Abstract
Introduction
Pheochromocytomas are neuroendocrine tumors of the adrenal glands. Up to 40% of the cases are caused by germline mutations in one of at least 15 susceptibility genes, making them the human neoplasms with the highest degree of heritability. Recurrent somatic alterations are found in about 50% of the more common sporadic tumors with NF1 being the most common mutated gene (20–25%). In many sporadic tumors, however, a genetic explanation is still lacking.
Materials and methods
We investigated the genomic landscape of sporadic pheochromocytomas with whole-exome sequencing of 16 paired tumor and normal DNA samples and extended confirmation analysis in 2 additional cohorts comprising a total of 80 sporadic pheochromocytomas.
Results
We discovered on average 33 non-silent somatic variants per tumor. One of the recurrently mutated genes was FGFR1, encoding the fibroblast growth factor receptor 1, which was recently revealed as an oncogene in pediatric brain tumors. Including a subsequent analysis of a larger cohort, activating FGFR1 mutations were detected in three of 80 sporadic pheochromocytomas (3.8%). Gene expression microarray profiling showed that these tumors clustered with NF1-, RET,- and HRAS-mutated pheochromocytomas, indicating activation of the MAPK and PI3K-AKT signal transduction pathways.
Conclusion
Besides RET and HRAS, FGFR1 is only the third protooncogene found to be recurrently mutated in pheochromocytomas. The results advance our biological understanding of pheochromocytoma and suggest that somatic FGFR1 activation is an important event in a subset of sporadic pheochromocytomas.
Electronic supplementary material
The online version of this article (10.1007/s00268-017-4320-0) contains supplementary material, which is available to authorized users.
Introduction
Pheochromocytomas (PCCs) are tumors arising from the neural crest-derived cells of the adrenal medulla, and paragangliomas (PGLs) are their extra-adrenal counterparts. They may cause hypertension due to overproduction of catecholamines, with symptoms including frequent episodes of headache, palpitations and sweating, and an increased risk of cardiovascular disease [1]. PCCs and PGLs have a highly diverse genetic background; up to 40% of the cases are caused by germline mutations in one of at least 15 so far identified susceptibility genes, making them the human neoplasms with the highest degree of heritability [1, 2]. More and more is also known regarding alterations in the sporadic tumors. During the past few years, somatic mutations have been revealed in several of the genes known from the hereditary cases, most frequently in NF1 [3, 4]. HRAS was found as the first gene with recurrent somatic mutations that is not associated with hereditary PCCs (6), and more recently, frequent somatic mutations were also discovered in ATRX [5, 6]. Initially, gene expression profiling showed that PCCs could be divided into at least two different clusters based on their expression signature [7]. The first cluster contains tumors with VHL, SDHx, and EPAS1 mutations and displays mRNA expression associated with the hypoxic response. The second cluster contains tumors with RET, NF1, TMEM127, and MAX mutations and is enriched for mRNA expression related to activation of kinase signaling cascades. Recently, third and fourth clusters have been described [8]. The third cluster contains tumors with somatic MAML3 fusion genes, CSDE1 and ATRX mutations leading to activation of the Wnt signaling. The fourth cluster contains genes known to be adrenal cortex markers (CYP11B2, CYP21A2, and STAR) and is associated with the presence of cortical cells and termed cortical admixture. Despite this remarkable progress in the field, a large portion of the sporadic tumors still remains without any known genetic driver event [9, 10]. We therefore hypothesized that there are additional targets of recurrent somatic mutations in PCCs. To examine this, we performed whole-exome sequencing of paired tumor and normal DNA from patients with sporadic PCC and followed up the results by studying whole-transcriptome gene expression.
Materials and methods
Patients and samples
The discovery cohort of this study consisted of 16 PCCs with corresponding peripheral blood samples from 16 patients operated at Linköping University Hospital, Sweden (Table S1). Findings were further investigated in 15 remaining PCCs from Sweden and Norway with known mutational status in PCC-associated genes (Table S1), followed by 49 additional sporadic PCCs from Hôpital de Brabois, Nancy, France (Table S2). All patients had been diagnosed with sporadic PCC due to absence of family history and syndromic presentation. Two sporadic PCCs and ten tumors from patients with hereditary syndromes were also available and tested (Table S2). Informed consent was obtained from all participants, and the local ethic committees approved the study.
Methods
All methods are described in the supplementary material.
Results
The mutation landscape of pheochromocytomas
We performed whole-exome sequencing of 16 tumors with paired constitutional DNA from patients with apparently sporadic disease (Table S1). Six of the cases had previously tested negative for mutations in known PCC-related genes [4, 9]. Sequencing generated an average depth of 119× for tumor samples and 97× for blood samples, with a depth of at least 10× in 97.9 and 97.5% of the exome, respectively. When investigating somatic mutations in the previously known genes, we detected truncating NF1 mutations in three of the tumors with previously unknown mutation status, as could be expected in the light of earlier studies [3, 4]. In addition, somatic mutations in the previously known genes VHL, RET, MAX, and HRAS were found in one tumor each (Table S3), in agreement with frequencies in previous reports (Table 1) [7, 10–13]. Despite the premises, we did not find any ATRX mutations [5, 6]. Moreover, we investigated germline alterations and detected a germline frameshift mutation in SDHB in one case for which the mutation status was previously unknown (Table S3). Germline missense variants were also found in EGLN2, KIF1Bβ, NF1, and SDHD, but bioinformatic analysis suggests that these are either benign or of unknown significance for the disease (Table S3).
Table 1.
Frequency of somatic mutations in pheochromocytomas
| Gene | NF1 (%) | KIF1Bβ (%) | ATRX (%) | HRAS (%) | VHL (%) | RET (%) | EPAS1 (%) | MAML3 (%) | CSDE1 (%) | MAX (%) | FGFR1 (%) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Literature | 9–48 [3, 4, 8, 41] | 1.5–29.6 [10, 41] | 5–12.6 [5, 6] | 6.5–10 [8, 13, 42, 43] | 1.4–7.9 [7, 8, 10, 42] | 2.2–5.6 [7, 8, 10, 42] | 2.2–5 [8, 10, 42] | 5 [8] | 2 [8] | 1.5 [10] | 0.5 [32] |
| Present study | 18.8 | 0 | 0 | 6.3 | 6.3 | 6.3 | 0 | n.d | n.d | 3.8 | 6.3 |
n.d not determined
We detected totally 542 non-silent somatic variants in the cohort of 16 tumors. The median number per tumor was 33 non-silent somatic variants, with a range from 16 to 60 per tumor. Most of the detected alterations were unique to one tumor, but some genes harbored non-silent somatic mutations in two or more samples, including the NF1 gene as could be expected. The recurrently altered genes also included FGFR1, in which activating point mutations were recently discovered in pediatric pilocytic astrocytoma [14].
Recurrent hotspot FGFR1 mutations
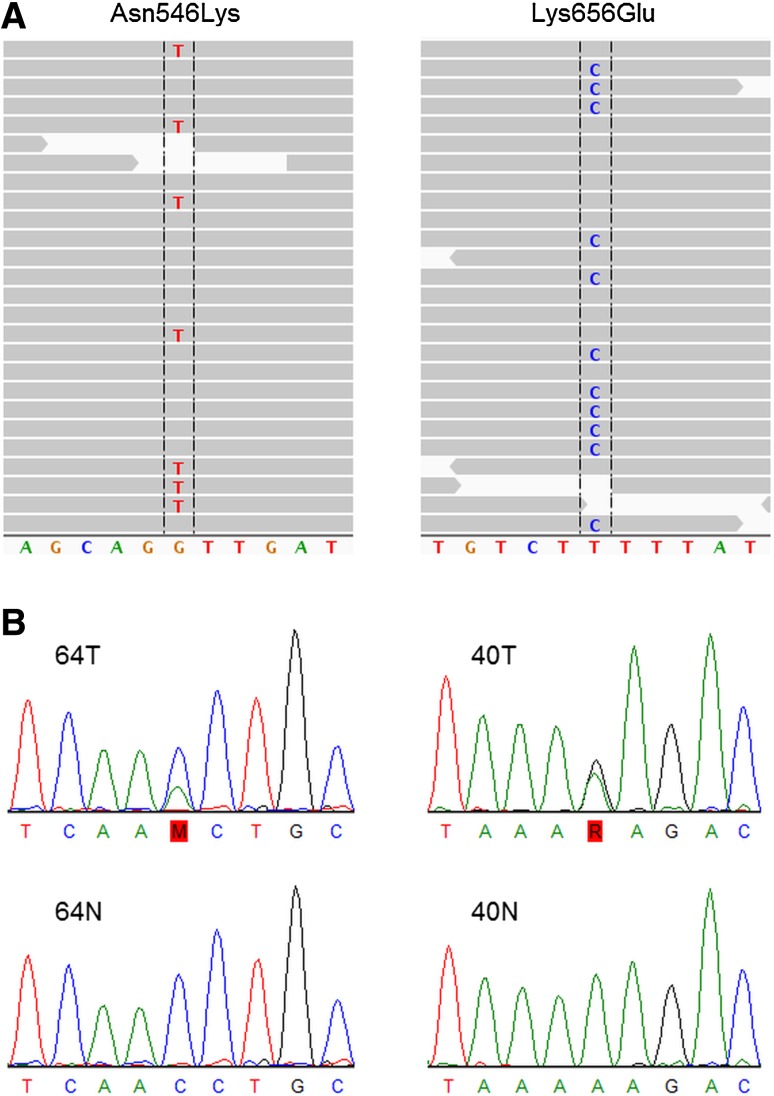
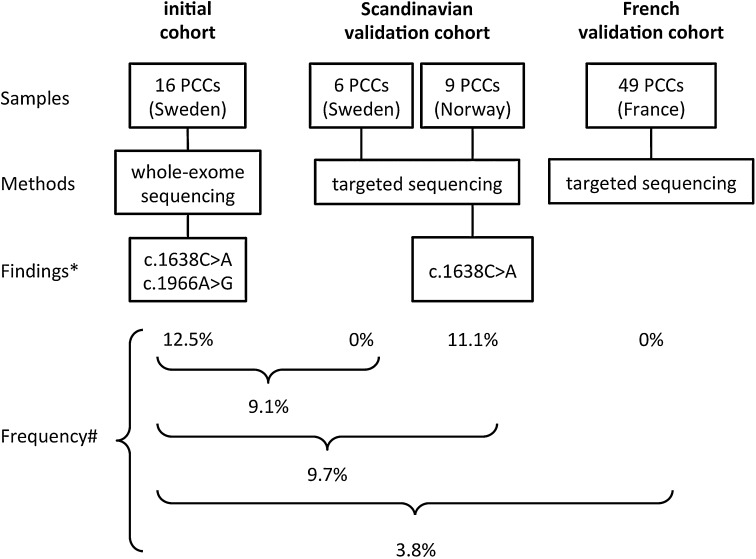
We detected two somatic FGFR1 mutations, Asn546Lys (c.1638C>A) and Lys656Glu (c.1966A>G) (Figs. 1 and 2). They exactly correspond to the two hotspot sites for activating mutations recently discovered in pediatric brain tumors [14]. To investigate the prevalence of FGFR1 mutations in sporadic PCCs, we performed Sanger sequencing of the three most common hotspots in FGFR1 (Fig. S1) in 15 remaining tumors from the Scandinavian cohort (Table S1) and detected one additional Asn546Lys mutation (Fig. 2). Subsequent analysis of 49 sporadic PCCs from France (Table S2) did not reveal any additional mutations (Fig. 2). In total, the combined cohort consisted of 80 apparently sporadic PCCs, of which three tumors (samples #40, #50, #64; 3.8%) carried FGFR1 mutations (Fig. 2). The cases with somatic FGFR1 mutations were two females and one male with a mean age of 66 years (Table 2). One of the FGFR1-mutated tumors also had a somatic MAX mutation, whereas the other two did not have any mutations in known PCC-associated genes (Fig. S2). Apart from the main cohort of sporadic PCCs, we tested also two sporadic PGLs and ten hereditary PCCs (four MEN2B, two VHL, three PGL1, and one PGL4), yet no FGFR1 mutations were detected. In addition, we searched samples with available RNA (Table S4) for a known fusion gene involving FGFR1, i.e., FGFR1-TACC1 [15, 16], which is known to play an oncogenic role in glioblastomas, but no gene fusions were found in PCCs.
Fig. 1.
Somatic hotspot mutations in FGFR1. a Overview of next-generation sequencing reads from the mutated sites in Integrative Genomics Viewer. Read bases that match the hg19 reference are displayed in gray, and mismatches are indicated with color coded alternate alleles (FGFR1 is oriented on the reverse strand; hence, the sequence is here the reverse complement to the transcribed sequence). b Validation of the mutations in the two tumor samples (64T and 40T) with Sanger sequencing (in the direction of transcription) and the corresponding sequences from blood samples
Fig. 2.
Frequency of FGFR1 mutations indifferent cohorts. Flow diagram simplifying the process of initial discovery and validation of somatic FGFR1 mutations in sporadic pheochromocytomas (PCCs). * With regard to somatic FGFR1 mutations; # of somatic FGFR1 mutations
Table 2.
Clinical and genetic data for cases with somatic FGFR1 mutations
| Case ID | Gender | Age (years) | Tumor size (mm) | Malignancy | Mutationa | Protein alteration | Copy number |
|---|---|---|---|---|---|---|---|
| 40 | Female | 63 | 32 | Benign | c.1638C>A | Asn546Lys | Gain |
| 50 | Female | 80 | 10 | Benign | c.1966A>G | Lys656Glu | Normal |
| 64 | Male | 56 | 32 | Benign | c.1638C>A | Asn546Lys | Normal |
aMutations were annotated according to the Ensembl transcript ENST00000447712
Copy number gains at the FGFR1 locus
Copy number analysis showed a gain of FGFR1 in four of 80 sporadic PCCs, one of which also had a FGFR1 mutation (Fig. S3). SNP microarray data from a previous study [4] were used to verify these findings where possible. For the sample with both FGFR1 copy gain and FGFR1 mutation, SNP microarray data were available and amplification of the FGFR1 locus was confirmed and microarray data showed that it was part of a ~2.5 Mb amplification unit on chromosome 8p (Fig. S4). Copy number loss in the FGFR1 region was seen in 11/91 sporadic PCCs (Table S4), none of which carried any FGFR1 mutations. For three of four cases with loss and where microarray data were also available, the loss could be seen to cover large parts of chromosome 8, suggesting that loss of FGFR1 in these cases is a passenger event (for the fourth sample, a loss was not detectable using the array technique, possibly because of a too low marker resolution).
Gene expression profiling of FGFR1-mutated tumors
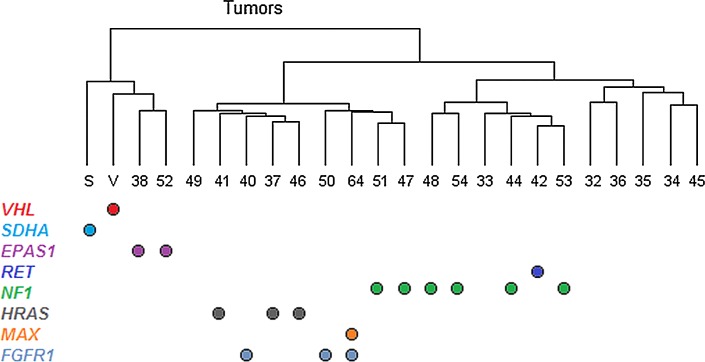
Global gene expression analysis showed that the three PCCs with FGFR1 mutations clustered in “cluster 2” together with tumors that contained mutations in RET, NF1, and HRAS. This indicates that FGFR1 mutations are associated with activation of kinase signal transduction pathways, including the MAPK and PI3K-AKT pathways [9]. Among all the genes that passed quality control, there were no genes that differed significantly in expression between tumors with and without FGFR1 mutations after correction for multiple testing. The reasons for this may be that the FGFR1-mutated tumors have a gene expression profile very similar to the other tumors in “cluster 2” and that the number of tested FGFR1-mutated tumors was too small to detect minor differences in expression patterns. There was no association between FGFR1 mutations and FGFR1 gene expression, as would be expected since the mutations are thought to have their effect on the protein level. However, copy gain of FGFR1 was associated with an enhanced FGFR1 gene expression (Fig. S5), suggesting a gene-dose effect.
Analysis of other fibroblast growth factor receptors
To further extend the knowledge of fibroblast growth factor receptors in PCCs, exons containing hotspot regions in FGFR2 and FGFR3 (Fig. S1) were investigated for mutations in all samples (sporadic and hereditary), but no mutations were detected. A very rare polymorphism in FGFR3 (rs17881656, Phe384Leu) was detected in three cases (two of which were hereditary) and was also present in normal DNA of the two patients where blood was available (Table S4). The variant was predicted as benign using the Polyphen-2, SIFT, and PROVEAN algorithms [17–19]. Allele frequency was then analyzed in a regional healthy control population (n = 739), as well as checked in the Swedish 1000 Genomes population (n = 1000; https://swegen-exac.nbis.se/) and the ExAC database (http://exac.broadinstitute.org/). The C-allele frequency was 1.6% in the PCC-cohort and 0.34, 0.4 and 0.31% in the regional control cohort, the Swedish 1000 Genome database and the ExAC database, respectively. Two-tailed Fisher’s exact test showed a borderline significant difference when compared to the Swedish control population (p = 0.049), the SweGen database (p = 0.07), and the ExAC database (p = 0.02). A rare germline variant in FGFR1 was detected in one case (c. 381T>G, Asp127Glu, rs750795714) and reported in only 2 out of 120,880 alleles in the ExAC database. It was outside of the mutational hotspots and was predicted as benign by PolyPhen. The known gene fusion involving FGFR3 (FGFR3-TACC3 [15, 16]) was also investigated, but no gene fusions were discovered. Available DNA microarray data from 21 tumors did not reveal any copy number alterations in any of the FGFR1-3 genes other than FGFR1. A summary of FGFR1-3 sequence variations, copy number alterations, and gene expression levels for all samples is given in Table S4.
Discussion
In this report, we present evidence that activating mutations in a gene encoding for fibroblast growth factor receptor 1, FGFR1, are important somatic events in some sporadic PCCs. FGFRs, which in humans include FGFR1 to FGFR4, are receptor tyrosine kinases that are involved in multiple processes during embryonic development, including mesenchymal–epithelial communication and formation of several organ systems [20, 21]. In the adult, fibroblast growth factor receptor signaling is involved in tissue homeostasis and regulates processes such as tissue repair, angiogenesis, and inflammation. Binding of FGFs to FGFRs induces receptor dimerization that activates the intracellular kinase domain and leads to transphosphorylation [21]. The phosphorylated tyrosine residues function as docking sites for adaptor proteins, for example FRS2, which can be phosphorylated by FGFRs. This leads to an activation of several signal transduction pathways, including the RAS-RAF-MAPK and PI3K-AKT pathways. The most established link between FGFRs and cancer is probably with bladder cancer, in which FGFR3 is one of the most commonly mutated genes [22] and the mutations are associated with a non-invasive behavior [23]. Mutations in FGFR2 and FGFR3 have also been reported in other tumor forms, whereas activation of FGFR1 has mainly been observed in the form of FGFR1 amplification in breast and lung cancer [20]. However, occasional point mutations and fusion genes involving FGFR1 have been observed in glioblastomas [24], which is in agreement with FGFR1 being the most abundant fibroblast growth factor receptor in the nervous system [25]. Most recently, frequent FGFR1 mutations in two hotspot sites were revealed in another brain tumor: pilocytic astrocytoma [14]. The two variants detected in this study, Asn546Lys and Lys656Glu, affect the kinase domain and have been shown to promote cell proliferation [26], and Asn546Lys also alter FGFR1 autophosphorylation [27]. Furthermore, overexpression of FGFR1 results in MAPK and AKT activation and neurite outgrowth in the rat pheochromocytoma cell line PC12 [25].
In this study, we discovered that the same FGFR1 hotspot residues as in certain brain tumors, Asn546 and Lys656 (also reported in 1 of 394 PCCs in the COSMIC database [28]), are recurrently mutated in PCCs. Interestingly, the first cohort investigated in this study, which consisted of Scandinavian cases, had a prevalence of FGFR1 mutations of almost 10%. However, further investigation in a French cohort did not reveal any additional mutations, resulting in a total frequency of 3.8% (Fig. 2) in the combined cohorts of 80 sporadic PCCs. This means that somatic FGFR1 mutations are a rare but recurrent event in PCCs (Table 1). The difference in the frequency of somatic FGFR1 mutations between PCCs from different countries may be due to random variation, or, alternatively, there could be regional differences in the frequency of mutations. Data on the ethnic background are not available in our cohort. Possibly, regional differences in the occurrence of certain germline variants could alter the predisposition to obtain somatic FGFR1 mutations. Such a phenomenon has previously been observed for a polymorphism that is associated with a hypermutability of the APC gene [29] involved in colorectal cancer. The significance of FGFR1 mutations in PCCs is also supported by data from the TCGA cohort of PCCs and PGLs (http://cancergenome.nih.gov/). Analyzed sequencing data accessed through cBioPortal [30, 31]) displayed a prevalence of Asn546Lys mutation (corresponding to the Asn577Lys in the cBioPortal) in 2/173 (1.2%) of the samples. Interestingly, Toledo et al. very recently found the Asn546Lys mutation in 1/43 PCCs/PGLs, but none in a 136 patient validation cohort supporting the role of FGFR1 mutation in PCCs/PGLs as a rare but recurrent event in three different cohorts at a frequency similar to several other PCC/PGL susceptibility genes [32]. In addition, we observed copy number gains in the FGFR1 region, which were associated with an increased FGFR1 gene expression, which is in agreement with an oncogenic role of FGFR1 in PCCs. Our findings constitute an additional PCC susceptibility gene associated with MAPK and AKT activation, and indeed, gene expression profiling showed that the FGFR1-mutated tumors clustered together with tumors having mutations in the NF1, RET, and HRAS genes (Fig. 3). Future studies in larger cohorts will be valuable to more accurately estimate the frequency and the potential clinical consequences of FGFR1 alterations in PCCs/PGLs. The potential association of the FGFR3 variant Phe384Leu (rs17881656) with PCCs warrants further examination.
Fig. 3.
Hierarchical clustering of tumors based on gene expression levels. Mutation status is indicated below the dendrogram, showing that tumors with FGFR1 mutations cluster together with those that have RET, NF1, or HRAS mutations. Whereas mutations in the known susceptibility genes were mutually exclusive, one mutation in the novel susceptibility gene, FGFR1, occurred in combination with a somatic MAX mutation
The whole-exome sequencing approach used in this study also detected a median of 33 non-silent somatic mutations in each tumor. Most of the altered genes detected here were only altered in a single sample, suggesting that there is a large diversity of somatic events occurring in PCCs. Many of these may be passenger events, and further studies with larger sample sizes will probably be required in order to identify additional true but infrequent drivers in the tumorigenesis [33]. In agreement with previous reports [34, 35], the exome sequencing technique was also useful in order to find a germline SDHB mutation in a patient with apparently sporadic PCCs, but we also detected numerous variants of unknown significance that may be difficult to interpret in a clinical setting. For gene and pathway discovery, however, next-generation sequencing and omics approaches have been incredibly successful [34, 36–38] and will likely result in the detection of additional PCC/PGL-associated genes in the near future. Indeed, recent exome sequencing initiatives published during the finishing of this work have revealed additional genes, including ATRX, MDH2, and KMT2D, to be potentially involved in PCC/PGL pathogenesis [5, 6, 39, 40], and FGFR1 can now be added to the list of PCC susceptibility genes. This finding advances our biological understanding of PCC development and possibly opens for novel therapeutic options involving FGFR inhibitors, which are currently being evaluated in other tumor forms [21].
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
The authors would like to acknowledge the support from FORSS (Medical Research Council of Southeast Sweden), Science for Life Laboratory, the National Genomics Infrastructure (NGI), Sweden, the Knut and Alice Wallenberg Foundation and UPPMAX for providing assistance in massive parallel sequencing and computational infrastructure. We also thank Miłosz Roliński, Ida Gustavsson, Cassandra Ekman, Annette Molbaek, and Åsa Schippert for valuable technical support. This work was supported by Grants from Linköping University, the Swedish Cancer Society, FORSS and LiU Cancer.
Compliance with ethical standards
Conflict of interest
The authors have no conflict of interest.
Footnotes
Electronic supplementary material
The online version of this article (10.1007/s00268-017-4320-0) contains supplementary material, which is available to authorized users.
Peter Söderkvist and Oliver Gimm have contributed equally to this manuscript.
References
- 1.Dahia PL. Pheochromocytoma and paraganglioma pathogenesis: learning from genetic heterogeneity. Nat Rev Cancer. 2014;14:108–119. doi: 10.1038/nrc3648. [DOI] [PubMed] [Google Scholar]
- 2.Favier J, Amar L, Gimenez-Roqueplo AP. Paraganglioma and phaeochromocytoma: from genetics to personalized medicine. Nat Rev Endocrinol. 2015;11:101–111. doi: 10.1038/nrendo.2014.188. [DOI] [PubMed] [Google Scholar]
- 3.Burnichon N, Buffet A, Parfait B, et al. Somatic NF1 inactivation is a frequent event in sporadic pheochromocytoma. Hum Mol Genet. 2012;21:5397–5405. doi: 10.1093/hmg/dds374. [DOI] [PubMed] [Google Scholar]
- 4.Welander J, Larsson C, Backdahl M, et al. Integrative genomics reveals frequent somatic NF1 mutations in sporadic pheochromocytomas. Hum Mol Genet. 2012;21:5406–5416. doi: 10.1093/hmg/dds402. [DOI] [PubMed] [Google Scholar]
- 5.Fishbein L, Khare S, Wubbenhorst B, et al. Whole-exome sequencing identifies somatic ATRX mutations in pheochromocytomas and paragangliomas. Nat Commun. 2015;6:6140. doi: 10.1038/ncomms7140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Flynn A, Benn D, Clifton-Bligh R, et al. The genomic landscape of phaeochromocytoma. J Pathol. 2015;236:78–89. doi: 10.1002/path.4503. [DOI] [PubMed] [Google Scholar]
- 7.Burnichon N, Vescovo L, Amar L, et al. Integrative genomic analysis reveals somatic mutations in pheochromocytoma and paraganglioma. Hum Mol Genet. 2011;20:3974–3985. doi: 10.1093/hmg/ddr324. [DOI] [PubMed] [Google Scholar]
- 8.Fishbein L, Leshchiner I, Walter V, et al. Comprehensive Molecular Characterization of Pheochromocytoma and Paraganglioma. Cancer Cell. 2017;31:181–193. doi: 10.1016/j.ccell.2017.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Welander J, Andreasson A, Brauckhoff M, et al. Frequent EPAS1/HIF2alpha exons 9 and 12 mutations in non-familial pheochromocytoma. Endocr Relat Cancer. 2014;21:495–504. doi: 10.1530/ERC-13-0384. [DOI] [PubMed] [Google Scholar]
- 10.Welander J, Andreasson A, Juhlin CC, et al. Rare germline mutations identified by targeted next-generation sequencing of susceptibility genes in pheochromocytoma and paraganglioma. J Clin Endocrinol Metab. 2014;99:E1352–1360. doi: 10.1210/jc.2013-4375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Crona J, Delgado Verdugo A, Maharjan R, et al. Somatic mutations in H-RAS in sporadic pheochromocytoma and paraganglioma identified by exome sequencing. J Clin Endocrinol Metab. 2013;98:E1266–1271. doi: 10.1210/jc.2012-4257. [DOI] [PubMed] [Google Scholar]
- 12.Burnichon N, Cascon A, Schiavi F, et al. MAX mutations cause hereditary and sporadic pheochromocytoma and paraganglioma. Clin Cancer Res. 2012;18:2828–2837. doi: 10.1158/1078-0432.CCR-12-0160. [DOI] [PubMed] [Google Scholar]
- 13.Oudijk L, de Krijger RR, Rapa I, et al. H-RAS mutations are restricted to sporadic pheochromocytomas lacking specific clinical or pathological features: data from a multi-institutional series. J Clin Endocrinol Metab. 2014;99:E1376–1380. doi: 10.1210/jc.2013-3879. [DOI] [PubMed] [Google Scholar]
- 14.Jones DT, Hutter B, Jager N, et al. Recurrent somatic alterations of FGFR1 and NTRK2 in pilocytic astrocytoma. Nat Genet. 2013;45:927–932. doi: 10.1038/ng.2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Singh D, Chan JM, Zoppoli P, et al. Transforming fusions of FGFR and TACC genes in human glioblastoma. Science. 2012;337:1231–1235. doi: 10.1126/science.1220834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Di Stefano AL, Fucci A, Frattini V, et al. Detection, Characterization, and Inhibition of FGFR-TACC Fusions in IDH Wild-type Glioma. Clin Cancer Res. 2015;21:3307–3317. doi: 10.1158/1078-0432.CCR-14-2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Adzhubei IA, Schmidt S, Peshkin L, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc. 2009;4:1073–1081. doi: 10.1038/nprot.2009.86. [DOI] [PubMed] [Google Scholar]
- 19.Choi Y, Sims GE, Murphy S, et al. Predicting the functional effect of amino acid substitutions and indels. PLoS One. 2012;7:e46688. doi: 10.1371/journal.pone.0046688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Haugsten EM, Wiedlocha A, Olsnes S, et al. Roles of fibroblast growth factor receptors in carcinogenesis. Mol Cancer Res. 2010;8:1439–1452. doi: 10.1158/1541-7786.MCR-10-0168. [DOI] [PubMed] [Google Scholar]
- 21.Turner N, Grose R. Fibroblast growth factor signalling: from development to cancer. Nat Rev Cancer. 2010;10:116–129. doi: 10.1038/nrc2780. [DOI] [PubMed] [Google Scholar]
- 22.Cheng L, Zhang S, Davidson DD, et al. Molecular determinants of tumor recurrence in the urinary bladder. Future Oncol. 2009;5:843–857. doi: 10.2217/fon.09.50. [DOI] [PubMed] [Google Scholar]
- 23.Hernandez S, Lopez-Knowles E, Lloreta J, et al. Prospective study of FGFR3 mutations as a prognostic factor in nonmuscle invasive urothelial bladder carcinomas. J Clin Oncol. 2006;24:3664–3671. doi: 10.1200/JCO.2005.05.1771. [DOI] [PubMed] [Google Scholar]
- 24.The_Cancer_Genome_Atlas_Research_Network Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hausott B, Schlick B, Vallant N, et al. Promotion of neurite outgrowth by fibroblast growth factor receptor 1 overexpression and lysosomal inhibition of receptor degradation in pheochromocytoma cells and adult sensory neurons. Neuroscience. 2008;153:461–473. doi: 10.1016/j.neuroscience.2008.01.083. [DOI] [PubMed] [Google Scholar]
- 26.Yoon K, Nery S, Rutlin ML, et al. Fibroblast growth factor receptor signaling promotes radial glial identity and interacts with Notch1 signaling in telencephalic progenitors. J Neurosci. 2004;24:9497–9506. doi: 10.1523/JNEUROSCI.0993-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lew ED, Furdui CM, Anderson KS, et al. The precise sequence of FGF receptor autophosphorylation is kinetically driven and is disrupted by oncogenic mutations. Sci Signal. 2009;2:6. doi: 10.1126/scisignal.2000021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Forbes SA, Bindal N, Bamford S, et al. COSMIC: mining complete cancer genomes in the Catalogue of Somatic Mutations in Cancer. Nucleic Acids Res. 2011;39:D945–950. doi: 10.1093/nar/gkq929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Laken SJ, Petersen GM, Gruber SB, et al. Familial colorectal cancer in Ashkenazim due to a hypermutable tract in APC. Nat Genet. 1997;17:79–83. doi: 10.1038/ng0997-79. [DOI] [PubMed] [Google Scholar]
- 30.Gao J, Aksoy BA, Dogrusoz U, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:l1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cerami E, Gao J, Dogrusoz U, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Toledo RA, Qin Y, Cheng ZM, et al. Recurrent mutations of chromatin-remodeling genes and kinase receptors in pheochromocytomas and paragangliomas. Clin Cancer Res. 2016;22:2301–2310. doi: 10.1158/1078-0432.CCR-15-1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lawrence MS, Stojanov P, Mermel CH, et al. Discovery and saturation analysis of cancer genes across 21 tumour types. Nature. 2014;505:495–501. doi: 10.1038/nature12912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Crona J, Verdugo AD, Granberg D, et al. Next-generation sequencing in the clinical genetic screening of patients with pheochromocytoma and paraganglioma. Endocr Connect. 2013;2:104–111. doi: 10.1530/EC-13-0009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McInerney-Leo AM, Marshall MS, Gardiner B, et al. Whole exome sequencing is an efficient and sensitive method for detection of germline mutations in patients with phaeochromcytomas and paragangliomas. Clin Endocrinol (Oxf) 2014;80:25–33. doi: 10.1111/cen.12331. [DOI] [PubMed] [Google Scholar]
- 36.Comino-Mendez I, Gracia-Aznarez FJ, Schiavi F, et al. Exome sequencing identifies MAX mutations as a cause of hereditary pheochromocytoma. Nat Genet. 2011;43:663–667. doi: 10.1038/ng.861. [DOI] [PubMed] [Google Scholar]
- 37.Letouze E, Martinelli C, Loriot C, et al. SDH mutations establish a hypermethylator phenotype in paraganglioma. Cancer Cell. 2013;23:739–752. doi: 10.1016/j.ccr.2013.04.018. [DOI] [PubMed] [Google Scholar]
- 38.Toledo RA, Qin Y, Srikantan S, et al. In vivo and in vitro oncogenic effects of HIF2A mutations in pheochromocytomas and paragangliomas. Endocr Relat Cancer. 2013;20:349–359. doi: 10.1530/ERC-13-0101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cascon A, Comino-Mendez I, Curras-Freixes M, et al. Whole-exome sequencing identifies MDH2 as a new familial paraganglioma gene. J Natl Cancer Inst. 2015 doi: 10.1093/jnci/djv053. [DOI] [PubMed] [Google Scholar]
- 40.Juhlin CC, Goh G, Healy JM, et al. Whole-exome sequencing characterizes the landscape of somatic mutations and copy number alterations in adrenocortical carcinoma. J Clin Endocrinol Metab. 2015;100:E493–502. doi: 10.1210/jc.2014-3282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Evenepoel L, Helaers R, Vroonen L, et al. KIF1B and NF1 are the most frequently mutated genes in paraganglioma and pheochromocytoma tumors. Endocr Relat Cancer. 2017;24:L57–L61. doi: 10.1530/ERC-17-0061. [DOI] [PubMed] [Google Scholar]
- 42.Crona J, Nordling M, Maharjan R, et al. Integrative genetic characterization and phenotype correlations in pheochromocytoma and paraganglioma tumours. PLoS One. 2014;9:e86756. doi: 10.1371/journal.pone.0086756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stenman A, Welander J, Gustavsson I, et al. HRAS mutation prevalence and associated expression patterns in pheochromocytoma. Genes Chromosomes Cancer. 2016;55:452–459. doi: 10.1002/gcc.22347. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.