ABSTRACT
RbdA is a positive regulator of biofilm dispersal of Pseudomonas aeruginosa. Its cytoplasmic region (cRbdA) comprises an N-terminal Per-ARNT-Sim (PAS) domain followed by a diguanylate cyclase (GGDEF) domain and an EAL domain, whose phosphodiesterase activity is allosterically stimulated by GTP binding to the GGDEF domain. We report crystal structures of cRbdA and of two binary complexes: one with GTP/Mg2+ bound to the GGDEF active site and one with the EAL domain bound to the c-di-GMP substrate. These structures unveil a 2-fold symmetric dimer stabilized by a closely packed N-terminal PAS domain and a noncanonical EAL dimer. The autoinhibitory switch is formed by an α-helix (S-helix) immediately N-terminal to the GGDEF domain that interacts with the EAL dimerization helix (α6-E) of the other EAL monomer and maintains the protein in a locked conformation. We propose that local conformational changes in cRbdA upon GTP binding lead to a structure with the PAS domain and S-helix shifted away from the GGDEF-EAL domains, as suggested by small-angle X-ray scattering (SAXS) experiments. Domain reorientation should be facilitated by the presence of an α-helical lever (H-helix) that tethers the GGDEF and EAL regions, allowing the EAL domain to rearrange into an active dimeric conformation.
IMPORTANCE Biofilm formation by bacterial pathogens increases resistance to antibiotics. RbdA positively regulates biofilm dispersal of Pseudomonas aeruginosa. The crystal structures of the cytoplasmic region of the RbdA protein presented here reveal that two evolutionarily conserved helices play an important role in regulating the activity of RbdA, with implications for other GGDEF-EAL dual domains that are abundant in the proteomes of several bacterial pathogens. Thus, this work may assist in the development of small molecules that promote bacterial biofilm dispersal.
KEYWORDS: cyclic di-GMP, PAS domain, GGDEF-EAL domain, Pseudomonas aeruginosa, biofilm, crystal structure, phosphodiesterase, diguanylate cyclase, allosteric control, GGDEF domain, allosteric
INTRODUCTION
Cytoplasmic second messengers provide an internal representation of external conditions encountered by bacterial cells. Original cues are amplified into an intracellular signal capable of eliciting various biochemical and metabolic changes. Bis-(3′,5′)-cyclic dimeric GMP (c-di-GMP) (1) is a second messenger involved in the regulation of several bacterial processes, such as motility, virulence, and biofilm formation (2, 3). The cellular concentration of c-di-GMP is controlled by enzymatic domains with opposing activities: GGDEF domain proteins with diguanylate cyclase (DGC) activity and EAL or HD-GYP domain proteins, which are c-di-GMP-specific phosphodiesterases (PDEs) (4–6). Synthesis of c-di-GMP from two GTP molecules results from the cooperative action of two GGDEF domains arranged in a 2-fold symmetric quaternary conformation such that their active (A) (half) sites face each other, and each half active site binds one GTP molecule. A second c-di-GMP binding site, termed the I site and containing an RXXD motif immediately preceding the GGDEF catalytic motif, functions in product inhibition of the DGC activity via a feedback inhibition mechanism (4, 7, 8). Conversely, hydrolysis of the c-di-GMP phosphodiester bond is catalyzed by EAL and HD-GYP domains, yielding 5′-pGpG or GMP. Active EAL domains characterized structurally so far also form 2-fold symmetric dimers through a conserved protein interface that involves two alpha helices (α5 and α6) and a regulatory loop from each monomer (loop 6; also named the β5-α5 loop) (9–12).
Many proteins containing either a GGDEF or EAL domain have been identified in pathogenic and nonpathogenic bacteria, where they regulate c-di-GMP levels and biofilm formation (13–19). Intriguingly, in several instances, these two enzymatic modules can be fused in the order GGDEF-EAL via a linker region. We refer to these as “GGDEF-EAL dual domains.” Several proteins containing a dual domain have either DGC activity, PDE activity, or no enzymatic activity at all, because one or the other enzymatic module has degenerate and hence inactive catalytic motifs (16, 20–24). For example, the FimX protein of Pseudomonas aeruginosa has inactive EAL and GGDEF domains and merely functions as a c-di-GMP cellular sensor (20). However, an increasing number of proteins with dual domains with both enzymatic activities and that can conditionally switch between DGC and PDE activities have been identified, including diguanylate cyclase 1 (dgc-1) from Gluconacetobacter xylinus (21, 25) and the GGDEF-EAL proteins BphG1 from Rhodobacter sphaeroides (26), ScrC from Vibrio parahaemolyticus (27), MSDGC1 from Mycobacterium smegmatis (28), Rv1354c from Mycobacterium tuberculosis (28), and CC3396 from Caulobacter crescentus (16). Given the genomic abundance of dual-domain proteins and the involvement of these proteins in signaling pathways of opportunistic bacterial pathogens, a better understanding of how their enzymatic activities are regulated, leading to a certain level of cellular c-di-GMP and a defined phenotype in terms of biofilm formation or dispersal, is crucial. Higher cellular c-di-GMP concentrations are associated with a sessile lifestyle and biofilm formation, while lower concentrations of the messenger correlate with motility and a planktonic lifestyle. For P. aeruginosa strain PAO1, 16 proteins containing a GGDEF-EAL dual domain have been identified (29). Most proteins containing either a GGDEF or EAL domain, as well as dual-domain proteins, have various regulatory and sensory domains located at their N termini. These sensor domains modulate activities in response to external stimuli (5, 17, 30), such as light or small ligands, including O2, NO, CO, or quorum-sensing molecules (reviewed in reference 3).
Pioneering studies were conducted on the MorA protein from P. aeruginosa (31, 32) and the LapD protein from Pseudomonas fluorescens (33). MorA is a membrane-bound regulator of flagellar development and biofilm formation (31) that comprises two functional DGC and PDE enzymatic domains. A crystal structure of the GGDEF-EAL dual domain of MorA revealed a two-lobe structure maintained by canonical dimeric interactions between the EAL domains, but with no interactions between the GGDEF domains, with their A sites facing away from each other, corresponding to an inactive DGC conformation (32). Interestingly, the linker region connecting the GGDEF domain to the EAL domain, which comprises 19 amino acids, folds into an α-helix that was christened H-helix, to emphasize its likelihood of functioning as a hinge region between the two enzymatic domains (32). LapD is a c-di-GMP sensor with a modular architecture encompassing a HAMP inside-out relay module and a GGDEF domain followed by an EAL domain, with the latter two being catalytically inactive due to the presence of degenerate sequences at their respective active sites (33). The c-di-GMP binding site lies in the EAL domain. In the absence of c-di-GMP, LapD is maintained in an off state by a helix (“signaling” helix, or S-helix) immediately N-terminal to the GGDEF domain that interacts with helix α6 of the EAL domain, restricting dinucleotide access to the EAL active site. c-di-GMP was proposed to release this autoinhibitory interaction and to lead to the formation of an EAL dimeric structure (33).
To gain further insight into how the enzymatic activity of dual-domain proteins in which both modules are potentially active is regulated, we chose to study the RbdA protein (PA0861) of P. aeruginosa, a medically important multidrug-resistant pathogen. The DipA (PA5017) (34) and RbdA (35) proteins have been shown to play important roles in regulating dispersion of P. aeruginosa. The RbdA protein was demonstrated to regulate the transition from a sessile to a motile lifestyle, possibly upon detection of hypoxic conditions, by hydrolyzing c-di-GMP (35). Moreover, the PDE activity of RbdA is allosterically stimulated when GTP binds to its GGDEF domain (35). Here we report the crystal structure of the cytoplasmic region of RbdA (cRbdA) in its free form at a resolution of 2.28 Å. This structure reveals the presence of a signaling helix (S-helix) that maintains the protein in an autoinhibited state in a manner reminiscent of LapD (33). We also report two binary complexes obtained by a brief soak of native crystals, namely, complexes of cRbdA with GTP/Mg2+ bound to the A site of the GGDEF domain, at 2.80-Å resolution, and of the c-di-GMP substrate bound to the EAL active site, at 3.31-Å resolution. Since the two binary complexes could be obtained only via a brief soak of cRbdA native crystals, not by cocrystallization of a preformed complex, the present crystallographic analysis precluded the observation of any large conformational changes that might be induced by GTP binding. Thus, we also studied the conformation of cRbdA in solution by using small-angle X-ray scattering (SAXS) and found evidence of large structural changes of cRbdA following GTP binding. As in the MorA protein (32), an α-helix between the GGDEF and EAL domains (H-helix) is likely to function as a hinge around which these two enzymatic domains can pivot. Together with pioneering work by other groups (32, 33), the present structures enlighten the regulation of c-di-GMP metabolism by bacterial proteins containing a dual domain and provide information for the design of small molecules with the aim of modulating the activity of these proteins. They illustrate the recycling of key functional structural elements to control the activity of dual domain-containing proteins.
RESULTS AND DISCUSSION
The EAL motif of RbdA is essential for downregulating biofilm formation.
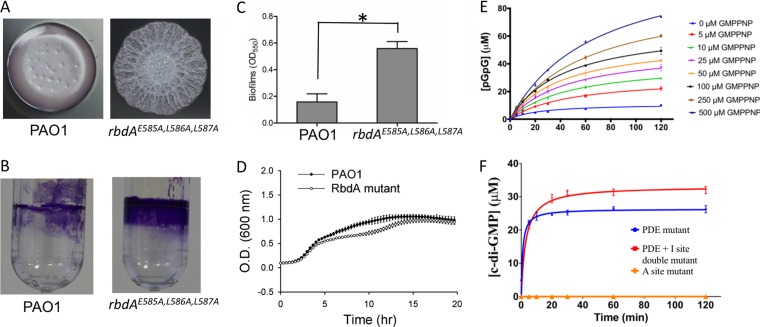
Higher levels of the second messenger c-di-GMP are associated with a sessile lifestyle, while lower levels of c-di-GMP, provoked by its breakdown by PDEs, is associated with biofilm dispersion and a planktonic bacterial lifestyle. In order to assess the role of the PDE domain of RbdA in promoting biofilm dispersal, we mutated the chromosomal copy of the rbdA gene to create the amino acid mutations E585A, L586A, and L587A in the PAO1 strain of P. aeruginosa (Fig. 1A) and found a 3-fold increase in biofilm formation for the corresponding triple mutant strain (Fig. 1B and C). The wrinkled colony morphology of the RbdA triple mutant strain is in agreement with the role of RbdA as a negative regulator of exopolysaccharide production proposed earlier (35). This result, which is also consistent with prior phenotypic analyses of PA14 and PAO1 transposon mutants (29, 35) in which the complete rbdA gene was inactivated, suggests that c-di-GMP breakdown by the EAL domain of RbdA is directly responsible for downregulating biofilm formation. However, we cannot rule out the possibility that the triple mutant is less stable than the wild-type (WT) RbdA protein and that the observed phenotype results from loss of the entire protein, not only from the “EAL” PDE enzymatic motif. Moreover, comparison over 20 h of the growth curves of P. aeruginosa PAO1 and bacteria containing the mutated chromosomal copy did not reveal significant changes (Fig. 1D).
FIG 1.
Activity of the RbdA protein. (A) Bacterial colony morphologies and biofilm phenotypes of P. aeruginosa PAO1 and the rbdAE585A,L586A,L587A chromosomal mutant. (B) Visualization of biofilm formation on polystyrene tubes. (C) Quantitative comparison of biofilm formation levels. The data shown are means of triplicate values. Standard deviations are shown by error bars (*, P < 0.05; two-tailed t test). (D) Comparative growth curves of P. aeruginosa PAO1 and the rbdAE585A,L586A,L587A chromosomal mutant (RbdA mutant). (E) Allosteric activation of PDE enzymatic activity of the cRbdA protein at 25°C in the presence of increasing concentrations of GMPPNP (ranging from 5 to 500 μM). (F) DGC enzymatic activity of RbdA. The PDE mutant bears the 585-ELL-587 → ALL single mutation, the PDE + I site double mutant has both 585-ELL-587 → ALL and 444-REGD-447 → AEGD mutations, and the A site mutant (453-GGDEF-457 → GGAAF) is the negative control. Reaction conditions are described in Materials and Methods.
Allosteric activation of PDE activity of cRbdA by addition of GMPPNP.
We also examined the allosteric activation of the PDE activity of cRbdA by addition of guanosine 5′-β-γ-imido triphosphate (GMPPNP), an isosteric analogue of GTP. We used GMPPNP instead of GTP to rule out the possibility that changes in the maximum velocity (Vmax) were due to an increased concentration of c-di-GMP substrate produced by the intrinsic DGC activity of cRbdA (see below). We found that GMPPNP is not converted into c-di-GMP by the GGDEF domain of cRbdA (see Fig. S1A in the supplemental material). Using GMPPNP concentrations ranging from 5 to 500 μM, we observed a gradual increase in Vmax of up to 10.5-fold at higher GMPPNP concentrations (Fig. 1E; Fig. S1B). This demonstrates that GMPPNP is an allosteric activator of the PDE activity of cRbdA (Fig. 1E). Moreover, this allosteric enhancement of PDE activity can be attributed to GMPPNP binding to the A site in the GGDEF region of cRbdA, because no enhancement was observed when an A-site mutant of cRbdA (which can no longer bind to either GMPPNP or GTP) was used (Fig. S1C).
Diguanylate cyclase activity of cRbdA.
Using a high-pressure liquid chromatography (HPLC)-based enzymatic assay, An et al. (35) reported that incubation of RbdA with GTP alone led to the formation of pGpG, suggesting that RbdA potentially has DGC activity. However, they noticed that c-di-GMP was rapidly degraded into pGpG by the PDE activity of this dual enzyme. In the present study, in order to eliminate product breakdown by the PDE activity of the wild-type RbdA EAL domain, we first used a cRbdA mutant devoid of PDE activity (Fig. 1F, “PDE mutant”) that shows DGC activity with a Vmax value of 22 μM/min, a Km value of 0.84 μM, and a kcat value of 26 min−1. Next, to also exclude possible feedback inhibition due to c-di-GMP product binding to the I site of the GGDEF domain, we used a double mutant (Fig. 1F, “PDE + I site double mutant”) in which both the EAL site and the I site were inactivated, leading to a slight increase in the catalytic activity (Vmax value of 33 μM/min, Km value of 3.3 μM, and kcat value of 33 min−1). A mutation in the A site completely abrogated DGC activity, demonstrating that cRbdA was responsible for the observed cyclase activity (and not a contaminant DGC that could have copurified during protein overexpression). Taken together, these data show that cRbdA has intrinsic GTP-stimulated PDE activity as well as DGC activity conferred by its GGDEF domain.
The cRbdA protein forms a stable dimer.
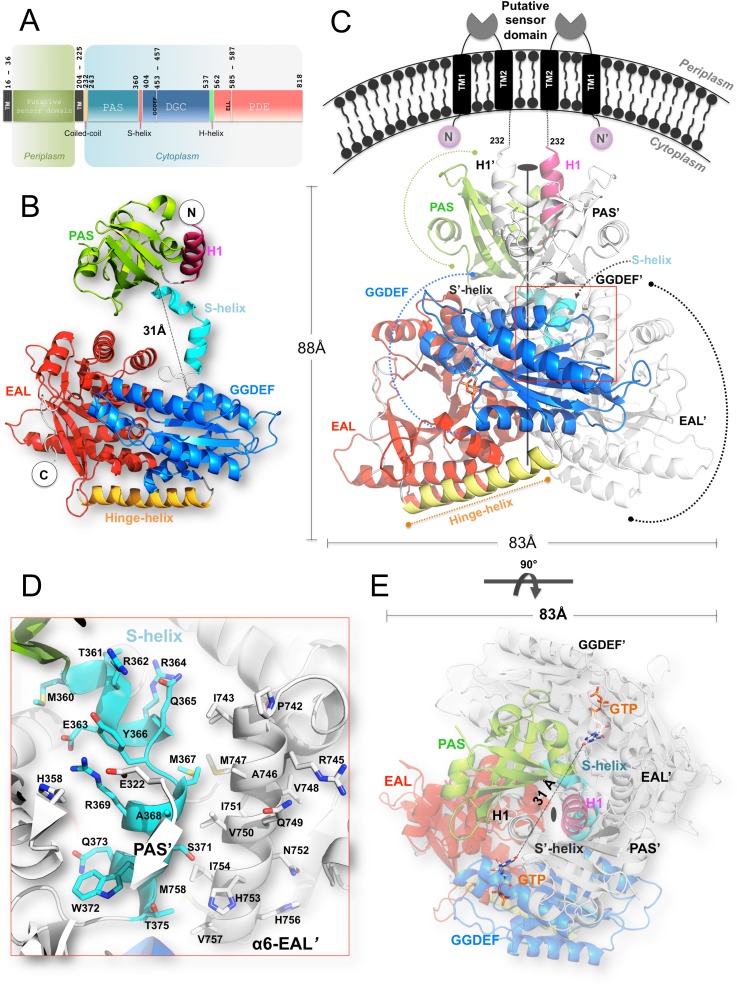
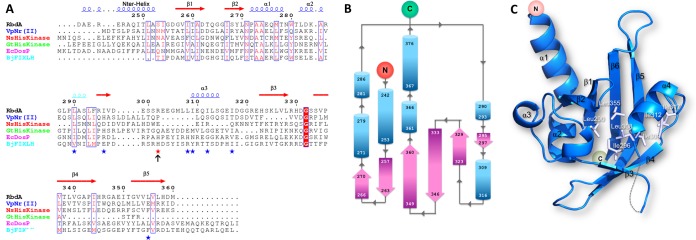
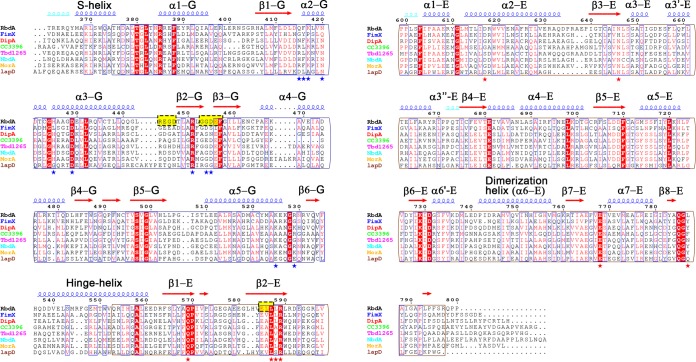
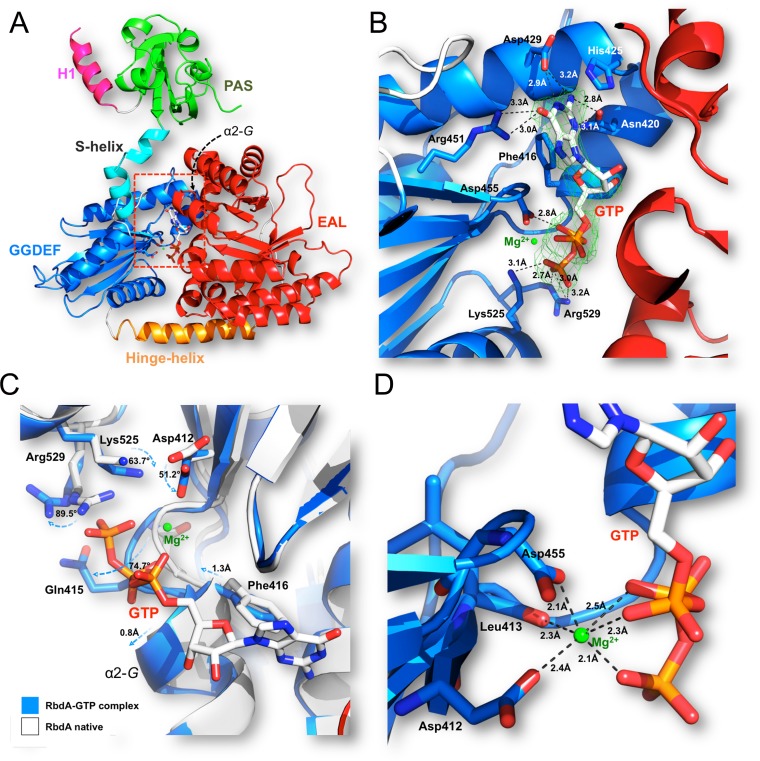
The RbdA protein contains two hydrophobic regions that anchor the protein to the P. aeruginosa inner membrane followed by a cytoplasmic region that comprises a Per-ARNT-Sim (PAS) domain followed by a GGDEF-EAL dual domain (Fig. 2A). Both the GGDEF and ELL catalytic amino acid motifs are present in the protein sequence, accounting for the observation that RbdA can be active as both a DGC and a PDE. The N-terminal domain of RbdA, which was omitted from the protein construct used for this study, is composed of ∼230 residues with low sequence identity to known protein structures. Two predicted transmembrane helical segments, at residues 16 to 36 (TM1) and 204 to 225 (TM2), surround a putative periplasmic sensor domain of hitherto unknown specificity and structure. The cRbdA fragment that was crystallized comprises the complete tripartite PAS, GGDEF, and EAL cytoplasmic domains of RbdA (Fig. 2B). The crystal asymmetric unit contains one cRbdA monomer (Fig. 2B), and a closely packed cRbdA dimer is generated through a crystallographic dyad, giving rise to a bilobe molecule with overall dimensions of 80 Å by 88 Å by 83 Å (Fig. 2C and E). Upon dimer formation, a total solvent-accessible surface area of 3,844 Å2 becomes buried at the monomer-monomer interface (Fig. 2C and D). A breakdown of buried surface areas for each of the three domains of the tripartite protein is given in Table S1. This large intermolecular interface is consistent with the observation that cRbdA forms stable dimers in solution at all tested concentrations, using both gel filtration chromatography and multiangle light scattering (MALS) (Fig. S2). A similar conclusion was reached using SAXS (see below). An assignment of the secondary structure elements of the cRbdA amino acid sequence, derived from the present crystallographic study, is displayed in Fig. 3A (PAS region), and a structure-based alignment with other bacterial GGDEF-EAL dual domains is shown in Fig. 4.
FIG 2.
Structure of the RbdA protein. (A) Domain organization of RbdA along its primary structure. Amino acid catalytic motifs and key structural features are indicated. (B) cRbdA monomer with each domain colored and labeled. The N and C termini are labeled. The PAS domain is colored green, the GGDEF domain cyan, the EAL domain red, and connecting segments crucial for protein dynamics (S-helix and H-helix) light blue and yellow, respectively. (C) cRbdA dimer. The position of the single dyad that runs through the crystallographic dimer is indicated. The region from positions 1 to 232 (not included in the cRbdA construct), leading to the periplasmic membrane, is represented schematically with dashed lines. (D) Magnified view of interactions between the S-helix and the dimerization helix α6-E′ from the EAL′ domain. (E) The cRbdA dimer is shown from a perpendicular direction, using the same color code.
FIG 3.
Structure of the PAS domain of RbdA. (A) Sequence comparisons of the PAS domains of RbdA of P. aeruginosa (accession number AAG04250.1), the two-component system sensor histidine kinase NRII of V. parahaemolyticus (accession number WP_049877818.1), the Nostoc histidine kinase NSHisKinase (accession number WP_010994604.1), the Geobacillus thermodenitrificans histidine kinase GtHiskinase (accession number WP_029761733.1), the oxygen sensor protein EcDosP of E. coli (accession number P76129.4), and FixL of Bradyrhizobium japonicum (accession number CAA40143.1). The histidine residue involved in heme chelation by FixL, which is not conserved in RbdA, is indicated with an arrow. Hydrophobic residues that form a possible ligand binding site of the PAS domain of cRbdA are indicated with stars. Secondary structure elements of the PAS domain of RbdA are displayed above the alignment. (B) Topology of the PAS domain. β-Strands are shown as purple arrows and α-helices as blue tubes, with residues at the extremities of each numbered. (C) Closeup view of the hydrophobic core housed inside the β-barrel of the PAS domain from RbdA. Hydrophobic residues forming a putative ligand binding site are depicted as white sticks and labeled.
FIG 4.
Structure-based amino acid sequence alignment of cRbdA and homologous domains from other GGDEF-EAL domain-containing bacterial proteins. Secondary structure elements are based on assignments for the cRbdA protein (this work). Strictly conserved residues are highlighted in red. The catalytic motifs of the DGC domain (A site or “GGDEF” motif and I site) and the PDE domain (“ELL” motif) are highlighted in yellow boxes. Key elements of the structure are indicated, including the H-helix (hinge-helix) and S-helix (present in RbdA and LapD but not in MorA; no structure is available for the dual domains of DipA, NbdA, CC3396, and Tbd1265). Accession numbers are as follows: Tbd1265, WP_011311777.1; LapD, WP_011331847.1; MorA, WP_073670889.1; FimX, WP_033999828.1; CC3396, WP_010921225.1; and NBDA, WP_048305406.1. Blue stars indicate GTP-interacting residues (GGDEF domain) and red stars c-di-GMP binding residues (EAL domain).
The PAS domain (residues 232 to 360).
Residues 233 to 240, which are linked to TM2 in the full-length protein, are disordered in the absence of the membrane anchoring regions that were not included in the cRbdA protein construct. The core structure of the PAS domain (Fig. 3) is composed of a six-stranded antiparallel β-sheet (with a flexible β-strand at the edge of the β-sheet and residues 301 to 306 being disordered) augmented by short α-helices. This topological arrangement (Fig. 3A and B) leads to the formation of a barrel-shaped module that houses a hydrophobic pocket in the interior of the PAS domain. A large number of aliphatic side chains, such as Leu290, Ile296, Leu308, Ile309, Ile312, Ile317, and Leu355, form a cavity that can bind small organic ligands (Fig. 3C). An automated database search for homologous three-dimensional (3D) structures returned the PAS domain from NRII, a protein involved in nitrogen regulation in Vibrio parahaemolyticus (PDB code 3B33), and PAS domains from sensor histidine kinases (PDB codes 3OLO and 5FQ1), with the highest Z-scores being >8.0. Note that these structures are devoid of heme, which is in line with the absence of a heme-coordinating histidine residue in the PAS domain sequence of RbdA (Fig. 3A). Accordingly, reconstitution experiments failed to give strong spectroscopic evidence for heme binding by the cRbdA protein, although weak binding of heme was observed when only the PAS domain was expressed (Fig. S3). The core region of the PAS domain (residues 255 to 360) is preceded by an α-helix (residues 242 to 253) (Fig. 3A and B) that is swapped with the symmetrically equivalent helix and inserts into the concave surface of the other PAS domain monomer (Fig. 2C). Interestingly, this swapped helix contains an exposed hydrophobic patch made by residues Ile249, Leu251, and Ile254 that forms interactions with the symmetry-equivalent residues (via the crystallographic dyad), leading to the formation of a short coiled-coil structure that runs in a direction roughly perpendicular to the cytoplasmic membrane (Fig. 2C). Thus, as seen for the PAS domain of Rhodobacter sphaeroides (PDB code 4L9E) (36), the role of the RbdA PAS domain may merely be limited to promoting protein oligomerization via its N-terminal helical extension. At the C-terminal side of the PAS domain, two α-helical segments (residues 360 to 380) project away from the core of the PAS domain and realize the connection with the GGDEF domain, such that the distance between the C terminus of the core PAS domain (taken as Met360) and the N terminus of the GGDEF domain (Thr380) is about 31 Å (Fig. 2B). As detailed below, this segment, which encompasses the S-helix, plays a key role in holding the bilobe structure in place by making contacts with the EAL domain of the partner molecule (Fig. 2D) (the domains from the other monomer are labeled with prime symbols [e.g., EAL′]).
The GGDEF domain and the complex with GTP/Mg2+.
The PDE activity of RbdA is enhanced in the presence of 50 μM GTP, leading to the complete hydrolysis of c-di-GMP after 30 min of incubation at 37°C, while only a small amount of substrate is hydrolyzed by RbdA in the absence of GTP (35). This observation is in line with our kinetic observation using the cRbdA protein in the presence of multiple concentrations of GMPPNP (Fig. 1E) and with published data (35). To obtain a molecular basis for this effect, we determined the structure of a binary complex between cRbdA and GTP via a brief soak of native crystals in the presence of Mg2+ (Fig. 5A and B; Table S2). A detailed view of the atomic interactions formed between GTP and residues from the A site of the GGDEF domain is displayed in Fig. 5B. Residue Asp455 from the GGDEF motif and Asp412 coordinate one Mg2+ ion, which neutralizes negative charges from the triphosphate group of GTP (Fig. 5C and D; Fig. S4). The N-2 atom of the guanine makes hydrogen bonds with the carbonyl oxygen of His425 and with the amide group of the amide side chain of Asn420. Overall, the bound GTP/Mg2+ closely overlaps the GTP/Mg2+ bound to the GGDEF domain of PleD (PDB code 2V0N) (8). To detect conformational changes in the RbdA protein that might be triggered by GTP binding, we superimposed the native/Hg structure with the GTP binary complex (Fig. 5C). Following superposition, the overall root mean square deviation (RMSD) between all main chain atoms is 0.48 Å. Compared to the native/Hg structure, the side chains of two basic residues, Lys525 (which makes a salt bridge with Asp412 in the native structure) and Arg529, are repositioned and neutralize the γ-phosphate group of GTP. Moreover, the N-terminal region of helix α2-G (Gln415-Phe416) undergoes a significant conformational change (RMSD of >2 Å) to accommodate the triphosphate group of GTP, with the main chain of Gln415 displaced by a distance of 2.8 Å and its side chain by 6.3 Å. It is possible that such local structural disturbances triggered by GTP binding propagate throughout the RbdA dimer, leading to larger conformational changes, such as whole-domain reorientations. However, these could not be observed using X-ray crystallography, possibly due to structural constraints on protein flexibility imposed by the crystal lattice. This hypothesis is consistent with the observation that longer soaks with GTP (typically >10 min) led to crystal lattice disruption and also with SAXS data demonstrating large conformational changes upon GTP binding, as detailed below. Binding of GTP to the A site of the GGDEF domain is in line with the work of An et al. (35), who observed that when the five signature motif residues GGDEF were mutated to alanine to generate the variant GGDEF-5A, the PDE activity of the mutant was similar to that of the WT enzyme, regardless of the presence or absence of GTP. In the present conformation of the cRbdA dimer, the two A sites (or GTP binding sites) are separated by a distance of ∼31 Å (Fig. 2E). In order to be active as a DGC, both GGDEF domains must adopt a specific 2-fold symmetrical orientation such that their respective half active sites are brought into close proximity (4, 7). We concluded that in the present orientation, the GGDEF domains of cRbdA do not function as a DGC but solely as GTP sensors and that GTP binding is conducive to an enhancement of PDE activity through an interaction between GTP and the A site of the GGDEF domains that triggers a range of conformational changes in the RbdA protein, both locally (Fig. 5C) and globally (see below).
FIG 5.
Interaction between the GGDEF domain of cRbdA and the GTP allosteric activator of PDE activity. (A) Overall view of the cRbdA monomer, highlighting the location of the bound GTP molecule within the GGDEF domain. (B and D) Closeup views showing the atomic interactions between GTP (sticks) and residues of the A site of the GGDEF domain (blue) (B) and the Mg (green sphere) coordination shell (D). A difference electron density map with Fo-Fc Fourier coefficients, with the GTP moiety omitted from the calculation, is displayed at a level of 3σ. (C) Conformational changes in RbdA triggered by GTP binding. An overlay of the native (blue) and GTP-bound (white) cRbdA structures is shown. Residues near the phosphate tail of GTP, including in helix α2-G, and undergoing large displacements between the GTP-free and bound states are shown as sticks and labeled. A list of atomic interactions between the GGDEF domain and GTP is given in Table S2 in the supplemental material, and contacts between the EAL domain of cRbdA and c-di-GMP are shown in Table S3.
The EAL domain and the complex with the c-di-GMP substrate.
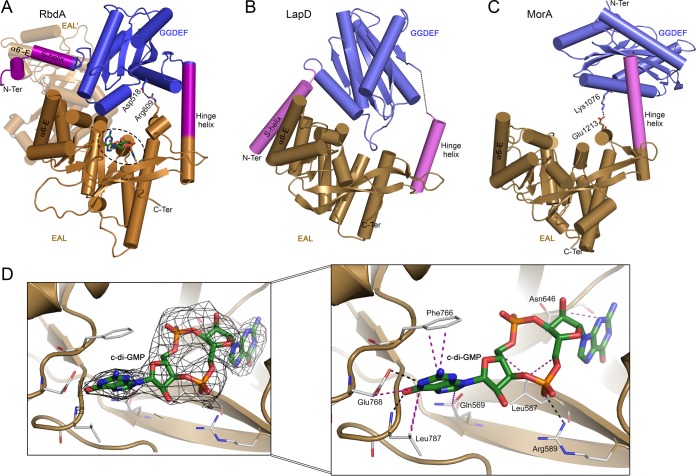
Following superpositioning of the structures of cRbdA bound to c-di-GMP and cRbdA, the overall RMSD between all main chain atoms is 0.56 Å. The connection between the GGDEF and EAL domains of cRbdA is made by an α-helix (residues 537 to 562) spanning a distance of 37 Å that we name the H-helix, to retain the nomenclature originally proposed for the dual domain-containing protein MorA from P. aeruginosa, in which this connecting helix is also present (32). Interestingly, a structurally equivalent helix is also located at the N-terminal end of the EAL domain of LapD, where it is named α0-E (33). Superpositioning of the GGDEF-EAL dual domains from MorA, LapD, and RbdA shows that the H-helix acts as a hinge region, allowing the GGDEF domain to adopt various orientations with respect to the EAL domain (Fig. 6A to C). The interdomain flexibility afforded by the presence of the H-helix potentially leads to the formation of alternative EAL-EAL′ and GGDEF-GGDEF′ dimeric assemblies while preserving the 2-fold symmetry of the whole RbdA dimer. The EAL domain of RbdA (residues 562 to 795) is formed by a (β/α)8 barrel capped by a lobe that comprises C-terminal residues 553 to 562 of the H-helix and helix α1-E, which forms part of the c-di-GMP substrate binding site. In the “closed conformation” of the cRbdA dimer captured here, the c-di-GMP binding site faces the A site from the GGDEF domain of the same monomer (Fig. 6A). Detailed views of the interactions between c-di-GMP and active site residues of the EAL domain are presented in Fig. 6D and Fig. S5. Even in the absence of Ca2+, which is commonly used to inhibit PDE activity, an intact c-di-GMP molecule is bound to the active site, which is in line with the observed weak intrinsic PDE activity of RbdA and with the brief soaking time used to obtain this complex. Overall, the orientation of c-di-GMP in the EAL active site is closely superimposable on that of the substrate in the isolated PDE of MorA bound to c-di-GMP (PDB code 4RNH) (32). Catalytic residues are located at the C-terminal end of the barrel, including residues that form the metal ion binding site and the evolutionarily conserved residues from loop 6 (D709FCAGMSS716). Although the complex was crystallized in the presence of Mg2+, no residual electron density indicative of a divalent metal is visible in the EAL active site. Systematic studies of dimerization by EAL domain-containing proteins showed that EAL domains form dimers through a conserved dimerization interface involving two α-helices (α5-E and α6-E) and loop 6 (Fig. 4) (2, 9–12). This mode of interaction is also found in the dimeric EAL proteins TBD1265 from Thiobacillus denitrificans (9), YahA (37), YkuI from Bacillus subtilis (11), RocR (12, 38, 39), and BlrP1 from Klebsiella pneumoniae (10), with the same structural elements involved in stabilizing the EAL-EAL interface (40). In the present quaternary conformation of the cRbdA dimer, the PAS′ and GGDEF′ domains occupy the space of the EAL dimeric partner (labeled EAL′), and disruption of the interatomic contacts established between the S-helix and helix α6-E′ and a large rotation of the GGDEF domain with respect to the EAL domain are required to allow the formation of a canonical and catalytically active EAL dimer. Moreover, an inward movement of loop 6 is also needed to avoid steric hindrance at the EAL-EAL canonical interface.
FIG 6.
Comparison of three dual domain-containing proteins, highlighting the range of relative orientations permitted by pivoting around the hinge helix. The EAL domain has the same orientation in panels A to C. (A) View of the GGDEF-EAL dual domain from RbdA, highlighting the hinge helix (magenta) that connects the EAL domain to the GGDEF domain and the S-helix (magenta) that contacts helix α6-E′ in trans from the partner EAL′ domain (shaded). The c-di-GMP moiety bound to the EAL domain is shown as green sticks (dashed circle). A salt bridge between Asp518 (GGDEF domain) and Arg609 (EAL domain) is also represented. (B) View of LapD (PDB code 3PJT). The S-helix contacts helix α6-E in cis from the same EAL monomer, resulting in an autoinhibited state. (C) View of MorA (PDB code 4LYK), which also possesses the H-helix, but with a flexible N-terminal segment (no visible S-helix in the available crystal structure). The Lys1076-Glu1213 salt bridge between the GGDEF and EAL domains is shown as sticks. (D) Difference electron density map with Fo-Fc Fourier coefficients, with c-di-GMP (sticks) omitted from the calculation. The inset shows a magnified view of the interactions established between the EAL domain of RbdA and c-di-GMP. Hydrogen bonds are shown as black dashed lines and van der Waals and stacking interactions as purple dashed lines.
Conformations adopted by cRbdA in solution.
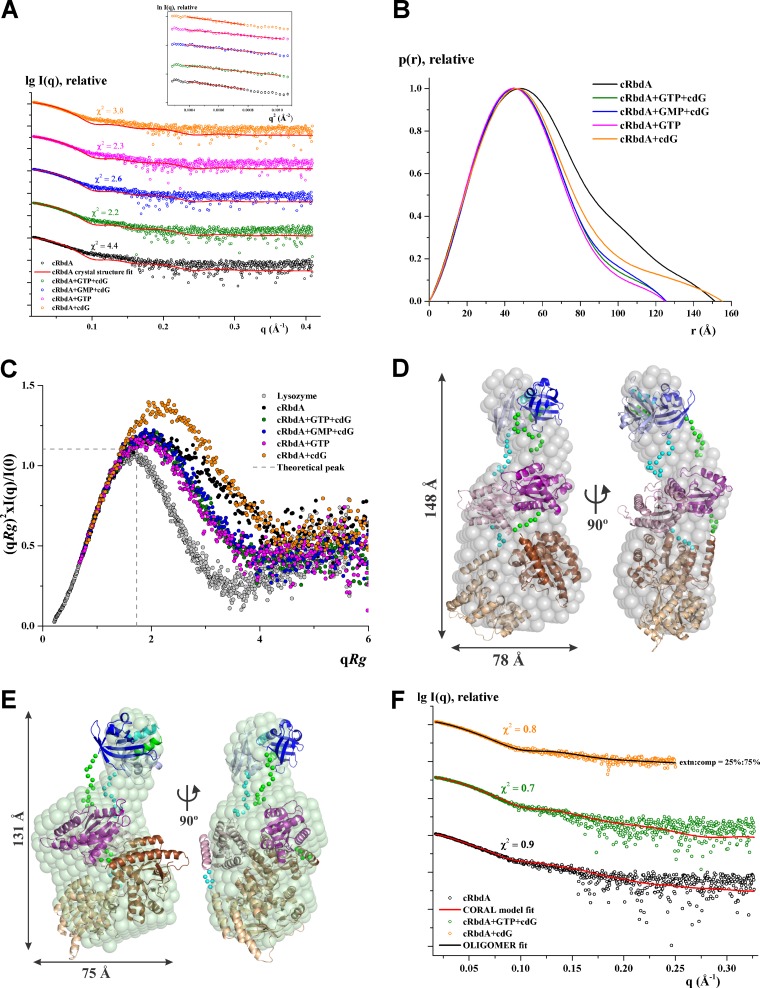
As detailed above, the crystal structure of cRbdA reveals a dimer locked in an autoinhibited conformation via an extensive set of interactions established in trans between the S-helix and the α6-E dimerization helix of the EAL′ domain (Fig. 2C), which is otherwise involved in EAL-EAL′ canonical dimer formation. In the closed conformation of the cRbdA dimer captured here (Fig. 6A), the c-di-GMP binding site of the EAL domain and the A site of the GGDEF domain of the same monomer face each other, largely occluded from the solvent, thereby restricting accessibility to their substrates. The observation of an autoinhibited conformation for cRbdA suggests that alternative dimeric structures are possible in solution, including one conformation where the EAL domains of each monomer assemble as a canonical dimer stabilized by isologous (symmetry-equivalent) interactions between helices α5-E and α6-E and loop 6 (35). In addition, we cannot rule out formation of a GGDEF dimer whereby both A sites would be brought into close proximity, leading to the activation of DGC activity, as suggested by the observation that RbdA can produce pGpG by using solely GTP as the substrate (35). We performed SAXS measurements to identify the range of conformations that cRbdA can adopt in solution. As shown in Fig. S6 and Table S4, cRbdA exists as a dimer in solution at all tested protein concentrations, and no aggregation is observed in the Guinier plot. The experimental radius of gyration (Rg) of cRbdA at 2.5 mg/ml is 45.4 ± 1.4 Å, and the maximum particle size (Dmax) is 151 ± 10 Å (Fig. 7A and B). Based on the Porod and excluded volumes and the volume of correlation, cRbdA adopts a dimeric state in solution at the concentrations used (Table S4). Rigid body modeling of cRbdA was performed by allowing flexibility for the regions connecting the individual domains and keeping the PAS dimer intact. The resulting molecular model is extended, has good agreement with the experimental scattering curve (χ2 = 0.9) (Fig. 7F), and fits nicely to the solution shape, with a normalized spatial distance (NSD) of 2.4 (Fig. 7D). Next, we performed an analysis of the conformations adopted by cRbdA in the presence of various ligands (Fig. 7A; Fig. S7). As shown in Fig. 7A and B, the experimental Rg value (46.2 ± 1.3 Å) of cRbdA remained unaffected by addition of c-di-GMP, within the error range, albeit with a different profile in the distance distribution plot (Fig. 7B). In contrast, addition of GTP led to a significant reduction of about 7 Å, with an Rg value of 38.5 ± 0.8 Å and a Dmax value of 125 ± 10 Å (Table S4), demonstrating that cRbdA converts to a more compact conformation in the presence of GTP. Interestingly, the presence of GTP and c-di-GMP (40.1 ± 0.8 Å) or GMP and c-di-GMP (40.7 ± 0.8 Å) also caused cRbdA to change to a more compact structure (Fig. 7A to C). An analysis of the flexibility of cRbdA alone or bound to GTP, GMP, or c-di-GMP by use of a normalized Kratky plot (Fig. 7C) demonstrated that cRbdA is intrinsically flexible, while cRbdA bound to either GTP, GTP and c-di-GMP, or GMP and c-di-GMP is less flexible. In comparison, cRbdA bound only to c-di-GMP showed more flexibility and appeared to be more extended, with a fit to the cRbdA extended conformation returning a χ2 value of 2.5. Analysis of mixtures of various conformational states indicated that cRbdA in the presence of c-di-GMP contains a 25% extended conformation (Rg = 46.2 Å) and a 75% compact conformation (Rg = 38.5 Å) (Fig. 7F), with a good agreement (χ2 = 0.8) with the experimental scattering curve. Low-resolution molecular shapes for cRbdA determined ab initio from the experimental data revealed a bilobe structure (Fig. 7D). The volume of the larger lobe can house the GGDEF-EAL dimer, while the smaller lobe is likely to correspond to the PAS domains. Remarkably, addition of GTP led to a change of the molecular shape suggestive of a reorganization of the dual domain that comprises the GGDEF-EAL dimer, while the PAS domains were shifted away from the dual domain (Fig. 7E). Given the relatively low resolution of the shapes determined by SAXS, however, several possible dimeric arrangements of the GGDEF-EAL dual domain, such as the one revealed by the cRbdA crystal structure or one where the two EAL monomers assemble as a canonical dimer, can be fit into the large lobe, with comparable discrepancies. Likewise, whether reorientation of the two GGDEF domains into an active DGC enzyme can also occur awaits further structural studies. In this respect, we note that the length of the H-helix is compatible with extensive domain reorientations leading to drastically distinct quaternary arrangements (Fig. 6).
FIG 7.
Solution X-ray scattering studies of cRbdA with ligands. (A) Experimental scattering patterns (○) and calculated scattering profiles (lines) of crystal structure dimers of cRbdA alone (black) and in complex with GTP and c-di-GMP (cdG), GMP and cdG, GTP, and cdG. (Inset) Guinier plots show linearity at all concentrations used, indicating no aggregation. The scattering profiles were offset, for clarity, by applying arbitrary scale factors. (B) Overlapping of pair-distance distribution function P(r) of cRbdA and its ligand complexes. cRbdA complexes with GTP and cdG, GMP and cdG, and GTP have similar profiles; however, cRbdA with cdG has an extended tail. (C) Normalized Kratky plot of cRbdA (black) compared to its complexes and the compact globular lysozyme (gray), with a peak (gray dashed line) representing the theoretical peak and assuming an ideal Guinier region of a globular particle. The scattering pattern of cRbdA and its complexes exhibits a broad bell-shaped profile shifted toward the right with respect to standard globular proteins, indicating the presence of motion in the protein. (D) Averaged and filtered envelope (gray) from 20 independent ab initio reconstructions created by use of DAMMIF superimposed onto a cartoon representation of the CORAL model with an extended conformation for cRbdA, with the PAS domain in blue, the GGDEF domain in purple, and the EAL domain in brown. The second subunit is colored in lighter shades. The linker regions modeled between the domains are shown as green and cyan spheres for the subunits. Front (left) and side (right) views are displayed. (E) Averaged and filtered ab initio low-resolution shape of cRbdA in the presence of GTP and c-di-GMP (green) superimposed on the compact conformation generated from CORAL. (F) Fitting of the CORAL model (red lines) to the experimental scattering patterns (○) for cRbdA alone (black) and with GTP and c-di-GMP (green). The theoretical scattering curve for the mixture of 75% compact and 25% extended conformations of cRbdA dimers (black line) calculated using the OLIGOMER program fits the experimental scattering pattern of cRbdA with cdG (orange), with a χ2 value of 0.8.
Model for allosteric activation of PDE activity of RbdA.
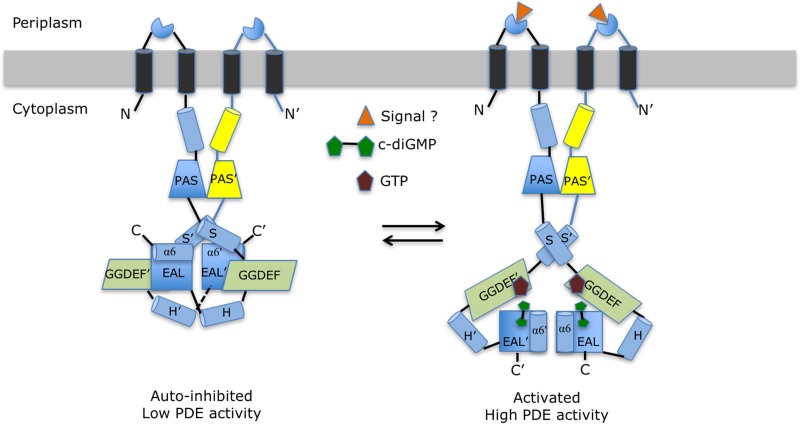
One main conclusion of solution studies conducted on cRbdA is that in the absence of ligand, the protein appears to be rather flexible but becomes more compact in the presence of GTP. Together with the observation of an apparently autoinhibited conformation in the crystal structure, these findings suggest the following model for the allosteric activation of the PDE activity of RbdA, which is consistent with available data (Fig. 8). In the simplest scenario, two conformational states adopted by the protein are associated with low and high PDE activities. The autoinhibited state is achieved via a set of interactions established in trans between helix α6-E′ and residues 361 to 365 from the S-helix, immediately N-terminal to the GGDEF domain, which locks the EAL domains in a noncanonical configuration. We also noted an evolutionarily conserved basic residue, Arg369 in RbdA (Fig. 2D and 4), that also projects from the S-helix but toward the PAS′ domain, and as a result is deeply buried in the protein interior at the dimer interface (Fig. 2D). Arg369 makes stacking interactions with the phenyl ring of Tyr366 and the imidazole ring of His358′ and one salt bridge with the carboxylic group of Glu322′ of the PAS′ domain (Fig. 2D). A preliminary mutagenesis study targeting Arg369 (Table S5) suggested that an Arg369Glu mutant partly released the autoinhibitory switch, leading to a higher PDE activity.
FIG 8.
Proposed allosteric mechanism regulating the PDE activity of RbdA. The left panel schematically represents the putative autoinhibited state observed in the cRbdA crystal structure. This resting state is stabilized by interactions between helix α6 from the EAL domain and the S′-helix immediately N-terminal to the GGDEF′ domain (and also between helix α6-E′ and the S-helix in the other monomer, via the dyad). These interactions lock both EAL domains of the RbdA dimer in a noncanonical configuration. Upon signal detection by either the putative periplasmic sensor domain (triangle) or GTP binding to the A sites of the GGDEF domains, local conformational changes (Fig. 5D) propagate throughout the protein and lead to the release of the autoinhibitory interactions between helix α6-E and the S′-helix. This is proposed to lead to the rearrangement of both EAL domains into a canonical dimer capable of hydrolyzing the incoming c-di-GMP substrate and to the observed higher PDE activity (Fig. 1D) conducive to biofilm dispersal (right panel).
Following GTP binding to the A site of the GGDEF domains (Fig. 5), local conformational changes as pictured in Fig. 5C for helix α2-G would propagate throughout the protein and lead to the release of the autoinhibitory interactions between α6-E′ and the S-helix, and possibly also exposure of Arg369 to the solvent, allowing the EAL domains to rearrange into a canonical dimer capable of hydrolyzing the incoming c-di-GMP substrate. An alternative but not mutually exclusive model for PDE activation involves detection of a signal by the putative periplasmic sensor domain, leading to a conformational rearrangement of the bundle of TM helices and of the S helices transmitted through the coiled-coil region N-terminal to the PAS domains. Again, the final PDE-active state is formed by the canonical EAL dimer, as displayed schematically in Fig. 8. Earlier studies (16) of the dual domain-containing protein CC3396 demonstrated that allosteric activation of PDE via GTP was derived from a reduction of the Km for c-di-GMP from 100 μM in the absence of GTP to 420 nM when GTP was present, and this large increase in affinity of the substrate is likely to derive from a better chemical complementarity afforded by structural changes induced in the c-di-GMP binding site upon canonical dimer formation. As discussed by Jenal and colleagues (16), why would the PDE activity be coupled to cellular levels of GTP? The cellular concentration of GTP is likely to reflect the overall cellular nutrient level, as it is also correlated with the level of the alarmone (p)ppGpp upon starvation of nutriments, including amino acids or nitrogen. When the GTP abundance is low, the PDE activity of RbdA is switched off to prevent exhaustion of the GTP pool, promoting biofilm formation. Conversely, in the presence of a large amount of GTP, the PDE activity is switched on, leading to biofilm dispersal. Importantly, we cannot exclude the existence of dimeric quaternary conformations for RbdA other than the two depicted schematically in Fig. 8, given our observation that a cRbdA double (I site and ELL) mutant can display DGC activity (Fig. 1F). Quaternary structures with the two GGDEF domains forming a dimer with both A sites facing each other may in principle be conducive to DGC activity, although their relevance is not established for RbdA, in the sense that this protein likely evolved to use its GGDEF domain as a GTP sensor, as outlined above, rather than for c-di-GMP synthesis. If this assumption is correct, quaternary conformations with A sites facing each other should represent only a minor population in the ensemble of possible protein structures present in equilibrium in solution. One interesting possibility is that RbdA also functions as a rheostat by integrating signals both from its putative periplasmic sensor domain and from the detection of intracellular GTP levels via its GGDEF domain, leading to a contextually optimal PDE activity. In this respect, we note an interesting overall mechanistic similarity between RbdA and the dimeric LapD sensor, which has a periplasmic output domain capable of binding a ligand (the LapG cysteine protease) and a cytoplasmic PDE domain able to sense c-di-GMP (33). For both RbdA and LapD, the autoinhibitory switch is made of an “S-helix” that locks the protein in a resting state via its interaction with the EAL domain, restricting accessibility to the EAL active site and preventing active dimer formation.
A large number of dual domain signaling systems fused to various sensor domains have now been identified (41–44). RbdA represents a useful model for studying how signals become integrated in the context of one protein with potential dual enzymatic activity. An important conclusion of the present work is that the apparent conservation of several regulatory structural elements, such as signaling (or S-helix) and lever (or H-helix) elements, across various bacterial proteins (Fig. 4 and 5) suggests that similar molecular mechanisms are at play in controlling c-di-GMP metabolism by GGDEF-EAL domain-containing dual proteins. Given the abundance of dual protein genes in the genomes of several major human pathogens, this observation offers attractive perspectives for better understanding the role of c-di-GMP in controlling biofilm formation and also for the design of molecules to interfere with this process.
MATERIALS AND METHODS
Bacterial strains, growth conditions, and biofilm formation and quantification.
Unless otherwise indicated, bacteria were grown at 37°C in lysogeny broth (LB). The wild-type P. aeruginosa PAO1 strain was obtained from the University of Washington. The rbdAE585A,L586A,L587A (rbdAAAA) mutant was created based on the PAO1 strain. Briefly, a DNA fragment of 1,518 bp containing an rbdAE585A,L586A,L587A gene construct was amplified by overlap extension PCR with P. aeruginosa PAO1 genomic DNA as the template. The PCR product was cloned into the vector pK18GT at BamHI and HindIII sites to generate the suicide plasmid pK18Gm-rbdAAAA. The suicide plasmid was transformed into Escherichia coli DH5α for subsequent conjugation with PAO1 by triparental mating with the helper plasmid pRK600. The transconjugants were selected on Pseudomonas isolation agar (PIA) medium plates (Difco) containing gentamicin (60 μg/ml). Colonies resulting from the first crossover events were streaked onto LB agar plates supplemented with 10% (wt/vol) sucrose, and sucrose-sensitive colonies were selected as positive colonies. Colonies that contained the correct mutations were identified by sequencing of the DNA fragment. The colony morphology assay was performed as follows. An overnight culture was diluted to an optical density at 600 nm (OD600) of 0.025 in T-broth (10 g/liter tryptone). Five microliters of diluted culture was spotted on a Congo red plate (10 g/liter tryptone, 40 μg/ml Congo red, 20 μg/ml Coomassie brilliant blue, 1% agar). The plates were incubated at room temperature for 3 days before photography was performed. For biofilm visualization, overnight cultures were diluted to an OD600 of 0.002 with fresh LB, and 500 μl of diluted culture was transferred to a 5-ml polystyrene tube and incubated at 37°C with shaking at 180 rpm for 24 h. Cultures were removed carefully. Biofilms were washed with water, stained with 1 ml 0.1% (wt/vol) crystal violet in each tube for 15 min, and then rinsed with water several times. The tubes were air dried for several hours before photography was performed. The biofilm assay was performed by diluting the bacterial culture at exponential phase to an OD600 of 0.05 with fresh LB broth. Subsequently, 100 μl of diluted culture was transferred to each well of a 96-well microplate and incubated at 37°C for 24 h. The biofilms were washed with water, stained with 125 μl 0.1% (wt/vol) crystal violet in each well for 15 min, and then rinsed with water. The plates were air dried for several hours. Biofilms were dissolved with 125 μl 30% (vol/vol) acetic acid in each well for 15 min, and the absorbance at 550 nm was measured. Each experiment was repeated at least three times in triplicate.
Growth curves.
OD measurements of bacterial cultures were performed in an Infinite 200 Pro microplate reader (Tecan). Absorbance was measured at a wavelength of 600 nm and a temperature of 37°C, and the mean for four readings was taken. Bacteria grown to an OD600 of 0.2 to 0.3 were diluted in fresh LB medium to an OD600 of 0.01. The culture was transferred to a Costar flat-bottomed 96-well plate with a lid and a volume of 200 μl per well. Plates were incubated for 67 cycles (about 22 h in total), and readings were taken after 1,000 s of shaking for 180 rpm and 30 s of static incubation in each cycle.
Cloning, expression, and purification of cRbdA.
A codon-optimized gene encoding the full-length (818 residues) wild-type protein RbdA (PA0861) from P. aeruginosa for expression in E. coli was purchased from Genscript. A fragment encoding residues 233 to 800 of the protein, which includes its entire PAS, GGDEF, and EAL domains (Fig. 2A), was subsequently cloned into the pNIC-CH2 expression vector (Novagen) with the coding sequence for a C-terminal hexahistidine tag (construct vc012 of the NTU Protein Production Platform; here referred to as cRbdA, with a calculated molecular mass of 64,439 Da). Successful recombinants were transformed into E. coli Rosetta(DE3) competent cells. Expression of the cRbdA protein was induced with 0.15 mM isopropyl-β-d-thiogalactopyranoside (IPTG; GoldBio) at 16°C for 16 to 20 h, following the addition of 4% (vol/vol) ethanol to the culture to enhance protein solubility. Cells were harvested by centrifugation and resuspended in buffer A [20 mM HEPES at pH 7.5, 500 mM NaCl, 10% (vol/vol) glycerol, and 0.5 mM tris(2-carboxyethyl)phosphine (TCEP)] supplemented with protease inhibitor cocktail set III, EDTA free (Calbiochem, Merck), and Benzonase nuclease (Sigma-Aldrich) prior to lysis using an LM20 Microfluidizer instrument (Microfluidics). The lysate was clarified by centrifugation at 48,000 × g for 40 min, and the supernatant was filtered through a 0.22-μm syringe filter before being loaded onto 2 ml of Ni-nitrilotriacetic acid (Ni-NTA) resin (Roche) preequilibrated with buffer A supplemented with 2 mM MgCl2 (buffer B). After washing with buffer B supplemented with 20 mM imidazole, bound proteins were eluted with buffer B containing 100 mM imidazole. The eluate was subjected to anion exchange using a HiTrap Q HP 5-ml column (GE Healthcare) equilibrated in buffer B, and cRbdA was eluted in a NaCl gradient ranging from 0 to 1 M NaCl. Finally, pooled fractions containing the cRbdA protein were concentrated and subjected to size exclusion chromatography (SEC) with a Superdex 200 16/600 column (GE Healthcare) preequilibrated in buffer B, with a 0.5-ml/min flow rate. Fractions containing cRbdA (including the C-terminal histidine tag), as confirmed by SDS-PAGE, were pooled and concentrated using Vivaspin concentrators (Sartorius). Concentrated proteins at 8 mg/ml were flash frozen in liquid N2 and stored at −80°C until use.
Cloning, expression, and purification of cRbdA mutants.
Synthetic genes were purchased from Bio Basic Asia Pacific (Singapore). We expressed the following four cRbdA mutants by using the method outlined above: (i) an A-site DGC-inactive mutant containing a 453-GGDEF-457 → GGAAF mutation, (ii) a PDE-inactive mutant containing a 585-ELL-587 → ALL mutation, (iii) an I-site mutant containing a 444-REGD-447 → AEGD mutation, and (iv) a double mutant with an inactive PDE and I site, with both 585-ELL-587 → ALL and 444-REGD-447 → AEGD mutations in order to measure unbiased DGC activity.
PDE activity measurement.
We measured the time course of pGpG formation without the allosteric activator GTP or in the presence of the isosteric guanosine 5′-(β-γ-imido)triphosphate (GMPPNP) molecule (a nonhydrolyzable GTP analogue). We found that GMPPNP is not converted to c-di-GMP by the DGC activity of cRbdA (see Fig. S1 in the supplemental material), thus ensuring that changes in Vmax are not due to an increased concentration of the c-di-GMP substrate produced by intrinsic cRbdA DGC activity. We used GMPPNP at concentrations of 0, 5, 10, 25, 50, 100, 250, or 500 μM, cRbdA at 1 μM, and c-di-GMP at 100 μM, and reactions were performed in duplicate. The hydrolysis of c-di-GMP was quenched by adding 0.1 M CaCl2 (to inhibit the EAL domain activity), and the amount of pGpG formed was resolved in a 4.6-mm by 250-mm by 5-μm C18 column (Agilent) in 20 mM triethylammonium bicarbonate, 9% (vol/vol) methanol, mounted on a Prominence HPLC (Shimadzu) and analyzed using Prism software (GraphPad, San Diego, CA).
DGC activity measurement.
We employed an HPLC-based assay analogous to that outlined above to analyze c-di-GMP formation by cRbdA. In order to eliminate potential feedback inhibition by c-di-GMP binding to the I site of the enzyme and also to prevent hydrolysis of c-di-GMP by the PDE activity of cRbdA, we used a cRbdA double mutant having both 585-ELL-587 → ALL and 444-REGD-447 → AEGD mutations to measure DGC activity. The A-site mutant of cRbdA (453-GGDEF-457 → GGAAF) was used as a negative control, and all reactions were done in triplicate.
Crystallization of cRbdA and crystallographic data collection.
The homogeneity and oligomeric state of cRbdA were assessed by multiangle light scattering in combination with size exclusion chromatography (SEC-MALS; Wyatt Technology) (Fig. S2). Crystallization conditions were screened by sitting-drop vapor diffusion in 96-well plates, using a mosquito LCP liquid handler (TTP Labtech). Rhombohedral crystals of the cRbdA proteins were obtained at 20°C by using 1 μl of protein at a concentration of 8 mg/ml and 1 μl of reservoir solution containing 0.1 M morpholineethanesulfonic acid (MES)-imidazole at pH 6.5 (obtained by mixing 30.6 ml of 1 M MES at pH 2.7 with 19.4 ml of 1 M imidazole at pH 9.9, as implemented in Morpheus “buffer system 1” [Molecular Dimensions]), 0.1 M carboxylic acid, 10% (wt/vol) polyethylene glycol 4000 (PEG 4000), and 20% (vol/vol) glycerol. Initial data sets to about 2.8- to 3.0-Å resolution could be collected with native crystals. Following a 5-min soak in reservoir solution containing 1 mM HgCl2, diffraction data were extended to 2.28-Å resolution (Table 1). The cRbdA-GTP and cRbdA–ci-di-GMP binary complexes were obtained by transferring native cRbdA crystals to a freshly prepared reservoir solution, supplemented with either 1 mM GTP and 2 mM MgCl2 or 1 mM c-di-GMP and 2 mM MgCl2, and soaking them for 4 to 5 min (Table 1). Longer soaking led to a severe deterioration of the diffraction quality of the crystals and to crystal breakage. Data were collected at the Australian Synchrotron (beamlines MXI and MXII; Clayton, Melbourne, Australia) and the Swiss Light Source (SLS) (beamline PXIII; Paul Scherrer Institute, Switzerland). Diffraction intensities were integrated with XDS (45) and scaled using SCALA (46) from the CCP4 suite (47). Data collection statistics are summarized in Table 1.
TABLE 1.
Data collection statistics
| Parameter | Value for data setc |
||
|---|---|---|---|
| Native, with HgCl2 | cRbdA-GTP/Mg2+ | cRbdA–c-di-GMP | |
| Soaking time (min)/concn (mM) | 5/1 | 4/1 | 5/1 |
| Beamline (synchrotron) | PXIII (SLS) | MXI (Australian Synchrotron) | PXIII (SLS) |
| Wavelength (Å) | 1.0000 | 0.9537 | 1.0000 |
| Resolution (Å) | 70.20–2.28 (2.40–2.28) | 47.90–2.80 (2.95–2.80) | 48.70–3.31 (3.49–3.31) |
| Space group | H32 | H32 | H32 |
| Cell parameters a, b, c (Å) | 134.71, 134.71, 210.45 | 134.73, 134.73, 209.95 | 134.88, 134.88, 211.22 |
| α/β/γ (°) | 90/90/120 | 90/90/120 | 90/90/120 |
| No. of measured reflections | 202,176 (29,411) | 193,115 (28,079) | 112,687 (16,413) |
| No. of unique reflections | 33,717 (4,851) | 18,306 (2,610) | 11,308 (1,624) |
| Redundancy | 6.0 (6.1) | 10.5 (10.8) | 10.0 (10.1) |
| <I/σ> | 10.7 (1.7) | 9.5 (2.5) | 11.3 (4.0) |
| Completeness (%) | 100.0 (100.0) | 100.0 (100.0) | 99.9 (99.6) |
| Rmergea (%) | 12.4 (106.0) | 27.6 (107.0) | 19.1 (61.8) |
| Rpimb (%) | 5.4 (45.4) | 8.9 (34.3) | 6.8 (20.4) |
| CC(1/2)d (%) | 99.7 (51.7) | 0.988 (0.705) | 99.1 (87.8) |
Rmerge = ∑|Ij − <I>|/∑Ij, where Ij is the intensity of an individual reflection and <I> is the average intensity of that reflection.
.
Values in parentheses indicate values in the highest-resolution shell.
CC(1/2), Pearson correlation coefficient.
Structure determination.
The asymmetric unit of the crystal comprises a single cRbdA monomer. The structure was determined using molecular replacement as implemented in the CCP4 package (47), leading to a partial model comprising only the GGDEF and EAL domains. Clear residual electron density was observed for large parts of the PAS domain as well as for segments linking the PAS domain to the GGDEF domain and the GGDEF domain to the EAL domain. The program BUCANEER (48) was then used to extend this partial model. Several rounds of manual model building were then performed using the program COOT (49), interspersed with refinement with the program BUSTER (50), leading to a model comprising residues 241 to 300 and 309 to 797 of the protein (Table 2). An anomalous Fourier map calculated with phases from the partial model revealed a major Hg site bound to Cys521 of the GGDEF domain (at a level of 6.85 σ) and two minor Hg sites, bound to Cys436 and Cys423, giving further confidence in the correctness of the structure determination. Interface areas and solvation energies were calculated by the protein interface, surface, and assembly service PISA (51). Figures were generated with the program PyMOL (Schrodinger Scientific). Structure-based alignment of cRbdA against homologs was generated with ESPript (52). Interface areas buried during dimer formation are listed in Table S1.
TABLE 2.
Refinement statistics
| Parameter | Value for data set |
||
|---|---|---|---|
| Native, with HgCl2 | cRbdA-GTP/Mg2+ | cRbdA–c-di-GMP | |
| Data range (Å) | 70.20–2.28 (2.35–2.28) | 27.55–2.80 (2.97–2.80) | 48.70–3.31 (3.63–3.31) |
| No. of reflections used for refinement | 33,717 (2,884) | 18,292 (2,774) | 11,285 (2,663) |
| No. of nonhydrogen protein atoms | 4,318 | 4,294 | 4,295 |
| Rworka (%) | 0.193 (0.224) | 0.172 (0.195) | 0.174 (0.167) |
| Rfreeb (%) | 0.229 (0.276) | 0.255 (0.259) | 0.259 (0.272) |
| CC Fo-Fc | 0.938 | 0.927 | 0.922 |
| RMSD | |||
| Bond length (Å) | 0.010 | 0.010 | 0.010 |
| Bond angle (°) | 1.11 | 1.19 | 1.21 |
| BWilson/Boverall (Å2) | 44.3/52.0 | 55.4/44.7 | 93.4/57.0 |
| Estimated coordinate error (Å)c | 0.19 | 0.36 | 0.53 |
| Mean B factor (Å2) | |||
| Protein | 38.8 | 45.0 | 58.1 |
| Ligands | |||
| c-di-GMP | 78.5 (46) | ||
| GTP | 47.2 (32) | ||
| Mg2+ | 18.9 (1) | ||
| Water molecules (no.) | 38.9 (303) | 40.0 (315) | 37.4 (162) |
| No. (%) of residues in Ramachandran plot | |||
| Most favored | 531 (97.1) | 518 (95.2) | 509 (93.6) |
| Allowed | 15 (2.7) | 22 (4.1) | 33 (6.1) |
| Disallowed | 1 (0.2) | 4 (0.7) | 2 (0.4) |
| PDB code | 5XGB | 5XGD | 5XGE |
Rwork = ∑||Fo| − |Fc||/∑|Fc|, where Fo denotes the observed structure factor amplitude and Fc denotes the structure factor amplitude calculated from the model.
Rfree was calculated in the same way as Rwork, but with 5% of randomly chosen reflections omitted from the refinement.
Estimated coordinate error is based on (dispersion precision indicator [DPI]) Rfree.
Small-angle X-ray scattering.
X-ray scattering data from wild-type cRbdA and its complexes with GTP, GMP, and c-di-GMP were collected on a Bruker Nanostar SAXS instrument equipped with a Metal-Jet X-ray source (Excillum, Germany) and a Våntec 2000 detector system. The scattering patterns were measured using a sample detector distance of 0.67 m and a wavelength (λ) of 1.3414 Å, covering the range of momentum transfer of 0.016 < q < 0.4 Å−1 [q = 4π sin(θ)/λ, where 2θ is the scattering angle]. To monitor for radiation damage, six 5-min exposures were collected for each protein sample, and no radiation effect was observed. cRbdA was measured at concentrations of 1.6, 2.5, and 3.6 mg/ml, while the ligand complexes were measured at 2.5 mg/ml. The data were normalized to the intensity of the transmitted beam, and the scattering of the buffer was subtracted. The difference curves were scaled for concentration, and the data processing steps were performed using the program package PRIMUS from the ATSAS package, version 2.7.1 (53). The forward scattering value I(0) and the radius of gyration (Rg) were computed using the Guinier approximation. The pair distribution functions were calculated by GNOM (54) and also provided the maximum particle size (Dmax), and the low-resolution shape of wild-type cRbdA was determined ab initio by use of DAMMIF (55). Qualitative particle motion was inferred by plotting the scattering patterns in the normalized Kratky plot [(qRg)2(I(q)/I(0)) versus qRg] (56). The theoretical scattering curves from atomic structures were generated and evaluated against experimental scattering curves by use of CRYSOL (57). Utilizing the intensities from each component, the volume fraction of each component could be determined by use of OLIGOMER (49). Rigid body modeling was performed using CORAL (58), by docking the individual domains of the high-resolution structures against the experimental data. The oligomeric state of the protein was confirmed from the molecular mass calculation, based on I(0), Porod volume (Vp), excluded volume (Vex), and volume of correlation (Vc) (59).
Accession number(s).
The structures from this study have been deposited in the Protein Data Bank under accession numbers 5XGB, 5XGD, and 5XGE.
Supplementary Material
ACKNOWLEDGMENTS
We thank the scientists and staff of the MXI and MXII beamlines (Australian Synchrotron, Clayton, Victoria, Australia) and the PXIII beamlines (Paul Scherrer Institute, Switzerland) for their expert assistance.
This work was supported by the Ministry of Education (MOE) Singapore, by AcRF tier 1 grant RG154/14 to the J.L. and S.A.R. laboratories and by an MOE tier 3 grant (grant MOE2012-T3-1-008) to G.G. Support from NRF and MOE Singapore under its RCE program is also acknowledged.
S.A.R., Z.-X.L., G.G., and J.L. conceived the experiments, C.L., Y.H.W., W.H.P., M.S.S.M., S.T.T., L.X., and J.L. performed the experiments, C.L., C.W.L., M.S.S.M., S.R., Z.-X.L., G.G., S.A.R., and J.L. analyzed the data, and J.L. wrote the manuscript, with input from all authors.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JB.00515-17.
REFERENCES
- 1.Ross P, Weinhouse H, Aloni Y, Michaeli D, Weinberger-Ohana P, Mayer R, Braun S, de Vroom E, van der Marel GA, van Boom JH, Benziman M. 1987. Regulation of cellulose synthesis in Acetobacter xylinum by cyclic diguanylic acid. Nature 325:279–281. https://www.nature.com/articles/325279a0. [DOI] [PubMed] [Google Scholar]
- 2.Hengge R. 2009. Principles of c-di-GMP signalling in bacteria. Nat Rev Microbiol 7:263–273. doi: 10.1038/nrmicro2109. [DOI] [PubMed] [Google Scholar]
- 3.Römling U, Galperin MY, Gomelsky M. 2013. Cyclic di-GMP: the first 25 years of a universal bacterial second messenger. Microbiol Mol Biol Rev 77:1–52. doi: 10.1128/MMBR.00043-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chan C, Paul R, Samoray D, Amiot NC, Giese B, Jenal U, Schirmer T. 2004. Structural basis of activity and allosteric control of diguanylate cyclase. Proc Natl Acad Sci U S A 101:17084–17089. doi: 10.1073/pnas.0406134101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chang AL, Tuckerman JR, Gonzalez G, Mayer R, Weinhouse H, Volman G, Amikam D, Benziman M, Gilles-Gonzalez M-A. 2001. Phosphodiesterase A1, a regulator of cellulose synthesis in Acetobacter xylinum is a heme-based sensor. Biochemistry 40:3420–3426. doi: 10.1021/bi0100236. [DOI] [PubMed] [Google Scholar]
- 6.Schirmer T, Jenal U. 2009. Structural and mechanistic determinants of c-di-GMP signalling. Nat Rev Microbiol 7:724–735. doi: 10.1038/nrmicro2203. [DOI] [PubMed] [Google Scholar]
- 7.Deepthi A, Liew CW, Liang ZX, Swaminathan K, Lescar J. 2014. Structure of a diguanylate cyclase from Thermotoga maritima: insights into activation, feedback inhibition and thermostability. PLoS One 9:e110912. doi: 10.1371/journal.pone.0110912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wassmann P, Chan C, Paul R, Beck A, Heerklotz H, Jenal U, Schirmer T. 2007. Structure of BeF3−-modified response regulator PleD: implications for diguanylate cyclase activation, catalysis, and feedback inhibition. Structure 15:915–927. doi: 10.1016/j.str.2007.06.016. [DOI] [PubMed] [Google Scholar]
- 9.Tchigvintsev A, Xu X, Singer A, Chang C, Brown G, Proudfoot M, Cui H, Flick R, Anderson WF, Joachimiak A, Galperin MY, Savchenko A, Yakunin AF. 2010. Structural insight into the mechanism of c-di-GMP hydrolysis by EAL domain phosphodiesterases. J Mol Biol 402:524–538. doi: 10.1016/j.jmb.2010.07.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barends TRM, Hartmann E, Griese JJ, Beitlich T, Kirienko NV, Ryjenkov DA, Reinstein DA, Shoeman RI, Gomelsky M, Schlichting I. 2009. Structure and mechanism of a bacterial light-regulated cyclic nucleotide phosphodiesterase. Nature 459:1015–1018. doi: 10.1038/nature07966. [DOI] [PubMed] [Google Scholar]
- 11.Minasov G, Padavattan S, Shuvalova L, Brunzelle JS, Miller DJ, Basler A, Massa C, Collart FR, Schirmer T, Anderson WF. 2009. Crystal structures of YkuI and its complex with second messenger cyclic di-GMP suggest catalytic mechanism of phosphodiester bond cleavage by EAL domains. J Biol Chem 284:13174–13184. doi: 10.1074/jbc.M808221200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen MW, Kotaka M, Vonrhein C, Bricogne G, Rao F, Chuah MLC, Svergun D, Schneider G, Liang ZX, Lescar J. 2012. Structural insights into the regulatory mechanism of the response regulator RocR from Pseudomonas aeruginosa in cyclic di-GMP signaling. J Bacteriol 184:4837–4846. doi: 10.1128/JB.00560-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ryjenkov DA, Tarutina M, Moskvin OV, Gomelsky M. 2005. Cyclic diguanylate is a ubiquitous signaling molecule in bacteria: insights into biochemistry of the GGDEF protein domain. J Bacteriol 187:1792–1798. doi: 10.1128/JB.187.5.1792-1798.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schmidt AJ, Ryjenkov DA, Gomelsky M. 2005. The ubiquitous protein domain EAL is a cyclic diguanylate-specific phosphodiesterase: enzymatically active and inactive EAL domains. J Bacteriol 187:4774–4781. doi: 10.1128/JB.187.14.4774-4781.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tamayo R, Tischler AD, Camilli A. 2005. The EAL domain protein VieA is a cyclic diguanylate phosphodiesterase. J Biol Chem 280:33324–33330. doi: 10.1074/jbc.M506500200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Christen M, Christen B, Folcher M, Schauerte A, Jenal U. 2005. Identification and characterization of a cyclic di-GMP-specific phosphodiesterase and its allosteric control by GTP. J Biol Chem 280:30829–30837. doi: 10.1074/jbc.M504429200. [DOI] [PubMed] [Google Scholar]
- 17.Galperin MY, Nikolskaya AN, Koonin EV. 2001. Novel domains of the prokaryotic two-component signal transduction systems. FEMS Microbiol Lett 203:11–21. doi: 10.1111/j.1574-6968.2001.tb10814.x. [DOI] [PubMed] [Google Scholar]
- 18.Hickman JW, Tifrea DF, Harwood CS. 2005. A chemosensory system that regulates biofilm formation through modulation of cyclic diguanylate levels. Proc Natl Acad Sci U S A 102:14422–14427. doi: 10.1073/pnas.0507170102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Paul R, Weiser S, Amiot NC, Chan C, Schirmer T, Giese B, Jenal U. 2004. Cell-cycle dependent dynamics localization of a bacterial response regulator with a novel diguanylate cyclase output domain. Genes Dev 18:715–727. doi: 10.1101/gad.289504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Navarro MVAS, De N, Bae N, Wang Q, Sondermann H. 2009. Structural analysis of the GGDEF-EAL domain-containing c-di-GMP receptor FimX. Structure 17:1104–1116. doi: 10.1016/j.str.2009.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tal R, Wong HC, Calhoon R, Gelfand D, Fear AL, Volman G, Mayer R, Ross P, Amikam D, Weinhouse H, Cohen A, Sapir S, Ohana P, Benziman M. 1998. Three cdg operons control cellular turnover of cyclic-di-GMP in Acetobacter xylinum: genetic organization and occurrence of conserved domains in isoenzymes. J Bacteriol 180:4416–4425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Weber H, Pesavento C, Possling A, Tischendorf G, Hengge R. 2006. Cyclic-di-GMP-mediated signalling within the sigma network of Escherichia coli. Mol Microbiol 62:1014–1034. doi: 10.1111/j.1365-2958.2006.05440.x. [DOI] [PubMed] [Google Scholar]
- 23.García B, Latasa C, Solano C, Portillo FG-D, Gamazo C, Lasa I. 2004. Role of the GGDEF protein family in Salmonella cellulose biosynthesis and biofilm formation. Mol Microbiol 54:264–277. doi: 10.1111/j.1365-2958.2004.04269.x. [DOI] [PubMed] [Google Scholar]
- 24.Kuchma SL, Brothers KM, Merritt JH, Liberati NT, Ausubel FM, O'Toole GA. 2007. BifA, a cyclic di-GMP phosphodiesterase, inversely regulates biofilm formation and swarming motility by Pseudomonas aeruginosa PA14. J Bacteriol 189:8165–8178. doi: 10.1128/JB.00586-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bae SO, Sugano Y, Ohi K, Shoda M. 2004. Features of bacterial cellulose synthesis in a mutant generated by disruption of the diguanylate cyclase 1 gene of Acetobacter xylinum BPR. Appl Microbiol Biotechnol 65:315–322. doi: 10.1007/s00253-004-1593-7. [DOI] [PubMed] [Google Scholar]
- 26.Tarutina M, Ryjenkov DA, Gomelsky M. 2006. An unorthodox bacteriophytochrome from Rhodobacter sphaeroides involved in turnover of the second messenger c-di-GMP. J Biol Chem 281:34751–34758. doi: 10.1074/jbc.M604819200. [DOI] [PubMed] [Google Scholar]
- 27.Boles BR, McCarter LL. 2002. Vibrio parahaemolyticus scrABC, a novel operon affecting swarming and capsular polysaccharide regulation. J Bacteriol 184:5946–5954. doi: 10.1128/JB.184.21.5946-5954.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kumar M, Chatterji D. 2008. Cyclic-di-GMP a second messenger required for long term survival, but not biofilm formation, in Mycobacterium smegmatis. Microbiology 154:2942–2955. doi: 10.1099/mic.0.2008/017806-0. [DOI] [PubMed] [Google Scholar]
- 29.Kulasakara H, Lee V, Brencic A, Liberati N, Urbach J, Miyata S, Lee DG, Neely AN, Hyodo M, Hayakawa Y, Ausubel FM, Lory S. 2006. Analysis of Pseudomonas aeruginosa diguanylate cyclases and phosphodiesterases reveals a role for bis-(3′-5′)-cyclic-GMP in virulence. Proc Natl Acad Sci U S A 103:2839–2844. doi: 10.1073/pnas.0511090103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Qi Y, Rao F, Luo Z, Liang Z-X. 2009. A flavin cofactor-binding PAS domain regulates c-di-GMP synthesis in AxDGC2 from Acetobacter xylinum. Biochemistry 48:10275–10285. doi: 10.1021/bi901121w. [DOI] [PubMed] [Google Scholar]
- 31.Choy W-K, Zou L, Syn CK-C, Zhang L-H, Swarup S. 2004. MorA defines a new class of regulators affecting flagellar development and biofilm formation in diverse Pseudomonas species. J Bacteriol 186:7221–7228. doi: 10.1128/JB.186.21.7221-7228.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Phippen CW, Mikolajek H, Schlaefli HG, Keevil CW, Webb JS, Tews I. 2014. Formation and dimerization of the phosphodiesterase active site of the Pseudomonas aeruginosa MorA, a bi-functional c-di-GMP regulator. FEBS Lett 588:4631–4636. doi: 10.1016/j.febslet.2014.11.002. [DOI] [PubMed] [Google Scholar]
- 33.Navarro MVAS, Newell PD, Krasteva PV, Chatterjee D, Madden DR, O'Toole GA, Sondermann H. 2011. Structural basis for c-di-GMP-mediated inside-out signaling controlling periplasmic proteolysis. PLoS Biol 9:e1000588. doi: 10.1371/journal.pbio.1000588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roy AB, Petrova OE, Sauer K. 2012. The phosphodiesterase DipA (PA5017) is essential for Pseudomonas aeruginosa biofilm dispersion. J Bacteriol 194:2904–2915. doi: 10.1128/JB.05346-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.An S, Wu J, Zhang LH. 2010. Modulation of Pseudomonas aeruginosa biofilm dispersal by a cyclic-di-GMP phosphodiesterase with a putative hypoxia-sensing domain. Appl Environ Microbiol 76:8160–8173. doi: 10.1128/AEM.01233-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Heintz U, Meinhart A, Winkler A. 2014. Multi-PAS domain-mediated protein oligomerization of PpsR from Rhodobacter sphaeroides. Acta Crystallogr D Biol Crystallogr 70:863–876. doi: 10.1107/S1399004713033634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sundriyal A, Massa C, Samoray D, Zehender F, Sharpe T, Jenal U, Schirmer T. 2014. Inherent regulation of EAL domain-catalyzed hydrolysis of second messenger cyclic di-GMP. J Biol Chem 289:6978–6990. doi: 10.1074/jbc.M113.516195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rao F, Qi Y, Chong HS, Kotaka M, Li B, Lescar J, Tang K, Liang Z-X. 2009. The functional role of a conserved loop in EAL domain-based c-di-GMP specific phosphodiesterase. J Bacteriol 191:4722–4731. doi: 10.1128/JB.00327-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rao F, Yang Y, Qi Y, Liang ZX. 2008. Catalytic mechanism of c-di-GMP specific phosphodiesterase: a study of the EAL domain-containing RocR from Pseudomonas aeruginosa. J Bacteriol 190:3622–3631. doi: 10.1128/JB.00165-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bellini D, Horrell S, Hutchin A, Phippen CW, Strange RW, Cai Y, Wagner A, Webb JS, Tews I, Walsh MA. 2017. Dimerisation-induced formation of the active site and the identification of three metal sites in EAL-phosphodiesterases. Sci Rep 7:42166. doi: 10.1038/srep42166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Barraud N, Schleheck D, Klebensberger J, Webb JS, Hassett DJ, Rice SA, Kjelleberg S. 2009. Nitric oxide signaling in Pseudomonas aeruginosa biofilms mediates phosphodiesterase activity, decreased cyclic di-GMP levels, and enhanced dispersal. J Bacteriol 191:7333–7342. doi: 10.1128/JB.00975-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ferreira RBR, Antunes LCM, Greenberg EP, McCarter LL. 2008. Vibrio parahaemolyticus ScrC modulates cyclic dimeric GMP regulation of gene expression relevant to growth on surfaces. J Bacteriol 190:851–860. doi: 10.1128/JB.01462-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.De N, Navarro MVAS, Raghavan RV, Sondermann H. 2009. Determinants for the activation and autoinhibition of the diguanylate cyclase response regulator WspR. J Mol Biol 393:619–633. doi: 10.1016/j.jmb.2009.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.De N, Pirruccello M, Krasteva PV, Bae N, Raghavan RV, Sondermann H. 2008. Phosphorylation-independent regulation of the diguanylate cyclase WspR. PLoS Biol 6:e67. doi: 10.1371/journal.pbio.0060067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kabsch W. 2010. Integration, scaling, space-group assignment and post-refinement. Acta Crystallogr D Biol Crystallogr 66:133–144. doi: 10.1107/S0907444909047374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Evans P. 2006. Scaling and assessment of data quality. Acta Crystallogr D Biol Crystallogr 62:72–82. doi: 10.1107/S0907444905036693. [DOI] [PubMed] [Google Scholar]
- 47.Acta Crystallographica Section D. 1994. The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr 50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 48.Cowtan K. 2006. The Buccaneer software for automated model building. 1. Tracing protein chains. Acta Crystallogr D Biol Crystallogr 62:1002–1011. doi: 10.1107/S0907444906022116. [DOI] [PubMed] [Google Scholar]
- 49.Emsley P, Lohkamp B, Scott WG, Cowtan K. 2010. Features and development of Coot. Acta Crystallogr D Biol Crystallogr 66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Smart OS, Womack TO, Flensburg C, Keller P, Paciorek W, Sharff A, Vonrhein C, Bricogne G. 2012. Exploiting structure similarity in refinement: automated NCS and target-structure restraints in BUSTER. Acta Crystallogr D Biol Crystallogr 68:368–380. doi: 10.1107/S0907444911056058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Krissinel E, Henrick K. 2007. Inference of macromolecular assemblies from crystalline state. J Mol Biol 372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 52.Robert X, Gouet P. 2014. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res 42:W320–W324. doi: 10.1093/nar/gku316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Konarev PV, Volkov VV, Sokolova AV, Koch MHJ, Svergun DI. 2003. PRIMUS: a Windows PC-based system for small-angle scattering data analysis. J Appl Crystallogr 36:1277–1282. doi: 10.1107/S0021889803012779. [DOI] [Google Scholar]
- 54.Svergun DI. 1992. Determination of the regularization parameter in indirect-transform methods using perceptual criteria. J Appl Crystallogr 25:495–503. doi: 10.1107/S0021889892001663. [DOI] [Google Scholar]
- 55.Franke D, Svergun DI. 2009. DAMMIF, a program for rapid ab-initio shape determination in small-angle scattering. J Appl Crystallogr 42:342–346. doi: 10.1107/S0021889809000338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Durand D, Vivès C, Cannella D, Pérez J, Pebay-Peyroula E, Vachette P, Fieschi F. 2010. NADPH oxidase activator p67phox behaves in solution as a multidomain protein with semi-flexible linkers. J Struct Biol 169:45–53. doi: 10.1016/j.jsb.2009.08.009. [DOI] [PubMed] [Google Scholar]
- 57.Svergun D, Barberato C, Koch MHJ. 1995. CRYSOL—a program to evaluate X-ray solution scattering of biological macromolecules from atomic coordinates. J Appl Crystallogr 28:768–773. doi: 10.1107/S0021889895007047. [DOI] [Google Scholar]
- 58.Petoukhov MV, Franke D, Shkumatov AV, Tria G, Kikhney AG, Gajda M, Gorba C, Mertens HDT, Konarev PV, Svergun DI. 2012. New developments in the ATSAS program package for small-angle scattering data analysis. J Appl Crystallogr 45:342–350. doi: 10.1107/S0021889812007662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mylonas E, Svergun DI. 2007. Accuracy of molecular mass determination of proteins in solution by small-angle X-ray scattering. J Appl Crystallogr 40:s245–s249. doi: 10.1107/S002188980700252X. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.