Abstract
Mitochondrial networks exhibit a variety of complex behaviors, including coordinated cell-wide oscillations of energy states as well as a phase transition (depolarization) in response to oxidative stress. Since functional and structural properties are often interwinded, here we characterized the structure of mitochondrial networks in mouse embryonic fibroblasts using network tools and percolation theory. Subsequently we perturbed the system either by promoting the fusion of mitochondrial segments or by inducing mitochondrial fission. Quantitative analysis of mitochondrial clusters revealed that structural parameters of healthy mitochondria laid in between the extremes of highly fragmented and completely fusioned networks. We confirmed our results by contrasting our empirical findings with the predictions of a recently described computational model of mitochondrial network emergence based on fission-fusion kinetics. Altogether these results offer not only an objective methodology to parametrize the complexity of this organelle but also support the idea that mitochondrial networks behave as critical systems and undergo structural phase transitions.
Introduction
Mitochondria arose around two billion years ago from the engulfment of an α-proteobacterium by a precursor of the modern eukaryotic cell1. Subsequent evolution shaped the relationship between mitochondria and their host cells, leading to a high degree of morphological and functional specialization in the organelle. Long known for their role in ATP production, mitochondria also participate in a myriad of essential cellular processes such as apoptosis, calcium buffering and phospholipid synthesis, among others2. In addition, mitochondria exhibit complex patterns including oscillatory dynamics, phase transitions and fractality3–5.
A typical mitochondrion comprises a network of tube-like structures, with fragments of all sizes (ranging from less than 1 μm to 15 μm or more)6. The current theoretical understanding proposes that mitochondrial morphology is maintained by two opposing processes, fission and fusion, which depending on their relative predominance determine the overall connectivity and structural properties of the network7. Although the structure of mitochondrial networks is generally described as complex, a quantitative description of such complexity is still lacking. Moreover, a recent model of mitochondrial dynamics suggested that mitochondrial networks are poissed at the critical point of a phase transition, albeit no connection between theory and phenomenology has been provided yet8.
In this work, we present a quantitative description of mitochondrial network structure using tools of network and percolation theory. To do that, we developed a pipeline to extract structural parameters from confocal images. Moreover, we evaluated how a recently published model of mitochondria fitted the data from real networks providing the missing link between theory and experiments.
Results
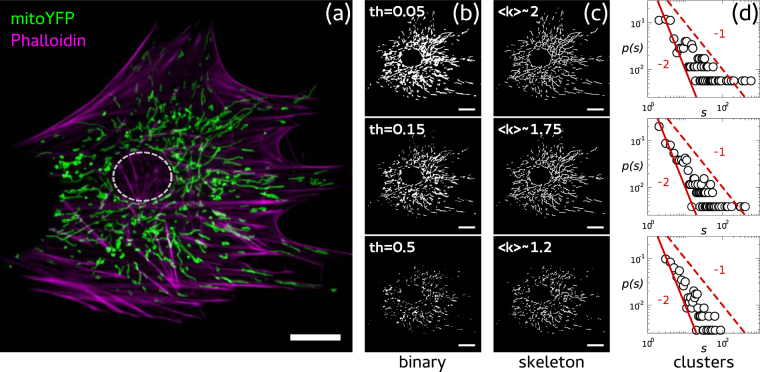
We developed a pipeline to quantify the structural complexity of mitochondrial networks from confocal microscopy images like the one presented in Fig. 1(a), where a mouse embryonic fibroblast (MEF) expressing a mitochondria-targeted yellow fluorescent protein (mYFP) is shown (see Methods). Our pipeline entailed three steps, the first of which was the convertion of a grayscale image into a binary image. To do that, we used the im2bw routine from Matlab (Natick, Massachusetts: The MathWorks Inc.). Figure 1(b) shows examples of binary images obtained from the same grayscale image (Fig. 1(a)) by choosing different threshold values. Second, binarized versions of the image were transformed into skeletons of uniform (one pixel) thickness, like the ones shown in Fig. 1(c), using the routine bwmorph from Matlab. These skeletons were composed of independent clusters of different sizes, each of them made up by either linear or branched segments. Following others9–11, we hypothesized that such skeletons constituted a good approximation of the structure of mitochondrial networks. Although mitochondrial networks are embeded in the 3-dimensional cellular volume, artefactual branching points in 2-dimensional reconstructions were negligible (see Supplementary Information). Finally, we extracted two different parameters from skeletons: cluster mass (s), by counting the number of pixels in each individual cluster, and pixel degree (k), by counting the nearest neighbors of each pixel (see Methods). Figure 1(d) shows cluster mass probability distribution functions corresponding to skeletons in Fig. 1(c). We saw that, while the connectivity decreased whit the threshold (as expected due to larger network fragmentation), p(s) exhibited a power law behavior that seemed to be robust against threshold variations.
Figure 1.
Network structure from mitochondrial images. (a) Confocal image of a MEF in which mitochondria are shown in green and the actin cytoskeleton in magenta. Image processing: (b) Grayscale images were converted to binary images by choosing the indicated threshold. (c) Detected lines in binary images were reduced to a single pixel width (skeleton). The mean degree 〈k〉 of each skeleton was computed by analysing the nearest neighbors of each pixel occupied. (d) Cluster mass distributions p(s) were computed by counting the number of pixels that made up each segment within clusters. Red curves represent power laws with exponents −1 (dashed) and −2 (continuous). Scale bars represent 10 μm.
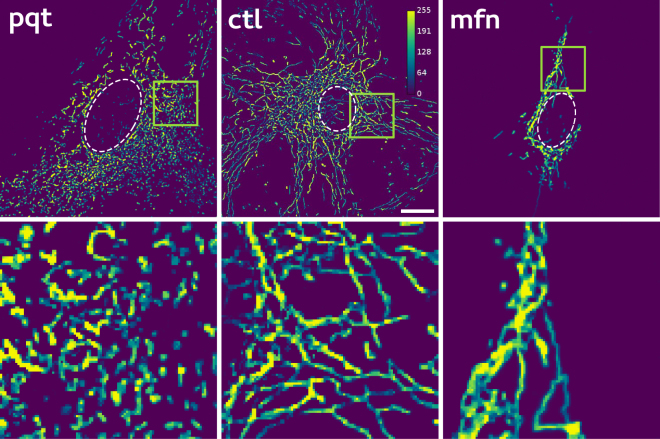
The long-tail behaviour displayed by p(s) was in accordance with recent work suggesting that mitochondrial networks operate near a percolation phase transition8. In order to test the existence of a structural transition, we perturbed the structure of mitochondrial networks in opposite directions, either by increasing mitochondrial fission using paraquat (pqt)12–15, or by promoting mitochondrial fusion by mitofusin 1 (mfn) over-expression16–19 (see Methods). Examples of morphological changes observed after applying the aforementioned treatments are presented in Fig. 2. A rapid qualitative inspection revealed that, compared to control (ctl) networks, mfn networks appeared as elongated interconnected strings, while in the case of the pqt networks, mitochondria seemed as independent small fragments. In fact, this qualitative visual inspection is the approach used routinely to evaluate mitochondrial morphology and status20–25, counting relative ratios of cells exhibiting a particular mitochondrial phenotype, different from the one found in control cells.
Figure 2.
Typical examples of mitochondrial network images obtained under different conditions. Confocal images of MEFs transfected with mYFP under control conditions (ctl), paraquat treatment (pqt) or mitofusin 1 over-expression (mfn). Pixel intensity is depicted using a pseudocolor (calibration bar). Insets highlight the effect asserted by each treatment on mitochondrial structure. Scale bars represent 10 μm.
Quantification of mitochondrial network properties
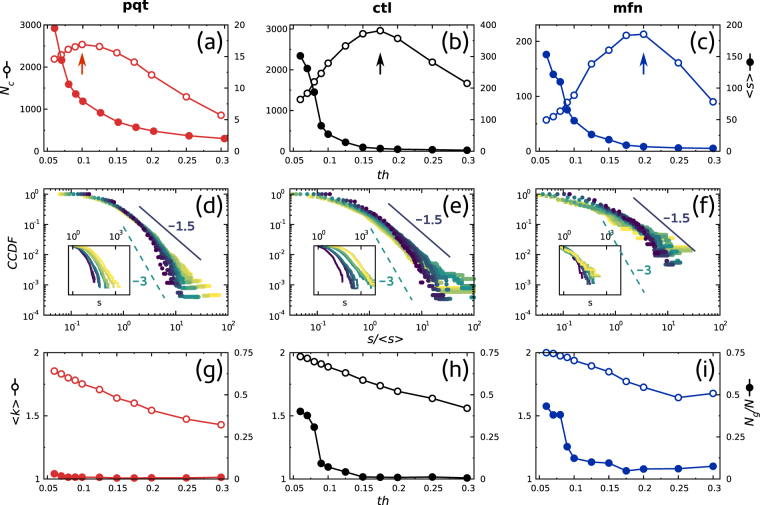
Although many methods have been proposed to extract networks from mitochondrial images, virtually all of them make use of an arbitrary thresholding step9–11,26–28. However, as illustrated in Fig. 1, the application of different intensity thresholds (th) resulted in different skeletons and hence different network architectures. We reasoned that a way to avoid this arbitrariness would be to compare the behavior of network parameters as a function of the threshold. As a proof of concept, we picked one network for each condition and computed basic structural parameters. The results obtained are shown in Fig. 3, where each column corresponds to one of the treatments: ctl in the middle, mfn on the right and pqt on the left. Similar results were obtained using other images (data not shown).
Figure 3.
Network parameters computed from single images. Center column corresponds to ctl, left column to pqt and right column to mfn. Top row panels (a–c) show the number of clusters (N c, empty symbols, left labels) and the mean cluster mass (〈s〉, filled symbols, right labels) as a function of the binarization threshold (th). Arrows point to th values at which the maximum number of clusters appears in each case. Middle row panels (d–f) show the cumulative distribution function of cluster mass as a function of th (colors denote different threshold values). Note that by normalizing each distribution using the mean cluster mass 〈s〉, all distributions collapse approximately to the same function. Power laws with exponents −1.5 and −3 are shown for reference purposes. Insets depict unnormalized distributions. Bottom row panels (g–i) depict the mean degree (〈k〉, empty symbols, left labels) and the normalized size of the largest cluster (N g/N, filled symbols, right labels) of the networks as a function of th.
As depicted in Fig. 3(a–c), we first looked at the number of clusters N c and the average cluster mass 〈s〉 of mitochondrial networks29. N c exhibited a non monotonic behavior in all cases, with a peak in the interval 0.1 ≤ th* ≤ 0.2 (Fig. 3(a–c), empty circles). This is expected, regardless of the nature or origin of the image, anytime a section is made cutting a rough landscape30,31. Interestingly, the peaks of pqt and mfn networks shifted to left and to right, respectively, from the peak of the ctl network, suggesting that the relative position of the peak could constitute a robust readout of network connectivity. Moreover, we envisioned that the position of the peak in N c could be useful as a less arbitrary criterion when the selection of a single threshold is needed.
On the other hand, 〈s〉 changed monotonically with the threshold in all cases (Fig. 3(a–c), solid circles). Regardless of the similar behavior, 〈s〉 values of the pqt network were much smaller than that of other networks, in accordance with the effect asserted by the compound on mitochondria. In the case of the mfn network, although its 〈s〉 values were within the same order of magnitude of those of the ctl network, it should be noted that mitofusin 1 over-expression reduced overall network size (see Fig. 2), making them proportionally bigger.
Long-tail distributions in Fig. 1(d) indicated that mitochondrial networks exhibited scale free properties, such as the coexistence of numerous small fragments with few massive clusters. To extract more precise information from mass distributions, we plotted the complementary cummulative distribution function (CCDF)
| 1 |
where p(s i) is the probability of finding a cluster of mass i in the network, which enhances the statistical significance of the high mass region32. Figure 3(d–f) show the CCDFs of cluster mass as a function of a range of threshold values. CCDFs describe how often the cluster mass is above a particular value s and we used them here to characterize the effect of the treatment on the giant cluster of the network. In addition, we found that the average cluster mass 〈s〉 can be used to normalize mass densities, eventually leading to a unique average representative distribution for any threshold value that followed approximately a power-law of the form
| 2 |
with 0.5 ≤ γ ≤ 2. This result allowed us to consider cluster mass distributions as roughly independent of the threshold value, which is a desirable property in any descriptive parameter used to quantify the structure of mitochondrial networks. More importantly, Fig. 3(d–f) suggest that perturbations in the fission/fusion balance altered mass frequencies and modified the mass of the giant cluster, causing a shift in the slope of the CCDF.
Figure 3(g–i) describe two additional topological quantities inspired in network theory, namely the average degree 〈k〉 (empty dots) and the normalized mass of the giant cluster N g/N (solid dots)33. Although 〈k〉 decreased monotonically in all cases, the relation 〈k〉pqt < 〈k〉ctl < 〈k〉mfn held for every particular threshold value, indicating once again that perturbing the fission/fusion balance drove the network connectivity to the extremes of disconnected fragments or fully connected clusters. Finally, when we computed N g/N across the threshold range we found that, despite the effect that the treatments asserted on network size, the ctl network displayed an intermediate behaviour compared to pqt and mfn networks.
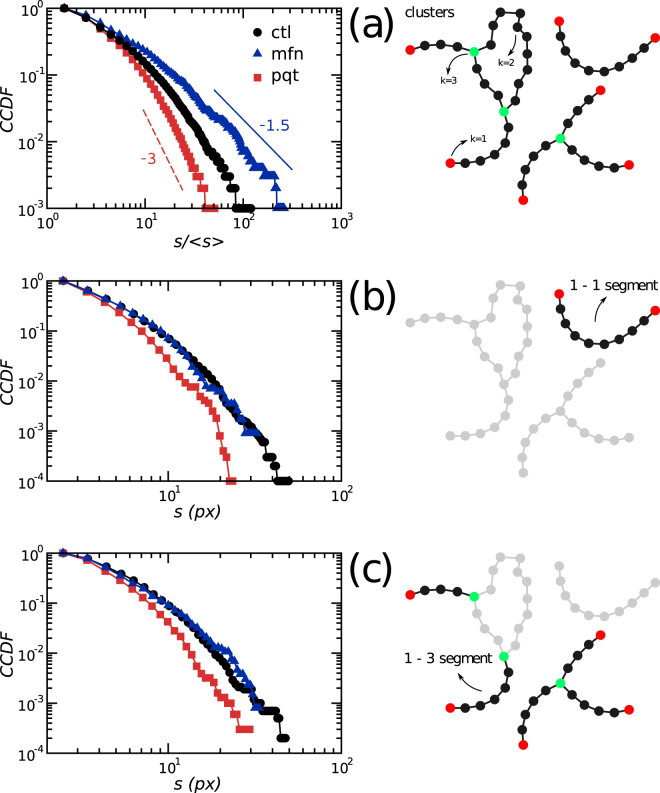
The results shown in Fig. 3 suggested that mitochondrial network structure changed in response to treatments in a rather predictable manner, with ctl networks always displaying an intermediate behavior. To gain insight into how the mitochondrial structure responds to perturbations we recomputed the CCDFs from Fig. 3(d–f) using all available data, by selecting a threshold value of 0.15. Similar results were obtained using thresholds in the range [0.1–0.2] (data not shown). Schemes in Fig. 4(a–c) define the elements measured in every network, namely clusters and segments, where 1-1 means a segment that connects two nodes with k = 1 and 1-3 refers to those segments connecting a node with k = 3 and a node with k = 1. Figure 4(a) shows that modifications of the fission/fusion balance changed the exponents of the power-law relation in the cluster mass distribution, adding support to our previous findings. Interestingly, mass distributions of 1-1 and 1-3 segments behaved similarly accross treatments, suggesting that differences between networks could not be attributed to changes in the mass of linear segments.
Figure 4.
Changes in mass distributions upon fission/fusion balance perturbation. (a) Cluster mass distributions followed a truncated power law. Shifts in the behavior upon treatment obeyed the relation γ mfn < γ ctl < γ pqt, where γ is the exponent of the CCDF. Power laws with exponents −1.5 and −3 are shown for reference purposes only. Mass distribution of open (b) and branched (c) segments showed exponential decays. Illustrations at the right side of each panel depict network elements measured.
Fission/fusion balance is linked to mitochondrial network complexity
In the previous section we have described a straightforward methodology and a set of structural parameters that allowed us to characterize basic features of mitochondrial networks. To gain a deeper understanding on the relationship between mitochondrial dynamics and network status we computed additional parameters in order to quantify the complexity of the different networks.
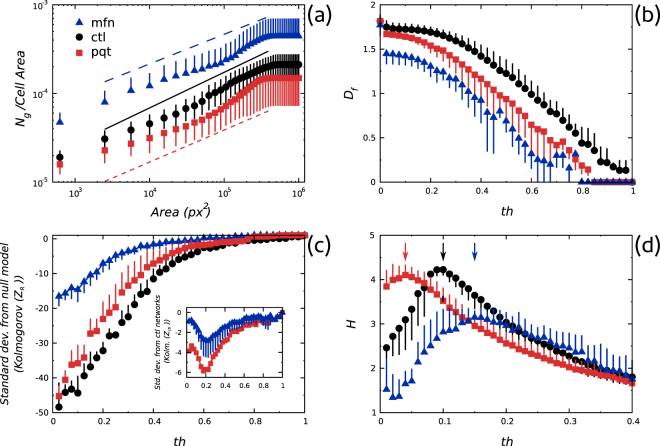
We investigated the scaling properties of mitochondrial networks by performing a finite size scaling analysis34,35. Results are summarized in Fig. 5(a) where the normalized mass of the giant cluster as a function of the area used to sample the image is shown. Fitted lines showed that the scaling behavior of pqt and mfn networks slightly deviated from the one displayed by ctl networks, suggesting that structural alterations caused by treatments affected the network scaling.
Figure 5.
Changes in mitochondrial network complexity. (a) Scaling of N g/N with the area used to sample the image. (b) Changes in fractal dimension upon paraquat treatment and mitofusin 1 over-expression. (c) Z-scores expressing the deviation of the observed structure from the random configuration in Kolmogorov complexity units. Inset shows the deviation of pqt and mfn network configurations from the ctl configuration in Kolmogorov complexity units. (d) Entropy of cluster mass distributions. Perturbations of the fission/fusion balance caused a shift in the critical threshold value th* (arrows).
Next, given the functional relevance of the spatial distribution of mitochondria36–39, we decided to quantify the fractal dimension D f of the networks40,41. As can be seen in Fig. 5(b), even though treatments had opposite effects on mitochondria, they both decreased the network D f, suggesting once again that alterations in the fission/fusion balance impacted on the space-filling properties of mitochondria.
These two observations prompted us to hypothesize that alterations in the fission/fusion balance tended to reduce network complexity. To test this hypothesis we computed the normalized Kolmogorov complexity42
| 3 |
where K is the Kolmogorov complexity of the network, μ R is the mean Kolmogorov complexity from 1000 randomized versions of the original skeleton and is the standard deviation from randomized versions of the original skeleton. As shown in Fig. 5(c), the distance between the observed network structure and the random configuration, in Kolmogorov complexity units, was maximal for ctl networks, strongly supporting the idea that perturbations to mitochondrial fission or fusion lowered network complexity.
Finally, we considered the possibility that changes in network parameters could be explained as shifts in the percolation regime of the networks, taking into account recent suggestions that healty mitochondrial networks are in a critical regime, characterized by the maximal heterogeneity in sizes of network subcomponents8. Specifically, we reasoned that the percolation transition threshold th* of the images could indicate the stage of the percolation process of the underlying networks, i.e., subcritical, critical or supercritical regime43. To test this, we computed the Shannon’s entropy of cluster masses44,45
| 4 |
where H j is the entropy at threshold j and is the fraction of the total mass of clusters of mass i at threshold j over the total mass of the network at threshold j. Plotting H j as a function of the threshold (Fig. 5(d)) we observed that each network type was characterized by a particular th* value. Moreover, the following relation was found , indicating that configurations found in pqt and mfn networks corresponded to subcritical and supercritical regimes, respectively.
In summary, our results showed that perturbations to fission/fusion kinetics gave rise to changes in connectivity patterns, altering the scaling properties and the percolation regime of mitochondrial networks.
Are mitochondrial networks poised at criticality
Structural and topological changes described in previous paragraphs were consistent with the idea that mitochondrial networks undergo a percolation transition29,46,47. This view propose that a steady-state mitochondrial network requires a proper balance of the two opposing tendencies, one towards fusing segments and one favoring fragmentation.
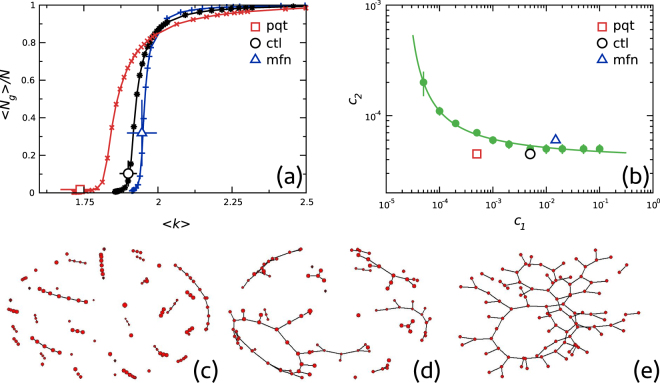
In an attempt to further test these interpretations we contrasted our experimental results with the predictions of a recently published mathematical model8 that contains explicit variables for relative fission/fusion rates c 1 and c 2 (see Methods). In order to proceed, a bootstrapping approach was required to extract model parameters from experimental data. First, the order parameter was defined as the ratio of the largest cluster 〈N g〉 over the network size N. Then, for a fixed value of c 1 both the order parameter (〈N g〉/N) and the average degree 〈k〉 as a function of c 2 were numerically computed. Next, for every value of c 1, 〈N g〉/N was plotted parametrically by varying c 2, as a function of 〈k〉 (See Methods).
Continuous lines in Fig. 6(a) are an example of those curves. It could be observed that, at least for the parameters region of interest, each point of the curves corresponded to a unique pair of values (c 1, c 2) (see Supplementary Information). This allowed us to roughly associate model parameters values to experimental data. The symbols in the figure correspond to an average of 〈N g〉/N and 〈k〉 over different network groups. The phase diagram in the (c 1, c 2) space is illustrated in Fig. 6(b). Filled symbols (with error bars) correspond to the maxima in the mean cluster size 〈s〉 (see Supplementary Information). The continuous line is a non linear fitting to the points and represents a reference to the eye for the location of the phase transition. Empty symbols correspond to parameter values extracted from experimental data shown in Fig. 6(a). Graphs on panels (c), (d) and (e) show examples of typical networks constructed with the model using the three derived empirical values (i.e., the points in (b) labeled pqt, ctl, mfn respectively). From Fig. 6 it can be concluded that ctl networks were in the vicinity of a percolation phase transition, while pqt and mfn network configurations corresponded to subcritical and supercritical regimes, respectively.
Figure 6.
Comparison of the present experimental results with those of Sukhorukov’ et al. model. (a) Model parameters extracted iteratively from experimental data. Open symbols and error bars correspond to means and standard deviations of all cells for the ctl, pqt and mfn groups (for a binarization threshold = 0.15). Each point of the extracted curves corresponds to a unique pair of values (c 1, c 2). (b) Phase diagram of the model in the (c 1, c 2) space. Filled symbols and the continuous line correspond to the location of the phase transition. The three open symbols (labeled pqt, ctl, mfn) correspond to the parameter values extracted from experimental data as shown in (a). Panels (c–e) show a graphical representation of the typical networks simulated using the three derived (c 1, c 2) values, corresponding to pqt, clt and mfn networks, respectively.
Discussion
Mitochondria organize as complex networks that display temporal and spatial coordination, pressumably by operating close to the edge of dynamic instability4. Following these ideas, in this work we explored the topological properties of mitochondria and we tested the hypothesis that mitochondrial networks are organized near the percolation transition43.
The main findings of the present study can be summarized in three aspects. First, we proposed a straightforward approach to quantify structural properties of mitochondrial networks. Applying the proposed method to control (ctl) and perturbed (pqt and mfn) networks, we identified regularities in the variations of connectivity patterns as well as changes in the behavior of the mass distribution of mitochondrial clusters. Moreover, data from additional experimental manipulations supported the conclusions obtained with paraquat treatment and mitofusin 1 over-expression (see Supplementary Information).
Second, by computing additional parameters we determined that the configuration found in control mitochondria is optimal. Specifically, we observed that promoting fission or fusion lowered the fractal dimension of the networks40, reducing the space-filling capacity of mitochondria. Additionally, we found that promoting either fission or fusion lowered the Kolmogorov complexity of mitochondrial networks42, increasing the randomness of their configuration.
Finally, we contrasted our empirical data with a recently published mathematical model8 finding that balanced fission/fusion dynamics lead to a network capable of a phase transition. Specifically, empirical network configurations were found at parameter regions predicted by the model, allowing us to classify network configurations in three different regimes, subcritical (pqt), critical (ctl) and supercritical (mfn), and adding support to the idea that, under normal physiological conditions, mitochondrial networks are poissed near a percolation transition point.
Methods
Cell culture
Mouse Embryonic Fibroblasts (MEFs) were obtained as described by Xu et al.48. Briefly, 13.5 days old mouse embryos were extracted from the mother uterus, rinsed with PBS and placed on a petri dish. The head and red organs were discarded and the remaining body was rinsed again with PBS and placed on a new petri dish. Using shaving blades, the tissue was chopped into pieces and trypsinized for 15 sec at 37 °C (PBS 10% Trypsin). Trypsin reaction was quenched with serum-containing media and the whole mixture was centrifuged for 5 min at 2000 rpm. The supernatant was discarded and the pellet resuspended in DMEM with 10% SFB, 1% GlutaMax and 1% Non-essential amino acids. Cell pellets from 4 embryos were seeded on 175 cm2 culture bottles and were allowed to grow for 48 h. C57BL6 mice were obtained from the Animal Facility of the Mercedes and Martín Ferreyra Medical Research Institute and National University of Córdoba (INIMEC-CONICET-UNC). All experimental protocols were approved by the Institutional Council of Animal Care (CICUAL-INIMEC-CONICET). All methods were carried out in accordance with the approved guidelines.
Mitochondrial network morphology manipulation and cell imaging
Mitochondria were visualized by lentiviral infection. Lentiviruses were produced as described by Baloh et al.49. Briefly, human embryonic kidney (HEK) 293 T cells were plated onto six-well plates and transfected with a polymerase-coding vector (REV), a packaging vector (8.71), an envelope vector (VSVG) and a shuttle vector encoding the mitochondrial-targeted yellow fluorescent protein (mitoYFP) using Lipofectamine 2000 (Invitrogen) reagent. Media was changed at 12 h and collected at 48 and 72 h, pooled and applied directly to MEFs cultures.
Manipulation of mitochondrial morphology was accomplished in two ways: (1) to induce mitochondrial fragmentation, mitoYFP-expressing MEF cultures were treated with 200 μM paraquat (Sigma-Aldrich) for 24 hours, and (2) to promote mitochondrial fusion, cultures were transfected (Lipofectamine 2000, Invitrogen) with a plasmid encoding the sequence of the human mitofusin 1 gene (MFN1, Addgene). Once ready, cells were fixed with 4% PFA in 4% sucrose-containing PBS. F-actin staining was performed on fixed cells using Alexa 546-phaloidin (Molecular Probes), following manufacturer’s protocol. In all cases, image acquisition was achieved using an Olympus IX81 inverted microscope equipped with a Disk Spinning Unit (DSU), epifluorescence illumination (150 W Xenon Lamp) and a microprocessor. MEFs were imaged using a 60x oil immersion objective, an ORCA AG (Hamamatsu) CCD camera and OSIS software.
Image analysis
All routines used for image processing and analysis were written in Matlab (The MathWorks, Natick, MA). As explained above, mitochondrial structures were extracted from micrographs of MEFs where mitochondria were fluorescently tagged. Individual 8-bit images were converted to binary (i.e., black & white) for different threshold values of intensity using the Matlab routine im2bw. For each threshold value (range 0–1) the skeleton (i.e., the image reduced to a trace of one- pixel thickness) was extracted using the Matlab routine bmorph. Subsequently, clusters were extracted using the Matlab routine bwlabel. The algorithm define a cluster as those pixels connected with at least one of the eight nearest neighbors. The degree was computed using a numerical routine that inspects the nearest neighbors of each pixel and decides if the site corresponds to a node of degree k = 1, k = 2 or k = 350. Ng/N scaling was computed by averaging the size of the biggest cluster in sections of the image of increasing area. It was determined that the reconstructed network topology was not affected by the issue of potential artifacts due to the 2D projection of the mitochondrial 3D structure (see Supplementary Information). The complementary cumulative distribution function is defined as
| 5 |
and was computed by calculating the fraction of clusters of mass higher than x, were x takes the value of all possible cluster masses. Fractal dimension (D f), Kolmogorov complexity and Entropy (H) were computed using the Matlab routines boxcount, kolmogorov and entropy, respectively.
Sukhorukov et al. Model
Numerical simulations were conducted using the model described in Sukhorukov et al.8. Briefly, the network structure emerges as the result of two fusion and two fission reactions between the tips of a set of L dimers. In the model, a dimer tip can connect (disconnect) to other dimer tips forming a network node, but at most three tips can be merged. In this way, the degree k of the nodes can take only the values k = 1 (isolated tip), k = 2 (two merged tips) and k = 3 (three merged tips); only two fusion processes are allowed: tip-to-tip (two nodes of degree k = 1 merge into a node of degree k = 2) and tip-to-side (a node of degree k = 1 and a node of degree k = 2 merge into a node of degree k = 3). To each fusion process there is an associated inverse (fission) one. The bias to each process can be written as rates of either fusion or fission51 represented as reaction processes on nodes X k:
| 6 |
| 7 |
where a 1 (b 1) is the reaction rate for tip-to-tip fusion (fission) process and a 2 (b 2) is the reaction rate for tip-to-side fusion (fission) process. The model can then be implemented as an agent based stochastic dynamics between a set of L reactant objects (dimers), submitted to the above described fusion and fission processes. The dynamics is simulated using Gillespie52 algorithm. Nodes participating in a particular event are chosen with equal probability within a list of the nodes with the same degree. Following ref.8 we assumed b 2 = (3/2)b 1 and varied the relative rates c i = a i/b i.
Sukhorukov et al. described in detail the steady state of the dynamics as a function of changes in c 1 and c 2 8. The system admits a plethora of network configurations in parameter space, including fragmented or hyperfused networks resulting from extreme values of fission and fusion activities as well as networks resembling those seen in healthy cells at intermediate values. In passing, notice that more recently this model was reformulated53 to include information on the microtubule cytoskeleton.
In the present work we performed simulations for L = 15000, roughly the estimated value for the average number of edges in control networks. In every simulation we run the algorithm 3L times after which we measured different quantities. This ensured that the distribution of nodes with degree k became stationary. For every set of values of (c 1, c 2) the procedure was repeated 100 times for different sequences of random numbers and the different quantities were averaged over this sample. The different quantities measured were: the average degree 〈k〉, the average fraction of nodes in the largest cluster 〈N g/N〉 and the average cluster size excluding the largest cluster 〈s〉 (see Supplementary Information), where averages were taken both over all nodes in the network and over different runs.
Data availability
Data are available upon request.
Electronic supplementary material
Acknowledgements
This work was partially supported by CONICET (Argentina) through Grants PIP 112-201101-00213, PIP 2015-01-00954 and PICT 2013-3142, and by SECyT (Universidad Nacional de Córdoba, Argentina). DRC thanks the support of Universidad Nacional de San Martín, Argentina.
Author Contributions
N.Z., E.Z., P.H. and D.C. conceived the experiments; N.Z. and E.Z. conducted the experiments; N.Z., E.Z., S.C., O.B. and D.C. analyzed the data. All authors reviewed the manuscript.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Nahuel Zamponi and Emiliano Zamponi contributed equally to this work.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-017-18351-5.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Nahuel Zamponi, Email: zamponi.n@gmail.com.
Pablo R. Helguera, Email: phelguera@immf.uncor.edu
References
- 1.Gray MW, Burger G, Lang BF. Mitochondrial evolution. Science. 1999;283:1476–1481. doi: 10.1126/science.283.5407.1476. [DOI] [PubMed] [Google Scholar]
- 2.Friedman JR, Nunnari J. Mitochondrial form and function. Nature. 2014;505:335–343. doi: 10.1038/nature12985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kurz FT, Derungs T, Aon MA, O’Rourke B, Armoundas AA. Mitochondrial networks in cardiac myocytes reveal dynamic coupling behavior. Biophysical Journal. 2015;108:1922–1933. doi: 10.1016/j.bpj.2015.01.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aon MA, Cortassa S, O’Rourke B. Percolation and criticality in a mitochondrial network. Proc Natl Acad Sci USA. 2004;101:4447–4452. doi: 10.1073/pnas.0307156101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lennon FE, et al. Unique fractal evaluation and therapeutic implications of mitochondrial morphology in malignant mesothelioma. Sci Rep. 2016;6:24578. doi: 10.1038/srep24578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Collins TJ, Berridge MJ, Lipp P, Bootman MD. Mitochondria are morphologically and functionally heterogeneous within cells. EMBO J. 2002;21:1616–1627. doi: 10.1093/emboj/21.7.1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen H, Chan DC. Mitochondrial dynamics in mammals. Curr Top Dev Biol. 2004;59:119–144. doi: 10.1016/S0070-2153(04)59005-1. [DOI] [PubMed] [Google Scholar]
- 8.Sukhorukov VM, Dikov D, Reichert AS, Meyer-Hermann M. Emergence of the mitochondrial reticulum from fission and fusion dynamics. PLoS Comput Biol. 2012;8:e1002745. doi: 10.1371/journal.pcbi.1002745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ouellet M, Guillebaud G, Gervais V, Lupien St-Pierre D, Germain M. A novel algorithm identifies stress-induced alterations in mitochondrial connectivity and inner membrane structure from confocal images. PLoS Comput Biol. 2017;13:e1005612. doi: 10.1371/journal.pcbi.1005612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nikolaisen J, et al. Automated quantification and integrative analysis of 2D and 3D mitochondrial shape and network properties. PLoS One. 2014;9:e101365. doi: 10.1371/journal.pone.0101365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shin JW, et al. Changes, and the Relevance Thereof, in Mitochondrial Morphology during Differentiation into Endothelial Cells. PLos One. 2016;11:e0161015. doi: 10.1371/journal.pone.0161015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang G-Y, Hirai K-I, Shimada H. Mitochondrial breakage Induced by the herbicide Paraquat in cultured human lung cells. J Electron Microsc (Tokyo) 1992;41:181–184. [PubMed] [Google Scholar]
- 13.Castello PR, Derek A, Patel D, Patel M. Mitochondria are a major source of Paraquat-induced reactive oxygen species production in the brain. J Biol Chem. 2007;282:14186–14193. doi: 10.1074/jbc.M700827200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cocheme HM, Murphy MP. Complex I is the major site of mitochondrial superoxide production by paraquat. J Biol Chem. 2008;283:1786–1798. doi: 10.1074/jbc.M708597200. [DOI] [PubMed] [Google Scholar]
- 15.Zhao G, et al. Crosstalk between Mitochondrial Fission and Oxidative Stress in Paraquat-Induced Apoptosis in Mouse Alveolar Type II Cells. Int J Biol Sci. 2017;13:888–900. doi: 10.7150/ijbs.18468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Santel A, Fuller MT. Control of mitochondrial morphology by a human mitofusin. J Cell Sci. 2001;114:867–874. doi: 10.1242/jcs.114.5.867. [DOI] [PubMed] [Google Scholar]
- 17.Legros, F., Lombès, A., Frachon, P. & Rojo, M. Mitochondrial fusion in human cells Is efficient, requires the inner membrane potential, and Is mediated by Mitofusins. Mol Biol Cell13, 4343–4354 (2002). [DOI] [PMC free article] [PubMed]
- 18.Rojo, M., Legros, F., Chateau, D. & Lombes, A. Membrane topology and mitochondrial targeting of mitofusins, ubiquitous mammalian homologs of the transmembrane GTPase Fzo. J Cell Sci115, 1663–1674 (2002). [DOI] [PubMed]
- 19.Santel A, et al. Mitofusin-1 protein is a generally expressed mediator of mitochondrial fusion in mammalian cells. J Cell Sci. 2003;116:2763–2774. doi: 10.1242/jcs.00479. [DOI] [PubMed] [Google Scholar]
- 20.Detmer, S. A. & Chan, D. C. Complementation between mouse Mfn1 and Mfn2 protects mitochondrial fusion defects caused by CMT2A disease mutations. J Cell Biol176, 405–414 (2007). [DOI] [PMC free article] [PubMed]
- 21.Loson OC, Song Z, Chen H, Chan DC. Fis1, Mff, MiD49, and MiD51 mediate Drp1 recruitment in mitochondrial fission. Mol Biol Cell. 2013;24:659–667. doi: 10.1091/mbc.E12-10-0721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li G, et al. Mitochondrial translocation and interaction of cofilin and Drp1 are required for erucin-induced mitochondrial fission and apoptosis. Oncotarget. 2015;30:1834–1849. doi: 10.18632/oncotarget.2795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li S, et al. Transient assembly of F-actin on the outer mitochondrial membrane contributes to mitochondrial fission. J Cell Biol. 2015;5:109–123. doi: 10.1083/jcb.201404050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moore AS, Wong YC, Simpson CL, Holzbaur EL. Dynamic actin cycling through mitochondrial subpopulations locally regulates the fission-fusion balance within mitochondrial networks. Nat Commun. 2016;30:12886. doi: 10.1038/ncomms12886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rehklau K, et al. Cofilin1-dependent actin dynamics control DRP1-mediated mitochondrial fission. Cell Death Dis. 2017;5:e3063. doi: 10.1038/cddis.2017.448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chevrollier, A. et al. Standardized mitochondrial analysis gives new insights into mitochondrial dynamics and OPA1 function. Int J Biochem Cell Biol44, 980–988 (2012). [DOI] [PubMed]
- 27.Vowinckel J, Hartl J, Butler R, Ralser M. MitoLoc: A method for the simultaneous quantification of mitochondrial network morphology and membrane potential in single cells. Mitochondrion. 2015;24:77–86. doi: 10.1016/j.mito.2015.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Peng JY, et al. Automatic morphological subtyping reveals new roles of caspases in mitochondrial dynamics. PLoS Comput Biol. 2011;7:e1002212. doi: 10.1371/journal.pcbi.1002212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stauffer, D. & Aharony, A. Introduction to Percolation Theory (CRC press, 1994).
- 30.Gallos LK, Barttfeld P, Havlin S, Sigman M, Makse HA. Collective behavior in the spatial spreading of obesity. Sci Rep. 2012;2:454. doi: 10.1038/srep00454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Makse HA, Havlin S, Stanley HE. Modelling urban growth patterns. Nature. 1995;377:608–612. doi: 10.1038/377608a0. [DOI] [Google Scholar]
- 32.Clauset A, Shalizi CR, Newman MEJ. Power-Law Distributions in Empirical Data. SIAM Rev. 2009;4:661–703. doi: 10.1137/070710111. [DOI] [Google Scholar]
- 33.Erdös P, Rényi A. On random graphs, I. Publicationes Mathematicae (Debrecen) 1959;6:290–297. [Google Scholar]
- 34.Andrea Cavagna A, et al. Scale-free correlations in starling flocks. Proc Natl Acad Sci USA. 2010;26:11865–11870. doi: 10.1073/pnas.1005766107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Attanasi A, et al. Finite-size scaling as a way to probe near-criticality in natural swarms. Phys Rev Lett. 2014;23:238102. doi: 10.1103/PhysRevLett.113.238102. [DOI] [PubMed] [Google Scholar]
- 36.Korobova F, Ramabhadran V, Higgs HN. An actin-dependent step in mitochondrial fission mediated by the ER-associated formin INF2. Science. 2013;339:6118. doi: 10.1126/science.1228360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang C, et al. Dynamic tubulation of mitochondria drives mitochondrial network formation. Cell Res. 2015;25:1108–1120. doi: 10.1038/cr.2015.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lewis SC, Uchiyama LF, Nunnari J. ER-mitochondria contacts couple mtDNA synthesis with mitochondrial division in human cells. Science. 2016;353:6296. doi: 10.1126/science.aaf5549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Valm AM, et al. Applying systems-level spectral imaging and analysis to reveal the organelle interactome. Nature. 2017;546:162–167. doi: 10.1038/nature22369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mandelbrot, B. B. The fractal geometry of nature. (W. H. Freeman and Company, New York, 1983).
- 41.Theiler J. Estimating fractal dimension. J Opt Soc Am A. 1990;7:1055. doi: 10.1364/JOSAA.7.001055. [DOI] [Google Scholar]
- 42.Li, M. & Vitanyi, P. An Introduction to Kolmogorov Complexity and its Applications (Springer, New York, 1st edition, 1993).
- 43.Stanley, H. E. Introduction to phase transitions and critical phenomena. (Oxford University Press, Oxford, 1971).
- 44.Molinero C, Murcio R, Arcaute E. The angular nature of road networks. Sci Rep. 2017;7:4312. doi: 10.1038/s41598-017-04477-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cover, T. M. & Thomas, J. A. Elements Of Information Theory. Wiley Series in Telecommunications and Signal Processing (Wiley, 2006).
- 46.Barrat, A., Barthelemy, M. & Vespignani, A. Dynamical processes on complex networks. Cambridge Univesity Press (2008).
- 47.Strogatz SH. Exploring complex networks. Nature. 2001;410:268–276. doi: 10.1038/35065725. [DOI] [PubMed] [Google Scholar]
- 48.Xu J. Preparation, culture, and immortalization of mouse embryonic fibroblasts. Curr Protoc Mol Biol. 2005;70:1–8. doi: 10.1002/0471142727.mb2801s70. [DOI] [PubMed] [Google Scholar]
- 49.Baloh RH, Schmidt RE, Pestronk A, Milbrandt J. Altered axonal mitochondrial transport in the pathogenesis of Charcot-Marie-Tooth disease from mitofusin 2 mutations. J Neurosci. 2007;27:422–30. doi: 10.1523/JNEUROSCI.4798-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dirnberger M, Kehl T, Neumann A. NEFI: Network Extraction From Images. Sci Rep. 2015;5:15669. doi: 10.1038/srep15669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu X, Weaver D, Shirihai O, Hajnoczky G. Mitochondrial “kiss-and-run”: interplay between mitochondrial motility and fusion–fission dynamics. EMBO J. 2009;28:3074–3089. doi: 10.1038/emboj.2009.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gillespie DT. Exact stochastic simulation of coupled chemical reactions. J. Phys. Chem. 1977;81:2340–2361. doi: 10.1021/j100540a008. [DOI] [Google Scholar]
- 53.Sukhorukov VM, Meyer-Hermann M. Structural heterogeneity of mitochondria induced by the microtubule cytoskeleton. Scientific Reports. 2015;5:13924. doi: 10.1038/srep13924. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data are available upon request.