Abstract
Scope
Soy flour diet (MS) prevented isoflavones from stimulating MCF-7 tumor growth in athymic nude mice, indicating that other bioactive compounds in soy can negate the estrogenic properties of isoflavones. The underlying signal transduction pathways to explain the protective effects of soy flour consumption were studied here.
Methods and results
Ovariectomized athymic nude mice inoculated with MCF-7 human breast cancer cells were fed either MS or purified isoflavone mix (MI), both with equivalent amounts of genistein. Positive controls received estradiol pellets and negative controls received sham pellets. GeneChip-Human-Genome-U133-Plus-2.0 Array platform was used to evaluate gene expressions, and results were analyzed using bioinformatics approaches. Tumors in MS-fed mice exhibited higher expression of tumor-growth-suppressing genes ATP2A3 and BLNK, and lower expression of oncogene MYC. Tumors in MI-fed mice expressed higher level of oncogene MYB and lower level of MHC-I and MHC-II, allowing tumor cells to escape immunosurveillance. MS-induced gene expression alterations were predictive of prolonged survival among estrogen-receptor-positive breast cancer patients, whilst MI-induced gene changes were predictive of shortened survival.
Conclusion
Our findings suggest dietary soy flour affects gene expression differently than purified isoflavones, which may explain why soy foods prevent isoflavones-induced stimulation of MCF-7 tumor growth in athymic nude mice.
Keywords: breast cancer, isoflavones, MCF-7, soy, whole-genome expression
Introduction
Soybeans have been incorporated into Asian diets for thousands of years, serving as good sources of protein. The most abundant biologically active compounds in soybeans are isoflavones, and they have been extensively studied due to their potential effects on promoting or inhibiting carcinogenesis [1, 2]. Breast cancer is the most common cancer among women, with estimated 232,670 new cases diagnosed in 2014 in the United States, of which 15% (40,000) resulted in deaths, making it the second most common cause for cancer deaths in women [3]. An association between high soy food intake and low breast cancer incidence has been reported in Asian American women who continued consuming moderate levels of soy on a daily basis compared with Asian American women who adopted Western dietary habits [4–6]. However, the causal link between soy intake and breast cancer remains to be established. Asian diets contained more vegetables, fruits and green tea, and less fat than Western diets; this has been correlated with lower breast cancer risk [7, 8] and higher survival rates among breast cancer patients [9]. Exposure to soy foods in early life appeared to be the key window of opportunity for breast cancer prevention [10]. However, starting isoflavones supplementation when an estrogen-receptor-positive (ER+) breast cancer is present may increase breast cancer risk [11, 12]. This is supported by the results obtained in a recent randomized placebo-controlled study in breast cancer patients who either received soy protein supplement or placebo for 7 to 30 days [13]. Microarray analysis indicated that soy supplementation lead to increased expression of several genes that induce cell proliferation, although Ki67 expression was not increased [13].
Preventive effects of soy food consumption could also be due to the complex compositional profile of bioactive compounds in soy diets other than isoflavones, referred to here as soy matrix effects. Soy foods consumed in traditional Asian diets such as tofu, soy milk and soy flour are minimally processed from whole soybeans; whilst Western soy products contain highly processed isoflavones extracts and soy protein isolates. We reported earlier that minimally processed soy flour and highly purified isoflavones with equivalent amounts of genistein influenced the growth of MCF-7 human breast cancer tumors differently in ovariectomized athymic nude mice [1]. Average tumor area in mice fed diet containing purified isoflavone mix (MI) was significantly larger (91 mm2) over an 11-week observation period than in mice fed soy flour diet containing isoflavones (MS) (40 mm2). The average tumor area regressed in the negative control (NC) group from 40 mm2 to 16 mm2. In contrast, the average tumor area of the positive control (PC) increased from 40 mm2 to 112 mm2. The molecular mechanisms driving the differences in MI and MS groups remained unknown.
A systems biology approach to study breast cancer was initiated over a decade ago [14]. It assembles models of biological systems from multiple systematic measurements and gives a holistic view of the underlying biological phenomena. Under the framework of systems biology, high-throughput genomic approaches, such as microarrays and next-generation sequencing, are powerful means to provide insight as to how dietary factors affect the development of breast cancer [15].
Our current work focused on elucidating whether dietary exposures to varying soy processing levels, but equivalent amounts of genistein (750 ppm aglycone, a dose which was observed to stimulate the growth of MCF-7 tumors in vivo [11]) influenced MCF-7 tumor whole-genome expression patterns differently. Compared with the previous study [1], we focused on four treatments, which were mixed isoflavones in soy flour (SF with MI, called MS in our study), purified isoflavone mix alone (MI), positive control (PC) that involved treating ovariectomized mice with estradiol (E2) and negative control (NC, no hormone or dietary treatment) groups, and applied whole-genome microarray expression analysis to the mRNA extracted from MCF-7 tumor in the four different treatment groups. The goal was to explain the underlying molecular mechanisms regulating the growth of MCF-7 breast cancer tumors in ovariectomized athymic nude mice through high-throughput data analysis approaches.
Materials and Methods
Animals and dietary treatments
The experimental design of the study, animals used, treatments and MCF-7 tumor collection were described in our previous study [1]. Briefly, ovariectomized athymic nude mice at 28 days of age were randomly assigned to four groups: soy flour (MS), purified isoflavone mix (MI), negative control (NC) and positive control (PC). The mixed isoflavone preparation contained predominantly glycosylated isoflavones comprised of 37.2 ± 1.3 % by weight genistein, of which 0.17 ± 0.004% was aglycone, and 15.8 ± 0.7% by weight daidzein, of which 0.19 ± 0.008% was the aglycone. The glycetein content and other constituents were not quantified. MS and MI groups contained equivalent amounts of genistein, and both PC and NC groups received no genistein. All mice were fed AIN93G diet, with corn oil substituted for soy oil. Mice in the PC group received 2 milligram estradiol (E2) pellets and mice in the NC group received sham pellets. AIN93G based MI and MS diets contained 750 ppm genistein aglycone and were individually formulated and balanced to contain equal amounts of protein, carbohydrate and fat. All mice were euthanized 12 weeks after inoculation of MCF-7 tumor cells and tumors were immediately frozen by submersion into liquid nitrogen. All of the experimental procedures were approved by the Illinois Institutional Animal Care and Use Committee (Protocol ID 13315) and were conducted in accordance with the regulations described in the Committee’s Manual.
RNA isolation and microarray quantification
GeneChip Human Genome U133 Plus 2.0 Array (Affymetrix, Santa Clara, CA), an oligonucleotide array for 22,680 genes containing 11 probe pairs per gene, was used. Each probe pair consisted of a Perfect Match (PM) and a Mismatch (MM), which were identical except for a single nucleotide change to the complement in the middle of the MM probe sequence that allowed for determination of nonspecific binding. Analyses were carried out at the W. M. Keck Center for Comparative and Functional Genomics at the University of Illinois, Urbana-Champaign. In each group, RNA was isolated from tumors in 3 separate mice. Target labeling was performed using total RNA for cDNA synthesis and preparing the biotinylated antisense cRNA using SuperScript II (Invitrogen Life Technologies, Rockville, MD) and EnzoBioArray High Yield RNA Transcript Labeling Kit (Enzo diagnostics, farmingdale, NY). Quality control, hybridization, washing, and scanning were performed as described in the GeneChip Expression Analysis Technical Manual (Affymetrix, Santa Clara, CA; Rev. 4). The microarray dataset was deposited in the National Center for Biotechnology Information (NCBI) Gene Expression Omnibus (GEO) database (http://www.ncbi.nlm.nih.gov/geo/) [16–18] with the accession number GSE63205.
Preprocessing of the microarray data
Quality assessment, summarization, normalization, and transformation of the microarray data were performed using the Bioconductor package for Affymetrix arrays with the R software [19]. Three widely used normalization methods were compared, including MAS5 [20], RMA [21] and GCRMA [22]. GCRMA was adopted based on the density plots, box-plots and principle component analysis (PCA) comparing raw data and the preprocessed data (File S1, Figure S1 – S3), which allowed comparisons between microarray experiments and the control of extraneous variation among experiments. Since microarray data are of high dimensionality, different groups are mixed in high dimensions due to the curse-of-dimensionality. PCA was applied to extract the top-three principal components for the purpose of comparative visualization in a three-dimensional subspace, which indicated the dissimilarity between PC and NC treatment groups and similarity shared by MI and MS treatment groups using the GCRMA method. A linear model was fit to the log2-intensity transformed intensity data for each probe ID using Limma package of bioconductor [19] in preparation for differential expression analysis of the data. The Empirical Bayes method [23] was subsequently used to borrow information across genes making these analyses even more stable. Present/Marginal/Absent (P/M/A) calls are good criteria to filter out the unexpressed probes [24]. The probes which were “Present” on at least 1 array or “Marginal” on at least 2 arrays were reserved for further analysis; 64.52% of the total probes were preserved after filtration.
Soy matrix effects on the whole-genome expression in ER+ breast cancer xenografted in ovariectomized athymic nude mice were studied in two different ways: 1) Tumor-growth-inhibiting effect uniquely possessed by MS and NC compared with PC; 2) Tumor-growth-promoting effect uniquely possessed by MI and PC compared with NC. Following this approach, six comparisons were made: MS versus PC (namely MSPC), NC versus PC (NCPC), MI versus PC (MIPC), MI versus NC (MINC), PC versus NC (PCNC) and MS versus PC (MSPC). To identify the significantly differentially expressed probes (DEPs) in each comparison, t-tests were conducted, and DEPs were then mapped to differentially expressed genes (DEGs). The p-values were adjusted with Benjamini and Hochberg’s false discovery rate (FDR) control [25] using multiple hypothesis tests to correct for multiple comparisons.
DEGs clustering
DEGs were selected using the FDR cutoff of less than or equal to 0.01 and log2-fold change of greater/less than or equal to 2. Those cutoff values were chosen based on the distribution of the volcano-plot on contrasts (File S1, Figure S4), which demonstrated the log-fold expression changes on the x-axis versus log-odds (negative log of the adjusted p-values with base 10) of differential expression on the y-axis. Six different sets of gene lists were further extracted using the Venn Diagram technique, which corresponded to the tumor growth inhibition (up-regulation, down-regulation and both directions) and tumor growth promotion (up-regulation, down-regulation and both directions). Lists of genes were clustered by hierarchical clustering method [26] after expression value of each probe ID being adjusted by row-mean values. Final clusters of genes were determined through comparisons of height cut versus dynamic tree cut method [27] with the following parameters: method = “hybrid”, minimum cluster size = 1 and sensitivity to cluster splitting at the level of 3.
Visualization of Kyoto Encyclopedia of Gene and Genomes (KEGG) pathways
KegArray software was utilized to explore the specific KEGG pathways involved with those DEGs that were identified. The online application KegArray software is available in KEGG [28, 29] (Kyoto Encyclopedia of Genes and Genomes) website at http://www.genome.jp/kegg/expression/.
Gene Ontology (GO) analysis
GOstats package [30] of bioconductor was used to process GO analysis with DEGs being identified earlier. Conditional hypergeometric tests, which removed the effects of child GO terms when testing parents GO terms, were conducted to identify the over-represented annotations in four venues: cellular component (CC), molecular function (MF), biological process (BP) and KEGG pathways. A cutoff value of the hypergeometric tests was set at FDR of less than or equal to 0.01 obtaining enriched CC, MF and BP, while FDR was less than or equal to 0.05 investigating enriched KEGG pathways, in order to address the problem of dramatically increased type I errors that occurred in the multiple comparison test.
Further, hundreds of over-represented GO terms (CC, MF and BP) which were identified by the hypergeometric tests were visualized and organized into gene networks in the aspect of tumor-growth-inhibiting and promoting effects using enrichment map software [31], which was implemented as a plug-in (http://baderlab.org/Software/EnrichmentMap) for the Cytoscape network visualization software [32].
Rank correlation test on the DEGs expression with breast cancer patient survival time
Spearman’s rank correlation tests were conducted between the DEGs expression level and the first distant metastatic event (distant metastasis free survival, DMFS) of breast cancer patients in three clinical datasets among 448 samples [33, 34]. Those clinical datasets (File S1, Table S1) were obtained from patients with ER+ breast cancer and treated with Tamoxifen monotherapy for five years. The DMFS of the patient samples ranged from 0 to 17 years (File S1, Figure S8), which was suitable for robust rank correlation analyses. P-values of less than 0.05 were used as the threshold to identify the significantly correlated DEGs.
Results
Differentially expressed probes (DEPs) between soy flour and purified isoflavone mix
Two main probe lists of interest were identified through Venn Diagrams (File S1, Figure S5). Ninety-three DEPs (0.27% of the total genes being investigated) modulated by MS were selected. They were identified as the genes overlapping between NCPC and MSPC out of overall overlapping genes among NCPC, MSPC and MIPC. Among those, 45 DEPs were up-regulated and 48 DEPs were down-regulated. Therefore, those DEPs that were shared by MS and NC compared with PC correlated with the prevention of proliferation of the MCF-7 tumors growth.
One-hundred and twenty-six DEPs (0.38% of the total genes being investigated) altered by MI were selected. They were identified as the genes overlapping between PCNC and MINC out of overall overlapping genes among PCNC, MSNC and MINC. Among those, 31 DEPs were up-regulated and 95 DEPs were down-regulated. Therefore, those DEPs that were shared by MI and PC compared with NC correlated with the stimulation of the MCF-7 tumors growth.
Detailed information of the above screened probe IDs are provided in the supplemental files (File S2 and File S3), which explicitly demonstrated their log fold-change expression value, FDR, probe set ID, gene title, gene symbol, ensembl, entrez ID, and relating BP, CC, MF in the GO, as well as KEGG pathway information. Up-regulation of ATP2A3, CLU, RUNX1T1, STAT1, and TNFRSF19 was shown to be associated with tumor-growth-inhibiting effect by MS. These genes were down-regulated in the MI group. Thus, they might explain the different effects of soy flour versus purified isoflavone mix on the growth of MCF-7 tumor in ovariectomized athymic nude mice.
DEG clusters linked to soy matrix modulation on breast cancer growth
DEGs clusters modulated by MS
We focused on 93 DEPs mapping to 77 distinct entrez gene identifiers for differentially expressed gene (DEGs) clustering. Eight clustering criteria were compared visually in the cluster dendrogram (File S1, Figure S6). Twenty clusters of DEGs were grouped across different patterns and visualized on the heat map as a color matrix (Figure 1a), and categorized in Table 1 by their cluster ID and gene symbol. DEG clusters modulated by MI. It was noted that 126 DEPs mapped to 100 distinct entrez gene identifiers, and thus we focused on those DEGs for clustering. Eight clustering criteria were compared visually in the cluster dendrogram (Figure 1b) and twenty-six clusters were categorized in Table 2 by their cluster ID and gene symbol.
Figure 1. DEGs expression clusters reflect soy matrix effects on MCF-7 tumor growth in ovariectomized athymic nude mice.
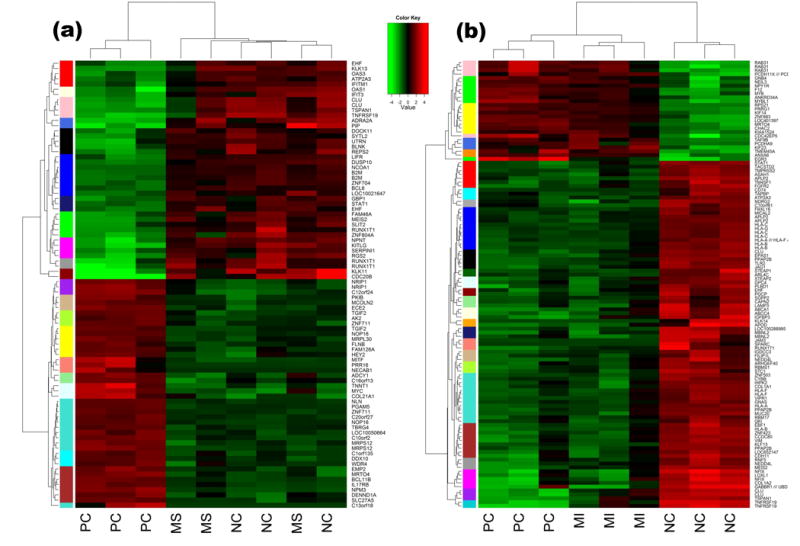
These clusters are demonstrated by a set of various sidebar colors on the heat map, in which green represents a low gene expression level, black represents medium expression and red denotes a high expression level. DEGs are in rows and experimental groups are in columns. a. DEGs expression clusters reflect MS negates MCF-7 cell growth. b. DEGs expression clusters reflect MI stimulates MCF-7 cell growth. MI, purified isoflavone mix diet; MS, soy flour diet; NC, negative control diet; PC, positive control diet.
Table 1. DEGs clusters modulated by dietary soy flour (MS) reduce MCF-7 tumor growth in ovariectomized athymic nude mice.
DEGs in the cluster from 1 to 10 are up-regulated. DEGs in the cluster from 11 to 20 are down-regulated.
| Cluster | Differentially Expressed Genes (DEGs) |
|---|---|
| 1 | EHF, KLK13, OAS3, ATP2A3, IFITM1 |
| 2 | OAS1, IFIT3 |
| 3 | CLU, TSPAN1, TNFRSF19 |
| 4 | ADRA2A, PIP |
| 5 | DOCK11, SYTL2, UTRN, BLNK, REPS2 |
| 6 | LIFR, DUSP10, NCOA1, B2M, ZNF704, BCL6, LOC100216479 |
| 7 | GBP1, STAT1, EHF |
| 8 | FAM46A, MEIS2, SLIT2, RUNX1T1, ZNF804A |
| 9 | NPNT, KITLG, SERPINI1, RGS2 |
| 10 | KLK11, CDC20B |
|
| |
| 11 | NRIP1, C12orf24 |
| 12 | PKIB, MCOLN2, ECE2 |
| 13 | TGIF2, AK2, ZNF711 |
| 14 | TGIF2, NOP16, MRPL30, FLNB, FAM126A, HEY2 |
| 15 | MITF, PRR16, NECAB1 |
| 16 | ADCY1, C16orf13 |
| 17 | TNNT1, MYC, COL21A1 |
| 18 | NLN, PGAM5, ZNF711, C20orf27, NOP16, TBRG4, LOC100506649, C10orf2, MRPS12, C13orf18 |
| 19 | C1orf135, DDX10, WDR4 |
| 20 | EMP2, MRTO4, BCL11B, IL17RB, NPM3, DENND1A, SLC27A5 |
Table 2. DEGs clusters modulated by purified isoflavone mix (MI) stimulate MCF-7 tumor growth in ovariectomized athymic nude mice.
DEGs in the cluster from 1 to 5 are up-regulated. DEGs in the cluster from 6 to 26 are down-regulated.
| Cluster | Differentially Expressed Genes (DEGs) |
|---|---|
| 1 | RAB31, PCDH11X /// PCDH11Y, CDC42EP5 |
| 2 | GNB4, NEIL3, NPY1R, F12, MYB, ANKRD34A, MYBL1, EGR3 |
| 3 | RPS21, PRRG1, KIF14, ZNF883, LOC401397, MRTO4, CHAC2, KIAA1524 |
| 4 | TAF9B, PCDHA9, KIF23 |
| 5 | TMEM45A, ANXA6 |
|
| |
| 6 | STAT1, TACSTD2, TMPRSS2, ASAH1, APLP2, TM4SF1, FGFR2 |
| 7 | CD74, TAPBP, ATP2A3 |
| 8 | NDRG2, C10orf81 |
| 9 | FBXL16, MICAL2, APLP2, HLA-G, HLA-C, HLA-A /// HLA-F /// HLA-J, HLA-B |
| 10 | CLU, EPAS1, PPAP2B, TLR2, JAG1 |
| 11 | STEAP1, ARL4C |
| 12 | STEAP2, GPC4, PLBD1 |
| 13 | EHF, PGCP |
| 14 | SGPP2, CAPN2, LAMP3 |
| 15 | ABCA1, ABCC4, IGFBP3 |
| 16 | KLK14, APOD |
| 17 | LOC100288985, MBNL2 |
| 18 | JAM3, SPARC, RUNX1T1 |
| 19 | IGDCC3, FILIP1L, NEDD4L |
| 20 | ARHGEF40, RBMS1, STC1 |
| 21 | ZNF503, CYBB, HIPK2, COL1A1, HLA-F, VIPR1, GNAS, HLA-A, PPAP2B, MUC20, RBM17, QKI |
| 22 | EBF1, HLA-B, ZNF423, CCDC80, VIM, KLF13, PPAP2B, LOC652147, CDH11 |
| 23 | RNF5, NEDD4L, MEIS2 |
| 24 | NFIX, LOXL1, COL1A2, GABBR1 /// UBD |
| 25 | CLU, TSPAN1 |
| 26 | TNFRSF19 |
Visualization of KEGG pathways regulated by soy matrix effects on breast cancer growth
Seventy-seven DEGs modulated by MS were mapped to 81 KEGG pathways. Visualization of KEGG pathways of interest was provided in the supplemental materials (File S4). Pathways in cancer contained the highest amount of DEGs being investigated, followed by cytokine-cytokine receptor interaction, osteoclast differentiation, proteoglycans in cancer, and JAK-STAT signaling pathway. Suppression of oncogene MYC was present in 44 pathways investigated, potentially leading to reduced cell proliferation and increased differentiation. Up-regulation of STAT1 was linked to 16 related pathways. STAT1 had both tumor suppressing and promoting properties. Down-regulation of ADCY1 was linked to 30 different pathways, such as estrogen signaling pathway, insulin secretion pathway, calcium signaling pathway, purine metabolism and bile secretion pathway. Up-regulation of BLNK was associated with regulation of actin cytoskeleton in the B cell receptor signaling pathway.
One-hundred DEGs regulated by MI were mapped to 127 KEGG pathways. Visualization of KEGG pathways of interest was provided in the supplemental materials (File S5). We observed down-regulation of MHC -I (HLA-A, HLA-B, HLA-C, HLA-F, HLA-G), TAPBP and MHC-II (CD74) in the highly correlated KEGG pathways. Since all these genes were involved in regulating immune responses, such as antigen processing and presentation, adhesion molecules (CAMs) and natural killer cell mediated cytotoxicity pathways, isoflavones were likely to suppress immune responses. Down-regulation of EPAS1, FGFR2, STAT1, and RUNX1T1 was seen in the pathways in cancer, suggesting that although isoflavones promoted the growth of MCF-7 tumors they inhibited pathways that induce angiogenesis and block differentiation. Down-regulation of GNAS was linked to 31 pathways, indicating changes in signal transduction, cell communication and endocrine functions. Down-regulation of TLR2 was associated with 16 pathways that were under categories of signal transduction, transport and catabolism, immune system, and cancer. Similarly, down-regulation of GNB4 was linked to 12 relating pathways under the categories of signaling transduction, immune system, nervous system and environmental adaptation.
Soy matrix modulation of breast cancer growth could be explained via enriched GO term regulation
Significantly over-represented GO terms were summarized in Figure 2. The tumor-growth-inhibiting MS up-regulated 117 Biological Process (BP) terms, which was over three times more than that were down-regulated. MS up-regulated and down-regulated number of Cellular Component (CC) terms similarly, as well as Molecular Function (MF) terms. GO terms were observed to be mainly down-regulated by MI which stimulated MCF-7 tumor growth. There were in total 204 down-regulated GO terms and 20 up-regulated terms by MI alone. Detailed GO analysis results were provided in the supplemental file (File S6).
Figure 2. Conditional gene ontology (GO) analysis adopted hypergeometric tests.
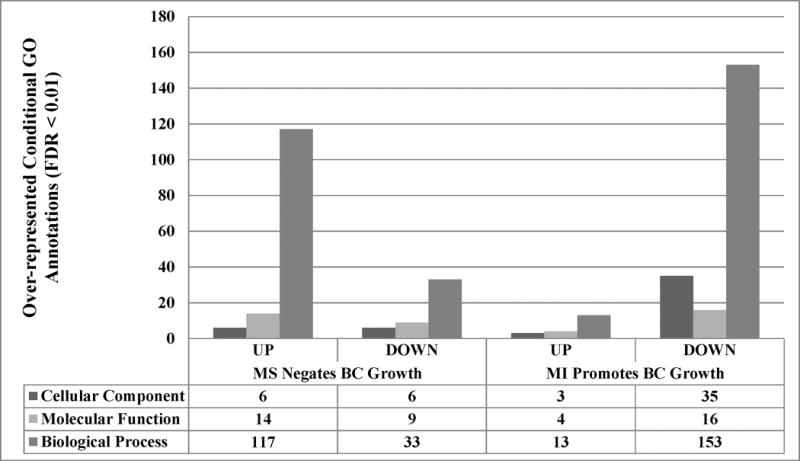
DEGs were filtered by the cutoff of FDR = 0.01 and |log (FC)| = 2. Further, FDR < 0.01 is considered as significant in the conditional hypergeometric test of GO annotations. MI, purified isoflavone mix diet; MS, soy flour diet.
Hundreds of identified over-represented GO terms were further interpreted and organized in the enrichment map. GO terms that were modulated by MS that inhibited tumor growth, were visualized in Figure 3. MS up-regulated 18 enriched GO term clusters and down-regulated 6 clusters, which were highlighted in the enrichment map. Several up-regulated enriched GO terms included immune responses, such as response to interferon, cell communication, negative regulation of signaling, and regulation of molecular function, along with others. All these GO terms belong to Biological Process (BP) category. Representative down-regulated enriched GO terms were nucleoside diphosphate biosynthetic process (BP), neuron differentiation (BP) and cholate-CoA ligase activity (molecular function, MF).
Figure 3. Enrichment map of GO terms distribution regulated by soy flour diet (MS) and reduced MCF-7 tumor growth in ovariectomized athymic nude mice.
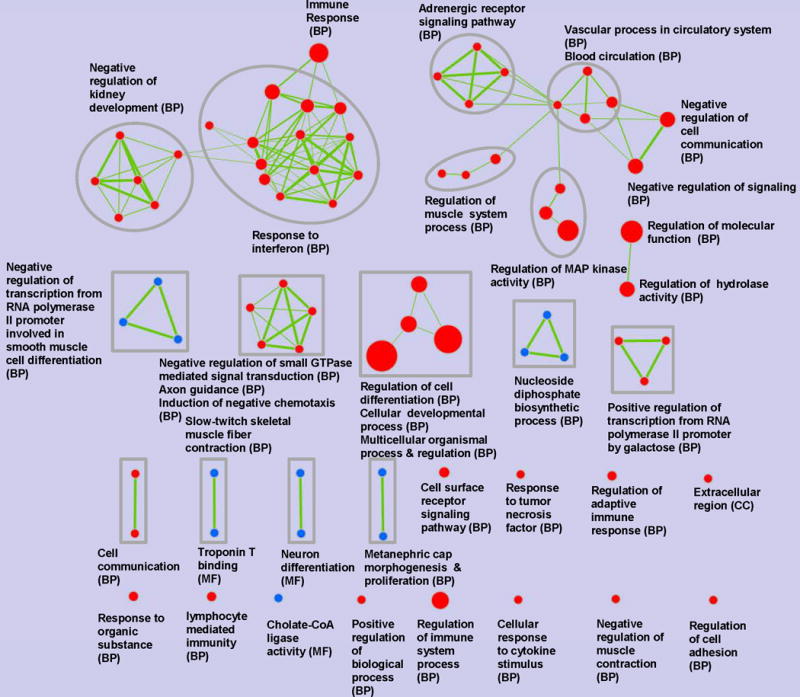
FDR Q-value cutoff ≤ 0.005 and Jaccard plus overlap combined coefficient cutoff ≥ 0.7. GO terms failed to pass the enrichment significance threshold are not shown. Nodes are colored according to the phenotype of the hypergeometric test of the GO terms: red denotes GO terms up-regulated by MS; blue denotes GO terms down-regulated by MS. The color intensity is proportional to the hypergeometric test significance (minus log FDR Q-value). Node size is proportional to the number of DEGs in corresponding GO term. Edge thickness is mapped to the overlap of DEGs between GO terms, which was calculated by the Jaccard plus overlap combined coefficient. Sub-networks (that is, clusters) were determined according to the corresponding functionally related GO terms, which were manually circled and assigned a label. The acronym in brackets represents the three main categories of GO: CC, MF and BP.
GO terms that were modulated by MI that promoted MCF-7 tumor growth, were shown in Figure 4. MI up-regulated 5 enriched GO term clusters and down-regulated 8 clusters, which were highlighted in the enrichment map. Note that 35 GO terms were clustered into 1 sub-network, which related to the down-regulation of immune response (BP) and endoplasmic reticulum (cellular component, CC). Each of their labels was shown in the zoom-in view on top of Figure 4. Other down-regulated enriched GO terms in the tumor-growth-promoting enrichment map contained cytokine production (BP), positive regulation of signal transduction (BP), negative regulation of cell-substrate adhesion (BP), platelet binding and activation (BP), identical protein binding (MF) and negative regulation of cell differentiation (BP). Representative up-regulated enriched GO terms were positive regulation of histone H3–K9 methylation (BP), F12 coagulation factor XII (BP), kinesin complex (CC), microtubule motor activity (MF), mitotic spindle elongation (BP) and endonucleolytic cleavage (BP).
Figure 4. Enrichment of GO terms distribution regulated by purified isoflavone mix diet (MI) and promoted MCF-7 tumor growth in ovariectomized athymic nude mice.
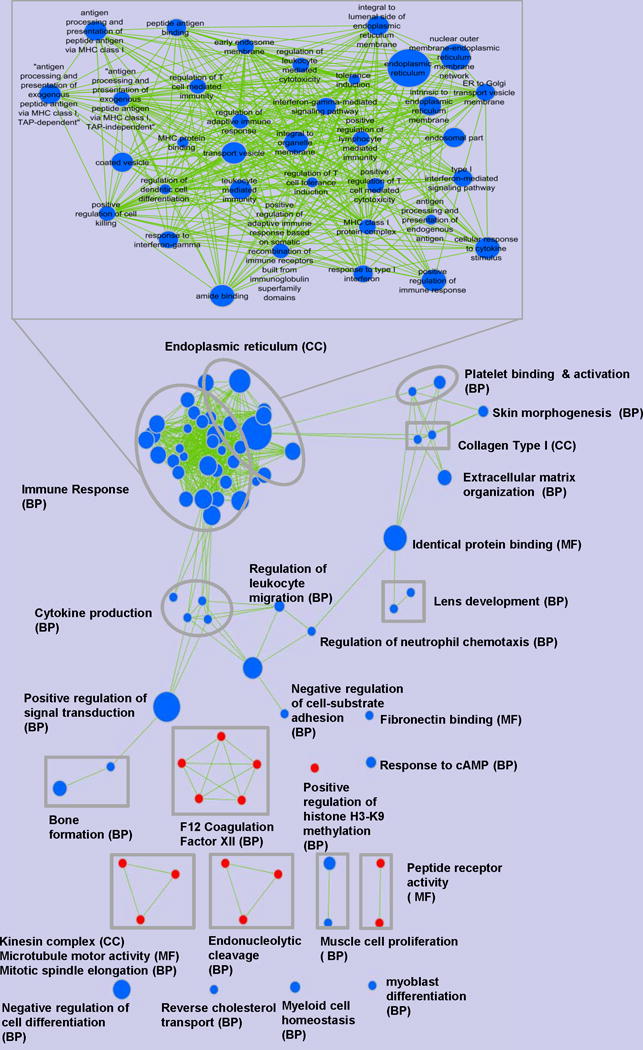
The same criteria adopted in Figure 3 apply here.
Consistent results regarding enriched KEGG pathways were obtained through conditional hypergeometric tests, along with KegArray Software visualization (refer to File S6 for integrated result). Under the FDR cutoff of less than or equal to 0.05, cytokine-cytokine receptor interaction, B cell receptor signaling pathway, JAK-STAT signaling pathway and osteoclast differentiation were up-regulated by MS. Basal transcription factors were up-regulated by MI. Further, antigen processing and presentation, cell adhesion molecules (CAMs), endocytosis, sphingolipid metabolism, natural killer cell mediated cytotoxicity, fat digestion and absorption, ABC transporters and bile secretion were down-regulated by MI.
Correlation analysis of gene expression level with breast cancer patient survival time based on soy matrix regulation
Significantly correlated DEGs, correlation coefficients and P-values of the Spearman’s rank correlation analyses are shown in the supplemental files (File S1, Table S2 and Table S3). MS primarily down-regulated DEGs (such as C1orf135, MRPS12 and TBRG4) that had expression levels negatively correlated with breast cancer patient survival time. Specifically, MS up-regulated DEGs (such as ADRA2A), which had expression levels positively correlated with breast cancer patient survival time (File S1, Table S2). MI primarily down-regulated DEGs (such as CD74, FRFR2, HLA-A/B/C/F/G, VIM and ZNF423) that had expression levels positively correlated with breast cancer patient survival time. MI also up-regulated DEGs (such as KIF23 and KIF14) with expression levels negatively correlated with breast cancer patient survival time (File S1, Table S3).
Discussion
We have previously shown that complex bioactive components in soy diets other than isoflavones (other bioactive compounds in soy are referred to here as soy matrix) modulated MCF-7 tumor growth in ovariectomized athymic nude mice [1]. Although MI was known to promote MCF-7 tumor growth, the MS diet containing an equal amount of genistein negated it. Previously we investigated 5 ER-related mRNA expression gene targets (pS2, PR, cyclin D1, bcl2 and aromatase) [1]. They were up-regulated by isoflavones in purified isoflavone mix (MI), but not by isoflavones in soy flour (MS), suggesting different levels of estrogenicity of diets containing equivalent amounts of genistein. However, other possible molecular mechanisms were not investigated. Our current study provides further evidence of differences between MS (traits shared by MS and NC comparing with PC) and MI (traits shared by MI and PC comparing with NC) by using an integrated MCF-7 tumor whole-genome expression, in which over 22,000 gene expression changes were investigated. Hierarchical clustering with DEGs (Figure 1) demonstrated that soy matrix modulated gene expression in MCF-7 breast cancer tumors differently than MI. The genes that we identified as DEGs are not just reflective of cell proliferation but also are reflective of biological pathways that if differentially expressed in normal cells and premalignant cells, increases the risk of these cells to undergo malignant transformation. Therefore, the DEGs are not necessarily caused by differences in cell proliferation but they are needed for the tumors to exhibit different growth or proliferation. By using bioinformatics tools to analyze extracted features from the heat map, our study highlights: 1) genes promoting or inhibiting tumor growth; 2) regulation of cancer cell signaling pathways; 3) differences in estrogen-responsive gene expression; 4) regulation of immune responses and 5) correlation of the DEGs expression level to survival time among ER+ breast cancer patients treated with Tamoxifen.
Inappropriate cell growth and division are restrained by some genes, which may help prevent the accumulation of mutations during cancer progression. Among them are genes that induce apoptosis. Genes that promote tumor growth often induce an increase in cell division, reduction in cell differentiation and inhibition of apoptosis. These genes have recently been linked to epigenomic programming of tumor initiating stem cells [35]. A large body of data [36, 37] has documented that activation of tumor-growth-promoting oncogenes and/or inactivation of tumor-growth-inhibiting genes function together to exacerbate breast cancer tumor progression. In contrast, inhibition of oncogenes and/or activation of genes that can inhibit tumor growth prevent progression of breast cancer. We identified ATP2A3, BLNK, CLU, RUNX1T1, STAT1, and TNFRSF19 as up-regulated by MS; however, it is unclear whether these genes can explain the protective effects of MS on MCF-7 tumor growth. For example, ATP2A3 has been reported to influence predisposition to cancer development by changing the cell and tissue environment [38]. Although STAT1 was acting as a tumor suppressor gene in some studies [39], several recent findings suggested that STAT1 acted as an oncogene and its high expression level was linked to resistance to cancer treatments and poor survival [40].
MS down-regulated MYC expression. It is well known that MYC is an estrogen-responsive gene [41], which participates in cell cycle regulation and MYC promotes proliferation of MCF-7 breast cancer cells [42]. MYC is down-regulated via the PI3K-AKT pathway [43]. We also observed that oncogene MYB was up-regulated by MI. Up-regulation of MYB was associated with ER+ breast cancer and reported to be critical for tumor cell growth, especially during an early period in mammary tumor development when it appeared to be imperative for tumor initiation [44]. A previous study demonstrated that MYB expression was induced by estrogen [41] in MCF-7 cells inoculated into ovariectomized athymic nude mice. Here, we confirm that MI mimicked the effects of E2 in affecting gene expression. Our findings suggest that MI stimulated MCF-7 tumor growth by increasing expression of tumor promoting genes.
MI down-regulated notch ligand JAG1 in the notch signaling pathway, which among many of its functions propels mammary stem cells to commit to differentiation [45]. Epigenetic alterations might also be affected by soy matrix through chromatin modification. Different levels of modulation were observed on transcription by soy matrix effects (File S6); for instance, MI up-regulated genes that regulate histone H3-H9 methylation. MS and MI had opposing effects on other cell survival pathways [46], including cell cycle/apoptosis, RAS, PI3K, STAT, MAPK and TGF-β pathways (File S4 and File S5). MI also suppressed genes that regulate apoptosis, such as CAPN2. This finding is consistent with previous studies showing that soy protein supplementation upregulated expression of the genes relating to cellular growth and proliferation, cell cycle, cell death and survival in breast cancer patients [13]. Collectively, we report that soy matrix did not induce similar adverse signaling changes, which were linked to increased cancer cell growth, to those induced by MI.
As mentioned earlier, complex bioactive components in soy may modulate the estrogenicity of diets containing equivalent amounts of genistein [1]. In a separate study [47], BALB/c mice fed MI diet had higher peak plasma levels of the active estrogenic aglycon form of genistein compared with mice fed MS. The results from the metabolism and deposition study of dietary isoflavones [47] indicate that MS diet reduced the aglycon content of genistein which may, in part, explain the lower estrogenic response in mice fed MS. The present study was designed to evaluate genome-wide effects and the results here provide further evidence that soy matrix significantly altered the estrogenicity of diet.
MS down-regulated the expression of estrogen-responsive genes, such as NRIP1 (RIP140) and IL17RB. RIP140 is required for normal mammary gland development, estrogen-dependent transcription regulation, cell proliferation in breast cancer, and functions as a co-activator for ERα-responsive genes. High RIP40 expression was linked to poor survival of breast cancer patients [48]. IL17BR was significantly increased by E2 in the MCF-7 cell line. Our data thus suggest that IL17BR is an estrogen-responsive gene, which is consistent with its positive correlation with ER status in primary breast cancer tumors [49]. BLNK is a potential tumor suppressor gene in breast cancer, since it inhibits MCF-7 cell growth and has been identified to be strongly repressed by E2 [50]. MS up-regulated BLNK expression. MI enhanced expression of estrogen-responsive genes, such as MYBL1 (B-MYB) and ERG3. B-MYB has been shown to promote cell survival by activating anti-apoptotic genes in breast cancer [51]. It was also shown to be strongly induced by E2 in the MCF-7 cell line [52] and was identified as one the overexpressed DEGs in breast cancer patients on soy protein supplementation diet compared with the placebo group [13]. ERG3 has been shown to be rapidly induced in estradiol-treated MCF-7 cells [53]. The transcription factor EGR3 is the bona fide target gene of ERα and involved in the estrogen-signaling pathway in breast cancer [53]. Our observations strongly suggest that MS negated the estrogenic properties of isoflavones by modulating estrogen-responsive gene expression.
Data shown in Figure 3 and Figure 4 demonstrated how immune responses were regulated by bioactive compounds found in soy matrix. Numerous immune response GO terms were up-regulated by MS. In contrast, MI caused a down-regulation of several immune response GO terms. DEGs cluster 9 (Table 2) that was modulated by MI contained many immune-related genes, such as HLA-G, HLA-C, HLA-A. Avoiding immune destruction by T and B lymphocytes, macrophages, and natural killer cells has been suggested to be an emerging hallmark of cancer [54]. In support of the notion that immune response is closely regulated in cancer, we observed that MS up-regulated lymphocyte mediated immunity (BP). On the other hand, MI down-regulated the positive regulation of lymphocyte mediated immunity (BP), positive regulation of macrophage cytokine production (BP) and natural killer cell mediated cytotoxicity (KEGG pathway). Previous studies have reported several T-cell mediated antitumor immune responses by isoflavones, including by genistein, in immunocompetent mice, rats and humans [55]. Since the results in our study were generated in athymic mice that are T-cell deficient, it is possible that T-cells are required for purified isoflavones to elicit antitumor immune responses and inhibit tumor growth. Further, our data indicate that soy matrix can provide antitumor immune responses in athymic nude mice that have B-cells and natural killer cells.
Previous studies indicated that reduced dietary fat consumption and increased fiber, vegetable, fruit and other nutrient intake associated with a plant-based, high-fiber diet improved overall survival rate of breast cancer patients [9]. Our study here elucidates how soy matrix effect might alter ER+ breast cancer patient survival time. Kinesins are a superfamily of motor proteins and they have been reported to be deregulated in different cancers. A recent study revealed that estrogen strongly induced the expression of 19 kinesin genes in ER+ breast cancer cells. Elevated kinesin gene expression was correlated with poor relapse-free survival of ER+ breast cancer patients [56]. KIF23 and KIF14 were up-regulated by MI in our study, and their expressions levels were negatively correlated with breast cancer patient survival time across all three clinical datasets being investigated. BRCA1 functions as an important tumor suppressor gene in breast cancer [57]. ZNF423 participates in the estrogen-dependent induction of BRCA1 expression, and an estrogen-inducible BRCA1 transcription factor, which is associated with decreased risk for breast cancer occurrence in selective estrogen receptor (SERM) therapy [58]. In the present study, MI down-regulated ZNF423 gene that is associated with longer survival times among ER+ breast cancer patients. Concurrently, gene expression of the major histocompatibility complex (MHC) family, such as HLA-A/B/C/F/G was suppressed by MI, which might lead to shorter breast cancer patient survival time. MHC molecules were of vital importance in regulating immune responses against cancer [59], and the expression level of MHC was positively correlated with patient survival time.
Collectively, this study is the first to elucidate the underlying molecular mechanisms modulated by the complex nature of the components in soy in a systematic fashion. In the future, studies will be conducted to characterize the composition of the bioactive components other than isoflavones in the soy flour diet, and their anticipated protective effects on breast cancer growth will be investigated. Further, studies will be designed to demonstrate that the genomic changes observed by us impact cancer cell proliferation and apoptosis. The results obtained in this study provide strong evidence that there is a difference in biological responses between consumption of soy flour versus purified isoflavone mix in the ovariectomized athymic nude mouse model of breast cancer.
Supplementary Material
File S1. Supplemental figures and tables in microarray expression analysis. (DOCX)
File S2. Differentially expressed genes (DEGs) list modulated by soy flour (MS) reduced MCF-7 tumor growth in ovariectomized athymic nude mice. Cutoff criteria are: FDR < 0.01 and |Log FC| < 2. (XLSX)
File S3. Differentially expressed genes (DEGs) list modulated by purified isoflavone mix (MI) stimulated MCF-7 tumor growth in ovariectomized athymic nude mice. Cutoff criteria are: FDR < 0.01 and |Log FC| < 2. (XLSX)
File S4. Visualization of KEGG pathways modulated by soy flour (MS) and reduced MCF-7 tumor growth in ovariectomized athymic nude mice. (DOCX)
File S5. Visualization of KEGG pathways modulated by purified isoflavone mix (MI) and stimulated MCF-7 tumor growth in ovariectomized athymic nude mice. (DOCX)
File S6. Over-represented gene ontology annotations include cellular component, molecular function, biological process and KEGG pathway modulated by soy flour (MS) and purified isoflavone mix (MI). (DOCX)
Acknowledgments
This work was supported by the NIH [P50AT006268] (WGH) from the National Center for Complementary and Alternative Medicines (NCCAM), the Office of Dietary Supplements (ODS) and the National Cancer Institute (NCI). Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the NCCAM, ODS, NCI or the National Institutes of Health. The views expressed in this paper do not necessarily reflect those of the U.S. Food and Drug Administration (FDA).
Abbreviations
- AIN93G
American Institute of Nutrition 93 Growth diet
- DEG
differentially expressed gene
- DEP
differentially expressed probe
- E2
17 β-estradiol
- ER+
estrogen-receptor-positive
- FDR
false discovery rate
- GO
gene ontology
- KEGG
Kyoto Encyclopedia of Gene and Genomes
- MI
purified isoflavone mix diet
- MS
soy flour diet
- NC
negative control diet
- PC
positive control diet
Footnotes
Author Contributions
WGH and YL conceived, designed the study, and wrote the manuscript and supplemental materials. YL analyzed the microarray data generated from W.M. Keck Center for Comparative and Functional Genomics at the University of Illinois. XW and JX performed correlation analysis of DEGs that YL identified with patient survival time using clinical data sets. SCF performed the composition analysis on the mixed isoflavone used to prepare MS and MI diets. YL, LH-C, YZ, XW, YP, JX, DRD and WGH gave conceptual advice and edited the manuscript.
Conflict of interest
The authors have declared no conflict of interest.
References
- 1.Allred CD, Allred KF, Ju YH, Goeppinger TS, et al. Soy processing influences growth of estrogen-dependent breast cancer tumors. Carcinogenesis. 2004;25:1649–1657. doi: 10.1093/carcin/bgh178. [DOI] [PubMed] [Google Scholar]
- 2.Helferich WG, Andrade JE, Hoagland MS. Phytoestrogens and breast cancer: a complex story. Inflammopharmacology. 2008;16:219–226. doi: 10.1007/s10787-008-8020-0. [DOI] [PubMed] [Google Scholar]
- 3.Siegel R, Ma J, Zou Z, Jemal A. Cancer statistics, 2014. CA Cancer J Clin. 2014;64:9–29. doi: 10.3322/caac.21208. [DOI] [PubMed] [Google Scholar]
- 4.Trock BJ, Hilakivi-Clarke L, Clarke R. Meta-analysis of soy intake and breast cancer risk. J Natl Cancer Inst. 2006;98:459–471. doi: 10.1093/jnci/djj102. [DOI] [PubMed] [Google Scholar]
- 5.Wu AH, Yu MC, Tseng CC, Pike MC. Epidemiology of soy exposures and breast cancer risk. Br J Cancer. 2008;98:9–14. doi: 10.1038/sj.bjc.6604145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hilakivi-Clarke L, Andrade JE, Helferich W. Is Soy Consumption Good or Bad for the Breast? J Nutr. 2010;140:2326s–2334s. doi: 10.3945/jn.110.124230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gandini S, Merzenich H, Robertson C, Boyle P. Meta-analysis of studies on breast cancer risk and diet: the role of fruit and vegetable consumption and the intake of associated micronutrients. Eur J Cancer. 2000;36:636–646. doi: 10.1016/s0959-8049(00)00022-8. [DOI] [PubMed] [Google Scholar]
- 8.Ogunleye AA, Xue F, Michels KB. Green tea consumption and breast cancer risk or recurrence: a meta-analysis. Breast Cancer Res Tr. 2010;119:477–484. doi: 10.1007/s10549-009-0415-0. [DOI] [PubMed] [Google Scholar]
- 9.McEligot AJ, Largent J, Ziogas A, Peel D, Anton-Culver H. Dietary fat, fiber, vegetable, and micronutrients are associated with overall survival in postmenopausal women diagnosed with breast cancer. Nutr Cancer. 2006;55:132–140. doi: 10.1207/s15327914nc5502_3. [DOI] [PubMed] [Google Scholar]
- 10.Messina M, Hilakivi-Clarke L. Early Intake Appears to Be the Key to the Proposed Protective Effects of Soy Intake Against Breast Cancer. Nutr Cancer. 2009;61:792–798. doi: 10.1080/01635580903285015. [DOI] [PubMed] [Google Scholar]
- 11.Hsieh CY, Santell RC, Haslam SZ, Helferich WG. Estrogenic effects of genistein on the growth of estrogen receptor-positive human breast cancer (MCF-7) cells in vitro and in vivo. Cancer Res. 1998;58:3833–3838. [PubMed] [Google Scholar]
- 12.Ju YH, Allred CD, Allred KF, Karko KL, et al. Physiological concentrations of dietary genistein dose-dependently stimulate growth of estrogen-dependent human breast cancer (MCF-7) tumors implanted in athymic nude mice. The Journal of nutrition. 2001;131:2957–2962. doi: 10.1093/jn/131.11.2957. [DOI] [PubMed] [Google Scholar]
- 13.Shike M, Doane AS, Russo L, Cabal R, et al. The effects of soy supplementation on gene expression in breast cancer: a randomized placebo-controlled study. J Natl Cancer Inst. 2014:106. doi: 10.1093/jnci/dju189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chuang HY, Hofree M, Ideker T. A decade of systems biology. Annu Rev Cell Dev Biol. 2010;26:721–744. doi: 10.1146/annurev-cellbio-100109-104122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sims AH. Bioinformatics and breast cancer: what can high-throughput genomic approaches actually tell us? J Clin Pathol. 2009;62:879–885. doi: 10.1136/jcp.2008.060376. [DOI] [PubMed] [Google Scholar]
- 16.Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Barrett T, Troup DB, Wilhite SE, Ledoux P, et al. NCBI GEO: archive for functional genomics data sets–10 years on. Nucleic Acids Res. 2011;39:D1005–1010. doi: 10.1093/nar/gkq1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barrett T, Wilhite SE, Ledoux P, Evangelista C, et al. NCBI GEO: archive for functional genomics data sets–update. Nucleic Acids Res. 2013;41:D991–995. doi: 10.1093/nar/gks1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gentleman R. Bioinformatics and computational biology solutions using R and Bioconductor. Springer Science+Business Media; New York: 2005. [Google Scholar]
- 20.Hubbell E, Liu WM, Mei R. Robust estimators for expression analysis. Bioinformatics. 2002;18:1585–1592. doi: 10.1093/bioinformatics/18.12.1585. [DOI] [PubMed] [Google Scholar]
- 21.Irizarry RA, Bolstad BM, Collin F, Cope LM, et al. Summaries of affymetrix GeneChip probe level data. Nucleic Acids Res. 2003:31. doi: 10.1093/nar/gng015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu ZJ, Irizarry RA, Gentleman R, Martinez-Murillo F, Spencer F. A model-based background adjustment for oligonucleotide expression arrays. J Am Stat Assoc. 2004;99:909–917. [Google Scholar]
- 23.Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3 doi: 10.2202/1544-6115.1027. Article3. [DOI] [PubMed] [Google Scholar]
- 24.Choe S, Boutros M, Michelson A, Church G, Halfon M. Preferred analysis methods for Affymetrix GeneChips revealed by a wholly defined control dataset. Genome Biol. 2005;6:R16. doi: 10.1186/gb-2005-6-2-r16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Benjamini Y, Hochberg Y. Controlling the False Discovery Rate - a Practical and Powerful Approach to Multiple Testing. J Roy Stat Soc B Met. 1995;57:289–300. [Google Scholar]
- 26.Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns (vol 95, pg 14863, 1998) Proc Natl Acad Sci U S A. 1999;96:10943–10943. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Langfelder P, Zhang B, Horvath S. Defining clusters from a hierarchical cluster tree: the Dynamic Tree Cut package for R. Bioinformatics. 2008;24:719–720. doi: 10.1093/bioinformatics/btm563. [DOI] [PubMed] [Google Scholar]
- 28.Ogata H, Goto S, Sato K, Fujibuchi W, et al. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 1999;27:29–34. doi: 10.1093/nar/27.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kanehisa M, Goto S, Sato Y, Furumichi M, Tanabe M. KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Res. 2012;40:D109–114. doi: 10.1093/nar/gkr988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Falcon S, Gentleman R. Using GOstats to test gene lists for GO term association. Bioinformatics. 2007;23:257–258. doi: 10.1093/bioinformatics/btl567. [DOI] [PubMed] [Google Scholar]
- 31.Merico D, Isserlin R, Stueker O, Emili A, Bader GD. Enrichment map: a network-based method for gene-set enrichment visualization and interpretation. PLoS One. 2010;5:e13984. doi: 10.1371/journal.pone.0013984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cline MS, Smoot M, Cerami E, Kuchinsky A, et al. Integration of biological networks and gene expression data using Cytoscape. Nat Protoc. 2007;2:2366–2382. doi: 10.1038/nprot.2007.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Loi S, Haibe-Kains B, Desmedt C, Wirapati P, et al. Predicting prognosis using molecular profiling in estrogen receptor-positive breast cancer treated with tamoxifen. BMC Genomics. 2008;9:239. doi: 10.1186/1471-2164-9-239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Symmans WF, Hatzis C, Sotiriou C, Andre F, et al. Genomic index of sensitivity to endocrine therapy for breast cancer. J Clin Oncol. 2010;28:4111–4119. doi: 10.1200/JCO.2010.28.4273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vicente-Duenas C, Romero-Camarero I, Cobaleda C, Sanchez-Garcia I. Function of oncogenes in cancer development: a changing paradigm. Embo J. 2013;32:1502–1513. doi: 10.1038/emboj.2013.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Osborne C, Wilson P, Tripathy D. Oncogenes and tumor suppressor genes in breast cancer: potential diagnostic and therapeutic applications. Oncologist. 2004;9:361–377. doi: 10.1634/theoncologist.9-4-361. [DOI] [PubMed] [Google Scholar]
- 37.Lee EY, Muller WJ. Oncogenes and tumor suppressor genes. Cold Spring Harb Perspect Biol. 2010;2:a003236. doi: 10.1101/cshperspect.a003236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Korosec B, Glavac D, Volavsek M, Ravnik-Glavac M. ATP2A3 gene is involved in cancer susceptibility. Cancer Genet Cytogen. 2009;188:88–94. doi: 10.1016/j.cancergencyto.2008.10.007. [DOI] [PubMed] [Google Scholar]
- 39.Chan SR, Vermi W, Luo J, Lucini L, et al. STAT1-deficient mice spontaneously develop estrogen receptor alpha-positive luminal mammary carcinomas. Breast Cancer Res. 2012;14:R16. doi: 10.1186/bcr3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Weichselbaum RR, Ishwaran H, Yoon T, Nuyten DSA, et al. An interferon-related gene signature for DNA damage resistance is a predictive marker for chemotherapy and radiation for breast cancer. Proc Natl Acad Sci U S A. 2008;105:18490–18495. doi: 10.1073/pnas.0809242105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Creighton CJ, Cordero KE, Larios JM, Miller RS, et al. Genes regulated by estrogen in breast tumor cells in vitro are similarly regulated in vivo in tumor xenografts and human breast tumors. Genome Biol. 2006;7:R28. doi: 10.1186/gb-2006-7-4-r28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hynes NE, Stoelzle T. Key signalling nodes in mammary gland development and cancer: Myc. Breast Cancer Res. 2009;11:210. doi: 10.1186/bcr2406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chang PH, Hwang-Verslues WW, Chang YC, Chen CC, et al. Activation of Robo1 signaling of breast cancer cells by Slit2 from stromal fibroblast restrains tumorigenesis via blocking PI3K/Akt/beta-catenin pathway. Cancer Res. 2012;72:4652–4661. doi: 10.1158/0008-5472.CAN-12-0877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yu Miao R, Drabsch Y, Cross RS, Cheasley D, et al. MYB Is Essential for Mammary Tumorigenesis. Cancer Res. 2011;71:7029–7037. doi: 10.1158/0008-5472.CAN-11-1015. [DOI] [PubMed] [Google Scholar]
- 45.Buckley NE, Nic An tSaoir CB, Blayney JK, Oram LC, et al. BRCA1 is a key regulator of breast differentiation through activation of Notch signalling with implications for anti-endocrine treatment of breast cancers. Nucleic Acids Res. 2013;41:8601–8614. doi: 10.1093/nar/gkt626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, et al. Cancer genome landscapes. Science (New York, N Y) 2013;339:1546–1558. doi: 10.1126/science.1235122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Allred CD, Twaddle NC, Allred KF, Goeppinger TS, et al. Soy processing affects metabolism and disposition of dietary isoflavones in ovariectomized BALB/c mice. J Agric Food Chem. 2005;53:8542–8550. doi: 10.1021/jf051246w. [DOI] [PubMed] [Google Scholar]
- 48.Rosell M, Nevedomskaya E, Stelloo S, Nautiyal J, et al. Complex Formation and Function of Estrogen Receptor alpha in Transcription Requires RIP140. Cancer Res. 2014;74:5469–5479. doi: 10.1158/0008-5472.CAN-13-3429. [DOI] [PubMed] [Google Scholar]
- 49.Wang Z, Dahiya S, Provencher H, Muir B, et al. The prognostic biomarkers HOXB13, IL17BR, and CHDH are regulated by estrogen in breast cancer. Clin Cancer Res. 2007;13:6327–6334. doi: 10.1158/1078-0432.CCR-07-0310. [DOI] [PubMed] [Google Scholar]
- 50.Casa AJ, Potter AS, Malik S, Lazard Z, et al. Estrogen and insulin-like growth factor-I (IGF-I) independently down-regulate critical repressors of breast cancer growth. Breast Cancer Res Tr. 2012;132:61–73. doi: 10.1007/s10549-011-1540-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sala A. B-MYB, a transcription factor implicated in regulating cell cycle, apoptosis and cancer. Eur J Cancer. 2005;41:2479–2484. doi: 10.1016/j.ejca.2005.08.004. [DOI] [PubMed] [Google Scholar]
- 52.Liu M, Fan J, Wang S, Wang Z, et al. Transcriptional profiling of Chinese medicinal formula Si-Wu-Tang on breast cancer cells reveals phytoestrogenic activity. BMC Complement Altern Med. 2013;13:11. doi: 10.1186/1472-6882-13-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Inoue A, Omoto Y, Yamaguchi Y, Kiyama R, Hayashi SI. Transcription factor EGR3 is involved in the estrogen-signaling pathway in breast cancer cells. J Mol Endocrinol. 2004;32:649–661. doi: 10.1677/jme.0.0320649. [DOI] [PubMed] [Google Scholar]
- 54.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 55.Masilamani M, Wei J, Sampson HA. Regulation of the immune response by soybean isoflavones. Immunol Res. 2012;54:95–110. doi: 10.1007/s12026-012-8331-5. [DOI] [PubMed] [Google Scholar]
- 56.Zou JX, Duan Z, Wang J, Sokolov A, et al. Kinesin family deregulation coordinated by bromodomain protein ANCCA and histone methyltransferase MLL for breast cancer cell growth survival and tamoxifen resistance. Mol Cancer Res. 2014;12:539–549. doi: 10.1158/1541-7786.MCR-13-0459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Silver DP, Livingston DM. Mechanisms of BRCA1 tumor suppression. Cancer Discov. 2012;2:679–684. doi: 10.1158/2159-8290.CD-12-0221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ingle JN, Liu M, Wickerham DL, Schaid DJ, et al. Selective estrogen receptor modulators and pharmacogenomic variation in ZNF423 regulation of BRCA1 expression: individualized breast cancer prevention. Cancer Discov. 2013;3:812–825. doi: 10.1158/2159-8290.CD-13-0038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Redondo M, Garcia J, Villar E, Rodrigo I, et al. Major histocompatibility complex status in breast carcinogenesis and relationship to apoptosis. Hum Pathol. 2003;34:1283–1289. doi: 10.1016/j.humpath.2003.06.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
File S1. Supplemental figures and tables in microarray expression analysis. (DOCX)
File S2. Differentially expressed genes (DEGs) list modulated by soy flour (MS) reduced MCF-7 tumor growth in ovariectomized athymic nude mice. Cutoff criteria are: FDR < 0.01 and |Log FC| < 2. (XLSX)
File S3. Differentially expressed genes (DEGs) list modulated by purified isoflavone mix (MI) stimulated MCF-7 tumor growth in ovariectomized athymic nude mice. Cutoff criteria are: FDR < 0.01 and |Log FC| < 2. (XLSX)
File S4. Visualization of KEGG pathways modulated by soy flour (MS) and reduced MCF-7 tumor growth in ovariectomized athymic nude mice. (DOCX)
File S5. Visualization of KEGG pathways modulated by purified isoflavone mix (MI) and stimulated MCF-7 tumor growth in ovariectomized athymic nude mice. (DOCX)
File S6. Over-represented gene ontology annotations include cellular component, molecular function, biological process and KEGG pathway modulated by soy flour (MS) and purified isoflavone mix (MI). (DOCX)


