Abstract
The facultative intracellular pathogen Listeria monocytogenes causes a severe food-borne infection in humans and animals. L. monocytogenes invasion factor InlB interacts with the tyrosine kinase c-Met via the N-terminal internalin domain. Previously, distinct variants of the InlB internalin domain (idInlB) have been described in L. monocytogenes field isolates. Three variants were used to restore full-length InlB expression in the L. monocytogenes strain EGDeΔinlB. Obtained isogenic L. monocytogenes strains were tested in the invasion assay and intravenous, intraperitoneal, and intragastric models of infection in mice. All idInlBs were functional, restored InlB activity as an invasion factor, and improved invasion of the parental strain EGDeΔinlB into human kidney HEK23 cells. Meanwhile, distinct idInlBs provided different mortality rates and bacterial loads in internal organs. When recombinant strains were compared, the variant designated idInlB14 decreased severity of disease caused by intravenous and intraperitoneal bacterial administration, whereas this variant improved intestine colonization and stimulated intragastric infection. Obtained results demonstrated that naturally occurring idInlBs differed in their impact on severity of L. monocytogenes infection in mice in dependence on the infection route.
1. Introduction
The Gram-positive bacterium Listeria monocytogenes is a causative agent of food-borne infection, listeriosis, in humans and many domestic and wild animals [1, 2]. Listeriosis is a rare but severe infection with lethality reaching 30% in humans [3, 4]. The most serious clinical manifestations of listeriosis are CNS disorders and fetus impairment in pregnant females.
The intestine is a port of bacterial entry in the course of food-borne listeriosis [1, 5]. L. monocytogenes crosses the intestinal barrier and colonizes the lamina propria from where it spreads to internal organs [5]. Systemic listeriosis includes colonization of the liver and spleen. If the infection is not controlled at this stage, a secondary bacteremia develops, and bacteria cross cerebral and/or maternal-fetal barriers to infect the brain and fetus [1, 2, 5, 6].
L. monocytogenes is a facultative intracellular pathogen that infects macrophages and a wide range of nonprofessional phagocytes [7]. Internalin (InlA) and InlB are major factors that provide active invasion in nonprofessional phagocytes [7, 8]. InlA and InlB belong to the internalin family. Internalin family proteins include an internalin domain, which consists of a central leucine rich repeat- (LRR-) domain flanked by amino-terminal N-cap and carboxy-terminal immunoglobulin-like (Ig-like) domains [9]. The internalin domain is directly involved in specific protein-protein interactions with mammalian cell surface receptors. InlB internalin domain specifically interacts with the eukaryotic receptor c-Met/HGFR (Hepatocyte Growth Factor Receptor) [10].
Mouse models are of key importance for studies of L. monocytogenes virulence. LD50 and bacterial counts in the liver and spleen are major ways used for assessment of L. monocytogenes strain virulence [11, 12]. Intravenous (i.v.) and intraperitoneal (i.p.) infection routes usually require a smaller infective dose and are used more often in L. monocytogenes research than intragastric (i.g.) or oral routes [13–15]. When several strains were compared, their relative virulence was dependent on the route of infection. Certain strains highly virulent when injected i.p. or i.p. are less virulent when applied orally or i.g., and vice versa [16, 17].
L. monocytogenes oral route infection in mice is dependent on InlB, which accelerates listerial invasion into M cells of ileal Peyer's patches (PPs) [18, 19]. Previously, we and others described isolation of L. monocytogenes from internal organs of wild small rodents belonging to the Murinae subfamily, which is closely related to Mus musculus. These data suggest that mouse intestine barrier crossing is effective enough for systemic infection to be established in the natural environment [20, 21]. Among 15 InlB variants found in L. monocytogenes strains, only two variants were revealed in L. monocytogenes isolated from wild small rodents [21, 22]. Existence of specific InlB variants was demonstrated for L. monocytogenes strains characterized by reduced virulence [23] or associated with cardiovascular disease [24].
To evaluate an impact of naturally occurring InlB variants on L. monocytogenes virulence, we used a set of previously described isogenic strains that carried distinct InlB internalin domains (idInlBs) [25]. Virulence of the recombinant strains was compared in mouse models. Distinct idInlBs provided different mortality rates and bacterial loads in internal organs, and the difference was dependent on the route of infection.
2. Materials and Methods
2.1. Bacterial Strains and Growth Conditions
Bacterial strains used in the work are listed in Table 1. L. monocytogenes were routinely cultivated in the Brain Heart Infusion (BHI, BD, USA) medium. To get a virulence regulon induction, L. monocytogenes was grown in BHI supplemented with 0,2% activated charcoal (Merck) [26]. E. coli used in cloning and expression procedures were cultivated in the LB medium (Sigma-Aldrich). All bacteria were grown at 37°C with agitation at 180 rpm. Antibiotics were added up to the following concentrations: erythromycin up to 10 and 300 μg ml−1 for L. monocytogenes and E. coli, respectively; ampicillin up to 100 μg ml−1; kanamycin up to 200 μg ml−1. All antibiotics were purchased from Sigma-Aldrich. To prepare a culture for infection, bacteria were grown up to mid-exponential phase, washed with PBS, aliquoted, and frozen in the presence of 10% glycerin. The concentration was determined by plating serial dilutions from the frozen culture the day before the experiment. Bacteria were thawed immediately before the experiment and resuspended in PBS up to the required concentration.
Table 1.
Strains and plasmids used in the work.
| Species/strain | Characteristics | Reference |
|---|---|---|
| L. monocytogenes | ||
| EGDe1 | Serovar 1/2a, type strain | [27] |
| EGDeΔinlB1 | EGDe derivative with inlB gene deletion | [28] |
| VIMHA004 | Serovar 4b, ST22, clinical isolate obtained from the stillborn, carried idInlB13 | [21] |
| VIMHA015 | Serovar 4b, ST12, Clinical isolate obtained from the stillborn, carried idInlB93 | [21] |
| VIMHA034 | Serovar 1/2a, ST3142, Clinical isolate obtained from the stillborn, carried idInlB143 | [21] |
| EGDeΔinlB::InlB1 | EGDeΔinlB supplemented with the pInlB1 plasmid | [25] |
| EGDeΔinlB::InlB9 | EGDeΔinlB supplemented with the pInlB9 plasmid | [25] |
| EGDeΔinlB::InlB14 | EGDeΔinlB supplemented with the pInlB14 plasmid | [25] |
| E. coli | ||
| JM109 | endA1 glnV44 thi-1 relA1 gyrA96 recA1 mcrB+Δ(lac-proAB) e14-[F′ traD36 proAB+lacIqlacZΔM15] hsdR17(rK-mK+) | Promega |
| BL21 (DE3) | F–ompT gal dcm lon hsdSB(rB-mB-) λ(DE3 [lacI lacUV5-T7 gene 1 ind1 sam7 nin5]) | NewEngland BioLabs |
| Plasmids | ||
| pTgem-easy | Cloning vector | Promega |
| pTRKH2 | Shuttle-vector | [29] |
| pInlAB | pTRKH2::PromoterinlAB::gwdInlB4 | [25] |
| pInlB1 | pInlAB::idInlB1 | [25] |
| pInlB9 | pInlAB::idInlB9 | [25] |
| pInlB14 | pInlAB::idInlB14 | [25] |
1The strain was generously provided by Professor J. A. Vazquez-Boland, University of Edinburgh. 2Sequence tyes (STs) are provided according to the MLST protocol by Ragon et al., 2008. 3idInlB is an internalin domain of InlB. 4gwdInlB is InlB GW-domain.
2.2. Mammalian Cells and Cultivation Conditions
Human embryonic kidney HEK-293 cells (the public collection of Gamaleya Center; http://virology.gamaleya.org/index.php?option=com_content&view=article&id=103:kollektsiya-kletochnykh-kultur&catid=91&Itemid=645) were used. Cells were grown in the DMEM medium supplemented with 10% FBS (fetal bovine serum) in the 5% CO2 atmosphere. In some experiments FBS was decreased up to 2%.
2.3. Recombinant Strains
Recombinant strain construction was described previously [25]. Shortly, the idInlB encoding DNA fragments obtained on lysates of L. monocytogenes strains VIMHA004, VIMHA015, and VIMHA034 (Table 1) with primers InlB3: 5′-CAAGCGGGATCCATCACCGTGCCAACGCC and InlB4: 5′-TATCCGGGATCCTGCTTCTACTTTTG were cloned into the derivative of the pTRKH2 vector [29] carrying the inlB gene, lacking the internalin domain encoding part. The obtained plasmids encoded the full-length InlB with different idInlBs. The plasmids were incorporated into the L. monocytogenes EGDeΔinlB strain by electroporation.
2.4. Cell Invasion Assay
Bacteria were prepared from the mid-exponential culture as it was described above. The “gentamycin assay” was performed as described previously [25]. Briefly, bacteria resuspended in DMEM were added with MOI 100 : 1 (bacteria to cell) to cells, incubated 1 h at 37°C in 5% CO2 atmosphere, and washed with PBS, and then the fresh medium supplemented with 100 μg ml−1 gentamycin (Fluka) was added. In 1 h, cells were washed and lysed with 1% Triton X-100. Serial dilutions of cell lysates were plated to count bacterial colonies. The invasion efficiency was determined as the ratio of intracellular bacteria to the number of applied bacteria.
2.5. Mice
Experiments on animals were conducted in accordance with the Russian Federation National Standard (GOST R52379-2005), directives of Ministry of Health of Russian Federation (number 753n from 26.08.2010; number 774н from 31.08.2010), and with the approval of the Biomedical Ethics Committee of Gamaleya Research Center of Epidemiology and Microbiology (number 93 from 22.10.2015). Animals were sacrificed using ether inhalation. Female BALB/c mice of 16–18 g, purchased from the nursery Stolbovaya (Moscow region, Russia), were kept at our breeding facilities in specific-pathogen-free conditions and used in all experiments.
2.6. Experimental Infection in Mice
For intravenous infection, mice were prewarmed with air heater and injected into the tail vein with bacteria suspended in 100 μl PBS. The doses are indicated in the text. For intraperitoneal infection, mice were injected intraperitoneally with bacteria suspended in 200 μl PBS. For intragastric infection, mice starved for 12 h were anesthetized by intraperitoneal injection of sodium pentobarbital (40 μg/g) and infected with bacteria suspended in 200 μl PBS by using the needle with the diameter 0.6 mm and the length 25 mm. The tip of the needle was cut and polished to smoothness to avoid stomach injury.
2.7. Mortality Rates and Bacterial Loads in Organs
To determine mortality rates, groups of 20 animals were infected i.v. with doses of bacteria higher or near LD50. The experiment was stopped as soon as 50% lethality was observed in at least one of the groups and all remained animals were euthanized.
Groups of 5 animals were used to determine bacterial loads in infected organs. Animals were euthanized with ether at time points specified in the text. The liver and the spleen were aseptically harvested and homogenized in 1 and 0,2 ml sterile PBS, respectively. The intestine was washed out with sterile PBS; visible Peyer's patches were separated and homogenized in 5 ml of sterile PBS. Small intestine was separately homogenized in 5 ml of sterile PBS. Tenfold serial dilutions of organ homogenates were plated on BHI agar supplemented with antibiotics if required. Plates were incubated at 37°C for 24 h The detection limit of this procedure was 10, 10, and 102 colony forming units (CFU) per liver, spleen, and intestine, respectively.
2.8. Statistics
All experiments were performed using duplicate samples. Results of repeated experiments were averaged. The one-tailed t-test was used for assessment of statistical significance.
3. Results
3.1. idInlB Variants Differentially Supported i.v. L. monocytogenes Infection
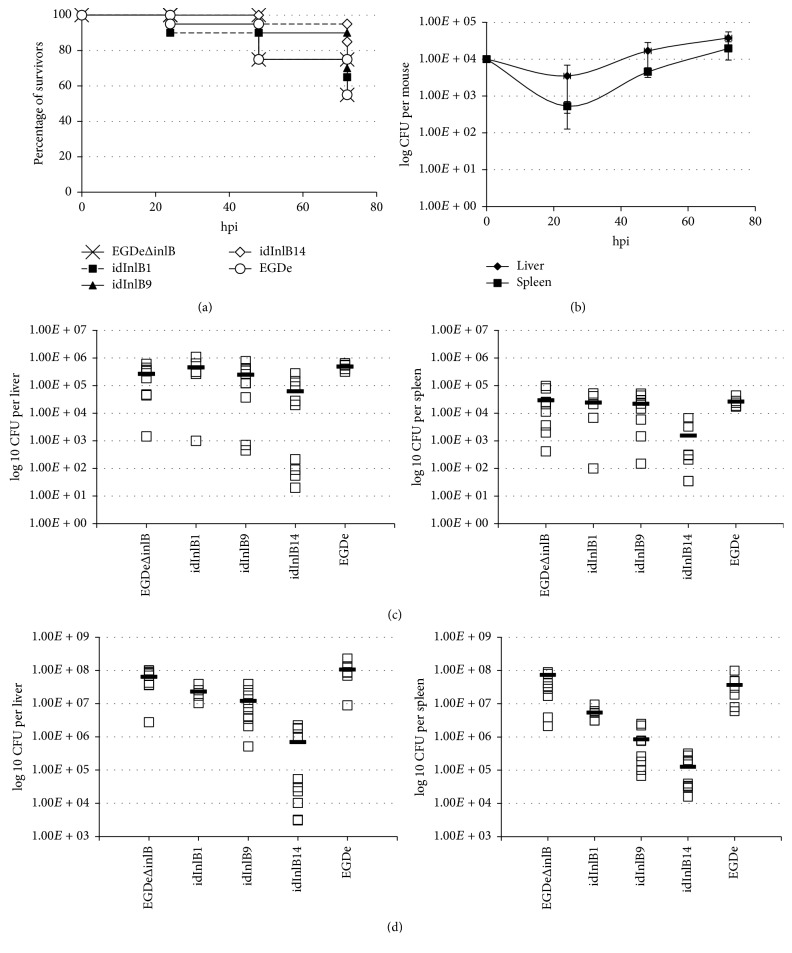
Firstly, we evaluated an impact of idInlB variants on the final outcome of intravenous (i.v.) infection. To compare mortality rates, 2 × 106 CFU of parental EGDeΔinlB, wild-type EGDe, or each recombinant strain was injected into the tail vein of female BALB/c mice. This bacterial load was near LD50 for the parental strain EGDeΔinlB (LD50 ≈ 1 × 106 in our experimental settings) and much higher than LD50 for the wild-type strain EGDe (LD50 ≈ 1,5 × 104). The 50% mortality was reached in 72 h after infection with both EGDeΔinlB and the wild-type strain EGDe (Figure 1) that was in line with the high infection dose. Mortality caused by all but one recombinant strains had similar rates. However, bacteria carrying idInlB14 caused noticeably lower mortality rates with only 5% mortality during the first three days (Figure 1(a)).
Figure 1.
Impact of idInlB variants on severity of intravenous infection in BALB/c mice. (a) Mortality caused by parental EGDeΔinlB, wild-type EGDe, and recombinant EGDeΔinlB::InlB1 (idInlB1), EGDeΔinlB::InlB9 (idInlB9), and EGDeΔinlB::InlB14 (idInlB14) strains. Groups of 20 mice were infected i.v. with 2 × 106 CFU as described in Materials and Methods. (b) Proliferation of EGDeΔinlB::InlB14 in the liver and spleen of mice i.v. infected with 1 × 104 CFU. Average and SD for groups of 5 mice are shown. (c) and (d) idInlB-dependent drop of bacterial loads in the liver and spleen 6 (c) and 24 (d) hpi. BALB/c mice were i.v. infected with 1 × 106 CFU. The data and average of two independent experiments performed on groups of 5 mice are shown.
To understand, whether the strain EGDeΔinlB::InlB14 was able to multiply within the host we followed bacterial loads in the liver and spleen of mice infected with 104 bacteria of this strain. A noticeable drop in bacterial loads was observed 24 hpi in comparison with the dose applied for infection: only (3,5 ± 2,3) × 103 CFU per mouse were found in the liver and (5,3 ± 1,8) × 102 CFU per mouse in the spleen (Figure 1(b)). However, after the initial drop, bacterial loads increased logarithmically suggesting that the strain is competent to multiply within the host.
To check whether the same drop happens with all other strains and at high infection doses and to establish at what time point it takes place, we compared bacterial loads in the liver and spleen for all strains 6 and 24 h after i.v. infection with 106 bacteria. By 6 hpi, the number of wild-type EGDe bacteria was higher than EGDeΔinlB in both the liver (4,92 × 105 versus 2,7 × 105 CFU per mouse, resp., p < 0,05) and the spleen (2,97 × 104 versus 2,4 × 104 CFU per mouse). The recombinant strain EGDeΔinlB::inlB9 behaved similarly to the parental, with average values of 2,47 × 105 and 2,20 × 104 CFU per liver and spleen, respectively. The strain EGDeΔinlB::inlB1 behaved more closely to the wild-type strain (4,57 × 105 and 2,4 × 104 CFU per liver and spleen, resp.). The strain EGDeΔinlB::inlB14 demonstrated the lowest loads in both the liver and spleen (6,25 × 104 and 1,25 × 103 CFU per liver and spleen, resp., p < 0,05).
By 24 hpi, EGDe and EGDeΔinlB accumulated in the liver to 1,08 × 108 and 6,47 × 107 CFU per mouse, respectively, while EGDeΔinlB::InlB1 and EGDeΔinlB::InlB9 accumulated to slightly lower values (2,32 × 107 and 1,22 × 107 CFU per mouse, resp., p < 0,05; Figure 1(d)). The difference between EGDeΔinlB and EGDeΔinlB::InlB14 reached 2 log10 (p < 0,01). In the spleen, all recombinant strains accumulated to values noticeably lower than control bacteria (p < 0,05). The difference between EGDeΔinlB::InlB14 and the parental strain in the spleen reached 2,76 log10 (7,42 × 107 and 1,27 × 105 CFU per mouse for EGDeΔinlB and EGDeΔinlB::InlB14, resp., p < 0,01).
Taken together, obtained results demonstrated that (i) InlB hyperproduction rather worsened than improved bacterial accumulation in the first 24 h; this effect was especially noticeable in the spleen 24 hpi; and (ii) InlB internalin domain variants differed in their ability to support i.v. infection with the idInlB14 demonstrating the worst results.
3.2. idInlBs Differed in Their Ability to Support i.p. Infection
In contrast with i.v. infection, all recombinant strains had improved bacterial loads in comparison with the parental strain EGDeΔinlB, when 106 bacteria were injected intraperitoneally (i.p.) and bacterial loads in the liver and spleen were detected 72 hpi (Figure 2; p < 0,05). However, pairwise strain comparison demonstrated that the EGDeΔinlB::InlB9 supplied noticeably higher infection levels than the variants EGDeΔinlB::InlB1 and EGDeΔinlB::InlB14. The effect was more pronounced in the liver: recombinant strain counts were higher comparatively to the parental (9306 ± 2342)-fold for EGDeΔinlB::InlB9 and (64 ± 18)- and (26 ± 6)-fold for EGDeΔinlB::InlB1 and EGDeΔinlB::InlB14, respectively. Loads of the wild-type EGDe strain were (4954 ± 1582)-higher than EGDeΔinlB. The same tendency was observed for colonization of the spleen: the counts of EGDeΔinlB::InlB9 and wild-type bacteria exceeded the counts of the parental strain (577 ± 97)- and (586 ± 70)-fold, respectively, while the counts of EGDeΔinlB::InlB1 and EGDeΔinlB::InlB14 exceeded parental (63 ± 8)- and (60 ± 11)-fold only.
Figure 2.
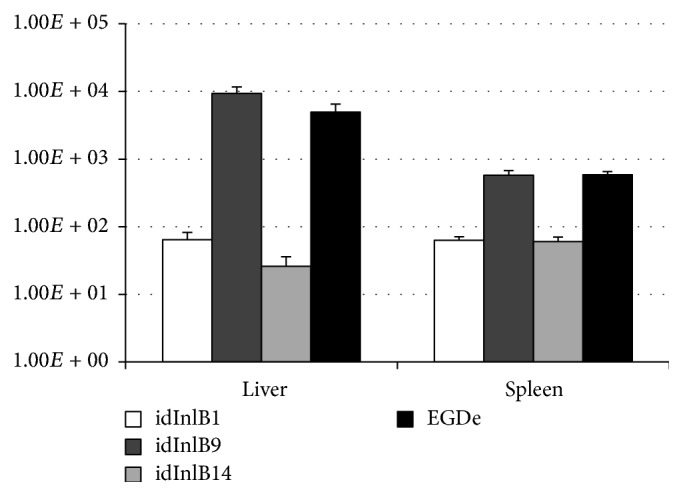
Impact of id InlB variants on severity of intraperitoneal infection in BALB/c mice. Results are shown as a relative increase of bacterial loads in the liver and spleen for the recombinant and wild-type strains comparatively with the strain EGDeΔinlB which lacked the inlB gene. The strains are designated as follows. EGDeΔinlB::InlB1, idInlB1; EGDeΔinlB::InlB9, idInlB9; EGDeΔinlB::InlB14, idInlB14; and EGDe, wild-type strain EGDe.
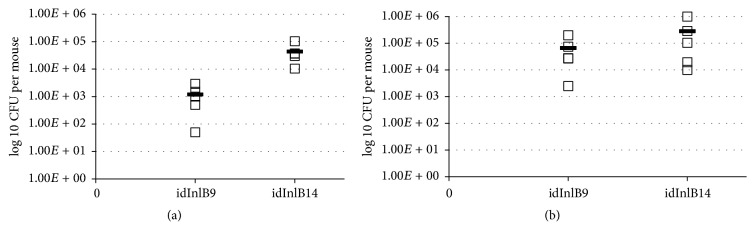
3.3. idInlB9 and idInlB14 Differentially Support Intestine Colonization
Previously, we have demonstrated that the idInlBs differed in their ability to support intragastric (i.g.) infection providing different bacterial loads in the liver but not in the spleen [25]. The idInlB14 gave rise to the highest loads in the liver 3 days after i.g. inoculation. In contrast, idInlB9, which as we demonstrated here provided high bacterial loads after i.p. injection, was low efficient in i.g. route infection [25].
The internal organ penetration after i.g. infection is not an instantaneous event as it happens after i.v. injection. Colonization of the intestine was shown to play an important role upon i.g. infection [30]. To check whether distinct idInlBs differentially affect gut colonization, EGDeΔinlB::InlB9 and EGDeΔinlB::InlB14 strains were applied i.g. in the dose of 108 CFU per mouse and the number of bacteria in the villous epithelium and Peyer's patches was counted 24 hpi. To count both intracellular and extracellular bacteria associated with the intestine, the intestine was washed thoroughly but it was not treated with gentamycin so both intracellular and surface attached bacteria could be counted. Counting of EGDeΔinlB::InlB14 bacteria was about 1,5log10 higher than EGDeΔinlB::InlB9 bacteria in Peyer's patches (Figure 3; p < 0,05). The difference between strains for villous epithelium was not so high, about 3-fold (not significant).
Figure 3.
Impact of id InlB variants 9 and 14 on colonization of the intestine. Bacterial loads in Peyer's patches (a) and villous epithelium (b) were determined for groups of 5 BALB/c mice infected intragastrically with 108 CFU of EGDeΔinlB::InlB9 (idInlB9) and EGDeΔinlB::InlB14 (idInlB14).
Taken together, obtained results demonstrated that InlB internalin domain variants differed in their ability to support infection in dependence on the route of bacterial injection under otherwise equal conditions. The idInlB14 was less effective when bacteria were injected i.v. while idInlB9 was less effective upon i.g. infection. The third variant idInlB1 demonstrated medium results in all tests.
3.4. All idInlBs Provided Invasion in Epithelial Cells
InlB promotes L. monocytogenes invasion in epithelial cells. To check whether idInlBs differ in their ability to support bacterial invasion in epithelial cells, human kidney carcinoma HEK293 cells were infected with recombinant and control bacteria (MOI 100 : 1, bacteria : cells). Relative invasion of EGDeΔinlB::InlB9 was 2,7-fold higher in HEK293 than EGDeΔinlB::InlB14 (Figure 4). However, the difference between recombinant strains was minor in comparison with invasion of the parental strain EGDeΔinlB that was about 2log10 lower for both cell lines. Therefore, all idInlBs were functional to support invasion in epithelial cells.
Figure 4.
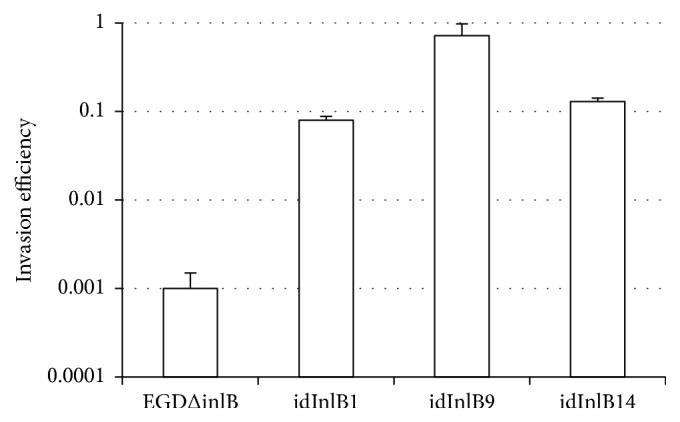
Impact of id InlB variants on invasion efficiency in human HEK293 epithelial cells. Cells were infected with MOI 100 and the invasion efficiency is shown as a ratio of intracellular bacteria to the number of bacteria used for infection. The average and SE of at least 3 independent experiments performed in duplicate are shown.
4. Discussion
Mouse models are of key importance for studies of listeriosis. L. monocytogenes strains differ considerably in their virulence [11, 12]. Moreover, an infection route affects severity of L. monocytogenes infection. The intragastric (i.g.) route is usually noticeably less effective than intravenous (i.v.) and intraperitoneal (i.p.) routes with LD50 differed by 4–6log10 [30, 31]. Still, other works reported a smaller difference within 1-2log10 or a better effectiveness of the i.g. infection when alternative L. monocytogenes strains were used [16, 17]. Isolation of L. monocytogenes from the internal organs of wild small rodents captured in the pristine habitats further supports the view that systemic L. monocytogenes infection in mice might take place via the oral route even at low doses of infection that might be expected in natural ecosystems [20, 22, 32].
Previously, we suggested that the sequence of the invasion factor InlB might be critical for L. monocytogenes spreading among small rodents in the pristine environment [21, 25]. Indeed, InlB is a key invasion factor responsible for the intestine barrier crossing in mice [19]. Among 15 InlB internalin domain variants described in L. monocytogenes isolated from different hosts, only two variants were discovered in strains isolated from wild small rodents [21, 25]. These are the InlB internalin domain variant 1 designated here the idInlB1, which was found in strains belonging to the phylogenetic lineage I, and the variant idInlB14 found in the lineage II strains. Comparison with available data established that idInlB14 was found in the widely used type strains 10403 and EGD but not in EGDe which was used in this work as a control and which carries the distinct idInlB [21, 25, 33]. Besides listed strains, idInlB14 was described by Témoin et al. in a group of strains that demonstrated low virulence in i.v. infected mice [23]. Complementation of low virulent strains with the plasmid encoding InlB from the strain EGDe improved virulence on the i.v. infection model [23]. Our data are in line with these results demonstrating that idInlB14 caused a decrease in bacterial counts after i.v. infection (see Figure 2). Meanwhile, the idInlB14 variant effectively supported the intragastric infection [25] and intestine colonization (see Figure 3).
5. Conclusions
Obtained results demonstrated that naturally occurring InlB internalin domain variants differed in their impact on the outcome of L. monocytogenes infection. The impact of distinct idInlBs was determined by the route of infection. The results supported the view that L. monocytogenes virulence on the models of intravenous and intraperitoneal mouse infection is not fully consistent with virulence on the intragastric route infection model. Particular idInlB variants might be responsible for this inconsistency.
Acknowledgments
The authors are thankful to Professor T. R. Klaenhammer and Professor J. A. Vazquez-Boland for providing the pTRKH2 vector and the L. monocytogenes EGDeΔinlB strain, respectively. The work was supported by RSF (Grant 16-15-00091).
Conflicts of Interest
The authors have no conflicts of interest to declare.
References
- 1.Vázquez-Boland J. A., Kuhn M., Berche P., et al. Listeria pathogenesis and molecular virulence determinants. Clinical Microbiology Reviews. 2001;14(3):584–640. doi: 10.1128/cmr.14.3.584-640.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Oevermann A., Zurbriggen A., Vandevelde M. Rhombencephalitis caused by listeria monocytogenes in humans and ruminants: A zoonosis on the rise? Interdisciplinary Perspectives on Infectious Diseases. 2010;2010 doi: 10.1155/2010/632513.632513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kathariou S. Listeria monocytogenes virulence and pathogenicity, a food safety perspective. Journal of Food Protection. 2002;65(11):1811–1829. doi: 10.4315/0362-028X-65.11.1811. [DOI] [PubMed] [Google Scholar]
- 4.De Noordhout C. M., Devleesschauwer B., Angulo F. J., et al. The global burden of listeriosis: a systematic review and meta-analysis. The Lancet Infectious Diseases. 2014;14(11):1073–1082. doi: 10.1016/s1473-3099(14)70870-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lecuit M. Understanding how Listeria monocytogenes targets and crosses host barriers. Clinical Microbiology and Infection. 2005;11(6):430–436. doi: 10.1111/j.1469-0691.2005.01146.x. [DOI] [PubMed] [Google Scholar]
- 6.Disson O., Lecuit M. Targeting of the central nervous system by Listeria monocytogenes. Virulence. 2012;3(2):213–221. doi: 10.4161/viru.19586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cossart P., Pizarro-Cerdá J., Lecuit M. Invasion of mammalian cells by Listeria monocytogenes: Functional mimicry to subvert cellular functions. Trends in Cell Biology. 2003;13(1):23–31. doi: 10.1016/S0962-8924(02)00006-5. [DOI] [PubMed] [Google Scholar]
- 8.Braun L., Dramsi S., Dehoux P., Bierne H., Lindahl G., Cossart P. InIB: An invasion protein of Listeria monocytogenes with a novel type of surface association. Molecular Microbiology. 1997;25(2):285–294. doi: 10.1046/j.1365-2958.1997.4621825.x. [DOI] [PubMed] [Google Scholar]
- 9.Schubert W.-D., Göbel G., Diepholz M., et al. Internalins from the human pathogen Listeria monocytogenes combine three distinct folds into a contiguous internalin domain. Journal of Molecular Biology. 2001;312(4):783–794. doi: 10.1006/jmbi.2001.4989. [DOI] [PubMed] [Google Scholar]
- 10.Shen Y., Naujokas M., Park M., Ireton K. InIB-dependent internalization of Listeria is mediated by the Met receptor tyrosine kinase. Cell. 2000;103(3):501–510. doi: 10.1016/S0092-8674(00)00141-0. [DOI] [PubMed] [Google Scholar]
- 11.Hoelzer K., Pouillot R., Dennis S. Animal models of listeriosis: A comparative review of the current state of the art and lessons learned. Veterinary Research. 2012;43(1, article no. 18) doi: 10.1186/1297-9716-43-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.D'Orazio S. E. F. Animal models for oral transmission of Listeria monocytogenes. Frontiers in Cellular and Infection Microbiology. 2014;4 doi: 10.3389/fcimb.2014.00015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Disson O., Nikitas G., Grayo S., Dussurget O., Cossart P., Lecuit M. Modeling human listeriosis in natural and genetically engineered animals. Nature Protocols. 2009;4(6):799–810. doi: 10.1038/nprot.2009.66. [DOI] [PubMed] [Google Scholar]
- 14.Cabanes D., Lecuit M., Cossart P. Animal models of Listeria infection. Current Protocols in Microbiology. 2008 doi: 10.1002/9780471729259.mc09b01s10. [DOI] [PubMed] [Google Scholar]
- 15.Ghanem E. N. B., Myers-Morales T., D’Orazio S. E. F. A mouse model of foodborne Listeria monocytogenes infection. Current Protocols in Microbiology. 2013 doi: 10.1002/9780471729259.mc09b03s31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pine L., Malcolm G. B., Plikaytis B. D. Listeria monocytogenes intragastric and intraperitoneal approximate 50% lethal doses for mice are comparable, but death occurs earlier by intragastric feeding. Infection and Immunity. 1990;58(9):2940–2945. doi: 10.1128/iai.58.9.2940-2945.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Barbour A. H., Rampling A., Hormaeche C. E. Comparison of the infectivity of isolates of Listeria monocytogenes following intragastric and intravenous inoculation in mice. Microbial Pathogenesis. 1996;20(4):247–253. doi: 10.1006/mpat.1996.0023. [DOI] [PubMed] [Google Scholar]
- 18.Pentecost M., Kumaran J., Ghosh P., Amieva M. R. Listeria monocytogenes internalin B activates junctional endocytosis to accelerate intestinal invasion. PLoS Pathogens. 2010;6(5):1–15. doi: 10.1371/journal.ppat.1000900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chiba S., Nagai T., Hayashi T., Baba Y., Nagai S., Koyasu S. Listerial invasion protein internalin B promotes entry into ileal Peyer's patches in vivo. Microbiology and Immunology. 2011;55(2):123–129. doi: 10.1111/j.1348-0421.2010.00292.x. [DOI] [PubMed] [Google Scholar]
- 20.Zaytseva E., Ermolaeva S., Somov G. P. Low genetic diversity and epidemiological significance of Listeria monocytogenes isolated from wild animals in the far east of Russia. Infection, Genetics and Evolution. 2007;7(6):736–742. doi: 10.1016/j.meegid.2007.07.006. [DOI] [PubMed] [Google Scholar]
- 21.Adgamov R., Zaytseva E., Thiberge J.-M., Brisse S., Ermolaeva S. Genetic Diversity in Microorganisms. In: Caliskan M., editor. InTEchOpen; 2012. pp. 235–250. [Google Scholar]
- 22.Voronina O. L., Ryzhova N. N., Kunda M. S., Kurnaeva M. A., Semenov A. N., Aksenova E. I. Diversity and pathogenic potential of listeria monocytogenes isolated from environmental sources in the russian federation. International Journal Of Modern Engineering Research. 2015;5:5–15. [Google Scholar]
- 23.Témoin S., Roche S. M., Grépinet O., Fardini Y., Velge P. Multiple point mutations in virulence genes explain the low virulence of Listeria monocytogenes field strains. Microbiology. 2008;154(3):939–948. doi: 10.1099/mic.0.2007/011106-0. [DOI] [PubMed] [Google Scholar]
- 24.Alonzo F., III, Bobo L. D., Skiest D. J., Freitag N. E. Evidence for subpopulations of Listeria monocytogenes with enhanced invasion of cardiac cells. Journal of Medical Microbiology. 2011;60(4):423–434. doi: 10.1099/jmm.0.027185-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sobyanin K., Sysolyatina E., Krivozubov M., Chalenko Y., Karyagina A., Ermolaeva S. Naturally occurring InlB variants that support intragastric Listeria monocytogenes infection in mice. FEMS Microbiology Letters. 2017 doi: 10.1093/femsle/fnx011. [DOI] [PubMed] [Google Scholar]
- 26.Ermolaeva S., Belyi Y., Tartakovskii I. Characteristics of induction of virulence factor expression by activated charcoal in Listeria monocytogenes. FEMS Microbiology Letters. 1999;174(1):137–141. doi: 10.1016/S0378-1097(99)00132-9. doi: 10.1016/S0378-1097(99)00132-9. [DOI] [PubMed] [Google Scholar]
- 27.Glaser P., Frangeul L., Buchrieser C., Rusniok C., Amend A., Baquero F. Comparative genomics of Listeria species. Science. 2001;294:849–852. doi: 10.1126/science.1063447. [DOI] [PubMed] [Google Scholar]
- 28.Kibardin A., Karpova T., Sapenko T., Vazquez-Boland J. A., Kiselev S., Ermolaeva S. Mammalian peptidoglycan recognition protein TagL inhibits Listeria monocytogenes invasion into epithelial cells. FEMS Immunology & Medical Microbiology. 2006;46(2):284–290. doi: 10.1111/j.1574-695X.2005.00038.x. [DOI] [PubMed] [Google Scholar]
- 29.O'Sullivan D. J., Klaenhammer T. R. High- and low-copy-number Lactococcus shuttle cloning vectors with features for clone screening. Gene. 1993;137(2):227–231. doi: 10.1016/0378-1119(93)90011-Q. [DOI] [PubMed] [Google Scholar]
- 30.Melton-Witt J. A., Rafelski S. M., Portnoy D. A., Bakardjiev A. I. Oral infection with signature-tagged Listeria monocytogenes reveals organ-specific growth and dissemination routes in guinea pigs. Infection and Immunity. 2012;80(2):720–732. doi: 10.1128/IAI.05958-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lecuit M., Vandormael-Pournin S., Lefort J., et al. A transgenic model for listeriosis: Role of internalin in crossing the intestinal barrier. Science. 2001;292(5522):1722–1725. doi: 10.1126/science.1059852. [DOI] [PubMed] [Google Scholar]
- 32.Gray M. L., Killinger A. H. Listeria monocytogenes and listeric infections. Bacteriological Reviews. 1966;30(2):309–382. doi: 10.1128/br.30.2.309-382.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bécavin C., Bouchier C., Lechat P., et al. Comparison of widely used Listeria monocytogenes strains EGD, 10403S, and EGD-e highlights genomic differences underlying variations in pathogenicity. mBio. 2014;5(2) doi: 10.1128/mBio.00969-14.e00969-14 [DOI] [PMC free article] [PubMed] [Google Scholar]