Abstract
Recent studies have revealed robust metabolic changes during cell differentiation. Mitochondria, the organelles where many vital metabolic reactions occur, may play an important role. Here, we report the involvement of SIRT3-regulated mitochondrial stress in osteoblast differentiation and bone formation. In both the osteoblast cell line MC3T3-E1 and primary calvarial osteoblasts, robust mitochondrial biogenesis and supercomplex formation were observed during differentiation, accompanied by increased ATP production and decreased mitochondrial stress. Inhibition of mitochondrial activity or an increase in mitochondrial superoxide production significantly suppressed osteoblast differentiation. During differentiation, SOD2 was specifically induced to eliminate excess mitochondrial superoxide and protein oxidation, whereas SIRT3 expression was increased to enhance SOD2 activity through deacetylation of K68. Both SOD2 and SIRT3 knockdown resulted in suppression of differentiation. Meanwhile, mice deficient in SIRT3 exhibited obvious osteopenia accompanied by osteoblast dysfunction, whereas overexpression of SOD2 or SIRT3 improved the differentiation capability of primary osteoblasts derived from SIRT3-deficient mice. These results suggest that SIRT3/SOD2 is required for regulating mitochondrial stress and plays a vital role in osteoblast differentiation and bone formation.
Bone health is determined by the homeostasis of osteoblast-mediated bone formation and osteoclast-mediated bone resorption, and osteoblast dysfunction can block bone formation and result in skeletal disorders such as osteoporosis.1, 2 Osteoblastic bone formation results from differentiation of primitive skeletal stem cells into osteoblast precursors that mature to become osteoblasts that will undergo differentiation to form the bone matrix and start mineralization.3 The differentiation process requires an elevated energy supply to fulfill altered cellular function and has been recognized to coordinate with energy production from mitochondria.4 Previous studies have revealed the critical roles of mitochondria in the differentiation of multiple cell types, such as cardiomyocytes,5 myoblasts,6 neural stem cells,7 dendritic cells,8 trophoblasts,9 and adipocytes;10 however, understanding of the precise involvement of mitochondria in osteoblast differentiation remains limited.
A recent study has shown that oxidative stress in osteoblasts plays a significant role in the pathogenesis of osteoporosis.11 Elevated levels of reactive oxygen species (ROS) have been shown to inhibit osteoblastic mineralization of MC3T3-E1 cells, and regulation of antioxidant systems affects bone formation.12 Because mitochondria are the major source of ROS as well as ATP production, manganese superoxide dismutase (SOD2) has naturally evolved to specifically localize in mitochondria to catalyze the dismutation of superoxide radicals into oxygen and hydrogen peroxide, thereby protecting cells against oxidative damage.13 Mitochondrial deacetylase Sirtuin 3 (SIRT3), a member of a conserved family of proteins that possess NAD-dependent deacetylase activity,14 has been suggested to coordinately deacetylate proteins involved in diverse pathways to regulate their activities in order to maintain mitochondrial function and metabolism under caloric restriction and stress.15, 16 Recent studies have revealed that SOD2 is a specific target of SIRT3. Through deacetylation at lysines 53, 68, and 89, SIRT3 effectively promotes SOD2 activity.17, 18 Increasing evidence has indicated that the SIRT3/SOD2 axis plays a vital role in neuron protection,19 liver health,20 aging, and carcinogenesis.21 Currently, studies of SIRT3/SOD2 in bone formation and homeostasis are quite limited, and a recent study showed that SIRT3 inhibited RANKL-mediated osteoclastogenesis,22 indicating a potential regulatory role of SIRT3 in bone health. However, the role of SIRT3 in osteogenesis remains unexplored. Herein, we investigated the involvement of SIRT3/SOD2 in osteoblast differentiation and bone formation. Our study demonstrates that SOD2 overexpression is specifically induced to maintain low mtROS, which coordinates robust mitochondrial biogenesis during osteoblast differentiation, and SIRT3 is induced to promote SOD2 activity. Loss of SIRT3 impaired mitochondrial homeostasis and function by increasing mtROS, thereby suppressing osteogenesis both in vitro and in vivo, indicating that SIRT3/SOD2 may be a potential target for promoting bone health.
Results
Osteogenic induction is accompanied by robust oxygen consumption
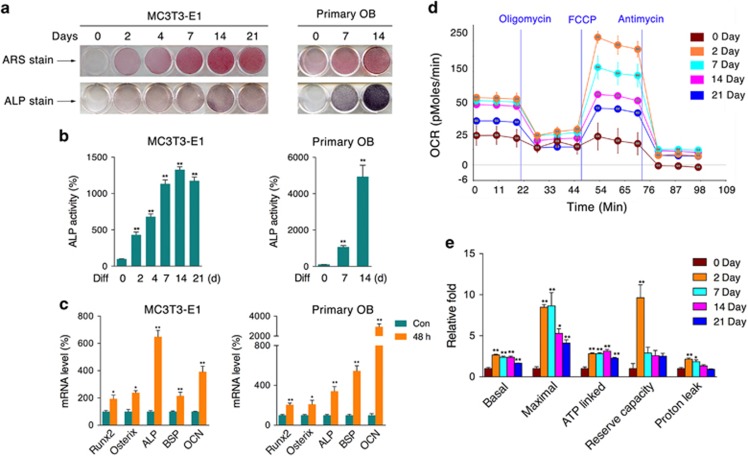
To initiate osteogenic induction, both MC3T3-E1 cells and primary osteoblasts were supplemented with conditioned medium for the indicated periods. Alizarin red S (ARS) and alkaline phosphatase (ALP) staining showed the capability of both cell types to differentiate and form mineralizing matrix (Figure 1a). ALP, as the by-product of osteoblast activity, was found to increase during the differentiation process (Figure 1b). Further mRNA analysis showed that the expression of osteogenic marker genes, including runt-related transcription factor 2 (Runx2), Osterix, ALP, bone sialoprotein (BSP), and osteocalcin (OCN), were significantly increased after 48 h of medium induction (Figure 1c). In addition, we observed that oxygen consumption during the differentiation periods from day 2 to day 21 was highly increased in MC3T3-E1 cells (Figures 1d and e), and a similar increase was also observed during the differentiation of primary osteoblasts (Supplementary Figure 1a), indicating that osteogenic differentiation is a process that requires enhanced mitochondrial function and energy supply.
Figure 1.
Increased mitochondrial oxygen consumption in osteoblast differentiation. MC3T3-E1 cells and calvaria-derived primary osteoblasts were induced to differentiate for the indicated periods of time. (a) ARS and ALP staining of differentiated cells. (b) ALP activity of cell homogenates. (c) After differentiation for 48 h, the relative mRNA levels of Runx2, Osterix, ALP, BSP, and OCN were determined by qRT-PCR. The oxygen consumption rate (OCR) of MC3T3-E1 cells at various time points was measured with an XF24 Extracellular Flux Analyzer (d) experiment program and (e) statistical analysis). Data are presented as the mean±S.E.M. from at least three independent experiments. *P<0.05, **P<0.01 versus relative control
Mitochondrial biogenesis is increased during osteogenic differentiation
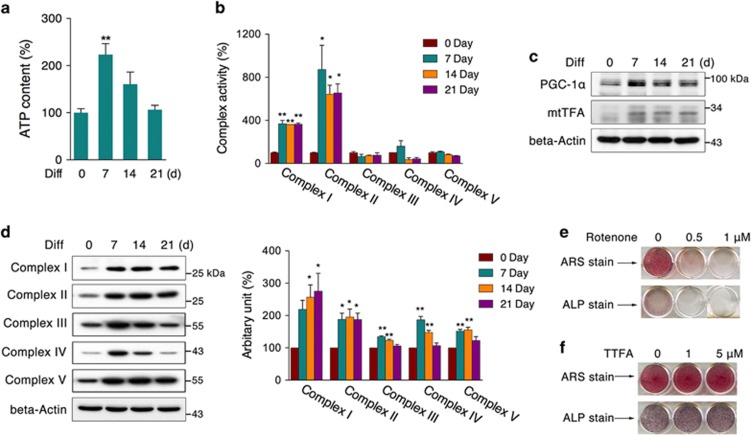
To further investigate the involvement of mitochondria in osteogenic differentiation, we measured ATP levels and mitochondrial complex activities and found that ATP content was significantly increased during the differentiation process (Figure 2a) and that complex I and II activities were robustly enhanced (Figure 2b), suggesting overall increased mitochondrial function. Further protein analysis showed that peroxisome proliferator-activated receptor γ coactivator-1α (PGC-1α) and mitochondrial transcription factor (mtTFA), the key regulating factors for mitochondrial biogenesis, were increased during differentiation (Figure 2c), accompanied by increased expression of mitochondrial complex subunits (Figure 2d). Consistently, differentiation of primary osteoblasts also showed increased mitochondrial biogenesis (Supplementary Figures 1b and c). Even though we found all five complex subunit protein expression increased in early differentiation (Figure 2d), it does not correlate with the observation that only complex I and II activities were increased (Figure 2b). Therefore, we further analyzed formation of mitochondrial supercomplexes using blue native polyacrylamide gel electrophoresis (BN-PAGE), and found supercomplex I + III + IV was largely increased at differentiation 7 and 21 days, whereas supercomplex I + III and III + IV showed no significant changes (Supplementary Figure 2). Thereby, we conclude that both increased mitochondrial biogenesis and mitochondrial supercomplex formation would contribute to improved mitochondrial function during the osteoblast differentiation. In addition, we used specific inhibitors to inhibit the activities of mitochondrial complexes I (rotenone) and II (TTFA) during differentiation, and only rotenone significantly suppressed the process (Figure 2e), whereas TTFA had no obvious effect (Figure 2f), as assessed by ARS and ALP staining. These data suggest that osteogenic differentiation is accompanied by mitochondrial biogenesis and relies on enhanced mitochondrial activity, especially complex I activity.
Figure 2.
Mitochondrial biogenesis during osteogenic differentiation. MC3T3-E1 cells were induced to differentiate for 7, 14, and 21 days. (a) ATP content, (b) mitochondrial complex activities, (c) protein levels of PGC-1α and mtTFA, and (d) mitochondrial complex subunits were tested. (e) Cells were induced to differentiate for 7 days with rotenone, and ARS and ALP staining were performed to evaluate differentiation capacity. (f) Cells were induced to differentiate for 7 days with TTFA, and ARS and ALP staining were performed. Data are presented as the mean±S.E.M. from at least three independent experiments. *P<0.05, **P<0.01 versus relative control
Antioxidant capacity is elevated during osteogenic differentiation
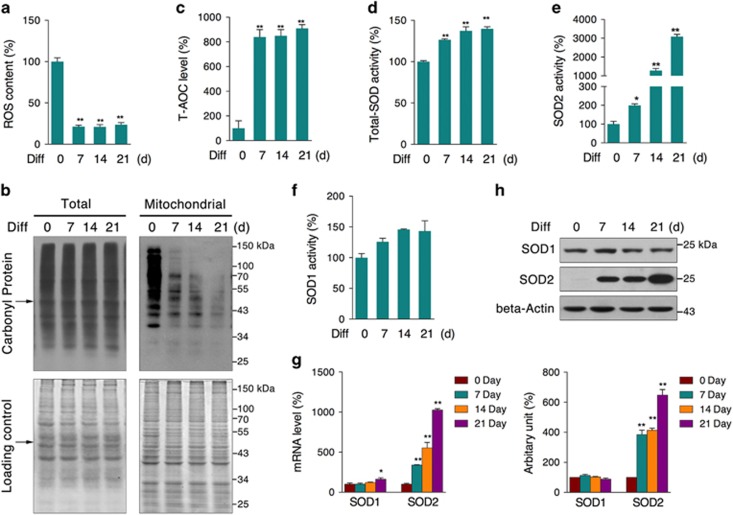
Mitochondria are the major source of endogenous ROS that can be molecular messengers as well as oxidative damage inducers. Although enhanced mitochondrial biogenesis and aerobic metabolism were observed during osteogenic differentiation, the level of its toxic by-products, ROS, was found to be unexpectedly decreased (Figure 3a). More interestingly, protein oxidation, especially the mitochondrial protein oxidation level, was dramatically decreased (Figure 3b). As expected, we observed increased levels of total antioxidant capacity (T-AOC) and total SOD activity (Figures 3c and d). Particularly, SOD2 activity was significantly increased (Figure 3e), whereas SOD1 activity was not affected (Figure 3f). Further analysis showed that SOD2 mRNA and protein were specifically increased during the differentiation process (Figures 3g and h). These data indicate that SOD2 may play a vital role in maintaining mitochondrial redox homeostasis during the differentiation process.
Figure 3.
Elevated antioxidative capacity during osteogenic differentiation. MC3T3-E1 cells were induced to differentiate for 7, 14, and 21 days, and cellular antioxidative capacity was evaluated by testing (a) cellular ROS content, (b) total and mitochondrial carbonyl protein levels, (c) T-AOC, (d) total-SOD activity, (e) SOD2 activity, (f) SOD1 activity, (g) mRNA levels of SOD1 and SOD2, and (h) protein levels of SOD1 and SOD2. Data are presented as the mean±S.E.M. from at least three independent experiments. *P<0.05, **P<0.01 versus relative control
SOD2 is vital to maintain osteogenic differentiation
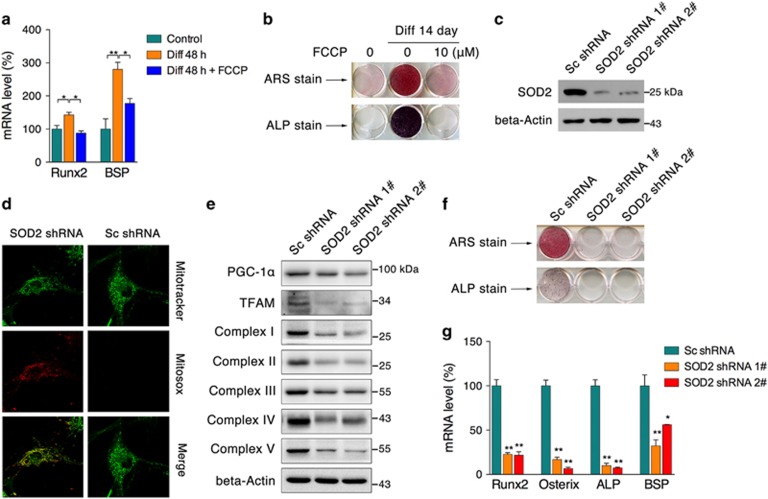
To confirm that the decrease of excessive ROS production is essential to osteogenic differentiation, we treated cells with sublethal doses of FCCP that can induce mitochondrial dysfunction and overgeneration of intracellular ROS. After osteogenic differentiation for 2 days, mRNA expression of Runx2 and BSP were suppressed in the presence of FCCP (Figure 4a). Meanwhile, the ALP and mineralization levels were also significantly reduced (Figure 4b), suggesting a principal role of maintaining redox homeostasis during differentiation. To obtain more insight into the role of SOD2 during differentiation, we generated SOD2-knockdown cells using specific shRNA constructs (Figure 4c) that induced mitochondrial ROS accumulation as expected (Figure 4d). In addition, mitochondrial complex subunits and the key transcription factors PGC-1α and mtTFA were all found to be downregulated in knockdown cells (Figure 4e), consistent with observations in erythroid cells,23 suggesting that SOD2 deficiency has more profound effects than ROS accumulation. Further osteogenic differentiation showed that SOD2 knockdown dramatically suppressed the process and lowered the osteoblast function, as evidenced by ARS and ALP staining (Figure 4f). qPCR analysis also revealed decreased osteoblast markers in knockdown cells subjected to induction for 48 h (Figure 4g). Given the previous observation that oxidative stress could trigger mitochondrial dysfunction and promote mitophagy,24 we propose that SOD2 deficiency associated with osteogenic arrest may attribute to oxidative stress-induced mitochondrial dysfunction.
Figure 4.
SOD2 is required for osteogenic differentiation. (a) MC3T3-E1 cells were induced to differentiate for 48 h with or without 10 μM FCCP, and mRNA levels of Runx2 and BSP were tested. (b) Cells were induced to differentiate for 14 days with or without 10 μM FCCP, and ARS and ALP staining was performed. (c) SOD2 protein content from cells that stably expressed SOD2 shRNA was tested by western blot. (d) Mitochondrial superoxide levels were visualized by fluorescence staining. (e) Mitochondrial biogenesis regulators and mitochondrial complex subunits were analyzed by western blot. (f) ARS and ALP staining of SOD2-knockdown cells differentiated for 7 days. (g) mRNA levels of Runx2, Osterix, ALP, and BSP from SOD2-knockdown cells differentiated for 2 days. Data are presented as the mean±S.E.M. from at least three independent experiments. *P<0.05, **P<0.01 versus relative control
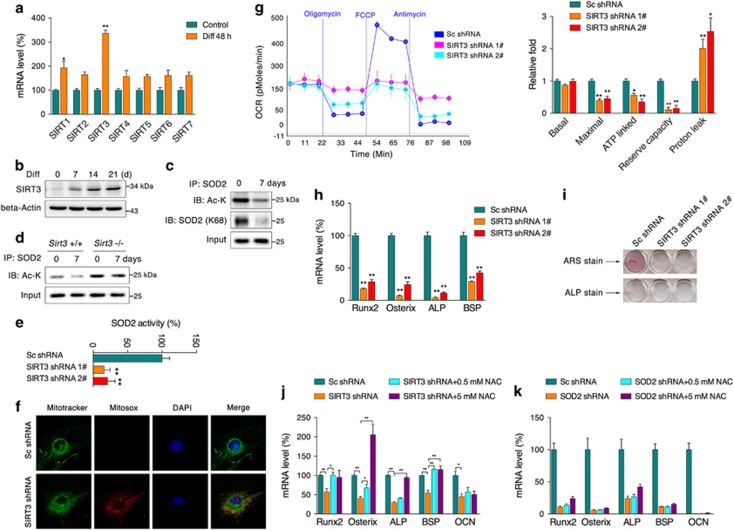
SIRT3 deacetylates SOD2 to maintain mitochondrial function and osteogenic differentiation
During osteogenic differentiation, mRNAs of the sirtuin family exhibited varying expression patterns. SIRT3 mRNA was more abundant than the other sirtuins after 2 days of differentiation (Figure 5a), and its protein expression level also continuously increased (Figure 5b), suggesting a significant activation of SIRT3 during differentiation. As an important sirtuin that is localized within mitochondria, SIRT3 has been noted to be an important regulator of basal ATP and overall energy homeostasis, and it has also been shown to deacetylate and activate SOD2,14, 17 mainly through lysine 68 (K68). We therefore assumed that SIRT3/SOD2 might regulate redox homeostasis and mitochondria function during differentiation. Immunoprecipitation analysis showed a significantly lower overall SOD2 acetylation and specific K68 acetylation levels after 7 days of induction, further supporting the observation of activated SIRT3 during differentiation (Figure 5c). Consistently, SOD2 immunoprecipitates collected after 7 days of differentiation of primary osteoblasts from wild-type and sirt3−/− mice showed that sirt3−/− osteoblasts had higher SOD2 acetylation level compared with wild type, and wild-type ostebolasts showed decreased SOD2 acetylation after 7 days of differentiation, whereas no difference was observed in sirt3−/− osteoblasts during the differentiation (Figure 5d). To further confirm whether deacetylation of SOD2 K68 by SIRT3 is involved in osteoblast differentiation, we constructed SOD2 K68 mutation. Both wild-type and K68 mutation SOD2 were overexpressed in stable SOD2-knockdown cells (Supplementary Figure 3a), wild-type SOD2 could sufficiently reduce overall ROS and mitochondrial superoxide levels (Supplementary Figures 3b and c), whereas SOD2 with K68 mutation failed to show significant improvement. Following overexpression of SOD2 constructs, cells were induced for 2 days of differentiation, and ALP activity was significantly increased by wild-type SOD2, whereas K68 mutation abolished the effect (Supplementary Figure 3d). Meanwhile, mRNA analysis of osteogenic markers showed similar changes as ALP activity (Supplementary Figure 3e). Above data indicate that SIRT3 deacetylating SOD2 at K68 contributes to osteoblast differentiation. However, whether K68 is the only target of SIRT3 on SOD2 during osteoblast differentiation still requires further investigation. Through specific shRNA constructs, we generated stable SIRT3-knockdown cells (Supplementary Figures 4a and b) that all showed significantly decreased SOD2 activity (Figures 5e) as well as decreased T-AOC (Supplementary Figure 4c). Fluorescence staining revealed robust mitochondrial superoxide accumulation, as expected (Figure 5f). Further energetic analysis showed that SIRT3-knockdown cells had reduced maximal respiration, ATP-linked respiration, and reserve capacity, as well as increased proton leakage (Figure 5g), suggesting significant mitochondrial dysfunction in SIRT3-knockdown cells; however, basal respiration was not affected. In addition, analysis of isolated mitochondria showed decreased mitochondrial complex I and II activities in knockdown cells (Supplementary Figure 4d). Mitochondrial DNA copy numbers and certain mitochondrial complex subunits were also found to be decreased, indicating suppressed mitochondrial biogenesis following suppression of SIRT3 (Supplementary Figures 4e and f). When subject to differentiation medium, knockdown cells showed decreased differentiation capability, as evidenced by decreased mRNA expression of osteogenic marker genes and diminished mineralization, (Figures 5h and i). In addition, to explore whether simply attenuating superoxide could promote the differentiation capability of SIRT3/SOD2-deficient cells, the free radical scavenger N-acetyl L-cysteine (NAC) was supplemented during differentiation. NAC could partially improve the expression of osteogenic markers in SIRT3-knockdown cells (Figure 5j), whereas no effects were observed in SOD2-knockdown cells (Figure 5k), indicating that the SIRT3/SOD2 axis is vital to maintaining mitochondrial function and osteogenic capacity and that SOD2 deficiency has more profound effects that could not be reversed by a simple free radical scavenger.
Figure 5.
SIRT3 deacetylates SOD2 to maintain mitochondrial function and osteogenic differentiation. (a) mRNA levels of sirtuin family members in MC3T3-E1 cells under differentiation conditions for 48 h. (b) SIRT3 protein expression in cells under differentiation conditions for the indicated period. (c) SOD2 acetylation and specific K68 acetylation after 7 days of differentiation was analyzed by immunoprecipitation. (d) SOD2 acetylation of primary osteoblasts from wild-type and Sirt3−/− mice after 7 days of differentiation was analyzed by immunoprecipitation. (e) SOD2 activity of SIRT3-knockdown cells. (f) Mitochondrial superoxide levels were visualized by fluorescence staining. (g) OCR analysis of SIRT3-knockdown cells. (h) mRNA levels of Runx2, Osterix, ALP, and BSP from SIRT3-knockdown cells that were differentiated for 2 days. (i) ARS and ALP staining of SIRT3-knockdown cells that were differentiated for 7 days. (j) mRNA levels of osteogenic markers in SIRT3-knockdown cells after differentiation for 2 days with or without NAC. (k) mRNA levels of osteogenic markers in SOD2-knockdown cells after differentiation for 2 days with or without NAC. Data are presented as the mean±S.E.M. from at least three independent experiments. *P<0.05, **P<0.01 versus relative control
SIRT3 knockout induces osteoporosis in mice
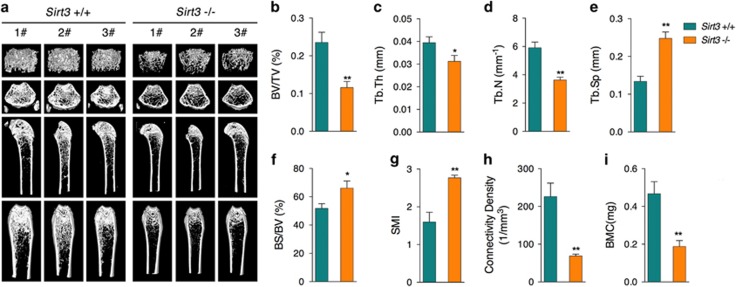
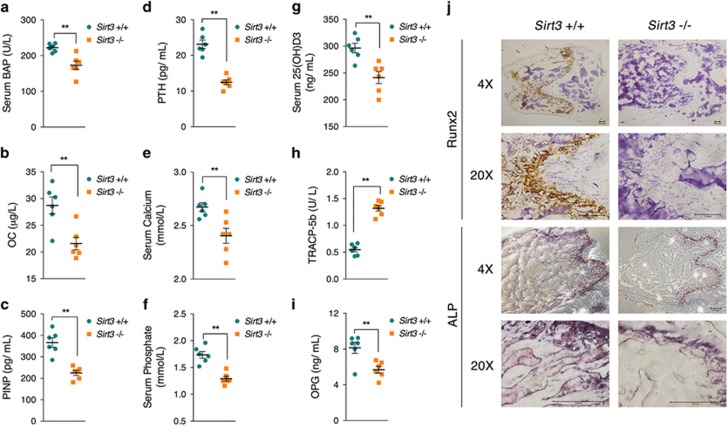
To further confirm the vital roles of SIRT3 in osteoblast function and bone formation in vivo, femur tissues from SIRT3-deficient (Sirt3−/−) mice were further analyzed. Microcomputed tomography (μCT) analysis clearly showed decreased bone volume per tissue volume (BV/TV; Figures 6a and b), trabecular thickness (Tb.Th; Figure 6c), and trabecular number (Tb.N; Figure 6d), as well as increased trabecular separation (Tb.Sp; Figure 6e) and specific bond surface (BS/BV; Figure 6f), indicating significant poor bone quality in Sirt3−/− mice. Structure model index (SMI) analysis revealed higher SMI values in Sirt3−/− mice compared with WT mice (Figure 6g), suggesting a higher proportion of rods to plates in trabecular bone and a potentially lower bone mechanical competence;25 this finding was further supported by the observation of decreased connectivity density in Sirt3−/− mice (Figure 6h). In addition, bone mineral content (BMC), the major feature of osteoporotic fractures, was also found to be significantly lower in Sirt3−/− mice (Figure 6i). Given these observations from the μCT analysis, we thus proposed that SIRT3 deficiency could contribute to bone loss, resulting in osteoporosis. To further explore the osteoblast function in Sirt3−/− mice, we collected serum samples from the mice. Bone alkaline phosphatase (BAP), the bone-specific isoform of alkaline phosphatase and a by-product of osteoblast activity, was found to be significantly decreased in Sirt3−/− mice (Figure 7a), accompanied by decreased osteocalcin (OC), a noncollagenous protein solely secreted by osteoblasts (Figure 7b). As a serum biomarker correlated with bone formation,26 procollagen type I N-terminal propeptide (PINP) was found to be dramatically decreased in Sirt3−/− mice (Figure 7c). Meanwhile, serum parathyroid hormone (PTH) (Figure 7d), calcium (Figure 7e), phosphate (Figure 7f), and 25(OH)D3 (Figure 7g) were all found to be decreased in Sirt3−/− mice. Moreover, Runx2, the leading factor that regulates osteoblast differentiation, was found dramatically decreased according to immunohistochemistry (IHC) analysis of femur sections (Figure 7j). A similar reduction in ALP expression (Figure 7j) was also observed. Taken together, these observations indicate that Sirt3−/− mice have dysregulated bone homeostasis that is partially attributed to osteoblast dysfunction. In addition, the osteoclast marker tartrate-resistant acid phosphatase 5b (TRACP-5b) was found to be increased in the serum of Sirt3−/− mice (Figure 7h), and a negative regulator of osteoclasts, osteoprotegerin (OPG), was found to be decreased (Figure 7i), suggesting that Sirt3−/− mice may have increased osteoclast activity that contributes to excessive bone resorption, consistent with recent findings showing that SIRT3 inhibited osteoclast differentiation by interfering with RANKL-induced expression of PGC-1β.22
Figure 6.
Decreased bone mass in Sirt3−/− mice. (a) Micro-CT images of the femurs of wild-type (Sirt3+/+) and SIRT3-knockout (Sirt3−/−) mice. In the analysis of the trabecular bone and architecture, the following parameters were calculated: (b) bone volume per tissue volume (BV/TV); (c) trabecular thickness (Tb.Th); (d) trabecular number (Tb.N); (e) trabecular spacing (Tb.Sp); (f) bone surface to bone volume (BS/BV); (g) structure model index (SMI); (h) connectivity density; and (i) bone mineral content (BMC). Data are presented as the means±S.E.M.; n=6 per group. *P<0.05, **P<0.01 versus relative control
Figure 7.
Serum and histochemical analyses of osteoblast function in Sirt3−/− mice. Serum was collected from wild-type and Sirt3−/− mice, and the levels of the following factors were analyzed: (a) bone alkaline phosphatase (BAP); (b) osteocalcin (OC); (c) procollagen type I N-terminal propeptide (P1NP); (d) parathyroid hormone (PTH); (e) calcium; (f) phosphate; (g) 25(OH)D3; (h) tartrate-resistant acid phosphatase 5b (TRACP-5b); and (i) osteoprotegerin (OPG). (j) Femur sections were prepared from wild-type and Sirt3−/− mice at 8 weeks and immunostained with an anti-Runx2 antibody and an ALP staining kit. Data are presented as the means±S.E.M.; n=6 per group. **P<0.01 versus relative control
Reestablishing the SIRT3/SOD2 axis promotes the differentiation ability of primary Sirt3−/− osteoblasts
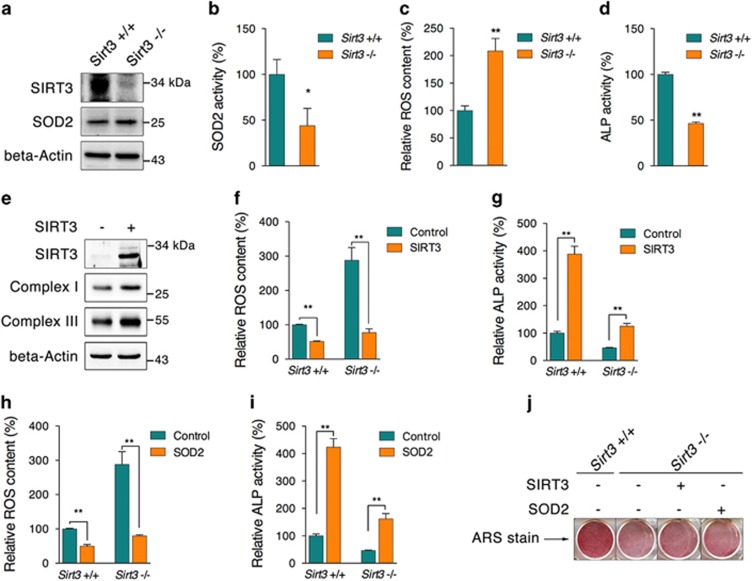
Following observations of bone structure and serum measurements, calvaria-derived primary osteoblasts from Sirt3−/− mice were further isolated and cultured. SIRT3 protein deficiency was confirmed in primary osteoblasts (Figure 8a). Although SOD2 content was not affected (Figure 8a), SOD2 activity was significantly decreased (Figure 8b), accompanied by increased ROS content (Figure 8c). Consistent with observations in MC3T3-E1 cells, impaired mitochondrial biogenesis and mitochondrial oxygen consumption capability were observed in the knockout osteoblasts (Supplementary Figures 5a and b). Meanwhile, ALP activity was found to be decreased, as expected (Figure 8d), and osteogenic markers were suppressed during differentiation (Supplementary Figure 5c). By overexpressing SIRT3 in knockout primary osteoblasts, we found that the protein contents of mitochondrial complex subunits were increased, indicating restored mitochondrial biogenesis (Figure 8e). Decreased ROS content and increased ALP activity were also observed following overexpression of SIRT3 in both wild-type and Sirt3-knockout osteoblasts (Figures 8f and g). In addition, similar improvements were observed by overexpressing SOD2 (Figures 8h and i). Eventually, ARS staining of osteoblast differentiation showed improved osteoblast function following either SIRT3 or SOD2 overexpression (Figure 8j). These data further support the vital role of the SIRT3/SOD2 axis in regulating osteoblast function and bone formation.
Figure 8.
Reestablishing the SIRT3/SOD2 axis improves primary Sirt3−/− osteoblast function. Calvaria-derived primary osteoblasts were isolated from neonatal wild-type and Sirt3−/− mice, and the (a) protein contents of SIRT3 and SOD2, (b) SOD2 activity, and (c) ROS content were analyzed. (d) ALP activity of primary osteoblasts after differentiation for 2 days. (e) SIRT3 adenovirus was applied to Sirt3−/− primary osteoblasts, and the protein contents of complexes I and III were analyzed by western blot. (f) ROS content of primary osteoblasts infected with SIRT3 adenovirus. (g) ALP activity of primary osteoblasts infected with SIRT3 adenovirus and after differentiation for 2 days. (h) ROS content of primary osteoblasts infected with SOD2 adenovirus. (i) ALP activity of primary osteoblasts infected with SOD2 adenovirus and after differentiation for 2 days. (j) ARS staining of primary osteoblasts infected with SIRT3 and SOD2 adenovirus after 7 days of differentiation. Data are presented as the mean±S.E.M. from at least three independent experiments. *P<0.05, **P<0.01 versus relative control
Discussion
Osteoblasts are the chief bone-making cells, and they produce a combination of extracellular proteins that constitute the major components of bone. Revealing the detailed mechanisms that regulate osteoblast differentiation and function is of great importance for clinical applications, such as the treatment of osteoporosis, a common skeletal disease associated with diminished osteoblast production and function.1 Mitochondria, the leading source of both ATP and ROS, have been indicated to participate in oxidative stress-induced osteoblast damage and dysfunction.27, 28 However, the role of mitochondria in osteoblast differentiation remains unexplored. In this study, we revealed the vital role of the SIRT3/SOD2 axis in regulating mitochondria and osteoblast function. Robust mitochondrial biogenesis and activity enhancement were required to support osteoblast differentiation, and the SIRT3/SOD2 axis was specifically activated to maintain the redox status of mitochondria to support mitochondrial function and osteoblast differentiation. Consistent with in vitro observations, Sirt3−/− mice exhibited reduced bone mass and impaired osteoblast function that was improved by reestablishing the SIRT3/SOD2 axis in vitro.
In addition to generating ATP, mitochondria are known to produce ROS through the electron transport chain. The popular ‘redox window’ hypothesis suggests that moderate levels of ROS are required for signal transduction to fulfill physiological cellular functions,29, 30 whereas excessive ROS levels can cause DNA damage and mitochondrial dysfunction, contributing to various pathological conditions.27, 31 In addition, the interaction between mitochondria and ROS has been well studied in the differentiation processes of various cell types, with the exception of osteoblast differentiation.5, 6, 8 In this study, we observed increased mitochondrial biogenesis and oxygen consumption during osteoblast differentiation that may naturally fulfill the energy need for producing a large amount of extracellular proteins. We measured the cellular ATP level to further support our conclusion. As directly measuring mitochondrial produced ATP is not feasible, the total cellular ATP was tested, and therefore the data could not directly correlate with mitochondrial activity. The increased ATP level at 7 days of differentiation may attribute to increased mitochondrial content and activity in early differentiation induction. Along the differentiation, utilization of ATP for phosphorylation and mineralization32, 33 is largely increased as well as enhanced ATP release from osteoblasts34 that may contribute to normalized cellular ATP level at late-stage differentiation. Meanwhile, instead of increased ROS production accompanying the enhanced oxygen consumption, the ROS level was dramatically decreased and remained low during differentiation, consistent with a previous study that exposure to nontoxic concentrations of hydrogen peroxide diminished osteoblast mineralization.12 We further found that inhibiting mitochondrial respiration with the complex I inhibitor rotenone or promoting ROS production with the mitochondrial oxidative phosphorylation uncoupler FCCP suppressed the osteoblast differentiation capacity. These data suggest that maintaining both the redox state and function of mitochondria is critical to preserve osteoblast function.
SOD is the primary defense enzyme that eliminates oxidative stress. We assumed that SOD was the major contributor to the decreased ROS levels during osteoblast differentiation. A previous study showed that coordinated regulation of mitochondrial biogenesis and antioxidant enzymes occurs synergistically during osteogenic differentiation of human mesenchymal stem cells (hMSCs).35 Interestingly, by investigating both SOD1 and SOD2, we found that only the mitochondria-specific SOD2 protein was significantly induced, accompanied by largely decreased mitochondrial protein oxidation, suggesting a unique role of SOD2 in osteogenic differentiation. Single-nucleotide polymorphism (SNP) mapping has suggested a strong association between SOD2 polymorphisms and osteoporosis in both Indian and Chinese populations.36, 37 Kobayashi et al. also reported that osteocytes of aged mice have lower SOD2 activity that is the cause of age-related bone loss.38 In addition, we further showed that SOD2 deficiency-induced mitochondrial dysfunction is the primary cause for osteoblast dysfunction, supporting a vital role of SOD2 in healthy bone. In addition to the expected elevated mitochondrial ROS levels and impaired mitochondrial function, SOD2 knockdown also induced significant loss of mitochondrial content, which could be a result of impaired mitochondrial biogenesis,39 or enhanced mitochondrial degradation by autophagy.40 These irreversibly damaged mitochondria could not be rescued by simple supplementation with the antioxidant NAC, providing the reason to the fact that supplementing SOD2-knockdown cells with NAC could not improve osteoblast differentiation.
Because of the vital role of SOD2 as an antioxidant in mitochondria, regulation of SOD2 has been widely studied. Multiple post-translational modifications of SOD2, including phosphorylation, nitration, glutathionylation, and methylation, have been identified.41 Recently, acetylation sites on SOD2 have been reported, and SIRT3 can specifically modify SOD2 to regulate its activity.17, 18 We found that Sirt3 mRNA was specifically induced during differentiation, followed by increased SIRT3 protein in mitochondria and a decreased SOD2 acetylation level. Although we showed that SIRT3 deacetylating SOD2 at K68 in osteoblasts, whether K68 is the only target of SIRT3 on SOD2 during osteoblasts differentiation still requires further investigation. In myoblasts, it has been demonstrated that SIRT3 positively regulated differentiation via its influence on the intracellular redox balance and mitochondrial homeostasis, such as mitochondrial biogenesis, ROS production, and SOD2 activity.42 In the current study, we found decreased SOD2 activity as well as increased mitochondrial superoxide in both Sirt3-knockdown MC3T3-E1 cells and Sirt3−/− primary osteoblasts, and both cell types showed impaired differentiation capability. Consistently, μCT analysis of Sirt3−/− mice indicated significant bone loss, and serum analysis showed that several osteoblast markers, including BAP, OC, and P1NP were decreased, suggesting that SIRT3 knockout induced significant osteoblast dysfunction that might be the major contributor to bone loss in mice. We also noticed an increase in the osteoclast marker TRACP-5b and a decrease in the osteoclast negative regulator OPG in the serum, indicating activated osteoclasts and bone resorption in Sirt3−/− mice. Bone mass is known to be maintained locally by the balance between osteoclastic bone resorption and osteoblastic bone formation. Although SIRT3 regulating SOD2 deacetylation and its activity has been indicated playing pivotal role in several biological activities, knowledge on the involvement of SIRT3/SOD2 in bone mass remains limited. Very recent study on osteoclasts revealed that SIRT3 could promote SOD2 activity to suppress osteoclastogensis, whereas loss of SIRT3 would decrease SOD2 activity and activate osteoclasts differentiation, leading to decreased bone volume43 that would also contribute to the decreased bone mass as we observed in Sirt3−/− mice. In the present study, we showed the same positive regulatory effect of SIRT3 on SOD2 activity and osteoblasts differentiation leading to decreased bone volume, and this is the same consequence as SIRT3 loss in osteoclasts. Taken together, all the observations indicate the key role of SIRT3/SOD2 in regulating bone mass through both osteoblasts and osteoclasts. These results suggest that systematically activating SIRT3 would be a potential therapeutic strategy for treating aging and disease-related osteoporosis.
In summary, this study reveals the indispensable role of mitochondria during osteoblast differentiation and demonstrates that SIRT3/SOD2 controls osteogenic differentiation and bone formation in regulating mitochondrial redox and function both in vitro and in vivo. Although further efforts should be made to explore the mechanisms underlying SIRT3/SOD2 activation and to determine the molecular correlation between osteoblasts and osteoclasts, manipulation of SIRT3/SOD2, which is confirmed to be a central regulator of mitochondrial adaptation in disease and aging, could provide novel therapeutics for age-related bone diseases.
Materials and methods
Reagents and antibodies
Cell culture medium, H2DCF-DA, MitoTracker Green, and MitoSOX Red were purchased from Life Technologies (San Diego, CA, USA). Assay kits for T-AOC, SOD, and ALP were purchased from Jiancheng Biochemical Inc., Ltd (Nanjing, China). The BCIP/NBT alkaline phosphatase color development kit was from Beyotime (Nanjing, China). Antibodies against SOD1, SOD2, mtTFA, PGC-1α, SIRT3, and acetyl-lysine were purchased from Santa Cruz Biotechnology (Santa Cruz, CA, USA). Antibodies against mitochondrial complexes I, II, III, IV, and V were purchased from Life Technologies. Antibodies against acetyl SOD2 K68 and Runx2 were purchased from Abcam (Cambridge, UK). Other reagents used in this study were purchased from Sigma (St. Louis, MO, USA).
Animals
Sirt3−/− mice in 129SV background (stock number 027975) were purchased from the Jackson Laboratory (Bar Harbor, ME, USA), and mice were backcrossed onto the C57BL/6 background for 10 generations. Wild-type C57BL/6 mice (Sirt3+/+) served as control. All animals were housed in a temperature (25–28 °C)- and humidity (60%)-controlled animal room and maintained on a 12 h light/12 h dark cycle with food and water provided during the experiments. Animal procedures were approved by Xi’an Jiaotong University Animal Care and Use Committee. All the methods were performed in accordance with approved guidelines, and all efforts were made to minimize the suffering and the number of animals used in this study.
Cell culture and in vitro differentiation
The clonal murine osteoblastic MC3T3-E1 cell line was cultured in α-minimum essential medium (MEM) supplemented with 10% (v/v) fetal bovine serum (FBS), 0.22% sodium bicarbonate, 100 U/ml penicillin, and 100 μg/ml streptomycin. Primary osteoblasts were obtained by enzyme digestion of calvaria from 2-day-old mice following a published protocol with minor modifications.44 Briefly, calvarial bones were isolated from the heads of killed neonatal mice and washed with phosphate-buffered saline (PBS). After removing the soft tissues, the calvaria were cut into pieces and incubated in 4 ml of digestion solution (0.25% trypsin with 0.02% EDTA) at 37 °C for 40 min with gentle shaking every 5 min. After digestion, the calvaria were cut into small pieces and suspended in complete cell culture medium. At 4 h after plating, the culture medium was supplemented to 8 ml, followed by static culture for 4 days. Primary osteoblasts growing out of the bone chips were trypsinized and transferred into a new dish for further experiments. All cells were maintained at 37 °C in a humidified atmosphere of 95% air and 5% CO2. For in vitro differentiation, when cells reached confluence, differentiation was initiated with 50 μg/ml ascorbic acid and 10 mM β-glycerophosphate in complete cell culture medium for the indicated periods, and differentiation medium was replaced every 3 days.
RNA isolation and real-time PCR
Total RNA was extracted using TRIzol (Invitrogen, Carlsbad, CA, USA) following the manufacturer’s protocol. Reverse transcription was performed using a PrimeScript RT-PCR Kit (Otsu, Shiga, Japan) followed by semiquantitative real-time PCR using specific primers. Data were normalized to the mRNA levels of actin that was used as a housekeeping gene, and were analyzed by the 2-ΔΔCT method. The final results are presented as a percentage of the control. Detailed primer sequences are provided in Supplementary Table 1.
Mineralization assay
Cells were cultured with α-MEM containing ascorbic acid and β-glycerophosphate in 12-well plates, and matrix mineralization was determined by the deposition of crystalline hydroxyapatite and visualized by ARS staining. After osteogenic differentiation, the cells were gently washed twice with PBS and fixed with ice-cold 70% ethanol for 30 min at 4 °C. Then, cells were stained with 5% ARS for 30 min at 37 °C. Cells were then washed with deionized water to remove excess dye, and the stained matrix was photographed.
ALP staining and activity assay
Cells were seeded into 12-well culture plates followed by differentiation for 2, 4, 7, 14, and 21 days. Cells were washed with PBS and fixed with 70% ethanol for 30 min at 4 °C. Then, ALP staining was performed with a BCIP/NBT alkaline phosphatase color development kit (Beyotime). For the ALP activity assay, the medium was removed, and the cells were washed and homogenized in ice-cold PBS. After centrifugation at 1000 × g for 10 min at 4 °C, the clear supernatant was used for measurement of the ALP activity and protein concentration. The ALP activity assay was performed with commercial kits following the manufacturer’s protocol (Jiancheng Biochemical).
Intracellular ROS determination
Intracellular generation of ROS was determined by the formation of fluorescent 2’,7’-dichlorofluorescein (DCF) upon oxidation of the non-fluorescent, reduced DCFH.45 After incubation with DCF for 30 min at 37 °C, the fluorescence intensity was observed with a fluorescence microscope and measured with a fluorescence spectrometer (FlexStation 3, Molecular Devices, Sunnyvale, CA, USA) at 485 nm excitation and 538 nm emission. Cellular oxidant levels were expressed as the relative DCF fluorescence per microgram of protein (BCA method). For observation of mitochondrial ROS, MitoSOX Red, a highly selective fluorescent probe, was used for the detection of superoxide generated within mitochondria. Cells were plated on glass coverslips in 12-well plates and stained with 2.5 μM MitoSOX Red diluted in serum-free culture medium for 20 min at 37 °C, accompanied by MitoTracker Green and nuclear staining. Then, coverslips were washed three times with PBS and mounted on the slides. The fluorescence images of mitochondria were visualized with confocal microscopy (Zeiss, Oberkochen, Germany).
Mitochondrial complex activity determination
Mitochondria in cultured cells were isolated as previously described.46 Briefly, cells were collected and resuspended in 1.0 ml of hypotonic buffer (10 mmol/l NaCl, 2.5 mmol/l MgCl2, 10 mmol/l Tris base, pH 7.5) and homogenized on ice with a glass homogenizer (Fisher Scientific, Pittsburgh, PA, USA). The homogenates were then centrifuged at 1300 × g for 5 min at 4 °C. The supernatant was centrifuged at 17 000 × g for 15 min at 4 °C, and the mitochondria pellet was resuspended in 100 μl of isotonic buffer (210 mmol/l mannitol, 70 mmol/l sucrose, 5 mmol/l Tris base, 1 mmol/l EDTA·2Na, pH 7.5). Assays for reduced nicotinamide adenine dinucleotide (NADH)-ubiquinone reductase (complex I), succinate-CoQ oxidoreductase (complex II), ubiquinol cytochrome c reductase (complex III), cytochrome c oxidase (complex IV), and Mg2+-ATPase (complex V) activities were performed according to methods previously described.47
Immunoblotting and immunoprecipitation
Cells were lysed with western and immunoprecipitation (IP) lysis buffers (Beyotime). The lysates were centrifuged at 13 000 × g for 6 min at 4 °C. The supernatants were collected and protein concentrations were determined with a BCA Protein Assay kit (Pierce, Rockford, IL, USA). Equal aliquots of protein samples were loaded onto 10% SDS-PAGE gels and transferred to nitrocellulose membranes. After blocking with 5% non-fat milk for 1 h, the membranes were incubated with primary antibodies at 4 °C overnight, after which they were incubated with horseradish peroxidase-conjugated secondary antibodies for 1 h at room temperature. Western blots were developed using an enhanced chemiluminescence (ECL) western blotting detection kit (Pierce) and quantified by scanning densitometry. To analyze protein interactions, cell lysates were incubated with Protein A/G agarose beads to perform a preclearing step to reduce nonspecific binding, followed by incubation with primary SOD2 antibody overnight at 4 °C, and the immune complexes were pulled down with fresh protein A/G agarose beads (Santa Cruz Biotechnology). The bound proteins were eluted in denaturing SDS sample buffer and analyzed by western blotting.
Protein carbonylation assay
Protein carbonyls in soluble proteins were assayed using an Oxyblot protein oxidation detection kit (Millipore, Temecula, CA, USA). Protein carbonyls were labeled with 2,4-dinitrophenylhydrazine and detected by western blot. As a loading control, equal amounts of the samples were subjected to 10% SDS-PAGE and stained with Coomassie brilliant blue. For determination of mitochondrial protein carbonyls, soluble mitochondrial proteins were isolated using the above method and analyzed.
Biochemical analysis
T-AOC and the activities of SOD1 and SOD2 were analyzed using Jiancheng Biochemical detection kits according to the corresponding kit protocols. ATP content was detected using an ATP bioluminescence assay kit (Sigma) as previously described.48
Oxygen consumption
MC3T3-E1 cells were seeded in XF 24-well microplates. After differentiation, oxygen consumption was measured with an XF24 Extracellular Flux Analyzer (Seahorse Bioscience, Billerica, MA, USA). In addition, 10 μM antimycin A, 6 μM FCCP, and 10 μM oligomycin were used as inhibitors, and the basal, maximal, ATP-linked, and non-mitochondrial respiration, as well as the reserve capacity and proton leakage, were determined and adjusted by protein concentration.49
Plasmids and lentivirus production
shRNAs containing a hairpin loop were synthesized and inserted into the pLKO.1-puro vector. The targeting sequences for SOD2 were 5′-GCTTACTACCTTCAGTATAAA-3′ and 5′-GCCACACATTAACGCGCAGAT-3′, and those for SIRT3 were 5′-GCCCAATGTCACTCACTACTT-3′ and 5′-AGACAGCTCCAACACGTTTAC-3′. Lentivirus was produced by transfecting 293T cells with the indicated expression plasmids, pCMV-VSV-G and pCMV-dR8.2. Viral supernatant was harvested 48 h after transfection. For viral infection, cells were incubated in medium containing virus and 8 μg/ml polybrene for 16 h. Cells were allowed to recover for 24 h before puromycin selection, and surviving pools were used for downstream analysis.
μCT-based analysis of bone structure
To determine the bone mineral density (BMD) and microarchitecture of femur trabeculae in mice, the left femur was scanned using a micro-CT scanner (GE eXplore Locus SP Micro-CT, GE Healthcare, Barrington, IL, USA). BMC, BS/BV, BV/TV, Tb.Th, Tb.N, Tb.Sp, SMI, and connectivity density were calculated from the region of interest (ROI). For cortical bone analysis, data, including total area (Tt.Ar), marrow area (Ma.Ar), and cortical area fraction (Ct.Ar/Tt.Ar), were taken from a 2-mm-long round region of the mid-diaphysis femur. All of the micro-CT data were calculated using MicroView v2.1.1 software and Advanced Bone Analysis application (GE Healthcare, IL, USA).
Immunohistochemistry
Femur tissues dissected from wild-type (Sirt3+/+) and Sirt3−/− mice were fixed using 10% formalin for 48 h and decalcified in 14% EDTA (pH 7.4) for 21 days at room temperature. After being frozen, sagittal-oriented sections were prepared for histological analysis, and ALP staining was performed with a TRAP/ALP Stain Kit (Wako, Osaka, Japan) according to the manufacturer’s protocol. A primary antibody that recognized mouse Runx2 (Abcam, 1 : 500, ab192256) was used for the IHC analysis.
Serum assay
Blood samples were allowed to clot undisturbed for 30 min at room temperature. Serum was separated through centrifugation at 3000 r.p.m. for 10 min. Serum calcium and phosphate levels were measured using an automated HITACHI 7600 clinical analyzer (HITACHI, Ltd, Tokyo, Japan). The serum levels of 25(OH)D3, PTH, P1NP, BAP, OC, OPG, and TRAP-5b were measured using ELISA assay kits following the manufacturer’s instructions.
Adenoviral infection
The cDNAs of mouse SOD2 and SIRT3 were cloned into pAd/CMV/V5-DEST expression vectors (Invitrogen) and transfected into the packaging cell line 293A, with an empty vector as a negative control. After amplification and titering, high-titer adenoviral stocks were collected. Then, calvaria-derived primary osteoblasts from Sirt3+/+ and Sirt3−/− mice were transfected with SOD2, SIRT3, or control adenoviral constructs. After 48 h, cell differentiation was induced for the indicated time periods, and cells were harvested for analysis.
Statistical analysis
Data are presented as the mean±S.E.M. from at least three independent experiments and were analyzed using GraphPad Prism-5 software (San Diego, CA, USA). Statistical significance was evaluated using one-way analysis of variance (ANOVA) followed by post hoc comparisons with Tukey’s HSD test. Differences between two groups were analyzed using Student’s t-test, and the level of significance was set at a value of P<0.05.
Acknowledgments
We thank Dr. Jun Wang at the Institute of Orthopedic Surgery, Xijing Hospital, Fourth Military Medical University, for his help on μCT-based analysis of bone structure. This study was supported by the National Basic Research Program (2015CB553602, 2014CB548200), the National Natural Science Foundation of China (81571050, 31570777, 91439113, 91649106).
Author contributions
ZHF, JKL and WLS designed the study, analyzed the data, wrote the manuscript, and directed the research. JG, ZHF, XQW, MQZ, and JX performed experiments. JL, SJH, LC, and KC conducted vector construction. JGL and ZFL directed animal study and primary cell culture.
Footnotes
Supplementary Information accompanies this paper on Cell Death and Differentiation website (http://www.nature.com/cdd)
Edited by RA Knight
The authors declare no conflict of interest.
Supplementary Material
References
- Long F. Building strong bones: molecular regulation of the osteoblast lineage. Nat Rev Mol Cell Biol 2011; 13: 27–38. [DOI] [PubMed] [Google Scholar]
- Croucher PI, McDonald MM, Martin TJ. Bone metastasis: the importance of the neighbourhood. Nat Rev Cancer 2016; 16: 373–386. [DOI] [PubMed] [Google Scholar]
- Kassem M, Bianco P. Skeletal stem cells in space and time. Cell 2015; 160: 17–19. [DOI] [PubMed] [Google Scholar]
- Kain KH, Popov VL, Herzog NK. Alterations in mitochondria and mtTFA in response to LPS-induced differentiation of B-cells. Bioch Biophys Acta 2000; 1494: 91–103. [DOI] [PubMed] [Google Scholar]
- San Martin N, Cervera AM, Cordova C, Covarello D, McCreath KJ, Galvez BG. Mitochondria determine the differentiation potential of cardiac mesoangioblasts. Stem Cells 2011; 29: 1064–1074. [DOI] [PubMed] [Google Scholar]
- Lee S, Tak E, Lee J, Rashid MA, Murphy MP, Ha J et al. Mitochondrial H2O2 generated from electron transport chain complex I stimulates muscle differentiation. Cell Res 2011; 21: 817–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xun Z, Lee DY, Lim J, Canaria CA, Barnebey A, Yanonne SM et al. Retinoic acid-induced differentiation increases the rate of oxygen consumption and enhances the spare respiratory capacity of mitochondria in SH-SY5Y cells. Mech Ageing Dev 2012; 133: 176–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Prete A, Zaccagnino P, Di Paola M, Saltarella M, Oliveros Celis C, Nico B et al. Role of mitochondria and reactive oxygen species in dendritic cell differentiation and functions. Free Radic Biol Med 2008; 44: 1443–1451. [DOI] [PubMed] [Google Scholar]
- Martinez F, Kiriakidou M, Strauss JF 3rd. Structural and functional changes in mitochondria associated with trophoblast differentiation: methods to isolate enriched preparations of syncytiotrophoblast mitochondria. Endocrinology 1997; 138: 2172–2183. [DOI] [PubMed] [Google Scholar]
- Tejerina S, De Pauw A, Vankoningsloo S, Houbion A, Renard P, De Longueville F et al. Mild mitochondrial uncoupling induces 3T3-L1 adipocyte de-differentiation by a PPARgamma-independent mechanism, whereas TNFalpha-induced de-differentiation is PPARgamma dependent. J Cell Sci 2009; 122(Pt 1): 145–155. [DOI] [PubMed] [Google Scholar]
- Rached MT, Kode A, Xu L, Yoshikawa Y, Paik JH, Depinho RA et al. FoxO1 is a positive regulator of bone formation by favoring protein synthesis and resistance to oxidative stress in osteoblasts. Cell Metab 2010; 11: 147–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arai M, Shibata Y, Pugdee K, Abiko Y, Ogata Y. Effects of reactive oxygen species (ROS) on antioxidant system and osteoblastic differentiation in MC3T3-E1 cells. IUBMB Life 2007; 59: 27–33. [DOI] [PubMed] [Google Scholar]
- Przedborski S, Schon EA. Loss of ROS—a radical response. Nat Genet 1998; 18: 99–100. [DOI] [PubMed] [Google Scholar]
- Ahn BH, Kim HS, Song S, Lee IH, Liu J, Vassilopoulos A et al. A role for the mitochondrial deacetylase Sirt3 in regulating energy homeostasis. Proc Natl Acad Sci USA 2008; 105: 14447–14452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebert AS, Dittenhafer-Reed KE, Yu W, Bailey DJ, Selen ES, Boersma MD et al. Calorie restriction and SIRT3 trigger global reprogramming of the mitochondrial protein acetylome. Mol Cell 2013; 49: 186–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai SH, Chen T, Wang YH, Zhu J, Luo P, Rao W et al. Sirt3 protects cortical neurons against oxidative stress via regulating mitochondrial Ca2+ and mitochondrial biogenesis. Int J Mol Sci 2014; 15: 14591–14609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Zhang J, Lin Y, Lei Q, Guan KL, Zhao S et al. Tumour suppressor SIRT3 deacetylates and activates manganese superoxide dismutase to scavenge ROS. EMBO Rep 2011; 12: 534–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu X, Brown K, Hirschey MD, Verdin E, Chen D. Calorie restriction reduces oxidative stress by SIRT3-mediated SOD2 activation. Cell Metab 2010; 12: 662–667. [DOI] [PubMed] [Google Scholar]
- Cheng A, Yang Y, Zhou Y, Maharana C, Lu D, Peng W et al. Mitochondrial SIRT3 mediates adaptive responses of neurons to exercise and metabolic and excitatory challenges. Cell Metab 2016; 23: 128–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pi H, Xu S, Reiter RJ, Guo P, Zhang L, Li Y et al. SIRT3-SOD2-mROS-dependent autophagy in cadmium-induced hepatotoxicity and salvage by melatonin. Autophagy 2015; 11: 1037–1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao R, Vassilopoulos A, Parisiadou L, Yan Y, Gius D. Regulation of MnSOD enzymatic activity by Sirt3 connects the mitochondrial acetylome signaling networks to aging and carcinogenesis. Antioxid Redox Signal 2014; 20: 1646–1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh JE, Shin JH, Jang ES, Park SJ, Park DR, Ko R et al. Sirtuin 3 (SIRT3) maintains bone homeostasis by regulating AMPK-PGC-1beta axis in mice. Sci Rep 2016; 6: 22511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin FM, Xu X, von Lohneysen K, Gilmartin TJ, Friedman JS. SOD2 deficient erythroid cells up-regulate transferrin receptor and down-regulate mitochondrial biogenesis and metabolism. PLoS ONE 2011; 6: e16894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piantadosi CA, Suliman HB. Redox regulation of mitochondrial biogenesis. Free Radic Biol Med 2012; 53: 2043–2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildebrand T, Ruegsegger P. Quantification of bone microarchitecture with the structure model index. Comput Methods Biomech Biomed Engin 1997; 1: 15–23. [DOI] [PubMed] [Google Scholar]
- Hale LV, Galvin RJ, Risteli J, Ma YL, Harvey AK, Yang X et al. PINP: a serum biomarker of bone formation in the rat. Bone 2007; 40: 1103–1109. [DOI] [PubMed] [Google Scholar]
- Ho WP, Chen TL, Chiu WT, Tai YT, Chen RM. Nitric oxide induces osteoblast apoptosis through a mitochondria-dependent pathway. Ann NY Acad Sci 2005; 1042: 460–470. [DOI] [PubMed] [Google Scholar]
- Choi EM, Suh KS, Lee YS. Liquiritigenin restores osteoblast damage through regulating oxidative stress and mitochondrial dysfunction. Phytother Res 2014; 28: 880–886. [DOI] [PubMed] [Google Scholar]
- Takano H, Zou Y, Hasegawa H, Akazawa H, Nagai T, Komuro I. Oxidative stress-induced signal transduction pathways in cardiac myocytes: involvement of ROS in heart diseases. Antioxid Redox Signal 2003; 5: 789–794. [DOI] [PubMed] [Google Scholar]
- Reczek CR, Chandel NS. ROS-dependent signal transduction. Curr Opin Cell Biol 2015; 33: 8–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urao N, Ushio-Fukai M. Redox regulation of stem/progenitor cells and bone marrow niche. Free Radic Biol Med 2013; 54: 26–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano Y, Addison WN, Kaartinen MT. ATP-mediated mineralization of MC3T3-E1 osteoblast cultures. Bone 2007; 41: 549–561. [DOI] [PubMed] [Google Scholar]
- Cutarelli A, Marini M, Tancredi V, D'Arcangelo G, Murdocca M, Frank C et al. Adenosine Triphosphate stimulates differentiation and mineralization in human osteoblast-like Saos-2 cells. Dev Growth Differ 2016; 58: 400–408. [DOI] [PubMed] [Google Scholar]
- Brandao-Burch A, Key ML, Patel JJ, Arnett TR, Orriss IR. The P2X7 receptor is an important regulator of extracellular ATP levels. Front Endocrinol 2012; 3: 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CT, Shih YR, Kuo TK, Lee OK, Wei YH. Coordinated changes of mitochondrial biogenesis and antioxidant enzymes during osteogenic differentiation of human mesenchymal stem cells. Stem Cells 2008; 26: 960–968. [DOI] [PubMed] [Google Scholar]
- Botre C, Shahu A, Adkar N, Shouche Y, Ghaskadbi S, Ashma R. Superoxide dismutase 2 polymorphisms and osteoporosis in Asian Indians: a genetic association analysis. Cell Mol Biol Lett 2015; 20: 685–697. [DOI] [PubMed] [Google Scholar]
- Deng FY, Lei SF, Chen XD, Tan LJ, Zhu XZ, Deng HW. An integrative study ascertained SOD2 as a susceptibility gene for osteoporosis in Chinese. J Bone Miner Res 2011; 26: 2695–2701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi K, Nojiri H, Saita Y, Morikawa D, Ozawa Y, Watanabe K et al. Mitochondrial superoxide in osteocytes perturbs canalicular networks in the setting of age-related osteoporosis. Sci Rep 2015; 5: 9148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos JM, Tewari S, Goldberg AF, Kowluru RA. Mitochondrial biogenesis and the development of diabetic retinopathy. Free Radic Biol Med 2011; 51: 1849–1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Nartiss Y, Steipe B, McQuibban GA, Kim PK. ROS-induced mitochondrial depolarization initiates PARK2/PARKIN-dependent mitochondrial degradation by autophagy. Autophagy 2012; 8: 1462–1476. [DOI] [PubMed] [Google Scholar]
- Candas D, Li JJ. MnSOD in oxidative stress response-potential regulation via mitochondrial protein influx. Antioxid Redox Signal 2014; 20: 1599–1617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdel Khalek W, Cortade F, Ollendorff V, Lapasset L, Tintignac L, Chabi B et al. SIRT3, a mitochondrial NAD(+)-dependent deacetylase, is involved in the regulation of myoblast differentiation. PLoS ONE 2014; 9: e114388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H, Lee YD, Kim HJ, Lee ZH, Kim HH. SOD2 and Sirt3 control osteoclastogenesis by regulating mitochondrial ROS. J Bone Miner Res 2017; 32: 397–406. [DOI] [PubMed] [Google Scholar]
- Jonason JH, O'Keefe RJ. Isolation and culture of neonatal mouse calvarial osteoblasts. Methods Mol Biol 2014; 1130: 295–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeBel CP, Ischiropoulos H, Bondy SC. Evaluation of the probe 2',7'-dichlorofluorescin as an indicator of reactive oxygen species formation and oxidative stress. Chem Res Toxicol 1992; 5: 227–231. [DOI] [PubMed] [Google Scholar]
- Hao J, Shen W, Yu G, Jia H, Li X, Feng Z et al. Hydroxytyrosol promotes mitochondrial biogenesis and mitochondrial function in 3T3-L1 adipocytes. J Nutr Biochem 2010; 21: 634–644. [DOI] [PubMed] [Google Scholar]
- Feng Z, Zou X, Jia H, Li X, Zhu Z, Liu X et al. Maternal docosahexaenoic acid feeding protects against impairment of learning and memory and oxidative stress in prenatally stressed rats: possible role of neuronal mitochondria metabolism. Antioxid Redox Signal 2012; 16: 275–289. [DOI] [PubMed] [Google Scholar]
- Zou X, Yan C, Shi Y, Cao K, Xu J, Wang X et al. Mitochondrial dysfunction in obesity-associated nonalcoholic fatty liver disease: the protective effects of pomegranate with its active component punicalagin. Antioxid Redox Signal 2014; 21: 1557–1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Li H, Zheng A, Yang L, Liu J, Chen C et al. Mitochondrial dysfunction-associated OPA1 cleavage contributes to muscle degeneration: preventative effect of hydroxytyrosol acetate. Cell Death Dis 2014; 5: e1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.