Philadelphia-negative neutrophilic leukemias—atypical chronic myeloid leukemia (aCML), chronic neutrophilic leukemia (CNL), MDS/MPNu (myelodysplastic/myeloproliferative neoplasm, unclassifiable), and MPNu (myeloproliferative neoplasm, unclassifiable)—are rare hematologic neoplasms characterized by leukocytosis, a hypercellular bone marrow comprised predominantly of granulocytic cells, absence of the Philadelphia chromosome (t(9;22); BCR-ABL1), and absence of PDGFRA/B or FGFR1 gene rearrangements. Occasional cases of CNL1 and MDS/MPNu,2 as well as a majority of aCML cases, exhibit non-specific cytogenetic abnormalities3 or the JAK2 V617F mutation,4 revealing their clonal nature. The primary genetic basis of these leukemias was unknown until recent work by Maxson et al.,5 which highlighted mutations in CSF3R as key leukemogenic drivers in ~ 60% of patients with aCML or CNL. Subsequent studies found CSF3R mutations in a majority of patients with CNL and only in a minority of patients with aCML,6 with parallel studies of aCML revealing the presence of recurrent SETBP1 and ETNK1 mutations.7,8 Despite these discoveries, knowledge of the full genetic landscape of Philadelphia-negative neutrophilic leukemias remains incomplete. To identify and characterize additional mutations contributing to the pathogenesis of these malignancies, we performed whole exome sequencing (WES) on 116 Philadelphia-negative neutrophilic leukemia samples (Supplementary Materials and Methods).
We identified 4/116 samples harboring mutations in CCND2, the gene encoding the cell cycle regulator cyclin D2. Of these four samples, one was pathologically confirmed as MPNu, while another was confirmed as MDS/MPNu (Supplementary Table 1). Interestingly, all mutated samples harbored variants in codon 281, with three samples possessing a P281S variant and the other carrying a P281L variant (Supplementary Table 1). These changes appeared to occur as heterozygous mutations and were present only in patients lacking CSF3R mutations, with one sample (13-00010) harboring a known pathogenic SETBP1 variant (I871T), and another (14-00247) possessing an SRSF2 P95H variant. Two out of four samples harbored a TET2 mutation (other observed mutations in Supplementary Table 2). All mutations with additional available DNA were confirmed by Sanger sequencing (Supplementary Figure 1).
To determine the potential incidence of CCND2 mutations in other hematologic malignancies, we interrogated cohorts of patients with AML (n = 239), BCR-ABL1-positive CML (CML; n = 44), chronic myelomonocytic leukemia (CMML; n = 24), B-cell acute lymphoblastic leukemia (B-ALL; n = 49), or T-ALL (n = 7) by whole exome sequencing for the presence of variants in CCND2 (Supplementary Table 2). We identified three AML patients harboring the same CCND2 variants (two P281S; one P281L), all confirmed as somatic (Supplementary Table 3). These findings are consistent with two recent reports of similar CCND2 mutations in AML.9,10 No mutations in CCND2 were detected in patients with CML, CMML, B-ALL or T-ALL (Supplementary Table 2). Taken together, these results suggest the presence of CCND2 mutations may unify a small but recurrent and selective subset of patients with the molecularly heterogeneous myeloid malignancies of Philadelphia-negative neutrophilic leukemias and AML.
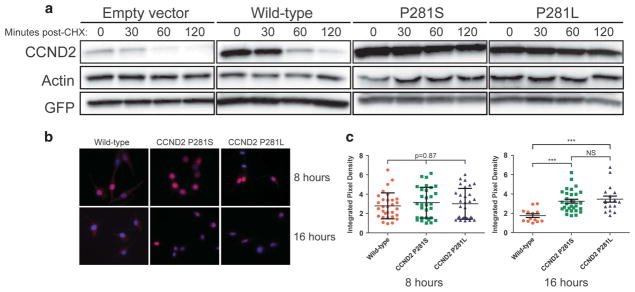
Previous work examining a missense variant in CCND2 (T280A), just one amino acid upstream of P281, demonstrated the variant to confer resistance to degradation.11 To assess whether the nearby variants identified in our patient cohort resulted in similar protein accumulation, NIH-3T3 cell lines stably overexpressing either empty vector, wild-type CCND2, or mutant CCND2 were treated with cycloheximide to block protein translation, and protein extracts from cells were analyzed by Western blotting (Figure 1a). While a rapid reduction in cyclin D2 protein was observed in cells overexpressing wild-type cyclin D2, this was not observed in cells overexpressing the mutant constructs. Therefore, CCND2 P281 variants result in the accumulation of degradation-resistant cyclin D2.
Figure 1.
CCND2 P281 variants are resistant to degradation and nuclear export. (a) Protein extracts from NIH-3T3 cells stably expressing each construct (top labels) were treated with cycloheximide (CHX) and were analyzed by western blotting using an antibody against CCND2. The wild-type CCND2 construct shows clear protein reduction by 60 min post-CHX treatment, while mutant constructs show relatively stable levels of protein, even by 120 min post-CHX treatment. (b) Representative immunofluorescence images of NIH-3T3 cells stably expressing either wild-type or CCND2 P281 variants. Cells were serum starved for 20 h in RPMI containing 0.1% FBS, reintroduced to serum, and then collected for immunofluorescence analysis at the specified time points post-serum reintroduction. Eight hours and 16 h post-reintroduction were chosen as collect time points because they represent time points at which the majority of wild-type CCND2 is nuclear and cytoplasmic, respectively. Red =CCND2, blue =DAPI. (c) Quantification of mean pixel density in the nucleus to establish nuclear localization of CCND2 constructs. At least ten cells were analyzed for each condition. ***P<0.0001 by one-way ANOVA and subsequent Tukey’s post hoc tests. Error bars show s.d.
Interestingly, the CCND2 T280A variant is also associated with constitutively nuclear localization of the protein.11 Because of the proximity of the CCND2 P281 variants to the previously reported T280A variant, we hypothesized that CCND2 P281 variants would be constitutively nuclear in their localization. While overexpressed wild-type CCND2 exhibited predominantly nuclear localization during G1 phase of the cell cycle and cytoplasmic staining during S phase, CCND2 P281 variant protein remained predominantly nuclear in its localization regardless of cell cycle phase (Figure 1b). Quantification of mean pixel density in the nucleus confirmed statistical significance of this difference (P<0.0001; Figure 1c).
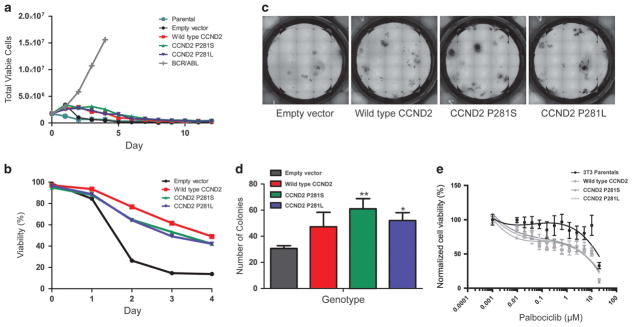
Variants in cyclin D1 have been reported at analogous residues to P281S and P281L of cyclin D2, exhibiting predominantly nuclear localization and oncogenic properties.12 We therefore assessed the transformative capacity of CCND2 P281 variants by generating Ba/F3 cell lines stably overexpressing either wild-type CCND2, CCND2 P281S or CCND2 P281L and removing IL-3 from their growth medium to assess for growth factor independence. As shown in Figure 2a, neither wild-type nor mutant CCND2 transformed Ba/F3 cells to IL-3-independent growth. However, cell lines that overexpressed CCND2 relative to cells expressing an empty control vector exhibited clear delays in cell death, suggesting that high levels of CCND2 expression can support enhanced cell survival (Figure 2b).
Figure 2.
Assessment of the oncogenicity and CDK4/6 inhibitor sensitivity of CCND2 P281 variants. (a) IL-3 withdrawal assay demonstrating the dependence of all CCND2-expressing lines on IL-3 for proliferation. (b) Normalized cell viability (relative to day 0 of IL-3 withdrawal) over time. Cells expressing CCND2 constructs show delayed decreases in viability relative to cells expressing an empty control vector. (c) Representative images of wells containing murine hematopoietic colonies grown in Methocult M3534 (supplemented with murine IL-3, IL-6 and SCF). Images were taken after 14 days of incubation. (d) Quantification of colony number, performed in triplicate, for murine hematopoietic cells expressing the labeled constructs. Colony number was compared between groups via one-way ANOVA followed by Tukey’s post hoc tests. **P<0.01 for comparison of colony number in empty vector-expressing cells versus the number in P281L-expressing cells. *P<0.05 for comparison of colony number in empty vector-expressing cells versus the number in P281L-expressing cells. No significant differences in colony numbers existed between wild-type CCND2-expressing cells and P281 variant-expressing cells. Error bars show s.d. (e) Sensitivity of CCND2-expressing NIH-3T3 cells to CDK4/6 inhibition. Cells expressing cyclin D2 constructs show increased sensitivity to palbociclib (a CDK4/6 inhibitor) relative to parental NIH-3T3 cells. Error bars show standard error of the mean for each data point.
Although CCND2 P281 variants did not confer growth factor independence in the Ba/F3 model, past work had demonstrated a role for CCND2 in cytokine-mediated proliferation of bone marrow progenitor cells.13 We thus examined whether expression of CCND2 P281 variants promoted the formation of mouse bone marrow progenitors in response to cytokines. Expression of CCND2 P281 variant constructs significantly enhanced the formation of colonies by mouse bone marrow progenitors relative to progenitors expressing an empty control vector, suggesting that enforced expression of these variants can enhance the proliferative potential of murine hematopoietic progenitors (Figures 2c and d). Notably, however, there were no significant differences in colony formation between wild-type CCND2-over-expressing cells and CCND2 variant-containing cells, presumably due to the wild-type CCND2 being overexpressed at levels that mimic the phenotype of the P281 variants (Figure 2d).
Cyclin D2 is synthesized in response to mitogens, whereupon it complexes with the cyclin-dependent kinases CDK4 and CDK6, leading to pRb inactivation and E2F-mediated upregulation of genes required for S phase entry.14 In recent years, inhibition of CDK4/6 has demonstrated effectiveness in various malignancies. Therefore, given the functional connection between cyclin D2 and CDK4/6, we hypothesized that cells expressing CCND2 P281 variants would be sensitive to CDK4/6 inhibition. While parental NIH-3T3 cells did not exhibit sensitivity to palbociclib, cell lines overexpressing either wild-type cyclin D2 or CCND2 P281 variants showed relatively increased sensitivity (Figure 2e). The results, therefore, indicate that leukemias that overexpress cyclin D2 may be susceptible to CDK4/6 inhibition.
The study herein provides evidence of CCND2 mutations in Philadelphia-negative neutrophilic leukemias that result in the overexpression and mislocalization of cyclin D2 protein, as well as increased survival and proliferation of hematopoietic cells. D-type cyclins are arbiters of cell cycle progression, especially between G1′ and S phase, and are frequently dysregulated in cancer. Of the D-type cyclins, cyclin D1 is the best-studied, with past work demonstrating its role in a variety of malignancies. While less is known about cyclin D2, its overexpression occurs in several cancer types.15–17 Recent work in T-cell acute lymphoblastic leukemias (T-ALL) has demonstrated leukemogenic chromosomal translocations involving cyclin D2 and the T-cell receptor,18 and over-expression of cyclin D2 has also been reported in chronic myeloid leukemia (CML) and Philadelphia-chromosome positive (Ph+)-ALL.19 However, to our knowledge, none of the aforementioned leukemias has been found to harbor CCND2 mutations, consistent with our analysis of CML, T-ALL and B-ALL samples (Supplementary Table 2).
While our work and the recent work of others suggests that the CCND2 variants contribute to protein degradation resistance, enhanced hematopoietic cell proliferation, and sensitivity to CDK4/6 inhibitors, additional experimental models will be required to further characterize the role of these variants in leukemogenesis. The pathogenic potential of CCND2 P281 variants may be manifested through promotion of genetic instability, thereby predisposing cells to develop additional deleterious mutations. Work examining the oncogenicity of the constitutively nuclear cyclin D1 variant T286A suggests that its improper nuclear accumulation during S phase triggers DNA damage and activation of the ATM-CHD2-p53 checkpoint pathway, leading to promotion of DNA re-replication.20 If CCND2 P281 variants do indeed increase genomic instability, one might expect that analysis of serial leukemia samples from patients with CCND2 mutations would reveal the presence of these mutations at or soon after diagnosis. The presence of these mutations in Philadelphia-negative neutrophilic leukemias, which are known to progress into a blastic ‘AML-like’ phase, may also support this hypothesis. Collectively, future studies that examine the combined effect of CCND2 mutations with other known deleterious mutations in these leukemias may provide additional insights into the relevance of cyclin D2 to leukemogenesis.
In summary, the present study provides novel insight into the genetics of myeloid malignancies, namely Philadelphia-negative neutrophilic leukemias. Our work suggests that in addition to the commonly recurring classes of genes that are frequently mutated in these malignancies, recurrent mutations in cyclin D2, and perhaps other cell cycle regulators, have biochemical and therapeutic consequences and may play important roles in the pathogenesis of these leukemias.
Supplementary Material
Acknowledgments
We thank the OHSU Massively Parallel Sequencing Shared Resource for their assistance with whole exome sequencing, and members of the Druker and Tyner laboratories for thoughtful and critical discussions. V.K. was supported by the Howard Hughes Institute Medical Research Fellows Program. C.A.E. and C.E.T. are funded through a Howard Hughes Medical Institute Investigator award to BJD JEM is supported by an ASH Scholar Award and the NCI (4R00CA190605-03). DKEV was supported by the National Science Foundation Graduate Research Program (DGE-1448072). JWT was supported by the Leukemia and Lymphoma Society, the V Foundation for Cancer Research, the Gabrielle’s Angel Foundation for Cancer Research, and the National Cancer Institute (5R00CA151457-04; 1R01CA183947-01). BJD is an Investigator of the Howard Hughes Medical Institute. This work was supported in part by an award from the St Baldrick’s Foundation (280290).
Footnotes
CONFLICT OF INTEREST
The authors declare no conflict of interest.
Supplementary Information accompanies this paper on the Leukemia website (http://www.nature.com/leu)
References
- 1.Matano S, Nakamura S, Kobayashi K, Yoshida T, Matsuda T, Sugimoto T. Deletion of the long arm of chromosome 20 in a patient with chronic neutrophilic leukemia: cytogenetic findings in chronic neutrophilic leukemia. Am J Hematol. 1997;54:72–75. doi: 10.1002/(sici)1096-8652(199701)54:1<72::aid-ajh11>3.0.co;2-c. [DOI] [PubMed] [Google Scholar]
- 2.DiNardo CD, Daver N, Jain N, Pemmaraju N, Bueso-Ramos C, Yin CC, et al. Myelodysplastic/myeloproliferative neoplasms, unclassifiable (MDS/MPN, U): natural history and clinical outcome by treatment strategy. Leukemia. 2014;28:958–961. doi: 10.1038/leu.2014.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hernandez JM, del Canizo MC, Cuneo A, Garcia JL, Gutierrez NC, Gonzalez M, et al. Clinical, hematological and cytogenetic characteristics of atypical chronic myeloid leukemia. Ann Oncol. 2000;11:441–444. doi: 10.1023/a:1008393002748. [DOI] [PubMed] [Google Scholar]
- 4.Baxter EJ, Scott LM, Campbell PJ, East C, Fourouclas N, Swanton S, et al. Acquired mutation of the tyrosine kinase JAK2 in human myeloproliferative disorders. Lancet. 2005;365:1054–1061. doi: 10.1016/S0140-6736(05)71142-9. [DOI] [PubMed] [Google Scholar]
- 5.Maxson JE, Gotlib J, Pollyea DA, Fleischman AG, Agarwal A, Eide CA, et al. Oncogenic CSF3R mutations in chronic neutrophilic leukemia and atypical CML. N Engl J Med. 2013;368:1781–1790. doi: 10.1056/NEJMoa1214514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pardanani A, Lasho TL, Laborde RR, Elliott M, Hanson CA, Knudson RA, et al. CSF3R T618I is a highly prevalent and specific mutation in chronic neutrophilic leukemia. Leukemia. 2013;27:1870–1873. doi: 10.1038/leu.2013.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Piazza R, Valletta S, Winkelmann N, Redaelli S, Spinelli R, Pirola A, et al. Recurrent SETBP1 mutations in atypical chronic myeloid leukemia. Nat Genet. 2013;45:18–24. doi: 10.1038/ng.2495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gambacorti-Passerini CB, Donadoni C, Parmiani A, Pirola A, Redaelli S, Signore G, et al. Recurrent ETNK1 mutations in atypical chronic myeloid leukemia. Blood. 2015;125:499–503. doi: 10.1182/blood-2014-06-579466. [DOI] [PubMed] [Google Scholar]
- 9.Eisfeld AK, Kohlschmidt J, Schwind S, Nicolet D, Blachly JS, Orwick S, et al. Mutations in the CCND1 and CCND2 genes are frequent events in adult patients with t(8;21)(q22;q22) acute myeloid leukemia. Leukemia. 2017;31:1278–1285. doi: 10.1038/leu.2016.332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Faber ZJ, Chen X, Gedman AL, Boggs K, Cheng J, Ma J, et al. The genomic landscape of core-binding factor acute myeloid leukemias. Nat Genet. 2016;48:1551–1556. doi: 10.1038/ng.3709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.He LM, Sartori DJ, Teta M, Opare-Addo LM, Rankin MM, Long SY, et al. Cyclin D2 protein stability is regulated in pancreatic beta-cells. Mol Endocrinol. 2009;23:1865–1875. doi: 10.1210/me.2009-0057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Alt JR, Cleveland JL, Hannink M, Diehl JA. Phosphorylation-dependent regulation of cyclin D1 nuclear export and cyclin D1–dependent cellular transformation. Genes Dev. 2000;14:3102–3114. doi: 10.1101/gad.854900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sasaki Y, Jensen CT, Karlsson S, Jacobsen SE. Enforced expression of cyclin D2 enhances the proliferative potential of myeloid progenitors, accelerates in vivo myeloid reconstitution, and promotes rescue of mice from lethal myeloablation. Blood. 2004;104:986–992. doi: 10.1182/blood-2003-07-2277. [DOI] [PubMed] [Google Scholar]
- 14.Deshpande A, Sicinski P, Hinds PW. Cyclins and cdks in development and cancer: a perspective. Oncogene. 2005;24:2909–2915. doi: 10.1038/sj.onc.1208618. [DOI] [PubMed] [Google Scholar]
- 15.Takano Y, Kato Y, Masuda M, Ohshima Y, Okayasu I. Cyclin D2, but not cyclin D1, overexpression closely correlates with gastric cancer progression and prognosis. J Pathol. 1999;189:194–200. doi: 10.1002/(SICI)1096-9896(199910)189:2<194::AID-PATH426>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 16.Delmer A, Ajchenbaum-Cymbalista F, Tang R, Ramond S, Faussat AM, Marie JP, et al. Overexpression of cyclin D2 in chronic B-cell malignancies. Blood. 1995;85:2870–2876. [PubMed] [Google Scholar]
- 17.Schmidt BA, Rose A, Steinhoff C, Strohmeyer T, Hartmann M, Ackermann R. Up-regulation of cyclin-dependent kinase 4/cyclin D2 expression but down-regulation of cyclin-dependent kinase 2/cyclin E in testicular germ cell tumors. Cancer Res. 2001;61:4214–4221. [PubMed] [Google Scholar]
- 18.Clappier E, Cuccuini W, Cayuela JM, Vecchione D, Baruchel A, Dombret H, et al. Cyclin D2 dysregulation by chromosomal translocations to TCR loci in T-cell acute lymphoblastic leukemias. Leukemia. 2006;20:82–86. doi: 10.1038/sj.leu.2404008. [DOI] [PubMed] [Google Scholar]
- 19.Deininger MW, Vieira SA, Parada Y, Banerji L, Lam EW, Peters G, et al. Direct relation between BCR-ABL tyrosine kinase activity and cyclin D2 expression in lymphoblasts. Cancer Res. 2001;61:8005–8013. [PubMed] [Google Scholar]
- 20.Aggarwal P, Lessie MD, Lin DI, Pontano L, Gladden AB, Nuskey B, et al. Nuclear accumulation of cyclin D1 during S phase inhibits Cul4-dependent Cdt1 proteolysis and triggers p53-dependent DNA rereplication. Genes Dev. 2007;21:2908–2922. doi: 10.1101/gad.1586007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.