Abstract
Background
Influenza A virus in swine herds represents a major problem for the swine industry and poses a constant threat for the emergence of novel pandemic viruses and the development of more effective influenza vaccines for pigs is desired. By optimizing the vector backbone and using a needle-free delivery method, we have recently demonstrated a polyvalent influenza DNA vaccine that induces a broad immune response, including both humoral and cellular immunity.
Objectives
To investigate the protection of our polyvalent influenza DNA vaccine approach in a pig challenge study.
Methods
By intradermal needle-free delivery to the skin, we immunized pigs with two different doses (500 μg and 800 μg) of an influenza DNA vaccine based on six genes of pandemic origin, including internally expressed matrix and nucleoprotein and externally expressed hemagglutinin and neuraminidase as previously demonstrated. Two weeks following immunization, the pigs were challenged with the 2009 pandemic H1N1 virus.
Results
When challenged with 2009 pandemic H1N1, 0/5 vaccinated pigs (800 μg DNA) became infected whereas 5/5 unvaccinated control pigs were infected. The pigs vaccinated with the low dose (500 μg DNA) were only partially protected. The DNA vaccine elicited binding-, hemagglutination inhibitory (HI) − as well as cross-reactive neutralizing antibody activity and neuraminidase inhibiting antibodies in the immunized pigs, in a dose-dependent manner.
Conclusion
The present data, together with the previously demonstrated immunogenicity of our influenza DNA vaccine, indicate that naked DNA vaccine technology provides a strong approach for the development of improved pig vaccines, applying realistic low doses of DNA and a convenient delivery method for mass vaccination.
Keywords: Challenge, DNA vaccine, H1N1pdm09, Needle-free immunization, Protection, Swine influenza
1. Introduction
Influenza A virus infections in swine herds constitute a well-known challenge to the swine industry. Reproductive problems together with weight loss and aggravation of secondary infections are characteristic of swine influenza and result in serious animal welfare problems and economic losses (Bennett et al., 1999, Olsen et al., 2006). The influenza infection in pigs resembles the infection in humans. The virus replicates in the epithelium of the entire respiratory tract but rarely infects other tissues (van der Laan et al., 2008). The disease lasts for 7–10 days seldom results in death of the animals (van der Laan et al., 2008). In addition, the tremendous genetic plasticity of the virus can result in transmission between animals as well as zoonotic transmission and adaptation to human hosts, resulting in novel pandemic influenza strains such as the pandemic 2009 H1N1 strain (Ito et al., 1998, Nelson and Vincent, 2015, Smith et al., 2009). A successful, more broadly protective vaccine for pigs against influenza A virus is very much desired, since it will improve the health in pig herds, limit the use of antibiotics and lower the risk of transmission to other species, such as humans. The current vaccines against influenza A virus for pigs are based on inactivated virus and only induce immunity against the virus strains included in the vaccines, thus providing only limited protection against the diverse spectrum of other circulating influenza strains (Sandbulte et al., 2015). DNA vaccine technology is already approved for use in pigs (Thacker et al., 2006) and has many advantages required for an effective influenza vaccine, such as rapid production, easy plasmid modification, a stable formulation and in vivo antigen expression leading to induction of both broad and long-lived cellular and humoral immunity (Kutzler and Weiner, 2008, Li and Petrovsky, 2015, Liu, 2011). The technique has previously been tested by us and others in pigs against influenza (Bragstad et al., 2013, Eriksson et al., 1998, Gorres et al., 2011, Heinen et al., 2002, Larsen et al., 2001, Macklin et al., 1998, Olsen, 2000). Several optimizations can be applied today to improve the production and immunogenicity of the vaccines. We and others have described improvements of influenza DNA vaccines, including optimizing the plasmid vector backbone (Borggren et al., 2015, Williams, 2013). and delivery of the vaccine intradermally with a convenient needle-free device developed for mass vaccination (Borggren et al., 2015, Martelli et al., 2007).
Recently, we reported a broad immune response induced in pigs by a DNA vaccine in vivo expressing six different genes of pandemic viral origin (Borggren et al., 2016, Borggren et al., 2015). The pandemic nature of the DNA genes makes them the ancestor of all subsequent strains and are naturally less glycosylated when expressed in vivo, compared to circulating virus strains with more glycosylation acquired by antigenic drift (Sriwilaijaroen and Suzuki, 2012, Wang et al., 2009, Wei et al., 2010). Consequently, a broader range of epitopes can be recognized by the DNA vaccine-induced response, thus producing a more cross-reactive immunity. Thus, our DNA vaccine induced both humoral and cellular immunity against virus strains homologous and heterologous to the DNA vaccine genes (Borggren et al., 2016). In the present study, we have evaluated the immunogenicity, dose-response, and protective effect of the same polyvalent DNA vaccine in an influenza-virus challenge study in pigs.
2. Materials and methods
2.1. Construction of DNA vaccines
The six influenza DNA vaccine genes have been described previously (Borggren et al., 2016, Borggren et al., 2015). Briefly, the 6 influenza DNA vaccine genes were designed from nucleotide sequences published in GenBank derived from only pandemic influenza strains; 1918 NP: A/Brevig Mission/1/18(H1N1), 1918 M: A/Brevig Mission/1/18(H1N1), 2009 HA: A/California/04/2009(H1N1)pdm09, 2009 NA: A/California/04/2009(H1N1)pdm09, 1968 HA: A/Aichi/2/1968(H3N2), 1968 NA: A/Aichi/2/1968(H3N2). The genes were synthesized and designed to include the appropriate restriction enzymes and the Kozak-sequence (GCCACC) upstream from the start codon, for efficient cloning and transcription into the expression vector. All genes were synthesized using only codons from highly expressed human or mammalian genes (codon optimized), except the M gene that was not codon optimized. The minimal NTC9385R plasmid, free of antibiotic resistance genes, was used as the expression vector backbone (Williams, 2013).
2.2. Animals and experimental design
Fifteen 5-to-6-week-old, recently weaned male pigs (Yorkshire x Landrace breed), tested influenza-negative by ID Screen® Influenza A Antibody Competition Multi-species ELISA (ID.VET, France), were procured from a commercial Spanish high-health herd free from Porcine Respiratory and Reproductive Syndrome (PRRS), Aujeszky’s disease, Pasteurella multocida and Brachyspira spp., but positive for Haemophilus parasuis, Mycoplasma hyopneumoniae and Actinobacillus pleuropneumoniae serotype 2. Prior to weaning, the pigs had been vaccinated against porcine circovirus type 2 (PCV2) and Mycoplasma hyopneumoniae. The pigs were randomly assigned to three groups of five animals (two vaccinated groups allocated in one box and one non-vaccinated group in another one). Boxes were subjected to negative pressure at the biosafety level 3 isolation facilities of the Centre de Recerca en Sanitat Animal (CReSA), Institut de Recerca i Tecnologia Agroalimentàries (IRTA), Spain. Pigs were allowed to acclimatize for one week before the initiation of the experiment.
With an interval of three weeks, two groups of pigs were vaccinated twice on the dorsal side of the back using the needle-free Intra-Dermal Application of Liquids (IDAL® MSD Animal Health) device (Ferrari et al., 2011, Visser et al., 1994). For use of the IDAL® device, the vaccine constructs were premixed at a 1:1 vol ratio with an α-tocopherol-based aqueous solution (Diluvac Forte®, MSD Animal Health) (Borggren et al., 2016, Borggren et al., 2015). Five pigs were immunized with 500 μg of DNA (83 μg per gene/plasmid) each (one shot of 200 μl (2.5 mg/ml Diluvac)) on the back of individual pigs). Five pigs received 800 μg of DNA (133 μg per gene/plasmid) each, distributed into four shot sites à 200 μl (1 mg/ml of Diluvac) on the back of individual pigs. Five pigs remained unvaccinated and constituted a non-immunized control group. Two weeks after the second vaccination, all pigs were challenged intranasally (i.n.) with 106 (TCID50)/pig of pandemic A/California/7/09 (H1N1)pdm09 applied in 1.5 ml into each nostril. All pigs were monitored daily for clinical signs of disease or any adverse vaccination-related effects. Rectal body temperatures were recorded daily starting from two days before challenge until the end of the experiment. Whole-blood samples were collected from the anterior vena cava of all pigs on days −36, −28, −21, −15, −7, 0, 7 and 13 post challenge (pc). Serum was isolated and stored at −20 °C for subsequent examination. On days 0, 3, 5, 7, 9 and 13 pc, nasal swab samples were collected in virus transport medium (phosphate-buffered saline (PBS) containing antimicrobial drugs (100 U/mL penicillin and 0.1 mg/ml streptomycin)) from all pigs to evaluate nasal virus shedding. Samples were stored at −80 °C until testing. Upon termination of the experiment, on day 13 pc, the pigs were euthanized by intravenous injection of a lethal dose of pentobarbital followed by exsanguination. Post mortem, gross-pathological evaluation was carried out and lung tissues (apical and middle lobes as well as other potential lobes if evidence of gross lesions) were taken and fixed by immersion in 10% buffered formalin. Lung tissues were subsequently embedded in paraffin, cut in 4 μm sections, stained with hematoxylin-eosin stain, and slides were observed under an optical microscope. Potential swine influenza-like lesions (broncho-interstitial pneumonia) were scored using a previously published work (Detmer et al., 2013).
The present study was approved by IRTA’s Ethics Committee for Animal Experimentation and the Animal Experimentation Commission from the Autonomous Community of Catalonia Government in compliance with the Directive, UE 63/2010 and the Spanish Legislation, RD 53/2013 and the Catalan Law 5/1995 and Decree 214/1997
2.3. Influenza virus detection
A quantitative reverse transcriptase polymerase chain reaction (qRT-PCR) assay was utilized to monitor viral loads in nasal swab samples (day 0, 3, 5, 7, 9 and 13 pc). RNA was extracted with a MagNA Pure LC Instrument applying the MagNA Pure LC Total Nucleic Acid Isolation Kit (Roche diagnostics). Primers and probes for the neuraminidase (NA) gene of challenge virus A/California/7/09 (H1N1)pdm09 were used to detect the challenge virus. The beta-actin housekeeping cellular gene was used as a control for correct sampling that should contain pig derived cell material in the swaps. Quantification of virus was performed by using a standard curve developed by serial dilutions of H1N1pdm09 virus with known TCID50/ml concentration, included with each RT-PCR assay. Assay sensitivity for detection of challenge virus was 15 TCID50/ml.
2.4. Enzyme-linked immunosorbent assay (ELISA)
An ELISA was conducted to measure influenza-specific IgG responses in the sera as previously described (Borggren et al., 2015). The influenza virus protein used for coating was hemagglutinin (HA) from A/California/04/09 (H1N1)pdm09 (Sino Biological Inc.). A horseradish peroxidase-conjugated anti-pig-IgG antibody (AbD Serotec) followed by TMB substrate (Kem-En-Tec Diagnostics) was used for detection. The binding antibody titers are expressed as the reciprocal of the sample dilution giving an optical density (OD) value of 1.0 (Bragstad et al., 2011).
2.5. Hemagglutination inhibition (HI) assay
The HI assay was performed according to the protocols of the WHO as previously described (Borggren et al., 2015) against virus strain A/California/07/09 (H1N1pdm).
2.6. Microneutralization assay (MN)
Development of neutralizing antibodies was determined according to the protocols of the WHO (Who, 2011). Viruses used were A/California/07/09 (H1N1pdm), A/swine/DK/102586/2007 (H1N1) and A/swine/DK/10496/2008 (H1N1), with 100 TCID50 as the inoculum. A/swine/DK/102586/2007 (H1N1) and A/swine/DK/10496/2008 (H1N1) both have an avian-like H1. The MN titers is defined as the reciprocal dilution giving 50% infection inhibition and is calculated as stated in the WHO protocol (Fenyö et al., 2009, Who, 2011) and Linear interpolation by Reed and Muench (Reed and Muench, 1938) is used to estimate titers if falling between two adjacent serum dilutions.
2.7. Neuraminidase inhibition (NAI) assay
The neuraminidase inhibition activity was determined using the NA-Fluor Influenza Neuraminidase Assay kit (Life technologies) according to the manufactures protocol, where serum and virus was allowed to incubate 30 min at 37 °C before adding the NA-Fluor™ Substrate. The two virus strains, A/California/07/09 (H1N1)pdm09 and A/swine/DK/10496/2008 (H1N1), were used at 1:10 dilution, determined to give a relative fluorescence unit (RFU) within the linear range. Serum from day −36, day 0 and day 13 pc were tested at 4-fold dilutions starting at 1:10 up to 1:1040. Non-linear regression, with log-transformed serum dilutions as the independent variable, was performed to calculate the serum dilution that would inhibit 50% NA activity (IC50) (Graph-Pad Prism software).
2.8. Statistical analysis
Differences between the groups were calculated using two-way ANOVA and Dunnett’s multiple comparison test, and correlations between different humoral immunity assays was performed using Spearman rank correlation (GraphPad Prism v.6, GraphPad software).
3. Results
3.1. Clinical evaluation
None of the pigs showed any signs of clinical disease during the experiment, and elevated body temperatures were not observed in any individual animal.
3.2. Pathological evaluation
Few pigs displayed mild pulmonary cranio-ventral consolidation (Table 1), potentially compatible with pulmonary collapse (non-virus related) or broncho-interstitial pneumonia (virus related). Histopathological evaluation revealed that pigs receiving the highest dose of DNA (800 μg) showed no lung lesions, thus no evidence of infection (5/5 scored 0). Those receiving the lower dose (500 μg) showed mild lung pathology (in 3/5 with a score of 0.5, indicating that only a few isolated airways were affected). Four out of five pigs from the unvaccinated group showed mild to moderate microscopic lung lesions consistent with broncho-interstitial pneumonia (1/5 scored 0; 1/5 scored 0.5; 2/5 scored 1.5 and 1/5 scored 2). The histopathological examination revealed that the lesions observed in three pigs were due to lung collapse, not related to the influenza infection.
Table 1.
Pathological findings.
| Group | Pig no. | Gross-pathological lesions | Scores–lungsA | Swine influenza virus-like lesions |
|---|---|---|---|---|
| 800 μg DNA/vacc. | 1 | No apparent lesions | 0 | None |
| 2 | Small consolidated areas (multifocal) in left median lobe | 0 | None | |
| 3 | Minimal consolidation foci in the left lobe | 0 | None | |
| 4 | No apparent lesions | 0 | None | |
| 5 | No apparent lesions | 0 | None | |
| 500 μg DNA/vacc. | 5 | Very minimal consolidation foci right cranial lobe and left median lobe | 0.5 | Very mild broncho-interstitial pneumonia |
| 7 | Small consolidated areas in right median lobe and cranial portion of right diaphragmatic lobe | 0.5 | Very mild broncho-interstitial pneumonia | |
| 8 | No apparent lesions | 0 | None | |
| 9 | Minimal consolidation areas/foci in the left medium and right cranial lobes | 0.5 | Very mild broncho-interstitial pneumonia | |
| 10 | No apparent lesions | 0 | None | |
| No vaccine | 11 | Lung consolidation in right median lobe and more areas in both cranial lobes | 1.5 | Mild-to-moderate broncho-interstitial pneumonia |
| 12 | Small consolidation areas in both median lobes | 2 | Moderate broncho-interstitial pneumonia | |
| 13 | Consolidation areas in right median and left median lobes and cranial areas of left diaphragmatic lobe | 0 | None | |
| 14 | Small areas of consolidation in the right apical lobe | 0.5 | Very mild broncho-interstitial pneumonia | |
| 15 | Very small areas of multifocal consolidation in the right apical lobe and moderate extensive areas in the left middle lobe | 1.5 | Mild-to-moderate broncho-interstitial pneumonia | |
Scoring system of lung lesions by influenza in pigs according to Detmer et al. (2013) 0, No airways affected; 0.5, Only a few isolated airways affected; 1, Localized cluster of affected airways (in 1 or 2 lobes); 1.5, Several airways affected throughout section plus minimal interstitial infiltrates; 2, Several airways affected throughout section plus mild to moderate interstitial infiltrates; 2.5, Several airways affected, often severely, plus moderate interstitial and alveolar infiltrates; 3, Many airways affected, often severely, plus moderate interstitial and alveolar infiltrates.
3.3. Antibody responses in DNA vaccinated pigs
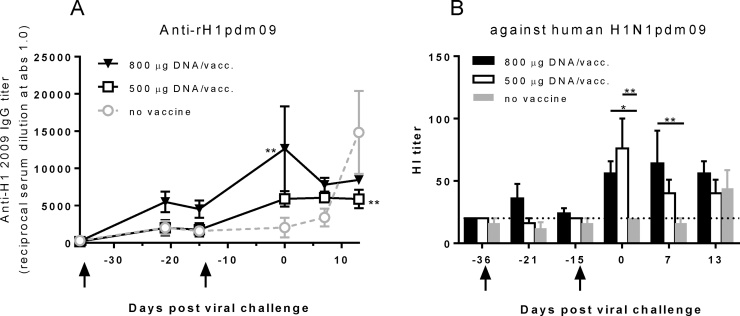
All vaccinated pigs developed antibodies to HA, NA, M, and NP proteins. Pigs receiving the highest dose of DNA (800 μg) developed influenza H1pdm09-specific binding antibodies by ELISA 14 days after the initial immunization (Fig. 1A). Following the second immunization, the antibody titers increased and pigs receiving the lower dose of DNA (500 μg) also developed antibodies. Control animals, not receiving any immunization, developed a detectable HA-specific antibody response only after viral challenge. Functionality of the HA binding antibodies was confirmed by the HI assay, demonstrating that DNA vaccination elicited an HI antibody response against the H1N1pdm09 virus (Fig. 1B). Both vaccinated groups showed significantly higher HI titers than the control group after the second immunization. After challenge, the vaccinated animals maintained their HI titers and the control animals demonstrated a response to the challenge virus two weeks after challenge. Binding antibody titers (ELISA) correlated with HI titers in the vaccinated animals (P < 0.0001, R = 0.69, Spearman rank correlation test).
Fig. 1.
HA-specific antibody responses following DNA vaccination and challenge. Pigs were vaccinated twice (arrows) i.d. with needle-free delivery with 800 μg (n = 5) or 500 μg (n = 5), or not DNA vaccinated at all (n = 5). Levels of IgG in the pre- and post-challenge sera were measured by ELISA against recombinant HA (H1N1pdm09) (A). Hemagglutination inhibition antibody responses in the pre- and post-challenge sera against H1N1pdm09 were measured (B). A Hi-titer cut-off of 20 IE/ml is indicated. Error bars indicate the mean ± SEM, and significant differences from the no-vaccine control group are indicated by **: p < 0.01; *: p < 0.05.
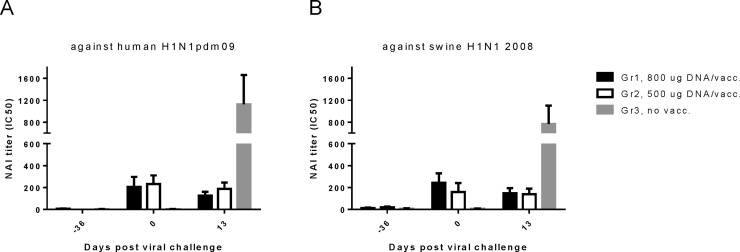
Neutralization (Fig. 2) and neuraminidase inhibition (NAI) (Fig. 3) assays supported the ELISA antibody response findings. Pigs given the highest dose of DNA showed robust levels of neutralizing antibodies against A/California/07/09 (H1N1)pdm09, A/swine/DK/102586/2007 (H1N1) and A/swine/DK/10496/2008 (H1N1). Both the high and low dose DNA vaccinations induced NAI antibodies against both A/California/07/09 (H1N1)pdm09 and A/swine/DK/10496/2008 (H1N1). After challenge, the unvaccinated pigs developed neutralizing antibodies as well as NAI antibodies, while the levels of the vaccinated animals remained unchanged. The ELISA IgG titers correlated with the neutralizing antibody titers in the vaccinated animals (P < 0.0001 R = 0.62, Spearman rank correlation test). The background values (day −36) in the MN assay differed in Fig. 2A–C, with a higher background pre-vaccination to the human H1N1pdm09. This could be due to the variation between the three different virus isolates in this assay. The higher neutralizing antibody titers against the human H1N1pdm09 compared to the two swine-virus isolates may be explained if H1N1pdm09 are easier to neutralize in in vitro settings. Alternatively, it is possible that some cross-reacting antibodies reacting with H1N1pdm09 in the MN assay were already present in the pigs. However, all pig sera were negative pre-vaccination in ELISA detecting binding antibodies to the H1N1pdm09. Thus, the background phenomenon in the MN assay was specific for the neutralization assay. Significant increase from this background value could be detected in Fig. 2A to monitor the effect of the vaccinations.
Fig. 2.
Cross-reactive anti-H1N1 neutralizing antibody responses in vaccinated pig sera. Pigs were vaccinated twice (arrows) i.d. with needle-free delivery with 800 μg (n = 5) or 500 μg (n = 5), or not DNA vaccinated at all (n = 5). The pre- and post-challenge pig sera were tested in a micro-neutralization (MN) assay. Neutralizing antibody titers, MN titers, were evaluated by the capacity of the sera to prevent the infection of MDCK cells by (A) H1N1pdm09 and (B) swine 2008 H1N1 virus isolates. The MN titer was defined as the reciprocal dilution providing 50% infection inhibition, calculated with a linear interpolation method (Reed and Muench, 1938). Serum samples with a titer below the detectable limit of the assay (lowest serum dilution tested was 1:20) were assigned a value of 10 for graphical representation and statistical analyses. Error bars indicate the mean ± SEM, and significant differences from the no-vaccine control group are indicated by **: p < 0.01; *: p < 0.05.
Fig. 3.
Cross-reactive anti-H1N1 neuraminidase inhibition (NAI) in vaccinated pig sera. Pigs were vaccinated twice (day −36 and day −14) i.d. with needle-free delivery with 800 μg (n = 5) or 500 μg (n = 5), or not DNA vaccinated at all (n = 5). The pre- and post-challenge pig sera were tested in an NAI assay. The NAI titers, or the serum dilutions that would inhibit 50% of neuraminidase activity (IC50), were tested against (A) H1N1pdm09 and (B) swine 2008 H1N1 isolates. Error bars indicate the mean ± SEM.
3.4. Detection of virus in nasal swabs
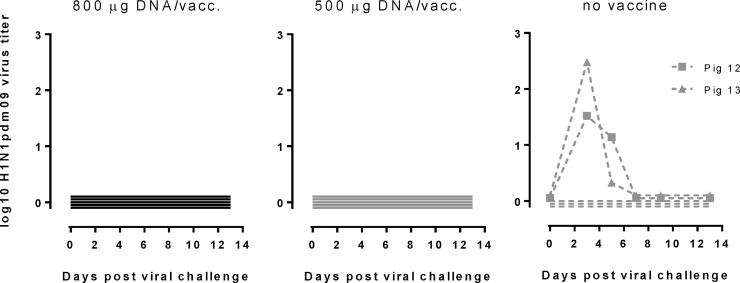
Influenza virus RNA was detected in nasal swabs in two out of five non-vaccinated pigs three days after challenge. Viral RNA could not be detected in any of the 10 vaccinated pigs (<15 TCID50/ml) (Fig. 4). The median peak viral load was 1.95 log TCID50/ml (range 1.61–2.29 log TCID50/ml; N = 2) for non-vaccinated control pigs, and they cleared the virus to undetectable levels at day 7 pc. Same results were obtained using qRT-PCR for the NA, PB1 and M viral genes (data not shown).
Fig. 4.
Protective efficacy of the influenza DNA vaccine. Vaccinated and control animals were challenged with pandemic H1N1 A/California/07/2009. Post-challenge viral loads were assessed for up to 13 days in nasal swabs. Data are expressed as the mean log10 virus titer ± SEM.
4. Discussion
Previously, we have shown that an optimized polyvalent influenza DNA vaccine induced cross-reactive humoral and cellular immune responses in growing pigs after needle-less intradermal application (Borggren et al., 2016). In the present study, we demonstrated that this DNA vaccine provided protection against influenza challenge in pigs. In the present study, we challenged our vaccine applying the commonly used animal model where each individual animal are infected with a standardized dose of challenge virus (Bragstad et al., 2013) as the alternative to larger studies using contact infection models. This enabled us to consider animal welfare recommendations by using a limited number of experimental animals and provided the opportunity to compare with the results of similar studies, e.g. (Bragstad et al., 2013, Busquets et al., 2010, Gorres et al., 2011). None of the vaccinated pigs shed virus. In addition, there was a total lack (800 μg group) or only minimal amounts (500 μg group) of microscopic lung lesions seen in the groups receiving the DNA immunization. The humoral response after viral challenge supported the capability of the vaccine to provide protection since the two vaccinated groups had no increase in antibody titers after challenge, indicating the presence of functional antibodies developed after vaccination. As observed in other influenza challenge studies (Busquets et al., 2010, Vergara-Alert et al., 2012), the challenged pigs did not show signs of clinical disease related to influenza infection. Gross and histopathological findings of broncho-interstitial pneumonia, indicative of influenza virus infection, were mainly observed in the unvaccinated pigs compared to the vaccinated pigs upon challenge, further supporting the protective capabilities of the DNA vaccine tested. The challenge virus H1N1pdm2009 turned out to constitute a relatively mild challenge when looking at the relatively low pathology scores. Yet, this virus is one of the major challenges in production pigs in Europe, and transmission between human and pigs were documented during the 2009 pandemic period (Nelson and Vincent, 2015, Rambaut and Holmes, 2009). Although it is the primary contemporary circulating influenza pathogen in industrialized pig production there are limited vaccine possibilities available for it. Therefore, we believe it represents a highly relevant virus for a novel vaccine study. Importantly, the absence of clinical signs of disease during the experiment supports previous findings (Borggren et al., 2016, Bragstad et al., 2013) which indicated that the vaccine did not show adverse effects in vaccinated animals.
Virus shedding was detected in two out of five unvaccinated control pigs, only. However, the development of antibody responses in all individual control pigs after challenge, demonstrated that all five control pigs were successfully infected. The amount of virus used for inoculation in the present study is comparable to that used successfully to induce effective infection in previous studies that also utilized intranasal inoculation (Bragstad et al., 2013, Trebbien et al., 2013). However, individual pig characteristics may have an effect on viral shedding, as seen also in humans (Lloyd-Smith et al., 2005, Skene et al., 2014). In addition, it remains possible that viral shedding may have occurred on days when nasal swab samples were not collected. Measurements of the housekeeping gene beta-actin in parallel to H1N1pdm09 virus in the qRT-PCR assay excluded differences in the sampled nasal swab material and/or RNA extraction (data not shown) as a source of variations in the nasal swab analyses.
All (5/5) non-vaccinated animals became infected, as evaluated by several or all criteria’s: lung pathology indicative of influenza infection in all but pig 13 (Table 1), that did however shed virus (Fig. 4) and/or viral shedding (Fig. 4, pig 12, 13), and importantly the development of specific antibodies in response to the challenge in all non-vaccinated animals (5/5). This was observed in all antibody tests applied, i.e. ELISA (Fig. 1A) and neutralizing antibody test (Fig. 2A) and HI test (Fig. 1B) and NA inhibition test (Fig. 3). In contrast, for the high vaccine dose (800 μg), none (0/5) got infected, as judged by all of the same measured criteria’s: no shedding (Fig. 4), no increase in specific binding antibodies (Fig. 1A (800 μg)), no increase in functional HAI antibodies (Fig. 1 B (800 μg)), no increase in MN titers (Fig. 2A (800 μg)), no increase in NAI (Fig. 3A (800 μg)), no viral shedding (Fig. 4) and no local lung pathology and no evidence of infection in the lung (Table 1). The titers in the vaccinated pigs increased due to the vaccinations from day −36 and −15 and until the day of challenge (day 0). However, from challenge to day 7 and 13 post challenge antibody titers remained either constant or decreased slightly (Fig. 1A + B) but in contrast to the non-challenged pigs there was no significant increase in the antibody titers. The slight drop in titers day 7 post challenge (Fig. 2A) could be due to antigen-antibody complexing or assay variation, but is not significant (Fig. 2A) and there was no change in titers from day of challenge (day 0) to day 13 post challenge, especially no significant increase from day of challenge as seen in the non-vaccinated pigs. Thus, 5/5 became infected in the non-vaccinated group versus 0/5 in the high dose (800 μg) vaccine group. This was statistically significant (p = 0.0079, Fishers exact test).
For the low vaccine group (500 μg), we did also not find any shedding of virus, no increase in specific binding antibodies (ELISA Fig. 1A (500 μg)), no increase in HI titers (Fig. 1B (500 μg)), no increase in NAI (Fig. 3A) (500 μg), no shedding (Table 1). However, we did see some local lung pathology in 3/5 pigs described as “very mild”, and a slight increase in Nab (Fig. 2A) although not statistically significant. Thus, the lower dose vaccine seemed to provide only partial protection. This could support a dose-related protection.
Recently, we reported an in-depth analysis of the immune response induced by this influenza DNA vaccine in a dose-response titration experiment, which included cross-reactive humoral and cellular dose-dependent vaccine responses (Borggren et al., 2016). Based on results from that study, the DNA vaccine doses used in the present study were chosen. The current results confirm and extend the prior results by demonstrating protection against influenza challenge by the induced response. Both doses of DNA vaccine given, 800 μg (133 μg per gene/plasmid) and 500 μg (83 μg per gene/plasmid), induced protection when pigs where challenged with pandemic H1N1. However, the induced humoral response in the higher dose 800 μg DNA group was broader in the in vitro neutralization analysis than the lower dose DNA group, which neutralized a virus homologous to the vaccine only. This observation indicates that a higher dose of vaccine is required to gain a broader cross-reactive humoral response. Thus, it is possible that a challenge with a more heterologous virus strain to the DNA vaccine would differentiate protection between the two DNA doses. However, it is promising for the DNA technology that even a low dose of 500 μg DNA, had a protective effect on a homologous virus strain, since the general low immunogenicity of DNA vaccines in larger animals (Ferraro et al., 2011) has previously been overcome with much larger doses of DNA (Enama et al., 2014). Previous challenge studies by us (Bragstad et al., 2013) and others (Gorres et al., 2011) have demonstrated protective effects with DNA vaccine doses of 2000 μg and up. It is tempting to speculate that the optimizations conducted for the DNA vaccine used herein (Borggren et al., 2015) have improved the immunogenicity in such a way that the potential to deliver substantially lower doses of DNA in the future seems realistic. This would be a central aspect for the practical use of a DNA vaccine in swine herds where vaccine costs are of utmost importance.
The demonstrated DNA dose-dependence was also reflected in the induced humoral response after immunizations. The higher DNA dose already demonstrated ELISA binding and HI titers after the first immunization, which were boosted after the second one. In contrast, the lower dose of 500 μg DNA seems to require two immunizations to induce a measurable humoral response. As for the commercial protein-based influenza vaccines the protective effect of the DNA most likely includes the vaccine-induced H1N1pdm09 IgG antibodies (Kutzler and Weiner, 2008, Pica and Palese, 2013, Van Reeth and Ma, 2013). In addition to the antibodies to HA, there were also antibodies to the NA surface protein, which neutralized the NA activity in the NAI assay. The relative or combined role of neutralizing anti-HA and anti-NA antibodies may be important (Pica and Palese, 2013) but will require further experiments to be defined.
Other potential mechanisms may be involved using DNA vaccines instead of protein vaccines, such as cellular immunity e.g. towards the more conserved antigens nucleoprotein (NP) and matrix (M) (Bragstad et al., 2011, Gorres et al., 2011, Liu, 2011, Sridhar et al., 2013). The cellular immunity was not assessed in this study, since our recent evaluation of vaccine-induced immunity already clearly demonstrated a strong cell-mediated immune response after DNA immunization (Borggren et al., 2016), including both IFN-γ-producing T cells against several different influenza specific proteins and functional proliferation following influenza stimulation. In addition, we have previously demonstrated protection against influenza induced by NP and M genes of 1918 origin alone in ferrets (Bragstad et al., 2011). However, in vivo expression of NP, M1 and M2 was confirmed by measuring antibody induction to these proteins in ELISA (data not shown).
Future studies including larger experiments using also contact-infected pigs are needed to address the potential impact of cross-reactive protection against infection of also other influenza strains. We have previously reported DNA-induced protective cross-reactive antibodies between different H3N2 influenza strains in pigs (Bragstad et al., 2013) and thus speculate that the present optimized DNA vaccine has the potential to induce broad protection.
Conflicts of interest
James Williams has an equity interest in Nature Technology Corporation. The other authors declare that there are no other conflicts of interest.
Acknowledgments
The technical assistance of Birgit Knudsen, Randi Thøgersen, Bente Andersen, Susanne Lopez Rasmussen, Lorena Córdoba, Mónica Pérez and the animal care staff at CReSA is gratefully acknowledged. This project has received funding from the European Union’s Seventh Framework Program for research, technological development and demonstration under grant agreement no. 602012 UNISEC consortium.
References
- Bennett R., Christiansen K., Clifton-Hadley R. Preliminary estimates of the direct costs associated with endemic diseases of livestock in Great Britain. Prev. Vet. Med. 1999;39:155–171. doi: 10.1016/s0167-5877(99)00003-3. [DOI] [PubMed] [Google Scholar]
- Borggren M., Nielsen J., Bragstad K., Karlsson I., Krog J.S., Williams J.A., Fomsgaard A. Vector optimization and needle-free intradermal application of a broadly protective polyvalent influenza A DNA vaccine for pigs and humans. Hum. Vaccines Immunother. 2015;11:1983–1990. doi: 10.1080/21645515.2015.1011987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borggren M., Nielsen J., Karlsson I., Dalgaard T.S., Trebbien R., Williams J.A., Fomsgaard A. A polyvalent influenza DNA vaccine applied by needle-free intradermal delivery induces cross-reactive humoral and cellular immune responses in pigs. Vaccine. 2016;34:3634–3640. doi: 10.1016/j.vaccine.2016.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bragstad K., Martel C.J., Thomsen J.S., Jensen K.L., Nielsen L.P., Aasted B., Fomsgaard A. Pandemic influenza 1918 H1N1 and 1968 H3N2 DNA vaccines induce cross-reactive immunity in ferrets against infection with viruses drifted for decades. Influenza Other Resp. Viruses. 2011;5:13–23. doi: 10.1111/j.1750-2659.2010.00177.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bragstad K., Vinner L., Hansen M.S., Nielsen J., Fomsgaard A. A polyvalent influenza A DNA vaccine induces heterologous immunity and protects pigs against pandemic A(H1N1)pdm09 virus infection. Vaccine. 2013;31:2281–2288. doi: 10.1016/j.vaccine.2013.02.061. [DOI] [PubMed] [Google Scholar]
- Busquets N., Segalés J., Córdoba L., Mussá T., Crisci E., Martín-Valls G.E., Simon-Grifé M., Pérez-Simó M., Pérez-Maíllo M., Núñez J.I., Abad F.X., Fraile L., Pina S., Majó N., Bensaid A., Domingo M., Montoya M. Experimental infection with H1N1 European swine influenza virus protects pigs from an infection with the 2009 pandemic H1N1 human influenza virus. Vet. Res. 2010;41:74–88. doi: 10.1051/vetres/2010046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Detmer S.E., Gunvaldsen R.E., Harding J.C. Comparison of influenza a virus infection in high- and low-birth-weight pigs using morphometric analysis. Influenza Other Resp. Viruses. 2013;7:2–9. doi: 10.1111/irv.12199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enama M.E., Ledgerwood J.E., Novik L., Nason M.C., Gordon I.J., Holman L.S., Bailer R.T., Roederer M., Koup R.A., Mascola J.R., Nabel G.J., Graham B.S. Phase I randomized clinical trial of VRC DNA and rAd5 HIV-1 vaccine delivery by intramuscular (IM), subcutaneous (SC) and intradermal (ID) administration (VRC 011) PLoS One. 2014:9. doi: 10.1371/journal.pone.0091366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriksson E., Yao F., Svensjö T., Winkler T., Slama J., Macklin M.D., Andree C., McGregor M., Hinshaw V., Swain W.F. In vivo gene transfer to skin and wound by microseeding. J. Surg. Res. 1998;78:85–91. doi: 10.1006/jsre.1998.5325. [DOI] [PubMed] [Google Scholar]
- Fenyö E.M., Heath A., Dispinseri S., Holmes H., Lusso P., Zolla-Pazner S., Donners H., Heyndrickx L., Alcami J., Bongertz V., Jassoy C., Malnati M., Montefiori D., Moog C., Morris L., Osmanov S., Polonis V., Sattentau Q., Schuitemaker H., Sutthent R., Wrin T., Scarlatti G. International network for comparison of HIV neutralization assays: the NeutNet report. PLoS One. 2009:4. doi: 10.1371/journal.pone.0004505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrari L., Borghetti P., Gozio S., De Angelis E., Ballotta L., Smeets J., Blanchaert A., Martelli P. Evaluation of the immune response induced by intradermal vaccination by using a needle-less system in comparison with the intramuscular route in conventional pigs. Res. Vet. Sci. 2011;90:64–71. doi: 10.1016/j.rvsc.2010.04.026. [DOI] [PubMed] [Google Scholar]
- Ferraro B., Morrow M.P., Hutnick N.A., Shin T.H., Lucke C.E., Weiner D.B. Clinical applications of DNA vaccines: current progress. Clin. Infect. Dis. 2011;53:296–302. doi: 10.1093/cid/cir334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorres J.P., Lager K.M., Kong W.P., Royals M., Todd J.P., Vincent A.L., Wei C.J., Loving C.L., Zanella E.L., Janke B., Kehrli M.E., Nabel G.J., Rao S.S. DNA vaccination elicits protective immune responses against pandemic and classic swine influenza viruses in pigs. Clin. Vaccine Immunol. 2011;18:1987–1995. doi: 10.1128/CVI.05171-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinen P.P., Rijsewijks F.A., de Boer-Luitjtze E.A., Bianchi A.T.J. Vaccination of pigs with a DNA construct expressing an influenza virus M2-nucleoprotein fusion protein exacerbates disease after challenge with influenza A virus. J. Gen. Virol. 2002;83:1851–1859. doi: 10.1099/0022-1317-83-8-1851. [DOI] [PubMed] [Google Scholar]
- Ito T., Couceiro J.N., Kelm S., Baum L.G., Krauss S., Castrucci M.R., Donatelli I., Kida H., Paulson J.C., Webster R.G., Kawaoka Y. Molecular basis for the generation in pigs of influenza A viruses with pandemic potential. J. Virol. 1998;72:7367–7373. doi: 10.1128/jvi.72.9.7367-7373.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutzler M.A., Weiner D.B. DNA vaccines: ready for prime time? Nat. Rev. Genet. 2008;9:776–788. doi: 10.1038/nrg2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen D.L., Karasin A., Olsen C.W. Immunization of pigs against influenza virus infection by DNA vaccine priming followed by killed-virus vaccine boosting. Vaccine. 2001;19:2842–2853. doi: 10.1016/s0264-410x(01)00014-7. [DOI] [PubMed] [Google Scholar]
- Li L., Petrovsky N. Molecular mechanisms for enhanced DNA vaccine immunogenicity. Expert Rev. Vaccines. 2015:1–17. doi: 10.1586/14760584.2016.1124762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M.A. DNA vaccines: an historical perspective and view to the future. Immunol. Rev. 2011;239:62–84. doi: 10.1111/j.1600-065X.2010.00980.x. [DOI] [PubMed] [Google Scholar]
- Lloyd-Smith J.O.O., Schreiber S.J.J., Kopp P.E.E., Getz W.M.M. Superspreading and the effect of individual variation on disease emergence. Nature. 2005;438:355–359. doi: 10.1038/nature04153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macklin M.D., McCabe D., McGregor M.W., Neumann V., Meyer T., Callan R., Hinshaw V.S., Swain W.F. Immunization of pigs with a particle-mediated DNA vaccine to influenza A virus protects against challenge with homologous virus. J. Virol. 1998;72:1491–1496. doi: 10.1128/jvi.72.2.1491-1496.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martelli P., Cordioli P., Alborali L.G., Gozio S., De Angelis E., Ferrari L., Lombardi G., Borghetti P. Protection and immune response in pigs intradermally vaccinated against porcine reproductive and respiratory syndrome (PRRS) and subsequently exposed to a heterologous European (Italian cluster) field strain. Vaccine. 2007;25:3400–3408. doi: 10.1016/j.vaccine.2006.12.050. [DOI] [PubMed] [Google Scholar]
- Nelson M.I., Vincent A.L. Reverse zoonosis of influenza to swine: new perspectives on the human-animal interface. Trends Microbiol. 2015;23:142–153. doi: 10.1016/j.tim.2014.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen C.W., Reeth K.V., Brown I. Diseases of Swine. 9th ed. 2006. Swine influenza. [Google Scholar]
- Olsen C.W. DNA vaccination against influenza viruses: a review with emphasis on equine and swine influenza. Vet. Microbiol. 2000:149–164. doi: 10.1016/s0378-1135(00)00175-9. [DOI] [PubMed] [Google Scholar]
- Pica N., Palese P. Toward a universal influenza virus vaccine: prospects and challenges. Annu. Rev. Med. 2013;64:189–202. doi: 10.1146/annurev-med-120611-145115. [DOI] [PubMed] [Google Scholar]
- Rambaut A., Holmes E. The early molecular epidemiology of the swine-origin A/H1N1 human influenza pandemic. PLoS Curr. 2009:1. doi: 10.1371/currents.RRN1003. (RRN1003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed L.J., Muench H. A simple method of estimating fifty percent endpoints. Am. J. Hyg. 1938;27:493–497. [Google Scholar]
- Sandbulte M., Spickler A., Zaabel P., Roth J. Optimal use of vaccines for control of influenza a virus in swine. Vaccines. 2015;3:22–73. doi: 10.3390/vaccines3010022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skene K.J., Paltiel A.D., Shim E., Galvani A.P. A marginal benefit approach for vaccinating influenza superspreaders. Med. Decis. Mak. 2014;34:536–549. doi: 10.1177/0272989X14523502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith G.J.D., Vijaykrishna D., Bahl J., Lycett S.J., Worobey M., Pybus O.G., Ma S.K., Cheung C.L., Raghwani J., Bhatt S., Peiris J.S.M., Guan Y., Rambaut A. Origins and evolutionary genomics of the 2009 swine-origin H1N1 influenza A epidemic. Nature. 2009;459:1122–1125. doi: 10.1038/nature08182. [DOI] [PubMed] [Google Scholar]
- Sridhar S., Begom S., Bermingham A., Hoschler K., Adamson W., Carman W., Bean T., Barclay W., Deeks J.J., Lalvani A. Cellular immune correlates of protection against symptomatic pandemic influenza. Nat. Med. 2013;19:1305–1312. doi: 10.1038/nm.3350. [DOI] [PubMed] [Google Scholar]
- Sriwilaijaroen N., Suzuki Y. Molecular basis of the structure and function of H1 hemagglutinin of influenza virus. Proc. Japan Acad. Ser. B. 2012;88:226–249. doi: 10.2183/pjab.88.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thacker E.L., Holtkamp D.J., Khan A.S., Brown P.A., Draghia-Akli R. Plasmid-mediated growth hormone-releasing hormone efficacy in reducing disease associated with Mycoplasma hyopneumoniae and porcine reproductive and respiratory syndrome virus infection. J. Anim. Sci. 2006;84:733–742. doi: 10.2527/2006.843733x. [DOI] [PubMed] [Google Scholar]
- Trebbien R., Bragstad K., Larsen L., Nielsen J., Bøtner A., Heegaard P.M., Fomsgaard A., Viuff B., Hjulsager C. Genetic and biological characterisation of an avian-like H1N2 swine influenza virus generated by reassortment of circulating avian-like H1N1 and H3N2 subtypes in Denmark. Virol. J. 2013;10:290. doi: 10.1186/1743-422X-10-290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Laan J.W., Herberts C., Lambkin-Williams R., Boyers A., Mann A.J., Oxford J. Animal models in influenza vaccine testing. Expert Rev. Vaccines. 2008;7:783–793. doi: 10.1586/14760584.7.6.783. [DOI] [PubMed] [Google Scholar]
- Van Reeth K., Ma W. Swine influenza virus vaccines: to change or not to change-that’s the question. Curr. Top. Microbiol. Immunol. 2013;370:173–200. doi: 10.1007/82_2012_266. [DOI] [PubMed] [Google Scholar]
- Vergara-Alert J., Argilaguet J.M., Busquets N., Ballester M., Martín-Valls G.E., Rivas R., López-Soria S., Solanes D., Majó N., Segalés J., Veljkovic V., Rodríguez F., Darji A. Conserved synthetic peptides from the hemagglutinin of influenza viruses induce broad humoral and T-cell responses in a pig model. PLoS One. 2012:7. doi: 10.1371/journal.pone.0040524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visser N., Egger W., Lutticken D. Intradermal application of Aujeszky’s disease virus strain Begonia with tocopherol-based adjuvant and a novel design injection device. Acta Vet. Hung. 1994;42:413–418. [PubMed] [Google Scholar]
- Wang C.-C., Chen J.-R., Tseng Y.-C., Hsu C.-H., Hung Y.-F., Chen S.-W., Chen C.-M., Khoo K.-H., Cheng T.-J., Cheng Y.-S.E., Jan J.-T., Wu C.-Y., Ma C., Wong C.-H. Glycans on influenza hemagglutinin affect receptor binding and immune response. Proc. Natl. Acad. Sci. 2009;106:18137–18142. doi: 10.1073/pnas.0909696106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei C.-J., Boyington J.C., Dai K., Houser K.V., Pearce M.B., Kong W.-P., Yang Z. -y., Tumpey T.M., Nabel G.J. Cross-Neutralization of 1918 and 2009 influenza viruses: role of glycans in viral evolution and vaccine design. Sci. Transl. Med. 2010;2 doi: 10.1126/scitranslmed.3000799. (24ra21-24ra21) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Who . World Heal. Organ.; 2011. Manual for the Laboratory Diagnosis and Virological Surveillance of Influenza; p. 153. [Google Scholar]
- Williams J.A. Vector design for improved DNA vaccine efficacy, safety and production. Vaccines. 2013;1:225–249. doi: 10.3390/vaccines1030225. [DOI] [PMC free article] [PubMed] [Google Scholar]