Abstract
Astrocyte has crucial roles in the central nervous system and accumulating evidence has shown its core function for brain complexity, plasticity and cognition. However, the essential key factors in the precise regulation of astrocytic differentiation remain largely uncharacterized. Here, we identified that RNF20, an E3 ligase of H2BK120 in the mammalian system, regulates astrocyte production from neural precursor cells. RNF20 deficiency by shRNA knockdown or deletion in conditional knockout mice impairs the astrocytic differentiation. Overexpression of RNF20 promotes astrocytic differentiation and can rescue the astrocyte production deficiency caused by RNF20 disruption. Furthermore, we demonstrate that RNF20 functions cooperatively with acetyltransferase MOF to promote astrocytic generation. RNF20-mediated H2Bub1 cooperating with MOF-mediated H4K16ac activates the transcription of Stat3. Together, these data indicate RNF20 is a critical regulator of astrocytic production, which may contribute to the understanding of neurological disorders with glial dysgenesis.
Cerebral cortical development is sequentially accomplished with neurogenesis and gliogenesis, and the latter orderly consists of astrocytic differentiation and oligodendrocyte production.1, 2 Astrocytic differentiation is an indispensable process whereby neural precursor cells generate astrocytes from embryonic day 16 after the peak of neurogenesis in the mouse brain.3, 4 Increasing evidence has shown that astrocytes play crucial roles in maintaining the normal cognitive network of the cerebral cortex, which contains neurotrophic support,5 neuronal migration,6, 7 synapse formation,8, 9 corpus callosum formation10 and mediation in the function of blood–brain barrier.11 Moreover, astrocyte dysfunction is associated with certain central nervous system disorders.8, 12, 13 However, the intracellular molecular regulatory mechanisms of astrocytic differentiation remain complicated and largely uncharacterized.
Astrocytic differentiation commences when neurogenesis is largely completed in the late embryonic stage.3 Astrocytes are derived from the common multipotent neural precursor cells as neurons.4, 14 Extracellular and intracellular signals cooperatively contribute to the lineage specification of neural precursor cells.3, 15 In mechanism, it has already been reported that Janus kinase-signal transducer and activator of transcription 3 (JAK–STAT3) signaling pathway plays a pivotal role in inducing neural precursor cells differentiating along astrocytic lineage.16, 17 The regulation in lineage specification is accompanied by epigenetic modifications all the way. At the early embroyonic stage, STAT3 is isolated to be accessible to the promoter of astrocyte-related gene, glial fibrillary acidic protein (GFAP), by DNA methylation on the promoter.18 As embryogenesis proceeds, activated STAT3 binds to the derepressed GFAP promoter to achieve asctrocytic potential.18 Then, the JAK–STAT3 pathway cooperates with multiple histone modifications to regulate astrocytic differentiation.19, 20 Furthermore, studies have revealed that the STAT3 level is also regulated by histone modifications.21, 22 Histone modifications maybe enhance or repress the JAK–STAT3 pathway during astrocytic differentiation.
During the process of neural precursor cells self-renewal and differentiation, a variety of epigenetic covalent modifications, such as methylation, acetylation and ubiquitination, dynamically occur on the tail of chromosomal histones. Histone modifications contribute to the fate determination of neural precursor cells and sequential generation of different cell types in the developing brain.20, 23, 24, 25, 26, 27 Furthermore, a part of studies is devoted to shed light on the connection between histone modifications and astrocytic differentiation.20, 28 Evidence has documented that dynamic changes in histone modifications induce transcription factors to be accessible to astrocyte-related gene promoter.28
Histones are subjected to be specifically catalyzed by enzymes on specific locus. RNF20, with a RING finger domain, known as an E3 ligase, catalyzes monoubiquitination of H2BK120 (H2Bub1, equivalent to H2BK123 in yeast) in vertebrates.29, 30, 31, 32 It has been reported that H2Bub1 is related to transcription activity and contributes to generate long transcripts by stimulating transcriptional elongation.33 Furthermore, H2Bub1 triggers following methylation of H3K4 and H3K79, which is also associated with transcriptional activation.34, 35, 36 Accordingly, RNF20 plays crucial roles in the stimulation of gene expression,32 regulation of meiotic recombination,37 suppression of tumorigenesis38 and control of cell size of yeast.30 Notably, previous study has highlighted the necessity of RNF20 in the execution of embryonic stem cells plasticity.39 Furthermore, based on the reported data sets,40 RNF20 is most highly expressed in astrocyte among various cell types in the cerebral cortex, suggesting that RNF20 may be involved in astrocyte production. However, the importance of RNF20 on astrocytic differentiation in the developing brain has never been reported.
Here, we demonstrate that RNF20 is a critical regulator of astrocytic differentiation. We have identified that RNF20 is required and sufficient for astrocytic differentiation. In mechanism, we discover that RNF20-mediated H2Bub1 in synergy with acetyltransferase MOF-mediated H4K16ac regulates STAT3 transcription. Our study suggests that RNF20 increases the expression of STAT3 and promotes astrocytic fate determination of neural precursor cells in the developing brain.
Results
RNF20 is abundantly expressed during the cortical astrocytic differentiation in the developing brain
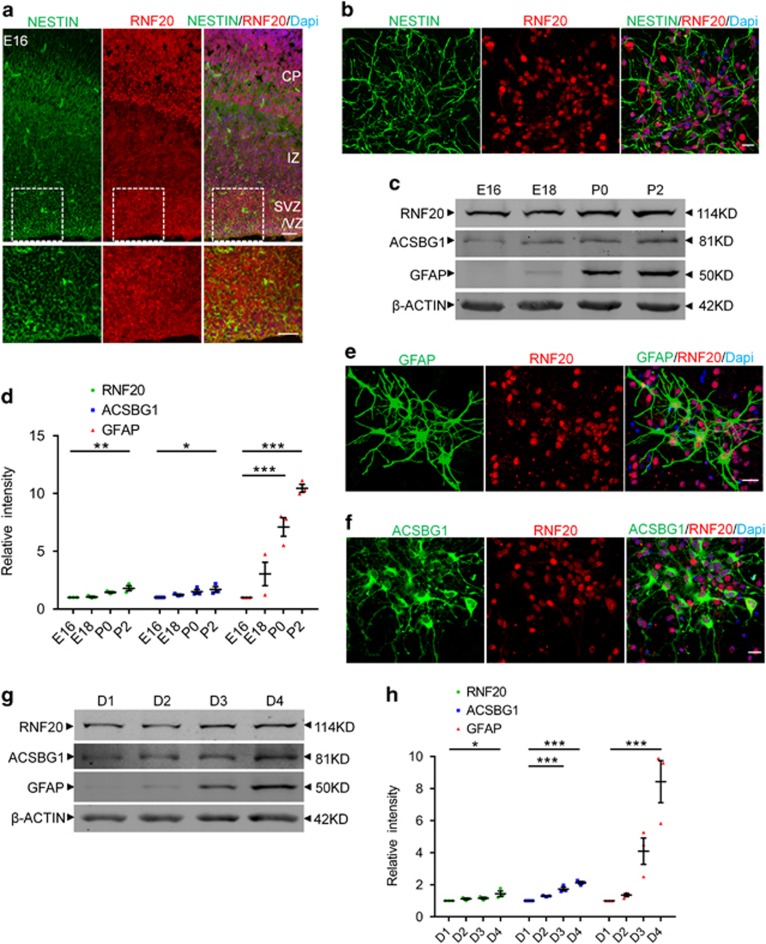
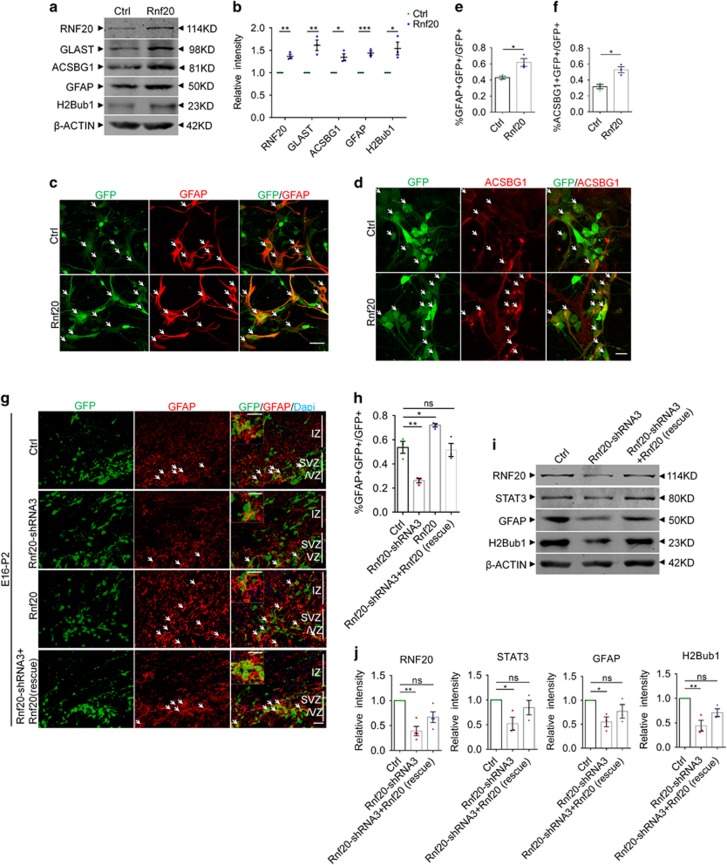
To investigate whether RNF20 plays a role in the astrocytic differentiation of the developing brain, we first analyzed the expression of RNF20 in the late embryonic brain. Immunostaining showed the abundant expression of RNF20 in the ventricular zone (VZ), subventricular zone (SVZ) and cortical plate (CP) of embryonic day (E) 16 cortex (Figure 1a). Importantly, RNF20 was prominently expressed in NESTIN-labeled neural precursor cells both in vivo and in vitro (Figures 1a and b). The protein expression pattern showed that expression of RNF20 increases from E16 to postnatal day (P) 2 in the developing brain (Figures 1c and d). Notably, the expression pattern of RNF20 is consistent with that of white matter astrocyte marker glial fibrillary acidic protein (GFAP) and gray matter astrocyte marker Acyl CoA Synthetase bubblegum family member 1 (ACSBG1)41 during the cerebral cortical astrocytic differentiation (Figures 1c and d). To identify whether RNF20 is expressed in astrocyte, we performed immunostaining in vitro. Results showed that RNF20 is highly expressed in GFAP and ACSBG1 labeled astrocyte (Figures 1e and f). RNF20 expression also showed an increased pattern during the differentiation of neural precursor cells in vitro (Figures 1g and h). These results suggest that RNF20 may participate in regulating the cortical astrocytic differentiation in the developing brain.
Figure 1.
Expression of RNF20 during the cortical astrocytic differentiation in the developing brain. (a) Immunostaining for RNF20 and NESTIN in E16 mouse cerebral cortex. In the VZ/SVZ, RNF20 is abundantly expressed in NESTIN-positive neural precursor cells. Enlarged images of the VZ/SVZ are shown in the lower panels. VZ, SVZ and CP stand for ventricular zone, subventricular zone and cortical plate, respectively. (b) RNF20 is colabeled with NESTIN in isolated E16 neural precursor cells in vitro. (c) Expression of RNF20, GFAP and ACSBG1 from E16, E18, P0 and P2 dorsal cortex is detected with indicated antibodies. (d) Quantification for protein levels of RNF20, ACSBG1 and GFAP in (c). Mean±S.E.M.; n=3. (e and f) RNF20 is colabeled with GFAP and ACSBG1 in astrocytes differentiated for 4 days from E16 neural precursor cells. (g) E16 primary neural precursor cells were differentiated for 1 day (D1), 2 days (D2), 3 days (D3) and 4 days (D4). Protein extracts were subjected to western blotting to detect the expression of RNF20, ACSBG1 and GFAP. (h) Quantification for protein levels of RNF20, ACSBG1 and GFAP in (g). Mean±S.E.M.; n=3. *P<0.05, **P<0.01, ***P<0.001, one-way ANOVA. Scale bar, 20 μm
RNF20 knockdown inhibits astrocytic differentiation from neural precursor cells
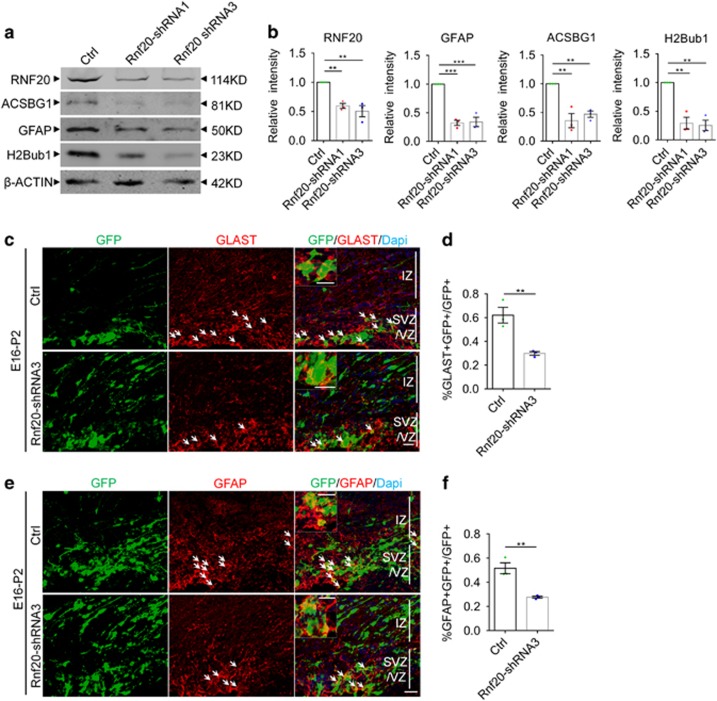
To determine the role of RNF20 in the cerebral cortical astrocytic differentiation, we designed four shRNAs targeting Rnf20. Results showed that all of the four shRNAs efficiently reduced the expression of RNF20 in N2a cells, while Rnf20-shRNA1 and Rnf20-shRNA3 had higher knockdown efficiency (Supplementary Figures S1a and b). We first examined the effects of RNF20 knockdown on differentiation of neural precursor cells from E16 cerebral cortex in vitro. A significant reduction was observed in the expression of astrocyte marker GFAP and ACSBG1 after RNF20 knockdown, accompanied by the reduction of H2Bub1 that is catalyzed by RNF20 (Figures 2a and b). We further knocked down RNF20 in neural precursor cells in vivo through in utero electroporation. We first used the astrocytic precursor marker glial high affinity glutamate transporter (GLAST)41 to investigate the specification of neural precursor cells along astrocytic lineage. We found that among the electroporated GFP-positive cells in VZ, SVZ and IZ, Rnf20-shRNA3 caused a decreased percentage of GLAST-positive astrocytic precursors (Figures 2c and d). Then, we used astrocyte marker GFAP to identify the astrocytic differentiation from neural precursor cells. Rnf20-shRNA3 also reduced the percentage of GFAP-positive astrocytes among the electroporated GFP-positive cells in VZ, SVZ and IZ in vivo (Figures 2e and f). Taken together, these results indicate that RNF20 knockdown inhibits the specification of neural precursor cells along astrocytic lineage and results in the decreased astrocytic differentiation.
Figure 2.
RNF20 knockdown reduces astrocyte production. (a) E16 neural precursor cells were isolated, infected and differentiated for 4 days. Western blotting showed the expression of RNF20, ACSBG1, GFAP and H2Bub1 with indicated antibodies. (b) Quantification for protein levels of RNF20, ACSBG1, GFAP and H2Bub1 in (a). Mean±S.E.M.; n=3. (c) Embryos from E16 pregnant mouse were electroporated with control (pSicoR-GFP) or Rnf20-shRNA3 plasmids. The percentage of GLAST-positive astrocytic precursor cells was investigated at P2. The arrows indicate GLAST-positive astrocytic precursor cells that were colabeled with GFP-positive cells. Enlarged images showed the colocalization of GLAST and GFP. (d) Quantification for the percentage of GLAST-positive astrocytic precursor cells in all of the GFP-positive cells in VZ, SVZ and IZ. Mean±S.E.M.; n=3. (e) E16 embryos were subjected to in utero electroporation and analyzed at P2. Immunostaining for GFAP was performed. The arrows indicate GFAP-positive astrocytes that were colabeled with GFP-positive cells. Enlarged images showed the colocalization of GFAP and GFP. (f) Quantification for the percentage of GFAP-positive astrocytes in all of the GFP-positive cells in VZ, SVZ and IZ. Mean±S.E.M.; n=3. **P<0.01, ***P<0.001, one-way ANOVA for (b), Student’s t-test for (d) and (f). Scale bar, 20 μm
Rnf20 conditional knockout leads to a continuous failure of astrocyte production
Knockdown experiments could not identify the effects of RNF20 on long-term astrocytic differentiation. To address this question, we use Rnf 20flox/flox mice crossing with hGFAP-Cre mice to generate Rnf 20flox/flox; hGFAP-Cre mice (referred to as hGFAP-Rnf 20−/−). Importantly, RNF20 was specifically knocked out in hGFAP promoter controlled precursor cells in this mutant model, which allows us to further confirm the role of RNF20 on astrocytic differentiation.
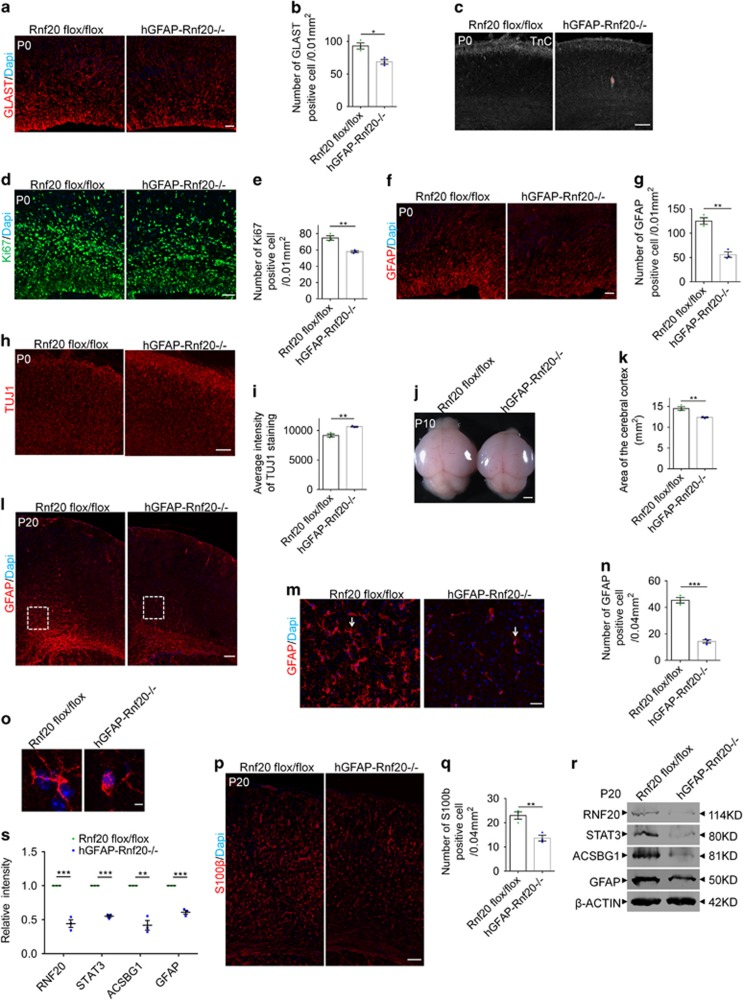
First, we found RNF20 was greatly reduced in the hGFAP-Rnf 20−/− brain at P0 (Figures 6c and d).Then, we studied the production of astrocytic precursors. Two astrocytic precursor markers GLAST and Tenascin C (TnC)41 showed moderate decreased production of astrocytic precursors in conditional knockout brain at P0 (Figures 3a–c). Immunostaining for Ki67 showed less proliferation of astrocytic precursors at P0 (Figures 3d and e). Furthermore, when we examined the astrocytic differentiation, we found that there was a decrease in the production of astrocytes immunostained by GFAP at P0 (Figures 3f and g). As compensation, a few more neurons were produced in conditional knockout brains at P0 (Figures 3h and i). To investigate the potential influence from apoptosis, we performed TUNEL staining and did not find significant differences between wild type and conditional knockout brains at P0 (Supplementary Figures S2a–c). Through continuing observation, we found that the area of the cerebral cortex in conditional knockout brains was smaller than that in wild type at P10 (Figures 3j and k).
Figure 3.
RNF20 deletion in hGFAP-Rnf 20−/− mice causes a persistent deficiency in astrocyte production. (a) Immunostaining showed the reduced expression of GLAST in dorsal cortices of hGFAP-Rnf 20−/− mice at P0. Scale bar, 20 μm. (b) Quantification for the number of GLAST-positive cell. Mean±S.E.M.; n=3. (c) Immunostaining showed the reduced expression of TnC in dorsal cortices of hGFAP-Rnf 20−/− mice at P0. Scale bar, 100 μm. (d) Immunostaining showed the reduced expression of Ki67 in dorsal cortices of hGFAP-Rnf 20−/− mice at P0. Scale bar, 50 μm. (e) Quantification for the number of Ki67-positive cell. Mean±S.E.M.; n=3. (f) Immunostaining showed the reduced expression of GFAP in dorsal cortices of hGFAP-Rnf 20−/− mice at P0. Scale bar, 20μm. (g) Quantification for the number of GFAP-positive cell. Mean±S.E.M.; n=3. (h) Immunostaining showed the increased expression of TUJ1 in dorsal cortices of hGFAP-Rnf 20−/− mice at P0. Scale bar, 100μm. (i) Quantification for the average intensity of TUJ1-positive cell. Mean±S.E.M.; n=3. (j) Morphological abnormality in size between wild type and hGFAP-Rnf 20−/− mice at P10. Scale bar, 1 mm. (k) Quantification for the area of cerebral cortex. Mean±S.E.M.; n=3. (l) The number of GFAP-positive astrocytes is dramatically reduced in the corpus collosum of the hGFAP-Rnf 20−/− mice at P20. Scale bar, 100 μm. (m) Higher magnification views from the dotted line area in (l) showed the reduction of GFAP expression in the cortex of the hGFAP-Rnf 20−/− mice. Scale bar, 20 μm. (n) Quantification for the number of GFAP-positive cell. Mean±S.E.M.; n=3. (o) Higher magnification for single GFAP-positive astrocyte labeled with arrows in (m) showed the morphological difference between wild type and hGFAP-Rnf 20−/− mice. Scale bar, 5 μm. (p) Immunostaining showed the reduction of S100β-positive astrocytes in the dorsal cortices of hGFAP-Rnf 20−/− mice at P20. Scale bar, 100 μm. (q) Quantification for the number of S100β-positive cell. Mean±S.E.M.; n=3. (r) Western blotting showed the reduced expression of RNF20, STAT3, ACSBG1 and GFAP in dorsal cortices of hGFAP-Rnf 20−/− mice at P20. (s) Levels of RNF20, STAT3, ACSBG1 and GFAP from (r) were quantified. Mean±S.E.M.; n=3. *P<0.05, **P<0.01, ***P<0.001, Student’s t-test
To address whether the astrocytic differentiation is delayed or prevented, we performed immunostaining at P20. We first found that RNF20 was expressed in GFAP-positive mature astrocyte in wild-type brain, but not in hGFAP-Rnf 20−/− brain (Supplementary Figure S2d). Western blotting also showed a clear reduced expression of RNF20 at P20 in conditional knockout brain cortex (Figures 3r and s).Then, immunostaining for GFAP in the white matter41 showed a persistent reduction at P20 in conditional knockout brain (Figures 3l–n). The astrocyte morphology in conditional knockout brains labeled by GFAP was less mature than that in wild type (Figures 3m and o). Meanwhile, immunostaining for S100β which labels mature astrocyte showed less astrocyte production across the cerebral cortex (Figures 3p and q). Also, western blotting further confirmed that expression of ACSBG1 and GFAP was reduced at P20 in hGFAP-Rnf 20−/− brain (Figures 3r and s). These findings indicate that conditional knockout of Rnf20 results in a persistent failure of astrocytic differentiation.
Deletion of RNF20 by Cre lentivirus in Rnf 20 flox/flox neural precursor cells impairs astrocyte production in vitro
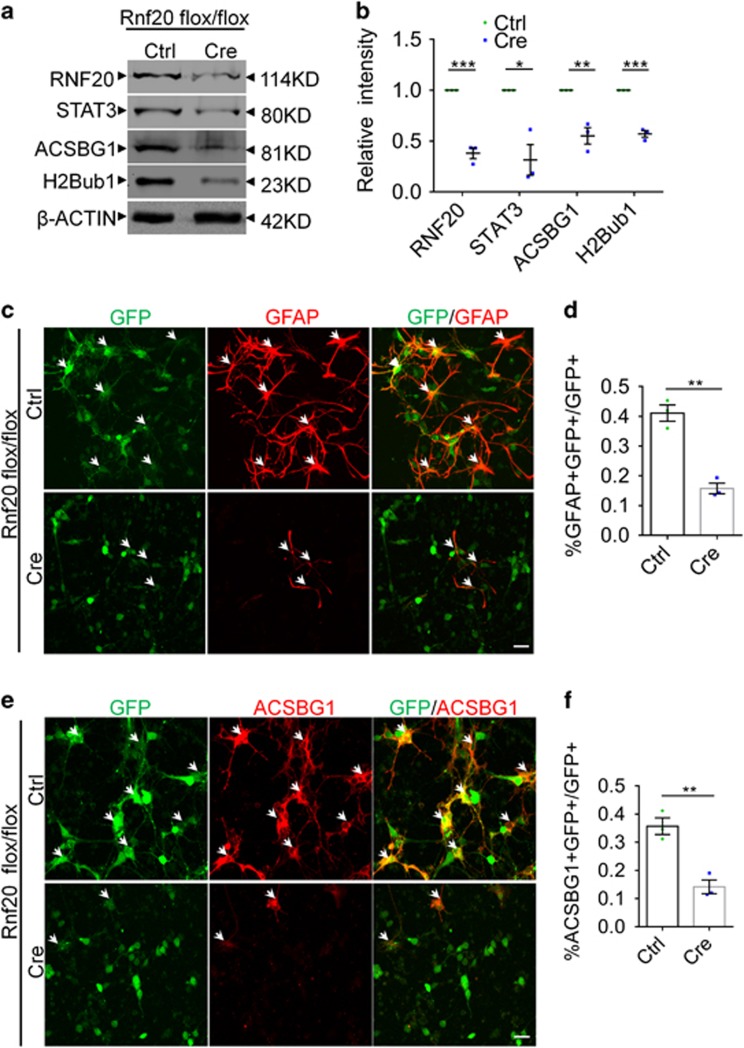
After identifying the requirement of RNF20 for astrocyte production in vivo, we further investigated the role of RNF20 in vitro. Control and Cre lentivirus were packaged and used to infect E16 Rnf 20flox/flox neural precursor cells. Western blotting showed high recombination efficiency of Cre recombinase (Figures 4a and b). Consistent with our results in vivo, RNF20 knockout in vitro also reduced the expression of ACSBG1 accompanied by the reduction of H2Bub1 (Figures 4a and b). Immunostaining illustrated that RNF20 knockout prevented neural precursor cells from differentiating into astrocytes labeled by GFAP and ACSBG1 (Figures 4c–f). Taken together, these results suggest that RNF20 is required for the astrocytic differentiation of neural precursor cells both in vivo and in vitro.
Figure 4.
Astrocyte production is inhibited in Cre lentivirus-mediated RNF20 conditional knockout in Rnf 20flox/flox precursor cells in vitro. (a) Western blotting showed the reduced expression of RNF20, STAT3, ACSBG1 and H2Bub1 in Cre lentivirus-infected Rnf 20flox/flox primary precursor cells. E16 Rnf 20flox/flox neural precursor cells were infected with control or Cre lentivirus and differentiated for 4 days in vitro. (b) Quantification for protein levels of RNF20, STAT3, ACSBG1 and H2Bub1 in (a). (c) Immunostaining showed the reduction of GFAP-positive astrocyte production in Cre lentivirus-infected Rnf 20flox/flox primary cells which were isolated at E16 and differentiated for 4 days in vitro. (d) The percentage of GFAP-positive astrocytes in GFP-positive cells was quantified. Mean±S.E.M.; n=3. (e) Immunostaining showed the reduction of ACSBG1-positive astrocyte production in Cre lentivirus-infected Rnf 20flox/flox primary cells which were isolated at E16 and differentiated for 4 days in vitro. (f) The percentage of ACSBG1-positive astrocytes in GFP-positive cells was quantified. Mean±S.E.M.; n=3. *P<0.05, **P<0.01, ***P<0.001, Student’s t-test. Scale bar, 20 μm
RNF20 is sufficient for astrocytic differentiation
As we have found out that RNF20 is required for astrocytic differentiation, then we wonder whether overexpression of RNF20 is able to promote astrocytic differentiation. We constructed RNF20 overexpression plasmid, both western blotting and immunostaining showed the efficiency of RNF20 overexpression (Supplementary Figures S3a and b). We found that overexpression of RNF20 in E16 neural precursor cells promoted the expression of astrocytic precursor marker GLAST, and leads to increased expression of astrocyte marker ACSBG1 and GFAP (Figures 5a and b). The increase of H2Bub1 suggests overexpression efficiency of RNF20 in vitro (Figures 5a and b). To confirm the effect of RNF20 on astrocytic fate, we conducted immunostaining in primary cells in vitro. We found that overexpression of RNF20 increased the percentage of colabeled GFAP and GFP-positive astrocyte (Figures 5c and e), and it had the same effect on ACSBG1-labeled astrocyte (Figures 5d and f). In utero electroporation showed that overexpression of RNF20 increased the percentage of GLAST-positive astrocytic precursors and the percentage of GFAP-positive astrocytes in vivo (Supplementary Figures S3c–f). We further observed that co-electroporated Rnf20-shRNA3 resistant-Rnf20 can restore astrocyte production defect caused by Rnf20-shRNA3 in vivo (Figures 5g and h). At the same time, overexpression of Rnf20-shRNA3 resistant-Rnf20 in E16 neural precursor cells can rescue GFAP expression level with a positive control of H2Bub1 in vitro (Figures 5i and j). Together, our results suggest that RNF20 is both required and instructive for astrocytic differentiation.
Figure 5.
RNF20 is sufficient for astrocytic differentiation. (a) E16 neural precursor cells were isolated, infected and differentiated for 4 days in vitro. Western blotting showed the expression of RNF20, GLAST, ACSBG1, GFAP and H2Bub1 with indicated antibodies. (b) Quantification for protein levels of RNF20, GLAST, ACSBG1, GFAP and H2Bub1 in (a). Mean±S.E.M.; n=3 (n=4 for RNF20). (c and d) Immunostaining showed that overexpression of RNF20 increased the production of GFAP- or ACSBG1-positive astrocytes. E16 neural precursor cells were isolated, infected and differentiated for 4 days in vitro. Scale bar in (c) 50μm, in (d) 20 μm. (e and f) The percentage of GFAP- or ACSBG1-positive cells in all the infected GFP-positive cells in (c) and (d) was quantified. Mean±S.E.M.; n=3. (g) Embryos from E16 pregnant mouse were electroporated with control, Rnf20-shRNA3, Rnf20 or co-electroporated with Rnf20-shRNA3 resistant-Rnf20 and Rnf20-shRNA3, and brains were harvested and analyzed at P2. Immunostaining for GFAP was performed. Enlarged images showed the colocalization of GFAP and GFP. Scale bar, 20 μm. (h) The percentage of GFAP-positive astrocytes in GFP-positive cells in VZ, SVZ and IZ was quantified. Mean± S.E.M.; n=3. (i) E16 neural precursor cells were infected with control or Rnf20-shRNA3 lentivirus, or co-infected with Rnf20-shRNA3 resistant-Rnf20 and Rnf20-shRNA3 lentivirus, and differentiated for 4 days in vitro. Western blotting showed the rescue experiments in vitro. Expression of RNF20, STAT3, GFAP and H2Bub1 were detected with indicated antibodies. (j) Quantification for protein levels of RNF20, STAT3, GFAP and H2Bub1 in (i). Mean±S.E.M.; n=3. *P<0.05, **P<0.01, ***P<0.001. NS, not significant; Student’s t-test for (b, e and f), one-way ANOVA for (h) and (j)
RNF20 promotes astrocyte production by enhancing the expression of STAT3
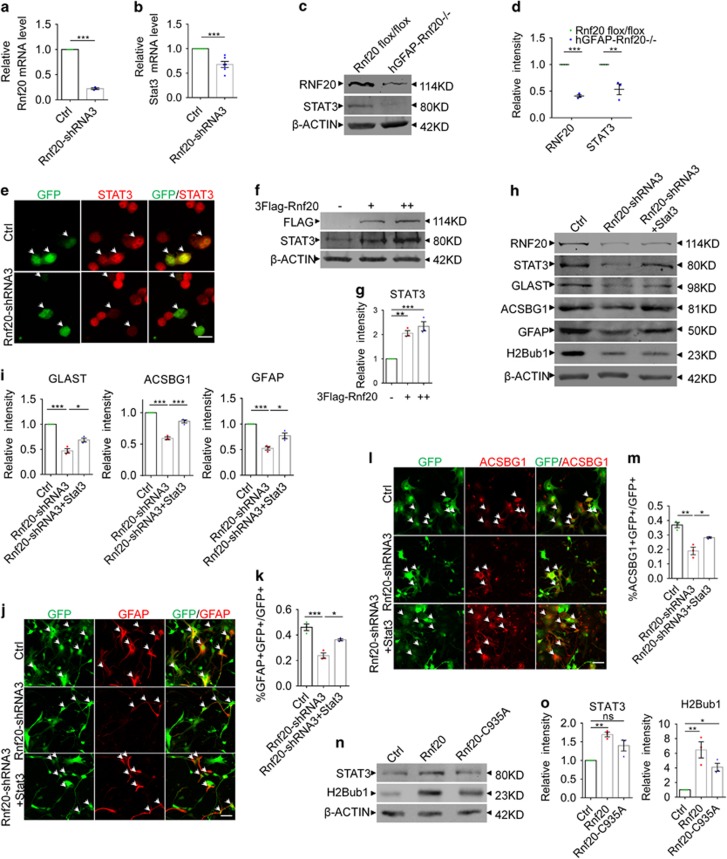
Previous studies have elucidated that transcription factor STAT3 can promote the expression of GFAP. Our discovery of RNF20 regulating astrocytic differentiation urged us to investigate whether STAT3 signaling is regulated by RNF20. Results from real-time PCR showed that RNF20 knockdown reduced the Stat3 mRNA level (Figures 6a and b). Western blotting showed that expression of STAT3 was reduced at P0 in the brain of hGFAP-Rnf 20−/− mice (Figures 6c and d), and a continuous reduction of STAT3 was observed at P20 (Figures 3r and s). Immunostaining demonstrated that lower immunoreactivity was detected in cells transfected with Rnf20-shRNA3 than in neighboring control cells, while there was no difference in the group transfected with control plasmid (Figure 6e). It turned out that overexpression of shRNA resistant-Rnf20 can restore the STAT3 expression level which was reduced in the RNF20 knockdown group (Figures 5i and j). We further found that STAT3 expression increased gradually in a RNF20 dose-dependent manner (Figures 6f and g). Finally, overexpression of STAT3 can partially restore the expression of astrocytic precursor marker GLAST, and astrocytic marker ACSBG1 and GFAP in the RNF20 knockdown group in vitro (Figures 6h and i). Immunostaining for GFAP (Figures 6j and k) and ACSBG1 (Figures 6l and m) in primary cells showed that STAT3 overexpression can partially rescue the astrocytic fate deficiency caused by RNF20 knockdown.
Figure 6.
RNF20 promotes the expression of STAT3. (a and b) Real-time PCR showed that RNF20 knockdown in E16 neural precursor cells reduced the relative mRNA level of STAT3 under differentiation condition for 4 days. Mean±S.E.M.; n=3 for Rnf20, n=6 for Stat3. (c) Western blotting showed the reduction of RNF20 and STAT3 in dorsal cortices of hGFAP-Rnf 20−/− mice at P0. (d) Protein levels of RNF20 and STAT3 in (c) were quantified. Mean±S.E.M.; n=3. (e) N2a cells were transfected with control or Rnf20-shRNA3 plasmids, and cultured for 48–72 h. Immunostaining showed Rnf20-shRNA3 reduces the immunoreactivity of STAT3. Scale bar, 20 μm. (f) Western blotting showed the increase of STAT3 expression is dependent on the dosage of RNF20. N2a cells were transfected with control or 3Flag-Rnf20 plasmids and cultured for 48–72 h. +, 1 μg; ++, 2 μg. (g) Protein levels of RNF20 and STAT3 in (f) were quantified. Mean±S.E.M.; n=3. (h) E16 neural precursor cells were infected with control or Rnf20-shRNA3 lentivirus, or co-infected with Rnf20-shRNA3 and Stat3 lentivirus, and cultured for 4 days under differentiation condition. Western blotting showed STAT3 can rescue the astrocyte production deficiency. RNF20, STAT3, GLAST, ACSBG1, GFAP and H2Bub1 were detected with indicated antibodies. (i) Protein levels of GLAST, ACSBG1 and GFAP in (h) were quantified. Mean±S.E.M.; n=3. (j and l) Immunostaining of GFAP and ACSBG1 showed that overexpression of STAT3 partially rescued the deficiency caused by Rnf20 knockdown. E16 neural precursor cells were isolated, infected and differentiated for 4 days. Scale bar, 50 μm. (k and m) Percentage of GFAP- or ACSBG1-positive cells was quantified. Mean±S.E.M.; n=3. (n) The mutant Rnf20-C935A failed to increase the expression of STAT3. N2a cells were transfected with control, Rnf20 or Rnf20-C935A plasmids, and cultured for 48–72 h. (o) Protein levels of STAT3 in (n) were quantified. Mean±S.E.M.; n=3. *P<0.05, **P<0.01, ***P<0.001, Student’s t-test for (a, b and d), one-way ANOVA for (g, i, k, m and o)
These results clearly demonstrate that RNF20 promotes the expression of STAT3. Previous studies have reported that RNF20 is an E3 ligase with a RING finger domain. Therefore, we introduced a mutation into the RING finger domain at the critical Cysteine 935. We found that, unlike overexpression of RNF20, RNF20-C935A mutant could not significantly increase the expression of STAT3 accompanied by partially decreased H2Bub1 (Figures 6n and o). According to these findings, it indicates that RNF20 regulates the expression of STAT3 associated with its E3 ligase function on H2B.
RNF20 is required for MOF recruitment to Stat3 gene locus
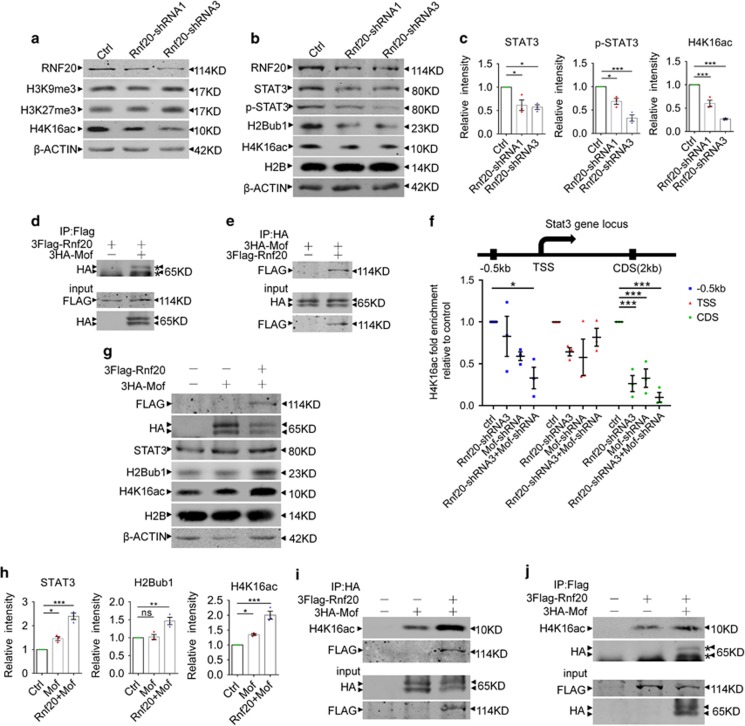
Previous studies have shown that extensive crosstalk exists among histone modifications. We wonder that whether H2Bub1 catalyzed by RNF20 further triggers some histone modification. To validate our hypothesis, we screened several histone modifications in RNF20 knockdown E16 neural precursor cells. We found that the H4K16 acetylation level (H4K16ac) was greatly reduced after RNF20 knockdown (Figures 7a and c). Consistent with our results, expression of both STAT3 and phosphorylated STAT3 was reduced (Figures 7b and c). These results can be repeated in N2a cells (Supplementary Figures S4a and b). Accordingly, RNF20 overexpression increased H2Bub1 and H4K16ac (Supplementary Figures S4c and d). The same change of H2Bub1 and H4K16ac reminds us a question: whether there is a connection between the two histone modifications (Figures 2b and 7c).
Figure 7.
RNF20 recruits MOF to Stat3 gene locus to acetylate H4K16. (a) Histone modifications screening after RNF20 knockdown via lentivirus infection in E16 primary neural precursor cells followed by 4 days differentiation. Histone modifications were detected with the indicated antibodies. (b) Western blotting showed the expression of RNF20, STAT3, p-STAT3, H2Bub1 and H4K16ac after lentivirus infection in E16 neural precursor cells followed by 4 days differentiation. (c) Protein levels of STAT3, p-STAT3 and H4K16ac in (b) were quantified. Mean±S.E.M.; n=3. (d and e) Co-IP assay was performed to examine the interaction between RNF20 and MOF. In (d), N2a cells were co-transfected with 3Flag-Rnf20 and control plasmids, or 3Flag-Rnf20 and 3HA-Mof plasmids. In (e), N2a cells were co-transfected with 3HA-Mof and control plasmids, or 3Flag-Rnf20 and 3HA-Mof plasmids. After 48–72 h, anti-Flag (d) or anti-HA (e) beads were used to pull down the proteins. Results were visualized by western blotting with the indicated antibodies. (f) Ch-IP assay showed knockdown of RNF20 promoted the reduction of H4K16ac caused by MOF knockdown at Stat3 coding region and the promoter. N2a cells were transfected with indicated plasmids and cultured for 48–72 h. (g) Western blotting showed that co-overexpression of 3Flag-Rnf20 and 3HA-Mof promotes more H4K16 acetylation and STAT3 expression than overexpression of single 3HA-Mof. N2a cells were transfected with indicated plasmids and cultured for 48–72 h. Results were visualized with indicated antibodies. (h) Protein levels of STAT3, H2Bub1 and H4K16ac in (g) were quantified. Mean±S.E.M.; n=3. (i and j) Co-IP assay showed that more H4K16ac was pulled down with anti-HA beads or anti-Flag beads in the 3Flag-Rnf20 and 3HA-Mof co-overexpressed group than in the 3HA-Mof or 3Flag-Rnf20 group. In (i and j), N2a cells were transfected with indicated plasmids and cultured for 48–72 h. Western blotting in (i) and (j) were performed with indicated antibodies. *P<0.05, **P<0.01, ***P<0.001, one-way ANOVA
To address this question, we cloned MOF acetylase which catalyzes H4K16 acetylation, and tested the interaction between the RNF20 E3 ligase and MOF acetylase. As illustrated by Co-IP experiment in N2a cells, Flag-tagged RNF20 can clearly pull down HA-tagged MOF (Figure 7d). Similarly, HA-tagged MOF doubtlessly pulls down Flag-tagged RNF20 (Figure 7e). Moreover, we obtained a better result in 293FT cells which have higher transfection efficiency (Supplementary Figures S4e and f). These results indicate that RNF20 strongly interacts with MOF.
As RNF20 knockdown reduced H4K16ac and RNF20 interacted with MOF, we reason that RNF20 is necessary for MOF recruitment to H4K16 at Stat3 gene locus. Chromatin immunoprecipitation (Ch-IP) assay demonstrated that either MOF or RNF20 suppression resulted in reduced H4K16ac mainly at the Stat3 coding region, and knockdown of RNF20 further promoted the reduction caused by MOF knockdown (Figure 7f). At the promoter of 0.5 kb upstream from Stat3 TSS (transcription start site), H4K16ac was reduced when RNF20 and MOF were both knocked down (Figure 7f). Accordingly, overexpression of RNF20 promoted the increase of H4K16ac caused by MOF overexpression at the Stat3 coding region (Supplementary Figure S4g). In protein level, co-overexpression of RNF20 and MOF further confirmed their association in H4K16ac. We identified that MOF overexpression can increase H4K16ac, and RNF20 grossly improves the activity of MOF acetylase (Figures 7g and h). Meanwhile, we found that the change of STAT3 expression coincides with that of H4K16ac (Figures 7g and h). However, H2Bub1 was increased only by RNF20 rather than MOF (Figures 7g and h). We further found that single RNF20 could increase H4K16ac probably by recruiting endogenous MOF (Supplementary Figures S4h and i). Co-IP assay further illuminated that more H4K16ac was pulled down with anti-HA beads in the 3Flag-Rnf20 and 3HA-Mof co-overexpressed group than in the 3HA-Mof and control plasmid co-overexpressed group (Figure 7i). Similarly, more H4k16ac was pulled down with anti-Flag beads in the co-overexpressed group than in the 3Flag-Rnf20 and control plasmid co-overexpressed group (Figure 7j). These findings indicate that RNF20 recruits MOF to acetylate H4K16 at Stat3 gene locus to promote Stat3 transcription.
Discussion
Astrocyte plays critical roles in brain network, while studies on astrocytic differentiation are far from matching with its significance. There are still some questions to be addressed during astrocytic differentiation. The differentiation process from neural precursor cells to astrocytes remains to be obscure, and more studies are needed to explain the process. We provide a perspective on histone modification to investigate the role of RNF20 in astrocytic differentiation. In consideration of the high expression level of RNF20 in astrocyte, we devoted to investigate its role in astrocytic differentiation. With in vivo experiments of RNF20 knockdown by in utero electroporation, we observed that depletion of RNF20 inhibited astrocyte production. In vivo and in vitro experiments of RNF20 conditional knockout further confirmed the phenotype. Our results demonstrated that RNF20 is required for astrocytic differentiation, and RNF20 overexpression promotes the process. Therefore, RNF20 is essential for the differentiation of neural precursor cells along astrocytic lineage.
Evidence has been accumulated to document that astrocyte plays critical roles in orchestrating neural circuits.5, 8, 12 Thus, it is equally pivotal to illuminate the mechanism underlying astrocyte production. Exploration in the molecular mechanism contributes to uncover potential possibilities in the pathology of central nervous system disorders. Studies have verified that JAK–STAT3 signaling pathway controls the astrocytic differentiation, whereas details in the regulation of pathways remain largely unknown. Here, we demonstrate that RNF20-mediated H2Bub1 coordinated with MOF-mediated H4K16ac to regulate the expression level of STAT3. Our findings first highlight the significance of these two histone modifications in astrocytic differentiation from neural precursor cells, which is coincident with cross-regulation among histone modifications.42
RNF20-mediated H2Bub1 triggers H3K4 and H3K79 methylation by stimulation of methyltransferase COMPASS and Dot1,36 and both of them promote active transcription. Additionally, we found another reliable way of RNF20-mediated H2Bub1 regulating transcriptional activation. H2Bub1 stimulated MOF to acetylate H4K16, and then cooperated with H4K16ac to promote the transcription of Stat3. Our study provides a new insight in the crosstalk between histone modifications. The crosstalk between H2Bub1 and H4K16ac indicates that modifications on different histone tails cooperate with each other to achieve specific gene transcription. Our findings suggest that RNF20 interacts with MOF to respectively catalyze their substrate histone in order to promote the transcription of Stat3. Our results drive and extend the study on H2Bub1 in transcription activation.
Astrocyte is an important component in maintaining high speed operation of the brain. Thus, abnormalities in astrocyte production are associated with a series of central nervous system disorders. Studies have paid attention to cognitive impairments caused by epigenetic dysfunction in neuron,43 while similar work needs to be done in astrocyte. Our study proves the significance of RNF20 in astrocytic differentiation as an epigenetic regulator, and explains the reason of high expression level of RNF20 in astrocyte. Taken together, disruption of RNF20 inhibits the differentiation of neural precursor cells along astrocytic lineage, and RNF20 is required and sufficient for astrocyte production through regulating Stat3 transcription by cooperation of H2Bub1 and H4K16ac. Our work was conducted at early astrocyte stages, and it is possible that RNF20 is also required for terminal astrocytic differentiation. In the future, extensive work needs to be done at late astrocyte stages to address this question. Our study provides new insights of RNF20 in astrocytic differentiation, and its regulatory mechanism may represent a new perspective of pathological study in central nervous system disorders.
Materials and methods
Animals
The Rnf 20flox/flox mice were generated according to the procedures as previously described.37 The hGFAP-Cre mice line was obtained from Jackson Laboratory (Bar Harbor, ME, USA). Homozygous Rnf 20flox/flox mice were mated to hGFAP-Cre mice to generate Rnf 20flox/+; hGFAP-Cre mice, and the F1 generation was crossed with homozygous Rnf 20flox/flox mice to generate Rnf 20flox/flox; hGFAP-Cre mice (referred to as hGFAP-Rnf 20−/−). Genotyping PCR primers of Rnf20 were as follows: forward, GCTGTAAGAGTTCTTAATGTATG, and reverse, GGCTTGTCACACAAGCATGAGCATC. The pregnant female mice of ICR line for in utero electroporation were purchased from Vital River Laboratories (Beijing, China). All of the animal experiments were conducted according to the Guide for the Care and Use of Laboratory Animals, and were approved by the Animal Committee of Institute of Zoology, Chinese Academy of Sciences.
Plasmid constructs
Sequences of the three small hairpin RNAs (Rnf20-shRNA1, 2, 4) targeting Rnf20 were obtained from Sigma (Clone ID of Rnf20-shRNA1: NM_182999.1-2559s1c1, Clone ID of Rnf20-shRNA2: NM_182999.1-580s1c1, Clone ID of Rnf20-shRNA4: NM_182999.1-1234s1c1). The Sequence of Rnf20-shRNA3 was obtained from previous study.44 The Rnf20-shRNAs were cloned into pSicoR-GFP vector. Flag-tagged full-length Rnf20, Rnf20-shRNA3 resistant-Rnf20 (rescue) and the mutated Rnf20-C935A were amplified by PCR, and cloned into pCDH vector. HA-taged Mof was cloned into pCDH vector. The target sequences of RNF20-shRNA were as follows:
Rnf20-shRNA1, 5′-GCGAATCAAGTCTAATCAG-3′
Rnf20-shRNA2, 5′-CGCATCATCCTTAAACGTT-3′
Rnf20-shRNA3, 5′-GGAGAGAGAACGAGAGAAA-3′
Rnf20-shRNA4, 5′-CGGAAGTTTGAGGAAATGA-3′.
Antibodies
The primary antibodies used in immunostaining were as follows: RNF20 (21615-1-AP; Proteintech, Rosemont, IL, USA), NESTIN (MAB353; Millipore, Darmstadt, Germany), ACSBG1 (ab118154; Abcam, Cambridge, UK), GFAP (G6171; Sigma, St. Louis, MO, USA), GFAP (Z0334 Dako, Santa Clara, CA, USA), GFAP (60190-1-Ig; Proteintech), GLAST (BMP009; MBL, Beijing, China), Tenascin C (110-68136; NB, Littleton, CO, USA), Ki67 (ab137876; Abcam), FLAG (F1804; Sigma), STAT3 (#9139; Cell Signaling Technology, Beverly, MA, USA). The primary antibodies used in western blotting were as follows: RNF20 (21615-1-AP; Proteintech), GLAST (20785-1-AP; Proteintech), ACSBG1 (ab118154; Abcam), GFAP (G6171; Sigma), STAT3 (#9139; Cell Signaling Technology), phospho-STAT3 (#9145; Cell Signaling Technology), Ubiquityl-Histone H2B (Lys 120) (#5546; Cell Signaling Technology), H4K16ac (07-329; Millipore), H3K9me3 (07-442; Millipore), Trimethyl-Histone H3 (Lys 27) (07-449; Millipore), H2B (ab1790; Abcam), FLAG (F1804; Sigma), HA (M20003; Abmart, Shanghai, China), β-ACTIN (20536-1-AP; Proteintech). Alexa-conjugated secondary antibodies (Jackson ImmunoResearch, West Grove, PA, USA) were used for immunostaining. IR Dye (LI-COR Biosciences, Lincoln, Nebraska, USA) 800 CW donkey anti-rabbit and 680 CW donkey anti-mouse secondary antibodies were used in western blotting.
Neural precursor cell culture
The isolation of cortical neural precursor cells from the dorsal cortex of E16 embryos was performed as follows. Cerebral cortices were isolated from E16 embryos, and then digested in papain at 37 °C for 5 min followed by three times quick washes with DMEM. Cortices were dissociated into single cells with a pipette in medium. The mixture was filtered with 70 μm filter membrane, and then centrifuged at 1000 r.p.m. for 5 min. The resuspended neural precursor cells were plated on poly-l-ornithine (Sigma, 10μg/ml) and laminin (Sigma, 5μg/ml) coated plates and cultured in proliferation medium overnight. The proliferation medium includes Neurobasal-A and DMEM/F12 (Invitrogen, Carlsbad, CA, USA), supplemented with 2% B27 (Invitrogen), 0.5% GlutaMAX, 5 ng/ml basic fibroblast growth factor (Invitrogen), 5 ng/ml epidermal growth factor (Invitrogen) and 1% penicillin/streptomycin (Invitrogen). After lentivirus infection, cells were cultured in differentiation medium for 4 days. The differentiation medium includes low-glucose DMEM (Gibco, Grand Island, NY, USA), supplemented with 2% B27, 1% fetal bovine serum (Invitrogen) and 1% penicillin/streptomycin (Invitrogen).
Lentivirus production and infection
Lentiviral DNA plasmids mixed with packaging plasmids were transfected into HEK293FT cells mediated by GenEscortI (Wisegen, Nanjing, China). The medium containing lentivius was harvest at 24, 48 and 72 h after tranfection. 293FT cells were used to detect the titer of lentivirus. For lentivirus infection, according to MOI (multiplicity of infection)=5, lentivirus was mixed with proliferation medium in the presence of 4μg/ml polybrene to infect neural precursor cells for 8 h. The neural precursor cells were differentiated in differentiation medium for 4 days. Further analysis, such as western blotting, immunostaining, and real-time PCR (RT-PCR) is performed at last.
Western blotting
Dorsal cortices or cultured cells were collected in lysis buffer RIPA (Solarbio, Beijing, China) containing 10 mM phenylmethanesulfonyl fluoride (PMSF) and cocktail, sonicated and cleared by centrifugation. Protein extracts were run on sodium dodecyl sulfate-polyacrylamide gels, and semi-dry transferred onto nitrocellulose membranes. The membranes were blocked in 5% defatted milk dissolved in PBST (PBS with 0.05% Tween-20) for 1 h at room temperature and then incubated with the primary antibodies overnight at 4 °C. Next day, the membranes were washed in PBST, incubated with the secondary antibodies for 1 h at room temperature and washed in PBST. ODYSSEY Sa Infrared Imaging System (LI-COR Biosciences) was used to scan the membranes.
In utero electroporation
In utero electroporation was performed as previously performed.45 Briefly, E16 pregnant mice were deeply anesthetized with sodium pentobarbital by intraperitoneal injection. Plasmids mixed with fast green (Sigma) were gently microinjected into the lateral ventricle of embryos. Embryonic brains were delivered five 50 ms pulses at 50V with a 950 ms interval using an electroporator (BTX ECM830) with 5 mm diameter electrodes.
Immunostaining
Embryonic brains were dissected, fixed in 4% paraformaldehyde for 24 h and dehydrated with 30% sucrose in PBS for 48 h at 4 °C. Fifteen micrometer cryosections were obtained with a freezing microtome (Leica, CM1950, Nussloch, Germany), and immunostained as the following procedures. Following fixation in 4% paraformaldehyde for 20 min, slices were washed in PBS containing 0.1% Triton X-100 (0.1% PBST, 1% PBST for protein in nucleus), blocked in 5% albumin bovine serum in 0.1% PBST for 1 h, and incubated with primary antibodies overnight at 4 °C. Slices were washed in 0.1% PBST, and incubated with secondary antibodies at room temperature for 1 h. Slices were washed with PBS containing 0.1% Triton X-100 three times prior to nuclear counterstaining with 4,6-diamidino-2-phenylindole (DAPI 2 μg/μl; Sigma). A Zeiss 780 laser scanning confocal microscope (Zeiss, Oberkochen, Germany) was used to capture the images. Immunostaining for cultured cells was performed as the same above procedures. As for the astroglial counts, the number of colabeled astrocytic marker with GFP-positive cells is divided by the number of all GFP-positive cells.
Co-immunoprecipitation
One microgram of each plasmid was co-transfected into N2A or HEK293FT cells in a six-well plate. Forty-eight hours later, cells were collected in lysis buffer RIPA (Solarbio) with cocktail and PMSF, sonicated and cleared by centrifugation. Six hundred micrograms protein extracts were incubated with 25 μl anti-Flag or anti-HA beads (MBL) overnight at 4 °C. Next, beads were washed with PBS containing 0.02% Tween for three times. Beads were resuspended with 40 μl 1 × SDS loading buffer and boiled for 10 min. The supernatant was subjected to western blotting.
Ch-IP assay
Cultured Cells were fixed with 2 ml fresh mixture of fixing solution (1 mM PH 8.0 EDTA, 0.1 M NaCl, 0.5 mM pH 8.0 EGTA, 50 mM pH 8.0 Hepes), 37% formaldehyde and PBS at the ratio of 0.7:0.3:10 at room temperature for 15 min. One hundred microliters 2.5 M glycine solution was added to terminate the crosslinking reaction at room temperature for 10 min. Cells were washed with cold PBS containing cocktail and PMSF on ice. Cells were collected with 200μl buffer 1 (0.05 M pH 7.5 Hepes-KOH, 0.14 M NaCl, 1 mM EDTA, 10% glycerol, 0.5% NP-40, 0.25% Triton X-100), rotated for 10 min at 4 °C and centrifuged at 4000 r.p.m. for 5 min. Precipitation was resuspended with 200μl buffer 2 (0.2 M NaCl, 1 mM EDTA, 0.5 mM EGTA, 0.01 M Tris PH 8.0), rotated for 10 min at 4 °C and centrifuged at 4000 r.p.m. for 5 min. Precipitation was resuspended with 650 μl buffer 3 (1 mM EDTA, 0.5 mM EGTA, 0.01 M Tris pH 8.0) and sonicated for 8 min with 8 s interval every 8 s. After 10 min centrifugation at 14 000 r.p.m., supernatant was collected and mixed with 350 μl buffer x (3% Triton X-100, 0.3% sodium deoxycholate, 3 mM EDTA, 3 mM PMSF, 1% cocktail). Half of the sample was incubated with beads coated with selected antibodies, and half of the sample with beads coated with IgG antibody at 4 °C overnight. Beads were washed with RIPA (50 mM pH 7.6 Hepes-KOH, 10 mM pH 8.0 EDTA, 0.7% Doc, 1% NP-40, 0.5 M LiCl, 1 mM PMSF) for six times, washed with TEN (10 mM Tris pH 8.0, 1 mM EDTA, 50 mM NaCl) and resuspended with 200 μl TES (50 mM Tris PH 8.0, 10 mM EDTA, 1% SDS). Beads were incubated at 65 °C for 15 min, and vibrated for 15 s every 2 min. Supernatant was transferred to a clean tube, and incubated at 65 °C overnight. Next, samples were used to extract genome DNA with Kit from TIANGEN (Beijing, China). Genome DNA was used to perform real-time PCR. The primers for real-time PCR were as follows: STAT3-CDS, 5′-GGCTTCTCCTTCTGGGTCTGG-3′
and 5′-ACCCATGATGTACCCTTCATT-3′
STAT3-ATG, 5′-AGACAGTCGAGACCCCTGACT-3′
and 5′-GGAAGCTGTCGCTGTACAGCT-3′
STAT3-0.5 kb, 5′-ACATTCCGCTAATGTCTCCTC-3′
and 5′-CTGTTCTGAGAGCTTTGTGC-3′.
Real-time PCR
The total RNA of cultured cells was extracted with Total RNA Kit (TIANGEN). The FastQuant RT Kit (TIANGEN) was used to perform the first-strand cDNA synthesis according to the manufacturer’s protocols. Quantitative Real-time PCR (qRT-PCR) was performed using the SuperReal PreMix Plus (SYBR Green I) Kit (TIANGEN). The primers used in the RT-PCR were as follows:
RNF20, 5′-TAAACGGAAGCTGAGAGAGGC-3′
and 5′-TGAGGAATGAGTGGCTAGGTC-3′
STAT3, 5′-GGCTTCTCCTTCTGGGTCTGG-3′
and 5′-ACCCATGATGTACCCTTCATT-3′.
Apoptosis assay
Apoptosis was analyzed with in situ cell death detection kit (Roche, Shanghai, China). Slices were fixed in 4% PFA for 20 min at room temperature, and then washed with PBS. Next, slices were blocked with 3% H2O2 in methanol for 10 min and washed with PBS. Then, slices were incubated in 0.1% Triton X-100 on ice for 2 min. Slices were incubated with TUNEL reaction mixture at 37 °C for 60 min. Images were captured with Zeiss 780 laser scanning confocal microscope.
Statistical analysis
The statistical analyses were performed by Student’s t-test between two groups and one-way ANOVA for more than two groups’ comparison. Quantitative data are shown as the mean±S.E.M. Statistical significant: NS, not significant, P>0.05, *P<0.05, ** P<0.01, *** P<0.001.
Acknowledgments
This work was supported by grants from the National Science Foundation of China (31730033 and 31621004), the National Key Basic Research Program of China (2015CB964500 and 2014CB964900) and K.C.Wong Education Foundation.
Author contributions
QL performed the research, analyzed the data and wrote the manuscript; WX constructed the plasmid of Mof and Mof-shRNA; WL provided Rnf 20flox/flox mice; and JJ designed the research and wrote the manuscript.
Footnotes
Supplementary Information accompanies this paper on Cell Death and Differentiation website (http://www.nature.com/cdd)
Edited by M Freeman
The authors declare no conflict of interest.
Supplementary Material
References
- Anderson DJ. Stem cells and pattern formation in the nervous system: the possible versus the actual. Neuron 2001; 30: 19–35. [DOI] [PubMed] [Google Scholar]
- McKay R. Stem cells in the central nervous system. Science 1997; 276: 66–71. [DOI] [PubMed] [Google Scholar]
- Barnabe-Heider F, Wasylnka JA, Fernandes KJ, Porsche C, Sendtner M, Kaplan DR et al. Evidence that embryonic neurons regulate the onset of cortical gliogenesis via cardiotrophin-1. Neuron 2005; 48: 253–65. [DOI] [PubMed] [Google Scholar]
- Qian X, Shen Q, Goderie SK, He W, Capela A, Davis AA et al. Timing of CNS cell generation: a programmed sequence of neuron and glial cell production from isolated murine cortical stem cells. Neuron 2000; 28: 69–80. [DOI] [PubMed] [Google Scholar]
- Liddelow S, Barres B. SnapShot: astrocytes in health and Disease. Cell 2015; 162: 1170–1170 e1. [DOI] [PubMed] [Google Scholar]
- Bolteus AJ, Bordey A. GABA release and uptake regulate neuronal precursor migration in the postnatal subventricular zone. J Neurosci 2004; 24: 7623–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko N, Marin O, Koike M, Hirota Y, Uchiyama Y, Wu JY et al. New neurons clear the path of astrocytic processes for their rapid migration in the adult brain. Neuron 2010; 67: 213–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barres BA. The mystery and magic of glia: a perspective on their roles in health and disease. Neuron 2008; 60: 430–40. [DOI] [PubMed] [Google Scholar]
- Christopherson KS, Ullian EM, Stokes CC, Mullowney CE, Hell JW, Agah A et al. Thrombospondins are astrocyte-secreted proteins that promote CNS synaptogenesis. Cell 2005; 120: 421–33. [DOI] [PubMed] [Google Scholar]
- Gobius I, Morcom L, Suarez R, Bunt J, Bukshpun P, Reardon W et al. Astroglial-mediated remodeling of the interhemispheric midline is required for the formation of the corpus callosum. Cell Rep 2016; 17: 735–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abbott NJ, Ronnback L, Hansson E. Astrocyte-endothelial interactions at the blood-brain barrier. Nat Rev Neurosci 2006; 7: 41–53. [DOI] [PubMed] [Google Scholar]
- Khakh BS, Sofroniew MV. Diversity of astrocyte functions and phenotypes in neural circuits. Nat Neurosci 2015; 18: 942–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloan SA, Barres BA. Mechanisms of astrocyte development and their contributions to neurodevelopmental disorders. Curr Opin Neurobiol 2014; 27: 75–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galileo DS, Gray GE, Owens GC, Majors J, Sanes JR. Neurons and glia arise from a common progenitor in chicken optic tectum: demonstration with two retroviruses and cell type-specific antibodies. Proc Natl Acad Sci USA 1990; 87: 458–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Namihira M, Nakashima K. Mechanisms of astrocytogenesis in the mammalian brain. Curr Opin Neurobiol 2013; 23: 921–7. [DOI] [PubMed] [Google Scholar]
- Bonni A, Sun Y, Nadal-Vicens M, Bhatt A, Frank DA, Rozovsky I et al. Regulation of gliogenesis in the central nervous system by the JAK-STAT signaling pathway. Science 1997; 278: 477–83. [DOI] [PubMed] [Google Scholar]
- Nakashima K, Yanagisawa M, Arakawa H, Kimura N, Hisatsune T, Kawabata M et al. Synergistic signaling in fetal brain by STAT3-Smad1 complex bridged by p300. Science 1999; 284: 479–82. [DOI] [PubMed] [Google Scholar]
- Takizawa T, Nakashima K, Namihira M, Ochiai W, Uemura A, Yanagisawa M et al. DNA methylation is a critical cell-intrinsic determinant of astrocyte differentiation in the fetal brain. Dev Cell 2001; 1: 749–58. [DOI] [PubMed] [Google Scholar]
- Asano H, Aonuma M, Sanosaka T, Kohyama J, Namihira M, Nakashima K. Astrocyte differentiation of neural precursor cells is enhanced by retinoic acid through a change in epigenetic modification. Stem Cells 2009; 27: 2744–52. [DOI] [PubMed] [Google Scholar]
- Cheng PY, Lin YP, Chen YL, Lee YC, Tai CC, Wang YT et al. Interplay between SIN3A and STAT3 mediates chromatin conformational changes and GFAP expression during cellular differentiation. PLoS ONE 2011; 6: e22018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patnala R, Arumugam TV, Gupta N, Dheen ST. HDAC inhibitor sodium butyrate-mediated epigenetic regulation enhances neuroprotective function of microglia during ischemic stroke. Mol Neurobiol 2016; 54: 6391–411. [DOI] [PubMed] [Google Scholar]
- Shen J, Jia W, Yu Y, Chen J, Cao X, Du Y et al. Pwp1 is required for the differentiation potential of mouse embryonic stem cells through regulating Stat3 signaling. Stem Cells 2015; 33: 661–73. [DOI] [PubMed] [Google Scholar]
- Benayoun BA, Pollina EA, Ucar D, Mahmoudi S, Karra K, Wong ED et al. H3K4me3 breadth is linked to cell identity and transcriptional consistency. Cell 2014; 158: 673–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juliandi B, Abematsu M, Nakashima K. Chromatin remodeling in neural stem cell differentiation. Curr Opin Neurobiol 2010; 20: 408–15. [DOI] [PubMed] [Google Scholar]
- Qiao Y, Yang X, Jing N. Epigenetic regulation of early neural fate commitment. Cell Mol Life Sci 2016; 73: 1399–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Y, Li M, Gu L, Chen X, Shi J, Zhang X et al. Chromatin remodeling during in vivo neural stem cells differentiating to neurons in early Drosophila embryos. Cell Death Differ 2016; 24: 409–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You L, Yan K, Zou J, Zhao H, Bertos NR, Park M et al. The lysine acetyltransferase activator Brpf1 governs dentate gyrus development through neural stem cells and progenitors. PLoS Genet 2015; 11: e1005034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song MR, Ghosh A. FGF2-induced chromatin remodeling regulates CNTF-mediated gene expression and astrocyte differentiation. Nat Neurosci 2004; 7: 229–35. [DOI] [PubMed] [Google Scholar]
- Thorne AW, Sautiere P, Briand G, Crane-Robinson C. The structure of ubiquitinated histone H2B. EMBO J 1987; 6: 1005–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang WW, Venkatasubrahmanyam S, Ianculescu AG, Tong A, Boone C, Madhani HD. A conserved RING finger protein required for histone H2B monoubiquitination and cell size control. Mol Cell 2003; 11: 261–6. [DOI] [PubMed] [Google Scholar]
- Wood A, Krogan NJ, Dover J, Schneider J, Heidt J, Boateng MA et al. Bre1, an E3 ubiquitin ligase required for recruitment and substrate selection of Rad6 at a promoter. Mol Cell 2003; 11: 267–74. [DOI] [PubMed] [Google Scholar]
- Zhu B, Zheng Y, Pham AD, Mandal SS, Erdjument-Bromage H, Tempst P et al. Monoubiquitination of human histone H2B: the factors involved and their roles in HOX gene regulation. Mol Cell 2005; 20: 601–11. [DOI] [PubMed] [Google Scholar]
- Pavri R, Zhu B, Li G, Trojer P, Mandal S, Shilatifard A et al. Histone H2B monoubiquitination functions cooperatively with FACT to regulate elongation by RNA polymerase II. Cell 2006; 125: 703–17. [DOI] [PubMed] [Google Scholar]
- Kim J, Guermah M, McGinty RK, Lee JS, Tang Z, Milne TA et al. RAD6-Mediated transcription-coupled H2B ubiquitylation directly stimulates H3K4 methylation in human cells. Cell 2009; 137: 459–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JS, Shukla A, Schneider J, Swanson SK, Washburn MP, Florens L et al. Histone crosstalk between H2B monoubiquitination and H3 methylation mediated by COMPASS. Cell 2007; 131: 1084–96. [DOI] [PubMed] [Google Scholar]
- Nakanishi S, Lee JS, Gardner KE, Gardner JM, Takahashi YH, Chandrasekharan MB et al. Histone H2BK123 monoubiquitination is the critical determinant for H3K4 and H3K79 trimethylation by COMPASS and Dot1. J Cell Biol 2009; 186: 371–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z, Song Z, Li G, Tu H, Liu W, Liu Y et al. H2B ubiquitination regulates meiotic recombination by promoting chromatin relaxation. Nucleic Acids Res 2016; 44: 9681–9697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shema E, Tirosh I, Aylon Y, Huang J, Ye C, Moskovits N et al. The histone H2B-specific ubiquitin ligase RNF20/hBRE1 acts as a putative tumor suppressor through selective regulation of gene expression. Genes Dev 2008; 22: 2664–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs G, Shema E, Vesterman R, Kotler E, Wolchinsky Z, Wilder S et al. RNF20 and USP44 regulate stem cell differentiation by modulating H2B monoubiquitylation. Mol Cell 2012; 46: 662–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Chen K, Sloan SA, Bennett ML, Scholze AR, O'Keeffe S et al. An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. J Neurosci 2014; 34: 11929–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Newbern JM, Wu Y, Morgan-Smith M, Zhong J, Charron J et al. MEK is a key regulator of gliogenesis in the developing brain. Neuron 2012; 75: 1035–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suganuma T, Workman JL. Crosstalk among histone modifications. Cell 2008; 135: 604–7. [DOI] [PubMed] [Google Scholar]
- Rudenko A, Tsai LH. Epigenetic modifications in the nervous system and their impact upon cognitive impairments. Neuropharmacology 2014; 80: 70–82. [DOI] [PubMed] [Google Scholar]
- Lee JH, Lee GY, Jang H, Choe SS, Koo SH, Kim JB. Ring finger protein20 regulates hepatic lipid metabolism through protein kinase A-dependent sterol regulatory element binding protein1c degradation. Hepatology 2014; 60: 844–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanada K, Gupta A, Tsai LH. Disabled-1-regulated adhesion of migrating neurons to radial glial fiber contributes to neuronal positioning during early corticogenesis. Neuron 2004; 42: 197–211. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.