Abstract
Protein ubiquitination plays a critical role in Toll-like receptor (TLR) signaling and innate immunity. Although several E3 ubiquitin ligases have been identified downstream of TLRs, the regulation of protein deubiquitination in TLR-triggered innate immune responses is poorly understood. We identified ubiquitin-specific protease 25 (USP25) as a regulator of TLR signaling. USP25 was recruited to the TLR4 signaling complex, and it associated with the adaptor proteins tumor necrosis factor receptor–associated factor 3 (TRAF3) and TRAF6 after stimulation of TLR4 with its ligand lipopolysaccharide (LPS). USP25 specifically reversed the Lys48-linked ubiquitination of TRAF3 that was mediated by the E3 ubiquitin ligase cIAP2 (cellular inhibitor of apoptosis 2). Deficiency in USP25 enhanced the extent of ubiquitination of TRAF3 and accelerated its degradation after TLR4 activation, which potentiated TLR4-induced activation of NF-κB (nuclear factor κB) and MAPK (mitogen-activated protein kinase) signaling, but inhibited activation of the transcription factor IRF3 (interferon regulatory factor 3). USP25-deficient mice exhibited increased susceptibility to LPS-induced septic shock compared to their wild-type counterparts, which was associated with enhanced production of proinflammatory cytokines and decreased production of interferon-α. Thus, by inhibiting the degradation of TRAF3 during TLR4 activation, USP25 enables a balanced innate immune response.
INTRODUCTION
Host pattern recognition receptors (PRRs) recognize structurally conserved pathogen-associated molecular patterns (PAMPs) from invading pathogens, which represents the first step in innate immune responses (1, 2). One major class of PRRs consists of the membrane-associated Toll-like receptors (TLRs), which exhibit well-established recognition of PAMPs (3). For example, TLR4 recognizes lipopolysaccharide (LPS), a common structure of the cell wall of Gram-negative bacteria (4), whereas TLR2, together with TLR1 or TLR6, is involved in the recognition of various microbial components, including lipoproteins with lipid chains covalently attached to conserved N-terminal cysteines that are found in Gram-positive bacteria (5, 6). TLR3 recognizes viral double-stranded RNA, whereas TLR7 and TLR8 recognize single-stranded viral RNA (7, 8), and TLR9 detects unmethylated CpG motifs present in bacterial and viral DNA (9). Upon binding to their ligands, TLRs initiate a series of signaling cascades that lead to activation of the transcription factors nuclear factor κB (NF-κB), interferon regulatory factor 3 (IRF3) and IRF7, as well as activator protein-1 (AP-1), which collaborate to induce transcription of a large number of target genes (3). These genes encode pro-inflammatory cytokines, chemokines, and type I interferons (IFNs), which collaborate to orchestrate innate immune responses (10, 11).
It is well recognized that TLR-triggered signaling depends on the adaptor proteins myeloid differentiation marker 88 (MyD88) and Toll–interleukin-1 (IL-1) receptor (TIR) domain–containing adaptor-inducing IFN-β (TRIF), which mediate distinct responses that are classified as MyD88- and TRIF-dependent pathways (3). TLR3-triggered signaling is exclusively dependent on TRIF, TLR4 triggers both MyD88- and TRIF-dependent signaling, whereas TLR2 signals only through MyD88. TLR-triggered TRIF and MyD88 signaling pathways depend on ubiquitination processes mediated by a number of E3 ubiquitin ligases, including tumor necrosis factor receptor–associated factor 3 (TRAF3), TRAF6, and cellular inhibitor of apoptosis 1 (cIAP1) and cIAP2 (collectively known as cIAP1/2) (3, 12). TRAF3 is an E3 ubiquitin ligase that mediates both MyD88- and TRIF-dependent pathways through distinct ubiquitination modifications (13–15). Upon activation of TLR3 or TLR4, TRIF-mediated signaling induces the Lys63 (K63)–linked ubiquitination of TRAF3, which is required for the TRAF family member–associated NF-κB activator–binding kinase (TBK1)–mediated activation of IRF3 and production of type I IFNs (16). In contrast, TLR2- or TLR4-initiated MyD88-dependent signaling triggers TRAF6- and cIAP1/2-mediated Lys48 (K48)–linked ubiquitination and degradation of TRAF3, which results in release of the TRAF6–cIAP1/2–IL-1 receptor–associated kinase (IRAK) signaling complex from the cell membrane to the cytosol and is required for transforming growth factor–β–activated kinase 1 (TAK1)–mediated activation of mitogen-activated protein kinases (MAPKs) and production of proinflammatory cytokines (15). Thus, in TLR4 signaling, activation of type I IFN and proinflammatory cytokine production are likely modulated by the type of ubiquitination of TRAF3 and its abundance in innate immune cells, the mechanism of which is not understood.
Similar to ubiquitination, the reverse process of protein deubiquitination is highly regulated. Deubiquitination is mediated by deubiquitinating enzymes (DUBs) and is implicated in numerous cellular functions, including cell cycle regulation, protein degradation, gene expression, and signal transduction (17, 18). About 100 putative DUBs are encoded by the human genome, and they belong to five different subfamilies, including ubiquitin-specific proteases (USPs), ubiquitin carboxy-terminal hydrolases, ovarian tumor (OTU) domain–containing proteases, Machado-Joseph disease proteases, and metalloproteases (19). Although several ubiquitin ligase enzymes regulate TLR-triggered signaling, only very few DUBs are known to regulate these pathways (20). A20 and cylindromatosis (turban tumor syndrome, also known as CYLD) are two OTU domain containing DUBs that inhibit TLR signaling by modulating the deubiquitination of TRAF6 or the inhibitor of κB (IκB) kinase (IKK) subunit IKKγ (also known as NEMO); however, their targets and the mechanisms involved are controversial (21–26). The K63-linked polyubiquitin chain on TRAF3 is cleaved by DUBA or OTUB1/2, and this process is required for the inhibition of TLR3-, TLR4-, or viral infection–induced production of type I IFNs (16, 27). However, how the K48-linked ubiquitination of TRAF3 is regulated by other DUBs and whether other DUBs are involved in TLR signaling remain unknown.
We previously found that USP25 dampens IL-17–mediated signaling by restricting the extent of the K63-linked ubiquitination of TRAF5 and TRAF6 by the IL-17 receptor (IL-17R) adaptor protein and E3 ubiquitin ligase Act1 (28). Here, we found that USP25 acted as a critical modulator of TLR4-dependent signaling. Deficiency in USP25 in mice potentiated LPS-induced degradative ubiquitination of TRAF3, which led to enhanced production of proinflammatory cytokines and impaired production of type I IFNs. USP25 physically interacted with TRAF3 after TLR4 activation and specifically removed K48-linked polyubiquitin chains from TRAF3, which maintained the cellular abundance of TRAF3 required for balanced TLR4-induced proinflammatory cytokine production and type I IFN responses. Our findings thus identify USP25 as a previously uncharacterized critical regulator of innate immune responses that prevents the degradation of TRAF3 during TLR4 activation.
RESULTS
LPS induces a physical association between USP25 and TRAF3
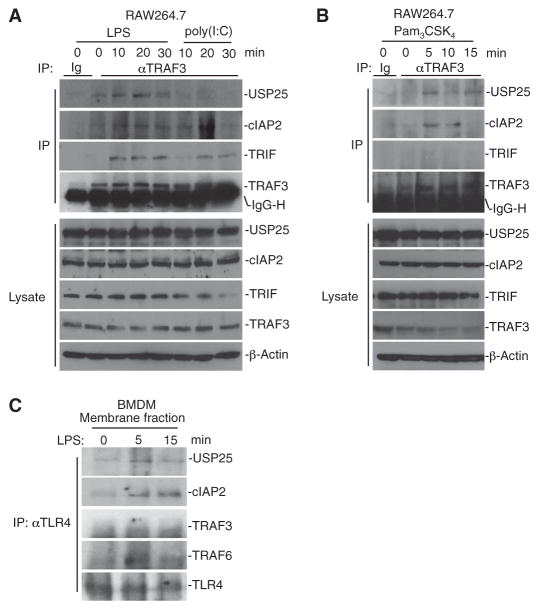
In our previous study, we demonstrated that USP25 interacts with TRAF3, TRAF5, and TRAF6 in an overexpression system in human embryonic kidney (HEK) 293T cells and that endogenous USP25-TRAF5 and USP25-TRAF6 interactions are promoted by IL-17 (28). However, the signals that stimulate an association between USP25 and TRAF3 are unknown. When we examined LPS-treated mouse bone marrow–derived macrophages (BMDMs) and bone marrow–derived dendritic cells (BMDCs), we found that the expression of Usp25 and the abundance of USP25 protein were substantially increased compared to those in untreated cells (fig. S1, A and B). Because TRAF3 plays an important role in TLR signaling, we examined whether USP25 interacted with TRAF3 in response to various TLR stimuli. We treated RAW264.7 cells (a mouse macrophage cell line) with poly(I:C) (a TLR3 ligand), LPS (a TLR4 ligand), or Pam3CSK4 (a TLR2 ligand) and performed immunoprecipitations and Western blotting analysis on the treated cells. We found that stimulation with LPS or Pam3CSK4, but not poly(I:C), promoted an association between USP25 and TRAF3 (Fig. 1, A and B). USP25 contains multiple domains, and we previously demonstrated that the ubiquitin-interacting motifs (UIMs) (residues 91 to 150) are required for its optimal association with TRAF6 and TRAF5 (28). However, here, our domain mapping analysis suggested that the peptidase (residues 151 to 654) and the coiled-coil domains (UCH-coil) (residues 655 to 1055) were essential for the USP25-TRAF3 association (fig. S1C). In addition, we observed a weak, but detectable, direct association between USP25 and TRAF3 in pull-down assays in vitro, whereas we did not detect a direct interaction between USP25 and TRAF6 (fig. S1D). When we subjected membrane fractions from LPS-treated BMDMs to immunoprecipitation with an antibody against TLR4 (anti-TLR4 antibody), we observed that USP25, TRAF3, and TRAF6 were co-immunoprecipitated (Fig. 1C), supporting the idea that LPS stimulates the formation of a membrane-bound TLR4 signaling complex consisting of USP25, TRAFs, and cIAPs. In addition, we found that USP25 interacted with MyD88, but not TRIF, in coimmunoprecipitation studies of transfected cells (fig. S1E) and that knockdown of TRAF3 or TRAF6 had no effect on the LPS-induced recruitment of USP25 to TLR4 (fig. S1F). These data suggest that LPS stimulates the recruitment of USP25 and TRAF3 to the TLR4 signaling complex in which USP25 interacts with TRAF3.
Fig. 1. LPS induces a physical association between USP25 and TRAF3.
(A and B) RAW264.7 cells were stimulated with LPS (10 μg/ml), poly(I:C) (50 μg/ml), or Pam3CSK4 (10 μg/ml) for the indicated times. Cells were then lysed, and cell lysates were subjected to immunoprecipitation (IP) with control immunoglobulin (Ig) or with antibody against TRAF3 (αTRAF3). The immuno-precipitates were analyzed by Western blotting with antibodies against the indicated proteins (top four blots in each panel). IgG-H indicates the IgG heavy chain. As controls, the abundances of the indicated proteins in the cell lysates were determined by Western blotting analysis with the appropriate specific antibodies (bottom panels). (C) USP25 is recruited to TLR4 after stimulation with LPS. BMDMs were left untreated or were stimulated with LPS (10 μg/ml) for the indicated times. Cell membrane fractions were prepared and resuspended in lysis buffer. The lysates were immunoprecipitated with anti-TLR4 antibody (αTLR4), and the immunoprecipitates were analyzed by Western blotting with antibodies against the indicated proteins. Data are representative of two independent experiments.
Usp25−/− mice are more susceptible than wild-type mice to LPS-induced septic shock
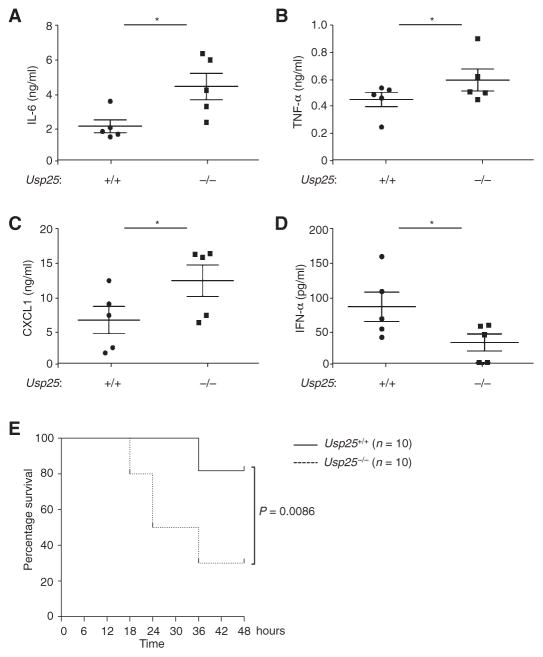
Next, we investigated the function of USP25 in TLR-stimulated innate immune responses in vivo. After challenge with LPS, Usp25−/− mice produced substantially increased amounts of tumor necrosis factor–α (TNF-α), IL-6, and the CXC chemokine CXCL1, but decreased amounts of IFN-α, in their sera than did their wild-type littermates (Fig. 2, A to D). In contrast, poly(I:C)-induced production of proinflammatory cytokines and IFN-α was comparable between the Usp25−/− mice and the wild-type controls (fig. S2). Moreover, Usp25−/− mice were more susceptible than were their wild-type littermates to LPS-induced septic shock (Fig. 2E). Together, these data suggest that USP25 differentially regulates TLR4 signaling in terms of the production of proinflammatory cytokines and type I IFNs in vivo.
Fig. 2. USP25 restricts LPS-induced septic shock as well as production of proinflammatory cytokines in vivo.
(A to D) Age- and sex-matched 7- to 8-week-old wild-type (WT) and Usp25−/− littermates (five mice each) were injected intraperitoneally with LPS. Two and a half hours later, the serum amounts of (A) IL-6, (B) TNF-α, (C) CXCL1, and (D) IFN-α were determined by enzyme-linked immunosorbent assay (ELISA). Data are means ± SD of three independent experiments. *P < 0.05 by t test. (E) USP25-deficient mice exhibit increased susceptibility to LPS-induced septic shock. WT and Usp25−/− littermates were treated with LPS as in (A). The survival of the mice was monitored over the next 48 hours. Data are combined from two independent experiments and are from a total of 10 mice of each genotype.
USP25 restricts proinflammatory cytokine production and promotes type I IFN production upon LPS stimulation
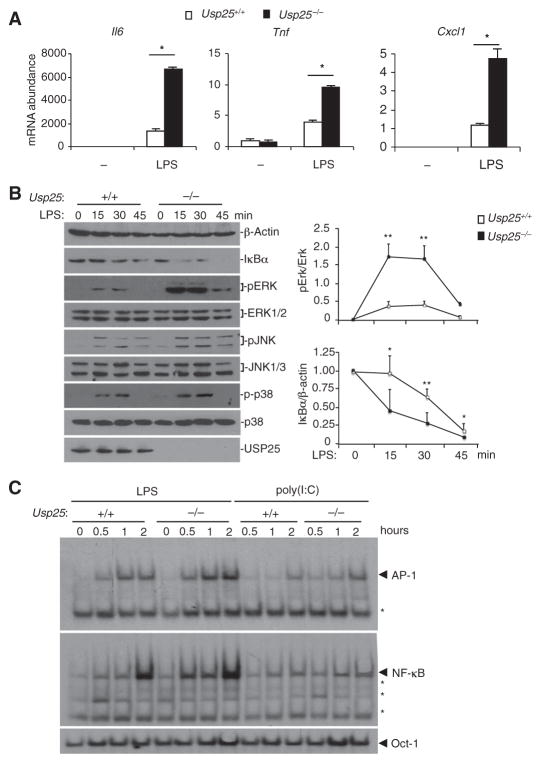
We next examined the effects of deficiency in USP25 on LPS-triggered signaling in mouse embryonic fibroblasts (MEFs). We found that LPS-induced expression of Il6, Tnf, and Cxcl1 was enhanced in Usp25−/−MEFs compared to that in wild-type MEFs (Fig. 3A). Consistent with the results of these gene expression studies, LPS-induced, but not poly(I:C)-induced, degradation of IκBα was accelerated, and the phosphorylation (and thus activation) of the MAPK extracellular signal–regulated kinase (ERK) was enhanced in Usp25−/− MEFs compared to that in wild-type MEFs (Fig. 3B and fig. S3). To further confirm that USP25 inhibited the LPS-induced activation of NF-κB and MAPKs, we performed electrophoretic mobility shift assays (EMSAs) with 32P-labeled consensus NF-κB and AP-1 (c-Jun–c-Fos) probes and showed that a deficiency in USP25 potentiated LPS-induced, but not poly(I:C)-induced, activation of the transcription factors NF-κB and AP-1 (Fig. 3C). These data suggest that USP25 inhibits TLR4-dependent, but not TLR3-dependent, signaling, thus inhibiting the activation of NF-κB and MAPKs in MEFs.
Fig. 3. USP25 inhibits TLR4-dependent activation of NF-κB and MAPKs in MEFs.
(A) USP25 inhibits the generation of proinflammatory cytokines in MEFs stimulated with LPS. WT or Usp25−/− MEFs were stimulated with LPS (1 μg/ml) for 3 hours before real-time reverse transcription polymerase chain reaction (RT-PCR) analysis was performed to determine the relative abundances of the indicated mRNAs. Data are means ± SD from three independent experiments. *P < 0.01 by t test. (B and C) Deficiency in USP25 potentiates LPS-induced activation of NF-κB and MAPKs in MEFs. (B) WT or Usp25−/− MEFs were stimulated with LPS (10 μg/ml), and the degradation of IκBα and phosphorylation of ERK, JNK (c-Jun N-terminal kinase), and p38 were analyzed by Western blotting with antibodies against the indicated proteins. Data are representative of three independent experiments. *P < 0.05; **P < 0.01 by analysis of variance (ANOVA). (C) WT or Usp25 −/− MEFs were stimulated with LPS (10 μg/ml) or poly(I:C) (50 μg/ml). Nuclear extracts were prepared and subjected to EMSA analysis with NF-κB, AP-1, and Oct-1 probes. Data are representative of three independent experiments. Asterisk indicates nonspecific bands.
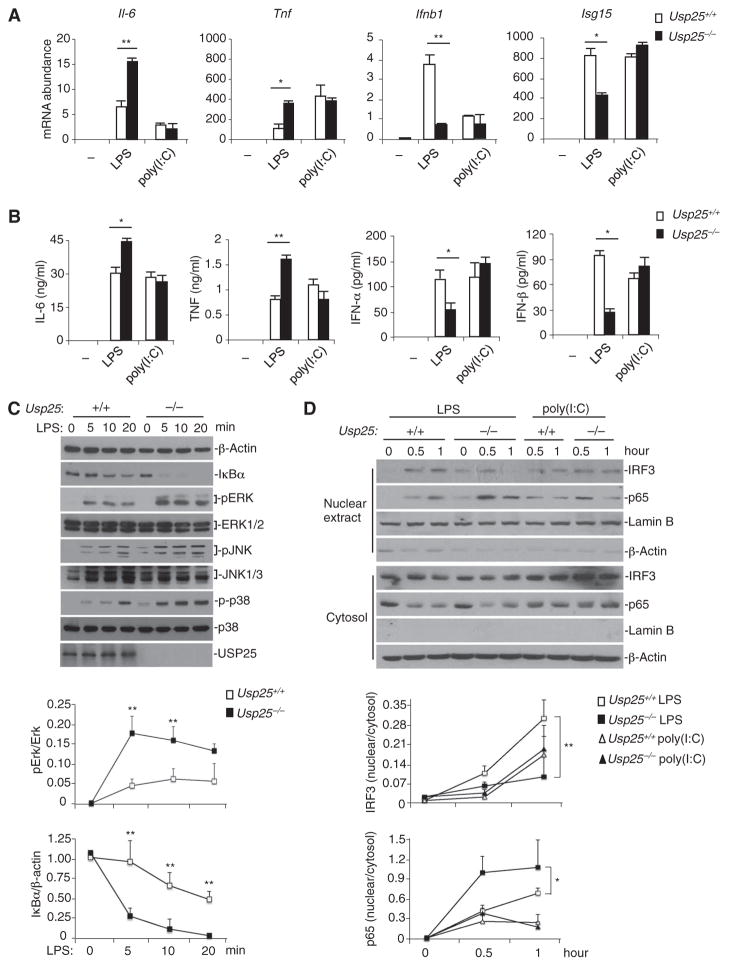
We further examined whether USP25 regulated LPS-induced signaling in other cell types. We isolated bone marrow cells from Usp25+/+ and Usp25−/− littermate mice, generated BMDMs and BMDCs through standard procedures, and stimulated the cells with LPS or poly(I:C). LPS-induced, but not poly(I:C)-induced, expression of Il6 and Tnf was increased in Usp25−/− BMDCs compared to that in wild-type cells (Fig. 4A). In the same experiments, we also found that LPS-induced, but not poly(I:C)-induced, expression of Ifnb1 and Isg15 (IFN-stimulated gene 15) was impaired in Usp25−/− BMDCs compared to that in wild-type BMDCs (Fig. 4A). Consistent with these observations, Usp25−/− BMDCs produced substantially increased amounts of proinflammatory cytokines and decreased amounts of type I IFNs compared to wild-type BMDCs in response to LPS (Fig. 4B). Similar results were obtained from experiments with wild-type and USP25-deficient BMDMs (fig. S4, A to C). Together, these data suggest that USP25 restricts the LPS-induced production of proinflammatory cytokines and promotes LPS-induced production of type I IFNs in various cell types.
Fig. 4. USP25 restricts TLR4-triggered proinflammatory signaling and promotes TLR4-induced type I IFN signaling.
(A and B) LPS-induced production of proinflammatory cytokines and type I IFNs is differentially regulated by USP25. WT and USP25-deficient BMDCs were stimulated with LPS (0.1 μg/ml) or poly(I:C) (25 μg/ml) (A) for 3 hours before real-time RT-PCR analysis of the abundance of the indicated mRNAs was performed or (B) for 12 hours before ELISA analysis for the indicated proteins was performed. Data are means ± SD from three independent experiments. *P < 0.05; **P < 0.01 by ANOVA. (C) LPS-induced activation of NF-κB and MAPKs is potentiated in USP25-deficient BMDCs. WT and Usp25−/− BMDCs were treated with LPS (10 μg/ml) for the indicated times. The degradation of IκBα and the activation of MAPKs were determined by Western blotting analysis with antibodies against the indicated proteins. (D) LPS-induced nuclear translocation of p65 is enhanced, whereas that of IRF3 is inhibited in USP25-deficient BMDCs. WT and Usp25−/− BMDCs were treated with LPS (10 μg/ml) or poly(I:C) (50 μg/ml) for the indicated times. Nuclear extracts and cytosolic fractions were prepared and analyzed by Western blotting with antibodies against the indicated proteins. Data are representative of at least three independent experiments. The intensities of the indicatedbands in (C) and (D) were quantified, and the ratios of the intensities of the corresponding bands were calculated and are shown in the graphs as means ± SD from three independent experiments. *P < 0.05; **P < 0.01 by ANOVA.
Because LPS-induced activation of the transcription factors NF-κB and IRF3 is responsible for the transcriptional activation of genes encoding proinflammatory cytokines and type I IFNs, respectively, we next examined the activation of NF-κB and IRF3 in wild-type and USP25-deficient BMDCs in response to LPS or poly(I:C). The activation of NF-κB and ERK was potentiated in LPS-treated, but not poly(I:C)-treated, Usp25−/− BMDCs compared to that in wild-type cells (Fig. 4C and fig. S4D). In addition, the translocation of the NF-κB subunit p65 from the cytosol to the nucleus was substantially promoted, whereas the translocation of IRF3 to the nucleus was strongly inhibited in Usp25−/− BMDCs compared to that in wild-type cells in response to LPS (Fig. 4D). Collectively, these data demonstrate that USP25 inhibits LPS-induced activation of NF-κB and promotes LPS-induced activation of IRF3, which is critical for the balanced production of proinflammatory cytokines and type I IFNs upon TLR4 activation.
The DUB activity of USP25 is required for regulation of TLR4 signaling
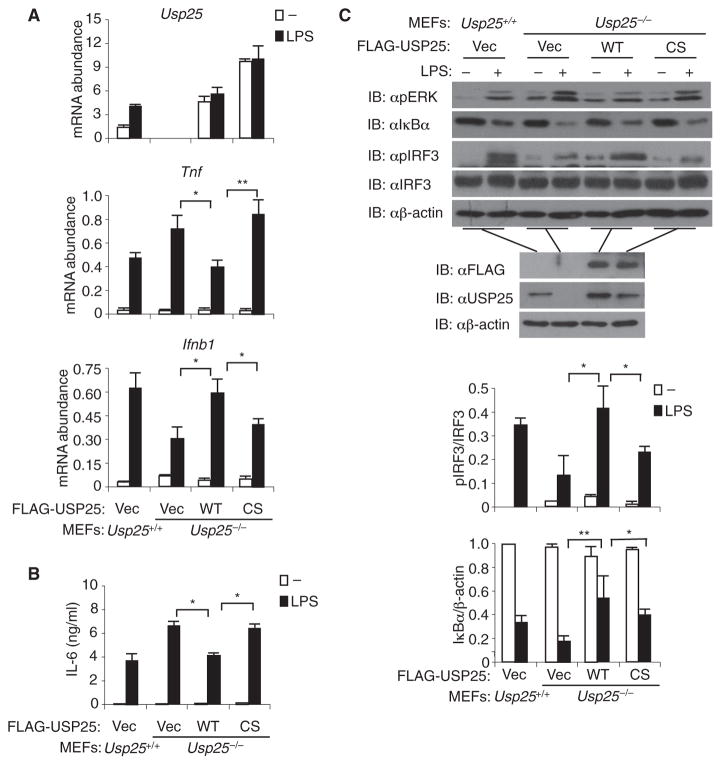
USP25 was originally identified as a deubiquitinase (DUB), and Cys178 of USP25 is critical for its DUB activity and is required for its regulation of IL-17–dependent signaling (29). We next examined whether the DUB activity of USP25 was required for regulating TLR4 signaling. Usp25−/− MEFs were reconstituted with wild-type USP25 or with the catalytically inactive mutant USP25(C178S), as previously described (28), which was confirmed at the mRNA level (Fig. 5A). Reconstitution with USP25, but not USP25(C178S), inhibited the LPS-induced expression of Tnf and production of IL-6 in Usp25−/− MEFs and restored the LPS-induced expression of Ifnb1 (Fig. 5, A and B). In similar experiments, reconstitution of Usp25−/− MEFs with either USP25 or USP25(C178S) had no effect on the poly(I:C)-induced expression of Tnf and Ifnb1 (fig. S5). Consistent with these observations, the LPS-induced activation of ERK and degradation of IκBα were inhibited, whereas the LPS-induced activation of IRF3 was restored, by reconstitution of Usp25−/−MEFs with USP25, but not USP25(C178S) (Fig. 5C). Together, these data suggest that the DUB activity of USP25 is required for its regulation of TLR4 signaling.
Fig. 5. The DUB activity of USP25 is required for the regulation of TLR4-induced generation of proinflammatory cytokines and type I IFNs.
(A to C) Usp25−/− MEFs were reconstituted with empty vector (Vec), FLAG-tagged WT USP25, or FLAG-tagged mutant USP25(C178S) (CS). (A) Reconstitution of Usp25−/−MEFs with USP25, but not USP25(C178S), alters the LPS-induced expression of Tnf and Ifnb1. The reconstituted cells were left untreated or were stimulated with LPS (1 μg/ml) for 6 hours before real-time RT-PCR analysis of the abundances of the indicated mRNAs was performed. (B) Reconstitution of Usp25−/− MEFs with USP25, but not USP25(C178S), inhibits LPS-induced production of IL-6 protein. The reconstituted cells were left untreated or were stimulated with LPS (1 μg/ml) for 24 hours before an ELISA assay to detect IL-6 was performed. Data are means ± SD from three independent experiments. *P < 0.05; **P < 0.01 by ANOVA. (C) LPS-induced activation of NF-κB and MAPKs is inhibited in Usp25−/− MEFs reconstituted with USP25, but not in Usp25−/− MEFs reconstituted with USP25(C178S). The indicated cells were left untreated or were stimulated with LPS (10 μg/ml) for 20 min before they were lysed, and the cell lysates were analyzed by Western blotting (IB) with the indicated antibodies (top five blots). Before stimulation, aliquots of cells were saved, and the abundances of the expressed proteins were examined by Western blotting with the indicated antibodies (bottom three blots). Data are representative of at least three independent experiments. The intensities of the indicated bands in (C) were quantified, and the ratios of the intensities of the corresponding bands were calculated and are shown in the bar graphs as means ± SD from three independent experiments. *P < 0.05; **P < 0.01 by ANOVA.
USP25 removes K48-linked ubiquitin chain from TRAF3
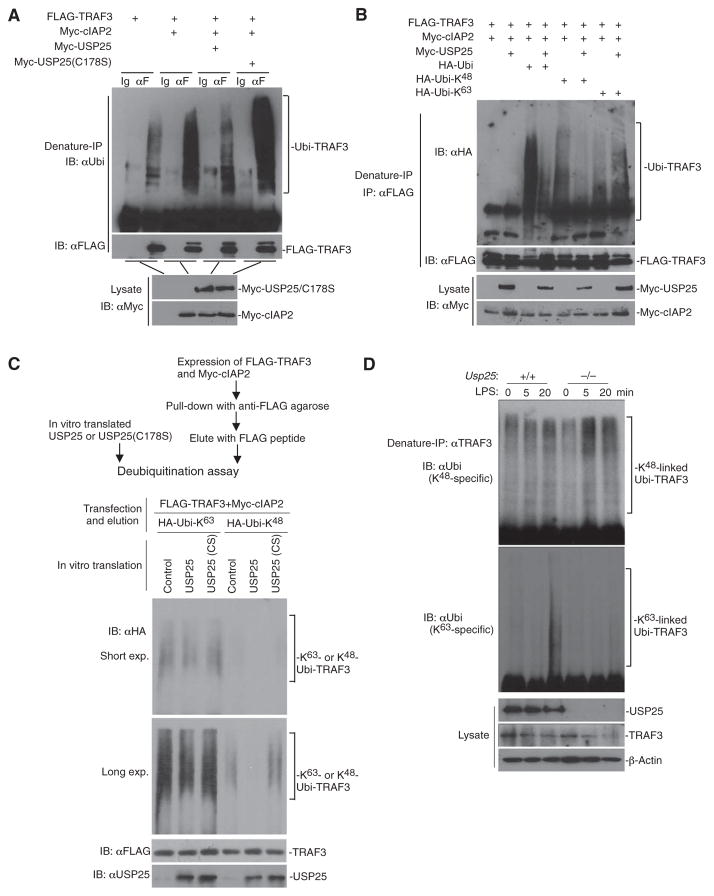
The LPS-stimulated activation of NF-κB signaling and induction of inflammatory-related gene expression are potentiated, whereas the activation of IRF3 and production of type I IFNs are diminished, in Traf3−/−BMDMs compared to wild-type BMDMs (14), which is similar to the phenotype that we observed in Usp25−/− mice and cells (Figs. 2 to 4). Because USP25 interacted with TRAF3 after stimulation with LPS and because the enzyme activity of USP25 was required for its regulation of TLR4 signaling (Figs. 1 and 5), we hypothesized that USP25 might target TRAF3 for deubiquitination in response to LPS. TRAF3 is an E3 ubiquitin ligase that catalyzes its self-ubiquitination, but it is also ubiquitinated by cIAPs upon LPS stimulation. However, we found that USP25 did not cleave the autoubiquitination chains from TRAF3 (fig. S6A). Instead, wild-type USP25, but not the inactive mutant USP25(C178S), removed the cIAP2-mediated polyubiquitin chains from TRAF3 (Fig. 6A). cIAP2 can mediate both K48-and K63-linked ubiquitination of TRAF3 (15, 30). We found that the cIAP2-mediated K48-linked polyubiquitin chain, but not the K63-linked species, was removed from TRAF3 and from its enzymatically inactive form TRAF3(RM) by USP25 in cells and in vitro (Fig. 6, B and C, and fig. S6B). These data suggest that USP25 specifically removes cIAP2-mediated, K48-linked polyubiquitin chains from TRAF3.
Fig. 6. USP25 cleaves the cIAP2-mediated K48-linked ubiquitin chain from TRAF3.
(A and B) USP25 cleaves K48-linked ubiquitin chains from TRAF3. HEK 293T cells were transfected with plasmids encoding the indicated constructs. Twenty hours later, cells were lysed and the cell lysates were denatured in 1% SDS and then heated to 95°C for 5 min. The denatured lysates were subjected to immunoprecipitation (denature-IP) with the control IgG (Ig) or an anti-FLAG antibody (αF). The immunoprecipitates were analyzed by Western blotting with antibodies against (A) ubiquitin (αUbi, top blot), (B) hemagglutinin (HA; αHA, top blot), or (A and B) FLAG (middle blots). The abundances of the indicated proteins in the cell lysates were determined by Western blotting analysis with an anti-Myc antibody (αMyc, bottom blots). (C) USP25 specifically cleaves K63-linked ubiquitin chains from TRAF3 in vitro. The strategy to obtain TRAF3 protein that was modified with either K63- or K48-linked ubiquitin chains and then combine them with USP25 or USP25(C178S) proteins is illustrated (also see Materials and Methods for further details). A deubiquitination assay was performed by mixing the indicated proteins followed by Western blotting analysis with antibodies against HA, FLAG, or USP25. exp., exposure. (D) Deficiency in USP25 results in increased K48-linked ubiquitination of TRAF3 after stimulation with LPS. WT and Usp25−/− BMDMs were treated with LPS (10 μg/ml) for the indicated times. The cells were lysed, and the cell lysates were subjected to denaturing IP with an anti-TRAF3 antibody. The immunoprecipitated samples were subjected to Western blotting analysis with antibodies specific for K48- or K63-linked ubiquitin (top two blots). The abundances of the indicated proteins in the cell lysates were determined by Western blotting analysis with specific antibodies against the indicated proteins (bottom blot). Data are representative of three (A and B) or two (C and D) independent experiments.
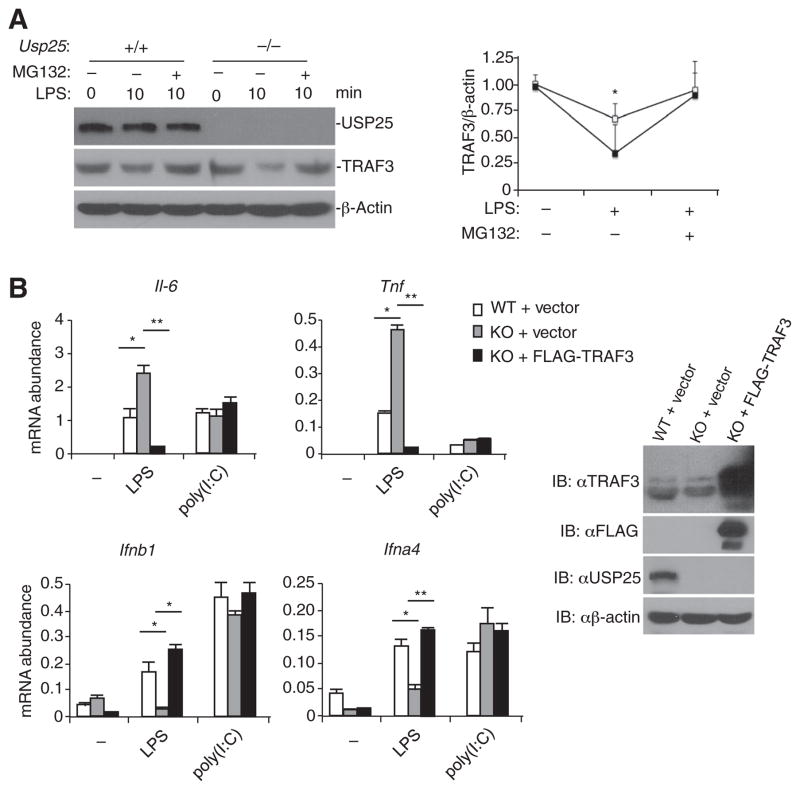
Because the activation of TLR4 results in the K48-linked ubiquitination and degradation of TRAF3 (15), we next determined the effects of a deficiency in USP25 on the LPS-induced ubiquitination and degradation of TRAF3. LPS induced the K48-linked ubiquitination and degradation of TRAF3 in wild-type BMDMs, and this was further enhanced in Usp25−/− BMDMs (Fig. 6D, top blot), whereas K63-linked ubiquitination of TRAF3 was diminished in Usp25−/−BMDMs after stimulation with LPS (Fig. 6D, middle blot), which was probably a result of the excessive degradation of TRAF3 in these cells and was responsible for the impaired production of IFN-α. In contrast, although USP25 specifically removed K63-linked ubiquitin chains from TRAF6 (fig. S7A) (28), the K63- and K48-linked ubiquitination of TRAF6 was comparable in wild-type and USP25-deficient BMDMs in response to LPS (fig. S7B). Treatment of cells with the proteasome inhibitor MG132 inhibited the LPS-stimulated degradation of TRAF3 in wild-type and Usp25−/−BMDMs (Fig. 7A). Furthermore, USP25-deficient MEFs reconstituted with TRAF3 showed substantially decreased LPS-induced expression of Il6 and Tnf compared to that in USP25-deficient cells transfected with empty plasmid, whereas reconstitution with TRAF3 restored the LPS-induced expression of Ifnb1 and Ifna4 compared to that in USP25-deficient MEFs transfected with empty plasmid (Fig. 7B). These results suggest that USP25 inhibits the TLR4-stimulated, K48-linked ubiquitination of TRAF3 to maintain the cellular abundance of TRAF3, which is responsible for the balanced production of proinflammatory cytokines and type I IFNs in response to LPS.
Fig. 7. Reconstitution of USP25-deficient MEFs with TRAF3 reverses the LPS-induced regulation of proinflammatory cytokine and type I IFN production.
(A) LPS-induced degradation of TRAF3 is inhibited by MG132. WT and Usp25−/− BMDMs were treated with MG132 for 1 hour before being stimulated with LPS (10 μg/ml) for the indicated times and then being analyzed by Western blotting for the indicated proteins. The graph shows the analysis of normalized TRAF3 abundances under the indicated conditions. (B) Reconstitution of USP25-deficient MEFs with TRAF3 inhibits the LPS-stimulated generation of proinflammatory cytokines and restores the generation of type I IFNs. Usp25−/− MEFs were transduced with either control retrovirus or retrovirus expressing FLAG-TRAF3, and the abundance of exogenous TRAF3 was determined by Western blotting analysis with anti-FLAG or anti-TRAF3 antibodies (right blots). The transduced cells were stimulated with LPS (1 μg/ml) or poly(I:C) (50 μg/ml) for 6 hours and then were analyzed by real-time PCR to determine the abundances of the indicated mRNAs (left panels). Data are representative of at least three independent experiments. Graphs show means ± SD of three independent experiments. *P < 0.05; **P < 0.01 by ANOVA.
DISCUSSION
TLR-stimulated signaling is critically regulated by ubiquitination and deubiquitination processes (31). Here, we identified USP25 as a critical modulator of TLR4-mediated, but not TLR3-mediated, signaling. A deficiency in USP25 potentiated LPS-induced, but not poly(I:C)-induced, activation of NF-κB and MAPKs, as well as the production of proinflammatory cytokines. LPS-induced, but not poly(I:C)-induced, activation of IRF3 and the production of type I IFNs were inhibited in Usp25−/− cells compared to that in wild-type cells. Consistent with these observations, compared to wild-type mice, Usp25−/− mice produced increased amounts of proinflammatory cytokines and decreased amounts of type I IFNs after challenge with LPS, but exhibited no differences in response to poly(I:C) challenge, and they exhibited increased susceptibility to LPS-induced septic shock than did their wild-type littermate controls. Together, these data demonstrate that USP25 differentially regulates the TLR4 signaling pathways that lead to the production of proinflammatory cytokines and type I IFNs.
TRAF3 is a critical adaptor protein that is required for TLR-triggered signaling. TRAF3 selectively activates the production of proinflammatory cytokines and type I IFNs through K48- or K63-linked ubiquitination events, respectively (15). The activation of TLR4 and TLR3 stimulates the K63-linked ubiquitination of TRAF3, which is required for the activation of IRF3 and the production of type I IFNs, and DUBA removes K63-linked ubiquitin chains from TRAF3, thereby inhibiting type I IFN responses (16). The activation of TLR4 also stimulates the K48-linked ubiquitination and subsequent degradation of TRAF3; however, the deubiquitinating enzyme(s) remains to be characterized. In our experiments, we found that USP25 specifically removed cIAP2-mediated K48-linked, but not K63-linked, ubiquitin chains from TRAF3 in cells and in vitro, even though USP25 can hydrolyze both K63- and K48-linked polyubiquitin chains into single ubiquitin molecules in vitro (29, 32). Consistent with these observations, LPS-stimulated, K48-linked ubiquitination of TRAF3 was further potentiated, and the degradation of TRAF3 was accelerated in Usp25−/− BMDMs compared to that in wild-type BMDMs, whereas reconstitution of USP25-deficient MEFs with TRAF3 inhibited the generation of proinflammatory cytokines and restored the generation of type I IFNs in response to LPS. Thus, our results suggest that USP25 cleaves K48-linked ubiquitin chain from TRAF3, which increases the cellular abundance of TRAF3 and accounts for the balanced production of proinflammatory cytokines and type I IFNs after activation of TLR4.
The E3 ubiquitin ligase and scaffold protein TRAF6 is an important mediator involved in various signaling pathways. We previously demonstrated that IL-17 stimulates the Act1-mediated addition of K63-linked ubiquitin chains to TRAF6, which is recognized and removed by USP25 (28). Here, we found that LPS also stimulated the K63-linked ubiquitination of TRAF6, which was not affected by a deficiency in USP25, although USP25 specifically cleaved K63-linked ubiquitin chain from TRAF6 in vitro. It is possible that different signals determine the distinct substrate specificities of USP25 in vivo, the exact mechanism of which requires further investigations.
Our data showed that USP25 was recruited to the TLR4 signaling complex independently of TRAF3 or TRAF6. Domain mapping analysis indicates that the UBA and UIM domains of USP25 interacted with TRAF6 (28), whereas the UCH-coil domains of USP25 interacted with and deubiquitinated TRAF3. One possible explanation for this difference is that USP25 is recruited to the TLR4-associated signaling complex through the recognition of, and interaction with, several molecules by its multiple domains. In addition, we observed that USP25 was recruited to the TLR4 signaling complex early after stimulation with LPS (5 to 10 min) and disassociated from TLR4 at later times (after 15 to 20 min). Upon binding to LPS, TLR4 is first delivered to lipid rafts and then to early endosomes to which TIRAP (TIR domain–containing adaptor protein)–MyD88–TRAF3 and TRAM (TRIF-related adaptor molecule)–TRIF–TRAF3 complexes are recruited, respectively (33–35). Therefore, we speculate that USP25 is probably recruited to LPS-stimulated MyD88-associated, but not TRIF-associated, signaling complexes. In this regard, we found that USP25 interacted with MyD88, but not TRIF, in coimmunoprecipitation studies of transfected cells. Whether and how the adaptor protein MyD88 or its upstream adaptor TIRAP specifies the recruitment of USP25 to the TLR4 signaling complex is unclear.
On the basis of our findings, we propose a working model of the role of USP25 in TLR-triggered signaling. In this model, TLR4 and TLR2 recruit TRAF6, TRAF3, cIAP1/2, and USP25 to form a signaling complex upon activation. In this complex, cIAP1/2 mediates the K48-linked ubiquitination and degradation of TRAF3, which leads to the disassociation of the signaling complex from cell membrane to cytosol, where it activates NF-κB and MAPK signaling, resulting in the production of proinflammatory cytokines. USP25 interacts with TRAF6 through its UBA-UIM domains, and it deubiquitinates K48-linked chains from TRAF3 through its UCH-coil domains, which helps to maintain the cellular abundance of TRAF3 by inhibiting its degradation. This form of regulation has two functional effects in innate immunity. First, USP25 restricts the excessive activation of proinflammatory responses, because TRAF3 has an inhibitory effect in this process. Second, in TLR4-triggered innate immune cells, USP25 also promotes the TRIF-dependent production of type I IFNs, a process in which TRAF3 is an essential stimulator. USP25 thereby maintains the appropriate TLR-triggered production of proinflammatory cytokines and type I IFNs.
Dysregulated ubiquitination of TLR-triggered signaling causes serious autoimmune diseases and tumorigenesis (36, 37). Our study provides insights into the dynamic and reversible nature of ubiquitin modification in the innate immune response. USP25 specifically cleaves K48-linked ubiquitin chains from TRAF3 after TLR4 activation. It is interesting that members of the large family of deubiquitinating enzymes have functional specificities; this feature may, in the future, provide new therapeutic targets for the treatment of immune diseases and cancers.
MATERIALS AND METHODS
Mice
Usp25−/− mice were generated as previously described (28). Mice were maintained in the specific pathogen–free facility of The University of Texas MD Anderson Cancer Center. Age- and sex-matched Usp25+/+ and Usp25−/− littermate mice were used for all experiments. All animal experiments were performed in accordance with protocols approved by the Institutional Animal Care and Use Committee of The University of Texas MD Anderson Cancer Center.
Antibodies and reagents
Mouse control IgG, horseradish peroxidase (HRP)–conjugated bovine anti-goat IgG, and HRP-conjugated anti-Myc, anti-ubiquitin, rabbit anti-HA, anti-IκBα, anti-ERK, anti-JNK, anti-p38, anti-TLR4, anti-TRAF6, anti-TRAF3, anti-cIAP2, anti-IRF3, anti-p65, and goat anti–lamin B antibodies were purchased from Santa Cruz Biotechnology; rabbit control IgG was purchased from Millipore; HRP-conjugated goat anti-mouse IgG and goat anti-rabbit IgG were from Thermo Scientific; mouse anti-FLAG and anti–β-actin antibodies were purchased from Sigma; anti–phosphorylated ERK(T202/Y204), anti–phosphorylated JNK(T183/Y185), and anti–phosphorylated p38(T180/Y182) antibodies were obtained from Cell Signaling Technology; and anti-TRIF antibody was purchased from Imgenex. Rabbit anti-USP25 antibody was described previously (29, 38) and was provided by G. Marfany (University of Barcelona, Barcelona, Spain). MG132, LPS, poly(I:C), and Pam3CSK4 were purchased from Sigma.
Cell culture
Primary MEFs were prepared from embryonic day 12.5 (E12.5) to E14.5 embryos and were cultured and transfected as previously described (28). Bone marrow cells were isolated from the femurs of the indicated mice. The cells were cultured in Dulbecco’s modified Eagle’s medium containing 20% fetal bovine serum, 1% streptomycin and penicillin, and 50 μM β-mercaptoethanol, with 20% L929 cell supernatant for BMDM differentiation or granulocyte-macrophage colony-stimulating factor (20 ng/ml) for BMDC differentiation. The medium was changed every 3 days. On day 7, BMDMs or BMDCs were harvested and incubated with fluorescently labeled antibodies against CD11b (BioLegend, 101206) or CD11c (BD Biosciences, 557401), respectively, and subjected to flow cytometric analysis to confirm purity. BMDMs and BMDCs (>95% purity) were used for subsequent analysis.
Constructs
Mammalian expression plasmids encoding FLAG- or Myc-tagged wild-type USP25 and its truncated mutants, TRAF6, TRAF3, and FLAG-USP25(C178S), were described previously (28). Plasmids encoding HA-ubiquitin, HA-ubiquitin (K48 only), and HA-ubiquitin (K63 only) were provided by Z. Chen (Southwestern Medical Center, Dallas), and plasmid encoding FLAG-TRAF3(RM) was provided by M. Karin (University of California San Diego, La Jolla).
Quantitative real-time PCR and ELISA
Cells treated with various stimuli were harvested in TRIzol (Invitrogen), and first-strand cDNA (complementary DNA) was synthesized with a reverse transcription kit (Invitrogen). Gene expression was examined with a Bio-Rad iCycler Optical System with an iQ SYBR Green Real-Time PCR kit (Bio-Rad Laboratories). Abundances of mRNAs of interest were normalized to that of β-actin. Gene-specific primers were previously described (28). The amounts of TNF-α, IL-6, and CXCL1 in cell culture media and sera were determined by ELISA with kits from BD Biosciences, whereas the amounts of IFN-β and IFN-α were measured by ELISA with kits from R&D Systems.
Isolation of cell membranes
Subcellular fractions were prepared as previously described (39). In brief, cells were resuspended and dounced in 1 ml of homogenization buffer [250 mM sucrose, 20 mM tris (pH 7.4), 2 mM MgCl2] on ice. The homogenate was centrifuged at 1000g for 10 min to remove crude nuclei and mitochondria (the P10 fraction). The supernatant (the S10 fraction) was further centrifuged at 20,000g for 1 hour at 4°C. The pellets containing membranes were lysed in lysis buffer [150 mM NaCl, 10 mM tris (pH 7.4), 1 mM EDTA, and 1% NP-40] for 10 min on ice, followed by centrifugation at 15,000g for 30 min at 4°C. The supernatants were then subjected to immunoprecipitation assays.
Nuclear extract preparation and EMSA
MEFs (2 × 106 cells) or BMDCs (5 × 106 cells) were lysed in lysis buffer [10 mM Hepes (pH 7.9), 10 mM KCl, 0.1 mM EDTA, and 0.4% NP-40] for 10 min on ice, followed by centrifugation at 15,000g for 1 min. The supernatants were saved as the cytosolic fraction. The pellets were washed once with lysis buffer and resuspended in 20 to 50 μl of extract buffer [20 mM Hepes (pH 7.9), 0.4 M NaCl, 1 mM EDTA] and shaken vigorously every 30 s for 15 min, followed by centrifugation at 15,000g for 10 min. The supernatant was subjected to EMSA with the 32P-radiolabeled oligo-nucleotide probes κB (Promega, E329A), AP-1 (Promega, E320A), or Oct-1 (Promega, E324A) or to Western blotting analysis.
Coimmunoprecipitations, Western blotting, and ubiquitination assays
Coimmunoprecipitations, Western blotting analysis, and ubiquitination assays were performed as previously described (28, 39, 40). Briefly, standard immunoprecipitations were performed under normal conditions in lysis buffer [20 mM tris (pH 7.4), 150 mM NaCl, 1 mM EDTA, 1% NP-40], and the immunoprecipitates were subjected to standard Western blotting analysis. For ubiquitination assays, the immunoprecipitates were reextracted in lysis buffer containing 1% SDS and denatured by heating for 5 min. The supernatants were diluted with regular lysis buffer until the concentration of SDS was decreased to 0.1%, and then, the samples were subjected to reimmunoprecipitation with the indicated antibodies. The immunoprecipitates were analyzed by Western blotting with antibodies specific for ubiquitin.
LPS- or poly(I:C)-induced cytokine production in mice
Age- and sex-matched Usp25+/+ and Usp25−/− mouse littermates were injected intraperitoneally with LPS (15 mg/kg) or poly(I:C) (5 μg/g). Survival was monitored every hour for 24 hours. Blood was obtained at 2.5 hours after challenge, and the concentrations of TNF-α, IL-6, CXCL1, and IFN-α in the sera were measured by ELISA.
Short hairpin RNAs
The short hairpin RNA (shRNA) lentiviral plasmid pLKO.1-shTRAF3 was purchased from Sigma (TRCN0000034220), and the plasmids pLKO.1-shTRAF6 and pLKO.1-shLuc were described previously (41) and were provided by L. Glimcher (Harvard Medical School, Boston, MA). HEK 293T cells were cotransfected with the appropriate lentiviral plasmid together with the packaging vectors pRRE, VSV-G, and RSV-Rev to obtain lentiviruses. Packaged viruses were then used to infect RAW264.7 cells, which were treated with polybrene (8 μg/ml) and then selected in the presence of puromycin (1 μg/ml) for 7 days. The puromycin-resistant cells were used for experiments.
In vitro deubiquitination assay
HEK 293T cells were cotransfected with plasmids encoding FLAG-TRAF3 and Myc-cIAP2 or FLAG-TRAF6 together with plasmids encoding HA-ubiqutin (K63 only) (HA-Ubi-K63) or HA-ubiqutin (K48 only) (HA-Ubi-K48). Twenty-four hours later, cells were lysed, and denatured immunoprecipitations were performed with anti-FLAG agarose. FLAG-tagged TRAF3 or TRAF6 modified with K63- or K48-linked ubiquitin was obtained by elution with the FLAG peptide. USP25 or USP25(C178S) proteins were generated with an in vitro transcription and translation kit (Promega). Deubiquitination experiments were performed by mixing the indicated proteins and incubating them at 30°C for 2 hours followed by incubation at 16°C overnight. Samples were then analyzed by Western blotting analysis with antibodies against HA, FLAG, or the indicated proteins.
Statistical analysis
Differences between experimental and control groups were determined by Student’s t test or by ANOVA analysis followed by the Bonferroni test. P values <0.05 were considered statistically significant. For animal survival analysis, the Kaplan-Meier method was adopted to generate graphs, and the survival curves were analyzed with log-rank analysis.
Supplementary Material
Acknowledgments
We thank M. Karin, X. Wang, Z. Chen, L. Glimcher, and H. Shu for the reagents and W. Peng, H. Hu, J. Jin, and members of the Dong lab for technical help.
Funding: This work was supported by grants from the NIH and the MD Anderson Cancer Center. B.Z. is an Odyssey Fellow supported by the CFP Foundation at the MD Anderson Cancer Center. C.D. is a Leukemia and Lymphoma Society Scholar and holds the Olga Keith and Harry Carothers Wiess Distinguished University Chair in Cancer Research at the MD Anderson Cancer Center. S.-C.S. was supported by NIH/National Institute of Allergy and Infectious Diseases grant R01 AI057555.
Footnotes
Author contributions: B.Z., Xikui Liu, and C.D. designed the experiments; B.Z., Xikui Liu, X.W., Xindong Liu, and H.L. performed the experiments; B.G.D. contributed reagents; B.Z., X. Lin, S.-C.S., and C.D. analyzed the data; and B.Z. and C.D. wrote the manuscript.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: Use of the Usp25−/− mice requires a material transfer agreement from The University of Texas MD Anderson Cancer Center.
REFERENCES AND NOTES
- 1.Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124:783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 2.Medzhitov R. Recognition of microorganisms and activation of the immune response. Nature. 2007;449:819–826. doi: 10.1038/nature06246. [DOI] [PubMed] [Google Scholar]
- 3.Kawai T, Akira S. The role of pattern-recognition receptors in innate immunity: Update on Toll-like receptors. Nat Immunol. 2010;11:373–384. doi: 10.1038/ni.1863. [DOI] [PubMed] [Google Scholar]
- 4.Poltorak A, He X, Smirnova I, Liu MY, Van Huffel C, Du X, Birdwell D, Alejos E, Silva M, Galanos C, Freudenberg M, Ricciardi-Castagnoli P, Layton B, Beutler B. Defective LPS signaling in C3H/HeJ and C57BL/10ScCr mice: Mutations in Tlr4 gene. Science. 1998;282:2085–2088. doi: 10.1126/science.282.5396.2085. [DOI] [PubMed] [Google Scholar]
- 5.Ozinsky A, Underhill DM, Fontenot JD, Hajjar AM, Smith KD, Wilson CB, Schroeder L, Aderem A. The repertoire for pattern recognition of pathogens by the innate immune system is defined by cooperation between Toll-like receptors. Proc Natl Acad Sci USA. 2000;97:13766–13771. doi: 10.1073/pnas.250476497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Alexopoulou L, Thomas V, Schnare M, Lobet Y, Anguita J, Schoen RT, Medzhitov R, Fikrig E, Flavell RA. Hyporesponsiveness to vaccination with Borrelia burgdorferi OspA in humans and in TLR1- and TLR2-deficient mice. Nat Med. 2002;8:878–884. doi: 10.1038/nm732. [DOI] [PubMed] [Google Scholar]
- 7.Alexopoulou L, Holt AC, Medzhitov R, Flavell RA. Recognition of double-stranded RNA and activation of NF-κB by Toll-like receptor 3. Nature. 2001;413:732–738. doi: 10.1038/35099560. [DOI] [PubMed] [Google Scholar]
- 8.Heil F, Hemmi H, Hochrein H, Ampenberger F, Kirschning C, Akira S, Lipford G, Wagner H, Bauer S. Species-specific recognition of single-stranded RNA via Toll-like receptor 7 and 8. Science. 2004;303:1526–1529. doi: 10.1126/science.1093620. [DOI] [PubMed] [Google Scholar]
- 9.Hemmi H, Takeuchi O, Kawai T, Kaisho T, Sato S, Sanjo H, Matsumoto M, Hoshino K, Wagner H, Takeda K, Akira S. A Toll-like receptor recognizes bacterial DNA. Nature. 2000;408:740–745. doi: 10.1038/35047123. [DOI] [PubMed] [Google Scholar]
- 10.Kawai T, Akira S. Toll-like receptors and their crosstalk with other innate receptors in infection and immunity. Immunity. 2011;34:637–650. doi: 10.1016/j.immuni.2011.05.006. [DOI] [PubMed] [Google Scholar]
- 11.Iwasaki A, Medzhitov R. Regulation of adaptive immunity by the innate immune system. Science. 2010;327:291–295. doi: 10.1126/science.1183021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Skaug B, Jiang X, Chen ZJ. The role of ubiquitin in NF-κB regulatory pathways. Annu Rev Biochem. 2009;78:769–796. doi: 10.1146/annurev.biochem.78.070907.102750. [DOI] [PubMed] [Google Scholar]
- 13.Häcker H, Redecke V, Blagoev B, Kratchmarova I, Hsu LC, Wang GG, Kamps MP, Raz E, Wagner H, Häcker G, Mann M, Karin M. Specificity in Toll-like receptor signalling through distinct effector functions of TRAF3 and TRAF6. Nature. 2006;439:204–207. doi: 10.1038/nature04369. [DOI] [PubMed] [Google Scholar]
- 14.Oganesyan G, Saha SK, Guo B, He JQ, Shahangian A, Zarnegar B, Perry A, Cheng G. Critical role of TRAF3 in the Toll-like receptor-dependent and -independent antiviral response. Nature. 2006;439:208–211. doi: 10.1038/nature04374. [DOI] [PubMed] [Google Scholar]
- 15.Tseng PH, Matsuzawa A, Zhang W, Mino T, Vignali DA, Karin M. Different modes of ubiquitination of the adaptor TRAF3 selectively activate the expression of type I interferons and proinflammatory cytokines. Nat Immunol. 2010;11:70–75. doi: 10.1038/ni.1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kayagaki N, Phung Q, Chan S, Chaudhari R, Quan C, O’Rourke KM, Eby M, Pietras E, Cheng G, Bazan JF, Zhang Z, Arnott D, Dixit VM. DUBA: A deubiquitinase that regulates type I interferon production. Science. 2007;318:1628–1632. doi: 10.1126/science.1145918. [DOI] [PubMed] [Google Scholar]
- 17.Komander D, Clague MJ, Urbé S. Breaking the chains: Structure and function of the deubiquitinases. Nat Rev Mol Cell Biol. 2009;10:550–563. doi: 10.1038/nrm2731. [DOI] [PubMed] [Google Scholar]
- 18.Reyes-Turcu FE, Ventii KH, Wilkinson KD. Regulation and cellular roles of ubiquitin-specific deubiquitinating enzymes. Annu Rev Biochem. 2009;78:363–397. doi: 10.1146/annurev.biochem.78.082307.091526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nijman SM, Luna-Vargas MP, Velds A, Brummelkamp TR, Dirac AM, Sixma TK, Bernards R. A genomic and functional inventory of deubiquitinating enzymes. Cell. 2005;123:773–786. doi: 10.1016/j.cell.2005.11.007. [DOI] [PubMed] [Google Scholar]
- 20.Sun SC. Deubiquitylation and regulation of the immune response. Nat Rev Immunol. 2008;8:501–511. doi: 10.1038/nri2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Boone DL, Turer EE, Lee EG, Ahmad RC, Wheeler MT, Tsui C, Hurley P, Chien M, Chai S, Hitotsumatsu O, McNally E, Pickart C, Ma A. The ubiquitin-modifying enzyme A20 is required for termination of Toll-like receptor responses. Nat Immunol. 2004;5:1052–1060. doi: 10.1038/ni1110. [DOI] [PubMed] [Google Scholar]
- 22.Shembade N, Ma A, Harhaj EW. Inhibition of NF-κB signaling by A20 through disruption of ubiquitin enzyme complexes. Science. 2010;327:1135–1139. doi: 10.1126/science.1182364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang YY, Li L, Han KJ, Zhai Z, Shu HB. A20 is a potent inhibitor of TLR3- and Sendai virus-induced activation of NF-κB and ISRE and IFN-β promoter. FEBS Lett. 2004;576:86–90. doi: 10.1016/j.febslet.2004.08.071. [DOI] [PubMed] [Google Scholar]
- 24.Wertz IE, O’Rourke KM, Zhou H, Eby M, Aravind L, Seshagiri S, Wu P, Wiesmann C, Baker R, Boone DL, Ma A, Koonin EV, Dixit VM. Deubiquitination and ubiquitin ligase domains of A20 downregulate NF-κB signalling. Nature. 2004;430:694–699. doi: 10.1038/nature02794. [DOI] [PubMed] [Google Scholar]
- 25.Zhang J, Stirling B, Temmerman ST, Ma CA, Fuss IJ, Derry JM, Jain A. Impaired regulation of NF-κB and increased susceptibility to colitis-associated tumorigenesis in CYLD-deficient mice. J Clin Invest. 2006;116:3042–3049. doi: 10.1172/JCI28746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang M, Wu X, Lee AJ, Jin W, Chang M, Wright A, Imaizumi T, Sun SC. Regulation of IκB kinase-related kinases and antiviral responses by tumor suppressor CYLD. J Biol Chem. 2008;283:18621–18626. doi: 10.1074/jbc.M801451200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li S, Zheng H, Mao AP, Zhong B, Li Y, Liu Y, Gao Y, Ran Y, Tien P, Shu HB. Regulation of virus-triggered signaling by OTUB1- and OTUB2-mediated deubiquitination of TRAF3 and TRAF6. J Biol Chem. 2010;285:4291–4297. doi: 10.1074/jbc.M109.074971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhong B, Liu X, Wang X, Chang SH, Liu X, Wang A, Reynolds JM, Dong C. Negative regulation of IL-17-mediated signaling and inflammation by the ubiquitin-specific protease USP25. Nat Immunol. 2012;13:1110–1117. doi: 10.1038/ni.2427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Denuc A, Bosch-Comas A, Gonzàlez-Duarte R, Marfany G. The UBA-UIM domains of the USP25 regulate the enzyme ubiquitination state and modulate substrate recognition. PLoS One. 2009;4:e5571. doi: 10.1371/journal.pone.0005571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mao AP, Li S, Zhong B, Li Y, Yan J, Li Q, Teng C, Shu HB. Virus-triggered ubiquitination of TRAF3/6 by cIAP1/2 is essential for induction of interferon-β (IFN-β) and cellular antiviral response. J Biol Chem. 2010;285:9470–9476. doi: 10.1074/jbc.M109.071043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.An H, Qian C, Cao X. Regulation of Toll-like receptor signaling in the innate immunity. Sci China Life Sci. 2010;53:34–43. doi: 10.1007/s11427-010-0011-x. [DOI] [PubMed] [Google Scholar]
- 32.Meulmeester E, Kunze M, Hsiao HH, Urlaub H, Melchior F. Mechanism and consequences for paralog-specific sumoylation of ubiquitin-specific protease 25. Mol Cell. 2008;30:610–619. doi: 10.1016/j.molcel.2008.03.021. [DOI] [PubMed] [Google Scholar]
- 33.Triantafilou M, Miyake K, Golenbock DT, Triantafilou K. Mediators of innate immune recognition of bacteria concentrate in lipid rafts and facilitate lipopolysaccharide-induced cell activation. J Cell Sci. 2002;115:2603–2611. doi: 10.1242/jcs.115.12.2603. [DOI] [PubMed] [Google Scholar]
- 34.Kagan JC, Su T, Horng T, Chow A, Akira S, Medzhitov R. TRAM couples endocytosis of Toll-like receptor 4 to the induction of interferon-β. Nat Immunol. 2008;9:361–368. doi: 10.1038/ni1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kagan JC. Signaling organelles of the innate immune system. Cell. 2012;151:1168–1178. doi: 10.1016/j.cell.2012.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Alameda JP, Moreno-Maldonado R, Navarro M, Bravo A, Ramírez A, Page A, Jorcano JL, Fernández-Aceñero MJ, Casanova ML. An inactivating CYLD mutation promotes skin tumor progression by conferring enhanced proliferative, survival and angiogenic properties to epidermal cancer cells. Oncogene. 2010;29:6522–6532. doi: 10.1038/onc.2010.378. [DOI] [PubMed] [Google Scholar]
- 37.Hymowitz SG, Wertz IE. A20: From ubiquitin editing to tumour suppression. Nat Rev Cancer. 2010;10:332–341. doi: 10.1038/nrc2775. [DOI] [PubMed] [Google Scholar]
- 38.Bosch-Comas A, Lindsten K, Gonzàlez-Duarte R, Masucci MG, Marfany G. The ubiquitin-specific protease USP25 interacts with three sarcomeric proteins. Cell Mol Life Sci. 2006;63:723–734. doi: 10.1007/s00018-005-5533-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhong B, Zhang L, Lei C, Li Y, Mao AP, Yang Y, Wang YY, Zhang XL, Shu HB. The ubiquitin ligase RNF5 regulates antiviral responses by mediating degradation of the adaptor protein MITA. Immunity. 2009;30:397–407. doi: 10.1016/j.immuni.2009.01.008. [DOI] [PubMed] [Google Scholar]
- 40.Chang M, Jin W, Sun SC. Peli1 facilitates TRIF-dependent Toll-like receptor signaling and proinflammatory cytokine production. Nat Immunol. 2009;10:1089–1095. doi: 10.1038/ni.1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Martinon F, Chen X, Lee AH, Glimcher LH. TLR activation of the transcription factor XBP1 regulates innate immune responses in macrophages. Nat Immunol. 2010;11:411–418. doi: 10.1038/ni.1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.