Abstract
Objective
Inadequate immunoregulation and elevated inflammation may be risk factors for posttraumatic stress disorder (PTSD), and microbial inputs are important determinants of immunoregulation; however, the association between the gut microbiota and PTSD is unknown. This study investigated the gut microbiome in a South African sample of PTSD-affected individuals and trauma-exposed (TE) controls, to identify potential differences in microbial diversity or microbial community structure.
Methods
The Clinician Administered Posttraumatic Stress Disorder Scale for DSM-5 (CAPS-5) was used to diagnose PTSD according to DSM-5 criteria. Microbial DNA was extracted from stool samples obtained from 18 individuals with PTSD and 12 TE control participants. Bacterial 16S ribosomal RNA (rRNA) gene V3/V4 amplicons were generated and sequenced. Microbial community structure, alpha-diversity, and beta-diversity were analyzed; random forest analysis was used to identify associations between bacterial taxa and PTSD.
Results
There were no differences between PTSD and TE control groups in alpha- or beta-diversity measures (e.g., alpha-diversity, Shannon index, t = 0.386, P = .70; beta diversity, based on analysis of similarities (ANOSIM), Bray Curtis test statistic = −0.033, P = .70); however, random forests analysis highlighted three phyla as important to distinguish PTSD status: Actinobacteria, Lentisphaerae, and Verrucomicrobia. Decreased total abundance of these taxa was associated with higher PTSD CAPS scores (r = −.387, P = .035).
Conclusions
In this exploratory study, measures of overall microbial diversity were similar among individuals with PTSD and TE controls; however, decreased total abundance of Actinobacteria, Lentisphaerae, and Verrucomicrobia was associated with PTSD status.
Keywords: childhood trauma, C-reactive protein, immunoregulation, inflammation, microbiome, posttraumatic stress disorder
INTRODUCTION
Violence and trauma are highly prevalent in South Africa (SA), with approximately 75% of the population experiencing at least one traumatic event, and more than half experiencing multiple traumatic events within their lifetime (1, 2). Interpersonal violence (e.g., physical and sexual assault, intimate partner violence) is the leading cause of injury in SA; a country where the homicide rate is seven times higher than the global average (3). The extent and severity of trauma exposure in SA has been found to contribute significantly to the overall burden of disease (4). Within the Diagnostic and Statistical Manual of Mental Disorders, Fifth Edition (DSM-5) (5), PTSD is classified as a trauma- and stress-related disorder that is characterized by the presence of symptoms in four diagnostic clusters (intrusion, avoidance, negative alterations in cognitions and mood, and alterations in arousal and reactivity) that significantly impair psychosocial functioning (5). Symptoms can persist for years following a traumatic event (6) and negatively affect quality of life (7, 8). Prevalence of PTSD in SA has been found to range between 2.3% and 19.9% (9, 10). In light of the substantial health and economic burden imposed by PTSD, research into the pathophysiology of the disease is imperative to both gain new insights into factors that contribute to the disease and to develop novel strategies for prevention and treatment.
Recent research has focused on the role of exaggerated inflammatory responses in the pathogenesis of PTSD. To this end, a subset of CD4+ T cells, the regulatory T cells (Tregs), have been found to be altered in PTSD-affected individuals (11, 12). Tregs play an important role in defense against inappropriate inflammatory responses, such as those observed in autoimmunity, allergy, and asthma (13). Reduced levels of Treg cells have been observed following exposure of human participants to a laboratory stressor (14), and in male and female refugees with chronic PTSD, relative to healthy controls (11, 12). Furthermore, reduced frequency of Tregs is associated with autoimmune diseases such as thyroiditis, inflammatory bowel disease, and rheumatoid arthritis (13), conditions for which individuals with PTSD show increased risk (15). Consistent with these findings, genome-wide association studies in PTSD cohorts revealed association with ANKRD55 (16), a gene associated with several autoimmune and inflammatory disorders, including multiple sclerosis (17, 18), type 2 diabetes mellitus (19), celiac disease (20), and rheumatoid arthritis (21). Additionally, Jergovic et al (22, 23) observed an altered Treg phenotype in male combat veterans with PTSD compared to healthy controls. PTSD has also been found to result in upregulation of interleukin (IL) 6 and proinflammatory cytokines, including interferon gamma (IFN-γ), IL-1β and tumor necrosis factor (TNF) (24–26). Elevated levels of C-reactive protein (CRP), a clinically used marker of inflammation, have also been observed in individuals with PTSD (27–29). Preexisting elevated CRP levels (30), or elevated IL-6 measured within 24 hours following trauma (31), have been found to predict post-deployment CAPS scores in war zone-deployed Marines or a diagnosis of PTSD in children six months following trauma, respectively.
An important factor determining immunoregulation, indicated by a balanced expansion of effector T cell populations and Tregs, is the human microbiome (32–34). The human microbiota comprises all the microorganisms (archaea, bacteria, eukaryotes, fungi, and viruses) harbored by the human body and the complete catalog of these microbial symbionts and their genes constitute the human microbiome (35). Research suggests that microbial inputs are essential for maintaining homeostasis and optimum health (36), controlling blood-brain barrier permeability (37), and regulating central nervous system (CNS) function (38). A complex, bidirectional system of communication exists between the gut microbiome, the gut, and the CNS (39). Data from animal studies indicate that environmental and gut microbial species elicit a significant impact on cognitive function, memory, and fundamental patterns of behaviour, such as social interaction and stress coping (40–43). Additionally, stress can influence the composition of the gut microbiota, and the bidirectional communication between microbiota and the CNS in turn influences stress reactivity (40, 41, 44). Alterations in microbiota have been shown to modulate plasticity-related (45–47), serotonergic (40, 45, 48, 49), and GABAergic (50–52) signaling systems in the CNS. Dysregulation of the gut microbiome (dysbiosis) therefore may influence risk of developing a disease, including stress- or trauma-related disorders (40, 41, 44).
Gut microbiota have also been found to play a role in programming of the hypothalamic–pituitary–adrenal (HPA) axis (one of the key regulators of the stress response system) (38, 53), with implications for stress-related disorders, including PTSD. Dysregulation of the HPA axis may contribute to the pathophysiology of PTSD (54). Glucocorticoid-mediated immunosuppression may result in the reduction of inflammatory responses in the short-term, but in the long-term can also lead to an imbalance in the homeostasis between pathobionts (resident microbes with pathogenic potential (55, 56), gut microbiota, and the mucosa. Indeed, glucocorticoids induce the expansion of pathobionts, such as Helicobacter spp., a gram-negative bacterium, shown to enhance chronic inflammatory diseases (57). Mice exposed to psychological stressors exhibit expansion of Helicobacter spp., as determined by absolute abundance, evaluated using real-time quantitative PCR (58), or relative abundance (40, 41), and this effect can be prevented by the administration of a glucocorticoid receptor antagonist (58). Helicobacter spp. induce colitis in IL-10−/− mice, which lack adequate immunoregulation, an effect that may be due to overactivation of host immune defenses (59–61). Immunization with a heat-killed preparation of an immunoregulatory bacterium that increases Treg and anti-inflammatory cytokines, such as IL-10 and transforming growth factor beta (TGFβ) (62), has recently been shown to prevent stress-induced increases in a PTSD-like syndrome in mice (40, 41), suggesting that the balance of proinflammatory and anti-inflammatory or immunoregulatory microbial inputs could contribute to the risk of developing a PTSD-like syndrome.
The present study investigated the gut microbiome profiles of a relatively homogeneous group of individuals of a unique South African mixed ancestry population1. Individuals diagnosed with PTSD were compared to individuals who were exposed to a traumatic event but did not develop PTSD, in order to identify microbial signatures associated with PTSD.
METHODS
Clinical and metabolic measures
All research participants provided written informed consent to take part in the study after the study procedures were explained in detail. The Health Research Ethics Committee 2 (HREC2) of Stellenbosch University approved the study. Participants were recruited through purposive sampling, utilizing various avenues including referrals from general and psychiatric hospitals and community clinics from Cape Town and surrounding areas as well as through print, radio and web advertisements. Samples included in this study were collected from August 2014 – February 2015.
Based on the MINI International Neuropsychiatric Interview, version 6.0 (MINI) (64) participants were excluded if they had bipolar or psychotic disorders or an alcohol or drug use disorder within the past six months. Other exclusionary criteria included: a neurological disorder, a diagnosis of metabolic syndrome (MetS), diarrhea within the past week, any antibiotic usage four weeks prior to stool sampling, or a diagnosis of inflammatory bowel disease (IBD), coeliac disease or irritable bowel syndrome (IBS). Written informed consent was obtained from all study participants. The study sample consisted of 18 PTSD and 12 trauma-exposed (TE) control participants of South African mixed ancestry matched for age, sex, time since index trauma and the number of traumatic event exposures. Demographic and clinical data were collected with structured demographic and medical history questionnaires designed for the SHARED ROOTS parent study. Posttraumatic stress disorder diagnosis was based solely on CAPS-5 severity scores, with those with a score of 23 or above being placed in the PTSD cohort. Current PTSD diagnosis and symptom severity were determined using the Clinician-Administered PTSD Scale for DSM-5 (CAPS-5) (65). The CAPS-5 is a structured diagnostic interview used to diagnose PTSD based on the Diagnostic and Statistical Manual of Mental Disorders, Fifth Edition (DSM-5) criteria and is the gold standard PTSD interview assessment. Plasma C-reactive protein (CRP) concentration was measured as a marker associated with inflammation. CRP assays were used to report CRP concentrations > 3.0 mg/dL; high-sensitivity CRP (hsCRP) assays were used to report CRP concentrations < 3.0 mg/dL. In 27 cases where these values were obtained using both assays, the values were highly correlated (Pearson correlation, r = .996; P < .001).
Adverse early life experience is an important determinant of risk for PTSD (66, 67). Consequently, we estimated adverse early life experience using the Childhood Trauma Questionnaire (CTQ). The CTQ (68) was used to screen for a history of child abuse and neglect. The CTQ consists of 28 self-report items used to calculate a total childhood trauma score by adding scores obtained on five trauma subscales (physical abuse, sexual abuse, emotional abuse, physical neglect and emotional neglect). Furthermore, prenatal maternal stress, compared to a low stress control condition, has been shown to affect the infant microbiota measured at 7, 14, 28, 80, and 110 days of age in infants, suggesting important effects of adverse early life experience on the gut microbiome (69).
Microbiome analyses
Microbial DNA was extracted from 1.4 ml stabilized stool (stool specimen homogenized in stool DNA stabilizing buffer) using the PSP® Spin Stool DNA Plus Kit (STRATEC Molecular, Birkenfeld, Germany) according to the manufacturer’s protocol 2 (“Isolation of total DNA from 1.4 ml stabilized stool homogenate with enrichment of bacterial DNA”). The 16S ribosomal RNA (rRNA) gene amplicons were generated for the V3 and V4 regions of the 16S rRNA bacterial gene, which were recommended by Klindworth et al. (70). Illumina adapter overhang nucleotide sequences were added to the gene-specific sequences. The full-length primer sequences targeting this region were
341 forward primer (5′-CCTACGGGNGGCWGCAG-3′) and
785 reverse primer (5′-GACTACHVGGGTATCTAATCC-3′)
Libraries were prepared using the 16S Metagenomic Sequencing Library Preparation Kit from Illumina, according to the manufacturer’s instructions. Libraries were sequenced using multiplexed Illumina HiSeq paired-end 100 base pair sequencing according to the manufacturer’s instructions. Base calling was performed and FASTQ sequence reads generated using Illumina Casava Pipeline 1.8.2. Initial quality assessment was based on data passing the Illumina Chastity filter. Subsequently, reads containing adaptors and/or PhiX control signal were removed. The second quality assessment was based on the remaining reads using the FASTQC quality control tool version 0.10.0.
Taxonomy from DNA sequences
The operational taxonomic unit (OTU) table was prepared using Quantitative Insights into Microbial Ecology (QIIME) v. 1.9 (71). Forward reads were demultiplexed using default parameters with a minimum quality score threshold set to 25. Following this step, 1,738,164 of 1,959,124 HiSeq reads passed quality control. These reads were assigned to OTUs using the closed-reference OTU picking method with Greengenes 97% reference database (Aug 13) (72).
Microbial diversity analysis
In microbiome studies, two commonly used measures of species diversity are α-diversity (assessing diversity within a sample) and β-diversity (assessing differences between samples, with greater β-diversity indicating greater dissimilarity). α- and β-diversity were analyzed using rarefied data, which corrects for differential sequencing depth among samples, using QIIME (71).
Comparison of taxa abundances
Operational taxonomic units of the rarefied data set were collapsed by taxonomic assignment and compared using QIIME (71). In addition, we used the R package VSURF (73) (Variable Selection using Random Forests) for feature selection on the 37 phyla. This method uses Random Forests, which are an ensemble approach from machine learning that rank the importance of features in terms of their ability to classify a variable of interest, while taking into account the complex interrelationships of the features (74). The VSURF function provides two sets of results: the “interpretation” subset of important variables that may include some redundancy, and the “prediction” subset that aims to eliminate redundancy while maintaining predictive accuracy. We then used marginal plots, partial plots and the find.interactions function of the R package RandomForestSRC (75) in order to interpret the findings of the variable selection process. Additionally, to interpret the findings, we also used Pearson’s correlation to evaluate the relationship between the random forest model outcomes and CAPs scores, CTQ scores, and other variables of potential interest.
For additional details of the clinical population, and detailed methods for microbiome analysis, see Supplemental methods (Supplemental Digital Content 1).
Results
Clinical and biological measures
A summary of key demographic and clinical data of the study participants is found in Table 1. Median CAPS-5 total scores were 33.5 for the PTSD group and 3.5 for the TE group (Table 1; P < .001; ranges: PTSD, 23–48, TE, 0–20). A difference in CTQ scores, with higher scores in the PTSD group, approached statistical significance (Table 1; P = .068). The type of traumatic event most frequently endorsed as the index trauma by the overall group was assault with a weapon (n = 7; 23.3%), followed by the sudden unexpected death of someone close to them (n = 6; 20.0%), physical assault (n = 5; 16.6%), and sexual assault (n = 4; 13.3%). Assault with a weapon (n = 4; 22.2%) and physical assault (n = 4; 22.2%) were the index traumas most frequently identified by those with PTSD, followed by the sudden unexpected death of someone close to them (n = 3; 16.7%), and sexual assault (n = 3; 16.7%); the index traumas most often experienced by individuals in the control group were assault with a weapon (n = 3; 25%) and the sudden unexpected death of someone close to them (n = 3; 25%). Six (33.3%) of those with PTSD were receiving concomitant psychotropic medications (three (16.7%) were on amitriptyline [5–25 mg] for sleep or neuralgia, one (5.6%) was on zolpidem [5 mg] for sleep, and two (11.1%) were on fluoxetine [20 mg] and citalopram [20 mg], respectively, for depression). Three individuals with PTSD (16.7%) and one control (8.3%) were on treatment for hypertension, and one control participant (8.3%) was receiving treatment for hypercholesterolemia. The only variables that differed significantly between PTSD participants and TE controls were mean systolic and diastolic blood pressure, with mean systolic and diastolic blood pressures lower in PTSD participants (Table 1).
Table 1.
Clinical and demographic variables of the study participants.
| Clinical/demographic variable | PTSD subjects (n = 18) | TE controls (n = 12) | P value |
|---|---|---|---|
| Age in years, mean (SD) | 42.0 (12.6) | 38.7 (11.7) | .52 |
| Female, n (%) | 14 (77.8) | 7 (58.3) | .26 |
| CAPS-5 total score, median (IQR) | 33.5 (30.0 – 36.7) | 3.5 (0 – 9.5) | <.001 |
| Time since index trauma (months), median (IQR) | 126 (39 – 231) | 48 (24 – 126) | .18 |
| Number of different types of traumatic experiences on LEC-5, mean (SD) | 6.3 (1.9) | 5.9 (3.9) | .75 |
| CTQ total score, median (IQR) | 54 (36.5; 81.5) | 38 (32.0; 44.0) | .068 |
| MINI current MDD, n (%) | 2 (11.1) | 0 (0) | .50 |
| MINI lifetime MDD, n (%) | 12 (66.7) | 6 (50.0) | .46 |
| MINI current comorbid anxiety disorder(s), n (%) | 6 (33.3) | 1 (8.3) | .19 |
| Current psychiatric medication, n (%) | 6 (33.3) | 0 (0) | .057 |
| Psychiatric medication current/lifetime, n (%) | 7 (38.9) | 3 (25.0) | .43 |
| Cigarette smoking (previous 6 months), n (%) | 9 (50.0) | 5 (41.7) | .87 |
| Alcohol use (previous 6 months), n (%) | 9 (50.0) | 7 (58.3) | .78 |
| Lifetime history of illicit substance use, n (%) | 3 (25.0) | 5 (41.7) | .21 |
| 1 CRP, median (IQR) | 1.4 (0.6 – 2.8) | 2.0 (1.0 – 6.2) | .11 |
| BMI, mean (SD) | 28.5 (7.0) | 28.6 (9.8) | .96 |
| Waist circumference (cm), mean (SD) | 90.7 (14.4) | 87.4 (16.3) | .57 |
| Mean systolic blood pressure (mmHg), mean (SD) | 120.3 (12.6) | 131.5 (14.0) | .035* |
| Mean diastolic blood pressure (mmHg), mean (SD) | 75.2 (7.47) | 83.7 (9.4) | .016* |
| Triglycerides (mmol/l), mean (SD) | 0.9 (0.3) | 1.0 (0.3) | .30 |
| HDL cholesterol (mmol/l), mean (SD) | 1.5 (0.4) | 1.6 (0.5) | .45 |
| Fasting glucose (mmol/l), median (IQR) | 5.0 (4.7; 5.3) | 5.0 (4.8; 5.2) | > .99 |
Abbreviations: BMI, body mass index; CAPS-5, Clinician Administered Posttraumatic Stress Disorder Scale for DSM-5; CRP, C-reactive protein; CTQ, Childhood Trauma Questionnaire; DSM-5 = Diagnostic and Statistical Manual of Mental Disorders, Fifth Edition; HDL, high density lipoprotein; IQR, interquartile range; LEC-5, Life Events Checklist for DSM-5; MDD, major depressive disorder; MINI, MINI International Neuropsychiatric Interview, version 6.0; SD, standard deviation.
CRP > 3.0 mg/L based on CRP assay; CRP < 3.0 mg/L based on hsCRP assay. Continuous data were summarised as means and standard deviations, if approximately normally distributed, and as medians and interquartile ranges (IQRs) if non-normally distributed. Differences between normally and non-normally distributed data were assessed using Student’s t-tests and Mann-Whitney U-tests, respectively. Categorical data were summarized as counts and percentages, and differences between groups were assessed using Chi-square or Fisher’s Exact tests, where appropriate.
P < 0.05, Student’s t-test.
Gut microbiome analyses
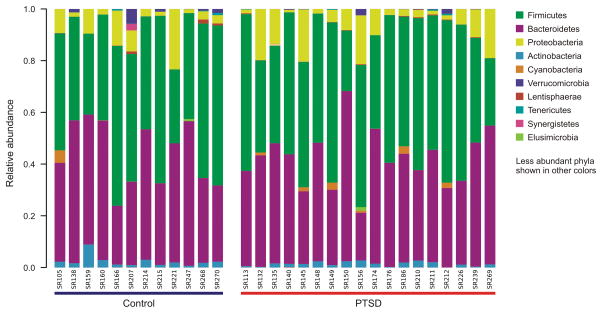
Of 1,959,124 reads resulting from sequencing, a total of 1,738,164 (88.7%) passed the QC filters applied to remove low quality reads. Of those reads, 1,690,568 (97.3%) aligned to the Greengenes database (Aug 13) with at least 97% similarity level. A total of 37 phyla were detected in the resulting data set. The top two phyla observed in all participants were Firmicutes and Bacteroidetes, followed by Proteobacteria and Actinobacteria, as expected. Figure 1 shows the ten most abundant phyla identified in PTSD participants and TE controls. No significant differences in relative abundances of individual taxa in PTSD participants and TE controls were observed (Figure 1, Supplemental Digital Content 1, Figure S1; Kruskal-Wallis tests with Bonferroni correction).
Figure 1.
Stacked bar chart indicating the relative abundances of the ten most abundant phyla detected in the gut microbiomes of PTSD participants and trauma-exposed (TE) controls.
There were also no significant differences in alpha (Supplemental Digital Content 1, Figure S2; Chao 1 (t = 0.832, P = .41), Observed species (t = 0.760, P = .45), Phylogenetic diversity (t = 0.510, P = .61), Shannon entropy (t = 0.386, P = .70), using data rarefied at 30,000 reads) or beta (Supplemental Digital Content 1, Figures S3 and S4) diversity between PTSD participants and TE controls. Specifically, analysis of similarities (ANOSIM) revealed that there were no differences between PTSD and TE control groups using the Bray-Curtis distance metric (test statistic = −0.033, P = .70), weighted UniFrac distance metric (test statistic = −0.016, P = .56), or unweighted UniFrac distance metric (test statistic = −0.013, P = .52).
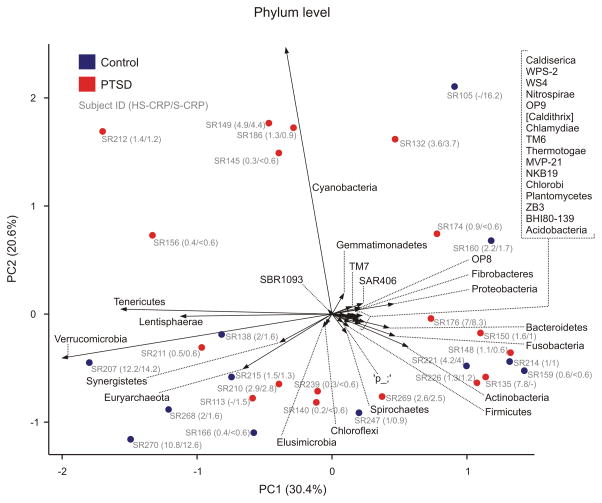
We created a biplot, projecting phylum level taxonomic information onto a PCoA plot, to determine which taxa drive sample distributions in PCoA space. Relative abundance of Cyanobacteria was found to be a major determinant of differences in bacterial community structure among samples (Figure 2). All Cyanobacteria belonged to order Gastranaerophilales (YS2/4C0d2), which has recently been defined as an order within the class of non-photosynthetic Melainabacteria (76); however, (77, 78) propose Melainabacteria as a separate phylum. Biplot analysis at the genus level revealed no clear condition-specific patterns (Supplemental Digital Content 1, Figure S5).
Figure 2.
Biplot illustrating phylum-level gut microbial community analysis (closed symbols) and composition analysis (vectors) using principal component analysis (PCA) of centre log ratio-transformed and standardized data from 18 PTSD participants (red symbols) and 12 trauma-exposed (TE) controls (blue symbols). The distance between symbols approximates the dissimilarity of their microbial communities, as measured by Euclidean distance. PCA axes 1 and 2 explain 30.4% and 20.6% of the variation, respectively. Vectors point in the direction of the greatest increase of values for the corresponding phylum across all PTSD and TE control participants; thirty-seven different phyla were detected. The angle between arrows indicates approximated correlation (>90° indicates a negative correlation). Gray text indicates subject identification number and plasma C-reactive protein (CRP) concentrations for individual participants as measured using high-sensitivity CRP (HS-CRP; CRP ≤ 3.0 mg/l) or sensitive (S-CRP; CRP > 3.0 mg/l) assays. Relative percent abundances of Cyanobacteria for individual PTSD participants were SR186, 2.87%, SR149, 2.78%, SR212, 2.14%, SR145, 1.67%, SR132, 1.23%, SR156, 0.23%, SR174, 0.06%, SR176, 0.01%, SR211, 0.01%, SR210, 0.0036%, SR135, 0.0035%. Relative percent abundances of Cyanobacteria for individual TE control participants were SR105, 4.65%, SR160, 0.11%, SR207, 0.027%, SR138, 0.014%, SR215, 0.0034%, SR214, 0.0037%. Cyanobacteria were undetectable in all other participants in the filtered dataset. Operational taxonomic units associated with the orders Haptophyceae, Streptophyta, Gloebacterales, Pseudanabaenales, and Synechococcales belonging to the Cyanobacterium phylum were observed in fewer than 20% of participants.
We used weighted gene co-expression network analysis (WGCNA) to investigate potential differences in microbial co-occurrence networks at the OTU level. To summarize the profiles of co-occurrence modules, we calculated the eigenvalue, which provides a mathematically optimal way of summarizing the co-occurrence patterns of all OTUs belonging to each module. To identify functional microbial communities (FMCs) or modules that were correlated with clinical traits, we used correlation tests to relate each eigenvalue to the clinical traits. Zhang and Horvath recommend selecting a soft thresholding power that satisfies a scale-free fit of R2 > .8 and a slope approximately equal to −1. As such, we selected β = 5 (R2 = 0.88, slope = −1.95) for construction of our adjacency matrix. The WGCNA analysis generated 22 different modules that were each arbitrarily assigned a unique color label. The bacterial dendrogram and module assignments are shown in Figure S6 (Supplemental Digital Content 1). The most highly connected nodes defined by scaled connectivity were dominated mainly by either the Bacteroidetes and Firmicutes phyla with either Lentisphaerae or Proteobacteria also showing the most connectivity for 2 modules (Supplemental Digital Content 1, Table S1). Module concept and descriptive statistics can be seen in Table S1 (Supplemental Digital Content 1). The cluster coefficient is a measure of localized network density or “cliquishness” within the modules. Network density is a measure of the total amount of possible connections that exist within a given module. Network centrality is a measure of the how much one individual node dominates the module’s connectedness. The p-value was generated from the Student’s t-test comparison between the eigenvalues of the PTSD participants and TE controls using Bonferroni correction. Finally the most connected node phylum was determined by the scaled connectivity coefficient. Analysis of the module eigenvalues determined that there were no differences between treatment groups for any of the 22 modules identified. Additionally, none of the traits of the participants that were regressed against the modules were significant when using Bonferroni correction.
The functional potential of microbial communities was investigated using PICRUSt (Supplemental Digital Content 1, Figure S7) (87). This analysis defined 328 functional groups; of these, 293 (89%) had positive values, while 35 (11%) had zero values for all samples. There were no significant differences between PTSD participants and TE controls for any functional groups, using Kruskal-Wallis one-way analysis of variance and Bonferroni correction to test for relative increases or relative decreases. While Kruskal-Wallis doesn’t explicitly account for compositionality, it can be useful for probing for obvious changes in metagenomics. A total of 5.5% of functional groups showed >50% relative increase in PTSD participants compared to TE controls, while a total of 2.4% of functional groups showed >50% relative decrease in PTSD participants compared to TE controls. The greatest percent increase (910%) was observed for photosynthesis – antenna proteins, which include proteins associated with the phycobilisome in Cyanobacteria and red algae, and may reflect increases in Cyanobacteria of the order Gastranaerophilales (YS2/4C0d2) in a subset of PTSD participants. Statistical comparison of this functional group in PTSD participants and TE controls revealed no significant difference (t = 3.86; P = 0.050; Bonferroni-adjusted P = 1.0).
Random Forest/VSURF
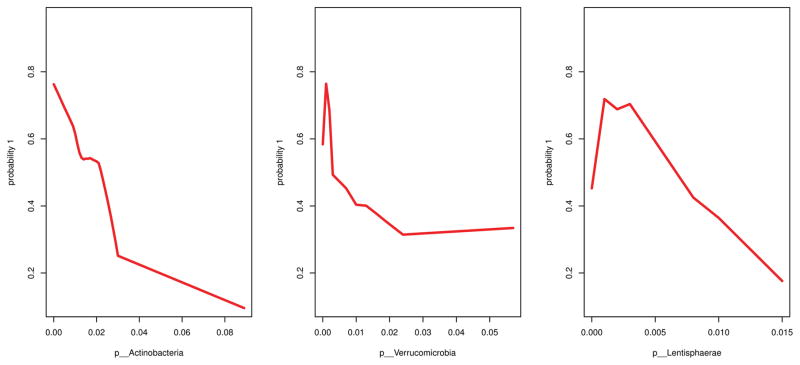
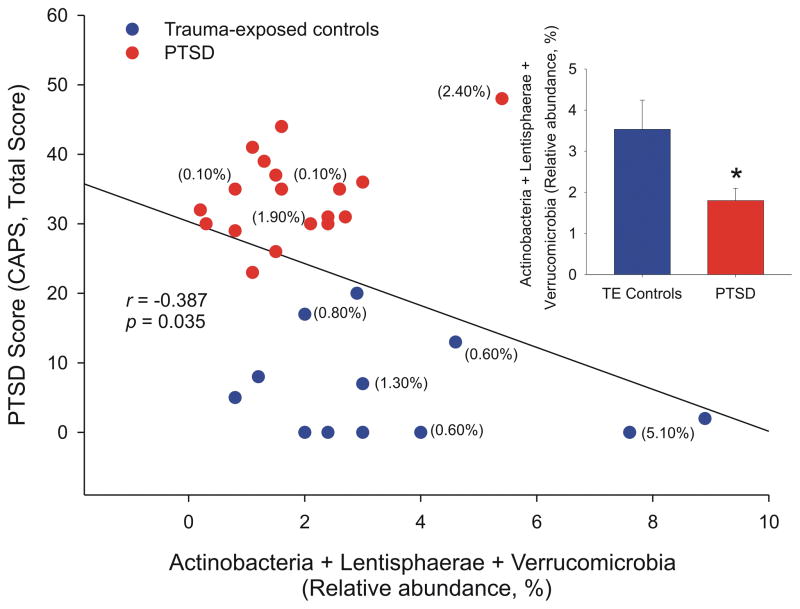
We used the R package VSURF (73) (Variable Selection using Random Forests) for feature selection on the 37 phyla. Random forest analysis identified three phyla (Actinobacteria, Lentisphaerae, and Verrucomicrobia) as highly important to classify PTSD versus TE controls. Of these, Lentisphaerae and Verrucomicrobia were correlated, and the more parsimonious set of results included only Actinobacteria and Verrucomicrobia. As per the random forest feature selection method that we used, we refer to this first set of three phyla as the “interpretation” set of results, and the subset of Actinobacteria and Verrucomicrobia as the “prediction” set. The Cohen’s d effect size for the interpretation model [Actinobacteria, Lentisphaerae, Verrucomicrobia] was 0.894; the Cohen’s d effect size for the prediction model [Actinobacteria, Verrucomicrobia] was 0.864. Thus, both models yield large effect sizes in terms of the taxonomic differences between individuals with PTSD and TE controls. The interpretation step of the variable selection function had an error rate of 30.7%; the prediction step had an error rate of 26.4%. The marginal plots, which show the unadjusted relationship between the predicted probability of PTSD and abundance of these taxa (Figure 3), and the partial plots (Figure S8), which show the adjusted relationship, were similar. For all three phyla, the general relationship was that higher abundance corresponded with a lower probability of PTSD. Similarly, lower total abundance of these taxa corresponded with higher PTSD CAPS score (Figure 4; Pearson’s r = −0.387; P = .035). There was no evidence of strong two-way interactions among the phyla. The prediction subset of results showed similar patterns (Figure S9).
Figure 3.
Marginal plots of the random forests predicted values for the estimated probability of posttraumatic stress disorder (PTSD) from the random forests versus the relative abundance of the three phyla identified as important for distinguishing PTSD status.
Figure 4.
Relationship between the random forests interpretation model, relative abundance of [Actinobacteria, Lentisphaerae, Verrucomicrobia] and posttraumatic stress disorder (PTSD) scores (Clinician Administered Posttraumatic Stress Disorder Scale for DSM-5 (CAPS), Total Score). Posttraumatic stress disorder was negatively correlated with the relative abundance of Actinobacteria, Lentisphaerae, and Verrucomicrobia phyla. In other words, PTSD diagnosis was associated with decreased abundance of these phyla (Pearson’s r = −.387; P = .035). Percentages in parentheses indicate the percent relative abundance of Akkermansia; Akkermansia was below the threshold of detection for all other participants. Sample sizes, PTSD participants, n = 18; TE controls, n = 12). *P < 0.05, Student’s t-test.
The relative abundance of [Actinobacteria, Verrucomicrobia], also was associated with childhood trauma scores (Childhood Trauma Questionnaire (CTQ), Total Score), with higher CTQ scores associated with lower total relative abundance (Figure S10; Pearson’s r = −0.375; P = .041). There was, however, no difference in CTQ scores in PTSD participants versus controls (Table 1; P = .068).
DISCUSSION
The present exploratory study evaluated microbial diversity and community structure in 18 PTSD participants and 12 TE controls. We found no significant differences in microbial community diversity or predicted functional capacity between PTSD and TE participants. However, random forests analysis highlighted three phyla, Actinobacteria, Lentisphaerae, and Verrucomicrobia, as important to distinguish PTSD participants, relative to TE controls. Decreased total abundance of these phyla was associated with higher PTSD CAPS scores. These results are interesting in part because they are consistent with predictions, based on theoretical grounds related to the hygiene hypothesis or the “Old Friends” hypothesis (32, 79–89), and studies using an animal model of PTSD (40, 41), leading to the hypothesis that decreased exposure to Actinobacteria and other anti-inflammatory/immunoregulatory “Old Friends” leads to increased vulnerability to PTSD (40). In this exploratory clinical study, the machine learning analysis is consistent with this hypothesis, with decreases in relative abundance of Actinobacteria, Lentisphaerae, and Verrucomicrobia in those with PTSD.
The Verrucomicrobia phylum was strongly represented by a single genus, Akkermansia, of which there is one species, muciniphila (on average, in participants with Verrucomicrobia, 61.9% of Verrucomicrobia belonged to the genus Akkermansia). A. muciniphila is thought to be potently anti-inflammatory in humans, and induces Treg cells (90). A. muciniphila has been reported to be reduced in a number of diseases or conditions associated with a failure of immunoregulation and/or increased inflammation, including type 1 and type 2 diabetes, obesity, inflammation and metabolic disorders during obesity, inflammatory bowel disease, appendicitis, atopic diseases, autism, and aging (for references, see (91)). The Actinobacteria phylum was strongly represented by the Collinsella genus (Collinsella represented 54.2% of Actinobacteria; expressed as mean percent of Actinobacteria among those participants with detectable Actinobacteria). Decreased relative abundance of Actinobacteria also has been described in individuals with major depressive disorder (92), while a recent study demonstrates a negative association between relative abundance of Actinobacteria and stress-induced increases in gut permeability (93). Conversely, a recent analysis of semi-supercentenarians (105–109 years of age), in comparison to adults, elderly individuals, and centenarians, found a higher prevalence of Akkermansia (Verrucomicrobia) as well as Eggerthella and Bifidobacterium (prominent genera of gut commensals belonging to the Actinobacterium phylum) in semi-supercentenarians (94). These data identify a clear hypothetical framework that can be investigated in future studies. It will be important to both replicate these findings in such studies, and to investigate the effects of the Actinobacterium, Lentisphaerae, and Verrucomicrobia phyla more in-depth.
These data are consistent with a previous clinical study of maternal prenatal stress, in which the relative abundances of Actinobacteria (including a consortium of Actinomycetaceae, Bifidobacterium, Collinsella, and Eggerthella) were low in infants whose mothers had experienced high cumulative stress, and the relative abundance of Akkermansia declined dramatically in the high cumulative stress group after the first month and remained low thereafter (69). It’s possible that, in the present study, maternal prenatal stress or adverse early life experience induced alterations in the relative abundances of Actinobacteria, Lentisphaerae, and Verrucomicrobia that persisted until the time of adult trauma and subsequent development and persistence of PTSD symptoms. Alternative explanations for these findings include current use of psychiatric medications by 6 of the 18 PTSD participants (33.3%), and the possibility that altered autonomic nervous system function (expected in some individuals with a history of early adversity (95)) may result in an altered environment for the gut microbes (96).
It is possible that elevated inflammation at the time of trauma exposure is critical for determining PTSD outcomes (30, 31). In support of the hypothesis that elevated inflammation before or immediately after trauma is an important factor determining development of PTSD symptoms following trauma exposure, pre-existing elevated CRP levels (30), or elevated IL-6 measured within 24 hours following trauma (31), have been found to predict subsequent PTSD symptoms. Studies in rodents are consistent with this hypothesis, as individual differences in stimulated IL-6 release prior to psychosocial stress predict subsequent vulnerability to anxiety and depression-like behavioral responses (97), and immunizations with an immunoregulatory bacterium that prevent stress-induced exaggeration of IL-6 release prevent development of a PTSD-like syndrome (40, 41).
Individual differences in the host immune response may play an important role in vulnerability to PTSD symptoms following trauma exposure. Consistent with this hypothesis, studies in rats show that glucocorticoids decrease IgA (which normally inhibits bacterial adherence to intestinal epithelial cells), increase bacterial adherence over two-fold, and increase bacterial translocation to mesenteric lymph nodes (98). Decreased immunoregulation, as evidenced by decreased frequency of Treg cells, or altered Treg function, may lead to overactive host immune defenses, increased gut permeability, colitis, and exaggerated PTSD symptoms following trauma exposure (11, 12, 14, 22, 23). Based on the current study, decreases in the relative abundances of Actinobacteria, Lentisphaerae, and Verrucomicrobia (including the prevalent human commensal, A. muciniphila) may contribute to decreased immunoregulation in PTSD.
Limitations
There are a number of important limitations of this study that deserve mention. The PTSD and TE control participants were comparable with regard to a number of clinical endpoints, including age, sex, time since index trauma, number of different types of traumatic experiences, current and lifetime depressive symptoms and comorbid anxiety disorders, smoking and alcohol use in the previous 6 months, lifetime history of illicit substance abuse, and symptoms of MetS. That being said, some members of the TE group reported subthreshold symptoms of PTSD, as well as clinically significant symptoms associated with other current psychiatric conditions. Nonetheless, there were no differences in plasma CRP concentrations between PTSD and TE control participants. Although elevated plasma concentrations of CRP, a biomarker of inflammation, have been observed in individuals with PTSD (27–29), symptoms not specifically evaluated with respect to CRP concentrations in the present study, such as re-experiencing symptoms, may account for this association (27). Previous studies have found that 23.0% of individuals in a community sample have elevated CRP (>3 mg/L) (28); a similar percentage of participants in the present study was found to have elevated plasma CRP concentrations (eight of 30 participants overall (26.7%), four of 18 PTSD participants (22.2%), and four of 12 TE controls (33.3%)). In contrast to previous studies with larger sample sizes, we were not able to detect increases in plasma CRP concentrations in PTSD participants, although previous studies did not use TE controls as a comparison group (28, 29). Indeed, trauma exposure per se might be a factor that contributed to elevated plasma CRP concentrations in individuals with PTSD in previous studies, as trauma exposure is associated with elevated CRP based on a transdiagnostic meta-analysis (99).
There are a number of other limitations of the current work. First, the study was of a cross-sectional nature. A longitudinal design will be required in order to determine the potential for causal effects of certain microbial profiles. Second, the sample sizes in this exploratory study were relatively small; consequently, the power of the study was not optimal. To guide design of future studies, we estimated power by Monte Carlo simulation as implemented in the HMP package in R, using the Dirichlet-multinomial parameters estimated from the data from this study. We performed 20,000 iterations for a range of read depths (10 to 100 thousand in increments of 10 thousand) and sample sizes (15 to 60 in increments of 15). Using these parameters, future studies would have >98% power to detect a difference between groups with 60 samples per group and using a Bonferroni correction for 20 comparisons (α = 0.0025; four clinical functional outcomes and five biological signatures). Third, some in the TE group exhibited posttraumatic symptoms and/or met criteria for other psychiatric conditions (e.g., major depressive disorder). Fourth, index traumas differed between study participants, and there was also a wide range of time since index trauma. Fifth, an additional potential limitation is that the CAPS-5 is a relatively new measure and guidance regarding criteria for case ascertainment or symptom severity cut-points is not widely available. Sixth, inclusion of functional metabolomics would inform potential mechanisms involved in any differences in microbial community structure or diversity. Finally, future studies should include non-trauma-exposed controls, in addition to TE controls, to explore the relationships among trauma exposure, the microbiota, and PTSD symptoms.
CONCLUSIONS
In this report, we tested the hypothesis of an association between the gut microbiome and PTSD. In our exploratory study, we were unable to detect differences in microbial community structure, or α- or β-diversity measures, however, random forests analysis identified a biological signature of vulnerability to developing PTSD; specifically, decreases in relative abundance of a consortium of three phyla with notable immunoregulation-promoting capabilities, including Actinobacteria, previously associated with lower stress-induced increases in gut permeability in humans. These phyla are biologically plausible in terms of their effects on PTSD, and they could form the a priori hypotheses for larger longitudinal studies, in which we could further evaluate both the combined and individual associations between these phyla and PTSD. Future studies addressing the limitations of this exploratory study will be required to validate these findings.
Supplementary Material
Acknowledgments
Conflicts of Interest and Source of Funding. LLH, SMJH, SMM, and SS are supported by the Medical Research Council of South Africa (MRC Flagship Grant RFA-UFSP-01-2013); SMJH, SMM, and SS are supported by the South African Research Chair in Posttraumatic Stress Disorder, funded by the Department of Science and Technology and the National Research Foundation. C.A.L. is currently receiving funding from the Alfred P. Sloan Foundation (G-2015-14165; G-2016-7077), Colorado Clinical & Translational Sciences Institute (CCTSI) (CNSTT-15-145), Colorado Department of Public Health and Environment (DCEED-3510), the National Institutes of Health (NIH) (R01 DA019921), the Department of Veterans Affairs Office of Research and Development (VA-ORD) (1 I21 RX002232-01), and the Office of Naval Research (N00014-15-1-2809). TTP is supported by the University of Maryland, Joint Institute for Food Safety and Applied Nutrition and the U.S. Food and Drug Administration through the cooperative agreement FDU.001418, the Veterans Affairs Administration through Merit Award MHBA-016-15S from CSR&D, and the Rocky Mountain MIRECC, Denver, CO. Christopher A. Lowry serves on the Scientific Advisory Board of Immodulon Therapeutics. For the remaining authors none were declared.
Abbreviations used in text
- BMI
body mass index
- CAPS-5
Clinician Administered Posttraumatic Stress Disorder Scale for DSM-5
- CTQ
Childhood Trauma Questionnaire
- CRP
C-reactive protein
- DSM-5
Diagnostic and Statistical Manual of Mental Disorders, Fifth Edition
- HDL
high density lipoprotein
- HPA
hypothalamic–pituitary–adrenal
- hsCRP
high sensitivity C-reactive protein
- IBD
inflammatory bowel disease
- IBS
irritable bowel syndrome
- IgA
immunoglobulin A
- IL
interleukin
- IFN-γ
interferon gamma
- IQR
interquartile range
- LEC-5
Life Events Checklist for DSM-5
- MDD
major depressive disorder
- MetS
metabolic syndrome
- MINI
MINI International Neuropsychiatric Interview, version 6.0
- OTUs
operational taxonomic units
- PICRUSt
phylogenetic investigation of communities by reconstruction of unobserved states
- PTSD
posttraumatic stress disorder, QIIME, Quantitative Insights into Microbial Ecology
- rRNA
ribosomal RNA
- SA
South Africa
- TE
trauma-exposed
- TGFβ
transforming growth factor β
- TNF
tumor necrosis factor
- Treg
regulatory T cell
- VSURF
Variable Selection using Random Forests
- WGCNA
weighted gene co-expression network analysis; weighted correlation network analysis
Footnotes
In South Africa, recognized population groups include Black African, Coloured, Indian or Asian, White, and Other. Coloured people, included in this study, constitute the largest population group in the Western Cape (63).
References
- 1.Williams SL, Williams DR, Stein DJ, Seedat S, Jackson PB, Moomal H. Multiple traumatic events and psychological distress: the South Africa stress and health study. J Trauma Stress. 2007;20(5):845–55. doi: 10.1002/jts.20252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kaminer D, Grimsrud A, Myer L, Stein DJ, Williams DR. Risk for post-traumatic stress disorder associated with different forms of interpersonal violence in South Africa. Soc Sci Med. 2008;67(10):1589–95. doi: 10.1016/j.socscimed.2008.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.World Health Organization. Violence Prevention. [4-3-2016];The Evidence. 2009 http://www.who.int/violence_injury_prevention/violence/4th_milestones_meeting/publications/en/
- 4.Norman R, Matzopoulos R, Groenewald P, Bradshaw D. The high burden of injuries in South Africa. Bull World Health Organ. 2007;85(9):695–702. doi: 10.2471/BLT.06.037184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.American Psychiatric Association. Diagnostic and statistical manual of mental disorders. 5. Washington, DC: 2013. [Google Scholar]
- 6.Bremner JD, Southwick SM, Darnell A, Charney DS. Chronic PTSD in Vietnam combat veterans: course of illness and substance abuse. Am J Psychiatry. 1996;153(3):369–75. doi: 10.1176/ajp.153.3.369. [DOI] [PubMed] [Google Scholar]
- 7.Mendlowicz MV, Stein MB. Quality of life in individuals with anxiety disorders. Am J Psychiatry. 2000;157(5):669–82. doi: 10.1176/appi.ajp.157.5.669. [DOI] [PubMed] [Google Scholar]
- 8.Zatzick DF, Marmar CR, Weiss DS, Browner WS, Metzler TJ, Golding JM, Stewart A, Schlenger WE, Wells KB. Posttraumatic stress disorder and functioning and quality of life outcomes in a nationally representative sample of male Vietnam veterans. Am J Psychiatry. 1997;154(12):1690–5. doi: 10.1176/ajp.154.12.1690. [DOI] [PubMed] [Google Scholar]
- 9.Atwoli L, Stein DJ, Koenen KC, McLaughlin KA. Epidemiology of posttraumatic stress disorder: prevalence, correlates and consequences. Curr Opin Psychiatry. 2015;28(4):307–11. doi: 10.1097/YCO.0000000000000167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Seedat SS. Post-traumatic stress disorder. South African Journal of Psychiatry. 2013:187–91. [Google Scholar]
- 11.Morath J, Gola H, Sommershof A, Hamuni G, Kolassa S, Catani C, Adenauer H, Ruf-Leuschner M, Schauer M, Elbert T, Groettrup M, Kolassa IT. The effect of trauma-focused therapy on the altered T cell distribution in individuals with PTSD: evidence from a randomized controlled trial. J Psychiatr Res. 2014;54:1–10. doi: 10.1016/j.jpsychires.2014.03.016. [DOI] [PubMed] [Google Scholar]
- 12.Sommershof A, Aichinger H, Engler H, Adenauer H, Catani C, Boneberg EM, Elbert T, Groettrup M, Kolassa IT. Substantial reduction of naive and regulatory T cells following traumatic stress. Brain Behav Immun. 2009;23(8):1117–24. doi: 10.1016/j.bbi.2009.07.003. [DOI] [PubMed] [Google Scholar]
- 13.Sakaguchi S, Yamaguchi T, Nomura T, Ono M. Regulatory T cells and immune tolerance. Cell. 2008;133(5):775–87. doi: 10.1016/j.cell.2008.05.009. [DOI] [PubMed] [Google Scholar]
- 14.Freier E, Weber CS, Nowottne U, Horn C, Bartels K, Meyer S, Hildebrandt Y, Luetkens T, Cao Y, Pabst C, Muzzulini J, Schnee B, Brunner-Weinzierl MC, Marangolo M, Bokemeyer C, Deter HC, Atanackovic D. Decrease of CD4(+)FOXP3(+) T regulatory cells in the peripheral blood of human subjects undergoing a mental stressor. Psychoneuroendocrinology. 2010;35(5):663–73. doi: 10.1016/j.psyneuen.2009.10.005. [DOI] [PubMed] [Google Scholar]
- 15.O’Donovan A, Cohen BE, Seal KH, Bertenthal D, Margaretten M, Nishimi K, Neylan TC. Elevated Risk for Autoimmune Disorders in Iraq and Afghanistan Veterans with Posttraumatic Stress Disorder. Biol Psychiatry. 2014;77(4):365–74. doi: 10.1016/j.biopsych.2014.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stein MB, Chen C-Y, Ursano RJ, Cai T, Gelernter J, Heeringa SG, Jain S, Jensen KP, Maihofer AX, Mitchell C, Nievergelt CM, Nock MK, Neale BM, Polimanti R, Ripke S, Sun X, Thomas ML, Wang Q, Ware EB, Borja S, Kessler RC, Smoller JW for the Army Study to Assess Risk and Resilience in Servicemembers (STARRS) Collaborators. Genome-wide association studies of posttraumatic stress disorder in 2 cohorts of US Army soldiers. JAMA Psychiatry. 2016:E1–E10. doi: 10.1001/jamapsychiatry.2016.0350. Published online May 11, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lill CM, Schjeide BM, Graetz C, Liu T, Damotte V, Akkad DA, Blaschke P, Gerdes LA, Kroner A, Luessi F, Cournu-Rebeix I, Hoffjan S, Winkelmann A, Touze E, Pico F, Corcia P, Otaegui D, Antiguedad A, Alcina A, Comabella M, Montalban X, Olascoaga J, Matesanz F, Dorner T, Li SC, Steinhagen-Thiessen E, Lindenberger U, Chan A, Rieckmann P, Hartung HP, Aktas O, Lohse P, Buttmann M, Kumpfel T, Kubisch C, Zettl UK, Epplen JT, Fontaine B, Zipp F, Vandenbroeck K, Bertram L. Genome-wide significant association of ANKRD55 rs6859219 and multiple sclerosis risk. J Med Genet. 2013;50(3):140–3. doi: 10.1136/jmedgenet-2012-101411. [DOI] [PubMed] [Google Scholar]
- 18.Alloza I, Otaegui D, de Lapuente AL, Antiguedad A, Varade J, Nunez C, Arroyo R, Urcelay E, Fernandez O, Leyva L, Fedetz M, Izquierdo G, Lucas M, Oliver-Martos B, Alcina A, Saiz A, Blanco Y, Comabella M, Montalban X, Olascoaga J, Matesanz F, Vandenbroeck K. ANKRD55 and DHCR7 are novel multiple sclerosis risk loci. Genes Immun. 2012;13(3):253–7. doi: 10.1038/gene.2011.81. [DOI] [PubMed] [Google Scholar]
- 19.Harder MN, Ribel-Madsen R, Justesen JM, Sparso T, Andersson EA, Grarup N, Jorgensen T, Linneberg A, Hansen T, Pedersen O. Type 2 diabetes risk alleles near BCAR1 and in ANK1 associate with decreased beta-cell function whereas risk alleles near ANKRD55 and GRB14 associate with decreased insulin sensitivity in the Danish Inter99 cohort. J Clin Endocrinol Metab. 2013;98(4):E801–E806. doi: 10.1210/jc.2012-4169. [DOI] [PubMed] [Google Scholar]
- 20.Zhernakova A, Stahl EA, Trynka G, Raychaudhuri S, Festen EA, Franke L, Westra HJ, Fehrmann RS, Kurreeman FA, Thomson B, Gupta N, Romanos J, McManus R, Ryan AW, Turner G, Brouwer E, Posthumus MD, Remmers EF, Tucci F, Toes R, Grandone E, Mazzilli MC, Rybak A, Cukrowska B, Coenen MJ, Radstake TR, van Riel PL, Li Y, de Bakker PI, Gregersen PK, Worthington J, Siminovitch KA, Klareskog L, Huizinga TW, Wijmenga C, Plenge RM. Meta-analysis of genome-wide association studies in celiac disease and rheumatoid arthritis identifies fourteen non-HLA shared loci. PLoS Genet. 2011;7(2):e1002004. doi: 10.1371/journal.pgen.1002004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Viatte S, Plant D, Bowes J, Lunt M, Eyre S, Barton A, Worthington J. Genetic markers of rheumatoid arthritis susceptibility in anti-citrullinated peptide antibody negative patients. Ann Rheum Dis. 2012;71(12):1984–90. doi: 10.1136/annrheumdis-2011-201225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jergovic M, Bendelja K, Vidovic A, Savic A, Vojvoda V, Aberle N, Rabatic S, Jovanovic T, Sabioncello A. Patients with posttraumatic stress disorder exhibit an altered phenotype of regulatory T cells. Allergy Asthma Clin Immunol. 2014;10(1):43. doi: 10.1186/1710-1492-10-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jergovic M, Bendelja K, Savic MA, Vojvoda V, Aberle N, Jovanovic T, Rabatic S, Sabioncello A, Vidovic A. Circulating levels of hormones, lipids, and immune mediators in post-traumatic stress disorder - a 3-month follow-up study. Front Psychiatry. 2015;6:49. doi: 10.3389/fpsyt.2015.00049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maes M, Lin AH, Delmeire L, Van GA, Kenis G, De JR, Bosmans E. Elevated serum interleukin-6 (IL-6) and IL-6 receptor concentrations in posttraumatic stress disorder following accidental man-made traumatic events. Biol Psychiatry. 1999;45(7):833–9. doi: 10.1016/s0006-3223(98)00131-0. [DOI] [PubMed] [Google Scholar]
- 25.Lindqvist D, Wolkowitz OM, Mellon S, Yehuda R, Flory JD, Henn-Haase C, Bierer LM, Abu-Amara D, Coy M, Neylan TC, Makotkine I, Reus VI, Yan X, Taylor NM, Marmar CR, Dhabhar FS. Proinflammatory milieu in combat-related PTSD is independent of depression and early life stress. Brain Behav Immun. 2014 doi: 10.1016/j.bbi.2014.06.003. [DOI] [PubMed] [Google Scholar]
- 26.Hoge EA, Brandstetter K, Moshier S, Pollack MH, Wong KK, Simon NM. Broad spectrum of cytokine abnormalities in panic disorder and posttraumatic stress disorder. Depress Anxiety. 2009;26(5):447–55. doi: 10.1002/da.20564. [DOI] [PubMed] [Google Scholar]
- 27.Heath NM, Chesney SA, Gerhart JI, Goldsmith RE, Luborsky JL, Stevens NR, Hobfoll SE. Interpersonal violence, PTSD, and inflammation: potential psychogenic pathways to higher C-reactive protein levels. Cytokine. 2013;63(2):172–8. doi: 10.1016/j.cyto.2013.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Spitzer C, Barnow S, Volzke H, Wallaschofski H, John U, Freyberger HJ, Lowe B, Grabe HJ. Association of posttraumatic stress disorder with low-grade elevation of C-reactive protein: evidence from the general population. J Psychiatr Res. 2010;44(1):15–21. doi: 10.1016/j.jpsychires.2009.06.002. [DOI] [PubMed] [Google Scholar]
- 29.Plantinga L, Bremner JD, Miller AH, Jones DP, Veledar E, Goldberg J, Vaccarino V. Association between posttraumatic stress disorder and inflammation: a twin study. Brain Behav Immun. 2013;30:125–32. doi: 10.1016/j.bbi.2013.01.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Eraly SA, Nievergelt CM, Maihofer AX, Barkauskas DA, Biswas N, Agorastos A, O’Connor DT, Baker DG. Assessment of plasma C-reactive protein as a biomarker of posttraumatic stress disorder risk. JAMA Psychiatry. 2014;71(4):423–31. doi: 10.1001/jamapsychiatry.2013.4374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pervanidou P, Kolaitis G, Charitaki S, Margeli A, Ferentinos S, Bakoula C, Lazaropoulou C, Papassotiriou I, Tsiantis J, Chrousos GP. Elevated morning serum interleukin (IL)-6 or evening salivary cortisol concentrations predict posttraumatic stress disorder in children and adolescents six months after a motor vehicle accident. Psychoneuroendocrinology. 2007;32(8–10):991–9. doi: 10.1016/j.psyneuen.2007.07.001. [DOI] [PubMed] [Google Scholar]
- 32.Rook GA, Raison CL, Lowry CA. Microbial ‘old friends’, immunoregulation and socioeconomic status. Clin Exp Immunol. 2014;177(1):1–12. doi: 10.1111/cei.12269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sefik E, Geva-Zatorsky N, Oh S, Konnikova L, Zemmour D, McGuire AM, Burzyn D, Ortiz-Lopez A, Lobera M, Yang J, Ghosh S, Earl A, Snapper SB, Jupp R, Kasper D, Mathis D, Benoist C. MUCOSAL IMMUNOLOGY. Individual intestinal symbionts induce a distinct population of RORgamma(+) regulatory T cells. Science. 2015;349(6251):993–7. doi: 10.1126/science.aaa9420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ohnmacht C, Park JH, Cording S, Wing JB, Atarashi K, Obata Y, Gaboriau-Routhiau V, Marques R, Dulauroy S, Fedoseeva M, Busslinger M, Cerf-Bensussan N, Boneca IG, Voehringer D, Hase K, Honda K, Sakaguchi S, Eberl G. MUCOSAL IMMUNOLOGY. The microbiota regulates type 2 immunity through RORgammat(+) T cells. Science. 2015;349(6251):989–93. doi: 10.1126/science.aac4263. [DOI] [PubMed] [Google Scholar]
- 35.Turnbaugh PJ, Ley RE, Hamady M, Fraser-Liggett CM, Knight R, Gordon JI. The human microbiome project. Nature. 2007;449(7164):804–10. doi: 10.1038/nature06244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gill SR, Pop M, Deboy RT, Eckburg PB, Turnbaugh PJ, Samuel BS, Gordon JI, Relman DA, Fraser-Liggett CM, Nelson KE. Metagenomic analysis of the human distal gut microbiome. Science. 2006;312(5778):1355–9. doi: 10.1126/science.1124234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Braniste V, Al-Asmakh M, Kowal C, Anuar F, Abbaspour A, Toth M, Korecka A, Bakocevic N, Guan NL, Kundu P, Gulyas B, Halldin C, Hultenby K, Nilsson H, Hebert H, Volpe BT, Diamond B, Pettersson S. The gut microbiota influences blood-brain barrier permeability in mice. Sci Transl Med. 2014;6(263):263ra158. doi: 10.1126/scitranslmed.3009759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.O’Mahony SM, Marchesi JR, Scully P, Codling C, Ceolho AM, Quigley EM, Cryan JF, Dinan TG. Early life stress alters behavior, immunity, and microbiota in rats: implications for irritable bowel syndrome and psychiatric illnesses. Biol Psychiatry. 2009;65(3):263–7. doi: 10.1016/j.biopsych.2008.06.026. [DOI] [PubMed] [Google Scholar]
- 39.Cryan JF, Dinan TG. Mind-altering microorganisms: the impact of the gut microbiota on brain and behaviour. Nat Rev Neurosci. 2012;13(10):701–12. doi: 10.1038/nrn3346. [DOI] [PubMed] [Google Scholar]
- 40.Reber SO, Siebler PH, Donner NC, Morton JT, Smith DG, Kopelman JM, Lowe KR, Wheeler KJ, Fox JH, Hassell JE, Jr, Greenwood BN, Jansch C, Lechner A, Schmidt D, Uschold-Schmidt N, Fuchsl AM, Langgartner D, Walker FR, Hale MW, Lopez PG, Van TW, Gonzalez A, Halweg-Edwards AL, Fleshner M, Raison CL, Rook GA, Peddada SD, Knight R, Lowry CA. Immunization with a heat-killed preparation of the environmental bacterium Mycobacterium vaccae promotes stress resilience in mice. Proc Natl Acad Sci U S A. 2016;113(22):E3130–E3139. doi: 10.1073/pnas.1600324113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reber SO, Langgartner D, Foertsch S, Postolache TT, Brenner LA, Guendel H, Lowry CA. Chronic subordinate colony housing paradigm: A mouse model for mechanisms of PTSD vulnerability, targeted prevention, and treatment-2016 Curt Richter Award Paper. Psychoneuroendocrinology. 2016;74:221–30. doi: 10.1016/j.psyneuen.2016.08.031. [DOI] [PubMed] [Google Scholar]
- 42.Dinan TG, Borre YE, Cryan JF. Genomics of schizophrenia: time to consider the gut microbiome? Mol Psychiatry. 2014;19(12):1252–7. doi: 10.1038/mp.2014.93. [DOI] [PubMed] [Google Scholar]
- 43.Jørgensen PB, Hansen JT, Krych L, Larsen C, Klein AB, Nielsen DS, Josefsen K, Hansen AK, Sorensen DB. A possible link between food and mood: dietary impact on gut microbiota and behavior in BALB/c mice. PLoS ONE. 2014;9(8):e103398. doi: 10.1371/journal.pone.0103398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Foster JA, McVey Neufeld KA. Gut-brain axis: how the microbiome influences anxiety and depression. Trends Neurosci. 2013;36(5):305–12. doi: 10.1016/j.tins.2013.01.005. [DOI] [PubMed] [Google Scholar]
- 45.Neufeld KM, Kang N, Bienenstock J, Foster JA. Reduced anxiety-like behavior and central neurochemical change in germ-free mice. Neurogastroenterol Motil. 2011;23(3):255–64. e119. doi: 10.1111/j.1365-2982.2010.01620.x. [DOI] [PubMed] [Google Scholar]
- 46.Bercik P, Denou E, Collins J, Jackson W, Lu J, Jury J, Deng Y, Blennerhassett P, Macri J, McCoy KD, Verdu EF, Collins SM. The intestinal microbiota affect central levels of brain-derived neurotropic factor and behavior in mice. Gastroenterology. 2011;141(2):599–609. doi: 10.1053/j.gastro.2011.04.052. [DOI] [PubMed] [Google Scholar]
- 47.Bercik P, Verdu EF, Foster JA, Macri J, Potter M, Huang X, Malinowski P, Jackson W, Blennerhassett P, Neufeld KA, Lu J, Khan WI, Corthesy-Theulaz I, Cherbut C, Bergonzelli GE, Collins SM. Chronic gastrointestinal inflammation induces anxiety-like behavior and alters central nervous system biochemistry in mice. Gastroenterology. 2010;139(6):2102–12. doi: 10.1053/j.gastro.2010.06.063. [DOI] [PubMed] [Google Scholar]
- 48.Heijtz RD, Wang S, Anuar F, Qian Y, Bjorkholm B, Samuelsson A, Hibberd ML, Forssberg H, Pettersson S. Normal gut microbiota modulates brain development and behavior. Proc Natl Acad Sci U S A. 2011;108(7):3047–52. doi: 10.1073/pnas.1010529108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Clarke G, Grenham S, Scully P, Fitzgerald P, Moloney RD, Shanahan F, Dinan TG, Cryan JF. The microbiome-gut-brain axis during early life regulates the hippocampal serotonergic system in a sex-dependent manner. Mol Psychiatry. 2013;18(6):666–73. doi: 10.1038/mp.2012.77. [DOI] [PubMed] [Google Scholar]
- 50.Bravo JA, Forsythe P, Chew MV, Escaravage E, Savignac HM, Dinan TG, Bienenstock J, Cryan JF. Ingestion of Lactobacillus strain regulates emotional behavior and central GABA receptor expression in a mouse via the vagus nerve. Proc Natl Acad Sci U S A. 2011;108(38):16050–5. doi: 10.1073/pnas.1102999108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Barrett E, Ross RP, O’Toole PW, Fitzgerald GF, Stanton C. gamma-Aminobutyric acid production by culturable bacteria from the human intestine. J Appl Microbiol. 2012;113(2):411–7. doi: 10.1111/j.1365-2672.2012.05344.x. [DOI] [PubMed] [Google Scholar]
- 52.Higuchi T, Hayashi H, Abe K. Exchange of glutamate and gamma-aminobutyrate in a Lactobacillus strain. J Bacteriol. 1997;179(10):3362–4. doi: 10.1128/jb.179.10.3362-3364.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Garcia-Rodenas CL, Bergonzelli GE, Nutten S, Schumann A, Cherbut C, Turini M, Ornstein K, Rochat F, Corthesy-Theulaz I. Nutritional approach to restore impaired intestinal barrier function and growth after neonatal stress in rats. J Pediatr Gastroenterol Nutr. 2006;43(1):16–24. doi: 10.1097/01.mpg.0000226376.95623.9f. [DOI] [PubMed] [Google Scholar]
- 54.Pace TW, Heim CM. A short review on the psychoneuroimmunology of posttraumatic stress disorder: from risk factors to medical comorbidities. Brain Behav Immun. 2011;25(1):6–13. doi: 10.1016/j.bbi.2010.10.003. [DOI] [PubMed] [Google Scholar]
- 55.Round JL, Mazmanian SK. The gut microbiota shapes intestinal immune responses during health and disease. Nat Rev Immunol. 2009;9(5):313–23. doi: 10.1038/nri2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chow J, Tang H, Mazmanian SK. Pathobionts of the gastrointestinal microbiota and inflammatory disease. Curr Opin Immunol. 2011;23(4):473–80. doi: 10.1016/j.coi.2011.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang CM, Kaltenboeck B. Exacerbation of chronic inflammatory diseases by infectious agents: Fact or fiction? World J Diabetes. 2010;1(2):27–35. doi: 10.4239/wjd.v1.i2.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Guo G, Jia KR, Shi Y, Liu XF, Liu KY, Qi W, Guo Y, Zhang WJ, Wang T, Xiao B, Zou QM. Psychological stress enhances the colonization of the stomach by Helicobacter pylori in the BALB/c mouse. Stress. 2009;12(6):478–85. doi: 10.3109/10253890802642188. [DOI] [PubMed] [Google Scholar]
- 59.Burich A, Hershberg R, Waggie K, Zeng W, Brabb T, Westrich G, Viney JL, Maggio-Price L. Helicobacter-induced inflammatory bowel disease in IL-10- and T cell-deficient mice. Am J Physiol Gastrointest Liver Physiol. 2001;281(3):G764–G778. doi: 10.1152/ajpgi.2001.281.3.G764. [DOI] [PubMed] [Google Scholar]
- 60.Kullberg MC, Ward JM, Gorelick PL, Caspar P, Hieny S, Cheever A, Jankovic D, Sher A. Helicobacter hepaticus triggers colitis in specific-pathogen-free interleukin-10 (IL-10)-deficient mice through an IL-12- and gamma interferon-dependent mechanism. Infect Immun. 1998;66(11):5157–66. doi: 10.1128/iai.66.11.5157-5166.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yang I, Eibach D, Kops F, Brenneke B, Woltemate S, Schulze J, Bleich A, Gruber AD, Muthupalani S, Fox JG, Josenhans C, Suerbaum S. Intestinal microbiota composition of interleukin-10 deficient C57BL/6J mice and susceptibility to Helicobacter hepaticus-induced colitis. PLoS ONE. 2013;8(8):e70783. doi: 10.1371/journal.pone.0070783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zuany-Amorim C, Sawicka E, Manlius C, Le Moine A, Brunet LR, Kemeny DM, Bowen G, Rook G, Walker C. Suppression of airway eosinophilia by killed Mycobacterium vaccae-induced allergen-specific regulatory T-cells. Nat Med. 2002;8(6):625–9. doi: 10.1038/nm0602-625. [DOI] [PubMed] [Google Scholar]
- 63.Census 2011 Census in brief. Pretoria: Statistics South Africa; 2012. [Google Scholar]
- 64.Sheehan DV, Lecrubier Y, Sheehan KH, Amorim P, Janavs J, Weiller E, Hergueta T, Baker R, Dunbar GC. The Mini-International Neuropsychiatric Interview (M.I.N.I): the development and validation of a structured diagnostic psychiatric interview for DSM-IV and ICD-10. J Clin Psychiatry. 1998;59(Suppl 20):22–33. [PubMed] [Google Scholar]
- 65.Weathers FW, Litz BT, Kean TM, Palmieri PA, Marx BP, Schnurr PP. The PTSD Checklist for DSM-5 (PCL-5) National Center for PTSD; 2013. [Google Scholar]
- 66.Widom CS. Posttraumatic stress disorder in abused and neglected children grown up. Am J Psychiatry. 1999;156(8):1223–9. doi: 10.1176/ajp.156.8.1223. [DOI] [PubMed] [Google Scholar]
- 67.Thayer Z, Barbosa-Leiker C, McDonell M, Nelson L, Buchwald D, Manson S. Early life trauma, post-traumatic stress disorder, and allostatic load in a sample of American Indian adults. Am J Hum Biol. 2016 doi: 10.1002/ajhb.22943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bernstein D, Fink L. Manual for the Childhood Trauma Questionnaire. New York: The Psychological Corporation; 1998. [Google Scholar]
- 69.Zijlmans MA, Korpela K, Riksen-Walraven JM, de Vos WM, de WC. Maternal prenatal stress is associated with the infant intestinal microbiota. Psychoneuroendocrinology. 2015;53:233–45. doi: 10.1016/j.psyneuen.2015.01.006. [DOI] [PubMed] [Google Scholar]
- 70.Klindworth A, Pruesse E, Schweer T, Peplies J, Quast C, Horn M, Glockner FO. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 2013;41(1):e1. doi: 10.1093/nar/gks808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7(5):335–6. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, Huber T, Dalevi D, Hu P, Andersen GL. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol. 2006;72(7):5069–72. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Genuer R, Poggi J-M, Tuleau-Malot C. VSURF: An R package for variable selection using random forests. The R Journal. 2015;7(2):19–33. [Google Scholar]
- 74.Breiman L. Random forests. Machine Learning. 2001;45:5–32. [Google Scholar]
- 75.Ishwaran H, Kogalur UB. [12-7-2016];Package “randomForestSRC”. 2016 http://www.rdocumentation.org/packages/randomForestSRC/functions/randomForestSRC_package.
- 76.Soo RM, Skennerton CT, Sekiguchi Y, Imelfort M, Paech SJ, Dennis PG, Steen JA, Parks DH, Tyson GW, Hugenholtz P. An expanded genomic representation of the phylum cyanobacteria. Genome Biol Evol. 2014;6(5):1031–45. doi: 10.1093/gbe/evu073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Di Rienzi SC, Sharon I, Wrighton KC, Koren O, Hug LA, Thomas BC, Goodrich JK, Bell JT, Spector TD, Banfield JF, Ley RE. The human gut and groundwater harbor non-photosynthetic bacteria belonging to a new candidate phylum sibling to Cyanobacteria. Elife. 2013;2:e01102. doi: 10.7554/eLife.01102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hug LA, Baker BJ, Anantharaman K, Brown CT, Probst AJ, Castelle CJ, Butterfield CN, Hersdorf AW, Amano Y, Ise K, Suzuki Y, Dudek N, Relman DA, Finstad KM, Amundson R, Thomas BC, Banfield JF. A new view of the tree of life. Nature Microbiology. 2016 doi: 10.1038/nmicrobiol.2016.48. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 79.Lowry CA, Smith DG, Siebler PH, Schmidt D, Stamper CE, Hassell JE, Jr, Yamashita PS, Fox JH, Reber SO, Brenner LA, Hoisington AJ, Postolache TT, Kinney KA, Marciani D, Hernandez M, Hemmings SMJ, Malan-Muller S, Wright KP, Knight R, Raison CL, Rook GAWR. The microbiota, immunoregulation and mental health: implications for public health. Current Environmental Health Reports. 2016;3:270–86. doi: 10.1007/s40572-016-0100-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Raison CL, Lowry CA, Rook GA. Inflammation, sanitation, and consternation: loss of contact with coevolved, tolerogenic microorganisms and the pathophysiology and treatment of major depression. Arch Gen Psychiatry. 2010;67(12):1211–24. doi: 10.1001/archgenpsychiatry.2010.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Rook GA, Lowry CA. The hygiene hypothesis and psychiatric disorders. Trends Immunol. 2008;29(4):150–8. doi: 10.1016/j.it.2008.01.002. [DOI] [PubMed] [Google Scholar]
- 82.Rook GA, Raison CL, Lowry CA. Can we vaccinate against depression? Drug Discov Today. 2012;17(9–10):451–8. doi: 10.1016/j.drudis.2012.03.018. [DOI] [PubMed] [Google Scholar]
- 83.Rook GA, Raison CL, Lowry CA. Microbiota, immunoregulatory old friends and psychiatric disorders. Adv Exp Med Biol. 2014;817:319–56. doi: 10.1007/978-1-4939-0897-4_15. [DOI] [PubMed] [Google Scholar]
- 84.Rook GA, Lowry CA, Raison CL. Hygiene and other early childhood influences on the subsequent function of the immune system. Brain Res. 2014;1617:47–62. doi: 10.1016/j.brainres.2014.04.004. [DOI] [PubMed] [Google Scholar]
- 85.Rook GA, Lowry CA, Raison CL. Microbial ‘Old Friends’, immunoregulation and stress resilience. Evol Med Public Health. 2013;2013(1):46–64. doi: 10.1093/emph/eot004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Rook GA, Raison CL, Lowry CA. Childhood microbial experience, immunoregulation, inflammation and adult susceptibility to psychosocial stressors and depression in rich and poor countries. Evol Med Public Health. 2013;2013(1):14–7. doi: 10.1093/emph/eos005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Rook GA, Lowry CA, Raison CL. Hygiene and other early childhood influences on the subsequent function of the immune system. Brain Res. 2015;1617:47–62. doi: 10.1016/j.brainres.2014.04.004. [DOI] [PubMed] [Google Scholar]
- 88.Rook GAW, Lowry CA. The hygiene hypothesis and affective and anxiety disorders. In: Rook GAW, editor. The Hygiene Hypothesis and Darwinian Medicine. Vol. 1. Basel: Birkhauser Publishing; 2009. [Google Scholar]
- 89.Rook GAW, Raison CL, Lowry CA. In: Microbial “Old Friends”, immunoregulation and psychiatric disorders. Heidt PJ, Midtvedt T, Rusch V, Versalovic J, editors. Vol. 26. Old Herborn University Press; 2013. pp. 61–90. Old Herborn University Monograph. [Google Scholar]
- 90.Shin NR, Lee JC, Lee HY, Kim MS, Whon TW, Lee MS, Bae JW. An increase in the Akkermansia spp. population induced by metformin treatment improves glucose homeostasis in diet-induced obese mice. Gut. 2014;63(5):727–35. doi: 10.1136/gutjnl-2012-303839. [DOI] [PubMed] [Google Scholar]
- 91.Gomez-Gallego C, Pohl S, Salminen S, de Vos WM, Kneifel W. Akkermansia muciniphila: a novel functional microbe with probiotic properties. Benef Microbes. 2016;7(4):571–84. doi: 10.3920/BM2016.0009. [DOI] [PubMed] [Google Scholar]
- 92.Jiang H, Ling Z, Zhang Y, Mao H, Ma Z, Yin Y, Wang W, Tang W, Tan Z, Shi J, Li L, Ruan B. Altered fecal microbiota composition in patients with major depressive disorder. Brain Behav Immun. 2015;48:186–94. doi: 10.1016/j.bbi.2015.03.016. [DOI] [PubMed] [Google Scholar]
- 93.Karl JP, Margolis LM, Madslien EH, Murphy NE, Castellani JW, Gundersen Y, Hoke AV, Levangie MW, Kumar R, Chakraborty N, Gautam A, Hammamieh R, Martini S, Montain SJ, Pasiakos SM. Changes in intestinal microbiota composition and metabolism coincide with increased intestinal permeability in young adults under prolonged physiologic stress. Am J Physiol Gastrointest Liver Physiol. 2017:ajpgi. doi: 10.1152/ajpgi.00066.2017. [DOI] [PubMed] [Google Scholar]
- 94.Biagi E, Franceschi C, Rampelli S, Severgnini M, Ostan R, Turroni S, Consolandi C, Quercia S, Scurti M, Monti D, Capri M, Brigidi P, Candela M. Gut Microbiota and Extreme Longevity. Curr Biol. 2016;26(11):1480–5. doi: 10.1016/j.cub.2016.04.016. [DOI] [PubMed] [Google Scholar]
- 95.De Bellis MD, Zisk A. The biological effects of childhood trauma. Child Adolesc Psychiatr Clin N Am. 2014;23(2):185–222. vii. doi: 10.1016/j.chc.2014.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Lyte M. Microbial endocrinology in the microbiome-gut-brain axis: how bacterial production and utilization of neurochemicals influence behavior. PLoS Pathog. 2013;9(11):e1003726. doi: 10.1371/journal.ppat.1003726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Hodes GE, Pfau ML, Leboeuf M, Golden SA, Christoffel DJ, Bregman D, Rebusi N, Heshmati M, Aleyasin H, Warren BL, Lebonte B, Horn S, Lapidus KA, Stelzhammer V, Wong EH, Bahn S, Krishnan V, Bolanos-Guzman CA, Murrough JW, Merad M, Russo SJ. Individual differences in the peripheral immune system promote resilience versus susceptibility to social stress. Proc Natl Acad Sci U S A. 2014;111(45):16136–41. doi: 10.1073/pnas.1415191111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Alverdy J, Aoys E. The effect of glucocorticoid administration on bacterial translocation. Evidence for an acquired mucosal immunodeficient state. Ann Surg. 1991;214(6):719–23. doi: 10.1097/00000658-199112000-00012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Tursich M, Neufeld RW, Frewen PA, Harricharan S, Kibler JL, Rhind SG, Lanius RA. Association of trauma exposure with proinflammatory activity: a transdiagnostic meta-analysis. Transl Psychiatry. 2014;4:e413. doi: 10.1038/tp.2014.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.