Abstract
Studying patterns of phenotypic variation among populations can shed light on the drivers of evolutionary processes. The house sparrow (Passer domesticus) is one of the world’s most ubiquitous bird species, as well as a successful invader. We investigated phenotypic variation in house sparrow populations across a climatic gradient and in relation to a possible scenario of an invasion. We measured variation in morphological, coloration, and behavioral traits (exploratory behavior and neophobia) and compared it to the neutral genetic variation. We found that sparrows were larger and darker in northern latitudes, in accordance with Bergmann’s and Gloger’s biogeographic rules. Morphology and behavior mostly differed between the southernmost populations and the other regions, supporting the possibility of an invasion. Genetic differentiation was low and diversity levels were similar across populations, indicating high gene flow. Nevertheless, the southernmost and northern populations differed genetically to some extent. Furthermore, genetic differentiation (F ST) was lower in comparison to phenotypic variation (P ST), indicating that the phenotypic variation is shaped by directional selection or by phenotypic plasticity. This study expands our knowledge on evolutionary mechanisms and biological invasions.
Introduction
The study of phenotypic variation among natural populations and its relation to ecology is one of the central concepts in the field of evolutionary biology1. Identifying patterns of variation can provide insight into the forces driving evolutionary processes, whether natural selection, phenotypic plasticity, genetic drift – or an interplay between more than one of these processes2,3. Latitudinal variation in morphology and color is often explained by two important biogeographic rules4,5. Bergmann’s rule predicts that larger body size should be advantageous to homoeothermic animals in colder climates (usually found in northern latitudes), due to lower surface-to-volume ratio allowing better heat conservation6,7. However, there is disagreement on the validity of this rule for birds (42% vs. 72% of bird species8,9). Gloger’s rule predicts that animals should be more heavily pigmented in humid than in arid habitats, supported by adaptive explanations (matching between background color and feathers which serves as camouflage, or higher resistance of dark feathers to bacteria, which thrive more in humid habitats10,11). The empirical evidence for this rule is convincing, as 96% of bird species conform to this prediction8.
Population genetics tools are essential for understanding evolutionary mechanisms12. Neutral genetic differentiation (measured, for example, by F ST index13) can indicate the level of gene flow between populations. In order to determine the evolutionary mechanism, genetic differentiation of quantitative traits (Q ST) is compared to the neutral genetic differentiation (Q ST-F ST comparison). Together, they point to the main evolutionary scenario: directional selection (Q ST > F ST), stabilizing selection (Q ST < F ST), or genetic drift (Q ST = F ST)14. Q ST is difficult to measure in wild populations, since it requires rearing of animals in common garden conditions; therefore, the phenotypic divergence (P ST) is commonly used as a proxy of Q ST 15–17.
Biological invasions threaten biodiversity and cause major economic losses worldwide18,19. Invasive species nevertheless provide an opportunity for studying evolutionary and ecological processes in real time20. Successful invaders must quickly adapt to novel environments and expand their range beyond the introduction site, eventually leading to phenotypic differentiation between populations21. For example, reduced levels of neophobia (the tendency to avoid novel objects or foods22) and increased exploratory behavior (searching behavior without immediate requirement23) may have an advantage in successful introductions24, and are therefore expected to characterize introduced populations25,26. However, it is not entirely understood how populations acquire such novel adjustments27. Reduction in genetic diversity is expected due to bottlenecks28,29, which may hinder adaptive evolution30. Furthermore, genetic evolution probably requires substantial time to operate31 (but see ref.32). Phenotypic plasticity, the ability of a genotype to express different phenotypes as a response to the environment, may therefore play an important role in the rapid habituation of introduced populations33.
The house sparrow (Passer domesticus) is one of the world’s most common bird species. Due to their commensal nature, and following numerous successful human-mediated introductions, house sparrows are abundant in almost every human-populated habitat34. Many well-documented introduction events exist around the world34–36. The exceptional success of the house sparrow as an invader, its great abundance, and the extensive knowledge on its biology and distribution, have made it a popular model in invasion biology34,37. One of the classic textbook study cases of rapid evolution in invasive populations was documented in house sparrows in North America38,39. A distinct pattern of variation in body size and color occurred within a time span of no longer than a century, corresponding to Bergmann’s and Gloger’s rules.
The house sparrow is native to Israel and abundant throughout the country. The local subspecies, P. d. biblicus, belongs to the Palearctic group36. However, in the southernmost part of Israel (Eilat area) sparrows appear to be smaller and brighter compared to other populations40,41. Thus, it was suggested that the southern populations may have been influenced by an Oriental subspecies, P. d. Indicus, introduced by ships via the Red Sea (which became possible from the 1950s, when the port of Eilat was established). At the same time, due to a steep climatic gradient in Israel from north to south42, a similar pattern of variation in both size and color is expected, in accordance with Bergmann’s and Gloger’s rules. Mean annual rainfall ranges between 25 mm in the south and 1,000 mm in the north, while mean annual temperature varies from 16 °C to 25 °C, respectively.
In contrast to other introductions of house sparrows, the assumed invasion in Israel is based on observational data alone and has not been quantitatively tested. In this study, we examined whether variation in morphology and color agrees with the expectations of the two mentioned biogeographic rules, and whether differentiation is greater between the southernmost region (Eilat) and other regions, in support of the invasion scenario. We measured behavioral variation between regions, since lower levels of neophobia and increased exploration are expected in the southernmost region, due to the potential advantage for invasive populations. We examined whether genetic structure occurs between populations, and searched for additional genetic “signatures” of invasion, such as reduced diversity. Finally, we examined whether the neutral genetic differentiation between populations, F ST, is significantly smaller than the differentiation of phenotypic traits, P ST, indicating local adaptation.
Results
Morphological Traits
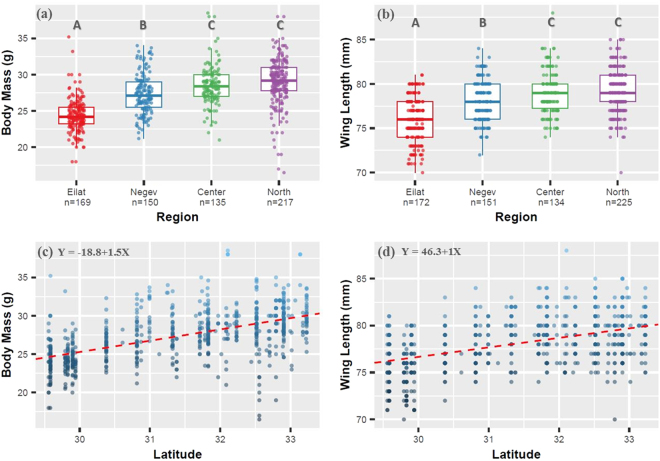
Five morphological traits were measured for sparrows from 18 populations and from museum specimens (N = 692; 398 males, 294 females; All mean values for each trait by region are listed in Supplementary Table S1). In one-way ANOVAs, four out of five traits differed among the four regions (from south to north: Eilat, Negev, Center, North) after Bonferroni correction for multiple comparisons: Body mass: F 3,667 = 98, P < 0.001; Wing length: F 3,678 = 65.5, P < 0.001 (Fig. 1a,b); Tarsus length: F 3,423 = 4.98, P < 0.05; Bill length: F 3,661 = 24.73, P < 0.001. Variation in bill width did not differ among regions (F 3,424 = 2.21, P = 0.43). In pairwise post-hoc Tukey tests we found a strong indication for morphological divergence (in all traits but tarsus length) between the region of Eilat and the more northern regions. Individuals from the area of Eilat were morphologically similar to the Oriental subspecies (P. d. indicus) in body size measurements36,43,44.
Figure 1.
(a,b) Results of one-way ANOVA for two morphological traits, body mass and wing length. Heavy line within each box represents the sample median. Lower and upper limits of each box represent the 25% and 75% quartiles, respectively. Limits of vertical lines (whiskers) represent the min and max values, excluding outliers. Letters above boxes (A, B, C) represent significant differences in post-hoc analyses. Eilat (A) significantly differs from both the Negev (B) and Center, North (C), while the Center does not differ from the North. (c,d) Results of regression analyses of two morphological traits, body mass and wing length, plotted against latitude.
We examined the contribution of latitude and climate features to the variation in morphology and color among populations. Precipitation and temperature PCAs were highly correlated with latitude (r = −0.93 for precipitation, r = 0.73 for temperature); therefore, in order to avoid the effects of multicollinearity, they were removed from following analyses. In a regression analysis with latitude as a predictor, all five traits showed a significant spatial pattern of becoming larger towards northern latitudes: Body Mass: F 1,669 = 282.5, r 2 = 0.296, P < 0.001; Wing length: F 2,679 = 95.8, r 2 = 0.218, P < 0.001 (Fig. 1c,d); Tarsus length: F 1,425 = 13.9, r 2 = 0.029, P < 0.001; Bill Length: F 1,663 = 58.05, r 2 = 0.079, P < 0.001; Bill width: F 1,426 = 5.4, r 2 = 0.02, P = 0.01. For traits which were sexually dimorphic (wing length and bill width), we conducted separate ANOVA and regression tests (results are given in Supplementary Table S2).
Color Brightness
Color brightness was analyzed for 380 males using a spectrophotometer on three parts of the body (belly, right and left cheeks). Mean values by region are listed in Supplementary Table S1.
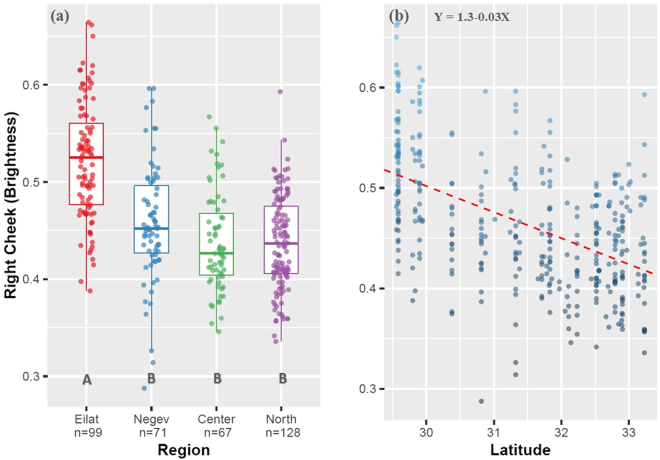
All three color measurements differed between regions: right cheek brightness (F 3,361 = 53.1, P < 0.001; Fig. 2a), left cheek (F 3,361 = 51.6, P < 0.001), and belly (Welch’s ANOVA: F 3,186 = 44.9, P < 0.001; Kruskal-Wallis χ2: 101.1, P < 0.001). In post-hoc analyses, the only significant difference in right cheek was between Eilat and the other three regions (P < 0.001). Left cheek differed between Eilat and the other regions (P < 0.001), and Center differed from the Negev region (P < 0.01). Belly brightness differed between Eilat and the North and Center (P < 0.001), but not from the Negev (P = 0.88), while the Negev differed from both the North and Center regions (P < 0.001). All three traits showed a significant spatial pattern of becoming darker towards the northern latitudes: right cheek (F 1,363 = 130.9, r 2 = 0.263, P < 0.001; Fig. 2b), left cheek (F 1,363 = 99.9, r 2 = 0.214, P < 0.001), belly brightness (F 1,365 = 135.2, r 2 = 0.268, P < 0.001).
Figure 2.
(a) Results of one-way ANOVA for right cheek brightness. Heavy line within each box represents the median. Limits of boxes represent the 25% and 75% quartiles, and limits of whiskers represent min and max values excluding outliers. Letters below boxes (A, B) represent significant differences in post-hoc analyses. Eilat (A) significantly differs from all other three regions, which are not different from one another (b) Results of regression analysis of right cheek brightness plotted against latitude.
Behavioral Assay
Behavioral assays of 81 male sparrows were scored for differences in exploratory behavior and neophobia between regions. Measures of exploratory behavior greatly varied among individuals (mean of hops ± SD: 44.6 ± 62.6; flight incidents: 8.7 ± 14.7; location changes: 13.7 ± 22.3; quarters visited: 2.1 ± 1.3; perches visited: 1.4 ± 1.8); however, we did not find significant variation among regions (χ2 = 0.69, df = 3, P = 0.87). Neophobia levels varied among regions (χ2 = 8.59, df = 3, P = 0.04; Fig. 3). Post-hoc analysis showed that the only significant pairwise difference was between Eilat and the North (P = 0.02). Sparrows in Eilat were less neophobic than those in the North.
Figure 3.
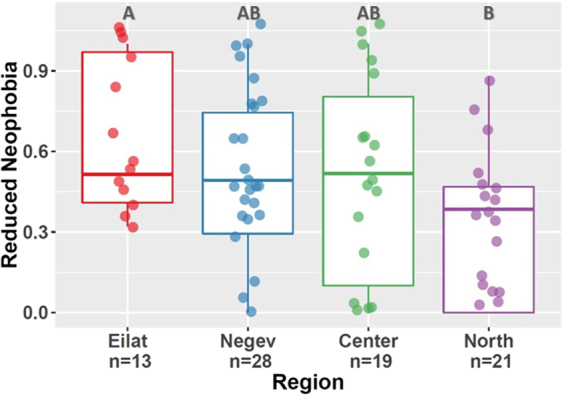
Variation in attributes of neophobia between regions. Higher score represents reduced neophobic response. Significant difference found in post-hoc Dunn’s Test between the region of Eilat and the North (represented by letters above boxes). Eilat (A) significantly differs from the North (B), but not from the Negev and Center (AB).
Genetic Analysis
We genotyped 267 sparrows (152 males, 115 females) from 14 populations, for 8 microsatellite loci. Overall, measures of genetic diversity did not differ among populations (Table 1). Expected heterozygosity (H E) was very similar in all 14 populations (0.74 – 0.79). Estimates of allelic (A R) and private allelic richness (A PR) were not lower in populations from the Eilat region than in all other populations. F IS estimates were mostly close to zero, and did not differ among populations (95% CIs comparisons). Heterozygosity excess as an indication of recent bottlenecks was non-significant in all populations but one (Hulda).
Table 1.
Summary statistics of genetic diversity attributes.
| Population | N | H E | F IS | A R | A PR | Heterozygosity excess |
|---|---|---|---|---|---|---|
| Eilot | 19 | 0.77 | 0.064 | 7.16 | 0.14 | NS |
| Elifaz Samar | 14 | 0.76 | 0.013 | 6.93 | 0.13 | NS |
| Yotvata Grofit | 28 | 0.79 | 0.088 | 7.21 | 0.21 | NS |
| Faran | 20 | 0.77 | 0.016 | 7.78 | 0.42 | NS |
| Idan | 19 | 0.77 | 0.113 | 7.09 | 0.08 | NS |
| Tlalim | 13 | 0.79 | 0.070 | 7.5 | 0.07 | NS |
| Gilat | 20 | 0.78 | 0.005 | 7.55 | 0.14 | NS |
| Avigdor | 15 | 0.77 | 0.048 | 7.09 | 0.02 | NS |
| Maale Hahamisha Ramat Rachel | 10 | 0.74 | −0.013 | 6.88 | 0.14 | NS |
| Hulda | 20 | 0.79 | 0.090 | 7.43 | 0.24 | 0.02 |
| Gaash | 19 | 0.78 | 0.006 | 7.2 | 0.12 | NS |
| Beit Alfa | 20 | 0.74 | 0.001 | 7.39 | 0.09 | NS |
| Ein Hamifratz | 20 | 0.76 | −0.004 | 7.65 | 0.35 | NS |
| Dafna Amir | 25 | 0.78 | 0.016 | 7.87 | 0.35 | NS |
N = individuals per population; H E = expected heterozygosity; F IS = inbreeding coefficient, high values indicate of high level of inbreeding; A R, A PR = allelic richness and private allelic richness, measured with HP-rare; Heterozygosity excess = results of 1-tail Wilcoxon rank test. Non-significant (NS), or the significant P-value for the population, measured with BOTTLENECK.
AMOVA revealed that most of the genetic variation (99.16%) was explained within populations, corresponding to a low global genetic differentiation over loci (F ST = 0.0084). Sex-specific genetic differentiation was similar for females (F ST = 0.012) and males (F ST = 0.010). Clustering results also revealed a lack of genetic structure, suggesting K = 1 as the most likely cluster (out of 14). Although global tests revealed an absence of genetic structure, pairwise comparisons between populations gave some significant F ST values, specifically between the southernmost population and the central and northern populations (Table 2). In support of these results, we also found a correlation between geographic distance and genetic differentiation among populations in a Mantel test (Z = 145.3, r = 0.37, P < 0.01), revealing a pattern of isolation by distance (IBD).
Table 2.
Pairwise geographic and genetic distances between 14 populations.
| 1 Eilot | 2 Elifaz Samar | 3 Yotvata Grofit | 4 Faran | 5 Idan | 6 Tlalim | 7 Gilat | 8 Avigdor | 9 Maale Hahamisha Ramat Rachel | 10 Hulda | 11 Gaash | 12 Beit Alfa | 13 Ein Hamifratz | 14 Dafna Amir | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | — | 23.7 | 36.3 | 89.9 | 139.8 | 157.3 | 195.1 | 236.6 | 248.3 | 249.1 | 294.0 | 328.3 | 368.4 | 409.5 |
| 2 | 0.0115 | — | 12.7 | 66.2 | 116.1 | 135.0 | 173.1 | 214.0 | 224.9 | 226.1 | 271.1 | 304.7 | 345.1 | 385.8 |
| 3 | −0.0005 | 0.0015 | — | 53.7 | 103.5 | 124.3 | 162.6 | 202.9 | 212.9 | 214.6 | 259.6 | 292.4 | 333.2 | 373.4 |
| 4 | 0.0007 | 0.0032 | 0.0052 | — | 49.9 | 77.2 | 115.2 | 152.7 | 159.9 | 163.0 | 208.0 | 238.7 | 280.2 | 319.7 |
| 5 | −0.0029 | 0.0065 | −0.0028 | 0.0075 | — | 52.1 | 82.2 | 111.5 | 112.6 | 118.7 | 163.0 | 189.4 | 232.4 | 270.1 |
| 6 | 0.0066 | −0.0046 | 0.0008 | 0.0104 | 0.0099 | — | 38.4 | 79.5 | 97.2 | 93.3 | 137.5 | 180.1 | 214.1 | 261.3 |
| 7 | 0.0188 | 0.0019 | 0.0054 | 0.0069 | 0.0092 | 0.0042 | — | 43.8 | 70.4 | 60.3 | 102.2 | 151.7 | 180.4 | 231.2 |
| 8 | 0.0103 | 0.0097 | 0.0055 | 0.0034 | 0.0038 | 0.0045 | −0.0021 | — | 36.9 | 18.6 | 58.5 | 110.6 | 136.6 | 188.6 |
| 9 | 0.0151 | 0.0150 | 0.0207 | 0.0023 | 0.0061 | 0.0272 | 0.0169 | 0.0044 | — | 21.8 | 53.2 | 83.0 | 120.3 | 164.1 |
| 10 | 0.0117 | 0.0022 | 0.0049 | 0.0083 | 0.0039 | 0.0001 | −0.0001 | 0.0014 | 0.0044 | — | 45.0 | 92.2 | 120.9 | 171.0 |
| 11 | 0.0269 | 0.0026 | 0.0124 | 0.0154 | 0.0068 | 0.0118 | −0.0027 | 0.0069 | 0.0238 | 0.0064 | — | 65.2 | 78.8 | 134.5 |
| 12 | 0.0283 | 0.0071 | 0.0164 | 0.0041 | 0.0150 | 0.0212 | 0.0015 | 0.0041 | 0.0199 | 0.0106 | 0.0069 | — | 53.1 | 81.5 |
| 13 | 0.0248 | −0.0043 | 0.0124 | 0.0094 | 0.0099 | 0.0122 | −0.0008 | 0.0093 | 0.0159 | 0.0102 | 0.0026 | −0.0008 | — | 62.4 |
| 14 | 0.0245 | 0.0069 | 0.0121 | 0.0066 | 0.0128 | 0.0066 | −0.0056 | 0.0031 | 0.0292 | 0.0069 | 0.0034 | −0.0002 | −0.0004 | — |
Pairwise geographic distances between 14 populations are presented above the diagonal and genetic differentiation values (F ST) appear below. A total of 18 significant F ST values are in bold. Nine out of 18 of the significant values are between populations from the region of Eilat (Eilot, Elifaz-Samar, Yotvata-Grofit) and populations from the Center and North regions.
Comparison of Phenotypic and Genetic Differentiation (PST - FST)
Lower 95% CI values of phenotypic differentiation among populations, P ST, were compared to the upper 95% CI of neutral genetic differentiation17 (0.016 for males and females; 0.024 for males only). P ST surpassed the global F ST value for six out of eight phenotypic traits (Table 3). The only morphological traits for which P ST was not larger than F ST were bill width, which also did not show significant variation between regions, and left cheek brightness. Critical values of c/h 2 were therefore estimated for all traits but bill width and left cheek brightness, with lower values indicating greater robustness for the comparison between phenotypic and genetic differentiation17. Tarsus length, followed by body mass, had the smallest critical c/h 2 and largest P ST value, while right cheek brightness had the largest critical c/h 2 value.
Table 3.
Summary of comparisons between phenotypic and genetic differentiation (P ST-F ST) among house sparrow populations in Israel.
| Trait | P ST | Lower 95% CI | Upper 95% CI | Critical c/h 2 | P ST-F ST |
|---|---|---|---|---|---|
| Body mass | 0.313 | 0.176 | 0.539 | 0.079 | P ST ≫ F ST |
| Wing length | 0.253 | 0.109 | 0.445 | 0.131 | P ST > F ST |
| Tarsus length | 0.443 | 0.292 | 0.680 | 0.046 | P ST ≫ F ST |
| Bill length | 0.130 | 0.054 | 0.296 | 0.171 | P ST > F ST |
| Bill width | 0.000 | 0.000 | 0.118 | — | P ST = F ST |
| Right cheek brightness | 0.113 | 0.025 | 0.288 | 0.959 | P ST > F ST |
| Left cheek brightness | 0.130 | 0.009 | 0.352 | — | P ST = F ST |
| Belly brightness | 0.379 | 0.204 | 0.662 | 0.124 | P ST ≫ F ST |
P ST value estimated for c = h 2. Critical c = h 2 calculated only for traits whose upper F ST CI > lower P ST CI. Upper F ST CI value used for the five morphological traits was 0.016 (males and females), and 0.024 for color brightness trait (male-only F ST).
Discussion
We examined patterns of phenotypic variation in morphology, plumage coloration, and behavior in house sparrows and compared them to patterns of neutral genetic variation. Our two main motivations were: first, to understand the evolutionary processes driving the phenotypic variation between populations; and second, to investigate the hypothesis that introduction of sparrows had occurred in the southernmost part of our research area.
We found evidence for spatial patterns of variation in morphology and color, which correspond to a climatic gradient and also support the possibility of introduction. Furthermore, inter-population behavioral differences present a pattern fitting an invasion scenario. Genetic differentiation and diversity analyses revealed that gene flow occurs between populations. Comparison of the phenotypic variation to genetic differentiation indicated that the variation cannot be explained by genetic drift alone. Overall, the results indicate that the observed variation between phenotypic traits derives from a process of local adaptation or phenotypic plasticity, and is in congruence with studies of house sparrow populations in both native and introduced ranges worldwide45–47.
Variation in all morphological measures coincides with the prediction of Bergmann’s rule, as sparrows become larger towards the northern localities. Although evidence for the validity of Bergmann’s rule across species and its adaptive advantage are equivocal, it is considered one of the central biogeographic principles. Thus, our results indicate a possible selective pressure leading to local adaptation. Additionally, four out of five measurements revealed significant differentiation among regions, with emphasized divergence between the southernmost populations and the other, more northern, regions. This provides empirical support for the assumption that foreign populations of a different, smaller subspecies (i.e, P. d. indicus), may occur in the Eilat region.
Interestingly, out of the five morphological traits, bill width and tarsus length demonstrated the weakest pattern of variation in relation to latitude and between regions. Both are skeletal measurements, and thus are less prone to environmental changes (however they may be affected by ontogenetic plasticity). In contrast, body mass, wing length, and bill length of house sparrows have been shown to respond to seasonal changes48,49, fluctuate between breeding and non-breeding seasons50, and body mass can vary on a diurnal basis46. This pattern may be indicative of a substantial plastic effect in shaping the morphological variation.
An increase in coloration along the north-south gradient was also evident, fitting the expectations of Gloger’s rule. Presuming an adaptive advantage for Gloger’s rule, the results indicate an adaptive basis for variation in plumage coloration of house sparrows. Complementary to variation in body size, differentiation between regions in color was also most significant between Eilat and the rest of the localities, with sparrows from Eilat being brighter, resembling the Oriental subspecies36.
Variation in exploratory behavior was evident among individuals, but we could not detect significant differences among regions. Conversely, significant but weak differences were detected in neophobic response between Eilat and the North region. Individuals from Eilat were less averse to novelty (visited decorated perches more) and spent more time in the center of the arena, as opposed to individuals from the North. Assuming that foreign populations occur in the southernmost region, this finding agrees with the hypothesis that assumes an advantage to being less neophobic in a novel habitat. The lack of difference in exploratory behavior among regions could perhaps be explained by the “adaptive flexibility hypothesis”25, according to which proxies of behavioral flexibility are expected only in the initial stage of introduction. Furthermore, exploratory behavior and reduced neophobia may induce costs, such as the risk of being revealed to predators or ingesting poisonous foods22. Consequently, cautiousness is likely to be favored once populations have become established.
Genetic differentiation between populations was low, demonstrating a lack of structure, indicating high gene flow among populations. We did find a pattern of isolation by distance, as a substantial portion of the significant pairwise differentiation values were between the southernmost populations, and some of the populations in the north and center. This agrees with the invasion scenario, as populations influenced by foreign immigrants are expected to genetically differ from their native counterparts. However, due to strong commensalism and the extremely sedentary nature of sparrows41,51, dispersal could be linked to human settlements, which are sparser in the southern, arid, area of Israel compared to the rest of the country. This alone could be a cause for some of the divergence found, independent of the introduction scenario. In contrast to the invasion scenario expectation, we found no evidence of a recent bottleneck52. However, this may be due to an admixture with the local subspecies53 or by a quick recovery from the bottleneck54. Altogether, inferring whether an introduction event occurred based on genetic diversity alone is at best partial, although it can provide additional support for more concrete data.
Most of the variation in phenotypic traits was greater than the neutral genetic differentiation (P ST > F ST), suggesting that phenotypic variation cannot be explained by genetic drift alone, but at least partially derives from natural selection. However, since the validity of the approximation of Q ST by P ST has been largely criticized55, the results should be interpreted cautiously. Contrary to Q ST, which measures additive genetic differentiation under common garden conditions56, P ST is calculated from phenotypic data alone, and thus it cannot distinguish between the contribution of plasticity and genetic evolution to the observed variation.
Our analyses of the genetic differentiation are based on a dataset of eight microsatellite loci. Although many studies have used comparable number of markers (6–14 microsatellite) to examine genetic differentiation and for F ST - P ST comparisons45–47,54,57–59, more markers (SNPs) may provide a better estimate for the genetic differentiation60. Increasing the number of markers would probably decrease the confidence limit, but will have lesser effect on the estimate. In support, a panel of 6736 variable SNPs and a panel of 14 microsatellites produced very similar F ST values in a study of house sparrow populations in Norway. However, the 95% confidence limit intervals for the microsatellite markers were wider61. That said, increasing the number of markers used may generate more reliable estimates of the genetic differentiation and effect our estimation of the level of adaptive evolution.
Since most of the documented phenotypic traits are plausibly affected by environmental conditions, some variation may be due to phenotypic plasticity. On the other hand, it is safe to assume that a genetic basis does exist for these morphological and color traits, as most of the discussed traits are heritable in house sparrows and other species62–64. Tarsus length, which is the least likely to be affected by phenotypic plasticity, had the smallest c/h 2 value, indicating that robustness of the P ST - F ST comparison was strongest for this trait.
Studying trait variation in a common species, such as the house sparrow, provides an excellent opportunity to investigate evolutionary and ecological questions. Our findings show that phenotypic variation over a climatic and latitudinal gradient agrees with recognized biogeographic rules, while also fitting the scenario of a possible introduction of foreign populations into the studied area. In order to further investigate the possibility of invasion, it may be informative to obtain genetic data from the original range of the subspecies in question, P. d. indicus. Comparing the low neutral genetic differentiation to the phenotypic variation indicates that random evolution by genetic drift is probably not the prime cause of the observed variation. Further investigation, which may include rearing of individuals from distant populations in common garden conditions, should determine whether variation has been shaped by a process of selection or by phenotypic plasticity. House sparrows have been experiencing severe population declines in vast parts of the world in recent decades65,66. Thus, expanding our current knowledge of its evolution and ecology may prove to be important for future conservation efforts for this ubiquitous and endearing bird.
Materials and Methods
Capture Sites and Sampling Procedures
A total of 486 adult house sparrows (260 males, 226 females) were caught using mist nets in 18 localities during 4/2013, and between 11/2014 and 6/2015, from north to south of Israel (~420 km). All capture sites were located near livestock farms in suburban and rural settlements (Table 4, Fig. 4). All birds were released back at the capture site after sampling. Following capture, we measured body mass with a digital scale (0.01 g accuracy), wing length with a ruler (1 mm), tarsus length, bill length and width with a digital caliper (0.01 mm). A small blood sample (20-50 µl) was collected from the brachial vein and stored in a 1.5 ml tube containing blood lysis buffer67 (0.1 M Tris pH 8; 0.1 M Ethylenediaminetetraacetic acid (EDTA); 0.01 M NaCl; 0.5% sodium dodecyl sulphate (SDS)). All males were measured using a spectrophotometer (i1Pro X-rite, Grand Rapids, Michigan; aperture of 4.5 mm diameter) for analysis of color brightness, on three parts of the body: belly (bright colored feathers under the bib), and right and left cheeks (white/grey colored area beneath the eye). Color data were obtained for males only, since plumage variation is expected to be more evident in males. GretagMacbeth’s Eye-One Share v1.4 software was used to translate the data to a 380–730 nm spectrum table (10 nm intervals) and converted to RGB values using a spectral calculator spreadsheet (© 2001–2016 Bruce Justin Lindbloom, http://www.brucelindbloom.com). We analyzed variation in one component of RGB (green), indicative of the relative brightness of the sample, transformed from a 0–255 scale to a scale of 0–1, where 0 = black (dark), 1 = white (bright). We also measured morphology and color traits in specimens collected over the past 80 years in Israel and vouchered at the Steinhardt Museum of Natural History, Tel Aviv University (SMNHTAU; 138 males and 68 females). Museum specimens were not included in the genetic analysis.
Table 4.
List of localities, coordinates and dates of sampling.
| Location | Region | Type | Date | Latitude (N) | Longitude (E) | Altitude (m) | |
|---|---|---|---|---|---|---|---|
| 1 | Eilot | Eilat | Petting zoo | 3/3/2015 | 29.5835 | 34.9604 | 89 |
| 2 | Elifaz | Eilat | Dairy farm | 17/4/2013 | 29.7928 | 35.0106 | 121 |
| 3 | Samar | Eilat | Dairy farm | 16/4/2013 | 29.8294 | 35.0238 | 92 |
| 4 | Yotvata | Eilat | Dairy farm | 2/3/2015 | 29.8995 | 35.0596 | 86 |
| 5 | Grofit | Eilat | Dairy farm | 15/4/2013 | 29.9393 | 35.0635 | 141 |
| 6 | Faran | Negev | Dairy farm | 4/3/2015 | 30.3776 | 35.1503 | 94 |
| 7 | Idan | Negev | Dairy farm | 22/2/2015 | 30.8140 | 35.2768 | −174 |
| 8 | Tlalim | Negev | Dairy farm | 23/2/2015 | 30.9929 | 34.7735 | 364 |
| 9 | Gilat | Negev | Dairy farm | 15/6/2015 | 31.3225 | 34.6507 | 138 |
| 10 | Avigdor | Center | Dairy farm | 9/11/2014 | 31.7094 | 34.7439 | 65 |
| 11 | Ramat Rachel | Center | Petting zoo | 26/3/2015 | 31.7410 | 35.2165 | 809 |
| 12 | Ma’ale Hahamisha | Center | Dairy farm | 28/12/2014 | 31.8197 | 35.1115 | 805 |
| 13 | Hulda | Center | Horse stable | 28/1/2015 | 31.8293 | 34.8818 | 121 |
| 14 | Gaash | Center | Petting zoo | 2/1/2015 | 32.2323 | 34.8252 | 23 |
| 15 | Beit Alfa | North | Dairy farm | 5/4/2015 | 32.5171 | 35.4315 | −85 |
| 16 | Ein Hamifratz | North | Dairy farm | 11/1/2015 | 32.9040 | 35.0972 | 6 |
| 17 | Amir | North | Dairy farm | 6/4/2015 | 33.1770 | 35.6236 | 76 |
| 18 | Dafna | North | Dairy farm | 6/4/2015 | 33.2299 | 35.6421 | 141 |
Locality numbers correspond to the numbers presented on the map in Fig. 4.
Figure 4.
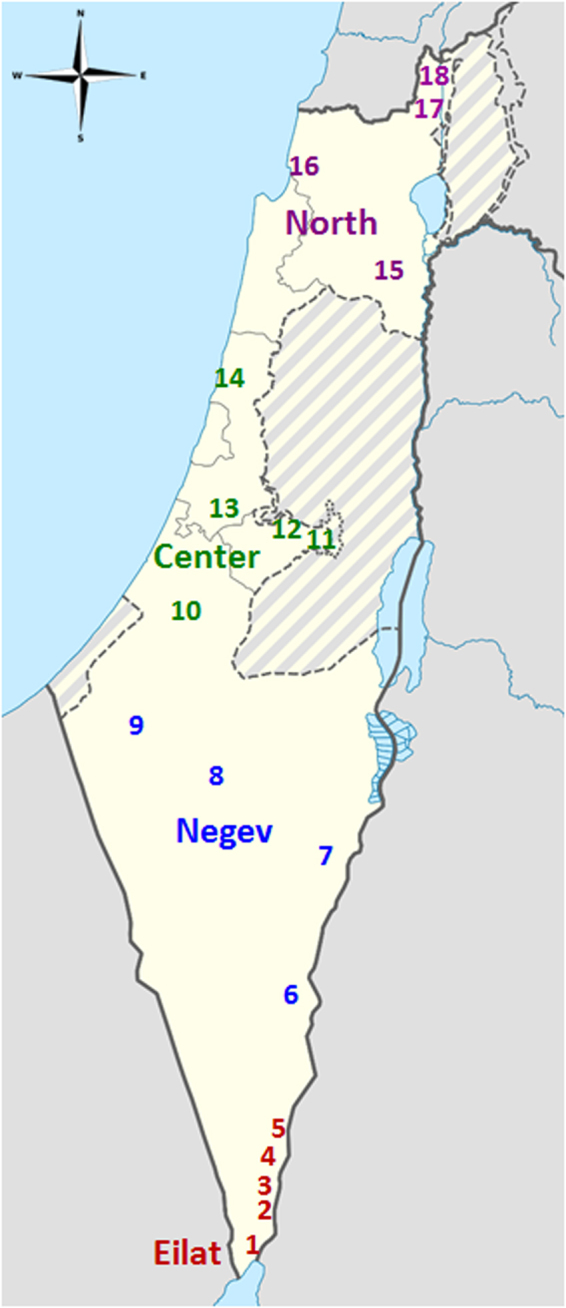
Map of sampling localities across Israel.
Behavioral Assay
A total of 94 males (2–9 at each locality, mean ± SD: 6.6 ± 2.2) were randomly selected for a novel environment test during 2014–2015. In order to reduce stress caused by handling68, measurements and blood samples were taken after the assay was completed for each bird. Variation in exploratory behavior is generally assessed by observing animals in an artificial arena, while neophobia is measured by the response to novel items or foods22. The novel environment setting and assay we used is based on a classic “open field” test, adapted from previous protocols69–71. The arena was located in a standard camping tent (3.0 m × 2.40 m, 1.90 m height). The tent floor was divided into four quadrats and six artificial wooden perches were placed inside (1.0 m height, 50 cm “branch”). In order to stimulate a novelty response, three perches were decorated with “novel” objects, presumed foreign to the natural habitat of the sparrows (see Supplementary Figure S4 for pictures of the tent and the novel items). In order to avoid possible behavioral variation caused by social context72, sparrows were released into the tent separately, and left to freely explore the arena for approximately 15 minutes, filmed throughout with a GoPro Hero3 + camera. Videos were later analyzed, blind to the capture localities. Out of the 94 males we were able to analyze only 81 due to technical problems (seven assays were removed since no camera recording was available; six others were removed due to strong wind conditions).
Exploratory behavior was measured according to the following variables, summed to a final score: (1) Proportion of the tent explored – number of quarters visited (0 to 4), and number of perches used (from 0 to 6); (2) Location changes (each movement between quarters/ from quarter to perch/ from perch to perch); (3) Number of hops; (4) Number of flight incidents.
Each variable received a score, normalized for the duration of each assay, and relative to the best result. For example, if a bird visited 6 perches during 10 minutes received the best result for this variable ( = 1), then a bird that visited only 3 perches during 20 minutes would receive a score of 0.25 out of 1. A separate score was given for neophobic behavior, measured according to the proportion of decorated perches visited (out of total time perched), and the proportion of time spent in the center of the arena as opposed to its corners.
Molecular Lab Procedures and Genotyping
Genomic DNA was extracted from blood samples of 267 sparrows captured in the field (152 males, 115 females; up to 20 representatives from each locality), according to the protocol of QIAGEN DNeasy blood & Tissue kit. DNA was amplified by polymerase chain reaction (PCR) for ten nuclear microsatellite markers: Pdo173, Pdo574, Pdo875, CAM01, CAM02, CAM05, CAM10, CAM15, CAM17, CAM2076. Forward primers were labeled with either 6-FAM, ROX, VIC or NED fluorescent dyes and PCR reactions were performed separately for each marker. PCR Products were combined into three multiplex mixes for genotyping (see Supplementary Table S3), and allele size scoring of the results was performed with GeneMarker v2.6.7 (SoftGenetics, LLC), verified and amended by eye.
Analysis of genetic data
Occurrences of linkage disequilibrium (LD) and deviation from Hardy-Weinberg equilibrium (HWE) were tested for each population, using Arlequin v3.5.2.277, as well as the presence of null alleles (checked with Cervus v3.0.778). Markers Pdo8 and CAM20 were excluded from the analysis due to significant deviation from HWE and high null allele percentage (37.6% for Pdo8, 20% for CAM20). Alleles per locus ranged from 4 to 28 for the eight remaining markers. Since sample sizes in some localities were too small to obtain meaningful results, they were merged with the closest geographic locality (between 4 to 12 km distance between two merged populations), forming a total of 14 genetic populations. Genetic structure of populations was evaluated using two methods: first, analysis of molecular variance, AMOVA79, implemented in Arlequin. This is a hierarchical analysis that partitions total variance into covariance components due to intra and inter-individual differences and inter-population differences. Populations were allocated into pre-defined groups (Eilat, Negev, Center, North). The second method used was Bayesian clustering implemented in STRUCTURE v2.3.480. STRUCTURE assigns samples into clusters (populations) using an admixture model, assuming correlation of allele frequencies, without prior knowledge of sample locality. We ran the clustering with a 20,000 burn-in period followed by 50,000 MCMC iterations for possible K = 1–14 populations, 10 times for each run (K). In order to infer which K was most likely, we ran the results in Structure Harvester81.
Genetic differentiation between populations was assessed by the F ST index13, estimated using Weir & Cockerham’s θ 82, and implemented in Arlequin. Pairwise F ST values were used to assess whether a pattern of isolation-by-distance (IBD) exists between populations, using the Isolation by Distance Web Service v3.2383. This application implements Mantel Test84 with 10,000 randomizations, for matrix correlation between genetic distance (F ST values) and geographic distance (in km, determined using Coordinate Distance Calculator; http://www.boulter.com/gps/distance).
Levels of genetic diversity within populations were estimated by allelic richness (AR) and private allelic richness (APR) in HP-Rare v1.185, compensating for differences in sample size. Variation in expected heterozygosity (H E) and F IS index86 were measured with the R package DiveRsity87. Detection of possible recent bottlenecks was achieved by measuring levels of heterozygosity excess, with BOTTLENECK v1.2.0288. We ran the software for Wilcoxon sign-rank test and mode-shift indicator89 with 1,000 replications under the two-phase model (T.P.M).
Statistical Analysis
All statistical analyses, unless noted otherwise, were performed in R v3.1.2 (R Development Core Team 2014).
Variation of phenotypic traits between regions
The sampled populations were separated into four regions (Eilat, Negev, Center, North). Variation in morphology, color, and behavior among regions was estimated for each trait separately using Analysis of Variance (ANOVA) and post-hoc Tukey tests. For traits that deviated from assumptions of normality and homoscedasticity we used two alternatives, Welch’s ANOVA, and a Kruskal-Wallis test, followed by Dunn’s post-hoc test.
Spatial morphological trends
We used linear regression analyses in order to examine the factors contributing to the variation in morphology and color (separately for each trait) with latitude and climate as predictor variables. For traits that deviated from the model’s normality assumption we applied square-root transformations.
Exact GPS coordinates (latitude, longitude and altitude) were obtained for all localities and approximated for museum specimens using GPS-coordinates website (© 2016 http://www.gps-coordinates.net). Arc GIS v10.3 (Esri Inc.) was used to produce climatic layers for all coordinates, extracted from the WorldClim90 database (www.worldclim.org). We ran two separate PCAs for temperature and precipitation (Temperature PC1 and Precipitation PC1, explaining 79% and 99.7% of the variation, respectively).
Phenotypic variation vs. genetic differentiation
Divergence of phenotypic traits between populations, P ST, was compared to the neutral genetic differentiation, F ST. Global F ST values and their 95% confidence intervals for males + females and for each sex separately were estimated with Arlequin.
| 1 |
The value of P ST, defined in eq. 1, was estimated for each morphological and coloration trait assuming that c = h 2, representing the ratio between variation caused by additive genetic effects (c), and heritability (h 2). Phenotypic variance between (σ2 B) and within (σ2 W) populations was estimated using the MCMCglmm package in R91. For this analysis we used a subset of the color and morphology dataset, including only individuals for which we had genetic data as well. We constructed linear mixed models for all phenotypic traits, using population (locality) as a random effect and sex as a fixed effect for morphological traits, and year of sampling as a fixed effect for color traits. The default priors of the R package were used (65,000 bootstrap iterations, 15,000 burn-in period, 50 = sampling interval). To account for the robustness of the comparison, for each trait where P ST exceeded F ST we calculated the critical c/h 2 value, for which the upper 95% CI of F ST equals the lower 95% CI of P ST 17.
Ethical Statement
All methods were carried out in accordance with the guidelines and regulations of the Israeli animal welfare law and the Israeli wildlife protection law. All capture and handling of birds, as well as all experiment protocols were approved and performed under permit (#L-14-062) granted by the Veterinary Service Center at the Sackler Faculty of Medicine, Tel Aviv University, according to the Israeli animal welfare law. The house sparrow is classified as a pest species under the Israeli wildlife protection law; therefore its capture for research purposes does not require an additional permit.
Data Availability
The datasets generated and analyzed during the current study are available from the corresponding author upon reasonable request.
Electronic supplementary material
Acknowledgements
We would like to thank lab members T. Magory Cohen and I. Goldshtein for their assistance during the field work and molecular lab work, and H. Friedman and D. Turjeman for their assistance in field work. We thank Y. Vortman for assistance with color analysis procedures, and I. Scharf for helpful comments on the manuscript. We thank the Steinhardt Museum of Natural History at Tel Aviv University for permission to use specimens from their collection for this research. Finally, we wish to thank all the farmers for allowing us to work on their properties.
Author Contributions
S.B.C. and R.D. both contributed to performing the research, analyzing the data, and writing the manuscript.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-017-18718-8.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Mayr, E. Animal species and evolution. (Harvard University Press, 1963).
- 2.Price TD, Qvarnstrom A, Irwin DE. The role of phenotypic plasticity in driving genetic evolution. Proc. R. Soc. Lond. B Biol. Sci. 2003;270:1433–1440. doi: 10.1098/rspb.2003.2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Crispo E. Modifying effects of phenotypic plasticity on interactions among natural selection, adaptation and gene flow. J. Evol. Biol. 2008;21:1460–1469. doi: 10.1111/j.1420-9101.2008.01592.x. [DOI] [PubMed] [Google Scholar]
- 4.Bergmann, C. Über die Verhältnisse der wärmeokönomie der Thiere zu ihrer Grösse. Göttinger Studien 595–708 (1847).
- 5.Gloger, C. L. Das Abändern der Vögel durch Einfluss des Klima’s (Breslau, August Schulz, 1833).
- 6.Mayr E. Geographical character gradients and climatic adaptation. Evolution. 1956;10:105–108. doi: 10.1111/j.1558-5646.1956.tb02836.x. [DOI] [Google Scholar]
- 7.Blackburn TM, Gaston KJ, Loder N. Geographic gradients in body size: a clarification of Bergmann’s rule. Divers. Distrib. 1999;5:165–174. doi: 10.1046/j.1472-4642.1999.00046.x. [DOI] [Google Scholar]
- 8.Zink R, Remsen J., Jr. Evolutionary processes and patterns of geographic variation in birds. Curr. Ornithol. 1986;4:1–69. [Google Scholar]
- 9.Meiri S, Dayan T. On the validity of Bergmann’s rule. J. Biogeogr. 2003;30:331–351. doi: 10.1046/j.1365-2699.2003.00837.x. [DOI] [Google Scholar]
- 10.Miller AH, Miller L. Geographic variation of the screech owls of the deserts of western North America. The Condor. 1951;53:161–177. doi: 10.2307/1364873. [DOI] [Google Scholar]
- 11.Burtt EH, Jr, Ichida JM. Gloger’s rule, feather-degrading bacteria, and color variation among song sparrows. The Condor. 2004;106:681–686. doi: 10.1650/7383. [DOI] [Google Scholar]
- 12.Hartl, D. L. & Clark, A. G. Principles of population genetics. Vol. 116 (Sinauer associates Sunderland, 1997).
- 13.Wright S. Isolation by distance. Genetics. 1943;28:114. doi: 10.1093/genetics/28.2.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Merilä J, Crnokrak P. Comparison of genetic differentiation at marker loci and quantitative traits. J. Evol. Biol. 2001;14:892–903. doi: 10.1046/j.1420-9101.2001.00348.x. [DOI] [Google Scholar]
- 15.Spitze K. Population structure in Daphnia obtusa: quantitative genetic and allozymic variation. Genetics. 1993;135:367–374. doi: 10.1093/genetics/135.2.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Leinonen T, O’Hara RB, Cano J, Merilä J. Comparative studies of quantitative trait and neutral marker divergence: a meta‐analysis. J. Evol. Biol. 2008;21:1–17. doi: 10.1111/j.1420-9101.2007.01445.x. [DOI] [PubMed] [Google Scholar]
- 17.Brommer J. Whither PST? The approximation of QST by PST in evolutionary and conservation biology. J. Evol. Biol. 2011;24:1160–1168. doi: 10.1111/j.1420-9101.2011.02268.x. [DOI] [PubMed] [Google Scholar]
- 18.Wilcove DS, Rothstein D, Dubow J, Phillips A, Losos E. Quantifying threats to imperiled species in the United States. Bioscience. 1998;48:607–615. doi: 10.2307/1313420. [DOI] [Google Scholar]
- 19.Pimentel, D. et al. Economic and environmental threats of alien plant, animal, and microbe invasions. Agr. Ecosyst. Environ. 84, 1–20 (2001).
- 20.Sax DF, et al. Ecological and evolutionary insights from species invasions. Trends Ecol. Evol. 2007;22:465–471. doi: 10.1016/j.tree.2007.06.009. [DOI] [PubMed] [Google Scholar]
- 21.Reznick DN, Ghalambor CK. The population ecology of contemporary adaptations: what empirical studies reveal about the conditions that promote adaptive evolution. Genetica. 2001;112:183–198. doi: 10.1023/A:1013352109042. [DOI] [PubMed] [Google Scholar]
- 22.Greenberg R, Mettke-Hofmann C. Ecological aspects of neophobia and neophilia in birds. Curr. Ornithol. 2001;16:119–178. [Google Scholar]
- 23.Immelmann, K. & Beer, C. A dictionary of ethology. (Harvard University Press, 1989).
- 24.Sol D, Timmermans S, Lefebvre L. Behavioural flexibility and invasion success in birds. Anim. Behav. 2002;63:495–502. doi: 10.1006/anbe.2001.1953. [DOI] [Google Scholar]
- 25.Wright TF, Eberhard J, Hobson E, Avery ML, Russello M. Behavioral flexibility and species invasions: the adaptive flexibility hypothesis. Ethol. Ecol. Evol. 2010;22:393–404. doi: 10.1080/03949370.2010.505580. [DOI] [Google Scholar]
- 26.Martin LB, Fitzgerald L. A taste for novelty in invading house sparrows. Passer domesticus. Behav. Ecol. 2005;16:702–707. doi: 10.1093/beheco/ari044. [DOI] [Google Scholar]
- 27.Sax DF, Brown JH. The paradox of invasion. Global Ecol. Biogeogr. 2000;9:363–371. doi: 10.1046/j.1365-2699.2000.00217.x. [DOI] [Google Scholar]
- 28.Barrett, S. & Richardson, B. In Ecology of Biological Invasions 21–33 (Cambridge University Press, 1986).
- 29.Baker AJ, Moeed A. Rapid genetic differentiation and founder effect in colonizing populations of common mynas (Acridotheres tristis) Evolution. 1987;41:525–538. doi: 10.1111/j.1558-5646.1987.tb05823.x. [DOI] [PubMed] [Google Scholar]
- 30.Willi Y, Van Buskirk J, Hoffmann AA. Limits to the adaptive potential of small populations. Annu. Rev. Ecol. Evol. Syst. 2006;37:433–458. doi: 10.1146/annurev.ecolsys.37.091305.110145. [DOI] [Google Scholar]
- 31.Slobodkin, L. B. Growth and regulation of animal populations. (Dover Publications, 1980).
- 32.Carroll SP, Hendry AP, Reznick DN, Fox CW. Evolution on ecological time-scales. Funct. Ecol. 2007;21:387–393. doi: 10.1111/j.1365-2435.2007.01289.x. [DOI] [Google Scholar]
- 33.Bradshaw AD. Evolutionary significance of phenotypic plasticity in plants. Adv. Genet. 1965;13:115–155. [Google Scholar]
- 34.Anderson, T. R. Biology of the Ubiquitous House Sparrow: From Genes to Populations. (Oxford University Press, 2006).
- 35.Robbins, C. S. Introduction, spread, and present abundance of the house sparrow in North America. Ornithol. Monogr. 3–9 (1973).
- 36.Summers-Smith, J. D. The sparrows: a study of the genus Passer. (T & AD Poyser, 1988).
- 37.Liebl AL, Schrey AW, Andrew SC, Sheldon EL, Griffith SC. Invasion genetics: Lessons from a ubiquitous bird, the house sparrow Passer domesticus. Curr. Zool. 2015;61:465–476. doi: 10.1093/czoolo/61.3.465. [DOI] [Google Scholar]
- 38.Johnston RF, Selander RK. House sparrows - rapid evolution of races in North America. Science. 1964;144:548. doi: 10.1126/science.144.3618.548. [DOI] [PubMed] [Google Scholar]
- 39.Johnston, R. F. & Selander, R. K. Evolution in the house sparrow. II. Adaptive differentiation in North American populations. Evolution 1–28 (1971). [DOI] [PubMed]
- 40.Hovel, H. Check-list of the Birds of Israel: With Sinai: an Annotated Check-list. (Tel Aviv University: Society for the Protection of Nature in Israel, 1987).
- 41.Shirihai, H. The birds of Israel. (Academic Press, 1996).
- 42.Jaffe, S. In The zoogeography of Israel (eds Yom-Tov, Yoram & Tchernov, E.) 79–94 (Springer Netherlands, 1988).
- 43.Gavrilov E. On hybridisation of the Indian and House Sparrows. Bull. Br. Ornithol. Club. 1965;85:112–114. [Google Scholar]
- 44.Saini HK, Dhindsa M, Toor H. Morphometric variation in an Indian population of the house sparrow, Passer domesticus indicus. Le Gerfaut. 1990;79:69–79. [Google Scholar]
- 45.Lima MR, et al. Genetic and morphometric divergence of an invasive bird: the introduced house sparrow (Passer domesticus) in Brazil. PLoS One. 2012;7:e53332. doi: 10.1371/journal.pone.0053332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kekkonen J, Jensen H, Brommer JE. Morphometric differentiation across House Sparrow Passer domesticus populations in Finland in comparison with the neutral expectation for divergence. Ibis. 2012;154:846–857. doi: 10.1111/j.1474-919X.2012.01252.x. [DOI] [Google Scholar]
- 47.Brommer J, Hanski I, Kekkonen J, Väisänen R. Size differentiation in Finnish house sparrows follows Bergmann’s rule with evidence of local adaptation. J. Evol. Biol. 2014;27:737–747. doi: 10.1111/jeb.12342. [DOI] [PubMed] [Google Scholar]
- 48.Packard GC. Seasonal variation in bill length of House Sparrows. Wilson Bull. 1967;79:345–346. [Google Scholar]
- 49.Folk C, Novotny I. Variation in body weight and wing length in the House Sparrow, Passer domesticus L., in the course of the year. Zool. Listy. 1970;19:333–342. [Google Scholar]
- 50.Schifferli L. Federgewicht des Haussperlings Passer domesticus im Jahresverlauf. Ornithol. Beob. 1981;78:113–118. [Google Scholar]
- 51.Kekkonen J, et al. Low genetic differentiation in a sedentary bird: house sparrow population genetics in a contiguous landscape. Heredity. 2011;106:183–190. doi: 10.1038/hdy.2010.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sirkkomaa S. Calculations on the decrease of genetic variation due to the founder effect. Hereditas. 1983;99:11–20. doi: 10.1111/j.1601-5223.1983.tb00729.x. [DOI] [PubMed] [Google Scholar]
- 53.Schrey AW, Liebl AL, Richards CL, Martin LB. Range expansion of house sparrows (Passer domesticus) in Kenya: evidence of genetic admixture and human-mediated dispersal. J. Hered. 2014;105:60–69. doi: 10.1093/jhered/est085. [DOI] [PubMed] [Google Scholar]
- 54.Schrey AW, et al. Broad-scale latitudinal patterns of genetic diversity among native European and introduced house sparrow (Passer domesticus) populations. Mol. Ecol. 2011;20:1133–1143. doi: 10.1111/j.1365-294X.2011.05001.x. [DOI] [PubMed] [Google Scholar]
- 55.Pujol B, Wilson A, Ross R, Pannell J. Are QST–FST comparisons for natural populations meaningful? Mol. Ecol. 2008;17:4782–4785. doi: 10.1111/j.1365-294X.2008.03958.x. [DOI] [PubMed] [Google Scholar]
- 56.McKay JK, Latta RG. Adaptive population divergence: markers, QTL and traits. Trends Ecol. Evol. 2002;17:285–291. doi: 10.1016/S0169-5347(02)02478-3. [DOI] [Google Scholar]
- 57.Lind MI, Ingvarsson PK, Johansson H, Hall D, Johansson F. Gene flow and selection on phenotypic plasticity in an island system of Rana temporaria. Evolution. 2011;65:684–697. doi: 10.1111/j.1558-5646.2010.01122.x. [DOI] [PubMed] [Google Scholar]
- 58.Rogell B, et al. Strong divergence in trait means but not in plasticity across hatchery and wild populations of sea-run brown trout Salmo trutta. Mol. Ecol. 2012;21:2963–2976. doi: 10.1111/j.1365-294X.2012.05590.x. [DOI] [PubMed] [Google Scholar]
- 59.Dor R, et al. Population genetics and morphological comparisons of migratory European (Hirundo rustica rustica) and sedentary East-Mediterranean (Hirundo rustica transitiva) Barn Swallows. J. Hered. 2012;103:55–63. doi: 10.1093/jhered/esr114. [DOI] [PubMed] [Google Scholar]
- 60.Evans DM, Cardon LR. Guidelines for genotyping in genomewide linkage studies: single-nucleotide–polymorphism maps versus microsatellite maps. The American Journal of Human Genetics. 2004;75:687–692. doi: 10.1086/424696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hagen IJ, et al. The easy road to genome-wide medium density SNP screening in a non-model species: development and application of a 10 K SNP-chip for the house sparrow (Passer domesticus) Mol. Ecol. Resour. 2013;13:429–439. doi: 10.1111/1755-0998.12088. [DOI] [PubMed] [Google Scholar]
- 62.Jensen H, et al. Sexual variation in heritability and genetic correlations of morphological traits in house sparrow (Passer domesticus) J. Evol. Biol. 2003;16:1296–1307. doi: 10.1046/j.1420-9101.2003.00614.x. [DOI] [PubMed] [Google Scholar]
- 63.Dor R, Lotem A. Heritability of nestling begging intensity in the House Sparrow (Passer domesticus) Evolution. 2009;63:738–748. doi: 10.1111/j.1558-5646.2008.00598.x. [DOI] [PubMed] [Google Scholar]
- 64.Garant D, Sheldon BC, Gustafsson L. Climatic and temporal effects on the expression of secondary sexual characters: genetic and environmental components. Evolution. 2004;58:634–644. doi: 10.1111/j.0014-3820.2004.tb01685.x. [DOI] [PubMed] [Google Scholar]
- 65.Hole DG, et al. Agriculture: Widespread local house-sparrow extinctions. Nature. 2002;418:931–932. doi: 10.1038/418931a. [DOI] [PubMed] [Google Scholar]
- 66.Daniels RR. Can we save the sparrow. Curr. Sci. 2008;95:1527–1528. [Google Scholar]
- 67.White, O. Mitochondrial DNA isolation. Molecular genetic analysis of populations, a practical approach (1992).
- 68.Cabanac M, Aizawa S. Fever and tachycardia in a bird (Gallus domesticus) after simple handling. Physiol. Behav. 2000;69:541–545. doi: 10.1016/S0031-9384(00)00227-4. [DOI] [PubMed] [Google Scholar]
- 69.Verbeek ME, Drent PJ, Wiepkema PR. Consistent individual differences in early exploratory behaviour of male great tits. Anim. Behav. 1994;48:1113–1121. doi: 10.1006/anbe.1994.1344. [DOI] [Google Scholar]
- 70.Liebl AL, Martin LB. Exploratory behaviour and stressor hyper-responsiveness facilitate range expansion of an introduced songbird. Proc. R. Soc. Lond. B Biol. Sci. 2012;279:4375–4381. doi: 10.1098/rspb.2012.1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Carvalho CF, et al. Personality traits are related to ecology across a biological invasion. Behav. Ecol. 2013;24:1081–1091. doi: 10.1093/beheco/art034. [DOI] [Google Scholar]
- 72.Stöwe M, et al. Novel object exploration in ravens (Corvus corax): effects of social relationships. Behav. Process. 2006;73:68–75. doi: 10.1016/j.beproc.2006.03.015. [DOI] [PubMed] [Google Scholar]
- 73.Neumann K, Wetton JH. Highly polymorphic microsatellites in the house sparrow Passer domesticus. Mol. Ecol. 1996;5:307–309. doi: 10.1111/j.1365-294X.1996.tb00319.x. [DOI] [PubMed] [Google Scholar]
- 74.Griffith SC, Stewart IR, Dawson DA, Owens IP, Burke T. Contrasting levels of extra-pair paternity in mainland and island populations of the house sparrow (Passer domesticus): is there an “island effect”? Biol. J. Linn. Soc. 1999;68:303–316. [Google Scholar]
- 75.Griffith SC, et al. Fourteen polymorphic microsatellite loci characterized in the house sparrow Passer domesticus (Passeridae, Aves) Mol. Ecol. Notes. 2007;7:333–336. doi: 10.1111/j.1471-8286.2006.01598.x. [DOI] [Google Scholar]
- 76.Dawson DA, et al. High-utility conserved avian microsatellite markers enable parentage and population studies across a wide range of species. BMC Genomics. 2013;14:176. doi: 10.1186/1471-2164-14-176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Excoffier L, Lischer HE. Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Resour. 2010;10:564–567. doi: 10.1111/j.1755-0998.2010.02847.x. [DOI] [PubMed] [Google Scholar]
- 78.Kalinowski ST, Taper ML, Marshall TC. Revising how the computer program CERVUS accommodates genotyping error increases success in paternity assignment. Mol. Ecol. 2007;16:1099–1106. doi: 10.1111/j.1365-294X.2007.03089.x. [DOI] [PubMed] [Google Scholar]
- 79.Excoffier L, Smouse PE, Quattro JM. Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics. 1992;131:479–491. doi: 10.1093/genetics/131.2.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Earl DA. Structure Harvester: a website and program for visualizing Structure output and implementing the Evanno method. Conserv. genet. resour. 2012;4:359–361. doi: 10.1007/s12686-011-9548-7. [DOI] [Google Scholar]
- 82.Weir BS, Cockerham CC. Estimating F-statistics for the analysis of population structure. Evolution. 1984;38:1358–1370. doi: 10.1111/j.1558-5646.1984.tb05657.x. [DOI] [PubMed] [Google Scholar]
- 83.Jensen JL, Bohonak AJ, Kelley ST. Isolation by distance, web service. BMC genetics. 2005;6:13. doi: 10.1186/1471-2156-6-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Mantel N. The detection of disease clustering and a generalized regression approach. Cancer Res. 1967;27:209–220. [PubMed] [Google Scholar]
- 85.Kalinowski S. T. hp‐rare 1.0: a computer program for performing rarefaction on measures of allelic richness. Mol. Ecol. Notes. 2005;5:187–189. doi: 10.1111/j.1471-8286.2004.00845.x. [DOI] [Google Scholar]
- 86.Weir, B. & Cockerham, C. Genetic data analysis II: Methods for discrete population genetic data. Sinauer Assoc. Inc., Sunderland, MA, USA (1996).
- 87.Keenan K, McGinnity P, Cross TF, Crozier WW, Prodöhl P. A. diveRsity: An R package for the estimation and exploration of population genetics parameters and their associated errors. Methods Ecol. Evol. 2013;4:782–788. doi: 10.1111/2041-210X.12067. [DOI] [Google Scholar]
- 88.Cornuet JM, Luikart G. Description and power analysis of two tests for detecting recent population bottlenecks from allele frequency data. Genetics. 1996;144:2001–2014. doi: 10.1093/genetics/144.4.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Luikart G, Allendorf F, Cornuet J, Sherwin W. Distortion of allele frequency distributions provides a test for recent population bottlenecks. J. Hered. 1998;89:238–247. doi: 10.1093/jhered/89.3.238. [DOI] [PubMed] [Google Scholar]
- 90.Hijmans RJ, Cameron SE, Parra JL, Jones PG, Jarvis A. Very high resolution interpolated climate surfaces for global land areas. Int. J. Climatol. 2005;25:1965–1978. doi: 10.1002/joc.1276. [DOI] [Google Scholar]
- 91.Hadfield JD. MCMC methods for multi-response generalized linear mixed models: the MCMCglmm R package. J. Stat. Softw. 2010;33:1–22. doi: 10.18637/jss.v033.i02. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated and analyzed during the current study are available from the corresponding author upon reasonable request.