Abstract
The polypeptide hormone prolactin (PRL) stimulates breast epithelial cell growth, differentiation, and motility through its cognate receptor, PRLr. PRLr is expressed in most breast cancers; however, its exact role remains elusive. Our laboratory previously described a novel mode of PRLr signaling in which Stat5a-mediated transcription is regulated through ligand-induced phosphorylation of the PRLr transactivation domain (TAD). Herein, we used a PRLr transactivation-deficient mutant (PRLrYDmut) to identify novel TAD-specific target genes. Microarray analysis identified 120 PRL-induced genes up-regulated by wild type but not PRLrYDmut. Compared with control, PRLr expression significantly induced expression of approximately 4700 PRL-induced genes, whereas PRLrYDmut ablated induction of all but 19 of these genes. Ingenuity pathway analysis found that the PRLr TAD most profoundly affected networks involving cancer and proliferation. In support of this, PRLrYDmut expression reduced anchorage-dependent and anchorage-independent growth. In addition, pathway analysis identified a link between the PRLr TAD and the estrogen and progesterone receptors (ERα/PR). Although neither ERα nor PR was identified as a PRL target gene, a TAD mutation significantly impaired ERα/PR expression and estrogen responsiveness. TMA analysis revealed a marked increase in nuclear, but not cytoplasmic, PRLr TAD phosphorylation as a function of neoplastic progression. We propose that PRLr TAD phosphorylation contributes to breast cancer pathogenesis, in part through regulation of ERα and PR, and has potential utility as a biomarker in this disease.
The hormone prolactin (PRL) is necessary for the normal growth, development, and function of the mammary gland. In addition to its role in normal cellular processes, there are abundant data at the cellular, epidemiological, and genetic levels that strongly implicate PRL in the pathogenesis of breast cancer. PRL promotes the growth of human breast cancer cells by acting as a potent mitogen and survival factor.1, 2, 3, 4, 5 Mice harboring a rat prl transgene develop mammary tumors that are both estrogen receptor (ERα) positive and ERα negative.6, 7 In addition, epidemiological studies demonstrate a positive correlation between circulating PRL levels and increased breast cancer risk8, 9, 10 and increased breast cancer metastasis.11 Thus, research aimed at elucidating the mechanisms of prolactin signaling and the modulating effects of this pathway may serve to uncover clinically viable therapies.12
PRL-mediated signaling requires the presence of its cell surface receptor, the prolactin receptor (PRLr). The engagement of PRL with its cognate receptor at the cell surface activates complex signaling networks that mediate development of the terminal ductal unit. This occurs, in part, through the activation of tyrosine kinases, such as Jak2 and Fyn/Src, that may rapidly phosphorylate the C-terminus of PRLr, serving to initiate activation of several signaling cascades.13
Given that elevated levels of serum PRL are associated with breast cancer risk,9, 14 the PRLr makes an attractive therapeutic target because it is detected in approximately 95% of breast cancers.4 Disruption of PRLr signaling in breast cancer cells causes cell growth inhibition and apoptosis induction.15 Furthermore, genetic ablation of the PRLr significantly delays SV40 large T-antigen–induced breast carcinomas in mice.16, 17, 18 Reports demonstrate that impaired turnover of PRLr contributes to a tumorigenic phenotype in vitro and in vivo by promoting high levels of constitutive signaling.19, 20, 21
Classically, it is thought that the PRLr and other transmembrane receptors function as static intermediaries to transmit information from outside the cell to associated messenger proteins that, in turn, translocate to the nucleus and directly affect gene expression.22 However, there are a multitude of reports that demonstrate the nuclear presence of both cell surface receptors and their ligands, thereby challenging this canonical model of transmembrane receptor signal transduction. These reports include, but are not limited to, members of the ErbB family of receptors,23, 24, 25, 26, 27 growth hormone receptor,28, 29 and fibroblast growth factor receptor.26, 30, 31, 32, 33 Data from these studies demonstrate that nuclear-localized transmembrane receptors contribute to transcriptional activation. Examples include nuclear epidermal growth factor receptor (EGFR)–mediated transactivation of Aurora A, iNOS, and COX2 genes34, 35, 36 and nuclear fibroblast growth factor receptor potentiation of c-jun.33 Moreover, preventing EGFR nuclear localization significantly decreases global EGF-induced transcription,37 strongly suggesting that the nuclear presence of a cell surface receptor is a necessary requisite in ligand-induced gene expression.
Interestingly, certain reports indicate that an increase in cell surface receptor nuclear translocation may contribute to tumorigenesis, perhaps by increasing pro-oncogenic gene expression. For instance, initial studies found EGFR in the nuclei of cells from various tissues, including thyroid cells, immortalized renal and ovarian epithelial cells, and primary tumors of the bladder and thyroid.38, 39 In subsequent reports, nuclear EGFR, but not cytoplasmic EGFR, positively correlated with tumor size and lymph node metastasis and negatively correlated with survival in breast cancer.23 A microarray study demonstrated that several genes, including COX2, were activated by EGFR, but not by nuclear localization–defective EGFR.37 Significantly, growth hormone receptor, which exhibits homology to the PRLr, can promote neoplastic changes in vitro and tumor growth in vivo when constitutively localized to the nucleus.29, 40 Nuclear-targeted growth hormone receptor also up-regulated the expression of proproliferative genes, including Survivin and Mybbp.29 Collectively, these studies suggest a pathological role for transmembrane receptor nuclear localization in human cancers. However, further characterization of the pathophysiological features of nuclear-localized cell surface receptors is required so that the potential therapeutic and prognostic value of this phenomenon can be unveiled.
Our previous work revealed that, on PRL stimulation, the PRLr translocates to the nucleus, where it engages in a transcriptional complex with Stat5a and high-mobility group N (HMGN) 2 to activate Stat5a-mediated gene expression.41 In addition, we identified a novel transactivation domain (TAD) within the PRLr and found that residues Y406 and D411 were necessary for PRLr-mediated transactivation. Expression of a transactivation-deficient mutant (Y406F/D411A; herein referred to as PRLrYDmut) significantly diminished both Gal4 reporter transactivation and Stat5a-mediated gene expression.41 Preliminary evidence also suggested that the transactivation properties of the PRLr correlate to a tumorigenic phenotype in vitro, as assessed by anchorage-independent growth.
Given the potential involvement of nuclear PRLr in transcriptional regulation and tumorigenesis, herein we examine how nuclear PRLr may regulate PRL-induced transcriptional events that contribute to a tumorigenic phenotype. Emerging evidence suggests that subcellular localization of proteins plays a critical pathological role and may have significant prognostic value.23, 42 Thus, we also investigated the expression and localization of Y406-phosphorylated (and, therefore, transcriptionally active) PRLr. The findings in this study do the following: i) demonstrate the importance of the PRLr TAD for global PRL-induced gene expression; ii) identify the role of the PRLr TAD in the expression of novel PRL-induced gene targets that are involved in proliferation, migration, and neoplastic progression; iii) find that the expression levels of ERα and progesterone receptor (PR) are regulated, in part, by the PRLr TAD; and iv) describe a significant correlation between nuclear Y406-phosphorylated PRLr and neoplastic progression in vivo. Together, these findings reveal a novel biological function and prognostic value for nuclear localized PRLr.
Materials and Methods
Cell Lines and Culture
T47D and MCF7 human breast cancer cell lines were obtained from ATCC (Manassas, VA) and cultured in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum, 100 U/mL penicillin, and 100 μg/mL streptomycin (Gibco, Grand Island, NY). The 293T/17 cells with stable integration of the SV40 large T antigen were supplied by Dr. Debu Chakravarti (Northwestern University, Chicago, IL) and were cultured in Dulbecco’s modified Eagle’s medium with 10% fetal bovine serum and 100 μg/mL streptomycin. All cells were incubated in a humidified 5% CO2/95% air atmosphere at 37°C. Estrogen receptor levels in MCF7 cell lines were previously reported by Saceda et al.43
Prolactin Treatment
Human recombinant PRL was a gift from Dr. Anthony Kossiakoff (University of Chicago, Chicago, IL). PRL was added to cells to yield a final concentration of 250 ng/mL.
Microarray Procedures and Data Analysis
To examine genome-wide changes in gene expression induced by PRL in the presence of wild-type (WT) or mutant PRLr, MCF7 cells stably infected with empty vector, WT PRLr, or PRLrYDmut retroviral constructs were generated, as previously described.41 Each cell line was plated at 70% confluency in six-well plates and 24 hours post-plating was serum starved for an additional 24 hours. Triplicate samples were either treated with 250 ng/mL PRL for 2 hours or left untreated. Total RNA was isolated from three independent cultures using RNeasy Mini Plus Kits (Qiagen, Valencia, CA). The integrity of RNA was determined using an Agilent Bioanalyzer (Agilent Technologies, Santa Clara, CA). Fluorescently labeled cDNA was prepared using TargetAmp aRNA Amplification Performance (Epicenter, Madison, WI), followed by hybridization to an Illumina HumanHT-12Version 3 microarray chip containing 44,000 probes (Illumina, San Diego, CA). Raw signal intensities of each probe were obtained using data analysis software (Beadstudio version 3.2; Illumina) and imported directly into the Partek Genomics Suite version 6.2 (Partek, Inc., St. Louis, MO) for statistical processing.
Microarray metadata complied with MIAME standards, and all samples were submitted to the National Center for Biotechnology Information Gene Expression Omnibus database for public access. Baseline statistical comparisons were performed using a two-way analysis of variance [where the interaction effect between receptor status (empty, PRLr, or PRLrYDmut) and treatment status (untreated versus PRL treatment)] was used to determine the P value. All outputs were filtered with a P < 0.05 cutoff ± 1.2-fold change cutoff. Gene lists were compiled by selecting two contrast groups (ie, WT + PRL versus MUT + PRL; WT versus E; empty + PRL versus E) and filtering out genes based on P < 0.05 and ±1.2 fold change cutoff. The WT + PRL versus MUT + PRL and empty + PRL versus E gene lists were then imported into Ingenuity Pathway Analysis (IPA) software, version 7.5 (Ingenuity Systems, Redwood City, CA), for network and pathway generation. In addition, gene lists were imported into Hierarchical Clustering Explorer 2.0 (http://www.cs.umd.edu/hcil/hce, last accessed August 2012). Heat maps were generated using hierarchical clustering of sample sets (eg, empty + PRL.A or empty + PRL.B) with average linkage methods and euclidean distance measure.
Western Blot Analysis
The T47D and MCF7 cells were plated in 10-mm dishes at 70% confluency. Cells were lysed in radioimmunoprecipitation assay buffer, subjected to 10% SDS-PAGE, and transferred to a polyvinylidene difluoride membrane and immunoblotted with the appropriate antibody (1 μg/mL). Blots were probed with anti-PRLr (Santa Cruz Biotechnology, Santa Cruz, CA), anti-V5 (Invitrogen, Carlsbad, CA), anti-GAPDH (Cell Signaling, Danvers, MA), anti-tubulin (Cell Signaling), or anti–phospho-Y406 PRLr antibodies (New England Biosystems, Ipswich, MA).
RT-PCR and Real-Time PCR
To validate microarray results, MCF7 transfectants were set up and treated as previously stated. To examine 17β estradiol (E2) induction of c-fos, PR, Myc (alias c-myc), and CCND1, MCF7 stable transfectants (PRLr overexpressing or knockdown cells) were removed from estrogen-containing media and cultured in steroid-free, phenol red–free media containing charcoal-stripped fetal bovine serum for 4 to 5 days before the start of experiments. Cells were plated in six-well plates at 70% confluency. The next day, cells were serum started for 24 hours and then treated with 10 nmol/L E2 for 1 hour. Total RNA was isolated using an RNA EZ miniprep kit (Qiagen), as previously reported.44 E2-induced gene expression dose-response curves for estrogen were previously optimized; thus, we chose the optimal concentration (10 nmol/L).45 cDNA was synthesized using qScript (Quanta Biosciences, Gaithersburg, MD). cDNA was diluted to 2.5 ng/μL (corresponding to RNA concentration). A total of 4 μL cDNA, 1 μL primers (2 μmol/L each), and 5 μL of two times Power SYBR MasterMix was used for real-time PCR in a 10-μL reaction volume performed in a 384-well plate. Real-time PCR was conducted on an ABI 7900HT thermocycler (Applied Biosystems, Foster City, CA). All real-time PCRs were run in triplicate. For RT-PCR, data were normalized to glyceraldehyde-3-phosphate dehydrogenase RNA or 18S ribosomal RNA (rRNA). The fold change for RT-PCR is represented as follows: 2–ΔΔCT[2–(CTtarget – CT18S rRNA)PRL – (CTtarget – CT18S rRNA)control] using empty vector, non-treated as a baseline. Primers used are listed in Table 1.
Table 1.
Primers Used for Microarray Validation
| Gene | Primer |
|---|---|
| 18S | |
| Forward | Olg 200: 5′-CCCCATGAACGAGGGAATT-3′ |
| Reverse | Olg 201: 5′-GGGACTTAATCAACGCAAGCTT-3′ |
| CEBPβ | |
| Forward | Olg 284: 5′-AGAACGAGCGGCTGCAGAAGA-3′ |
| Reverse | Olg 285: 5′-CAAGTTCCGCAGGGTGCTGA-3′ |
| ERα | |
| Forward | Olg 331: 5′-GGAAGCTACTGTTTGCTCCTAACTTG-3′ |
| Reverse | Olg 332: 5′-AGATCTCCACCATGCCCTCTAC-3′ |
| PR | |
| Forward | Olg 345: 5′-TCAGTGGGCAGATGCTGTATTT-3′ |
| Reverse | Olg 346: 5′-GCCACATGGTAAGGCATAATGA-3′ |
| RARA | |
| Forward | Olg 444: 5′-GCCGCCCCCACATGTT-3′ |
| Reverse | Olg 445: 5′-TGATGCTTCGCAGGTCAGTAA-3′ |
| RASD | |
| Forward | Olg 446: 5′-CGGCTCAGGCAGCAGATC-3′ |
| Reverse | Olg 447: 5′-CGTTCTCCTTGGTTTTGTTCTTG-3′ |
| ZYX | |
| Forward | Olg 458: 5′-AGGACATGGAGCATCCTCAGA-3′ |
| Reverse | Olg 459: 5′-GCATCGGCCGCAGAGTT-3′ |
| MIDN | |
| Forward | Olg 462: 5′-ACAAAGACACCCGGCTCAGT-3′ |
| Reverse | Olg 463: 5′-GGTCAGCTTGCTGCCATCA-3′ |
| ERRFI1 | |
| Forward | Olg 466: 5′-GTCATGCCCCCGACACA-3′ |
| Reverse | Olg 467: 5′-GCAGTGCTTTGCTGCTGACA-3′ |
| SRF | |
| Forward | Olg 470: 5′-CCTTTGCCACCCGAAAACT-3′ |
| Reverse | Olg 471: 5′-AGTGCCTTGCCGGTCTCA-3′ |
| MYADM | |
| Forward | Olg 474: 5′-ATGCCACCGCCCTTGTT-3′ |
| Reverse | Olg 475: 5′-GCCGCCATACTTCTCATCGA-3′ |
| CCND1 | |
| Forward | Olg 310: 5′-CCGTCCATGCGGAAGATC-3′ |
| Reverse | Olg 311: 5′-GAAGACCTCCTCCTCGCACTT-3′ |
| c-fos | |
| Forward | 5′-GGGCAAGGTGGAACAGTTATC-3′ |
| Reverse | 5′-CCGCTTGGAGTGTATCAGTCA-3′ |
| c-myc | |
| Forward | Olg 314: 5′-GGATTTTTTTCGGGTAGTGGAA-3′ |
| Reverse | Olg 315: 5′-TTCCTGTTGGTGAAGCTAACGTT-3′ |
Primers were obtained using Primer Express 3.1 software (Applied Biosystems). Most primer pairs are located at exon-exon junctions, except the intronless 18S rRNA. Primers were tested by dissociation curve analysis to ensure amplification of a single amplicon. All primers are located inside the coding sequence.
Olg, oligonucleotide.
Retroviral Production
To generate overexpression cell lines, PRLr or PRLrYDmut in pTracer was amplified through a PCR and ligated into the retroviral pBabe–green fluorescent protein vector using EcoRI and SalI, as previously described.41 Knockdown cell lines were generated using a predesigned HMGN2 short hairpin RNA (shRNA) pRFP retroviral vector (Origene, Rockville, MD); PRLr shRNA was designed and cloned into a pRFP vector [sequences: HMGN2, 5′-GTGTCAGGCAATCTGGACT-TTCCAGTGAT-3′; short hairpin PRLr (shPRLr), 5′-CAA-CTGCATAACCTTTACACT-3′]. Recombinant retroviruses for pBabe, PRLr, PRLrYDmut, Scrambled shRNA, and shPRLr were generated using transfection of retroviral vectors with Lipofectamine 2000 (Invitrogen, Carlsbad, CA) in 293T/17 cells, along with pVSVG and pGalPol vectors. Viral supernatant was collected 48 hours after transfection and added to the appropriate cell lines, along with 8 μg/mL polybrene. Cells were spin infected at 500 × g at 32°C for 2 hours. Infected cells were enriched by fluorescence-activated cell sorting for green fluorescent protein (overexpression cell lines) or 2 μg/mL puromycin selection (knockdown cell lines). After puromycin selection, all cells were grown in a maintenance dose of 1 μg/mL puromycin. Puromycin was removed 24 hours before experimental setup. For rescue cell lines, MCF7 overexpression cell lines (empty, PRLr, or PRLrYDmut) were subsequently infected with non-specific short hairpin (shNS) retrovirus or shPRLr retrovirus and selected with 2 μg/mL puromycin.
siRNA-Mediated Knockdown
Small-interfering RNA (siRNA) knockdown experiments were conducted as previously reported.46 The siRNAs against PRLr were custom made from Ambion (Austin, TX), using the following sequence: 5′-GCAGUUU-CUGGGAUGAACUdTdT-3′.
Soft Agar Colony Formation
Soft agar assays were performed as previously described.47 Briefly, 1 mL of 0.6% agarose/growth media mixture was added to each well of a six-well plate. A total of 3 × 105 cells were used to generate a single-cell suspension in 0.3% agarose/growth media. A 500-μL suspension was added to each well. Once solidified, 1 mL of growth media, supplemented with 250 ng/mL PRL, was added to each well. New media supplemented with PRL were added every 2 to 3 days. Pictures were taken on day 20. Colony size and number were quantified using ImageJ software version 1.46 (cutoff value for colony size, ≥50 μm) (NIH, Bethesda, MD).
Proliferation
T47D cells [small-interfering control (sicontrol) or siPRLr transfected] or MCF7 transfectants were plated in 96-well plates and cultured in growth medium for 24 hours. Each well was pulsed with 0.5 Ci of tritiated thymidine (48 Ci/mmol; Amersham Pharmacia Biotech, Piscataway, NJ) for 4 to 6 hours. Cells were harvested onto the membrane with Filtermate Harvester (Perkin Elmer, Waltham, MA) before analysis with a MicroBeta TriLux Scintillation Counter (Perkin Elmer).
ERE Luciferase
MCF7 stable transfectants were grown in estrogen-free media for 4 to 5 days before the start of the experiment. Luciferase assays using the estrogen responsiveness element (ERE)–luciferase reporter construct were conducted as follows: MCF7-transfectants cells were seeded into a 48-well plate at a density of 2 × 104. Cells were serum started for 24 hours and then transfected with 250 ng of luciferase reporter and 2.5 ng of renilla reporter, the Fugene HD transfection reagent (Promega, Madison, WI). At 24 hours after transfection, cells were treated with 10 nmol/L E2 for 6 hours. Cells were then using Dual Reporter Luciferase reagents (Promega) and read by a Victor Microplate Reader (Perkin Elmer). All transfections were performed in triplicate, and each individual sample was read in duplicate. Reporter Luciferase reagents (Promega) were read by a Victor Microplate Reader. All transfections were performed in triplicate or quadruplicate, and each individual sample was read in duplicate. The ratio of luciferase/renilla was calculated, and the results are reported as fold change compared with untreated MCF7-empty.
TMA Data
Breast tissues were obtained from the Tissue Core of the Northwestern University Breast Cancer Program in the form of a TMA. All tissues were stripped of all patient identifiers before use and, thus, were anonymous to the investigators. The TMA consisted of 15 patient-matched normal, tumor, and lymph node metastasis tissues from patients with breast cancer and six patient-matched normal and tumor tissue samples from patients with breast cancer. Within this cohort, there were 8 ER+/Her2− patient specimens and 13 ER−/Her2− patient specimens. The presence of tumor and the integrity of each tissue were confirmed by H&E staining. Before analysis by immunohistochemistry (IHC), the TMA was incubated at 65°C for 10 minutes, before deparaffinization in xylene and rehydration through graded ethanol, followed by antigen retrieval in citrate buffer (Zymed 00-5001, pH 6.0) at 95°C for 20 minutes. The slides were blocked in the blocking buffer (2.5% bovine serum albumin and 0.1% Triton X-100) for 10 minutes and incubated with a phospho-Y406 PRLr-specific antibody (New England Peptide, Ipswich) (1:10 dilution for immunofluorescence; 1:20 dilution for IHC) overnight at 4°C. Antigen-antibody complexes were detected for immunofluorescence by incubation with an Alexa 488 Fluor–conjugated secondary antibody (1:100 dilution) for 1 hour. For IHC, detection of an antibody-antigen complex used a horseradish peroxidase–conjugated secondary antibody, followed by diaminobenzidine labeling, as previously described.48 Visual scoring of the immunofluorescence-labeled images was performed as previously described,48 with modification. Scoring was restricted to assessment of label intensity on a 0 to 3 scale (0, absent; 1, dim; 2, bright; and 3, very bright). In a second cohort, 7 ER+ tumors and 26 ER− tumors were analyzed for phospho-Y406 staining.
Statistical Analysis
Statistical analysis was performed using one- or two-way analysis of variance with post hoc Bonferroni on GraphPad Prism version 4 (GraphPad Software, La Jolla, CA). The results are shown as the means with error bars depicting ± SEM. P < 0.05 was considered statistically significant. All experiments were performed at least three times, unless otherwise indicated.
Results
Expression of the PRLrYDmut Impairs Global PRL-Induced Gene Expression
Given our previous data that expression of a transactivation-deficient PRLr impaired PRL-induced CISH expression,41 we hypothesized that PRLrYDmut would impair global PRL-induced gene expression. To test this hypothesis, we used MCF7-PRLr, MCF7-PRLrYDmut, or MCF7-empty vector stable transfectants (herein referred to as PRLr, PRLrYDmut, and empty, respectively), as described in our previous report.41 MCF7 breast cancer cells were selected for their moderate level of endogenous PRLr expression and moderate response to PRL stimulation, which was significantly increased on ectopic expression of WT PRLr.46, 49 As shown in Figure 1A, immunoblot analysis of MCF7 stable transfectants demonstrated equal levels of ectopic PRLr expression using either an anti-V5 antibody to detect epitope-tagged PRLr or an anti-PRLr antibody. To demonstrate that stable cell lines expressed physiological levels of PRLr, endogenous PRLr levels in T47D breast cancer cells were assessed in conjunction with stable cell lines (Figure 1A). In addition, immunoblot analysis using an anti–phospho-Y406 PRLr antibody revealed that PRLrYDmut was not Y406 phosphorylated (Figure 1B).41
Figure 1.
Mutation of the prolactin receptor transactivation domain prevents its phosphorylation and impairs global PRL-induced gene expression. A: Western blot analysis demonstrating PRLr or PRLrYDmut overexpression in MCF7 stable cell lines using an anti-PRLr or an anti-V5 antibody (to detect only epitope-tagged PRLr). Comparison to endogenous PRLr in T47D cells demonstrates that stable cell lines express physiological levels of PRLr. B: Mutation of the PRLr TAD (PRLrYDmut) impairs phosphorylation of Y406. MCF7 cells were serum starved, treated with PRL, lysed, and subjected to immunoprecipitation (IP) analysis using an anti-V5 antibody. Eluted samples were analyzed by using Western blot analysis with an anti–phospho-Y406 PRLr antibody. Blots were stripped and reprobed for anti-V5 to show proper pull down. C: Heat maps of genes identified in microarray analysis were generated with unsupervised hierarchical clustering. MCF7 stable cells were serum starved for 24 hours, then treated with PRL for 2 hours. Data reveal a global view of PRL-induced genes up-regulated in MCF7 PRLr-expressing cells compared with MCF7 transactivation-deficient PRLrYDmut-expressing cells. D: Heat map of genes differentially regulated by PRL. In C and D, the biological replicates of WT, WT + PRL, and MUT + PRL group together, whereas replicates of MUT are grouped with empty control samples. Labels specify samples/treatments (empty, empty vector; WT, PRLr; MUT, PRLrYDmut; +P, PRL treatment) and biological repeats (ie, empty.A, empty.B, and empty.C). The scale is provided, where green represents down-regulated genes and red represents up-regulated genes. E: Venn diagram of PRL-induced genes up-regulated by PRLr or PRLrYDmut. Gene lists comparing WT + PRL versus empty + PRL or MUT + PRL versus empty + PRL (1.2-fold up-regulation, P < 0.05) were generated. F: Venn diagram of transcripts regulated by PRLr expression or PRL treatment. Gene lists comparing PRLr versus empty or empty + PRL versus empty (1.2-fold up-regulation, P < 0.05) were generated. The results demonstrate many genes regulated by PRLr expression and PRL treatment and 486 genes that are shared between both groups. G and H: Real-time PCR demonstrates that IER3 (G) and CCND1 (H) are up-regulated in a PRL-induced manner by expression of PRLr, but not PRLrYDmut. Ctrl, control; mut, prolactin receptor harboring Y406F/D411A mutations; PY406, phosphorylation on PRLr residue tyrosine 406. ***P < 0.001.
The requirement of the PRLr TAD for global PRL-induced gene expression was examined by microarray analysis of PRL-stimulated MCF7 stable transfectants (2 hours, PRL treatment). Heat maps were compiled using hierarchical clustering from gene lists generated by analysis of PRL-regulated genes up- or down-regulated 1.2-fold by PRLr versus PRLrYDmut and empty + PRL versus empty (P < 0.05). These analyses demonstrated 120 (PRLr + PRL versus PRLrYDmut + PRL) and 947 (empty + PRL versus empty) differentially regulated genes (Figure 1, C and D). Tables 2 and 3 list the top up- and down-regulated genes in PRLr versus PRLrYDmut analysis, and Tables 4 and 5 show the top up- and down-regulated PRL-induced genes, respectively. The complete list of genes is shown in Supplemental Table S1. Interestingly, when analyzing differential PRLr-induced gene expression in PRLr versus PRLrYDmut, we found only two down-regulated genes, supporting the hypothesis that the PRLr TAD was important for transcriptional regulation.
Table 2.
List of Top 75 Genes Up-Regulated by PRLr, but Not PRLrYDmut
| Gene symbol | P value (WT + PRL vs MUT + PRL) | Fold change (WT + PRL vs MUT + PRL) |
|---|---|---|
| RASD1 | 1.08 × 10−7 | 2.08403 |
| ACTG2 | 2.06 × 10−6 | 2.06483 |
| IER5L | 5.81 × 10−5 | 1.98411 |
| FOSB | 1.63 × 10−6 | 1.98166 |
| CTGF | 1.71 × 10−7 | 1.97731 |
| RARA | 2.93 × 10−6 | 1.92939 |
| ZYX | 7.44 × 10−7 | 1.92049 |
| PHLDA2 | 3.19 × 10−5 | 1.8765 |
| MIDN | 8.01 × 10−7 | 1.86918 |
| METRNL | 3.40 × 10−5 | 1.8273 |
| ZMYND19 | 5.64 × 10−6 | 1.80599 |
| PPRC1 | 2.45 × 10−5 | 1.79972 |
| SRF | 6.53 × 10−7 | 1.79468 |
| NT5DC3 | 5.14 × 10−6 | 1.7858 |
| ERRFI1 | 1.43 × 10−5 | 1.78173 |
| ZYX | 5.35 × 10−5 | 1.76553 |
| VCL | 3.66 × 10−5 | 1.76477 |
| CDC42BPB | 0.00024143 | 1.75944 |
| IRX5 | 5.10 × 10−6 | 1.75459 |
| CCDC85B | 6.23 × 10−7 | 1.75194 |
| DOK7 | 2.81 × 10−6 | 1.74303 |
| AKAP8L | 1.71 × 10−7 | 1.70602 |
| CSNK1D | 1.45 × 10−6 | 1.69846 |
| FHL2 | 4.77 × 10−7 | 1.69067 |
| MYADM | 5.53 × 10−7 | 1.68906 |
| JUNB | 2.93 × 10−6 | 1.68347 |
| FKSG30 | 0.000136381 | 1.68143 |
| CEBPB | 7.28 × 10−5 | 1.66644 |
| VCL | 1.94 × 10−5 | 1.65654 |
| PIM3 | 0.000163325 | 1.64606 |
| FHL2 | 3.10 × 10−6 | 1.62748 |
| IRF2BP2 | 1.00 × 10−5 | 1.62036 |
| EGR1 | 0.000603453 | 1.61063 |
| KRT80 | 7.42 × 10−5 | 1.59395 |
| CCNY | 5.71 × 10−7 | 1.59178 |
| PHLDA1 | 9.31 × 10−9 | 1.57512 |
| KCNF1 | 0.000154103 | 1.56515 |
| METRNL | 6.36 × 10−5 | 1.54002 |
| HOXC8 | 1.84 × 10−6 | 1.53373 |
| PINX1 | 1.23 × 10−5 | 1.53056 |
| OSGIN1 | 4.87 × 10−5 | 1.52242 |
| MYADM | 6.66 × 10−5 | 1.51566 |
| MED10 | 0.000979181 | 1.50776 |
| ETS2 | 1.55 × 10−5 | 1.505 |
| SMTN | 2.68 × 10−6 | 1.50201 |
| TTLL3 | 0.000165101 | 1.49711 |
| EGR3 | 6.95 × 10−5 | 1.4918 |
| PDGFB | 9.28 × 10−5 | 1.4864 |
| RBM38 | 6.14 × 10−6 | 1.48294 |
| 0.000311265 | 1.46592 | |
| DAPK3 | 1.85 × 10−5 | 1.46258 |
| EGR2 | 3.05 × 10−6 | 1.43838 |
| CRTC2 | 0.000456804 | 1.4357 |
| PLEKHG2 | 4.41 × 10−5 | 1.43032 |
| AKIRIN1 | 0.000200831 | 1.42602 |
| XIST | 0.00189227 | 1.41457 |
| LRRC45 | 7.23 × 10−6 | 1.41242 |
| PHF17 | 7.79 × 10−5 | 1.40478 |
| SH3BP1 | 2.70 × 10−5 | 1.40251 |
| IL5RA | 0.000536148 | 1.40106 |
| CISH | 0.00021071 | 1.39824 |
| JMJD6 | 0.000208113 | 1.39622 |
| EGR4 | 1.81 × 10−6 | 1.38468 |
| TRIM41 | 1.62 × 10−6 | 1.38151 |
| ENTPD4 | 5.14 × 10−5 | 1.37924 |
| ADAM15 | 2.70 × 10−5 | 1.36798 |
| FLJ12949 | 0.000412296 | 1.36246 |
| CGI-96 | 0.000364017 | 1.35468 |
| ZNF326 | 1.40 × 10−5 | 1.35227 |
| DCAF15 | 0.000985216 | 1.34618 |
| HYAL2 | 8.82 × 10−5 | 1.3417 |
| NAV2 | 0.000767715 | 1.33526 |
| TNFSF13B | 0.00122398 | 1.33072 |
| IKZF3 | 0.00165765 | 1.32625 |
| CDK5R1 | 0.000193887 | 1.32588 |
Microarray values were analyzed in Partek Genomics Suite. To generate the list, the following criteria were used: Contrast PRLr + PRL versus PRLrYDmut + PRL (fold change, 1.2; P < 0.05). The full list of genes that were up- or down-regulated in this analysis can be found in Supplemental Table S1.
Table 3.
Genes Identified to Be Down-Regulated by PRLr but Not PRLrYDmut
| Gene symbol | P value (WT + PRL vs MUT + PRL) | Fold change (WT + PRL vs MUT + PRL) |
|---|---|---|
| S100A14 | 0.00244896 | −1.48855 |
| TRQ1 | 0.00694574 | −1.31522 |
Microarray values were analyzed in Partek Genomics Suite. To generate the list, the following criteria were used: Contrast PRLr + PRL versus PRLrYDmut + PRL (fold change, −1.2; P < 0.05). The full list of genes that were up- or down-regulated in this analysis can be found in Supplemental Table S1.
Table 4.
List of Top 75 Genes Up-Regulated by PRL Treatment
| Column ID | P value (E + PRL vs E) | Fold change (E + PRL vs E) |
|---|---|---|
| MYADM | 5.61 × 10−7 | 3.39825 |
| TNFRSF12A | 8.74 × 10−5 | 3.04048 |
| SRF | 5.86 × 10−6 | 2.70749 |
| ACTG2 | 0.000236 | 2.61423 |
| FHL2 | 3.96 × 10−6 | 2.4949 |
| KRT80 | 3.64 × 10−5 | 2.45465 |
| IER3 | 0.000551 | 2.41686 |
| VCL | 8.89 × 10−5 | 2.39255 |
| PHLDA2 | 0.000907 | 2.38364 |
| TUFT1 | 0.000781 | 2.35088 |
| SPATA2L | 0.002807 | 2.32634 |
| MYADM | 0.000117 | 2.24982 |
| FHL2 | 1.69 × 10−6 | 2.22291 |
| RND3 | 1.19 × 10−5 | 2.20115 |
| RARA | 0.000791 | 2.16631 |
| IER5L | 0.006757 | 2.16468 |
| SOX18 | 0.000219 | 2.1109 |
| MIDN | 9.43 × 10−5 | 2.09812 |
| ZYX | 0.000448 | 2.07648 |
| RRS1 | 0.00289 | 2.05318 |
| VASN | 0.001214 | 2.05121 |
| FAM89A | 0.002013 | 2.01716 |
| PPRC1 | 0.000593 | 2.00695 |
| METRNL | 0.003639 | 1.98267 |
| MAP6D1 | 0.003279 | 1.97972 |
| PLD6 | 0.004202 | 1.97447 |
| F2RL1 | 0.000201 | 1.97006 |
| AMY1A | 0.025242 | 1.94975 |
| NIP7 | 0.000781 | 1.9489 |
| CEBPB | 0.000818 | 1.93153 |
| PFKFB3 | 0.000625 | 1.91705 |
| RASD1 | 0.003702 | 1.91148 |
| LYAR | 0.001735 | 1.90932 |
| FJX1 | 0.000274 | 1.8845 |
| PAQR4 | 0.003875 | 1.86233 |
| STC2 | 0.001385 | 1.8623 |
| HSPC111 | 0.000765 | 1.86223 |
| SURF6 | 0.002016 | 1.85036 |
| LYAR | 0.007156 | 1.84848 |
| MARS2 | 0.000352 | 1.84408 |
| BAG3 | 0.005463 | 1.83933 |
| TRQ1 | 3.92 × 10−6 | 1.83782 |
| HPS6 | 0.00679 | 1.8351 |
| ZBTB9 | 0.003265 | 1.83483 |
| TIGD5 | 0.007036 | 1.83323 |
| CHSY1 | 0.002988 | 1.82873 |
| ISG20L1 | 0.020593 | 1.8287 |
| MED10 | 0.000918 | 1.82562 |
| NOP16 | 0.000846 | 1.81675 |
| TRIB1 | 0.010389 | 1.81587 |
| BRIX1 | 0.000918 | 1.80872 |
| BRIX1 | 0.000399 | 1.80792 |
| LAD1 | 0.014801 | 1.78896 |
| EHD1 | 0.026665 | 1.78436 |
| AXUD1 | 0.028168 | 1.77712 |
| NT5DC3 | 0.000906 | 1.77692 |
| NXT1 | 0.00217 | 1.77464 |
| EGR3 | 0.002321 | 1.7677 |
| AMD1 | 0.000217 | 1.76583 |
| MSX1 | 0.003956 | 1.75815 |
| URB2 | 0.000284 | 1.74167 |
| GFOD1 | 0.011517 | 1.74014 |
| OKL38 | 0.018381 | 1.73959 |
| CDR2 | 0.001357 | 1.73377 |
| RPUSD1 | 0.002498 | 1.73329 |
| SLC10A3 | 0.035118 | 1.72924 |
| ZMYND19 | 0.001026 | 1.72882 |
| GPATCH4 | 0.001869 | 1.72762 |
| PRMT6 | 0.001279 | 1.72567 |
| FAM131A | 0.002641 | 1.72494 |
| ETF1 | 0.020531 | 1.72486 |
| NOL6 | 0.031402 | 1.72195 |
| CNN2 | 0.004733 | 1.71051 |
| DEXI | 0.006981 | 1.70672 |
| BRPF1 | 0.004484 | 1.70664 |
Microarray values were analyzed in Partek Genomics Suite. To generate the list, the following criteria were used: Contrast empty + PRL versus empty (fold change, 1.2; P < 0.05). The full list of genes that were up- or down-regulated in this analysis can be found in Supplemental Table S2.
Table 5.
List of Top 43 Genes Down-Regulated by PRL Treatment
| Gene symbol | P value (E + PRL vs E) | Fold change (E + PRL vs E) |
|---|---|---|
| BCL6 | 0.008276 | −3.08845 |
| HBP1 | 0.002612 | −1.72044 |
| CROP | 0.009112 | −1.65935 |
| CDK5RAP3 | 0.001033 | −1.62475 |
| FBXO32 | 0.019224 | −1.57676 |
| YPEL2 | 0.032369 | −1.54352 |
| TSC22D3 | 0.028116 | −1.54264 |
| TXNIP | 0.046969 | −1.48875 |
| CYTH2 | 0.004058 | −1.44713 |
| ZNF627 | 0.027492 | −1.42244 |
| NPDC1 | 0.037997 | −1.41707 |
| GOLGA8B | 0.045398 | −1.4126 |
| SFRS18 | 0.041675 | −1.4106 |
| EFNA1 | 0.033587 | −1.39753 |
| ARRDC3 | 0.029103 | −1.39268 |
| FLJ45244 | 0.016318 | −1.36964 |
| GRB7 | 0.032281 | −1.36174 |
| MRRF | 0.033059 | −1.36159 |
| WSB1 | 0.020118 | −1.36044 |
| FLJ38717 | 0.017432 | −1.36011 |
| PAQR6 | 0.00055 | −1.35852 |
| TPM2 | 0.001857 | −1.35103 |
| DOC2A | 0.015327 | −1.30835 |
| SNHG7 | 0.007631 | −1.30588 |
| CSAD | 0.04618 | −1.29469 |
| MXD3 | 0.000178 | −1.28976 |
| TTLL3 | 0.038842 | −1.28717 |
| TMBIM1 | 0.04702 | −1.27126 |
| NPHP3 | 0.025933 | −1.2635 |
| HOXA11AS | 0.034316 | −1.25319 |
| FBXO32 | 0.038922 | −1.24535 |
| GRB7 | 0.022039 | −1.23197 |
| HSF4 | 0.000446 | −1.22095 |
| PH-4 | 0.000155 | −1.22047 |
| TBC1D17 | 0.031072 | −1.21674 |
| DFNB31 | 0.009157 | −1.21327 |
| IDUA | 0.000806 | −1.21304 |
| SNORA7A | 0.007839 | −1.21012 |
| PRR22 | 8.91 × 10−5 | −1.20667 |
| FLJ40473 | 0.018857 | −1.2045 |
| ANO9 | 0.013677 | −1.20402 |
| FRAT1 | 0.00489 | −1.20336 |
| GPR19 | 0.045708 | −1.20036 |
Microarray values were analyzed in Partek Genomics Suite. To generate the list, the following criteria were used: Contrast empty + PRL versus empty (fold change, −1.2; P < 0.05). The full list of genes that were up- or down-regulated in this analysis can be found in Supplemental Table S2.
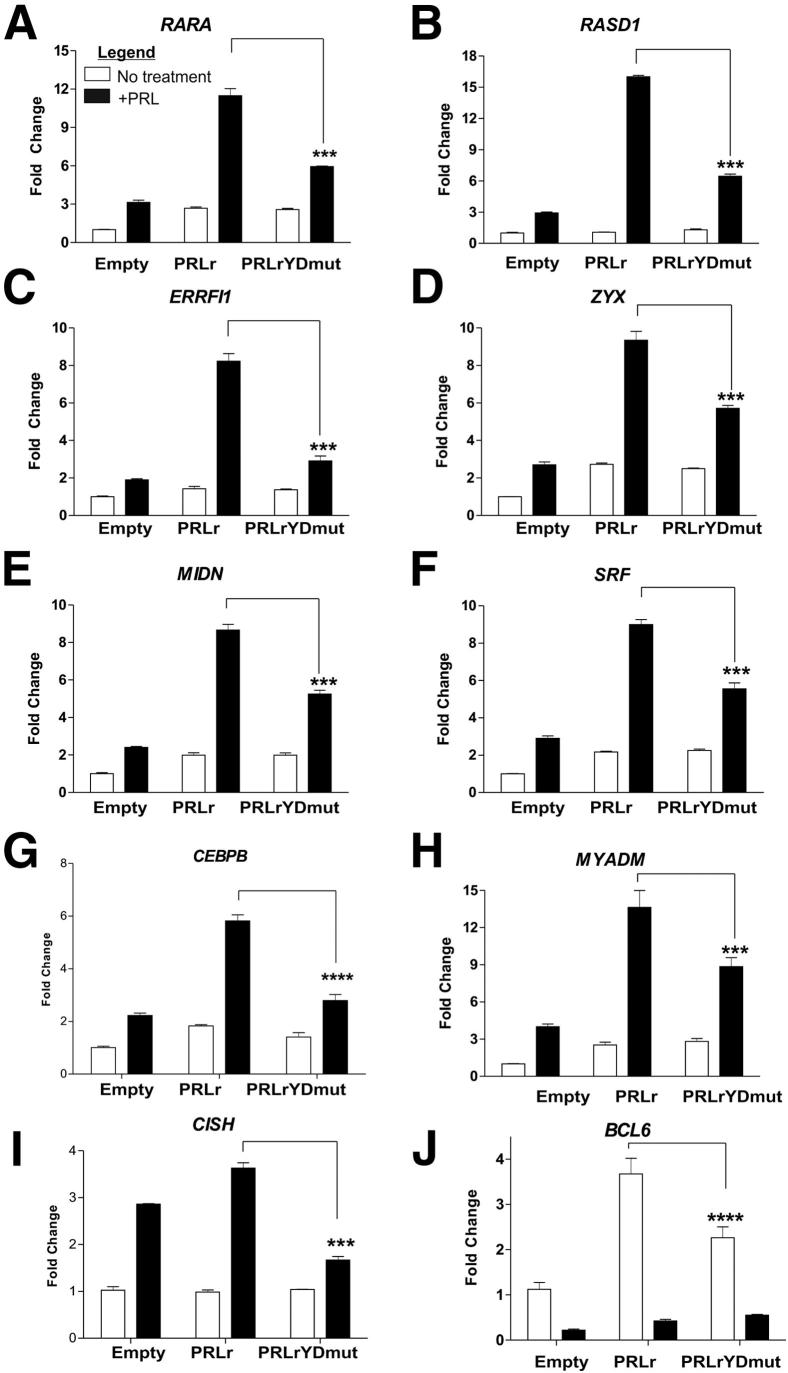
Select genes from Tables 2 and 3 were validated by quantitative PCR (qPCR). For gene validation studies, we focused on genes identified in analysis of PRLr + PRL versus PRLrYDmut + PRL, given that our main objective was to find those genes that required the PRLr TAD. However, we specifically chose those genes that also demonstrated a robust PRL response within this list. As shown in Figure 2, A–H, on PRL treatment, all validated genes were increased or decreased by twofold or greater. These results, in concordance with microarray analysis, demonstrated that mutation of the PRLr TAD reduced PRL-induced gene expression (Figure 2, A–H). As a negative control, we measured the expression of a non-PRL-responsive gene, FAS, and observed no significant differences between control, PRLr, and PRLrYDmut (Supplemental Figure S1). We chose not to further filter the list shown in Tables 4 and 5 by comparing it with the genes identified in our analysis of empty + PRL versus empty. This was because of the fact that, in previous studies, we found that MCF7 cells required exogenously expressed PRLr for appreciable levels of PRL-induced gene expression.49
Figure 2.
Validation of genes identified by microarray analysis. Real-time PCR demonstrates that RARA (A), RASD1 (B), ERRFI1 (C), ZYX (D), MIDN (E), SRF (F), CEBPβ (G), MYADM (H), and CISH (I) are up-regulated in a PRL-induced manner by expression of PRLr but not PRLrYDmut. Furthermore, because BCL6 was identified as the top down-regulated gene by PRL, J shows that basal levels of BCL6 are up-regulated by the PRLr but not PRLrYDmut and, thus, the percentage down-regulation of BCL6 is significantly decreased by the expression of PRLrYDmut. Results are representative of three separate experiments with SEM. All P values were calculated using one-way analysis of variance with a Bonferroni multiple comparisons test. Calculated P values are highlighted in the figure. ***P < 0.001, ****P < 0.0001.
We previously demonstrated that the PRLr TAD was necessary for recruitment of HMGN2 to the transcriptional complex and, in turn, HMGN2 was necessary for PRL-mediated transcription of CISH.41 Given this, we reasoned that knockdown of HMGN2 would significantly reduce the transcription of genes identified in microarray analysis. shRNA-mediated knockdown of HMGN2 was, therefore, used to test this hypothesis. Depletion of HMGN2 in MCF7 cells significantly impaired PRL-mediated transcription of myeloid-associated differentiation factor (MYADM), retinoic acid receptor (RARA), serum response factor (SRF), zyxin (ZYX), and ERBB receptor feedback inhibitor 1 (ERRFI1) (Supplemental Figure S2). in a similar manner to that of PRLrYDmut expression (Figure 2, D–F).
We next determined how many genes were significantly increased in both PRL-treated PRLr-expressing versus empty vector-expressing and PRLrYDmut-expressing versus empty vector-expressing cells using comparative analysis. Gene lists were generated in Partek Genomics Suite by comparing PRLr + PRL versus empty + PRL or PRLrYDmut + PRL versus empty + PRL (1.2-fold up-regulation, P < 0.05). Unexpectedly, we found that 4705 transcripts were significantly increased in PRLr-expressing cells, whereas in PRLrYDmut-expressing cells, all but 19 of these up-regulated transcripts were lost (Figure 1E; the complete gene list is shown in Supplemental Tables S2 and S3).
We also queried genes that were regulated by either the PRLr or PRL alone. This query identified a total of 5831 genes regulated by the PRLr (Supplemental Table S4) and 947 genes regulated by PRL (Supplemental Table S2). A total of 486 identified genes were shared by both groups (Supplemental Table S5). The top identified genes included those identified from the previous analysis (Figure 1C). We additionally identified genes that were not found in our previous analyses (PRLr + PRL versus PRLrYDmut + PRL). Two of these genes, immediate early response factor 3 (IER3) and cyclin D1 (CCND1), were subsequently validated by qPCR. As shown in Figure 1, G and H, PRL induced expression of both genes, whereas PRLrYDmut expression markedly decreased induction compared with PRLr. Taken together, these data strongly suggested that a subset of PRL-induced genes might require the transactivation activity of the PRLr for proper expression. Furthermore, the Venn diagram in Figure 1F suggests that subsets of genes may exhibit an independent response to either PRL or nuclear PRLr (a full list of overlapping genes is given in Supplemental Table S6).
PRLr Depletion or Mutation of the PRLr TAD Results in Impaired Tumorigenic Potential in Vitro
In previous reports, overexpression of the PRLr increased tumor cell growth potential.50 Furthermore, the expression of a degradation-resistant PRLr construct accelerated proliferation and increased invasive growth.20 To determine whether the PRLr TAD might be an integral part of PRLr-mediated proliferation and tumorigenesis, gene lists comparing differential regulation of PRLr- with PRLrYDmut-expressing cells were uploaded into IPA. The analysis revealed that the top molecular and cellular functions affected by the PRLr TAD were cellular growth/proliferation and gene expression (Table 6). Moreover, cancer was identified as the top disorder affected by the PRLr TAD.
Table 6.
List of Top Networks Affected by Mutation of the PRLr TAD
| Variable | P value | No. of molecules |
|---|---|---|
| Top diseases and disorders | ||
| Cancer | 1.23 × 10−4 to 4.42 × 10−2 | 13 |
| Reproductive system disease | 1.03 × 10−3 to 4.41 × 10−2 | 9 |
| Top molecular and cellular functions | ||
| Cellular growth and proliferation | 1.38 × 10−4 to 3.80 × 10−2 | 10 |
| Gene expression | 1.84 × 10−4 to 3.42 × 10−2 | 7 |
The gene list shown in Supplemental Table S1, for PRL-specific genes differentially regulated between PRLr and PRLrYDmut, was uploaded into IPA. A core IPA was run using this gene list (and filters for breast cancer were applied). This table illustrates the top diseases and top molecular and cellular functions identified using this analysis.
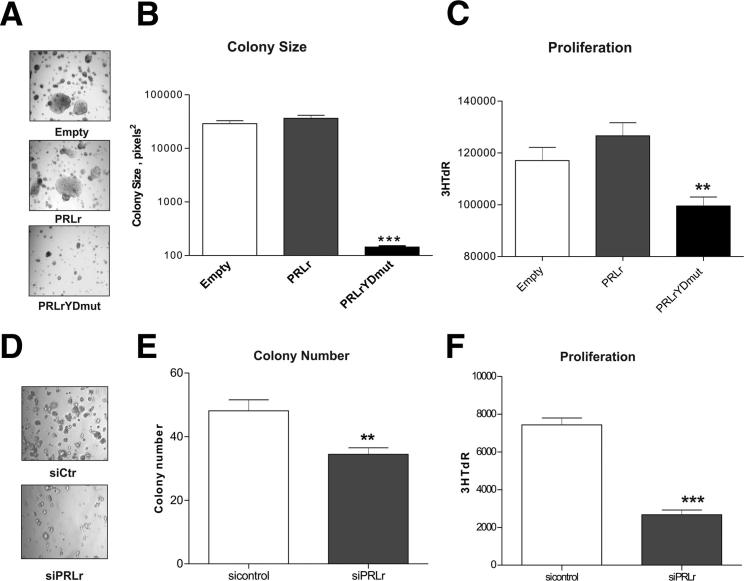
We next examined the functional effects of PRLr and PRLrYDmut expression based on our findings that PRLr enhanced the transcription of genes involved in proliferation/tumorigenesis. First, a soft agar assay was used to determine whether the transactivation function of the PRLr was relevant to tumorigenic growth in vitro. The expression of the PRLr, but not the PRLrYDmut, previously induced soft agar colony formation in the partially transformed MCF10AΔp53 cell line.41 Correspondingly, as shown in Figure 3, A and B, PRLrYDmut demonstrated a dominant negative effect by impairing colony growth in ER+ MCF7 human breast cancer cells. In addition, the proliferation of MCF7 stable transfectants was measured. The expression of PRLrYDmut significantly decreased proliferation, as measured by tritiated thymidine incorporation (Figure 3C).
Figure 3.
PRLrYDmut expression or PRLr depletion impairs anchorage-dependent and anchorage-independent growth in breast cancer cells. A: MCF7 stable cells were grown on soft agar for 3 weeks, and at day 20 images were taken under phase-contrast microscopy. B: Quantification of colony area; analysis was performed using ImageJ software colony cutoff, with a cutoff value of ≥50 μm. All conditions were performed in triplicate. C: Proliferation of MCF7 stable transfectants, as determined by the uptake of tritiated thymidine, as indicated by 3HTdR count on the y axis. Results were obtained two times, where the mean ± SEM is representative of 24 replicate measurements from one experiment. D: T47D cells transfected with the indicated siRNA cells were grown on soft agar for 2 weeks, and pictures were taken at day 13 under phase-contrast microscopy. E: Quantification of colony number. Analysis was performed using ImageJ software. All experiments were performed in triplicate. F: Proliferation of T47D cells transfected with the indicated siRNAs. Results were determined by the uptake of tritiated thymidine, shown by scintillation count on the y axis. Results were performed two times, where the mean ± SEM is representative of 24 replicate measurements from one experiment. 3HTdR, tritiated thymidine; empty, empty vector control; sicontrol, nonsilencing siRNA; siPRLr, siRNA targeted against PRLr. **P < 0.01, ***P < 0.001.
To complement these data, we examined the functional effects of siRNA-mediated PRLr knockdown on T47D breast cancer cells, an ER+ cell line known to express high levels of the PRLr. Knockdown assessed by Western blot analysis demonstrated approximately 90% efficiency (see Supplemental Figure S3). Colony formation was significantly reduced in cells harboring siRNA against the PRLr (Figure 3, D and E). In previous studies, down-regulation of the PRLr resulted in slower cell growth in normal growth conditions.21 Similarly, siRNA targeted against the PRLr inhibited cell proliferation (Figure 3F). Taken as a whole, these results suggested that the PRLr, and specifically the PRLr TAD, contributed to both anchorage-dependent and anchorage-independent growth of human breast cancer cells.
Mutation of the PRLr TAD or Depletion of the PRLr Results in Impaired ERα and PR Levels
Genes differentially regulated by both the PRLr TAD and PRL were used to generate interaction networks using IPA (Figure 4, A and B, respectively). Both ERα and PR were prominently situated in both the network highlighting the function of the PRLr TAD (Figure 4A) and the network highlighting PRL-specific effects (Figure 4B). Subsets of genes in these networks have been validated (Figure 1, G and H, and Figure 2), including RARA, CEBPβ, IER3, CCND1, and BCL6. In previous reports, PRL acted in synergy with the steroid hormones estrogen and progesterone to promote/increase mammary growth, development, and differentiation.51 Furthermore, co-expression of the PRLr with the estrogen receptor (ERα) and the progesterone receptor (PR) was observed in many breast tumors.50, 52, 53 Previous reports demonstrated that PRL up-regulated PR50 and ERα in breast cancer cells54, 55; this induction was observed only after prolonged PRL stimulation (approximately 24 to 72 hours). We, therefore, examined the effects of prolonged PRL exposure on ERα/PR mRNA levels in PRLr or PRLrYDmut-expressing MCF7 cells. Although prolonged PRL stimulation (48 hours) did not alter ERα levels (data not shown), it resulted in a modest increase in PR transcription (Supplemental Figure S4). Moreover, PRLr, but not PRLrYDmut, expression potentiated this effect (Supplemental Figure S4). Collectively, these data suggested that the PRLr-specific modulation of ERα and PR transcription might occur through an indirect mechanism.
Figure 4.
IPA identifies that the ER and PR are affected by both the PRLr TAD and PRL. A and B: Gene lists generated by Partek Genomics Suite; PRLr + PRL versus PRLrYDmut + PRL (A) and empty + PRL versus empty (B) were uploaded into IPA. The top interaction networks generated by IPA are shown for genes affected by the PRLr TAD (A) and PRL treatment (B), where red shows up-regulated genes; green, down-regulated genes; and gray, integral protein complexes that are affected by these changes. Empty, empty vector control; sicontrol, nonsilencing siRNA; siPRLr, siRNA targeted against PRLr.
By using gene lists generated from the microarray analysis, we further examined the effects of the PRLr on ER/PR expression. We found that both ER and PR were identified as significantly up-regulated in untreated PRLr-expressing cells compared with control (1.6- and 1.4-fold, respectively; Table 7), whereas neither ER nor PR were present in the gene list comparing untreated PRLrYDmut-expressing cells with control (cutoff ± 1.2-fold change, P < 0.05). Given this, we analyzed ERα and PR expression via qPCR in both PRLr and PRLrYDmut-expressing cells (normal growth conditions). As expected, both ERα and PR expression levels were decreased in response to PRLrYDmut expression compared with PRLr (Figure 5, A and B).
Table 7.
Estrogen and Progesterone Receptor Expression Is Up-Regulated in Untreated PRLr-Expressing Cells
| Gene symbol | P value | Fold up-regulated (WT vs E) |
|---|---|---|
| ESR1 | 0.005595 | 1.65559 |
| PGR | 0.00217 | 1.42978 |
By using the list generated in Supplemental Table S5, for WT versus empty, the fold change and P values are highlighted for ESR1 and PGR. Partek Genomics Suite was also used to analyze MUT versus empty; however, neither ESR1 nor PGR passed the applied filters (±1.2-fold change, P < 0.05).
Figure 5.
Mutation of the PRLr TAD or PRLr depletion impairs expression of ERα and PR. A and B: Real-time PCR performed on cDNA collected from MCF7 stable cells to measure ERα (A) or PR (B) mRNA levels. Results are representative of two separate experiments. The values presented are the mean ± SEM from replicate measurements within one experiment. C: Western blot analysis demonstrating PRLr knockdown in MCF7 shPRLr cells versus shNS control cells. D and E: Real-time PCR performed on cDNA collected from MCF7 shNS or shPRLr cells to measure ERα (D) or PR (E) mRNA levels. Results are representative of three separate experiments. The values presented are the mean ± SEM from replicate measurements within one experiment. Empty, empty vector control, expressing MCF7 cells. *P < 0.05, **P < 0.01, and ***P < 0.001.
To validate these results, ERα/PR transcript levels were assessed on PRLr depletion. As shown in Figure 5C, shRNA-mediated knockdown of the PRLr in MCF7 cells significantly reduced PRLr protein levels compared with control shRNA (cells termed shPRLr and shNS, respectively). PRLr depletion also significantly reduced ERα (Figure 5D) and PR transcript levels (Figure 5E). To complement shRNA data, siRNA against the PRLr was also used. This also resulted in an appreciable reduction of ERα (Supplemental Figure S5A) and PR (Supplemental Figure S5B) transcript levels. Finally, we questioned whether reintroduction of the PRLr could rescue the decreased expression of ERα and PR seen in PRLr knockdown cell lines. To do this, we reintroduced PRLr or PRLrYDmut into shPRLr MCF7 cells. Because shRNA targeted the PRLr 3′-untranslated region, it enabled the reintroduction of PRLr or PRLrYDmut, which lacked the PRLr 3′-untranslated region. By using these rescue cell lines, the expression of ERα and PR in MCF7 was assessed. Reintroduction of PRLr or PRLrYDmut restored PRLr levels to slightly higher than physiological levels (Supplemental Figure S5C). Interestingly, the introduction of PRLr into shPRLr cells rescued ERα and PR levels to approximately 25% to 30%, whereas expression of PRLrYDmut did not increase ERα to higher than shPRLr levels (Supplemental Figure S5, D and E). Collectively, these data strongly suggested that PRLr TAD was critical for proper expression of ERα and PR.
Mutation of the PRLr TAD or Depletion of the PRLr Results in Decreased Estrogen Responsiveness
We next questioned if diminished ERα levels in PRLrYDmut-expressing cells would correlate to decreased estrogen responsiveness by using a luciferase reporter construct driven by multiple estrogen response elements (Figure 6A). MCF7 stable cell lines were transiently transfected with the ERE-luciferase reporter, and transcriptional activity was measured in response to 10 nmol/L E2. Interestingly, PRLr expression promoted a significant increase in E2-driven luciferase activity, whereas PRLrYDmut expression yielded levels similar to control (Figure 6B). As a second measure of estrogen responsiveness, the ER-regulated genes c-fos, PR, c-myc, and CCND1 were analyzed by qPCR. MCF7 stable transfectants were depleted of estrogen for 4 days, followed by the addition of E2 (1 hour). PRLrYDmut expression significantly decreased E2-driven transcription (Figure 6, C–E and Supplemental Figure S6A). We did not, however, observe an increase in transcription with PRLr expression, which might be due to saturation of E2-induced transcription.
Figure 6.
PRLrYDmut expression or PRLr depletion decreases estrogen responsiveness. A: Schematic of ERE-luciferase reporter. B: MCF7 stable cells were transfected with an ERE-luciferase reporter and a control reporter for 24 hours, serum starved for an additional 24 hours, and treated with E2 for 6 hours. Results are presented as fold change relative to non-treated empty vector control. Values are from a representative experiment repeated three separate times, in which SEM represents the error of three replicates performed within one experiment. C–E: Real-time PCR performed on cDNA collected from E2-treated MCF7 stable cell pools to determine c-fos (C), PR (D), and c-myc (E) mRNA induction. MCF7 transfectants were serum starved for 24 hours before 1 hour of E2 treatment. Values were normalized to 18S RNA, and fold change was calculated by comparing each value with the non-treated control sample; values are representative of three separate experiments with SEM (F–H). Real-time PCR performed on cDNA collected from E2-treated MCF7 stable knockdown cell lines to determine c-fos (F), PR (G), and c-myc (H) mRNA induction. All P values in A–H were calculated using one-way analysis of variance with a Bonferroni multiple comparisons test. Calculated P values are highlighted in the figure. Empty, empty vector control, expressing MCF7 cells. *P < 0.05, **P < 0.01, and ***P < 0.001.
To corroborate these data, we additionally measured E2-driven transcription in MCF7 shNS or shPRLr cells. Depletion of the PRLr completely inhibited E2-mediated induction of c-fos, PR, c-myc (Figure 6, F–H), and CCND1 (Supplemental Figure S6B). Taken together, our data suggested that the PRLr was critical for estrogen responsiveness in breast cancer cells, in part through its TAD.
Y406 Phosphorylation Is Up-Regulated in Malignant Breast Tissue
To investigate the correlation between the PRLr and the steroid hormone receptors, we queried Oncomine Research, a cancer profiling database (http://www.oncomine.org, last accessed June 30, 2012). Database analysis using Oncomine Research revealed several studies that found a significant increase in PRLr expression in breast cancer compared with normal breast tissue (Table 8 and Figure 7A).56, 57, 58 This analysis also revealed that the PRLr was expressed at higher levels in ER+ and PR+ breast cancers compared with ER−/PR− breast cancers59, 60, 61, 62 (Table 8 and Figure 7, B and C).
Table 8.
PRLr Expression in Breast Carcinoma
| Source | Tissue | P value | Fold change (PRLr up-regulation) |
|---|---|---|---|
| PRLr expression in breast cancer vs normal | |||
| 56 | Ductal breast cancer vs norm | 1.44 × 10−16 | 2.945 |
| 57 | Invasive ductal breast carcinoma vs norm | 3.03 × 10−10 | 2.95 |
| 58 | Ductal breast carcinoma vs norm | 2.39 × 10−6 | 2.008 |
| PRLr expression in ER+ vs ER− breast tumors | |||
| 59 | Ductal breast carcinoma | 1.47 × 10−10 | 2.932 |
| 60 | Invasive ductal breast carcinoma | 2.51 × 10−12 | 2.203 |
| PRLr expression in PR+ vs PR− breast tumors | |||
| 61 | Breast carcinoma | 1.42 × 10−6 | 1.759 |
| 62 | Breast carcinoma | 6.69 × 10−6 | 1.336 |
To determine the clinical relevance of PRLr expression in human mammary carcinoma, we used the Oncomine cancer microarray database to analyze the expression profile of PRLr in human cancers. The table lists four independent studies in which PRLr was up-regulated in ductal or invasive breast cancer versus normal tissue (study: PRLr expression in breast cancer). In addition, three studies demonstrated higher PRLr expression in ER+ versus ER− breast tumors (study: PRLr expression in ER+ versus ER− breast cancers), and two studies demonstrated higher PRLr expression in PR+ versus PR− breast cancers. The type of study, tissue type, fold up-regulation (of PRLr), and corresponding P values are listed. In addition, all references are listed in the table.
Figure 7.
PRLr Y406 phosphorylation is increased as a function of neoplastic progression. A–C: Data analysis performed using the Oncomine database. Representative graphs demonstrating that the PRLr is overexpressed in ductal breast cancer compared with normal breast tissue (A),56 the PRLr is more highly expressed in ER+ versus ER− breast tumors (B),59 and the PRLr is more highly overexpressed in PR+ versus PR− breast tumors (C).61D: Representative anti–phospho-Y406 PRLr IHC from patient-matched normal, primary tumor, and lymph node metastasis breast tissue. The photomicrograph demonstrates the observed increase in PRLr Y406 phosphorylation in malignant tissue, with significant accumulation in the cell nucleus. Original magnification, ×400. E: Quantification of nuclear PRLr Y406 phosphorylation. F: Quantification of cytoplasmic PRLr Y406 phosphorylation. The results are shown as the mean ± SEM, and P values were determined using a one-way analysis of variance. G: Values from TMA analysis were stratified into ER− and ER+ groups for analysis, and a two-way analysis of variance was performed to analyze the effects of both ER status and tumor status. Statistical analysis demonstrated a significant increase in Y406 phosphorylation as a function of tumor status and ER positivity, although no individual groups were significantly different from each other, as analyzed by the post hoc Bonferroni test. Normal, normal breast tissue; Tumor, primary breast tumor; LN Met, lymph node metastasis. *P < 0.05, **P < 0.01, and ***P < 0.001.
Because microarray analysis and RT-qPCR data both suggested that the PRLr TAD might play a functional role in PRLr-driven tumorigenesis, we next assessed the clinical relevance of an active PRLr TAD (via Y406 phosphorylation). To that end, we performed IHC with a phospho-specific anti-PRLr antibody on a breast cancer progression TMA. This antibody was characterized in our previous work.41 The TMA consisted of 15 patient-matched normal, tumor, and lymph node metastasis tissue and six patient-matched normal and tumor tissue samples that were taken from both ER+/Her2− and ER−/Her2− patients with breast cancer (as described in Materials and Methods). Notably, PRLr Y406 phosphorylation was primarily observed in the nucleus of tissue samples, consistent with the hypothesis that phosphorylation activated the nuclear transactivation properties of the PRLr. Visual scoring of PRLr Y406 phosphorylation revealed a significant increase in nuclear intensity of primary breast tumor and lymph node metastasis samples compared with normal samples (Figure 7, D and E). Moreover, the staining intensity increased as a function of neoplastic progression. No significant differences, however, were observed in cytoplasmic intensity of PRLr Y406 phosphorylation (Figure 7F). Given the identified association between the PRLr Y406 phosphorylation and ER expression, samples from the TMA were stratified into ER− or ER+ cohorts. Although no individual tumor groups were significantly different from each other in ER− and ER+ cohorts, a global increase in phospho-Y406 staining was observed between the ER− and ER+ groups (Figure 7G). Taken together, these data supported a pathological role for PRLr Y406 phosphorylation in human breast cancer and suggested that assessment of PRLr Y406 phosphorylation in the nucleus of breast cancer tissue samples might have prognostic utility.
Discussion
Although overexpression of the PRLr in breast cancers has been previously reported,4 our understanding of the specific pathways and transcriptional axes that account for this phenomenon remains insufficient. Herein, we reveal associations between the PRLr TAD and the following: i) global PRL-induced gene expression, ii) ERα and PR transcription, and iii) neoplastic progression. Given this, the data presented herein have a direct and significant correlation to the pathogenesis of both ERα/PR-positive tumors, in which the PRLr may potentially function as a key regulator that contributes to steroid hormone receptor expression in breast cancers.
Microarray analysis confirmed the hypothesis that the PRLr TAD contributes to PRL-induced gene expression. Although several genes identified in microarray analysis have previously been identified as PRL-specific target genes, such as CCND163, 64 and C/EBPβ,65 we also identified several novel PRL-induced genes, such as dexamethasone-induced Ras-related protein 1 (RASD1), MYADM, and ZYX. These novel PRL-induced target genes are significant for the following reasons: i) RASD1 functions as a tumor suppressor gene by inhibiting cell growth,66 ii) MYADM is important for the organization of lamellipodium extension and cell motility,67 and iii) reports suggest that ZYX is a critical regulator of DNA damage–induced cell death induction.68 The role of such genes in normal and malignant processes within the mammary gland requires further characterization, but it will ultimately contribute to the overall knowledge of the complex PRL/PRLr signaling network.
IPA of microarray data revealed cellular growth/proliferation and gene expression as the top molecular functions affected by the PRLr TAD. In addition, IPA identified cancer as the top disorder associated with PRLr TAD function. These results are particularly relevant because subsets of genes identified by microarray analysis are implicated in proliferation and migration and may play a role in breast cancer pathogenesis. For instance, the cell cycle protein cyclin D1 is up-regulated in approximately 50% of breast tumors.69, 70 Similarly, the transcription factor, C/EBPβ, is implicated in the pathogenesis of breast cancer,71 because an elevation in C/EBPβ expression is associated with metastatic breast cancer, a high tumor grade, and an overall poorer prognosis.72 SRF regulates transcription of the oncogene, c-fos, which is critical for the growth of MCF7 breast cancer cells.73 Targeting RARA via its ligand lowers the threshold of taxol-induced cell killing,74 and loss of ERRFI1 is associated with ErbB2/HER-2 oncogenic potency, leading to herceptin resistance.75 The notion that the PRLr, but not the transactivation-deficient PRLrYDmut, up-regulates genes involved in a proproliferative/migratory phenotype is reinforced by our observation that PRLrYDmut expression decreases proliferation and soft agar colony formation in vitro. Together, these data suggest that the PRLr TAD plays a key role in both PRL-induced gene expression and cellular functions that contribute to a tumorigenic phenotype.
Surprisingly, although a subset overlap existed when comparing PRL with PRLr regulated genes, many genes were regulated by either PRL (n = 383) or PRLr (n = 4683). These data are comparable with early reports that PRL and PRLr may have independent functions in the nucleus.76 Differential gene regulation could result from constitutive cytoplasmic-to-nuclear shuttling of Stat5a77 or HMGN2,78 which can, in turn, enable the binding of PRLr to its target genes. Jak2 has also been in the nucleus,79, 80 and could potentially promote the phosphorylation of the PRLr TAD directly in the nucleus. Interestingly, Jak2 also promotes H3Y41 phosphorylation, preventing binding of HP1α and, thus, promoting an open chromatin state.80 Another possibility is that the autocrine/paracrine production of endogenous PRL could promote nuclear localization of PRLr independently of exogenous PRL.5
Although we observed that a functional PRLr TAD is required for the expression of ERα, neither ERα nor PR was identified as an immediate early-response gene after PRL stimulation. We propose two scenarios to explain how the PRLr TAD may be regulating the transcription of these genes. The first scenario is indirect regulation. In the rat corpus luteum, PRL stimulates ERα transcription through the activation of Stat5a. These reports demonstrate that constitutively active PRLr enhances transcription of ERα, as measured by an ER-luciferase reporter containing the proximal ERα promoter.81, 82 Furthermore, mutation of Stat5a binding within the proximal promoter of rat ERα prevents the PRL-mediated induction of an ER-luciferase reporter. PRL also increases ERα transcription after 12 hours, and Jak2 tyrosine kinase is required for this increase.81, 82 Herein, we demonstrate that PRL-mediated induction of PR gene expression occurs only after long-term treatment (ie, 48-hour treatment; Supplemental Figure S4). Although the previously described studies suggest that Stat5a is in some way required for ERα transcription, in all of these reports induction of transcription occurred only after prolonged PRL stimulation. In addition, Stat5a binding sites have not been identified within the human ERα proximal promoter. Thus, the possibility cannot be excluded that the reported results were due to the PRL-specific induction of a subset of genes whose protein products, in turn, served to regulate ERα transcription. A second possibility for PRLr TAD-mediated regulation of ER/PR is coregulation. For instance, Stat5a may regulate ERα in coordination with another transcription factor. An example of this is in the case of PRL-mediated β-casein transcriptional activation, which requires Stat5a, C/EBPβ, and the glucocorticoid receptor. However, if this is the case, it remains to be seen why PRL does not promote immediate early induction of ER.
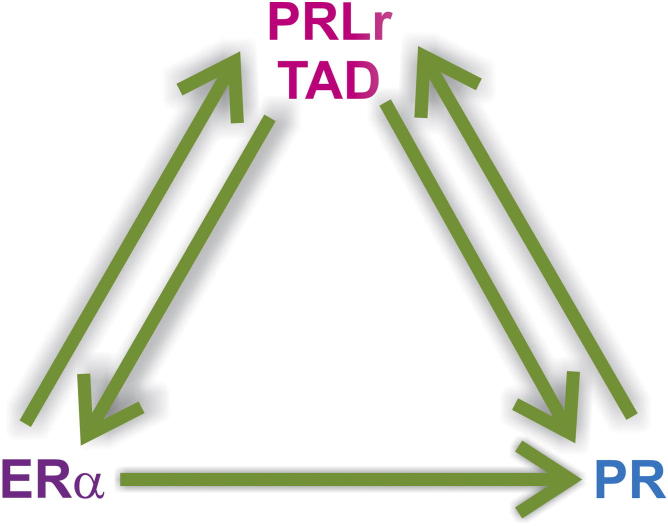
PRL and steroid hormones function synergistically in the mammary gland to regulate growth and differentiation. Progesterone and estrogen can increase PRLr expression,83, 84, 85, 86 and in both cases, the coordinated interactions of ERα or PR with C/EBPβ on the PRLr promoter were responsible for the up-regulation of PRLr expression. In addition, PRL can amplify the expression of its own receptor.87 We report herein that the PRLr TAD mutation decreases expression of ER and PR specifically in the presence of serum, which possesses hormones (ie, lactogens, estrogen, and progesterone).88 It is, therefore, conceivable that the indirect action of PRLr on ER/PR expression is a product of cross talk between prolactin, steroid hormones, and their respective receptors. In serum-free conditions, mutation of the PRLr TAD decreases PR expression only after treatment with estrogen. This further supports a mechanism of cross talk between the PRL/PRLr and steroid hormones/receptors affecting ER-mediated gene expression. Given that the PRLr TAD may play a role in ERα and PR expression and ER and/or PR regulate PRLr expression, we propose a model in which levels of ERα, PR, and PRLr expression are controlled by a tripartite regulatory loop. This proposed model suggests that levels of either the PRLr or PRLr TAD phosphorylation contribute to the balance of ERα/PR/PRLr expression and synergy (Figure 8).
Figure 8.
Correlation between ERα, PR, and PRLr expression results from a tripartite regulatory loop. Model demonstrating the relationship between PRLr, ERα, and PR. Herein, perturbation of the PRLr, PRLr Y406 phosphorylation, ERα, or PR level disrupts the balance in expression of all three receptors.
The relevance of our observations in MCF7 breast cancer cells was substantiated by TMA, in which up-regulation of PRLr Y406 phosphorylation occurs as a function of neoplastic progression in breast cancer. We only observe increased PRLr Y406 phosphorylation in the nuclear compartment, with no significant differences in the cytoplasm. This finding is particularly significant because the subcellular localization of translocated receptors may be of clinical importance. For example, the expression of EGFR does not yet provide predictive value for the overall survival of patients with breast cancer. However, IHC analysis of 130 breast carcinomas by Lo et al23 reveals a significant inverse correlation between nuclear EGFR expression and breast cancer patient survival, whereas no such correlation is identified with cytoplasmic EGFR. Collectively, these reports emphasize the importance of examining phosphorylation status and subcellular localization of translocated receptors as prognostic markers in breast cancer pathogenesis.
Co-expression of the PRLr with ERα and PR is observed in many breast tumors,50, 52, 53, 89 and prolonged PRL exposure increases ERα and PR levels in the ER/PR-positive breast cancer cell lines, T47D and MCF7.50, 55 These observations suggest that the transcriptional cross regulation of PRLr, ERα, and PR may contribute to their co-expression in breast cancer. However, although others have suggested that PRL may stimulate/contribute to ERα and PR transcription,54, 55 to our knowledge, this is the first report to determine the requirement for PRLr Y406 phosphorylation in steroid hormone receptor transcription. Thus, because the quantification and inhibition of both ER and ErbB2 are useful in the stratification and therapy of breast cancer,90 similar strategies may be of utility for the PRLr.
Acknowledgments
We thank Drs. Debu Chakravarti (Northwestern University) for the use of reagents for retroviral experiments, Anthony Kossiakoff (University of Chicago) for providing human recombinant PRL, Donald McDonald (Duke University) for supplying the three times ERE-luciferase reporter, Feng Fang (Northwestern University) for his guidance in cloning and microarray experimental design, and Joe Devaney and Monica Hubal (Children’s National Medical Center) for their assistance in microarray analysis.
Footnotes
Supported by a predoctoral traineeship award (DOD BC073401) and grants from Avon and the Lynn Sage Cancer Research Foundations.
Current address of A.A.F., George Washington University School of Medicine and Health Sciences, Center for Genetic Medicine Research, Children’s National Medical Center, Washington, DC.
Supplemental material for this article can be found at http://dx.doi.org/10.1016/j.ajpath.2012.09.021.
Supplemental Data
Validation of genes identified by microarray analysis. Negative control is used; real-time PCR demonstrates that FAS is not affected by PRL treatment of PRLr/PRLrYDmut expression.
HMGN2 knockdown impairs gene expression in a similar manner to expression of PRLrYDmut. A–E: qPCR was performed on control of HMGN2 knockdown cell lines (shNS or shHMGN2) expressing cell lines treated with PRL (1 hour). Results demonstrated impaired PRL-induced gene expression with depletion of HMGN2. shHMGN2, MCF7 HMGN2 shRNA-expressing cells; shNS, MCF7 control shRNA-expressing cells. **P < 0.01, ***P < 0.001.
Efficient siRNA-mediated knockdown of the PRLr. Western blot analysis demonstrating PRLr knockdown in T47D cells transfected with siPRLr, sicontrol as a negative control (Ctr), and small-interfering glyceraldehyde-3-phosphate dehydrogenase (GAPDH) as a positive control. Cells were harvested at 48 hours after transfection.
Prolonged PRL induces PR gene expression. Real-time PCR was performed on cDNA collected from MCF7 stable cell lines with expression of the indicated constructs. MCF7 cells were grown in charcoal-stripped, phenol red–free media for 24 hours, followed by treatment of PRL for 48 hours. Data are representative of two separate experiments, in which the mean ± SEM represents the error of replicate measurements from a single experiment. Empty, empty vector. **P < 0.01, ***P < 0.001.
PRLr depletion impairs expression of ERα and PR. A and B: Real-time PCR performed on cDNA collected from T47D cells transfected with the indicated siRNAs to measure ERα (A) or PR (B) mRNA levels. Results are representative of three separate experiments. The values presented are the mean ± SEM from replicate measurements within one experiment. C: Real-time PCR performed on cDNA collected from MCF7 rescue cell lines, demonstrating successful PRLr rescue by PRLr or PRLrYDmut. Results were normalized to PRLr levels in control cells expressing shNS, along with an empty vector control. D and E: Real-time PCR performed on cDNA collected from MCF7 rescue cells to measure ERα (D) or PR (E) mRNA levels. Results are shown as percentage expression compared with shNS control cells. EV, empty vector; siPRLr, siRNA targeted against PRLr. **P < 0.01, ***P < 0.001.
PRLrYDmut expression or PRLr depletion decreases estrogen-responsiveness. A: Real-time PCR performed on cDNA collected from E2-treated MCF7 stable cell pools to determine CCND1 mRNA induction. MCF7 transfectants were serum starved for 24 hours, before 1 hour of E2 treatment. B: Real-time PCR performed on cDNA collected from E2-treated MCF7 stable knockdown cell lines to determine CCND1 mRNA induction. Values were normalized to 18S RNA, and fold change was calculated by comparing each value with the non-treated control sample and are representative of three separate experiments with SEM. All P values were calculated using one-way analysis of variance with a Bonferroni multiple-comparison test. Calculated P values are highlighted in the figure. Empty, empty vector control, expressing MCF7 cells. **P < 0.01, ***P < 0.001.
References
- 1.Das R., Vonderhaar B.K. Prolactin as a mitogen in mammary cells. J Mammary Gland Biol Neoplasia. 1997;2:29–39. doi: 10.1023/a:1026369412612. [DOI] [PubMed] [Google Scholar]
- 2.Perks C.M., Keith A.J., Goodhew K.L., Savage P.B., Winters Z.E., Holly J.M. Prolactin acts as a potent survival factor for human breast cancer cell lines. Br J Cancer. 2004;91:305–311. doi: 10.1038/sj.bjc.6601947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Maus M.V., Reilly S.C., Clevenger C.V. Prolactin as a chemoattractant for human breast carcinoma. Endocrinology. 1999;140:5447–5450. doi: 10.1210/endo.140.11.7245. [DOI] [PubMed] [Google Scholar]
- 4.Reynolds C., Montone K.T., Powell C.M., Tomaszewski J.E., Clevenger C.V. Expression of prolactin and its receptor in human breast carcinoma. Endocrinology. 1997;138:5555–5560. doi: 10.1210/endo.138.12.5605. [DOI] [PubMed] [Google Scholar]
- 5.Clevenger C.V., Plank T.L. Prolactin as an autocrine/paracrine factor in breast tissue. J Mammary Gland Biol Neoplasia. 1997;2:59–68. doi: 10.1023/a:1026325630359. [DOI] [PubMed] [Google Scholar]
- 6.Wennbo H., Gebre-Medhin M., Gritli-Linde A., Ohlsson C., Isaksson O.G., Tornell J. Activation of the prolactin receptor but not the growth hormone receptor is important for induction of mammary tumors in transgenic mice. J Clin Invest. 1997;100:2744–2751. doi: 10.1172/JCI119820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rose-Hellekant T.A., Arendt L.M., Schroeder M.D., Gilchrist K., Sandgren E.P., Schuler L.A. Prolactin induces ERalpha-positive and ERalpha-negative mammary cancer in transgenic mice. Oncogene. 2003;22:4664–4674. doi: 10.1038/sj.onc.1206619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hankinson S.E., Willett W.C., Michaud D.S., Manson J.E., Colditz G.A., Longcope C., Rosner B., Speizer F.E. Plasma prolactin levels and subsequent risk of breast cancer in postmenopausal women. J Natl Cancer Inst. 1999;91:629–634. doi: 10.1093/jnci/91.7.629. [DOI] [PubMed] [Google Scholar]
- 9.Tworoger S.S., Sluss P., Hankinson S.E. Association between plasma prolactin concentrations and risk of breast cancer among predominately premenopausal women. Cancer Res. 2006;66:2476–2482. doi: 10.1158/0008-5472.CAN-05-3369. [DOI] [PubMed] [Google Scholar]
- 10.Tworoger S.S., Eliassen A.H., Sluss P., Hankinson S.E. A prospective study of plasma prolactin concentrations and risk of premenopausal and postmenopausal breast cancer. J Clin Oncol. 2007;25:1482–1488. doi: 10.1200/JCO.2006.07.6356. [DOI] [PubMed] [Google Scholar]
- 11.Mujagic Z., Mujagic H. Importance of serum prolactin determination in metastatic breast cancer patients. Croat Med J. 2004;45:176–180. [PubMed] [Google Scholar]
- 12.Anderson E., Ferguson J.E., Morten H., Shalet S.M., Robinson E.L., Howell A. Serum immunoreactive and bioactive lactogenic hormones in advanced breast cancer patients treated with bromocriptine and octreotide. Eur J Cancer. 1993;29A:209–217. doi: 10.1016/0959-8049(93)90178-i. [DOI] [PubMed] [Google Scholar]
- 13.DaSilva L., Howard O.M., Rui H., Kirken R.A., Farrar W.L. Growth signaling and JAK2 association mediated by membrane-proximal cytoplasmic regions of prolactin receptors. J Biol Chem. 1994;269:18267–18270. [PubMed] [Google Scholar]
- 14.Clevenger C.V., Furth P.A., Hankinson S.E., Schuler L.A. The role of prolactin in mammary carcinoma. Endocr Rev. 2003;24:1–27. doi: 10.1210/er.2001-0036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fuh G., Wells J.A. Prolactin receptor antagonists that inhibit the growth of breast cancer cell lines. J Biol Chem. 1995;270:13133–13137. doi: 10.1074/jbc.270.22.13133. [DOI] [PubMed] [Google Scholar]
- 16.Bogorad R.L., Courtillot C., Mestayer C., Bernichtein S., Harutyunyan L., Jomain J.B., Bachelot A., Kuttenn F., Kelly P.A., Goffin V., Touraine P. Identification of a gain-of-function mutation of the prolactin receptor in women with benign breast tumors. Proc Natl Acad Sci U S A. 2008;105:14533–14538. doi: 10.1073/pnas.0800685105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Canbay E., Degerli N., Gulluoglu B.M., Kaya H., Sen M., Bardakci F. Could prolactin receptor gene polymorphism play a role in pathogenesis of breast carcinoma? Curr Med Res Opin. 2004;20:533–540. doi: 10.1185/030079904125003232. [DOI] [PubMed] [Google Scholar]
- 18.Oakes S.R., Robertson F.G., Kench J.G., Gardiner-Garden M., Wand M.P., Green J.E., Ormandy C.J. Loss of mammary epithelial prolactin receptor delays tumor formation by reducing cell proliferation in low-grade preinvasive lesions. Oncogene. 2007;26:543–553. doi: 10.1038/sj.onc.1209838. [DOI] [PubMed] [Google Scholar]
- 19.Li Y., Clevenger C.V., Minkovsky N., Kumar K.G., Raghunath P.N., Tomaszewski J.E., Spiegelman V.S., Fuchs S.Y. Stabilization of prolactin receptor in breast cancer cells. Oncogene. 2006;25:1896–1902. doi: 10.1038/sj.onc.1209214. [DOI] [PubMed] [Google Scholar]
- 20.Plotnikov A., Varghese B., Tran T.H., Liu C., Rui H., Fuchs S.Y. Impaired turnover of prolactin receptor contributes to transformation of human breast cells. Cancer Res. 2009;69:3165–3172. doi: 10.1158/0008-5472.CAN-08-4033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Swaminathan G., Varghese B., Fuchs S.Y. Regulation of prolactin receptor levels and activity in breast cancer. J Mammary Gland Biol Neoplasia. 2008;13:81–91. doi: 10.1007/s10911-008-9068-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Watson C.J., Miller W.R. Elevated levels of members of the STAT family of transcription factors in breast carcinoma nuclear extracts. Br J Cancer. 1995;71:840–844. doi: 10.1038/bjc.1995.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lo H.W., Xia W., Wei Y., Ali-Seyed M., Huang S.F., Hung M.C. Novel prognostic value of nuclear epidermal growth factor receptor in breast cancer. Cancer Res. 2005;65:338–348. [PubMed] [Google Scholar]
- 24.Lin S.Y., Makino K., Xia W., Matin A., Wen Y., Kwong K.Y., Bourguignon L., Hung M.C. Nuclear localization of EGF receptor and its potential new role as a transcription factor. Nat Cell Biol. 2001;3:802–808. doi: 10.1038/ncb0901-802. [DOI] [PubMed] [Google Scholar]
- 25.Komuro A., Nagai M., Navin N.E., Sudol M. WW domain-containing protein YAP associates with ErbB-4 and acts as a co-transcriptional activator for the carboxyl-terminal fragment of ErbB-4 that translocates to the nucleus. J Biol Chem. 2003;278:33334–33341. doi: 10.1074/jbc.M305597200. [DOI] [PubMed] [Google Scholar]
- 26.Ni C.Y., Murphy M.P., Golde T.E., Carpenter G. γ-Secretase cleavage and nuclear localization of ErbB-4 receptor tyrosine kinase. Science. 2001;294:2179–2181. doi: 10.1126/science.1065412. [DOI] [PubMed] [Google Scholar]
- 27.Offterdinger M., Schofer C., Weipoltshammer K., Grunt T.W. c-erbB-3: a nuclear protein in mammary epithelial cells. J Cell Biol. 2002;157:929–939. doi: 10.1083/jcb.200109033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lobie P.E., Wood T.J., Chen C.M., Waters M.J., Norstedt G. Nuclear translocation and anchorage of the growth hormone receptor. J Biol Chem. 1994;269:31735–31746. [PubMed] [Google Scholar]
- 29.Conway-Campbell B.L., Wooh J.W., Brooks A.J., Gordon D., Brown R.J., Lichanska A.M., Chin H.S., Barton C.L., Boyle G.M., Parsons P.G., Jans D.A., Waters M.J. Nuclear targeting of the growth hormone receptor results in dysregulation of cell proliferation and tumorigenesis. Proc Natl Acad Sci U S A. 2007;104:13331–13336. doi: 10.1073/pnas.0600181104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marti U., Wells A. The nuclear accumulation of a variant epidermal growth factor receptor (EGFR) lacking the transmembrane domain requires coexpression of a full-length EGFR. Mol Cell Biol Res Commun. 2000;3:8–14. doi: 10.1006/mcbr.2000.0177. [DOI] [PubMed] [Google Scholar]
- 31.Feng S., Xu J., Wang F., Kan M., McKeehan W.L. Nuclear localization of a complex of fibroblast growth factor(FGF)-1 and an NH2-terminal fragment of FGF receptor isoforms R4 and R1alpha in human liver cells. Biochim Biophys Acta. 1996;1310:67–73. doi: 10.1016/0167-4889(95)00146-8. [DOI] [PubMed] [Google Scholar]
- 32.Johnston C.L., Cox H.C., Gomm J.J., Coombes R.C. Fibroblast growth factor receptors (FGFRs) localize in different cellular compartments: a splice variant of FGFR-3 localizes to the nucleus. J Biol Chem. 1995;270:30643–30650. doi: 10.1074/jbc.270.51.30643. [DOI] [PubMed] [Google Scholar]
- 33.Reilly J.F., Maher P.A. Importin β–mediated nuclear import of fibroblast growth factor receptor: role in cell proliferation. J Cell Biol. 2001;152:1307–1312. doi: 10.1083/jcb.152.6.1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hung L.Y., Tseng J.T., Lee Y.C., Xia W., Wang Y.N., Wu M.L., Chuang Y.H., Lai C.H., Chang W.C. Nuclear epidermal growth factor receptor (EGFR) interacts with signal transducer and activator of transcription 5 (STAT5) in activating Aurora-A gene expression. Nucleic Acids Res. 2008;36:4337–4351. doi: 10.1093/nar/gkn417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lo H.W., Hung M.C. Nuclear EGFR signalling network in cancers: linking EGFR pathway to cell cycle progression, nitric oxide pathway and patient survival. Br J Cancer. 2006;94:184–188. doi: 10.1038/sj.bjc.6602941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lo H.W., Cao X., Zhu H., Ali-Osman F. Cyclooxygenase-2 is a novel transcriptional target of the nuclear EGFR-STAT3 and EGFRvIII-STAT3 signaling axes. Mol Cancer Res. 2010;8:232–245. doi: 10.1158/1541-7786.MCR-09-0391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lo H.W., Cao X., Zhu H., Ali-Osman F. Cyclooxygenase-2 is a novel transcriptional target of the nuclear EGFR-STAT3 and EGFRvIII-STAT3 signaling axes. Mol Cancer Res. 2010;8:232–245. doi: 10.1158/1541-7786.MCR-09-0391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Marti U., Ruchti C., Kampf J., Thomas G.A., Williams E.D., Peter H.J., Gerber H., Burgi U. Nuclear localization of epidermal growth factor and epidermal growth factor receptors in human thyroid tissues. Thyroid. 2001;11:137–145. doi: 10.1089/105072501300042785. [DOI] [PubMed] [Google Scholar]
- 39.Lipponen P., Eskelinen M. Expression of epidermal growth factor receptor in bladder cancer as related to established prognostic factors, oncoprotein (c-erbB-2, p53) expression and long-term prognosis. Br J Cancer. 1994;69:1120–1125. doi: 10.1038/bjc.1994.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lincoln D.T., Sinowatz F., Temmim-Baker L., Baker H.I., Kolle S., Waters M.J. Growth hormone receptor expression in the nucleus and cytoplasm of normal and neoplastic cells. Histochem Cell Biol. 1998;109:141–159. doi: 10.1007/s004180050212. [DOI] [PubMed] [Google Scholar]
- 41.Fiorillo A.A., Medler T.R., Feeney Y.B., Liu Y., Tommerdahl K.L., Clevenger C.V. HMGN2 inducibly binds a novel transactivation domain in nuclear PRLr to coordinate Stat5a-mediated transcription. Mol Endocrinol. 2011;25:1550–1564. doi: 10.1210/me.2011-0106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Clarke R.B. p27KIP1 phosphorylation by PKB/Akt leads to poor breast cancer prognosis. Breast Cancer Res. 2003;5:162–163. doi: 10.1186/bcr596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Saceda M., Lippman M.E., Chambon P., Lindsey R.L., Ponglikitmongkol M., Puente M., Martin M.B. Regulation of the estrogen receptor in MCF-7 cells by estradiol. Mol Endocrinol. 1988;2:1157–1162. doi: 10.1210/mend-2-12-1157. [DOI] [PubMed] [Google Scholar]
- 44.Fang F., Rycyzyn M.A., Clevenger C.V. Role of c-Myb during prolactin-induced signal transducer and activator of transcription 5a signaling in breast cancer cells. Endocrinology. 2009;150:1597–1606. doi: 10.1210/en.2008-1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cho H.S., Ng P.A., Katzenellenbogen B.S. Differential regulation of gene expression by estrogen in estrogen growth-independent and -dependent MCF-7 human breast cancer cell sublines. Mol Endocrinol. 1991;5:1323–1330. doi: 10.1210/mend-5-9-1323. [DOI] [PubMed] [Google Scholar]
- 46.Fang F., Zheng J., Galbaugh T.L., Fiorillo A.A., Hjort E.E., Zeng X., Clevenger C.V. Cyclophilin B as a co-regulator of prolactin-induced gene expression and function in breast cancer cells. J Mol Endocrinol. 2010;44:319–329. doi: 10.1677/JME-09-0140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Galbaugh T., Feeney Y.B., Clevenger C.V. Prolactin receptor-integrin cross-talk mediated by SIRPalpha in breast cancer cells. Mol Cancer Res. 2010;8:1413–1424. doi: 10.1158/1541-7786.MCR-10-0130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.McHale K., Tomaszewski J.E., Puthiyaveettil R., Livolsi V.A., Clevenger C.V. Altered expression of prolactin receptor-associated signaling proteins in human breast carcinoma. Mod Pathol. 2008;21:565–571. doi: 10.1038/modpathol.2008.7. [DOI] [PubMed] [Google Scholar]
- 49.Fang F., Antico G., Zheng J., Clevenger C.V. Quantification of PRL/Stat5 signaling with a novel pGL4-CISH reporter. BMC Biotechnol. 2008;8:11. doi: 10.1186/1472-6750-8-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ormandy C.J., Hall R.E., Manning D.L., Robertson J.F., Blamey R.W., Kelly P.A., Nicholson R.I., Sutherland R.L. Coexpression and cross-regulation of the prolactin receptor and sex steroid hormone receptors in breast cancer. J Clin Endocrinol Metab. 1997;82:3692–3699. doi: 10.1210/jcem.82.11.4361. [DOI] [PubMed] [Google Scholar]
- 51.Horseman N.D. Prolactin and mammary gland development. J Mammary Gland Biol Neoplasia. 1999;4:79–88. doi: 10.1023/a:1018708704335. [DOI] [PubMed] [Google Scholar]
- 52.Murphy L.J., Murphy L.C., Vrhovsek E., Sutherland R.L., Lazarus L. Correlation of lactogenic receptor concentration in human breast cancer with estrogen receptor concentration. Cancer Res. 1984;44:1963–1968. [PubMed] [Google Scholar]
- 53.Grimm S.L., Seagroves T.N., Kabotyanski E.B., Hovey R.C., Vonderhaar B.K., Lydon J.P., Miyoshi K., Hennighausen L., Ormandy C.J., Lee A.V., Stull M.A., Wood T.L., Rosen J.M. Disruption of steroid and prolactin receptor patterning in the mammary gland correlates with a block in lobuloalveolar development. Mol Endocrinol. 2002;16:2675–2691. doi: 10.1210/me.2002-0239. [DOI] [PubMed] [Google Scholar]
- 54.Shafie S., Brooks S.C. Effect of prolactin on growth and the estrogen receptor level of human breast cancer cells (MCF-7) Cancer Res. 1977;37:792–799. [PubMed] [Google Scholar]
- 55.Gutzman J.H., Miller K.K., Schuler L.A. Endogenous human prolactin and not exogenous human prolactin induces estrogen receptor alpha and prolactin receptor expression and increases estrogen responsiveness in breast cancer cells. J Steroid Biochem Mol Biol. 2004;88:69–77. doi: 10.1016/j.jsbmb.2003.10.008. [DOI] [PubMed] [Google Scholar]
- 56.Sorlie T., Tibshirani R., Parker J., Hastie T., Marron J.S., Nobel A., Deng S., Johnsen H., Pesich R., Geisler S., Demeter J., Perou C.M., Lonning P.E., Brown P.O., Borresen-Dale A.L., Botstein D. Repeated observation of breast tumor subtypes in independent gene expression data sets. Proc Natl Acad Sci U S A. 2003;100:8418–8423. doi: 10.1073/pnas.0932692100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhao H., Langerod A., Ji Y., Nowels K.W., Nesland J.M., Tibshirani R., Bukholm I.K., Karesen R., Botstein D., Borresen-Dale A.L., Jeffrey S.S. Different gene expression patterns in invasive lobular and ductal carcinomas of the breast. Mol Biol Cell. 2004;15:2523–2536. doi: 10.1091/mbc.E03-11-0786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Richardson A.L., Wang Z.C., De Nicolo A., Lu X., Brown M., Miron A., Liao X., Iglehart J.D., Livingston D.M., Ganesan S. X chromosomal abnormalities in basal-like human breast cancer. Cancer Cell. 2006;9:121–132. doi: 10.1016/j.ccr.2006.01.013. [DOI] [PubMed] [Google Scholar]
- 59.Lu X., Wang Z.C., Iglehart J.D., Zhang X., Richardson A.L. Predicting features of breast cancer with gene expression patterns. Breast Cancer Res Treat. 2008;108:191–201. doi: 10.1007/s10549-007-9596-6. [DOI] [PubMed] [Google Scholar]
- 60.Ginestier C., Cervera N., Finetti P., Esteyries S., Esterni B., Adelaide J., Xerri L., Viens P., Jacquemier J., Charafe-Jauffret E., Chaffanet M., Birnbaum D., Bertucci F. Prognosis and gene expression profiling of 20q13-amplified breast cancers. Clin Cancer Res. 2006;12:4533–4544. doi: 10.1158/1078-0432.CCR-05-2339. [DOI] [PubMed] [Google Scholar]
- 61.Minn A.J., Gupta G.P., Siegel P.M., Bos P.D., Shu W., Giri D.D., Viale A., Olshen A.B., Gerald W.L., Massague J. Genes that mediate breast cancer metastasis to lung. Nature. 2005;436:518–524. doi: 10.1038/nature03799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ivshina A.V., George J., Senko O., Mow B., Putti T.C., Smeds J., Lindahl T., Pawitan Y., Hall P., Nordgren H., Wong J.E., Liu E.T., Bergh J., Kuznetsov V.A., Miller L.D. Genetic reclassification of histologic grade delineates new clinical subtypes of breast cancer. Cancer Res. 2006;66:10292–10301. doi: 10.1158/0008-5472.CAN-05-4414. [DOI] [PubMed] [Google Scholar]
- 63.Brockman J.L., Schroeder M.D., Schuler L.A. PRL activates the cyclin D1 promoter via the Jak2/Stat pathway. Mol Endocrinol. 2002;16:774–784. doi: 10.1210/mend.16.4.0817. [DOI] [PubMed] [Google Scholar]
- 64.Brockman J.L., Schuler L.A. Prolactin signals via Stat5 and Oct-1 to the proximal cyclin D1 promoter. Mol Cell Endocrinol. 2005;239:45–53. doi: 10.1016/j.mce.2005.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Nanbu-Wakao R., Fujitani Y., Masuho Y., Muramatu M., Wakao H. Prolactin enhances CCAAT enhancer-binding protein-beta (C/EBP beta) and peroxisome proliferator-activated receptor gamma (PPAR gamma) messenger RNA expression and stimulates adipogenic conversion of NIH-3T3 cells. Mol Endocrinol. 2000;14:307–316. doi: 10.1210/mend.14.2.0420. [DOI] [PubMed] [Google Scholar]
- 66.Vaidyanathan G., Cismowski M.J., Wang G., Vincent T.S., Brown K.D., Lanier S.M. The Ras-related protein AGS1/RASD1 suppresses cell growth. Oncogene. 2004;23:5858–5863. doi: 10.1038/sj.onc.1207774. [DOI] [PubMed] [Google Scholar]
- 67.Aranda J.F., Reglero-Real N., Kremer L., Marcos-Ramiro B., Ruiz-Sáenz A., Calvo M., Enrich C., Correas I., Millán J., Alonso M.A. MYADM regulates Rac1 targeting to ordered membranes required for cell spreading and migration. Mol Biol Cell. 2011;22:1252–1262. doi: 10.1091/mbc.E10-11-0910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Crone J., Glas C., Schultheiss K., Moehlenbrink J., Krieghoff-Henning E., Hofmann T.G. Zyxin is a critical regulator of the apoptotic HIPK2-p53 signaling axis. Cancer Res. 2011;71:2350–2359. doi: 10.1158/0008-5472.CAN-10-3486. [DOI] [PubMed] [Google Scholar]
- 69.Bartkova J., Lukas J., Muller H., Lutzhoft D., Strauss M., Bartek J. Cyclin D1 protein expression and function in human breast cancer. Int J Cancer. 1994;57:353–361. doi: 10.1002/ijc.2910570311. [DOI] [PubMed] [Google Scholar]
- 70.Gillett C., Fantl V., Smith R., Fisher C., Bartek J., Dickson C., Barnes D., Peters G. Amplification and overexpression of cyclin D1 in breast cancer detected by immunohistochemical staining. Cancer Res. 1994;54:1812–1817. [PubMed] [Google Scholar]
- 71.Grimm S.L., Rosen J.M. The role of C/EBPbeta in mammary gland development and breast cancer. J Mammary Gland Biol Neoplasia. 2003;8:191–204. doi: 10.1023/a:1025900908026. [DOI] [PubMed] [Google Scholar]
- 72.Zahnow C.A. CCAAT/enhancer-binding protein beta: its role in breast cancer and associations with receptor tyrosine kinases. Expert Rev Mol Med. 2009;11:e12. doi: 10.1017/S1462399409001033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lu C., Shen Q., DuPre E., Kim H., Hilsenbeck S., Brown P.H. cFos is critical for MCF-7 breast cancer cell growth. Oncogene. 2005;24:6516–6524. doi: 10.1038/sj.onc.1208905. [DOI] [PubMed] [Google Scholar]
- 74.Wang Q., Yang W., Uytingco M.S., Christakos S., Wieder R. 1,25-Dihydroxyvitamin D3 and all-trans-retinoic acid sensitize breast cancer cells to chemotherapy-induced cell death. Cancer Res. 2000;60:2040–2048. [PubMed] [Google Scholar]
- 75.Anastasi S., Sala G., Huiping C., Caprini E., Russo G., Iacovelli S., Lucini F., Ingvarsson S., Segatto O. Loss of RALT/MIG-6 expression in ERBB2-amplified breast carcinomas enhances ErbB-2 oncogenic potency and favors resistance to Herceptin. Oncogene. 2005;24:4540–4548. doi: 10.1038/sj.onc.1208658. [DOI] [PubMed] [Google Scholar]
- 76.Clevenger C.V., Altmann S.W., Prystowsky M.B. Requirement of nuclear prolactin for interleukin-2–stimulated proliferation of T lymphocytes. Science. 1991;253:77–79. doi: 10.1126/science.2063207. [DOI] [PubMed] [Google Scholar]
- 77.Iyer J., Reich N.C. Constitutive nuclear import of latent and activated STAT5a by its coiled coil domain. FASEB J. 2008;22:391–400. doi: 10.1096/fj.07-8965com. [DOI] [PubMed] [Google Scholar]
- 78.Louie D.F., Gloor K.K., Galasinski S.C., Resing K.A., Ahn N.G. Phosphorylation and subcellular redistribution of high mobility group proteins 14 and 17, analyzed by mass spectrometry. Protein Sci. 2000;9:170–179. doi: 10.1110/ps.9.1.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Nilsson J., Bjursell G., Kannius-Janson M. Nuclear Jak2 and transcription factor NF1-C2: a novel mechanism of prolactin signaling in mammary epithelial cells. Mol Cell Biol. 2006;26:5663–5674. doi: 10.1128/MCB.02095-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Dawson M.A., Bannister A.J., Gottgens B., Foster S.D., Bartke T., Green A.R., Kouzarides T. JAK2 phosphorylates histone H3Y41 and excludes HP1alpha from chromatin. Nature. 2009;461:819–822. doi: 10.1038/nature08448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Frasor J., Park K., Byers M., Telleria C., Kitamura T., Yu-Lee L.Y., Djiane J., Park-Sarge O.K., Gibori G. Differential roles for signal transducers and activators of transcription 5a and 5b in PRL stimulation of ERalpha and ERbeta transcription. Mol Endocrinol. 2001;15:2172–2181. doi: 10.1210/mend.15.12.0745. [DOI] [PubMed] [Google Scholar]
- 82.Frasor J., Barkai U., Zhong L., Fazleabas A.T., Gibori G. PRL-induced ERalpha gene expression is mediated by Janus kinase 2 (Jak2) while signal transducer and activator of transcription 5b (Stat5b) phosphorylation involves Jak2 and a second tyrosine kinase. Mol Endocrinol. 2001;15:1941–1952. doi: 10.1210/mend.15.11.0722. [DOI] [PubMed] [Google Scholar]
- 83.Dong J., Tsai-Morris C.H., Dufau M.L. A novel estradiol/estrogen receptor alpha-dependent transcriptional mechanism controls expression of the human prolactin receptor. J Biol Chem. 2006;281:18825–18836. doi: 10.1074/jbc.M512826200. [DOI] [PubMed] [Google Scholar]
- 84.Hovey R.C., Trott J.F., Ginsburg E., Goldhar A., Sasaki M.M., Fountain S.J., Sundararajan K., Vonderhaar B.K. Transcriptional and spatiotemporal regulation of prolactin receptor mRNA and cooperativity with progesterone receptor function during ductal branch growth in the mammary gland. Dev Dyn. 2001;222:192–205. doi: 10.1002/dvdy.1179. [DOI] [PubMed] [Google Scholar]
- 85.Petz L.N., Nardulli A.M. Sp1 binding sites and an estrogen response element half-site are involved in regulation of the human progesterone receptor A promoter. Mol Endocrinol. 2000;14:972–985. doi: 10.1210/mend.14.7.0493. [DOI] [PubMed] [Google Scholar]
- 86.Schultz J.R., Petz L.N., Nardulli A.M. Estrogen receptor alpha and Sp1 regulate progesterone receptor gene expression. Mol Cell Endocrinol. 2003;201:165–175. doi: 10.1016/s0303-7207(02)00415-x. [DOI] [PubMed] [Google Scholar]
- 87.Djiane J., Durand P. Prolactin-progesterone antagonism in self regulation of prolactin receptors in the mammary gland. Nature. 1977;266:641–643. doi: 10.1038/266641a0. [DOI] [PubMed] [Google Scholar]
- 88.Briand P., Lykkesfeldt A.E. Effect of estrogen and antiestrogen on the human breast cancer cell line MCF-7 adapted to growth at low serum concentration. Cancer Res. 1984;44:1114–1119. [PubMed] [Google Scholar]
- 89.Bonneterre J., Peyrat J.P., Beuscart R., Demaille A. Biological and clinical aspects of prolactin receptors (PRL-R) in human breast cancer. J Steroid Biochem Mol Biol. 1990;37:977–981. doi: 10.1016/0960-0760(90)90453-r. [DOI] [PubMed] [Google Scholar]
- 90.Fisher B., Redmond C., Fisher E.R., Caplan R. Relative worth of estrogen or progesterone receptor and pathologic characteristics of differentiation as indicators of prognosis in node negative breast cancer patients: findings from National Surgical Adjuvant Breast and Bowel Project Protocol B-06. J Clin Oncol. 1988;6:1076–1087. doi: 10.1200/JCO.1988.6.7.1076. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Validation of genes identified by microarray analysis. Negative control is used; real-time PCR demonstrates that FAS is not affected by PRL treatment of PRLr/PRLrYDmut expression.
HMGN2 knockdown impairs gene expression in a similar manner to expression of PRLrYDmut. A–E: qPCR was performed on control of HMGN2 knockdown cell lines (shNS or shHMGN2) expressing cell lines treated with PRL (1 hour). Results demonstrated impaired PRL-induced gene expression with depletion of HMGN2. shHMGN2, MCF7 HMGN2 shRNA-expressing cells; shNS, MCF7 control shRNA-expressing cells. **P < 0.01, ***P < 0.001.
Efficient siRNA-mediated knockdown of the PRLr. Western blot analysis demonstrating PRLr knockdown in T47D cells transfected with siPRLr, sicontrol as a negative control (Ctr), and small-interfering glyceraldehyde-3-phosphate dehydrogenase (GAPDH) as a positive control. Cells were harvested at 48 hours after transfection.
Prolonged PRL induces PR gene expression. Real-time PCR was performed on cDNA collected from MCF7 stable cell lines with expression of the indicated constructs. MCF7 cells were grown in charcoal-stripped, phenol red–free media for 24 hours, followed by treatment of PRL for 48 hours. Data are representative of two separate experiments, in which the mean ± SEM represents the error of replicate measurements from a single experiment. Empty, empty vector. **P < 0.01, ***P < 0.001.
PRLr depletion impairs expression of ERα and PR. A and B: Real-time PCR performed on cDNA collected from T47D cells transfected with the indicated siRNAs to measure ERα (A) or PR (B) mRNA levels. Results are representative of three separate experiments. The values presented are the mean ± SEM from replicate measurements within one experiment. C: Real-time PCR performed on cDNA collected from MCF7 rescue cell lines, demonstrating successful PRLr rescue by PRLr or PRLrYDmut. Results were normalized to PRLr levels in control cells expressing shNS, along with an empty vector control. D and E: Real-time PCR performed on cDNA collected from MCF7 rescue cells to measure ERα (D) or PR (E) mRNA levels. Results are shown as percentage expression compared with shNS control cells. EV, empty vector; siPRLr, siRNA targeted against PRLr. **P < 0.01, ***P < 0.001.
PRLrYDmut expression or PRLr depletion decreases estrogen-responsiveness. A: Real-time PCR performed on cDNA collected from E2-treated MCF7 stable cell pools to determine CCND1 mRNA induction. MCF7 transfectants were serum starved for 24 hours, before 1 hour of E2 treatment. B: Real-time PCR performed on cDNA collected from E2-treated MCF7 stable knockdown cell lines to determine CCND1 mRNA induction. Values were normalized to 18S RNA, and fold change was calculated by comparing each value with the non-treated control sample and are representative of three separate experiments with SEM. All P values were calculated using one-way analysis of variance with a Bonferroni multiple-comparison test. Calculated P values are highlighted in the figure. Empty, empty vector control, expressing MCF7 cells. **P < 0.01, ***P < 0.001.