Abstract
Currently, oncology testing includes molecular studies and cytogenetic analysis to detect genetic aberrations of clinical significance. Next-generation sequencing (NGS) allows rapid analysis of multiple genes for clinically actionable somatic variants. The WUCaMP assay uses targeted capture for NGS analysis of 25 cancer-associated genes to detect mutations at actionable loci. We present clinical validation of the assay and a detailed framework for design and validation of similar clinical assays. Deep sequencing of 78 tumor specimens (≥1000× average unique coverage across the capture region) achieved high sensitivity for detecting somatic variants at low allele fraction (AF). Validation revealed sensitivities and specificities of 100% for detection of single-nucleotide variants (SNVs) within coding regions, compared with SNP array sequence data (95% CI = 83.4–100.0 for sensitivity and 94.2–100.0 for specificity) or whole-genome sequencing (95% CI = 89.1–100.0 for sensitivity and 99.9–100.0 for specificity) of HapMap samples. Sensitivity for detecting variants at an observed 10% AF was 100% (95% CI = 93.2–100.0) in HapMap mixes. Analysis of 15 masked specimens harboring clinically reported variants yielded concordant calls for 13/13 variants at AF of ≥15%. The WUCaMP assay is a robust and sensitive method to detect somatic variants of clinical significance in molecular oncology laboratories, with reduced time and cost of genetic analysis allowing for strategic patient management.
Traditional approaches to the genetic characterization of clinical oncology specimens include cytogenetic analysis, fluorescence in situ hybridization (FISH), and molecular studies of single genes. These methodologies are complementary to each other and generate information of diagnostic and prognostic relevance. However, as new insight is gained into the complexities of cancer at the molecular level, the need emerges to obtain a more detailed cancer genetic profile for improved patient management. As illustrated by recent studies, identifying DNA mutations in cancer may aid in understanding clonal evolution,1 risk stratification,2 and therapeutic strategies.3, 4 With the advent of next-generation sequencing (NGS), a more complete biological characterization of a tumor can be attained at the molecular level.5
Increased access to sequencing technology and a decrease in the associated costs have made it possible for clinical laboratories to develop testing strategies using NGS. Clinical tests may be targeted to a panel of genes relevant to a given phenotype or disease, or may be more broad in scope (eg, whole-exome or whole-genome analyses). To date, most NGS clinical testing has focused on the detection of constitutional rather than somatic sequence variation, such as that reported in neuromuscular disease, mitochondrial disorders, familial cancer syndromes, cardiomyopathy, ciliopathies, and familial hypercholesterolemia.6, 7, 8, 9, 10, 11, 12 Nonetheless, there is an increasing role for NGS testing to direct the management of oncology patients. For example, KRAS mutations in codons 12 and 13 are observed in approximately 40% of colorectal cancer cases,13 and these correlate with a poor response to anti-EGFR antibody therapy.14 Likewise, the detection of an EGFR exon 19 or 21 mutation in non–small cell lung cancer is correlated with sensitivity to EGFR tyrosine kinase inhibitors, including gefitinib and erlotinib.15 Concomitant NGS analysis of a select set of genes with relevance across a broad scope of cancers increases the likelihood of detecting rare but clinically actionable variants (such as KIT mutations, which are present in <10% of thymic carcinomas16), is an aid in selecting therapeutics for tumors harboring multiple genetic changes (such as combination therapies used for synergistic suppression), and allows for a tailored treatment regimen and more personalized patient care.17 Unlike single-gene tests, applied chiefly to cancer types commonly harboring mutations in one gene, the use of NGS-based testing of multiple oncology targets also allows for detection of rare and at times unexpected genetic variation. Moreover, the additive cost of performing multiple single-gene assays quickly exceeds that of multiplex testing but provides less in terms of cumulative information.
The emerging use of NGS approaches in clinical laboratories has brought increased interest in the development of guidelines to ensure that NGS testing to direct patient care is performed to the same rigorous standards as other clinical tests focused on the analysis of nucleic acids, such as DNA sequence analysis by Sanger methodology, DNA copy number analysis by microarray analysis, and detection of chromosome aberrations by interphase FISH. To that end, several organizations have promulgated guidelines for clinical NGS analysis. The College of American Pathologists (CAP) has released a checklist covering NGS.18 Although the checklist addresses both the technical and bioinformatics components of NGS, it is structured as a series of requirements, with little guidance as to how the requirements should be met in routine clinical practice. The Next Generation Sequencing–Standardization of Clinical Testing (Nex-StoCT) working group facilitated by the U.S. Centers for Disease Control and Prevention (CDC) recently provided a much more detailed document covering the validation of clinical NGS tests,19 as has the New York State Department of Health (http://www.wadsworth.org/labcert/TestApproval/forms/NextGenSeq_ONCO_Guidelines.pdf, last accessed November 7, 2013). The recommendations from both groups are comprehensive and cover both the laboratory-based and bioinformatics components of NGS, but again there is little detail as to how the requirements should be met in actual practice. Similarly, the Clinical and Laboratory Standards Institute (CLSI) will soon offer descriptive guidance on the implementation of clinical NGS (projected for the first quarter of 2014 as document MM09-A2, an update of the currently available document on nucleic acid sequencing20), but does not address issues such as validation or quality control (QC) directly. The U.S. Food and Drug Administration (FDA) is likewise aware of the need for increased oversight of NGS as a laboratory-developed test (http://www.fda.gov/MedicalDevices/NewsEvents/WorkshopsConferences/ucm255327.htm, last accessed October 30, 2013); to date, however, the FDA has issued no specific regulatory guidance.
Thus, although several organizations have recognized the need for formalized guidelines under which clinical NGS can be performed, there remains a gap between the general requirements that have been published and detailed and focused information regarding how those requirements should be satisfied in routine practice. It is understandable that clinical NGS laboratories may be struggling with questions of what constitutes an appropriate approach to these requirements, including documentation of the analytical wet-bench process used to generate NGS data and of the bioinformatics used to support the analysis, interpretation, as well as reporting of NGS-based results as required by the various regulatory bodies.18, 19, 20 Different laboratories address the regulatory requirements in various ways, and the discrete validation paradigms pursued by each laboratory need to be disseminated, so that their strengths and weaknesses can be evaluated. To this end, here we report the validation performed at Genomics and Pathology Services at Washington University for an NGS test designed to detect sequence variation within a targeted panel of actionable oncology genes (Figure 1), a validation process designed to meet the regulatory guidelines that have thus far been published.
Figure 1.
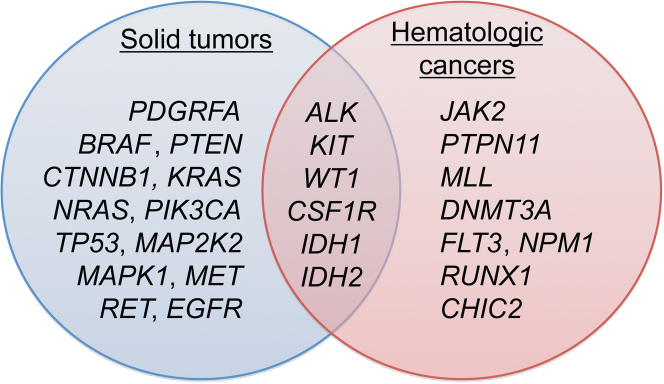
The Washington University Cancer Mutation Profiling (WUCaMP) gene set includes NGS analysis of 25 genes with relevance across multiple tumor types. As supplements to the set, ALK and MLL are assessed by FISH for rearrangements. The choice of genes for the set was based on direct clinical actionability of the target mutations, as determined by consensus between pathologists and oncologists at our institution.
Materials and Methods
WUCaMP Assay Design
The Washington University Cancer Mutation Profiling (WUCaMP) gene set targets oncology genes containing known, clinically important variants. Genes were selected based on the presence of described mutations with an established role for targeted therapy (or that affect response to targeted therapy), an established role in current treatment paradigms, and a record of reimbursement when sequenced individually. A series of meetings with medical oncologists across a range of subspecialties was used to limit the number of genes in the set to those viewed as having the most clinical utility at that time.
Genes
For 25 genes (BRAF, CHIC2, CSF1R, CTNNB1, DNMT3A, EGFR, FLT3, IDH1, IDH2, JAK2, KIT, KRAS, MAP2K2, MAPK1, MET, NPM1, NRAS, PDGFRA, PIK3CA, PTEN, PTPN11, RET, RUNX1, TP53, and WT1), all exons and 200 bp flanking each exon were sequenced by NGS (Figure 1). The capture region (Supplemental Table S1) totals approximately 300 kb. Interphase FISH for ALK and MLL (to detect rearrangements) and for EGFR (to detect amplification) supplements the sequence information derived from the 25 genes. (We note that MLL has been reclassified as KMT2A, but for consistency with current clinical practice here we use the MLL gene symbol.)
Target Selection
Illumina (San Diego, CA) sequencing libraries were enriched for the targeted 25 genes using solution-phase biotinylated cRNA capture reagents (SureSelect; Agilent Technologies, Santa Clara, CA). The design used Agilent’s proprietary algorithm and synthetic process to generate the custom capture reagents for the panel. Targeted regions were based on custom coordinates for annotated exons of the 25 genes from NCBI RefSeq release 45, incorporating 200 bp flanking each exon (to ensure high coverage at exon boundaries) and 1000 bp upstream and downstream of each gene. The resulting capture region comprised approximately 20% coding and 80% noncoding sequence. The UCSC Genome Browser (http://genome.ucsc.edu, last accessed August 23, 2013) mappability data (both ENCODE/CRG alignability and ENCODE/OpenChrom uniqueness) were used during assay design to ensure the uniqueness of the regions selected for capture and to make sure that areas with possible homology to pseudogenes would not prohibit mapping and variant detection. Mappability scores, WUCaMP assay mapping quality data by gene and exon, and location of homologous sequences are presented in Supplemental Tables S2 and S3. PIK3CA exons 10 to 14 exhibited the greatest homology to another genomic location (a pseudogene on chromosome 22; Supplemental Table S3) and showed as high as 33% of reads with a mapping quality score of <50 (Supplemental Table S2). However, these exons achieved high coverage despite potential ambiguous mapping of some paired reads; lack of interference by the homologous region was confirmed by successful mapping and detection of simulated mutations in these exons in silico (data not shown).
Sample Collection and DNA Extraction
The full WUCaMP workflow is diagrammed in Figure 2. DNA was extracted from FFPE and/or fresh-frozen tissues using a QIAamp DNeasy blood and tissue kit (Qiagen, Valencia, CA). DNA was extracted from cell lines from the HapMap Project21 and from blood and bone marrow using a QIAamp DNA blood mini kit (Qiagen). Sample quality was assessed using gel electrophoresis, Qubit fluorometer (Life Technologies, Carlsbad, CA), and NanoDrop spectrophotometer (Thermo Fisher Scientific, Waltham, MA) readings. Requirements for acceptable genomic DNA were as follows: total mass ≥750 ng (by Qubit fluorometry), absorbance ratio A260/A280 at ≥1.7 and ≤2.1, and absorbance ratio A260/A230 at ≥0.7. In our clinical experience with this assay, extracted DNA from approximately 30% of specimens falls below a 750-ng threshold. We have recently demonstrated identical analytical sensitivity and specificity with DNA inputs as low as 200 ng (R.D. Head et al, unpublished data); in our clinical experience, yields fall below a 200-ng threshold for approximately 13% of specimens on which extraction is performed.
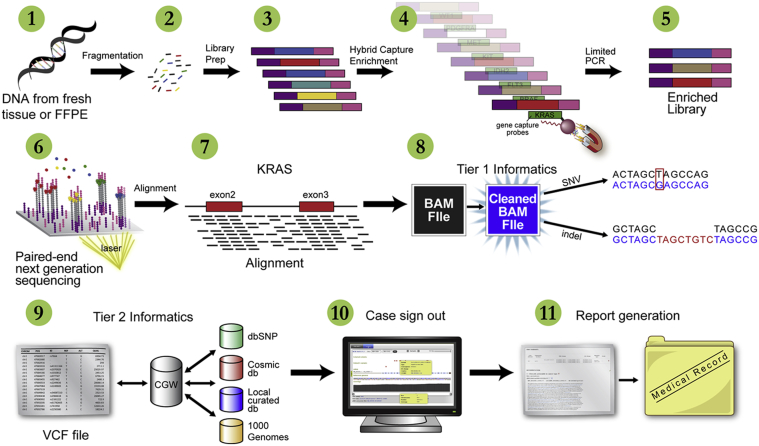
Figure 2.
Schematic view of the WUCaMP assay workflow. DNA is extracted from tumor tissue (1) derived from fresh or FFPE specimens and fragmented by sonication (2). Libraries are prepared and amplified via limited-cycle PCR (3) and enriched for WUCaMP genes by fluid phase hybridization to custom cRNA capture reagents (4). The hybridized product is amplified (5) and sequenced on an Illumina HiSeq 2000 or Illumina MiSeq instrument (6). Paired-end reads are aligned to the genome (7), PCR duplicates are removed (8), and variant calls are made (9). Variants are annotated and classified by our internally developed CGW application, using publicly available and proprietary databases, and the case is reviewed and interpreted by a clinical genomicist for sign-out in CGW (10). A report is then issued to the medical record (11).
Library Preparation and Amplification, Targeted Capture, and Illumina-Based Sequencing
Genomic DNA (750 to 1000 ng) was fragmented using a Covaris S220 series sonicator (Covaris, Woburn, MA) and QC was performed using an Agilent Bioanalyzer 2100 (Agilent Technologies) to ensure an average fragment size of 160 to 230 bp. Fragmentation was followed by end repair, A-tailing, and sequencing adapter ligation (which included a unique nucleic acid barcode, or index) using an Agilent SureSelect library kit. The adapter-ligated DNA was amplified via selective, limited-cycle PCR for a total of seven cycles. Prepared library (500 ng) was hybridized for 24 to 72 hours to WUCaMP custom capture baits (Agilent Technologies). The hybridized product was amplified for 14 PCR cycles using Agilent post-capture primers and a custom indexing primer. QC was performed on the amplified product using an Agilent Bioanalyzer HS chip to ensure that the final library fragment size ranged from 260 to 600 bp; the product was quantified using an Invitrogen Quant-iT dsDNA HS assay (Life Technologies) to ensure a yield of ≥5 μmol/L for sequencing. Exome sequencing was performed using targeted solution-phase enrichment of whole-genome shotgun sequencing libraries with SureSelect Human ALL Exon V3 biotinylated cRNA capture reagents (Agilent Technologies) according to the manufacturer’s instructions.
For paired-end 101-bp sequencing on the Illumina HiSeq 2000 instrument, captured libraries were denatured and loaded onto an Illumina cBot instrument at 12 to 16 pmol/L for cluster generation according to the manufacturer’s instructions. Up to 20 WUCaMP libraries were sequenced per HiSeq lane. A PhiX control (Illumina) was added to lane 8 of each flowcell. For paired-end 150-bp sequencing, captured libraries were denatured and loaded onto an Illumina MiSeq instrument at 8 pmol/L for on-board cluster generation and sequencing according to the manufacturer’s instructions. Up to four WUCaMP libraries were sequenced at a time per MiSeq instrument with version 2 upgrade, 2012. A denatured PhiX control (11 pmol/L) was added to each sample at 1% as a control.
Bioinformatics Pipeline and Data Analysis
For analysis and interpretation, we used the following software packages (all accessed September 6, 2011): Novoalign (version 2.07.11, for alignment to the reference human genome; Novocraft Technologies, Selangor, Malaysia), SAMtools22 (version 0.1.18-1), picard tools (version 1.53, to remove PCR duplicates; http://picard.sourceforge.net), vcftools (version 0.1.6, to merge VCF files; http://vcftools.sourceforge.net), BEDTools23 (to compare the VCF files), the Genome Analysis Toolkit24, 25 [GATK version 1.2, for local realignment and base quality-score recalibration, and to call single-nucleotide variants (SNVs) and small indel variants], Integrative Genomics Viewer26 (IGV version 2.0.16 or later, for visualization), IGV-tools27 (version 1.5.15, to index VCF files), and our internally developed Clinical Genomicist Workstation application (CGW version 1.0, for visualization and interpretation). Software parameters and commands are available on request.
All analyses were based on the human reference sequence UCSC build hg19 (NCBI build 37.2). SNVs with a unique coverage depth of <50× or a strand bias of >100 by Fisher’s exact test were excluded from analyses. The coverage-depth threshold of 50× was determined empirically to minimize false-positive calls while maintaining high sensitivity for the range of DNA input amounts expected in the assay. Variants were reported according to Human Genome Variation Society nomenclature (http://www.hgvs.org/mutnomen, last accessed September 6, 2011) and were classified into eight categories, based on clinical actionability and previously reported data in the literature. Variations found in dbSNP (version 132; http://www.ncbi.nlm.nih.gov/projects/SNP) that have >5% minor allele frequency in at least one population or that were reported by the 1000 Genomes Project28 (http://www.1000genomes.org) from the initial discovery phase, all pilot phases, or the production phase were classified as known polymorphisms. Database versions current as of September 6, 2011 were used for the launch of the assay.
Additional data analysis was performed using Microsoft Excel 2010 software and the split–apply–combine strategy29 and R.30 Confidence intervals were calculated online (http://www.vassarstats.net/clin1.html, last accessed October 4, 2013). Graphing was performed using R software version 2.15.1 and ggplot2 for R.31 Sanger sequence data were visualized and analysis was aided using Mutation Surveyor software (SoftGenetics, State College, PA).
Validation Specimens and Assessment of Analytical Performance Characteristics
For validation of the WUCaMP assay, we tested 78 tumor specimens, including 67 solid tumors derived from formalin fixed, paraffin embedded (FFPE) samples and 11 hematologic malignancy (acute myeloid leukemia) specimens derived from involved blood or bone marrow, and three HapMap cell-line DNA samples. Analyses were performed in a masked fashion for 15 of the 78 tumor specimens, as described below. Extracted DNA was prepared and sequenced multiple times for some specimens, to assess reproducibility (38 specimens were sequenced twice and 1 specimen was sequenced four times), yielding a total of 119 tumor specimen sequencing data sets from solid-tumor and hematologic specimens. For 16 of the FFPE specimens, matched fresh-frozen tissue was also tested for comparison. In addition to the 78 validation tumor specimens, after launch of the assay 51 routine clinical NGS specimens were tested in parallel by Sanger sequencing in a comparative analysis (described below).
Quality and depth-of-coverage metrics were measured across all 119 tumor specimen data sets, to establish an acceptable reference range for key measures (including percentage of reads mapped to the reference sequence, percentage of reads mapped to the target region, and number of unique on-target reads).
Analytical performance characteristics were assessed using HapMap and clinical specimens. For accuracy and to calculate analytic sensitivity and specificity, variant calls made by our pipeline for HapMap specimen NA19240 were compared with publicly available data on that specimen from two orthogonal platforms, the Illumina Omni 2.5M microarray (ftp://ftp.1000genomes.ebi.ac.uk/vol1/ftp/technical/working/20101206_hapmap_omni_results)32 and whole-genome sequence data from Complete Genomics (ftp://ftp2.completegenomics.com/YRI_trio/ASM_Build37_2.0.0/NA19240).33 To assess precision, including repeatability and reproducibility of the assay, we measured concordance between repeat runs, between different lanes of the Illumina HiSeq instrument, between instruments, and between samples prepared by different technicians.
Accuracy was also assessed using parallel Sanger testing in routine clinical NGS samples. PCR amplicons (368 primer pairs, synthesized by Integrated DNA Technologies, Coralville, IA; sequences are available on request) were developed to span the entire coding region, nearly 60 kb, of the WUCaMP gene set (with the exception of exon 1 of WT1, for technical reasons). The targeted amplicon size was 250 to 500 bp, and exons larger than the targeted amplicon size were covered by multiple amplicons with 50- to 100-bp overlap, to ensure complete high-quality coverage. Small exons separated by short intronic regions were covered by a single amplicon if possible. All primers were ordered with the appropriate sense and antisense M13 tail sequences, to ensure full-length Sanger reads; when used to verify NGS results, all amplicons were sequenced with bidirectional coverage or in duplicate with unidirectional coverage.
The limit of detection of the assay was assessed using preanalytic and in silico mixtures of two HapMap samples, NA18507 and NA19129, to validate the detection of variants at low variant allele fractions (VAFs) and coverage depths. The two samples were first sequenced individually to establish a gold-standard set of variants using two established variant calling tools, SAMtools,22 and GATK.24, 25 For NA18507 and NA19129, respectively, 277 and 305 calls were concordant between the two variant callers and present at a unique coverage depth of >50×, including 33 and 36 variants, respectively, in coding regions. Positions with low coverage (<50×) and positions at which the two variant callers were discordant were masked during analysis of HapMap mixes, to avoid including true variants missed during the gold-standard analysis.
Sensitivity for variants at low allele fraction (AF) was first assessed using two preanalytical mixed samples of NA18507 and NA19129. DNA from each sample was mixed to achieve proportions of 50% and 20% of NA19129 by mass, and 1000 ng of the mix was used for library generation and sequencing, as described above, to achieve a mean unique coverage of approximately 600×. Sensitivity was assessed for NA18507 variants in the 50% mix and for NA19129 variants in the 20% mix, both across the entire target region and in coding regions and adjacent splice sites only. False-positive calls were defined as those variants identified in the mix that were not included in the gold-standard set for either sample (after excluding positions with either discordant calls or coverage of <50× in the gold-standard set).
Low VAF detection was also assessed as a function of coverage, using a set of mixed samples derived in silico from NA18507 and NA19129. Mapped reads from the individual data sets were sampled and combined to generate synthetic data sets with 1000× unique coverage and mix proportions of 50%, 20%, 10%, and 2% of NA18507, with NA19129 for the remaining proportion in each; these mixed samples were expected to have VAFs of 25%, 10%, 5%, and 1% at positions with heterozygous variants unique to NA18507. The mixed data sets were also downsampled at random to achieve mean coverages of 750×, 500×, and 250×, for a total of 16 mixed data sets with a range of mix proportion and coverage levels.
To demonstrate the ability of the assay to detect clinically relevant somatic variation, we sequenced masked clinical specimens previously analyzed by Sanger sequencing or fragment analysis in Clinical Laboratory Improvement Amendments (CLIA)–certified, CAP-accredited clinical diagnostic laboratories. Identified variants were reported according to HGVS nomenclature, using genomic, coding DNA, and protein reference sequences; read alignments and variant call information were also reviewed in the Integrative Genomics Viewer26, 27 to visually confirm the presence of the variant identified by CGW.
Multiplexed Sequencing Validation
We assessed the frequency of contamination that occurs during library preparation by preparing a library from each of two human genomic DNA samples side by side, each with a unique index. Each indexed library was sequenced individually on three separate lanes. The frequency at which a valid index other than the index used for the sample was identified in the sequenced reads was calculated. Reads with an invalid index (ie, reads with an index sequence that was not matched to any of the 96 indices used in our laboratory) were discarded from the analysis. To assess for possible crossover between indexed samples on the instrument during multiplex sequencing, libraries from HapMap genomic DNA samples NA18507 and NA19240 were generated side by side, enriched for exome targets as described above, and pooled in the same lane for multiplex sequencing on a HiSeq 2000 instrument. All positions in the targeted regions of our panel with unique homozygous nonreference genotypes in NA19240 relative to NA18507 were identified (n = 15), and the base counts at those positions in sequence data from NA18507 were generated. The converse analysis was also performed, querying positions in the data from NA19240 for which NA18507 had unique homozygous nonreference genotypes (n = 11). A similar analysis was performed for whole-exome data.
QC
Data collected during the assay validation phase were applied toward the development of QC metrics for use in routine clinical specimen testing. These data were derived from the 78 tumor specimens described above, which were considered representative of the type to be encountered in our standard workflow. A QC report consists of three parts.
Level 1. Specimen-level sequencing metrics are parameters indicating overall sequencing quality and coverage for the specimen, including total reads, percent mapped, number of on-target reads, percentage of reads on target, percentage of unique on-target reads (ie, not potential PCR duplicates), and percentage of targeted positions with ≥50×, ≥400×, and ≥1000× unique coverage.
Level 2. The exon-level coverage metric comprises a list of exons in the capture region that did not achieve 50× unique coverage at 95% or more of positions (including gene name and exon number). Low coverage of such exons is declared in the clinical report.
Level 3. The clinically actionable variants metric comprises a list of curated nucleotide positions with known clinically actionable variants that did not achieve 50× unique coverage.
Results
Overview
WUCaMP is a high-coverage, NGS-based clinical test for detection of somatic mutations in cancer. DNA is extracted from tumors, fragmented, amplified via limited-cycle PCR, and subjected to solution-phase enrichment of the exons and flanking intronic sequence of 25 target genes (totaling ∼300 kb), using custom-designed biotinylated cRNA capture reagents (SureSelect; Agilent Technologies). Paired-end, 101-bp or 150-bp sequencing is performed on an Illumina HiSeq 2000 or an Illumina MiSeq instrument, respectively. In routine practice, data are analyzed to generate an automated draft clinical report, which is edited and finalized by a Washington University clinical genomicist and returned to the ordering physician. Interphase FISH for ALK, MLL, and EGFR accompanies the NGS analysis for detection of gene rearrangements and copy number variation at these loci.
Selection of tumor tissue for analysis by a Board-certified anatomical pathologist and great depth of coverage are integral to the WUCaMP assay. Detection of somatic variants in cancer is most straightforward when only tumor cell DNA is sampled, such as in acute leukemias with high blast percentages. In solid tumors, however, the malignant cells are always surrounded by supporting stromal cells, inflammatory cells, and benign parenchymal tissue elements, resulting in the generation of sequence derived from both tumor and nontumor DNA that includes both constitutional and somatic variants. For the WUCaMP assay, a pathologist reviews H&E-stained slides cut from tumor specimens and marks the areas of greatest tumor cellularity and viability, to minimize contamination from nontumor cell DNA. Even when areas of very high tumor cellularity are sampled, tumor heterogeneity can result in a range of observed AFs. The deep coverage, averaging approximately 1018× unique coverage across the WUCaMP assay capture region for 78 validation tumor specimens (Figure 3), allows detection of variants at low AF resulting from low tumor cellularity or tumor heterogeneity, as described below.
Figure 3.
Distribution of unique coverage depth across the full WUCaMP capture region. The percentage of targeted WUCaMP positions (including both coding and flanking intronic sequence) that achieve unique coverage depth on the HiSeq instrument greater than or equal to that shown on the x axis is plotted. Rectangles (dashed lines) indicate the unique coverage depth achieved at 95% of positions and at 50% of positions (median unique coverage). On the y-axis scale, 1.0 indicates 100%.
Analysis and interpretation of sequence data and variants detected in the 25 WUCaMP genes are managed by CGW, our custom Web-based application. The CGW application manages order intake, including case number, patient and specimen details, and nucleic acid index; facilitates the bioinformatics analyses for base calling, read mapping, and variant calling using various tools and filters; incorporates genomic annotation, including dbSNP and the Catalogue Of Somatic Mutations In Cancer (COSMIC) database (http://cancer.sanger.ac.uk/cancergenome/projects/cosmic, last accessed October 30, 2013); applies custom clinical-grade annotations curated by Genomics and Pathology Services; provisionally classifies each variant into one of the eight variant levels; and inserts appropriate curated interpretations into a draft clinical report in the context of the constellation of variants and clinical indication. A clinical genomicist reviews specimen-level and exon-level QC data, inspects variants and position-level coverage data in the context of annotation resources using the CGW gene viewer, assesses the medical relevance of the identified variants with assistance from the available curated interpretations, and edits and finalizes the clinical report for sign-out.
To validate the WUCaMP assay, we assessed analytical performance characteristics on 81 unique validation samples (78 tumor specimens and 3 HapMap21 DNA cell lines) and performed a complete clinical validation for the detection of SNVs.
Establishment of Quality Metrics and Reportable Range
We measured quality and depth-of-coverage metrics across all clinical validation specimen data sets to establish acceptable run-level QC parameters, including the percentage of reads mapped to the reference sequence, the percentage of mapped reads that mapped specifically to the target region, and the number of unique on-target reads (Table 1). On the Illumina HiSeq instrument, the average total number of reads across 99 FFPE validation tumor specimen data sets was 13.9 million (SD = 4.8 million). An average of 98.6% (SD = 1.5%) of the bases were mapped, and an average of 42.7% (SD = 11.8%) of the mapped reads were on-target (ie, within the ∼300-kb capture region). On average, validation specimens yielded 2.9 million unique on-target reads (SD = 1.0 million), representing approximately 21% of the average total reads per case.
Table 1.
Average Quality and Depth-of-Coverage Metrics across Validation Samples on Illumina Platforms
| Instrument | Sample type and number | Total reads (no.) | Mapped (%) | On target (%) | On-target reads (no.) | On target, unique (%) | On-target reads, unique (no.) |
|---|---|---|---|---|---|---|---|
| HiSeq | FFPE (n = 99) | 13.9 × 106 ± 4.8 × 106 | 98.6 ± 1.5 | 42.7 ± 11.8 | 6.0 × 106 ± 3.0 × 106 | 56.2 ± 20.3 | 2.9 × 106 ± 1.0 × 106 |
| HiSeq | Fresh frozen and cell line (n = 64) | 13.5 × 106 ± 4.5 × 106 | 99.1 ± 0.5 | 42.6 ± 8.6 | 5.77 × 106 ± 2.6 × 106 | 67.7 ± 11.9 | 3.7 × 106 ± 1.3 × 106 |
| MiSeq | FFPE (n = 4) | 6.1 × 106 ± 0.2 × 106 | 96.3 ± 1.0 | 47.6 ± 7.6 | 2.8 × 106 ± 0.5 × 106 | 80.5 ± 2.6 | 2.3 × 106 ± 0.4 × 106 |
Data are expressed as means ± SD.
The read and coverage statistics were similar for sequencing runs of libraries prepared from fresh-frozen tissue specimens, blood or bone marrow specimens, or cell lines. Similar numbers of on-target reads were achieved from formalin-fixed and unfixed sources of DNA. These data indicate that formalin-fixed tissues are suitable for NGS-based analyses, as documented elsewhere.34
The percentage of base positions at which high-quality unique on-target reads were achieved at depths of 50×, 400×, and 1000× was calculated (Table 2). For an average FFPE specimen sequenced on the HiSeq instrument, 96.9% (SD = 1.3%) of positions were covered at a depth of ≥50×, 82.6% (SD = 12.2%) at a depth of ≥400×, and 46.2% (SD = 23.0%) at a depth of ≥1000×. With the MiSeq instrument, distribution of coverage was nearly identical to that of the HiSeq.
Table 2.
Percent of Capture Region That Met Unique On-Target Coverage Depth Thresholds on Illumina Platforms
| Instrument | Sample type and number | Capture region that met threshold (%) |
||
|---|---|---|---|---|
| ≥50× | ≥400× | ≥1000× | ||
| HiSeq | FFPE (n = 99) | 96.9 ± 1.3 | 82.6 ± 12.2 | 46.2 ± 23.0 |
| HiSeq | Fresh frozen and cell line (n = 64) | 97.4 ± 0.6 | 86.7 ± 3.5 | 52.4 ± 24.3 |
| MiSeq | FFPE (n = 4) | 95.1 ± 0.4 | 82.5 ± 1.2 | 48.4 ± 1.3 |
Quality metric data obtained from the assay validation phase were used to establish a QC report for use in the standard workflow for clinical specimen testing. This allowed for an evaluation of the data quality generated from each clinical sample in comparison with the aggregate validation data metrics. Indicators including specimen-level sequencing metrics, exon-level coverage metrics, and coverage at clinically actionable nucleotide positions were assessed for each clinical sample. If the quality of a given clinical sample was deemed inadequate (because of low coverage), the assay was repeated to improve this outcome.
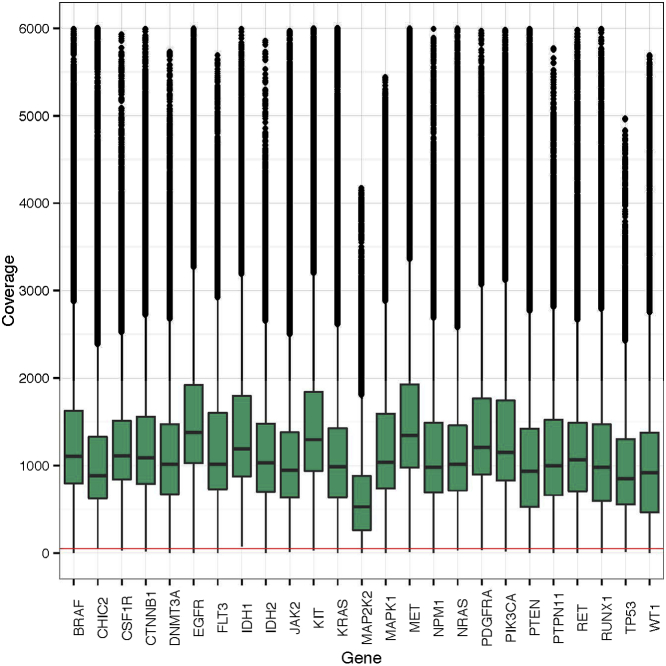
To determine the reportable range, defined as the region of the genome in which sequence of an acceptable quality can be derived, average depth of unique coverage for each base position in each exon of the 25 WUCaMP genes was determined for all validation tumor specimen data sets (n = 119). High coverage on a gene-by-gene (Figure 4) and an exon-by-exon (Figure 5) basis was present throughout the 25-gene capture region. A limited number of exons (largely the GC-rich first exon of several genes; Supplemental Tables S2 and S3) did not achieve 50× unique coverage at 95% of exonic base-pair positions across validation samples. Those regions included exon 1 in BRAF, EGFR, FLT3, IDH2, KRAS, MAP2K2, MAPK1, MLL, PIK3CA, PTPN11, RET, and WT1, and also exons 2 and 8 in the DNMT3A gene, representing 14 of the 393 exons (3.6%) contained in our target capture region.
Figure 4.
Distribution of unique coverage depth across the 25 genes in the WUCaMP panel. Unique coverage data across 119 validation tumor specimen data sets from HiSeq sequencing are plotted by gene. Each box represents the interquartile range, with the midline as the median unique coverage; whiskers represent exon coverage for a given sequencing run within 2 SD of the median. Outlier exons for a sequencing run are plotted as independent dots.
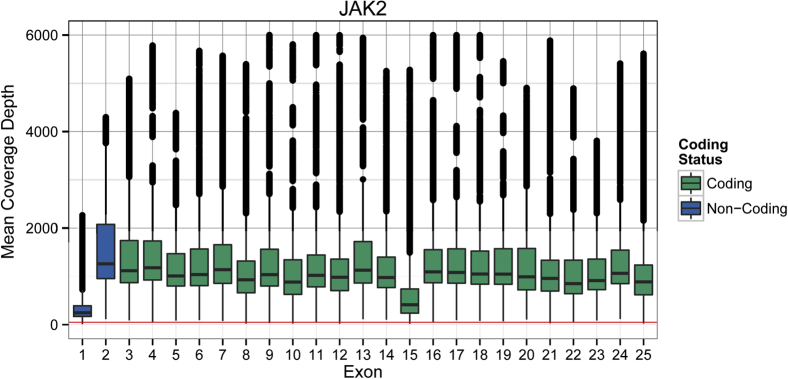
Figure 5.
Distribution of unique coverage depth across exons in JAK2, one gene of the WUCaMP panel. Unique coverage data across 119 validation tumor specimen data sets from HiSeq sequencing are plotted by exon. Box–whisker plots are defined as for Figure 4, except that unique coverage level is considered by position rather than averaged across an exon. The red horizontal line near the x axis indicates 50× unique coverage. JAK2 coverage was slightly below average, relative to other WUCaMP genes (data not shown).
Accuracy and Analytical Sensitivity and Specificity
Demonstrating accuracy across the large number of clinically important positions sequenced in the approximately 300-kb WUCaMP capture region was not feasible using clinical cancer specimens. Unlike those of clinical specimens, the genomes of cell lines from the HapMap Project have been thoroughly characterized by a number of genotyping and sequencing methods, providing ample reference data to validate SNV detection over a range of coverages and sequence contexts. SNV calls made by our pipeline for HapMap cell line NA19240 were therefore compared with publicly available data on NA19240 from two orthogonal platforms, the Illumina Omni 2.5M microarray (Table 3) and whole-genome sequence data from Complete Genomics (Table 4), the latter of which allowed for comparison of the majority of sites in our capture region.
Table 3.
Minimum SNV Analytic Sensitivity, Specificity, and Positive Predictive Value Calculated for WUCaMP Illumina Platforms versus Positions Genotyped by the Illumina Omni 2.5M Array
| Instrument | Region assessed | Sensitivity (%) [TP/(TP+FN)] | 95% CI | Specificity (%) [TN/(TN+FP)] | 95% CI | PPV (%)[TP/(TP+FP)] | 95% CI |
|---|---|---|---|---|---|---|---|
| HiSeq | Entire capture region | 95.9 (118/123) | 90.3–98.5 | 99.6 (273/274) | 97.7–99.9 | 99.2 (118/119) | 94.7–99.9 |
| Coding region only | 100.0 (25/25) | 83.4–100.0 | 100.0 (79/79) | 94.2–100.0 | 100.0 (25/25) | 83.4–100.0 | |
| MiSeq | Entire capture region | 95.9 (118/123) | 90.3–98.5 | 100.0 (274/274) | 98.3–100.0 | 100.0 (118/118) | 96.1–100.0 |
| Coding region only | 100.0 (25/25) | 83.4–100.0 | 100.0 (79/79) | 94.2–100.0 | 100.0 (25/25) | 83.4–100.0 |
FN, false negative; FP, false positive; PPV, positive predictive value; TN, true negative; TP, true positive.
Table 4.
Minimum SNV Analytic Sensitivity, Specificity, and Positive Predictive Value of WUCaMP on Illumina Platforms versus Complete Genomics Whole Genome Sequence
| Instrument | Region assessed [size (bp)] | Sensitivity (%) [TP/(TP+FN)] | 95% CI | Specificity (%) [TN/(TN+FP)] | 95% CI | PPV (%)[TP/(TP+FP)] | 95% CI |
|---|---|---|---|---|---|---|---|
| HiSeq | Entire capture region (306,336) | 98.3 (297/302) | 95.9–99.4 | 100.0 (288,262/288,276) | 99.9–100.0 | 95.5 (299/313) | 92.4–97.4 |
| HiSeq | Coding region only (59,490) | 100.0 (40/40) | 89.1–100.0 | 100.0 (57,645/57,645) | 99.9–100.0 | 100.0 (40/40) | 89.1–100.0 |
| MiSeq | Entire capture region (306,336) | 98.3 (297/302) | 95.9–99.4 | 100.0 (288,575/288,580) | 99.9–100.0 | 98.3 (297/302) | 95.9–99.4 |
| MiSeq | Coding region only (59,490) | 100.0 (40/40) | 89.1–100.0 | 100.0 (57,700/57,700) | 99.9–100.0 | 100.0 (40/40) | 89.1–100.0 |
Unfiltered coding region variant calls from the WUCaMP pipeline are presented in Supplemental Table S4. Reference base calls and homozygous and heterozygous nonreference SNV calls were extracted from the Omni 2.5M microarray and Complete Genomics data on NA19240 and were compared with SNV calls made by our WUCaMP pipeline for eight independent NA19240 library preparations sequenced across three separate HiSeq runs. Minimum sensitivity, specificity, and positive predictive value (PPV) calculated across these eight preparations from the Omni 2.5M and Complete Genomics comparisons are reported in Tables 3 and 4, respectively. For the latter comparison, a total of 288,955 sites were compared, 302 of which were called nonreference (homozygous or heterozygous) by Complete Genomics. The sensitivity of WUCaMP across our capture region, as assessed using Complete Genomics as the gold standard, was ≥98.3% [95% confidence interval (95% CI) = 95.9–99.4] across runs, and the specificity was 100.0% (95% CI = 99.9–100.0). The PPV across the capture region ranged from 95.5% (95% CI = 92.4–97.4) to 97.7% (95% CI = 95.1–99.0) across runs.
Because only approximately 20% of our capture region includes coding sequence, which generally has higher coverage (because of greater sequence complexity and more complete tiling with capture baits) and contains the vast majority of the clinically relevant loci, we also assessed these metrics limited to the coding region sequence. This analysis showed coding region sensitivity, specificity, and PPV were 100% (95% CI = 89.1–100.0 for sensitivity and PPV and 95% CI = 99.9–100.0 for specificity) when using Complete Genomics as the gold standard, with >57,600 sites compared (Table 4).
To extend the assessment of the accuracy of the assay to routine clinical samples, a concurrent testing strategy was used for 51 WUCaMP cases. Coding regions containing hotspots for mutations associated with clinical outcomes in certain cancers were PCR-amplified and sequenced by the Sanger method. This analysis identified 56 coding variants, including germline polymorphisms and somatic variants (Table 5). All 56 coding variants were also identified by NGS in the WUCaMP assay, indicating that the WUCaMP assay is at least as sensitive as Sanger sequencing. Of the variant calls made by NGS, seven were not made by Sanger sequencing. Of these, no result was obtained for six variant positions, because of amplification failure (likely attributable to degraded DNA derived from older FFPE tissues); the seventh was likely below the limit of detection of Sanger sequencing (a SNV at an estimated AF of 20%).
Table 5.
Sanger Correlation Results
| Sanger panel | Genes and exons covered | Cases tested (no.) | Codingvariants identified by Sanger (no.) | Sanger coding variants supported by NGS (no.) |
|---|---|---|---|---|
| Breast |
KIT exon 11 PIK3CA exons 10, 21 TP53 exons 5, 6, 7, 8 |
2 | 1 | 1 |
| Colon/gastrointestinal |
BRAF exon 15 CTNNB1 exon 3 KIT exon 11 KRAS exons 2, 3 PIK3CA exons 10, 21 TP53 exons 5, 6, 7 |
2 | 0 | 0 |
| Lung |
BRAF exon 15 EGFR exons 18, 19, 20, 21 KIT exon 11 KRAS exons 2, 3 PIK3CA exons 10, 21 MET exon 14 |
36 | 43 | 43 |
| Cholangiocarcinoma |
BRAF exon 15 IDH1 exon 4 IDH2 exon 4 KIT exon 11 KRAS exon 2 PIK3CA exon 10 TP53 exons 5, 6, 7, 8 |
1 | 0 | 0 |
| Hematologic |
BRAF exon 15 IDH1 exon 4 IDH2 exon 4 JAK2 exon 14 KIT exons 8, 11, 17 PIK3CA exon 10 TP53 exons 5, 6, 7, 8 |
2 | 2 | 2 |
| Pancreatic |
BRAF exon 15 CTNNB1 exon 3 KIT exon 11 KRAS exon 2 PIK3CA exons 5, 21 TP53 exons 4, 5, 6, 7 |
9 | 10 | 10 |
| Total | 52∗ | 56 | 56 |
Of the 51 cases, 1 was tested for two Sanger panels.
Precision and Reproducibility
To assess precision, including repeatability and reproducibility of the assay, HapMap cell line NA19240 was subjected to sequencing and analyses as described under Materials and Methods on repeat runs that differed in the technician performing library preparation, HiSeq lane number, instrument, or instrument run (Table 6). We calculated average coding region variant-call concordance rates of 100.0 ± 0.0% between samples prepared by different technicians, and 98.7 ± 1.2% between samples prepared by the same technician on different days. Inter-run reproducibility, in which the same library preparation was sequenced on two separate runs of the same sequencing instrument, yielded a coding region variant-call concordance of 98.8 ± 1.6%. Coding region concordance was 99.2 ± 1.2% when a single library preparation was run on two different instruments. Concordance in the coding regions fell short of 100% in some comparisons because of a single position that was not covered at the 50× depth required to make a call in our pipeline in one of the sequenced specimens in the pairwise comparison. This position was identified in dbSNP as rs12872889, in the coding portion of exon 1 of the FLT3 gene.
Table 6.
Reproducibility and Repeatability of the WUCaMP Assay on the Illumina HiSeq System
| Comparison | Capture region |
Coding region |
Comparisons (no.) | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Variants |
Concordance |
Variants |
Concordance |
||||||
| Mean no. | SD | % | SD | Mean no. | SD | % | SD | ||
| Intratechnician repeatability∗ | 353 | 1.4 | 96.6 | 1.3 | 43 | 0.5 | 98.7 | 1.2 | 9 |
| Intertechnician reproducibility† | 353 | 1.4 | 97.3 | 0.4 | 43 | 0.5 | 100.0 | 0.0 | 4 |
| Interlane reproducibility‡ | 357 | 6.3 | 95.1 | 1.9 | 43 | 0.5 | 98.8 | 1.6 | 2 |
| Inter-run reproducibility§ | 352 | 0.0 | 97.2 | 1.6 | 43 | 0.5 | 98.8 | 1.6 | 2 |
| Interinstrument reproducibility¶ | 346 | 3.7 | 96.9 | 1.1 | 43 | 0.4 | 99.2 | 1.2 | 6 |
The same technician performed multiple library preparations from the same specimen on different days; HiSeq lane also varied.
Different technicians performed library preparations on the same specimen, which were sequenced together in the same lane.
The same library preparation was sequenced in two different lanes on the same HiSeq run.
The same library preparation was sequenced on two separate HiSeq runs.
The same library preparation was sequenced on two different instruments.
Limit of Detection
The sensitivity for detecting variants at low AF that occur because of tumor heterogeneity or stromal contamination was assessed using preanalytic and in silico mixtures of two HapMap samples. DNAs from HapMap samples NA18507 and NA19129 were mixed at 1:1 and 4:1 ratios and were sequenced as described under Materials and Methods. Variants in each sample that were heterozygous and unique to that sample relative to the other (as described under Materials and Methods) were used to determine sensitivity, because these positions were expected to have VAFs of 25% and 10%, respectively, for the 50% and 20% mixes.
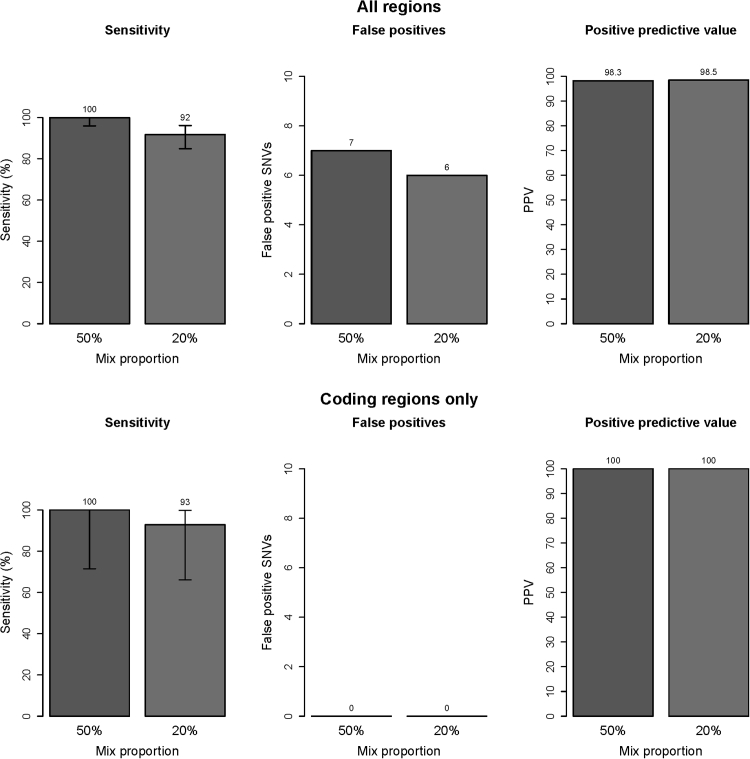
Review of the VAFs at heterozygous positions unique to NA18507 (n = 95, of which 11 were coding) revealed the mean observed VAF to be 23.9% in the 50% mix and the mean VAF of NA19129-specific variants to be 13.1% in the 20% mix, demonstrating good agreement with the expected values. For the 50% mix, the sensitivity of detection for all NA18507 heterozygous unique variants meeting the validated quality criteria was 100%, both across the full capture region (n = 90; 95% CI = 94.9–100.0) and in coding regions (n = 11; 95% CI = 67.9–100.0) (Figure 6). For the 20% mix, the sensitivity of detection for all heterozygous NA19129 variants meeting the quality criteria (n = 109, including 14 coding; 13.1% mean observed VAF) was 92% (95% CI = 84.5–95.9) and 93% (95% CI = 64.2–99.6), respectively, across the capture region and in coding regions (Figure 6). Considering only variants with observed VAFs of ≥10% (n = 67), the sensitivity was 100% (95% CI = 93.2–100.0). Seven and six false-positive calls were detected in the 50% and 20% NA19129 mixes, respectively, yielding PPVs of 98.3% (95% CI = 96.3–99.2) and 98.5% (95% CI = 96.5–99.3) across the full capture region. No false-positive calls were detected in the analysis of coding regions in either mix for PPVs of 100% (95% CI = 90.6–100.0 for the 50% mix and 95% CI = 90.4–100.0 for the 20% mix) (Figure 6).
Figure 6.
Low VAF detection. For all target regions (top row) and for coding regions only (bottom row), sensitivity, false positives, and PPV are presented for one sample with a 50% mix proportion and a second sample with a 20% mix proportion. Error bars indicate the 95% binomial confidence interval for each point estimate. Top row: n = 109 variants (50% mix); n = 95 variants (20% mix). Bottom row: n = 11 variants (50% mix); n = 14 variants (20% mix). PPV = TP/(TP+FP).
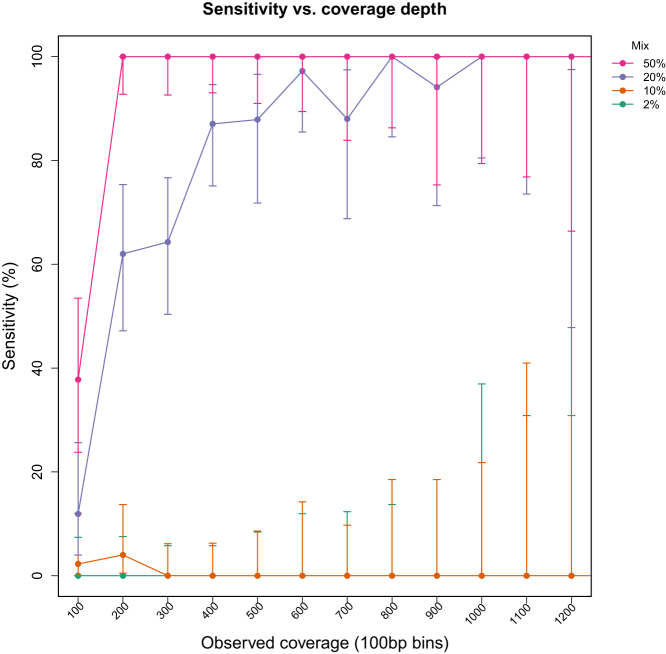
The sensitivity for detecting variants at low AF was also assessed as a function of coverage depth using in silico mixing of NA18507 and NA19129 sequencing reads and random downsampling, as described under Materials and Methods. Synthetic mixed samples were generated from mapped read data from NA18507 and NA19129 in proportions of 50%, 20%, 10%, and 2%, which resulted in mean observed VAFs of 23.8%, 10.6%, 5.8%, and 1.1% for NA18507 variants that were heterozygous and unique (n = 95). The performance of the variant calling pipeline for each mix, with variants binned into groups based on unique coverage levels (0 to 100×, 101 to 200×, and so on) at the variant position, is presented in Figure 7. Sensitivity was calculated based on the proportion of variants in a given mix and coverage bin that were detected. For variants with 25% AF (50% mix), the sensitivity was 38% (95% CI = 23.8–53.5) and 100% (95% CI = 92.7–100.0), respectively, at 100× and 200× unique coverage. For variants with 10% AF (20% mix), the sensitivity was 16% (95% CI = 4.0–25.6) at 100×, reaching approximately 90% (95% CI = 75.1–96.6) at 400× and 100% (95% CI = 79.4–100.0) at 1000× unique coverage (Figure 7). Sensitivity for variants with 5% and 1% AF (10% and 2% mixes, respectively) was essentially 0% at all coverage levels.
Figure 7.
Sensitivity for low VAF detection as a function of coverage depth. Synthetic mixed samples were generated from two individual HapMap samples in silico, with mix proportions of 50%, 20%, 10%, and 2% and mean coverage levels across the entire target region of 1000×, 750×, 500×, and 250×. Each mixed sample had 95 heterozygous variants unique to the minor sample present at mean observed VAFs of 23.8%, 10.6%, 5.8%, and 1.1%, respectively. Data indicate the sensitivity (percent detected) for variants with observed coverage in bins of 100. Error bars indicate the 95% binomial confidence interval for each point estimate.
Multiplexed Sequencing Validation
Because combining multiple samples in a single sequencing run introduces the possibility of cross-sample contamination, we examined the fidelity of indexed library sequencing by determining the frequency of spurious indices (ie, index sequences other than those expected) after multiplex sequencing. We first assessed the frequency of contamination that occurs during library preparation by preparing libraries from two human genomic DNA samples side by side and sequencing in separate lanes, as described under Materials and Methods. A total of 95 and 94 spurious indices were identified in samples 1 and 2, respectively, and the frequency of reads with the most commonly encountered spurious index was 0.0074% and 0.1822% in samples 1 and 2, respectively (Table 7). The most frequent spurious index in sample 1 was the index used for library preparation for sample 2 and vice versa, reflecting contamination during library preparation, most likely due to aerosol formation. However, the frequency of detection of spurious indices in this experiment did not exceed 0.2% of the reads, which is far below the limit of detection of the assay.
Table 7.
Evaluation of Index Contamination during Library Preparation
| Lane | Sample 1 |
Sample 2 |
||||
|---|---|---|---|---|---|---|
| Frequency of correct index (%) | Frequency of most frequent spurious index (%) | Spurious indices detected (no.) | Frequency of correct index (%) | Frequency of most frequent spurious index (%) | Spurious indices detected (no.) | |
| 1 | 99.994 | 0.0033 | 93 | 99.808 | 0.1822 | 94 |
| 2 | 99.994 | 0.0036 | 94 | 99.809 | 0.1817 | 94 |
| 3 | 99.985 | 0.0074 | 95 | 99.809 | 0.1808 | 91 |
In addition, to investigate crossover between indexed samples on the instrument during multiplex sequencing, we assessed base calls in multiplexed HapMap samples at positions with unique homozygous nonreference genotypes in one sample relative to the other, as described under Materials and Methods. Because these positions are homozygous for the reference allele in one HapMap sample and homozygous nonreference in the other, all observed nonreference bases in the index with the homozygous reference genotype represent either sequencing errors or contamination from the other HapMap sample. The analysis of positions homozygous for the reference allele in NA19240 resulted in 0/2726 (0%) reads that matched NA18507 (Table 8). In the converse analysis, 1/1586 reads (0.06%) matched the other sample. An extended analysis across the entire exome yielded a 0.03% rate of potential sample crossover at homozygous reference positions in both HapMap samples. This resulted in an estimated on-instrument crosstalk frequency of 0.03% to 0.06%. The total error rate, as estimated by fraction of any nonreference bases called at such positions, was 0.04% to 0.06% across the WUCaMP capture region and 0.3% to 0.4% across the exome.
Table 8.
Evaluation of Index Cross-Talk between Two HapMap Samples, by Region Analyzed
| Metric | Positions homozygous reference in NA19240 and homozygous nonreference in NA18507 | Positions homozygous reference in NA18507 and homozygous nonreference in NA19240 |
|---|---|---|
| WUCaMP target region | ||
| Total positions (no.) | 15 | 11 |
| Nonreference calls matching other HapMap [n/N (%)] | 0/2726 (0) | 1/1587 (0.06) |
| Other nonreference calls [n/N (%)] | 1/2726 (0.04) | 0/1587 (0) |
| Fraction of any nonreference bases to total bases (%) | 0.04 | 0.06 |
| Whole exome | ||
| Total positions (no.) | 8866 | 8769 |
| Nonreference calls matching other HapMap [n/N (%)] | 549/1,683,767 (0.03) | 462/1,627,991 (0.03) |
| Other nonreference calls [n/N (%)] | 4462/1,683,767 (0.27) | 6161/1,627,991 (0.38) |
| Fraction of any nonreference bases to total bases (%) | 0.30 | 0.41 |
Detection of Pathogenic Variants in Known Clinical Standards
The sensitivity of the WUCaMP assay to detect pathogenic tumor mutations was demonstrated by assaying 15 masked clinical specimens known to harbor one or more pathogenic SNVs in the capture region, based on prior Sanger sequencing or other genotyping assay performed in a CLIA-licensed clinical laboratory. All SNVs (13/13 on the HiSeq instrument and 4/4 on the MiSeq) were detected using our pipeline when the AF of these variants was ≥15% (Table 9). Three SNVs present at <5% AF that were detected by FLT3 tyrosine kinase domain genotyping were not detected by our pipeline (as expected, based on our reported sensitivity of <10% VAF).
Table 9.
Confirmation of Known Clinically Important Mutations by WUCaMP on Illumina Sequencing Instruments
| Instrument and sample ID | Expected mutation [reported allele fraction (%)] | NGS assay result? | Observed allele fraction by NGS (%) |
|---|---|---|---|
| HiSeq | |||
| 4V00Oz | IDH1 R132S | Yes | 38.8 |
| 20 | FLT3-TKD D835 (15) | Yes | 20.2 |
| 3Y056A | IDH2 R140Q | Yes | 41.8 |
| 22 | FLT3-TKD D835 (2.9) | No | 0.3 |
| 21 | FLT3-TKD D835 (43) | Yes | 30.2 |
| 3Y0563 | IDH2 R140Q | Yes | 29.2 |
| 23 | FLT3-TKD D835 (2.5) | No | 1.2 |
| 24 | FLT3-TKD D835 (3.6) | No | 2.8 |
| 3Y055Y | IDH1 R132C | Yes | 43.2 |
| 3Y055Y | NRAS G12D | Yes | 37.7 |
| 3y0562 | IDH1 R132C | Yes | 31.4 |
| 28 | RET V804M | Yes | 51 |
| 38 | JAK2 V617F | Yes | 19.1 |
| 35 | JAK2 V617F | Yes | 43.8 |
| 40 | JAK2 V617F | Yes | 87.2 |
| 32 | RET C634G | Yes | 50.8 |
| MiSeq | |||
| 20 | FLT3-TKD D835 (15) | Yes | 23.4∗ |
| 3y0562 | IDH1 R132C | Yes | 34.1∗ |
| 28 | RET V804M | Yes | 47.5∗ |
| 35 | JAK2 V617F | Yes | 41.4∗ |
Mean observed allele fraction across two sequencing runs.
Discussion
Our comprehensive approach for the validation of SNVs in the WUCaMP assay included measuring analytic sensitivity and specificity for inherited variants across the entire targeted region in HapMap samples and for variants at low AF using synthetic mixtures of these samples to simulate the heterogeneity expected in cancer specimens. We also measured reproducibility between and within runs, instruments, and technologists, and determined QC metrics based on well in excess of 100 sequencing data sets derived from validation tumor specimens. Finally, we called variants in a masked fashion for 15 specimens with known cancer-associated variants.
Analytic sensitivity and specificity for inherited SNPs across the target region were evaluated using a well-characterized HapMap sample (NA19240) that made it possible to use orthogonal sequence and genotype data generated previously by Complete Genomics whole-genome sequencing (302 SNVs and nearly 300,000 reference sites) and the Illumina Omni 2.5M SNP array (123 SNVs), which showed technical sensitivity and specificity of 100% by both comparisons within the coding regions of the genes in the WUCaMP assay. Our dilution experiments showed 100% sensitivity for variants an observed VAF of ≥10% in all targeted regions and also in coding regions only, while maintaining a PPV of >98% overall and a PPV of 100% in coding regions. These results have been confirmed by a more comprehensive set of dilution experiments that explored a wider range of mix proportions and SNV detection software.35
We also showed that analytical sensitivity and specificity of the NGS assay obtained on the MiSeq instrument were similar to those obtained on the HiSeq 2000. Finally, testing of 15 masked specimens previously tested in accredited clinical laboratories showed that our NGS assay correctly identified 100% of pathogenic mutations with a VAF of ≥15%. It is worth noting that no false-positive coding calls were observed, which is especially important when genetic testing is performed to direct the adjuvant therapy of cancer patients, because in this setting the absence of mutations in some genes has an equally important role in the selection of treatment regimen as the presence of mutations in other genes. Use of the WUCaMP assay for analysis of the three other classes of sequence changes (ie, copy number variants, structural rearrangements, and small insertions and deletions) will require a similar validation paradigm. At present, efforts are ongoing to assess the ability of the WUCaMP assay to detect these additional types of genetic aberration.36
The utility of the genes included in the WUCaMP assay has been well documented in prior clinical studies, and treatment decisions can be directly affected by the detection of certain mutations in the WUCaMP gene set. For example, there are certain mutations in the kinase domains of genes that predict response to specific targeted therapies: EGFR and ERBB2 mutations in non–small cell lung cancer,37, 38 JAK2 mutations in myeloproliferative disorders,39 BRAF mutations in melanoma,40 ALK mutations in neuroblastoma,41 and KIT and PDGFRA mutations in gastrointestinal stromal tumors.42 In contrast, other mutations predict resistance to therapy; for example, detection of codon 12 mutations in KRAS predicts resistance to treatment with EGFR tyrosine kinase inhibitors in lung cancer43 and resistance to treatment with anti-EGFR monoclonal antibodies in colorectal cancer.44 Unexpected findings, such as the discovery of BRAF V600E in an ovarian cancer, with subsequent response to treatment with a BRAF inhibitor,45 support the utility of sequencing genes that are not strictly characteristic for the patient’s tumor type. Thus, the WUCaMP assay that we have validated here provides actionable information to direct patient care in routine practice.
Compared with genetic tests designed to detect inherited (constitutional) mutations, clinical molecular oncology tests have additional complexities because they target acquired (somatic) mutations. Tumor samples are an admixture of cell types, tumor viability varies within the tissue samples, and tumors harbor intrinsic genetic heterogeneity—all of which can affect test results. Analysis of FFPE specimens is further complicated by the chemical effects of formalin fixation on nucleic acids, which introduce crosslinks that can inhibit the enzymatic steps of DNA library preparation and also can cause direct DNA damage.34, 46 Nonetheless, these challenges can be overcome through the use of NGS technologies, which have been already used to sequence cancer genomes, including acute myeloid leukemia,47 small- and non-small cell lung cancer,48, 49 and melanoma,50 among others. Our own demonstration that read and coverage statistics for fresh tissues are similar to those of FFPE is also paramount to validation of the WUCaMP assay for use in routine clinical practice, because for the vast majority of solid-tumor specimens only FFPE tissue is available for testing.
Based on our in silico sensitivity analysis, 85% and 100% of variants with 10% AF were detected at 400× and 1000× unique coverage levels, respectively. Thus, the high average coverage level achieved by the WUCaMP assay (>1000×) is crucial for the detection of variants at low AF. However, even at this depth, somatic variants present at AFs of <10% are missed by the current implementation of our informatics pipeline. Such low VAFs are known to occur in cancer specimens, and their detection may be possible with other software and ultimately with technical advances in NGS library preparation and analysis.35 Currently, we use a workaround whereby individual base counts at hotspot positions recurrently mutated in cancer (eg, KRAS codon 12 or BRAF V600) are generated for review independently of the variant calls returned by the informatics pipeline, allowing identification of variants below the 10% threshold of automated detection.
In routine clinical practice, WUCaMP NGS assay QC is performed at multiple points in the workflow. First, a surgical pathologist performs histopathological examination of archival H&E-stained tissue sections. A region of tumor cellularity of ≥20% is required; the cutoff is selected based on our estimated VAF limit of detection. This step not only minimizes the chance of obtaining false-negative results due to the presence of variants below the level of detection, but also avoids performing unnecessary testing. Second, a clinical genomicist reviews QC parameters of DNA extraction, library preparation, and targeted capture to evaluate DNA quality and library product quantity and fragment size. Third, a clinical genomicist evaluates three levels of sequencing run QC metrics, benchmarked to reference standards obtained from our 119 validation tumor specimen data sets.
The level 1 metrics concern read and coverage depth, including the number of unique on-target reads achieved across the 300-kb capture region and the percentage of positions that reach unique coverage thresholds of 50×, 400×, and 1000×. In routine clinical use, specimens that fail to match the range of coverage observed during validation require particular scrutiny of the remaining QC measures. The level 2 metric is a measure of which exons failed to achieve 50× unique coverage at 95% of exonic positions; this is frequently the first exon of genes, which are often GC-rich and therefore difficult to amplify by PCR and/or to capture by fluid-phase targeted hybrid capture.51 Of the 14 exons that failed this level of QC on an average validation run, 2 exons are entirely noncoding; according to the COSMIC database, no recurrent mutations are reported in the remaining 12 exons. It is unlikely, therefore, that a clinically important mutation would be missed because of the lower coverage in these regions. For such failed exons, orthogonal testing is not performed; instead, low coverage of the exon is declared in the clinical report. Finally, the level 3 metric verifies that all curated actionable mutation positions have achieved a unique coverage depth of ≥50×.
We have considered the utility of verifying NGS somatic variant findings by Sanger sequencing, and indeed performed targeted Sanger sequencing in parallel with the WUCaMP assay on 51 patient specimens after launch of the test. It is important to emphasize that this analysis included verification of both wild-type as well as SNV sequence calls, to monitor the occurrence of false negatives as well as false positives. An approach focused on verification of identified SNVs alone would have represented a form of discrepant analysis (also known as review bias), which is well established as an intrinsically biased approach that generates significant overestimates of test performance metrics, including sensitivity and specificity.52, 53 In our analysis, all SNVs identified by Sanger sequencing were also called by the WUCaMP assay, whereas a single high-quality, low-AF SNV identified by the WUCaMP assay was not called by Sanger sequencing. This finding, although unsurprising in light of the limitations of Sanger sequencing, underscores the shortcomings of verifying somatic NGS calls using an approach with inherently inferior sensitivity. Given the high sensitivity and specificity of the WUCaMP assay for variants at observed AFs of 10% to 100%, targeted verification of positive and negative findings by Sanger sequencing on every clinical case would not improve the accuracy of test results.
To make review of the NGS data from the WUCaMP assay efficient and accurate, we have curated all published variants of the genes in the assay associated with treatment or clinical outcomes into a database that links the variant with an annotated interpretation based on the medical literature. The need for this functionality, together with the requirement for software that can integrate publicly available NGS analysis tools and NGS viewers, led us to develop the CGW comprehensive software application. In general terms, CGW filters and annotates the variant calls to remove those reported in dbSNP with a VAF of >5%. It then marks variants that are reported in COSMIC and attaches the appropriate genomic, coding, and protein nomenclature and curated clinical interpretations from the internally developed medical annotation database noted above (with visualization of the identified variants and hyperlinks to relevant databases) before generating a draft report, all of which significantly reduces the time and effort required to review data-heavy NGS results. The WUCaMP report classifies variants into levels based on the potential actionability of the variant, a classification that is tailored to the type of the variant in the setting of the specific tumor type. SNPs not reported in COSMIC are classified as known polymorphisms, and non-SNP variants lacking evidence of actionability or prognostic value from the literature are classified as variants of unknown significance.
Proficiency testing for NGS is currently unavailable. However, reference materials for NGS proficiency testing are being developed by CAP, the National Institute of Standards and Technology (NIST), and the CDC, with plans for pilot offerings in 2013 and full implementation in 2014. In the interim, we have developed our own alternative performance assessment strategy in accord with CAP recommendations, as follows. The alternative assessment (AA) is performed biannually, and two samples are analyzed in each AA cycle. One sample is a tissue specimen processed starting from the DNA isolation step, to evaluate the wet-bench procedure and the bioinformatic analysis; the second sample is an NGS sequence file, to evaluate the bioinformatic analysis only. The cases used for AA are either obtained from other CLIA-licensed laboratories or from clinical cases previously processed and signed out in our laboratory; the cases cover both disease-associated and naturally occurring sequence variations across the genomic regions targeted by our test (as recommended19). Each clinical genomicist (either a pathologist with subspecialty certification in Molecular Genetic Pathology from the American Board of Pathology or a laboratory director certified by the American Board of Medical Genetics in Clinical Molecular Genetics and/or Clinical Cytogenetics, or by the American Board of Clinical Chemistry) performs a masked independent review of the two cases and signs them out in the laboratory information management system. Three types of comparisons are then performed. First, the QC metrics of library preparation and sequencing are compared with those of the original run. Second, the variant calls are compared with those of the original run. Third, the variant classification and interpretation are compared between the reviewing clinical genomicists and the original report. Any significant differences observed for the QC metrics, variant calls, or classification between the AA and the original analysis are rigorously assessed until the source of the discrepancy is identified and resolved. It is worth noting that our AA approach is somewhat similar to the CAP proficiency testing challenges for chromosomal microarray analysis, in which DNA is provided to the laboratory to test from the wet-bench procedure through bioinformatic analysis and subsequent interpretation, with additional challenge questions on paper used to further gauge interpretative components of array testing.
As with any genetic test, there is a risk of detecting secondary or incidental findings. DNA sequence analysis of tumor tissue may identify variants linked to familial cancer syndromes in addition to somatically acquired mutations. Correct classification of these variants can be difficult, because there is overlap between mutations described as somatic changes and those linked to familial cancer syndromes.48, 50, 54, 55 Although it is possible to distinguish between a germline and a somatic change by paired analysis of germline and tumor tissue, current laboratory workflows and economic realities limit NGS to the tumor specimen itself. In the context of the WUCaMP assay, whenever a clinically significant variant is suspected to be constitutional, the ordering clinician is informed, formal medical genetic evaluation is recommended, and, if clinically indicated, a constitutional sample (such as a buccal swab or peripheral blood specimen) is obtained, to establish the correct classification of the problematic variant by Sanger sequencing.
Here, we have described the validation of an NGS-based clinical oncology test for the detection of somatic variation in actionable gene targets. The validation process we describe for SNVs addresses recommendations of Gargis et al,19 the NGS checklist requirements of CAP,18 and NGS regulations of the New York State Department of Health. As detailed above, our validation approach for independent confirmation of SNVs meets these requirements, inasmuch as our approach includes comparison of our results with publicly available whole-genome sequence data, SNV calls extracted from publicly available microarray data, and data from Sanger sequencing performed in our laboratory. Likewise, we have independently confirmed well in excess of 10 SNVs from patient samples in which NGS detected a 1-bp change.
There is admittedly some ambiguity as to the precise intent of some of the recommendations of the regulatory entities. For example, the New York State Department of Health regulations require that a minimum of 10 positive patient samples “for each type of variant in each target area” must be independently confirmed. It is reasonable to interpret the phrase “each type of variant” as referring to the class of variant (namely, SNVs as opposed to indels, structural variants, or copy number variants), rather than to the functional consequence of the variant (splice site, versus coding region synonymous, missense, or nonsense) and to interpret the phrase “target area” as referring to the entire 300-kb capture region of our assay, rather than to specific mutation hotspots, exons, or genes, as we did for the validation of our assay. Nonetheless, other interpretations are possible that would clearly require different validation paradigms in routine clinical practice. Because the New York State Department of Health requirements seem to be the most rigorous yet published, there is no doubt that the ambiguities must be addressed. Thus, the type of regulatory uncertainty highlighted by our validation approach for SNVs in tumor samples will likely be of interest to many entities as they develop guidelines for NGS-based clinical testing that address the technical and bioinformatic features of hybrid capture-based methods, as well as amplification-based methods.
The various features of our validation approach for SNVs in tumor samples evaluated by NGS should prove useful to clinical laboratories developing similar approaches to test tissue samples to guide patient management in the emerging paradigm of precision medicine (or personalized medicine), whether at the time of diagnosis or at relapse. Similarly, our validation approach should also prove useful for clinical laboratories developing validation approaches for the other classes of somatic variation, including indels, copy number variation, and structural variation.
Acknowledgment
We thank Manoj Tyagi for assistance with data analysis.
Footnotes
See related Commentary on page7.
Supported by Genomics and Pathology Services, Washington University School of Medicine, St. Louis, MO.
C.E.C., H.A.-K., and A.J.B. contributed equally to this work.
Supplemental material for this article can be found at http://dx.doi.org/10.1016/j.jmoldx.2013.10.002.
Supplemental Data
References
- 1.Walter M.J., Shen D., Ding L., Shao J., Koboldt D.C., Chen K., Larson D.E., McLellan M.D., Dooling D., Abbott R., Fulton R., Magrini V., Schmidt H., Kalicki-Veizer J., O’Laughlin M., Fan X., Grillot M., Witowski S., Heath S., Frater J.L., Eades W., Tomasson M., Westervelt P., DiPersio J.F., Link D.C., Mardis E.R., Ley T.J., Wilson R.K., Graubert T.A. Clonal architecture of secondary acute myeloid leukemia. N Engl J Med. 2012;366:1090–1098. doi: 10.1056/NEJMoa1106968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Patel J.P., Gönen M., Figueroa M.E., Fernandez H., Sun Z., Racevskis J., Van Vlierberghe P., Dolgalev I., Thomas S., Aminova O., Huberman K., Cheng J., Viale A., Socci N.D., Heguy A., Cherry A., Vance G., Higgins R.R., Ketterling R.P., Gallagher R.E., Litzow M., van den Brink M.R., Lazarus H.M., Rowe J.M., Luger S., Ferrando A., Paietta E., Tallman M.S., Melnick A., Abdel-Wahab O., Levine R.L. Prognostic relevance of integrated genetic profiling in acute myeloid leukemia. N Engl J Med. 2012;366:1079–1089. doi: 10.1056/NEJMoa1112304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kalari K.R., Rossell D., Necela B.M., Asmann Y.W., Nair A., Baheti S., Kachergus J.M., Younkin C.S., Baker T., Carr J.M., Tang X., Walsh M.P., Chai H.S., Sun Z., Hart S.N., Leontovich A.A., Hossain A., Kocher J.P., Perez E.A., Reisman D.N., Fields A.P., Thompson E.A. Deep sequence analysis of non-small cell lung cancer: integrated analysis of gene expression, alternative splicing, and single nucleotide variations in lung adenocarcinomas with and without oncogenic KRAS mutations. Front Oncol. 2012;2:12. doi: 10.3389/fonc.2012.00012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen J., Huang X.F., Katsifis A. Activation of signal pathways and the resistance to anti-EGFR treatment in colorectal cancer. J Cell Biochem. 2010;111:1082–1086. doi: 10.1002/jcb.22905. [DOI] [PubMed] [Google Scholar]
- 5.Duncavage E.J., Abel H.J., Szankasi P., Kelley T.W., Pfeifer J.D. Targeted next generation sequencing of clinically significant gene mutations and translocations in leukemia. Mod Pathol. 2012;25:795–804. doi: 10.1038/modpathol.2012.29. [DOI] [PubMed] [Google Scholar]
- 6.Vasli N., Böhm J., Le Gras S., Muller J., Pizot C., Jost B., Echaniz-Laguna A., Laugel V., Tranchant C., Bernard R., Plewniak F., Vicaire S., Levy N., Chelly J., Mandel J.L., Biancalana V., Laporte J. Next generation sequencing for molecular diagnosis of neuromuscular diseases. Acta Neuropathol. 2012;124:273–283. doi: 10.1007/s00401-012-0982-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Calvo S.E., Compton A.G., Hershman S.G., Lim S.C., Lieber D.S., Tucker E.J., Laskowski A., Garone C., Liu S., Jaffe D.B., Christodoulou J., Fletcher J.M., Bruno D.L., Goldblatt J., Dimauro S., Thorburn D.R., Mootha V.K. Molecular diagnosis of infantile mitochondrial disease with targeted next-generation sequencing. Sci Transl Med. 2012;4:118ra10. doi: 10.1126/scitranslmed.3003310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pritchard C.C., Smith C., Salipante S.J., Lee M.K., Thornton A.M., Nord A.S., Gulden C., Kupfer S.S., Swisher E.M., Bennett R.L., Novetsky A.P., Jarvik G.P., Olopade O.I., Goodfellow P.J., King M.C., Tait J.F., Walsh T. ColoSeq provides comprehensive Lynch and polyposis syndrome mutational analysis using massively parallel sequencing. J Mol Diagn. 2012;14:357–366. doi: 10.1016/j.jmoldx.2012.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Voelkerding K.V., Dames S., Durtschi J.D. Next generation sequencing for clinical diagnostics–principles and application to targeted resequencing for hypertrophic cardiomyopathy: a paper from the 2009 William Beaumont Hospital Symposium on Molecular Pathology. J Mol Diagn. 2010;12:539–551. doi: 10.2353/jmoldx.2010.100043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sikkema-Raddatz B., Johansson L.F., de Boer E.N., Almomani R., Boven L.G., van den Berg M.P., van Spaendonck-Zwarts K.Y., van Tintelen J.P., Sijmons R.H., Jongbloed J.D., Sinke R.J. Targeted next-generation sequencing can replace Sanger sequencing in clinical diagnostics. Hum Mutat. 2013;34:1035–1042. doi: 10.1002/humu.22332. [DOI] [PubMed] [Google Scholar]
- 11.Berg J.S., Evans J.P., Leigh M.W., Omran H., Bizon C., Mane K., Knowles M.R., Weck K.E., Zariwala M.A. Next generation massively parallel sequencing of targeted exomes to identify genetic mutations in primary ciliary dyskinesia: implications for application to clinical testing. Genet Med. 2011;13:218–229. doi: 10.1097/GIM.0b013e318203cff2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vandrovcova J., Thomas E.R.A., Atanur S.S., Norsworthy P.J., Neuwirth C., Tan Y., Kasperaviciute D., Biggs J., Game L., Mueller M., Soutar A.K., Aitman T.J. The use of next-generation sequencing in clinical diagnosis of familial hypercholesterolemia. Genet Med. 2013 doi: 10.1038/gim.2013.55. http://dx.doi.org/10.1038/gim.2013.55 [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 13.Neumann J., Zeindl-Eberhart E., Kirchner T., Jung A. Frequency and type of KRAS mutations in routine diagnostic analysis of metastatic colorectal cancer. Pathol Res Pract. 2009;205:858–862. doi: 10.1016/j.prp.2009.07.010. [DOI] [PubMed] [Google Scholar]
- 14.Dienstmann R., Vilar E., Tabernero J. Molecular predictors of response to chemotherapy in colorectal cancer. Cancer J. 2011;17:114–126. doi: 10.1097/PPO.0b013e318212f844. [DOI] [PubMed] [Google Scholar]
- 15.Ladanyi M., Pao W. Lung adenocarcinoma: guiding EGFR-targeted therapy and beyond. Mod Pathol. 2008;21(Suppl 2):S16–S22. doi: 10.1038/modpathol.3801018. [DOI] [PubMed] [Google Scholar]
- 16.Girard N. Thymic tumors: relevant molecular data in the clinic. J Thorac Oncol. 2010;5(10 Suppl 4):S291–S295. doi: 10.1097/JTO.0b013e3181f209b9. [DOI] [PubMed] [Google Scholar]
- 17.Godley L.A. Profiles in leukemia. N Engl J Med. 2012;366:1152–1153. doi: 10.1056/NEJMe1200409. [DOI] [PubMed] [Google Scholar]
- 18.College of American Pathologists: Molecular Pathology Checklist. Northfield, IL, College of American Pathologists, 2012
- 19.Gargis A.S., Kalman L., Berry M.W., Bick D.P., Dimmock D.P., Hambuch T. Assuring the quality of next-generation sequencing in clinical laboratory practice. Nat Biotechnol. 2012;30:1033–1036. doi: 10.1038/nbt.2403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.National Committee for Clinical Laboratory Standards (NCCLS) Clinical and Laboratory Standards Institute; Wayne, PA: 2004. Nucleic acid sequencing methods in diagnostic laboratory medicine; approved guideline. CLSI (NCCLS) document MM09-A. [Google Scholar]
- 21.International HapMap Consortium. Frazer K.A., Ballinger D.G., Cox D.R., Hinds D.A., Stuve L.L., Gibbs R.A. A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449:851–861. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R., 1000 Genome Project Data Processing Subgroup The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Quinlan A.R., Hall I.M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., Kernytsky A., Garimella K., Altshuler D., Gabriel S., Daly M., DePristo M.A. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.DePristo M.A., Banks E., Poplin R., Garimella K.V., Maguire J.R., Hartl C., Philippakis A.A., del Angel G., Rivas M.A., Hanna M., McKenna A., Fennell T.J., Kernytsky A.M., Sivachenko A.Y., Cibulskis K., Gabriel S.B., Altshuler D., Daly M.J. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Robinson J.T., Thorvaldsdóttir H., Winckler W., Guttman M., Lander E.S., Getz G., Mesirov J.P. Integrative genomics viewer. Nat Biotechnol. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thorvaldsdóttir H., Robinson J.T., Mesirov J.P. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform. 2013;14:178–192. doi: 10.1093/bib/bbs017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.1000 Genomes Project Consortium. Abecasis G.R., Altshuler D., Auton A., Brooks L.D., Durbin R.M., Gibbs R.A., Hurles M.E., McVean G.A. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wickham H. The split-apply-combine strategy for data analysis. J Stat Software. 2011;40:1–29. [Google Scholar]
- 30.R Development Core Team . R Foundation for Statistical Computing; Vienna, Austria: 2012. R: A language and environment for statistical computing. [Google Scholar]
- 31.Wickham H. ed 2. Springer; New York: 2009. ggplot: Elegant Graphics for Data Analysis. [Google Scholar]
- 32.1000 Genomes Project Consortium. Abecasis G.R., Altshuler D., Auton A., Brooks L.D., Durbin R.M., Gibbs R.A., Hurles M.E., McVean G.A. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Drmanac R., Sparks A.B., Callow M.J., Halpern A.L., Burns N.L., Kermani B.G. Human genome sequencing using unchained base reads on self-assembling DNA nanoarrays. Science. 2010;327:78–81. doi: 10.1126/science.1181498. [DOI] [PubMed] [Google Scholar]
- 34.Spencer D.H., Sehn J.K., Abel H.J., Watson M.A., Pfeifer J.D., Duncavage E.J. Comparison of clinical targeted next-generation sequence data from formalin-fixed and fresh-frozen tissue specimens. J Mol Diagn. 2013;15:623–633. doi: 10.1016/j.jmoldx.2013.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Spencer D.H., Tyagi M., Vallania F., Bredemeyer A., Pfeifer J.D., Mitra R.D., Duncavage E.J. Performance of common analysis methods for detecting low-frequency single nucleotide variants in targeted next-generation sequence data. J Mol Diagn. 2014;16:75–88. doi: 10.1016/j.jmoldx.2013.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Spencer D.H., Abel H.J., Lockwood C.M., Payton J.E., Szankasi P., Kelley T.W., Kulkarni S., Pfeifer J.D., Duncavage E.J. Detection of FLT3 internal tandem duplication in targeted, short-read-length, next-generation sequencing data. J Mol Diagn. 2013;15:81–93. doi: 10.1016/j.jmoldx.2012.08.001. [DOI] [PubMed] [Google Scholar]
- 37.Paez J.G., Jänne P.A., Lee J.C., Tracy S., Greulich H., Gabriel S., Herman P., Kaye F.J., Lindeman N., Boggon T.J., Naoki K., Sasaki H., Fujii Y., Eck M.J., Sellers W.R., Johnson B.E., Meyerson M. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004;304:1497–1500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- 38.Stephens P., Hunter C., Bignell G., Edkins S., Davies H., Teague J. Lung cancer: intragenic ERBB2 kinase mutations in tumours. Nature. 2004;431:525–526. doi: 10.1038/431525b. [DOI] [PubMed] [Google Scholar]
- 39.Kralovics R., Passamonti F., Buser A.S., Teo S., Tiedt R., Passweg J.R., Tichelli A., Cazzola M., Skoda R.C. A gain-of-function mutation of JAK2 in myeloproliferative disorders. N Engl J Med. 2005;352:1779–1790. doi: 10.1056/NEJMoa051113. [DOI] [PubMed] [Google Scholar]
- 40.Davies H., Bignell G.R., Cox C., Stephens P., Edkins S., Clegg S. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949–954. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- 41.Janoueix-Lerosey I., Lequin D., Brugières L., Ribeiro A., de Pontual L., Combaret V., Raynal V., Puisieux A., Schleiermacher G., Pierron G., Valteau-Couanet D., Frebourg T., Michon J., Lyonnet S., Amiel J., Delattre O. Somatic and germline activating mutations of the ALK kinase receptor in neuroblastoma. Nature. 2008;455:967–970. doi: 10.1038/nature07398. [DOI] [PubMed] [Google Scholar]
- 42.Hirota S., Isozaki K., Moriyama Y., Hashimoto K., Nishida T., Ishiguro S., Kawano K., Hanada M., Kurata A., Takeda M., Tunio G.M., Matsuzawa Y., Kanakura Y., Shinomura Y., Kitamura Y. Gain-of-function mutations of c-kit in human gastrointestinal stromal tumors. Science. 1998;279:577–580. doi: 10.1126/science.279.5350.577. [DOI] [PubMed] [Google Scholar]
- 43.Pao W., Wang T.Y., Riely G.J., Miller V.A., Pan Q., Ladanyi M., Zakowski M.F., Heelan R.T., Kris M.G., Varmus H.E. KRAS mutations and primary resistance of lung adenocarcinomas to gefitinib or erlotinib. PLoS Med. 2005;2:e17. doi: 10.1371/journal.pmed.0020017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Karapetis C.S., Khambata-Ford S., Jonker D.J., O’Callaghan C.J., Tu D., Tebbutt N.C., Simes R.J., Chalchal H., Shapiro J.D., Robitaille S., Price T.J., Shepherd L., Au H.J., Langer C., Moore M.J., Zalcberg J.R. K-ras mutations and benefit from cetuximab in advanced colorectal cancer. N Engl J Med. 2008;359:1757–1765. doi: 10.1056/NEJMoa0804385. [DOI] [PubMed] [Google Scholar]
- 45.Falchook G.S., Long G.V., Kurzrock R., Kim K.B., Arkenau T.H., Brown M.P., Hamid O., Infante J.R., Millward M., Pavlick A.C., O’Day S.J., Blackman S.C., Curtis C.M., Lebowitz P., Ma B., Ouellet D., Kefford R.F. Dabrafenib in patients with melanoma, untreated brain metastases, and other solid tumours: a phase 1 dose-escalation trial. Lancet. 2012;379:1893–1901. doi: 10.1016/S0140-6736(12)60398-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gilbert M.T., Haselkorn T., Bunce M., Sanchez J.J., Lucas S.B., Jewell L.D., Van Marck E., Worobey M. The isolation of nucleic acids from fixed, paraffin-embedded tissues-which methods are useful when? PLoS One. 2007;2:e537. doi: 10.1371/journal.pone.0000537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ley T.J., Mardis E.R., Ding L., Fulton B., McLellan M.D., Chen K. DNA sequencing of a cytogenetically normal acute myeloid leukaemia genome. Nature. 2008;456:66–72. doi: 10.1038/nature07485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pleasance E.D., Stephens P.J., O’Meara S., McBride D.J., Meynert A., Jones D. A small-cell lung cancer genome with complex signatures of tobacco exposure. Nature. 2010;463:184–190. doi: 10.1038/nature08629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Govindan R., Ding L., Griffith M., Subramanian J., Dees N.D., Kanchi K.L., Maher C.A., Fulton R., Fulton L., Wallis J., Chen K., Walker J., McDonald S., Bose R., Ornitz D., Xiong D., You M., Dooling D.J., Watson M., Mardis E.R., Wilson R.K. Genomic landscape of non-small cell lung cancer in smokers and never-smokers. Cell. 2012;150:1121–1134. doi: 10.1016/j.cell.2012.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pleasance E.D., Cheetham R.K., Stephens P.J., McBride D.J., Humphray S.J., Greenman C.D. A comprehensive catalogue of somatic mutations from a human cancer genome. Nature. 2010;463:191–196. doi: 10.1038/nature08658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Aird D., Ross M.G., Chen W.S., Danielsson M., Fennell T., Russ C., Jaffe D.B., Nusbaum C., Gnirke A. Analyzing and minimizing PCR amplification bias in Illumina sequencing libraries. Genome Biol. 2011;12:R18. doi: 10.1186/gb-2011-12-2-r18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hadgu A. Discrepant analysis is an inappropriate and unscientific method. J Clin Microbiol. 2000;38:4301–4302. doi: 10.1128/jcm.38.11.4301-4302.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lipman H.B., Astles J.R. Quantifying the bias associated with use of discrepant analysis. Clin Chem. 1998;44:108–115. [PubMed] [Google Scholar]
- 54.Shah S.P., Morin R.D., Khattra J., Prentice L., Pugh T., Burleigh A., Delaney A., Gelmon K., Guliany R., Senz J., Steidl C., Holt R.A., Jones S., Sun M., Leung G., Moore R., Severson T., Taylor G.A., Teschendorff A.E., Tse K., Turashvili G., Varhol R., Warren R.L., Watson P., Zhao Y., Caldas C., Huntsman D., Hirst M., Marra M.A., Aparicio S. Mutational evolution in a lobular breast tumour profiled at single nucleotide resolution. Nature. 2009;461:809–813. doi: 10.1038/nature08489. [DOI] [PubMed] [Google Scholar]
- 55.Mardis E.R., Ding L., Dooling D.J., Larson D.E., McLellan M.D., Chen K. Recurring mutations found by sequencing an acute myeloid leukemia genome. N Engl J Med. 2009;361:1058–1066. doi: 10.1056/NEJMoa0903840. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.