Abstract
Escherichia coli can hardly grow anaerobically on glycerol without exogenous electron acceptor. The formate-consuming methanogen Methanobacterium formicicum plays a role as a living electron acceptor in glycerol fermentation of E. coli. Wild-type and mutant E. coli strains were screened for succinate production using glycerol in a co-culture with M. formicicum. Subsequently, E. coli was adapted to glycerol fermentation over 39 rounds (273 days) by successive co-culture with M. formicicum. The adapted E. coli (19.9 mM) produced twice as much succinate as non-adapted E. coli (9.7 mM) and 62% more methane. This study demonstrated improved succinate production from waste glycerol using an adapted wild-type strain of E. coli with wild-type M. formicicum, which is more useful than genetically modified strains. Crude glycerol, an economical feedstock, was used for the cultivation. Furthermore, the increase in methane production by M. formicicum during co-culture with adapted E. coli illustrated the possibility of energy-saving effects for the fermentation process.
Electronic supplementary material
The online version of this article (10.1007/s10295-017-1994-0) contains supplementary material, which is available to authorized users.
Keywords: Long-term adaptation, Escherichia coli, Methanobacterium formicicum, Succinate, Crude glycerol
Introduction
Crude glycerol is an excellent feedstock candidate that is discarded as waste from biodiesel production [4, 5]. The waste glycerol from biodiesel production accounts for approximately 10% (w/w), or approximately 14 million tons [1, 19]. The bioconversion of glycerol to chemical building blocks is important to support the biofuel industry, as well as to lower production costs for succinate. Succinate is a multi-purpose platform chemical that can be produced from renewable biomass by microbes [8, 20, 25]. The global succinate market has experienced steady growth and reached 157.2 million USD and 58.5 kilotons in 2015 [9].
Various groups studied the succinate production from glycerol. The microbes known to produce succinate from glycerol are Anaerobiospirillum succiniciproducens [6], Pasteurellaceae family species and Mannheimia succiniciproducens [17], Actinobacillus succinogenes [24], Yarrowia lipolytica [29], Corynebacterium glutamicum [8], and Escherichia coli. Several studies have investigated the succinate production from glycerol using E. coli strains. Dharmadi et al. [4] focused on the pH-dependent mechanism of the E. coli fermentation of glycerol. They found that the production of CO2 from formate was required for increased glycerol consumption and succinate production. Blankschien et al. [2] improved succinate production by blocking the synthesis of competing by-products and the expression of Lactococcuslactis pyruvate carboxylase, which drives the generation of succinate from pyruvate production. Zhang et al. [30] engineered three gene mutations (pck*, ptsI −, pflB −) in E. coli ATCC 8739. The redirection of carbon flow in the engineered genes resulted in the maximum succinate yield. Soellner et al. [18] constructed a double mutant of E. coli (∆pykA, ∆pykF), from which a fast-growing strain was selected. In the selected strain, the third mutation in PEP carboxylase was found. Most recently, Li et al. [7] engineered an E. coli strain (ldhA −, pflB −, pck*) and performed two-stage fermentation that lead to an enhanced succinate production. In addition, A. succinogenes also enhanced succinate production from glycerol in the presence of dimethyl sulfoxide (DMSO) under controlled continuous microaerobic culture [16].
The mainstream approach of genetic engineering has generally adopted strategies for glucose fermentation, i.e., the elimination of competing pathways with the adjustment of the redox-balance and strengthening of the C3 to C4 branch, combined with process engineering to overcome the intrinsic redox imbalance [23]. E. coli strains rarely grow with glycerol in anaerobic conditions in the absence of an external electron acceptor: glycerol is imported by glycerol facilitator (GlpF), activated by glycerol kinase (GlpK) with ATP consumption, and oxidized to dihydroxyacetone phosphate (DHAP), whereby menaquinone (MQ) is reduced to menaquinol (MQH2) (Fig. S1). MQH2 emerges as every glycerol utilized, which must be recycled, therefore anaerobic growth on glycerol requires additional electron acceptors such as nitrate, DMSO, trimethylamine N-oxide (TMAO), or fumarate [22], and the amount of endogenous fumarate is not sufficient to recycle MQH2 to MQ. To overcome the redox imbalance of glycerol fermentation, Richter and Gescher [12] introduced the co-culture of E. coli and Methanobacterium formicicum, which uses formate in addition to H2–CO2 as its energy sources [15]. Glycerol fermentation and succinate production were higher in the co-cultures than in E. coli monocultures [12].
Our study screened co-cultures of several strains of E. coli (wild-type and genetically modified strains) with M. formicicum. We then adapted the E. coli to co-culture with M. formicicum in glycerol fermentation for 273 days. The long-term adapted E. coli developed in the present study demonstrated approximately twofold higher succinate levels than the non-adapted E. coli during crude glycerol fermentation.
Materials and methods
Strains and culture
E. coli K-12 strain MG1655 was used as the wild-type, and E. coli K-12 BW25113 gene knockout mutants were purchased from the National BioResource Project (National Institute of Genetics, Japan). M. formicicum JF-1 was obtained from the Leibniz Institute German Type Culture Collection (DSMZ, Germany). E. coli cells were anaerobically grown at 37 °C in Luria Broth (Affymetrix inc., USA), and kanamycin (30 µg/mL) was included for mutant E. coli strains. M. formicicum was anaerobically cultivated at 37 °C in DSMZ 119 medium. E. coli and M. formicicum were cultivated up to OD600 1.20 and 0.27, respectively.
The adaptation medium contained 3 mM KH2PO4, 1 mM K2HPO4, 4 mM NH4Cl, 5 mM KCl, 6 mM NaCl, 1 mM MgCl2, 21 mM HCO3Na, 5 mM CO3Na2, 0.2 mM of sodium ascorbate, 5.1 mM CaCl2, 10 mL NB trace mineral solution [3], 1.0 mL selenite-tungstate solution (13 mM NaOH, 17 μM Na2SeO3, and 12 μM Na2WO4), 10 mL vitamin solution (DSMZ, media 141), 0.1% (w/v) yeast extract, 1 mM cysteine, and 2 µM resazurin. The pH value was adjusted to 7.0. To adapt E. coli on glycerol fermentation with M. formicicum, the co-cultivation of E. coli and M. formicicum was continuously sub-cultured until the 39th round, where each co-cultivation took 7 days. The co-culture was performed in 100 mL adaptation medium with 70 mM glycerol in 250-mL rubber-stoppered infusion bottles and cultivated anaerobically under a sterile 80% H2 + 20% CO2 gas mixture at 37 °C. Twenty percent of the co-culture pre-stage was inoculated into fresh medium. To this, additional 20% M. formicicum (v/v) that was cultured in DSMZ 119 medium was inoculated.
The crude glycerol fermentation medium contained 1.5 mM KH2PO4, 2.3 mM K2HPO4, 9.4 mM NH4Cl, 2 mM MgSO4, 2 mM CaCl2, 38.8 mM NaCl, 0.01 mM FeSO4, 20 mM HCO3Na, trace element solution SL-10 (DSMZ, media 320), 10 mL vitamin solution (DSMZ, media 141), 0.1% (w/v) yeast extract, 0.2% (w/v) casitone, 1.7 mM cysteine, 1.3 mM Na2S, and 2 µM resazurin. Ten percent E. coli (v/v) and 30% M. formicicum (v/v) were inoculated in the medium with 80 mM crude glycerol (AEKYUNG PETROCHEMICAL CO. LTD., Korea) (Table S1), and cultivated at 37 °C for 4 days anaerobically under a sterile 80% N2 + 20% CO2 gas mixture. DMSO (50 mM) was used to test the effect of an electron acceptor.
HPLC analysis
Substrates and products in the supernatant of 1-mL cultures were analyzed using a HPLC Hitachi LaChrom Elite system (Hitachi High Technologies, Japan), consisting of an L-2130 pump, an L-2350 column oven, and an L-2200 auto-sampler. Ten-microliter samples were injected and separated using an Aminex HPX-87H ion-exclusion column (300 mm × 7.8 mm i.d., Bio-Rad, USA). The mobile phase was 4 mM H2SO4, which was pumped at a constant flow rate of 0.55 mL/min. The quantitative determination of substances was carried out using an L-2490 refractive index detector and an L-2400 UV detector (210 nm).
Methane determination by GC
One-milliliter sample from the air space of culture was analyzed using a 6500GC System (YL Instruments, Korea). Gas samples were injected and separated using a Carboxen 1006 PLOT column (30 m × 0.53 mm i.d., Sigma-Aldrich Co. LLC., USA). The quantitative determination of methane was carried out using a flame ionization detector (YL Instruments, Korea).
Cell growth analysis
Cell density of mixed M. formicicum and E. coli was determined at 600 nm wavelength using a UV/VIS spectrophotometer (X-ma1200, Human Corporation, Korea). The proliferation of M. formicicum and E. coli cells were quantitated by quantitative real time PCR (qRT-PCR), as described previously [28]. The M. formicicum primers, forward (5′- CGWAG GGAAG CTGTT AAGT-3′) and reverse (5′- TACCG TCGTC CACTC CTT-3′), and E. coli K-12 primers, forward (5′- ACTCC TACGG GAGGC AG-3′) and reverse (5′- GACTA CCAGG GTATC TAATC C-3′), were obtained from Cosmo Genetech (Korea); product sizes were 343 and 468 bp, respectively. Standard curves for the qRT-PCR were obtained by using plasmids that included partial 16S rRNA genes of M. formicicum M.o.H. and E. coli K-12, which were provided by the Environmental Bioprocess Engineering Laboratory (POSTECH, Korea). For standard curves of M. formicicum and E. coli, 16S rRNA gene copy numbers ranged from 2.6 × 109 to 2.6 × 102 and from 2.5 × 109 to 2.5 × 102, respectively. Logarithmic values of different 16S rRNA gene amounts were plotted against the threshold cycle (CT) number from each result. The linear range of the standard curve was selected based on the R 2 value of slopes, which were 0.9964 and 0.9945 for M. formicicum and E. coli, respectively. The average slope and average intercept were calculated, and the resulting equation was used to quantify 16S rRNA gene abundance in samples. CT values of each sample were compared to the corresponding standard curve. Genomic DNA was extracted using NucleoSpin Microbial DNA kits (Macherey–Nagel, Germany) and used as a template for qRT-PCR. Total reaction volume was 20 μL included 400 nM each primer in SensiFAST™ SYBR No-ROX Mix (Bioline, USA). The qRT-PCR analysis used 40 cycles of denaturation at 95 °C for 10 s, annealing at 60 °C for 20 s, and extension at 72 °C for 20 s and was performed in a Corbett Research Rotor-Gen RG-3000A (Qiagen, Germany) and the Rotor-Gene software, version 6.1.93.
Statistics
Statistical analyses were performed using PASW Statistics 18. Unpaired two-tailed student’s t tests were performed to analyze the data. Statistical significance was defined as P < 0.05.
Results and discussion
Co-culture of wild-type or mutant E. coli with M. formicicum
In the co-culture of wild-type E. coli with M. formicicum, glycerol consumption and succinate production were highly improved by 12-fold and 8-fold, respectively, in comparison with the single cultivation (Table 1). Accordingly, other fermentation products were also increased, but formate was used up by M. formicicum (Table 1). To select the most suitable E. coli strain for succinate production during co-fermentation, mutant strains with specific gene deletions (pflB, adhE, pta, or ackA) involved in each competitive pathway against succinate production were cultivated with M. formicicum under the conditions of glycerol fermentation without exogenous electron acceptors (Table S2). The pflB (pyruvate formate lyase) mutant, for which all pathways other than that of succinate production were blocked, served as a negative control. The adhE (alcohol dehydrogenase) mutant did not grow at all in either single- or co-culture, indicating that ethanol production is an unavoidable step. In pta (phosphate acetyltransferase) and ackA (acetate kinase) mutants of E. coli, the acetate production was blocked, with the co-cultures of M. formicicum designed to produce succinate without both formate and acetate. In the co-culture, the pta or ackA E. coli mutants could grow to some extent, but the glycerol consumption was low, and the succinate production level did not exceed half that of wild-type E. coli co-culture (Table S2).
Table 1.
Glycerol fermentation in single- and co-culture of Escherichia coli with Methanobacterium formicicum
| Consumed (mM) | Produced (mM) | Cell number (mL−1) | |||||
|---|---|---|---|---|---|---|---|
| Glycerol | Succinate | Formate | Acetate | Ethanol | E. coli | M. formicicum | |
| Single culture | |||||||
| E. coli | 4.6 ± 1.6 | 1.0 ± 0.1 | 10.7 ± 3.5 | 1.7 ± 0.9 | 9.6 ± 2.0 | 3.4 × 108 ± 0.4 × 108 | ND |
| Co-culture | |||||||
| E. coli
M. formicicum |
53 ± 12.8a | 8.0 ± 0.7a | 0 | 7.3 ± 1.0 | 47.2 ± 16.1 | 1.1 × 109 ± 0.2 × 109 | 2.1 × 108 ± 1.2 × 108 |
Product analysis and cell growth were determined after 7 days of fermentation. Values report means ± standard deviations for three replicates
aValue means a significant difference between single- and co-culture (unpaired samples t test, P < 0.05). ND, not determined
Collectively, co-culture of E. coli mutants with M. formicicum did not efficaciously improve succinate production. Among E. coli strains, we found that wild-type was the best strain for co-culturing with M. formicicum. For this reason, wild-type E. coli was performed into long-term adaptation for succinate production. Moreover, the use of wild-type (non-GMO) microbes is an incomparably large advantage for industrial applications.
Crude glycerol fermentation by long-term adapted or non-adapted E. coli
Without an exogenous electron acceptor, E. coli hardly ferment glycerol. The co-cultivation of E. coli with M. formicicum allows E. coli to promote glycerol fermentation (Table 1), as reported by Richter and Gescher [12]. During co-cultivation for 7 days, E. coli fermented 53 mM of glycerol and produced 8 mM of succinate, while formate was completely consumed by M. formicicum (Table 1). This co-cultivation of E. coli with M. formicicum was adapted via 39 successive rounds. The adaptation lasted 273 days in total, in which each co-cultivation was carried out in batch culture for 1 week, and successively inoculated.
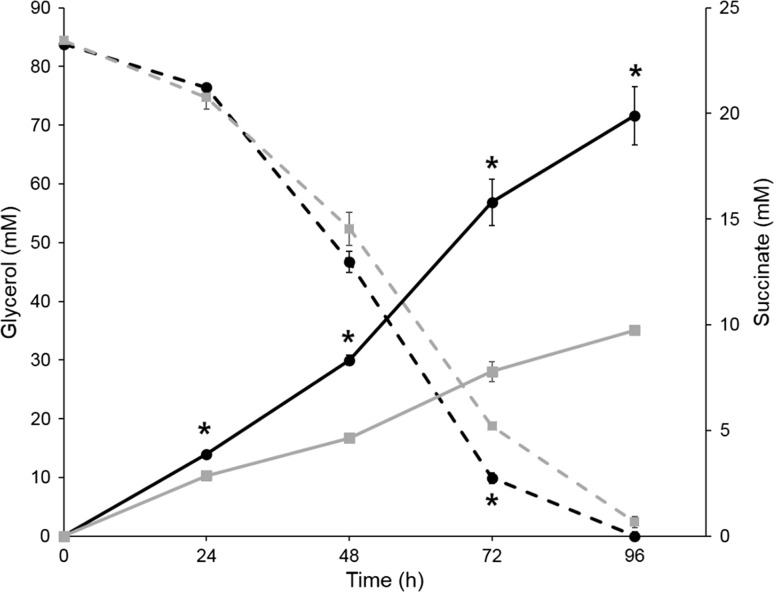
The E. coli adapted to glycerol fermentation exhibited a two-fold increase in succinate production (19.9 mM) over non-adapted E. coli (9.7 mM succinate) during crude glycerol fermentation for 96 h (4 days) (Fig. 1; Table 2). Twenty-four percent of the PEP (from 83.8 mM glycerol) was metabolized and reduced to succinate in the adapted E. coli co-culture, whereas only 12% of the PEP (from 82 mM glycerol) was reduced to succinate in the non-adapted E. coli co-culture (Fig. 1; Table 2; Fig. S1). Methane production was higher in the adapted co-culture (206978 ppm) than in the non-adapted (127552 ppm) (Table 2). Methane production is often followed by improved growth or substrate consumption rates of the primary carbon source consumers and methane can easily be collected for use as energy in fermentation processes [12, 27]. The adapted E. coli (14.0 mM) also exhibited improved succinate production in the presence of DMSO compared with non-adapted E. coli (7.2 mM), but co-culture with M. formicicum produced even more succinate (19.9 mM) (Table 2). Co-culture with M. formicicum permitted a higher crude glycerol consumption by E. coli than that of culture with DMSO, which indicated that formate consumption by M. formicicum, a living electron acceptor, is more advantageous than for the supply of the electron acceptor DMSO (Table 2). All product analysis data collected over the course of fermentation, including the pH values, are shown in Tables S3 and S4.
Fig. 1.
Succinate production from crude glycerol fermentation during co-culture of Escherichia coli with Methanobacterium formicicum. Solid lines represent succinate production, and dotted lines represent glycerol consumption by adapted (black circles) and non-adapted (gray squares) E. coli. *Value for the adapted E. coli was significantly different from that for the non-adapted E. coli (unpaired samples t test, P < 0.05). Plotted points report the means, and error bars report the standard deviations for independent samples taken in triplicate
Table 2.
Fermentative characteristics by adapted Escherichia coli on crude glycerol
| 39th E. coli
M. formicicum |
1st E. coli
M. formicicum |
39th E. coli
DMSO |
1st E. coli
DMSO |
39th E. coli
M. formicicum DMSO |
1st E. coli
M. formicicum DMSO |
|
|---|---|---|---|---|---|---|
| Fermentation | ||||||
| Consumption (mM) | ||||||
| Glycerol | 83.8 ± 0.8 | 82.0 ± 1.0 | 46.3 ± 1.1 | 54.1 ± 1.1 | 78.5 ± 1.2 | 81.0 ± 1.4 |
| Production (mM) | ||||||
| Succinate | 19.9 ± 1.4a | 9.7 ± 0.2 | 14.0 ± 0.2a | 7.2 ± 0 | 19.8 ± 0.5a | 12.6 ± 0.4 |
| Acetate | 6.1 ± 1.8 | 5.3 ± 0.8 | 2.7 ± 0a | 1.5 ± 0.1 | 7.0 ± 0.4 | 4.6 ± 1.4 |
| Ethanol | 56.9 ± 3.5a | 74.8 ± 3.0 | 30.8 ± 0.1a | 52.2 ± 1.6 | 47.3 ± 2.0a | 67.5 ± 5.7 |
| Formate | 0 | 0 | 7.6 ± 0.3a | 14.4 ± 1.2 | 0 | 0 |
| Methane (ppm) | 206977.7 ± 72056.8 | 127552.0 ± 11659.79 | 105.7 ± 81.5 | 528.0 ± 454.5 | 122956.7 ± 84541.3 | 62341.3 ± 3672.3 |
| Growth | ||||||
| Cell density (OD600) | 1.33 ± 0.03 | 1.31 ± 0.02 | 1.01 ± 0.01a | 1.10 ± 0 | 1.32 ± 0.02 | 1.41 ± 0.03 |
| E. coli cell number (mL−1) | 8.9 × 108 ± 7.2 × 107 | 8.5 × 108 ± 1.5 × 108 | 6.3 × 108 ± 7.0 × 107a | 7.8 × 108 ± 5.5 × 107 | 8.6 × 108 ± 4.0 × 107 | 9.5 × 108 ± 5.5 × 107 |
| M. formicicum cell number (mL−1) | 7.8 × 107 ± 1.3 × 107 | 1.1 × 108 ± 1.7 × 107 | ND | ND | 6.3 × 107 ± 4.1 × 106 | 5.6 × 107 ± 1.2 × 107 |
E. coli was adapted to Methanobacterium formicicum by 39 successive rounds of cultivation on glycerol. The adapted (39th-round) or non-adapted (1st-round) E. coli was cultivated for 96 h on crude glycerol with M. formicicum or DMSO
Values report means ± standard deviations for three replicates
ND not determined, 39th E. coli adapted E. coli, 1st E. coli non-adapted E. coli
aValue means a significant difference between co-culture groups of adapted and non-adapted E. coli (unpaired samples t test, P < 0.05)
Under anaerobic conditions, E. coli cannot grow with glycerol as its sole carbon and energy source due to the metabolic dilemma of redox-balancing and energy acquisition (Fig S1). During conversion of glycerol to PEP, MQH2 and NADH2 are generated. For redox balancing, PEP could be reduced to succinate whereby MQH2 and NADH2 would be re-oxidized, but no ATP is generated in this pathway. For energy acquisition, PEP should be also degraded over pyruvate to acetate, formate, ethanol, or lactate. NADH2 is re-oxidized in the ethanol or lactate production, but this pathway requires additional electron acceptors like fumarate, DMSO, TMAO, or nitrate, of which reduction is coupled with oxidation of MQH2 [22]. Therefore, glycerol fermentation by E. coli alone was very slow and showed low levels of products (Table 1). The interspecies transfer of formate from E. coli to M. formicicum and consumption of formate by M. formicicum improved the glycerol fermentation by E. coli. Formate is derived from pyruvate in nonoxidative cleavage by PFL (pyruvate formate lyase), and reducing equivalents of the reaction remain in the formate [13]. Therefore, formate metabolism is a critical step for adjusting redox balance in fermentation [14]. In the absence of an exogenous electron acceptor, the formate channel FocA exports formate. As the external pH decreases, formate is re-imported by FocA, undergoes disproportionation into CO2 and H2 by cytoplasmic orientated formate hydrogenlyase (FHL), and the excess redox equivalents are released as H2 [11]. FHL complex is composed of formate dehydrogenase H (FDH-H) [HCOO− → CO2 + H+ + 2e−, E′0 = − 432 mV] and hydrogenase 3 [2H+ + 2e− → H2, E′0 = − 414 mV] [10, 21]. Moreover, as the affinity of FDH-H of FHL to formate is very low (K m = 26 mM) [14], therefore the FHL reaction was not sufficient to solve the redox imbalance of glycerol fermentation. M. formicicum also possesses a FocA-similar formate channel FdhC [26]. Therefore, in co-culture, formate exported by E. coli is imported into M. formicicum and is quickly used, which re-adjusts the equilibrium in the direction of fermentation. M. formicicum uses both H2 and formate as electron donors [15].
In conclusion, this study successfully adapted an E. coli strain for succinate production from waste glycerol by 39 successive rounds (273 days) of co-culture of E. coli and M. formicicum. The adapted E. coli produced twice amount of succinate in co-culture in comparison with the non-adapted E. coli, and the methane production by M. formicicum increased by 62%, whereas the glycerol consumption and cell growth were not increased, and the ethanol production decreased by 24%. We, therefore, speculated that the formate transfer from E. coli to M. formicicum became more efficient during the adaptation process, whereby the reduction step to ethanol production was decreased and the C4-branch enzymes, including PEP carboxylase, malate dehydrogenase, fumarase, and fumarate reductase, were upregulated. The basis of phenotypic changes should be further investigated by genome and transcriptome analyses.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgments
This research was supported by the Basic Research Laboratory (BRL) through a National Research Foundation (NRF) grant funded by the government of Korea (MSIP) (2015R1A4A1041997). We are grateful to Prof. Seokhwan Hwang (POSTECH, Korea) for supplying plasmids for qRT-PCR of M. formicicum and E. coli.
Footnotes
Electronic supplementary material
The online version of this article (10.1007/s10295-017-1994-0) contains supplementary material, which is available to authorized users.
References
- 1.Anand P, Saxena RK. A comparative study of solvent-assisted pretreatment of biodiesel derived crude glycerol on growth and 1,3-propanediol production from Citrobacter freundii. New Biotechnol. 2012;29:199–205. doi: 10.1016/j.nbt.2011.05.010. [DOI] [PubMed] [Google Scholar]
- 2.Blankschien MD, Clomburg JM, Gonzalez R. Metabolic engineering of Escherichia coli for the production of succinate from glycerol. Metab Eng. 2010;12:409–419. doi: 10.1016/j.ymben.2010.06.002. [DOI] [PubMed] [Google Scholar]
- 3.Coppi MV, Leang C, Sandler SJ, Lovley DR. Development of a genetic system for Geobacter sulfurreducens. Appl Environ Microbiol. 2001;67:3180–3187. doi: 10.1128/AEM.67.7.3180-3187.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dharmadi Y, Murarka A, Gonzalez R. Anaerobic fermentation of glycerol by Escherichia coli: a new platform for metabolic engineering. Biotechnol Bioeng. 2006;94:821–829. doi: 10.1002/bit.21025. [DOI] [PubMed] [Google Scholar]
- 5.Kenar JA. Glycerol as a platform chemical: sweet opportunities on the horizon? Lipid Technol. 2007;19:249–253. doi: 10.1002/lite.200700079. [DOI] [Google Scholar]
- 6.Lee PC, Lee WG, Lee SY, Chang HN. Succinic acid production with reduced by-product formation in the fermentation of Anaerobiospirillum succiniciproducens using glycerol as a carbon source. Biotechnol Bioeng. 2001;72:41–48. doi: 10.1002/1097-0290(20010105)72:1<41::AID-BIT6>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 7.Li Q, Wu H, Li Z, Ye Q. Enhanced succinate production from glycerol by engineered Escherichia coli strains. Bioresour Technol. 2016;218:217–223. doi: 10.1016/j.biortech.2016.06.090. [DOI] [PubMed] [Google Scholar]
- 8.Litsanov B, Brocker M, Oldiges M, Bott M. Succinic acid. In: Bisaria VS, Kondo A, editors. Bioprocessing of renewable resources to commodity bioproducts. New Jersey: Wiley; 2014. pp. 435–472. [Google Scholar]
- 9.Markets and Markets (2016) Top Market Reports. In: Succinic acid market by type (bio-based, petro-based), application (polyurethane, resins, coatings and pigments, pharmaceuticals, plasticizers, food and beverage, PBS/PBST, solvents and lubricants, de-icer solutions, personal care, and others), and by region—global forecast to 2021. Available via marketsandmarkets.com. http://www.marketsandmarkets.com/Market-Reports/succinic-acid-market-402.html. Accessed 1 Jul 2017
- 10.McDowall JS, Murphy BJ, Haumann M, Palmer T, Armstrong FA, Sargent F. Bacterial formate hydrogenlyase complex. Proc Natl Acad Sci USA. 2014;111:E3948–E3956. doi: 10.1073/pnas.1407927111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pinske C, Sawers RG. Anaerobic formate and hydrogen metabolism. EcoSal Plus. 2016 doi: 10.1128/ecosalplus.esp-0011-2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Richter K, Gescher J. Accelerated glycerol fermentation in Escherichia coli using methanogenic formate consumption. Bioresour Technol. 2014;162:389–391. doi: 10.1016/j.biortech.2014.04.011. [DOI] [PubMed] [Google Scholar]
- 13.Sawers RG, Clark DP. Fermentative pyruvate and acetyl-coenzyme a metabolism. EcoSal Plus. 2004 doi: 10.1128/ecosalplus.3.5.3. [DOI] [PubMed] [Google Scholar]
- 14.Sawers RG. Formate and its role in hydrogen production in Escherichia coli. Biochem Soc Trans. 2005;33:42–46. doi: 10.1042/BST0330042. [DOI] [PubMed] [Google Scholar]
- 15.Schauer NL, Ferry JG. Metabolism of formate in Methanobacterium formicicum. J Bacteriol. 1980;142:800–807. doi: 10.1128/jb.142.3.800-807.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schindler BD, Joshi RV, Vieille C. Respiratory glycerol metabolism of Actinobacillus succinogenes 130Z for succinate production. J Ind Microbiol Biotechnol. 2014;41:1339–1352. doi: 10.1007/s10295-014-1480-x. [DOI] [PubMed] [Google Scholar]
- 17.Scholten E, Dägele D. Succinic acid production by a newly isolated bacterium. Biotechnol Lett. 2008;30:2143–2146. doi: 10.1007/s10529-008-9806-2. [DOI] [PubMed] [Google Scholar]
- 18.Soellner S, Rahnert M, Siemann-Herzberg M, Takors R, Altenbuchner J. Evolution of pyruvate kinase-deficient Escherichia coli mutants enables glycerol-based cell growth and succinate production. J Appl Microbiol. 2013;115:1368–1378. doi: 10.1111/jam.12333. [DOI] [PubMed] [Google Scholar]
- 19.Stasiak-Różańska L, Błażejak S, Gientka I, Bzducha-Wróbel A, Lipińska E. Utilization of a waste glycerol fraction using and reusing immobilized Gluconobacter oxydans ATCC 621 cell extract. Electron J Biotechnol. 2017;27:44–48. doi: 10.1016/j.ejbt.2017.03.003. [DOI] [Google Scholar]
- 20.Thakker C, Martı´nez I, San KY, Bennett GN. Succinate production in Escherichia coli. Biotechnol J. 2012;7:213–224. doi: 10.1002/biot.201100061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thauer RK, Jungermann K, Decker K. Energy conservation in chemotrophic anaerobic bacteria. Bacteriol Rev. 1977;41:100–180. doi: 10.1128/br.41.1.100-180.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Unden G, Bongaerts J. Alternative respiratory pathways of Escherichia coli: energetics and transcriptional regulation in response to electron acceptors. Biochim Biophys Acta. 1997;1320:217–234. doi: 10.1016/S0005-2728(97)00034-0. [DOI] [PubMed] [Google Scholar]
- 23.Unden G, Strecker A, Kleefeld A, Kim OB. C4-Dicarboxylate utilization in aerobic and anaerobic growth. EcoSal Plus. 2016 doi: 10.1128/ecosalplus.esp-0021-2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vlysidis A, Binns M, Webb C, Theodoropoulos C. Glycerol utilisation for the production of chemicals: conversion to succinic acid, a combined experimental and computational study. Biochem Eng J. 2011;58–59:1–11. doi: 10.1016/j.bej.2011.07.004. [DOI] [Google Scholar]
- 25.Werpy T, Petersen G. Top value added chemicals from biomass. Washington DC: US Department of Energy; 2004. [Google Scholar]
- 26.White WB, Ferry JG. Identification of formate dehydrogenase-specific messenger-RNA species and nucleotide-sequence of the fdhC gene of Methanobacterium formicicum. J Bacteriol. 1992;174:4997–5004. doi: 10.1128/jb.174.15.4997-5004.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang ST, Tang IC. Methanogenesis from lactate by a co-culture of Clostridium formicoaceticum and Methanosarcina mazei. Appl Microbiol Biotechnol. 1991;35:119–123. doi: 10.1007/BF00180648. [DOI] [Google Scholar]
- 28.Yu Y, Lee C, Kim J, Hwang S. Group-specific primer and probe sets to detect methanogenic communities using quantitative real-time polymerase chain reaction. Biotechnol Bioeng. 2005;89:670–679. doi: 10.1002/bit.20347. [DOI] [PubMed] [Google Scholar]
- 29.Yuzbashev TV, Yuzbasheva EY, Sobolevskaya TI, Laptev IA, Vybornaya TV, Larina AS, Matsui K, Fukui K, Sineoky SP. Production of succinic acid at low pH by a recombinant strain of the aerobic yeast Yarrowia lipolytica. Biotechnol Bioeng. 2010;107:673–682. doi: 10.1002/bit.22859. [DOI] [PubMed] [Google Scholar]
- 30.Zhang X, Shanmugam KT, Ingram LO. Fermentation of glycerol to succinate by engineered strains of Escherichia coli. Appl Environ Microbiol. 2010;76:2397–2401. doi: 10.1128/AEM.02902-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.