Abstract
The significance of mitochondrial metabolism in cancer cells has recently been gaining attention. Among other findings, One-carbon folate metabolism has been reported to be closely associated with cellular characteristics in cancer. To study molecular targets for efficient cancer therapy, we investigated the association between the expressions of genes that code enzymes involved in one-carbon metabolism and survival rate of patients with adenocarcinomas of the colorectum and lung. Patients with high expression of genes that control the metabolic cycle of tetrahydrofolate (THF) in mitochondria, SHMT2, MTHFD2, and ALDH1L2, have a shorter overall survival rate compared with patients with low expression of these genes. Our results revealed that these genes could be novel and more promising anticancer targets than dihydrofolate reductase (DHFR), the current target of drug therapy linked with folate metabolism, suggesting the rationale of drug discovery in cancer medicine.
Introduction
Folate is very important in bioorganic systems, although it cannot be synthesized in humans1. It is acquired from food and converted to tetrahydrofolate [tetrahydrofolic acid (THF)], which is the reactant in the metabolic pathway of the folate cycle. The folate cycle has a crucial role in supplying one-carbon (C1) groups (e.g., methyl, methylene, and methenyl groups) that are transferred to biomolecules, such as amino acids and nucleotides, which explains why the metabolism of the folate cycle is also known as C1 metabolism2–9. C1 metabolism plays a role in DNA synthesis and repair via production of nucleic acid components5,6. In the first step of C1 metabolism, some of the folate is converted to an intermediate metabolite, dihydrofolate (DHF), by a reduction reaction catalyzed by an enzyme, dihydrofolate reductase (DHFR), and the rest is directly converted to THF. THF is extensively used as a source of intermediates in other reaction pathways involved in C1 metabolism7–10. DHF is also converted to THF with time.
Recently, C1 metabolism has received much attention as a target of cancer therapy, and many studies have reported it to be a promising target for cancer treatment11–13. Many drugs have been developed to target folate metabolism by inhibiting DHFR. However, folate metabolism is absolutely imperative not only for cancer cells but also for normal cells. Folate deficiency in humans results in the reduction of DNA synthesis and methylation11,12. Therefore, inhibition of DHFR is known to cause toxicity in normal cells as well as cancer cells.
In contrast, mitochondrial metabolism has received much attention as a potential target for cancer therapy7,13. We focused on the reactions among THF, 5,10-methylene THF (CH2-THF), and 10-formyl THF (CHO-THF) in C1 metabolism, which occur in the cytoplasm and mitochondria (Fig. 1). If folate metabolism in the mitochondria is responsible for poor prognosis in cancer patients, then the selective targeting of this pathway could be effective with few side effects because of the parallel pathway in the cytoplasm.
Figure 1.
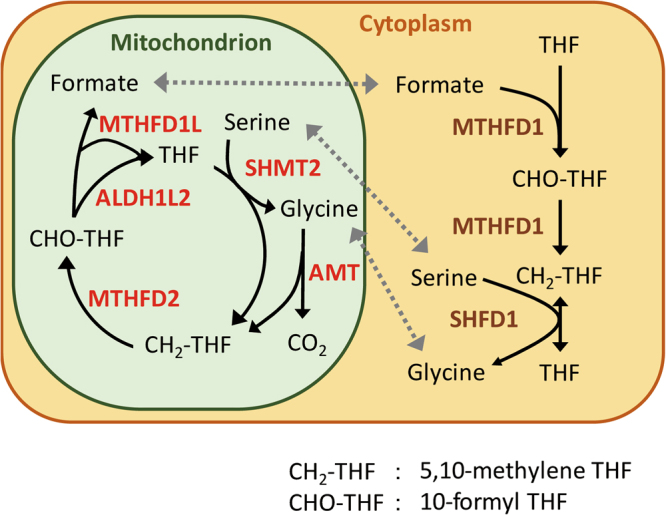
Schematic diagram of the metabolic cycle of THF and the C1 metabolic pathway.
Herein we have performed computational analysis to investigate the association between transcriptome profiles of genes involved in C1 metabolism in patients with colorectal cancer (CRC) and lung adenocarcinoma (LA) and the overall survival (OS) rate of patients. We have previously reported the usefulness of transcriptome analysis in examining cellular metabolism and its close correlation with information gained from metabolome analysis14.
It is noteworthy that there was no strong association between the gene expression of DHFR and the OS of patients (Fig. 2). In contrast, we could identify other mitochondrial genes involved in C1 metabolism that are strongly associated with the prognosis in cancer patients, which we believe have the potential to be more efficient anticancer targets than DHFR, which is the anticancer target of folate metabolism.
Figure 2.
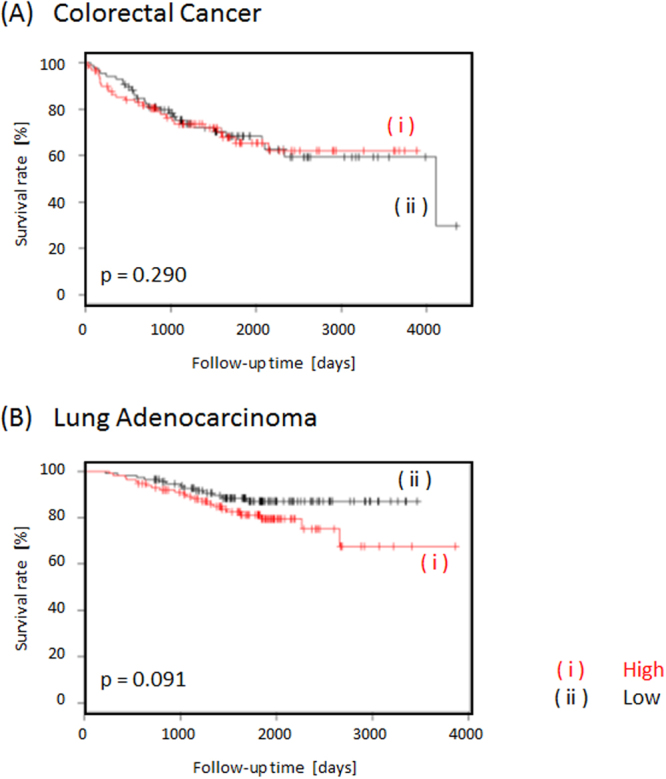
Kaplan–Meier curves of overall survival for (A) colorectal cancer and (B) lung adenocarcinoma according to the expression of DHFR, which is the conventional anticancer target of folate metabolism.
Results
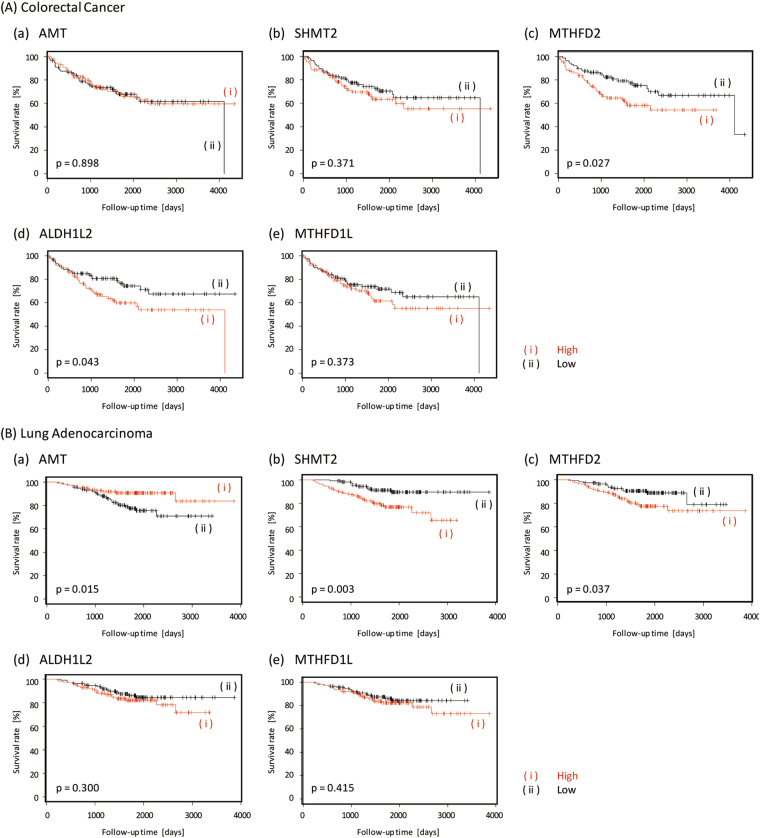
Unexpectedly, the expression of DHFR, the conventional target for folate metabolism in cancer, did not dictate the OS in patients with CRC or LA. We thus searched for better anticancer targets of C1 metabolism and then performed correlation analysis between the expressions of genes involved in C1 metabolism and prognosis in patients with CRC and LA. In this study, we focused on five genes expressed in the mitochondria (ATM, SHMT2, MTHFD2, ALDH1L2, and MTHFD1L). The Kaplan–Meier curves for these genes in patients with CRC and LA are shown in Fig. 3. Although the expression of ATM did not affect the prognosis in patients with CRC, high expression of the gene was associated with a better prognosis in patients with LA (p = 0.015). In contrast, the high expression of SHMT2 was associated with a significantly worse prognosis in patients with LA (p = 0.003), although the prognosis was not different in patients with CRC. The high expression of MTHFD2 was associated with poor survival rates in patients with CRC (p = 0.027) and LA (p = 0.037). Similar to SHMT2, the high expression of ALDH1L2 was associated with a significantly worse prognosis in patients with LA (p = 0.043), although the prognosis was not different in patients with CRC. The expression of MTHFD1L was not associated with patient prognosis. With the exception of AMT, we found that the high expression of four genes is associated globally with worse prognoses in patients with CRC and LA.
Figure 3.
Kaplan–Meier curves of overall survival for (A) colorectal cancer and (B) lung adenocarcinoma according to the expression of genes involved in mitochondrial-specific folate metabolism (a) ATM, (b) SHMT2, (c) MTHFD2, (d) ALDH1L2, and (e) MTHFD1L.
From these results, we focused on the THF cycle in the mitochondria. We performed combination analyses to investigate the correlation among SHMT2, MTHFD2, and ALDH1L2, which all are involved in the THF cycle described above. Figure 4A (CRC) and 4B (LA) show the Kaplan–Meier curves for the combination of SHMT2 with MTHFD2. Patients with high expressions of SHMT2 and MTHFD2 had worse prognoses compared to those with the other expression patterns of the enzymes in CRC (p = 0.030) and LA (p < 0.002). As shown in Supplementary Table 1, this behavior has a poor association with the age and sex of patients with CRC and LA. Additionally, according to Cox proportional hazards analysis and the likelihood ratio test, the p values were 0.032 and 0.034 for CRC, respectively, and 0.003 and 0.002 for LA, respectively. These results suggested that the prognosis could be improved in both cancers by decreasing the expression of either of the enzymes, SHMT2 or MTHFD2. In addition, patients with CRC and LA having high expressions of SHMT2, MTHFD2, and ALDH1L2 had worse prognoses (Fig. 4C) (CRC), 4D (LA). Survival rate analysis also revealed that patients with high expression of all three enzymes had worse prognoses than the other patients with CRC (p = 0.030) and LA (p = 0.037). In this case, we performed the same statistical analyses. As shown in Supplementary Table 2, although there was bias in the distribution of sex between both groups, the poor association with patient age also was obtained for CRC and LA. According to Cox proportional hazards analysis and the likelihood ratio test, the p values were 0.027 and 0.033 for CRC, respectively, and 0.025 and 0.031 for LA, respectively.
Figure 4.
Correlation analysis of SHMT2, MTHFD2, and ALDH1L2, which are the enzymes involved in the THF cycle in the mitochondria. The Kaplan–Meier curves of the combination of the expression of SHMT2 and MTHFD2 for (A) colorectal cancer and (B) lung adenocarcinoma and of the combination of the expression of SHMT2, MTHFD2, and ALDH1L2 for (C) colorectal cancer and (D) lung adenocarcinoma.
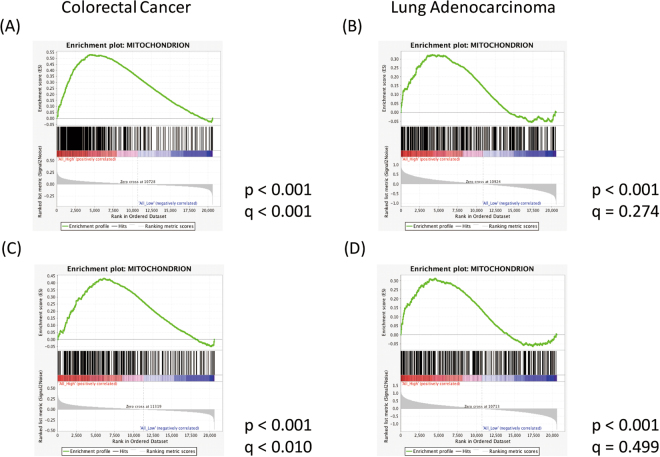
In addition, gene set enrichment analysis (GSEA) for genes involved in mitochondrial metabolism was performed to investigate whether the association between the THF cycle and cancer prognosis could be explained by the activity of the mitochondrial C1 metabolism. GSEA analyses revealed that, in CRC and LA, patients with high expression of SHMT2 and MTHFD2 had a high expression of genes associated with mitochondrial metabolism (Fig. 5A,B). Similarly, Fig. 5C and D show the GSEA results between all high- and low-expression groups for SHMT2, MTHFD2, and ALDH1L2. These figures also show the same tendency. These GSEA analyses revealed an association between the activities of the mitochondrial metabolism and the THF cycle.
Figure 5.
(A,B) GSEA analyses between the high-expression group and low-expression group in SHMT2 and MTHFD2 for colorectal cancer and lung adenocarcinoma, respectively. (C,D) Similarly, GSEA analyses were conducted between all high- and low-expression groups in SHMT2, MTHFD2, and ALDH1L2.
We then investigated the association between the expression of genes involved in folic acid metabolism in the cytoplasm and prognosis. Intriguingly, the expression of SHMT1, MTHFD1, and ALDH1L1 had no influence on the survival rate of patients with CRC and LA (Supplementary Figure 1), suggesting it is not the folate metabolism per se but the THF cycle in the mitochondria that confers poor prognosis in cancers.
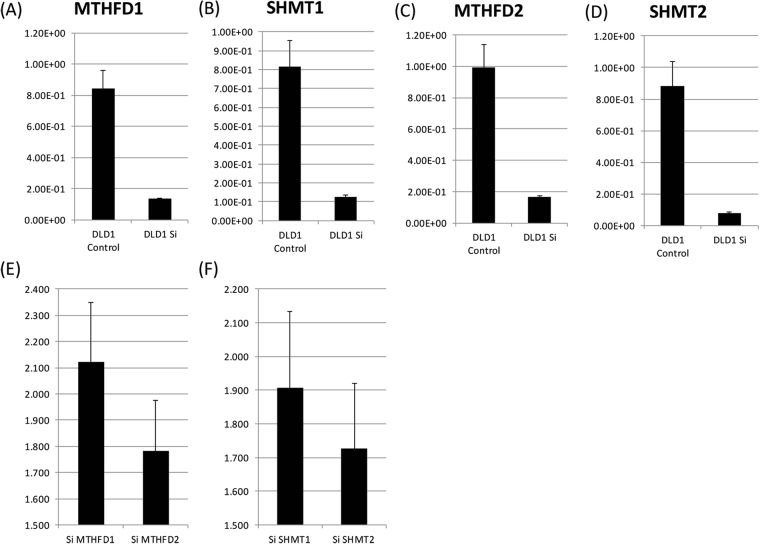
Additionally, we have performed some siRNA knockdown experiments for cytoplasm enzyme (MTHFD1 and SHMT1) and C1 metabolic enzymes (MTHFD2 and SHMT2) at the in vitro level. As shown in Fig. 6A–D, the expression level of these mRNAs were downregulated by siRNA. Then, siRNA knockdown of SHMT2 and MTHFD2 resulted in more apparent inhibitory effect than SHMT1 and MTHFD1 (Fig. 6E,F). Moreover, we checked the expression of MTHFD2, SHMT2, and DHFR using the RefExA database. Although MTHFD2 and SHMT2 were highly expressed in colon cancer cells, DHFR had a very low or undetectable expression in colon cancer cells (Supplementary Figure 2). These data further suggest mitochondrial folate metabolism to be a promising target for cancer treatment.
Figure 6.
(A–D) Gene expression analysis for SHMT1, MTHFD1, SHMT2, and MTHFD2, under siRNA transfected condition. MTT assay under siRNA transfected conditions. (E) siMTHFD1 and siMTHFD2, and (F) SHMT1 and siSHMT 2.
Discussion
Our study unexpectedly revealed that the expression of DHFR, the current anticancer target of folic acid metabolism, had no influence on the prognosis in patients with CRC or LA. Instead, we observed that the expression of genes controlling THF metabolism was associated with cancer prognosis, suggesting these genes to be more effective targets for cancer therapy than DHFR. Furthermore, we identified that the expression of genes was involved with only mitochondrial THF metabolism and not cytoplasmic THF metabolism, which was associated with a worse prognosis. Knockdown of the aforementioned enzymes in cancer cells confirmed that the mitochondrial enzymes involved in THF metabolism were crucial for cancer proliferation, but the cytoplasmic enzymes involved in THF metabolism were dispensable for cellular proliferation.
Specifically, we identified SHMT2 and MTHFD2 to be the most promising anticancer targets of folate metabolism from the correlational analyses between gene expression and survival. This may be because these enzymes are a crucial part of the THF cycle in mitochondria, and interference with their activities is predicted to create a bottleneck in the reaction cycle (Supplementary Figure 3).
Specific targeting of the mitochondrial folate metabolism would spare the physiological folate metabolism for occurring in the cytoplasm and thus may be associated with minimal side effects.
We also demonstrated the usefulness of correlational analysis between gene expression patterns and survival in cancers to identify novel targets of cancer metabolism. The novel targets identified by such analyses were validated by in vitro studies, which confirmed the usefulness of the correlational analysis in identifying potential targets.
We revealed novel anticancer targets of folate metabolism, which we predict will be associated with minimal toxicity given sparing of the physiological folate metabolism in the cytoplasm. Specifically, we predicted that SHMT2 and MTFD2 have the potential to be more effective anticancer targets than DHFR, which is the current anticancer target of folate metabolism.
Materials and Methods
Microarray Data of Gene Expression in Patients with CRC and LA
The GSE17536 database15 from Gene Expression Omnibus at the National Center for Biotechnology Information was used to analyze the effects of gene expression on the OS of patients (n = 177) with CRC, and the GSE31210 database16 was used for patients with LA (n = 226). These databases were generated using the Affymetrix Human Genome U133 Plus 2.0 Array. In these databases, the level of C1 expression is obtained with multiple probes for some genes. We selected the gene expression data for statistical analysis for probes that showed the widest variance of the expression for a particular gene and divided the expression data into low- and high-expression groups at the median. The GSE17536 and GSE31210 databases have data for patients of a broad generation aged around 60 years (range, 26–92 years and 30–76 years, respectively). The GSE17536 database reports gene expression for Stages I, II, III, and IV cancer. In contrast, the GSE31210 database has expression data for patients with only Stages I and II disease.
Analysis of the Association between Gene Expression and Survival Rate
We focused on DHFR, the current anticancer target of folate metabolism, and five mitochondrial-specific genes involved in folate metabolism: ATM, SHMT2, MTHFD2, ALDH1L2, and MTHFD1L. For all of these genes, Kaplan–Meier curves for patients with CRC and LA was generated. Subsequently, combination analyses were performed to investigate the correlations among SHMT2, MTHFD2, and ALDH1L2, which are part of the THF cycle in mitochondria. In these analyses, we compared patients in the high-expression groups to those in the other groups. All Kaplan–Meier curves were generated using R package version 3.0.217 with the survival package18.
Cell Culture and MTT Assay
CRC cell line DLD1 was cultured in Dulbecco’s modified Eagle’s medium containing 10% fetal bovine serum. SHMT1, MTHFD1, SHMT2, and MTHFD2 knockdown was performed using siRNA oligonucleotides synthesized from Sigma–Aldrich Corp. (St. Louis, MO, USA). After 36 h of culture from seeding, siRNAs were transfected into DLD1 cells at 20 nM final concentration with lipofectamine RNAiMax (Life Technologies, Carlsbad, CA, USA) with a forward transfection method according to the manufacturer’s protocol. MTT assay was performed using MTT (Nakarai Tesque) according to the manufacturer’s protocol.
Gene Expression Analysis
Gene expression level was analyzed using the RefExA database (http://sbmdb.genome.rcast.u-tokyo.ac.jp/refexa/main_search.jsp).
Electronic supplementary material
Acknowledgements
We would like to thank the members of our laboratories for their fruitful discussion and Drs. Masaaki Miyo and Kozo Noguchi for their support of statistical analysis. This work received the following financial support: grants-in-aid for Scientific Research and P-Direct Grants from the Ministry of Education, Culture, Sports, Science, and Technology (MK, YD, MM, and HI); a grant-in-aid from the Ministry of Health, Labour, and Welfare (MK, HI, and MM); a grant from the Kobayashi Cancer Research Foundation (HI); a grant from the Princess Takamatsu Cancer Research Fund, Japan (HI); a grant from the National Institute of Biomedical Innovation (MK, HI, and MM) and a grant from the Osaka University Drug Discovery Funds (NN, JK, KK, MK, MM, and HI). Partial support was received from Takeda Science and Medical Research Foundation through institutional endowments (MM and HI), Princess Takamatsu Cancer Research Fund (MM and HI), Kobayashi Cancer Research Foundation (MM and HI), Suzuken Memorial Foundation (MK), Pancreas Research Foundation of Japan (KK) and the Nakatomi Foundation (MK).
Author Contributions
J.K., M.K., H.C., A.A., K.K., and N.N. performed some computational analyses. J.K., M.K., H.C., A.A., D.S., T.K., T.S., Y.D., M.M., and H.I. designed the analyses. J.K., M.K., and H.C. drafted the article.
Footnotes
Jun Koseki and Masamitsu Konno contributed equally to this work.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-017-18456-x.
Competing financial interests: Institutional endowments were received partially from Taiho Pharmaceutical Co., Ltd.; Evidence Based Medical (EBM) Research Center; Unitech Co., Ltd.; Yakult Honsha Co., Ltd.; Chugai Co., Ltd.; and Merck Co., Ltd. These funders had no role in the main experimental equipment, supplies expenses, study design, data collection and analysis, decision to publish, or preparation of this manuscript. The authors declare no competing financial interests.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Masaki Mori, Email: mmori@gesurg.med.osaka-u.ac.jp.
Hideshi Ishii, Email: hishii@gesurg.med.osaka-u.ac.jp.
References
- 1.Stokstad ELR, Koch J. Folic acid metabolism. Physiol. Rev. 1967;47:83–116. doi: 10.1152/physrev.1967.47.1.83. [DOI] [PubMed] [Google Scholar]
- 2.Coon MJ, Robinson WG. Amino acid metabolism. Annu. Rev. Biochem. 1958;27:561–612. doi: 10.1146/annurev.bi.27.070158.003021. [DOI] [PubMed] [Google Scholar]
- 3.Jaenicke L. Vitamin and coenzyme function: Vitamin B12 and Folic Acid. Annu. Rev. Biochem. 1964;326:287–316. doi: 10.1146/annurev.bi.33.070164.001443. [DOI] [PubMed] [Google Scholar]
- 4.Brown, G. M. Biogenesis and Metabolism of Folic Acid. (ed. Greenberg, D. M.) Chapter 24, 383–410, Metabolic Pathways, Vol. IV (Academic Press, New York. 1970).
- 5.Duthie SJ, et al. Folate, DNA stability and colo-rectal neoplasia. Poc. Nutr. Soc. 2004;63:571–578. doi: 10.1079/PNS2004. [DOI] [PubMed] [Google Scholar]
- 6.Jabrin S, Ravanel S, Gambonnet B, Douce R, Rébeillé F. One-carbon metabolism in plants. Regulation of tetrahydrofolate synthesis during germination and seedling development. Plant Physiol. 2003;131:1431–1439. doi: 10.1104/pp.016915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Christensen KE, MacKenzie RE. Mitochondrial one-carbon metabolism is adapted to the specific needs of yeast, plants and mammals. BioEssays. 2006;28:595–605. doi: 10.1002/bies.20420. [DOI] [PubMed] [Google Scholar]
- 8.Stover PJ, Field MS. Trafficking of intracellular folates. Adv. Nutr. 2011;2:325–331. doi: 10.3945/an.111.000596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Momb J, et al. Deletion of Mthfd1l causes embryonic lethality and neural tube and craniofacial defects in mice. Proc. Natl. Acad. Soc. USA. 2013;110:549–554. doi: 10.1073/pnas.1211199110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hsiao T-H, et al. Ethanol-induced upregulation of 10-formyltetrahydrofolate dehydrogenase helps relieve ethanol-induced oxidative stress. Mol. Cell. Biol. 2014;34:498–509. doi: 10.1128/MCB.01427-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dominguez-Salas P, Cox SE, Prentice AM, Hennig BJ, Moore SE. Maternal nutritional status, C1 metabolism and offspring DNA methylation: a review of current evidence in human subjects. Proc. Nutr. Soc. 2012;71:154–165. doi: 10.1017/S0029665111003338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Williams EA. Folate, colorectal cancer and the involvement of DNA methylation. Proc. Nutr. Soc. 2012;71:592–597. doi: 10.1017/S0029665112000717. [DOI] [PubMed] [Google Scholar]
- 13.Nilsson R, et al. Metabolic enzyme expression highlights a key role for MTHFD2 and the mitochondrial folate pathway in cancer. Nat. Commun. 2014;5:3128. doi: 10.1038/ncomms4128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Koseki J, et al. A trans-omics mathematical analysis reveals novel functions of the ornithine metabolic pathway in cancer stem cells. Sci. Rep. 2016;6:20726. doi: 10.1038/srep20726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Smith JJ, et al. Experimentally derived metastasis gene expression profile predicts recurrence and death in patients with colon cancer. Gastroenterology. 2010;138:958–968. doi: 10.1053/j.gastro.2009.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Okayama H, et al. Identification of genes upregulated in ALK-positive and EGFR/KRAS/ALK-negative lung adenocarcinomas. Cancer Res. 2012;72:100–111. doi: 10.1158/0008-5472.CAN-11-1403. [DOI] [PubMed] [Google Scholar]
- 17.Team R. C. A language and environment for statistical computing. (R Foundation for Statistical Computing, Vienna, Austria 2013).
- 18.Therneau, T. M. & Grambsch, P. M. Modeling Survival Data. Extending the Cox Model. (N.Y. Springer, 2000).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.