Abstract
The chemokine CCL17, mainly produced by dendritic cells (DCs) in the immune system, is involved in the pathogenesis of various inflammatory diseases. As a ligand of CCR4, CCL17 induces chemotaxis and facilitates T cell-DC interactions. We report the identification of two novel RNA aptamers, which were validated in vitro and in vivo for their capability to neutralize CCL17. Both aptamers efficiently inhibited the directed migration of the CCR4+ lymphoma line BW5147.3 toward CCL17 in a dose-dependent manner. To study the effect of these aptamers in vivo, we used a murine model of contact hypersensitivity. Systemic application of the aptamers significantly prevented ear swelling and T cell infiltration into the ears of sensitized mice after challenge with the contact sensitizer. The results of this proof-of-principle study establish aptamers as potent inhibitors of CCL17-mediated chemotaxis. Potentially, CCL17-specific aptamers may be used therapeutically in humans to treat or prevent allergic and inflammatory diseases.
Keywords: CCL17, TARC, CCL22, chemokine, aptamer, SELEX, contact hypersensitivity, allergy, cell migration, skin homing
Graphical Abstract
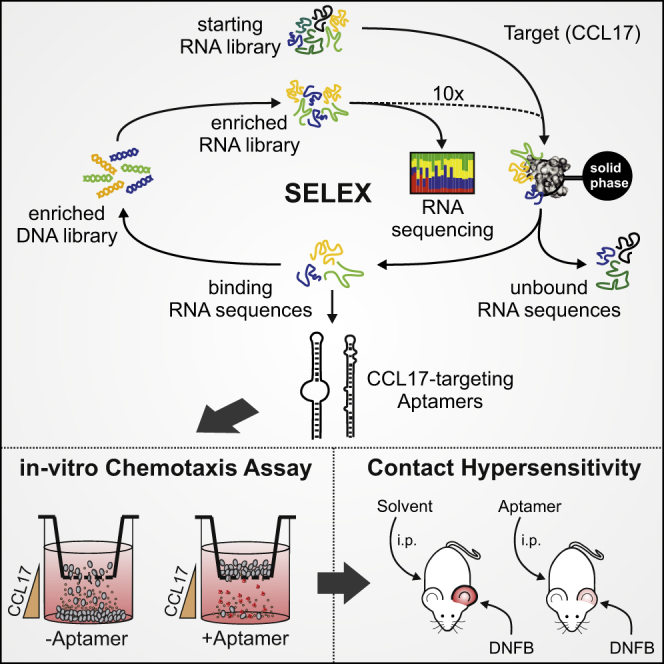
The chemokine CCL17 promotes T cell attraction to peripheral tissues and thereby exacerbates inflammatory reactions. Using SELEX, Fülle et al. selected high-affinity aptamers, which neutralize CCL17 in murine contact hypersensitivity. Because of their small size and structural properties, such aptamers may represent new versatile drugs for allergic skin diseases.
Introduction
Chemokines are essential to direct appropriate trafficking of leukocytes within the organism during development, homeostasis, and the orchestration of immune responses. In addition to cell migration, they promote integrin-dependent cell adhesion and confer activation signals. Because chemokines and their receptors are also crucial mediators of inflammatory diseases, they represent promising targets for therapeutic approaches.1 Here, we describe the identification of RNA aptamers able to neutralize the chemokine CCL17 with the intention to develop a new class of drugs for treatment of allergic and inflammatory diseases.
The chemokine superfamily comprises 48 or 40 small chemotactic proteins in human and mouse, respectively, which interact with 1 or several of the 20 known chemokine receptors belonging to the G-protein-coupled seven-transmembrane receptor family.2, 3 Because several chemokines may bind to the same receptor and some chemokines stimulate more than one chemokine receptor, there is a significant promiscuity in this system. This applies particularly to inflammatory chemokines, whereas homeostatic chemokines are evolutionarily conserved and less redundant.3 The chemokine CCL17 (formerly called thymus and activation regulated chemokine [TARC]), a ligand of CCR4, was originally identified in the thymus4 and later detected in a subset of conventional dendritic cells (cDCs) in the gut-associated lymphoid tissue, as well as in inflamed skin and lymph nodes (LNs).5, 6 CCL17 not only facilitates T cell-DC interactions,7 but also enables emigration of Langerhans cells (LCs) from the skin by enhancing the responsiveness to CCR7 and CXCR4 ligands.8 In addition, CCL17 is produced by alternatively activated macrophages together with CCL22, the other known CCR4 ligand.9 CCL17 and CCL22 have dual functions, acting as homeostatic as well as inflammatory chemokines.3, 10 In mouse models, CCL17 was shown to promote various allergic and inflammatory diseases, such as contact hypersensitivity (CHS),6, 11 atopic dermatitis (AD),8, 12 inflammatory bowel disease,13 and atherosclerosis.14 In humans, CCL17 serves as a biomarker of atopic diseases because serum levels of CCL17 are strongly enhanced in patients suffering from atopic dermatitis, allergic rhinitis, or asthma.15, 16, 17 In addition, CCL17 serves as a valuable biomarker for treatment response in patients suffering from Hodgkin’s lymphoma.18 Recently, CCL17 was also shown to promote inflammatory pain in mouse models of collagen- or GM-CSF-induced arthritis in a prostaglandin-dependent manner.19
In contrast to the pro-inflammatory action of CCL17, data reported in the literature associate CCL22 mainly with the recruitment of regulatory T cells (Tregs) and inhibition of anti-tumor immune responses.20, 21 Recently, CCL17 and CCL22 were demonstrated to transduce distinct signaling outputs via CCR4, in particular regarding receptor internalization and β-arrestin binding, a phenomenon called biased agonism.1, 22, 23 The two CCR4 ligands also appear to bind to different regions of the CCR4 protein.21 Thus, it is possible that CCR4 antagonists differentially affect the function of CCL17 and CCL22 depending on the site of receptor interaction.22
In this study, we used the aptamer technology to identify novel CCL17-neutralizing nucleic acid molecules. Aptamers are short, synthetic RNA or DNA oligonucleotides, which can be selected to specifically bind proteins and inhibit their biological functions.24, 25 An aptamer specific for the vascular endothelial growth factor (pegaptanib)26 has been successfully used in the clinic for treatment of the wet form of age-related macular degeneration.27 In addition, several other aptamers are currently in clinical trials,28, 29 and Spiegelmers (L-RNA aptamers) specific for the chemokines CXCL12 or CCL2 were described for potential treatment of chronic lymphatic leukemia or diabetic nephropathy, respectively.30, 31 To isolate aptamers with specificity for CCL17, we employed a SELEX (systematic evolution of ligands by exponential enrichment) procedure using a 2′-deoxy-2′-fluoro pyrimidine-bearing RNA library.32 We identified two CCL17-specific aptamers that efficiently inhibit cell migration in vitro and in vivo. Thus, we validated that the application of aptamer-based inhibitors of CCL17 represents a promising strategy for systemic or local prevention of allergic and inflammatory reactions.
Results
The Identification of CCL17-Binding Aptamers
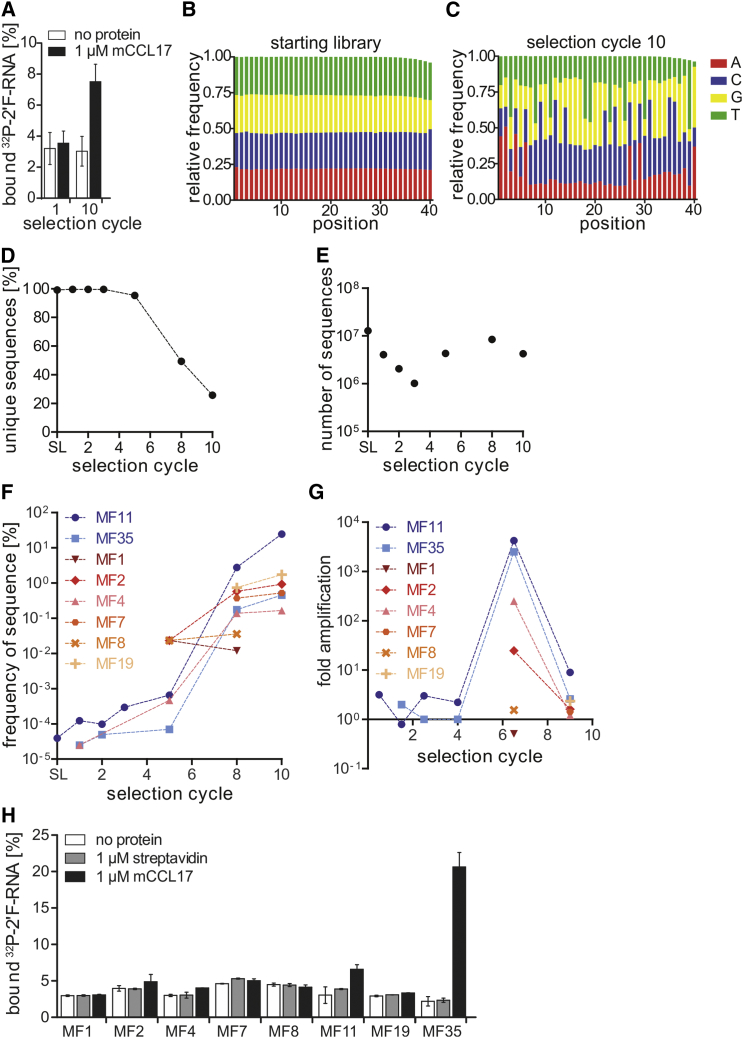
Aptamers binding to CCL17 were identified by SELEX. As nucleic acid library we employed a 2′-deoxy-2′-fluoro pyrimidine-modified RNA (2′F-RNA) library.32 This type of nucleic acid library has an enhanced stability toward nuclease degradation; thus, aptamers derived from these can be applied in vivo without laborious stabilization efforts.33 We performed 10 selection cycles with increasing stringency (Table S1) and analyzed the obtained library for CCL17 binding by filter retention analysis. This analysis revealed an increased binding of the RNA library after 10 selection cycles compared with the library from the first selection cycle (Figure 1A). The enriched library was cloned, and the containing sequences were analyzed by Sanger sequencing (Table S2). These data revealed that the library from selection cycle 10 is heterogeneous. We subsequently performed next-generation sequencing (NGS) to get an in-depth view on the population of the starting library and the libraries obtained from selection cycles 1, 2, 3, 5, 8, and 10. We analyzed the frequency of each sequence found by Sanger sequencing (Table S2). Furthermore, the distribution of the four nucleotides in the random region of the sequenced libraries was calculated (Figures 1B and 1C; Figure S1). The NGS data revealed that the enrichment of CCL17 binding RNA species occurred after selection cycle 5, whereas a shift in nucleotide distribution can be observed in selection cycle 8 and became more pronounced in selection cycle 10 (Figures 1B and 1C; Figure S1). In line, the number of unique sequences starts to decline in selection cycle 5 but is still high in selection cycle 10 (>25%) (Figure 1D). The total number of sequence reads, however, stays constant between 1 and 10 million for every selection cycle (Figure 1E). The strongest enrichment of CCL17 binding sequences was detected between selection cycles 5 and 8 (Figures 1F and 1G). We have chosen some of the most abundant sequences and performed a filter retention analysis to determine CCL17 binding species. These data revealed MF11 and MF35 as putative aptamers, whereas all other sequences tested were found not to interact with CCL17 (Figure 1H). Noteworthy is that MF11 represents the most dominant sequence in the libraries from selection cycles 8 and 10, whereas MF35 has a similar frequency as other sequences that have been found not to bind to CCL17. This enrichment profile indicates that the selection conditions may not have been optimal. Nevertheless, the combination of NGS and filter retention analyses allowed the assignment of novel aptamers that recognize CCL17. Analysis of NGS data was accomplished with the software tool COMPAS34, 35 (see also Supplemental Information).
Figure 1.
Filter Retention Assay and Next-Generation Sequencing Results of the CCL17 Selection
(A) 2′F-RNA selection cycles 1 and 10 were analyzed by radioactive filter retention assay (n = 3, mean ± SD). (B and C) Nucleotide distribution at the different positions of the random region in the starting library (B) and the final selection cycle 10 (C). (D) Frequency of unique sequences in all selection cycles and the starting library. The frequency was calculated by dividing the overall number of sequences by the number of unique sequences. (E) Number of sequence reads in the next-generation sequencing analysis per selection cycle and starting library. (F) Frequency of binding and non-binding sequences in all selection cycles and the starting library. Missing data points indicate that the sequence could not be detected in the next-generation sequencing data of the respective round. Blue sequences are binding ones; non-binding sequences are depicted in shades of red and yellow. (G) Fold amplification of binding and non-binding sequences at different stages during the SELEX procedure. Calculation of the fold amplification was performed by dividing the frequency of the sequence of interest in the respective selection cycle by its frequency in the previous cycle. The data points are depicted on the x axis in the middle of the two selection cycles from which the fold amplification value was calculated. Missing data points indicate that the sequence could not be detected in the next-generation sequencing data of the respective round. (H) 2′F-RNA sequences identified by Sanger sequencing were analyzed by radioactive filter retention assay (n = 2–3, mean ± SD).
MF11 and MF35 Specifically Inhibit CCL17-Dependent Cell Migration
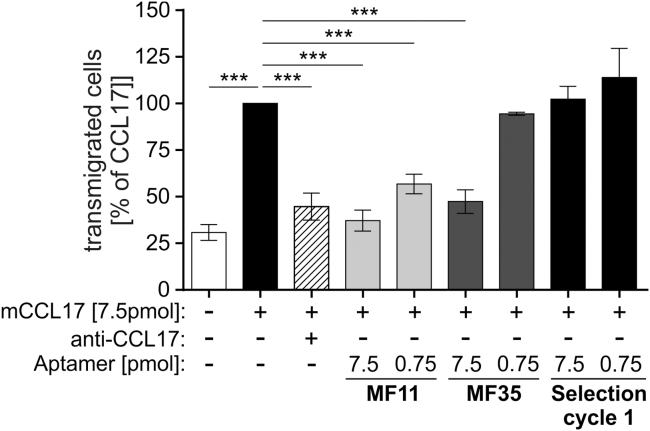
To test whether MF11 and MF35 are able to inhibit the chemotactic activity of CCL17, we employed an in vitro transwell migration assay using the thymoma line BW5147.3, which expresses CCR4 and CCR8, and shows chemotactic migration toward CCL17.5 For optimization of the assay system, we first titrated the minimal concentration of recombinant murine CCL17 (mCCL17) required to achieve a robust migration of BW5147.3 cells. At a concentration of 100 ng/mL mCCL17, equivalent to 7.5 pmol mCCL17/well, migration of the cells was somewhat more than half maximal (Figure S2A). For all further in vitro transwell assays, this concentration was chosen to achieve a high sensitivity of the assay for inhibition of CCL17-dependent chemotaxis. As an internal standard, the number of cells that migrated toward mCCL17 without any further additions was set to 100%. Medium only was used as negative control. As a positive control for CCL17 inhibition, we first employed a commercially available CCL17-specific neutralizing antibody at a molar ratio of 2:1 or 1:1 (antibody/mCCL17), which was added to the lower chamber together with mCCL17 (Figure S2B). The antibody inhibited the CCL17-dependent cell migration significantly at a 1:1 molar ratio, and no cell migration was detectable at a 2:1 ratio. To test the inhibitory capacity of aptamers MF11 and MF35, they were first added at molar ratios of 1:1 (7.5 pmol/well) and at an aptamer/CCL17 ratio of 1:10 (0.75 pmol/well), and compared with the non-enriched library from selection cycle 1 of the SELEX procedure. Both MF11 and MF35 significantly inhibited transmigration of the cells at doses of 7.5 pmol/well, and MF11 even showed significant inhibition at 0.75 pmol/well (Figure 2). Thus, the two newly identified CCL17-specific aptamers efficiently inhibited CCL17-dependent cell migration.
Figure 2.
Aptamer-Dependent Inhibition of CCL17-Dependent Cell Migration In Vitro
Migration of BW5147.3 cells toward 100 ng/mL (7.5 pmol/well) mCCL17 was measured in a transwell system. The full-size and unmodified aptamers MF11 and MF35, as well as the RNA pool from selection cycle 1, were tested at equimolar concentrations (7.5 pmol) and at an aptamer/CCL17 ratio of 1:10 (0.75 pmol). As a control, 7.5 pmol of an anti-CCL17 monoclonal antibody (1.88 μg/mL) was added to the lower compartment. After 2 hr, transmigrated cells in the lower compartment were counted and calculated as percent of CCL17 alone (n = 3–5, mean ± SEM). Statistical significance was tested by one-way ANOVA with Bonferroni post hoc test for multiple comparisons (***p < 0.001).
Truncation and Modification of the Aptamers
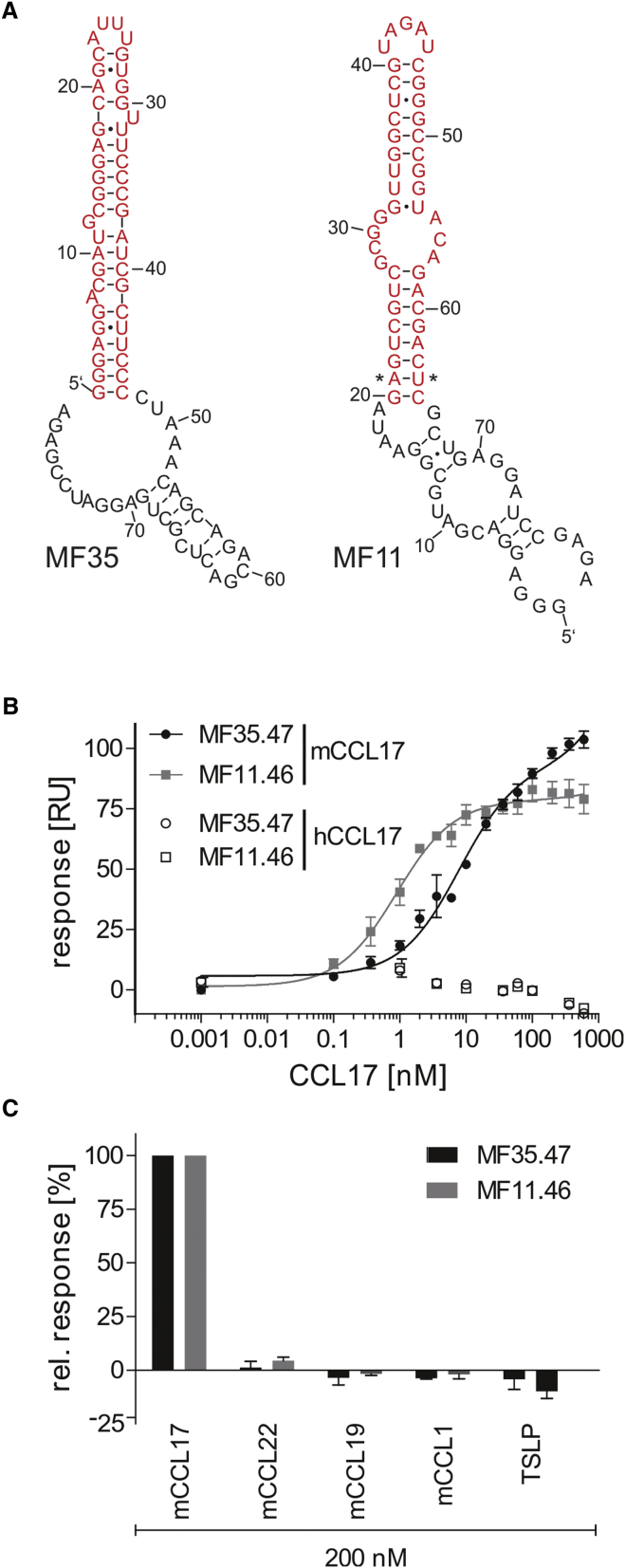
MF11 and MF35 were truncated to facilitate chemical synthesis of the aptamers for in vivo applications. Based on the prediction of the secondary structure of both aptamers by mfold,36 we shortened both aptamers accordingly to keep the extended hairpin structure (Figure 3A). This truncation yielded MF11.46 and MF35.47, both of which bear less than 50 nucleotides and, thus, are suitable for solid-phase synthesis. The truncated variants were analyzed for the interaction with mCCL17 using surface plasmon resonance (SPR). To achieve this, we immobilized biotinylated versions of both aptamers on the sensor surface and injected increasing concentrations of mCCL17. As control, we immobilized a non-binding scrambled version of MF35.47, named control aptamer in the following, and recorded the difference in response between the aptamer and the control. The response values of the equilibrium phase were then plotted against the injected concentration to determine the respective dissociation constant. These experiments revealed dissociation constants of 0.9 ± 0.1 and 7.5 ± 1.5 nM for MF11.46 and MF35.47, respectively (Figure 3B). The same experiment was done using human CCL17 (hCCL17), but no concentration-dependent interaction was observed (Figure 3B). Further, we investigated a series of other chemokines at concentrations of 200 nM, 27- to 200-fold higher than the observed KD values for mCCL17. To these chemokines, no binding was detectable, indicating a high specificity of the two aptamers (Figure 3C).
Figure 3.
Aptamer Sequences and Surface Plasmon Resonance Analysis of Aptamer Affinity and Specificity
(A) Structures and sequences of MF35 and MF11, as predicted by mfold.36 The truncated versions of the aptamers are depicted in red. The asterisks (*) in MF11 point out a base pair that has been exchanged to GC in MF11.46 to facilitate in vitro transcription. (B) Surface plasmon resonance measurement using CCL17 as analyte and 2′F-RNA aptamers as immobilized ligands. MF35.47 and MF11.46 bind to murine CCL17 with a dissociation constant (KD) of 7.5 ± 1.5 and 0.9 ± 0.1 nM, respectively (n = 2, mean ± SD). (C) Surface plasmon resonance measurement using chemokines as analytes and 2′F-RNA aptamers as immobilized ligands. 200 nM (∼27- to 200-fold of KD) of different chemokines was tested for their binding to MF35.47 and MF11.46 (n = 2, mean ± SD).
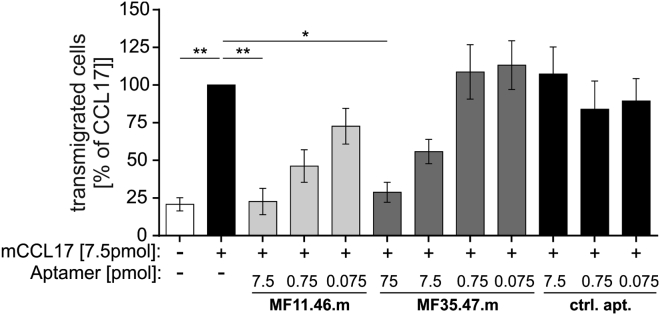
It has been shown that the half-life of aptamers in vivo can be enhanced by the addition of polyethyleneglycol (PEG) tails and cap structures at the 5′- and 3′- terminal ends. These groups supposedly prolong aptamer resistance toward exonucleases and the rapid renal clearance.33 Therefore, both aptamers were modified with a 3′-dT-cap structure and a 20 kDa PEG tail at the 5′ end (MF11.46.m, MF35.47.m) (Figure S3). To ascertain that the modified aptamers were still functional, they were tested in the transwell assay in comparison with the equally modified, non-binding control aptamer at doses of 7.5, 0.75, and 0.075 pmol/well. The truncated and modified versions of both aptamers inhibited CCL17-dependent chemotaxis of the BW5147.3 cells similarly as observed for the parental full-length aptamers (Figure 4). We also determined the IC50 for both modified aptamers in direct comparison with the CCL17-neutralizing antibody MAB529 using the same transwell assay (Figure S4). Whereas MF35.47 and MAB529 had a very similar IC50 with 2.9 and 3.3 pmol, respectively, that of MF11.46.m was about 8-fold lower (0.42 pmol), indicating a very high efficiency of MF11.46.m in vitro. In addition, we determined the serum stability of the non-modified and modified aptamers, demonstrating that all were stable for at least 10 days in human serum, whereas a reduction of RNA was observed in mouse serum over this time period. In addition, the modification slightly increased the stability of both aptamers in mouse serum after 2–3 days (Figure S5).
Figure 4.
MF11.46.m and MF35.47.m Efficiently Inhibit CCL17-Dependent Cell Migration In Vitro
The ability of the shortened and modified (5′-PEGylated, 3′-capped) aptamers to inhibit CCL17-dependent migration of BW5147.3 was measured in a transwell system. MF11.46.m and MF35.47.m were tested at a molar ratio of 1:1 (7.5 pmol), 1:10 (0.75 pmol), and 1:100 (0.075 pmol) (aptamer:mCCL17). An equally modified scrambled control aptamer served as a negative control. After 2 hr, cells in the lower compartment were counted and calculated as percent of migration toward CCL17 alone (n = 3, mean ± SEM). Statistical significance was tested by one-way ANOVA with Bonferroni post hoc test for multiple comparisons (**p < 0.01; *p < 0.05).
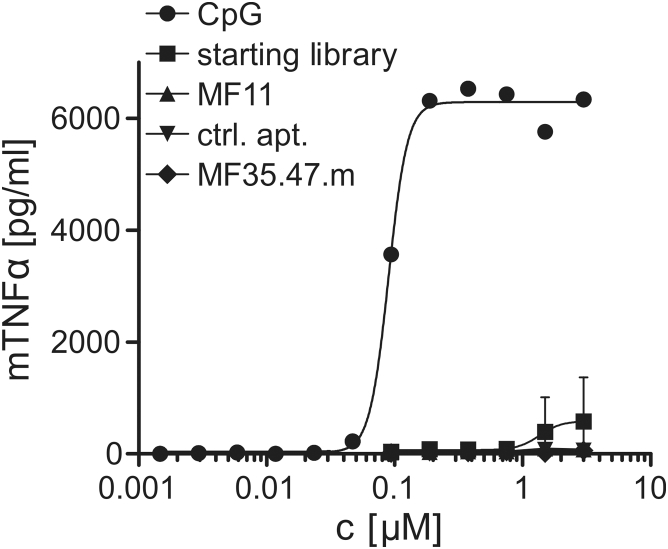
A general concern regarding the use of DNA or RNA oligonucleotides for application in vivo is their potential ability to activate cell surface or intracellular pattern recognition receptors (PRRs), such as Toll-like receptors (TLRs) 3, 7, and 9 or retinoic acid inducible gene I (RIG-I)-like receptors.37, 38 To test for such immunostimulatory activity of the CCL17-specific aptamers, we added the starting library, the full-length MF11, MF35.47.m, and the modified scrambled control aptamer to cultures of immortalized murine embryonic stem cell-derived macrophages at concentrations of 0.094 to 3 μM and compared with immunostimulatory CpG DNA oligonucleotides (1.46 nM to 3 μM range). As readout for PRR activation, the concentration of tumor necrosis factor alpha (TNF-α) in the supernatant was measured after 24 hr. It showed a minor TNF-α release after treatment of the cells with the starting library, but very low to no secretion upon addition of the aptamers MF11 and MF35.47.m, or the control aptamer (Figure 5). Therefore, the CCL17-specific aptamers do not appear to be immunostimulatory.
Figure 5.
MF11.46.m and MF35.47.m Do Not Induce TNF-α Secretion in Macrophages
Murine embryonic stem cell-derived macrophages were incubated with increasing concentrations of TLR9 ligand CpG oligodeoxynucleotide (ODN) 1826 type B, the starting library, or aptamers MF11, MF35.47.m, and control aptamer for 24 hr. TNF-α levels in the supernatants were determined by homogeneous time-resolved fluorescence (HTRF) assay (n = 4, mean ± SD).
Establishment of an In Vivo Assay System for CCL17-Dependent Cell Migration
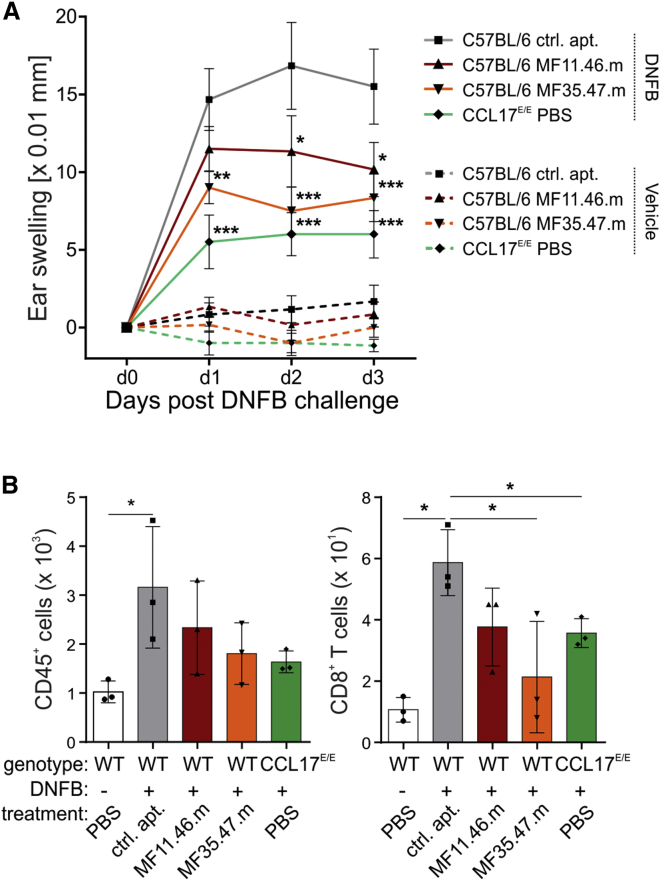
We next aimed to demonstrate the effectiveness of the aptamers to inhibit CCL17 function in vivo. As demonstrated previously, CCL17-deficient (CCL17E/E) mice are protected against CHS induced by classical contact sensitizers, such as 2,4-dinitro-1-fluorobenzene (DNFB).6 In this short-term assay, mice are exposed epicutaneously to DNFB at the belly to sensitize the T cell response and are several days later challenged with DNFB on the ear.39 Here, we measured the ear-swelling response of primed wild-type (WT) and CCL17E/E mice challenged on the right ear with DNFB on days 1, 2, and 3 after challenge (Figure S6A). As a control, the swelling of the left ear of each animal that was exposed to the vehicle (acetone/olive oil) only is depicted. We could confirm that the CHS response of CCL17E/E mice to DNFB is significantly reduced compared with WT mice on days 1 and 2 after challenge. In addition, flow cytometric analysis of DNFB-treated ears at day 4 after challenge revealed significantly reduced numbers of infiltrating CD45+ leukocytes and CD8+ T cells in CCL17E/E mice compared with WT mice (Figures S6B, left and right, respectively, and S7). Based on this, we tested whether systemic treatment of the mice with the in vitro validated CCL17-specific neutralizing antibody either immediately before sensitization or before challenge inhibits the ear-swelling response. For this purpose, mice received 200 μg of anti-CCL17 or isotype control antibody at day −5 or day 0 of the CHS protocol. Only if the antibodies were injected at the time of elicitation did we observe a notable reduction in the ear-swelling response, although this was not statistically significant (Figure S8). We therefore decided to test the efficacy of the CCL17-specific aptamers during the elicitation phase rather than the earlier phase of sensitization. Because the half-life of the aptamers is likely to be much shorter than that of the antibodies, we performed two injections of 5 nmol aptamer/mouse 1 hr before and 12 hr after challenge with DNFB. As depicted in Figure 6A, both CCL17-specific aptamers significantly inhibited the ear-swelling response compared with the control aptamer, with MF35.47.m being even more efficient than MF11.46.m. This was somewhat unexpected because MF11.46.m inhibited the in vitro migratory response with an approximately 10-fold higher efficiency than MF35.47.m. As an additional readout for the CHS response, we also analyzed the leukocytic infiltrate in the ears by flow cytometry at day 4 after challenge. Interestingly, we noticed a clear reduction in the number of CD45+ leukocytes in the ears of the MF11.46.m- and MF35.47.m-treated mice compared with ears of mice treated with the control aptamer (Figure 6B, left). This reduction was almost as strong as that observed in CCL17E/E mice. With regard to the infiltration of CD8+ T cells, the main mediators of the CHS response,40 treatment with MF11.46.m resulted in a partial and MF35.47.m in a significant reduction of the CD8+ T cell number that was as pronounced as that observed in the CCL17-deficient mice (Figure 6B, right).
Figure 6.
Reduced Ear Swelling and Leukocyte Infiltration into the Ears following Application of CCL17-Specific Aptamers
C57BL/6 WT and CCL17E/E mice were sensitized with DNFB on day −8 and day −7. On day 0, WT mice were injected i.p. with 5 nmol MF11.46.m and MF35.47.m, or the control aptamer 1 hr before and 12 hr after DNFB challenge. CCL17E/E mice received PBS only. (A) Ear swelling of WT and CCL17E/E mice 24 (day 1), 48 (day 2), and 72 hr (day 3) after application of DNFB (solid lines) or vehicle (dashed lines) (n = 6 per group, mean ± SEM). One representative of three experiments is shown. Data were tested for statistical significance by two-way ANOVA with Bonferroni post hoc test for multiple comparisons (***p < 0.001). (B) Flow cytometric analysis of the immune cell infiltrate. At day 4, ears were digested and the isolated cells subjected to staining for flow cytometry. Absolute numbers of CD45+ and CD3+CD8+ cells were determined by flow cytometry (n = 3 per group, mean ± SD). Data were tested for statistical significance by Student’s t test compared with the control aptamer-treated group (*p < 0.05).
To obtain a better estimate of the dose of aptamer required for efficient inhibition of the CHS response, we also tested the efficacy of applying two injections of 1 nmol aptamer using the same experimental setup as in Figure 6. Although some inhibition of ear swelling could be observed, the differences were not significant compared with the control aptamer (Figure S9). Using the shorter protocol, sensitizing mice at days −5 and −4, we also tested MF35.47.m for inhibition of the ear-swelling response at doses of 1 and 10 nmol/injection (two injections/mouse: 1 hr before and 12 hr after challenge). Again, at 1 nmol/injection, only a slight, non-significant inhibition was observed, whereas injection of 10 nmol resulted in a significant inhibition that was now similar to that seen in CCL17E/E mice (Figures S10A and S10B). In conclusion, MF35.47.m, and to a lesser extent MF11.46.m, proved to be highly efficient in blocking CCL17 in vivo.
Discussion
The selective blockade of chemokine-chemokine receptor interactions is considered a very promising approach for the inhibition of inflammatory or allergic responses, as well as for cancer therapy. Because of the large promiscuity within the chemokine family,3 chemokine receptors often appear to be the more suitable targets for drug development. In the case of CCR4, both small-molecule inhibitors41 and neutralizing monoclonal antibodies have been generated and tested in clinical trials.17, 42, 43, 44 In line with previous findings in mice that CCR4 is expressed on Tregs,45 and that anti-CCL22 antibodies reduced Treg recruitment into ovarian tumors,20 the humanized CCR4-specific monoclonal antibodies mogamulizumab and mAb156743, 44 displayed anti-tumor activity.17, 42 Furthermore, some patients treated with mogamulizumab developed a severe skin rash, which has also been attributed to the depletion of Tregs.17, 46 This effect strongly contrasts with the fact that CCL17 is a well-known biomarker of atopic diseases, and that deficiency of CCL17 prevents allergic skin inflammation and many other inflammatory diseases in mouse models.6, 8, 13, 14, 19 Recently, the phenomenon of biased agonism, based on differential activation of arrestin signaling, was reported for the two CCR4 ligands (for review, see Solari and Pease1), indicating that blockade or deficiency of CCR4 is likely to have a different outcome than selective inhibition of either CCL17 or CCL22. Based on the data available from CCL17-deficient mice, we hypothesize that CCL17 exerts a pro-inflammatory function, whereas CCL22 may be mainly responsible for Treg recruitment and feedback inhibition of CCR4 activity. Supporting this theory and in line with the phenotypic changes in CCL17-deficient mice,6, 8, 13, 14, 19 CCL17-specific neutralizing antibodies were able to inhibit atherosclerosis in a mouse model.14 In contrast, simultaneous application of anti-CCL17 and anti-CCL22 antibodies during the elicitation phase of CHS did not inhibit the ear-swelling response47 or attraction of activated T cells to the skin.48 In line, CCR4-knockout mice develop enhanced CHS responses,49 and CCR4-deficient T cells showed no impairment in skin homing, whereas additional treatment with CCL27-specific antibodies significantly inhibited skin homing in a CHS model, indicating a cooperative function of CCR4 and CCR10.50 In addition to the possibility of biased signaling through CCR4, it can also not be excluded at present that CCL17 interacts with an additional, as yet unknown chemokine receptor.
To specifically target CCL17 as a pro-inflammatory chemokine, we here aimed to explore whether high-affinity RNA aptamers with specificity for CCL17 can be identified and are able to efficiently inhibit allergic skin reactions in vivo. In comparison with neutralizing antibodies, aptamers are more amenable for GMP production because they can be chemically synthesized and modified without the need for large-scale cell culture.29 Furthermore, aptamers with an appropriately modified ribonucleotide backbone are resistant to nuclease degradation and immunologically inert. Thus, the half-life of aptamers for therapeutic application, such as pegaptanib or others currently used in clinical trials, is between 12 hr and 10 days.33 In particular, with regard to inhibition of inflammatory reactions in the skin, the smaller size and secondary structure of aptamers may facilitate their entry into the dermal and epidermal compartment, and potentially also permit epicutaneous application. As demonstrated in this study, the aptamers MF11.46 and MF35.47 possessed a very high affinity for murine CCL17 of 0.9 and 7.5 nM, respectively. In addition, both aptamers very efficiently inhibited the chemotactic activity of mCCL17 in the in vitro transwell assay. Here, MF11.46 showed good inhibition at a molarity 10-fold lower than that of mCCL17 itself, indicating a so far unexplained dominant-negative activity of this aptamer. In the in vivo CHS assay, however, MF11.46 unexpectedly turned out to be less efficient than MF35.47. This may be explained by a shorter in vivo half-life of MF11.46 or a potential absorbance of MF11.46 by certain tissues in the intact organism. It should be noted in this context that MF11 was the most strongly enriched sequence during the SELEX as determined by NGS, presumably because of its very high affinity for mCCL17. Nevertheless, the less abundant sequence MF35 also identified by NGS later turned out to be more efficient than MF11 under in vivo conditions.
CHS represents a well-studied mouse model of human allergic contact dermatitis, which is induced by epicutaneous application of contact sensitizers, such as DNFB.39, 51 During the sensitization phase, the innate immune system is activated through release of danger-associated molecular patterns, and in addition, endogenous peptides are chemically modified by the contact sensitizer inducing an adaptive immune response.51 Interferon-γ-producing antigen-specific CD8+ T cells were shown to be the main mediators of CHS.39 Recent work by Natsuaki et al.52 demonstrated the existence of perivascular clusters of macrophages and DCs in the skin, to which CD8+ T cells were attracted after DNFB challenge in a chemokine-dependent manner. Because the influx of CD8+ T cells to the inflamed ears was specifically reduced after treatment with the CCL17-specific aptamers described in this study, it is possible that they inhibit this important step of extravasation and DC-T cell interactions.
After this proof-of-principle demonstration of the efficiency of CCL17-specific aptamers for inhibition of CHS reactions during the elicitation phase, future experiments will be designed to assess the duration of inhibition, the possibility to achieve inhibition by an epicutaneous application regimen, and the potency of the aptamers to ameliorate other inflammatory reactions. For these preclinical investigations, the availability of aptamers specific for murine CCL17 in combination with a variety of CCL17-dependent murine disease models will be highly advantageous.
Recently, Abboud et al.53 identified the compound GPN279 as a small-molecule inhibitor of both murine and human CCL17 with an IC50 of approximately 5 μM for inhibition of Ca2+ flux in vitro. They also showed that application of 350 μmol GPN279/kg body weight was sufficient to significantly inhibit leukocyte inflammation in a mouse model of airway inflammation. These doses are about 1,000-fold higher compared with the dosing of MF11.46 and MF35.47 in this study. Therefore, the CCL17-specific aptamers described here appear to be more efficient inhibitors than GPN279. Despite the structural similarity of mouse and human CCL17, MF35.47 and MF11.46, however, do not bind to human CCL17. To predict a potential binding site of MF35.47 and/or MF11.46 to murine CCL17, we investigated which amino acids differ between the human and mouse CCL17 proteins based on the crystal structure of human CCL17 (Figure S11).54 Obviously, the greatest differences are present in the C-terminal α helix of CCL17, whereas the N-terminal dimerization domain is largely conserved. Therefore, it is possible that the aptamers mainly interact with the C-terminal part and/or the adjacent receptor binding loops, similar to what has been described for binding of the NOX-A36 aptamer to CCL2 on the basis of a crystal structure of the aptamer-chemokine complex.31 The NOX-A36 aptamer is already in phase 2 clinical trials and appears to be a promising drug for treatment of diabetic nephropathies.55 Because we were very successful in isolating high-affinity aptamers for murine CCL17, and at least two aptamers directed against human chemokines are already in clinical trials, it appears highly likely that aptamers specific for human CCL17 can also be developed in the future and may facilitate the development of novel drugs for treatment of atopic and inflammatory diseases.
Materials and Methods
Selection of CCL17-Binding Aptamers
The library template DNA was purchased from Ella Biotech (Martinsried, Germany) and is composed of a randomized 40-nt region flanked by the primer binding sites (5′-GGGGGAATTCTAATACGACTCACTATAGGGAGGACGATGCGG-N40-CAGACGACTCGCTGAGGATCCGAGA-3′). The start of the 2′F-RNA sequence is underlined. The 2′F-RNA starting library was prepared by T7 in vitro transcription for 4 hr at 37°C. 2′F pyrimidines were incorporated by the T7(Y639F) RNA polymerase mutant. The selection procedure was started by incubation of 1 nmol starting library with 80 μL of CCL17 beads (∼1 μg of mCCL17) in a total volume of 300 μL of selection buffer (PBS [pH 7.4], 1 mM MgCl2, 1 mM CaCl2) for 30 min at 37°C. The beads were washed twice with selection buffer and the retained 2′F-RNA eluted in 55 μL of ddH2O for 5 min at 80°C. 50 μL of 2′F-RNA was reverse transcribed and amplified using the forward primer 5′-GGGGGAATTCTAATACGACTCACTATAGGGAGGACGATGCGG-3′ and reverse primer 5′-TCTCGGATCCTCAGCGAGTCGTC-3′. The 2′F-RNA for the subsequent selection cycles was prepared by T7 in vitro transcription for 20 min at 37°C. From the second selection cycle, counter selection was done by pre-incubation of the enriched 2′F-RNA with 80 μL of uncoupled beads. To gradually increase the selection pressure, we increased the number of washing steps by two in every second selection cycle, ending with 8. To identify individual aptamer sequences, the reverse transcribed library of the 10th selection cycle was ligated into pCR2.1-TOPO vectors in accordance with the manufacturer’s guidelines (TOPO-TA cloning kit; Invitrogen) and sequenced by GATC Biotech (Köln, Germany).
Transwell Migration Assay
BW5147.3 thymic lymphoma cells (ATCC TIB-47) were incubated for 2 hr at 37°C in starvation medium (RPMI + 0.5% FCS, P/S) and subsequently adjusted to 1 × 106 cells/mL. Lower chambers of transwell plates (5-μm pores; Sigma-Aldrich, St. Louis, MO, USA) were filled with 600 μL of starvation media with or without supplementation of 100 ng/mL (≥7.5 pmol) recombinant murine CCL17 (BioLegend, San Diego, CA, USA). As a positive control, the CCL17 neutralizing antibody MAB529 (Biotechne, Wiesbaden, Germany) was used at 1.88 μg/mL (≥7.5 pmol). Inhibitory function of CCL17-specific aptamers was tested at different concentrations as indicated in the figures. To exclude possible unspecific effects of the RNA aptamers, we used a scrambled version of MF35.47.m as a control. Upper chambers were loaded with 100 μL of the cell suspension (1 × 105 cells). After allowing the cells to migrate for 2 hr at 37°C, the transmigrated cells in the lower chamber were harvested and their absolute number determined using flow cytometry (FACSCanto; BD Biosciences, Heidelberg, Germany). Results are shown as percent of migration normalized to migration toward mCCL17 alone. All conditions were tested in duplicates or triplicates.
Mice
Animals were bred under specific pathogen-free conditions in the animal facility of the LIMES Institute, University of Bonn. All experiments were performed using female 8- to 12-week-old WT (C57BL/6J-RCCHsd; Envigo, Rossdorf, Germany) or CCL17E/E mice. CCL17E/E mice were generated as previously described6 and backcrossed into the C57BL/6J-RCCHsd background. All experiments were approved by the government of North Rhine-Westphalia (Az. 87-51.04.2010.A260 and 84-02.04.2016.A226).
Contact Hypersensitivity
For sensitization, mice were anesthetized with isoflurane and shaved before the abdominal skin was topically treated with 70 μL of 0.25% w/v DNFB (Sigma-Aldrich) in acetone/olive oil (5:1) on day −8 or day −5 as indicated in the figure legends. Mice were single housed for the duration of the experiment to avoid cross-contamination of the animals. The next day, the abdomen was treated with another 70 μL of 0.25% w/v DNFB. At day 0, mice were anesthetized and the baseline thickness of the ears was determined using a thickness-gauge caliper. Aptamers were administered i.p. in PBS 1 hr before and 12 hr after DNFB challenge. Control animals received i.p. injections of the solvent. For challenge, dorsal and ventral sides of the right ear were treated epicutaneously with 10 μL of 0.3% w/v DNFB in acetone/olive oil. As negative control, dorsal and ventral sides of the left ear were treated with 10 μL of acetone/olive oil only. Ear swelling was assessed 24 (day 1), 48 (day 2), and 72 hr (day 3) later. Absolute swelling of the ears at indicated time points was calculated by subtracting baseline thickness.
Statistical Analysis
Data were analyzed with GraphPad Prism 6 (GraphPad Software, San Diego, CA, USA) using one-way or two-way ANOVA with Bonferroni post hoc test for multiple comparisons. The level of significance was denoted as *p < 0.05, **p < 0.01, and ***p < 0.001 as indicated in the figure legends.
For description of additional methods, see Supplemental Materials and Methods.
Author Contributions
L.F., N.S., M.F., F.G., F.P., H.J.S., F.V.O., S.K.H., A.B.E., O.S., J.S., M. Blank, C.G., and K.H. designed, performed, and analyzed experiments. M. Beyer, H.W., E.L., and J.L.S. designed experiments and helped with critical advice and discussion. G.M. and I.F. designed experiments, supervised the study, and wrote the manuscript.
Conflicts of Interest
The authors declare no competing financial interest.
Acknowledgments
We are grateful to Martina Schunk, Philip Hatzfeld, and Björn Zapke for technical help. This work was supported by the Deutsche Forschungsgemeinschaft through SFB704 (projects A1, A25, and Z4) and GRK 2168. M. Beyer, E.L., J.L.S., and I.F. are members of the Bonn Cluster of Excellence “ImmunoSensation.”
Footnotes
Supplemental Information includes Supplemental Materials and Methods, eleven figures, and two tables and can be found with this article online at https://doi.org/10.1016/j.ymthe.2017.10.005.
Contributor Information
Günter Mayer, Email: gmayer@uni-bonn.de.
Irmgard Förster, Email: irmgard.foerster@uni-bonn.de.
Supplemental Information
References
- 1.Solari R., Pease J.E. Targeting chemokine receptors in disease--a case study of CCR4. Eur. J. Pharmacol. 2015;763(Pt B):169–177. doi: 10.1016/j.ejphar.2015.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zlotnik A., Yoshie O. Chemokines: a new classification system and their role in immunity. Immunity. 2000;12:121–127. doi: 10.1016/s1074-7613(00)80165-x. [DOI] [PubMed] [Google Scholar]
- 3.Zlotnik A., Yoshie O. The chemokine superfamily revisited. Immunity. 2012;36:705–716. doi: 10.1016/j.immuni.2012.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Imai T., Yoshida T., Baba M., Nishimura M., Kakizaki M., Yoshie O. Molecular cloning of a novel T cell-directed CC chemokine expressed in thymus by signal sequence trap using Epstein-Barr virus vector. J. Biol. Chem. 1996;271:21514–21521. doi: 10.1074/jbc.271.35.21514. [DOI] [PubMed] [Google Scholar]
- 5.Lieberam I., Förster I. The murine beta-chemokine TARC is expressed by subsets of dendritic cells and attracts primed CD4+ T cells. Eur. J. Immunol. 1999;29:2684–2694. doi: 10.1002/(SICI)1521-4141(199909)29:09<2684::AID-IMMU2684>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 6.Alferink J., Lieberam I., Reindl W., Behrens A., Weiss S., Hüser N., Gerauer K., Ross R., Reske-Kunz A.B., Ahmad-Nejad P. Compartmentalized production of CCL17 in vivo: strong inducibility in peripheral dendritic cells contrasts selective absence from the spleen. J. Exp. Med. 2003;197:585–599. doi: 10.1084/jem.20021859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Semmling V., Lukacs-Kornek V., Thaiss C.A., Quast T., Hochheiser K., Panzer U., Rossjohn J., Perlmutter P., Cao J., Godfrey D.I. Alternative cross-priming through CCL17-CCR4-mediated attraction of CTLs toward NKT cell-licensed DCs. Nat. Immunol. 2010;11:313–320. doi: 10.1038/ni.1848. [DOI] [PubMed] [Google Scholar]
- 8.Stutte S., Quast T., Gerbitzki N., Savinko T., Novak N., Reifenberger J., Homey B., Kolanus W., Alenius H., Förster I. Requirement of CCL17 for CCR7- and CXCR4-dependent migration of cutaneous dendritic cells. Proc. Natl. Acad. Sci. USA. 2010;107:8736–8741. doi: 10.1073/pnas.0906126107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mantovani A., Sica A., Sozzani S., Allavena P., Vecchi A., Locati M. The chemokine system in diverse forms of macrophage activation and polarization. Trends Immunol. 2004;25:677–686. doi: 10.1016/j.it.2004.09.015. [DOI] [PubMed] [Google Scholar]
- 10.Griffith J.W., Sokol C.L., Luster A.D. Chemokines and chemokine receptors: positioning cells for host defense and immunity. Annu. Rev. Immunol. 2014;32:659–702. doi: 10.1146/annurev-immunol-032713-120145. [DOI] [PubMed] [Google Scholar]
- 11.Kitajima M., Ziegler S.F. Cutting edge: identification of the thymic stromal lymphopoietin-responsive dendritic cell subset critical for initiation of type 2 contact hypersensitivity. J. Immunol. 2013;191:4903–4907. doi: 10.4049/jimmunol.1302175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stutte S., Gerbitzki N., Novak N., Förster I. Expression and function of CCL17 in atopic dermatitis. In: Esparza-Gordillo J., editor. Atopic Dermatitis – Disease Etiology and Clinical Management. InTech; 2012. pp. 81–104. [Google Scholar]
- 13.Heiseke A.F., Faul A.C., Lehr H.A., Förster I., Schmid R.M., Krug A.B., Reindl W. CCL17 promotes intestinal inflammation in mice and counteracts regulatory T cell-mediated protection from colitis. Gastroenterology. 2012;142:335–345. doi: 10.1053/j.gastro.2011.10.027. [DOI] [PubMed] [Google Scholar]
- 14.Weber C., Meiler S., Döring Y., Koch M., Drechsler M., Megens R.T., Rowinska Z., Bidzhekov K., Fecher C., Ribechini E. CCL17-expressing dendritic cells drive atherosclerosis by restraining regulatory T cell homeostasis in mice. J. Clin. Invest. 2011;121:2898–2910. doi: 10.1172/JCI44925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kataoka Y. Thymus and activation-regulated chemokine as a clinical biomarker in atopic dermatitis. J. Dermatol. 2014;41:221–229. doi: 10.1111/1346-8138.12440. [DOI] [PubMed] [Google Scholar]
- 16.Hartl D., Lee C.G., Da Silva C.A., Chupp G.L., Elias J.A. Novel biomarkers in asthma: chemokines and chitinase-like proteins. Curr. Opin. Allergy Clin. Immunol. 2009;9:60–66. doi: 10.1097/ACI.0b013e32831f8ee0. [DOI] [PubMed] [Google Scholar]
- 17.Yoshie O., Matsushima K. CCR4 and its ligands: from bench to bedside. Int. Immunol. 2015;27:11–20. doi: 10.1093/intimm/dxu079. [DOI] [PubMed] [Google Scholar]
- 18.Sauer M., Plütschow A., Jachimowicz R.D., Kleefisch D., Reiners K.S., Ponader S., Engert A., von Strandmann E.P. Baseline serum TARC levels predict therapy outcome in patients with Hodgkin lymphoma. Am. J. Hematol. 2013;88:113–115. doi: 10.1002/ajh.23361. [DOI] [PubMed] [Google Scholar]
- 19.Achuthan A., Cook A.D., Lee M.-C., Saleh R., Khiew H.-W., Chang M.W.N., Louis C., Fleetwood A.J., Lacey D.C., Christensen A.D. Granulocyte macrophage colony-stimulating factor induces CCL17 production via IRF4 to mediate inflammation. J. Clin. Invest. 2016;126:3453–3466. doi: 10.1172/JCI87828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Curiel T.J., Coukos G., Zou L., Alvarez X., Cheng P., Mottram P., Evdemon-Hogan M., Conejo-Garcia J.R., Zhang L., Burow M. Specific recruitment of regulatory T cells in ovarian carcinoma fosters immune privilege and predicts reduced survival. Nat. Med. 2004;10:942–949. doi: 10.1038/nm1093. [DOI] [PubMed] [Google Scholar]
- 21.Viney J.M., Andrew D.P., Phillips R.M., Meiser A., Patel P., Lennartz-Walker M., Cousins D.J., Barton N.P., Hall D.A., Pease J.E. Distinct conformations of the chemokine receptor CCR4 with implications for its targeting in allergy. J. Immunol. 2014;192:3419–3427. doi: 10.4049/jimmunol.1300232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ajram L., Begg M., Slack R., Cryan J., Hall D., Hodgson S., Ford A., Barnes A., Swieboda D., Mousnier A., Solari R. Internalization of the chemokine receptor CCR4 can be evoked by orthosteric and allosteric receptor antagonists. Eur. J. Pharmacol. 2014;729:75–85. doi: 10.1016/j.ejphar.2014.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mariani M., Lang R., Binda E., Panina-Bordignon P., D’Ambrosio D. Dominance of CCL22 over CCL17 in induction of chemokine receptor CCR4 desensitization and internalization on human Th2 cells. Eur. J. Immunol. 2004;34:231–240. doi: 10.1002/eji.200324429. [DOI] [PubMed] [Google Scholar]
- 24.Zhou J., Rossi J. Aptamers as targeted therapeutics: current potential and challenges. Nat. Rev. Drug Discov. 2017;16:181–202. doi: 10.1038/nrd.2016.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Famulok M., Mayer G. Aptamers and SELEX in chemistry & biology. Chem. Biol. 2014;21:1055–1058. doi: 10.1016/j.chembiol.2014.08.003. [DOI] [PubMed] [Google Scholar]
- 26.Ruckman J., Green L.S., Beeson J., Waugh S., Gillette W.L., Henninger D.D., Claesson-Welsh L., Janjić N. 2′-Fluoropyrimidine RNA-based aptamers to the 165-amino acid form of vascular endothelial growth factor (VEGF165). Inhibition of receptor binding and VEGF-induced vascular permeability through interactions requiring the exon 7-encoded domain. J. Biol. Chem. 1998;273:20556–20567. doi: 10.1074/jbc.273.32.20556. [DOI] [PubMed] [Google Scholar]
- 27.Gragoudas E.S., Adamis A.P., Cunningham E.T., Jr., Feinsod M., Guyer D.R., VEGF Inhibition Study in Ocular Neovascularization Clinical Trial Group Pegaptanib for neovascular age-related macular degeneration. N. Engl. J. Med. 2004;351:2805–2816. doi: 10.1056/NEJMoa042760. [DOI] [PubMed] [Google Scholar]
- 28.Sundaram P., Kurniawan H., Byrne M.E., Wower J. Therapeutic RNA aptamers in clinical trials. Eur. J. Pharm. Sci. 2013;48:259–271. doi: 10.1016/j.ejps.2012.10.014. [DOI] [PubMed] [Google Scholar]
- 29.Lee J.W., Kim H.J., Heo K. Therapeutic aptamers: developmental potential as anticancer drugs. BMB Rep. 2015;48:234–237. doi: 10.5483/BMBRep.2015.48.4.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hoellenriegel J., Zboralski D., Maasch C., Rosin N.Y., Wierda W.G., Keating M.J., Kruschinski A., Burger J.A. The Spiegelmer NOX-A12, a novel CXCL12 inhibitor, interferes with chronic lymphocytic leukemia cell motility and causes chemosensitization. Blood. 2014;123:1032–1039. doi: 10.1182/blood-2013-03-493924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Oberthür D., Achenbach J., Gabdulkhakov A., Buchner K., Maasch C., Falke S., Rehders D., Klussmann S., Betzel C. Crystal structure of a mirror-image L-RNA aptamer (Spiegelmer) in complex with the natural L-protein target CCL2. Nat. Commun. 2015;6:6923. doi: 10.1038/ncomms7923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mi J., Liu Y., Rabbani Z.N., Yang Z., Urban J.H., Sullenger B.A., Clary B.M. In vivo selection of tumor-targeting RNA motifs. Nat. Chem. Biol. 2010;6:22–24. doi: 10.1038/nchembio.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Keefe A.D., Pai S., Ellington A. Aptamers as therapeutics. Nat. Rev. Drug Discov. 2010;9:537–550. doi: 10.1038/nrd3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Blank M. Next-generation analysis of deep sequencing data: bringing light into the black box of SELEX experiments. Methods Mol. Biol. 2016;1380:85–95. doi: 10.1007/978-1-4939-3197-2_7. [DOI] [PubMed] [Google Scholar]
- 35.Blind M., Blank M. Aptamer selection technology and recent advances. Mol. Ther. Nucleic Acids. 2015;4:e223. doi: 10.1038/mtna.2014.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Paludan S.R., Bowie A.G. Immune sensing of DNA. Immunity. 2013;38:870–880. doi: 10.1016/j.immuni.2013.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.O’Neill L.A.J., Golenbock D., Bowie A.G. The history of Toll-like receptors - redefining innate immunity. Nat. Rev. Immunol. 2013;13:453–460. doi: 10.1038/nri3446. [DOI] [PubMed] [Google Scholar]
- 39.Honda T., Egawa G., Grabbe S., Kabashima K. Update of immune events in the murine contact hypersensitivity model: toward the understanding of allergic contact dermatitis. J. Invest. Dermatol. 2013;133:303–315. doi: 10.1038/jid.2012.284. [DOI] [PubMed] [Google Scholar]
- 40.Xu H., DiIulio N.A., Fairchild R.L. T cell populations primed by hapten sensitization in contact sensitivity are distinguished by polarized patterns of cytokine production: interferon gamma-producing (Tc1) effector CD8+ T cells and interleukin (Il) 4/Il-10-producing (Th2) negative regulatory CD4+ T cells. J. Exp. Med. 1996;183:1001–1012. doi: 10.1084/jem.183.3.1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pere H., Montier Y., Bayry J., Quintin-Colonna F., Merillon N., Dransart E., Badoual C., Gey A., Ravel P., Marcheteau E. A CCR4 antagonist combined with vaccines induces antigen-specific CD8+ T cells and tumor immunity against self antigens. Blood. 2011;118:4853–4862. doi: 10.1182/blood-2011-01-329656. [DOI] [PubMed] [Google Scholar]
- 42.Bayry J., Tartour E., Tough D.F. Targeting CCR4 as an emerging strategy for cancer therapy and vaccines. Trends Pharmacol. Sci. 2014;35:163–165. doi: 10.1016/j.tips.2014.02.003. [DOI] [PubMed] [Google Scholar]
- 43.Ishida T., Joh T., Uike N., Yamamoto K., Utsunomiya A., Yoshida S., Saburi Y., Miyamoto T., Takemoto S., Suzushima H. Defucosylated anti-CCR4 monoclonal antibody (KW-0761) for relapsed adult T-cell leukemia-lymphoma: a multicenter phase II study. J. Clin. Oncol. 2012;30:837–842. doi: 10.1200/JCO.2011.37.3472. [DOI] [PubMed] [Google Scholar]
- 44.Chang D.-K., Sui J., Geng S., Muvaffak A., Bai M., Fuhlbrigge R.C., Lo A., Yammanuru A., Hubbard L., Sheehan J. Humanization of an anti-CCR4 antibody that kills cutaneous T-cell lymphoma cells and abrogates suppression by T-regulatory cells. Mol. Cancer Ther. 2012;11:2451–2461. doi: 10.1158/1535-7163.MCT-12-0278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Iellem A., Mariani M., Lang R., Recalde H., Panina-Bordignon P., Sinigaglia F., D’Ambrosio D. Unique chemotactic response profile and specific expression of chemokine receptors CCR4 and CCR8 by CD4(+)CD25(+) regulatory T cells. J. Exp. Med. 2001;194:847–853. doi: 10.1084/jem.194.6.847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ishida T., Ito A., Sato F., Kusumoto S., Iida S., Inagaki H., Morita A., Akinaga S., Ueda R. Stevens-Johnson syndrome associated with mogamulizumab treatment of adult T-cell leukemia/ lymphoma. Cancer Sci. 2013;104:647–650. doi: 10.1111/cas.12116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mirshahpanah P., Li Y.Y.Y., Burkhardt N., Asadullah K., Zollner T.M. CCR4 and CCR10 ligands play additive roles in mouse contact hypersensitivity. Exp. Dermatol. 2008;17:30–34. doi: 10.1111/j.1600-0625.2007.00630.x. [DOI] [PubMed] [Google Scholar]
- 48.Wang X., Fujita M., Prado R., Tousson A., Hsu H.C., Schottelius A., Kelly D.R., Yang P.A., Wu Q., Chen J. Visualizing CD4 T-cell migration into inflamed skin and its inhibition by CCR4/CCR10 blockades using in vivo imaging model. Br. J. Dermatol. 2010;162:487–496. doi: 10.1111/j.1365-2133.2009.09552.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lehtimäki S., Tillander S., Puustinen A., Matikainen S., Nyman T., Fyhrquist N., Savinko T., Majuri M.L., Wolff H., Alenius H., Lauerma A. Absence of CCR4 exacerbates skin inflammation in an oxazolone-induced contact hypersensitivity model. J. Invest. Dermatol. 2010;130:2743–2751. doi: 10.1038/jid.2010.208. [DOI] [PubMed] [Google Scholar]
- 50.Reiss Y., Proudfoot A.E., Power C.A., Campbell J.J., Butcher E.C. CC chemokine receptor (CCR)4 and the CCR10 ligand cutaneous T cell-attracting chemokine (CTACK) in lymphocyte trafficking to inflamed skin. J. Exp. Med. 2001;194:1541–1547. doi: 10.1084/jem.194.10.1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kaplan D.H., Igyártó B.Z., Gaspari A.A. Early immune events in the induction of allergic contact dermatitis. Nat. Rev. Immunol. 2012;12:114–124. doi: 10.1038/nri3150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Natsuaki Y., Egawa G., Nakamizo S., Ono S., Hanakawa S., Okada T., Kusuba N., Otsuka A., Kitoh A., Honda T. Perivascular leukocyte clusters are essential for efficient activation of effector T cells in the skin. Nat. Immunol. 2014;15:1064–1069. doi: 10.1038/ni.2992. [DOI] [PubMed] [Google Scholar]
- 53.Abboud D., Daubeuf F., Do Q.T., Utard V., Villa P., Haiech J., Bonnet D., Hibert M., Bernard P., Galzi J.L., Frossard N. A strategy to discover decoy chemokine ligands with an anti-inflammatory activity. Sci. Rep. 2015;5:14746. doi: 10.1038/srep14746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Asojo O.A., Boulègue C., Hoover D.M., Lu W., Lubkowski J. Structures of thymus and activation-regulated chemokine (TARC) Acta Crystallogr. D Biol. Crystallogr. 2003;59:1165–1173. doi: 10.1107/s0907444903009454. [DOI] [PubMed] [Google Scholar]
- 55.NoxxonPharma. (2012). NOXXON initiates phase IIa of anti-CCL2/MCP-1 Spiegelmer® NOX-E36 for treatment of diabetic nephropathy. June 19, 2012. https://www.noxxon.com/downloads/pressrel/2012-06-19_NOXXON_Initiates_Phase_IIa_of_Spiegelmer_NOX-E36.pdf.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.