Summary
In this issue of Cancer Discovery, Shi and colleagues add further insight into the role of exon 3 MEK1 mutations in BRAF inhibitor resistance by demonstrating the presence of P124SMEK1 and I111SMEK1 mutations concurrently with V600E/KBRAF mutations at baseline in 16% of melanoma specimens. Although the presence of P124SMEK1 or I111SMEK1 mutations did not predict for resistance, and these alleles were not selected for upon BRAF inhibition, other exon 3 MEK1 mutations, such as C121S, did convey resistance, suggesting a role for defined exon 3 MEK1 mutations in acquired BRAF inhibitor resistance.
The discovery that 50% of all cutaneous melanomas harbor activating mutations in the serine/threonine kinase BRAF has revolutionized expectations for melanoma therapy. In the recent phase III randomized clinical trial, 53% of patients selected on the basis of their melanomas harboring a position-600 mutation showed good levels of response to the small-molecule BRAF inhibitor vemurafenib [Zelboraf, PLX4032; Genentech (1)]. Despite these promising results, and the unprecedented hope this offers to patients with melanoma, significant hurdles remain. We do not yet fully understand why nearly one half of all patients with BRAF-mutant melanoma do not show a response to BRAF inhibitors, as measured by Response Evaluation Criteria in Solid Tumors (RECIST) criteria. Among the patients with BRAF-mutant melanoma whose tumors do respond well to treatment initially, most responses tend to be short-lived, with resistance emerging in nearly every case. Updated data from the phase II clinical trial of vemurafenib shows an average progression-free survival of 7 months, with an overall survival of 16 months. It is clear that both intrinsic and acquired resistance to BRAF inhibitors is common and efforts are underway to identify these resistance mechanisms and to develop mitigation strategies.
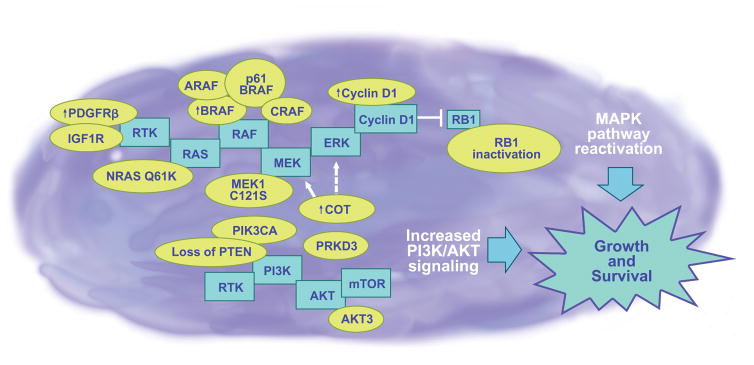
A large number of potential BRAF inhibitor resistance mechanisms have now been described. The identified mechanisms differ from those implicated in the acquired resistance to other targeted therapy agents, such as imatinib in chronic myeloid leukemia and erlotinib in non–small cell lung cancer, in that they do not involve secondary kinase mutations at the “gatekeeper” site. Instead, a diverse array of acquired resistance mechanisms have been reported, including mutations in NRAS, upregulated expression of a number of receptor tyrosine kinases (IGF1R, PDGFR-β), truncations of BRAF, amplification of BRAF, and increased expression of kinases known to activate the mitogen-activated protein kinase (MAPK) signaling pathway, particularly COT and CRAF [Fig. 1 (2)].
Figure 1.
Various mechanisms (shown in yellow balloons) have been defined that can lead to BRAF inhibitor resistance in baseline and disease-progressed BRAF-mutated melanomas and include PDGFRβ and IGF1R receptor tyrosine kinase signaling, secondary NRAS mutations, V600EBRAF amplification, V600EBRAF p61 splice variant, RAF isoform signal switching, C121SMEK1 mutation, COT amplification, increased AKT activity, loss of PTEN, PRKD3, amplified cyclin D1, and RB1 inactivation. The central theme of BRAF inhibitor escape is the reactivation of MAP kinase and increased PI3K/AKT/mTOR signaling (skeleton pathways shown in blue boxes), which leads to melanoma growth and survival. As a further complication to the incredibly diverse resistance landscape, it has been demonstrated that multiple mutations, and possibly others that have yet to be identified, can occur within the same melanoma tumor or cell line, resulting in intratumor heterogeneity. It is expected that the complexity of mutations that can occur within one tumor will ultimately redirect our strategies towards individualized therapy.
Melanomas are well known to be “addicted” to signaling through the Ras/Raf/MEK/ERK MAPK pathway, with the majority of melanoma driver oncogenes reported thus far known to activate this signal transduction cascade. Even among those melanomas lacking the obvious MAPK pathway activators such as oncogenic BRAF and NRAS, constitutive levels of MEK/ERK signaling are still required. It therefore comes as little surprise that melanomas possess multiple means to reactivate MAPK signaling and that many of the acquired BRAF inhibitor resistance mechanisms reported so far, including most of those that have been convincingly validated in clinical specimens, are MAPK dependent. The fact that near total MAPK pathway blockade is required for BRAF inhibitor efficacy in patients with melanoma suggests that even modest increases in the level of signaling through the Ras/Raf/MEK/ERK pathway are sufficient to convey drug resistance, and there is now experimental evidence to support this contention (3).
In this issue of Cancer Discovery, Shi and colleagues (4) present new data exploring the potential role of exon 3 MEK1 mutations in intrinsic and acquired BRAF inhibitor resistance. This work builds on a previous study in which investigators implicate the role of an acquired exon 3 C121SMEK1 mutation in one patient with melanoma who did not respond to vemurafenib therapy (5). Using a series of matched (pre- and posttreatment) samples from 31 patients treated with either vemurafenib or dabrafenib (GSK2118436), the authors performed whole-exome sequencing and demonstrated that 5 of 31 specimens harbored either P124SMEK1 or I111SMEK1 mutations concurrently with V600E/KBRAF mutations. These mutations were somatic, and no equivalent mutations in MEK2 were noted in the pretreatment samples. Unexpectedly, failure of BRAF inhibitor therapy did not select for exon 3 MEK1 mutations, and the only patients with MEK1 mutations in their melanomas upon disease progression already harbored these at baseline.
Of clinical significance, the presence of either the P124SMEK1 or I111SMEK1 mutations was not predictive of up-front resistance to BRAF inhibitor therapy, with 4 of the 5 patients with concurrent BRAF and MEK1 mutations displaying objective responses in the tumors biopsied and 3 of these patients ultimately achieving an overall partial response (as measured by RECIST criteria). Because these exon 3 MEK1 mutations did not appear to predict for intrinsic BRAF inhibitor resistance, the authors next performed in vitro studies and observed that lentiviral-mediated introduction of either wild-type MEK1, P124SMEK1, or I111SMEK1 did not increase activation of extracellular signal-regulated kinase (ERK) in melanoma cells that were V600E/KBRAF mutant. In contrast, introduction of the P124SMEK1 mutant into human cells (HEK293) lacking oncogenic BRAF increased phospho-ERK signaling, suggesting that the oncogenic BRAF in melanoma cells was dominant over the 2 exon 3 MEK1 mutants with regard to ERK activation. Although it is not yet clear why mutant BRAF would be more effective at activating ERK than P124SMEK1 or I111SMEK1, melanoma cells are known to have impaired feedback inhibition in the Ras/Raf/MEK/ERK signaling pathway, suggesting that inhibition/activation at different levels of the signaling cascade may not be equivalent (6).
Consistent with the identified MEK1 mutants having little influence upon phospho-ERK levels, further studies by Shi and colleagues (4) showed neither of the MEK1 mutants to convey vemurafenib resistance in either growth or clonogenic assays. Not all exon 3 MEK1 mutants identified from patients with melanoma were similarly inactive with regards to vemurafenib resistance. In agreement with previously published work, Shi and colleagues (4) provided in vitro data demonstrating the ability of the C121SMEK1 mutation to restore levels of phospho-ERK signaling in the face of BRAF inhibition leading to a decrease in the sensitivity to vemurafenib (5). It therefore seems that although some mutations in MEK1 may play a role in BRAF inhibitor resistance, the baseline presence of a MEK1 mutation is not necessarily predictive of a diminished drug response. Further study of a larger series of pre- and posttreatment biopsies is therefore required to determine the spectrum of MEK1 mutations that have utility as predictive biomarkers.
The evolving experience of targeted therapy in melanoma and other tumors shows single-agent inhibitor strategies to be largely ineffective at delivering durable antitumor responses. Although combination therapy strategies may yet prove to be curative in genetically defined cases, much is still to be done in terms of developing combination therapies and matching these with tumor genotypes. In line with the observation that restoration of MAPK signaling mediates resistance to vemurafenib, Shi and colleagues (4) demonstrated that “vertical inhibition” of the MAPK pathway through combined vemurafenib and MEK inhibitor (AZD6244) treatment led to a strong degree of synergy in melanoma cell lines with V600EBRAF mutations. This observation, which corroborates earlier studies in which the authors demonstrated that combined BRAF and MEK inhibition delays the onset of BRAF inhibitor resistance, is currently the subject of an eagerly awaited phase II clinical trial [dabrafenib plus trametinib (GSK1120212); ref.¤7, 8]. The combination of a BRAF plus a MEK inhibitor is likely to prove particularly beneficial in preventing the paradoxical MAPK signaling-driven development of squamous tumors that often occur in patients with melanoma receiving BRAF inhibitor therapy (9).
Reactivation of MAPK signaling is not the only mechanism used by melanoma cells to escape from BRAF inhibition, and it has been suggested that resistance may also be managed through the dual targeting of mutant BRAF and components of the PI3K/AKT/mTOR pathway (2, 3). At this juncture, we do not have good biomarkers for segregating V600EBRAF mutant melanomas into those that would benefit from the BRAF and MEK inhibitor combination versus those that would respond better to the BRAF and PI3K/AKT inhibitor combination. Ultimately, however, these issues may be difficult to resolve. There is a growing realization that tumors are much more heterogeneous than previously suspected, and it is expected that this will complicate our efforts to develop patient-specific combination therapies. A recent whole-exome sequencing analysis of multiple specimens taken from the same patient with renal cell carcinoma highlighted the nature of intratumoral diversity and showed that two thirds of the mutations found in one specimen were not detected in other tumor biopsies from the same patient (10). Most remarkably of all, a favorable prognosis gene signature and an unfavorable prognosis gene signature were found in different regions of the same tumor nodule (10). With this in mind, it is likely that multiple BRAF inhibitor resistance mechanisms could coexist within the same patient’s melanoma and require different management strategies. There is already evidence from the whole-genome sequencing of cell lines that melanoma is a tumor with a very high mutational load (11). This mutational complexity is likely to drive the high levels of intratumoral heterogeneity already known to exist in melanoma and lead to the development of evolutionarily diverse tumors within the same patient. Understanding this heterogeneity and developing strategies to manage multiple coexistent therapeutic escape routes will prove critical in our efforts to develop combination therapies that deliver durable responses to patients with disseminated melanoma.
Footnotes
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Authors’ Contributions
Conception and design: K.S.M. Smalley
Writing, review, and/or revision of the manuscript: K.H.T. Paraiso, K.S.M. Smalley
Administrative, technical, or material support: K.H.T. Paraiso
References
- 1.Chapman PB, Hauschild A, Robert C, Haanen JB, Ascierto P, Larkin J, et al. Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N Engl J Med. 2011;364:2507–16. doi: 10.1056/NEJMoa1103782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fedorenko IV, Paraiso KH, Smalley KS. Acquired and intrinsic BRAF inhibitor resistance in BRAF V600E mutant melanoma. Biochem Pharmacol. 2011;82:201–9. doi: 10.1016/j.bcp.2011.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Su F, Bradley WD, Wang QQ, Yang H, Xu LZ, Higgins B, et al. Resistance to selective BRAF inhibition can be mediated by modest upstream pathway activation. Cancer Res. 2012;72:969–78. doi: 10.1158/0008-5472.CAN-11-1875. [DOI] [PubMed] [Google Scholar]
- 4.Shi H, Moriceau G, Kong X, Koya RC, Nazarian R, Pupo GM, et al. Preexisting MEK1 exon 3 mutations in V600E/KBRAF melanomas do not confer resistance to BRAF inhibitors. Cancer Discov. 2012;5:414–24. doi: 10.1158/2159-8290.CD-12-0022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wagle N, Emery C, Berger MF, Davis MJ, Sawyer A, Pochanard P, et al. Dissecting therapeutic resistance to RAF inhibition in melanoma by tumor genomic profiling. J Clin Oncol. 2011;29:3085–96. doi: 10.1200/JCO.2010.33.2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pratilas CA, Taylor BS, Ye Q, Viale A, Sander C, Solit DB, et al. (V600E)BRAF is associated with disabled feedback inhibition of RAF-MEK signaling and elevated transcriptional output of the pathway. Proc Natl Acad Sci U S A. 2009;106:4519–24. doi: 10.1073/pnas.0900780106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Paraiso KH, Fedorenko IV, Cantini LP, Munko AC, Hall M, Sondak VK, et al. Recovery of phospho-ERK activity allows melanoma cells to escape from BRAF inhibitor therapy. Br J Cancer. 2010;102:1724–30. doi: 10.1038/sj.bjc.6605714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Infante JR, Falchook GS, Lawrence DA, Weber J, Kefford R, Bendell J, et al. Phase I/II Study of the oral MEK1/2 inhibitor GSK1120212 dosed in combination with the oral BRAF inhibitor GSK2118436. J Clin Oncol. 2011;29:CRA8503. [Google Scholar]
- 9.Su F, Viros A, Milagre C, Trunzer K, Bollag G, Spleiss O, et al. RAS mutations in cutaneous squamous-cell carcinomas in patients treated with BRAF inhibitors. N Engl J Med. 2012;366:207–15. doi: 10.1056/NEJMoa1105358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gerlinger M, Rowan AJ, Horswell S, Larkin J, Endesfelder D, Gronroos E, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med. 2012;366:883–92. doi: 10.1056/NEJMoa1113205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pleasance ED, Cheetham RK, Stephens PJ, McBride DJ, Humphray SJ, Greenman CD, et al. A comprehensive catalogue of somatic mutations from a human cancer genome. Nature. 2010;463:191–6. doi: 10.1038/nature08658. [DOI] [PMC free article] [PubMed] [Google Scholar]