Abstract
RNA‐binding proteins (RBPs) and noncoding (nc)RNAs (such as microRNAs, long ncRNAs, and others) cooperate within a post‐transcriptional network to regulate the expression of genes required for many aspects of cancer behavior including its sensitivity to chemotherapy. Here, using an RBP‐centric approach, we explore the current knowledge surrounding contributers to post‐transcriptional gene regulation (PTGR) in ovarian cancer and identify commonalities that hint at the existence of an evolutionarily conserved core PTGR network. This network regulates survival and chemotherapy resistance in the contemporary context of the cancer cell. There is emerging evidence that cancers become dependent on PTGR factors for their survival. Further understanding of this network may identify innovative therapeutic targets as well as yield crucial insights into the hard‐wiring of many malignancies, including ovarian cancer. WIREs RNA 2018, 9:e1432. doi: 10.1002/wrna.1432
This article is categorized under:
-
1
RNA Interactions with Proteins and Other Molecules > Protein–RNA Interactions: Functional Implications
-
2
Translation > Translation Mechanisms
-
3
RNA in Disease and Development > RNA in Disease
INTRODUCTION
Epithelial ovarian cancer (EOC) is diagnosed in approximately 239,000 women every year worldwide. It is the most lethal of gynecological cancers; only 45% of those with the disease remain alive 5 years after their diagnosis.1 The most significant contributor to its high mortality is that, owing to its subtle presenting symptoms, EOC is predominantly diagnosed at an advanced stage but also has a high rate of recurrence. Initial treatment comprised of surgery and chemotherapy is usually effective at inducing remission but, in 70–80% patients, the cancer recurs within 2 years. At this point re‐treatment with chemotherapy becomes increasingly futile as the cancer cells become more ‘chemoresistant.’2
Unlike many cancers with an average of 30–60 mutations each, EOC has a relatively bland mutational profile. According to The Cancer Genome Atlas (TCGA) Research Network data only nine nonsynonymous gene mutations are present in high‐grade serous ovarian cancer (HGSOC), the most common histological subtype. The two most frequent are TP53 and BRCA mutations, present in 96 and 22%, respectively.3 The other mutations are present in only 2–6% cases. Although poly(adenosine diphosphate (ADP)‐ribose) polymerase (PARP) inhibitors have transformed the management of those with hereditary BRCA mutations and p53‐targeted therapies are in clinical trials, the low penetrance of the others limits the utility of mutation‐driven approaches.4, 5 HGSOC has a high degree of chromosomal instability (CIN), attributed to mutations and promoter methylations in DNA homologous repair genes. High CIN is a predictor of greater sensitivity to DNA‐damaging chemotherapies like platinum agents.3, 6, 7 Tumors like EOC with low mutational burden show limited response to immune checkpoint inhibitors, such as those designed to target PD1 and PDL1.8 This is borne out in current clinical practice where studies of checkpoint inhibitors as monotherapies in EOC have shown limited efficacy.9 For these reasons, cytotoxic chemotherapy remains the mainstay of treatment for non‐BRCA mutated ovarian cancer patients, in the primary (newly diagnosed) context and at relapse.
CHEMOTHERAPY RESISTANCE
Two chemotherapies predominate in the clinical management of ovarian cancer: the pseudoalkylating agent carboplatin and the microtubule spindle poison paclitaxel. Carboplatin is derived from cisplatin and functions by inducing cell damaging cross‐links between DNA, RNA, and protein in rapidly dividing cells. Paclitaxel functions by binding β tubulin microtubules and preventing spindle remodeling during cell division, resulting in mitotic arrest.10 Resistance mechanisms to paclitaxel include point mutations or alterations in the expression of β‐tubulin isotypes such as upregulation of the β3‐tubulin (TUBB3) isoform and overexpression of the efflux protein P‐glycoprotein 1 (encoded by the gene MDR1) that exports paclitaxel from the cell.11, 12 For carboplatin (and other platinum agents such as cisplatin) whose cellular uptake is reliant on heavy metal pathways, resistance mechanisms include down‐regulation of the copper channel influx transporters CTR1 and CTR2, upregulation of the efflux transporter ATP7B, increased DNA damage repair, and enhanced survival signaling.13, 14 It is likely that at diagnosis ovarian cancer contains a proportion of innately resistant cells that gain clonal dominance with each recurrence and utilize multiple resistance mechanisms.15 Therapies to successfully reverse resistance remain elusive and, for patients with recurrent disease, death from progressive disease is unfortunately inevitable.
There is now an accumulating body of evidence that therapeutic opportunities lie within the realm of post‐transcriptional biology.
POST‐TRANSCRIPTIONAL GENE REGULATION (PTGR)
The former central dogma of molecular biology was that nucleic acids regulate protein in an irreversibly linear sequence.16 However, comparisons between gene and protein expression within cells and more recently within HGSOC itself have revealed that the ratio of mRNA to protein is not 1:1 and the abundance of a particular cellular protein cannot necessarily be predicted from the copy number of its encoding gene.17, 18 It is now understood that post‐transcriptional factors bind mRNAs and modify (and in some cases dominate) the amount of protein they can generate.19 These regulatory factors have been identified as noncoding RNAs (ncRNAs) and RNA‐binding proteins (RBPs). Both influence protein expression by controlling the maturation, modification (including stability and translation efficiency), subcellular transport, and/or degradation of their target mRNAs.20 While it was once assumed these two classes of PTGR factors worked in isolation they are now known to act co‐operatively. Pier Paolo Pandolfi was the first to describe a competing endogenous (ce)RNA network, initially around microRNAs but later including other ncRNAs, that collectively coordinates gene expression.21 Although current descriptions of the ceRNA network include RBPs, we will refer here to this cooperation between RBPs and ncRNAs as the ‘PTGR network.’ In cancer, it is apparent that a PTGR network is co‐opted to regulate the expression of genes that participate in pathological processes such as cell migration, invasion, and metastasis. Advances in computational biology have allowed the collation of expression profiling datasets in common cancers and their correlation with known, validated interactions to generate putative ceRNA network maps. In EOC these maps have focused around microRNAs, lncRNAs and mRNAs but are yet to include RBPs.22, 23 Here we discuss RBPs that have been identified and known to be deregulated in ovarian cancer and their interactions with other PTGR factors.
RNA Binding Proteins
From the moment mRNA transcripts are synthesized they become bound to RBPs that are involved in every stage of their lifespan.20 Although RBPs can be found bound at many sites along an mRNA transcript, they tend to bind within the untranslated regions (UTRs) of mRNA transcripts. Some bind to conserved sequences [such as adenylate‐uridylate (AU)‐rich elements], others recognize structural conformations like stem loops or hairpin bends.24, 25 RBPs have RNA binding domains, such as RNA recognition motifs (RRM) and hnRNP K‐homology domains.26 While the first RBPs to be identified were members of the canonical eIF4F cap‐binding complex, other RBPs have since been characterized, including those with noncanonical roles in specialized protein synthesis.27 There are now an estimated 1542 genes encoding RBPs.28 This has led to comparisons of the differential levels and patterns of RBP expression between normal and cancer cells. Cellular levels of RBP genes are significantly higher than ncRNAs suggesting RBPs have a more prominent role in PTGR.29 Although RBPs function in the synthesis of all proteins, they have an important role in cellular stress by selectively modulating gene expression to provide a stress response. Thus, RBPs are important for disease prevention in normal cells but their deregulation can contribute to many pathological conditions such as inflammation and cancer.30 For this reason, and because they bind multiple mRNAs within a single physiological or pathological pathway, RBPs have been identified as potential drug targets.19 Recent advances in drug discovery have led to an expanded repertoire of what is considered ‘druggable’ and now includes gene depletion by anti‐sense oligonucleotides and RNA intereference as well as inhibitors of protein–protein, and protein–RNA interactions in addition to classical kinase inhibitors.31 This, combined with preclinical evidence of efficacy but minimal toxicity from serendipitous RBP inhibitors like the eIF4E inhibitor ribavirin, has highlighted the feasibility of targeting RBPs.32 However, RBPs do not act alone and there is accumulating evidence of interactions with other PTGR network factors to be considered (Figure 1).
Figure 1.
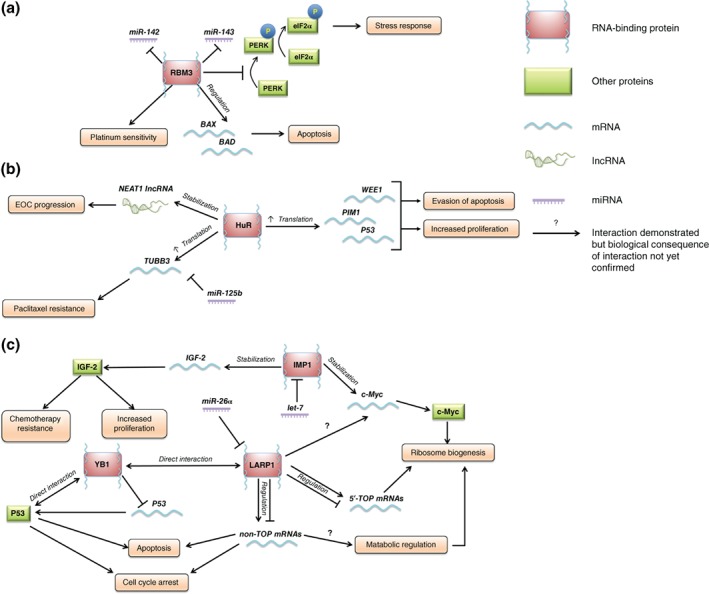
(a)–(c) RNA‐binding proteins influence epithelial ovarian cancer (EOC) progression through complex networks with mRNAs, noncoding RNAs, and other proteins. (a) RNA‐binding motif protein 3 (RBM3) regulates platinum sensitivity and patient survival through regulation of mRNAs involved in apoptosis and the stress response. (b) HuR exerts an oncogenic effect through stabilization and therefore increased translation of a range of mRNAs. (c) RNA‐binding proteins, such as YB1, LARP1, and IMP1 may converge on multiple subsets of mRNAs and signaling pathways as part of a network that drives progression of EOC and/or resistance to chemotherapy.
Noncoding RNAs
It is estimated that <2% of the human genome is stably transcribed into protein, the rest is believed to transcribe mRNAs that do not encode proteins and are termed noncoding RNAs (ncRNAs). NcRNAs are classified by size into long noncoding RNAs (lncRNAs) of over 200 and small RNAs with fewer than 200 nucleotides. While some ncRNAs like transfer, ribosomal and spliceosomal RNAs have established cellular housekeeping functions and are constitutively expressed, others including lncRNAs, microRNAs (miRNAs or miRs), small interfering RNAs (siRNAs), and PIWI‐interacting RNAs (piRNAs) were initially perceived as transcriptional ‘junk.’ However, as detailed in a number of reviews, they are now recognized as having important regulatory roles in gene expression.33, 34, 35, 36 New classes of small RNAs have more recently been defined such as transfer RNA‐related fragments (tRFs), pseudogenes, and circular RNAs (circRNAs) although these have yet to be fully characterized in EOC.37, 38, 39
Long Noncoding RNAs
LncRNAs range from 200 to 100,000 nucleotides in length. They have some overlapping characteristics with mRNA being transcribed by RNA Polymerase II and are (usually) 5′ capped and polyadenylated. However, unlike mRNAs, they do not have a typical open reading frame, are usually shorter, and are expressed at lower levels.40 Importantly, lncRNAs are processed and regulated differentially and appear to have expression patterns that are specific to subcellular site, cell type, developmental stage, or disease. Approximately 23,000 lncRNAs have been described so far, of which only a few have been shown to have important roles in cellular regulation and fewer still have been linked to cancer.41 LncRNAs act by multiple mechanisms such as altering chromatin remodeling, binding and regulating transcription factors and acting as protein–protein interaction scaffolds. Through these mechanisms, lncRNAs fine‐tune protein expression and in cancers act as oncogenes, tumor suppressors, or as both depending on the circumstances. Intriguingly, some lncRNAs have been shown to act as microRNA sponges and carry microRNA response elements (MREs) to bind and sequester miRNAs away from their intended mRNA targets.42 LncRNAs have also been identified as RBP‐sponges, an area of discovery in its infancy as data from Photoactivatable Ribonucleoside‐Enhanced Crosslinking and Immunoprecipitation (PAR‐CLIP), individual‐nucleotide resolution UV crosslinking and immunoprecipitation (iCLIP), and other RBP‐immobilization methods become available.43 Those such as MALAT1, HOST2, PVT1 NEAT1, and HOTAIR have been identified in EOC and both PVT1 and HOTAIR have been linked to platinum resistance by regulating apoptosis factors and via NF‐κB activation, respectively.44, 45, 46
MicroRNAs
MicroRNAs (miRs) are a class of endogenous, small noncoding RNAs of approximately 22 nucleotides in length that bind and repress translation. It is estimated that each microRNA regulates around 100 genes and that, as PTGR factors, microRNAs fine‐tune protein expression.47 They are transcribed from intronic, exonic, or intragenic regions of protein encoding ‘host’ genes, in parallel with host gene expression. A ‘cluster’ of multiple microRNAs can be transcribed from a single gene. After undergoing a series of processing steps, mature single‐stranded microRNA is integrated with Argonaute (Ago) into an RNA induced silencing complex (RISC) which then binds its targets by partial base pairing between the microRNA 5′ ‘seed region’ and the 3′ MRE in the target mRNA. Depending on the complementarity between these two sequences, the target mRNA is either degraded by Ago‐mediated cleavage or attenuated. The latter occurs at translation initiation by the RISC hindering assembly of the eIF4F complex or preventing the recruitment of ribosome components.48 Since microRNAs were first discovered, the number to have been identified in human cells has now reached 2500 and it is estimated that around 60% all genes are associated with miRNAs.49, 50, 51, 52 For this reason, they are involved in almost all biological processes. In cancer, in which microRNAs are generally downregulated, they contribute to many aspects of the disease, from its initial formation to its chemotherapy responsiveness.53, 54, 55 This is partially because microRNA clusters are often located within genomic regions amplified, deleted, hyper‐, or hypomethylated in cancer.56 This is particularly relevant in EOC with its high genomic instability.57 Of the 34 miRNAs currently known to be deregulated in cancer, 17 have been associated with HGSOC.58, 59 Those linked to cisplatin resistance include Let‐7 family members let‐7e and 7i and miR‐214 and miR‐30a‐5p, whereas miR‐663, miR‐622, and miR‐130b regulate paclitaxel resistance via the expression of p53 network genes and MDR1 respectively.60, 61, 62, 63 In addition, miR‐200c is associated with reversal of resistance via its regulation of TUBB3. Although these studies identify putative target mRNAs that could explain the resistance phenotype, it is highly likely that there are hundreds of other genes regulated by each microRNA that contribute incrementally toward chemotherapy resistance.
As summarized by Ciafrè and Galardi,64 RBPs work collaboratively with miRNAs, either by attracting the miRNA‐RISC complex to degrade mRNAs or, if the RBP‐binding or ‘USER’ site on the target mRNA is close or overlapping the MRE, by preventing the interaction between a miRNA and its target mRNA resulting in translation derepression. RBPs can compete with microRNAs by binding and modifying the structure of the target mRNA or by sequestering it away from microRNAs to another location within the cell.
PATHOLOGICAL RBPS AND THEIR PTGR NETWORKS IN OVARIAN CANCER
The relationships that exist between RBPs and microRNAs, lncRNAs and other ncRNAs have yet to be fully characterized but this is becoming achievable using state‐of‐the‐art computational models like SimiRa (http://vsicb‐simira.helmholtz‐muenchen.de), CircInteractome (circinteractome.nia.nih.gov), doRiNA (dorina.mdc‐berlin.de.), and StarBase (starbase.sysu.edu.cn) that integrate and interrogate data from publically available RNA‐Seq and CLIP datasets to generate interaction maps and/or provide functional categorisations.65 The RBPs described below have been previously been characterized in the context of EOC and any known interactions with ncRNAs are outlined (Table 1).
Table 1.
RNA‐Binding Proteins (RBPs) Described in Epithelial Ovarian Cancer and Their Known Interactions with ncRNAs
| RBP | Abbreviation | Family | Biological Role | Role in Epithelial Ovarian Cancer (EOC) | RBP–ncRNA Interaction |
|---|---|---|---|---|---|
| RNA‐binding motif protein 3 | RBM3 | Glycine‐rich RNA binding protein (GRP)66 | Regulates global protein synthesis under normal physiological conditions and in response to cold temperature and low oxygen tension114 | Elevated mRNA and protein levels are associated with a favorable prognosis and sensitivity to platinum‐based chemotherapy through post‐transcriptional regulation of the apoptosis mediators BCL2, BAX, and genes involved in DNA integrity as well as impairment of DNA damage repair following chemotherapy67 | RBM3 regulates the expression of temperature sensitive miR‐142‐5p and miR‐143 to attenuate pathological hyperthermia and downregulates miR‐143‐mediated nitric oxide‐induced apoptosis by interfering with p38 MAPK kinase signaling in human SH‐SY5Y neuroblastoma cells115 |
| ELAV‐like protein 1 | HuR | Embryonic lethal abnormal visual system (ELAV)116, 117 | HuR positively regulates stability and/or translation of ZEB2 mRNA which plays a key role in ovarian cancer progression, where concomitant high cytoplasmic HuR and nuclear ZEB2 correlates with unfavorable prognosis120 |
|
|
| IGF2 mRNA‐binding protein 1 | IGF2BP1/IMP1 | IGF2 mRNA‐binding protein (IMP)121 | Stabilizes mRNAs that drive drug resistance such as c‐myc and MDR191, 92, 93 | IMP1 is a downstream target gene of let‐7 miRNA. Let‐7 negatively regulates IMP1 expression,95, 96 increasing the sensitivity of resistant ovarian cancer to Taxanes92 | |
| Y‐box binding protein 1 | YBX1/YB‐1 | Cold‐shock domain containing proteins99 | Transcription, translation, DNA damage repair,127, 128 mRNA stability and translational regulation in the cytoplasm129, 130, 131 | Nuclear expression positively correlates with poor prognosis132 and cisplatin resistance133 | miR‐190a negatively regulates mRNA and protein levels of YB‐1 in prostate cancer103 |
| La‐related protein 1 | LARP1 | La‐related protein (LARP)104 | Regulates the stability and/or translation of mRNAs required for ribosome biogenesis and cell survival and proliferation106, 107 | Promotes EOC progression and resistance to chemotherapy through post‐transcriptional regulation of cell survival mRNAs such as BCL2 and BIK108, 109 |
RNA‐Binding Motif Protein 3
RNA‐binding motif protein 3 (RBM3) is a member of the glycine‐rich RNA binding protein (GRP) family. Transcript levels of RBM3 are elevated in cell lines immediately following cold shock (when taken from 37 to 32°C) and RBM3 is often co‐expressed with another GRP family member, cold‐inducible RNA‐binding protein.66 Levels of RBM3 have subsequently shown to be elevated across multiple cancers, generally in association with a favorable outcome. In the context of EOC, mRNA and protein levels of RRM3 were correlated with outcome in tissue collected from 163 and 151 patients with EOC respectively. Messenger RNA levels of RBM3 were found to be an independent predictor of better relapse‐free and overall survival while presence (vs absence) of RBM3 protein was strongly associated with prolonged overall survival. Higher RBM3 (mRNA or protein) was associated with increased platinum‐sensitivity, attributed to its interactome of apoptosis regulating mRNAs such as BCL‐2, BAX as well as genes involves in DNA integrity; upregulation of RBM3 impaired DNA damage repair after chemotherapy and improved its cytotoxic effect.67 RBM3 also inhibits PERK phosphorylation and prevents cell death from endoplasmic reticulum (ER) stress.68 In addition, RBM regulates the biogenesis of temperature sensitive miRNAs such as miR‐142 and miR‐143 but has not yet been associated with any oncomiRs or lncRNAs.69
HuR
The ubiquitously expressed mRNA‐binding protein HuR (ELAVL1) is one of the four membered embryonic lethal abnormal visual (ELAV) system family of proteins expressed in human cells.70 In embryos, these evolutionarily conserved proteins are involved in neuronal development. In normal adult cells HuR is expressed at low levels and shuttles from the nucleus into the cytoplasm in response to stress where it stabilizes and/or promotes the translation of mRNAs containing AU‐rich elements within their 3′ UTRs.71 In cancers, elevated levels of cytoplasmic HuR positively correlate with treatment resistance and adverse survival outcome, attributed to its target mRNAs such as PIM1, the DNA damage‐regulated G2 checkpoint gene Wee1, the TNF‐related apoptosis‐inducing ligand (TRAIL) component DR4 and the cell survival regulator TP53.72, 73, 74, 75, 76, 77, 78 In addition, HuR is linked to paclitaxel resistance as it binds and stabilizes the mRNA encoding TUBB3 in competition with miR‐200c. When HuR is nuclear, miR‐200c binds the 3′ UTR of cytoplasmic TUBB3 inhibiting it and conferring a good prognosis, but when HuR is cytoplasmic it competes with miR‐200c and the expression of TUBB3 is derepressed.79 Similarly, HuR has been shown to compete with miR‐125b in binding the 3′ UTR of TP53 during DNA damage preventing miR‐125b from reaching its MRE site.80 Control of the shuttling of HuR between the cytoplasm and the nucleus is a crucial component of its regulation. In a recent study using murine macrophages, HuR was shown to be bound and PARylated by the DNA damage‐repair protein PARP1 following immune stimulation. This post‐translational modification (the addition of a polymeric poly‐ADP ribose chain) of HuR appears to stabilize its interaction with target mRNAs in the cytoplasm.81
HuR interacts with lncRNAs such as nuclear enriched abundant transcript 1 (NEAT1). NEAT1 is up‐regulated in EOC where it is associated with more advanced disease and poorer prognosis.82 In ovarian cancer cell lines, ectopic expression of NEAT1 promotes cell proliferation and invasion. Work by Chai et al. in OVCAR3 ovarian cells showed that HuR binds and stabilizes NEAT1 and increases its expression. However, the microRNA miR124‐3p competes with HuR to destabilize NEAT1.83 In other cancers, HuR has been shown to bind the lncRNAs HOX transcript antisense intergenic RNA (HOTAIR) and metastasis‐associated lung adenocarcinoma transcript‐1 (MALAT1) enhancing their activity as microRNA sponges.84, 85 HOTAIR is upregulated in patients with EOC where it drives proliferation, migration, and invasion and acts as a sponge for miR‐373, thereby derepressing the Ras oncogene family member Rab22a.86 Levels of MALAT1 have been shown to be elevated in a number of cancers including EOC and, in SKOV3 cells, MALAT1 drives proliferation, invasion, and tumorigenicity.87 The mechanism of action for MALAT1 in EOC has remained elusive although it has been shown, in endometrial cancer, to sponge miR‐200c thus derepressing TUBB3 expression.88 More recent studies in EOC lines have demonstrated MALAT1 acts as a sponge to miR‐506 and derepresses the apoptosis inhibitor iASPP.89 It is therefore likely that, in EOC, HuR drives oncogenic behavior and chemoresistance through direct interactions with its target mRNAs and indirectly via its PTGR network.
IGF2 mRNA‐Binding Protein 1 (IGF2BP1/IMP1)
The IMP family is comprised of IMP 1–3 and is so named because its members bind to the 5′‐UTR of insulin‐like growth factor 2 (IGF2) mRNA. They are important for embryogenesis and (apart from IMP2) are expressed at low levels in adult cells but all three paralogues are highly expressed in cancers where levels are associated with poor prognosis.90 IMP1 modulates the turnover of target mRNAs during stress by trafficking mRNAs to cytosolic mRNP complexes, presumably to protect them from degradation. In addition to its eponymous target IGF2, IMP1 stabilizes c‐myc as well as the Adenosine triphosphate (ATP)‐dependent efflux pump MDR1 and thus has been implicated in ovarian chemoresistance.91, 92, 93 RNAi knockdown of IMP‐1 in cell lines from several types of cancers reduces c‐Myc levels, inhibits cell proliferation, and triggers apoptosis.94 IMP1 is negatively regulated by the microRNA let7 and cooperates within an ‘oncogenic triangle’ between IMP1 and two other let‐7 targets (the RBP) LIN28B and the transcriptional regulator HMGA2.95, 96 Although an inhibitory interaction between IMP1 and lncMyoD has been postulated in skeletal muscle cells, there are currently no documented interactions between IMP1 and lncRNAs in cancer.97
Y‐Box Binding Protein 1 (YBX1/YB1)
YB‐1 is a member of an evolutionarily conserved family of cold‐shock domain containing proteins.98, 99 YB‐1 is essential for normal embryogenesis and levels of the protein are high in embryos but decline in subsequent stages of development.100 Initially classified as a transcription factor, YB‐1 was also shown to have RNA‐binding capability by virtue of two RNA‐binding motifs contained within its cold shock domain. It has both nuclear and cytosolic localization and elevated expression of YB‐1 has been observed in many cancers, including EOC where higher levels are correlated with adverse survival outcome. YB‐1 null cells have enhanced sensitivity to multiple stresses including from genotoxic drugs such as cisplatin. The mechanism behind the YB‐1‐regulated stress response is not yet fully understood and may have transcriptional and post‐transcriptional components. The protein undergoes nuclear accumulation under stress. YB‐1 binds CCAAT elements (Y‐box) in the promoter region of MD1 and nuclear levels of YB‐1 correlate with expression of P glycoprotein.101 Reduction of YB‐1 in colorectal cancer cell lines causes induction of p53‐dependent cell death and TP53 levels rise after YB‐1 repression. YB‐1 has also been shown to directly bind p53 protein at the site of DNA damage.102 As yet YB‐1 has not been associated with ncRNAs in EOC. In prostate cancer, miR‐190a has been shown to directly bind and repress YB‐1. In advanced prostate cancer miR‐190a expression is reduced resulting in YB‐1 derepression and activation of androgen‐receptor cell signaling.103
La‐Related Protein 1
La‐related protein 1 belongs to a seven‐member family of RBPs called the LARPs. It is highly evolutionarily conserved with orthologues present in all metazoan species.104 In Drosophila, LARP1 is required for normal embryonic development as well as spermatocyte formation.105 In normal human cells LARP1 is predominantly cytoplasmic and expressed at low levels where it acts downstream of mTORC1 to bind and regulate the translation of mRNA transcripts required for ribosome manufacture (biogenesis) and cell proliferation.106, 107 These are TOP mRNAs characterized by a consensus 5′ terminal oligopyrimidine (TOP) sequence. Levels of LARP1 protein are elevated in a number of epithelial malignancies including EOC and confers adverse survival outcome. LARP1 is required for stress response; cells depleted for LARP1 have heightened sensitivity to genotoxic agents like cisplatin and paclitaxel, as well as other stresses such as hypoxia and glucose starvation. In cancer cell lines, LARP1 has been shown to bind an interactome of approximately 3000 mRNAs that is enriched for transcripts encoding cell survival and RNA biogenesis proteins. In vitro and in vivo inhibition of LARP1 induces cell death in part through its post‐transcriptional regulation of the apoptosis mediators BCL2 and BIK.108, 109 Although no consensus binding sequences within its targets have yet been identified, LARP1 binds these mRNAs via its C‐terminal ‘DM15 repeat region’ that adopts a HEAT domain‐like configuration.110 Although it has not been associated with ncRNAs in EOC, in breast and prostate cancer LARP1 has recently been shown to be one of several targets of miR‐26a.111, 112 In endothelial cells, the lncRNA TGFB2‐OT1 acts as a sponge to miR‐4459 resulting in derepressed expression of LARP1.113
SUMMARY AND FUTURE PERSPECTIVES
It is evident that, as with other cancers, a PTGR network exists in EOC that regulates the malignant characteristics of the disease. There are notable similarities between the RBPs described here. They are all oncofetal genes required for embryonic development that are mobilized during stress in normal adult cells but are highly expressed in cancer where they drive cell migration, invasion, and tumorigenesis. They are also evolutionarily ancient. RBM3 and YB1 are ancestral cold‐shock genes, LARP1, HuR, and IMP1 belong to gene families that predate eukaryotes. Unlike ncRNAs that have tumor site, type and stage specificity, the RBPs described here have activities that are consistent across multiple cancer types. The majority are proto‐oncogenic and their inhibition induces apoptotic cell death. This indicates not only that these RBPs are influential ‘nodes’ within the PTGR network but also that they have a crucial function in maintaining cancer cell survival.
There is much overlap in the target mRNAs described here. They encode proteins controlling ATPase efflux pumps, copper uptake proteins, tubulin isoforms, and apoptotic regulators implying they belong to a core set of mRNAs that are themselves evolutionarily conserved. As these proteins also control the uptake and efflux of chemotherapy drugs, they are correspondingly and coincidentally associated with treatment resistance. The possibility of the existence of a core set of PTGR targets indicates that analysis of the interactomes of more cancer RBPs (once they are known) may reveal more overlapping candidates. While fully annotating such a complex network may seem daunting, the commonalities between those factors known so far suggests they exist within a single coordinated stress response. This implies that a similar post‐transcriptional response is deployed whatever form of stress the cell is exposed to. If this were proven to be the case, there would be huge therapeutic advantage in targeting a PTGR node when genomic heterogeneity so limits the success of conventional targeted therapies.
Like transcription factors, the nonenzymatic nature of RBPs have earned them the reputation of being ‘undruggable’ and knockdown approaches using RNAi or antisense have been preferred. An example is therapeutic RNAi against HuR which causes enhanced apoptosis and chemosensitivity in EOC cell lines and xenograft models but has yet to reach the clinic.134 As it is likely that each RBP is deployed in many different contexts in the normal and cancer cell, a preferable approach may prove to be the blockade of the site of interaction between an RBP and its core target gene(s) or ncRNAs. Fortunately, improvements in drug design have heralded the development of molecules capable of disrupting protein:protein as well as protein:RNA interactions.135 To embark on such a drug discovery program for RBPs described here will require a clear understanding of the direct interactome of each RBP, the site of these interactions and their impact on the wider PTGR network. Currently, although the RBPs described here have identifiable RRM, it cannot be assumed that these are the site of interactions with the core mRNAs regulating survival and chemosensitivity. For example, the DM15 region of LARP1, now known to bind cell survival transcripts (such as BCL2 and BIK) is C‐terminally placed and somewhat remote from the N‐terminal La‐RRM (La motif) which is the de facto site of mRNA binding in other LARP family members.104
As with other cancer types, a comprehensive annotation of the PTGR network in EOC is in its infancy. Although many cancer RBPs have been identified from high throughput screens, few have undergone functional analysis and their target mRNA interactomes are undocumented. This prevents their incorporation into PTGR algorithms and thus limits our understanding of the roles played by RBPs in post‐transcriptional gene networks. As our knowledge of these networks unfolds, it is likely that opportunities for developing new inhibitors will be revealed. Also, the discovery that some RBPs are subject to PARylation and other post‐translational modifications may provide insights into their contribution to the clinical responses that are currently observed with targeted cancer therapies. Here a biased, RBP‐centric review of the published literature surrounding PTGR in ovarian cancer gives fascinating insights into an ancient network that dominates multiple aspects of cancer behavior including chemotherapy resistance.
Conflict of interest: The authors have declared no conflicts of interest for this article.
REFERENCES
- 1. Cancer Research Statistics . Available at: http://www.cancerresearchuk.org/health‐professional/cancer‐statistics/statistics‐by‐cancer‐type/ovarian‐cancer. (Accessed March 25, 2017).
- 2. Jayson GC, Kohn EC, Kitchener HC, Ledermann JA. Ovarian cancer. Lancet 2014, 384:1376–1388. [DOI] [PubMed] [Google Scholar]
- 3. Cancer Genome Atlas Research Network . Integrated genomic analyses of ovarian carcinoma. Nature 2011, 7353:609–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Konecny GE, Kristeleit RS. PARP inhibitors for BRCA1/2‐mutated and sporadic ovarian cancer: current practice and future directions. Br J Cancer 2016, 115:1157–1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bykov VJ, Wiman KG. Mutant p53 reactivation by small molecules makes its way to the clinic. FEBS Lett 2014, 588:2622–2627. [DOI] [PubMed] [Google Scholar]
- 6. Cope L, Wu RC, Shih IM, Wang TL. High level of chromosomal aberration in ovarian cancer genome correlates with poor clinical outcome. Gynecol Oncol 2013, 128:500–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. McClelland SE, Burrell RA, Swanton C. Chromosomal instability: a composite phenotype that influences sensitivity to chemotherapy. Cell Cycle 2009, 8:3262–3266. [DOI] [PubMed] [Google Scholar]
- 8. Rizvi NA, Hellmann MD, Snyder A, Kvistborg P, Makarov V, Havel JJ, Lee W, Yuan J, Wong P, Ho TS, et al. Cancer immunology. Mutational landscape determines sensitivity to PD‐1 blockade in non‐small cell lung cancer. Science 2015, 348:124–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hardwick N, Frankel PH, Cristea M. New approaches for immune directed treatment for ovarian cancer. Curr Treat Options Oncol 2016, 17:14. [DOI] [PubMed] [Google Scholar]
- 10. Jordan MA, Toso RJ, Thrower D, Wilson L. Mechanism of mitotic block and inhibition of cell proliferation by taxol at low concentrations. Proc Natl Acad Sci U S A 1993, 90:9552–9556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kavallaris M, Kuo DY, Burkhart CA, Regl DL, Norris MD, Haber M, Horwitz SB. Taxol‐resistant epithelial ovarian tumors are associated with altered expression of specific beta‐tubulin isotypes. J Clin Invest 1997, 100:1282–1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Penson RT, Oliva E, Skates SJ, Glyptis T, Fuller AF Jr, Goodman A, Seiden MV. Expression of multidrug resistance‐1 protein inversely correlates with paclitaxel response and survival in ovarian cancer patients: a study in serial samples. Gynecol Oncol 2004, 93:98–106. [DOI] [PubMed] [Google Scholar]
- 13. Kilari D, Guancial E, Kim ES. Role of copper transporters in platinum resistance. World J Clin Oncol 2016, 7:106–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Galluzzi L, Senovilla L, Vitale I, Michels J, Martins I, Kepp O, Castedo M, Kroemer G. Molecular mechanisms of cisplatin resistance. Oncogene 2012, 31:1869–1883. [DOI] [PubMed] [Google Scholar]
- 15. Castellarin M, Milne K, Zeng T, Tse K, Mayo M, Zhao Y, Webb JR, Watson PH, Nelson BH, Holt RA. Clonal evolution of high‐grade serous ovarian carcinoma from primary to recurrent disease. J Pathol 2013, 229:515–524. [DOI] [PubMed] [Google Scholar]
- 16. Crick F. Central dogma of molecular biology. Nature 1970, 227:561–563. [DOI] [PubMed] [Google Scholar]
- 17. Zhang H, Liu T, Zhang Z, Payne SH, Zhang B, McDermott JE, Zhou JY, Petyuk VA, Chen L, Ray D, et al., et al. Integrated proteogenomic characterization of human high‐grade serous ovarian cancer. Cell 2016, 166:755–765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Schwanhäusser B, Busse D, Li N, Dittmar G, Schuchhardt J, Wolf J, Chen W, Selbach M. Global quantification of mammalian gene expression control. Nature 2011, 473:337–342. [DOI] [PubMed] [Google Scholar]
- 19. Blagden SP, Willis AE. The biological and therapeutic relevance of mRNA translation in cancer. Nat Rev Clin Oncol 2011, 8:280–291. [DOI] [PubMed] [Google Scholar]
- 20. Moore MJ. From birth to death: the complex lives of eukaryotic mRNAs. Science 2005, 309:1514–1518. [DOI] [PubMed] [Google Scholar]
- 21. Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell 2011, 146:353–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zhang S, Ng MK. Gene‐microRNA network module analysis for ovarian cancer. BMC Syst Biol 2016, 10(suppl 4):117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zhou M, Wang X, Shi H, Cheng L, Wang Z, Zhao H, Yang L, Sun J. Characterization of long non‐coding RNA‐associated ceRNA network to reveal potential prognostic lncRNA biomarkers in human ovarian cancer. Oncotarget 2016, 7:12598–12611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Chen CY, Shyu AB. AU‐rich elements: characterization and importance in mRNA degradation. Trends Biochem Sci 1995, 20:465–470. [DOI] [PubMed] [Google Scholar]
- 25. Li X, Kazan H, Lipshitz HD, Morris QD. Finding the target sites of RNA‐binding proteins. WIREs RNA 2014, 5:111–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lunde BM, Moore C, Varani G. RNA‐binding proteins: modular design for efficient function. Nat Rev Mol Cell Biol 2007, 8:479–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sonenberg N. ATP/Mg++‐dependent cross‐linking of cap binding proteins to the 5′ end of eukaryotic mRNA. Nucleic Acids Res 1981, 9:1643–1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Gerstberger S, Hafner M, Tuschl T. A census of human RNA‐binding proteins. Nat Rev Genet 2014, 15:829–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kechavarzi B, Janga SC. Dissecting the expression landscape of RNA‐binding proteins in human cancers. Genome Biol 2014, 15:R14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Morris AR, Mukherjee N, Keene JD. Systematic analysis of posttranscriptional gene expression. WIREs Syst Biol Med 2010, 2:162–180. [DOI] [PubMed] [Google Scholar]
- 31. Makley LN, Gestwicki JE. Expanding the number of “druggable” targets: non‐enzymes and protein‐protein interactions. Chem Biol Drug Des 2013, 81:22–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Assouline S, Culjkovic B, Cocolakis E, Rousseau C, Beslu N, Amri A, Caplan S, Leber B, Roy DC, Miller WH Jr, et al. Molecular targeting of the oncogene eIF4E in acute myeloid leukemia (AML): a proof‐of‐principle clinical trial with ribavirin. Blood 2009, 114:257–260. [DOI] [PubMed] [Google Scholar]
- 33. Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell 2009, 136:629–641. [DOI] [PubMed] [Google Scholar]
- 34. Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell 2009, 136:215–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Carthew RW, Sontheimer EJ. Origins and mechanisms of miRNAs and siRNAs. Cell 2009, 136:642–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Thomson T, Lin H. The biogenesis and function of PIWI proteins and piRNAs: progress and prospect. Annu Rev Cell Dev Biol 2009, 25:355–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Fu Y, Lee I, Lee YS, Bao X. Small non‐coding transfer RNA‐derived RNA fragments (tRFs): their biogenesis, function and implication in human diseases. Genomics Inform 2015, 13:94–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Johnsson P, Morris KV, Grandér D. Pseudogenes: a novel source of trans‐acting antisense RNAs. Methods Mol Biol 2014, 1167:213–226. [DOI] [PubMed] [Google Scholar]
- 39. Chen LL. The biogenesis and emerging roles of circular RNAs. Nat Rev Mol Cell Biol 2016, 17:205–211. [DOI] [PubMed] [Google Scholar]
- 40. Quinn JJ, Chang HY. Unique features of long non‐coding RNA biogenesis and function. Nat Rev Genet 2016, 17:47–62. [DOI] [PubMed] [Google Scholar]
- 41. Bartonicek N, Maag JL, Dinger ME. Long noncoding RNAs in cancer: mechanisms of action and technological advancements. Mol Cancer 2016, 15:43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Thomson DW, Dinger ME. Endogenous microRNA sponges: evidence and controversy. Nat Rev Genet 2016, 17:272–283. [DOI] [PubMed] [Google Scholar]
- 43. Yoon JH, Gorospe M. Cross‐linking immunoprecipitation and qPCR (CLIP‐qPCR) analysis to map interactions between longnoncoding RNAs and RNA‐binding proteins. Methods Mol Biol 2016, 1402:11–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Zhao M, Qiu Y, Yang B, Sun L, Hei K, Du X, Li Y. Long non‐coding RNAs involved in gynecological cancer. Int J Gynecol Cancer 2014, 24:1140–1145. [DOI] [PubMed] [Google Scholar]
- 45. Liu E, Liu Z, Zhou Y, Mi R, Wang D. Overexpression of long non‐coding RNA PVT1 in ovarian cancer cells promotes cisplatin resistance by regulating apoptotic pathways. Int J Clin Exp Med 2015, 8:20565–20572. [PMC free article] [PubMed] [Google Scholar]
- 46. Özeş AR, Miller DF, Özeş ON, Fang F, Liu Y, Matei D, Huang T, Nephew KP. NF‐κB‐HOTAIR axis links DNA damage response, chemoresistance and cellular senescence in ovarian cancer. Oncogene 2016, 35:5350–5361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Brennecke J, Stark A, Russell RB, Cohen SM. Principles of microRNA–target recognition. PLoS Biol 2005, 3:e85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Iwakawa HO, Tomari Y. The functions of microRNAs: mRNA decay and translational repression. Trends Cell Biol 2015, 25:651–665. [DOI] [PubMed] [Google Scholar]
- 49. Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin‐4 encodes small RNAs with antisense complementarity to lin‐14. Cell 1993, 75:843–854. [DOI] [PubMed] [Google Scholar]
- 50. Lagos‐Quintana M, Rauhut R, Lendeckel W, Tuschl T. Identification of novel genes coding for small expressed RNAs. Science 2001, 294:853–858. [DOI] [PubMed] [Google Scholar]
- 51. Friedländer MR, Lizano E, Houben AJ, Bezdan D, Báñez‐Coronel M, Kudla G, Mateu‐Huertas E, Kagerbauer B, González J, Chen KC, et al. Evidence for the biogenesis of more than 1,000 novel human microRNAs. Genome Biol 2014, 15:R57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Gunaratne PH, Creighton CJ, Watson M, Tennakoon JB. Large‐scale integration of microRNA and gene expression data for identification of enriched microRNA‐mRNA associations in biological systems. Methods Mol Biol 2010, 667:297–315. [DOI] [PubMed] [Google Scholar]
- 53. Nana‐Sinkam SP, Croce CM. MicroRNA regulation of tumorigenesis, cancer progression and interpatient heterogeneity: towards clinical use. Genome Biol 2014, 15:445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. An X, Sarmiento C, Tan T, Zhu H. Regulation of multidrug resistance by microRNAs in anti‐cancer therapy. Acta Pharm Sin B 2017, 7:38–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Lu J, Getz G, Miska EA, Alvarez‐Saavedra E, Lamb J, Peck D, Sweet‐Cordero A, Ebet BL, Mak RH, Ferrando AA, et al. MicroRNA expression profiles classify human cancers. Nature 2005, 435:834–838. [DOI] [PubMed] [Google Scholar]
- 56. Montano M. MicroRNAs: miRRORS of health and disease. Transl Res 2011, 157:157–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Kan CW, Howell VM, Hahn MA, Marsh DJ. Genomic alterations as mediators of miRNA dysregulation in ovarian cancer. Genes Chromosomes Cancer 2015, 54:1–19. [DOI] [PubMed] [Google Scholar]
- 58. Davidson B, Tropé CG, Reich R. The clinical and diagnostic role of microRNAs in ovarian carcinoma. Gynecol Oncol 2014, 133:640–646. [DOI] [PubMed] [Google Scholar]
- 59. Zhang S, Lu Z, Unruh AK, Ivan C, Baggerly KA, Calin GA, Li Z, Bast RC Jr, Le XF. Clinically relevant microRNAs in ovarian cancer. Mol Cancer Res 2015, 13:393–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Cai J, Yang C, Yang Q, Ding H, Jia J, Guo J, Wang J, Wang Z. Deregulation of let‐7e in epithelial ovarian cancer promotes the development of resistance to cisplatin. Oncogenesis 2013, 2:e75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Liu J, Wu X, Liu H, Liang Y, Gao X, Cai Z, Wang W, Zhang H. Expression of microRNA‐30a‐5p in drug‐resistant and drug‐sensitive ovarian cancer cell lines. Oncol Lett 2016, 12:2065–2070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Kim YW, Kim EY, Jeon D, Liu JL, Kim HS, Choi JW, Ahn WS. Differential microRNA expression signatures and cell type‐specific association with taxol resistance in ovarian cancer cells. Drug Des Devel Ther 2014, 8:293–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Zong C, Wang J, Shi TM. MicroRNA 130b enhances drug resistance in human ovarian cancer cells. Tumour Biol 2014, 35:12151–12156. [DOI] [PubMed] [Google Scholar]
- 64. Ciafrè SA, Galardi S. microRNAs and RNA‐binding proteins: a complex network of interactions and reciprocal regulations in cancer. RNA Biol 2013, 10:935–942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Preusse M, Marr C, Saunders S, Maticzka D, Lickert H, Backofen R, Theis F. SimiRa: a tool to identify coregulation between microRNAs and RNA‐binding proteins. RNA Biol 2015, 12:998–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Danno S, Nishiyama H, Higashitsuji H, Yokoi H, Xue JH, Itoh K, Matsuda T, Fujita J. Increased transcript level of RBM3, a member of the glycine‐rich RNA‐binding protein family, in human cells in response to cold stress. Biochem Biophys Res Commun 1997, 236:804–807. [DOI] [PubMed] [Google Scholar]
- 67. Ehlén Å, Nodin B, Rexhepaj E, Brändstedt J, Uhlén M, Alvarado‐Kristensson M, Pontén F, Brennan DJ, Jirström K. RBM3‐regulated genes promote DNA integrity and affect clinical outcome in epithelial ovarian cancer. Transl Oncol 2011, 4:212–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Zhu X, Zelmer A, Kapfhammer JP, Wellmann S. Cold‐inducible RBM3 inhibits PERK phosphorylation through cooperation with NF90 to protect cells from endoplasmic reticulum stress. FASEB J 2016, 30:624–634. [DOI] [PubMed] [Google Scholar]
- 69. Wong JJ, Au AY, Gao D, Pinello N, Kwok CT, Thoeng A, Lau KA, Gordon JE, Schmitz U, Feng Y, et al. RBM3 regulates temperature sensitive miR‐142‐5p and miR‐143 (thermomiRs), which target immune genes and control fever. Nucleic Acids Res 2016, 44:2888–2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Hinman MN, Lou H. Diverse molecular functions of Hu proteins. Cell Mol Life Sci 2008, 65:3168–3181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Lal A, Kawai T, Yang X, Mazan‐Mamczarz K, Gorospe M. Antiapoptotic function of RNA‐binding protein HuR effected through prothymosin alpha. EMBO J 2005, 24:1852–1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Denkert C, Weichert W, Pest S, Koch I, Licht D, Köbel M, Reles A, Sehouli J, Dietel M, Hauptmann S. Overexpression of the embryonic‐lethal abnormal vision‐like protein HuR in ovarian carcinoma is a prognostic factor and is associated with increased cyclooxygenase 2 expression. Cancer Res 2004, 64:189–195. [DOI] [PubMed] [Google Scholar]
- 73. Filippova N, Yang X, Wang Y, Gillespie GY, Langford C, King PH, Wheeler C, Nabors LB. The RNA‐binding protein HuR promotes glioma growth and treatment resistance. Mol Cancer Res 2011, 9:648–659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Zhu Z, Wang B, Bi J, Zhang C, Guo Y, Chu H, Liang X, Zhong C, Wang J. Cytoplasmic HuR expression correlates with P‐gp, HER‐2 positivity, and poor outcome in breast cancer. Tumour Biol 2013, 34:2299–2308. [DOI] [PubMed] [Google Scholar]
- 75. Blanco FF, Jimbo M, Wulfkuhle J, Gallagher I, Deng J, Enyenihi L, Meisner‐Kober N, Londin E, Rigoutsos I, Sawicki JA, et al. The mRNA‐binding protein HuR promotes hypoxia‐induced chemoresistance through posttranscriptional regulation of the proto‐oncogene PIM1 in pancreatic cancer cells. Oncogene 2016, 35:2529–2541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Lal S, Burkhart RA, Beeharry N, Bhattacharjee V, Londin ER, Cozzitorto JA, Romeo C, Jimbo M, Norris ZA, Yeo CJ, et al. HuR posttranscriptionally regulates WEE1: implications for the DNA damage response in pancreatic cancer cells. Cancer Res 2014, 74:1128–1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Romeo C, Weber MC, Zarei M, DeCicco D, Chand SN, Lobo AD, Winter JM, Sawicki JA, Sachs JN, Meisner‐Kober N, et al. HuR contributes to TRAIL resistance by restricting death receptor 4 expression in pancreatic cancer cells. Mol Cancer Res 2016, 14:599–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Mazan‐Mamczarz K, Galbán S, López de Silanes I, Martindale JL, Atasoy U, Keene JD, Gorospe M. RNA‐binding protein HuR enhances p53 translation in response to ultraviolet light irradiation. Proc Natl Acad Sci USA 2003, 100:8354–8359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Prislei S, Martinelli E, Mariani M, Raspaglio G, Sieber S, Ferrandina G, Shahabi S, Scambia G, Ferlini C. MiR‐200c and HuR in ovarian cancer. BMC Cancer 2013, 13:72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Ahuja D, Goyal A, Ray PS. Interplay between RNA‐binding protein HuR and microRNA‐125b regulates p53 mRNA translation in response to genotoxic stress. RNA Biol 2016, 13:1152–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Ke Y, Han Y, Guo X, Wen J, Wang K, Jiang X, Tian X, Ba X, Boldogh I, Zeng X. PARP1 promotes gene expression at the post‐transcriptiona level by modulating the RNA‐binding protein HuR. Nat Commun 2017, 8:14632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Li Z, Wei D, Yang C, Sun H, Lu T, Kuang D. Overexpression of long noncoding RNA, NEAT1 promotes cell proliferation, invasion and migration in endometrial endometrioid adenocarcinoma. Biomed Pharmacother 2016, 84:244–251. [DOI] [PubMed] [Google Scholar]
- 83. Chai Y, Liu J, Zhang Z, Liu L. HuR‐regulated lncRNA NEAT1 stability in tumorigenesis and progression of ovarian cancer. Cancer Med 2016, 5:1588–1598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Xu CZ, Jiang C, Wu Q, Liu L, Yan X, Shi R. A feed‐forward regulatory loop between HuR and the long noncoding RNA HOTAIR promotes head and neck squamous cell carcinoma progression and metastasis. Cell Physiol Biochem 2016, 40:1039–1051. [DOI] [PubMed] [Google Scholar]
- 85. Latorre E, Carelli S, Raimondi I, D'Agostino V, Castiglioni I, Zucal C, Moro G, Luciani A, Ghilardi G, Monti E, et al. The ribonucleic complex HuR‐MALAT1 represses CD133 expression and suppresses epithelial‐mesenchymal transition in breast cancer. Cancer Res 2016, 76:2626–2636. [DOI] [PubMed] [Google Scholar]
- 86. Zhang Z, Cheng J, Wu Y, Qiu J, Sun Y, Tong X. LncRNA HOTAIR controls the expression of Rab22a by sponging miR‐373 in ovarian cancer. Mol Med Rep 2016, 14:2465–2472. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 87. Zou A, Liu R, Wu X. Long non‐coding RNA MALAT1 is up‐regulated in ovarian cancer tissue and promotes SK‐OV‐3 cell proliferation and invasion. Neoplasma 2016, 63:865–872. [DOI] [PubMed] [Google Scholar]
- 88. Lei R, Xue M, Zhang L, Lin Z. Long noncoding RNA MALAT1‐regulated microRNA 506 modulates ovarian cancer growth by targeting iASPP. Onco Targets Ther 2016, 10:35–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Li Q, Zhang C, Chen R, Xiong H, Qiu F, Liu S, Zhang M, Wang F, Wang Y, Zhou X, et al. Disrupting MALAT1/miR‐200c sponge decreases invasion and migration in endometrioid endometrial carcinoma. Cancer Lett 2016, 383:28–40. [DOI] [PubMed] [Google Scholar]
- 90. Bell JL, Wächter K, Mühleck B, Pazaitis N, Köhn M, Lederer M, Hüttelmaier S. Insulin‐like growth factor 2 mRNA‐binding proteins (IGF2BPs): post‐transcriptional drivers of cancer progression? Cell Mol Life Sci 2013, 70:2657–2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Boyerinas B, Park SM, Shomron N, Hedegaard MM, Vinther J, Andersen JS, Feig C, Xu J, Burge CB, Peter ME. Identification of let‐7‐regulated oncofetal genes. Cancer Res 2008, 68:2587–2591. [DOI] [PubMed] [Google Scholar]
- 92. Boyerinas B, Park SM, Murmann AE, Gwin K, Montag AG, Zillhardt M, Hua YJ, Lengyel E, Peter ME. Let‐7 modulates acquired resistance of ovarian cancer to Taxanes via IMP‐1‐mediated stabilization of multidrug resistance 1. Int J Cancer 2012, 130:1787–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Weidensdorfer D, Stöhr N, Baude A, Lederer M, Köhn M, Schierhorn A, Buchmeier S, Wahle E, Hüttelmaier S. Control of c‐myc mRNA stability by IGF2BP1‐associated cytoplasmic RNPs. RNA 2009, 15:104–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Mahapatra L, Mao C, Andruska N, Zhang C, Shapiro DJ. High‐throughput fluorescence anisotropy screen for inhibitors of the oncogenic mRNA binding protein, IMP‐1. J Biomol Screen 2014, 19:427–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Busch B, Bley N, Müller S, Glaß M, Misiak D, Lederer M, Vetter M, Strauß HG, Thomssen C, Hüttelmaier S. The oncogenic triangle of HMGA2, LIN28B and IGF2BP1 antagonizes tumor‐suppressive actions of the let‐7 family. Nucleic Acids Res 2016, 44:3845–3864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Jønson L, Christiansen J, Hansen TV, Vikeså J, Yamamoto Y, Nielsen FC. IMP3 RNP safe houses prevent miRNA‐directed HMGA2 mRNA decay in cancer and development. Cell Rep 2014, 7:539–551. [DOI] [PubMed] [Google Scholar]
- 97. Gong C, Li Z, Ramanujan K, Clay I, Zhang Y, Lemire‐Brachat S, Glass DJ. A long non‐coding RNA, LncMyoD, regulates skeletal muscle differentiation by blocking IMP2‐mediated mRNA translation. Dev Cell 2015, 34:181–191. [DOI] [PubMed] [Google Scholar]
- 98. Kohno K, Izumi H, Uchiumi T, Ashizuka M, Kuwano M. The pleiotropic functions of the Y‐box‐binding protein, YB‐1. Bioessays 2003, 25:691–698. [DOI] [PubMed] [Google Scholar]
- 99. Lu ZH, Books JT, Ley TJ. YB‐1 is important for late‐stage embryonic development, optimal cellular stress responses, and the prevention of premature senescence. Mol Cell Biol 2005, 25:4625–4637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Inoue I, Matsumoto K, Yu Y, Bay BH. Surmounting chemoresistance by targeting the Y‐box binding protein‐1. Anat Rec 2012, 295:215–222. [DOI] [PubMed] [Google Scholar]
- 101. Saji H, Toi M, Saji S, Koike M, Kohno K, Kuwano M. Nuclear expression of YB‐1 protein correlates with P‐glycoprotein expression in human breast carcinoma. Cancer Lett 2003, 190:191–197. [DOI] [PubMed] [Google Scholar]
- 102. Lasham A, Moloney S, Hale T, Homer C, Zhang YF, Murison JG, Braithwaite AW, Watson J. The Y‐box‐binding protein, YB1, is a potential negative regulator of the p53 tumor suppressor. J Biol Chem 2003, 278:35516–35523. [DOI] [PubMed] [Google Scholar]
- 103. Xu S, Wang T, Song W, Jiang T, Zhang F, Yin Y, Jiang SW, Wu K, Yu Z, Wang C, et al. The inhibitory effects of AR/miR‐190a/YB‐1 negative feedback loop on prostate cancer and underlying mechanism. Sci Rep 2015, 5:13528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Stavraka C, Blagden S. The La‐related proteins, a family with connections to cancer. Biomolecules 2015, 5:2701–2722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Blagden SP, Gatt MK, Archambault V, Lada K, Ichihara K, Lilley KS, Inoue YH, Glover DM. Drosophila Larp associates with poly(A)‐binding protein and is required for male fertility and syncytial embryo development. Dev Biol 2009, 334:186–197. [DOI] [PubMed] [Google Scholar]
- 106. Tcherkezian J, Cargnello M, Romeo Y, Huttlin EL, Lavoie G, Gygi SP, Roux PP. Proteomic analysis of cap‐dependent translation identifies LARP1 as a key regulator of 5′TOP mRNA translation. Genes Dev 2014, 28:357–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Fonseca BD, Zakaria C, Jia JJ, Graber TE, Svitkin Y, Tahmasebi S, Healy D, Hoang HD, Jensen JM, Diao IT, et al. La‐related protein 1 (LARP1) represses terminal oligopyrimidine (TOP) mRNA translation downstream of mTOR complex 1 (mTORC1). J Biol Chem 2015, 290:15996–16020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Hopkins TG, Mura M, Al‐Ashtal HA, Lahr RM, Abd‐Latip N, Sweeney K, Lu H, Weir J, El‐Bahrawy M, Steel JH, et al. The RNA‐binding protein LARP1 is a post‐transcriptional regulator of survival and tumorigenesis in ovarian cancer. Nucleic Acids Res 2016, 44:1227–1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Mura M, Hopkins TG, Michael T, Abd‐Latip N, Weir J, Aboagye E, Mauri F, Jameson C, Sturge J, Gabra H, et al. LARP1 post‐transcriptionally regulates mTOR and contributes to cancer progression. Oncogene 2015, 34:5025–5036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Lahr RM, Mack SM, Héroux A, Blagden SP, Bousquet‐Antonelli C, Deragon JM, Berman AJ. The La‐related protein 1‐specific domain repurposes HEAT‐like repeats to directly bind a 5′TOP sequence. Nucleic Acids Res 2015, 43:8077–8088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Kato M, Goto Y, Matsushita R, Kurozumi A, Fukumoto I, Nishikawa R, Sakamoto S, Enokida H, Nakagawa M, Ichikawa T, et al. MicroRNA‐26a/b directly regulate La‐related protein 1 and inhibit cancer cell invasion in prostate cancer. Int J Oncol 2015, 47:710–718. [DOI] [PubMed] [Google Scholar]
- 112. Castellano L, Dabrowska A, Pellegrino L, Ottaviani S, Cathcart P, Frampton AE, Krell J, Stebbing J. Sustained expression of miR‐26a promotes chromosomal instability and tumorigenesis through regulation of CHFR. Nucleic Acids Res 2017, 45:4401–4412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Huang S, Lu W, Ge D, Meng N, Li Y, Su L, Zhang S, Zhang Y, Zhao B, Miao J. A new microRNA signal pathway regulated by long noncoding RNA TGFB2‐OT1 in autophagy and inflammation of vascular endothelial cells. Autophagy 2015, 11:2172–2183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Dresios J, Aschrafi A, Owens GC, Vanderklish PW, Edelman GM, Mauro VP. Cold stress‐induced protein Rbm3 binds 60S ribosomal subunits, alters microRNA levels, and enhances global protein synthesis. Proc Natl Acad Sci U S A 2005, 102:1865–1870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Ma WJ, Cheng S, Campbell C, Wright A, Furneaux H. Cloning and characterization of HuR, a ubiquitously expressed Elav‐like protein. J Biol Chem 1996, 5:8144–8151. [DOI] [PubMed] [Google Scholar]
- 116. Yang HJ, Ju F, Guo XX, Ma SP, Wang L, Cheng BF, Zhuang RJ, Zhang BB, Shi X, Feng ZW, et al. RNA‐binding protein RBM3 prevents NO‐induced apoptosis in human neuroblastoma cells by modulating p38 signaling and miR‐143. Sci Rep 2017, 7:41738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Myer VE, Fan XC, Steitz JA. Identification of HuR as a protein implicated in AUUUA‐mediated mRNA decay. EMBO J 1997, 16:2130–2139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Galbán S, Kuwano Y, Pullmann R Jr, Martindale JL, Kim HH, Lal A, Abdelmohsen K, Yang X, Dang Y, Liu JO, et al. RNA‐binding proteins HuR and PTB promote the translation of hypoxia‐inducible factor 1alpha. Mol Cell Biol 2008, 28:93–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Katsanou V, Milatos S, Yiakouvaki A, Sgantzis N, Kotsoni A, Alexiou M, Harokopos V, Aidinis V, Hemberger M, Kontoyiannis DL. The RNA‐binding protein Elavl1/HuR is essential for placental branching morphogenesis and embryonic development. Mol Cell Biol 2009, 29:2762–2776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Silvia P, Enrica M, Gian FZ, Marco P, Flavia F, Marisa M, Simona M, Giuseppina R, Giovanni S, Cristiano F. Role and prognostic significance of the epithelial‐mesenchymal transition factor ZEB2 in ovarian cancer. Oncotarget 2015, 6:18966–18979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Nielsen J, Christiansen J, Lykke‐Andersen J, Johnsen AH, Wewer UM, Nielsen FC. A family of insulin‐like growth factor II mRNA‐binding proteins represses translation in late development. Mol Cell Biol 1999, 19:1262–1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Zhang HL, Eom T, Oleynikov Y, Shenoy SM, Liebelt DA, Dictenberg JB, Singer RH, Bassell GJ. Neurotrophin‐induced transport of a beta‐actin mRNP complex increases beta‐actin levels and stimulates growth cone motility. Neuron 2001, 31:261–275. [DOI] [PubMed] [Google Scholar]
- 123. Yaniv K, Fainsod A, Kalcheim C, Yisraeli JK. The RNA‐binding protein Vg1 RBP is required for cell migration during early neural development. Development 2003, 130:5649–5661. [DOI] [PubMed] [Google Scholar]
- 124. Farina KL, Huttelmaier S, Musunuru K, Darnell R, Singer RH. Two ZBP1 KH domains facilitate beta‐actin mRNA localization, granule formation, and cytoskeletal attachment. J Cell Biol 2003, 160:77–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Nishino J, Kim S, Zhu Y, Morrison SJ. A network of heterochronic genes including Imp1 regulates temporal changes in stem cell properties. Elife 2013, 2:e00924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Stohr N, Lederer M, Reinke C, Meyer S, Hatzfeld M, Singer RH, Huttelmaier S. ZBP1 regulates mRNA stability during cellular stress. J Cell Biol 2006, 175:527–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Mossink MH, van Zon A, Scheper RJ, Sonneveld P, Wiemer EA. Vaults: a ribonucleoprotein particle involved in drug resistance? Oncogene 2003, 22:7458–7467. [DOI] [PubMed] [Google Scholar]
- 128. Koike K, Uchiumi T, Ohga T, Toh S, Wada M, Kohno K, Kuwano M. Nuclear translocation of the Y‐box binding protein by ultraviolet irradiation. FEBS Lett 1997, 417:390–394. [DOI] [PubMed] [Google Scholar]
- 129. Evdokimova V, Ruzanov P, Imataka H, Raught B, Svitkin Y, Ovchinnikov LP, Sonenberg N. The major mRNA‐associated protein YB‐1 is a potent 5′ cap‐dependent mRNA stabilizer. EMBO J 2001, 20:5491–5502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Ashizuka M, Fukuda T, Nakamura T, Shirasuna K, Iwai K, Izumi H, Kohno K, Kuwano M, Uchiumi T. Novel translational control through an iron‐responsive element by interaction of multifunctional protein YB‐1 and IRP2. Mol Cell Biol 2002, 22:6375–6383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Fukuda T, Ashizuka M, Nakamura T, Shibahara K, Maeda K, Izumi H, Kohno K, Kuwano M, Uchiumi T. Characterization of the 5′‐untranslated region of YB‐1 mRNA and autoregulation of translation by YB‐1 protein. Nucleic Acids Res 2004, 32:611–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Kamura T, Yahata H, Amada S, Ogawa S, Sonoda T, Kobayashi H, Mitsumoto M, Kohno K, Kuwano M, Nakano H. Is nuclear expression of Y box‐binding protein‐1 a new prognostic factor in ovarian serous adenocarcinoma? Cancer 1999, 85:2450–2454. [DOI] [PubMed] [Google Scholar]
- 133. Hideaki Y, Hiroaki K, Toshiharu K, Satoshi A, Toshio H, Kimitoshi K, Michihiko K, Hitoo N. Increased nuclear localization of transcription factor YB‐1 in acquired cisplatin‐resistant ovarian cancer. J Cancer Res Clin Oncol 2002, 128:621–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Huang YH, Peng W, Furuuchi N, Gerhart J, Rhodes K, Mukherjee N, Jimbo M, Gonye GE, Brody JR, Getts RC, et al. Delivery of therapeutics targeting the mRNA‐binding protein HuR using 3DNA nanocarriers suppresses ovarian tumor growth. Cancer Res 2016, 76:1549–1559. [DOI] [PubMed] [Google Scholar]
- 135. Yan C, Higgins PJ. Drugging the undruggable: transcription therapy for cancer. Biochim Biophys Acta 2013, 1835:76–85. [DOI] [PMC free article] [PubMed] [Google Scholar]


