociation Constants for Rev WT
| DExD-Box Protein | Kd (nM) |
|---|---|
| DDX21 | 21 +/− 7 |
| DDX21 DExD | 60 +/− 9 |
| DDX21 (183–620) | 48 +/− 27 |
| DDX3 | 122 +/− 15 |
| DDX3 DExD | 200 +/− 83 |
| DDX1 | 41 +/− 11 |
| DDX1 DExD* | 79 +/− 7 |
| DDX1 SPRY* | n.d. |
Data from Hammond et al, 2017
Figure 5.
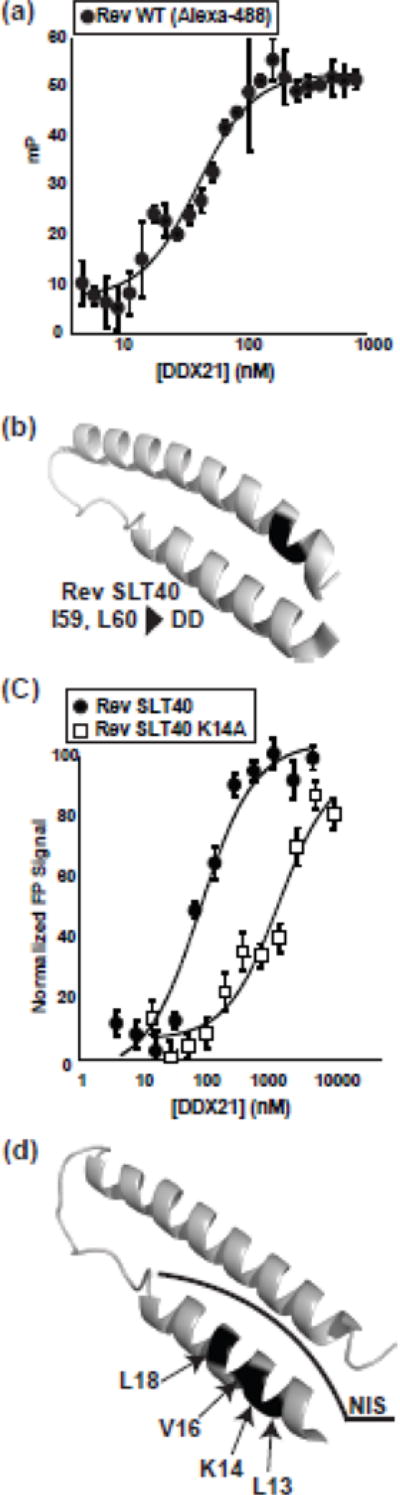
DDX proteins bind to Rev in a similar manner. a) A representative fluorescence polarization experiment with fluorescently labelled Rev and increasing concentrations of DDX21. For line fit, see materials and methods. b) Tertiary structure of Rev (pdb: 4PMI) with SLT40 mutations (I59D, I60D) colored black. c) Fluorescence binding data for Rev SLT40 and Rev SLT40 K14A) d) Tertiary structure of Rev (pdb: 4PMI) with residues 13, 14, 16 and 18 labelled and colored black. NIS region is marked by a black line.
Rev interaction with DDX21 is affected by four amino acids located within the NIS
Work by Fang et al demonstrated that a segment of Rev, the Nuclear Diffusion Inhibitory Signal (NIS), was sufficient to interact with DDX1 in vivo (diagramed in Figures 1C and 5d)[15, 40]. Further, mutations made in this region disrupt Rev-DDX1 interaction in vivo [13] and in vitro (Table 4). Because this region is also required for higher-order Rev multimerization, mutations that inhibit Rev-DDX interaction have been shown to also inhibit Rev-Rev interaction[2, 9, 41]. This has led to two possible models to explain binding defects of these mutants. Either the NIS region is directly responsible for binding to DDX proteins and/or DDX proteins may require Rev dimerization to bind.
Table 4.
Protein Equilibrium Dissociation Constants for Rev & Mutants
| DDX21 bound | |
|---|---|
| Rev Construct | Kd (nM) |
| Rev WT | 21 +/− 7 |
| Rev SLT40 | 49 +/− 17 |
| Rev SLT40 E10A | 29 +/− 17 |
| Rev SLT40 D11A | 42 +/− 19 |
| Rev SLT40 L12A | 240 +/− 23 |
| Rev SLT40 L13A | >1110 |
| Rev SLT40 K14A | >2714 |
| Rev SLT40 V16A | >1119 |
| Rev SLT40 V16D | >1225 |
| Rev SLT40 R17A | 89 +/− 55 |
| Rev SLT40 L18A | >630 |
| Rev SLT40 I19A | 315 +/− 250 |
| Rev SLT40 K20A | 79 +/− 19 |
| Rev SLT40 F21A | 210 +/− 154 |
| Rev SLT40 L22A | 297 +/− 258 |
| Rev SLT40 Y23A | 38 +/− 30 |
| Rev SLT40 Q24A | 54 +/− 30 |
| DDX1 bound | |
| Rev Construct | Kd (nM) |
| Rev WT | 41 +/− 11 |
| Rev V16D | >500 nM |
To distinguish which of these models is operative, a previously identified monomer-only Rev mutant, SLT40, was tested for binding to DDX21[42]. Here residues I59 and L60, positions located in one part of the Rev oligomerization domains, but outside the NIS, have been mutated to aspartic acids, leading to a defect in higher order Rev oligomerization (Figure 5b). Rev-SLT40 binds to DDX21 at near-WT affinity (Kd of 49 +/− 17) (Figure 5C, Table 4), which is consistent with a binding model where monomeric Rev binds to DDX21, with the Rev NIS acting as a DDX binding platform.
To further characterize the interaction between DDX21 and Rev, a single alanine scan of the monomeric SLT40 Rev NIS was performed (Table 4). While most alanine position mutations showed small to no binding defect, four positions, L13, K14, V16, and L18 showed substantial and reproducible defects. Of these, K14A displayed the largest binding defect with a greater than 100-fold decrease in binding affinity (Figures 5c, 5d and Table 4). These data taken together strongly implicate the NIS region in direct binding to DDX21, and by extension, to other DDX proteins.
DDX protein interaction profile with Rev in vivo
Multiple studies have investigated the interaction of DEAD proteins and Rev-containing complexes (RCCs) in vivo[10, 12, 13, 20–22]. For example, one study demonstrated that DDX21 or DDX3 interact with RCCs in vivo and suggested this interaction was differentially RNA dependent[10]. In another series of experiments, DDX1 interaction with RCCs was shown to be disrupted by a V16D mutation[13]. To more clearly compare the interactions of all three DEAD-Box proteins with RCCs, and the importance of RNA in this interaction, co-IP experiments were performed.
For this work, a stable HeLa cell line containing a chromosomally integrated doxycycline-inducible Gag-RRE expression construct, termed T-Rex-GagRRE HeLa, was established (Figure 6a). These cells were transduced with a lentiviral vector encoding Rev-GFP-V5. In some cases cells were co-transduced with vectors encoding either DDX1 or DDX21 to increase intracellular concentrations of these helicases; endogenous levels of DDX3 were deemed sufficient for this work. The expression of the GagRRE mRNA was induced in the various cell populations with doxycycline, and cells were then harvested. IPs then were performed using antibodies against DDX21, DDX1 and DDX3, and Western blotting was performed to assess helicase IP efficiency and co-IP of Rev.
Figure 6.
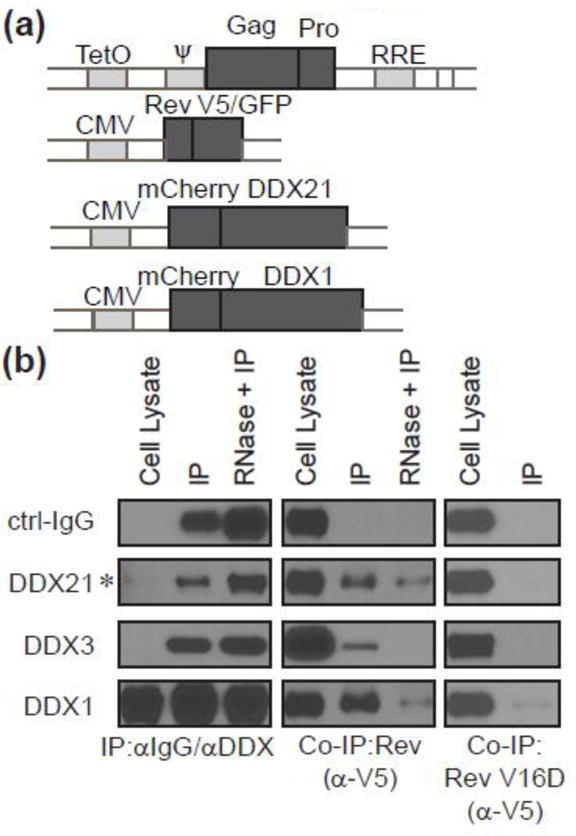
Co-IP between Rev and DEAD proteins. (a) Schematic diagrams of the plasmids used for cell line engineering and lentivirus production. Top: plasmid for Flp recombinase-mediated integration and its expression is under the control of a CMV/Tetracycline-regulated promoter. Flp-In T-Rex HeLa cells were transfected with this plasmid and selected with Hygromycin to establish stable T-Rex-GagRRE HeLa cells. The bottom three diagrams represent the lentivectors containing indicated proteins. (b) Western blot analysis of co-IP between Rev and DEAD proteins. To examine the interactions of DEAD helicases and Rev, T-Rex-GagRRE HeLa cells were transduced with lentiviruses of Rev-GFP-V5 for DDX3 IP, or cotransduced with lentiviruses of mCherry-DDX1-V5, or mCherry-DDX21-V5 for 24 hr followed by Doxycycline addition for another 24 hr for DDX1 or DDX21 IP. Cell lysates with and without RNase treatment were incubated with indicated antibodies, and then analyzed by western blotting with indicated. Cells expressing Rev(V16D)-GFP-V5 were analyzed with Rev-GFP-V5 in parallel. Asterisk indicates overexposed samples for better visualization.
Each DDX-specific antibody efficiently immunoprecipitated its cognate target protein (Figure 6b). When samples were probed for the presence of Rev, all DDX-specific antibodies showed significantly higher co-IP signal than with the negative control mouse IgG (Figure 6b). To assess the role of RNA in the DDX/RCC interactions, some samples were incubated with RNase A prior to IP (Figure 6b). For DDX21 and DDX1, the co-immunoprecipitated Rev levels decreased when compared to RNAse untreated samples, but was still clearly detected. Co-IP of Rev was not detected for DDX3 in the presence of RNAse A. To further investigate the role of direct protein binding on RCC/DDX association, Rev V16D was tested for the ability to associate with each DDX protein complex. In each case, Rev showed an absence or marked decrease in IP with DEAD-box proteins, as would be expected if in vivo association were due to a direct interaction (Figure 6b, fourth row of panels).
These data indicate that these three tested DEAD-box proteins can robustly interact with RCCs in vivo, and while RNA may enhance this interaction, it does not seem strictly required for DDX1 and DDX21 binding. Further, mutations that inhibit the interaction between DDX proteins and Rev in vitro also inhibit the interaction between DDX proteins and RCCs in vivo.
DDX21 binds more tightly to the RRE than DDX1
We previously demonstrated that DDX1 interacts with the full length RRE with an apparent Kd of ~250 nM using electrophoretic gel shift assays (EMSA) and radiolabeled RNA[13]. Further, when these assays were conducted at higher current, the RNA-protein complex dissociated, revealing a DDX1-dependent RRE structural transition[24]. To test the apparent RNA binding affinity of DDX21, EMSA experiments were again performed under high current conditions. As seen with DDX1, DDX21 binding resulted in the RNA signal shifting to the well (Figure 7a). However, DDX21 bound to the RRE RNA with 5–10 fold higher affinity when compared to DDX1 (apparent Kd of 37 +/− 2 nM). Further, no additional DDX21-dependent species were resolved under high current conditions.
Figure 7.
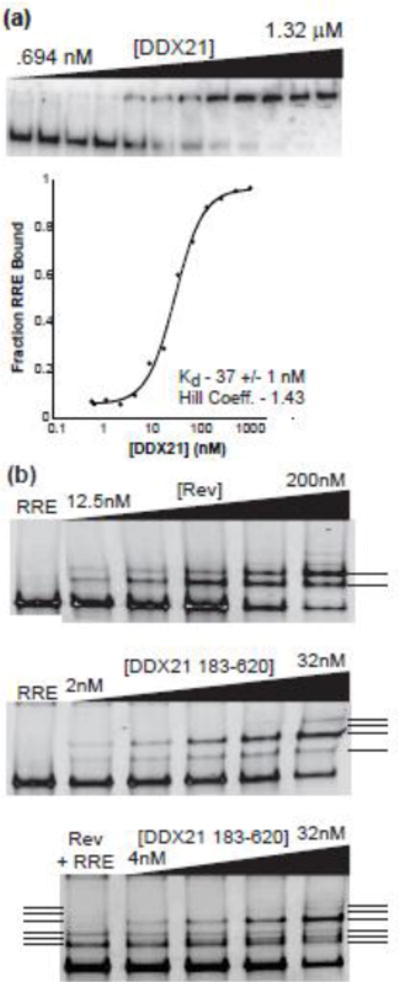
DDX21 and Rev bind independently to the RRE. a) Electrophoretic gel mobility shift titration of DDX21 into radiolabeled RRE RNA (top panel), and quantified fraction bound graph (on bottom). See materials and methods for line fit of bottom panel. Kd is noted on bottom. b) EMSA titration of Rev (top panel), DDX21(83–620) (middle panel), and DX21(83–620) in the presence of 25 nM Rev (bottom panel) into RRE RNA; RRE band imaged via ethidium bromide staining and U.V. fluorescence. Rev-dependent species are marked by a solid line, and DDX21(83–620)-dependent species by dotted lines.
Because DDX21 association results in a “well-shift” in radioactive EMSA assays, a DDX21 construct was created composed of the conserved DEAD and Helicase C domains required for ATP-dependent helicase activity in DDX proteins, as well as preserving the conserved Rev binding site. The construct, labelled DDX21(183–620), shows an efficient ATP stimulation of helicase activity, though the residual ATP-independent helicase activity was no longer present (Table 2). DDX21(183–620) resolves into distinct protein-bound RRE species when using a previously described modified EMSA assay where the RNA is visualized via UV fluorescence (Figure 6b, middle panel)[43]. Upon the addition of Rev, previously described single-step oligomeric RRE/Rev species can be resolved (Figure 7b, top panel). A similar titration of DDX21(183–620) results in the formation of two distinct species at lower concentrations, and a third at higher concentrations (Figure 7b, middle panel).
To test the ability of Rev, DDX21(183–620) and the RRE to form a tripartite complex, an EMSA was performed with constant concentrations of Rev and RRE, and increasing concentrations of DDX21(183–620). Surprisingly, instead of the predicted formation of a three-member complex, only a combination of the previously described species was observed (Figure 7b, bottom panel). This would indicate that either the tripartite species concentration is so small it cannot be imaged using this technique, or that the complex is too unstable for evaluation using EMSAs, or that a ternary complex does not form.
DDX21 enhances Rev oligomerization on the RRE in parallel to DDX1
A hallmark of the Rev/RRE assembly pathway is the initial high affinity binding of a single Rev protein to stem IIB, and subsequent oligomerization of Rev along the RRE utilizing both RNA and protein binding domains[2, 3, 9, 44, 45]. EMSA experiments have been used historically to discern the number of Rev monomers bound to the RRE at any given concentration[3, 45]. However, our previous work demonstrated that this data is actually a convolution of both RRE structural transitions and Rev binding events[24]. Therefore, only an assay that directly measures the binding of Rev to the RRE can be used to assess effects of DDX proteins on Rev binding and subsequent oligomerization on the RRE. One such method uses total internal reflection fluorescence (TIRF) microscopy to visualize Rev association with the RRE at single molecule resolution[14, 23, 24, 46]. In this experiment, a full length RRE molecule is immobilized on a quartz surface via biotin-streptavidin attachment, utilizing a biotinylated oligonucleotide that is hybridized at the 3′ end of the RRE. Then, Rev labeled at a specific cysteine residue with Alexa Fluor 555 (A555), is allowed to interact with the RRE and is excited through the evanescent field of a 532 nm laser. A representative fluorescence intensity versus time trace showing Rev molecules interacting with a single immobilized RRE molecule is presented in Figure 8a. Rev associates with and dissociates from the RRE several times during the observation period (Figure 8a). A single Rev monomer emits a fluorescent intensity of ~200 a.u. in our experimental conditions, so one RRE-bound Rev molecule has an intensity of 200 a.u.
Figure 8.
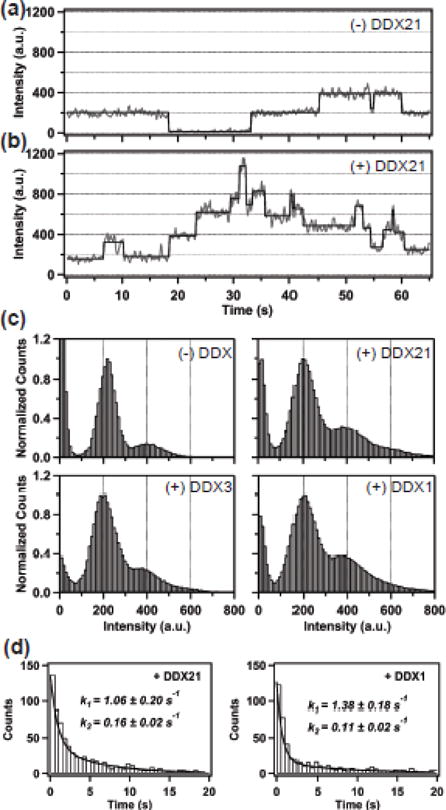
Assembly of Rev on the RRE monitored by single-molecule fluorescence spectroscopy. (a) Rev-RRE assembly in the absence of any DDX protein. Representative time trace (grey) of A555 emission over time, showing individual Rev monomer binding and dissociation events. The emission intensity of a single Rev monomer is ~200 units. The black lines are fits to a hidden Markov model. (b) Rev-RRE assembly in the presence of 150 nM DDX21. Same format as in (a). More Rev monomers assemble on the RRE in the presence of DDX21. (c) Normalized intensity histograms compiled from more than 100 traces, representing individual Rev-RRE stoichiometry states as separate peaks in the absence and presence of 150 nM DDX proteins, as indicated. (d) Dwell time histograms for the first Rev monomer binding to the RRE in the presence of DDX21 (left) and DDX1 (right). Histograms were fitted to a biexponential function, indicated by the solid line. The fitted rate constants and their associated uncertainties are indicated.
A histogram of all binding events in each experiment can be compiled and the distribution of Rev oligomerization states can be assessed (Figure 8c, top left panel). At the concentrations used in this experiment (1 nM Rev), Rev binds to the RRE primarily as a monomer (a dominant signal at 200 a.u.), with a minor peak corresponding to two bound Rev monomers. As demonstrated previously, the presence of DDX1 increases the proportion of fluorescent signal at higher intensity values, indicating an increase in the number of Rev monomers associated with each RRE molecule (Figure 8c, bottom right panel)[46]. Recently, we showed that DDX1 acts through the RRE RNA to promote Rev-RRE assembly [23, 24]. This ability of DDX1 to promote oligomerization of Rev on the RRE may be a general property shared by other DEAD-box helicases, or this activity might reflect the helicase activities and substrate preferences of a particular helicase. Indeed, when DDX21 was added to a Rev/RRE assembly assay, enhanced oligomerization behavior of Rev was again observed, similar to that in the presence of DDX1 (Figure 8b and 8c top right panel). Surprisingly, the presence of DDX3 had little effect on Rev oligomerization on the RRE (Figure 8c bottom left). This indicates that DDX3 doesn’t promote Rev oligomerization on the RRE as efficiently as DDX1 or DDX21.
We also performed a dwell time analysis for the first Rev monomer binding to the RRE in the presence of DDX21 or DDX1. In both cases, we observed similar bi-exponential kinetic behavior: rate constants 1.06 ± 0.20 s−1 and 0.16 ± 0.06 s−1 in the presence of DDX21 (Figure 8d left) and 1.38 ± 0.18 s−1 and 0.11 ± 0.02 s−1 in the presence of DDX1 (Figure 8d right). In both cases, the fast phase is predominant, whereas the slow phase corresponds to the Rev binding rate observed in the absence of any DDX protein (0.09 ± 0.01 s−1), agreeing with previous results [23]. These results indicate that DDX1 and DDX21 both accelerate binding of the first Rev monomer to the RRE and likely act by a similar mechanism.
Discussion
DDX21 is a multifunctional enzyme
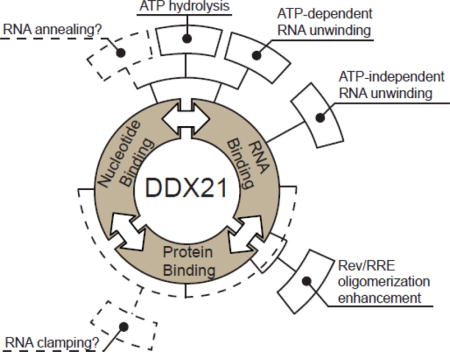
DEAD-box helicases are multifunctional proteins that can directly bind to RNA and nucleoside triphosphates through their DEAD-box homology domains, and bind to additional factors through accessory domains[47]. DDX21, a protein first isolated from Hela cell nuclear extracts, was characterized previously, exhibiting ATPase/dATPase and RNA unwinding activities [48]. Here we characterized the recombinant, untagged DDX21 protein, focusing on RNA-stimulated ATPase activity, RNA helicase activity, and direct RNA and accessory protein binding. Figure 9 diagrams a summary of the binding and enzymatic activities of DDX21 in this work, as well as indicating hypothetical activities hinted by our research. These data indicate the measured ATPase and helicase enzymatic activities of DDX21, like DDX1 and DDX3, fall within the range of activities generally observed for DEAD-box helicases. Surprisingly, even in the absence of ATP, some basal helicase activity was observed for DDX21. It is unclear what the mechanism of an ATP-independent helicase activity might be, or indeed if it has a physiological relevance outside this artificial context, but it is worth noting that this activity was not present in the DEAD-core DDX21(183–620), although ~80% of the ATP-dependent helicase activity of the full length enzyme was retained (Table 2). This might indicate a synergistic activity between the DEAD core and accessory domains during RNA remodeling.
Figure 9.
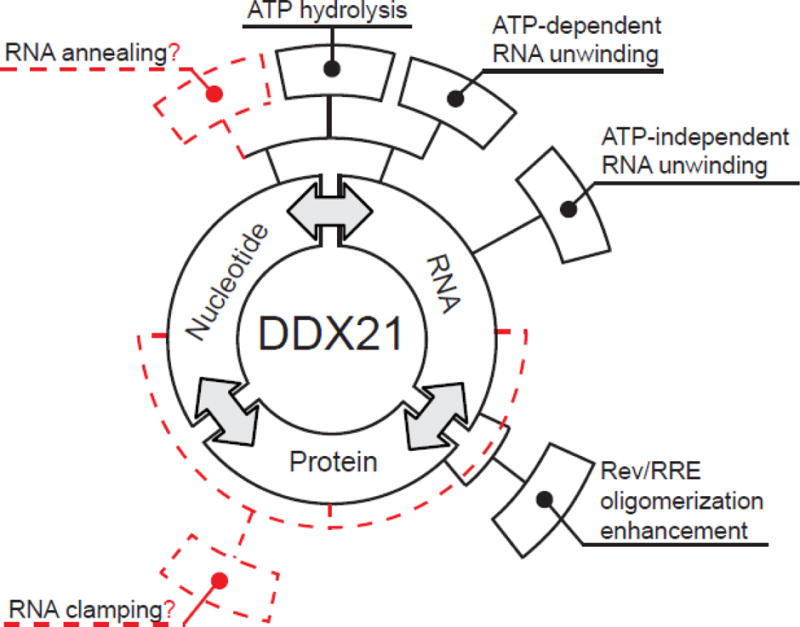
Binding and enzymatic activities of DDX21. Figure adapted from Putnam and Jankowsky, 2013. DDX21 enzyme has RNA, nucleotide and protein binding activities (first level of boxes in black). One binding activity is often affected by other binding activities (annotated by double-headed arrows), and can combine to create enzymatic-effects (annotated by boxes in second layer). Often, multiple binding activities are required for single enzymatic activities(solid and dashed lines from first to second tier). Solid black boxes in the second ring are enzymatic or functional activities measured in this study, while dashed red boxes represent activities only suggested by this or other studies.
DDX21 and the Rev/RRE components
The Rev binding activities of DDX21, DDX3 and DDX1 are strikingly similar. Each was previously shown to interact with RCCs in vivo [10, 13, 15, 40], and work here has demonstrated the ability of each of these proteins to interact directly with Rev through their respective DEAD domains. This conserved mode of binding would likely explain why Rev is able to interact intracellularly with a large number of DEAD-box proteins[10, 20]. The data presented here indicate a direct role for the Rev NIS in this protein-protein interaction. Of the residues shown to have an effect on binding, only L13 and V16 show high conservation among HIV strains, while position 14 shows a high preference for basic amino acids and position 18 for nonpolar amino acids [45]. Further, the V16D mutations in Rev result in a binding defect both DDX21 and DDX1 in vitro and in vivo, implicating a direct protein-protein interaction between Rev and DEAD-box proteins intracellularly.
DDX21 binds to the RRE with a higher affinity than that reported for DDX1. However, ATPase assays suggest that RRE-derived RNAs are not better substrates for activity than generic oligo-ribonucleotides, indicating that while the RRE is itself a substrate for RNA-dependent ATPase activity, it is not necessarily a preferred one.
Titration of excess Rev protein into DDX21 ATPase and RNA unwinding assays showed a dose-dependent inhibition of DDX21 activity (Table 2, Figure 4). This inhibition is not significant in the presence of stoichiometric amount of Rev protein, and supplying excess RNA reduces the apparent inhibition, as does addition of an RNA-binding defective mutant of Rev. Based on the observations above, we propose that both ATPase and ATP-dependent RNA unwinding activities of DDX21 are inhibited by Rev, and this inhibition is linked to Rev’s RNA binding properties. A possible explanation would be that Rev, at the concentrations used here, titrates RNA substrates away from DDX21 binding. However, data indicating DDX21’s ATP-independent RNA helicase activity is not affected by Rev would suggest a mechanism more complicated than simple titration. Indeed, the data presented here would be consistent with an RNA clamping model, where hydrolysis byproducts are not released, but some background helicase activity is observed due to stable DDX/RNA binding and induction of strand displacement in the presence of Rev[49]. However, it seems unlikely that the in vivo concentration of Rev would be high enough for total nuclear DDX21 population ATPase inhibition [50, 51].
DDX1 and DDX21 have similar activities
Our results indicate that DDX1 and DDX21 (and unlike DDX3) have very similar biochemical properties, binding to Rev with similar affinities through their conserved DEAD domains, reorganizing RNA architecture in the absence of ATP and enhancing Rev oligomerization on the RRE. Previous work in vivo indicated that while these proteins may have similar roles in the HIV lifecycle, they are not merely redundant members of an export mechanism, as the knockdown of either has a significant effect on HIV release from the cell, though they may be involved in the same mechanism[10, 13]. These overlaps in function may reflect a direct interaction between DDX1 and DDX21 in the cell, and indeed, these proteins have been implicated as playing a part in the double stranded RNA pathway together, and were hypothesized to directly interact with each other to do so[31]. Future studies investigating the role each of these proteins play in the RRE export process, as well as their native cellular interaction and functions together, is a natural extension of the present work.
DDX21, DDX1 and DDX3 in the Rev/RRE assembly pathway
We previously hypothesized that all that would be required for the stimulation of Rev binding to the RRE is a functionally conserved DEAD-helicase protein capable of binding RNA (as demonstrated for DDX1)[24]. To test this, Rev oligomerization assays were performed using TIRF microscopy and indicated that DDX1 and DDX21 had nearly identical effects on both Rev oligomerization and Rev’s initial RRE on-rate. Somewhat surprising was the relative lack of effect DDX3 had in this assay. This data taken with in vivo IP experiments presented here, as well as from other papers, would indicate that while DDX3 is capable of interacting with RCCs in vivo, its mechanistic role is distinct from that of DDX1 and DDX21[10, 16–19, 21, 22]. Indeed, DDX3 is somewhat unique in that it is capable of efficiently interacting with a variety of nucleic acid partners, including NTPs, dNTPs, DNA and RNA[35]. This broad specificity likely reflects its previously implied roles in HIV translation and nuclear pore export, but unfortunately does not explain the mechanistic role it might be playing in Rev-related processes.
It remains unclear what roles DEAD-box proteins play in the HIV lifecycle. This is partly due to the fact that their mechanistic roles in native cellular processes are still not well understood, and the authentic substrates for the HIV-specific functions for helicases are not yet defined. HIV represents a somewhat simplified platform with which to understand the enzymatic and functional roles these helicases perform in vivo, and in this work we more clearly define the biochemical activities of DDX21, DDX1 and DDX3. However, while we have RNA, nucleotide, and protein binding data for these three DDXs, the critical step to connect these activities with specific steps in the HIV life cycle remains unclear. Our work here and previously now provides a number of tools to study the role of DEAD-box helicases in the Rev/RRE pathway in vivo and the HIV lifecycle.
Materials and Methods
Plasmid construction for protein purification
RRE, DDX1 and Rev S10C constructs are described previously[14, 24, 41]. RevSLT40 and subsequent alanine mutants described above were created using Quick Change II site directed mutagenesis (Agilent) according to manufacturer protocol. DDX3, DDX21 and subdomain constructs were created using Gateway Cloning Technology™ (Invitrogen). DDX21 (HsCD00042538) and DDX3 (HsCD00040249) containing constructs in pDONR221 were purchased from DNASU. A DDX3 DEAD domain comprised of aa 211–403, a DDX21 DEAD domains of aa 217–396 and DDX21(183–620) were PCR amplified and recombined into fresh pDONR221 according to manufacturer protocol (Invitrogen). All DDX proteins and subdomains were then recombined into pDest566 according to manufacturer’s protocol. Final plasmids were named pMBPDDX3, pMBPDDX3DEAD, pMBPDDX21, pMBPDDX21DEAD and pMBPDDX21183-620. Each construct contained, from the N-terminus, coding regions for a 6X His-tag, an MBP tag, a TEV cleavage site and ended in the DDX protein of interest. All plasmids were verified via sequencing reactions (Eurofins Operon). Gateway vectors were a generous gift of the Dominic Esposito Lab at the National Cancer Institute.
Plasmid construction for human cell in vivo protein expression
Rev WT, V16D, and EGFP were amplified by PCR, and digested with NheI and XhoI, XhoI and SacII, respectively. The digested fragments were then cloned into pcDNA3.1 (Invitogen) containing V5 epitope tag. The whole cassette of Rev-EGFP-V5 was then digested with NheI and PmeI, and ligated to lentiviral vector pCDHblast MCSNard OST-LMNA (with CMV promoter, a gift from Tom Misteli) (Addgene plasmid # 22661) where OST-LMNA was removed by BamHI and NotI, then replaced with oligos containing NheI and BstZ17 I. Constructs containing DDX1 or DDX21, which were amplified by PCR, digested with NheI and XhoI, and inserted into pcDNA3.1 containing mCherry and V5 tag at N-terminus and C-terminus of multiple cloning sites (unpublished), respectively. The cassettes of mCherry-DDX1-V5 or mCherry-DDX21-V5 were then amplified by PCR, digested with BamHI and SalI, then cloned into lentiviral vector (with CMV promoter, a gift from Eric Campeau) (Addgene plasmid #17448).
Flp-In-T-Rex-GagRRE HeLa cell production
GagRRE was amplified by PCR from pCMV-GagRRE[52], digested with EcoRV and AleI, and cloned into pcDNA5-FRT-TO-6xMS2 (unpublished) with AleI site to generate pcDNA-FRT-TO-GagRRE-6xMS2. This plasmid was co-transfected with pOG44 into Flp-In T-Rex HeLa cells (Invitrogen), a gift from Don Cleveland, and subsequently selected with hygromycin to establish a stable cell line per manufacturer’s instructions. Cells were maintained in Dulbecco’s Modified Eagle Medium (DMEM, Gibco), 10% Tetracycline-free FCS (Clontech), 2 mM L-glutamine (Gibco), 100 U ml-1 penicillin and 100 μg/ml streptomycin (Gibco) at 37°C in 5% CO2 incubator.
DDX and Rev Protein Purification
All DDX1 (including subdomain constructs) and Rev (including mutant constructs) proteins were purified as previously described[13, 24]. DDX3, DDX21 and sub-domain proteins were expressed in Rosetta pLysS cells (Novagen, MA), grown in LB broth to an OD600 of 0.6 at 37°C. Temperature was then adjusted to 25°C and protein expression induced by the addition of 1mM IPTG for 12–18 hours. Cells were harvested by centrifugation and stored at −20°C until ready for use, or immediately used in protein purification. Cell pellets were resuspended and lysed in B-Per reagent (Pierce Biotechnology, MA) supplemented with lysozyme (0.1mg/ml) at room temperature for 20 min. Cell debris was removed by centrifugation, and the resulting supernatant was adjust to 0.5M NaCl and polyethyleneimine (Sigma, MO) added to a final concentration of 0.025%. Sample was again centrifuged to remove precipitated nucleic acid, and resulting supernatant applied to HisTrap column (GE healthcare) using a AKTA FLPC (GE healthcare). After washing the column with wash buffer (20mM Tris, pH 7.8 and 0.5M NaCl) DDX proteins were eluted using 0–0.25M imidazole gradients, and protein presence in each fraction assayed via SDS PAGE followed by coomassie staining. Pooled fractions of proteins were dialyzed against Heparin Column buffer A (20mM Tris 7.8, 75mM NaCl, 1mM EDTA, 1mM TCEP and 2% glycerol) and further purified on a 5ml HiTrap Heparin Column (GE healthcare) and eluted using a 75mM to 2M NaCL gradient. Purified DDX proteins were dialyzed against Heparin Buffer A, and cleaved by Tev protease (1:20 mass ratio) overnight. The resultant DDX proteins were further purified from the HisMBP tag and Tev protease via Heparin column as described above.
RNA production
Stem IIB, poly(U)21, FAM-labelled 10mer, Dab-labelled 26mer, and 14mer RNA oligomers were purchased from Thermo Fisher. Full length RRE RNAs used for EMSA or TIRF experiments were produced as previously described[24, 53, 54].
ATPase activity assay
DDX21 ATPase activity was measured using an NADH coupled ATPase assay as previously described[36]. Reaction mixtures were monitored for the depletion of NADH by measuring Abs(340) with an Envision 2104 Multi Plate Reader (Perkin Elmer). 50ul reactions contained 10μM DDX21, 20mM Tris pH7.5, 60mM NaCl, 55mM KCl, 2mM MgCl2, 5% Glycerol and 0.1mg/ml BSA. The reactions with either increasing amounts of ATP or RNA were carried out at room temperature in UV-transparent 96 well plates (Grenier Bio-one), and the hydrolysis rates analyzed using Michaelis-Menten equations. Because it is difficult to accurately measure the path length of a 50μl reaction volume, a concentration dependent standard curve of NADH was generated in the same 50μl buffer to convert OD340 to an NADH molar value, resulting in a value of 1abs unit equaling 0.98±0.006mM NADH. Pyruvate Kinase, Lactate dehydrogenase, and ATP were purchased from Sigma. Phosphoenolpyruvate was purchased from Santa Cruz Biotechnology.
Fluorescence based helicase assay
DDX21 RNA unwinding activity assay was adapted from previous work and was measured in real time using an Envision 2104 Multilabel Plate Reader (Perkin Elmer)[39]. 50 mL reaction mixtures consisted of a 20nM pre-formed RNA duplex (FAM-labelled 10mer and DAB labelled 26mer), 100 nM 14mer RNA competitor, 2μM DDX21 or DDX21(183–620), 1X helicase buffer (20mM Tris pH7.5, 60mM NaCl, 55mM KCl, 2mM MgCl2 and 0.67U/μL RNaseOUT), and 0 or 1 mM ATP. The resulting Fluorescence data was fitted as previously described using Igor (Wavemetrics) to the following equation
where F(t) is fluorescent signal at any given time (t), F0 is initial fluorescent signal, F∞ is maximal fluorescent signal and kunw is the unwinding rate constant [39].
Fluorescence polarization assay
Fluorescence polarization binding assays were conducted as previously described[13, 24]. All Rev constructs were fluorescently labelled as previously described using maleimide-alexa555 (Invitrogen)[24]. Reaction conditions contained 15 nM Rev-alexa555 (or mutants). Samples were prepared in 96-well black plate (Grenier) and polarization was measured using 2014 Envision multi-label Reader. The average polarization values were calculated from 7 consecutive readings of each plate. Equilibrium Kd values were determined by fitting to quadratic equations using Igor (Wavemetrics) as described previously[13].
Co-IP assay and western blotting
For production of DEAD-box and Rev producing cells, Flp-In-T-Rex-GagRRE-HeLa cells (~1.5×106) were seeded and transduced with various lentiviruses as indicated in the text and figure 6. After 24 hours Doxycycline was added to 1 μg/ml. Cells were grown for an additional 24 hours at 37°C to induce the expression of GagRRE mRNA and then harvested using trypsin digestion. For IP, cell pellets were resuspended with IP buffer [50 mM Tris (pH 7.4), 75 mM NaCl, 1mM MgCl2, 1% Nonidet P-40, 1 mM DTT, 1 mM phenylmethylsulfonyl fluoride and complete protease inhibitor cocktail (EDTA-free, Roche Diagnostics)] (2). Where indicated, cell lysates were treated with 50 μg/mL RNase A (Amresco) at 37°C for 30 min. Lysates were cleared by centrifugation at 10,000 × g for 10 min at 4°C. An aliquot of the supernatant was saved for Western blotting (“input”), while the remainder was used for a 30 min IP incubation with the following antibodies: Rabbit IgG (Cell Signaling Technology), rabbit α-DDX21 IgG (10528-1-AP, Proteintech, Rosemond, IL), mouse α-DDX3 IgG (ab50703, Abcam), rabbit α-DDX1 IgG (11357-1-AP, Proteintech). The lysate-antibody mixtures were then incubated with protein-G Dynabeads (Life Technologies) for 30 min. Beads were washed three times with wash buffer [50 mM Tris (pH 7.4), 75 mM NaCl, 1mM MgCl2, 0.05% Triton X-100] and bound proteins were eluted with SDS sample buffer. Protein samples were resolved by SDS-PAGE and transferred to nitrocellulose membranes (Amersham Biosciences). Mouse a-V5 antibody (R960-25, Life Technologies) was used to detect Rev-GFP-V5 (WT or V16D), DDX21 and DDX1, while DDX3 was detected using the α-DDX3 antibody noted above.
Electromobility shift assay (EMSA)
Radioactive EMSA assays (including radiolabeling of RRE and buffer preparation) were performed as previously described[24]. UV-visualized EMSA assays were adapted from previous protocols as follows[43]. 20 μL reaction mixtures containing varying DDX21 or Rev concentrations, 12.5nM RRE RNA, 1X reaction buffer (50mM Tris pH 7.5, 150mM KCl, 10% glycerol, 1% Triton X-100, 0.1 mg/mL BSA and 0.67U/μL RNaseOUT) and DEPC-treated H2O were incubated at room temperature for 30 minutes to come to binding equilibrium. Reactions were then loaded onto a 10% TBE polyacrylamide gel (Invitrogen) and run for 2 hours at 200V. Gels were soaked in SYBRGold Stain (Invitrogen) for 30 minutes and RNA visualized using a BioRad Gel Doc EZ Imager™. Image contrast was adjusted slightly using photoshop (Adobe®).
Total internal reflection fluorescence (TIRF) microscopy of Rev oligomerization
The full-length RRE (351 nt) containing extensions at both the 3′ and 5′ ends was transcribed in vitro from an appropriate DNA template using T7 RNA polymerase as previously described[23]. A biotinylated DNA oligonucleotide (28 nt) was hybridized at the 3′ end of the RRE transcript for surface-immobilization. 500 pM of RRE-DNA hybrid was immobilized on a streptavidin-coated quartz surface, as described elsewhere [14, 23, 46]. The quartz surface was passivated with polyethylene glycol to inhibit nonspecific adsorption of Rev or DDX proteins, as described [55]. Rev was labeled with A555 at a single N-terminal cysteine, as described[14]. 1 nm A555-Rev with or without 150 nM DDX protein, in 50 mM Hepes buffer (pH7.5) containing 2 mM Trolox, 150 mm KCl, 10 mM K2SO4, 2 mM MgCl2, 2 mM DTT, and oxygen scavenging system (50 μg/ml glucose oxidase, 10 μg/ml catalase, and 2–5% wt/vol glucose)[55], was introduced into the surrounding solution. The A555 fluorophore was excited using a 532 nm laser in a custom-built prism-based TIRF microscope. Emission from A555 was collected through a water immersion objective and recorded on an intensified CCD camera with 100 ms time resolution [56, 57]. A single-molecule data acquisition package (downloaded from https://physics.illinois.edu/cplc/software/) was used to record data. Fluorescence intensity traces from individual Rev-RRE complexes were extracted from the CCD camera movie files and processed using a custom program written in Matlab. All intensity time traces were corrected for background and smoothed using 3-point linear averaging. Multiple time traces (>100) were used to generate fluorescence intensity histograms using Matlab and Igor Pro software (WaveMetrics). Individual time traces were fitted using Hidden Markov modeling (McKinney et. al) to determined the dwell times spent in one state prior to transition to another state. The information from multiple times traces was compiled in the form of dwell time histograms, which were fitted with exponential functions to determine the rate constants for transitions from one state to another, using Igor Pro software (WaveMetrics).
Research Highlights.
DDX21 has been implicated in HIV-1 Rev-dependent nuclear export.
DDX21 is an RNA-dependent ATPase, with both ATP-dependent and ATP-independent helicase activities.
DDX21 ATP-dependent activities (ATPase and RNA helicase) are inhibited by the presence of Rev, and this inhibition is dependent on Rev binding to RNA substrates.
DEAD-box proteins bind to Rev through their conserved N-terminal DEAD-domain, and Rev through its NIS domain.
DDX21 is able to stimulate Rev binding to the RRE in a manner similar to that described for DDX1, while DDX3 cannot.
Acknowledgments
Funding for this work was provided by the National Institute of General Medical Sciences (grant P50 GM082545; W. Sundquist PI, to J.R.W, D.P.M. and L.G.), the American Cancer Society (fellowship award 121633-PF-11-222-01-RMC to J.A.H) and California HIV/AIDS Research Program (fellowship award F12-SRI-210 to R.L.; fellowship award F12-SRI-203 to L.Z.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Fornerod M, Ohno M, Yoshida M, Mattaj IW. CRM1 is an export receptor for leucine-rich nuclear export signals. Cell. 1997;90:1051–60. doi: 10.1016/s0092-8674(00)80371-2. [DOI] [PubMed] [Google Scholar]
- 2.Jain C, Belasco JG. Structural model for the cooperative assembly of HIV-1 Rev multimers on the RRE as deduced from analysis of assembly-defective mutants. Mol Cell. 2001;7:603–14. doi: 10.1016/s1097-2765(01)00207-6. [DOI] [PubMed] [Google Scholar]
- 3.Daugherty MD, D’Orso I, Frankel AD. A solution to limited genomic capacity: using adaptable binding surfaces to assemble the functional HIV Rev oligomer on RNA. Mol Cell. 2008;31:824–34. doi: 10.1016/j.molcel.2008.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jayaraman B, Crosby DC, Homer C, Ribeiro I, Mavor D, Frankel AD. RNA-directed remodeling of the HIV-1 protein Rev orchestrates assembly of the Rev-Rev response element complex. Elife. 2014;3:e04120. doi: 10.7554/eLife.04120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.DiMattia MA, Watts NR, Stahl SJ, Rader C, Wingfield PT, Stuart DI, et al. Implications of the HIV-1 Rev dimer structure at 3.2 A resolution for multimeric binding to the Rev response element. Proc Natl Acad Sci U S A. 2010;107:5810–4. doi: 10.1073/pnas.0914946107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Malim MH, Hauber J, Le SY, Maizel JV, Cullen BR. The HIV-1 rev trans-activator acts through a structured target sequence to activate nuclear export of unspliced viral mRNA. Nature. 1989;338:254–7. doi: 10.1038/338254a0. [DOI] [PubMed] [Google Scholar]
- 7.Malim MH, Cullen BR. HIV-1 structural gene expression requires the binding of multiple Rev monomers to the viral RRE: implications for HIV-1 latency. Cell. 1991;65:241–8. doi: 10.1016/0092-8674(91)90158-u. [DOI] [PubMed] [Google Scholar]
- 8.Tiley LS, Malim MH, Tewary HK, Stockley PG, Cullen BR. Identification of a high-affinity RNA-binding site for the human immunodeficiency virus type 1 Rev protein. Proc Natl Acad Sci U S A. 1992;89:758–62. doi: 10.1073/pnas.89.2.758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jain C, Belasco JG. A structural model for the HIV-1 Rev-RRE complex deduced from altered-specificity rev variants isolated by a rapid genetic strategy. Cell. 1996;87:115–25. doi: 10.1016/s0092-8674(00)81328-8. [DOI] [PubMed] [Google Scholar]
- 10.Naji S, Ambrus G, Cimermancic P, Reyes JR, Johnson JR, Filbrandt R, et al. Host cell interactome of HIV-1 Rev includes RNA helicases involved in multiple facets of virus production. Mol Cell Proteomics. 2012;11 doi: 10.1074/mcp.M111.015313. M111 015313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Popow J, Jurkin J, Schleiffer A, Martinez J. Analysis of orthologous groups reveals archease and DDX1 as tRNA splicing factors. Nature. 2014;511:104–7. doi: 10.1038/nature13284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lin MH, Sivakumaran H, Jones A, Li D, Harper C, Wei T, et al. A HIV-1 Tat mutant protein disrupts HIV-1 Rev function by targeting the DEAD-box RNA helicase DDX1. Retrovirology. 2014;11:121. doi: 10.1186/s12977-014-0121-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Edgcomb SP, Carmel AB, Naji S, Ambrus-Aikelin G, Reyes JR, Saphire AC, et al. DDX1 is an RNA-dependent ATPase involved in HIV-1 Rev function and virus replication. J Mol Biol. 2012;415:61–74. doi: 10.1016/j.jmb.2011.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Robertson-Anderson RM, Wang J, Edgcomb SP, Carmel AB, Williamson JR, Millar DP. Single-molecule studies reveal that DEAD box protein DDX1 promotes oligomerization of HIV-1 Rev on the Rev response element. J Mol Biol. 2011;410:959–71. doi: 10.1016/j.jmb.2011.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fang J, Acheampong E, Dave R, Wang F, Mukhtar M, Pomerantz RJ. The RNA helicase DDX1 is involved in restricted HIV-1 Rev function in human astrocytes. Virology. 2005;336:299–307. doi: 10.1016/j.virol.2005.03.017. [DOI] [PubMed] [Google Scholar]
- 16.Brai A, Fazi R, Tintori C, Zamperini C, Bugli F, Sanguinetti M, et al. Human DDX3 protein is a valuable target to develop broad spectrum antiviral agents. Proc Natl Acad Sci U S A. 2016;113:5388–93. doi: 10.1073/pnas.1522987113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Frohlich A, Rojas-Araya B, Pereira-Montecinos C, Dellarossa A, Toro-Ascuy D, Prades-Perez Y, et al. DEAD-box RNA helicase DDX3 connects CRM1-dependent nuclear export and translation of the HIV-1 unspliced mRNA through its N-terminal domain. Biochimica et biophysica acta. 2016;1859:719–30. doi: 10.1016/j.bbagrm.2016.03.009. [DOI] [PubMed] [Google Scholar]
- 18.Soto-Rifo R, Rubilar PS, Ohlmann T. The DEAD-box helicase DDX3 substitutes for the cap-binding protein eIF4E to promote compartmentalized translation initiation of the HIV-1 genomic RNA. Nucleic acids research. 2013;41:6286–99. doi: 10.1093/nar/gkt306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yasuda-Inoue M, Kuroki M, Ariumi Y. DDX3 RNA helicase is required for HIV-1 Tat function. Biochem Biophys Res Commun. 2013;441:607–11. doi: 10.1016/j.bbrc.2013.10.107. [DOI] [PubMed] [Google Scholar]
- 20.Yasuda-Inoue M, Kuroki M, Ariumi Y. Distinct DDX DEAD-box RNA helicases cooperate to modulate the HIV-1 Rev function. Biochem Biophys Res Commun. 2013;434:803–8. doi: 10.1016/j.bbrc.2013.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lai MC, Wang SW, Cheng L, Tarn WY, Tsai SJ, Sun HS. Human DDX3 interacts with the HIV-1 Tat protein to facilitate viral mRNA translation. PloS one. 2013;8:e68665. doi: 10.1371/journal.pone.0068665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ishaq M, Hu J, Wu X, Fu Q, Yang Y, Liu Q, et al. Knockdown of cellular RNA helicase DDX3 by short hairpin RNAs suppresses HIV-1 viral replication without inducing apoptosis. Mol Biotechnol. 2008;39:231–8. doi: 10.1007/s12033-008-9040-0. [DOI] [PubMed] [Google Scholar]
- 23.Lamichhane R, Hammond JA, Pauszek RF, 3rd, Anderson RM, Pedron I, van der Schans E, et al. A DEAD-box protein acts through RNA to promote HIV-1 Rev-RRE assembly. Nucleic acids research. 2017 doi: 10.1093/nar/gkx206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hammond JA, Lamichhane R, Millar DP, Williamson JR. A DEAD-Box Helicase Mediates an RNA Structural Transition in the HIV-1 Rev Response Element. J Mol Biol. 2017;429:697–714. doi: 10.1016/j.jmb.2017.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang Y, Baysac KC, Yee LF, Saporita AJ, Weber JD. Elevated DDX21 regulates c-Jun activity and rRNA processing in human breast cancers. Breast cancer research : BCR. 2014;16:449. doi: 10.1186/s13058-014-0449-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jung Y, Lee S, Choi HS, Kim SN, Lee E, Shin Y, et al. Clinical validation of colorectal cancer biomarkers identified from bioinformatics analysis of public expression data. Clin Cancer Res. 2011;17:700–9. doi: 10.1158/1078-0432.CCR-10-1300. [DOI] [PubMed] [Google Scholar]
- 27.Henning D, So RB, Jin R, Lau LF, Valdez BC. Silencing of RNA helicase II/Gualpha inhibits mammalian ribosomal RNA production. The Journal of biological chemistry. 2003;278:52307–14. doi: 10.1074/jbc.M310846200. [DOI] [PubMed] [Google Scholar]
- 28.Calo E, Flynn RA, Martin L, Spitale RC, Chang HY, Wysocka J. RNA helicase DDX21 coordinates transcription and ribosomal RNA processing. Nature. 2015;518:249–53. doi: 10.1038/nature13923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sloan KE, Leisegang MS, Doebele C, Ramirez AS, Simm S, Safferthal C, et al. The association of late-acting snoRNPs with human pre-ribosomal complexes requires the RNA helicase DDX21. Nucleic acids research. 2015;43:553–64. doi: 10.1093/nar/gku1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen G, Liu CH, Zhou L, Krug RM. Cellular DDX21 RNA helicase inhibits influenza A virus replication but is counteracted by the viral NS1 protein. Cell host & microbe. 2014;15:484–93. doi: 10.1016/j.chom.2014.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang Z, Kim T, Bao M, Facchinetti V, Jung SY, Ghaffari AA, et al. DDX1, DDX21, and DHX36 helicases form a complex with the adaptor molecule TRIF to sense dsRNA in dendritic cells. Immunity. 2011;34:866–78. doi: 10.1016/j.immuni.2011.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Linder P, Jankowsky E. From unwinding to clamping - the DEAD box RNA helicase family. Nat Rev Mol Cell Biol. 2011;12:505–16. doi: 10.1038/nrm3154. [DOI] [PubMed] [Google Scholar]
- 33.Valdez BC, Henning D, Perumal K, Busch H. RNA-unwinding and RNA-folding activities of RNA helicase II/Gu-two activities in separate domains of the same protein. European journal of biochemistry/FEBS. 1997;250:800–7. doi: 10.1111/j.1432-1033.1997.00800.x. [DOI] [PubMed] [Google Scholar]
- 34.Valdez BC. Structural domains involved in the RNA folding activity of RNA helicase II/Gu protein. European journal of biochemistry/FEBS. 2000;267:6395–402. doi: 10.1046/j.1432-1327.2000.01727.x. [DOI] [PubMed] [Google Scholar]
- 35.Garbelli A, Beermann S, Di Cicco G, Dietrich U, Maga G. A motif unique to the human DEAD-box protein DDX3 is important for nucleic acid binding, ATP hydrolysis, RNA/DNA unwinding and HIV-1 replication. PloS one. 2011;6:e19810. doi: 10.1371/journal.pone.0019810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Norby JG. Coupled assay of Na+,K+−ATPase activity. Methods Enzymol. 1988;156:116–9. doi: 10.1016/0076-6879(88)56014-7. [DOI] [PubMed] [Google Scholar]
- 37.Lee CG, Hurwitz J. A new RNA helicase isolated from HeLa cells that catalytically translocates in the 3′ to 5′ direction. The Journal of biological chemistry. 1992;267:4398–407. [PubMed] [Google Scholar]
- 38.Noble CG, Song H. MLN51 stimulates the RNA-helicase activity of eIF4AIII. PloS one. 2007;2:e303. doi: 10.1371/journal.pone.0000303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bizebard T, Dreyfus M. A FRET-based, continuous assay for the helicase activity of DEAD-box proteins. Methods Mol Biol. 2015;1259:199–209. doi: 10.1007/978-1-4939-2214-7_13. [DOI] [PubMed] [Google Scholar]
- 40.Fang J, Kubota S, Yang B, Zhou N, Zhang H, Godbout R, et al. A DEAD box protein facilitates HIV-1 replication as a cellular co-factor of Rev. Virology. 2004;330:471–80. doi: 10.1016/j.virol.2004.09.039. [DOI] [PubMed] [Google Scholar]
- 41.Edgcomb SP, Aschrafi A, Kompfner E, Williamson JR, Gerace L, Hennig M. Protein structure and oligomerization are important for the formation of export-competent HIV-1 Rev-RRE complexes. Protein Sci. 2008;17:420–30. doi: 10.1110/ps.073246608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Thomas SL, Oft M, Jaksche H, Casari G, Heger P, Dobrovnik M, et al. Functional analysis of the human immunodeficiency virus type 1 Rev protein oligomerization interface. J Virol. 1998;72:2935–44. doi: 10.1128/jvi.72.4.2935-2944.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fang X, Wang J, O’Carroll IP, Mitchell M, Zuo X, Wang Y, et al. An unusual topological structure of the HIV-1 Rev response element. Cell. 2013;155:594–605. doi: 10.1016/j.cell.2013.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Daugherty MD, Booth DS, Jayaraman B, Cheng Y, Frankel AD. HIV Rev response element (RRE) directs assembly of the Rev homooligomer into discrete asymmetric complexes. Proc Natl Acad Sci U S A. 2010;107:12481–6. doi: 10.1073/pnas.1007022107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Daugherty MD, Liu B, Frankel AD. Structural basis for cooperative RNA binding and export complex assembly by HIV Rev. Nat Struct Mol Biol. 2010;17:1337–42. doi: 10.1038/nsmb.1902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pond SJ, Ridgeway WK, Robertson R, Wang J, Millar DP. HIV-1 Rev protein assembles on viral RNA one molecule at a time. Proc Natl Acad Sci U S A. 2009;106:1404–8. doi: 10.1073/pnas.0807388106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Putnam AA, Jankowsky E. DEAD-box helicases as integrators of RNA, nucleotide and protein binding. Biochimica et biophysica acta. 2013;1829:884–93. doi: 10.1016/j.bbagrm.2013.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Flores-Rozas H, Hurwitz J. Characterization of a new RNA helicase from nuclear extracts of HeLa cells which translocates in the 5′ to 3′ direction. The Journal of biological chemistry. 1993;268:21372–83. [PubMed] [Google Scholar]
- 49.Nielsen KH, Chamieh H, Andersen CB, Fredslund F, Hamborg K, Le Hir H, et al. Mechanism of ATP turnover inhibition in the EJC. RNA. 2009;15:67–75. doi: 10.1261/rna.1283109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pomerantz RJ, Seshamma T, Trono D. Efficient replication of human immunodeficiency virus type 1 requires a threshold level of Rev: potential implications for latency. J Virol. 1992;66:1809–13. doi: 10.1128/jvi.66.3.1809-1813.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Beck M, Schmidt A, Malmstroem J, Claassen M, Ori A, Szymborska A, et al. The quantitative proteome of a human cell line. Mol Syst Biol. 2011;7:549. doi: 10.1038/msb.2011.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wodrich H, Bohne J, Gumz E, Welker R, Krausslich HG. A new RNA element located in the coding region of a murine endogenous retrovirus can functionally replace the Rev/Rev-responsive element system in human immunodeficiency virus type 1 Gag expression. J Virol. 2001;75:10670–82. doi: 10.1128/JVI.75.22.10670-10682.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hammond JA, Rambo RP, Kieft JS. Multi-domain packing in the aminoacylatable 3′ end of a plant viral RNA. J Mol Biol. 2010;399:450–63. doi: 10.1016/j.jmb.2010.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hammond JA, Rambo RP, Filbin ME, Kieft JS. Comparison and functional implications of the 3D architectures of viral tRNA-like structures. RNA. 2009;15:294–307. doi: 10.1261/rna.1360709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lamichhane R, Daubner GM, Thomas-Crusells J, Auweter SD, Manatschal C, Austin KS, et al. RNA looping by PTB: Evidence using FRET and NMR spectroscopy for a role in splicing repression. Proc Natl Acad Sci U S A. 2010;107:4105–10. doi: 10.1073/pnas.0907072107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lamichhane R, Berezhna SY, Gill JP, Van der Schans E, Millar DP. Dynamics of site switching in DNA polymerase. J Am Chem Soc. 2013;135:4735–42. doi: 10.1021/ja311641b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Berezhna SY, Gill JP, Lamichhane R, Millar DP. Single-molecule Forster resonance energy transfer reveals an innate fidelity checkpoint in DNA polymerase I. J Am Chem Soc. 2012;134:11261–8. doi: 10.1021/ja3038273. [DOI] [PMC free article] [PubMed] [Google Scholar]


