Abstract
Aberrant expression and regulation of miRNAs have been implicated in multiple stages of tumorigenic processes. The current study was designed to explore the biological function and epigenetic regulation of miR-34a in human cholangiocarcinoma (CCA). Our data show that the expression of miR-34a is decreased significantly in CCA cells compared with non-neoplastic biliary epithelial cells. Forced overexpression of miR-34a in CCA cells inhibited their proliferation and clonogenic capacity in vitro, and suppressed tumor xenograft growth in severe combined immunodeficiency mice. We identified three key components of the Notch pathway, Notch1, Notch2, and Jagged 1, as direct targets of miR-34a. Our further studies show that down-regulation of miR-34a is caused by Enhancer of zeste homolog 2 (EZH2)-mediated H3 lysine 27 trimethylation as well as DNA methylation. Accordingly, treatment with the EZH2 inhibitor, selective S-adenosyl-methionine-competitive small-molecule (GSK126), or the DNA methylation inhibitor, 5-Aza-2′-deoxycytidine, partially restored miR-34a levels in human CCA cells. Immunohistochemical staining and Western blot analyses showed increased EZH2 expression in human CCA tissues and cell lines. We observed that GSK126 significantly reduced CCA cell growth in vitro and intrahepatic metastasis in vivo. Our findings provide novel evidence that miR-34a expression is silenced epigenetically by EZH2 and DNA methylation, which promotes CCA cell growth through activation of the Notch pathway. Consequently, these signaling cascades may represent potential therapeutic targets for effective treatment of human CCA.
Cholangiocarcinoma (CCA) is a malignant epithelial cancer of the biliary tree with poor prognosis.1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12 Despite an increasing incidence and mortality rate of CCA worldwide, therapeutic options for patients still are limited and ineffective. The molecular pathogenesis underlying CCA development involves genetic and epigenetic changes, alterations of oncogenic and tumor-suppressive pathways, contribution of the tumor stroma compartment, and aberrant expression of miRNAs.
The Notch signaling pathway is implicated critically in the development and progression of CCA.13, 14, 15, 16, 17, 18, 19 It consists of Notch receptors (Notch1 to 4) and two types of ligands, Jagged (Jag 1 and 2) and Delta-like (Dll 1, 3, and 4) family. The interaction of Notch receptors and ligands leads to proteolytic cleavage of Notch receptor by γ-secretase and thereby translocation of Notch intracellular domain into the nucleus, which activates the expression of target genes. Enhanced Notch1 expression has been reported in human CCA tissue and cell lines.20 In animal models of cholangiocarcinogenesis, mice with constitutive activation of the Notch1 intracellular domain in hepatoblasts (N1IC;AlbCre) developed intrahepatic CCA through up-regulation of cyclin E.15 Similarly, mice with overexpression of the Notch2 intracellular domain in liver (N2IC;AlbCre) developed mixed hepatocellular-CCA during treatment with the hepatic carcinogen diethylnitrosamine.18 In both studies, the Notch pathway activated a subset of liver progenitor cells (Sox9 and K19-positive cells) and promoted carcinogenesis. Furthermore, up-regulation of the Notch1 intracellular domain by inducible nitric oxide synthase also has been reported in human CCA. These findings document an important role of Notch signing in CCA.20
miRNAs are small noncoding RNAs (approximately 22 nucleotides in length) that typically repress the expression of target genes through mRNA degradation or inhibition of translation.21, 22 They are known to target mRNAs through complementary binding at 3′untranslated regions (UTRs) with their seed sequences (usually two to seven nucleotides). Studies in recent years have shown that miRNAs have diverse biological and pathophysiological functions including regulation of carcinogenesis.23, 24 For example, many tumor-suppressor miRNAs are silenced epigenetically by DNA methylation and/or histone modification, although these alterations also can activate the expression of oncogenes, leading to cancer cell proliferation, inhibition of apoptosis, and cell-cycle progression.23, 24 Recently, studies from several groups have reported dysregulation of several miRNAs in CCA and their contributions to cholangiocarcinogenesis.25 In this regard, epigenetic modifications underlying down-regulation of miRNAs is being recognized as an important mechanism for CCA development and progression. Thus, miRNAs and epigenetic molecules represent important therapeutic targets to be evaluated for future treatment of CCA.
Enhancer of zeste homolog 2 (EZH2) is a histone methyltransferase that catalyzes histone H3 lysine 27 (H3K27) trimethylation and thus regulates gene expression epigenetically.26, 27 It is the catalytic subunit of polycomb repressor complex 2, which mediates silencing of target genes via local chromatin reorganization. Studies have shown that EZH2 plays a critical role in cancer initiation, progression, and metastasis, as well as in cancer stem cell biology, although the biological effects and molecular mechanisms of EZH2 depend on the tissue context and specific cancer types.26, 27 Given the documented oncogenic functions of EZH2 in a number of cancer types, targeting EZH2 with specific inhibitors has been considered a promising anticancer therapy.26, 27 Notably, increased expression of EZH2 has been found in several types of cancers and correlates with tumor aggressiveness and poor prognosis.28, 29, 30, 31 However, little is known about the role of EZH2 in the epigenetic regulation of miRNAs and its mechanism of action in CCA.
In the present study, we aimed to determine the biological function and epigenetic regulation of miR-34a in human CCA. We report for the first time that miR-34a is a key tumor-suppressive miRNA that is silenced epigenetically by EZH2-mediated H3K27me3 and DNA methylation in CCA. Our data provide novel evidence that miR-34a suppresses CCA cell growth in vitro and in vivo through targeting Notch pathway–related genes (Notch1, Notch2, and Jagged1). Furthermore, we show that the expression of EZH2 is increased markedly in human CCA tissues and cell lines. We have found that pharmacologic inhibition of EZH2 restores miR-34a expression and inhibits CCA cell growth in vitro and inhibits intrahepatic tumor metastasis in vivo. Our findings suggest the possibility of targeting miR-34a and EZH2, in addition to Notch signaling, for effective treatment of human CCA.
Materials and Methods
Reagents and Antibodies
EZH2 inhibitor, GSK126, was purchased from Cellagen Technology (San Diego, CA) and Sellekchem (Houston, TX). 5-Aza-2′-deoxycytidine (5-Aza-CdR) was obtained from Sigma-Aldrich (St. Louis, MO). Water soluble tetrazolium salts-1 (WST1) assay reagent was purchased from Roche (Indianapolis, IN). The antibodies against EZH2, histone H3, Notch1, Notch2, and Jagged1 were purchased from Cell Signaling Technology (Beverly, CA). The anti-H3K27me3 antibody was purchased from Abcam (Cambridge, MA). The antibodies against DNA methyltransferase (DNMT)1 and DNMT3a were obtained from Santa Cruz Biotechnology (Santa Cruz, CA). The anti–β-actin was obtained from Sigma-Aldrich.
Cell Culture
Human CCA cells including CCLP1, SG231, HUCCT1, and TFK1 were cultured according to our methods as described previously.32 Briefly, the CCLP1 cells were cultured in Dulbecco's modified Eagle's medium (Invitrogen, Carlsbad, CA) containing 10% fetal bovine serum (Sigma-Aldrich) and antibiotics (100 U/mL penicillin and 100 μg/mL streptomycin). The SG231 cells were cultured in Opti-MEM medium (Invitrogen, Carlsbad, CA) containing 5% fetal bovine serum and antibiotics. The HUCCT1 and TFK1 cells were cultured in RPMI-1640 medium containing 10% fetal bovine serum and antibiotics. The immortalized human cholangiocyte H69 cells were provided by Dr. Gregory J. Gores (Mayo Clinic College of Medicine, Rochester, MN) and cultured in Bronchial Epithelial Cell Basal Medium (Lonza, Basel, Switzerland) supplemented with growth factors (BEGM SingleQuot Kit; Lonza, Walkersville, MD) and 10% fetal bovine serum. All cells were cultured in a humidified 5% CO2 incubator at 37°C.
Transient Transfection
CCLP1 and SG231 cells were cultured in a 6-well plate and transfected with miR-34a mimic/inhibitor or its negative control RNA (Qiagen, Valencia, CA) using Oligofectamine reagent (Invitrogen) according to the manufacturer's instructions. After 48 hours, the cells were collected for Western blot and quantitative RT-PCR (RT-qPCR) analysis.
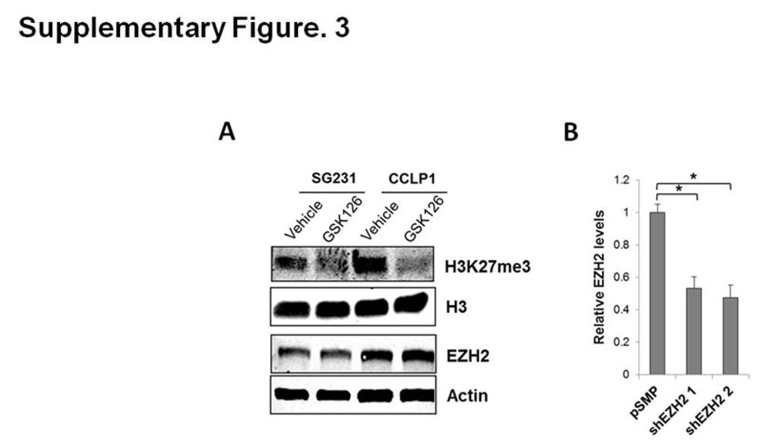
For EZH2 shRNA transfection, the cells were transfected with two different EZH2 shRNAs (OriGene Technologies, Rockville, MD) or its control vector (pSMP; OriGene) using Lipofectamine and Plus reagents (Invitrogen) according to the manufacturer's instructions. After 48 hours of transfection, the cells were collected for RT-qPCR analysis.
Cell Proliferation Assay
The growth of CCA cells was measured by cell counting or WST-1 assay. The cells (1 × 104 per well) were seeded in 12-well plates in triplicate and transfected with miR-34a mimic or scramble control. On the indicated day, the cells were detached using 0.05% trypsin-EDTA and suspended in 1 mL of culture medium. After staining cells with a 1:1 ratio of Trypan blue (Invitrogen), the number of cells was counted under the microscope. For WST-1 assay, the cells (5 × 103 per well) were seeded in 96-well plates at least in triplicate and treated with vehicle control or N-[N-(3,5-Difluorophenacetyl)-L-alanyl]-S-phenylglycine t-butyl ester (DAPT)/GSK126. At the indicated time point, the culture medium was removed and cells were incubated with 90 μL of serum-free medium containing 10 μL of WST-1 reagents for 2 hours at 37°C. The absorbance of each sample was measured at 450 nm by using an automatic enzyme-linked immunosorbent assay plate reader (VersaMax Microplate Reader; Molecular Devices, Sunnyvale, CA).
Colony Forming Assay
The cells (1 × 103) were plated in 10-cm dishes and cultured for 14 days to allow colony formation. The colonies were fixed with 100% methanol and then stained with 0.1% crystal violet solution. For GSK126/DAPT treatment, the cells (5 × 103) were plated in 10-cm dishes and treated with the indicated doses of GSK126/DAPT or vehicle. After 14 days of cell culture, colony formation was determined by crystal violet staining.
RT-qPCR
Total RNA from cells was extracted with TRIzol reagent (Invitrogen) and reverse-transcribed with iScript Supermix (Bio-Rad, Hercules, CA). After normalization of cDNA concentration, RT-qPCR was performed in triplicate using SsoAdvanced SYBR green supermix on a C1000 thermal cycler (Bio-Rad). Primer sequences used in this study are detailed in Table 1. Results were analyzed using CFX Manager Software version 3.1 (Bio-Rad) and normalized to actin or glyceraldehyde-3-phosphate dehydrogenase. For detection of miR-34a, reverse transcription was performed using the miScript II RT Kit (Qiagen) and RT-qPCR was performed with the miScript SYBR Green PCR Kit (Qiagen). MiR-34a miScript Primer Assays (Qiagen) were used to determine the expression of miR-34a. RNA U6 small nuclear 2 (Qiagen) was used as internal control.
Table 1.
Primer Sequences
| Target | Forward primer | Reverse primer |
|---|---|---|
| Notch1 | 5′-ATGTGTTCTCGGAGTGTGTATG-3′ | 5′-AGGGACCAAGAACTTGTATAACC-3′ |
| Jag1 | 5′-GGGAACCCGATCAAGGAAAT-3′ | 5′-CGCCTCTGAACTCTTACTTCTG-3′ |
| Notch2 | 5′-AATCCCTGACTCCAGAACG-3′ | 5′-TGGTAGACCAAGTCTGTGATGAT-3′ |
| EZH2 | 5′-TTGTTGGCGGAAGCGTGTAAAATC-3′ | 5′-TCCCTAGTCCCGCGCAATGAGC-3′ |
| Actin | 5′-AGAAAATCTGGCACCACACC-3′ | 5′-GTCTCAAACATGATCTGGG-3′ |
| Chip primer 1 (P1) | 5′-ATAATGGTTGGGGCAGGAGG-3′ | 5′-TAAAGGTTCCCAGAGACA-3′ |
| Chip primer 2 (P2) | 5′-GAAAGGGTGGGAAATGCAGG-3′ | 5′-GGGTGGGAAGGGACATGG-3′ |
| MiR-34a gene methylated primer | 5′-GGTTTTGGGTAGGCGCGTTTC-3′ | 5′-TCCTCATCCCCTTCACCGCCG-3′ |
| MiR-34a gene unmethylated primer | 5′-IIGGTTTTGGGTAGGTGTGTTTT-3′ | 5′-AATCCTCATCCCCTTCACCACCA-3′ |
Western Blot
Western blot was performed as we described previously.33 Briefly, the cell lysates were sonicated and centrifuged at 12,000 × g for 15 minutes to obtain supernatants. The protein concentration was measured using a Bio-Rad Protein Assay Kit. Equal amounts of protein samples were separated on SDS-PAGE (Bio-Rad) and transferred to a nitrocellulose membrane (Bio-Rad). After blocking the membranes with 5% skim milk in 0.1% Tween 20 in phosphate-buffered saline for 1 hour at room temperature, the membranes were incubated with primary antibodies overnight at 4°C. On the following day, the membranes were incubated with IRDye800-conjugated secondary antibodies and developed using the LI-COR Odyssey Imaging system (LI-COR Biosciences, Lincoln, NE).
Construction of 3′UTR Luciferase Reporter Plasmids and Luciferase Reporter Activity Assay
Notch1 (SC215771), Jagged1 (HmiT004470), and Notch2 (HmiT011875) 3′-UTR reporter plasmids were obtained from Origene (Rockville, MD) and Genecopoeia (Rockville, MD). Mutation of the putative miR-34a target sites at the Notch1, Jag1, and Notch2 3′UTRs was achieved using the QuikChange II site-directed mutagenesis kit (Agilent Technologies, Santa Clara, CA). The mutagenic primers were as follows: Notch1 forward, 5′-GTCAGCCCAGGCATTATCATTCCCCAGAAAAGGGTAGGATGCC-3′; reverse, 5′-GGCATCCTACCCTTTTCTGGGGAATGATAATGCCTGGGCTGAC-3′; Jag1 forward, 5′-TTGATTTCCTCACTTAAGGCAGGTAATATACTCTATGGCAAATCTAAACAGTGATC-3′; reverse, 5′-GATCACTGTTTAGATTTGCCATAGAGTATATTACCTGCCTTAAGTGAGGAAATCAA-3′; and Notch2 forward, 5′-GAGATCAGTAAAAAGTTTGAAAGGTAATATTGTCCTCCTCATCACTGAAACCTGTTG-3′; reverse, 5′-CAACAGGTTTCAGTGATGAGGAGGACAATATTACCTTTCAAACTTTTTACTGATCTC-3′. The accuracy of the mutant constructs was verified by DNA sequencing.
SG231 cells were co-transfected with miR-34a mimic/negative control and Notch1, Jagged1, or Notch2 wild-type/mutant 3′-UTR reporter plasmid. After 24 hours of transfection, the cell lysates were collected and luciferase activity was measured using a dual-luciferase reporter assay system (Promega, Madison, WI) in a Centro XS3 LB 960 microplate fluorescence reader (Berthold Technologies, Ontario, Canada). The luciferase values were normalized to protein concentration.
Bisulfite Conversion and Methylation-Specific PCR
Genomic DNA was isolated from cells using the DNeasy Blood and Tissue Kit (Qiagen) according to the manufacturer's instructions. Extracted genomic DNA (1 to 2 μg per sample) was treated with bisulfite using the EzDNA Methylation-Gold kit (Zymo Research, Orange, CA) and then methylation-specific PCR (MSP) was performed using the ZymoTaq Premix (Zymo Research). The PCR conditions were as follows: 95°C for 10 minutes followed by 40 cycles of 95°C for 30 seconds, 56°C for 30 seconds, and 72°C for 30 seconds. Final extension was performed at 72°C for 7 minutes. The PCR products were separated in a 2% agarose gel and identified by ethidium bromide staining. The primer sequences for MSP are described in Table 1.
Establishment of a Stable Cell Line
SG231 cells were transfected with miR-34a expression vector (HmiR0005-MR03; Genecopoeia) or control vector (CmiR0001-MR03; Genecopoeia) using Lipofectamine and Plus reagent (Invitrogen). After 72 hours of transfection, the medium was replaced with fresh medium containing increasing concentrations of puromycin (from 0.2 to 1 μg/mL; Invitrogen) for selection. The selection medium was changed every 2 to 3 days and viable cells were subcultured with selection medium. Transfection efficiency was monitored by green fluorescent protein signals under a fluorescent microscope and the expression of miR-34a was confirmed by RT-qPCR analysis.
Chromatin Immunoprecipitation Assay
The chromatin immunoprecipitation (ChIP) assay was performed with the Simplechip Enzymatic Chromatin IP kit (Cell Signaling Technology, Billerica, MA) according to the manufacturer's instructions. Briefly, the cross-linked chromatin was sonicated and subjected to immunoprecipitation with 8 μg of anti-H3K27me3 (Abcam)/anti-EZH2 (Abcam) or mouse/rabbit IgG control. Purified ChIP DNA was obtained and amplified by real-time quantitative PCR using specific primers detecting the CpG-enriched upstream region of human miR-34a. The primer sequences are shown in Table 1.
Immunohistochemistry
Immunohistochemistry of EZH2 was performed in the formalin-fixed, paraffin-embedded tissue specimens surgically resected from CCA patients according to the approval of the Institutional Review Board. The tissue sections were deparaffinized and subjected to heat retrieval at 95°C for 40 minutes and cooling down to room temperature for 20 minutes. After washing the sections with deionized water, endogenous peroxidase activity was quenched by incubation with 3% H2O2 for 5 minutes, followed by buffer wash (TWB945; Biocare Medical). The sections were incubated with avidin (AB972; Biocare Medical, Pike Lane Concord, CA) for 10 minutes and biotin for 10 minutes. Nonspecific binding was blocked by Sniper (BS966; Biocare Medical) for 10 minutes. The sections were incubated with EZH2 primary antibody at a 1:50 dilution for 60 minutes at room temperature. After repeated washes, the sections were incubated with horseradish-peroxidase–conjugated secondary antibody for 30 minutes and horseradish-peroxidase–labeled micropolymer (mach-4; Biocare Medical) for 30 minutes at room temperature. The 3,3′-diaminobenzidine was added and color was developed for 1 minute. Finally, the slides were counterstained with hematoxylin.
CCA Xenograft Model
We used 5- or 6-week-old male nonobese diabetic CB17-prkdc/severe combined immunodeficiency (SCID) mice purchased from The Jackson Laboratory (Bar Harbor, ME). Scramble control miRNA or miR-34a–overexpressed SG231 cells (2 × 107 cells/50 μL) were mixed with BD Matrigel matrix high concentration (BD Biosciences, San Jose, CA) in a total volume of 100 μL for a 1:1 ratio, and then cell suspension mixture (1 × 106 cells/10 μL) was injected directly into the livers of SCID mice (n = 6) using a Hamilton syringe (model 75 N SYR; Hamilton Co., Reno, NV). The mice were observed over 6 weeks for tumor formation. At sacrifice, the livers were dissected surgically and the tumor volume was calculated by the following formula: Tumor volume = (π × length × width2)/6.
For EZH2 inhibitor study, CCLP1 cells (2 × 107 cells/50 μL) were mixed with BD Matrigel matrix high concentration in a total volume of 100 μL for a 1:1 ratio. Cell suspension mixture (1 × 106 cells/10 μL) was inoculated directly into the liver of SCID mice (n = 6) using a Hamilton syringe. After 3 weeks, the mice were randomized into two groups and treated with vehicle control (20% Captisol) or GSK126 [150 mg/kg diluted with dimethyl sulfoxide and 20% modified cyclodextrin (Captisol, San Diego, CA)]) by intraperitoneal injection twice a week for an additional 5 weeks. At the end of the experiments (8 weeks), the liver tumors were recovered and the tumor volume was calculated. All animal studies were conducted according to the protocol approved by the Tulane Institutional Animal Care and Use Committee.
Statistical Analysis
Statistical analysis was performed using Graphad Prism version 5 (GraphPad Software, La Jolla, CA). The tests included the t-test, one-way analysis of variance along with Bonferroni adjustment, the Wilcoxon signed-rank test, and the Fisher exact test. The RT-qPCR data are expressed as means ± SEM, whereas all other quantitative data are expressed as means ± SD. A P value <0.05 was considered statistically significant.
Results
MiR-34a Suppresses CCA Cell Growth in Vitro
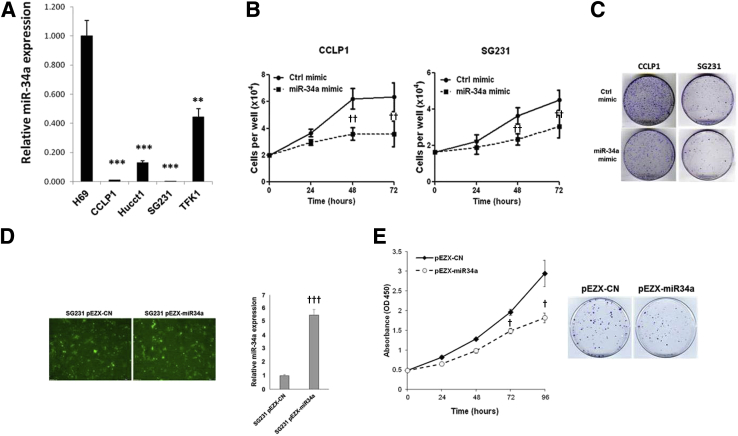
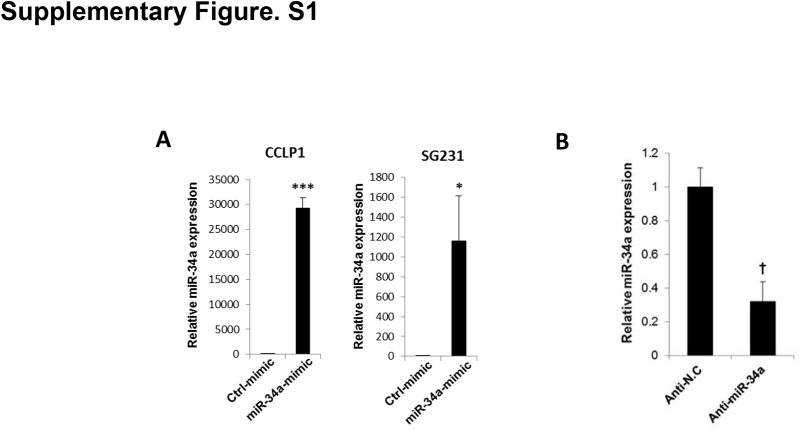
We first evaluated miR-34a expression levels in human CCA cell lines (CCLP1, HUCCT1, SG231, and TFK1) by RT-qPCR analysis. Our data showed that miR-34a was down-regulated significantly in human CCA cells compared with the non-neoplastic human cholangiocyte cell line (H69) (Figure 1A). To investigate the functional role of miR-34a in CCA progression, we transfected CCLP1 and SG231 cells with miR-34a mimic (because these cells express the lowest level of miR-34a among the four CCA cell lines); RT-qPCR analysis confirmed increased miR-34a in cells transfected with the miR-34a mimic compared with cells transfected with scramble control miRNA (Supplemental Figure S1A). We observed that miR-34a mimic significantly inhibited the growth and colony formation of CCLP1 and SG231 cells (Figure 1, B and C). As a parallel approach, we also stably transfected SG231 cells with the miR-34a expression or control vector (containing green fluorescent protein); the transfection efficiency was monitored by fluorescence microscopy for green fluorescent protein and by RT-qPCR analysis for miR-34a (an approximately fivefold increase of miR-34a was observed in miR-34a stable cells) (Figure 1D). We observed that the miR-34a stable cells showed decreased cell proliferation and colony formation compared with scramble control cells (Figure 1E). Together, these findings show that miR-34a mimics treatment or overexpression inhibits human CCA cell growth and decreases their colonogenic capacity in vitro.
Figure 1.
MiR-34a regulates proliferation and colony formation of human cholangiocarcinoma cells in vitro. A: The levels of miR-34a in nonmalignant human cholangiocyte cell (H69) and cholangiocarcinoma cells (CCLP1, SG231, HUCCT1, and TFK1) by quantitative RT-PCR (RT-qPCR) analysis. B: The growth of cells (CCLP1 and SG231) was measured by counting at the indicated time point after transfection of miR-34a mimic or scramble control. C: Representative images of colony formation. CCLP1 and SG231 cells were transfected with miR-34a mimic or scramble control and cultured for 10 days, and stained with crystal violet. D: Successful overexpression of miR-34a was confirmed by green fluorescent protein (GFP) expression under fluorescence microscope and RT-qPCR in SG231 cells with stable miR-34a overexpression. E: The proliferation and colony formation for control (pEZX-CN) and miR-34a–overexpressed (pEZX-miR34a) SG231 cells was determined by WST1 assay and crystal violet staining, respectively. Data are expressed as means ± SEM (A and D); data are expressed as means ± SD (B and E). ∗∗P < 0.01, ∗∗∗P < 0.001 versus H69; †P < 0.05, ††P < 0.01, †††P < 0.001 versus control at each time point. Original magnification: ×100 (D). Ctrl, control.
MiR-34a Inhibits Notch Pathway by Targeting Notch1, Notch2, and Jagged1
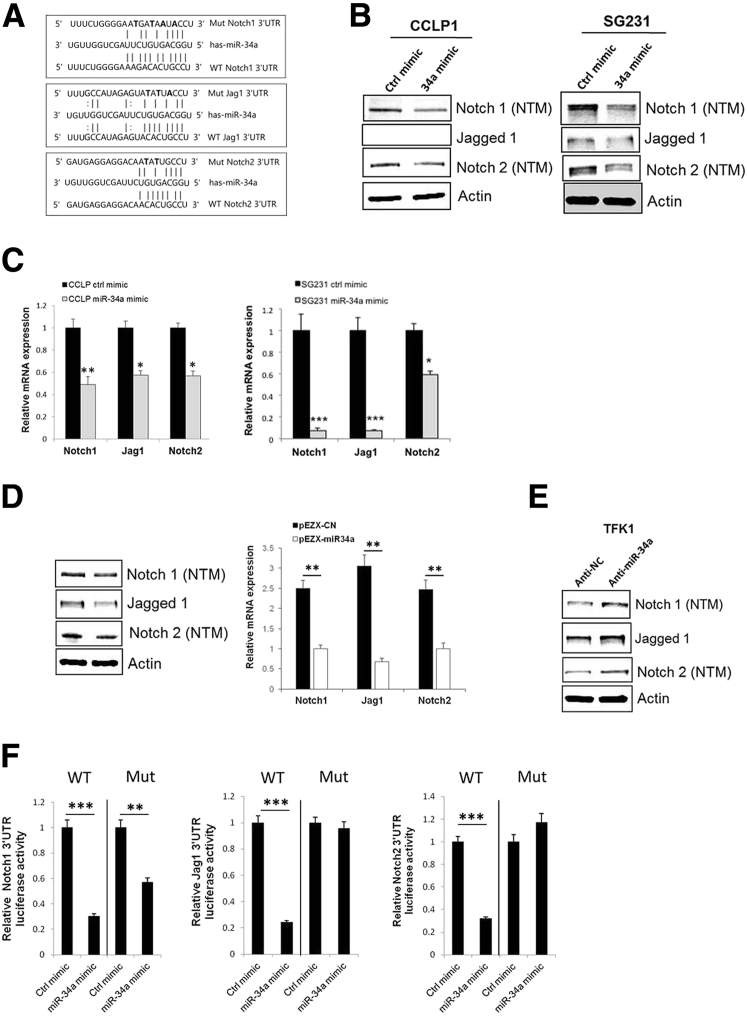
We next used the miRNA target prediction resources (http://microRNA.org and http://www.targetscan.org, last accessed March 4, 2016) to identify potential targets of miR-34a. Our analyses led to the identification of putative miR-34a binding sites at 3′UTRs of three genes in the Notch pathway, including Notch1, Jagged1, and Notch2 (Figure 2A). This finding is noteworthy given that Notch pathway activation is critically implicated in cholangiocarcinogenesis.13, 15, 17, 18 We observed that treatment with miR-34a mimic significantly decreased the expression levels of Notch1, Jag1, and Notch2 in both CCLP1 and SG231 cells, as shown by Western blot and RT-qPCR analysis (Figure 2, B and C) (Jag1 protein was not detectable in CCLP1 cells). Similarly, we noted decreased levels of Notch1, Jag1, and Notch2 in miR-34a stably overexpressed cells compared with scramble control miRNA-expressed cells (Figure 2D). Because the TFK1 cells express a relatively high level of miR-34a among the four CCA cell lines used in this study, we transfected the TFK1 cells with the miR-34a inhibitor or scramble control miRNA. RT-qPCR analysis showed that miR-34a inhibitor transfection led to an approximately 75% reduction of miR-34a expression in these cells (Supplemental Figure S1B). We observed that inhibition of miR-34a increased the expression of Notch1, Jag1, and Notch2 in the TFK1 cells (Figure 2E). These findings provide novel evidence that miR-34a regulates three key molecules of the Notch pathway in human CCA cells.
Figure 2.
MiR-34a inhibits the Notch pathway by targeting Notch1, Notch2, and Jag1. A: Putative miR-34a binding sites in the 3′untranslated region (UTR) of Notch1, Jag1, and Notch2 mRNA. 3′UTR fragments of Notch1, Jag1, and Notch2 containing wild-type or mutated (three mutated nucleotides are in bold) miR-34a binding site. B: Western blot analysis for Notch1 (transmembrane/intracellular region: notch transmembrane region), Notch2, and Jag1 in CCLP1 and SG231 cells transfected with miR-34a mimic or scramble control (Jag1 protein was not detectable in CCLP1 cells). C: Relative Notch1, Jag1, and Notch2 mRNA expression in CCLP1 and SG231 cells transfected with miR-34a mimic or scramble control. D: Western blot and quantitative RT-PCR analysis for Notch1, Jag1, and Notch 2 in control (pEZX-CN) and miR-34a–overexpressed (pEZX-miR34a) SG231 cells. E: Western blot analysis for Notch1, Jag1, and Notch2 in TFK1 cells transfected with anti–miR-34a and scramble control. F: Luciferase reporter activity in SG231 cells transfected with wild-type (WT) or mutant (Mut) Notch1, Jag1, or Notch2 3′UTR plasmids and treated with miR-34a mimic or scramble control miRNA. Data are expressed as means ± SEM (C and D); data are expressed as means ± SD (F). ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001 versus control mimic or plasmid transfected cells. Anti-NC, antisense miRNA negative control; ctrl, control.
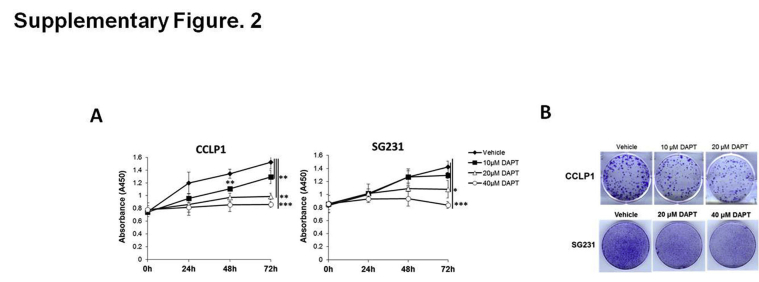
To validate miR-34a targeting of Notch1, Jag1, and Notch2 in CCA cells, we transfected SG231 cells with wild-type or mutant Notch1, Jag1, or Notch2 3′UTR luciferase reporter constructs; the transfected cells then were treated with miR-34a mimic or scramble control miRNA to determine luciferase reporter activity. Our data showed that miR-34a mimic treatment significantly decreased Notch1, Jag1, and Notch2 3′UTR luciferase reporter activities and that the effects were abolished by mutation of the miR-34a binding site in the respective 3′UTRs (Figure 2F). These findings established Notch1, Jag1, and Notch2 as direct targets of miR-34a in human CCA cells. The role of Notch signaling in human CCA cells used in our system was confirmed by the observation that treatment with the γ-secretase inhibitor, DAPT (which prevents proteolytic cleavage of the Notch intracellular domain), inhibited the growth and colony formation of CCLP1 and SG231 cells (Supplemental Figure S2).
Overexpression of miR-34a Inhibits CCA Growth in Vivo
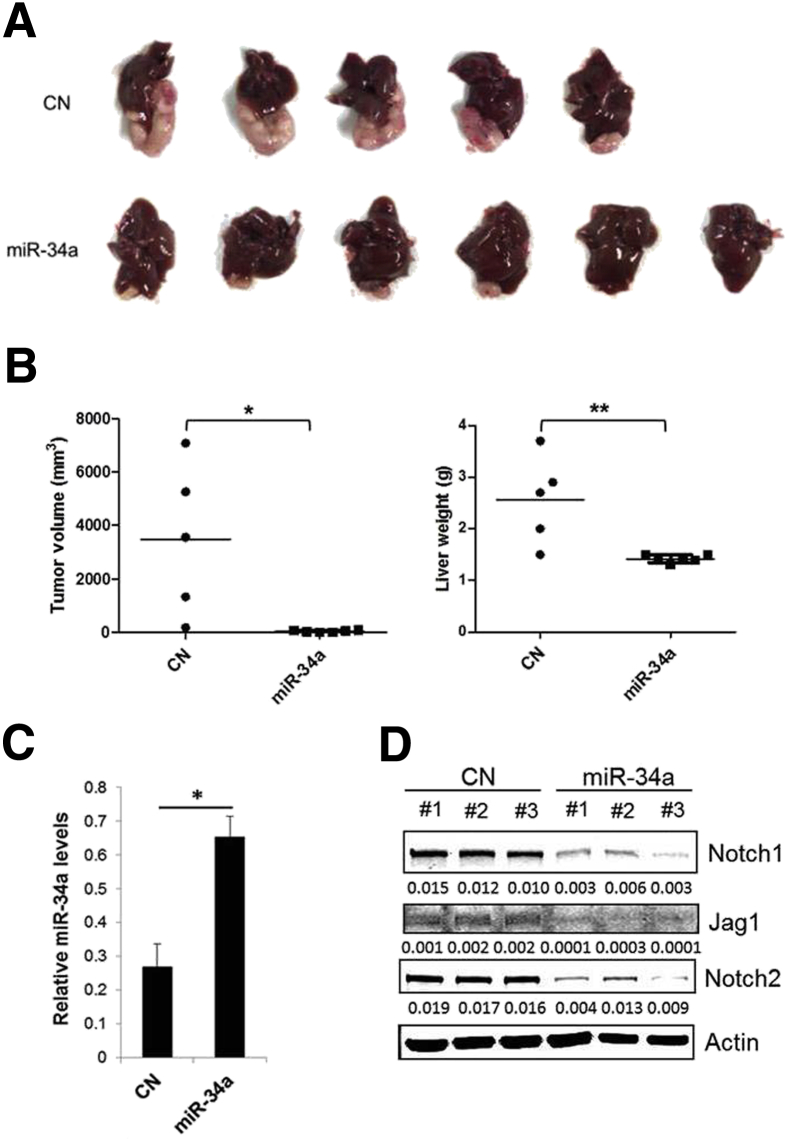
Given the antitumor effect of miR-34a on CCA cells in vitro, we further examined whether overexpression of miR-34a would inhibit tumor growth in a CCA xenograft model. To this end, SG231 cells stably expressing miR-34a or scramble control miRNA were inoculated directly into the livers of SCID mice and the animals were observed for 6 weeks to monitor tumor growth. The miR-34a–overexpressed cells formed smaller-sized tumors in the liver compared with the scramble control cells (Figure 3A). The miR-34a–overexpressed tumors showed significantly decreased tumor volume, and hence decreased liver weight, compared with the scramble control tumors (P < 0.05 and P < 0.01) (Figure 3B). No intrahepatic metastasis was observed in the livers injected with either control or miR-34a–overexpressed cells. An increased miR-34a level in miR-34a–overexpressed tumors was confirmed by RT-qPCR analysis (Figure 3C). We observed that the miR-34a–overexpressed tumors showed reduced expression of Notch1, Jag1, and Notch2, as determined by Western blot analysis (Figure 3D). Thus far, our data provide novel evidence that miR-34a suppresses CCA growth by targeting the Notch pathway in vitro and in vivo.
Figure 3.
Overexpression of miR-34a inhibits cholangiocarcinoma growth in vivo. SG231 cells stably expressing miR-34a or scramble control miRNA (1.0 × 106) were mixed in Matrigel solution and inoculated directly into the livers of severe combined immunodeficiency (SCID) mice (intrahepatic injection). We initially started with six mice per group, but one mouse from the control group died during the injection. After 6 weeks, the mice were sacrificed and monitored for tumor growth. A: Representative images of xenograft tumors recovered from five mice for control and six mice for miR-34a–overexpressing cells. B: Tumor volume and liver weight. C: The levels of miR-34a in xenograft tumor tissues as determined by quantitative RT-PCR. Data are expressed as means ± SEM. D: Western blot analysis for Notch1, Jag1, and Notch2 in control and miR-34a–overexpressed tumors. ∗P < 0.05, ∗∗P < 0.01. CN, scramble control.
EZH2 Epigenetically Silences miR-34a Expression through H3K27 Trimethylation in CCA Cells
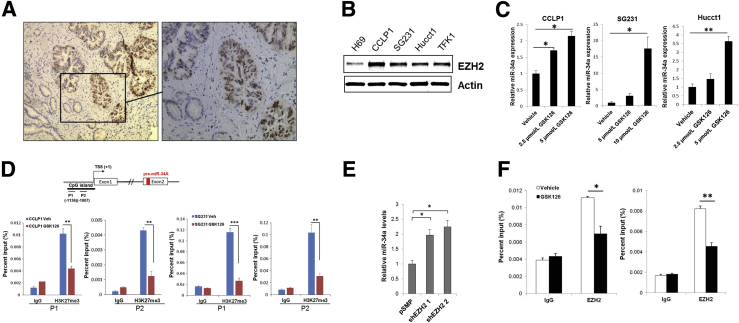
We next explored the mechanism that mediates down-regulation of miR-34a in CCA cells. Intriguingly, a recent study reported that H3K27 trimethylation (H3K27me3) is enriched in the promoter region of the miR-34a gene.34 Given that H3K27 trimethylation is mediated by the histone methyltransferase EZH2, we evaluated the expression of EZH2 in human CCA tissues by immunohistochemical staining. Remarkably, we observed that EZH2 is expressed highly in human CCA tissues compared with the non-neoplastic peribiliary glands (Figure 4A). Nuclear expression of EZH2 was observed in 12 of 15 cases of human CCA (80%), although it was not observed in the non-neoplastic peribiliary glands in all 15 cases (P < 0.01) (Table 2). Accordingly, increased EZH2 expression also was observed in human CCA cell lines (CCLP1, HUCCT1, SG231, and TFK1) compared with the non-neoplastic human cholangiocytes (H69) (Figure 4B). To determine whether EZH2 may contribute to the suppression of miR-34a via H3K27 trimethylation, we assessed the effect of GSK126, a selective EZH2 inhibitor, on miR-34a expression. We observed that GSK126 treatment reduced the global level of H3K27me3, without alteration of EZH2 expression (Supplemental Figure S3A). Notably, GSK126 treatment led to a significant increase in miR-34a expression in CCLP1, SG231, and HUCCT1 cells (Figure 4C). Moreover, ChIP–quantitative PCR assay with an anti-H3K27me3 antibody showed that the enrichment of H3K27me3 at the pre-miR-34a gene promoter region (P1; -1136 bp, P2; -1007 bp) was decreased significantly by GSK126 in both CCLP1 and SG231 cells (Figure 4D). These findings support the regulation of miR-34a expression by EZH2-mediated H3K27me3. Consistent with the findings in GSK126-treated cells, knockdown of EZH2 with two different shRNAs resulted in an increase of miR-34a levels (Figure 4E and Supplemental Figure S3B). ChIP–quantitative PCR with an anti-EZH2 antibody showed that GSK126 treatment significantly reduced the association of EZH2 with the pre–miR-34a gene promoter (Figure 4F). Collectively, these data provide novel evidence that EZH2 epigenetically represses miR-34a expression through H3K27 trimethylation in CCA cells.
Figure 4.
EZH2 represses miR-34a expression through H3K27 trimethylation in cholangiocarcinoma (CCA) cells. A: Representative immunohistochemistry for EZH2 in human CCA tissue. The brown color indicates positive signals; nuclei were counterstained as blue. The boxed area in the left panel is shown at higher magnification in the right panel. B: Western blot analysis for EZH2 in nonmalignant human cholangiocyte cell (H69) and CCA cells (CCLP1, SG231, HUCCT1, and TFK1). C: The levels of miR-34a in CCA cells with/without GSK126 treatment for 72 hours, as determined by quantitative RT-PCR (RT-qPCR). D: Chromatin immunoprecipitation (ChIP) assay. The chromatin extracted from CCLP1 and SG231 cells treated with or without GSK126 was subjected to immunoprecipitation with H3K27me3 antibody, and the precipitated DNA was subjected to RT-qPCR analysis using two sets of primers to amplify the miR-34a promoter region, as shown in the schematic diagram. Normal mouse IgG was used as the negative control. E: RT-qPCR analysis for miR-34a in SG231 cells transfected with two individual EZH2 shRNA or control vector (pSMP). F: ChIP assay with EZH2 antibody followed by RT-qPCR analysis in SG231 cells with or without GSK126 treatment. Data are expressed as means ± SD (D and F); data are expressed as means ± SEM (C and E). ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001. Original magnification: ×100 (A, left panel); ×200 (A, right panel). TSS, transcription start site.
Table 2.
Graphic Presentation of EZH2 Staining Intensity in Human Cholangiocarcinoma Tissue
| Staining intensity | Non-neoplastic, n (%) | Cholangiocarcinoma, n (%) |
|---|---|---|
| 0 | 15/15 (100) | 2/15 (13.3) |
| 0.5 | 0/0 (0) | 5/15 (33.3) |
| 1 | 0/0 (0) | 2/15 (13.3) |
| 2 | 0/0 (0) | 2/15 (13.3) |
| 3 | 0/0 (0) | 4/15 (26.7) |
n = 15. P < 0.01 (Wilcoxon signed-rank test).
The EZH2 Inhibitor, GSK126, Inhibits CCA Cell Growth and Metastasis in Vivo
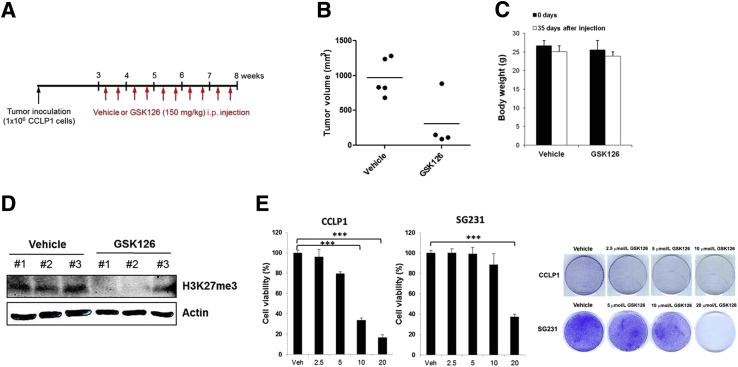
Our data presented thus far indicate that pharmacologic and genetic inhibition of EZH2 in CCA cells restores miR-34a expression through H3K27 trimethylation. We next used a mouse model of CCA to assess the therapeutic efficacy of EZH2 inhibition on CCA progression in vivo. For this purpose, CCLP1 cells mixed with Matrigel solution were injected directly into the livers of SCID mice and the animals were observed for 3 weeks to allow tumor establishment. The mice then were assigned randomly to two groups and treated with 150 mg/kg GSK126 or vehicle control (20% Captisol) by intraperitoneal injection twice a week for an additional 5 weeks (Figure 5A). We observed that GSK126 treatment decreased the growth of inoculated CCA cells. The average tumor volume of GSK126-treated mice is lower than that of vehicle-treated mice (Figure 5B). Notably, GSK126 treatment significantly reduced intrahepatic metastasis [which was defined by visible tumor nodules (>1 mm) in the uninjected liver lobes] (Figure 5B). The body weight of the mice did not differ significantly between the vehicle- and GSK126-treated groups, indicating that GSK126 treatment was tolerated in those animals (Figure 5C). The liver tumor tissues from the GSK126-treated group appeared to have decreased H3K27 trimethylation compared with the tumor tissues from the vehicle-treated group (Figure 5D). We observed that the effect of GSK126 in mice somewhat varied depending on the individual animals.
Figure 5.
Effect of EZH2 inhibitor, GSK126, on cholangiocarcinoma cell growth in vivo and in vitro. A: Experimental design of tumor xenograft and GSK126 intraperitoneal injection in SCID mice. CCLP1 cells (1.0 × 106) were mixed in Matrigel solution and inoculated into severe combined immunodeficiency (SCID) mice (initially started with six mice per group) via intrahepatic injection and the mice were treated with vehicle (20% Captisol) or 150 mg/kg GSK126 twice a week for 5 weeks. B: Tumor volume of vehicle and GSK126-treated mice. After injection of CCLP1 cells into the left liver lobe in the mice, intrahepatic metastasis was determined at 8 weeks by observation of the tumor nodule on the uninjected liver lobes. All of the vehicle-treated mice (five mice) had intrahepatic metastasis, whereas one of four GSK126-treated mice had intrahepatic metastasis (P < 0.05, Fisher exact test). C: Body weight changes of the mice. D: The level of trimethylated H3K27 in liver tumor tissues from mice treated with vehicle or GSK126 (only three liver tumor samples were available for Western blot because of the very small tumor size in the GSK126-treated group). E: The growth of CCLP1 and SG231 cells was determined by WST1 assay after treatment of GSK126 or vehicle for 72 hours. Colony-forming assay in CCLP1 and SG231 cells with or without GSK126 treatment for 10 days. Data are expressed as means ± SD. ∗∗∗P < 0.001. Veh, vehicle.
We also evaluated the effect of GSK126 on CCA cell growth in vitro. CCLP1 and SG231 cells were treated with different doses of GSK126 and the cells were analyzed for proliferation and colony formation. We observed that treatment with GSK126 significantly reduced CCA cell proliferation and colony formation (Figure 5E). Taken together, our results suggest that EZH2 inhibition decreased CCA growth and metastasis, at least in part, through epigenetic activation of miR-34a.
MiR-34a Expression Also Is Regulated by CpG Methylation
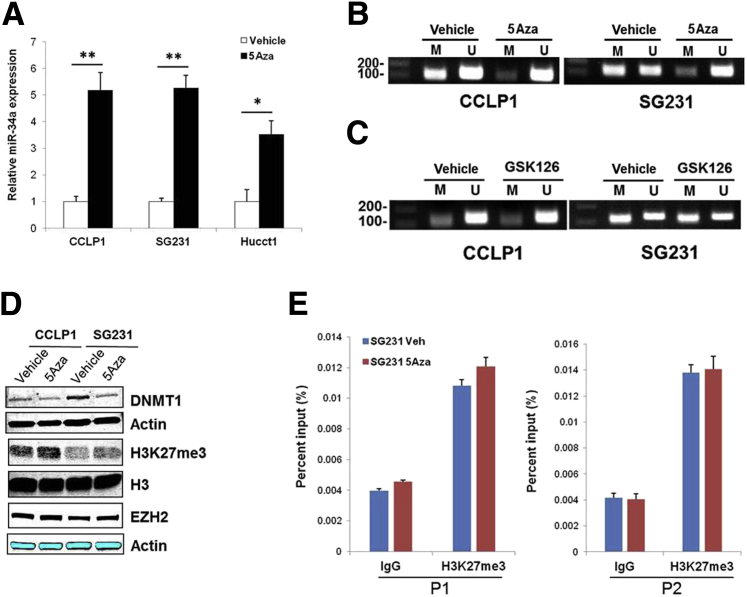
Previous studies have reported that miR-34a gene promoter contains CpG islands and its expression is silenced by aberrant CpG methylation in several types of cancer.35, 36 Thus, we further examined whether CpG methylation also might contribute to miR-34a down-regulation in CCA cells, in addition to EZH2-mediated H3K27 trimethylation. Specifically, the CCLP1, SG231, and HUCCT1 cells were treated with 5-Aza-CdR, an inhibitor of DNA methyltransferases, and miR-34a levels were determined by RT-qPCR analysis. miR-34a expression was increased significantly by 5-Aza-CdR treatment compared with vehicle control (Figure 6A). Moreover, MSP was performed using primers specific for methylated or unmethylated sites in CpG islands of miR-34a promoter (Table 1). The MSP analysis showed that 5-Aza-CdR treatment caused demethylation of the miR-34a promoter region, suggesting regulation of miR-34a expression by DNA methylation (Figure 6B).
Figure 6.
EZH2-mediated H3K27 trimethylation and DNA methylation independently silence miR-34a expression. A: Relative miR-34a expression in cholangiocarcinoma (CCA) cells treated with 3 μmol/L 5-Aza-2′-deoxycytidine (5-Aza-CdR) or vehicle for 72 hours, as determined by quantitative RT-PCR (RT-qPCR) analysis. B: Methylation-specific PCR (MSP) of genomic DNA from CCLP1 and SG231 cells treated with or without 3 μmol/L 5-Aza-CdR treatment. C: MSP analysis in CCLP1 and SG231 cells treated with 2.5 or 10 μmol/L GSK126. D: Western blot analysis for DNMT1, H3K27 trimethylation (H3K27me3), and EZH2 in CCLP1 and SG231 cells with or without GSK126 treatment. E: Chromatin immunoprecipitation (ChIP) assay. The chromatin extracted from SG231 cells treated with 5-Aza-CdR or vehicle was subjected to immunoprecipitation with H3K27me3 antibody, and the precipitated DNA was subjected to RT-qPCR analysis using two sets of primers to amplify the miR-34a promoter region. Data are expressed as means ± SEM. ∗P < 0.05, ∗∗P < 0.01. 5Aza, 5-Aza-CdR; M, methylation-specific primer to the miR-34a promoter sequence; U, unmethylation-specific primer to the miR-34a promoter sequence.
Our data presented hitherto suggest that repression of miR-34a in CCA cells is associated with both EZH2-mediated H3K27me3 and DNA methylation. Because histone and DNA methylation are known to control the epigenetic mechanism of gene silencing cooperatively,37, 38 we further explored whether EZH2-dependent H3K27 trimethylation and DNA methylation may regulate miR-34a expression independently or coordinately. We treated CCA cells with the EZH2 inhibitor GSK126 and the cells were analyzed for CpG methylation of the miR-34a gene promoter by MSP analysis. Our data showed that the methylation status of CpG islands in the miR-34a gene promoter was not altered by GSK126 treatment (Figure 6C). On the other hand, we observed that although 5-Aza-CdR treatment reduced DNMT1 expression, it did not alter the levels of H3K27me3 and EZH2 in CCA cells (Figure 6D). ChIP assay using an anti-H3K27me3 antibody showed enhancement of H3K27me3 at the miR-34a promoter region (P1, -1136 bp; P2, -1007 bp); however, such H3K27me3 enrichment was unaffected by 5-Aza-CdR treatment (Figure 6E). Therefore, EZH2-mediated H3K27me3 and DNA methylation appear to silence miR-34a expression epigenetically through independent mechanisms in human CCA cells.
Discussion
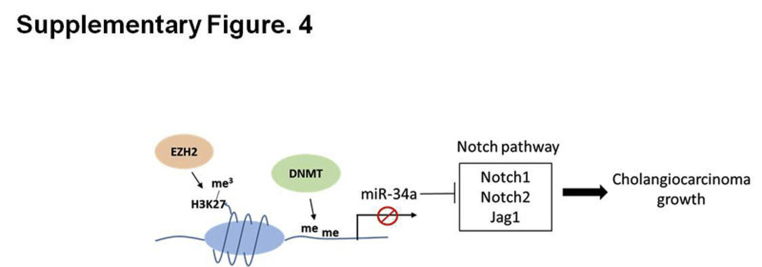
The present study provides novel evidence that miR-34a is silenced epigenetically by EZH2-mediated H3K27 trimethylation and DNA methylation in human CCA cells. Our data show that miR-34a is an important tumor-suppressive miRNA that effectively inhibits CCA cell growth in vitro and in vivo through targeting the Notch pathway receptors and ligand. We observed that pharmacologic inhibition of the Notch pathway recapitulates the effect of miR-34a on CCA growth. Moreover, we show that EZH2 is highly expressed in human CCA tissues compared with non-neoplastic peribiliary glands, and that inhibition of EZH2 restored miR-34a expression and thus inhibits CCA growth and intrahepatic metastasis. Our data presented in this article show a novel epigenetic mechanism that silences miR-34a expression in CCA cells and identifies Notch signaling as a direct target of miR-34a (Supplemental Figure S4). Our findings provide important preclinical evidence for targeting miR-34a and related molecules (eg, miR-34a mimic and EZH2/Notch inhibitors) for future treatment of human CCA.
Deregulation of miRNAs, which can be caused by genetic/epigenetic modification and transcriptional regulation, has been involved in cancer development and progression.24 A number of tumor-suppressive miRNAs that are silenced aberrantly are known to prevent cancer development/progression by negatively regulating proto-oncogenes and/or genes involved in apoptosis or cell differentiation.39, 40 Among these miRNAs, miR-34a expression is reported to be inactivated by aberrant DNA methylation and this is implicated in tumor suppression,35, 41 although the regulation and function of miR-34a in CCA has not been fully understood. Our data in this study show that miR-34a is silenced epigenetically by EZH2-mediated H3K27me3 and DNA methylation in human CCA cells. Consistent with these findings, we observed that restoration of miR-34a in human CCA cells led to inhibition of cell proliferation and colony formation in vitro. Furthermore, our data show that miR-34a significantly decreased CCA growth in SCID mice. These findings suggest that miR-34a replacement therapy may represent an effective strategy for CCA treatment.
Epigenetic silencing of miR-34a has been found in several types of cancer such as breast, kidney, lung, and prostate cancers, as a result of aberrant DNA methylation of its promoter region.35, 42, 43, 44 Consistent with previous studies, our findings in the current article showed that treatment of the DNA demethylating agent 5-Aza-CdR caused demethylation of the miR-34a gene promoter region (as determined by methylation-specific PCR) and restored miR-34a expression in CCA cells.34, 36 EZH2 is a histone methyltransferase that mediates epigenetic gene silencing through H3K27 trimethylation. Accumulating studies have identified several miRNAs that are repressed by EZH2 in cancers.45, 46, 47 Here, we show that EZH2 epigenetically silences miR-34a expression by promoting H3K27 trimethylation, and that inhibition of EZH2 up-regulated miR-34a expression in CCA cells. Although DNA methylation and H3K27 trimethylation have been linked to establish and maintain gene silencing,38 our data showed no apparent epigenetic cross-talk between DNA methylation and H3K27 trimethylation in silencing of miR-34a expression in CCA.
Given the evidence for oncogenic functions of EZH2 in many types of cancer, targeting EZH2 with specific inhibitors has been considered a compelling anticancer therapy.26, 48 Because the expression of EZH2 is increased markedly in human CCA tissue and cells as shown in the current study, we have evaluated the antitumor effect of EZH2 inhibitor, GSK126, on CCA growth in vitro and in vivo. Notably, treatment of CCA cells with GSK126 significantly decreased tumor cell growth and colony formation in vitro. Intraperitoneal injection of GSK126 in tumor-bearing SCID mice also inhibited tumor growth and intrahepatic metastasis in SCID mice. Based on our observation that GSK126 restored miR-34a expression in CCA cells, inhibition of EZH2 may suppress CCA progression at least partially through up-regulation of miR-34a.
Compelling studies have documented the oncogenic roles of the Notch pathway in the development and progression of CCA.13, 14, 15, 16, 17, 18, 19 However, treatment with anti-Notch receptors/ligands antibodies or γ-secretase inhibitor has shown variable effects on tumor growth in animal models of cholangiocarcinogenesis.15, 16, 49, 50 In the current study, we identified Notch1, Jag1, and Notch2 as target genes of miR-34a in human CCA cells. We observed that inhibition of the Notch pathway by either miR-34a overexpression or γ-secretase inhibitor (DAPT) significantly reduced human CCA cell growth and clonogenic capacity. These findings not only validate the role of miR-34a–regulated Notch signaling in cholangiocarcinogenesis, but also suggest an intriguing possibility of targeting miR-34a in combination with anti-Notch therapies for effective treatment of human CCA.
In conclusion, the current study establishes miR-34a as an important tumor suppressor in human CCA. Our findings disclose a novel epigenetic regulatory mechanism that mediates miR-34a gene silencing. The functional and mechanistic studies described in this study show that miR-34a inhibits CCA growth by targeting the Notch pathway. Our findings suggest that restoration of endogenous miR-34a expression by targeting epigenetic machinery (such as the EZH2 inhibitor) or delivery of exogenous miR-34a mimic may represent novel therapeutic strategies for the treatment of human CCA.
Footnotes
Supported by NIH grant R01CA102325 (T.W.).
Disclosures: None declared.
Supplemental material for this article can be found at http://dx.doi.org/10.1016/j.ajpath.2017.06.014.
Supplemental Data
Supplementary Figure S1.
MiR-34a levels in cholangiocarcinoma cells transfected with miR-34a mimic/inhibitor or scramble control. A: The expression of miR-34a in CCLP1 and SG231 cells transfected with miR-34a mimic or scramble control. B: The levels of miR-34a in TFK1 cells transfected with miR-34a inhibitor or scramble control. The miR-34a level was measured by quantitative RT-PCR analysis. RNA U6 small nuclear 2 was used as internal control. Data are expressed as means ± SEM. ∗P < 0.05, ∗∗∗P < 0.001 versus ctrl-mimic transfected cells; †P < 0.05 versus anti-NC. Anti-NC, antisense-negative control; Ctrl, control.
Supplementary Figure S2.
Inhibition of Notch pathway decreases cholangiocarcinoma growth and colony formation. A: The growth of cells was measured by WST1 assay after DAPT or vehicle treatment at 24, 48, and 72 hours. B: Colony-forming assay in CCLP1 and SG231 cells with/without DAPT treatment for 10 days. Data are expressed as means ± SD. ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001.
Supplementary Figure S3.
Effect of EZH2 inhibition on H3K27me and EZH2 levels. A: Western blot for H3K27me and EZH2 in CCA cells treated with vehicle or GSK126. B: SG231 cells were transfected with two individual EZH2 shRNAs or control vector (pSMP) for 48 hours and then EZH2 mRNA levels were determined by quantitative RT-PCR analysis. Actin was used as an internal control. Data are expressed as means ± SEM. ∗P < 0.05.
Supplementary Figure S4.
Schematic diagram of the mechanism involving epigenetic silencing of miR-34a and its impact on cholangiocarcinoma growth. me, methylation.
References
- 1.Bridgewater J., Galle P.R., Khan S.A., Llovet J.M., Park J.W., Patel T., Pawlik T.M., Gores G.J. Guidelines for the diagnosis and management of intrahepatic cholangiocarcinoma. J Hepatol. 2014;60:1268–1289. doi: 10.1016/j.jhep.2014.01.021. [DOI] [PubMed] [Google Scholar]
- 2.Razumilava N., Gores G.J. Cholangiocarcinoma. Lancet. 2014;383:2168–2179. doi: 10.1016/S0140-6736(13)61903-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rizvi S., Gores G.J. Pathogenesis, diagnosis, and management of cholangiocarcinoma. Gastroenterology. 2013;145:1215–1229. doi: 10.1053/j.gastro.2013.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Patel T. Cholangiocarcinoma–controversies and challenges. Nat Rev Gastroenterol Hepatol. 2011;8:189–200. doi: 10.1038/nrgastro.2011.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gatto M., Bragazzi M.C., Semeraro R., Napoli C., Gentile R., Torrice A., Gaudio E., Alvaro D. Cholangiocarcinoma: update and future perspectives. Dig Liver Dis. 2010;42:253–260. doi: 10.1016/j.dld.2009.12.008. [DOI] [PubMed] [Google Scholar]
- 6.Khan S.A., Davidson B.R., Goldin R.D., Heaton N., Karani J., Pereira S.P., Rosenberg W.M., Tait P., Taylor-Robinson S.D., Thillainayagam A.V., Thomas H.C., Wasan H. Guidelines for the diagnosis and treatment of cholangiocarcinoma: an update. Gut. 2012;61:1657–1669. doi: 10.1136/gutjnl-2011-301748. [DOI] [PubMed] [Google Scholar]
- 7.Francis H., Alpini G., DeMorrow S. Recent advances in the regulation of cholangiocarcinoma growth. Am J Physiol Gastrointest Liver Physiol. 2010;299:G1–G9. doi: 10.1152/ajpgi.00114.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Everhart J.E., Ruhl C.E. Burden of digestive diseases in the United States part III: liver, biliary tract, and pancreas. Gastroenterology. 2009;136:1134–1144. doi: 10.1053/j.gastro.2009.02.038. [DOI] [PubMed] [Google Scholar]
- 9.Tyson G.L., El-Serag H.B. Risk factors for cholangiocarcinoma. Hepatology. 2011;54:173–184. doi: 10.1002/hep.24351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sirica A.E., Dumur C.I., Campbell D.J., Almenara J.A., Ogunwobi O.O., Dewitt J.L. Intrahepatic cholangiocarcinoma progression: prognostic factors and basic mechanisms. Clin Gastroenterol Hepatol. 2009;7:S68–S78. doi: 10.1016/j.cgh.2009.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Palmer W.C., Patel T. Are common factors involved in the pathogenesis of primary liver cancers? A meta-analysis of risk factors for intrahepatic cholangiocarcinoma. J Hepatol. 2012;57:69–76. doi: 10.1016/j.jhep.2012.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sia D., Tovar V., Moeini A., Llovet J.M. Intrahepatic cholangiocarcinoma: pathogenesis and rationale for molecular therapies. Oncogene. 2013;32:4861–4870. doi: 10.1038/onc.2012.617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Geisler F., Strazzabosco M. Emerging roles of Notch signaling in liver disease. Hepatology. 2015;61:382–392. doi: 10.1002/hep.27268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Morell C.M., Strazzabosco M. Notch signaling and new therapeutic options in liver disease. J Hepatol. 2014;60:885–890. doi: 10.1016/j.jhep.2013.11.028. [DOI] [PubMed] [Google Scholar]
- 15.Zender S., Nickeleit I., Wuestefeld T., Sorensen I., Dauch D., Bozko P., El-Khatib M., Geffers R., Bektas H., Manns M.P., Gossler A., Wilkens L., Plentz R., Zender L., Malek N.P. A critical role for notch signaling in the formation of cholangiocellular carcinomas. Cancer Cell. 2013;23:784–795. doi: 10.1016/j.ccr.2013.04.019. [DOI] [PubMed] [Google Scholar]
- 16.Huntzicker E.G., Hotzel K., Choy L., Che L., Ross J., Pau G., Sharma N., Siebel C.W., Chen X., French D.M. Differential effects of targeting Notch receptors in a mouse model of liver cancer. Hepatology. 2015;61:942–952. doi: 10.1002/hep.27566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sekiya S., Suzuki A. Intrahepatic cholangiocarcinoma can arise from Notch-mediated conversion of hepatocytes. J Clin Invest. 2012;122:3914–3918. doi: 10.1172/JCI63065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dill M.T., Tornillo L., Fritzius T., Terracciano L., Semela D., Bettler B., Heim M.H., Tchorz J.S. Constitutive Notch2 signaling induces hepatic tumors in mice. Hepatology. 2013;57:1607–1619. doi: 10.1002/hep.26165. [DOI] [PubMed] [Google Scholar]
- 19.Razumilava N., Gores G.J. Notch-driven carcinogenesis: the merging of hepatocellular cancer and cholangiocarcinoma into a common molecular liver cancer subtype. J Hepatol. 2013;58:1244–1245. doi: 10.1016/j.jhep.2013.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ishimura N., Bronk S.F., Gores G.J. Inducible nitric oxide synthase up-regulates Notch-1 in mouse cholangiocytes: implications for carcinogenesis. Gastroenterology. 2005;128:1354–1368. doi: 10.1053/j.gastro.2005.01.055. [DOI] [PubMed] [Google Scholar]
- 21.Fabian M.R., Sonenberg N., Filipowicz W. Regulation of mRNA translation and stability by microRNAs. Annu Rev Biochem. 2010;79:351–379. doi: 10.1146/annurev-biochem-060308-103103. [DOI] [PubMed] [Google Scholar]
- 22.Filipowicz W., Bhattacharyya S.N., Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet. 2008;9:102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- 23.Garofalo M., Croce C.M. microRNAs: master regulators as potential therapeutics in cancer. Annu Rev Pharmacol Toxicol. 2011;51:25–43. doi: 10.1146/annurev-pharmtox-010510-100517. [DOI] [PubMed] [Google Scholar]
- 24.Croce C.M. Causes and consequences of microRNA dysregulation in cancer. Nat Rev Genet. 2009;10:704–714. doi: 10.1038/nrg2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pisarello M.J., Loarca L., Ivanics T., Morton L., LaRusso N. MicroRNAs in the cholangiopathies: pathogenesis, diagnosis, and treatment. J Clin Med. 2015;4:1688–1712. doi: 10.3390/jcm4091688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim K.H., Roberts C.W. Targeting EZH2 in cancer. Nat Med. 2016;22:128–134. doi: 10.1038/nm.4036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kondo Y. Targeting histone methyltransferase EZH2 as cancer treatment. J Biochem. 2014;156:249–257. doi: 10.1093/jb/mvu054. [DOI] [PubMed] [Google Scholar]
- 28.Varambally S., Dhanasekaran S.M., Zhou M., Barrette T.R., Kumar-Sinha C., Sanda M.G., Ghosh D., Pienta K.J., Sewalt R.G., Otte A.P., Rubin M.A., Chinnaiyan A.M. The polycomb group protein EZH2 is involved in progression of prostate cancer. Nature. 2002;419:624–629. doi: 10.1038/nature01075. [DOI] [PubMed] [Google Scholar]
- 29.Kleer C.G., Cao Q., Varambally S., Shen R., Ota I., Tomlins S.A., Ghosh D., Sewalt R.G., Otte A.P., Hayes D.F., Sabel M.S., Livant D., Weiss S.J., Rubin M.A., Chinnaiyan A.M. EZH2 is a marker of aggressive breast cancer and promotes neoplastic transformation of breast epithelial cells. Proc Natl Acad Sci U S A. 2003;100:11606–11611. doi: 10.1073/pnas.1933744100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bachmann I.M., Halvorsen O.J., Collett K., Stefansson I.M., Straume O., Haukaas S.A., Salvesen H.B., Otte A.P., Akslen L.A. EZH2 expression is associated with high proliferation rate and aggressive tumor subgroups in cutaneous melanoma and cancers of the endometrium, prostate, and breast. J Clin Oncol. 2006;24:268–273. doi: 10.1200/JCO.2005.01.5180. [DOI] [PubMed] [Google Scholar]
- 31.Sasaki M., Ikeda H., Itatsu K., Yamaguchi J., Sawada S., Minato H., Ohta T., Nakanuma Y. The overexpression of polycomb group proteins Bmi1 and EZH2 is associated with the progression and aggressive biological behavior of hepatocellular carcinoma. Lab Invest. 2008;88:873–882. doi: 10.1038/labinvest.2008.52. [DOI] [PubMed] [Google Scholar]
- 32.Zhang J., Han C., Wu T. MicroRNA-26a promotes cholangiocarcinoma growth by activating beta-catenin. Gastroenterology. 2012;143:246–256.e8. doi: 10.1053/j.gastro.2012.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Song K., Han C., Zhang J., Lu D., Dash S., Feitelson M., Lim K., Wu T. Epigenetic regulation of microRNA-122 by peroxisome proliferator activated receptor-gamma and hepatitis b virus X protein in hepatocellular carcinoma cells. Hepatology. 2013;58:1681–1692. doi: 10.1002/hep.26514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu Y.W., Sun M., Xia R., Zhang E.B., Liu X.H., Zhang Z.H., Xu T.P., De W., Liu B.R., Wang Z.X. LincHOTAIR epigenetically silences miR34a by binding to PRC2 to promote the epithelial-to-mesenchymal transition in human gastric cancer. Cell Death Dis. 2015;6:e1802. doi: 10.1038/cddis.2015.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lodygin D., Tarasov V., Epanchintsev A., Berking C., Knyazeva T., Korner H., Knyazev P., Diebold J., Hermeking H. Inactivation of miR-34a by aberrant CpG methylation in multiple types of cancer. Cell Cycle. 2008;7:2591–2600. doi: 10.4161/cc.7.16.6533. [DOI] [PubMed] [Google Scholar]
- 36.Vogt M., Munding J., Gruner M., Liffers S.T., Verdoodt B., Hauk J., Steinstraesser L., Tannapfel A., Hermeking H. Frequent concomitant inactivation of miR-34a and miR-34b/c by CpG methylation in colorectal, pancreatic, mammary, ovarian, urothelial, and renal cell carcinomas and soft tissue sarcomas. Virchows Arch. 2011;458:313–322. doi: 10.1007/s00428-010-1030-5. [DOI] [PubMed] [Google Scholar]
- 37.Cedar H., Bergman Y. Linking DNA methylation and histone modification: patterns and paradigms. Nat Rev Genet. 2009;10:295–304. doi: 10.1038/nrg2540. [DOI] [PubMed] [Google Scholar]
- 38.Vire E., Brenner C., Deplus R., Blanchon L., Fraga M., Didelot C., Morey L., Van Eynde A., Bernard D., Vanderwinden J.M., Bollen M., Esteller M., Di Croce L., de Launoit Y., Fuks F. The polycomb group protein EZH2 directly controls DNA methylation. Nature. 2006;439:871–874. doi: 10.1038/nature04431. [DOI] [PubMed] [Google Scholar]
- 39.Zhang B., Pan X., Cobb G.P., Anderson T.A. microRNAs as oncogenes and tumor suppressors. Dev Biol. 2007;302:1–12. doi: 10.1016/j.ydbio.2006.08.028. [DOI] [PubMed] [Google Scholar]
- 40.Hammond S.M. MicroRNAs as tumor suppressors. Nat Genet. 2007;39:582–583. doi: 10.1038/ng0507-582. [DOI] [PubMed] [Google Scholar]
- 41.Misso G., Di Martino M.T., De Rosa G., Farooqi A.A., Lombardi A., Campani V., Zarone M.R., Gulla A., Tagliaferri P., Tassone P., Caraglia M. Mir-34: a new weapon against cancer? Mol Ther Nucleic Acids. 2014;3:e194. doi: 10.1038/mtna.2014.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shen Z., Zhou C., Li J., Ye D., Li Q., Wang J., Cui X., Chen X., Bao T., Duan S. Promoter hypermethylation of miR-34a contributes to the risk, progression, metastasis and poor survival of laryngeal squamous cell carcinoma. Gene. 2016;593:272–276. doi: 10.1016/j.gene.2016.07.047. [DOI] [PubMed] [Google Scholar]
- 43.Xie K., Liu J., Chen J., Dong J., Ma H., Liu Y., Hu Z. Methylation-associated silencing of microRNA-34b in hepatocellular carcinoma cancer. Gene. 2014;543:101–107. doi: 10.1016/j.gene.2014.03.059. [DOI] [PubMed] [Google Scholar]
- 44.Cui X., Zhao Z., Liu D., Guo T., Li S., Hu J., Liu C., Yang L., Cao Y., Jiang J., Liang W., Liu W., Li S., Wang L., Wang L., Gu W., Wu C., Chen Y., Li F. Inactivation of miR-34a by aberrant CpG methylation in Kazakh patients with esophageal carcinoma. J Exp Clin Cancer Res. 2014;33:20. doi: 10.1186/1756-9966-33-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cao Q., Mani R.S., Ateeq B., Dhanasekaran S.M., Asangani I.A., Prensner J.R., Kim J.H., Brenner J.C., Jing X., Cao X., Wang R., Li Y., Dahiya A., Wang L., Pandhi M., Lonigro R.J., Wu Y.M., Tomlins S.A., Palanisamy N., Qin Z., Yu J., Maher C.A., Varambally S., Chinnaiyan A.M. Coordinated regulation of polycomb group complexes through microRNAs in cancer. Cancer Cell. 2011;20:187–199. doi: 10.1016/j.ccr.2011.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang L., Zhang X., Jia L.T., Hu S.J., Zhao J., Yang J.D., Wen W.H., Wang Z., Wang T., Zhao J., Wang R.A., Meng Y.L., Nie Y.Z., Dou K.F., Chen S.Y., Yao L.B., Fan D.M., Zhang R., Yang A.G. c-Myc-mediated epigenetic silencing of microRNA-101 contributes to dysregulation of multiple pathways in hepatocellular carcinoma. Hepatology. 2014;59:1850–1863. doi: 10.1002/hep.26720. [DOI] [PubMed] [Google Scholar]
- 47.Wang Y., Wang M., Wei W., Han D., Chen X., Hu Q., Yu T., Liu N., You Y., Zhang J. Disruption of the EZH2/miRNA/beta-catenin signaling suppresses aerobic glycolysis in glioma. Oncotarget. 2016;7:49450–49458. doi: 10.18632/oncotarget.10370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yamaguchi H., Hung M.C. Regulation and role of EZH2 in cancer. Cancer Res Treat. 2014;46:209–222. doi: 10.4143/crt.2014.46.3.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Villanueva A., Alsinet C., Yanger K., Hoshida Y., Zong Y., Toffanin S., Rodriguez-Carunchio L., Sole M., Thung S., Stanger B.Z., Llovet J.M. Notch signaling is activated in human hepatocellular carcinoma and induces tumor formation in mice. Gastroenterology. 2012;143:1660–1669.e7. doi: 10.1053/j.gastro.2012.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Viatour P., Ehmer U., Saddic L.A., Dorrell C., Andersen J.B., Lin C., Zmoos A.F., Mazur P.K., Schaffer B.E., Ostermeier A., Vogel H., Sylvester K.G., Thorgeirsson S.S., Grompe M., Sage J. Notch signaling inhibits hepatocellular carcinoma following inactivation of the RB pathway. J Exp Med. 2011;208:1963–1976. doi: 10.1084/jem.20110198. [DOI] [PMC free article] [PubMed] [Google Scholar]