Abstract
Duchenne muscular dystrophy (DMD) is a severe, progressive muscle wasting disorder caused by reading frame disrupting mutations in the DMD gene. Exon skipping is a therapeutic approach for DMD. It employs antisense oligonucleotides (AONs) to restore the disrupted open reading frame, allowing the production of shorter, but partly functional dystrophin protein as seen in less severely affected Becker muscular dystrophy patients. To be effective, AONs need to be delivered and effectively taken up by the target cells, which can be accomplished by the conjugation of tissue-homing peptides. We performed phage display screens using a cyclic peptide library combined with next generation sequencing analyses to identify candidate muscle-homing peptides. Conjugation of the lead peptide to 2′-O-methyl phosphorothioate AONs enabled a significant, 2-fold increase in delivery and exon skipping in all analyzed skeletal and cardiac muscle of mdx mice and appeared well tolerated. While selected as a muscle-homing peptide, uptake was increased in liver and kidney as well. The homing capacity of the peptide may have been overruled by the natural biodistribution of the AON. Nonetheless, our results suggest that the identified peptide has the potential to facilitate delivery of AONs and perhaps other compounds to skeletal and cardiac muscle.
Keywords: phage display, 2′-O-methyl phosphorothioate, dystrophin, antisense oligonucleotides, exon skipping, cyclic peptide, 7-mer
Upon systemic administration of antisense oligonucleotides (AONs), most AONs end up in liver and/or kidney and are lost for targeting other organs. Jirka et al. (2017) show a valuable approach to identify muscle-homing peptides that leads to 2-fold improvement of delivery and bioactivity of systemically administrated AONs in muscle.
Introduction
Duchenne muscular dystrophy (DMD) is a severe, progressive, X-linked muscle wasting disorder that affects 1 in 5,000 newborn boys worldwide.1, 2 In general, DMD patients are diagnosed before the age of 5 years, become wheelchair dependent around the age of 12 years, need assisted ventilation around 20 years of age, and currently have a life expectancy of ∼30 years in the Western world.2 DMD is caused by out-of-frame or nonsense mutations in the DMD gene that lead to truncated, non-functional dystrophin proteins. Dystrophin provides stability to the muscle fibers upon contraction.3 Lacking dystrophin, muscle fibers are easily and continuously damaged, and eventually are replaced by non-functional fibrotic and adipose tissues. In contrast, Becker muscular dystrophy (BMD) is a muscle wasting disorder caused by mutations in the same gene, but here mutations maintain the open reading frame and allow production of an internally deleted, but partially functional dystrophin protein. The phenotype of patients with BMD is milder and less progressive, and patients have generally near-normal life expectancies.2 Restoration of the reading frame in DMD patients would in theory allow the production of a shorter, but partly functional dystrophin protein as seen in patients with BMD.4, 5 This restoration can be achieved with antisense oligonucleotides (AONs) that recognize specific exons during pre-mRNA splicing and induce skipping of target exons.6, 7, 8, 9 For DMD, the most advanced exon-skipping AON chemistries are the negatively charged 2′-O-methyl phosphorothioate (2OMePS) and the neutral phosphorodiamidate morpholino oligomers (PMOs).10 The first exon-skipping drug eteplirsen (a PMO marketed as EXONdys 51) was recently approved by the US Food and Drug Administration (FDA) (https://www.fda.gov/NewsEvents/Newsroom/PressAnnouncements/ucm521263.htm).
Delivery Challenges for AON
Exon-skipping AONs need to be delivered and taken up adequately by the target tissue and enter the target tissue cells to be effective. Furthermore, when taken up by endocytosis, they have to escape from the endosomes and reach the nucleus where splicing takes place. Because the human body consists of 30%–40% muscle, body-wide treatment is necessary for DMD. This appeared feasible for AONs, as observed in preclinical animal studies for both 2OMePS and PMO AONs; however, in humans, it remains challenging to reach a significant clinical benefit for DMD patients.6, 9, 10, 11, 12, 13 Studies in animal models have revealed that large portions of the AON end up in the liver and/or kidney, while uptake and consequently exon-skipping levels in skeletal muscle and heart are low and very low, respectively, when using 2OMePS and PMO AON doses comparable with those used in humans in clinical trials.14, 15, 16 Obviously, improved AON delivery to skeletal and cardiac muscle is anticipated to enhance therapeutic effects of AONs.
Improving AON Delivery by Formulations
To enhance the delivery of 2OMePS AONs, polyethylenimine (PEI) and poly(ethylene glycol) (PEG) copolymers alone, combined with the cell-penetrating TAT peptide (GRKKRRQRRRPQ), adsorbed colloidal gold (CG), or a combination have been investigated. The PEI-PEG copolymer combined with TAT was most potent and resulted in 6-fold increased dystrophin-positive fibers and up to 30% of dystrophin expression compared to wild-type levels upon intramuscular administration in tibialis anterior muscle of mdx mice.17 However, these polyplexes have a positive charge that limits their biodistribution due to nonspecific binding to target unrelated components. Later the encapsulation of these polyplexes in biodegradable poly(lactide-coglycolide) (PLGA) nanospheres was investigated to improve the strategy. Nevertheless, upon intramuscular administration, no improvement in dystrophin levels was observed compared with the unencapsulated polyplexes.18 2OMePS AONs adsorbed onto poly(methyl methacrylate) (PMMA)/N-isopropyl-acrylamide+ (NIPAM) nanoparticles (ZM2) have been investigated as well. Intraperitoneal administration of these nanoparticles resulted in 20% exon 23 skipping levels and up to 40% dystrophin-positive muscle fibers in mdx mice.19 Later, Bassi et al.20 showed that this was persistent for more than 90 days. A next generation of this approach involves ZM4 nanoparticles; however, effective delivery to muscle remains to be evaluated.21
Small-sized polyethylenimine (PEI)-conjugated pluronic copolymers (PCMs) have been evaluated to improve the uptake of PMOs. Intravenous administration resulted in increased exon 23 skipping levels and, on average, 15% dystrophin-positive muscle fibers (particularly in cardiac muscle tissue) when combining the PMO with PCMs compared with the PMO alone (<5%) in mdx mice.22 Nonetheless, the overall percentages of exon skipping and dystrophin-positive fibers remained low.
Improving AON Delivery by Conjugation
For PMOs, cell-penetrating peptides (CPPs) have been studied most intensively. CPPs are short cationic peptides designed to transport drug into cells. Moulton et al.23 were one of the first to describe the use of arginine-rich peptides to enhance the delivery and uptake of PMOs in muscle for DMD (PPMOs). Unfortunately, these PPMOs were poorly tolerated in non-human primates because of kidney toxicity.24 Throughout the years various improvements have been made regarding arginine-rich CPPs, RXR4, B-peptide (RXRRBR)2, and the more recently developed Pip peptides.25, 26 The most potent Pip peptides come from the Pip5 and Pip6 series. Conjugates resulted in vivo, upon systemic administration, in high levels of exon 23 skipping and dystrophin protein production in skeletal and cardiac muscle. Further evaluation of these conjugates showed improvement of muscle strength and cardiac function in exercised mdx mice.27, 28, 29 The newly developed CPPs appear to be well tolerated in mdx mice. Nevertheless, they contain many arginine residues, making it questionable whether they are not toxic in higher animals than mice, as shown in the early studies of Moulton et al.23
Despite the fact that the well-studied CPPs have the potential to improve the delivery of PMOs, they are not suitable for delivery of 2OMePS. The cationic nature of CPPs has the tendency to strongly form aggregates when combined with the anionic 2OMePS AON backbone. For the 2OMePS AON chemistry, we therefore chose to use (non-highly cationic) tissue-homing peptides, identified using phage display technology, a well-described technique to identify target-specific peptides, antibodies, and proteins.30 Phages (so-called bacteriophages) are viruses that consist of DNA or RNA within a protein coat. A phage library is constructed by fusing a foreign peptide or protein with one of the protein coat genes in such a way that these are expressed on the surface of the phage. Phage libraries consist of millions or billions of uniquely constructed phages from which affinity selection takes place, a process called biopanning (incubate phage library with target, wash away non-binders, isolate binding phages).30 This approach can be cumbersome, and it is well-known that many false-positive peptides (parasite sequences, e.g., have an amplification bias or bind to plastic) have been identified.31 However, analyzing the outcome of phage display experiments with next generation sequencing (NGS) improves the chance of success greatly.32 NGS allows us to use just a single screening round, preventing parasitic peptide sequences to dominate the outcome and making identification of parasitic peptide sequences easier and more reliable. The first encouraging results were reported using a short, linear 7-mer homing peptide selected from in vivo phage display biopanning experiments toward skeletal and cardiac muscle using the Ph.D.-7 phage display library.33 This library expresses a few copies of a linear 7-mer peptide at the N terminus of the PIII protein of the phage. In addition, a cyclic 7-mer peptide library, Ph.D.-C7C, is available. This C7C-peptide library shares its features with the linear library but expresses peptides that are cyclized by disulfide bridges between two cysteine residues that are positioned at each end of the random 7-mer peptide. We reasoned that the conformational restriction by cyclization would lead to higher affinity binding, and thus to potentially more efficient targeting peptides.
Here, we explored the identification of muscle-homing peptides with new phage display peptide library screens, in vitro and in vivo, using the Ph.D.-C7C peptide library combined with NGS analyses. This allowed the identification of a cyclic peptide (CyPep10), which, upon conjugation to a 2OMePS AON, resulted in a significant, 2-fold increase in delivery and exon 23 skipping in all analyzed skeletal muscles and heart of the mdx mouse model and appeared to be well tolerated. Although this peptide was selected for being muscle specific, a more general increased delivery of the conjugate throughout tissues was seen. Nonetheless, results suggest that the identified peptide has the potential to facilitate targeted delivery of AONs and possible other compounds to skeletal and cardiac muscle for DMD.
Results
Candidate Peptide Identification
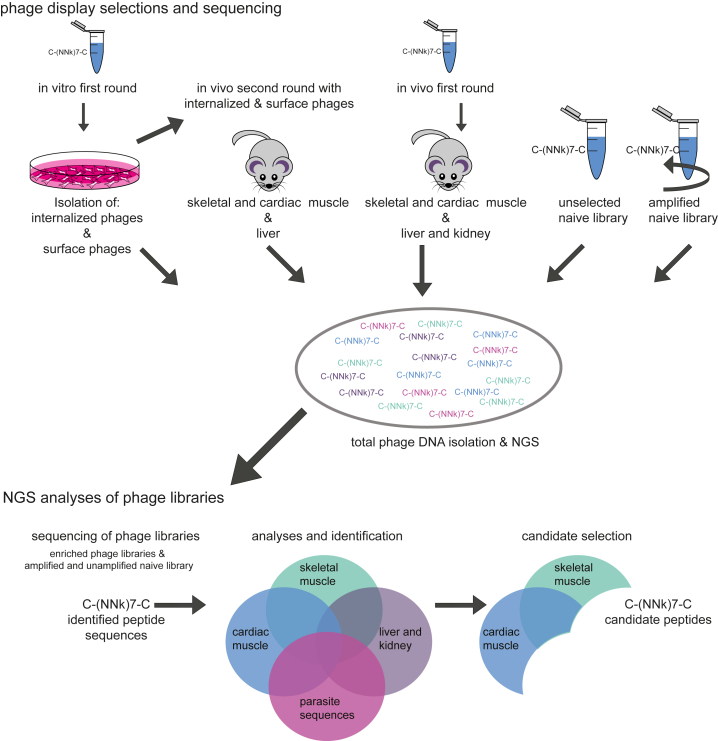
To identify peptides that enhance the delivery of AONs to skeletal and cardiac muscle, we used the Ph.D.-C7C phage display peptide library. A schematic outline of the screening procedure is given in Figure 1. We performed a first-round in vitro screening using human control myotubes in which internalized and surface-bound phages were isolated and amplified. Subsequently, the naive library was also used for a first-round in vivo enriched internalized phage fraction, and the enriched surface-bound phage fraction was used for a second in vivo screening round in mdx mice, a mouse model for DMD. Gastrocnemius, quadriceps, and heart were isolated for positive phage selection. Liver and kidney were isolated for negative phage selection. All enriched libraries from the biopanning selections, and the naive library with and without one round of bacterial amplification, were sequenced using a published NGS approach32 with further adaptations and improvements to increase efficiency. First, we barcoded each of the libraries using sample-specific barcodes in the reverse primer, allowing all enriched libraries to be pooled in one lane and the two naive libraries to be pooled in a second lane. Results are summarized in Table S1 and Figure S1.
Figure 1.
Phage Display Selections and Sequencing
A schematic overview of the phage display selection experiments and candidate peptide identification. “k” represents an admixture of 50% T and 50% G.
The enriched phage libraries were first filtered for parasitic sequences with a propagation advantage to identify candidate peptides. Sequences were considered parasitic when the frequency count in the naive amplified library minus the frequency count in the unamplified naive library was greater than 2. Second, candidate peptides were selected based on the following criteria: (1) the candidate peptide sequence has a relatively low frequency count or is absent in liver and kidney, and (2) the candidate peptide sequence has either a high frequency count in skeletal muscle and low frequency count in cardiac muscle or vice versa, or (3) the candidate peptide sequence has a high frequency count in both skeletal and cardiac muscle (Figure 1). Lastly, we took the charge of the peptides into account: candidate peptide sequences with more than two positively charged residues (arginines or lysines) were removed from the candidate list because they were expected to likely form aggregates when attempting to couple them to negatively charged AONs.
In Vitro Evaluation of the Peptides
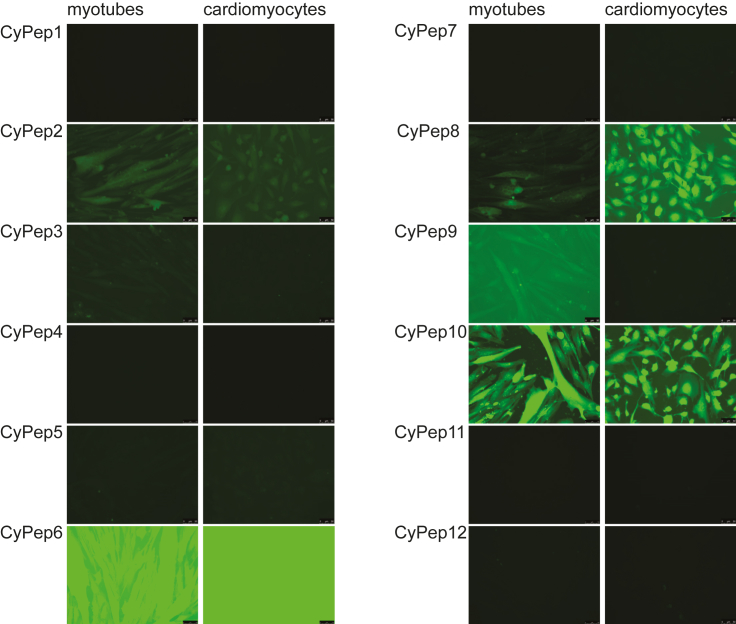
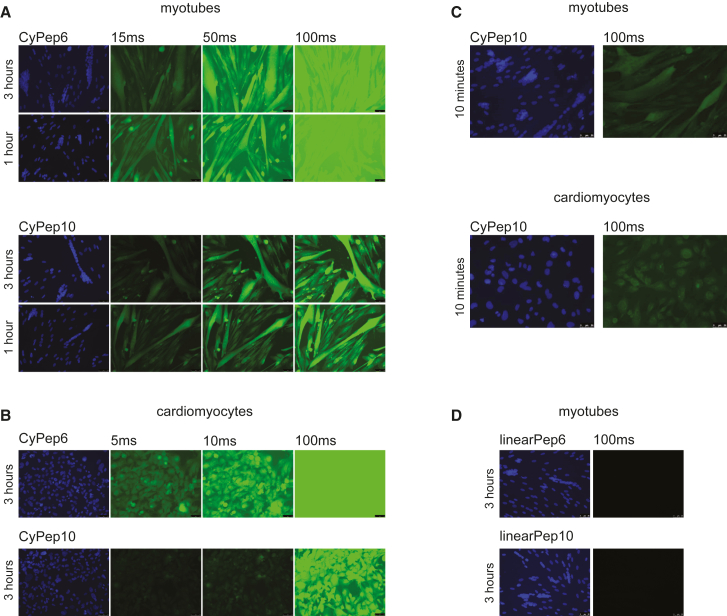
For in vitro evaluation of uptake by skeletal and cardiac cells, we synthesized the best 12 candidate peptides equipped with a fluorescein isothiocyanate (FITC) label (Table 1). Human control myotubes or human cardiomyocytes cultured on cover glass slips were incubated for 3 hr with 2.25 μM candidate peptides. After thorough washing, slides were fixed with methanol and embedded in mounting media containing DAPI to stain the nuclei. Most of the peptides were not taken up based on the absence of fluorescence in the cells. Cells incubated with CyPep2, CyPep8, or CyPep9 were weakly fluorescent, whereas incubation with CyPep6 (CLNSLFGSC) and CyPep10 (CQLFPLFRC) resulted in bright fluorescence throughout the cells with fluorescence also observed in the nuclei (Figure 2). When we repeated the experiment using different incubation times, clear fluorescence of CyPep6 and CyPep10 could be seen in human control myotubes after 1 hr of incubation (Figure 3A). In cardiomyocytes, higher fluorescence intensity was observed for CyPep6 compared with Cypep10 after 3 hr of incubation (Figure 3B). Interestingly, incubation with CyPep10 resulted in detectable levels of fluorescence in human control myotubes and human cardiomyocyte cultures already after an incubation of 10 min (Figure 3C), something we did not observe for CyPep6 (data not shown).
Table 1.
Amino Acid Sequence of the Cyclic Peptide Candidates
| Peptide | Sequences | Group |
|---|---|---|
| CyPep1 | CQVRSNTTC | muscle |
| CyPep2 | CSLFKNFRC | muscle/heart |
| CyPep3 | CRADFYTTC | heart |
| CyPep4 | CRENTNHTC | heart |
| CyPep5 | CWNEDHTWC | muscle/heart |
| CyPep6 | CLNSLFGSC | muscle/heart |
| CyPep7 | CLLGHTNNC | muscle |
| CyPep8 | CFSHTYRVC | muscle |
| CyPep9 | CTYSPTEVC | heart |
| CyPep10 | CQLFPLFRC | muscle/heart |
| CyPep11 | CTLQDQATC | heart |
| CyPep12 | CMQHSMRVC | muscle/heart |
Figure 2.
In Vitro Evaluation of Fluorescently Labeled Cyclic Peptides
Human control myotubes and cardiomyocytes were incubated with 2.25 μM FITC-labeled CyPeps for 3 hr. Slides were embedded in mounting media containing DAPI to stain nuclei. Fluorescence intensities were analyzed with fluorescence microscopy at 20× magnification; representative pictures are shown.
Figure 3.
In Vitro Evaluation of CyPep6 and CyPep10
CyPep6 and CyPep10 were incubated at a dose of 2.25 μM; slides were embedded in mounting media containing DAPI and analyzed with fluorescence microscopy at 20× magnification. (A) Human control myotubes incubated with CyPep6 or CyPep10 for 3 or 1 hr. (B) Human cardiomyocytes incubated with CyPep6 or CyPep10 for 3 hr. (C) Human control myotubes or cardiomyocytes incubated with CyPep10 for 10 min. (D) Human control myotubes incubated with linearPep6 or linearPep10 for 3 hr. Representative pictures are shown.
To study whether the cyclization of the peptide is crucial for the observed results, we synthesized FITC-labeled linear forms of CyPep6 and CyPep10 with Cys→Ala substitutions. No fluorescence was observed for both peptides, suggesting that not only the sequence, but also the cyclization is responsible for the positive fluorescence observed (Figure 3D).
Peptide Conjugation Does Not Impair Exon Skipping In Vitro and In Vivo
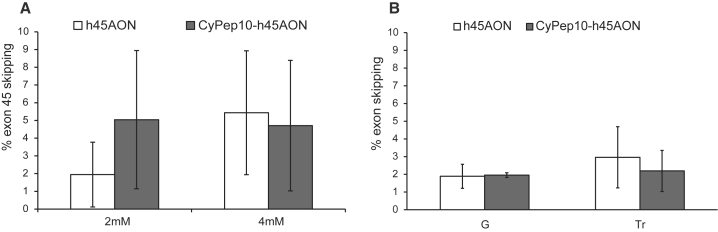
We then conjugated CyPep10 to a human AON targeting human dystrophin exon 45 (h45AON). To test whether the conjugation of the peptide impaired the exon-skipping efficiency of the AON, we incubated human control myotubes with 2 or 4 μM h45AON or CyPep10-h45AON for 96 hr without a transfection reagent. Subsequently, the cells were washed, followed by RNA isolation and nested RT-PCR. Exon-skipping levels were determined semiquantitatively using a lab-on a-chip-analyzer as described before by Spitali et al.34 This revealed no differences in exon-skipping levels (Figure 4A).
Figure 4.
Evaluation of CyPep10-AON Conjugate
CyPep10 was conjugated to an AON targeting human dystrophin exon 45 (h45AON and CyPep10-h45AON) to evaluate whether the conjugation of a cyclic peptide has any influence on the exon-skipping functionality of the AON. (A) Human control myotubes incubated with 2 or 4 μM AON without any transfection reagent for 96 hr. Results show the average exon-skipping levels of two independent experiments in duplo. (B) IM injection in two hDMD mice (pretreated with cardiotoxin) with 20 μg of h45AON or molar equivalent of CyPep10-h45AON for 2 consecutive days. Results represent an average of two independent experiments in duplo. Bars represent mean ± SD. G, gastrocnemius; Tr, triceps.
We also assessed whether the conjugation of the peptide affected the exon-skipping efficiency of the AON in vivo. Hereto, we injected the CyPep10-h45AON in hDMD mice, a mouse model with the human DMD gene integrated in the mouse genome.35 This mouse model has healthy muscle, because human dystrophin compensates for the lack of mouse dystrophin. We pretreated gastrocnemius and triceps muscles intramuscularly (IM) with cardiotoxin injections to induce muscle necrosis and enhance AON uptake.15 Two days later, muscles were injected with 20 μg of h45AON or a molar equivalent of CyPep10-h45AON contralateral for 2 consecutive days. One week after the last injection, RNA was isolated, nested RT-PCR was performed, and exon-skipping levels were determined semiquantitatively using a lab-on a-chip-analyzer.34 No differences in exon-skipping levels were found, showing that the conjugation of a cyclic peptide to an AON also has no negative influence on its exon-skipping ability in vivo (Figure 4B).
Conjugation of CyPep10 to AONs Increases Uptake and Exon-Skipping Levels after Systemic Delivery in mdx Mice
We evaluated CyPep6 and CyPep10 for their ability to enhance delivery and efficacy of AONs in skeletal and cardiac muscle in mdx mice upon systemic administration. Mdx mice (C57Bl/10ScSn-DMDmdx/J) have a point mutation in exon 23 of the mouse Dmd gene, which leads to a premature stop codon, resulting in an absence of the dystrophin protein.36 Skipping exon 23 in the mdx mouse Dmd pre-mRNA bypasses the mutation, maintaining the reading frame, and results in a shorter but (partly) functional protein. CyPep6 and CyPep10 were conjugated to a 2OMePS AON targeting the exon 23/intron 23 splice site (23AON).37 We systemically treated 4-week-old mdx mice (four to five per group) subcutaneously (s.c.) four times per week with 50 mg/kg 23AON, a molar equivalent of CyPep6-23AON or CyPep10-23AON, or saline for 8 weeks. Animals were sacrificed 1 week after the last injection.
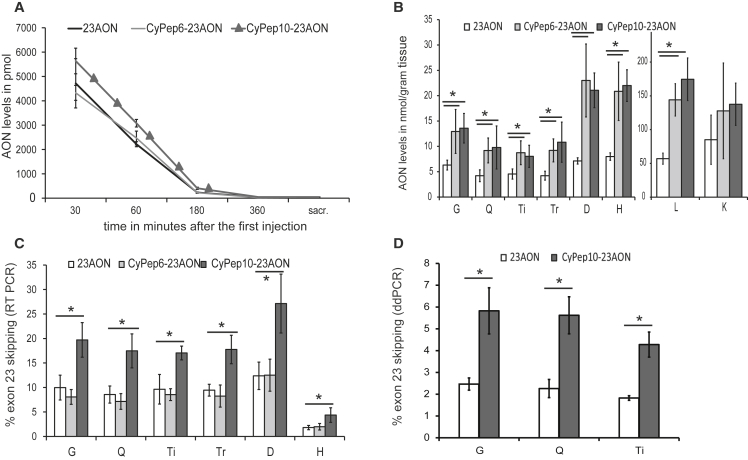
Evaluation of AON levels in plasma, obtained at several time points (sparse sampling approach) after the first injection, revealed increased levels of CyPep10-23AON in plasma for the first 3 hr after injection, but not for CyPep6-23AON (Figure 5A). In tissues, a 2-fold (on average) increase of CyPep6- and CyPep10-conjugated 23AON was observed in gastrocnemius, quadriceps, tibialis anterior, and triceps muscle, and 3- and 2.8-fold increases were observed in diaphragm and heart, respectively. Unexpectedly, 2.5- and 3-fold increases were found in liver for CyPep6- and CyPep10-conjugated 23AON. A less prominent and non-significant 1.5-fold increase in kidney was seen for both conjugated 23AONs (Figure 5B).
Figure 5.
Systemic Evaluation of CyPep6- and CyPep10-Conjugated AON
Subcutaneous administration of 4-week-old mdx mice (four to five per group), four times per week, with 50 mg/kg 23AON, a molar equivalent of CyPep6-23AON, CyPep10-23AON, or saline for 9 weeks. One week after the last injection tissues were isolated. (A) After the first injection blood samples were taken at several time points and at sacrifice to determine AON levels in plasma. (B) A hybridization-ligation assay was used to determine AON levels in tissue. (C) RNA was isolated and exon 23 skipping levels were evaluated by single RT-PCR and semiquantitatively determined by lab-on-a-chip analysis. (D) Exon-skipping levels in gastrocnemius, quadriceps, and triceps muscle were quantified using ddPCR for CyPep10-23AON- and 23AON-treated mice. Bars represent mean ± SD. A one-way ANOVA with a Bonferroni post hoc test was used to determine significance (*p < 0.05). D, diaphragm; G, gastrocnemius; H, heart; L, liver; K, kidney; Q, quadriceps; Ti, tibialis anterior; Tr, triceps.
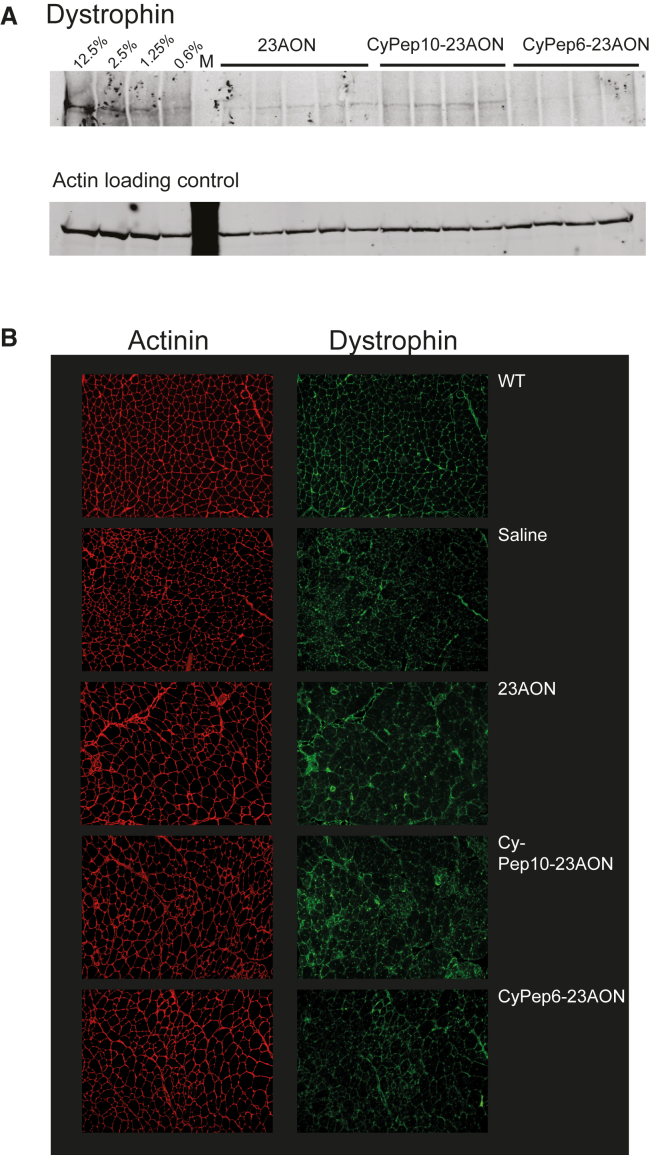
Exon-skipping levels in skeletal and cardiac muscle, determined with single-round RT-PCR, showed a significant, 2-fold increase in exon 23 skipping for CyPep10-23AON compared to the unconjugated 23AON (Figure 5C). Surprisingly, CyPep6 conjugation did not show any improvement in exon-skipping levels. Dystrophin could be detected in quadriceps of all AON (conjugated and unconjugated)-treated mice by western blot analysis. Upon visual inspection, a slight increase seems to be present for mice treated with CyPep10-23AON (Figure 6A), but levels were too low for quantification. We also performed dystrophin immunostaining on muscle sections of the triceps muscles. This revealed slight increases in dystrophin staining for all AON (conjugated and unconjugated)-treated mice compared with saline-treated mice and appears to be more prominent for CyPep10-23AON-treated mice (Figure 6B).
Figure 6.
Dystrophin Analysis
(A and B) Dystrophin protein levels have been determined (A) in quadriceps muscle by western blot and (B) by immunofluorescent staining of dystrophin in triceps muscle. The first four lanes in (A) named 12.5%–0.6% are diluted control protein lysates. M, marker.
We replicated the RT-PCR results for CyPep10-23AON for gastrocnemius, quadriceps, and triceps muscles using a highly quantitative method, i.e., digital droplet PCR (ddPCR).38 These results showed a 2.4-fold increase, on average, in exon 23 skipping levels for CyPep10-23AON compared with the unconjugated 23AON. This supports our findings from the RT-PCR (Figure 5D).
Lastly, in samples obtained just prior to sacrifice, we evaluated blood and plasma parameters for liver and kidney damage and function to determine any possible toxicity of the conjugates used. Hemoglobin levels, urea (a marker for kidney function), and alkaline phosphatase (ALP; a marker for hepatobiliary function) showed no significant differences between groups and were within the normal range for mdx mice. Levels of glutamate pyruvate transaminase (GPT) and glutamic oxaloacetic pyruvate transaminase (GOT), markers for liver and muscle damage, and creatine kinase (CK), a marker for muscle damage, were evaluated. No significant differences were observed for GPT. GOT levels were slightly lower, but not significantly, for CyPep10-23AON. CK was significantly decreased in CyPep10-23AON-treated mice compared with the 23AON, but not compared with saline-treated mice because of the high variation observed in this group (Figure S3).
Investigation of the Uptake Mechanism Points toward Receptor-Mediated Uptake
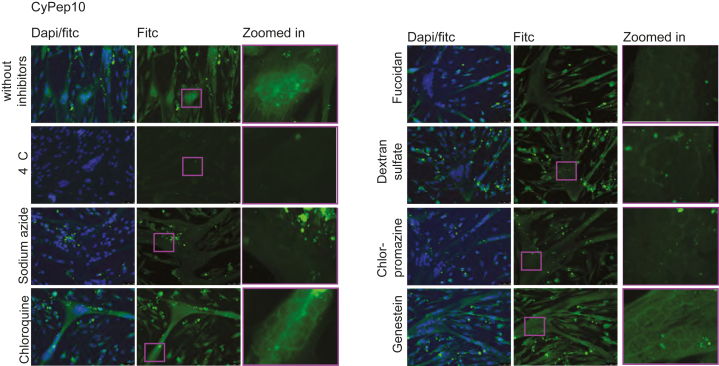
Encouraged by the positive results for CyPep10, we set out to study the possible uptake mechanisms for this peptide. Human control myotubes were incubated with a number of pharmacological inhibitors (Table 2) for 3.5 hr, where 2.25 μM FITC-CyPep10 was added after 30 min. CyPep10 showed a clear energy-dependent uptake, because no fluorescence was observed at 4°C, and reduced fluorescence was observed after pre-incubation with sodium azide. We speculate that there is no endosomal entrapment of FITC-CyPep10 based on the finding that incubation with 5 μg/mL chloroquine did not result in increased fluorescence and no punctate fluorescence, which is indicative for endosomal entrapment, is observed. With fucoidan (75 μg/mL) and dextran sulfate (2 μg/mL) blocking uptake by scavenger receptors, we observed less fluorescence, suggesting that scavenger receptors are involved in the uptake of FITC-labeled CyPep10. Incubation with chlorpromazine (2.5 μM) resulted also in less fluorescence, suggesting that clathrin-mediated uptake is involved. In contrast, incubation with genistein (200 μM), an inhibitor of caveolin-mediated uptake, did not show any difference in fluorescence compared with CyPep10 alone. Combined, these results point toward an energy-dependent uptake of CyPep10 via scavenger receptors accumulated in clathrin-coated pits of the muscle cell membrane (Figure 7).
Table 2.
Pharmacological Inhibitors
| Condition | Concentration | Mechanism |
|---|---|---|
| 4°C | – | inhibition of energy-dependent uptake |
| Sodium azide | 7.7 μM | inhibitor of ATPase (energy-dependent uptake) |
| Chloroquine | 5 μg/mL | causes swelling and disruption of endocytic vesicles, facilitating endosomal escape |
| Chlorpromazine | 2.5 μM | inhibitor of clathrin-mediated uptake |
| Fucoidan | 75 μg/mL | inhibitor of scavenger receptor-mediated uptake |
| Dextran sulfate | 2 μg/mL | inhibitor of scavenger receptor-mediated uptake |
| Genistein | 200 μM | inhibitor of caveolin-mediated uptake |
Figure 7.
Uptake Mechanism of CyPep10
Human control myotubes were incubated with pharmacological inhibitors for 30 min prior to a 3-hr incubation with CyPep10 in the presence of the inhibitors. Representative pictures are shown. Scale bar, 50 μm.
Discussion
Exon skipping is a therapeutic approach for DMD. Improved delivery of AON to the target tissue is anticipated to increase the therapeutic effect. Achieving this in a safe and effective way is challenging. Most progress to increase the delivery and efficacy has been achieved by the conjugation of arginine-rich CPPs to non-anionic AONs such as peptide nucleic acid (PNA) and PMO. It was found that some of these highly cationic peptides were poorly tolerated in higher animals because of kidney toxicity.24 A different approach is the use of tissue-homing peptides, which are not necessarily highly cationic. In 1999, Samoylova and Smith39 described a phage, expressing the 7-mer peptide ASSLNIA, which showed increased fluorescence in murine tissues compared with control phages upon histochemical analyses. Unfortunately, the peptide alone was unable to increase delivery or exon-skipping levels in mdx mice when conjugated to a PMO.40 More recently, Gao et al.41 reported the M12 peptide RRQPPRSISSHP, which, upon conjugation to a PMO, resulted in improved but variable exon-skipping levels in mdx mice in skeletal muscle, but not heart. Previously we described the 7-mer P4 peptide LGAQSNF identified through phage display studies, which, upon conjugation to a 2OMePS AON, resulted in significantly increased exon-skipping levels in diaphragm and cardiac muscle tissue of mdx mice.33 Taken together, tissue-homing peptide conjugation to AONs appears to be a promising strategy.
We believe the approach we have used here (Figure 1) further facilitates the identification of effective muscle-homing peptides from phage display studies for several reasons. First, as already outlined by other studies,32, 42, 43, 44 the use of NGS enables the study of large numbers of sequences. By sequencing the naive library before and after one round of bacterial amplification, one can identify library-specific peptide sequences that are overrepresented or have an amplification bias, the so-called parasite sequences.32, 44 With our approach, we assessed that a little more than 1.9 million unique sequences in the naive amplified library are potentially parasitic sequences (around 21% of the total library). In the enriched libraries, ∼11% of the unique sequences were represented by these parasitic sequences. However, even with NGS, only a fraction of possible sequences in the enriched libraries are sequenced (for comparison, the naive library contains 1.28 × 109 unique sequences, whereas we sequenced approximately 1–2.5 × 106 sequences of the enriched libraries). Nevertheless, these millions of sequences are a vast improvement over the ∼50–100 phage clones generally sequenced with Sanger sequencing.33 Furthermore, many sequences in the enriched library will be present only once or twice, with the more abundant sequences primarily being of interest. As such, NGS will generally provide sufficient depth to identify interesting candidates. In addition, the databases Pepbank45 and Sarotup31 can be used to filter known parasitic sequences or target-unrelated sequences, but are mainly applicable to linear peptides and do not provide sufficient information about cyclic peptides. Second, combining a first-round in vitro screening with a second selection round in vivo screening, we aimed to increase the potential binding of peptides toward muscle, because peptides with no affinity toward muscle would be deselected in the first round. By isolating phages from liver and kidney, we were able to filter peptides that predominately were taken up by these tissues (e.g., target-unrelated sequences).
The 12 selected candidate peptides contained a group of 7 that were identified in either skeletal or cardiac muscle (data not shown), and a group of 5 that were identified in both skeletal and cardiac muscle. In vitro evaluation of these candidates in human control myotubes and cardiomyocyte cultures left us with two peptides for further study: CyPep6 and CyPep10, both identified in skeletal and cardiac muscle. Interestingly, of the peptides selected based on presence in skeletal muscle or cardiac muscle, only three out of seven showed some degree of fluorescence. While in the group selected based on presence in skeletal and cardiac muscles, two out of three showed bright fluorescence and one showed some fluorescence. This suggests that for future studies it may be more efficient to focus only on peptides enriched in both skeletal and cardiac muscle tissue.
Because this was the first time cyclic peptides were conjugated to 2OMePS AONs for DMD, we investigated the influence of such conjugation on exon-skipping efficiency in vitro (human control myotubes) and in vivo (IM administration in hDMD mice) in a humanized setting. No clear differences were observed in the efficiency of conjugated and unconjugated AON, suggesting that the conjugation has no influence on the exon-skipping efficiency of the AON. We can only speculate why in these settings no improved efficiency is seen for the conjugate. It should be kept in mind that the peptides have been selected based on phage display data for their ability to reach muscle fibers after systemic administration. As such, it is not a given that they also facilitate uptake in cultured cells in vitro or by muscle fibers after local injection.
Systemic administration of CyPep6- and CyPep10-conjugated 23AON in mdx mice revealed a 2-fold increase of AON levels in skeletal muscle and around a 3-fold increase in diaphragm muscle and heart for both peptide conjugates over the unconjugated 23AON. This resulted in a significant 2-fold increase in exon skipping in all tissues analyzed for CyPep10-23AON, but surprisingly not for CyPep6-23AON (Figure 5). A potential explanation for the discrepancy between CyPep6-23AON tissue levels and activity may lie in the fact that the peptides were isolated from muscle and heart tissue homogenates, and not selected for their capacity to actually enter the tissue or tissue cells. It is therefore possible that CyPep6-23AON conjugate is trapped somewhere in the endothelium or interstitium. Judged on the fluorescence observed in vitro, CyPep6 outperformed CyPep10 for both cell lines at time points 1 and 2 hr, except for the 10-min incubation time point (Figure 3). CyPep6 appears to be taken up by cells efficiently, however not as fast as CyPep10. This could indicate that different mechanistic uptake pathways are involved and that fast uptake in vitro might lead to a better outcome in vivo. If CyPep6 alone does enter tissue and tissue cells, conjugation to the larger AON may have negatively affected the tissue or cellular uptake in vivo. The fact that CyPep10-23AON did show increased exon skip levels suggests that this peptide conjugate is taken up by the target cells efficiently. It is also possible that different physicochemical properties underlie these differences, which will be addressed in future experiments.
Unfortunately, dystrophin levels were very low and could not be accurately quantified. It is known that dystrophin levels accumulate for 12–24 weeks after treatment initiation,46 so the time of sacrifice (deemed optimal for analysis of AON and exon-skipping levels) is probably not optimal for assessing dystrophin levels. Results of dystrophin analyses in immunostained muscle sections are in line with the western blot results. Taken together these results are encouraging, but future experiments will have to reveal whether longer-term treatment with CyPep10-AON has a true impact on the level of dystrophin restoration.
Compared with unconjugated AON, we observed for CyPep10-23AON similar uptake in kidney, but a 2.5-fold increase in AON levels in liver. No clear indication is found in our NGS data that could explain the increased uptake in liver. CyPep10 showed a 4-fold increase in frequency count in the second round in vivo in quadriceps and heart compared with the first round in vitro (internalized phage fraction). Frequency counts in liver were 2.5 times lower in the second selection round in vivo compared with quadriceps and heart. After the first selection round in vivo, the counts in liver were 6.5 times lower compared with heart and 1.5 times lower compared with kidney. Thus the kidney and liver levels of CyPep10-AON conjugate may seem surprising given that the peptide was selected based on low uptake in these organs. However, one should not forget that the identified peptide was conjugated, in multiple copies, to a phage, while now it is singly conjugated to a 2OMePS AON, which is by itself taken up efficiently by liver and kidney. It is not unlikely that this is further facilitated by peptide conjugation and/or that increased levels are due to the increased plasma half-life.
We finally investigated the mechanism by which FITC-labeled CyPep10 is taken up. Results clearly show that uptake of CyPep10 by muscle cells is energy dependent, because incubation at 4°C or with an ATP inhibitor almost completely prevented uptake. The involvement of scavenger receptors accumulated in clathrin-coated pits is also implied, because uptake was inhibited by inhibitors of scavenger receptors and clathrin-mediated uptake. Clathrin-mediated uptake is one of the main and best studied uptake pathways.47 It requires strong receptor-ligand binding and clustering in clathrin-coated pits in an energy-dependent way. The clathrin-coated pits invaginate and pinch off from the membrane into clathrin-coated vesicles, which later on form early and late endosomes. Subsequently, ligand and receptors are sorted and transported to their appropriated cellular destination, such as Golgi apparatus, liposomes, nucleus, or back to the cell membrane. Endosomes appear to play a role after the peptide enters the cell in clathrin-coated vesicles. This is supported by the fact that pre-incubation with chloroquine, an inhibitor of endosomal entrapment, did not result in a clear increase in fluorescence. In line, no clear indications were found that CyPep10 is taken up or trapped in endosomes because no punctate fluorescence was observed in the cytoplasm of the cells.
In conclusion, we have identified CyPep10 from phage display studies as a potential candidate for AON delivery. Although this peptide was selected as muscle specific, upon conjugation to the AON, the conjugate showed general improved tissue levels in a mouse model for DMD. Extended studies will be needed to assess whether long-term treatment leads to beneficial effects on muscle quality and function and to assess safety in more detail.
Materials and Methods
Animal Care
All experiments were approved by the animal experimental commission (DEC) of the Leiden University Medical Center (LUMC) and performed according to Dutch regulation for animal experimentation. Mice were housed with 12-hr light-dark cycles in individually ventilated cages and had access to standard chow and water ad libitum. In all studies mice of mixed gender were used.
Cell Culture
All cells were cultured in an incubator at 37°C and 5% CO2. All experiments have been performed in live cells.
Human control myoblasts (7304-1 cells, kindly provided by Dr. Vincent Mouly,48 only used for phage display biopanning) were grown in NutMix F-10 (HAM) medium supplemented with GlutaMAX-I, 20% fetal bovine serum (FBS), and 1% penicillin/streptomycin (P/S) (all from GIBCO-BRL, the Netherlands) in flasks coated with purified bovine dermal collagen (collagen) for cell culture (Nutacon, the Netherlands). Cells were plated on collagen-coated Petri dishes and grown to 90% confluence before switching to differentiation medium (Dulbecco’s medium [DMEM], without phenol red, with 2% FBS, 1% P/S, 2% GlutaMAX-I, and 1% glucose; all from GIBCO-BRL, the Netherlands). Cells were allowed to differentiate for 7–14 days.
Human control myoblasts (Km155.c25 cells, kindly provided by Dr. Vincent Mouly, only used for the in vitro evaluation of labeled peptides and conjugates) were grown in skeletal muscle cell growth medium (Promocell, C-23160) supplemented with an extra 15% FBS and 50 μg/mL gentamicin (PAA Laboratories, USA) in uncoated flasks until 70%–80% confluence was reached. Cells were plated in a six-well plate with 0.5% gelatin-coated cover glass slips (Sigma-Aldrich, the Netherlands), at a density of 1 × 105 cells per well, 48 hr prior to differentiation. Reaching 90% confluence, medium was switched to differentiation medium (DMEM [without phenol red] with 2% FBS, 50 μg/mL gentamicin, 2% GlutaMAX-I, and 1% glucose). Cells were allowed to differentiate for 3–5 days.
Immortalized human cardiomyocytes from a healthy individual (Applied Biological Materials, Canada) were grown in Prigrow I medium supplemented with 10% FBS and 1% P/S in collagen-coated flasks. Cells were plated in collagen-coated cover glass slips in six-well plates and grown until confluence prior to experiments.
Phage Display Peptide Library
The Ph.D.-C7C Phage Display Peptide Library kit (New England Biolabs [NEB], USA) was used in the experiments. This library is generated from a degenerate (NNK)7 oligonucleotide library (K represents an admixture of 50% T and 50% G) and two cysteine residues that are positioned at each end of the random 7-mer peptide.
In Vitro Biopanning
In vitro biopanning was performed as previously described by ’t Hoen et al.32 Differentiated human control myoblasts cells were washed three times with PBS and incubated with DMEM supplemented with 0.1% BSA for 1 hr at 37°C, 5% CO2. Cells were washed with PBS and incubated with 2 × 1011 phages from the Ph.D.-C7C Phage Display Peptide Library kit (NEB) in 3 mL of DMEM medium for 1 hr at 37°C, while shaking at 70 rounds per minute. After incubation, the cells were gently washed six times by incubating with 5 mL of ice-cold DMEM containing 0.1% BSA, for 5 min. Subsequently, the cells were incubated for 10 min on ice with 3 mL of 0.1 M HCl (pH 2.2) to elute cell-surface-bound phages, which was neutralized by addition of 0.6 mL of 0.5 M Tris. To recover the cell-associated phages, we lysed cells for 1 hr on ice in 3 mL of 30 mM Tris-HCl, 1 mM EDTA [pH 8]. Phages from each fraction were titrated and amplified according to the manufacturer’s instruction (NEB).
In Vivo Biopanning
In total, three mdx mice (C57Bl/10ScSn-DMDmdx /J) were injected intravenously (i.v.) with 2 × 1011 phages either from the first-round in vitro cell-surface-bound phages, in vitro internalized phages (i.e., second selection round in vivo), or the naive Ph.D.-C7C library (i.e., first in vivo selection round). Phages were circulated for 1 hr, after which mice were under anesthesia perfused with PBS. Quadriceps muscles, heart, and liver were isolated from mice injected with phages from the in vitro selection. Gastrocnemius and quadriceps muscles, heart, liver, and kidney were isolated from the mouse injected with the naive library. Tissues were homogenized in Tris-buffered saline (TBS) buffer using a MagNalyzer according to manufacturer’s instruction (Roche Diagnostics, the Netherlands). Phages were titrated and amplified according to manufacturer’s instruction (NEB) (from here on referred to as enriched phage library).
DNA Isolation and NGS
Total phage DNA was isolated from all enriched phage libraries, the naive unselected library, and the naive library after a single round of bacterial amplification. From each enriched phage library, 2 × 1011 phage particles were added to 500 μL of lysogeny broth (LB) growth media in a 1.5-mL tube. The phages were precipitated with 200 μL of PEG 8000/NaCl (Sigma-Aldrich) for 3–4 hr at room temperature. Phages were pelleted, and DNA was isolated according to the manufacturer’s instruction. The final pellet (phage DNA) was dissolved in Milli-Q water, and DNA concentration was determined by NanoDrop (Thermo Fisher Scientific, the Netherlands). Phage DNA was amplified by PCR using the following primers (* is a phosphorothioate bond): forward: 5′-AAT GAT ACG GCG ACC ACC GAG ATC TAC ACT TCC TTT AGT GGT ACC TTT CTA TTC TC*A-3′; reverse: 5′-CAA GCA GAA GAC GGC ATA CGA GAT CGG BARCODE TCT ATG GGA TTT TGC TAA ACA ACT TT*C-3′.
The PCR primers used to amplify the phage DNA contain a subsequence that recognized the sequence flanking the 27-nt-long unknown insert sequence (including the sequences encoding for the two cysteines), the adapters necessary for binding to the Illumina flow cell and a unique barcode (underlined) for every enriched phage library. The PCR protocol applied was the following: 1 ng of phage DNA was incubated with 2.625 U high-fidelity Taq polymerase (Roche Diagnostics), 20 pM primers in 1× high-fidelity PCR buffer containing 15 mM MgCl2, and amplified for 20 cycles, each consisting of an incubation for 30 s at 94°C, 30 s at 67°C, and 30 s at 72°C. The PCR was stopped in exponential phase to mitigate PCR-induced sequence biases. The final PCR product was purified with the QIAquick PCR purification kit (QIAGEN, USA). Concentrations, as well as the correct length of the PCR products, were established with an Agilent 2100 Bioanalyzer DNA 1000 assay (Agilent Technologies, USA). All PCR products from the enriched phage libraries were combined in a single lane. Phage fractions from the naive unselected library (with and without amplification) were combined together in another lane of the Illumina flow cell. Both pools were subjected to solid-phase amplification in the cluster station following manufacturer’s specification (Illumina, USA). Up to 50 cycles of single end sequencing (the minimal amount required for single end sequencing in the department due to presence of unrelated samples in the other lanes) were performed using a custom sequencing primer that started exactly at the first position of the unknown insert sequence (5′-ACA CTT CCT TTA GTG GTA CCT TTC TAT TCT CAC TC*T-3′). Sequencing was performed with the Illumina HiSeq 2000 with a v3 flow cell and reagents (Illumina, USA).
NGS Analyses
The Illumina CASAVA 1.8.2 software was used to extract fastq files from Illumina BCL files and to split the data based on the individual sample barcodes. For further analyses, sequences were filtered out if they did not fulfill the following criteria: sequences should start with GCT TGT followed by (NNK)7 and end with TGC GGT GGA GGT, with N being any nucleotide and K being G or T. Subsequently, sequences were translated to amino acid sequences with a custom perl script using conventional amino acid codon tables. When the stop codon TAG was encountered, this was changed to a CAG codon according to manufacturer’s instruction (NEB). An overview of the coverage is shown in Table S1 and Figure 1. All sequenced phage library data were normalized by a square root transformation on the number of counts in the library, a commonly applied data transformation to stabilize the variance in count data.35 Subsequently, parasitic sequences were excluded. Parasitic sequences were defined as sequences for which the frequency count in the naive amplified library minus the frequency count in the unamplified naive library was greater than 2. Next, two separate analyses were performed. First, sequences with a frequency count higher than 2 in liver and/or kidney were removed from the enriched skeletal and cardiac muscle libraries. Sequences in the skeletal and cardiac muscle libraries were, per individual enriched library, ranked by frequency count and interesting candidates divided in two groups, i.e., “skeletal muscle” and “cardiac muscle.” Second, the threshold for liver and kidney was ignored, and skeletal and cardiac muscle libraries were ranked based on frequency count. Peptide sequences with higher frequency counts in liver and/or kidney compared with skeletal or cardiac muscle were removed. Interesting candidates were selected and combined in the group “cardiac muscle and skeletal muscle.” A final list of 25 candidate peptides was created (7 peptides for skeletal muscle; 7 peptides for cardiac muscle, and 9 peptides for skeletal and cardiac muscle groups). Finally, sequences with more than two positively charged amino acids (arginines or lysines) were removed from the final candidate list. Of this list, the best 12 candidate peptides were selected for further evaluation (3 from the skeletal muscle group, 4 from the cardiac muscle group, and 5 from the skeletal and cardiac muscle group).
Fluorescently Labeled Peptides
Fluorescently labeled cyclic and linear (i.e., with Cys→Ala substitution) peptides were obtained from Pepscan (the Netherlands); C terminus amidated peptides containing an Ahx (6-aminohexanoic acid) spacer attached to the N terminus were equipped with an FITC label at the Ahx amine group. Cyclic peptides were obtained by internal disulfide formation.
In Vitro Evaluation of FITC-Labeled Peptides
Positive/Negative Screening
FITC-labeled peptides were dissolved in water and, if necessary, acetic acid was used to help dissolve the peptide (final acetic acid concentration in experiment <0.2% v/v.) Final peptide concentrations were determined by spectrophotometric analysis at 490 nm (pH 7.5; ∼20 times diluted in 1 M Tris-HCl buffer [pH 7.5]).
Human control myotubes and immortalized human cardiomyocytes were washed twice with PBS and incubated with 2.25 μM FITC-labeled peptides in serum-free media for 3 hr at 37°C and 5% CO2. Cells were washed three times with PBS and fixed with cold methanol (−20°C) for 5 min (human control myotubes) or 10 min (human cardiomyocytes). Subsequently, the glass slides were shortly air-dried and embedded on microscope slides with Vectashield hard set containing DAPI mounting media (Vector Laboratories, UK). After drying for 30 min, slides were analyzed with fluorescence microscopy, 20× magnification (Leica DM5500 B) using a CCD camera (Leica DFC 360 FX).
Mechanistic Uptake Experiments
Human control myotubes in six-well plate on gelatin-coated cover glass slips (as previous described) were incubated for 30 min at 37°C and 5% CO2 with 7.7 μM sodium azide (NaN3; Riedel-de Haën, Germany), 5 μg/mL chloroquine diphosphate (Sigma-Aldrich), 75 μg/mL fucoidan (from Fucus vesiculosus; Sigma-Aldrich), 2 μg/mL dextran sulfate sodium salt (Sigma-Aldrich), 2.5 μM chlorpromazine (Sigma-Aldrich), and 200 μM genistein (Sigma-Aldrich), or were kept at 4°C prior to the addition of 2.25 μM FITC-labeled peptide and subsequently incubated for another 3 hr at 37°C and 5% CO2 or 4°C in the presence of the inhibitors (one inhibitor per well). Cells were washed and slides were analyzed as previously described for positive/negative screening of the peptides.
AON and CyPep-AON Conjugates
General
The 5′-carboxylate linker phosphoramidite was purchased from Link Technologies (UK). All solvents and reagents were obtained from Sigma-Aldrich (the Netherlands) or Acros (Belgium) and used as received unless indicated otherwise. Cyclic peptides used for the conjugation were synthesized by Bachem (Switzerland). Observed molecular weights were corrected for reference standard values.
AON Synthesis
2OMePS AONs modified with a 5′-carboxylate linker were prepared through standard phosphoramidite chemistry protocols, using a 2-chlorotrityl (Clt)-protected amidite for the last coupling (15 equivalent grade [eq], 20 min modified coupling conditions) and final removal of the Clt group. Cleavage/deprotection (0.1 M NaOH in MeOH/H2O 4/1 [v/v], 18 hr, 55°C), addition of NaCl and desalting by FPLC, and subsequent lyophilization yielded the desired crude AON, which was of sufficient purity to use in the next steps. AONs sequences are as follows: h45AON, 5′-UGCCGCUGCCCAAUGCCAUCCUG-3′; 23AON, 5′-GGCCAAACCUCGGCUUACCU-3′.
CyPep10-h45AON Synthesis
The 5′-carboxylate-modified h45AON (1 μmol) was added to a solution of O-(1H-6-chlorobenzotriazole-1-yl)-1,1,3,3-tetramethyluronium hexafluorophosphate (HCTU; 2.3 eq) and hydroxybenzotriazole (HOBt; 2 eq) in DMSO (0.4 mL) for preactivation by shaking for 3 min at room temperature (RT). CyPep10 (2 μmol and 2.3 eq N,N-diisopropylethylamine [DiPEA] in 0.1 mL of dimethylformamide [DMF]) was added, and the reaction mixture was shaken for 1 hr at RT. Reversed phase high-performance liquid chromatography (RP-HPLC) purification was followed by salt exchange using a small excess of NaCl. Excess salt was removed by FPLC, and the conjugate was evaporated to dryness three times from Milli-Q, yielding CyPep10-h45AON (0.3 μmol [31%]).
CyPep6-23AON and CyPep10-23AON Synthesis
Both conjugates were obtained through a similar procedure as described for CyPep10-h45AON, in larger scale from six separate pooled syntheses: CyPep6-23AON (38 μmol [37%]) and CyPep10-23AON (38 μmol [36%]).
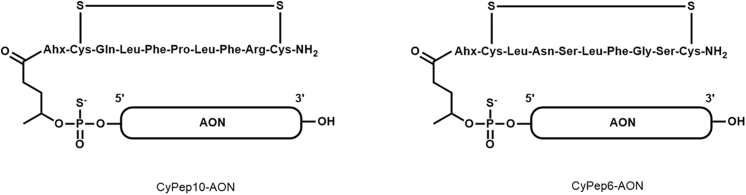
A representative illustration of the CyPep-AONs is given in Figure 8 and a representative chromatogram in Figure S2. The molecular weight data of the CyPep-AON conjugates are given in Table S3. All conjugates had a purity >85%.
Figure 8.
CyPep-AON Conjugates
The chemical structures of the conjugates.
In Vitro Evaluation of Peptide-Conjugated AON
Human control myotubes (differentiated for 3–5 days) were incubated with 2 or 4 μM AON or peptide-conjugated AON (CyPep-h45AON) for 96 hr in differentiation medium without a transfection reagent. After incubation, cells were washed three times with PBS and RNA was isolated by adding 500 μL of TriPure (Roche Diagnostics) to each well to lyse the cells. This was followed by chloroform extraction in a 1:5 ratio on ice for 5 min. After centrifugation (4°C, 15 min, 15,400 relative centrifugal force [rcf]) the upper aqueous phase was precipitated for 30 min on ice with equal volume of isopropanol. Subsequently, RNA was pelleted down by centrifugation (4°C, 15 min, 15,400 rcf) and the pellet was washed with 70% ethanol. The final pellet (RNA) was dissolved in Milli-Q water. For cDNA synthesis, half of the RNA was used in a 20 μL reaction with a specific reverse primer in exon 48 and transcriptor reverse transcriptase (Roche Diagnostics) for 30 min at 55°C and 5 min at 85°C to terminate the reaction. For PCR analysis, 3 μL of cDNA was incubated with 0.625 U of AmpliTaq polymerase (Roche Diagnostics), 10 pM primers (reverse primer in exon 48 and a forward primer in exon 43), one-time SuperTaq PCR buffer (Enzyme Technologies, UK), and amplified for 20 cycles, each consisting of an incubation for 40 s at 94°C, 40 s at 60°C, and 80 s at 72°C. This PCR was followed by a nested PCR in which 1.5 μL of the first PCR was incubated with 1.25 U of AmpliTaq polymerase (Roche Diagnostics), 20 pM primers (reverse primer in exon 47 and a forward primer in exon 44), and one-time SuperTaq PCR buffer (Enzyme Technologies, UK) amplified for 32 cycles, each consisting of an incubation for 40 s at 94°C, 40 s at 60°C, and 60 s at 72°C. Exon-skipping levels were semiquantitatively determined as the percentages of the total (wild-type and skipped) product with the Agilent 2100 Bioanalyzer.
In Vivo Evaluation of Peptide-Conjugated AONs
hDMD Mice
Four hDMD mice were injected in the gastrocnemius and triceps muscles with cardiotoxin 2 days prior to injection with 20 μg of h45AON or equimolar CyPep10-h45AON contralateral for 2 consecutive days. One week after the last injection the mice were sacrificed; quadriceps (non-injected control), gastrocnemius, and triceps muscles were isolated to determine exon skip levels as described for the in vitro evaluation of peptide-conjugated AON.
Mdx Mice
Four-week-old (four to five mice per group, mixed male/female [m/f]) mdx mice (C57Bl/10ScSn-DMDmdx/J) were s.c. injected with 50 mg/kg AON, molar equivalent of CyPep-AON, or saline four times per week for 8 weeks. After the first injection, blood was collected via the tail vein to determine AON levels in plasma at 30 min, 1 hr, 3 hr, and 6 hr via sparse sampling approach. One week after the last injection, blood was collected via the tail vein to determine AON in plasma and assess plasma levels of markers for liver and kidney function and damage. Subsequently, mice were anesthetized, sacrificed by perfusion with PBS, and gastrocnemius, quadriceps, tibialis anterior, triceps, diaphragm, heart, kidney, and liver were isolated to determine exon skip, AON, and dystrophin levels in tissue.
Determination of In Vivo Exon Skip Levels
Skeletal and cardiac muscles were homogenized in TriPure buffer (Roche Diagnostics) using the MagNaLyzer and MagNaLyzer green beads (Roche Diagnostics). Total RNA was isolated as described for the in vitro evaluation of peptide-conjugated AON.
RT-PCR
cDNA was generated using 400 ng of RNA in a 20 μL reaction with random hexamer primers and transcriptor reverse transcriptase (Roche Diagnostics) for 45 min at 42°C. For PCR analysis, 1.5 μL of cDNA was incubated with 1.25 U of Taq polymerase (Roche Diagnostics), 20 pM primers (reverse primer in exon 24, forward primer in exon 22), and one-time SuperTaq PCR buffer (Enzyme Technologies, UK) and amplified for 30 cycles, each consisting of an incubation for 30 s at 94°C, 30 s at 60°C, and 30 s at 72°C. Exon-skipping levels were semiquantitatively determined as the percentages of the total (wild-type and skipped) product with the Agilent 2100 Bioanalyzer (lab-on-a-chip analyzer) as described previously by Spitali et al.34
ddPCR
cDNA was generated in 20 μL reactions, using 1,000 ng of total RNA with random hexamer primers (Roche Diagnostics) and transcriptor reverse transcriptase (Roche Diagnostics) according to the manufacturer’s instructions. ddPCR was performed in duplicate as previously described38 on 0.5 μL of cDNA using a TaqMan assay spanning the exon 22–23 junction to detect the non-skipped fragment and an assay spanning the exon 22–24 junction to detect the skipped fragment (sequences are listed in Table S2). The concentration (in copies/μL sample mix) of the skipped assay and non-skipped assay was used to calculate the percentage of exon 23 skip [copies/μL skipped/(copies/μL skipped + copies/μL non-skipped)*100].
Determination of AON Levels in Plasma and Tissue
For measuring the concentration of (CyPep)-23AON in plasma and tissue samples, a hybridization-ligation assay based on one previously published was used49 following adaptations previously described.50 Tissues were homogenized in 100 mM Tris-HCl (pH 8.5), 200 mM NaCl, 0.2% SDS, 5 mM EDTA, and 2 mg/mL proteinase K using MagNaLyzer green bead tubes in a MagNaLyzer (Roche Diagnostics). Samples were diluted 600 and 6,000 times (muscle) or 6,000 and 60,000 (liver and kidney) in pooled control mdx tissue in PBS. Calibration curves of the analyzed 23AON prepared in 60 times pooled control mouse mdx tissue in PBS were included. All analyses were performed in duplicate. For plasma, the samples were diluted as follows: t = 30 min and 1 hr 10,000 and 100,000 times, t = 3 hr 1,000 and 10,000 times, t = 6 hr and sacrifice 100 and 1,000 times, all in pooled control plasma of mdx mice in PBS. Calibration curves of the analyzed 23AON prepared in 100 times pooled control plasma of mdx mice in PBS were included.
Determination of Protein Levels by Western Blot
Quadriceps muscle samples were homogenized in MagNaLyser green bead tubes with a MagNaLyser (Roche Diagnostics, the Netherlands). Samples were homogenized for 20 s at speed 7,000 (two to five rounds) in 1 mL of 125 mM Tris-HCl (pH 6.8) buffer supplemented with 20% (w/v) SDS. Protein concentrations were determined by the bicinchoninic acid (BCA) protein assay kit (Thermo Fisher Scientific, the Netherlands) using BSA as a standard according to manufacturer’s instruction. Prior to loading, a sample volume (for muscle tissue and reference samples) representing 25 μg of total protein was supplemented with loading buffer consisting of 125 mM Tris-HCl (pH 6.8), 20% (v/v) glycerol, 5% (v/v) β-mercaptoethanol, and 0.0008% (w/v) bromophenol blue, to reach a final volume of 20 μL. Subsequently, this was incubated for 5 min at 95°C. Reference concentration samples were made by diluting total protein from wild-type in mdx lysates of the same muscle type. Samples were loaded on Criterion XT Tris acetate (polyacrylamide) gels containing 18 slots, with a linear resolving gel gradient of 3%–8% (Bio-Rad Laboratories, the Netherlands). Gels were run for 1 hr at 75 V (∼0.07 A), followed by a 2-hr incubation at 150 V (∼0.12 A), on ice, using XT Tricine as running buffer (Bio-Rad Laboratories, the Netherlands). The gel was blotted on a nitrocellulose membrane (Bio-Rad Laboratories, the Netherlands) using the ready-to-use Trans-Blot Turbo transfer packs and the Trans-Blot Turbo transfer system from Bio-Rad at 2.5 A (∼25 V) for 10 min (standard Bio-Rad protocol for high molecular weight proteins). The membrane was blocked for 1 hr with 5% (w/v) non-fat dried milk powder (ELK Campina Melkunie, the Netherlands) in a TBS buffer (10 mM Tris-HCl [pH 8.0] and 0.15 M NaCl). Membranes were washed three times for 10 min with TBS-T buffer (TBS buffer with 0.005% [v/v] Tween 20) and incubated with primary antibody for dystrophin (GTX15277, 1:2,000; Gene Tex, USA) and primary antibody for loading control, alpha-actinin (AB72592, 1:7,500; Abcam, UK), in TBS buffer overnight at room temperature with gentle agitation. Membranes were washed three times for 15 min in TBS-T buffer and incubated with secondary antibody IRDye 800 CW for dystrophin (1:5,000, IgG; Li-Cor, NE, USA) and IRDye 680TL for alpha-actinin (1:10,000; Li-Cor) in TBS buffer for 1 hr. Membranes were washed two times for 15 min in TBS-T buffer followed by a final washing step of 15 min in TBS buffer. Membranes were analyzed with the Odyssey system and software (Li-Cor).
Immunofluorescent Staining of Muscle Sections
Left triceps muscles were snap frozen in 2-methylbutane (Sigma-Aldrich) cooled in liquid nitrogen. Sections of 8 μm were made on SuperFrost Ultra Plus slides (Thermo Fisher Scientific, Germany) with a Leica CM3050 S cryostat (Leica Microsystems, the Netherlands). Slides with frozen muscle sections were warmed up for 30 min at room temperature, fixated in ice-cold acetone for 5 min, and air-dried for 30 min. A circle was drawn around each muscle section with a DAKO pen (Dako Denmark, Denmark) and washed with PBS. Subsequently, slides were stained overnight at 4°C with primary antibody (Dystrophin antibody, 1:50 [Santa Cruz, Germany]; sc-7461; Laminin antibody, 1:100 [Abcam, UK]; ab11575) in blocking buffer (PBS/0.05% Tween/5% horse serum). The next day slides were washed three times with PBS and incubated with labeled secondary antibody for 60 min at room temperature in the dark (Alexa 594 red [for laminin], 1:1,000 [Life Technology, the Netherlands]; Alexa 488 green [for dystrophin], 1:1,000 [Life Technology, the Netherlands]). Subsequently, slides were washed three times with PBS and embedded in ProLong Gold Antifade Mounting medium. Slides were dried overnight at 4°C and analyzed using a BZ-X700 fluorescent microscope (Keyence, Japan), 10× magnification.
Safety Evaluation
Blood was collected in lithium-heparin-coated microvettes CB300 (Sarstedt, the Netherlands). Hemoglobin (HB), urea, ALP, glutamate pyruvate transaminase (GPT), glutamic oxaloacetic transaminase (GOT), and CK levels were determined using Reflotron strips (Roche Diagnostics) in the Reflotron Plus machine (Roche Diagnostics) as previously described.33
Statistical Analysis
A one-way ANOVA with a post hoc test (Bonferroni) was used to determine significant differences in exon-skipping levels, AON levels, and plasma protein levels. Results were deemed significantly different when p < 0.05.
Author Contributions
Conceptualization and Methodology: S.M.G.J., P.A.C.t.H., V.D.P., P.C.d.V., B.A., A.M.A.-R.; Formal Analyses: S.M.G.J.; Investigation: S.M.G.J., C.T.L.-d.W., R.C.V.; Project Administration and Writing – Original Draft: S.M.G.J.; Writing – Review and Editing: P.A.C.t.H., P.C.d.V., A.M.A.-R.; Supervision: P.C.d.V., A.M.A.-R.; Funding Acquisition: A.M.A.-R.
Conflicts of Interest
A.M.A.-R. is a co-inventor of patents of the LUMC on exon skipping, licensed by LUMC to Prosensa Therapeutics, and is entitled to a share or royalties. S.M.G.J., A.M.A.-R., P.A.C.t.H., B.A., and P.C.d.V. are co-inventors on a patent application that includes the peptides reported in this work. A.M.A.-R. also declares being an ad hoc consultant for Global Guidepoint, GLC consulting, Deerfield Institute, Bioclinica, Grunenthal, Summit PLC, PTC Therapeutics, and BioMarin and is a member of the scientific advisory board of ProQR and Philae Pharmaceuticals. Remuneration for these activities goes to LUMC. P.C.d.V., R.C.V., B.A., and V.D.P. are employees of BioMarin Nederland BV.
Acknowledgments
We hereby thank the following people for their technical or intellectual input: M. van Putten, J.W. Boertje-van der Meulen, M. Hulsker, L. van Vliet, M. Overzier, K. Putker, A. Harder (student) employed by the LUMC (Leiden, the Netherlands); and D. Muilwijk, K.H. Pang, R. Vermue, N.A. Datson, and J.C.T. van Deutekom employed by BioMarin Nederland BV (Leiden, the Netherlands). Studies were financed by the Dutch Ministry of Economic Affairs (IOP-Genomics grant IGE7001 to P.A.C.t.H.) and a grant from the Prinses Beatrix Spierfonds (W-OR13-06 to A.M.A.-R.).
Footnotes
Supplemental Information includes three figures and three tables and can be found with this article online at https://doi.org/10.1016/j.ymthe.2017.10.004.
Supplemental Information
References
- 1.Moat S.J., Bradley D.M., Salmon R., Clarke A., Hartley L. Newborn bloodspot screening for Duchenne muscular dystrophy: 21 years experience in Wales (UK) Eur. J. Hum. Genet. 2013;21:1049–1053. doi: 10.1038/ejhg.2012.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Emery A.E. The muscular dystrophies. Lancet. 2002;359:687–695. doi: 10.1016/S0140-6736(02)07815-7. [DOI] [PubMed] [Google Scholar]
- 3.Blake D.J., Weir A., Newey S.E., Davies K.E. Function and genetics of dystrophin and dystrophin-related proteins in muscle. Physiol. Rev. 2002;82:291–329. doi: 10.1152/physrev.00028.2001. [DOI] [PubMed] [Google Scholar]
- 4.Monaco A.P., Bertelson C.J., Liechti-Gallati S., Moser H., Kunkel L.M. An explanation for the phenotypic differences between patients bearing partial deletions of the DMD locus. Genomics. 1988;2:90–95. doi: 10.1016/0888-7543(88)90113-9. [DOI] [PubMed] [Google Scholar]
- 5.Muntoni F., Torelli S., Ferlini A. Dystrophin and mutations: one gene, several proteins, multiple phenotypes. Lancet Neurol. 2003;2:731–740. doi: 10.1016/s1474-4422(03)00585-4. [DOI] [PubMed] [Google Scholar]
- 6.Aartsma-Rus A., Janson A.A., Kaman W.E., Bremmer-Bout M., den Dunnen J.T., Baas F., van Ommen G.J., van Deutekom J.C. Therapeutic antisense-induced exon skipping in cultured muscle cells from six different DMD patients. Hum. Mol. Genet. 2003;12:907–914. doi: 10.1093/hmg/ddg100. [DOI] [PubMed] [Google Scholar]
- 7.Aartsma-Rus A. Antisense-mediated modulation of splicing: therapeutic implications for Duchenne muscular dystrophy. RNA Biol. 2010;7:453–461. doi: 10.4161/rna.7.4.12264. [DOI] [PubMed] [Google Scholar]
- 8.Mendell J.R., Goemans N., Lowes L.P., Alfano L.N., Berry K., Shao J., Kaye E.M., Mercuri E., Eteplirsen Study Group and Telethon Foundation DMD Italian Network Longitudinal effect of eteplirsen versus historical control on ambulation in Duchenne muscular dystrophy. Ann. Neurol. 2016;79:257–271. doi: 10.1002/ana.24555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mendell J.R., Rodino-Klapac L.R., Sahenk Z., Roush K., Bird L., Lowes L.P., Alfano L., Gomez A.M., Lewis S., Kota J., Eteplirsen Study Group Eteplirsen for the treatment of Duchenne muscular dystrophy. Ann. Neurol. 2013;74:637–647. doi: 10.1002/ana.23982. [DOI] [PubMed] [Google Scholar]
- 10.Aartsma-Rus A. Dystrophin analysis in clinical trials. J. Neuromuscul. Dis. 2014;1:41–53. [PubMed] [Google Scholar]
- 11.Flanigan K.M., Voit T., Rosales X.Q., Servais L., Kraus J.E., Wardell C., Morgan A., Dorricott S., Nakielny J., Quarcoo N. Pharmacokinetics and safety of single doses of drisapersen in non-ambulant subjects with Duchenne muscular dystrophy: results of a double-blind randomized clinical trial. Neuromuscul. Disord. 2014;24:16–24. doi: 10.1016/j.nmd.2013.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Voit T., Topaloglu H., Straub V., Muntoni F., Deconinck N., Campion G., De Kimpe S.J., Eagle M., Guglieri M., Hood S. Safety and efficacy of drisapersen for the treatment of Duchenne muscular dystrophy (DEMAND II): an exploratory, randomised, placebo-controlled phase 2 study. Lancet Neurol. 2014;13:987–996. doi: 10.1016/S1474-4422(14)70195-4. [DOI] [PubMed] [Google Scholar]
- 13.Fall A.M., Johnsen R., Honeyman K., Iversen P., Fletcher S., Wilton S.D. Induction of revertant fibres in the mdx mouse using antisense oligonucleotides. Genet. Vaccines Ther. 2006;4:3. doi: 10.1186/1479-0556-4-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heemskerk H.A., de Winter C.L., de Kimpe S.J., van Kuik-Romeijn P., Heuvelmans N., Platenburg G.J., van Ommen G.J., van Deutekom J.C., Aartsma-Rus A. In vivo comparison of 2′-O-methyl phosphorothioate and morpholino antisense oligonucleotides for Duchenne muscular dystrophy exon skipping. J. Gene Med. 2009;11:257–266. doi: 10.1002/jgm.1288. [DOI] [PubMed] [Google Scholar]
- 15.Heemskerk H., de Winter C., van Kuik P., Heuvelmans N., Sabatelli P., Rimessi P., Braghetta P., van Ommen G.J., de Kimpe S., Ferlini A. Preclinical PK and PD studies on 2′-O-methyl-phosphorothioate RNA antisense oligonucleotides in the mdx mouse model. Mol. Ther. 2010;18:1210–1217. doi: 10.1038/mt.2010.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu B., Xiao B., Cloer C., Shaban M., Sali A., Lu P., Li J., Nagaraju K., Xiao X., Lu Q.L. One-year treatment of morpholino antisense oligomer improves skeletal and cardiac muscle functions in dystrophic mdx mice. Mol. Ther. 2011;19:576–583. doi: 10.1038/mt.2010.288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sirsi S.R., Schray R.C., Guan X., Lykens N.M., Williams J.H., Erney M.L., Lutz G.J. Functionalized PEG-PEI copolymers complexed to exon-skipping oligonucleotides improve dystrophin expression in mdx mice. Hum. Gene Ther. 2008;19:795–806. doi: 10.1089/hum.2007.129. [DOI] [PubMed] [Google Scholar]
- 18.Sirsi S.R., Schray R.C., Wheatley M.A., Lutz G.J. Formulation of polylactide-co-glycolic acid nanospheres for encapsulation and sustained release of poly(ethylene imine)-poly(ethylene glycol) copolymers complexed to oligonucleotides. J. Nanobiotechnology. 2009;7:1. doi: 10.1186/1477-3155-7-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ferlini A., Sabatelli P., Fabris M., Bassi E., Falzarano S., Vattemi G., Perrone D., Gualandi F., Maraldi N.M., Merlini L. Dystrophin restoration in skeletal, heart and skin arrector pili smooth muscle of mdx mice by ZM2 NP-AON complexes. Gene Ther. 2010;17:432–438. doi: 10.1038/gt.2009.145. [DOI] [PubMed] [Google Scholar]
- 20.Bassi E., Falzarano S., Fabris M., Gualandi F., Merlini L., Vattemi G., Perrone D., Marchesi E., Sabatelli P., Sparnacci K. Persistent dystrophin protein restoration 90 days after a course of intraperitoneally administered naked 2'OMePS AON and ZM2 NP-AON complexes in mdx mice. J. Biomed. Biotechnol. 2012;2012:897076. doi: 10.1155/2012/897076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Falzarano M.S., Bassi E., Passarelli C., Braghetta P., Ferlini A. Biodistribution studies of polymeric nanoparticles for drug delivery in mice. Hum. Gene Ther. 2014;25:927–928. doi: 10.1089/hum.2014.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang M., Wu B., Lu P., Cloer C., Tucker J.D., Lu Q. Polyethylenimine-modified pluronics (PCMs) improve morpholino oligomer delivery in cell culture and dystrophic mdx mice. Mol. Ther. 2013;21:210–216. doi: 10.1038/mt.2012.236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moulton H.M., Nelson M.H., Hatlevig S.A., Reddy M.T., Iversen P.L. Cellular uptake of antisense morpholino oligomers conjugated to arginine-rich peptides. Bioconjug. Chem. 2004;15:290–299. doi: 10.1021/bc034221g. [DOI] [PubMed] [Google Scholar]
- 24.Moulton H.M., Moulton J.D. Morpholinos and their peptide conjugates: therapeutic promise and challenge for Duchenne muscular dystrophy. Biochim. Biophys. Acta. 2010;1798:2296–2303. doi: 10.1016/j.bbamem.2010.02.012. [DOI] [PubMed] [Google Scholar]
- 25.Betts C.A., Wood M.J. Cell penetrating peptide delivery of splice directing oligonucleotides as a treatment for Duchenne muscular dystrophy. Curr. Pharm. Des. 2013;19:2948–2962. doi: 10.2174/1381612811319160009. [DOI] [PubMed] [Google Scholar]
- 26.Betts C., Saleh A.F., Arzumanov A.A., Hammond S.M., Godfrey C., Coursindel T., Gait M.J., Wood M.J. Pip6-PMO, a new generation of peptide-oligonucleotide conjugates with improved cardiac exon skipping activity for DMD treatment. Mol. Ther. Nucleic Acids. 2012;1:e38. doi: 10.1038/mtna.2012.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Betts C.A., Saleh A.F., Carr C.A., Hammond S.M., Coenen-Stass A.M., Godfrey C., McClorey G., Varela M.A., Roberts T.C., Clarke K. Prevention of exercised induced cardiomyopathy following Pip-PMO treatment in dystrophic mdx mice. Sci. Rep. 2015;5:8986. doi: 10.1038/srep08986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yin H., Saleh A.F., Betts C., Camelliti P., Seow Y., Ashraf S., Arzumanov A., Hammond S., Merritt T., Gait M.J., Wood M.J. Pip5 transduction peptides direct high efficiency oligonucleotide-mediated dystrophin exon skipping in heart and phenotypic correction in mdx mice. Mol. Ther. 2011;19:1295–1303. doi: 10.1038/mt.2011.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lehto T., Castillo Alverez A., Gauck S., Gait M.J., Coursindel T., Wood M.J., Lebleu B., Boisguerin P. Cellular trafficking determines the exon skipping activity of Pip6a-PMO in mdx skeletal and cardiac muscle cells. Nucleic Acids Res. 2014;42:3207–3217. doi: 10.1093/nar/gkt1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Smith G.P. Filamentous fusion phage: novel expression vectors that display cloned antigens on the virion surface. Science. 1985;228:1315–1317. doi: 10.1126/science.4001944. [DOI] [PubMed] [Google Scholar]
- 31.Huang J., Ru B., Li S., Lin H., Guo F.B. SAROTUP: scanner and reporter of target-unrelated peptides. J. Biomed. Biotechnol. 2010;2010:101932. doi: 10.1155/2010/101932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.’t Hoen P.A., Jirka S.M., Ten Broeke B.R., Schultes E.A., Aguilera B., Pang K.H., Heemskerk H., Aartsma-Rus A., van Ommen G.J., den Dunnen J.T. Phage display screening without repetitious selection rounds. Anal. Biochem. 2012;421:622–631. doi: 10.1016/j.ab.2011.11.005. [DOI] [PubMed] [Google Scholar]
- 33.Jirka S.M., Heemskerk H., Tanganyika-de Winter C.L., Muilwijk D., Pang K.H., de Visser P.C., Janson A., Karnaoukh T.G., Vermue R., ’t Hoen P.A. Peptide conjugation of 2′-O-methyl phosphorothioate antisense oligonucleotides enhances cardiac uptake and exon skipping in mdx mice. Nucleic Acid Ther. 2014;24:25–36. doi: 10.1089/nat.2013.0448. [DOI] [PubMed] [Google Scholar]
- 34.Spitali P., Heemskerk H., Vossen R.H., Ferlini A., den Dunnen J.T., ’t Hoen P.A., Aartsma-Rus A. Accurate quantification of dystrophin mRNA and exon skipping levels in duchenne muscular dystrophy. Lab. Invest. 2010;90:1396–1402. doi: 10.1038/labinvest.2010.98. [DOI] [PubMed] [Google Scholar]
- 35.’t Hoen P.A., de Meijer E.J., Boer J.M., Vossen R.H., Turk R., Maatman R.G., Davies K.E., van Ommen G.J., van Deutekom J.C., den Dunnen J.T. Generation and characterization of transgenic mice with the full-length human DMD gene. J. Biol. Chem. 2008;283:5899–5907. doi: 10.1074/jbc.M709410200. [DOI] [PubMed] [Google Scholar]
- 36.Sicinski P., Geng Y., Ryder-Cook A.S., Barnard E.A., Darlison M.G., Barnard P.J. The molecular basis of muscular dystrophy in the mdx mouse: a point mutation. Science. 1989;244:1578–1580. doi: 10.1126/science.2662404. [DOI] [PubMed] [Google Scholar]
- 37.Mann C.J., Honeyman K., McClorey G., Fletcher S., Wilton S.D. Improved antisense oligonucleotide induced exon skipping in the mdx mouse model of muscular dystrophy. J. Gene Med. 2002;4:644–654. doi: 10.1002/jgm.295. [DOI] [PubMed] [Google Scholar]
- 38.Verheul R.C., van Deutekom J.C., Datson N.A. Digital droplet PCR for the absolute quantification of exon skipping induced by antisense oligonucleotides in (pre-)clinical development for Duchenne muscular dystrophy. PLoS ONE. 2016;11:e0162467. doi: 10.1371/journal.pone.0162467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Samoylova T.I., Smith B.F. Elucidation of muscle-binding peptides by phage display screening. Muscle Nerve. 1999;22:460–466. doi: 10.1002/(sici)1097-4598(199904)22:4<460::aid-mus6>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 40.Yin H., Moulton H.M., Betts C., Seow Y., Boutilier J., Iverson P.L., Wood M.J. A fusion peptide directs enhanced systemic dystrophin exon skipping and functional restoration in dystrophin-deficient mdx mice. Hum. Mol. Genet. 2009;18:4405–4414. doi: 10.1093/hmg/ddp395. [DOI] [PubMed] [Google Scholar]
- 41.Gao X., Zhao J., Han G., Zhang Y., Dong X., Cao L., Wang Q., Moulton H.M., Yin H. Effective dystrophin restoration by a novel muscle-homing peptide-morpholino conjugate in dystrophin-deficient mdx mice. Mol. Ther. 2014;22:1333–1341. doi: 10.1038/mt.2014.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dias-Neto E., Nunes D.N., Giordano R.J., Sun J., Botz G.H., Yang K., Setubal J.C., Pasqualini R., Arap W. Next-generation phage display: integrating and comparing available molecular tools to enable cost-effective high-throughput analysis. PLoS ONE. 2009;4:e8338. doi: 10.1371/journal.pone.0008338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McLaughlin M.E., Sidhu S.S. Engineering and analysis of peptide-recognition domain specificities by phage display and deep sequencing. Methods Enzymol. 2013;523:327–349. doi: 10.1016/B978-0-12-394292-0.00015-1. [DOI] [PubMed] [Google Scholar]
- 44.Matochko W.L., Cory Li S., Tang S.K., Derda R. Prospective identification of parasitic sequences in phage display screens. Nucleic Acids Res. 2014;42:1784–1798. doi: 10.1093/nar/gkt1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shtatland T., Guettler D., Kossodo M., Pivovarov M., Weissleder R. PepBank—a database of peptides based on sequence text mining and public peptide data sources. BMC Bioinformatics. 2007;8:280. doi: 10.1186/1471-2105-8-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Verhaart I.E., van Vliet-van den Dool L., Sipkens J.A., de Kimpe S.J., Kolfschoten I.G., van Deutekom J.C., Liefaard L., Ridings J.E., Hood S.R., Aartsma-Rus A. The dynamics of compound, transcript, and protein effects after treatment with 2OMePS antisense oligonucleotides in mdx mice. Mol. Ther. Nucleic Acids. 2014;3:e148. doi: 10.1038/mtna.2014.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McMahon H.T., Boucrot E. Molecular mechanism and physiological functions of clathrin-mediated endocytosis. Nat. Rev. Mol. Cell Biol. 2011;12:517–533. doi: 10.1038/nrm3151. [DOI] [PubMed] [Google Scholar]
- 48.Zhu C.H., Mouly V., Cooper R.N., Mamchaoui K., Bigot A., Shay J.W., Di Santo J.P., Butler-Browne G.S., Wright W.E. Cellular senescence in human myoblasts is overcome by human telomerase reverse transcriptase and cyclin-dependent kinase 4: consequences in aging muscle and therapeutic strategies for muscular dystrophies. Aging Cell. 2007;6:515–523. doi: 10.1111/j.1474-9726.2007.00306.x. [DOI] [PubMed] [Google Scholar]
- 49.Yu R.Z., Baker B., Chappell A., Geary R.S., Cheung E., Levin A.A. Development of an ultrasensitive noncompetitive hybridization-ligation enzyme-linked immunosorbent assay for the determination of phosphorothioate oligodeoxynucleotide in plasma. Anal Biochem. 2002;304:19–25. doi: 10.1006/abio.2002.5576. [DOI] [PubMed] [Google Scholar]
- 50.Jirka S.M., Tanganyika-de Winter C.L., Boertje-van der Meulen J.W., van Putten M., Hiller M., Vermue R., de Visser P.C., Aartsma-Rus A. Evaluation of 2'-deoxy-2'-fluoro antisense oligonucleotides for exon skipping in Duchenne muscular dystrophy. Mol Ther Nucleic Acids. 2015;4:e265. doi: 10.1038/mtna.2015.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.