Abstract
Background
Adverse viral and medication effects on adipose tissue contribute to the development of metabolic disease in HIV-infected persons, but T cells also have a central role modulating local inflammation and adipocyte function. We sought to characterize potentially pro-inflammatory T cell populations in adipose tissue among persons on long-term antiretroviral therapy, and assess whether adipose tissue CD8+ T cells represent an expanded, oligoclonal population.
Methods
We recruited 10 HIV-infected, overweight and obese adults on efavirenz, tenofovir, and emtricitabine for >4 years with consistent viral suppression. We collected fasting blood and subcutaneous abdominal adipose tissue to measure the percentage of CD4+ and CD8+ T cells expressing activation, exhaustion, late-differentiation/senescence, and memory surface markers. We performed T cell receptor (TCR) sequencing on sorted CD8+ cells. We compared the proportion of each T cell subset, and the TCR repertoire diversity, in blood versus adipose tissue.
Results
Adipose tissue had a higher percentage of CD3+CD8+ T cells compared to blood (61.0% vs. 51.7%, P<0.01), and was enriched for both activated CD8+HLA-DR+ T cells (5.5% vs. 0.9%, P<0.01) and late-differentiated CD8+CD57+ T cells (37.4% vs. 22.7%, P<0.01). Adipose tissue CD8+ T cells displayed distinct TCRβ V and J gene usage, and the Shannon’s Entropy index, a measure of overall TCRβ repertoire diversity, was lower compared to blood (4.39 vs. 4.46; P=0.05).
Conclusions
Adipose tissue is enriched for activated and late-differentiated CD8+ T cells with distinct TCR usage. These cells may contribute to tissue inflammation and impaired adipocyte fitness in HIV-infected persons.
Keywords: HIV, obesity, metabolic disease, adipose tissue, T cell receptor, clonality
Introduction
HIV-infected individuals are at increased risk of developing dyslipidemia and insulin resistance, which appears to be due, in part, to adverse changes in adipocyte metabolic fitness.1-7 Exposure to HIV viral proteins and antiretroviral therapy (ART) agents depletes adipocyte mitochondrial DNA and electron transport chain proteins, which is accompanied by impaired fatty acid metabolism, altered adipokine secretion, and reduced insulin sensitivity.8-15 Adipose tissue from HIV-infected persons also has lower expression of key adipocyte regulatory proteins involved in maturation and control of lipid storage and release.16-23
The stromal vascular fraction of adipose tissue contains a diverse mix of immune cells forming a complex paracrine signaling milieu, which modulates local inflammation and adipocyte function. Studies in animal models and HIV-negative persons demonstrate the central role of T cells in regulating local inflammation, adipocyte gene expression, energy uptake, and metabolic homeostasis in subcutaneous and visceral fat.24-31 CD8+ T cell infiltration into adipose tissue is an early and necessary step preceding the recruitment of M1-phenotype (tumor necrosis factor-α [TNF-α], interleukin [IL]-6, IL-12, and IL-23-producing) macrophages, which act via adipocyte surface receptors and other mechanisms to inhibit insulin signaling by reducing expression of insulin receptor substrate-1 (IRS-1), phosphoinositide 3-kinase p85α, and glucose transporter type 4 (GLUT4).24,28,30,32-36 Furthermore, spectrotype analyses of adipose tissue CD8+ T cells in the setting of obesity have described a conserved repertoire of T cell receptor (TCR) Vα and Vβ chain families as compared to cells in lean tissue, suggesting that these CD8+ T cells may undergo oligoclonal expansion in adipose tissue.24,37
Alterations in adipose tissue T cell distributions are also observed in the setting of HIV infection. Adipose tissue from HIV-infected persons has a lower percentage of CD4+ memory T cells and higher CD8+ memory T cells compared to HIV-negative persons.36 Furthermore, both the CD4+ and CD8+ memory T cells predominantly express the CD69 activation marker, and the median copy number of latent HIV DNA in adipose tissue CD4+ T cells is similar to the median copy number in circulating CD4+ T cells.35,36 Co-culture of preadipocytes with CD4+ T cells harboring latent HIV virus in vitro increases preadipocyte IL-6 expression nearly 3-fold, and this finding may reflect an in vivo process contributing to adipose tissue inflammation in HIV-infected persons.36 Similar changes are observed in studies of simian immunodeficiency virus (SIV), where SIV infection is accompanied by a significantly higher proportion of activated CD4+ and CD8+ cells in both subcutaneous and visceral adipose tissue compared to uninfected animals, and the mean level of SIV RNA in adipose tissue is similar to CD4+ T cells in circulation and in mesenteric lymph nodes.35
In this study, we sought to extend our understanding of adipose tissue immunology in HIV-infected persons, and assess whether adipose tissue is enriched for CD4+ and CD8+ T cells expressing activation, exhaustion, late-differentiation/senescence, and memory surface markers compared to blood. Furthermore, we compare the relative CD8+ T cell oligoclonality and TCR Vβ gene family usage, and the frequency of latently HIV-infected CD4+ T cells, in adipose tissue compared to blood among HIV-infected adults on long-term ART with sustained viral suppression.
Methods
Participants were recruited from an existing cohort enrolled to study the effects of HIV infection and obesity on metabolic disease, which has been previously described.38,39 All subjects had received efavirenz, tenofovir, and emtricitabine (i.e., the combination pill Atripla) for a minimum of 4 years, and had consistent HIV-1 RNA measurements <50 copies/ml for at least the prior 3 years (single viral ‘blips’ of less than 1000 copies/ml followed by measurements <50 copies/ml and no documentation of missed doses or poor adherence were allowed). Additional inclusion criteria were a blood CD4+ T cell count >350 cells/μl at the time of enrollment, no use of any anti-diabetic or statin (i.e., HMG CoA reductase inhibitor) medication, no self-reported heavy alcohol (defined as >11 drinks/week) or cocaine/amphetamine use, no active infectious conditions aside from HIV, and no previously diagnosed diabetes, cardiovascular, or rheumatologic disease.
All visits occurred in the Vanderbilt Clinical Research Center between 8 and 11 am. Subjects fasted for a minimum of 8 hours and blood was collected for mononuclear cell separation prior to other procedures. Subcutaneous abdominal adipose tissue biopsies were collected approximately 3 cm to the right of the umbilicus after anesthetizing the skin with lidocaine and infiltrating 10 ml of sterile saline and lidocaine as tumescent fluid. We collected 3-5 grams of adipose tissue using a 2.7 mm blunt, side-ported liposuction catheter (Tulip CellFriendly™ GEMS system Carraway Harvester, Tulip Medical Products, San Diego, CA) designed for the extraction of viable adipocytes and stromal vascular cells during cosmetic adipose tissue transfer procedures.40 Tissue was recovered in droplets of generally less than 3 mm diameter, limiting the need to mechanically mince the sample, and repeatedly rinsed with saline to eliminate contamination by blood. The adipose tissue was placed in a gentleMACS™ Dissociator (Miltenyi Biotec, Bergisch Gladbach, Germany) followed by incubation with collagenase. Mononuclear cells were separated by Ficoll-Paque Plus density gradient and cryopreserved in FBS with 10% DMSO.
Blood and adipose tissue mononuclear cell aliquots were thawed and stained for CD3, CD4, CD8, activation (CD38, HLA-DR, and CD69), exhaustion (PD1), late-differentiation/senescence (CD57), and memory (CD45RO) surface markers. Cells were run on a Fortessa (Becton Dickson Biosciences, San Jose, CA) flow cytometer, and CD4+ and CD8+ T cells were FACS sorted for subsequent assays. The percentage of each T cell subset was assessed using FACSDiva software (Becton Dickson Biosciences).
We purified genomic DNA from sorted CD4+ and CD8+ T cells using DNA IQ System (Promega, Madison, WI). DNA from adipose tissue and blood CD4+ T cells was used to compare the size of the latent HIV reservoir using the QX200 droplet digital PCR (ddPCR) PCR system and polymerase kit (Bio-Rad Laboratories, Hercules, CA) with primers and probes specific for the HIV LTR and gag genes. Reactions were partitioned into oil droplets using an automated droplet generator. We quantified the positive droplets that contained amplified HIV sequences (QuantaSoft software, Bio-Rad Laboratories, Hercules, CA); the number of HIV-1 gag or LTR copies was normalized with the RNaseP housekeeping gene, and then extrapolated to calculate the mean number of HIV copies per million CD4+ T cells.
DNA from adipose tissue and blood CD8+ T cells was used for bulk TCRβ CDR3 region amplification and sequencing using the ImmunoSEQ assay (Adaptive Biotechnologies, Seattle, WA).41,42 In this method, bias-controlled V and J gene primers are used to amplify rearranged V(D)J segments for sequencing. Nonproductive sequences were identified by non-templated insertions or deletions, which produced frameshifts or premature stop codons and were excluded from analysis. Extracted DNA from a minimum of 300,000 sorted CD8+ T cells was sequenced for all blood samples. Extracted DNA was sequenced from the seven adipose tissue samples that yielded more than 1000 sorted CD8+ T cells (range 1480-24,890 cells); the remaining 3 samples yielded CD8+ T cell counts below the recommended 1000 cell limit of the ImmunoSEQ assay. Two adipose tissue samples yielded less than 100 productive TCRβ sequence reads and were excluded, yielding a final sample of 5 paired adipose and blood TCRβ repertoires. The blood CD8+ TCRβ repertoire from subject 4 had a lower clonality score (0.021) compared to other subjects (range 0.079-0.226). Therefore, a second DNA sample from subject 4 was sequenced, which confirmed both the clonality score and the identity and frequency of the most prevalent sequences.
All subjects provided written informed consent and study procedures were in accordance with the human experimentation ethical standards of the Vanderbilt Institutional Review Board and with the Helsinki Declaration of 1975, as revised in 2000.
Statistical analysis
We compared the percentage of CD4+ and CD8+ T cells expressing each surface marker in paired blood and adipose tissue samples using Wilcoxon signed-rank tests. Raw amino acid and nucleotide data was exported from the Adaptive Biosciences ImmunoSEQ analyzer and the TCRβ V and J sequences were aligned using the international ImMunoGeneTics information system (IMGT).43 We compared the CD8+ TCRβ repertoire diversity in adipose tissue versus blood using the tcR R statistics package.44 Proportional downsampling and bootstrapping were used to account for read count distribution differences between tissue compartments and between patients. Frequencies of TCRβ V and J gene usage, and V-J gene pairing, were analyzed using the open-source VDJtools software package (https://github.com/mikessh/vdjtools).45 Analysis of variance (ANOVA) was used for comparisons between adipose and blood samples, and between subjects.
Results
The demographic and clinical characteristics of the 10 subjects are shown in Table 1. Six subjects were female, 7 were non-white, median age was 46 years (interquartile range [IQR] 39, 51), median blood CD4+ T cell count was 819 cells/μl (IQR 697, 1113), and median duration of ART was 9.6 years (IQR 6.8, 14.3). All subjects were overweight or obese (BMI >25.0 kg/m2), and median BMI was 32.8 kg/m2 (IQR 28.5, 36.5). None of the subjects were diabetic according to hemoglobin A1c (HbA1c) criteria (i.e., ≥6.5%), however 5 subjects were pre-diabetic (HbA1c 5.7-6.4%).
Table 1.
Description of study subjects
| Subject number | Sex | Age | Race | CD4+ count, cells/μl | ART regimen | Duration of ART, years | BMI, kg/m2 | Hemoglobin A1c, % |
|---|---|---|---|---|---|---|---|---|
| 1 | F | 44 | Black | 1311 | EFV/TDF/FTC | 4.6 | 36.4 | 5.4 |
| 2 | M | 53 | White | 713 | EFV/TDF/FTC | 8.1 | 34.8 | 5.5 |
| 3 | M | 34 | Black | 1053 | EFV/TDF/FTC | 7.0 | 37.0 | 5.7 |
| 4 | F | 40 | Black | 779 | EFV/TDF/FTC | 7.6 | 34.0 | 5.8 |
| 5 | F | 49 | Black | 502 | EFV/TDF/FTC | 25.6 | 27.2 | 5.6 |
| 6 | F | 47 | White | 1292 | EFV/TDF/FTC | 18.0 | 28.9 | 5.7 |
| 7 | M | 51 | Black | 859 | EFV/TDF/FTC | 13.1 | 41.2 | 5.9 |
| 8 | F | 37 | Black | 1025 | EFV/TDF/FTC | 12.1 | 27.7 | 6.4 |
| 9 | F | 45 | Black | 755 | EFV/TDF/FTC | 11.1 | 28.8 | 4.8 |
| 10 | M | 51 | White | 647 | EFV/TDF/FTC | 6.2 | 31.7 | 5.4 |
Adipose tissue contained a lower percentage of CD4+ T cells, and a higher percentage of CD8+ cells, in comparison to blood (Table 2; P<0.01 for both). The percentage of CD8+ T cells expressing the HLA-DR activation marker in adipose tissue was over 6-fold higher compared to those in blood (5.5% vs. 0.9%, P<0.01). Similarly, the percentage of CD8+ T cells expressing CD57, a marker of late differentiation or replicative senescence, was significantly higher in adipose tissue than in blood (37.4% vs. 22.7%, P<0.01). Adipose tissue was also enriched for CD4+ T cells expressing CD57 (5.7% vs. 2.2%, P<0.01) and HLA-DR (3.5% vs. 1.0%, P<0.01), though at lower absolute levels as compared to the CD8+ T cells. Adipose tissue CD4+ T cell expression of the TCR-coupled CD69 activation marker was over 5-fold higher compare to blood, but among memory CD4+ CD45RO+ T cells the CD69 expression in adipose tissue was 20-fold higher (5.9% vs. 0.3%, P=0.04).
Table 2.
Surface marker expression on CD4+ and CD8+ T cells from paired blood and adipose tissue (n=10)
| Comparison of median percentage in blood vs. adipose tissue | |||
|---|---|---|---|
| T cell subset | Median in Blood (IQR) | Median in Adipose Tissue (IQR) | P-value† |
| CD4+ %* | 42.6 (35.1, 45.8) | 33.1 (28.3, 38.9) | <0.01 |
| CD4+ PD1+% (exhaustion) | 24.6 (20.2, 27.0) | 24.9 (17.7, 30.5) | 0.86 |
| CD4+ CD57+% (senescence) | 2.2 (1.4, 6.6) | 5.7 (3.3, 14.9) | <0.01 |
| CD4+ CD38+% (activation) | 4.0 (1.9, 6.3) | 2.7 (2.0, 4.2) | 0.44 |
| CD4+ DR+% (activation) | 1.0 (0.6, 1.6) | 3.5 (1.5, 5.6) | <0.01 |
| CD4+ CD69+% (activation) | 0.3 (0.2, 0.3) | 1.3 (0.3, 4.5) | 0.02 |
| CD4+ CD45RO+% (memory) | 51.3 (47.6, 62.7) | 53.7 (47.6, 69.5) | 0.96 |
| CD4+ CD45RO+ cells expressing CD69 (%) | 0.3 (0.3, 0.5) | 5.9 (0.3, 6.3) | 0.04 |
| CD8+ %* | 51.7 (42.7, 59.6) | 61.0 (52.8, 67.2) | <0.01 |
| CD8+ PD1+% (exhaustion) | 20.6 (11.4, 27.7) | 20.0 (13.4, 25.1) | 0.89 |
| CD8+ CD57+% (senescence) | 22.7 (15.2, 26.7) | 37.4 (27.6, 45.1) | <0.01 |
| CD8+ CD38+% (activation) | 0.5 (0.5, 0.6) | 0.9 (0.5, 1.1) | 0.01 |
| CD8+ DR+% (activation) | 0.9 (0.8, 1.2) | 5.5 (3.2, 10.3) | <0.01 |
| CD8+ CD69+% (activation) | 0.5 (0.3, 0.8) | 1.1 (0.53, 1.4) | 0.15 |
| CD8+ CD45RO+% (memory) | 31.3 (18.1, 55.9) | 32.6 (20.8, 54.6) | 0.31 |
Represents the percentage of CD3+ T cells that expressed CD4 or CD8. All other values represent the percentage of the CD4+ or CD8+ population expressing indicated surface markers.
Wilcoxon Signed Rank Test – a bold p-value indicates the median of differences between paired blood and adipose tissue is not equal to zero with a significance level of 0.05.
Latent HIV DNA was detected in the two adipose tissue samples with more than 5000 sorted CD4+ T cells, but not in samples with fewer CD4+ cells; in both subjects the copy number was higher in adipose tissue CD4+ cells compared to blood (3.50 vs 3.14, and 3.46 vs. 3.22, log DNA copies/million CD4+ cells).
Five adipose tissue samples yielded sufficient CD8+ TCRβ sequences (range 319 to 4,548 productive templates) for analysis. The 10 most prevalent TCRβ clones comprised a larger percentage of total clones in adipose tissue compared to paired blood in all 5 subjects (25% vs. 16% of total repertoire, respectively; P=0.04, Figure 1), and the Shannon’s Entropy index, a measure of overall repertoire diversity, was lower in adipose tissue compared to blood (4.39 vs. 4.46, respectively; P=0.05). Notably, we did not observe the same TCRβ sequences, also referred to as public clonotypes (i.e., specific rearrangements observed in multiple individuals), occurring at greater than 0.9% frequency in adipose tissue among any two study subjects. We graphed V and J gene pairing in adipose tissue versus blood using Circos plots (Figure 2). These plots demonstrate visual differences in gene usage and V-J gene pairing between blood and adipose tissue, but these differences were not statistically significant between the five subjects (P=0.12).
Figure 1.
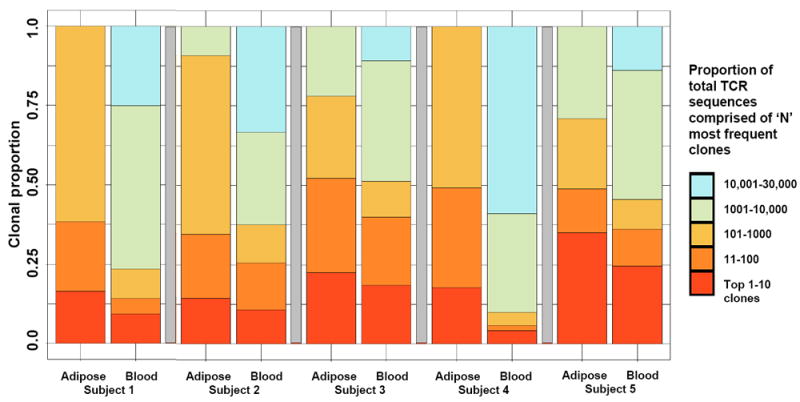
Greater clonality of the CD8+ T cell receptor β repertoire in adipose tissue compared to blood
Colored bars represent the relative proportion of all productive CD8+ T cell receptor (TCR)β sequences from each subject’s paired adipose tissue and blood that comprised the 10 most prevalent clones (red bar), the 100 most prevalent clones (dark orange bar), etc. Proportional downsampling and bootstrapping were used to account for read count distribution differences between tissue compartments. The 10 most prevalent TCRβ clones comprised a larger percentage of total clones in adipose tissue compared to paired blood in all 5 subjects (25% vs. 16% of total repertoire, respectively; P=0.04), and the Shannon’s Entropy index, a measure of overall repertoire diversity, was lower in adipose tissue compared to blood (4.39 vs. 4.46, respectively; P=0.05).
Figure 2.
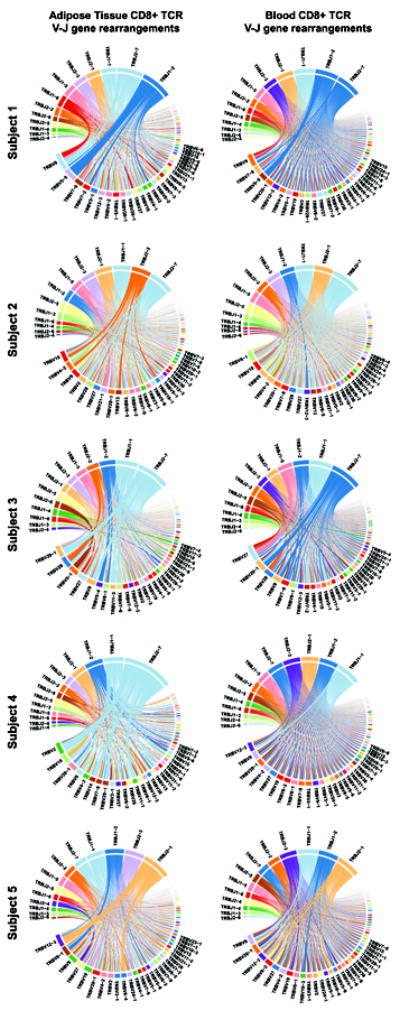
Distinct CD8+ T cell receptor β V and J gene arrangement distributions in adipose tissue compared to blood
Circos plots showing the combinations of V and J genes used in the rearranged T cell receptor (TCR)β expressed by CD8+ T cells in adipose tissue and matched blood samples. For each subject, the left-sided plot represents sorted adipose tissue CD8+ T cells and the right-sided plot represents sorted blood CD8+ T cells. The upper section of each plot depicts TCRβ (abbreviated TRB) J genes and the lower section depicts V genes. The arc length of each colored block denotes the relative frequency at which the gene family was identified. Rearrangement of a J gene with a V gene segment in a clonal TCR sequence is represented by a ribbon carrying the color of the J gene family to the color of the V gene family participating in the pairing. The width of the ribbon corresponds to the frequency at which each particular V-J rearrangement occurs in the TCR repertoire.
Discussion
In this study, we found the subcutaneous adipose tissue of overweight and obese HIV patients on long-term ART is enriched for both activated and late-differentiated CD4+ and CD8+ T cells compared to blood, and the overall population of CD8+ cells in adipose tissue demonstrates greater clonality and a higher fraction of the repertoire devoted to the most prevalent TCRβ sequences. The increased proportion of antigen-stimulated activated, late-differentiated, and memory CD8+ T cells in adipose tissue, as opposed to naïve cells, would be expected to confer a more clonal TCRβ repertoire. However, the finding that the most prevalent V and J gene pairings appears to differ between tissue compartments suggests the adipose tissue TCRβ repertoire does not simply represent a concentration of the most common blood sequences, as might be observed if activated and memory T cells collected in adipose tissue in a random manner.
This is the first study, to our knowledge, to use TCR sequencing to compare clone repertoires in blood and adipose tissue compartments from HIV-infected persons. While CD8+ TCRβ clonality was greater in every adipose tissue sample compared to the matched blood, there was minimal shared sequence homology between subjects, indicating a lack of public clonotypes characteristic of conserved antigenic stimuli. This finding could indicate a ‘founder effect’ in which certain adipose tissue CD8+ T cells undergo oligoclonal expansion in the presence of local stimuli, such as IL-2, and fail to egress and equilibrate with the blood. Alternately, this finding could also reflect a homing of specific CD8+ cells to adipose tissue and expansion in response to a unique antigen, or neo-antigen, as might result from oxidative damage or other protein modifications.
While prior studies have shown higher CD4+ and CD8+ T cell expression of HLA-DR in the adipose tissue of SIV-infected macaques compared to blood, we are not aware of prior studies in HIV-infected humans or animal models showing an enrichment of CD57+ T cells. Expression of CD57, a terminally sulfated glycan carbohydrate epitope, on T cells is considered a marker of late differentiation and reduced replicative capacity, or an inability to proliferate in response to antigen-stimulation and increased susceptibility to activation-induced apoptosis.46-48 A higher percentage of CD57+ T cells is implicated in several inflammatory diseases, 49-51 and CD57 expression on CD4+ and CD8+ T cells in the blood is higher in HIV-infected persons compared to HIV-negative.48,52,53 We observed a far higher percentage of CD8+ T cells expressing CD57 in adipose tissue (37.4%) compared to CD4+ T cells expressing CD57 (5.7%). CD8+ T cells expressing CD57 have higher interferon-γ and TNF-α production after TCR stimulation compared to CD8+CD57- T cells, and demonstrate a distinct gene expression profile characterized by greater cytotoxic effector potential (e.g., production of perforin, granzymes, and granulysin), greater adhesion molecule expression, and reduced chemokine receptor expression.54,55 Granulysin is one of the most overexpressed genes in CD8+CD57+ compared to CD8+CD57- T cells and a central component in the granules of cytotoxic T cells and NK cells, which serves as a potent immune cell chemoattractant and proinflammatory activator.56-58 A central question for further research is whether the percentage of CD57+ T cells in adipose tissue is higher in HIV-infected persons compared to HIV-negative, and whether these cells are expressing a cytokine profile that contributes to a local inflammatory environment and changes in adipocyte function.
Our study findings contribute to the limited literature on adipose tissue T cells in HIV-infected persons or SIV-infected macaques.35,36,59,60 A study combining subcutaneous and visceral adipose tissue from live donors and cadavers found a higher proportion of memory (CD45RO+) CD8+ T cells and lower memory CD4+ T cells in the adipose tissue of HIV-infected subjects as compared to HIV-negative controls, and the memory cells predominantly expressed the CD69 activation marker suggesting the possible presence of receptor stimulation in the local environment.36 Two studies of cynomolgus macaques found SIV infection was associated with a higher percentage of CD8+ T cells and lower CD4+ T cells in both the subcutaneous and visceral adipose tissue compared to uninfected animals.35,59 The percentage of activated (HLA-DR+) CD4+ and CD8+ T cells, and the density of activated macrophages, was significantly higher in subcutaneous and visceral adipose tissue of SIV-infected animals compared to uninfected controls.35 Adipocytes from SIV-infected macaques also have lower CCAAT enhancer–binding protein-α (C/EBPα; a master transcription factor controlling adipogenesis), C/EBPβ, leptin, and GLUT4 expression compared to uninfected animals, but higher expressions of IL-2, IL-7, and CCL19, three cytokines that may contribute to the homing and survival of T cells in adipose tissue.59
Irrespective of HIV infection, the presence of activated and memory CD4+ and CD8+ T cells in adipose tissue contributes to local inflammation and reduced adipocyte insulin sensitivity and energy storage. Infiltration of several T cell subtypes, including CD4+ Th1-type cells,26,31,37,61 CD8+ cytotoxic T cells,24,37,62 and natural killer T cells,63,64 promotes the recruitment of M1-phenotype macrophages and other innate immune cells. These cells secrete inflammatory cytokines and chemokines that inhibit adipocyte insulin signaling through changes in gene transcription and the phosphorylation of IRS-1.28-30 Furthermore, in vitro studies have found obese fat independently activates CD8+ T cells and induces proliferation, while lean fat has little effect.24 Analyses of adipose tissue CD8+ T cells in the setting of obesity have also identified a predominance of memory cells and a relatively conserved repertoire of T cell receptor Vα and Vβ chain families, indicating these cells may undergo oligoclonal expansion.24,26,37 Lastly, experimental models depleting CD8+ T cells in adipose tissue result in reduced macrophage density and improved insulin sensitivity, suggesting that immunotherapy could have a future application in the management of metabolic disease.24,26,65,66
Further studies are needed to understand the etiology of the oligoclonal expansion of CD8+ T cells in adipose tissue. As noted above, this could result from a TCR specificity-independent mechanism in which random activated and memory T cells enter the adipose tissue, adopt a tissue-resident phenotype inhibiting egress to the blood, and then expand in response to local IL-2 or other cytokines. It may also represent a TCR-specific accumulation in response to an antigenic stimuli that is present in the adipose tissue of HIV-infected persons but is not homologous or highly conserved; one example might be diverse proteins or lipids altered by local inflammation or oxidative damage. Notably, animal studies have also identified a predominance of specific TCR Vα and Vβ chain families in the adipose tissue of obese animals (though not using the sequencing approach we employed).24,26,37 While these prior studies and our results do not resolve the etiology of the clonal CD8+ T cell expansion, the similar findings in animal models of obesity suggest the stimuli originates in the adipose tissue and is not specific to HIV-infected persons.
The primary limitations of our study were the small sample size, quantity of adipose tissue T cells collected, and the technical challenge of eliminating peripheral blood contamination from the liposuction aspirate. Our group collected only 3-5 grams of adipose tissue from each participant, and the yield of sorted CD4+ and CD8+ T cells was insufficient to perform TCRβ sequencing or latent HIV-quantification on all subjects. Liposuction requires the infiltration of lidocaine and saline (i.e., tumescent fluid) prior to tissue aspiration, and the mechanical trauma leads to contamination of this fluid with blood. While our adipose tissue aspirates were repeatedly rinsed prior to enzymatic and mechanical digestion, we observed markedly lower absolute expression of CD69 on CD4+ and CD8+ memory T cells compared to prior reports from HIV-infected humans and SIV-infected macaques (though we did find 20-fold higher CD69 expression on our adipose CD4+ memory cells compared to blood).35,36,59 In the absence of problems with antibody-binding, which we do not suspect, this lower absolute CD69 expression on memory CD4+ T cells suggests a degree of contamination by peripheral T cells. Such contamination would have biased our comparisons of T cell subset percentages blood versus adipose tissue towards the null, and therefore the true differences between compartments may actually be more pronounced than we report. Going forward, our group has developed a protocol for multiple adipose tissue saline baths immediately after aspiration, and we are investigating candidate surface markers indicative of T cells present in adipose tissue (i.e., a ‘tissue resident’ phenotype), which could be used as cell-sorting criterion.
Our study only sequenced CD8+ cell TCRs, which was based on prior studies demonstrating restricted CD8+ cell TCR gene families in mouse models of obesity and insulin resistance,24,26,37 and to allow for latent HIV quantification on the CD4+ cells. Larger volumes of tissue aspiration in future studies will permit sequencing of both the CD8+ and CD4+ TCR repertoire. To reduce potential confounding from different ART regimens, our study only enrolled subjects with long-term viral suppression on EFV/TDF/FTC, but consequently we could not assess the effect of unsuppressed viremia or other ART medications on T cell profiles. Finally, larger studies enrolling subjects with a wider range of insulin resistance will be needed to assess the relationship between adipose tissue T cell populations and glucose tolerance and metabolic disease more broadly.
In summary, our study demonstrates that subcutaneous adipose tissue from HIV patients is enriched for activated and late-differentiated CD8+ T cells, and is characterized by a disproportionate expansion of specific CD8+ T cell clones compared to blood. We also demonstrate the feasibility of utilizing liposuction to collect and extract adipose tissue T cells, and the use of TCRβ sequencing to characterize receptor repertoire and gene segment usage in this cell population. Based on our findings, further studies are needed to understand the etiology of the observed oligoclonal expansion and its relationship to adipocyte function and metabolic disease in HIV-infected persons.
Acknowledgments
The authors thank the participants in the Adiposity and Immune Activation Cohort study.
Source of Funding: This work was supported by NIAID grant K23 AI100700, NIDDK grant R01 DK112262, the NIH-funded Vanderbilt Clinical and Translational Science award from NCRR/NIH grant UL1 RR024975, the NIH-funded Tennessee Center for AIDS Research grant P30 AI110527, National Institute of General Medical Sciences awards T32 GM007347 and T32 GM008320, and National Heart, Lung, and Blood Institute award T32 HL069765. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Prior presentations: These data were presented at the International Workshops on Co-morbidities and Adverse Drug Reactions in HIV, New York, NY, September 12-13 2016, and the Conference on Retroviruses and Opportunistic Infections, Boston, MA, February 13-16, 2017.
Disclosures: No authors report any conflicts of interest or relevant financial relationships
References
- 1.Brown TT, Cole SR, Li X, et al. Antiretroviral therapy and the prevalence and incidence of diabetes mellitus in the multicenter AIDS cohort study. Arch Intern Med. 2005;165(10):1179–1184. doi: 10.1001/archinte.165.10.1179. [DOI] [PubMed] [Google Scholar]
- 2.De Wit S, Sabin CA, Weber R, et al. Incidence and risk factors for new-onset diabetes in HIV-infected patients: the Data Collection on Adverse Events of Anti-HIV Drugs (D:A:D) study. Diabetes Care. 2008;31(6):1224–1229. doi: 10.2337/dc07-2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Capeau J, Bouteloup V, Katlama C, et al. Ten-year diabetes incidence in 1046 HIV-infected patients started on a combination antiretroviral treatment. AIDS. 2012;26(3):303–314. doi: 10.1097/QAD.0b013e32834e8776. [DOI] [PubMed] [Google Scholar]
- 4.Leroyer S, Vatier C, Kadiri S, et al. Glyceroneogenesis is inhibited through HIV protease inhibitor-induced inflammation in human subcutaneous but not visceral adipose tissue. J Lipid Res. 2011;52(2):207–220. doi: 10.1194/jlr.M000869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gallego-Escuredo JM, Villarroya J, Domingo P, et al. Differentially altered molecular signature of visceral adipose tissue in HIV-1-associated lipodystrophy. J Acquir Immune Defic Syndr. 2013;64(2):142–148. doi: 10.1097/QAI.0b013e31829bdb67. [DOI] [PubMed] [Google Scholar]
- 6.Ben-Romano R, Rudich A, Torok D, et al. Agent and cell-type specificity in the induction of insulin resistance by HIV protease inhibitors. AIDS. 2003;17(1):23–32. doi: 10.1097/00002030-200301030-00005. [DOI] [PubMed] [Google Scholar]
- 7.Rudich A, Ben-Romano R, Etzion S, Bashan N. Cellular mechanisms of insulin resistance, lipodystrophy and atherosclerosis induced by HIV protease inhibitors. Acta Physiol Scand. 2005;183(1):75–88. doi: 10.1111/j.1365-201X.2004.01383.x. [DOI] [PubMed] [Google Scholar]
- 8.Brinkman K, Smeitink JA, Romijn JA, Reiss P. Mitochondrial toxicity induced by nucleoside-analogue reverse-transcriptase inhibitors is a key factor in the pathogenesis of antiretroviral-therapy-related lipodystrophy. Lancet. 1999;354(9184):1112–1115. doi: 10.1016/S0140-6736(99)06102-4. [DOI] [PubMed] [Google Scholar]
- 9.Lewis W, Day BJ, Copeland WC. Mitochondrial toxicity of NRTI antiviral drugs: an integrated cellular perspective. Nat Rev Drug Discov. 2003;2(10):812–822. doi: 10.1038/nrd1201. [DOI] [PubMed] [Google Scholar]
- 10.Caron M, Auclair M, Lagathu C, et al. The HIV-1 nucleoside reverse transcriptase inhibitors stavudine and zidovudine alter adipocyte functions in vitro. AIDS. 2004;18(16):2127–2136. doi: 10.1097/00002030-200411050-00004. [DOI] [PubMed] [Google Scholar]
- 11.Caron M, Auclairt M, Vissian A, Vigouroux C, Capeau J. Contribution of mitochondrial dysfunction and oxidative stress to cellular premature senescence induced by antiretroviral thymidine analogues. Antivir Ther. 2008;13(1):27–38. [PubMed] [Google Scholar]
- 12.Feeney ER, Mallon PW. Impact of mitochondrial toxicity of HIV-1 antiretroviral drugs on lipodystrophy and metabolic dysregulation. Curr Pharm Des. 2010;16(30):3339–3351. doi: 10.2174/138161210793563482. [DOI] [PubMed] [Google Scholar]
- 13.Galluzzi L, Pinti M, Troiano L, et al. Changes in mitochondrial RNA production in cells treated with nucleoside analogues. Antivir Ther. 2005;10(1):191–195. [PubMed] [Google Scholar]
- 14.Cote HC. Possible ways nucleoside analogues can affect mitochondrial DNA content and gene expression during HIV therapy. Antivir Ther. 2005;10(Suppl 2):M3–11. [PubMed] [Google Scholar]
- 15.Jones SP, Qazi N, Morelese J, et al. Assessment of adipokine expression and mitochondrial toxicity in HIV patients with lipoatrophy on stavudine- and zidovudine-containing regimens. J Acquir Immune Defic Syndr. 2005;40(5):565–572. doi: 10.1097/01.qai.0000187443.30838.3e. [DOI] [PubMed] [Google Scholar]
- 16.Shikuma CM, Gangcuangco LM, Killebrew DA, et al. The role of HIV and monocytes/macrophages in adipose tissue biology. J Acquir Immune Defic Syndr. 2014;65(2):151–159. doi: 10.1097/01.qai.0000435599.27727.6c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Casteilla L, Cousin B, Carmona M. PPARs and Adipose Cell Plasticity. PPAR Res. 2007;2007:68202. doi: 10.1155/2007/68202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Caron M, Vigouroux C, Bastard JP, Capeau J. Antiretroviral-related adipocyte dysfunction and lipodystrophy in HIV-infected patients: Alteration of the PPARgamma-dependent pathways. PPAR Res. 2009;2009:507141. doi: 10.1155/2009/507141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mandard S, Patsouris D. Nuclear control of the inflammatory response in mammals by peroxisome proliferator-activated receptors. PPAR Res. 2013;2013:613864. doi: 10.1155/2013/613864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kintscher U, Law RE. PPARgamma-mediated insulin sensitization: the importance of fat versus muscle. Am J Physiol Endocrinol Metab. 2005;288(2):E287–291. doi: 10.1152/ajpendo.00440.2004. [DOI] [PubMed] [Google Scholar]
- 21.Medina-Gomez G, Gray SL, Yetukuri L, et al. PPAR gamma 2 prevents lipotoxicity by controlling adipose tissue expandability and peripheral lipid metabolism. PLoS Genet. 2007;3(4):e64. doi: 10.1371/journal.pgen.0030064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Semple RK, Chatterjee VK, O’Rahilly S. PPAR gamma and human metabolic disease. J Clin Invest. 2006;116(3):581–589. doi: 10.1172/JCI28003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Garrabou G, Lopez S, Moren C, et al. Mitochondrial damage in adipose tissue of untreated HIV-infected patients. AIDS. 2011;25(2):165–170. doi: 10.1097/QAD.0b013e3283423219. [DOI] [PubMed] [Google Scholar]
- 24.Nishimura S, Manabe I, Nagasaki M, et al. CD8+ effector T cells contribute to macrophage recruitment and adipose tissue inflammation in obesity. Nat Med. 2009;15(8):914–920. doi: 10.1038/nm.1964. [DOI] [PubMed] [Google Scholar]
- 25.Feuerer M, Herrero L, Cipolletta D, et al. Lean, but not obese, fat is enriched for a unique population of regulatory T cells that affect metabolic parameters. Nat Med. 2009;15(8):930–939. doi: 10.1038/nm.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Winer S, Chan Y, Paltser G, et al. Normalization of obesity-associated insulin resistance through immunotherapy. Nat Med. 2009;15(8):921–929. doi: 10.1038/nm.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Winer DA, Winer S, Shen L, et al. B cells promote insulin resistance through modulation of T cells and production of pathogenic IgG antibodies. Nat Med. 2011;17(5):610–617. doi: 10.1038/nm.2353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hotamisligil GS, Peraldi P, Budavari A, Ellis R, White MF, Spiegelman BM. IRS-1-mediated inhibition of insulin receptor tyrosine kinase activity in TNF-alpha- and obesity-induced insulin resistance. Science. 1996;271(5249):665–668. doi: 10.1126/science.271.5249.665. [DOI] [PubMed] [Google Scholar]
- 29.Senn JJ, Klover PJ, Nowak IA, Mooney RA. Interleukin-6 induces cellular insulin resistance in hepatocytes. Diabetes. 2002;51(12):3391–3399. doi: 10.2337/diabetes.51.12.3391. [DOI] [PubMed] [Google Scholar]
- 30.Lumeng CN, Bodzin JL, Saltiel AR. Obesity induces a phenotypic switch in adipose tissue macrophage polarization. J Clin Invest. 2007;117(1):175–184. doi: 10.1172/JCI29881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kintscher U, Hartge M, Hess K, et al. T-lymphocyte infiltration in visceral adipose tissue: a primary event in adipose tissue inflammation and the development of obesity-mediated insulin resistance. Arterioscler Thromb Vasc Biol. 2008;28(7):1304–1310. doi: 10.1161/ATVBAHA.108.165100. [DOI] [PubMed] [Google Scholar]
- 32.Weisberg SP, McCann D, Desai M, Rosenbaum M, Leibel RL, Ferrante AW., Jr Obesity is associated with macrophage accumulation in adipose tissue. J Clin Invest. 2003;112(12):1796–1808. doi: 10.1172/JCI19246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gao D, Madi M, Ding C, et al. Interleukin-1beta mediates macrophage-induced impairment of insulin signaling in human primary adipocytes. Am J Physiol Endocrinol Metab. 2014;307(3):E289–304. doi: 10.1152/ajpendo.00430.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lumeng CN, Deyoung SM, Saltiel AR. Macrophages block insulin action in adipocytes by altering expression of signaling and glucose transport proteins. Am J Physiol Endocrinol Metab. 2007;292(1):E166–174. doi: 10.1152/ajpendo.00284.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Damouche A, Lazure T, Avettand-Fenoel V, et al. Adipose Tissue Is a Neglected Viral Reservoir and an Inflammatory Site during Chronic HIV and SIV Infection. PLoS Pathog. 2015;11(9):e1005153. doi: 10.1371/journal.ppat.1005153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Couturier J, Suliburk JW, Brown JM, et al. Human adipose tissue as a reservoir for memory CD4+ T cells and HIV. AIDS. 2015;29(6):667–674. doi: 10.1097/QAD.0000000000000599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang H, Youm YH, Vandanmagsar B, et al. Obesity increases the production of proinflammatory mediators from adipose tissue T cells and compromises TCR repertoire diversity: implications for systemic inflammation and insulin resistance. J Immunol. 2010;185(3):1836–1845. doi: 10.4049/jimmunol.1000021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Koethe JR, Grome H, Jenkins CA, Kalams SA, Sterling TR. The metabolic and cardiovascular consequences of obesity in persons with HIV on long-term antiretroviral therapy. AIDS. 2016;30(1):83–91. doi: 10.1097/QAD.0000000000000893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Koethe JR, Jenkins CA, Petucci C, Culver J, Shepherd BE, Sterling TR. Superior Glucose Tolerance and Metabolomic Profiles, Independent of Adiposity, in HIV-Infected Women Compared With Men on Antiretroviral Therapy. Medicine (Baltimore) 2016;95(19):e3634. doi: 10.1097/MD.0000000000003634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Alexander RW, Harrell DB. Autologous fat grafting: use of closed syringe microcannula system for enhanced autologous structural grafting. Clinical, cosmetic and investigational dermatology. 2013;6:91–102. doi: 10.2147/CCID.S40575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Robins HS, Campregher PV, Srivastava SK, et al. Comprehensive assessment of T-cell receptor beta-chain diversity in alphabeta T cells. Blood. 2009;114(19):4099–4107. doi: 10.1182/blood-2009-04-217604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.DeWitt WS, Emerson RO, Lindau P, et al. Dynamics of the cytotoxic T cell response to a model of acute viral infection. J Virol. 2015;89(8):4517–4526. doi: 10.1128/JVI.03474-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li S, Lefranc MP, Miles JJ, et al. IMGT/HighV QUEST paradigm for T cell receptor IMGT clonotype diversity and next generation repertoire immunoprofiling. Nature communications. 2013;4:2333. doi: 10.1038/ncomms3333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nazarov VI, Pogorelyy MV, Komech EA, et al. tcR: an R package for T cell receptor repertoire advanced data analysis. BMC Bioinformatics. 2015;16:175. doi: 10.1186/s12859-015-0613-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shugay M, Bagaev DV, Turchaninova MA, et al. VDJtools: Unifying Post-analysis of T Cell Receptor Repertoires. PLoS Comput Biol. 2015;11(11):e1004503. doi: 10.1371/journal.pcbi.1004503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Focosi D, Bestagno M, Burrone O, Petrini M. CD57+ T lymphocytes and functional immune deficiency. J Leukoc Biol. 2010;87(1):107–116. doi: 10.1189/jlb.0809566. [DOI] [PubMed] [Google Scholar]
- 47.Brenchley JM, Karandikar NJ, Betts MR, et al. Expression of CD57 defines replicative senescence and antigen-induced apoptotic death of CD8+ T cells. Blood. 2003;101(7):2711–2720. doi: 10.1182/blood-2002-07-2103. [DOI] [PubMed] [Google Scholar]
- 48.Palmer BE, Blyveis N, Fontenot AP, Wilson CC. Functional and phenotypic characterization of CD57+CD4+ T cells and their association with HIV-1-induced T cell dysfunction. J Immunol. 2005;175(12):8415–8423. doi: 10.4049/jimmunol.175.12.8415. [DOI] [PubMed] [Google Scholar]
- 49.Maeda T, Yamada H, Nagamine R, et al. Involvement of CD4+, CD57+ T cells in the disease activity of rheumatoid arthritis. Arthritis Rheum. 2002;46(2):379–384. doi: 10.1002/art.10133. [DOI] [PubMed] [Google Scholar]
- 50.Palmer BE, Mack DG, Martin AK, Maier LA, Fontenot AP. CD57 expression correlates with alveolitis severity in subjects with beryllium-induced disease. J Allergy Clin Immunol. 2007;120(1):184–191. doi: 10.1016/j.jaci.2007.03.009. [DOI] [PubMed] [Google Scholar]
- 51.Hoffmann J, Fiser K, Weaver J, et al. High-throughput 13-parameter immunophenotyping identifies shifts in the circulating T-cell compartment following reperfusion in patients with acute myocardial infarction. PLoS One. 2012;7(10):e47155. doi: 10.1371/journal.pone.0047155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Papagno L, Spina CA, Marchant A, et al. Immune activation and CD8+ T-cell differentiation towards senescence in HIV-1 infection. PLoS Biol. 2004;2(2):E20. doi: 10.1371/journal.pbio.0020020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lieberman J, Trimble LA, Friedman RS, et al. Expansion of CD57 and CD62L-CD45RA+ CD8 T lymphocytes correlates with reduced viral plasma RNA after primary HIV infection. AIDS. 1999;13(8):891–899. doi: 10.1097/00002030-199905280-00004. [DOI] [PubMed] [Google Scholar]
- 54.Le Priol Y, Puthier D, Lecureuil C, et al. High cytotoxic and specific migratory potencies of senescent CD8+ CD57+ cells in HIV-infected and uninfected individuals. J Immunol. 2006;177(8):5145–5154. doi: 10.4049/jimmunol.177.8.5145. [DOI] [PubMed] [Google Scholar]
- 55.Tae Yu H, Youn JC, Lee J, et al. Characterization of CD8(+)CD57(+) T cells in patients with acute myocardial infarction. Cell Mol Immunol. 2015;12(4):466–473. doi: 10.1038/cmi.2014.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pena SV, Hanson DA, Carr BA, Goralski TJ, Krensky AM. Processing, subcellular localization, and function of 519 (granulysin), a human late T cell activation molecule with homology to small, lytic, granule proteins. J Immunol. 1997;158(6):2680–2688. [PubMed] [Google Scholar]
- 57.Jongstra J, Schall TJ, Dyer BJ, et al. The isolation and sequence of a novel gene from a human functional T cell line. J Exp Med. 1987;165(3):601–614. doi: 10.1084/jem.165.3.601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Deng A, Chen S, Li Q, Lyu SC, Clayberger C, Krensky AM. Granulysin, a cytolytic molecule, is also a chemoattractant and proinflammatory activator. J Immunol. 2005;174(9):5243–5248. doi: 10.4049/jimmunol.174.9.5243. [DOI] [PubMed] [Google Scholar]
- 59.Couturier J, Agarwal N, Nehete PN, et al. Infectious SIV resides in adipose tissue and induces metabolic defects in chronically infected rhesus macaques. Retrovirology. 2016;13:30. doi: 10.1186/s12977-016-0260-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pallikkuth S, Mohan M. Adipose Tissue: Sanctuary for HIV/SIV Persistence and Replication. Trends Microbiol. 2015;23(12):748–750. doi: 10.1016/j.tim.2015.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Duffaut C, Zakaroff-Girard A, Bourlier V, et al. Interplay between human adipocytes and T lymphocytes in obesity: CCL20 as an adipochemokine and T lymphocytes as lipogenic modulators. Arterioscler Thromb Vasc Biol. 2009;29(10):1608–1614. doi: 10.1161/ATVBAHA.109.192583. [DOI] [PubMed] [Google Scholar]
- 62.Rausch ME, Weisberg S, Vardhana P, Tortoriello DV. Obesity in C57BL/6J mice is characterized by adipose tissue hypoxia and cytotoxic T-cell infiltration. Int J Obes (Lond) 2008;32(3):451–463. doi: 10.1038/sj.ijo.0803744. [DOI] [PubMed] [Google Scholar]
- 63.Ohmura K, Ishimori N, Ohmura Y, et al. Natural killer T cells are involved in adipose tissues inflammation and glucose intolerance in diet-induced obese mice. Arterioscler Thromb Vasc Biol. 2010;30(2):193–199. doi: 10.1161/ATVBAHA.109.198614. [DOI] [PubMed] [Google Scholar]
- 64.Mantell BS, Stefanovic-Racic M, Yang X, Dedousis N, Sipula IJ, O’Doherty RM. Mice lacking NKT cells but with a complete complement of CD8+ T-cells are not protected against the metabolic abnormalities of diet-induced obesity. PLoS One. 2011;6(6):e19831. doi: 10.1371/journal.pone.0019831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rocha VZ, Folco EJ, Sukhova G, et al. Interferon-gamma, a Th1 cytokine, regulates fat inflammation: a role for adaptive immunity in obesity. Circ Res. 2008;103(5):467–476. doi: 10.1161/CIRCRESAHA.108.177105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Stolarczyk E, Vong CT, Perucha E, et al. Improved insulin sensitivity despite increased visceral adiposity in mice deficient for the immune cell transcription factor T-bet. Cell Metab. 2013;17(4):520–533. doi: 10.1016/j.cmet.2013.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]


