Abstract
Background
The project entitled Surveillance of Antibiotic Use and Resistance in Intensive Care Units (SARI) was initiated in Germany in 2000. In this article, we describe developments in antibiotic use and resistance rates in the participating intensive care units over the years 2001–2015.
Methods
The intensive care units supplied monthly figures on patient days, antibiotic use (in defined daily doses, DDD), and resistance data for 13 pathogens. The density of antibiotic use per 1000 patient days was calculated on the basis of antibiotic use, DDD, and patient days, and the resistance density per 1000 patient days was calculated from the number of resistant pathogens.
Results
In the years 2001–2015, data on 2 920 068 patient days were collected in 77 intensive care units. The average overall antibiotic use rose by 19% over this period, with a marked increase in the density of carbapenem use (from 76 to 250 DDD per 1000 patient days, +230%) and piperacillin-tazobactam use (from 42 to 146 DDD per 1000 patient days, +247%). The proportion of Escherichia coli and Klebsiella pneumoniae isolates that were resistant to third-generation cephalosporins increased markedly initially, then remained stable over the remainder of the observation period. The proportion of methicillin-resistant Staphylococcus aureus was stable over the entire period. The rates of vancomycin resistance among Enterococcus faecium isolates and imipenem resistance among gram-negative pathogens increased from 2.3% to 13.3% and from 0.1% to 0.3%, respectively.
Conclusion
The resistance density of gram-negative multiresistant pathogens in the participating intensive care units increased markedly. The rise in imipenem-resistant pathogens arouses particular concern. The increased use of broad-spectrum/reserve antibiotics may well have contributed to this development. Efforts to use antibiotics rationally, e.g., with the support of multidisciplinary “antibiotic stewardship” teams, are therefore vitally important. As participation in SARI is voluntary, these surveillance data cannot be considered representative of Germany as a whole.
Worldwide, the consumption of antibiotics has increased substantially in the past decades (1). Increased use of antibiotics promotes—in addition to other factors (2)—the selection and spread of antibiotic-resistant or multiresistant pathogens, with the result that the treatment of infections caused by these pathogens becomes more difficult (3). The problem is concentrated in intensive care units (ICUs) (4), where often multimorbid patients generally present with a higher risk for nosocomial infections (5) and infections with multiresistant pathogens can lead to additional complications, prolonged hospital stays, and higher healthcare costs (6– 9).
In February 2000 the project for the surveillance of antibiotic use and resistance in intensive care units (SARI) in Germany was initiated for the purpose of benchmarking (10, 11). After a one-year pilot phase, SARI has continuously captured antibiotic use densities and resistance data for selected pathogens on ICUs in Germany.
This article aims to describe the development of antibiotic resistance and changes in resistance rates in the past 15 years in this cohort of ICUs in Germany.
Methods
Participation in SARI is voluntary. The methods are explained in greater detail in the eMethods section and have already been described elsewhere (10– 12) (http://sari.eu-burden.info/down/protokoll.pdf). In sum, participating ICUs report on a monthly basis the number of patient days, use (in g) of all orally or parenterally administered antibiotics, and resistance rates of the following pathogens:
Staphylococcus (S.) aureus
Streptococcus pneumoniae
Coagulase-negative staphylococci (CNS)
Enterococcus faecalis
Enterococcus faecium
Escherichia (E.) coli
Klebsiella (K.) pneumoniae
Enterobacter cloacae
Serratia marcescens
Citrobacter spp.
Pseudomonas aeruginosa
Stenotrophomonas maltophilia
Acinetobacter (A.) baumannii.
Resistance testing can be performed according to German industry standard (DIN) 58940, the CLSI (Clinical & Laboratory Standards Institute), or EUCAST (European Committee on Antimicrobial Susceptibility Testing). Copy strains—that is, isolates detected within 30 days from a patient with an identical antibiogram—were not included in the analysis. The frequency of taking specimens from patients is the prerogative of the clinician in the respective ICU; tests performed exclusively for the purpose of screening were not considered. We did not collect data on the total number specimens sampled, the location of the specimen sampling, nor on whether an infection or colonization was present or whether the pathogen was acquired in an outpatient or inpatient setting.
From the antibiotics use, the defined daily doses (DDD), and the patient days, the antibiotic use density was calculated as follows: (antibiotic use in g/DDD in g) × 1000 patient days. The resistance rate of a pathogen is calculated from the number of resistant isolates of a species against a specific antibiotic, divided by the number of all pathogens tested against this antibiotic × 100. The antibiotic resistance density results from the number of resistant pathogens/1000 patient days.
Statistical analysis
Pooled mean values, medians, and interquartile ranges (25th and 75th percentile) of antibiotic use density, resistance rates, and antibiotic resistance density were calculated from the reported data for the period 2001–2015. Since not all ICUs participated in SARI for the entire duration of the study, a sensitivity analysis was used to calculate the antibiotic use density, resistance rates, and resistance density for only those ICUs that had reported data continuously from 2001 to 2015 (core cohort). At the time points 2001 and 2015, the results of the core cohort were compared with those of the total cohort, in order to identify possible differences in trends in antibiotic use and resistance rates. We used the Wilcoxon test to calculate for both cohorts on the basis of the inpatient ward whether the use of antibiotics and the resistance rates of the analyzed pathogens changed between 2001 and 2015. We used SAS 9.4 (SAS Institute, Cary, NC, USA) to evaluate the data.
Results
In 2001–2015 data were collected in 44 hospitals in 13 federal states on 77 ICUs with a total of 2 920 068 patient days (etable 1). The median size of the hospitals was 572 (interquartile range 411–1008) beds, and the median size of the ICUs was 12 (10– 16) beds. 45% of ICUs were managed in an interdisciplinary way, 25% specalized in internal medicine, and 30% were surgical.
eTable 1. Baseline characteristics of intensive care units participating in the surveillance project since 2001*.
| Variable |
2001–2015 Total cohort (N = 77 ICUs) |
2001–2015 Core cohort (N =20 ICUs) |
| Hospitals, No | 44 | 10 |
| Participating ICUs, No | 77 | 20 |
| No of ICU beds, median (IQR) | 12 (10– 16) | 12 (10– 15) |
| Type of ICU | ||
| Interdisciplinary, n (%) | 35 (45) | 6 (30) |
| Medical, n (%) | 19 (25) | 7 (35) |
| Surgical, n (%) | 23 (30) | 7 (35) |
| Data collection of ICUs in months, median (IQR) | 89 (60–156) | 180 (164–180) |
| No of hospital beds, median (IQR) | 572 (411–1008) | 956 (308–1484) |
| Medical care level of hospital | ||
| Maximum care: university medical center, n (%) | 8 (18) | 3 (30) |
| Maximum care: other, n (%) | 8 (18) | 3 (30) |
| Secondary care with specialty focus, n (%) | 12 (27) | 1 (10) |
| Secondary care hospital, n (%) | 1 (2) | – |
| Standard care, n (%) | 13 (30) | 2 (20) |
| Basic care, n (%) | 2 (5) | 1 (10) |
| Patient days in total | 2 920 068 | 1 229 428 |
* The total cohort includes all ICUs that ever submitted data to SARI (N = 77). The core cohort includes those ICUs, that continuously submitted data from 2001 through 2015 (N = 20, core cohort). The data of the total cohort were included in the main analysis in the manuscript. The data of the core cohort were used for the sensitivity analysis and compared with those of the total cohort.
ICU: intensive care unit; IQR: interquartile range; SARI: Surveillance der Antibiotika-Anwendung und der bakteriellen Resistenzen auf deutschen Intensivstationen [surveillance of antibiotic use and resistance in intensive care units]
Antibiotic use
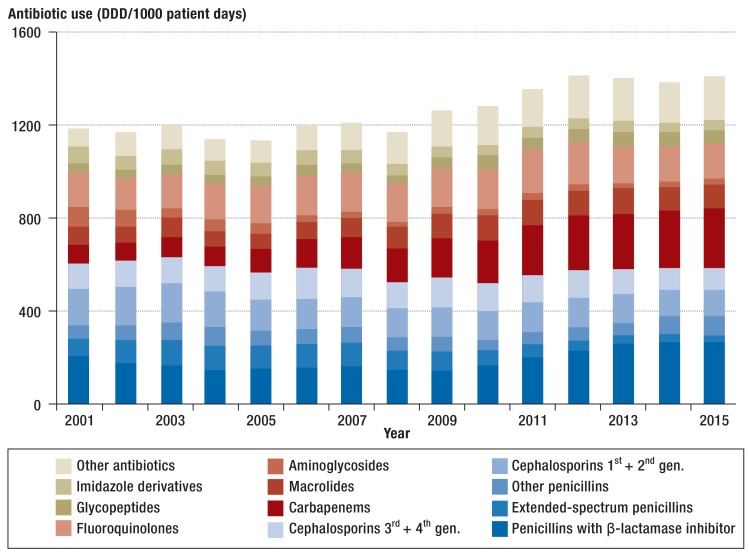
The total consumption of antibiotics over the study period increased by 19%, from 1180 DDD/1000 patient days to 1407 DDD/1000 patient days in 2015 (Figure 1, Table 1). The antibiotic use density, however, varied between ICUs: for example, the median in 2015 was 1330 DDD/1000 patient days and the interquartile range 1145–1605 DDD/1000 patient days. The five groups of antibiotics used most often in 2015 were penicillins with ß-lactamase inhibitors (262 DDD/1000 patient days), carbapenems (250 DDD/1000 patient days), fluoroquinolones (157 DDD/1000 patient days), macrolides (104 DDD/1000 patient days), and third-generation cephalosporins (91 DDD/1000 patient days), which accounted for 61% of the total use.
Figure 1.
Trends in antibiotic use density (defined daily doses/1000 patient days) in intensive care units (N = 77) in Germany from 2001 to 2015.
DDD, defined daily dose (www. whocc.no/atc_ddd_index); gen: generation
Table 1. Antibiotic use density (defined daily doses/1000 patient days) for selected antibiotics in intensive care units (N = 77) in 2001 versus 2015.
| ATC code | Antibiotic/group |
2001 DDD/1000 PD |
2015 DDD/1000 PD |
Change 2001–2015 (%) |
P value* |
| J01 | All excl sulbactam | 1180 | 1407 | 19 | 0.028 |
| J01CA | Extended-spectrum penicillins | 74 | 36 | –52 | 0.007 |
| J01CR | Combination of penicillins with BLI | 206 | 262 | 28 | 0.015 |
| – Piperacillin/ Tazobactam |
42 | 146 | 247 | <0.001 | |
| J01DD | 3rd generation cephalosporins | 106 | 91 | –14 | 0.069 |
| – Ceftazidime | 30 | 24 | –20 | 0.076 | |
| J01DH | Carbapenems | 76 | 250 | 230 | <0.001 |
| – Imipenem – Meropenem |
47 29 |
37 211 |
–23 638 |
0.015 <0.001 |
|
| J01XA | Glycopeptides | 38 | 57 | 48 | 0.537 |
| J01MA | Fluoroquinolones | 151 | 157 | 4 | 0.440 |
| J01FA | Macrolides | 77 | 104 | 36 | 0.036 |
| J01G | Aminoglycosides | 86 | 22 | –75 | <0.001 |
| J01XD | Imidazole derivatives | 70 | 42 | –40 | 0.001 |
| J01XX | Other antibiotics | 8 | 85 | 928 | <0.001 |
| J01XX | – Fosfomycin | 4 | 13 | 204 | 0.041 |
| J01XX | – Linezolid | 0 | 38 | – | <0.001 |
| J01AA12 | – Tigecyclin | 0 | 15 | – | <0.001 |
| J01XX09 | – Daptomycin | 0 | 18 | – | <0.001 |
* Wilcoxon test
ATC: anatomic-therapeutic-chemical classification system; DDD: defined daily dose; PD: patient days; excl: excluding; BLI: ß-lactamase inhibitor; extended spectrum penicillins: ampicillin, amoxicillin, mezlocillin, piperacillin; ß-lactamase resistant penicillins: flucloxacillin, oxacillin; penicillins with BLI: amoxicillin-clavulanic acid, ampicillin/sulbactam, piperacillin-tazobactam; 3 rd generation cephalosporins: cefotaxime, ceftazidim, ceftriaxone, cefixime; carbapenems: imipenem, meropenem, ertapenem; glycopeptides: vancomycin, teicoplanin; fluoroquinolones: ciprofloxacin, ofloxacin, levofloxacin, moxifloxacin, norfloxacin; macrolides: erythromycin, roxithromycin, clarithromycin, azithromycin; aminoglycosides: gentamicin, streptomycin, tobramycin, neomycin, amikacin, netilmicin; imidazoles: metronidazole
Among the classes of antibiotics, the use of carbapenems increased most, from 76 DDD/1000 patient days in 2001 to 250 DDD/1000 patient days in 2015 +230%) (Table 1 and eTable 2). This increase is mainly accounted for by the use of meropenem (+638%). Increases were also noted in the use of penicillins with ß-lactamase inhibitors (+28%), glycopeptides (+48%), macrolides (+36%), and other antibiotics (+928%). The notable increase in the use of other antibiotics is mainly due to linezolid, tigecycline, and daptomycin—substances that were not, or had only just become, available in 2001. In the group of penicillins with ß-lactamase inhibitors, the greatest increase was seen for piperacillin/tazobactam (+247%). Over the observation period, the use of first and second generation cephalosporins decreased (-29%), as did that of aminoglycosides (-75%) and imidazoles (-40%) (Figure 1, Table 1, and eTable 2).
eTable 2. Development of antibiotic use density (defined daily doses/1000 patient days) in intensive care units (N = 77) from 2001 to 2015.
| ATC code | Antibiotic/group | 2001 | 2002 | 2003 | 2004 | 2005 | 2006 | 2007 | 2008 | 2009 | 2010 | 2011 | 2012 | 2013 | 2014 | 2015 |
| J01 | Total incl sulbactam | 1225 | 1296 | 1346 | 1291 | 1283 | 1383 | 1378 | 1311 | 1424 | 1396 | 1436 | 1455 | 1402 | 1380 | 1412 |
| J01 | Total excl sulbactam | 1180 | 1166 | 1200 | 1138 | 1132 | 1193 | 1203 | 1167 | 1258 | 1279 | 1353 | 1412 | 1399 | 1380 | 1407 |
| J01CE | β-Lactamase susceptible penicillins | 20 | 21 | 25 | 30 | 21 | 23 | 20 | 17 | 19 | 20 | 19 | 18 | 15 | 23 | 23 |
| J01CA | Extended spectrum penicillins | 74 | 101 | 111 | 102 | 99 | 101 | 103 | 82 | 89 | 67 | 61 | 48 | 33 | 36 | 36 |
| J01CF | β-Lactamase-resistant penicillins | 38 | 40 | 48 | 50 | 44 | 37 | 45 | 41 | 42 | 25 | 32 | 34 | 34 | 50 | 56 |
| J01CG | β-Lactamase inhibitor (BLI) | 44 | 129 | 147 | 153 | 151 | 190 | 175 | 145 | 166 | 117 | 83 | 43 | 3 | 0 | 5 |
| J01CR | Combination of penicillins with BLI | 206 | 174 | 161 | 146 | 151 | 158 | 160 | 145 | 136 | 161 | 194 | 227 | 263 | 268 | 262 |
| J01CR25 | – Piperacillin/tazobactam | 42 | 33 | 26 | 24 | 39 | 40 | 42 | 45 | 50 | 71 | 87 | 105 | 122 | 136 | 146 |
| J01DB | 1st generation cephalosporins | 47 | 47 | 42 | 39 | 33 | 39 | 33 | 28 | 34 | 29 | 35 | 38 | 36 | 38 | 33 |
| J01DC | 2nd generation cephalosporins | 113 | 118 | 130 | 112 | 96 | 94 | 98 | 95 | 95 | 93 | 93 | 88 | 91 | 75 | 80 |
| J01DD | 3rd generation cephalosporins | 106 | 109 | 108 | 107 | 109 | 124 | 112 | 112 | 123 | 118 | 115 | 109 | 98 | 91 | 91 |
| J01DE | 4th generation cephalosporins | 2 | 3 | 2 | 6 | 12 | 11 | 10 | 4 | 4 | 4 | 5 | 15 | 13 | 5 | 6 |
| J01DH | Carbapenems | 76 | 79 | 88 | 82 | 96 | 120 | 133 | 143 | 168 | 184 | 214 | 231 | 233 | 246 | 250 |
| J01DH02 | – Meropenem | 29 | 36 | 51 | 47 | 47 | 60 | 62 | 72 | 85 | 105 | 147 | 162 | 175 | 200 | 211 |
| J01DH21 | – Imipenem | 47 | 43 | 38 | 35 | 46 | 53 | 63 | 62 | 74 | 73 | 62 | 65 | 53 | 43 | 37 |
| J01XA | Glycopeptides | 38 | 36 | 38 | 35 | 39 | 40 | 38 | 36 | 46 | 62 | 54 | 57 | 59 | 62 | 57 |
| J01MA | Fluoroquinolones | 151 | 140 | 145 | 155 | 166 | 172 | 169 | 165 | 171 | 172 | 185 | 184 | 160 | 152 | 157 |
| J01E | Sulfonamides and trimethoprim | 29 | 36 | 38 | 17 | 20 | 17 | 20 | 26 | 25 | 22 | 20 | 34 | 32 | 41 | 40 |
| J01AA | Tetracyclines | 8 | 9 | 8 | 12 | 10 | 10 | 11 | 8 | 12 | 14 | 8 | 10 | 8 | 9 | 9 |
| J01FA | Macrolides | 77 | 70 | 80 | 72 | 70 | 75 | 83 | 89 | 106 | 108 | 111 | 106 | 110 | 99 | 104 |
| J01FF | Lincosamines | 25 | 25 | 26 | 24 | 26 | 25 | 21 | 24 | 25 | 23 | 27 | 23 | 22 | 23 | 24 |
| J01G | Aminoglycosides | 86 | 68 | 45 | 46 | 41 | 30 | 28 | 25 | 27 | 28 | 27 | 26 | 24 | 24 | 22 |
| J01GB03 | – Gentamicin | 38 | 28 | 22 | 22 | 23 | 17 | 17 | 16 | 12 | 13 | 12 | 10 | 10 | 12 | 11 |
| J01XB | Polymyxines | 2 | 3 | 2 | 2 | 0 | 0 | 0 | 0 | 8 | 9 | 12 | 13 | 15 | 1 | 2 |
| J01XD | Imidazole derivatives | 70 | 59 | 68 | 64 | 59 | 67 | 60 | 51 | 48 | 46 | 47 | 48 | 46 | 42 | 42 |
| J01A | Tuberculostatic drugs | 5 | 6 | 10 | 6 | 7 | 5 | 7 | 16 | 14 | 11 | 15 | 14 | 16 | 23 | 27 |
| J01XX | Other antibiotics | 8 | 23 | 23 | 31 | 33 | 44 | 52 | 59 | 67 | 82 | 79 | 90 | 90 | 72 | 85 |
| J01XX | – Fosfomycin | 4 | 10 | 7 | 8 | 6 | 6 | 5 | 8 | 9 | 13 | 4 | 10 | 11 | 9 | 13 |
| J01XX | – Linezolid | 0 | 10 | 13 | 20 | 24 | 31 | 27 | 32 | 35 | 35 | 45 | 42 | 42 | 33 | 38 |
| J01AA12 | – Tigecyclin | 0 | 0 | 0 | 0 | 0 | 4 | 14 | 13 | 16 | 22 | 18 | 17 | 18 | 14 | 15 |
| J01XX09 | – Daptomycin | 0 | 0 | 0 | 0 | 0 | 0 | 5 | 3 | 4 | 10 | 9 | 19 | 16 | 15 | 18 |
| Diverse | 3 | 4 | 4 | 3 | 5 | 5 | 3 | 2 | 2 | 3 | 2 | 1 | 2 | 1 | 1 |
DDD: defined daily doses (www.whocc.no/atc_ddd_index); total incl sulbactam: all antibiotics including sulbactam as individual preparation; total excl sulbactam: all antibiotics excluding sulbactam as individual preparation; ß-lactamase susceptible penicillins: benzylpenicillin, phenoxymethylpenicillin; extended spectrum penicillins: ampicillin, amoxicillin, mezlocillin, piperacillin; ß-lactamase-resistant penicillins: flucloxacillin, oxacillin; penicillin with ß-lactamase inhibitor (BLI): amoxicillin-clavulanic acid, ampicillin/sulbactam, piperacillin-tazobactam; 1st generation cephalosporins: cefazolin, cephalexin; 2nd generation cephalosporins: cefuroxime, cefotiam, cefaclor; 3rd generation cephalosporins: cefotaxime, ceftazidime, ceftriaxone, cefixime; carbapenems: imipenem, meropenem, ertapenem; glycopeptides: vancomycin, teicoplanin; fluoroquinolones: ciprofloxacin, ofloxacin, levofloxacin, moxifloxacin, norfloxacin; trimethoprim-sulfonamide: sulfamethoxazol, trimethoprim; tetracyclines: doxycyclin, minocyclin; macrolides: erythromycin, roxithromycin, clarithromycin, azithromycin; aminoglycosides: gentamicin, streptomycin, tobramycin, neomycin, amikacin, netilmicin; imidazoles: metronidazole; others: fosfomycin, linezolid, daptomycin, tigecyclin; diverse: antibiotics with fewer than 10 DDD/1000 patient days: monobactams, streptogramins, amphenicols, steroids, paromomycin, taurolidin, atovaquone, nitrofurantoin; ATC: anatomic-therapeutic-chemical classification system
Antibiotic resistance
In the period from 2001 through 2015, a total of 263 639 isolates were tested (138 686 gram-positive and 124 953 gram-negative), which corresponds to a copy-strain adjusted isolation rate of 90 isolates per 1000 patient days for the 13 pathogens. From 2001 through 2010, almost 60% of laboratories used the DIN 58940 and 40% the CSLI standard (etable 3) every year. From 2010 onwards, the first laboratories switched to the EUCAST standard. In 2013, 40% used DIN 58940, 39% used the CLSI standard, and 21% the EUCAST standard.
eTable 3. Microbiological tests/standards used to determine bacterial resistance*.
|
Microbiological tests used (monthly figures by number of ICUs), n (%) |
|||
| Year | DIN | EUCAST | CLSI |
| 2001 | 233 (60) | 0 | 158 (40) |
| 2002 | 251 (60) | 0 | 168 (40) |
| 2003 | 276 (61) | 0 | 177 (39) |
| 2004 | 251 (54) | 0 | 216 (46) |
| 2005 | 279 (56) | 0 | 221 (44) |
| 2006 | 303 (56) | 0 | 239 (44) |
| 2007 | 330 (61) | 0 | 215 (39) |
| 2008 | 318 (62) | 0 | 192 (38) |
| 2009 | 306 (57) | 0 | 228 (53) |
| 2010 | 293 (58) | 18 (4) | 193 (38) |
| 2011 | 265 (51) | 77 (15) | 180 (34) |
| 2012 | 258 (41) | 131 (21) | 242 (38) |
| 2013 | 246 (40) | 126 (21) | 241 (39) |
| From 2014 | No data reported | ||
* Of laboratories participating in SARI.
CLSI: Clinical & Laboratory Standards Institute; DIN: Deutsches Institut für Normung [German standards institute]; EUCAST: European Committee on Antimicrobial Susceptibility Testing; ICU: intensive care unit; SARI: Surveillance der Antibiotika-Anwendung und der bakteriellen Resistenzen auf deutschen Intensivstationen [surveillance of antibiotic use and resistance in intensive care units]
Since 2001 the number of isolates increased by 22%, with the greatest increase observed in E. coli (+87%), Enterococcus species (+65%), and K. pneumoniae (+63%), whereas the greatest decrease was observed among A. baumannii isolates (–72%). The decrease in A. baumannii isolates was constant over the entire study period, but this effect may have been affected by additional species differentiation that was introduced by some laboratories from 2012 onwards see ( section).
The most commonly identified gram-positive pathogens were S. aureus (n = 54 320), Enterococcus faecalis (n = 26 578), and Enterococcus faecium (n = 17 813); the most common gram-negative pathogens were E. coli (n = 44 809), Pseudomonas aeruginosa (n = 27 216), and K. pneumoniae (n = 17 529). Trends in selected resistance patterns for four gram-positive and five gram-negative pathogens are shown in Table 2 and eTable 4.
Table 2. Development of selected resistance rates (pooled means in %) and resistance densities (per 1000 patient days) in intensive care units (N = 77) in 2001 versus 2015.
| Resistance rates |
2001 Pooled mean |
2015 Pooled mean |
Change 2001 to 2015 (%) |
P value* |
| Staphylococcus aureus (oxacillin) | 26.0 | 22.7 | –13 | 0.255 |
| Enterococcus faecium (vancomycin) | 2.3 | 13.3 | 470 | <0.001 |
|
Klebsiella pneumoniae (cefotax./ceftr./cefta.) |
4.5 | 15.7 | 247 | <0.001 |
| Klebsiella pneumoniae (imipenem) | 0.4 | 1.6 | 269 | 0.005 |
| Acinetobacter baumannii (imipenem) | 1.1 | 42.8 | 3620 | <0.001 |
| Escherichia coli (cefotax./ceftr./cefta.) | 1.3 | 16.3 | 1113 | <0.001 |
| Escherichia coli (imipenem) | 0.1 | 0.3 | 292 | 0.196 |
| Resistance density (per 1000 patient days) | ||||
| Staphylococcus aureus (oxacillin) | 4.2 | 3.5 | –16 | 0.985 |
| Enterococcus faecium (vancomycin) | 0.1 | 1.1 | 1416 | <0.001 |
|
Klebsiella pneumoniae (cefotax./ceftr./cefta.) |
0.2 | 1.4 | 451 | <0.001 |
| Acinetobacter baumannii (imipenem) | 0.0 | 0.3 | 917 | <0.001 |
| Escherichia coli (cefotax./ceftr./cefta.) | 0.2 | 3.7 | 2270 | <0.001 |
| Escherichia coli (imipenem) | 0.0 | 0.1 | 711 | 0.170 |
* Wilcoxon test
Cefotax./ceftr./cefta.: cefotaxime/ceftriaxone/ceftazidime
eTable 4. Development of selected resistance rates (pooled mean values in %) in intensive care units (N = 77) in 2001–2015.
| Pathogen | Antibiotic | 2001 | 2002 | 2003 | 2004 | 2005 | 2006 | 2007 | 2008 | 2009 | 2010 | 2011 | 2012 | 2013 | 2014 | 2015 |
| Gram-positive pathogens | ||||||||||||||||
| Staphylococcus aureus | Oxacillin | 26.0 | 22.4 | 20.9 | 19.5 | 22.6 | 22.5 | 21.5 | 22.4 | 22.6 | 24.5 | 27.2 | 25.0 | 22.6 | 23.6 | 22.7 |
| Ciprofloxacin/Ofloxacin | 28.2 | 24.1 | 22.9 | 24.3 | 28.9 | 29.7 | 30.9 | 34.0 | 35.9 | 34.4 | 35.2 | 35.7 | 31.9 | 31.3 | 29.9 | |
| Vancomycin | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.1 | 0.1 | 0.4 | 0.1 | 0.0 | |
| Linezolid | 0.0 | 0.0 | 0.0 | 0.2 | 0.1 | 0.0 | 0.1 | 0.0 | 0.0 | 0.0 | 0.0 | 0.1 | 0.1 | 0.2 | 0.0 | |
| Enterococcus faecalis | Ciprofloxacin | 38.7 | 63.9 | 66.7 | 62.2 | 56.4 | 60.6 | 66.6 | 65.1 | 66.8 | 69.6 | 72.6 | 46.2 | 50.1 | 48.0 | 43.0 |
| Ampicillin | 0.0 | 0.0 | 0.0 | 1.8 | 2.7 | 1.2 | 1.2 | 1.1 | 1.2 | 1.9 | 2.2 | 2.5 | 1.3 | 0.7 | 0.5 | |
| Vancomycin | 0.1 | 0.2 | 0.0 | 0.0 | 0.2 | 0.0 | 0.1 | 0.2 | 0.0 | 0.3 | 0.2 | 0.3 | 0.5 | 0.4 | 0.0 | |
| Coagulase-negative streptococci | Vancomycin | 0.1 | 0.0 | 0.1 | 0.3 | 0.0 | 0.2 | 0.1 | 0.3 | 0.0 | 0.1 | 0.1 | 0.2 | 0.3 | 0.5 | 0.1 |
| Teicoplanin | 0.7 | 0.2 | 1.0 | 0.5 | 0.8 | 0.4 | 1.0 | 0.5 | 0.7 | 3.0 | 3.8 | 3.8 | 8.0 | 5.2 | 7.4 | |
| Enterococcus faecium | Ciprofloxacin | 93.8 | 73.3 | 77.4 | 81.5 | 80.9 | 88.7 | 90.2 | 89.9 | 93.4 | 94.2 | 93.6 | 92.7 | 93.4 | 93.4 | 91.0 |
| Vancomycin | 2.3 | 1.2 | 1.2 | 5.4 | 5.4 | 2.9 | 3.4 | 8.0 | 7.1 | 7.3 | 11.7 | 10.8 | 12.8 | 14.3 | 13.3 | |
| Linezolid | 0.0 | 0.0 | 0.0 | 0.3 | 3.1 | 0.2 | 0.7 | 0.5 | 0.5 | 0.2 | 0.9 | 0.9 | 1.1 | 1.0 | 1.6 | |
| Gram-negative pathogens | ||||||||||||||||
| Escherichia coli | Piperacillin/β-lactamase inhibitor | 5.8 | 5.6 | 4.5 | 6.5 | 9.7 | 10.9 | 14.4 | 13.9 | 17.8 | 18.5 | 27.9 | 17.0 | 13.3 | 11.5 | 12.3 |
| Ciprofloxacin | 8.3 | 11.9 | 14.1 | 16.5 | 18.2 | 16.7 | 21.1 | 24.4 | 24.5 | 25.2 | 27.8 | 25.5 | 25.3 | 25.6 | 26.0 | |
| Cefota./ceftr./cefta. | 1.3 | 1.8 | 2.8 | 3.8 | 3.6 | 5.0 | 10.6 | 9.9 | 12.5 | 11.9 | 17.0 | 13.6 | 15.9 | 17.7 | 16.3 | |
| Imipenem | 0.1 | 0.2 | 0.1 | 0.0 | 0.0 | 0.1 | 0.6 | 0.2 | 0.1 | 0.2 | 0.1 | 0.2 | 0.2 | 0.3 | 0.3 | |
| Pseudomonas aeruginosa | Piperacillin/tazobactam | 22.5 | 24.7 | 21.2 | 21.0 | 18.7 | 18.8 | 19.8 | 19.0 | 18.3 | 23.5 | 28.4 | 19.4 | 27.2 | 26.5 | 25.6 |
| Ciprofloxacin | 19.7 | 18.4 | 15.6 | 19.5 | 17.4 | 20.8 | 19.4 | 16.5 | 18.9 | 16.9 | 25.2 | 21.1 | 22.1 | 21.3 | 18.4 | |
| Ceftazidim | 14.3 | 18.5 | 17.4 | 18.7 | 20.5 | 33.4 | 32.5 | 32.6 | 32.4 | 29.1 | 24.1 | 17.3 | 19.6 | 18.4 | 17.1 | |
| Imipenem | 24.0 | 22.8 | 23.5 | 23.8 | 22.0 | 26.5 | 28.9 | 24.5 | 29.6 | 32.7 | 32.4 | 29.6 | 31.8 | 30.3 | 31.8 | |
| Meropenem | 8.9 | 14.4 | 9.0 | 14.0 | 14.1 | 17.7 | 17.8 | 16.9 | 19.8 | 22.8 | 23.1 | 19.3 | 22.4 | 18.6 | 20.1 | |
| Amikacin | 8.6 | 12.6 | 7.5 | 5.6 | 3.4 | 3.3 | 5.2 | 4.5 | 2.9 | 6.4 | 12.7 | 8.9 | 7.9 | 7.8 | 5.3 | |
| Klebsiella pneumoniae | Piperacillin/β-lactamase inhibitor | 8.6 | 11.9 | 8.2 | 11.0 | 12.9 | 9.1 | 14.5 | 15.8 | 15.1 | 23.1 | 21.3 | 16.9 | 19.6 | 20.3 | 16.4 |
| Ciprofloxacin | 5.1 | 9.9 | 4.2 | 8.4 | 5.9 | 5.9 | 9.3 | 12.2 | 8.6 | 18.4 | 16.7 | 16.3 | 15.8 | 15.4 | 15.9 | |
| Cefotax./ceftr./cefta. | 4.5 | 10.8 | 6.0 | 6.0 | 6.9 | 6.3 | 10.2 | 12.5 | 10.9 | 20.1 | 19.5 | 12.2 | 16.6 | 16.5 | 15.7 | |
| Imipenem | 0.4 | 0.2 | 0.3 | 0.3 | 0.0 | 0.3 | 0.4 | 1.3 | 0.0 | 0.7 | 1.5 | 0.8 | 1.4 | 1.7 | 1.5 | |
| Enterobacter cloacae | Ciprofloxacin | 4.6 | 5.6 | 10.7 | 6.5 | 5.3 | 6.2 | 9.0 | 6.4 | 6.0 | 6.9 | 8.3 | 8.1 | 7.1 | 7.1 | 7.4 |
| Cefotax./ceftr./cefta. | 39.5 | 69.4 | 63.4 | 33.1 | 31.4 | 35.6 | 37.2 | 34.6 | 38.6 | 39.8 | 38.5 | 34.8 | 36.3 | 36.8 | 31.7 | |
| Imipenem | 0.1 | 0.0 | 0.6 | 0.0 | 0.7 | 0.9 | 0.7 | 0.3 | 1.0 | 1.0 | 0.9 | 0.2 | 0.7 | 1.1 | 0.9 | |
| Acinetobacter baumannii | Ceftazidime | 28.1 | 13.1 | 15.7 | 14.6 | 16.3 | 27.9 | 21.3 | 12.8 | 22.8 | 35.9 | 29.4 | 24.3 | 33.1 | 51.4 | 61.8 |
| Imipenem | 1.1 | 0.7 | 1.0 | 2.5 | 6.2 | 19.4 | 9.0 | 3.9 | 19.0 | 14.4 | 10.9 | 21.0 | 11.7 | 27.0 | 42.8 | |
Cefotax./Ceftr./Cefta.: cefotaxime/ceftriaxone/ceftazidime
Gram-positive pathogens
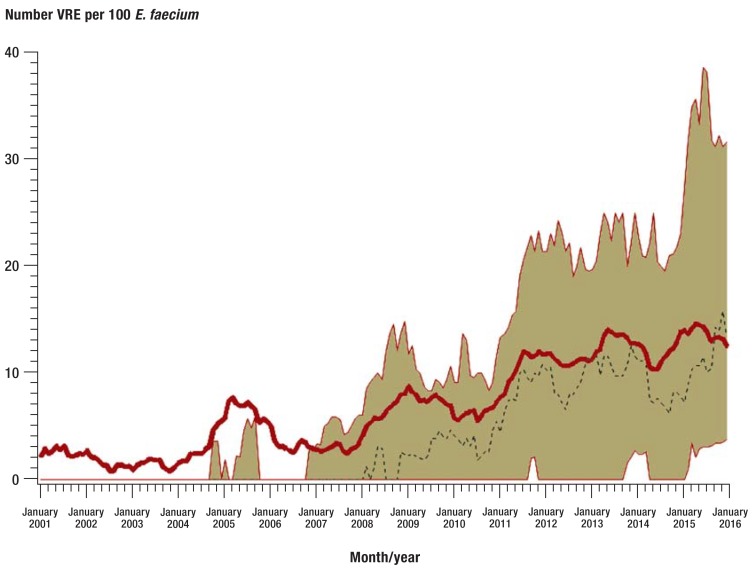
According to SARI, the proportion of methicillin-resistant S. aureus (MRSA) has stabilized in recent years (etable 4). However, in 2015, almost 23% of all S. aureus strains were still resistant to oxacillin (table 2). Of note is the increase in vancomycin-resistant E. faecium (VRE) isolates. In 2001, only individual VRE-isolates were confirmed, whereas in 2015 more than 75% of all ICUs participating in SARI were affected; the resistance rate was 13.3%, which translates into a resistance density of 1.1 VRE/1000 patient days (Table 2 and eTable 4, Figure 2). A new observation among this pathogen was an increase in linezolid-resistant isolates, to 1.6% (n = 28/1776 isolates) in 2015 (etable 4).
Figure 2.
Trends in the resistance rate of vancomycin-resistant E. faecium (VRE) in SARI intensive care units (N = 77) from 2001 to 2015. Values are smoothed and show 12-month moving averages. The bold red line shows the pooled mean value of all SARI ICUs, the beige area marks the interquartile range (25th and 75th percentile), and the dotted black line the median.
E. faecium: Enterococcus faecium; SARI: Surveillance der Antibiotika-Anwendung und der bakteriellen Resistenzen auf deutschen Intensivstationen [surveillance of antibiotic use and resistance in intensive care units]; ICU: intensive care unit
Gram-negative pathogens
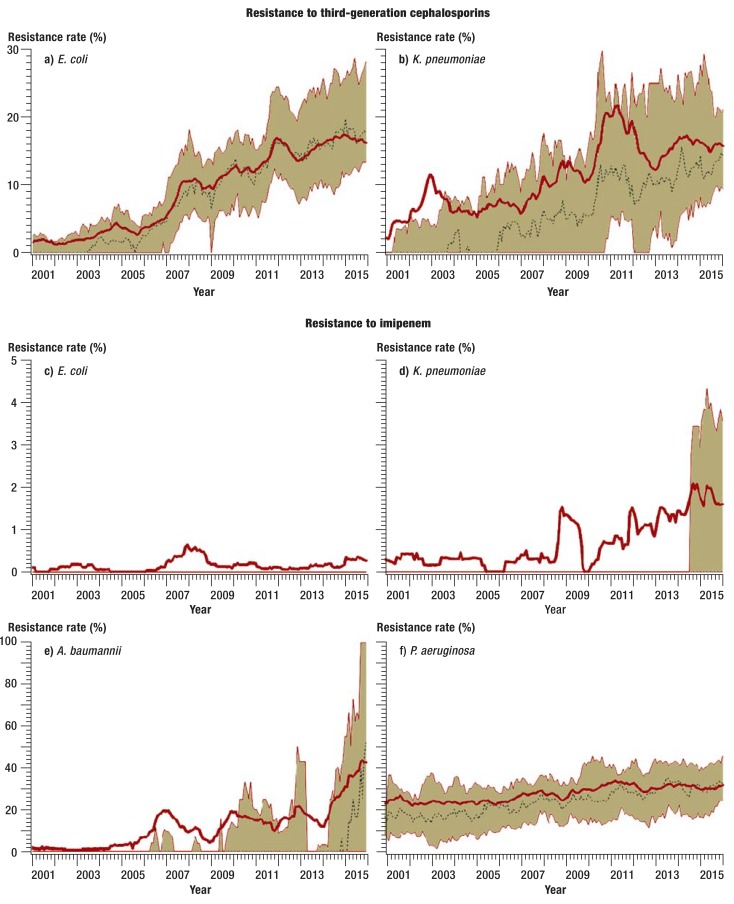
Between 2001 and 2011, the proportion of E. coli and K. pneumoniae isolates with resistance to third-generation cephalosporins increased notably (efigure). Since then the resistance rate has stabilized, and in 2015 it was 16.3% and 15.7%, respectively (table 2). Resistance rates to ciprofloxacin in these two pathogens increased over the entire study period from 8.3% to 26% (E. coli) and from 5.1% to 15.9% (K. pneumoniae) (etable 4). In recent years, the proportion of imipenem-resistant K. pneumoniae isolates has also risen. At the start of the study period, such isolates were seen only in individual cases. In 2015, by contrast, they were confirmed in more than 25% of all participating ICUs. The resistance rate was 1.6% (eFigure, Table 2).
eFigure.
Trends in resistance rates to third-generation cephalosporins and imipenem of selected gram-negative pathogens in SARI intensive care units (ICUs) from 2001 to 2015. a+b) resistance to third-generation cephalosporins; c–f) resistance to imipenem. Values are smoothed and reflect 12-month moving averages. The bold red line shows the pooled mean value for all SARI ICUs, the beige area marks the interquartile range (25th and 75th percentile), the dotted black line shows the median. A. baumannii: Acinetobacter baumannii; E. coli: Escherichia coli; K. pneumoniae: Klebsiella pneumoniae; P. aeruginosa: Pseudomonas aeruginosa; SARI: Surveillance der Antibiotika-Anwendung und der bakteriellen Resistenzen auf deutschen Intensivstationen [surveillance of antibiotic use and resistance in intensive care units]
The increase in A. baumannii isolates with resistance to imipenem was particularly pronounced. The resistance rate has more than doubled over recent years and was 43% in 2015 (eFigure, Table 2).
Resistance density
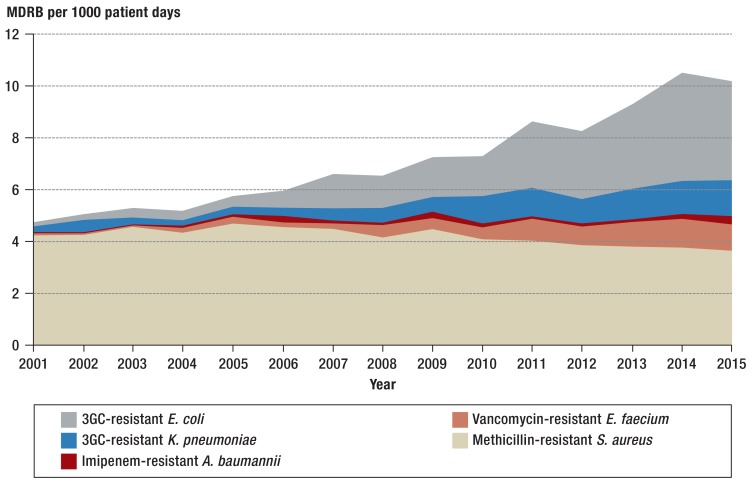
Even though the resistance density is considered a measure for the actual resistance burden, the resistance density for MRSA was stable from 2001 to 2015, whereas it notably increased in VRE (from 0.1/1000 patient days to 1.1/1000 patient days) and imipenem-resistant A. baumannii (from 0.03/1000 patient days to 0.3/1000 patient days) (Table 2, Figure 3). Since 2001 the resistance density of multiresistant gram-negative pathogens has altogether increased substantially and accounted for 54% of the resistance burden in the ICUs participating in SARI by 2015.
Figure 3.
Trends in the incidence density of resistant pathogens in intensive care units (N = 77) from 2001 to 2015
MDRB: multidrug resistant bacteria; 3GC: 3rd generation cephalosporins;
E. coli: Escherichia coli; K: pneumoniae: Klebsiella pneumoniae; A. baumannii: Acinetobacter baumannii; E. faecium: Enterococcus faecium; S. aureus: Staphylococcus aureus
Discussion
Since the start of the SARI project more than 15 years ago, the use of antibiotics in intensive care units participating in SARI has risen by 19%. This rise is mainly due to the increased use since 2009 of piperacillin/tazobactam, carbapenems, and glycopeptides. Studies from France, Norway, and Switzerland similarly observed an increase in antibiotic use in intensive care units, especially of reserve antibiotics or broad-spectrum antibiotics (13– 15). Because of different study periods and possible differences in the study populations, a direct comparison of the results is feasible only to a limited degree.
Regarding the development of resistance in gram-positive pathogens, the increase of VRE in SARI ICUs is notable, and the incipient confirmations of linezolid-resistant E. faecium isolates further restrict therapeutic options (16). The development and selection of VRE can be explained primarily by the use of different antibiotics and the resultant selection pressure on the Enterococcus species that naturally occurs in the gastrointestinal tract (17– 19). In addition to vancomycin (19), ceftriaxone (20) and antibiotics with anaerobic activity—for example, metronidazole or piperacillin/tazobactam—seem to have an important role in the selection process (18, 21, 22). According to SARI, the rise in VRE started in 2007. Simultaneously, the use of glycopeptide antibiotics and piperacillin/tazobactam increased. By contrast, we did not identify an increase in the use of third-generation cephalosporins and imidazole derivatives in the study period. In terms of the spread of VRE in hospitals, however, other factors also have an important role. Because of their high environmental tenacity, VRE can survive for long periods on inanimate surfaces, which facilitates transmission between patients, especially if hygiene measures are not strictly adhered to (19, 23).
In addition to VRE, according to the European Centre for Disease Prevention and Control (ECDC), another cause for concern is the increase of multiresistant gram-negative pathogens in invasive infections (24). Europe-wide between 2011 and 2014, the mean proportion of VRE increased from 6.2% to 7.9%, whereas the proportion of E. coli and K. pneumoniae isolates with resistance to third-generation cephalosporins increased from 9.6% to 12% and from 23.6% to 28%, respectively. Furthermore, the proportion of imipenem-resistant K. pneumoniae isolates increased from 6% to 7.3% (24). Systematic reviews and meta-analyses indicate that the case fatality rate in infections with VRE (25), E. coli and K. pneumoniae with resistance to third-generation cephalosporins (26), and K. pneumoniae with resistance to imipenem (27) is significantly raised compared with infections with susceptible pathogens.
In SARI ICUs, resistant gram-negative pathogens have gained in importance. Except for P. aeruginosa, they were identified in individual cases only at the start of the study period. In the meantime, resistant gram-negative pathogens have become responsible for half of the resistance burden in SARI ICUs. The proportion of E. coli and K. pneumoniae isolates with resistance to third-generation cephalosporins in SARI ICUs has not risen further in the past 3–4 years after a notable increase between 2001 and 2011, but the resistance rate in both pathogens in 2015 was still high, above 15%. One explanation of this stagnation may be the fact that because of increasing resistance against third-generation cephalosporins, carbapenems had to be used increasingly in empiric and definitive antibiotic therapy (12). This facilitated the selection of imipenem-resistant K. pneumoniae and A. baumannii isolates that are difficult to treat and, in the worst case scenario, are associated with hospital outbreaks (28– 31). On SARI ICUs, imipenem-resistant A. baumannii isolates have been identified regularly since 2005, whereas imipenem-resistant E. coli isolates have hardly been identified at all, and K. pneumoniae isolates only since 2014. A look back, however, shows that the development of resistance of the same pathogens against third-generation cephalosporins started in a similar way 15 years ago, and in the meantime this has become an ongoing problem in Germany’s ICUs. The fact that the proportion of resistant Enterobactericeae seems to increase even in the general population (32) makes it clear that it is not enough to study resistance patterns only in hospitals.
Intervention options
The rising prevalence of multiresistant pathogens presents an enormous challenge to medical professionals. For this reason, the use of classes of antibiotics that can still be used to treat the relevant pathogens—for example, in empiric antibiotic therapy—is increasingly necessary. This is especially the case for the increasingly older and comorbid patients in ICUs (4, 33). In order to slow down the rise and spread of multiresistant pathogens in intensive care units, strict adherence to hygiene measures is required, as is rational use of antibiotics, for example, with the support of multidisciplinary “antibiotic stewardship” teams (3, 34, 35). Furthermore, surveillance of antibiotic consumption and antibiotic resistance according to § 23 paragraph 4 of the German Protection Against Infection Act (Infektionsschutzgesetz, IfSG) can contribute to optimizing the use of antibiotics and observing the further spread of multiresistant pathogens (36). Ultimately, a multidisciplinary and cross-sectoral approach (One Health concept) is needed to stop the spread of multiresistant pathogens in general; in additional to human and veterinary medicine, animal husbandry, agriculture, and the environment will have to be included in this concept (37).
Limitations
Because of the ecological study design, the results should be interpreted with caution, and the following limitations should be borne in mind:
• (Participation in SARI is voluntary. For this reason, it is not clear to which extent the results are generalizable to all of Germany’s ICUs. Because ICUs are largely heterogeneous in terms of patient populations, size, and hospitals’ different levels of medical care, we cannot assume that the results are representative for the whole of Germany. As the proportion of university hospitals and maximum care hospitals is very high in our sample, antibiotic use and resistance rates may have been overestimated.
• (Only 20 of the 77 ICUs provided data continuously over the entire study period. However, the sensitivity analysis (etable 5) does not give any indication that antibiotic use and resistance rates in the total cohort and the core cohort developed materially differently.
eTable 5. Sensitivity analysis: antibiotic use (defined daily doses), resistance rate (%), and resistance density (per 1000 person days), 2001 versus 2015*1.
| 2001 | 2001 | 2015 | 2015 | 2015 | |||
| Cohort (ICU) | Pooled mean | Median (IQR) | Pooled mean | Median (IQR) | % increase vs 2001 | P value*2 | |
| Antibiotic use in DDD | |||||||
| Total excl sulbactam | Core(20) | 1222 | 1166 (1008–1360) | 1477 | 1486 (1213–1613) | 21 | 0.038 |
| Total (77) | 1180 | 1162 (953–1434) | 1407 | 1322 (1146–1605) | 19 | 0.028 | |
| Extended-spectrum penicillins | Core(20) | 64 | 63 (25–110) | 38 | 28 (25–50) | −42 | 0.135 |
| Total (77) | 74 | 72 (24–123) | 36 | 27 (13–56) | −52 | 0.007 | |
| Penicillins with β-lactamase- inhibitor without Pseudomonas effectiveness | Core(20) | 204 | 182 (73–291) | 254 | 235 (211–316) | 25 | 0.106 |
| Total (77) | 206 | 209 (86–268) | 262 | 246 (188–346) | 28 | 0.015 | |
| Piperacillin/tazobactam | Core(20) | 42 | 37 (0–70) | 143 | 150 (126–179) | 243 | 0.000 |
| Total (77) | 42 | 32 (0–64) | 146 | 150 (102–179) | 247 | 0.000 | |
| 3rd generation cephalosporins | Core(20) | 100 | 101 (79–140) | 96 | 95 (59–132) | −4 | 0.345 |
| Total (77) | 106 | 104 (77–141) | 91 | 79 (52–124) | −14 | 0.069 | |
| Carbapenems | Core(20) | 89 | 86 (48–125) | 293 | 254 (180–489) | 231 | 0.000 |
| Total (77) | 76 | 67 (42–114) | 250 | 189 (123–373) | 230 | 0.000 | |
| Glycopeptides | Core(20) | 42 | 25 (21–57) | 72 | 54 (26–106) | 72 | 0.113 |
| Total (77) | 38 | 26 (18–55) | 57 | 29 (15–834) | 48 | 0.537 | |
| Fluoroquinolones | Core(20) | 158 | 131 (68–196) | 142 | 132 (106–201) | −10 | 0.927 |
| Total (77) | 151 | 131 (85–192) | 157 | 148 (107–213) | 4 | 0.440 | |
| Macrolides | Core(20) | 93 | 75 (20–108) | 99 | 98 (57–153) | 7 | 0.315 |
| Total (77) | 77 | 64 (19–101) | 104 | 96 (55–145) | 36 | 0.036 | |
| Aminoglycosides | Core(20) | 109 | 77 (39–112) | 24 | 14 (6–35) | −78 | 0.000 |
| Total (77) | 86 | 70 (28–102) | 22 | 12 (6–34) | −75 | 0.000 | |
| Imidazole derivatives | Core(20) | 76 | 100 (21–131) | 50 | 30 (16–46) | −34 | 0.094 |
| Total (77) | 70 | 71 (41–105) | 42 | 31 (18–52) | −40 | 0.001 | |
| Other antibiotics | Core(20) | 11 | 4 (0–15) | 89 | 78 (54–115) | 734 | 0.000 |
| Total (77) | 8 | 2 (0–13) | 85 | 63 (34–124) | 928 | 0.000 | |
| Piperacillin and piperacillin/tazobactam | Core(20) | 68 | 72 (25–107) | 143 | 151 (126–180) | 111 | 0.000 |
| Total (77) | 62 | 59 (34–88) | 146 | 152 (102–179) | 134 | 0.000 | |
| Resistance rate (%) | |||||||
|
Oxacillin-resistant Staphylococcus aureus |
Core(20) | 31.0 | 20.7 (9.7–41.2) | 21.5 | 21.4 (12.7–31) | −31 | 0.987 |
| Total (77) | 26.0 | 17.5 (8.6–32.3) | 22.7 | 25.5 (15.4–36.7) | −13 | 0.255 | |
|
Vancomycin-resistant Enterococcus faecium |
Core(20) | 1.0 | 0 (0–0) | 16.5 | 24.7 (10.8–37.7) | 1559 | 0.000 |
| Total (77) | 2.3 | 0 (0–0) | 13.3 | 14 (3.8–33.3) | 470 | 0.000 | |
|
Cefotax./Ceftr./Cefta.-resistant Klebsiella pneumoniae |
Core(20) | 3.0 | 0 (0–4.2) | 17.2 | 16.3 (11.6–23.2) | 472 | 0.000 |
| Total (77) | 4.5 | 0 (0–4) | 15.7 | 14.1 (9.3–21.1) | 247 | 0.000 | |
|
Imipenem-resistant Klebsiella pneumoniae |
Core(20) | 0.8 | 0 (0–0) | 3.0 | 1.1 (0–4.5) | 296 | 0.024 |
| Total (77) | 0.4 | 0 (0–0) | 1.6 | 0 (0–3.6) | 269 | 0.005 | |
|
Imipenem-resistant Acinetobacter baumannii |
Core(20) | 0.0 | 0 (0–0) | 53.7 | 54.6 (0–100) | – | 0.000 |
| Total (77) | 1.1 | 0 (0–0) | 42.8 | 52.3 (0–100) | 3620 | 0.000 | |
|
Cefotax./ceftr./cefta.-resistant Escherichia coli |
Core(20) | 0.9 | 0 (0–0) | 19.9 | 18.6 (14.9–28.2) | 2121 | 0.000 |
| Total (77) | 1.3 | 0 (0–1.5) | 16.3 | 17.6 (13.3–28.2) | 1113 | 0.000 | |
|
Imipenem-resistant Escherichia coli |
Core(20) | 0.0 | 0 (0–0) | 0.0 | 0 (0–0) | – | 1.000 |
| Total (77) | 0.1 | 0 (0–0) | 0.3 | 0 (0–0) | 292 | 0.196 | |
| Resistance density (per 1000 patient days) | |||||||
|
Oxacillin-resistant Staphylococcus aureus |
Core(20) | 5.3 | 2.1 (0.9–4.3) | 2.0 | 1.8 (0.8–2.3) | −62 | 0.542 |
| Total (77) | 4.2 | 2.3 (1–4.2) | 3.5 | 2.3 (1.3–4.3) | −16 | 0.985 | |
|
Vancomycin-resistant Enterococcus faecium |
Core(20) | 0.0 | 0 (0–0) | 1.0 | 0.8 (0.2–1.5) | 3549 | 0.000 |
| Total (77) | 0.1 | 0 (0–0) | 1.1 | 0.7 (0.2–1.7) | 1416 | 0.000 | |
|
Cefotax./ceftr./cefta.-resistant Klebsiella pneumoniae |
Core(20) | 0.2 | 0 (0–0.3) | 1.1 | 1 (0.7–1.5) | 577 | 0.000 |
| Total (77) | 0.2 | 0 (0–0.2) | 1.4 | 1 (0.4–1.7) | 451 | 0.000 | |
|
Imipenem-resistant Acinetobacter baumannii |
Core(20) | 0.0 | 0 (0–0) | 0.4 | 0 (0–0.5) | – | 0.001 |
| Total (77) | 0.0 | 0 (0–0) | 0.3 | 0 (0–0.3) | 917 | 0.000 | |
|
Cefotax./ceftr./cefta.-resistant Escherichia coli |
Core(20) | 0.1 | 0 (0–0) | 2.3 | 2.5 (1.5–3) | 2342 | 0.000 |
| Total (77) | 0.2 | 0 (0–0.2) | 3.7 | 2.6 (1.4–3.9) | 2270 | 0.000 | |
|
Imipenem-resistant Escherichia coli |
Core(20) | 0.0 | 0 (0–0) | 0.0 | 0 (0–0) | – | 1.000 |
| Total (77) | 0.0 | 0 (0–0) | 0.1 | 0 (0–0) | 711 | 0.170 | |
*1 For the intensive care units that continuously participated in SARI from 2001 through 2015 (N = 20, core cohort) compared with total cohort (N = 77)
*2 Wilcoxon test
Cefotax./ceftr./cefta.: cefotaxime/ceftriaxone/ceftazidime; DDD: defined daily doses; total: total cohort (N = 77 ICUs); ICU: intensive care unit; IQR: interquartile range; core: core cohort(N = 20 ICUs); SARI: Surveillance der Antibiotika-Anwendung und der bakteriellen Resistenzen auf deutschen Intensivstationen [surveillance of antibiotic use and resistance in intensive care units]
• (Resistance testing was done by different laboratories and followed different standards (DIN 58940, CLSI, and EUCAST). These standards partly differ in terms of their threshold values for the categories “susceptible,” “intermediate,” or “resistant” (to a particular antibiotic). If laboratories swap CLSI for EUCAST, the resistance rate of some pathogens to certain antibiotics may rise (38). Furthermore, threshold values also changed over the study period within certain testing methods—for example, CLSI 2009–2011. This poses an additional obstacle to the interpretation of the resistance rate (38– 40). Even though in our data, the effect of such changes cannot be identified, an (additional) rise in resistance rates from 2011 onwards seems to be plausible.
• (As SARI did not collect data on the collection sites where pathogens were isolated, the proportions of infections and colonization cannot be calculated.
• (The DDD used to describe antibiotic use does not necessarily reflect the recommended daily dose (RDD) or the prescribed daily dose (PDD) in Germany. For this reason, antibiotic use may have been overestimated—for example, when the prescribed dose was higher than the defined daily dose in some ß-lactam antibiotics—or underestimated—for example, when a reduced dose was given to patients with kidney failure.
In spite of these limitations, the present data can help to better assess trends in antibiotic consumption and resistance patterns in Germany’s ICUs and to develop measures to combat the development of resistance.
Supplementary Material
Methods
Participation in the surveillance of antibiotic use and bacterial resistance in German intensive care wards (SARI) is voluntary. The methods are explained in the relevant study protocol (http://sari.eu-burden.info/down/protokoll.pdf) and have been described in detail elsewhere (10–12). Interested, non-pediatric intensive care units (ICUs)—independently of the type of hospital— in Germany have been able to enroll since 2001. The following criteria have to be met in order to be able to participate:
Ideally, ICUs are already participating in a module of the hospital infection surveillance system KISS (Krankenhaus-Infektions-Surveillance-System), because important data, such as the size of the hospital, the type of ICU, and the number of ICU beds, would have already been collected. In the context of quality control measures, participating laboratories sent bacterial isolates to the central study laboratory, which re-tested resistance patterns and undertook proficiency tests (12). The present study included all ICUs that provided data on antibiotic use and pathogens’ resistance patterns between 2001 and 2015.
Antibiotic use
Data on antibiotic use were collected via the pharmacies in participating hospitals. The documented use of all oral and parenteral antibiotics in a ward is calculated in defined daily doses (DDD, in g) per antibiotic. The defined daily dose is a mathematical variable defined by the World Health Organization (WHO), which corresponds to the assumed mean daily maintenance dose for the main indication of a medical drug in adults, and which enables international comparison of antibiotic use data (e1). From antibiotic use, the defined daily dose, and the number of patient days, the antibiotic use density is calculated by using the following formula: (antibiotic use in g/defined daily dose in g) × 1000 patient days.
Resistance data
The ICUs participating in SARI reported the numbers of confirmed isolates of the following 13 pathogens:
Since 2012, some laboratories have conducted further species identification within the A. baumannii complex (A. baumannii [sensu stricto], A. calcoaceticus, A. pittii, and A. nosocomialis), which means that the isolates previously attributed to the A. baumannii complex (sensu stricto) are detected less often (e2).
The frequency with which specimens are taken is the clinicians’ prerogative in the relevant ICU; SARI does not consider mere screening investigations. We did not collect data on the number of specimens sampled, the collection site, nor on whether infection or colonization was present, and whether the pathogen had been community-acquired or hospital-acquired.
In addition to the isolates, the laboratories responsible for the ICUs report on a monthly basis the number of isolates tested for specific antibiotics and resistant isolates. The antibiotics that are to be tested per species are defined in the study protocol (http://sari.eu-burden.info/down/protokoll.pdf). Resistance is tested for by following the German industry standard (DIN) 58940, CLSI (Clinical & Laboratory Standards Institute), or EUCAST (European Committee on Antimicrobial Susceptibility Testing). Until the 31 December 2013, information about the test method employed was reliably documented. Copy strains—that is, isolates detected within 30 days from a patient with an identical antibiogram—were not included in the evaluation. An isolate is considered non-identical if at least one of the antibiotics that were predefined for each species deviated in terms of the classification R (resistant), S (susceptible), or I (intermediate)—for example, from R or S to I.
A pathogen’s resistance rate is calculated from the number of resistant isolates of a species to a certain antibiotic, divided by the number of all pathogens tested against this antibiotic × 100. The resistance density results from the number of resistant pathogens/1000 patient days.
Feedback
Participating ICUs received annual feedback on their own antibiotic use density compared with that of all participating ICUs (reference values), as well as on their own resistance rate and resistance density compared with the reference values of those ICUs using the same testing methods.
Statistical analysis
From the data, we calculated pooled means, medians, and interquartile ranges (IQR, 25th and 75th percentile) for the antibiotic use density, resistance rate, and resistance density for 2001–2015. Not all ICUs participated in SARI for the entire study period. For this reason, we calculated in a sensitivity analysis the antibiotic use density, resistance rate, and resistance density of commonly used antibiotics and selected pathogens only for those ICUs that reported data continuously from 2001 through 2015 (core cohort). We compared the results of the core cohort and the total cohort at 2001 and 2015, in order to identify possible differences in the development of antibiotic use and resistance rates. We used the Wilcoxon test to find out for both cohorts, whether antibiotic use and resistance rates of the analyzed pathogens changed from 2001 to 2015. Trends over time in the resistance rates of selected pathogens were displayed graphically as a moving average over 12 months (smoothed graph). The reference values for antibiotic use density and resistance rates (pooled data from 2001–2015) that were calculated in the context of the SARI project are in the public domain at sari.eu-burden.info. We used SAS 9.4 (SAS Institute Inc., Cary, NC, USA) to analyze our data.
Sensitivity analysis
In total, 20 of the 77 ICUs continuously provided data for the entire study period. Hospitals in the core cohort (20 ICUs) had a greater median number of beds than the total cohort (77 ICUs); the larger proportion consisted of hospitals offering maximum medical care (etable 1). Trends in antibiotic use in the core and total cohorts are comparable (etable 4). The use of all antibiotics increased from 2001 to 2015 in the core cohort by a mean of 21% (p = 0.04) and in the total cohort by 19% (p = 0.03). The use of carbapenems (+231% core cohort versus +230% total cohort), piperacillin/tazobactam (+243% versus +247%), or other antibiotics (+734% versus +928) increased to a comparable degree in both cohorts, and the use of aminoglycosides (–78% versus –75%) decreased to a comparable degree in both cohorts (p <0.001 for all values). Trends in the resistance rate in the core cohort and total cohort are similar (etable 5). Slight differences were seen in the resistance rate of vancomycin-resistant E. faecium (VRE): the resistance rate of VRE increased in the core cohort from 1% to 16.5% (+1560%; p <0.001) and in the total cohort from 2.3% to 13.3% (+470%; p <0.001).
A named person is nominated to have responsibility for the project.
Resistance testing is done by using German industry standards (DIN 58940), CLSI (Clinical & Laboratory Standards Institute), or EUCAST (European Committee on Antimicrobial Susceptibility Testing).
Patient days, antibiotic use, and resistance rates of selected pathogens are reported on a monthly basis to the study center at Charité Berlin.
Staphylococcus aureus
Streptococcus pneumoniae
Coagulase-negative staphylococci
Enterococcus faecalis
Enterococcus faecium
Escherichia coli
Klebsiella pneumoniae
Enterobacter cloacae
Serratia marcescens
Citrobacter spp.
Pseudomonas aeruginosa
Stenotrophomonas maltophilia
Acinetobacter (A.) baumannii.
Key Messages.
The large increase in vancomycin-resistant enterococci and the increase in resistance to impenem should prompt a strengthening of attention given to these subjects nationwide, as well as a demand for and implementation of relevant prevention measures.
In this context, rapid diagnostic evaluation and targeted therapy are of great importance, as are effective measures for preventing the spread of multiresistant pathogens.
Specialists working in “antibiotic stewardship” and prevention of infection should provide regular advice at least in all hospitals with intensive care units.
The rise in the use of broad-spectrum and reserve antibiotics should provide an impetus for optimizing rational antibiotic use in hospitals.
Owing to the study design—that is, voluntary participation of the intensive care units and the composition of the group under study over time—we cannot claim that the study results are representative for Germany.
Acknowledgments
Acknowledgment
Translated from the original German by Birte Twisselmann, PhD.
We thank all participants in the SARI project for their support.
Funding
The SARI project received funding from the German Federal Ministry of Education and Research (BMBF) until the end of 2006.
Footnotes
Conflict of interest statement
The authors declare that no conflict of interest exists.
References
- 1.Van Boeckel TP, Gandra S, Ashok A, et al. Global antibiotic consumption 2000 to 2010: an analysis of national pharmaceutical sales data. Lancet Infect Dis. 2014;14:742–750. doi: 10.1016/S1473-3099(14)70780-7. [DOI] [PubMed] [Google Scholar]
- 2.Holmes AH, Moore LS, Sundsfjord A, et al. Understanding the mechanisms and drivers of antimicrobial resistance. Lancet. 2016;387:176–187. doi: 10.1016/S0140-6736(15)00473-0. [DOI] [PubMed] [Google Scholar]
- 3.Laxminarayan R, Duse A, Wattal C, et al. Antibiotic resistance-the need for global solutions. Lancet Infect Dis. 2013;13:1057–1098. doi: 10.1016/S1473-3099(13)70318-9. [DOI] [PubMed] [Google Scholar]
- 4.Sader HS, Farrell DJ, Flamm RK, Jones RN. Antimicrobial susceptibility of Gram-negative organisms isolated from patients hospitalized in intensive care units in United States and European hospitals (2009-2011) Diagn Microbiol Infect Dis. 2014;78:443–448. doi: 10.1016/j.diagmicrobio.2013.11.025. [DOI] [PubMed] [Google Scholar]
- 5.Vincent JL, Rello J, Marshall J, et al. International study of the prevalence and outcomes of infection in intensive care units. JAMA. 2009;302:2323–2329. doi: 10.1001/jama.2009.1754. [DOI] [PubMed] [Google Scholar]
- 6.Neidell MJ, Cohen B, Furuya Y, et al. Costs of healthcare- and community-associated infections with antimicrobial-resistant versus antimicrobial-susceptible organisms. Clin Infect Dis. 2012;55:807–815. doi: 10.1093/cid/cis552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cheah AL, Spelman T, Liew D, et al. Enterococcal bacteraemia: factors influencing mortality, length of stay and costs of hospitalization. Clin Microbiol Infect. 2013;19:E181–E189. doi: 10.1111/1469-0691.12132. [DOI] [PubMed] [Google Scholar]
- 8.Shorr AF. Review of studies of the impact on Gram-negative bacterial resistance on outcomes in the intensive care unit. Crit Care Med. 2009;37:1463–1469. doi: 10.1097/CCM.0b013e31819ced02. [DOI] [PubMed] [Google Scholar]
- 9.Stewardson AJ, Allignol A, Beyersmann J, et al. The health and economic burden of bloodstream infections caused by antimicrobial-susceptible and non-susceptible Enterobacteriaceae and Staphylococcus aureus in European hospitals, 2010 and 2011: a multicentre retrospective cohort study. Euro Surveill. 2016;21 doi: 10.2807/1560-7917.ES.2016.21.33.30319. pii 30319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Meyer E, Jonas D, Schwab F, Rueden H, Gastmeier P, Daschner FD. Design of a surveillance system of antibiotic use and bacterial resistance in German intensive care units (SARI) Infection. 2003;31:208–215. doi: 10.1007/s15010-003-3201-7. [DOI] [PubMed] [Google Scholar]
- 11.Meyer E, Schwab F, Jonas D, Rueden H, Gastmeier P, Daschner FD. Surveillance of antimicrobial use and antimicrobial resistance in intensive care units (SARI): 1 antimicrobial use in German intensive care units. Intensive Care Med. 2004;30:1089–1096. doi: 10.1007/s00134-004-2266-9. [DOI] [PubMed] [Google Scholar]
- 12.Meyer E, Schwab F, Schroeren-Boersch B, Gastmeier P. Dramatic increase of third-generation cephalosporin-resistant E coli in German intensive care units: secular trends in antibiotic drug use and bacterial resistance, 2001 to 2008. Crit Care. 2010;14 doi: 10.1186/cc9062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Haug JB, Berild D, Walberg M, Reikvam A. Increased antibiotic use in Norwegian hospitals despite a low antibiotic resistance rate. J Antimicrob Chemother. 2011;66:2643–2646. doi: 10.1093/jac/dkr361. [DOI] [PubMed] [Google Scholar]
- 14.Pluss-Suard C, Pannatier A, Kronenberg A, Muhlemann K, Zanetti G. Hospital antibiotic consumption in Switzerland: comparison of a multicultural country with Europe. J Hosp Infect. 2011;79:166–171. doi: 10.1016/j.jhin.2011.05.028. [DOI] [PubMed] [Google Scholar]
- 15.Dumartin C, L‘Heriteau F, Pefau M, et al. Antibiotic use in 530 French hospitals: results from a surveillance network at hospital and ward levels in 2007. J Antimicrob Chemother. 2010;65:2028–2036. doi: 10.1093/jac/dkq228. [DOI] [PubMed] [Google Scholar]
- 16.O‘Driscoll T, Crank CW. Vancomycin-resistant enterococcal infections: epidemiology, clinical manifestations, and optimal management. Infect Drug Resist. 2015;8:217–230. doi: 10.2147/IDR.S54125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eckburg PB, Bik EM, Bernstein CN, et al. Diversity of the human intestinal microbial flora. Science. 2005;308:1635–1638. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Donskey CJ, Chowdhry TK, Hecker MT, et al. Effect of antibiotic therapy on the density of vancomycin-resistant enterococci in the stool of colonized patients. N Engl J Med. 2000;343:1925–1932. doi: 10.1056/NEJM200012283432604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Arias CA, Murray BE. The rise of the Enterococcus: beyond vancomycin resistance. Nat Rev Microbiol. 2012;10:266–278. doi: 10.1038/nrmicro2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McKinnell JA, Kunz DF, Chamot E, et al. Association between vancomycin-resistant Enterococci bacteremia and ceftriaxone usage. Infect Control Hosp Epidemiol. 2012;33:718–724. doi: 10.1086/666331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McKinnell JA, Kunz DF, Moser SA, et al. Patient-level analysis of incident vancomycin-resistant enterococci colonization and antibiotic days of therapy. Epidemiol Infect. 2016;144:1748–1755. doi: 10.1017/S0950268815003118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stiefel U, Paterson DL, Pultz NJ, Gordon SM, Aron DC, Donskey CJ. Effect of the increasing use of piperacillin/tazobactam on the incidence of vancomycin-resistant enterococci in four academic medical centers. Infect Control Hosp Epidemiol. 2004;25:380–383. doi: 10.1086/502409. [DOI] [PubMed] [Google Scholar]
- 23.Mutters NT, Mersch-Sundermann V, Mutters R, Brandt C, Schneider-Brachert W, Frank U. Control of the spread of vancomycin-resistant enterococci in hospitals: epidemiology and clinical relevance. Dtsch Arztebl Int. 2013;110:725–731. doi: 10.3238/arztebl.2013.0725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.European Centre for Disease Prevention and Control. Antimicrobial resistance surveillance in Europe 2014. Annual report of the european antimicrobial resistance Surveillance Network (EARS-Net). Stockholm: ECDC 2015. www.ecdc.europa.eu/en/publications/Publications/antimicrobial-resistance-europe-2014.pdf (last accessed on 24 October 2016) [Google Scholar]
- 25.DiazGranados CA, Zimmer SM, Klein M, Jernigan JA. Comparison of mortality associated with vancomycin-resistant and vancomycin-susceptible enterococcal bloodstream infections: a meta-analysis. Clin Infect Dis. 2005;41:327–333. doi: 10.1086/430909. [DOI] [PubMed] [Google Scholar]
- 26.World Health Organization (WHO) Antimicrobial resistance: global report on surveillance Geneva: WHO 2014. apps.who.int/iris/bitstream/10665/112642/1/9789241564748_eng.pdf (last accessed on 1 June 2017) [Google Scholar]
- 27.Xu L, Sun X, Ma X. Systematic review and meta-analysis of mortality of patients infected with carbapenem-resistant Klebsiella pneumoniae. Ann Clin Microbiol Antimicrob. 2017;16 doi: 10.1186/s12941-017-0191-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nordmann P, Cuzon G, Naas T. The real threat of Klebsiella pneumoniae carbapenemase-producing bacteria. Lancet Infect Dis. 2009;9:228–236. doi: 10.1016/S1473-3099(09)70054-4. [DOI] [PubMed] [Google Scholar]
- 29.Maragakis LL, Perl TM. Acinetobacter baumannii: epidemiology, antimicrobial resistance, and treatment options. Clin Infect Dis. 2008;46:1254–1263. doi: 10.1086/529198. [DOI] [PubMed] [Google Scholar]
- 30.Campos AC, Albiero J, Ecker AB, et al. Outbreak of Klebsiella pneumoniae carbapenemase-producing K pneumoniae: a systematic review. Am J Infect Control. 2016;44:1374–1380. doi: 10.1016/j.ajic.2016.03.022. [DOI] [PubMed] [Google Scholar]
- 31.Munoz-Price LS, Poirel L, Bonomo RA, et al. Clinical epidemiology of the global expansion of Klebsiella pneumoniae carbapenemases. Lancet Infect Dis. 2013;13:785–796. doi: 10.1016/S1473-3099(13)70190-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Woerther PL, Burdet C, Chachaty E, Andremont A. Trends in human fecal carriage of extended-spectrum beta-lactamases in the community: toward the globalization of CTX-M. Clin Microbiol Rev. 2013;26:744–758. doi: 10.1128/CMR.00023-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.MacVane SH. Antimicrobial resistance in the intensive care unit: a focus on gram-negative bacterial infections. J Intensive Care Med. 2017;32:25–37. doi: 10.1177/0885066615619895. [DOI] [PubMed] [Google Scholar]
- 34.Goff DA, Kullar R, Goldstein EJ, et al. A global call from five countries to collaborate in antibiotic stewardship: united we succeed, divided we might fail. Lancet Infect Dis. 2017;17:e56–e63. doi: 10.1016/S1473-3099(16)30386-3. [DOI] [PubMed] [Google Scholar]
- 35.Schuts EC, Hulscher ME, Mouton JW, et al. Current evidence on hospital antimicrobial stewardship objectives: a systematic review and meta-analysis. Lancet Infect Dis. 2016;16:847–856. doi: 10.1016/S1473-3099(16)00065-7. [DOI] [PubMed] [Google Scholar]
- 36.Schweickert B, Kern WV, de With K, et al. [Surveillance of antibiotic consumption: clarification of the “definition of data on the nature and extent of antibiotic consumption in hospitals according to section sign 23 paragraph 4 sentence 2 of the IfSG“] Bundesgesundheitsblatt Gesundheitsforschung Gesundheitsschutz. 2013;56:903–912. doi: 10.1007/s00103-013-1764-8. [DOI] [PubMed] [Google Scholar]
- 37.Niedrig M, Eckmanns T, Wieler L. One-Health-Konzept: Eine Antwort auf resistente Bakterien? Dtsch Arztebl. 2017;114 [Google Scholar]
- 38.Hombach M, Bloemberg GV, Bottger EC. Effects of clinical breakpoint changes in CLSI guidelines 2010/2011 and EUCAST guidelines 2011 on antibiotic susceptibility test reporting of gram-negative bacilli. J Antimicrob Chemother. 2012;67:622–632. doi: 10.1093/jac/dkr524. [DOI] [PubMed] [Google Scholar]
- 39.Rodloff A, Bauer T, Ewig S, Kujath P, Muller E. Susceptible, intermediate, and resistant—the intensity of antibiotic action. Dtsch Arztebl Int. 2008;105:657–662. doi: 10.3238/arztebl.2008.0657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stokkou S, Tammer I, Zibolka S, Grabau C, Geginat G. Impact of minimal inhibitory concentration breakpoints on local cumulative bacterial susceptibility data and antibiotic consumption. BMC Res Notes. 2014;7 603 doi: 10.1186/1756-0500-7-603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- E1.World Health Organization (WHO) DDD: definition and general considerations. WHO. www.whocc.no/ddd/definition_and_general_considera (last accessed on 8 November 2016) [Google Scholar]
- E2.Robert Koch-Institut. Acinetobacter baumannii - ein Krankenhauskeim mit beunruhigendem Entwicklungspotenzial. Epidemiologisches Bulletin 2013: 32. www.rki.de/DE/Content/Infekt/EpidBull/Archiv/2013/Ausgaben/32_13.pdf?__blob=publicationFile (last accessed on 1 August 2017) [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Methods
Participation in the surveillance of antibiotic use and bacterial resistance in German intensive care wards (SARI) is voluntary. The methods are explained in the relevant study protocol (http://sari.eu-burden.info/down/protokoll.pdf) and have been described in detail elsewhere (10–12). Interested, non-pediatric intensive care units (ICUs)—independently of the type of hospital— in Germany have been able to enroll since 2001. The following criteria have to be met in order to be able to participate:
Ideally, ICUs are already participating in a module of the hospital infection surveillance system KISS (Krankenhaus-Infektions-Surveillance-System), because important data, such as the size of the hospital, the type of ICU, and the number of ICU beds, would have already been collected. In the context of quality control measures, participating laboratories sent bacterial isolates to the central study laboratory, which re-tested resistance patterns and undertook proficiency tests (12). The present study included all ICUs that provided data on antibiotic use and pathogens’ resistance patterns between 2001 and 2015.
Antibiotic use
Data on antibiotic use were collected via the pharmacies in participating hospitals. The documented use of all oral and parenteral antibiotics in a ward is calculated in defined daily doses (DDD, in g) per antibiotic. The defined daily dose is a mathematical variable defined by the World Health Organization (WHO), which corresponds to the assumed mean daily maintenance dose for the main indication of a medical drug in adults, and which enables international comparison of antibiotic use data (e1). From antibiotic use, the defined daily dose, and the number of patient days, the antibiotic use density is calculated by using the following formula: (antibiotic use in g/defined daily dose in g) × 1000 patient days.
Resistance data
The ICUs participating in SARI reported the numbers of confirmed isolates of the following 13 pathogens:
Since 2012, some laboratories have conducted further species identification within the A. baumannii complex (A. baumannii [sensu stricto], A. calcoaceticus, A. pittii, and A. nosocomialis), which means that the isolates previously attributed to the A. baumannii complex (sensu stricto) are detected less often (e2).
The frequency with which specimens are taken is the clinicians’ prerogative in the relevant ICU; SARI does not consider mere screening investigations. We did not collect data on the number of specimens sampled, the collection site, nor on whether infection or colonization was present, and whether the pathogen had been community-acquired or hospital-acquired.
In addition to the isolates, the laboratories responsible for the ICUs report on a monthly basis the number of isolates tested for specific antibiotics and resistant isolates. The antibiotics that are to be tested per species are defined in the study protocol (http://sari.eu-burden.info/down/protokoll.pdf). Resistance is tested for by following the German industry standard (DIN) 58940, CLSI (Clinical & Laboratory Standards Institute), or EUCAST (European Committee on Antimicrobial Susceptibility Testing). Until the 31 December 2013, information about the test method employed was reliably documented. Copy strains—that is, isolates detected within 30 days from a patient with an identical antibiogram—were not included in the evaluation. An isolate is considered non-identical if at least one of the antibiotics that were predefined for each species deviated in terms of the classification R (resistant), S (susceptible), or I (intermediate)—for example, from R or S to I.
A pathogen’s resistance rate is calculated from the number of resistant isolates of a species to a certain antibiotic, divided by the number of all pathogens tested against this antibiotic × 100. The resistance density results from the number of resistant pathogens/1000 patient days.
Feedback
Participating ICUs received annual feedback on their own antibiotic use density compared with that of all participating ICUs (reference values), as well as on their own resistance rate and resistance density compared with the reference values of those ICUs using the same testing methods.
Statistical analysis
From the data, we calculated pooled means, medians, and interquartile ranges (IQR, 25th and 75th percentile) for the antibiotic use density, resistance rate, and resistance density for 2001–2015. Not all ICUs participated in SARI for the entire study period. For this reason, we calculated in a sensitivity analysis the antibiotic use density, resistance rate, and resistance density of commonly used antibiotics and selected pathogens only for those ICUs that reported data continuously from 2001 through 2015 (core cohort). We compared the results of the core cohort and the total cohort at 2001 and 2015, in order to identify possible differences in the development of antibiotic use and resistance rates. We used the Wilcoxon test to find out for both cohorts, whether antibiotic use and resistance rates of the analyzed pathogens changed from 2001 to 2015. Trends over time in the resistance rates of selected pathogens were displayed graphically as a moving average over 12 months (smoothed graph). The reference values for antibiotic use density and resistance rates (pooled data from 2001–2015) that were calculated in the context of the SARI project are in the public domain at sari.eu-burden.info. We used SAS 9.4 (SAS Institute Inc., Cary, NC, USA) to analyze our data.
Sensitivity analysis
In total, 20 of the 77 ICUs continuously provided data for the entire study period. Hospitals in the core cohort (20 ICUs) had a greater median number of beds than the total cohort (77 ICUs); the larger proportion consisted of hospitals offering maximum medical care (etable 1). Trends in antibiotic use in the core and total cohorts are comparable (etable 4). The use of all antibiotics increased from 2001 to 2015 in the core cohort by a mean of 21% (p = 0.04) and in the total cohort by 19% (p = 0.03). The use of carbapenems (+231% core cohort versus +230% total cohort), piperacillin/tazobactam (+243% versus +247%), or other antibiotics (+734% versus +928) increased to a comparable degree in both cohorts, and the use of aminoglycosides (–78% versus –75%) decreased to a comparable degree in both cohorts (p <0.001 for all values). Trends in the resistance rate in the core cohort and total cohort are similar (etable 5). Slight differences were seen in the resistance rate of vancomycin-resistant E. faecium (VRE): the resistance rate of VRE increased in the core cohort from 1% to 16.5% (+1560%; p <0.001) and in the total cohort from 2.3% to 13.3% (+470%; p <0.001).
A named person is nominated to have responsibility for the project.
Resistance testing is done by using German industry standards (DIN 58940), CLSI (Clinical & Laboratory Standards Institute), or EUCAST (European Committee on Antimicrobial Susceptibility Testing).
Patient days, antibiotic use, and resistance rates of selected pathogens are reported on a monthly basis to the study center at Charité Berlin.
Staphylococcus aureus
Streptococcus pneumoniae
Coagulase-negative staphylococci
Enterococcus faecalis
Enterococcus faecium
Escherichia coli
Klebsiella pneumoniae
Enterobacter cloacae
Serratia marcescens
Citrobacter spp.
Pseudomonas aeruginosa
Stenotrophomonas maltophilia
Acinetobacter (A.) baumannii.