Abstract
Papain-like cysteine proteases (PLCPs) are a class of proteolytic enzymes involved in many plant processes. Compared with the extensive research in Arabidopsis thaliana, little is known in castor bean (Ricinus communis) and physic nut (Jatropha curcas), two Euphorbiaceous plants without any recent whole-genome duplication. In this study, a total of 26 or 23 PLCP genes were identified from the genomes of castor bean and physic nut respectively, which can be divided into nine subfamilies based on the phylogenetic analysis: RD21, CEP, XCP, XBCP3, THI, SAG12, RD19, ALP and CTB. Although most of them harbor orthologs in Arabidopsis, several members in subfamilies RD21, CEP, XBCP3 and SAG12 form new groups or subgroups as observed in other species, suggesting specific gene loss occurred in Arabidopsis. Recent gene duplicates were also identified in these two species, but they are limited to the SAG12 subfamily and were all derived from local duplication. Expression profiling revealed diverse patterns of different family members over various tissues. Furthermore, the evolution characteristics of PLCP genes were also compared and discussed. Our findings provide a useful reference to characterize PLCP genes and investigate the family evolution in Euphorbiaceae and species beyond.
Introduction
Castor bean (Ricinus communis L., 2n = 20) and physic nut (Jatropha curcas L., 2n = 22) are two economically important species that belong to the spurge family, Euphorbiaceae1,2. Castor bean, a perennial shrub of the monotypic Ricinus genus, is indigenous to Africa. The oil produced in castor seeds, mainly composed of the unusual hydroxylated fatty acid ricinoleic acid, is widely used for industrial, medicinal and cosmetic purposes, having prompted its domestication in many tropical, subtropical and warm temperate regions around the world3. Physic nut, also known Barbados nut, purging nut or jatropha, is a semi-evergreen shrub or small tree originated from central America, and now is widely cultivated in tropical and subtropical regions around the world4. Physic nut is a potential energy plant with the high oil content (up to 50%) in seeds and the fossil fuel-like oil composition that can be easily processed into bio-diesel5. The draft genome of castor bean was reported in 2010, which consists of 25,878 scaffolds spanning approximate 400 Mb6. The genome size of physic nut was estimated to be 350 Mb and two genome assemblies have been available7,8. The more complete assembly is about 320 Mb consisting of 23,125 scaffolds, and the number of putative protein-encoding genes of 27,172 is slightly smaller than 31,221 in castor bean6,8. Most importantly, except for the ancient so-called γ whole-genome duplication (WGD) event shared by core eudicots including Arabidopsis thaliana and poplar (Populus trichocarpa), comparative genomics analysis indicated that both castor bean and physic nut didn’t experience additional recent WGD6,8,9. From this perspective, analysis of certain gene families in castor bean and physic nut may provide insights into lineage-specific evolution in high plants especially in core eudicots.
Papain-like cysteine proteases (PLCPs) are a class of proteolytic enzymes that are associated with plant growth, development, protein storage and mobilization, organ senescence, abscission, seed germination, immunity and stress response10–14. Featuring a catalytic cysteine as a nucleophile during proteolysis, PLCPs are classed as the family C1A of clan CA and thus are also known as C1A cysteine proteases15. PLCPs are produced as preproproteases which usually include a signal peptide, an auto-inhibitory pro-domain and a mature protease domain16. The signal peptide ensures that the proprotease enters the endomembrane system, whereas the pro-domain prevents premature activation of the protease. Thereby, the protease precursors are usually inactive or weakly active. To become active, PLCPs need to be processed either by self-processing or with the aid of processing enzymes, which depends on the pH, the action of other proteases and protease inhibitors, and the cellular or extracellular environment10,17. In animals, PLCPs are often called cathepsins, and PLCPs in plants are named cathepsin L-, B-, H-, or F-like based on sequence similarity. Furthermore, the L-like can be subclassed into five phylogenetic subgroups (i.e. A–E)18. Although several properties of individual PLCPs have been reported in a wide range of plant species19–21, the genome-wide analysis is still limited to several species such as Arabidopsis, poplar and rubber (Hevea brasiliensis) which were proven to have undergone two or one recent doubling events respectively2,16,22–25. In Arabidopsis, 31 PLCPs were recently divided into nine subfamilies based on the phylogenetic analysis17: Subfamily 9 (CTB3-like or CTB) contains three cathepsin B-like PLCPs; Subfamily 8 (ALP-like or ALP) contains two cathepsin H-like PLCPs with the vacuolar-targeting NPIR motif at the N-terminal; Subfamily 7 (RD19A-like or RD19) contains four cathepsin F-like PLCPs; Subfamily 6 (SAG12-like or SAG12) contains six members; both Subfamily 5 (THI1-like or THI) and Subfamily 4 (XBCP3-like or XBCP3) contain a single member; Subfamily 3 (XCP2-like or XCP) contains two members; Subfamily 2 (CEP1-like or CEP) contains three members with the endoplasmic reticulum (ER)-localizing KDEL motif at the C-terminal; Subfamily 1 (RD21A-like or RD21) contains nine members (see Supplementary Table S1).
In this study, genome-wide identification of castor bean and physic nut PLCP family genes was carried out. Their gene structures were manually curated through aligning transcriptome data to the gene-encoding scaffolds. Furthermore, the sequence feature, evolutionary relationship and expression pattern were also investigated and compared.
Results
Characterization of 26 PLCP genes in castor bean
The initial search of the castor bean genome resulted in 28 loci putatively encoding PLCP homologs. All of them were predicted by the genome annotation6, however, two loci (i.e. 29900.t000066 and 29675.t000004) were shown to encode pseudogenes and were not further analyzed. The remaining 26 loci are distributed across 16 scaffolds. Most scaffolds were found to harbor a single PLCP gene, however, six of them were shown to contain more than one, i.e., scaffold30170 (4), scaffold29646 (3), scaffold29900 (3), scaffold30131 (2), scaffold28962 (2) and scaffold29910 (2) (Table 1).
Table 1.
List of 26 RcPLCP genes identified in this study.
| Gene name | Locus ID | Protein ID | Scaffold | Predicted position | Identified position | EST hits | ASa | AA | MW (KDa) | pI | GRAVY | iPSORTb | At_orthologc | Jc_orthologc |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| RcRD21A | 30170.t000243 | 30170.m013831 | scaffold30170 | 1214556–1211809 | 1214722–1211316 | 16 | Yes | 469 | 52.11 | 5.39 | −0.492 | S | AtRD21A | JcRD21A |
| RcRD21B | 29801.t000069 | 29801.m003124 | scaffold29801 | 415594–412915 | 415851–412454 | 10 | — | 471 | 52.18 | 5.71 | −0.470 | S | — | JcRD21B |
| RcRD21C | 29970.t000002 | 29970.m000973 | scaffold29970 | 17319–21021 | 17149–21809 | 0 | — | 383 | 42.96 | 6.56 | −0.419 | M | AtRDL1 | JcRD21C |
| RcCEP1 | 30147.t000097 | 30147.m013826 | scaffold30147 | 2830277–2828642 | 2830593–2828056 | 3 | Yes | 360 | 40.11 | 5.97 | −0.571 | S | AtCEP1 | JcCEP1 |
| RcCEP2 | 29929.t000288 | 29929.m004785 | scaffold29929 | 1643832–1642090 | 1643874–1641686 | 0 | Yes | 359 | 40.39 | 5.87 | −0.603 | S | — | JcCEP2 |
| RcXCP1 | 30162.t000046 | 30162.m001301 | scaffold30162 | 1780890–1779633 | 1781121–1779118 | 0 | — | 349 | 39.01 | 5.44 | −0.390 | S | AtXCP1 | JcXCP1 |
| RcXCP2 | 30170.t000213 | 30170.m013801 | scaffold30170 | 4386293–4387733 | 4386238–4387922 | 1 | — | 349 | 39.03 | 5.25 | −0.412 | S | AtXCP2 | JcXCP2 |
| RcXBCP3 | 30170.t000524 | 30170.m014112 | scaffold30170 | 2901779–2899595 | 2901979–2898207 | 1 | — | 466 | 51.59 | 7.43 | −0.296 | S | AtXBCP3 | JcXBCP3 |
| RcXBCP3L | 29381.t000001 | 29381.m000072 | scaffold29381 | 7721–11783 | 7678–12160 | 13 | — | 501 | 55.85 | 5.05 | −0.432 | S | — | JcXBCP3L |
| RcTHI1 | 29646.t000057 | 29646.m001109 | scaffold29646 | 344102–345635 | 343762–345957 | 1 | Yes | 347 | 38.63 | 5.48 | −0.439 | S | AtTHI1 | JcTHI1 |
| RcSAG12H1 | 30131.t000408 | 30131.m007257 | scaffold30131 | 2504598–2505766 | 2504200–2506430 | 0 | — | 362 | 41.11 | 6.17 | −0.412 | S | AtSAG12 | JcSAG12H3 |
| RcSAG12H2 | 28962.t000017 | 28962.m000448 | scaffold28962 | 92991–94101 | 92944–94101 | 0 | — | 340 | 37.20 | 5.22 | −0.441 | S | AtSAG12 | JcSAG12H4 |
| RcSAG12H3 | 28962.t000018 | 28962.m000449 | scaffold28962 | 96412–97522 | 96181–97735 | 0 | Yes | 340 | 37.46 | 5.16 | −0.424 | S | AtSAG12 | JcSAG12H4 |
| RcSAG12H4 | 29646.t000033 | 29646.m001085 | scaffold29646 | 207629–208856 | 207526–209015 | 0 | — | 349 | 38.55 | 9.33 | −0.347 | S | AtSAG12 | — |
| RcSAG12H5 | 29646.t000034 | 29646.m001086 | scaffold29646 | 211653–212894 | 211496–213036 | 0 | — | 342 | 38.06 | 8.59 | −0.417 | S | AtSAG12 | — |
| RcSAG12H6 | 29900.t000065 | 29900.m001603 | scaffold29900 | 407069–405926 | 407069–405639 | 0 | — | 344 | 38.12 | 5.13 | −0.428 | S | AtSAG12 | JcSAG12H7 |
| RcSAG12H7 | 29910.t000015 | 29910.m000924 | scaffold29910 | 208698–206791 | 208876–206709 | 0 | — | 341 | 37.41 | 4.86 | −0.458 | S | AtSAG12 | JcSAG12H8 |
| RcSAG12H8 | 29910.t000014 | 29910.m000923 | scaffold29910 | 204533–202640 | 204533–202640 | 0 | — | 342 | 37.40 | 4.71 | −0.439 | S | AtSAG12 | JcSAG12H8 |
| RcPAP1 | 29900.t000078 | 29900.m001616 | scaffold29900 | 487805–488928 | 487718–489114 | 0 | — | 340 | 37.64 | 6.33 | −0.325 | S | AtPAP1 | — |
| RcPAP2 | 29900.t000077 | 29900.m001615 | scaffold29900 | 483849–485660 | 483819–485847 | 1 | — | 343 | 37.89 | 4.87 | −0.343 | S | AtPAP1 | — |
| RcPAP3 | 29827.t000145 | 29827.m002672 | scaffold29827 | 836744–835493 | 836744–835443 | 0 | — | 342 | 37.91 | 5.07 | −0.371 | M | AtPAP1 | — |
| RcRD19A | 30131.t000249 | 30131.m007098 | scaffold30131 | 1514779–1516461 | 1514766–1516903 | 8 | Yes | 373 | 41.07 | 5.95 | −0.312 | S | AtRD19A | JcRD19A |
| RcRD19B | 30170.t000534 | 30170.m014122 | scaffold30170 | 2964658–2966204 | 2964578–2966528 | 68 | Yes | 366 | 40.38 | 5.68 | −0.249 | S | AtRD19C | JcRD19B |
| RcRD19C | 28462.t000004 | 28462.m000130 | scaffold28462 | 46401–48802 | 46261–49312 | 17 | Yes | 381 | 41.73 | 5.87 | −0.037 | S | AtRD19D | JcRD19C |
| RcALP1 | 29739.t000193 | 29739.m003757 | scaffold29739 | 1197717–1200174 | 1197581–1200666 | 3 | Yes | 358 | 39.21 | 5.88 | −0.185 | S | AtALP | JcALP1 |
| RcCTB1 | 30076.t000074 | 30076.m004510 | scaffold30076 | 399770–402553 | 399629–403162 | 2 | Yes | 359 | 39.84 | 5.90 | −0.200 | S | AtCTB2 | JcCTB1 |
a“Yes” represents genes containing alternative splicing isoforms; b“S, M and C” represent signal peptide, mitochondrial targeting peptide or chloroplast transit peptide, respectively; cThe best ortholog hit.
Except for RcCEP1 (GenBank accession number AF050756)26, homology analysis showed that no full-length cDNA sequences of the other 25 RcPLCP genes were reported in any public database (as of Dec 2016). Nevertheless, 13 members had EST (expressed sequence tag) hits in GenBank and RcRD19B was found to harbor the maximal hit of 68 ESTs. Moreover, the expression of other genes was supported by RNA sequencing reads derived from various tissue transcriptomes, i.e. leaf, flower, endosperm and seed27–29. Except for RcSAG12H8, the transcription regions of all other RcPLCP genes were successfully extended based on the read alignment (Table 1).
Since the gene models released in castor bean were the result of an automatic annotation6, an expert revision of their gene structures was conducted via mapping ESTs and RNA sequencing reads to the identified scaffolds. Interesting, six out of the 26 annotated gene models were proved to be inaccurate. The locus 29970.t000002 (RcRD21C) was predicted to encode 366 residues (29970.m000973), and it represents only the 3′ sequence of the gene which encodes 383 residues (see Supplementary File S1). The locus 30162.t000046 (RcXCP1) was predicted to contain three introns putatively encoding 324 residues (30162.m001301), however, hundreds of RNA sequencing reads indicated that the “GAAA” sequence in the first exon was absent from the genome assembly. Thereby, this locus promises to harbor two introns encoding 349 residues (see Supplementary File S2). The locus 30170.t000524 (RcXBCP3) was predicted to contain four introns putatively encoding 422 residues (30170.m014112), however, read alignment indicated that this locus harbors five introns putatively encoding 466 residues (see Supplementary File S3). The locus 29381.t000001 (RcXBCP3L) was predicted to encode 417 residues (29381.m000072), however, read alignment indicated that this locus encodes 501 residues (see Supplementary File S4). The locus 29827.t000145 (RcPAP3) was predicted to contain two introns putatively encoding 321 residues (29827.m002672), however, read alignment indicated that this locus harbors one intron putatively encoding 342 residues (see Supplementary File S5). The locus 30076.t000074 (RcCTB1) was predicted to encode 376 residues (30076.m004510), however, read alignment indicated that this locus encodes 359 residues (see Supplementary File S6). Additionally, 10 genes (i.e. RcRD21A, RcCEP1, RcCEP2, RcTHI1, RcSAG12H3, RcRD19A, RcRD19B, RcRD19C, RcALP1 and RcCTB1) were shown to have alternative splicing isoforms (Table 1).
Four gene pairs (i.e. paralogs) can be defined as tandem duplicates for their close organization on same scaffolds and high sequence identity, i.e., 97.4% between RcSAG12H7 (29910.t000015) and RcSAG12H8 (29910.t000014), 96.8% between RcSAG12H2 (28962.t000017) and RcSAG12H3 (28962.t000018), 87.5% between RcSAG12H4 (29646.t000033) and RcSAG12H5 (29646.t000034), and 74.1% between RcPAP1 (29900.t000078) and RcPAP2 (29900.t000077). However, whether RcPAP3 is a proximal duplicate of RcPAP1 or RcPAP2 still needs to be confirmed, since the 25,878 assembled scaffolds have not been anchored to the chromosomes yet6.
Characterization of 23 PLCP genes in physic nut
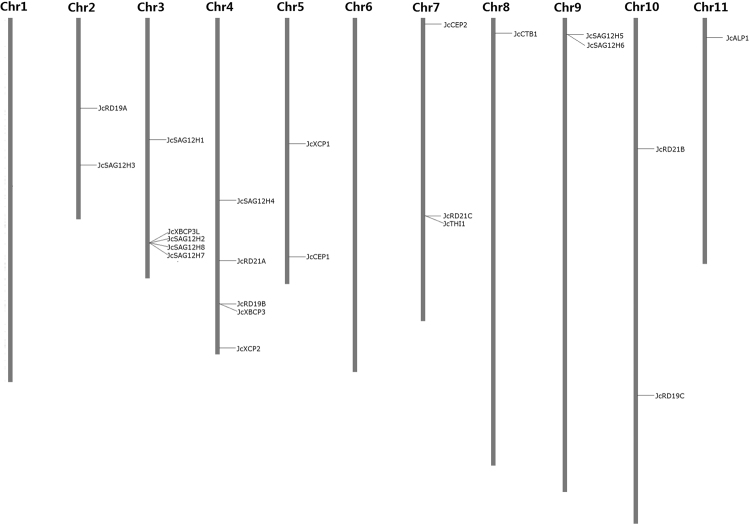
After discarding four pseudogenes (i.e. JCGZ_22119, JCGZ_05109 and two unpredicted loci on scaffold170), a total of 23 PLCP-encoding loci were identified from the physic nut genome. Among them, 22 loci were predicted by the automatic genome annotation8, whereas one more locus putatively encoding a SAG12 subfamily member was identified from scaffold684 (i.e. JcSAG12H8, see Supplementary File S7). These loci are distributed across 17 scaffolds. Like observed in castor bean, most scaffolds contain one PLCP gene, and four scaffolds encoding more than one are as follows: scaffold684 (4), scaffold341 (2), scaffold159 (2) and scaffold872 (2) (Table 2). When taking the linkage map with 1208 markers8 into account, these scaffolds can be further anchored to nine chromosomes (Chrs), i.e., Chr2 (scaffold84 and scaffold96), Chr3 (scaffold26 and scaffold684), Chr4 (scaffold5, scaffold46, scaffold159 and scaffold221), Chr5 (scaffold3 and scaffold328), Chr7 (scaffold341 and scaffold502), Chr8 (scaffold392), Chr9 (scaffold872), Chr10 (scaffold7 and scaffold464) and Chr11 (scaffold211). The distribution of JcPLCP genes looks uneven: Chromosomes 3 and 4 contain the most of five PLCP genes, followed by Chromosome 7 with three genes (Fig. 1).
Table 2.
List of 23 JcPLCP genes identified in this study.
| Gene name | Locus ID | Scaffold | Predicted position | Identified position | Chr | EST hits | ASa | AA | MW (KDa) | pI | GRAVY | iPSORTb | At_orthologc | Rc_orthologc |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| JcRD21A | JCGZ_16099 | scaffold46 | 3722788–3725887 | 3726134–3722381 | 4 | 53 | — | 466 | 51.79 | 5.28 | −0.515 | S | AtRD21A | RcRD21A |
| JcRD21B | JCGZ_22120 | scaffold7 | 104053–105002 | 103886–106508 | 10 | 6 | — | 475 | 52.68 | 5.39 | −0.504 | S | — | RcRD21B |
| JcRD21C | JCGZ_12447 | scaffold341 | 1511831–1514897 | 1511754–1515287 | 7 | 3 | Yes | 366 | 41.12 | 5.40 | −0.372 | S | AtRDL1 | RcRD21C |
| JcCEP1 | JCGZ_11373 | scaffold328 | 482830–484642 | 484817–482666 | 5 | 30 | — | 360 | 40.17 | 5.69 | −0.552 | S | AtCEP1 | RcCEP1 |
| JcCEP2 | JCGZ_17869 | scaffold502 | 2577993–2579143 | 2579634–2577807 | 7 | 36 | — | 358 | 39.94 | 6.19 | −0.548 | S | — | RcCEP2 |
| JcXCP1 | JCGZ_10746 | scaffold3 | 258651–259875 | 258620–260019 | 5 | 0 | Yes | 349 | 39.05 | 5.59 | −0.361 | S | AtXCP1 | RcXCP1 |
| JcXCP2 | JCGZ_08041 | scaffold221 | 310092–311414 | 311442–309951 | 4 | 0 | — | 350 | 39.18 | 5.40 | −0.367 | S | AtXCP2 | RcXCP2 |
| JcXBCP3 | JCGZ_04495 | scaffold159 | 1388654–1391224 | 1391430–1388201 | 4 | 0 | Yes | 441 | 48.82 | 6.06 | −0.333 | S | AtXBCP3 | RcXBCP3 |
| JcXBCP3L | JCGZ_21572 | scaffold684 | 2356094–2358380 | 2355980–2358669 | 3 | 3 | — | 524 | 58.04 | 5.24 | −0.369 | C | — | RcXBCP3L |
| JcTHI1 | JCGZ_12228 | scaffold341 | 82305–83446 | 83709–81708 | 7 | 6 | Yes | 347 | 38.94 | 8.43 | −0.615 | S | AtTHI1 | RcTHI1 |
| JcSAG12H1 | JCGZ_09604 | scaffold26 | 600732–601907 | 602051–600546 | 3 | 0 | — | 345 | 38.26 | 7.99 | −0.438 | S | AtSAG12 | — |
| JcSAG12H2 | JCGZ_21557 | scaffold684 | 2264217–2264672 | 2264714–2263227 | 3 | 0 | — | 345 | 38.40 | 8.56 | −0.377 | S | AtSAG12 | — |
| JcSAG12H3 | JCGZ_24483 | scaffold84 | 187704–188806 | 189021–187389 | 2 | 0 | — | 339 | 37.60 | 5.94 | −0.424 | S | AtSAG12 | RcSAG12H1 |
| JcSAG12H4 | JCGZ_17185 | scaffold5 | 199542–200754 | 199432–201896 | 4 | 0 | — | 340 | 37.42 | 5.13 | −0.413 | S | AtSAG12 | RcSAG12H2 |
| JcSAG12H5 | JCGZ_25371 | scaffold872 | 469319–470468 | 472278–473649 | 9 | 0 | — | 340 | 37.59 | 5.01 | −0.424 | S | AtSAG12 | RcSAG12H2 |
| JcSAG12H6 | JCGZ_25372 | scaffold872 | 472278–473548 | 469201–470468 | 9 | 0 | — | 340 | 37.28 | 6.90 | −0.426 | S | AtSAG12 | RcSAG12H2 |
| JcSAG12H7 | JCGZ_21549 | scaffold684 | 2211946–2213074 | 2213483–2211423 | 3 | 0 | — | 344 | 37.98 | 4.73 | −0.372 | S | AtSAG12 | RcSAG12H6 |
| JcSAG12H8 | — | scaffold684 | — | 2215711–2214317 | 3 | 0 | — | 342 | 37.48 | 4.97 | −0.413 | S | AtSAG12 | RcSAG12H7 |
| JcRD19A | JCGZ_26761 | scaffold96 | 231461–233265 | 233346–230988 | 2 | 67 | — | 370 | 40.56 | 5.95 | −0.246 | S | AtRD19A | RcRD19A |
| JcRD19B | JCGZ_04503 | scaffold159 | 1432232–1434263 | 1432160–1434615 | 4 | 11 | — | 368 | 40.92 | 5.81 | −0.338 | S | AtRD19C | RcRD19B |
| JcRD19C | JCGZ_16165 | scaffold464 | 314963–316717 | 316756–314735 | 10 | 11 | — | 383 | 42.10 | 6.03 | −0.106 | S | AtRD19D | RcRD19C |
| JcALP1 | JCGZ_07488 | scaffold211 | 2991934–2994769 | 2994891–2991702 | 11 | 51 | Yes | 358 | 39.44 | 5.88 | −0.147 | S | AtALP | RcALP1 |
| JcCTB1 | JCGZ_14145 | scaffold392 | 1649834–1653016 | 1653334–1649409 | 8 | 7 | — | 358 | 39.71 | 6.07 | −0.230 | S | AtCTB2 | RcCTB1 |
a“Yes” represents genes containing alternative splicing isoforms; b“S, M and C” represent signal peptide, mitochondrial targeting peptide or chloroplast transit peptide, respectively; cThe best ortholog hit.
Figure 1.
Chromosomal distribution of 23 JcPLCP genes. The eleven linkage groups or chromosomes were constructed with 1208 DNA markers, where the chromosome number is indicated at the top.
As of Dec 2016, although no full-length cDNA sequences were available for all physic nut PLCP family genes, 12 members were found to have EST hits in GenBank and JcRD19A harbored the maximum of 67 hits. Moreover, the expression of other members was supported by RNA sequencing reads derived from transcriptomes of various tissues including callus, root, leaf, inflorescence meristem, flower, embryo and seed7,8,30–34. Based on the read alignment, the transcription regions of all JcPLCP genes were extended and seven predicted gene models were optimized (Table 2). The locus JCGZ_22120 (JcRD21B) was predicted to encode 242 residues, and it represents only the 5′ sequence of the gene which encodes 471 residues (see Supplementary File S8). The locus JCGZ_17869 (JcCEP2) was predicted to encode 269 residues, and it represents only the 3′ sequence of the gene which encodes 358 residues (see Supplementary File S9). The locus JCGZ_21572 (JcXBCP3L) was predicted to encode 508 residues, and it represents only the 3′ sequence of the gene which encodes 524 residues (see Supplementary File S10). The locus JCGZ_12228 (JcTHI1) was predicted to encode 315 residues, and it represents only the 3′ sequence of the gene which encodes 347 residues (see Supplementary File S11). The locus JCGZ_09604 (JcSAG12H1) was predicted to encode 311 residues, and it represents only the 3′ sequence of the gene which encodes 345 residues (see Supplementary File S12). The locus JCGZ_21557 (JcSAG12H2) was predicted to encode 155 residues, and it represents only the 5′ sequence of the gene which encodes 345 residues (see Supplementary File S13). The locus JCGZ_21549 (JcSAG12H7) was predicted to encode 324 residues, and it represents only the 3′ sequence of the gene which encodes 344 residues (see Supplementary File S14). Additionally, five genes (i.e. JcRD21C, JcXCP1, JcXBCP3, JcTHI1 and JcALP1) were shown to have alternative splicing isoforms (Table 2).
Several gene pairs were shown to exhibit high sequence identity, i.e., 97.8% between JcSAG12H1 and JcSAG12H2, 88.6% between JcSAG12H5 and JcSAG12H6, 76.4% between JcSAG12H7 and JcSAG12H8, 67.2% between JcSAG12H8 and JcSAG12H1, 66.8% between JcSAG12H8 and JcSAG12H2, 65.4% between JcSAG12H7 and JcSAG12H1, 65.2% between JcSAG12H7 and JcSAG12H2. JcSAG12H2/7/8 and JcSAG12H5/6 can be defined as tandem duplicates for their adjacent organization on same scaffolds, whereas JcSAG12H1 can be defined as the recent proximal duplicate of JcSAG12H2 for their distribution on two distinct scaffolds of Chromosome 3 (Table 2 and Fig. 1).
Phylogenetic analysis of castor bean and physic nut PLCPs
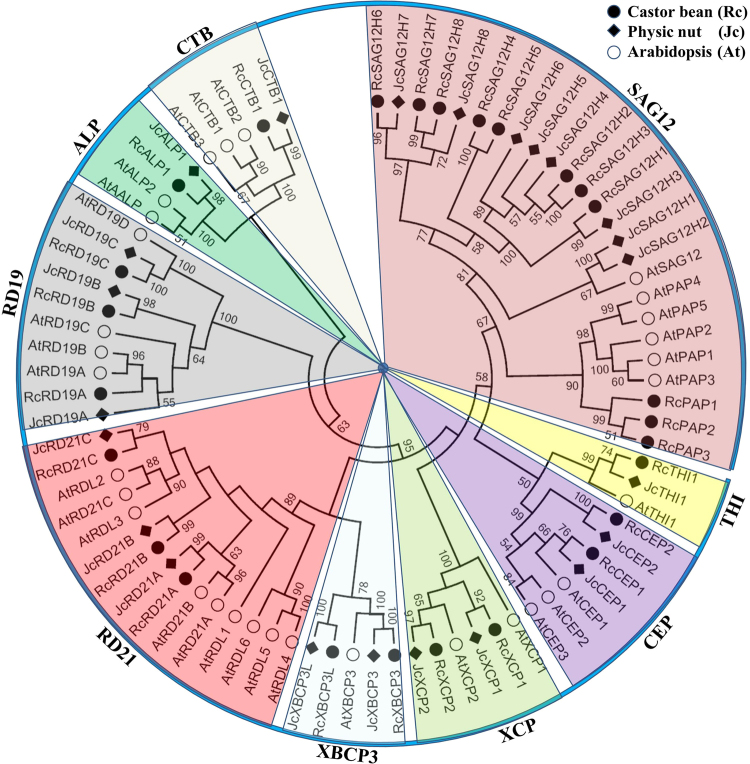
According to the reciprocal BLASTP analysis, 26 RcPLCPs have 20 or 14 orthologous groups (OGs) in physic nut and Arabidopsis, respectively (Table 1), and 23 JcPLCPs have 19 or 13 OGs in castor bean and Arabidopsis, respectively (Table 2), suggesting gene-specific duplication and/or loss occurred. To reveal the evolutionary relationship, we constructed a phylogenetic tree using 80 PLCPs from castor bean, physic nut as well as the extensively studied Arabidopsis. Results showed that castor bean and physic nut PLCPs can be divided into nine subfamilies as described in Arabidopsis, i.e., RD21, CEP, XCP, XBCP3, THI, SAG12, RD19, ALP and CTB (Figs 2 and 3). Furthermore, RcRD21B/JcRD21B, RcCEP2/JcCEP2 and two XBCP3 members (named RcXBCP3L/JcXBCP3L), which have no orthologs in Arabidopsis, were found to form new groups (Tables 1 and 2, and Fig. 2).
Figure 2.
Phylogenetic analysis of castor bean, physic nut and Arabidopsis PLCPs. Sequence alignment and construction of the phylogenetic tree were performed using MUSCLE or MEGA6, respectively. The subfamily names are indicated next to their cluster and the distance scale denotes the number of amino acid substitutions per site.
Figure 3.
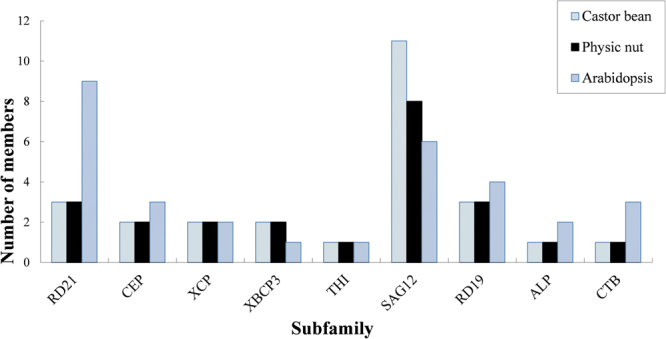
Distribution of castor, physic nut and Arabidopsis PLCP genes in subfamilies.
As shown in Fig. 2, a large number of AtPLCPs were grouped in pairs, reflecting the occurrence of two recent WGD events22. These gene pairs are widely distributed in different subfamilies, only excluding subfamilies XBCP3 and THI with a single member. In contrast, few gene pairs were found in castor bean and physic nut, which are limited to the SAG12 subfamily. In Arabidopsis, the SAG12 subfamily is composed of six members, which can be further divided into two groups named SAG12 and PAP. The PAP group was also present in castor bean but absent from physic nut, suggesting specific gene loss in the latter. Compared with Arabidopsis that contains a single SAG12 group member, both castor bean and physic nut contain eight members, which were shown to form four subgroups (Fig. 2).
Gene structure, sequence feature and conserved motifs
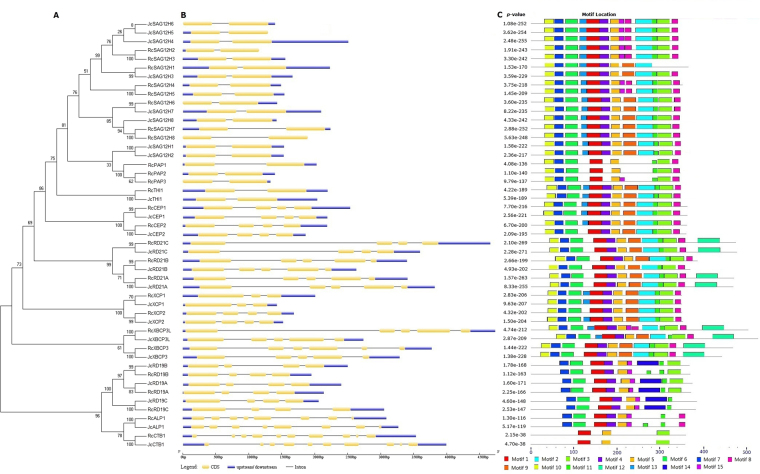
The exon-intron structure of castor bean and physic nut PLCP genes was investigated based on optimized gene models, which are supported by ESTs and/or RNA sequencing reads. As shown in Fig. 4B, these genes harbor at least one intron, varying from one to ten as observed in Arabidopsis. Although genes in different subfamilies were shown to harbor distinct exon-intron structures, the structure is usually conserved within the subfamily and between orthologs across three compared species. Without any exception, genes in the ALP subfamily all contain seven introns. Except for AtSAG12 containing two introns, other members in subfamilies SAG12 and THI feature one intron. Most genes in subfamilies CEP, XCP and RD19 contain three introns, whereas RcXCP1, JcXCP1, AtCEP1, AtCEP2, AtCEP3, AtRD19A and AtRD19B contain two introns instead. Genes in the XBCP3 subfamily usually contain four introns, while RcXBCP3 contains five introns instead. Genes in the CTB subfamily usually contain ten introns, however, AtCTB1 harbors nine introns instead. Compared with the ORF (open reading frame) length (1023–1506 bp with the average of 1119 bp in castor bean, 1020–1575 bp with the average of 1135 bp in physic nut, and 1026–1392 bp with the average of 1118 bp in Arabidopsis), the gene size (from start to stop codons) of each gene is relatively more variant (1111–4315 bp with the average of 1933 bp in castor bean, 1103–3183 bp with the average of 1860 bp in physic nut, and 1137–2471 bp with the average of 1603 bp in Arabidopsis) (Fig. 4B and Supplementary Table S1).
Figure 4.
Structural features of castor and physic nut PLCP genes. (A) An unrooted phylogenetic tree constructed using MEGA6. (B) A graphic representation of exon-intron structures displayed using GSDS. (C) Distribution of 15 conserved motifs.
Sequence analysis showed that the deduced PLCP proteins were predicted to harbor an average MW (molecular weight) of 41 kDa and pI (isoelectric point) value of 4.7. Since the predicted GRAVY (grand average of hydropathicity) values were all shown to be less than 0 (varying from −0.04 to −0.62), these proteins are more likely to be hydrophilic. According to the subcellular localization analysis, a hydrophobic signal peptide was also found at the N-terminal of each protein (Tables 1 and 2, and Supplementary Table S1), where JcALP1 and RcALP1 include the NPIR motif for the vacuolar localization as observed in AtALP35. Except for JcCEP2 that harbors a RDEL motif at the C-terminal, RcCEP1, RcCEP2 and JcCEP1 contain a KDEL motif for the ER retention26,36,37.
Motif compositions among different Rc/JcPLCPs were also investigated and the results were shown in Fig. 3 and Supplementary Fig. S1. Among the 15 motifs identified using MEME, Motifs 1–11 and 13 are broadly distributed. Motif 7 includes the ERFNIN consensus sequence. This motif as well as Motifs 10 and 6 are characterized as the well-studied Inhibitor_I29 (PF08246), which is the core of the auto-inhibitory pro-domain10,16. Motifs 1, 4, 5, 9, 15, 2, 11, 3 and 8 are characterized as the Peptidase_C1 domain (PF00112), where Motifs 1, 11 and 3 include the Cys, His or Asn active site respectively10,11. Motifs 14 and 15 are also part of the Peptidase_C1 domain, where Motif 14 is specific to the RD19 subfamily and Motif 15 is only found in subfamilies SAG12, XBCP3, RD19 and ALP. Motif 13 is the link of the Inhibitor_I29 domain and the Peptidase_C1 domain, which was shown to contain the cleavage site for generation of a mature enzyme21,38. Motif 12, which is limited to RD21 and XBCP3 subfamilies, is characterized as the well-studied GRAN domain (PF00396) (Fig. 3C).
Expression patterns of RcPLCP genes in various tissues
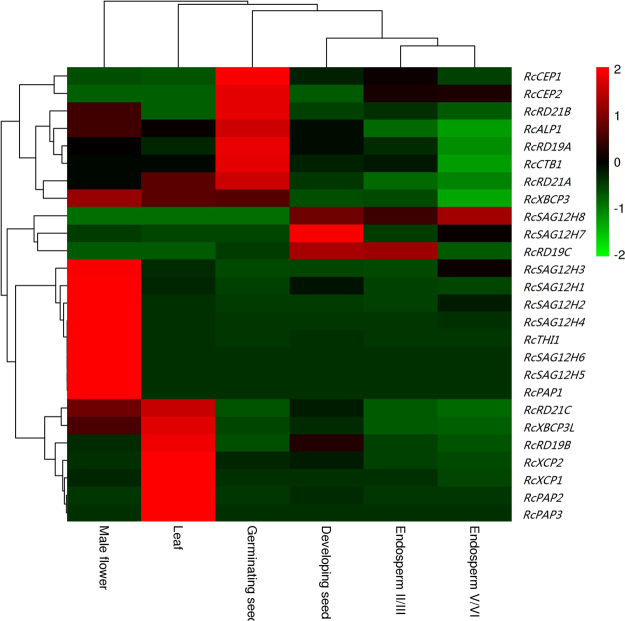
Transcriptional profiling revealed that 26 RcPLCP genes were expressed in at least one of the tested tissues or developing stages of a certain tissue, i.e., 20 in leaf, 23 in male flower, 19 in endosperm II/III, 16 in endosperm V/VI, 21 in developing seed and 17 in germinating seed. According to the FPKM value, the total transcripts of RcPLCP genes were most abundant in male flower, followed by germinating seed (Group I); moderate in endosperm II/III, leaf and developing seed (Group II, occupying 21–27% of Group I); and, considerably low in endosperm V/VI (Group III, occupying 6–7% of Group I). In male flower, the unique member of the THI subfamily contributes the major transcripts, occupying about 72% of the total PLCP transcripts; by contrast, the second highly abundant RD21 subfamily occupies only 10%. In leaf, subfamilies RD19 and RD21 occupy about 69% of the total PLCP transcripts. In endosperm and seed, the CEP subfamily usually contributes the major PLCP transcripts, though the RD19 subfamily plays a more important role in developing seed. In endosperm V/VI, the CEP subfamily occupies about 88% of the total PLCP transcripts. In endosperm II/III, the CEP subfamily occupies about 57% of the total PLCP transcripts, and the less abundant subfamilies RD19 and RD21 occupy about 19% or 13% respectively. In germinating seed, the CEP subfamily occupies about 51% of the total PLCP transcripts, and the less abundant subfamilies RD21 and RD19 occupy about 25% or 11% respectively. In developing seed, three highly abundant subfamilies RD19, CEP and RD21 occupy about 35%, 28% or 17% of the total PLCP transcripts respectively (Fig. 5).
Figure 5.
Transcriptional profiling of 26 RcPLCP genes over various tissues. Color scale denotes FPKM normalized log10 transformed counts.
Although the transcript level is diverse, most RcPLCP genes were shown to express in all examined tissues. RcCEP1 represents the most expressed gene in endosperm II/III, endosperm V/VI, developing seed and germinating seed, occupying about 46%, 41%, 43% or 27% of the total PLCP transcripts in each sample respectively; by contrast, its transcript level in leaf is extremely low. Compared with RcCEP1, the transcript level of RcCEP2 is comparable in endosperm V/VI, but is relatively lower in other tissues. RcTHI1 represents the most expressed gene in flower; RcRD19B and RcRD21A represent the most expressed genes in leaf, occupying about 29% or 25% of the total PLCP transcripts respectively. Nevertheless, the expression of RcRD19B was not detected in endosperm V/VI; RcSAG12H5, RcSAG12H6 and RcPAP1 seem to be flower-specific; and, RcPAP3 seems to be leaf-specific (Fig. 5).
Based on the expression pattern across various tissues, 26 RcPLCP genes were clustered into four groups: Group 1 prefers to express in germinating seed, including 2 CEPs (RcCEP1 and RcCEP2), 2 RD21s (RcRD21B and RcRD21A), RcRD19A, RcALP1, RcCTB1 and RcXBCP3; Group 2 prefers to express in developing seed and endosperm, including RcRD19C, 2 SAG12s (RcSAG12H7 and RcSAG12H8); Group 3 prefers to express in male flower, including RcTHI1, 7 SAG12s (RcSAG12H1, RcSAG12H2, RcSAG12H3, RcSAG12H4, RcSAG12H5, RcSAG12H6 and RcPAP1); and, Group 4 prefers to express in leaf, including 2 XCPs (RcXCP1 and RcXCP2), 2 SAG12s (RcPAP2 and RcPAP3), RcRD21C, RcXBCP3L and RcRD19B.
Expression patterns of JcPLCP genes in various tissues
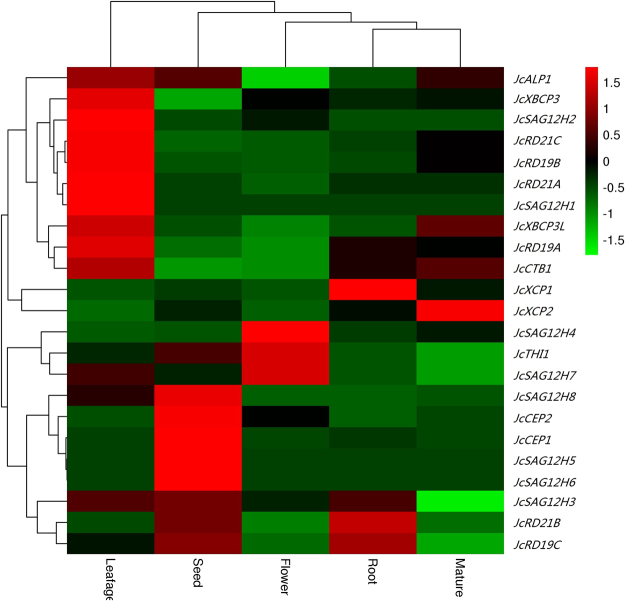
As shown in Fig. 6, transcriptional profiling supported the expression of all JcPLCP genes in at least one of the examined tissues, i.e., 21 in root, 19 in flower bud, 23 in seed, 21 in leafage and 18 in mature leaf. The total transcripts were most abundant in leafage (Group I); moderate in seed, mature leaf and root (Group II, occupying 37–47% of Group I); and, considerably low in flower bud (Group III, occupying about 13% of Group I). In most tissues, RD19 and RD21 subfamilies contribute the major PLCP transcripts, e.g., 72% in leafage, 62% in flower, and 54% in root. By contrast, subfamilies RD19 and ALP occupy 55% of the total PLCP transcripts in mature leaf, and the CEP subfamily occupies 55% of the total PLCP transcripts in seed. Several important JcPLCP genes were also identified for a certain tissue. JcRD19A presents the most expressed gene in flower bud and root, both occupying about 30% of the total PLCP transcripts. JcCEP1 presents the most expressed gene in seed, occupying about 55% of the total PLCP transcripts, though its expression level is relatively low in flower bud, leafage and mature leaf. In leafage, the transcript level of JcRD21A, JcRD19A, JcRD19B and JcALP1 is considerably high, where JcALP1 also presents the most expressed gene in mature leaf (Fig. 6). These genes were also clustered into four groups based on the tissue-specific expression pattern: Group 1 prefers to express in leafage, including 2 RD21s (JcRD21A and RcRD21C), 2 RD19s (JcRD19A and RcRD19B), 2 XBCP3 (JcXBCP3 and JcXBCP3L), 2 SAG12 (JcSAG12H1 and JcSAG12H2), JcALP1 and JcCTB1; Group 2 prefers to express in root (JcXCP1) or mature leaf (JcXCP2); Group 3 prefers to express in flower bud, including JcTHI1, 2 SAG12s (JcSAG12H4 and JcSAG12H7); and, Group 4 prefers to express in seed, including 2 CEP (JcXCP1 and JcXCP2), 4 SAG12s (JcSAG12H3, JcSAG12H5, JcSAG12H6, and JcSAG12H8), JcRD21B and JcRD19C.
Figure 6.
Transcriptional profiling of 23 JcPLCP genes over various tissues. Color scale denotes FPKM normalized log10 transformed counts.
Discussion
Small number but high diversity of PLCP family genes in castor bean and physic nut
WGDs are widespread and play an important role in the origin and diversification of the angiosperms39,40. Arabidopsis, an annual herb with a relatively short life cycle and small size, services as a popular model species for the study of plant biology and genetics. The relatively small diploid genome (approximately 135 Mb) made it the first plant to be sequenced, completed in December of the year 200041. However, analysis of the Arabidopsis genome has revealed several unexpected secrets. During the last 120 million years, Arabidopsis was shown to have experienced three WGDs known as γ, β and α22,42. The γ event occurred at about 117 million years ago (Mya)9, which is shared by all core eudicots, e.g. castor bean, physic nut, rubber, cassava (Manihot esculenta), poplar, papaya (Carica papaya), cacao (Theobroma cacao) and grapevine (Vitis vinifera)6,8,24,25,43–47. The β event occurred at 61–65 Mya, shortly after its divergence with its close species papaya44, which is shared by the Cleomaceae plants48. The α event is Brassicaceae-specific, occurred within a window of 23–50 Mya22,49. Following these WGD events, the ancestral A. thaliana genome was hugely rearranged and gene copies have been massively lost, and almost half of the genome was lost since its divergence with A. lyrata at about 10–13 Mya50. The genome-wide analysis indicated that the Arabidopsis PLCP gene family is comprised of 31 members that can be divided into nine subfamilies based on sequence similarity17. As shown in Fig. 2, a relatively high number of Arabidopsis PLCP gene pairs were identified in most subfamilies. The 18 duplicates were shown to be resulted from different modes of gene duplication, i.e., γ (2), β (2), α (5), tandem (5), proximal (1) and transposed (3)51 (see Supplementary Table S1).
Despite containing more or comparable protein-coding loci than Arabidopsis (i.e. 27,416 in TAIR10), our genome-wide survey revealed that castor bean (i.e. 31,221) and physic nut (i.e. 27,172) encode relatively less PLCP genes, i.e. 26 or 23, respectively. The number occupies 0.08% of the total loci in both species, which is relatively smaller than 0.11% in Arabidopsis. The expression of all these genes was supported by available EST and/or RNA sequencing reads, suggesting that they may have function in these two species. Moreover, all PLCP genes in physic nut can be anchored to nine out of the 11 chromosomes based on available DNA markers8. Based on the phylogenetic analysis, these genes can be assigned to nine previously described subfamilies (i.e. RD21, CEP, XCP, XBCP3, THI, SAG12, RD19, ALP and CTB)17.
Except for the SAG12 subfamily, one-to-one orthologous relationships were found between castor bean and physic nut, and conserved synteny between these two species can be clearly observed. Despite the castor bean genome is highly fragmented, we are able to anchor 25 out of 26 RcPLCP genes to eight physic nut chromosomes based on the synteny analysis (Supplementary Fig. S2). By contrast, gene-specific expansion was observed in the SAG12 subfamily, which can be further divided into two groups (i.e., SAG12 and PAP). The PAP group was shown to be lost in physic nut, but highly expanded in castor bean (i.e. 3 members) as observed in Arabidopsis (i.e. 5 members)17. The SAG12 group is comprised of a single gene with two introns in Arabidopsis, whereas eight members with a single intron were found in both castor bean and physic nut. As shown in Fig. 2, the SAG12 group is obviously clustered into four subgroups: Subgroup 1 (Ia) includes JcSAG12H1–2 and AtSAG12; Subgroup 2 (Ib) includes JcSAG12H3–6 and RcSAG12H1–3; Subgroup 3 (Ic) includes RcSAG12H4–5; and Subgroup 4 (Id) includes JcSAG12H7–8 and RcSAG12H6–8. Since members of all four subgroups can be found in other plant species (see below), castor bean and physic nut promise to have lost Ic or Ia subgroup members, respectively; and, the unique AtSAG12 is more likely to be the result of massive gene loss after WGDs. As for other subfamilies, most members in castor bean and physic nut were shown to harbor one to three orthologs in Arabidopsis, however, the orthologs of RcRD21B/JcRD21B, RcCEP2/JcCEP2 and RcXBCP3L/JcXBCP3L have also been lost in Arabidopsis. Thereby, it is probably safe to say that the ancestral Euphorbiaceae genome contained 20 PLCP family genes, i.e., three RD21s, two CEPs, two XCPs, two XBCP3s, one THI, five SAG12s, three RD19s, one ALP and one CTB.
Evolution of the PLCP gene family in castor bean and physic nut
As discussed above, the PLCP family genes in castor bean and physic nut promise to evolve from 20 ancestors, and gene-specific expansion and/or loss was shown to be restricted to the SAG12 subfamily. Although the exon-intron pattern between orthologs is highly conserved, RcXBCP3 has obtained an additional intron close to the 3′ untranslated region (Fig. 4). As expected, the sequence length and nucleotide substitution of introns are relatively more variable than that in exons between orthologs. Nevertheless, as shown in Fig. 2, the origin and evolution of subfamily members still needs to be resolved. The available genomes of several representative plants allow us to discuss this issue. When taking advantage of the castor bean, physic nut and Arabidopsis PLCP genes to trace their orthologs in these plants, we are able to find one RD21 and three CTBs in a single celled green alga, Chlamydomonas reinhardtii 52; one RD21, one RD19, one ALP and one CTB in spikemoss (Selaginella moellendorffii), an ancient vascular species first appeared at about 400 Mya53; one RD21, one CEP, two XCP, one XBCP, four SAG12s, one RD19, one RD19, one ALP and one CTB in Amborella trichopoda, a sister species to all other flowering plants54; two RD21s, one CEP, two XCPs, one XBCP3, four SAG12s, two RD19s, one RD19, one ALP and one CTB in rice (Oryza sativa), a model species of monocotyledons55; one RD21, one CEP, two XCPs, two XBCP3 (including one XBCP3L), one THI, four SAG12s, one RD19, one RD19, one ALP and one CTB in Aquilegia coerulea, a basal species of most eudicot clade56.
As shown in Fig. 2, the RD21 subfamily contains five OGs: Group I includes RcRD21A, JcRD21A, AtRD21A and AtRD21B; Group II includes RcRD21B and JcRD21B; Group III includes RcRD21C, JcRD21C and AtRDL1; Group IV includes AtRD21C, AtRDL2 and AtRDL3; and, Group V includes AtRDL4, AtRDL5 and AtRDL6 (Supplementary Table S2). Group I members are widely distributed, which can be traced back to C. reinhardtii 52. The divergence of other groups is more likely to occur in the common ancestor of core eudicots, which was proven to experience the whole-genome triplication γ event9. Plant species not having undergone any recent WGD were found to contain one RD21A ortholog, one RD21B ortholog and one RD21C ortholog, e.g. papaya, cacao and grapevine43–45. And species such as poplar and cassava that have experienced one recent WGD24,47 contain one or two orthologs for RD21A, RD21B and RD21C, respectively. However, Group II is more likely to be lost in Brassicaceae, e.g. A. thaliana, A. lyrata, Brassica rapa and B. oleracea 41,50,57,58. In contrast, Groups IV and V were shown to be restricted to Brassicaceous plants probably resulted from the β event or fast evolution, though gene-specific expansion and/or loss were found in B. rapa and B. oleracea (Supplementary Table S2).
The CTB subfamily can also be traced back to C. reinhardtii 52, and a single member was found in most plant species, especially those not having experienced recent WGDs. In contrast, gene expansion was found in C. reinhardtii, poplar, cassava and Brassicaceous plants, resulted from recent WGDs and local duplication24,38,45,50. In A. thaliana, AtCTB3 was produced from AtCTB2 via the α WGD, which is shared by A. lyrata, B. rapa and B. oleracea; AtCTB1 was produced from AtCTB2 via tandem duplication, which is only shared by A. lyrata 41,50,57,58 (Supplementary Tables S1 and 2).
The CEP subfamily contains two OGs: Group I includes RcCEP1, JcCEP1, AtCEP1, AtCEP2 and AtCEP3; and, Group II includes RcCEP2 and JcCEP2. Group I members are relatively primitive, which can be traced back to A. trichopoda 54. This group was shown to be highly expanded through WGD and local duplication in Brassicaceous plants. By contrast, Group II is more likely to be derived from the γ event, and gene-specific loss occurred in Brassicaceous plants41,50,57,58 (Supplementary Table S2).
The XCP subfamily also includes two OGs: Group I includes RcXCP1, JcXCP1 and AtXCP1; and, Group II includes RcXCP2, JcXCP2 and AtXCP2. Like the CEP subfamily, Group I of the XCP subfamily can also be traced back to A. trichopoda 54, while Group II is more likely to be resulted from the γ event (Supplementary Tables S1 and 2).
The XBCP3 subfamily contains two OGs: Group I includes RcXBCP3, JcXBCP3 and AtXBCP3, which can be traced back to the ancestral angiosperm; and, Group II includes RcXBCP3L and JcXBCP3L, which can be traced back to A. coerulea, though specific gene loss occurred in Brassicaceous plants (Supplementary Table S2). Interesting, Group II is highly expanded in poplar, resulted from the recent WGD and local duplication24 (Supplementary Table S2).
The THI subfamily usually contains a single member, which can be traced back to the ancestor of eudicots. However, specific gene loss was found in poplar24 (Supplementary Table S2).
The RD19 subfamily contains three OGs: Group I includes RcRD19A, JcRD19A, AtRD19A and AtRD19B; Group II includes RcRD19B, JcRD19B and AtRD19C; and, Group III includes RcRD19C, JcRD19C and AtRD19D. Groups I and III can be traced back to spikemoss53, whereas Group II is more likely to be resulted from the γ event (Supplementary Tables S1 and 2). In grapevine, Group II is highly expanded through local duplication43.
The SAG12 subfamily contains two main groups, i.e. SAG12 and PAP. The origin of the PAP group is still not clear, since it was only found in castor bean, cassava as well as Brassicaceous plants (Supplementary Table S2), which belong to two distinct plant families. In Brassicaceae, the PAP group is highly expanded via the α WGD, tandem duplication and transposed duplication51. The SAG12 group can be further divided into four orthologous subgroups: Ia can be traced back to A. trichopoda, and has been lost in castor bean, papaya, cacao and grapevine; Ib is more likely to appear in the common ancestor of core eudicots along with the γ event, and has been lost in Brassicaceous plants; Ic may also appear along with the γ event, and has been lost in physic nut, cacao as well as Brassicaceous plants; and, Id is more likely to be Euphorbiaceae-specific14,21 (Supplementary Table S2).
In addition to gene copies and exon-intron structures, expression divergence was also observed between orthologs/paralogs. The transcript level of several OGs such as RcRD21A/JcRD21A/AtRD21A, RcRD19A/JcRD19A/AtRD19A, RcRD19B/JcRD19B/ AtRD19C, RcALP1/JcALP1/AtALP and RcCTB1/JcCTB1/AtCTB3 is highly abundant in leaf, flower and seed. In contrast, the paralogs of AtRD21A, AtRD19A, AtALP and AtCTB3, i.e., AtRD21B, AtRD19B, AtALP2 and AtCTB1/AtCTB2, are considerably less expressed, though they are also constitutively expressed in these tissues59. As for two OGs of subfamilies CEP and XCP, which were generated along with the γ event, expression divergence is even more obvious. Among them, RcCEP1/JcCEP1 and RcXCP2/JcXCP2 have become the dominated isoforms in all tested tissues (Figs 5 and 6). Similar expression pattern can also be observed for their orthologs in Arabidopsis, e.g. AtXCP1 and AtXCP2 59. It is worth noting that, RcTHI1 represents the most abundant gene in the male flower of castor bean, which is in accord with the expression pattern of its ortholog in Arabidopsis (i.e. AtTHI1); by contrast, the expression level of its ortholog in physic nut (i.e. JcTHI1) is considerably low in flower bud (Figs 5 and 6).
Conclusions
The first genome-wide analysis of PLCP family genes in castor bean and physic nut was performed in the present study, resulting in 26 or 23 members respectively. The phylogenetic analysis assigned them into nine subfamilies. Novel groups or subgroups, which are absent from Arabidopsis, were identified in RD21, CEP, XBCP3 and SAG12 subfamilies. Their orthologs are widely distributed in core eudicots, suggesting gene-specific loss occurred in Arabidopsis and other Brassicaceous plants. Moreover, the evolution characteristics of castor bean and physic nut PLCP family genes were also compared and discussed. Our findings provide a useful reference to characterize PLCP genes and analyze the family evolution in Euphorbiaceous plants and other species.
Materials and Methods
Identification and manual curation of PLCP genes in castor bean and physic nut
The genomic data for castor bean6 and physic nut8 were downloaded from Phytozome v1160 or NCBI (http://www.ncbi.nlm.nih.gov/), respectively. 31 Arabidopsis PLCPs obtained from TAIR1061 were used as queries for the homologous search. Sequences with an E-value of less than 1e−5 in the TBLASTN search62 were collected, and the positive genomic sequences were predicted using GeneMark.hmm63. The predicted gene models were further confirmed with transcriptome data available in NCBI, including cDNAs, ESTs and RNA sequencing reads. The presence of the Peptidase_C1 domain in deduced proteins was checked using SMART64. Gene expression annotation and definition of tandem/proximal duplications were performed as described before14,46. The alternative splicing (AS) isoforms were identified using Cufflinks (v2.2.1)65. The BRH (Best Reciprocal Hits) method66 was used to define orthologs across species, and the systematic name was assigned based on the best Arabidopsis ortholog.
Multiple sequence alignment and phylogenetic analysis
Sequence alignment of full-length PLCPs was conducted using MUSCLE67. The unrooted phylogenetic tree based on the alignment was constructed using MEGA668 with the maximum likelihood method, where the bootstrap was set to 1,000 replicates. Gene structures were displayed using GSDS (http://gsds.cbi.pku.edu.cn/).
Analysis of sequence feature and conserved motifs
Protein properties were determined using the online tool ProtParam (http://web.expasy.org/protparam/) and subcellular localization was predicted using iPSORT (http://ipsort.hgc.jp/). Conserved motifs were analyzed using the MEME Suite (http://meme-suite.org/tools/meme) as described before14.
Gene expression analysis
Global gene expression profiles over various tissues were examined based on Illumina RNA sequencing data available in NCBI SRA, i.e., root (SRX750579), leafage (SRX750580), mature leaf (SRX1097498), flower bud (SRX1037655) and seed (SRX750581) in physic nut; expanding leaf (ERX021378), male developing flower (ERX021379), developing endosperm II/III (ERX021375), developing endosperm V/VI (ERX021376), developing seed (SRX485027) and germinating seed (ERX021377) in castor bean. Read mapping was performed using Bowtie 269, and the FPKM (fragments per kilobase of exon per million fragments mapped)70 was used to determine the gene expression level as described before46.
Electronic supplementary material
Acknowledgements
This work was supported by the National Natural Science Foundation of China (31700580, 31371556).
Author Contributions
Z.Z. conceived and directed the study. Z.Z., Q.H., G.X. and L.Y. performed the experiments and analysis. Z.Z. wrote the paper. All authors approved the manuscript.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-017-18760-6.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Zeng C, et al. Conservation and divergence of microRNAs and their functions in Euphorbiaceous plants. Nucleic Acids Res. 2010;38:981–995. doi: 10.1093/nar/gkp1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zou Z, et al. Genome-wide identification of Jatropha curcas aquaporin genes and the comparative analysis provides insights into the gene family expansion and evolution in Hevea brasiliensis. Front. Plant Sci. 2016;7:395. doi: 10.3389/fpls.2016.00395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rivarola M, et al. Castor bean organelle genome sequencing and worldwide genetic diversity analysis. PLoS One. 2011;6:e21743. doi: 10.1371/journal.pone.0021743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Montes Osorio LR, et al. High level of molecular and phenotypic biodiversity in Jatropha curcas from Central America compared to Africa, Asia and South America. BMC Plant Biol. 2014;14:77. doi: 10.1186/1471-2229-14-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fairless D. Biofuel: the little shrub that could—maybe. Nature. 2007;449:652–655. doi: 10.1038/449652a. [DOI] [PubMed] [Google Scholar]
- 6.Chan AP, et al. Draft genome sequence of the oilseed species Ricinus communis. Nat. Biotechnol. 2010;28:951–956. doi: 10.1038/nbt.1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sato S, et al. Sequence analysis of the genome of an oil-bearing tree, Jatropha curcas L. DNA Res. 2011;18:65–76. doi: 10.1093/dnares/dsq030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wu P, et al. Integrated genome sequence and linkage map of physic nut (Jatropha curcas L.), a biodiesel plant. Plant J. 2015;81:810–821. doi: 10.1111/tpj.12761. [DOI] [PubMed] [Google Scholar]
- 9.Jiao Y, et al. A genome triplication associated with early diversification of the core eudicots. Genome Biol. 2012;13:R3. doi: 10.1186/gb-2012-13-1-r3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.van der Hoorn RA. Plant proteases: from phenotypes to molecular mechanisms. Annu. Rev. Plant Biol. 2008;59:191–223. doi: 10.1146/annurev.arplant.59.032607.092835. [DOI] [PubMed] [Google Scholar]
- 11.Diaz-Mendoza M, Velasco-Arroyo B, González-Melendi P, Martínez M, Díaz I. C1A cysteine protease-cystatin interactions in leaf senescence. J. Exp. Bot. 2014;65:3825–3833. doi: 10.1093/jxb/eru043. [DOI] [PubMed] [Google Scholar]
- 12.Lu H, et al. Subfamily-specific fluorescent probes for cysteine proteases display dynamic protease activities during seed germination. Plant Physiol. 2015;168:1462–1475. doi: 10.1104/pp.114.254466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Misas-Villamil JC, van der Hoorn RA, Doehlemann G. Papain-like cysteine proteases as hubs in plant immunity. New Phytol. 2016;212:902–907. doi: 10.1111/nph.14117. [DOI] [PubMed] [Google Scholar]
- 14.Zou Z, Xie GS, Yang LF. Papain-like cysteine protease encoding genes in rubber (Hevea brasiliensis): comparative genomics, phylogenetic and transcriptional profiling analysis. Planta. 2017;246:999–1018. doi: 10.1007/s00425-017-2739-z. [DOI] [PubMed] [Google Scholar]
- 15.Rawlings ND, Barrett AJ, Finn R. Twenty years of the MEROPS database of proteolytic enzymes, their substrates and inhibitors. Nucleic Acids Res. 2016;44:D343–350. doi: 10.1093/nar/gkv1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Beers EP, Jones AM, Dickerman AW. The S8 serine, C1A cysteine and A1 aspartic protease families in Arabidopsis. Phytochemistry. 2004;65:43–58. doi: 10.1016/j.phytochem.2003.09.005. [DOI] [PubMed] [Google Scholar]
- 17.Richau KH, et al. Subclassification and biochemical analysis of plant papain-like cysteine proteases displays subfamily-specific characteristics. Plant Physiol. 2012;158:1583–1599. doi: 10.1104/pp.112.194001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Martínez M, Diaz I. The origin and evolution of plant cystatins and their target cysteine proteinases indicate a complex functional relationship. BMC Evol. Biol. 2008;8:198. doi: 10.1186/1471-2148-8-198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Drenth J, Jansonius JN, Koekoek R, Swen HM, Wolthers BG. Structure of papain. Nature. 1968;218:929–932. doi: 10.1038/218929a0. [DOI] [PubMed] [Google Scholar]
- 20.Than ME, et al. The 2.0 A crystal structure and substrate specificity of the KDEL-tailed cysteine endopeptidase functioning in programmed cell death of Ricinus communis endosperm. J. Mol. Biol. 2004;336:1103–1116. doi: 10.1016/j.jmb.2003.12.075. [DOI] [PubMed] [Google Scholar]
- 21.Zou Z, Liu J, Yang L, Xie G. Survey of the rubber tree genome reveals a high number of cysteine protease-encoding genes homologous to Arabidopsis SAG12. PLoS One. 2017;12:e0171725. doi: 10.1371/journal.pone.0171725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bowers JE, Chapman BA, Rong J, Paterson AH. Unravelling angiosperm genome evolution by phylogenetic analysis of chromosomal duplication events. Nature. 2003;422:433–438. doi: 10.1038/nature01521. [DOI] [PubMed] [Google Scholar]
- 23.García-Lorenzo M, Sjödin A, Jansson S, Funk C. Protease gene families in Populus and Arabidopsis. BMC Plant Biol. 2006;6:30. doi: 10.1186/1471-2229-6-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tuskan GA, et al. The genome of black cottonwood, Populus trichocarpa (Torr. & Gray) Science. 2006;313:1596–1604. doi: 10.1126/science.1128691. [DOI] [PubMed] [Google Scholar]
- 25.Tang C, et al. The rubber tree genome reveals new insights into rubber production and species adaptation. Nat Plants. 2016;2:16073. doi: 10.1038/nplants.2016.73. [DOI] [PubMed] [Google Scholar]
- 26.Schmid M, Simpson D, Kalousek F, Gietl C. A cysteine endopeptidase with a C-terminal KDEL motif isolated from castor bean endosperm is a marker enzyme for the ricinosome, a putative lytic compartment. Planta. 1998;206:466–475. doi: 10.1007/s004250050423. [DOI] [PubMed] [Google Scholar]
- 27.Troncoso-Ponce MA, et al. Comparative deep transcriptional profiling of four developing oilseeds. Plant J. 2011;68:1014–1027. doi: 10.1111/j.1365-313X.2011.04751.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brown AP, et al. Tissue-specific whole transcriptome sequencing in castor, directed at understanding triacylglycerol lipid biosynthetic pathways. PLoS One. 2012;7:e30100. doi: 10.1371/journal.pone.0030100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xu W, Dai M, Li F, Liu A. Genomic imprinting, methylation and parent-of-origin effects in reciprocal hybrid endosperm of castor bean. Nucleic Acids Res. 2014;42:6987–6998. doi: 10.1093/nar/gku375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.King AJ, Li Y, Graham IA. Profiling the developing Jatropha curcas L. seed transcriptome by pyrosequencing. BioEnergy Res. 2011;4:211–221. doi: 10.1007/s12155-011-9114-x. [DOI] [Google Scholar]
- 31.Natarajan P, Parani M. De novo assembly and transcriptome analysis of five major tissues of Jatropha curcas L. using GS FLX titanium platform of 454 pyrosequencing. BMC Genomics. 2011;12:191. doi: 10.1186/1471-2164-12-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang H, Zou Z, Wang S, Gong M. Global analysis of transcriptome responses and gene expression profiles to cold stress of Jatropha curcas L. PLoS One. 2013;8:e82817. doi: 10.1371/journal.pone.0082817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Juntawong P, et al. Elucidation of the molecular responses to waterlogging in Jatropha roots by transcriptome profiling. Front. Plant Sci. 2014;5:658. doi: 10.3389/fpls.2014.00658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pan BZ, Chen MS, Ni J, Xu ZF. Transcriptome of the inflorescence meristems of the biofuel plant Jatropha curcas treated with cytokinin. BMC Genomics. 2014;15:974. doi: 10.1186/1471-2164-15-974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ahmed SU, et al. The plant vacuolar sorting receptor AtELP is involved in transport of NH(2)-terminal propeptide-containing vacuolar proteins in Arabidopsis thaliana. J. Cell Biol. 2000;149:1335–1344. doi: 10.1083/jcb.149.7.1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Napier RM, Fowke LC, Hawes C, Lewis M, Pelham HR. Immunological evidence that plants use both HDEL and KDEL for targeting proteins to the endoplasmic reticulum. J. Cell Sci. 1992;102:261–271. doi: 10.1242/jcs.102.2.261. [DOI] [PubMed] [Google Scholar]
- 37.Hierl G, Höwing T, Isono E, Lottspeich F, Gietl C. Ex vivo processing for maturation of Arabidopsis KDEL-tailed cysteine endopeptidase 2 (AtCEP2) pro-enzyme and its storage in endoplasmic reticulum derived organelles. Plant Mol. Biol. 2014;84:605–620. doi: 10.1007/s11103-013-0157-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen GH, et al. Molecular characterization of a senescence-associated gene encoding cysteine proteinase and its gene expression during leaf senescence in sweet potato. Plant Cell Physiol. 2002;43:984–991. doi: 10.1093/pcp/pcf125. [DOI] [PubMed] [Google Scholar]
- 39.De Bodt S, Maere S. & Van de Peer, Y. Genome duplication and the origin of angiosperms. Trends Ecol. Evol. 2005;20:591–597. doi: 10.1016/j.tree.2005.07.008. [DOI] [PubMed] [Google Scholar]
- 40.Soltis DE, et al. Polyploidy and angiosperm diversification. Am. J. Bot. 2009;96:336–348. doi: 10.3732/ajb.0800079. [DOI] [PubMed] [Google Scholar]
- 41.Arabidopsis Genome Initiative Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2000;408:796–815. doi: 10.1038/35048692. [DOI] [PubMed] [Google Scholar]
- 42.Jiao Y, et al. Ancestral polyploidy in seed plants and angiosperms. Nature. 2011;473:97–100. doi: 10.1038/nature09916. [DOI] [PubMed] [Google Scholar]
- 43.Jaillon O, et al. The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature. 2007;449:463–467. doi: 10.1038/nature06148. [DOI] [PubMed] [Google Scholar]
- 44.Ming R, et al. The draft genome of the transgenic tropical fruit tree papaya (Carica papaya Linnaeus) Nature. 2008;452:991–996. doi: 10.1038/nature06856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Motamayor JC, et al. The genome sequence of the most widely cultivated cacao type and its use to identify candidate genes regulating pod color. Genome Biol. 2013;14:r53. doi: 10.1186/gb-2013-14-6-r53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zou Z, et al. Genome-wide identification of rubber tree (Hevea brasiliensis Muell. Arg.) aquaporin genes and their response to ethephon stimulation in the laticifer, a rubber-producing tissue. BMC genomics. 2015;16:1001. doi: 10.1186/s12864-015-2152-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bredeson JV, et al. Sequencing wild and cultivated cassava and related species reveals extensive interspecific hybridization and genetic diversity. Nat. Biotechnol. 2016;34:562–570. doi: 10.1038/nbt.3535. [DOI] [PubMed] [Google Scholar]
- 48.Barker MS, Vogel H, Schranz ME. Paleopolyploidy in the Brassicales: analyses of the Cleome transcriptome elucidate the history of genome duplications in Arabidopsis and other Brassicales. Genome Biol. Evol. 2009;1:391–399. doi: 10.1093/gbe/evp040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vanneste K, Baele G, Maere S, Van de Peer Y. Analysis of 41 plant genomes supports a wave of successful genome duplications in association with the Cretaceous-Paleogene boundary. Genome Res. 2014;24:1334–1347. doi: 10.1101/gr.168997.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hu TT, et al. The Arabidopsis lyrata genome sequence and the basis of rapid genome size change. Nat. Genet. 2011;43:476–481. doi: 10.1038/ng.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang Y, Tan X, Paterson AH. Different patterns of gene structure divergence following gene duplication in Arabidopsis. BMC Genomics. 2013;14:652. doi: 10.1186/1471-2164-14-652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Merchant SS, et al. The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science. 2007;318:245–250. doi: 10.1126/science.1143609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Banks JA, et al. The Selaginella genome identifies genetic changes associated with the evolution of vascular plants. Science. 2011;332:960–963. doi: 10.1126/science.1203810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Amborella Genome Project The Amborella genome and the evolution of flowering plants. Science. 2013;342:1241089. doi: 10.1126/science.1241089. [DOI] [PubMed] [Google Scholar]
- 55.Ouyang S, et al. The TIGR Rice Genome Annotation Resource: improvements and new features. Nucleic Acids Res. 2007;35:D883–887. doi: 10.1093/nar/gkl976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kramer EM. Aquilegia: a new model for plant development, ecology, and evolution. Annu. Rev. Plant Biol. 2009;60:261–277. doi: 10.1146/annurev.arplant.043008.092051. [DOI] [PubMed] [Google Scholar]
- 57.Wang X, et al. The genome of the mesopolyploid crop species Brassica rapa. Nat. Genet. 2011;43:1035–1039. doi: 10.1038/ng.919. [DOI] [PubMed] [Google Scholar]
- 58.Liu S, et al. The Brassica oleracea genome reveals the asymmetrical evolution of polyploid genomes. Nat. Commun. 2014;5:3930. doi: 10.1038/ncomms4930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zimmermann P, Hirsch-Hoffmann M, Hennig L, Gruissem W. GENEVESTIGATOR. Arabidopsis microarray database and analysis toolbox. Plant Physiol. 2004;136:2621–2632. doi: 10.1104/pp.104.046367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Goodstein DM, et al. Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res. 2012;40:D1178–1186. doi: 10.1093/nar/gkr944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lamesch P, et al. The Arabidopsis Information Resource (TAIR): improved gene annotation and new tools. Nucleic Acids Res. 2012;40:D1202–1210. doi: 10.1093/nar/gkr1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Altschul SF, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3401. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lomsadze A, Ter-Hovhannisyan V, Chernoff Y, Borodovsky M. Gene identification in novel eukaryotic genomes by self-training algorithm. Nucleic Acids Res. 2005;33:6494–6506. doi: 10.1093/nar/gki937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Letunic I, Doerks T, Bork P. SMART: recent updates, new developments and status in 2015. Nucleic Acids Res. 2015;43:D257–260. doi: 10.1093/nar/gku949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Trapnell C, et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 2012;7:562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Moreno-Hagelsieb G, Latimer K. Choosing BLAST options for better detection of orthologs as reciprocal best hits. Bioinformatics. 2008;24:319–324. doi: 10.1093/bioinformatics/btm585. [DOI] [PubMed] [Google Scholar]
- 67.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-seq. Nat. Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.