Summary
Intracellular protein gradients underlie essential cellular and developmental processes, but the mechanisms by which they are established are incompletely understood. During the asymmetric division of the C. elegans zygote, the RNA-binding protein MEX-5 forms an anterior-rich cytoplasmic gradient that causes the RNA-binding protein POS-1 to form an opposing, posterior-rich gradient. We demonstrate that the polo-like kinase PLK-1 mediates the repulsive coupling between MEX-5 and POS-1 by increasing the mobility of POS-1 in the anterior. PLK-1 is enriched in the anterior cytoplasm and phosphorylates POS-1, which is both necessary and sufficient to increase POS-1 mobility. Regulation of POS-1 mobility depends on both the interaction between PLK-1 and MEX-5 and between MEX-5 and RNA, suggesting that MEX-5 may recruit PLK-1 to RNA in the anterior. The low concentration of MEX-5/PLK-1 in the posterior cytoplasm provides a permissive environment for the retention of POS-1, which depends on POS-1 RNA-binding. Our findings describe a novel reaction/diffusion mechanism in which the asymmetric distribution of cytoplasmic PLK-1 couples two RNA-binding protein gradients, thereby partitioning the cytoplasm.
ETOC
The mechanisms that polarize the cytoplasm are not understood. HAN et al. show that the polo-like kinase PLK-1 regulates cytoplasmic polarization in the C. elegans zygote. PLK-1 acts with the RNA-binding protein MEX-5 to locally control the mobility of POS-1, thereby driving the formation of a POS-1 concentration gradient.
Introduction
Protein concentration gradients encode spatial information that is necessary for a wide range of cellular and developmental processes. The central role morphogen gradients play in patterning cell identity across tissues has motivated decades of research into how gradients are established over long length scales [1]. It had been assumed that gradients could not be maintained within a single cell because of the dominant effects of diffusion at short length scales. However, initial theoretical work showed that if a kinase and phosphatase are spatially separated within a cell, a gradient in the phosphorylation state of their substrate could arise [2]. This idea was extended by the observation that if phosphorylation substantially changes a substrate’s diffusion coefficient, a protein concentration gradient will arise with high concentrations in the region of low protein diffusivity [3]. Recent findings have demonstrated that such reaction/diffusion mechanisms generate intracellular gradients that underlie fundamental cellular processes including organization of the mitotic spindle, control of the timing and position of cell division, and asymmetric cell division [4–7]. Therefore, characterization of the mechanisms that locally control protein diffusivity is central to understanding how intracellular gradients are sculpted.
The segregation of cytoplasmic fate determinants during the asymmetric division of the C. elegans zygote provides a classic and particularly striking example of how gradients can spatially pattern cells. Shortly following fertilization, the Tandem CCCH Zinc Finger (TZF) RNA-binding proteins MEX-5 and MEX-6 (MEX-5/6 hereafter) segregate over the course of ~10 minutes to the anterior cytoplasm in response to the posterior kinase PAR-1 [8–11]. PAR-1 phosphorylates MEX-5 and increases its mobility in the posterior, leading to the preferential retention of MEX-5 in the anterior [12–14], likely through the formation of slow-diffusing MEX-5/RNA complexes [13]. The MEX-5/6 gradients, in turn, stimulate the redistribution of germ plasm RNA-binding proteins to the posterior cytoplasm [9]. For example, MEX-5/6 act through unknown mechanisms to increase the mobility of the TZF RNA-binding proteins POS-1 and PIE-1 in the anterior, which leads to their progressive redistribution to the posterior [15, 16]. The coupling between the MEX-5/6 and PIE-1 and POS-1 gradients raise two important questions. First, how do RNA-binding proteins such as MEX-5/6 regulate the diffusion, and therefore the localization, of other RNA-binding proteins? Second, how can MEX-5/6 target specific RNA-binding proteins such as POS-1 and PIE-1 among the many cytoplasmic RNA-binding proteins?
MEX-5/6 were recently shown to control the segregation of germ granules (P granules in C. elegans) through a competitive RNA-binding mechanism. P granules are micron-sized, non-membranous assemblages composed of RNA and RNA-binding proteins [17, 18]. MEX-5/6 compete with the P granule components PGL-3 and MEG-3 for RNA, such that there is a local depletion of available RNA in the anterior cytoplasm [19, 20]. As a consequence, P granules are destabilized/disassembled in the anterior and stabilized/assembled in the posterior cytoplasm. It has not been determined whether similar mechanisms underlie the segregation of other germ plasm RNA-binding proteins.
In this study, we provide evidence that MEX-5 acts through the polo-like kinase PLK-1 to control the segregation of POS-1. We show that POS-1 segregation depends on both the interaction of MEX-5 with RNA and the interaction between MEX-5 and PLK-1, which has previously been shown to lead to the accumulation of PLK-1 in the anterior cytoplasm [21–23]. We show that PLK-1 phosphorylates POS-1 and acts to increase POS-1 mobility in the anterior. Additionally, we show that the retention of POS-1 in the posterior depends on POS-1 RNA-binding, suggesting that PLK-1 phosphorylation may inhibit the formation of slow-diffusing POS-1/RNA complexes. Our findings elucidate a novel reaction/diffusion mechanism by which PLK-1 kinase couples two RNA-binding protein concentration gradients, leading to the partitioning of the cytoplasm of the C. elegans zygote.
Results
MEX-5 RNA-binding is required for POS-1 and PIE-1 segregation
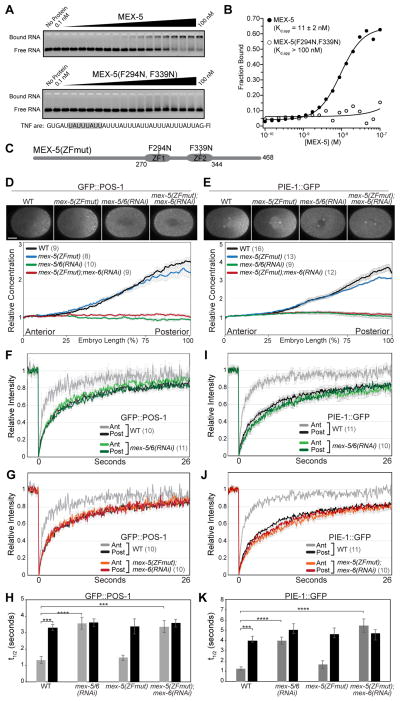
To begin to characterize the mechanisms underlying POS-1 and PIE-1 segregation, we asked whether MEX-5 RNA-binding is required for its ability to control POS-1 and PIE-1 localization. The mammalian TZF protein Tis11d binds RNA through conserved aromatic residues within its ZF domains (Figure S1A) that form hydrophobic stacking interactions with RNA [24, 25]. Using an in vitro electrophoretic mobility shift assay (EMSA), we find that mutations at the corresponding residues (F294N;F339N) in MEX-5 reduce the apparent affinity (Kd,app) of recombinant MBP-tagged MEX-5(aa236–350) for TNF-ARE RNA [10] from 11 nM ± 2 nM to greater than 100 nM (Figures 1A and 1B). We used CRISPR/Cas9 to edit the endogenous MEX-5 locus to generate mex-5(F294N;F339N), hereafter referred to as mex-5(ZFmut). mex-5(ZFmut) embryos are viable (Figure S1B) and segregate POS-1 and PIE-1 normally (Figures 1C–1E, and S1G–S1I). We therefore analyzed the function of MEX-5(ZFmut) in embryos depleted of the partially redundant MEX-5 homologue MEX-6 [9] using mex-6(RNAi), which specifically depletes MEX-6 and not MEX-5 or MEX-5(ZFmut) (Figure S1C). In the presence of MEX-5, MEX-6 is dispensable for the segregation of GFP::MEX-5, GFP::MEX-5(ZFmut) (Figure S1D), GFP::POS-1 (Figures 5B and 5C) and PIE-1::GFP (Figure S1E) [9]. Like mex-5/6(RNAi) embryos, mex-5(ZFmut);mex-6(RNAi) embryos are inviable (Figure S1B) and, as described below, exhibit severe defects in GFP::POS-1 and PIE-1::GFP segregation.
Figure 1. MEX-5 RNA-binding is required for GFP::POS-1 and PIE-1::GFP segregation.
A. EMSA comparing the affinity of MBP-tagged MEX-5(aa236–350) (top) and MBP-tagged MEX-5(aa236–350; F294N, F339N) (bottom) for fluorescein labeled TNF-ARE RNA from the TNFα 3′UTR [10]. The target RNA is shown at the bottom with a MEX-5 binding site highlighted in grey. B. The fraction of bound RNA is plotted as a function of MEX-5 concentration. Kd,app was determined by fitting the data to the Hill equation [10] and is the average of at least three experiments. Error in the key is the standard deviation. Little or no binding of MEX-5(aa236–350; F294N, F339N) was observed at the highest concentration tested (100 nM), so we estimate the Kd,app to be greater than 100 nM. C. Schematic of MEX-5(ZFmut) indicating the position of the TZF RNA-binding domains (ZF1 and ZF2) and the F294N and F339N substitutions. D, E. Top: GFP::POS-1 (D) and PIE-1::GFP (E) localization in the indicated embryos. For this and subsequent figures, images are acquired at NEBD at the cell midplane and embryos are oriented with the anterior to the left and the posterior to the right. Scale bar is 10 μm. Bottom: Relative GFP::POS-1 and PIE-1::GFP concentration along the A/P axis in embryos of the indicated genotype (normalized to the concentration at the anterior end). F–G, I–J. Mean FRAP recovery curves for GFP::POS-1 (F and G) and PIE-1::GFP (I and J) in the anterior and posterior of embryos of the indicated genotype. The same WT data are presented in the top and bottom graphs (error bars are only shown in the top graph for visual clarity). Photobleaching was performed at time = 0. H, K. Mean half-time of recovery (t1/2) for the indicated FRAP experiments. A = Anterior, P = Posterior. In this and subsequent figures, p values are: * <0.05; ** <0.01; *** <0.001; **** <0.0001; n.s. = not significant (>0.05). For all graphs, error bars represent SEM and the number of embryos analyzed are in parentheses. Throughout this figure, WT embryos were treated with a control RNAi (empty L4440 vector). See also Figure S1, S2 and Table S1.
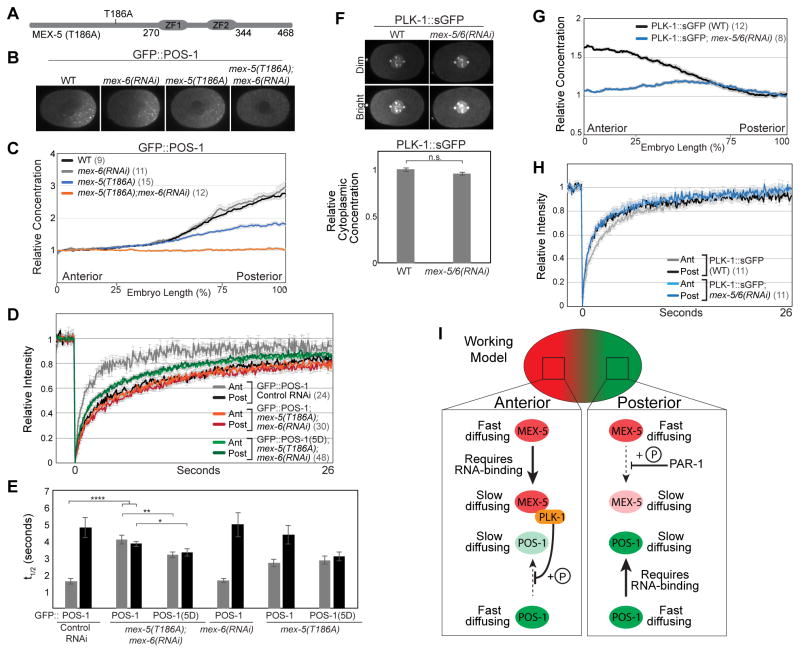
Figure 5. Characterization of mex-5(T186A);mex-6(RNAi) and PLK-1::sGFP.
A. Schematic of MEX-5(T186A). B–D. GFP::POS-1 localization (B), relative concentration along the A/P axis (C), and FRAP curves (D) in embryos of the indicated genotype. GFP::POS-1 FRAP curves are similar in control RNAi and mex-6(RNAi) embryos (see Figure S5A). E. Mean t1/2 for the indicated FRAP experiments. F. PLK-1::sGFP localization in WT and mex-5/6(RNAi) embryos. Top: Duplicate images with different intensity normalizations (dim and bright). The centrosomal and chromosomal signals in the “bright” images are saturated. Bottom: the concentration of PLK-1::sGFP in the cytoplasm is not affected by mex-5/6(RNAi). G. Relative concentration of PLK-1::sGFP along the A/P axis (normalized to the posterior end). H. FRAP curves for PLK-1::sGFP in wild-type and mex-5/6(RNAi) embryos. I. Working model. MEX-5 (red) and POS-1 (green) exist in fast-diffusing complexes and slow-diffusing complexes. The interaction of both MEX-5 and POS-1 with slow-diffusing complexes depends on their ability to bind RNA. PAR-1 phosphorylation of MEX-5 (depicted as “+P”) inhibits the formation of slow-diffusing MEX-5 complexes in the posterior, leading to their enrichment in the anterior. PLK-1 phosphorylation of POS-1 inhibits the formation of slow-diffusing POS-1 complexes in the anterior where PLK-1 likely accumulates in slow-diffusing RNA complexes due to its interaction with MEX-5, which was shown by yeast two-hybrid [23]. As a consequence, slow-diffusing POS-1 complexes accumulate in the posterior, giving rise to the POS-1 concentration gradient. PLK-1 is depicted as inhibiting POS-1 association with slow-diffusing complexes for simplicity, but could also promote POS-1 dissociation from slow-diffusing complexes. See also Figure S3, S5 and Table S1.
To monitor GFP::POS-1 and PIE-1::GFP mobility, we used a fluorescence recovery after photobleaching (FRAP) assay. Consistent with previous studies [15, 16], we find that the diffusivity of both GFP::POS-1 and PIE-1::GFP are significantly higher in the anterior than the posterior of wild-type embryos (Figures 1F, 1H, 1I, 1K and Table S1). In contrast, GFP::POS-1 and PIE-1::GFP exhibit slow mobility throughout the cytoplasm of mex-5(ZFmut);mex-6(RNAi) embryos and mex-5/6 (RNAi) embryos (Figures 1F – 1K; Table S1), and apparently as a consequence, fail to segregate (Figures 1C–1E, S1F and S1G) [15]. MEX-5 and MEX-5(ZFmut) are expressed at similar levels (Figure S1C), indicating that decreased MEX-5 concentration does not cause the defects in GFP::POS-1 and PIE-1::GFP dynamics in mex-5(ZFmut);mex-6(RNAi) embryos. Additionally, MEX-5/6 increase PIE-1::GFP mobility in both par-3 and par-1 mutant embryos in which MEX-5/6 are uniformly distributed [15], indicating that the failure to increase GFP::POS-1 and PIE-1::GFP mobility in mex-5(ZFmut);mex-6(RNAi) is not due to the reduced segregation of MEX-5 RNA-binding mutants, per se (Figure S1D) [13, 19]. Therefore, we conclude that MEX-5 RNA-binding is required for MEX-5 to stimulate increased GFP::POS-1 and PIE-1::GFP mobility in the anterior. MEX-5 RNA-binding is also required to suppress MEG-3 granule assembly in the anterior [19], suggesting that MEX-5 RNA-binding may be a common requirement for germ plasm segregation. We note that although MEG-3/4 are important for P granule segregation, they do not contribute to PIE-1 [26] or POS-1 [27] segregation (Figure S2).
RNA-binding mediates POS-1 and PIE-1 retention in the posterior
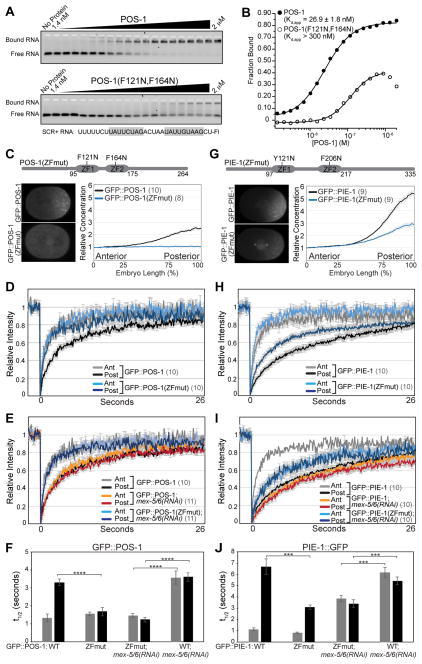
Having demonstrated that MEX-5 RNA-binding is required to increase POS-1 and PIE-1 mobility, we next asked whether POS-1 or PIE-1 RNA-binding is required for their segregation. We hypothesized that interaction with RNA could retard POS-1 and PIE-1 mobility, as was previously shown for MEX-5 [13]. To test this hypothesis, we introduced mutations in the POS-1 ZF domains (F121N;F164N) and in the PIE-1 ZF domains (Y121N;F206N) that are predicted to disrupt the hydrophobic stacking interactions necessary to bind RNA (Figure S1A). In vitro, the affinity of POS-1(F121N;F164N) for a fragment of the GLP-1 RNA [28] is significantly lower (>300 nM) than wild-type POS-1 (26.9 nM ± 1.8 nM) (Figures 2A and 2B). POS-1 and PIE-1 alleles containing these mutations (GFP::POS-1(ZFmut) and GFP::PIE-1(ZFmut) hereafter) were expressed in embryos from single-copy transgenes [29] at levels similar to wild-type GFP::POS-1 and GFP::PIE-1 transgenes (Figure S3).
Figure 2. RNA-binding mediates POS-1 and PIE-1 retention in the posterior cytoplasm.
A EMSA comparing the affinity of MBP-tagged POS-1(aa80–180) and MBP-tagged POS-1(aa80–180; F121N, F164N) for fluorescein labeled SCR+ RNA from the glp-1 3′UTR [28]. The target RNA is shown at the bottom with the POS-1 binding sites highlighted in grey. B. The fraction of bound RNA plotted as a function of POS-1 concentration. Kd,app was determined as in Figure 1B. We did not observe saturation of POS-1(aa80–180; F121N, F164N) binding at the highest concentration tested (2 μM). We approximate the Kd,app to be greater than 300 nM, which shows approximately half maximal binding observed in the wild-type POS-1(aa80–180) EMSA. C, G. Top: Schematic of POS-1 (C) and PIE-1 (G) indicating the position of the TZF RNA-binding domains (ZF1 and ZF2) and the ZFmut substitutions. Bottom: Images of embryos expressing the indicated GFP::POS-1 and GFP::PIE-1 transgenes and quantification of their relative concentration along the A/P axis. D–E, H–I. FRAP recovery curves for the indicated GFP::POS-1 (D and E) and GFP::PIE-1 (H and I) transgenes. The wild-type GFP::POS-1 and GFP::PIE-1 recovery curves are also displayed in Figure 1. F, J. Mean t1/2 for the indicated FRAP experiments. See also Figure S3 and Table S1.
GFP::POS-1 mobility is significantly lower in the posterior than in the anterior, leading to the formation of a ~3-fold posterior-rich concentration gradient. In contrast, the mobility of GFP::POS-1(ZFmut) is significantly higher than GFP::POS-1 in the posterior, and as a result GFP::POS-1(ZFmut) fails to segregate (Figures 2C – 2D, 2F and Table S1). We note that the GFP::POS-1(ZFmut) does not localize to posterior granules in the zygote, suggesting RNA-binding contributes to the recruitment of POS-1 into granules. GFP::PIE-1(ZFmut) exhibited similar, although less severe, defects in its dynamics. The mobility of GFP::PIE-1(ZFmut) is significantly faster than GFP::PIE-1 in the posterior (Figures 2H, 2J and Table S1) resulting in a reduced differential in mobility between the anterior and posterior. As a consequence, GFP::PIE-1(ZFmut) forms a concentration gradient that is ~50% shallower than that of GFP::PIE-1 (Figure 2G). Previous work demonstrated that in addition to the ZF domains, the C-terminus of PIE-1 contributes to its segregation [30], and we speculate that the C-terminus may mediate additional interactions that contribute to PIE-1 retention in the posterior. Because we have not determined the affinity of PIE-1(ZFmut) for RNA, another possibility is that PIE-1(ZFmut) may retain affinity for RNA.
The above findings suggest that the association of POS-1 and PIE-1 with RNA retards their mobility in the posterior and that RNA-bound MEX-5/6 may negatively regulate their association with RNA in the anterior. One prediction of this model is that the increased mobility exhibited by POS-1 and PIE-1 RNA-binding mutants should be independent of MEX-5/6. Indeed, the mobility of GFP::POS-1(ZFmut) is significantly higher than GFP::POS-1 in mex-5/6(RNAi) embryos (Figures 2E, 2F and Table S1). Additionally, the mobility of GFP::PIE-1(ZFmut) in mex-5/6(RNAi) embryos is intermediate between the mobility of GFP::PIE-1 in the anterior and posterior of wild-type embryos (Figures 2I, 2J and Table S1). We conclude that RNA-binding is required for GFP::POS-1 retention and contributes significantly to GFP::PIE-1 retention in the posterior cytoplasm. We propose that MEX-5/6 activity in the anterior inhibits the association of these two proteins with slow-diffusing RNA complexes. Because additional unknown mechanisms appear to contribute to PIE-1 retention in the posterior, we focused our subsequent analysis on POS-1.
The POS-1 C-terminus is required for segregation
To determine the regions of POS-1 required for its segregation, we analyzed the localization of a series of GFP::POS-1 truncations in the regions flanking the centrally positioned ZF domains (aa95–175) (Figure S4A). We find that GFP::POS-1 N-terminal truncations (GFP::POS-1(aa30–264), GFP::POS-1(aa60–264) and GFP::POS-1(aa90–264)) form normal posterior-rich gradients (Figures S4A and S4B). Depletion of endogenous POS-1 by RNAi did not affect the GFP::POS-1(aa90–264) gradient (Figures S4C and S4D), indicating that its segregation does not depend on interactions with endogenous POS-1. Similarly, GFP::POS-1(aa1–239) also segregates to the posterior, forming a gradient that is slightly weaker than the GFP::POS-1 gradient (Figures S4A and S4B). In contrast, GFP::POS-1(aa1–199) is symmetrically distributed throughout the cytoplasm of the polarized zygote (Figures S4A and S4B). These data indicate that GFP::POS-1 segregation requires the C-terminus but not the N-terminus of POS-1 and suggest that residues aa199–239 are particularly important. Interestingly, this region is conserved within nematode POS-1 homologs, but is absent from other nematode TZF proteins [31].
Predicted PLK-1 phosphorylation sites are required for POS-1 segregation
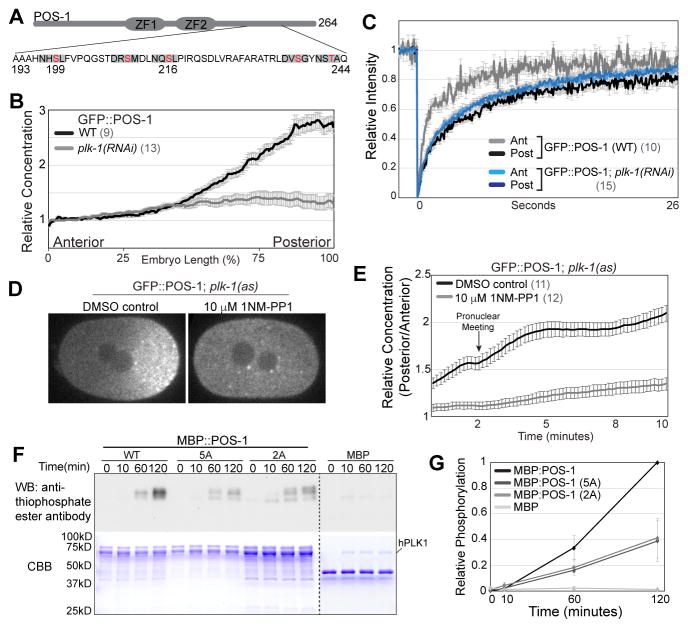
Strikingly, there are five potential polo-like kinase phosphorylation sites (D/E/N X S/T) within or immediately adjacent to the region aa199–239 of POS-1 (Figure 3A) [32]. Two of the sites, Ser199 and Ser216, have a Leu in the +1 position and therefore match the preferred PLK-1 consensus phosphorylation site [33]. PLK-1 and PLK-2 have been shown to interact with MEX-5/6 by yeast two-hybrid, to accumulate in the anterior cytoplasm in a MEX-5/6-dependent manner and to be required for MEX-5/6 to stimulate the segregation of PIE-1 to the posterior [23]. We hypothesized that PLK-1 and/or PLK-2 might phosphorylate POS-1 and thereby control POS-1 mobility. As an initial test of this hypothesis, we characterized POS-1 dynamics in plk-1(RNAi) embryos. GFP::POS-1 was symmetrically distributed in 10 out of 13 plk-1(RNAi) embryos examined (Figure 3B), and exhibited uniformly slow mobility in plk-1(RNAi) embryos (Figure 3C), indicating that PLK-1 is required to increase POS-1 mobility in the anterior. Interestingly, defects in PIE-1 segregation were previously observed in plk-1/2(RNAi) but not plk-1(RNAi) [23], which taken together with our findings, suggests that low levels of PLK activity may be sufficient for PIE-1 segregation but not for POS-1 segregation.
Figure 3. PLK-1 kinase activity is required for POS-1 segregation.
A. Schematic of POS-1 with predicted PLK-1 phosphorylation sites within the region aa193 – 244. Residues in red were mutated to either Alanine (5A) or Aspartic Acid (5D). Grey boxes indicate the D/E/N X S/T Φ motif. B. Relative GFP::POS-1 concentration along the A/P axis in WT (control RNAi) and plk-1(RNAi) embryos C. FRAP recovery curves for GFP::POS-1 in WT (control RNAi) and plk-1(RNAi) embryos. D. Images of GFP::POS-1; plk-1(as) embryos treated with DMSO control or 1NM-PP1. E. Posterior/anterior ratio of GFP::POS-1 concentration in DMSO control and 1NM-PP1 treated plk-1(as) embryos from 2 minutes before pronuclear meeting to 8 minutes after pronuclear meeting. F. In vitro kinase assay with hPLK1 and the indicated substrates. Top: phosphorylation was monitored using a thiophosphate ester antibody, which recognizes alkylated ATP-γS. Bottom: Coomassie stained gel. G. Quantification of in vitro kinase assays (n=4). Background was defined as the the values before kinase addition (t=0) for each reaction. Values within each experiment were normalized to the final MBP:POS-1 time point. See also Figure S4 and Table S1.
To determine whether PLK-1 kinase activity is required in the zygote to control POS-1 mobility, we generated an analog sensitive allele of PLK-1 (plk-1(as) hereafter) by introducing the C52V and L115G mutations [34] at the endogenous PLK-1 locus. GFP::POS-1;plk-1(as) embryos were permeabilized [35] and treated with either DMSO control or 10 μM 1NM-PP1, which inhibits analog sensitive kinases [36]. 12/13 embryos treated with 1NM-PP1 exhibited delayed nuclear envelope breakdown and failed cytokinesis, indicating that PLK-1 was efficiently inhibited under these conditions [37]. Importantly, GFP::POS-1 segregation was dramatically reduced in 1NM-PP1 treated embryos relative to control embryos (Figures 3D – 3E), indicating that PLK-1 kinase activity is required for GFP::POS-1 segregation.
Using in vitro kinase assays, we find that human PLK1 phosphorylates full length recombinant MBP:POS-1 but not MBP (Figures 3F – 3G). Alanine substitutions at all five potential PLK-1 phosphorylation sites within the region aa199–242 (MBP:POS-1(5A)) significantly reduced hPLK1 phosphorylation of POS-1 (Figures 3F – 3G). Alanine substitutions at Ser199 and Ser216 reduced phosphorylation to a similar extent (Figures 3F – 3G), suggesting these two residues are the primary hPLK1 phosphorylation sites within this region of POS-1. The remaining phosphorylation of MBP:POS-1(5A) by hPLK1 suggests the presence of additional PLK-1 phosphorylation site(s) in POS-1.
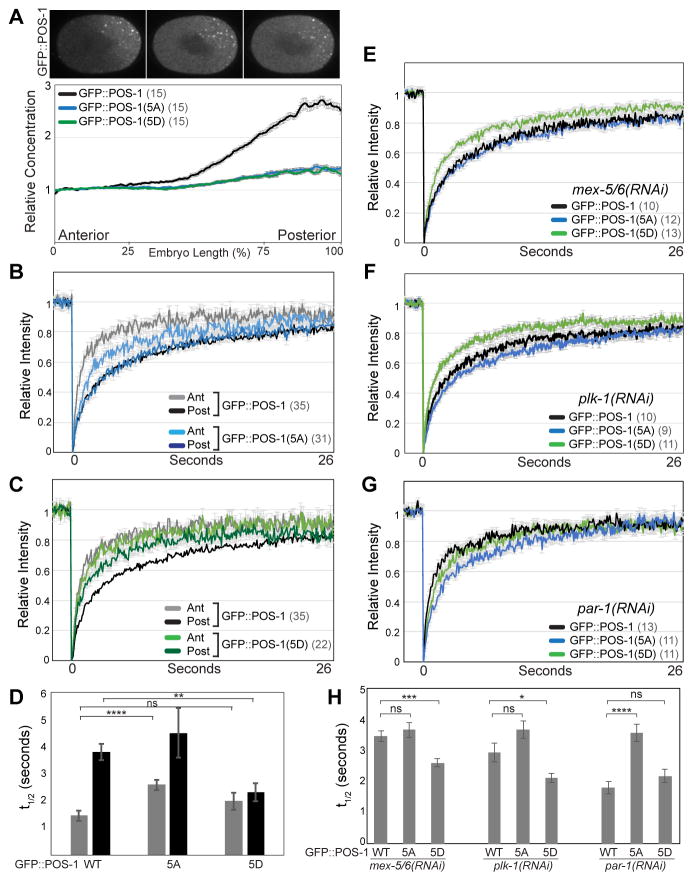
We next expressed GFP::POS-1 transgenes in which these five predicted PLK-1 phosphorylation sites were mutated to either non-phosphorylatible (GFP::POS-1(5A)) or phosphomimetic (GFP::POS-1(5D)) residues. GFP::POS-1(5A) forms a significantly weaker gradient than GFP::POS-1 (Figure 4A). Similarly, alanine substitutions at either Ser199 or Ser216 significantly reduce POS-1 segregation (Figure S4E), suggesting that these residues are the primary sites of POS-1 regulation by PLK-1. Importantly, GFP::POS-1(5A) is significantly less mobile than GFP::POS-1 in the anterior (Figures 4B and 4D and Table S1), consistent with the idea that phosphorylation increases POS-1 mobility in the anterior.
Figure 4. PLK-1 phosphorylation controls POS-1 mobility.
A. Top: Images of embryos expressing the indicated GFP::POS-1 alleles. Bottom: relative concentration of the indicated GFP::POS-1 alleles along the A/P axis. B, C. FRAP recovery curves for the indicated GFP::POS-1 alleles. D, H. Mean t1/2 for the indicated FRAP experiments. E–G. FRAP recovery curves for the indicated GFP::POS-1 alleles in mex-5/6(RNAi), plk-1(RNAi) and par-1(RNAi) embryos. As with all other FRAP experiments in this study, FRAP was performed simultaneously in the anterior and posterior cytoplasm. Because GFP::POS-1 mobility is uniform in mex-5/6(RNAi), plk-1(RNAi) and in par-1(RNAi) embryos, the anterior and posterior recovery curves for each genotype were averaged together. See also Figure S3 and Table S1.
Like GFP::POS-1(5A), GFP::POS-1(5D) forms a significantly weaker gradient than GFP::POS-1 (Figure 4A). However, in contrast to GFP::POS-1(5A), GFP::POS-1(5D) is significantly more mobile than GFP::POS-1 in the posterior (Figure 4C and 4D and Table S1), indicating that the phosphomimetic substitutions disrupt the retention of POS-1 in the posterior. These data are consistent with a model in which the differential in POS-1 mobility along the A/P axis result from a cycle of phosphorylation/dephosphorylation that increases POS-1 mobility in the anterior and decreases POS-1 mobility in the posterior. We note that both GFP::POS-1(5A) and GFP::POS-1(5D) exhibit some differential in mobility between the anterior and posterior (Figures 4B – 4D), raising the possibility that additional PLK-1 phosphorylation site(s) may contribute to the regulation of POS-1 mobility.
We next sought to determine whether the phosphorylation status of POS-1 regulates POS-1 mobility downstream of MEX-5/6 and PLK-1 function. In mex-5/6(RNAi) and plk-1(RNAi) embryos, the mobility of GFP::POS-1(5D) is significantly higher than either GFP::POS-1 or GFP::POS-1(5A) (Figures 4E, 4F, 4H and Table S1). Additionally, in par-1(RNAi) embryos in which MEX-5/6 are active throughout the cytoplasm, the mobility of GFP::POS-1(5A) is significantly lower than either GFP::POS-1 or GFP::POS-1(5D) (Figures 4G, 4H and Table S1). We conclude that phosphorylation of POS-1 by PLK-1 is both necessary and sufficient to increase the mobility of POS-1 downstream of MEX-5/6 and PLK-1 activity.
Interaction between MEX-5/6 and PLK-1 is required for GFP::POS-1 segregation
MBK-2 kinase phosphorylates MEX-5 T186 (and presumably MEX-6 at the analogous residue, T190), priming MEX-5/6 for Polo-box domain (PBD)-dependent binding of PLK-1 [23]. Consistent with the requirement for MBK-2 in the segregation of POS-1 [38, 39], we find that GFP::POS-1 mobility is uniformly slow in mbk-2(RNAi) embryos (Table S1). Using GFP::MEX-5 transgenes, Nishi et al showed that the mex-5(T186A) mutation does not affect MEX-5 gradient formation, but reduces embryonic viability and disrupts PIE-1 degradation in somatic cells, a MEX-5/6 dependent function [23, 40]. To test whether the interaction between PLK-1 and MEX-5 is required for the control of POS-1 dynamics, we introduced the T186A mutation at the endogenous mex-5 locus and analyzed GFP::POS-1 dynamics (Figure 5A). mex-5(T186A) mutant embryos are viable and exhibit defects in GFP::POS-1 dynamics consistent with a partial reduction in MEX-5/6 activity (Figures 5B, 5C, S5B, S5C and Table S1). Like mex-5/6(RNAi) embryos, mex-5(T186A);mex-6(RNAi) embryos are inviable and fail to segregate GFP::POS-1 (Figures 5B and 5C) because GFP::POS-1 mobility is uniformly slow (Figures 5D, 5E and Table S1). Furthermore, the mobility of GFP::POS-1(5D) is significantly faster than wild-type GFP::POS-1 in mex-5(T186A);mex-6(RNAi) embryos (Figures 5D, 5E and Table S1). These findings suggest that the interaction between MEX-5/6 and PLK-1 is required for the control of POS-1 mobility.
Previous studies have demonstrated that PLK-1 forms an anterior-rich cytoplasmic gradient in the polarized zygote [21–23]. We quantified the strength of the PLK-1 gradient using a strain in which PLK-1 was tagged at the endogenous locus with superfolder GFP [41] and found that PLK-1::sGFP forms a ~1.6 fold anterior-rich cytoplasmic gradient that depends on MEX-5/6 [23] (Figures 5F and 5G). The mobility of PLK-1::sGFP in wild-type embryos is slower in the anterior than the posterior cytoplasm (Figures 5H and Table S1). Depletion of MEX-5/6 increases the mobility of PLK-1::sGFP in the anterior cytoplasm (Figure 5H and Table S1), indicating that MEX-5/6 recruit PLK-1 into slow-diffusing complexes in the anterior cytoplasm. Importantly, the overall concentration of cytoplasmic PLK-1::sGFP is not affected by MEX-5/6 depletion (Figure 5F), supporting the conclusion that PLK-1 depends on its interaction with MEX-5/6 in order to control POS-1 mobility.
Discussion
We propose the following working model for POS-1 segregation (Figure 5I). Both MEX-5 and POS-1 can associate with slow-diffusing complexes in a manner that depends on their ability to bind RNA. PLK-1 interacts with MEX-5 and is recruited into slow-diffusing complexes in the anterior cytoplasm [12–14, 23], positioning PLK-1 to phosphorylate POS-1 and inhibit the formation of stable, slow-diffusing POS-1/RNA complexes in the anterior. Because the formation of slow-diffusing MEX-5 complexes requires MEX-5 RNA-binding [13], one possibility is that MEX-5/6 recruit PLK-1 to RNA in the anterior cytoplasm. In the posterior, the relatively low concentration of MEX-5/6 and PLK-1 provides a permissive environment for POS-1 to bind slow-diffusing RNA complexes, which retards POS-1 diffusion and results in the preferential retention of POS-1 in the posterior cytoplasm. Our model for POS-1 segregation is similar in many respects to that for MEX-5 segregation. MEX-5 mobility is controlled by PAR-1 kinase, which phosphorylates MEX-5 and is required to increase MEX-5 mobility in the posterior cytoplasm [12, 13], likely by decreasing the association of MEX-5 with slow-diffusing RNA complexes. The uniformly distributed phosphatase PP2A acts to reduce MEX-5 mobility and allow its retention in the anterior cytoplasm [13]. We predict the existence of an analogous cytoplasmic phosphatase(s) that dephosphorylates POS-1, thereby enabling its retention in the posterior.
Segregation of other germ plasm proteins in response to MEX-5/6
It was recently demonstrated that MEX-5/6 suppress P granule assembly/stability in the anterior cytoplasm through a competitive RNA-binding mechanism [19, 20]. In vitro, MEX-5/6 can compete with the P granule RNA-binding proteins MEG-3 and PGL-3 for RNA and thereby suppress the assembly of MEG-3 and PGL-3 into phase-separated liquid droplets. In vivo, suppression of MEG-3 phase separation in the anterior by MEX-5 depends on MEX-5 interaction with RNA and can be overcome by increasing the concentration of RNA in the embryo [19]. These observations indicate that MEX-5/6 can prevent the assembly/stability of P granules in the anterior by depleting the available pool of RNA required to assemble P granules. In contrast, competition between MEX-5/6 and POS-1 for RNA does not appear to underlie POS-1 responsiveness to MEX-5/6. First, the observation that GFP::POS-1(5A) decreases POS-1 mobility in the anterior suggests that the retention of POS-1 in the anterior is not limited by the availability of slow-diffusing RNAs. Second, our current model is that MEX-5 accumulates in the anterior through interaction with RNA [13], which a competitive binding model predicts would be sufficient to stimulate POS-1 segregation. However, in plk-1 and mbk-2 mutant embryos, MEX-5/6 segregates normally but does not drive POS-1 segregation [23, 38, 39]. The dynamics of P granules in plk-1/2 embryos have not been reported, and it will be interesting to learn whether PLK-1/2 contribute to P granule segregation or if they selectively target a subset of germ plasm components such as POS-1.
The mechanisms controlling the segregation of PIE-1 have not been fully elucidated. Nishi et al demonstrated PLK-1/2 are required for PIE-1 segregation [23], suggesting that the mechanisms controlling POS-1 segregation could also drive PIE-1 segregation. Importantly, however, the mechanisms that drive POS-1 and PIE-1 segregation appear to differ in at least two respects. First, our observation that PIE-1(ZFmut) exhibits only a partial defect in posterior retention indicates the existence of additional, unknown mechanisms that mediate PIE-1 retention. Second, in mbk-2 mutants, an unknown mechanism stimulates PIE-1 but not POS-1 segregation during mitosis [38]. One possible explanation for the segregation of PIE-1 in mbk-2 mutants could be a low level of residual MEX-5/6 interaction with PLK-1/2 that is sufficient to drive PIE-1, but not POS-1, segregation. Alternatively, MEX-5/6 may act through unknown, PLK-independent mechanisms that specifically regulate PIE-1 segregation. In the future, it will be important to determine to what extent the multiple predicted PLK-1/2 phosphorylation sites on PIE-1 regulate its dynamics.
Mechanisms linking the PLK-1 and MEX-5/6 gradients to POS-1 segregation
How does PLK-1 phosphorylation increase POS-1 diffusivity? The simplest model is that phosphorylation of POS-1 reduces its affinity for RNA, thereby inhibiting POS-1 association with slow-diffusing RNA complexes. Alternatively, phosphorylation may regulate POS-1 interactions with unknown factors that are required in addition to RNA-binding for the formation of slow-diffusing complexes. Consistent with the latter possibility, POS-1 lacking the C-terminus binds RNA with high affinity in vitro [28, 42], but exhibits uniformly fast mobility in vivo (GFP::POS-1(1–199)) (data not shown), suggesting that RNA-binding is required but not sufficient to retard POS-1 mobility. Understanding how the C-terminus contributes to the formation of slow-diffusing POS-1 complexes will provide a foundation for understanding how PLK-1 phosphorylation inhibits their formation.
Polo-like kinases control a wide range of processes during the cell cycle and are therefore subject to layers of regulation that ensure they are only active at the appropriate place and time [43]. The activation of polo-like kinases is often controlled by interaction of the PBD domain with binding partners, which relieves auto-inhibition of the kinase domain [43]. Consistent with such a mechanism controlling PLK-1 activity in the zygotic cytoplasm, Nishi et al found that PLK-1 interaction with MEX-5/6 increases PLK-1 kinase activity in vitro [23]. In addition, our finding that MEX-5 RNA-binding is required for control of POS-1 mobility suggests that PLK-1 must both interact with MEX-5/6 and be recruited to slow-diffusing complexes in order to inhibit POS-1 retention. In this respect, MEX-5/6 could be viewed as targeting factors that position PLK-1 in the cytoplasm where it can most efficiently control POS-1 segregation. Because depletion of MEX-5/6 only increases PLK-1::sGFP mobility in the anterior and not the posterior (Figure 5H), we predict that the gradient in “active” cytoplasmic PLK-1/MEX-5/6 complexes may be substantially stronger than the overall cytoplasmic PLK-1 gradient.
The posterior kinase PAR-1 phosphorylates MEX-5 and likely decreases the association of MEX-5 with slow-diffusing RNA complexes [13]. Taken together with the current finding that MEX-5/6 RNA-binding is required for control of POS-1 mobility, PAR-1 phosphorylation would be predicted to not only decrease MEX-5 concentration in the posterior, but to also prevent MEX-5/6 from targeting PLK-1 to POS-1 in the posterior cytoplasm. Consistent with this prediction, previous analysis of GFP::PIE-1 mobility showed that MEX-5/6 are more active in par-1 mutant embryos than in par-3 mutant embryos, which retain PAR-1 activity [15].
Our findings have three important implications for understanding how the zygotic cytoplasm in partitioned. First, only proteins whose mobility is increased by PLK-1 phosphorylation would be sensitive to the mechanisms described here, which could explain why only specific RNA-binding proteins redistribute to the posterior in response to MEX-5/6. Second, we propose that the combined requirement for PLK-1 interaction with MEX-5/6 and MEX-5/6 interaction with RNA converts their gradual concentration gradients into much stronger, switch-like activity gradients. The formation of such a switch-like activity gradient may explain the apparent amplification of the relatively weak PLK-1 concentration gradient as it gives rise to the significantly stronger POS-1 gradient. Third, we propose that the reaction/diffusion mechanisms by which PAR-1 patterns the MEX-5 gradient significantly impacts the subsequent reaction/diffusion mechanisms that pattern the POS-1 gradient. Because a series of reaction/diffusion mechanisms are thought to underlie the dramatic spatial reorganization of the zygote [7, 44], a full understanding of this process will likely require an integrated analysis of the mechanisms that amplify and dampen polarity cues as they propagate within the cortex and into the cytoplasm.
STAR METHODS
Contact for Reagent and Resource Sharing
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Erik Griffin (erik.e.griffin@dartmouth.edu).
Experimental Model and Subject Details
C. elegans hermaphrodite strains were maintained on Nematode Growth Medium (NGM) plates containing 3 g/L NaCl, 2.5 g/L peptone and 17 g/L agar supplemented with 1 mM CaCl2, 1 mM MgSO4, 1 mM KPO4 and 5 mg/L Cholesterol with E. coli OP50 as a source of food [46]. All strains were derived from the Bristol N2 wild-type strain and are listed in the Key Resources table. All strains were maintained at 20°C with the exception of transgenic strains, which were maintained at 25°C. Because the gene products analyzed in this study are maternally deposited in the zygote, we refer to the progeny of mothers homozygous for an indicated mutation as mutant embryos. For example, mex-5(ZFmut) mutant embryos are the progeny of mothers homozygous for the mex-5(ZFmut) mutation. RNAi treated embryos are the progeny of mothers exposed to dsRNA using the feeding protocol [47]. Briefly, HT115 bacteria (CGC) were transformed with an RNAi feeding plasmid derived from L4440 [47], grown in liquid culture and 110 μL were seeded on NGM plates containing 1 mM isopropyl-β-D-thiogalactoside and 25 μg/ml carbenicillin and allowed to dry overnight at room temperature. mex-5/6, par-1 and mbk-2 RNAi were performed by placing L4 animals on RNAi plates for 24 hours at 25°C. plk-1 RNAi was performed by placing late L3 animals on RNAi plates for 28 hours at 25°C.
Method Details
Molecular biology and transgenes
Single-copy transgenes were generated using the MosSCI approach [29]. All transgenes were expressed using the mex-5 promoter region [48] amplified from genomic DNA by PCR using the oligonucleotides BH0234 and BH0235. The mex-5 promoter region and PCR amplified GFP (using oligos BH0236 and BH0237 for POS-1 and BH0236 and BH0240 for PIE-1) and the POS-1 ORF + 3′UTR (oligos BH0238 and BH0239) or PIE-1 ORF + 3′UTR (oligos BH0241 and BH0242) were inserted into pCFJ350 (Addgene) [29] by Gibson assembly using the In-Fusion HD Cloning system (TaKaRa) to generate the MosSCI vectors pBH55 (GFP::POS-1) and pBH56 (GFP::PIE-1). pBH57 (GFP::POS-1(F121N, F164N)), pBH58 (GFP::PIE-1(Y121N, F206N)), pBH66 - pBH70 (GFP::POS-1 truncation alleles) and pBH74 – pBH78 (GFP::POS-1 with single alanine substitutions) were derived from pBH55. To generate strains carrying single-copy insertions, EG8079 oxTi179 II; unc-119(ed3) III (CGC, RRID:WB-STRAIN:EG8079) hermaphrodites were injected with 50 ng/μl MosSCI plasmids and two sgRNA plasmids, pXW7.01 and pXW7.02 (kind gift from Katya Voronina, University of Montana), that were used to excise the ttTi5605 Mos1 transposon. To clone GFP::POS-1 with mutations at the five predicted PLK-1 phosphorylation sites (S199, S210, S216, S237, T242), gBlock gene fragments bearing either 5A or 5D mutations were ordered from IDT (Coralville, IA) and were inserted into pBH55 to generate pBH71 and pBH72, respectively. The GFP::POS-1 transgene (EGD224) rescued pos-1(zu148) (CGC, RRID:WB-STRAINLJJ462) homozygous mutants to 100% embryonic viability, indicating that the GFP::POS-1 transgene is functional. GFP::POS-1 was expressed from transgenes because insertion of GFP at the endogenous pos-1 locus resulted in >90% sterility. GFP::PIE-1(ZFmut) was expressed from a transgene because we were unable to introduce the Y121N;F206N mutations at the PIE-1 locus, presumably because this allele is toxic.
Gene editing
With the exception of the PLK-1::sGFP strain, gene editing was performed using the dpy-10 Co-CRISPR/Cas9 approach [49]. All gene edits were confirmed by sequencing. To generate the MEX-5 F294N and F339N mutations, we used sgRNAs vectors pBH46 and pBH47 (derived from pRB1017 [49]) to create double strand breaks flanking the MEX-5 zinc fingers. The donor template was a PCR product amplified from a plasmid containing the MEX-5 F294N and F339N mutations using the oligonucleotides BH0144 and BH0145. The gain-of-function dpy-10(cn64) allele was used as a co-conversion marker with the gRNA plasmid pJA58 and the ssODN BH0271 (IDT) as the repair template [49]. Our injection mix included 50 ng/μL pDD162 that expresses Cas9 [50], 40 ng/μL sgRNA plasmids, 35 ng/μL PCR homology repair template, 15 ng/μL pJA58 and 300 nM dpy-10(cn64) ssODN homology repair template.
To introduce the MEX-5 T186A mutation at the endogenous mex-5 locus and to introduce the C52V and L115G mutations at the endogenous plk-1 locus (to generate the plk-1(as) allele), we assembled microinjection mixes consisting of 200 ng/μL crRNAs, 1.3 μg/μL tracrRNA (Dharmacon-GE Lifesciences), 100 ng/μL ssODN or 500 ng/μL PCR template and 0.67 μg/μL Cas9 protein (Dharmacon), as described [51]. These mixes were injected into the strain EGD224. For the MEX-5 T186A mutation, crRNAs BH0280 and BH0281 were used and the ssODN BH0282 served as the repair template (IDT). For PLK-1 C52V and L115G mutations, crRNAs BH0300 and BH0301 were used to create flanking double strand breaks and a gBlock (IDT) containing the C52V and L115G mutations amplified with oligos BH0303 and BH0304 served as the repair template. Unlike mex-5(zu199) animals [9], both mex-5(T186A) and mex-5(ZFmut) are viable and can be maintained as homozygous mutants, indicating that these mutants retain some MEX-5 function. Homozygous plk-1 (as) worms display no defects in embryonic viability.
The PLK-1::sGFP strain [41] was generated by Dhanya Cheereambathur (Desai lab) using the CRISPR –Cas9 approach described in [50]. This allele includes the GGRAGSG linker between PLK-1 and sGFP. The PLK-1::sGFP strain is 98.9% embryonic viable (n=446 embryos) at 20°C. We were unable to analyze homozygous mex-5(egx1[F294N, F339N]); plk-1(it17[plk-1::sGFP::loxp) embryos as these alleles are synthetically lethal. By immunofluorescence, PLK-1 was reported to localize to centrosomes and to metaphase chromosomes [37]. Similarly, PLK-1::sGFP is intensely concentrated on centrosomes and on metaphase chromosomes in the zygote. In addition, we see relatively weak PLK-1::sGFP signal on chromosomes and the nuclear envelope beginning during pronuclear centration (the images in the Figure 5F were taken at nuclear envelope breakdown).
Inhibition of plk-1(as)
The egg shell of GFP::POS-1; plk-1(as) embryos were permeabilized by treatment with perm-1(RNAi) [35] using the RNAi feeding method. perm-1(RNAi) bacteria were diluted with control L4440 (RNAi) bacteria (dilutions ranged from 1:10 to 1:20) and seeded on RNAi plates. GFP::POS-1 worms were incubated on these plates for 24 hours at 25°C. Embryos were dissected from mothers in 3 μL of L15 embryonic growth media [52]. Then, roughly one hundred 20 micron polysterene beads in 1 μL L15 embryonic growth media were added followed by 4 μL L15 embryonic growth media containing 20 μM 1NM-PP1 (Cayman Chemical) or DMSO control. Imaging began between 3 and 4 minutes from the time of drug addition. Only embryos that had exited meiosis and initiated polarization (as evidenced by smoothening of the posterior cortex) and were imaged for at least 2 minutes prior to pronuclear meeting were analyzed.
Microscopy
Images were collected on a Marianas spinning disk confocal microscope controlled by the Slidebook software package (Intelligent Imaging Innovations, Denver, CO) and built around a Zeiss Axio Observer Z.1 equipped with a Zeiss Plan-Apochromat 63×/1.4NA oil immersion objective, a CSU-X1 spinning disk (Yokogawa, Tokyo, Japan), an Evolve 512X512 EMCCD camera (Photometrics, Tucson, AZ) and a 50mW 488nm solid state laser. For all acquisitions, the camera intensification was 300 (out of 1000) and the gain was 1. Time lapse images (Figures S1F and S1G) were collected at the embryo midplane at 10 second intervals with 60% laser power and 300 msec exposures. Movies were temporally registered at pronuclear meeting. Images for gradient quantification were collected at nuclear envelope breakdown at the cell midplane with 60% laser power and 600 msec exposures.
FRAP experiments were performed using a Phasor photomanipulation unit (Intelligent Imaging Innovations), which delivered 488nm light simultaneously to two 4 μm diameter circular ROIs (one in the anterior and one in the posterior cytoplasm). Photobleaching lasted for 90 msec and images were collected at 93.1 msec per frame for 300 frames.
Kinase Assay
MBP:POS-1, MBP:POS-1(5A), MBP:POS-1(2A) and MBP were expressed in BL21(DE3)pLysS cells (Fisher Scientific). MBP was expressed from the pMAL-c2X vector that was modified to include three additional STOP codons to prevent translation read through of the single STOP codon in the original vector. Bacteria were induced with 0.5 mM IPTG and cultured overnight at 16°C. 100 μM Zn(OAc)2 was added at the time of induction. Bacterial pellets were lysed in Lysis Buffer (50 mM Tris-HCl, pH 8.0, 200 mM NaCl, 2 mM DTT, 100 μM Zn(OAc)2, 1 mM NaF, 2 mM Na3VO4, 1 μg/mL Pepstatin A, 1 mM PMSF, and supplemented by cOmplete EDTA-free Protease Inhibitor cocktail (Sigma-Aldrich)) by sonication (power 3.5, 15 pulses per round, 3 rounds), clarified by centrifugation at 17,000g for 30 minutes and bound in batch to Amylose Resin (NEB) for 2 hours at 4° C. Columns were washed with Wash Buffer (10 mM Tris-HCl, pH 8.0, 200 mM NaCl, 2 mM DTT, 100 μM Zn(OAc)2) and eluted with Wash Buffer + 10 mM Maltose.
Kinase assays were performed at 37°C by diluting protein (~0.2μg) into kinase reaction buffer (40 mM MOPS, pH 7.0, 100 μM Zn(OAc)2, 10 mM MgCl2) containing 10 mM ATP-γS (Abcam) and 21 ng hPLK1 (EMD Millipore). Samples were taken at the indicated time points and quenched with 20 mM EDTA. P-nitrobenzyl mesylate (Abcam) was added to 2.5 mM and samples were incubated for 2 hours at room temperature to alkylate ATP-γS, generating a thiophosphate ester. Kinase reactions were analyzed by Western Blot using 1:5000 thiophosphate ester specific primary antibody (Abcam) and 1:10,000 peroxidase-conjugated AffiniPure Goat anti-rabbit IgG secondary antibody (Jackson ImmunoResearch). Blots were developed with the Clarity Western ECL Substrate (Bio-Rad) and imaged with the ChemiDoc XRS system (Bio-Rad) with exposure time of 20 seconds. In parallel, reaction samples were run on SDS-PAGE gels and stained with Coomassie Brilliant Blue to quantify substrate levels.
Embryonic viability assays
For viability assays, late L4 larvae were grown for 24 hours at 20° C to generate staged adults. Ten adults were each picked to individual seeded NGM plates and allowed to lay eggs for two hours. Embryonic viability was calculated as the percentage of total eggs laid that hatched into L1s.
Western blotting
To quantify the expression levels of MEX-5 and MEX-5(ZFmut), 50 young adults were collected for each strain in 50 μl M9 buffer. 4× Laemmli sample buffer (Bio-Rad) was added to 1×, and samples were lysed by three freeze/thaw cycles on dry ice and in a 42° C water bath. DTT was then added to a final concentration of 200 mM before boiling at 98° C for 5 min. Equal volumes of each sample were loaded onto Any kD mini-PROTEAN TGX Gels (Bio-Rad). Samples were transferred to Immobilon-P transfer membrane (EMD Millipore) and probed with 1:500 guinea pig anti-MEX-5 primary antibody [13] and 1:10,000 peroxidase-conjugated AffiniPure Goat anti-guinea pig IgG secondary antibody (Jackson ImmunoResearch). The blot was then stripped with Restore PLUS Western Blot Stripping Buffer (Thermo Fisher Scientific) and probed with 1:600 mouse monoclonal anti-α-tubulin primary antibody DM1A (Sigma-Aldrich) and 1:5000 peroxidase-conjugated AffiniPure Goat anti-mouse IgG secondary antibody (Jackson Immuno Research). All antibodies were dissolved in TBST containing 5% milk. Blots were developed with the Clarity Western ECL Substrate (Bio-Rad) and imaged with the ChemiDoc XRS system (Bio-Rad) with an exposure time of 20 seconds.
Protein Purification for RNA binding assays
Recombinant MBP-tagged MEX-5 was purified as described previously [10] with some modification. Briefly, plasmids encoding wild-type MEX-5 TZF domains (pBH97: pHMTc-MEX-5(aa236–350)) or MEX-5(ZFmut) TZF domains (pBH98: pHMTc-MEX-5(aa236–350; F294N, F339N)) were transformed into E. coli strain BL21. Protein expression was induced for three hours in mid-log phase with 1 mM IPTG. Cells were disrupted in lysis buffer (200 mM NaCl, 50 mM Tris-Cl, pH 8.0, 100 μM Zn(OAc)2, 2 mM DTT), clarified extract was loaded onto an amylose affinity column (New England Biolabs), washed extensively, then eluted with lysis buffer supplemented with 10 mM maltose. Fractions containing the protein were dialyzed into low salt Q column buffer (20 mM NaCl, 50 mM Tris-Cl, pH 8.8, 100 μM Zn(OAc)2, 2 mM DTT) overnight, and then loaded onto a HiTrap Q column (GE HealthCare). The protein was eluted across a 20 mM to 1 M NaCl gradient. Fractions containing the protein were combined and dialyzed into low salt S column buffer (20 mM NaCl, 50 mM MOPS, pH 6.0, 100 μM Zn(OAc)2, 2 mM DTT) overnight. The dialyzed sample was loaded onto a HiTrap S column (GE HealthCare), and fractions containing the protein were collected across a gradient from 20 mM to 1M NaCl. The eluted protein was dialyzed into storage buffer (25 mM NaCl, 25 mM Tris-Cl, pH 8.0, 100 μM Zn(OAc)2, 2 mM DTT) overnight and stored at 4°C until use. The protein concentration was estimated from its UV absorbance at 280 nm and overall purity was estimated by SDS-PAGE. We note that the presence of apparent proteolysis products that co-purify with both MEX-5 variants.
Recombinant MBP-tagged POS-1 was purified as described previously [42]. In brief, the plasmid encoding wild-type POS-1 TZF domains (pHMTc-POS-1(aa80–180) [42]) and POS-1(ZFmut) TZF domains (pBH96: pHMTc-POS-1(aa80–180; F121N, F164N)) were transformed into BL21, the protein was induced, cells were lysed, and the protein purified over an amylose column as described for MEX-5 above. Fractions containing the protein were combined and dialyzed into low salt S buffer overnight. The protein was loaded over a HiTrap S column. For POS-1(aa80–180), the flow through was collected, then dialyzed into low salt Q buffer overnight. The protein was then purified over a HiTrap Q column as described for MEX-5 above. POS-1(aa80–180; F121N, F164N) protein did not flow through the HiTrap S column, presumably due to its reduced RNA-binding activity and concomitant reduction in associated negative charge. Therefore, this protein was eluted from the column across a gradient from 20 mM to 1 M NaCl. Both variants were dialyzed in storage buffer and stored as MEX-5 above. The concentration of each was estimated from its UV absorbance at 280 nm, and purity assessed by SDS-PAGE.
EMSA
Electrophoretic mobility shift assays were performed as previously described [42]. Trace FAM-labeled RNA oligonucleotides (2.9 nM, IDT) corresponding to fragments of the glp-1 3′UTR (SCR+: 5′-UUUUU CUUAU UCUAG ACUAA UAUUG UAAGC U-Fl-3′) or the TNFα 3′UTR (TNF-ARE: 5′-GUGAU UAUUU AUUAU UUAUU UAUUA UUUAU UUAUU AG-Fl-3′) were equilibrated with varying concentrations of purified recombinant POS-1 or MEX-5, respectively, in binding buffer (50 mM Tris, pH 8.0, 100 mM NaCl, 0.01 mg/mL tRNA, 0.01% (v/v) IGEPAL, and 100 μM ZnOAc2) for at least 3 hours. Prior to loading onto a 5% polyacrylamide 1XTB slab gel, 10 μL of 0.005% (w/v) bromocresol green in 30% glycerol was added to 100 μL of each sample. The gels were run at 120V in a 4°C room for 90 minutes, then analyzed on a Fuji FLA-9000 laser imager to detect bound and free RNA. The fraction of bound RNA was quantified using MultiGauge software (Fuji) and fit to the Hill Equation using Igor Pro (WaveMetrics) to determine the apparent binding constant, as previously described [10].
Quantification and Statistical Analysis
For quantification of the mean intensity in the anterior and posterior cytoplasm (Figures 1F, 1G and 3E), the mean fluorescence intensities in the anterior half, the posterior half, and throughout the zygote were determined at each time point. The background signal from outside the embryo was subtracted and fluorescence intensities were normalized to the initial total value for each embryo. For line scan analysis, fluorescence intensities were averaged for 15-pixel line scans at both 25% and 75% embryo length along the D/V axis for each embryo. The fluorescence intensities along the A/P axis were normalized to the average of the first 10 pixels of the end with low concentration (the posterior for PLK-1 and the anterior for POS-1 and PIE-1). To average the fluorescence values among multiple embryos that differ slightly in length, the signal at 201 regularly spaced points along the A/P axis was estimated using linear interpolation with the interp1 function in Matlab (version R2015a, Mathworks, Natick, MA) [15].
FRAP curves were corrected for photobleaching during recovery by normalizing to a fluorescence intensity within a 4μm diameter circle outside of bleach region. A region of the same size outside of the embryo was selected as the background. The fluorescence intensity immediately following photobleaching was set to zero, and the background-subtracted intensities were normalized to the average of the pre-bleach values. The final relative intensity was then averaged among the indicated number of embryos. t1/2 and percent recovery values were calculated in Prism6 (GraphPad Software Inc, La Jolla, CA) by fitting to a one-phase association non-linear regression curve. Because FRAP assays were performed simultaneously in the anterior and posterior cytoplasm, statistical comparisons between the anterior and posterior cytoplasm were performed using a paired student t-test with Welch’s correction. Statistical comparisons between embryos with different genotypes were performed using an unpaired student t-test with Welch’s correction.
Supplementary Material
Highlights.
POS-1 segregation requires the interaction of MEX-5 with RNA and PLK-1
PLK-1 phosphorylation increases POS-1 mobility in the anterior cytoplasm
RNA-binding mediates the retention of POS-1 in the posterior
Acknowledgments
We thank Sharon Bickel, Bing He, Katya Voronina and members of the Griffin lab for comments on the manuscript. We thank Dhanya Cheerambathur and Arshad Desai (UCSD) for the PLK-1::sGFP strain, Craig Mello (UMass) for the PIE-1::GFP strain, Katya Voronina (U. Montana) and Josh Arribere and Andy Fire (Stanford) for sgRNA plasmids. Some C. elegans strains were provided by the Caenorhabditis Genetics Center, which is funded by the NIH Office of Research Infrastructure Programs (P40 OD010440). Work in the Griffin lab is supported by NIH Grant R01GM110194 to E.E.G.. Work in the Ryder lab is supported by NIH Grant R01GM117237 to S.P.R.
Footnotes
Author Contributions
B.H and E.G. designed the project, performed experiments, analyzed results and wrote the manuscript. K.R.A. performed the EMSA experiments. S.P.R analyzed the EMSA data. M.R. performed the experiments in the Figure S1F and S1G. X.F. generated the strains in Figure S4E.
Declaration of Interests
The authors declare no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Wartlick O, Kicheva A, Gonzalez-Gaitan M. Morphogen gradient formation. Cold Spring Harbor Perspectives in Biology. 2009;1:a001255. doi: 10.1101/cshperspect.a001255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brown GC, Kholodenko BN. Spatial gradients of cellular phospho-proteins. FEBS Letters. 1999;457:452–454. doi: 10.1016/s0014-5793(99)01058-3. [DOI] [PubMed] [Google Scholar]
- 3.Lipkow K, Odde DJ. Model for protein concentration gradients in the cytoplasm. Cellular and Molecular Bioengineering. 2008;1:84–92. doi: 10.1007/s12195-008-0008-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kiekebusch D, Thanbichler M. Spatiotemporal organization of microbial cells by protein concentration gradients. Trends in Microbiology. 2014;22:65–73. doi: 10.1016/j.tim.2013.11.005. [DOI] [PubMed] [Google Scholar]
- 5.Fuller BG. Self-organization of intracellular gradients during mitosis. Cell Division. 2010;5:1–21. doi: 10.1186/1747-1028-5-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Howard M. How to build a robust intracellular concentration gradient. Trends Cell Biology. 2012;22:311–317. doi: 10.1016/j.tcb.2012.03.002. [DOI] [PubMed] [Google Scholar]
- 7.Griffin EE. Cytoplasmic localization and asymmetric division in the early embryo of Caenorhabditis elegans. Wiley Interdisciplinary Reviews Developmental Biology. 2015;4:267–282. doi: 10.1002/wdev.177. [DOI] [PubMed] [Google Scholar]
- 8.Guo S, Kemphues KJ. par-1, a gene required for establishing polarity in C. elegans embryos, encodes a putative Ser/Thr kinase that is asymmetrically distributed. Cell. 1995;81:611–620. doi: 10.1016/0092-8674(95)90082-9. [DOI] [PubMed] [Google Scholar]
- 9.Schubert CM, Lin R, de Vries CJ, Plasterk RH, Priess JR. MEX-5 and MEX-6 function to establish soma/germline asymmetry in early C. elegans embryos. Molecular Cell. 2000;5:671–682. doi: 10.1016/s1097-2765(00)80246-4. [DOI] [PubMed] [Google Scholar]
- 10.Pagano JM, Farley BM, Mccoig LM, Ryder SP. Molecular basis of RNA recognition by the embryonic polarity determinant MEX-5. Journal of Biological Chemistry. 2007;282:8883–8894. doi: 10.1074/jbc.M700079200. [DOI] [PubMed] [Google Scholar]
- 11.Cuenca AA, Schetter A, Aceto D, Kemphues K, Seydoux G. Polarization of the C. elegans zygote proceeds via distinct establishment and maintenance phases. Development. 2003;130:1255–1265. doi: 10.1242/dev.00284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tenlen JR, Molk JN, London N, Page BD, Priess JR. MEX-5 asymmetry in one-cell C. elegans embryos requires PAR-4- and PAR-1-dependent phosphorylation. Development. 2008;135:3665–3675. doi: 10.1242/dev.027060. [DOI] [PubMed] [Google Scholar]
- 13.Griffin EE, Odde DJ, Seydoux G. Regulation of the MEX-5 gradient by a spatially segregated kinase/phosphatase cycle. Cell. 2011;146:955–968. doi: 10.1016/j.cell.2011.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Daniels BR, Dobrowsky TM, Perkins EM, Sun SX, Wirtz D. MEX-5 enrichment in the C. elegans early embryo mediated by differential diffusion. Development. 2010;137:2579–2585. doi: 10.1242/dev.051326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu Y, Zhang H, Griffin EE. Coupling between cytoplasmic concentration gradients through local control of protein mobility in the Caenorhabditis elegans zygote. Molecular Biology of the Cell. 2015;26:2963–2970. doi: 10.1091/mbc.E15-05-0302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Daniels BR, Perkins EM, Dobrowsky TM, Sun SX, Wirtz D. Asymmetric enrichment of PIE-1 in the Caenorhabditis elegans zygote mediated by binary counterdiffusion. Journal of Cell Biology. 2009;184:473–479. doi: 10.1083/jcb.200809077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Voronina E. The diverse functions of germline P-granules in Caenorhabditis elegans. Molecular Reproduction and Development. 2013;80:624– 631. doi: 10.1002/mrd.22136. [DOI] [PubMed] [Google Scholar]
- 18.Updike D, Strome S. P granule assembly and function in Caenorhabditis elegans germ cells. Journal of Andrology. 2010;31:53–60. doi: 10.2164/jandrol.109.008292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Smith J, Calidas D, Schmidt H, Lu T, Rasoloson D, Seydoux G. Spatial patterning of P granules by RNA-induced phase separation of the intrinsically-disordered protein MEG-3. Elife. 2016;5:e21337. doi: 10.7554/eLife.21337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Saha S, Weber CA, Nousch M, Adame-Arana O, Hoege C, Hein MY, Osborne-Nishimura E, Mahamid J, Jahnel M, Jawerth L, et al. Polar positioning of phase-separated liquid compartments in cells regulated by an mRNA competition mechanism. Cell. 2016;166:1572–1584. doi: 10.1016/j.cell.2016.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rivers DM, Moreno S, Abraham M, Ahringer J. PAR proteins direct asymmetry of the cell cycle regulators Polo-like kinase and Cdc25. Journal of Cell Biology. 2008;180:877–885. doi: 10.1083/jcb.200710018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Budirahardja Y, Gonczy P. PLK-1 asymmetry contributes to asynchronous cell division of C. elegans embryos. Development. 2008;135:1303–1313. doi: 10.1242/dev.019075. [DOI] [PubMed] [Google Scholar]
- 23.Nishi Y, Rogers E, Robertson SM, Lin R. Polo kinases regulate C. elegans embryonic polarity via binding to DYRK2-primed MEX-5 and MEX-6. Development. 2008;135:687–697. doi: 10.1242/dev.013425. [DOI] [PubMed] [Google Scholar]
- 24.Hudson BP, Martinez-Yamout MA, Dyson HJ, Wright PE. Recognition of the mRNA AU-rich element by the zinc finger domain of TIS11d. Nature Structure and Molecular Biology. 2004;11:257–264. doi: 10.1038/nsmb738. [DOI] [PubMed] [Google Scholar]
- 25.Lai WS, Kennington EA, Blackshear PJ. Interactions of CCCH zinc finger proteins with mRNA: non-binding tristetraprolin mutants exert an inhibitory effect on degradation of AU-rich element-containing mRNAs. Journal of Biological Chemistry. 2002;277:9606–9613. doi: 10.1074/jbc.M110395200. [DOI] [PubMed] [Google Scholar]
- 26.Wang JT, Smith J, Chen B-C, Schmidt H, Rasoloson D, Alexandre AP, Lambrus BG, Calidas D, Betzig E, Seydoux G. Regulation of RNA granule dynamics by phosphorylation of serine-rich, intrinsically disordered proteins in C. elegans. eLife. 2014;3:e04591. doi: 10.7554/eLife.04591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen JX, Cipriani PG, Mecenas D, Polanowska J, Piano F, Gunsalus KC, Selbach M. In vivo interaction proteomics in Caenorhabditis elegans embryos provides new insights into P Granule dynamics. Molecular and Cellular Proteomics. 2016;15:1642–1657. doi: 10.1074/mcp.M115.053975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Farley BM, Ryder SP. POS-1 and GLD-1 repress glp-1 translation through a conserved binding site cluster. Molecular Biology of the Cell. 2012;23:4473–4483. doi: 10.1091/mbc.E12-03-0216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Frokjaer-Jensen C, Davis MW, Ailion M, Jorgensen EM. Improved Mos1-mediated transgenesis in C. elegans. Nature Methods. 2012;9:117–118. doi: 10.1038/nmeth.1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reese K, Dunn M, Waddle J, Seydoux G. Asymmetric segregation of PIE-1 in C. elegans Is mediated by two complementary mechanisms that act through separate PIE-1 protein domains. Molecular Cell. 2000;6:445–455. doi: 10.1016/s1097-2765(00)00043-5. [DOI] [PubMed] [Google Scholar]
- 31.Konwerski J, Senchuk M, Petty E, Lahaie D, Schisa JA. Cloning and expression analysis of pos-1 in the nematodes Caenorhabditis briggsae and Caenorhabditis remanei. Developmental Dynamics. 2005;233:1006–1012. doi: 10.1002/dvdy.20421. [DOI] [PubMed] [Google Scholar]
- 32.Liu Z, Ren J, Cao J, He J, Yao X, Jin C, Xue Y. Systematic analysis of the Plk-mediated phosphoregulation in eukaryotes. Briefings in Bioinformatics. 2013;14:344–360. doi: 10.1093/bib/bbs041. [DOI] [PubMed] [Google Scholar]
- 33.Santamaria A, Wang B, Elowe S, Malik R, Zhang F, Bauer M, Schmidt A, Sillje HH, Korner R, Nigg EA. The Plk1-dependent phosphoproteome of the early mitotic spindle. Molecular and Cellular Proteomics. 2011;10:M110.004457. doi: 10.1074/mcp.M110.004457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Woodruff JB, Wueseke O, Viscardi V, Mahamid J, Ochoa SD, Bunkenborg J, Widlund PO, Pozniakovsky A, Zanin E, Bahmanyar S, et al. Centrosomes. Regulated assembly of a supramolecular centrosome scaffold in vitro. Science. 2015;348:808–812. doi: 10.1126/science.aaa3923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Carvalho A, Olson SK, Gutierrez E, Zhang K, Noble LB, Zanin E, Desai A, Groisman A, Oegema K. Acute drug treatment in the early C. elegans embryo. PLoS ONE. 2011;6:e24656. doi: 10.1371/journal.pone.0024656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bishop AC, Ubersax JA, Petsch DT, Matheos DP, Gray NS, Blethrow J, Shimizu E, Tsien JZ, Schultz PG, Rose MD, et al. A chemical switch for inhibitor-sensitive alleles of any protein kinase. Nature. 2000;407:395–401. doi: 10.1038/35030148. [DOI] [PubMed] [Google Scholar]
- 37.Chase D, Serafinas C, Ashcroft N, Kosinski M, Longo D, Ferris DK, Golden A. The polo-like kinase PLK-1 is required for nuclear envelope breakdown and the completion of meiosis in Caenorhabditis elegans. Genesis. 2000;26:26–41. doi: 10.1002/(sici)1526-968x(200001)26:1<26::aid-gene6>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
- 38.Pellettieri J, Reinke V, Kim SK, Seydoux G. Coordinate activation of maternal protein degradation during the egg-to-embryo transition in C. elegans. Developmental Cell. 2003;5:451–462. doi: 10.1016/s1534-5807(03)00231-4. [DOI] [PubMed] [Google Scholar]
- 39.Pang KM, Ishidate T, Nakamura K, Shirayama M, Trzepacz C, Schubert CM, Priess JR, Mello CC. The minibrain kinase homolog, mbk-2, is required for spindle positioning and asymmetric cell division in early C. elegans embryos. Developmental Biology. 2004;265:127–139. doi: 10.1016/j.ydbio.2003.09.024. [DOI] [PubMed] [Google Scholar]
- 40.DeRenzo C, Reese KJ, Seydoux G. Exclusion of germ plasm proteins from somatic lineages by cullin-dependent degradation. Nature. 2003;424:685–689. doi: 10.1038/nature01887.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Martino L, Morchoisne-Bolhy S, Cheerambathur DK, Van Hove L, Dumont J, Joly N, Desai A, Doye V, Pintard P. Channel nucleoporins recruit PLK-1 to nuclear pore complexes to direct nuclear envelope breakdown in C. elegans. Developmental Cell. 2017;43:157–171. doi: 10.1016/j.devcel.2017.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Farley BM, Pagano JM, Ryder SP. RNA target specificity of the embryonic cell fate determinant POS-1. RNA. 2008;14:2685–2697. doi: 10.1261/rna.1256708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zitouni S, Nabais C, Jana SC, Guerrero A, Bettencourt-Dias M. Polo-like kinases: structural variations lead to multiple functions. Nature Reviews Molecular Cell Biology. 2014;15:433–452. doi: 10.1038/nrm3819. [DOI] [PubMed] [Google Scholar]
- 44.Hoege C, Hyman AA. Principles of PAR polarity in Caenorhabditis elegans embryos. Nature Reviews Molecular Cell Biology. 2013;14:315–322. doi: 10.1038/nrm3558. [DOI] [PubMed] [Google Scholar]
- 45.Kim H, Ishidate T, Ghanta KS, Seth M, Conte D, Shirayama M, Mello CC. A Co-CRISPR strategy for efficient genome editing in Caenorhabditis elegans. Genetics. 2014;197:1069–1080. doi: 10.1534/genetics.114.166389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Timmons L, Fire A. Specific interference by ingested dsRNA. Nature. 1998;395:854. doi: 10.1038/27579. [DOI] [PubMed] [Google Scholar]
- 48.Merritt C, Rasoloson D, Ko D, Seydoux G. 3′ UTRs are the primary regulators of gene expression in the C. elegans germline. Current Biology. 2008;18:1476–1482. doi: 10.1016/j.cub.2008.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Arribere JA, Bell RT, Fu BXH, Artiles KL, Hartman PS, Fire AZ. Efficient marker-free recovery of custom genetic modifications with CRISPR/Cas9 in Caenorhabditis elegans. Genetics. 2014;198:837–846. doi: 10.1534/genetics.114.169730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dickinson DJ, Ward JD, Reiner DJ, Goldstein B. Engineering the Caenorhabditis elegans genome using Cas9-triggered homologous recombination. Nature Methods. 2013;10:1028–1034. doi: 10.1038/nmeth.2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Paix A, Folkmann A, Rasoloson D, Seydoux G. High efficiency, homology-directed genome editing in Caenorhabditis elegans using CRISPR-Cas9 ribonucleoprotein complexes. Genetics. 2015;201:47–54. doi: 10.1534/genetics.115.179382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Strange K, Christensen M, Morrison R. Primary culture of Caenorhabditis elegans developing embryo cells for electrophysiological, cell biological and molecular studies. Nature Protocols. 2007;2:1003–1012. doi: 10.1038/nprot.2007.143. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.