Abstract
The cytoplasmic tails of some coronavirus (CoV) spike (S) proteins contain an endoplasmic reticulum retrieval signal (ERRS) that can retrieve S proteins from the Golgi to the endoplasmic reticulum (ER); this process is thought to accumulate S proteins at the CoV budding site, the ER-Golgi intermediate compartment (ERGIC), and to facilitate S protein incorporation into virions. However, we showed previously that porcine epidemic diarrhoea CoV S proteins lacking the ERRS were efficiently incorporated into virions, similar to the original virus. Thus, the precise role of the ERRS in virus assembly remains unclear. Here, the roles of the S protein ERRS in severe acute respiratory syndrome CoV (SARS-CoV) intracellular trafficking and S incorporation into virus-like particles (VLPs) are described. Intracellular trafficking and indirect immunofluorescence analysis suggested that when M protein was present, wild-type S protein (wtS) could be retained in the pre- and post-medial Golgi compartments intracellularly and co-localized with M protein in the Golgi. In contrast, mutant S protein lacking the ERRS was distributed throughout the ER and only partially co-localized with M protein. Moreover, the intracellular accumulation of mutant S protein, particularly at the post-medial Golgi compartment, was significantly reduced compared with wtS. A VLP assay suggested that wtS that reached the post-medial compartment could be returned to the ERGIC for subsequent incorporation into VLPs, while mutant S protein could not. These results suggest that the ERRS of SARS-CoV contributes to intracellular S protein accumulation specifically in the post-medial Golgi compartment and to S protein incorporation into VLPs.
Keywords: severe acute respiratory syndrome coronavirus, spike protein, ER retrieval signal, trafficking, virus assembly, virus-like particles
Introduction
Severe acute respiratory syndrome (SARS) is a life-threatening disease caused by SARS coronavirus (SARS-CoV) (Drosten et al., 2003; Fouchier et al., 2003; Ksiazek et al., 2003; Rota et al., 2003). SARS-CoV belongs to the order Nidovirales and family Coronaviridae, which are enveloped viruses with a large positive-sense ssRNA genome. CoVs can be genetically and antigenically divided into four genera: Alpha-, Beta-, Gamma- and Deltacoronaviruses (de Groot et al., 2012; Woo et al., 2012). SARS-CoV belongs to the genus Betacoronavirus. All CoV virions possess three envelope proteins, two major glycoproteins, membrane (M) proteins, spike (S) proteins and minor small envelope (E) proteins that are incorporated into the CoV envelope (Cavanagh, 1981; Hogue & Brian, 1986; King & Brian, 1982; Masters, 2006; Wege et al., 1979). Some CoVs contain an additional enveloped protein, haemagglutinin-esterase (HE) (King et al., 1985; Lissenberg et al., 2005). M and E proteins play important roles in virus assembly and budding (Baudoux et al., 1998; DeDiego et al., 2007; Fischer et al., 1998; Kuo & Masters, 2002; Vennema et al., 1996), while S proteins are responsible for receptor binding and viral entry (Heald-Sargent & Gallagher, 2012; Li, 2013).
A characteristic feature of CoVs is their ability to assemble at and bud into the lumen of the endoplasmic reticulum (ER)-Golgi intermediate compartment (ERGIC) (Krijnse-Locker et al., 1994; Tooze et al., 1984). For efficient virus assembly, viral protein interaction and trafficking near the ERGIC is required. M proteins play a key role in protein interaction because the incorporation of nucleocapsid (N), E and S proteins into virions is mediated by M proteins at the budding site (Kuo & Masters, 2002; Narayanan et al., 2000; Nguyen & Hogue, 1997; Opstelten et al., 1995). Furthermore, M protein is indispensable for virus-like particle (VLP) formation (Baudoux et al., 1998; Hsieh et al., 2005; Huang et al., 2004; Siu et al., 2008; Vennema et al., 1996). In protein trafficking, three envelope (membrane) proteins must be transported and retained near the ERGIC because membrane proteins generally reach the plasma membrane through the secretory pathway. In fact, M and E proteins from avian infectious bronchitis virus (IBV) and mouse hepatitis virus (MHV) possess intrinsic intracellular retention signals to ensure they accumulate at or near the budding site (Corse & Machamer, 2000; Klumperman et al., 1994; Lim & Liu, 2001; Machamer et al., 1990).
S protein forms a homotrimer that is highly glycosylated and comprises S1 and S2 domains. The S2 domain can be divided into three domains: a large ectodomain, a transmembrane domain and a cytoplasmic tail (CT) (Fig. 1a). Two distinct retention signals are found in the CT (Fig. 1b). One is an ER retrieval signal (ERRS) that can be further classified into dilysine (KKxx-COOH) or dibasic (KxHxx-COOH) motifs (Lontok et al., 2004; McBride et al., 2007). The other retention signal is a tyrosine-dependent localization signal (YxxI or YxxF motif) (Schwegmann-Wessels et al., 2004; Winter et al., 2008). The retention mechanism of tyrosine-dependent localization signals is unclear, while that of ERRSs is well established. Since the ERRS can bind directly to coatomer complex I (COPI), S proteins containing an ERRS are recruited into COPI vesicles and retrieved from the Golgi to the ER in retrograde (Cosson & Letourneur, 1994; Lee et al., 2004). The repeated cycling of S proteins between the ER and the Golgi leads to S protein intracellular retention.
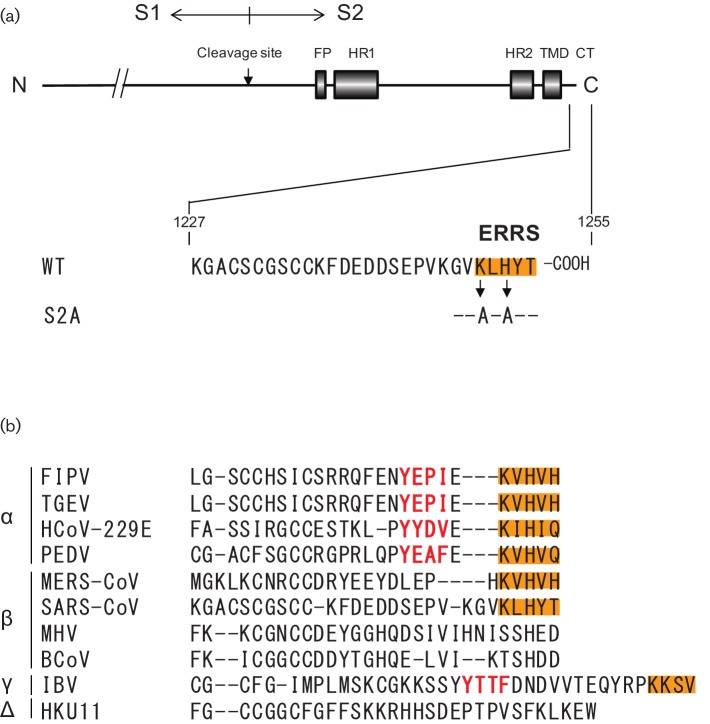
Fig. 1.
Schematic of wtS and S2A proteins of SARS-CoV and the comparison of CoV S proteins C-terminal ends. (a) SARS-CoV S protein consists of 1255 aa and contains two α-helical heptad repeats (HR1 and HR2) (Bosch et al., 2004; Matsuyama et al., 2005; Ujike et al., 2008), a putative fusion peptide (FP), transmembrane domain (TMD) and trypsin cleavage site. Part of the amino acid sequences of the CT are indicated and the ERRS (orange box) are located from amino acid positions 1251–1255. (b) Comparison of the C-terminal ends of 10 CoV-S proteins that included Alpha-, Beta-, Gamma- and Deltacoronaviruses. The amino acid sequence alignment was performed by the clustalw program. Tyrosine-dependent localization signals/internalization signals (Yxxθ motif, where θ can be F, I, L, M or V) are shown in red. The orange box indicates ERRS (KxHxx- or KKxx-motif).
However, the inherent retention potency of ERRS varies significantly among CoV genera or species (Fig. 1b). The S proteins of transmissible gastroenteritis virus (TGEV) and IBV have both an ERRS and a tyrosine-dependent localization signal and are primarily retained intracellularly. In contrast, the S proteins of SARS-CoV possess only an ERRS and cannot be retained intracellularly, resulting in the release of S protein into the plasma membrane. However, when co-expressed with M protein, S and M protein interaction near the budding site led to S protein intracellular retention, but mutant S proteins lacking the ERRS are transported to the plasma membrane (McBride et al., 2007). The authors of this work also show that, while the recombinant CT of wild-type S protein (wtS), but not S2A, could bind directly to COPI, both S protein tails exhibited an identical ability to interact with M protein in an in vitro binding assay (McBride et al., 2007). These results suggest that, although the ERRS of SARS-CoV S proteins is weak so that S proteins alone are eventually released into the plasma membrane, the ERRS can provide sufficient opportunities to interact with M proteins at the budding site by repeated cycling between the Golgi and ER, resulting in intracellular retention. These results indicate that the ERRS plays an important role in S protein trafficking and accumulation near the budding site.
Although protein trafficking and intracellular accumulation mediated by ERRSs is thought to be important for efficient virion assembly, the function of ERRS in this process has been largely neglected, as only a few studies on its role in virion assembly have been reported. Recombinant IBV lacking dilysine ERRS reduced growth only at later time-points post-infection (Youn et al., 2005), while the growth of murine adapted porcine epidemic diarrhea coronavirus (PEDV) lacking dibasic ERRS remained unchanged compared with original PEDV (Shirato et al., 2011), suggesting that in the whole virion the ERRS of both IBV and PEDV had marginal or little effect on viral replication. Moreover, since early release of S proteins lacking ERRS to the plasma membrane would fail to efficiently retain S proteins near the budding site, mutant virus was expected to contain less S protein in virions. However, the extent of S protein incorporation into virions did not differ between mutant and original PEDV (Shirato et al., 2011). Thus, the role of the ERRS of CoV S protein in virus assembly remains unclear.
In this study, we generated a SARS-CoV S protein lacking a dibasic ERRS and studied the role of the ERRS in S protein intracellular trafficking and incorporation into VLPs.
Results
Expression of M protein led to different intracellular trafficking between wild-type S and S2A proteins
The S protein of SARS-CoV consists of 1255 aa and the dibasic ERRS in the CT are located from 1251 to 1255 aa positions at the C-terminal end (Fig. 1a).
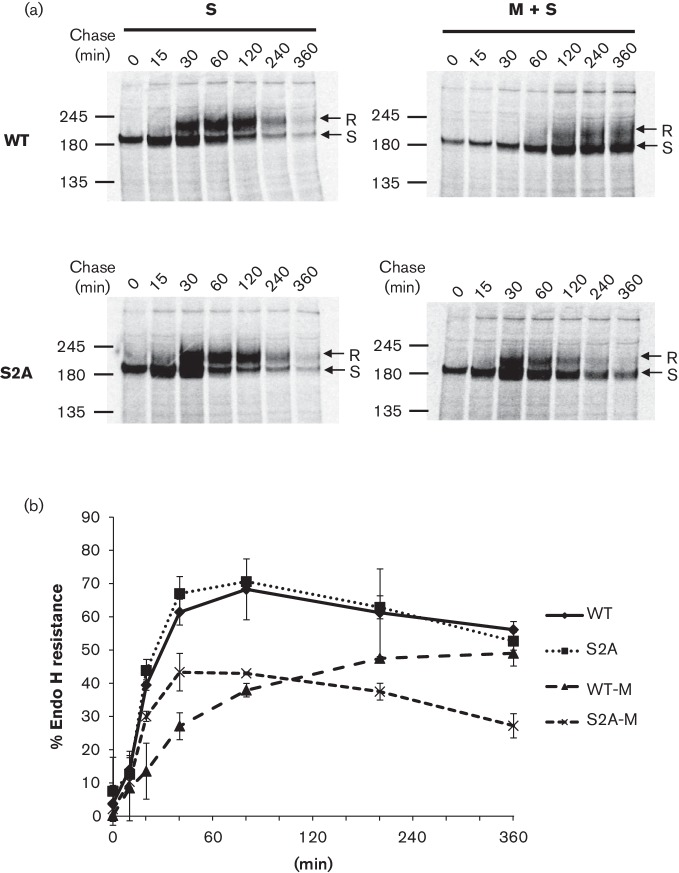
In previous studies, pulse-chase experiments showed small differences in the trafficking rate between wtS and mutant S proteins lacking ERRS when expressed alone. In contrast, when S protein was co-expressed with M protein, indirect immunofluorescence microscopy (IF) showed that wtS was retained intracellularly and strongly co-localized with M protein at the Golgi. Mutant S protein, on the other hand, did not co-localize with M protein, resulting in mutant S protein release to the plasma membrane (McBride et al., 2007). This suggested that the intracellular trafficking between wtS and mutant S proteins drastically changed when M protein was present. To further study S protein intracellular trafficking in the presence of M protein, we introduced the same mutation into the cDNA of the wtS gene (called S2A) (Fig. 1a) and observed the cellular localization of wtS and S2A by a pulse-chase experiment. 293T cells transiently transfected with pCAGGS-wtS or -S2A with or without pCAGGS-M were labelled 40 h post-transfection with 35S methionine/cysteine for 20 min and chased for 0 or 60 min. S protein was immunoprecipitated and analysed by SDS-PAGE. Since the carbohydrate chains of S proteins that are transported to the medial Golgi become resistant to Endo H digestion, S proteins were digested with or without Endo H to evaluate their localization. Both S proteins expressed independently (without M protein) showed a single band sensitive to Endo H without chase, and two bands under the 60 min chase, with the upper band showing resistance to Endo H. This suggests that both proteins reached the medial Golgi similarly after a 60 min chase (Fig. 2a, left). On the other hand, when M was co-expressed, wtS proteins were not processed to Endo H resistance after a 60 min chase whereas S2A proteins were processed (Fig. 2a, right), suggesting that wtS protein had reduced intracellular trafficking from the ER to the medial-Golgi and could be retained intracellularly at the pre-medial Golgi compartment under the 60 min chase.
Fig. 2.
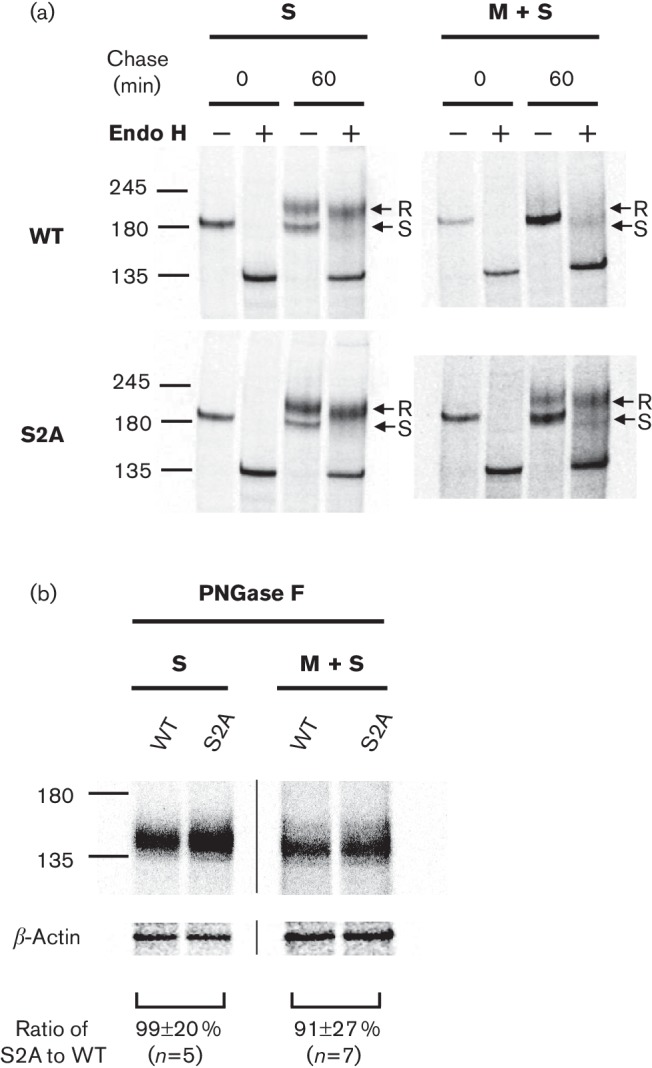
Glycosylation profile and comparison of wtS and S2A expression in the presence or absence of M protein. (a) 293T cells expressing wtS or S2A with or without co-expression of M protein were metabolically labelled at 40 h post-transfection for 20 min and then chased for 0 or 60 min. Cellular S proteins were immunoprecipitated with anti-Sect-2 and immunocomplexes were mock-treated (−) or treated with Endo H (+). Arrows indicate the Endo H-resistant (R) and -sensitive (S) forms. (b) Immunoprecipitated S proteins were treated with PNGase F and band densities from the same gels were compared, and standardized to the density of a β-actin loading control. Results are shown as means±sd from independent experiments (N= number of independent experiments). The values −245', ‘180' and ‘135' represents molecular mass (kDa).
Since the band profile between wtS and S2A was different when M protein was present, both proteins were digested with peptide-N-glycosidase F (PNGase F) (cleaves all carbohydrate chains to obtain a single band) and band densities were compared from the same gel to evaluate the expression of wtS and S2A. We observed no significant differences of their band densities between wtS and S2A proteins under 60 min chase in the presence or absence of M proteins (Fig. 2b). However, the detection sensitivity of both S proteins was reduced when M protein was co-expressed.
In the presence of M protein, the ERRS of SARS-CoV can reduce S protein intracellular trafficking and mediate S protein accumulation at pre- and post-medial Golgi compartments
To further explore the subcellular localization and trafficking of S protein from ER to Golgi, 293T cells expressing wtS or S2A with or without M protein were labelled for 20 min and chased for various times over a 6 h period. Both S proteins, when expressed independently, showed upper bands, corresponding to the Endo H-resistant form, primarily after the 30 min chase. The amount of the resistant form increased until 120 min chase, and then total band densities gradually decreased until 360 min chase (Fig. 3a, left). The kinetics of resistance to Endo H digestion showed no significant differences between wtS and S2A (Fig. 3b). On the other hand, when M protein was co-expressed, the appearance of the resistant form of wtS was delayed until 120 min chase and the intensities of both sensitive and resistant forms were not remarkably reduced from 120–360 min. In contrast, the resistant form of S2A was detected after 30 min chase, similar to that without M protein, and was significantly reduced until 360 min chase, while some amount of the sensitive form was still detected (Fig. 3a, right). The kinetics of wtS resistance to Endo H digestion were remarkably slower than that of S2A in the presence of M protein (Fig. 3b). These data suggested that, when M protein was co-expressed, wtS could accumulate at the pre- and post-medial Golgi compartments for 120–360 min, while S2A lacking ERRS could be partially retained at the pre-medial Golgi compartment but failed to accumulate at the post-medial Golgi compartment at 360 min. Thus, the ERRS of S protein plays an important role in reducing S protein trafficking and accumulation in the pre- and especially post-medial Golgi compartments, likely by repeated cycling between the Golgi and ER via the ERRS.
Fig. 3.
Kinetics of intracellular wtS and S2A trafficking in the presence or absence of M protein. (a) 293T cells expressing S protein were labelled and chased for the indicated times. S protein was immunoprecipitated with anti-Sect-2 and analysed by SDS-PAGE. Arrows indicate the Endo H-resistant (R) and -sensitive (S) forms. (b) The band densities corresponding to wtS and S2A were quantified to evaluate intracellular trafficking from ER to medial-Golgi. Results are shown as means±sd from three independent experiments. The values ‘245', ‘180' and ‘135' represents molecular mass (kDa).
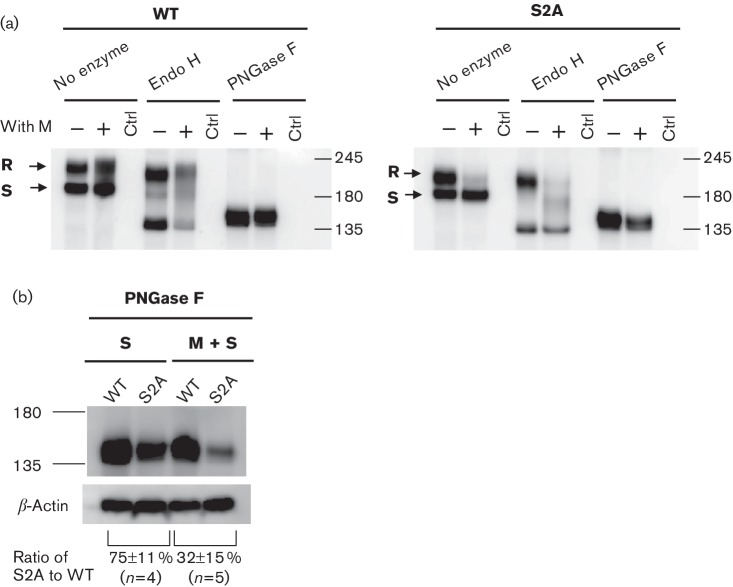
S2A lacking the ERRS significantly reduced the cellular accumulation of S protein at steady state
To support the above data, we evaluated the cellular accumulation and subcellular localization of wtS and S2A proteins at steady state by immunoprecipitation and immunoblotting. This experiment was performed at 66 h post-transfection, because our goal was to determine the role of the ERRS in S protein incorporation into VLPs and we knew that VLPs were recovered at 66 h post-transfection. 293T cells transiently expressing wtS or S2A with or without M protein were lysed and proteins were immunoprecipitated with mouse mAb SKOT-3 and then digested with or without Endo H or PNGase F. Proteins were separated by SDS-PAGE, transferred to a PVDF membrane and then incubated with rabbit antiserum. As expected, the band profile of the two S proteins did not differ significantly in the absence of M protein at steady state (Fig. 4a). On the other hand, when M protein was co-expressed, the Endo H-sensitive and -resistant forms of wtS were detected, while Endo H-sensitive and a small amount of resistant form of S2A were detected at steady state. This suggested once again the importance of ERRS in cellular accumulation of S protein at the pre- and particularly post-medial Golgi compartments in the presence of M protein. To evaluate total accumulation levels, both proteins were digested with PNGase F, and their band densities were compared on the same gel. The total expression of S2A was reduced by 25 % without M protein compared with that of wtS protein, while drastically declined by 67 % when M protein was co-expressed (Fig. 4b).
Fig. 4.
Steady state accumulation of wtS and S2A in the presence or absence of M protein. (a) 293T cells expressing S proteins were lysed at 66 h post-transfection and S proteins were immunoprecipitated with anti-SKOT-3. Immunocomplexes were mock-treated (−) or treated with Endo H or PNGase F (+). Proteins were analysed by SDS-PAGE, transferred to membrane and immunoblotted with S1-2. Since the expression of S2A in the presence of M protein was significantly reduced, the volume of the sample loaded into the SDS-PAGE gel was doubled. (b) Band densities corresponding to wtS and S2A treated with PNGase F were compared, and standardized to the density of a β-actin loading control. Results are shown as means±sd from independent experiments (N= number of independent experiments). The values ‘245', ‘180' and ‘135' represents molecular mass (kDa).
WtS was retained at the Golgi intracellularly and strongly co-localized with M protein, whereas S2A was distributed through the ER and only partially co-localized with M protein
A previous IF study showed that wtS was retained at the Golgi and strongly co-localized with M protein, while S2A did not co-localize with M protein, resulting in release to the plasma membrane where the carbohydrate chains of S protein acquired a Endo H-resistant form (McBride et al., 2007). However, in this study, a small amount of Endo H-resistant form of S2A protein was detected at steady state. To clarify this difference, we performed IF studies by immunofluorescence and confocal laser microscopy. COS-7 cells transiently expressing wtS or S2A in the presence or absence of M protein were double-stained on the cell surface or internally with rabbit anti-M serum and SKOT-3 at 40 h post-transfection, since cell detachment and morphological changes were observed at 66 h post-transfection. When independently expressed, both wtS and S2A were detected on their cell surface similarly and showed similar internal staining patterns (Fig. 5a). Confocal microscopy showed that both proteins largely overlapped with ER-membrane marker calnexin and partially with Golgi-marker 1.4-galactosyltransferase throughout the secretory pathway (Fig. 5b). On the other hand, when M protein was co-expressed, the majority of wtS was retained intracellularly, with little detected on the cell surface (Fig. 5a), and it was strongly co-localized with Golgi-marker and M protein (Fig. 5b). In contrast, the majority of S2A was distributed throughout the cytoplasm, and a few of them were detected on the cell surface (Fig. 5a). S2A was only partially co-localized with Golgi-marker and M protein, while it was largely co-localized with ER-marker (Fig. 5b). Pulse-chase experiments in addition to this IF study suggested that, in the presence of M proteins, wtS was retained at the pre- and post-medial Golgi intracellularly and co-localized with M protein at the Golgi. Conversely, S2A was only partially co-localized with M protein and largely distributed through the ER. Since a few S2A proteins were detected on the cell surface and a small amount of Endo H-resistant form of S2A was detected at steady state in the presence of M protein, S2A protein at or up to the post-medial Golgi compartment seemed to degrade rapidly.
Fig. 5.
Subcellular localization of wtS and S2A in the presence or absence of M protein. (a) COS-7 cells expressing S protein with or without M protein were double-stained on the cell surface or internally with rabbit anti-M serum and SKOT-3 and with secondary antibodies. (b) To determine the subcellular localization of S proteins, the cells were stained with ER-membrane marker anti-calnexin or co-transfected with pAcGFP1-Golgi Vector, which expressed, Golgi-marker 1.4-galactosyltransferase fused to eGFP.
Intracellular accumulation of S proteins by ERRS contributed to the incorporation of S proteins into VLPs
To determine the extent of wtS and S2A protein incorporation into VLPs, a VLP incorporation assay was performed as described previously with some modifications (Ujike et al., 2012). 293T cells co-transfected with 0.75–2.25 µg pCAGGS-SAR-CoV S or S2A plasmids with a constant amount of pCAGGS-SARS-CoV E, M and N plasmids were labelled at 48 h post-transfection with 35S methionine/cysteine for 18 h, and VLPs released into the supernatant were analysed by SDS-PAGE (Fig. 6a, right). Cellular S protein was detected as described above and cellular N and M proteins were detected by immunoblotting using anti-N and anti-M rabbit antiserum, respectively (Fig. 6a, left). To determine relative amounts of cellular S protein and S protein incorporation into VLPs, band densities of Endo H-sensitive and -resistant forms were quantified and standardized to the density of M protein (Fig. 6b). Similar to experiments when M and S protein were co-expressed, cellular wtS showed both forms, while S2A predominantly showed the sensitive form (Fig. 6a, left). Although gels showed that both wtS and S2A protein levels have a tendency to increase dose-dependently with the transfected S plasmid, the Endo H-sensitive form of cellular wtS appeared to be saturated (Fig. 6b, left; open bars in wtS). In VLPs, both Endo H-sensitive and -resistant forms of wtS proteins were successfully incorporated whereas the Endo H-sensitive form of S2A was primarily incorporated in a dose-dependent fashion (Fig. 6b, right). Since the Endo H-resistant form of wtS was incorporated into VLPs, it seems likely that wtS that passed through the ERGIC became Endo H-resistant in the medial Golgi compartment; however, a portion of ERRS-harbouring wtS could return to the ERGIC for subsequent incorporation into VLPs (Endo H-resistant form). In contrast, S2A lacking the ERRS failed to accumulate in the medial Golgi compartment, resulting in little incorporation of the Endo H-resistant form of S protein into VLPs. This result suggested that intracellular accumulation of S protein by ERRS contributed to the incorporation of S protein, particularly at the post-medial Golgi compartment (Endo H-resistant form), into VLPs.
Fig. 6.
Incorporation of wtS and S2A proteins into VLPs and cell attachment experiment (a) 293T cells were co-transfected with 0.75–2.25 µg pCAGGS-S and a constant amount of pCAGGS-M, N and E. At 48 h post-transfection, cells were metabolically labelled for 18 h and released VLPs were analysed by SDS-PAGE (right). ellular S proteins were detected as described in Fig. 4 and cellular M and N proteins were immunoblotted using anti-M or -N rabbit serum. (b) To determine relative amounts of cellular S protein and S protein incorporation into VLPs, band densities corresponding to sensitive (open bar) and resistant forms (filled bar) were quantified and standardized to the density of M protein. Band densities corresponding to wtS proteins (0.7 µg plasmid transfected) in cells or VLPs were set to 1.0. Results are shown as means±sd from three independent experiments. (c) VLPs containing wtS and S2A proteins or VLPs without S proteins were absorbed to 293T cells expressing ACE2 on ice for 3 h, and N proteins of VLPs before and after cell attachment were detected by immunoblotting with anti-N rabbit serum and S pro teins of VLPs before were detected as described in Fig. 6(a). The density of N protein after cell attachment was normalized to that of N protein (filled bar) and total S protein (open bar) (right). The results are shown as the mean±sd from three independent experiments. The values ‘245', ‘180' and ‘135' represents molecular mass (kDa).
Finally, since wtS-VLP contains both Endo H-sensitive and -resistant forms, but S2A-VLP contains primarily the sensitive form, the cell attachment ability between wtS- and S2A-VLPs was compared. WtS- and S2A-VLPs (1.5 µg pCAGGS-SARS-CoV S or S2A was co-transfected with E, M and N plasmids), or VLP without S protein, were concentrated and added to 293T cells transiently expressing ACE2, and incubated on ice for 3 h. Cells were washed with ice-cold 0.2 % BSA-PBS three times. N proteins of VLPs before and after cell attachment were detected by immunoblotting with anti-SARS-CoV N antiserum. After cell attachment, N proteins were not detected in VLPs lacking S protein, while they were detected in wtS- and S2A-VLPs to different extents (Fig. 6c, left). In contrast, unfortunately, 35S lablled S proteins of VLPs could be deteced before cell attachment, but they could not after cell attachment. Only wtS of VLPs could be barely detected by immunoblotting with S1-2 antiserum (data not shown). To standardize VLP attachment, the band densities of N proteins in VLPs after cell attachment were quantified and standardized to the density of N and S proteins of VLPs before attachment (Fig. 6c). Although cell attachment by S2A-VLPs was significantly reduced compared with wtS-VLPs in the N protein standardization (Fig. 6c, right, filled bar), it was comparable to that in the S protein standardization (open bar). Thus, the reduced cell attachment ability of S2A-VLPs is likely due to low-level incorporation of S2A protein into VLPs.
Discussion
This study indicates that the ERRS of SARS-CoV S protein plays an important role in intracellular accumulation of the S protein, particularly at the post-medial Golgi compartments (Endo H-resistant form) when M proteins were co-expressed. Furthermore, it showed that their accumulation by ERRS would contribute to the incorporation of S protein at the post-medial Golgi compartment into VLPs.
The retention potency of the ERRS of CoV S proteins was significantly different among CoV genera or species (Ujike & Taguchi, 2015). The ERRS of SARS-CoV was too weak to retain S proteins intracellularly. Previous pulse-chase experiments and IF studies showed that, when expressed independently, wtS and S2A proteins displayed small differences in intracellular trafficking through the Golgi over 40 min, and that both wtS and S2A proteins were detected on the cell surface and displayed similar subcellular localization in HeLa cells (McBride et al., 2007). These results are consistent with ours, though cell type (293T and COS-7 cells) and experiment duration (6 h) were different.
On the other hand, when M proteins were co-expressed, the behaviour of S proteins was significantly altered. Previous IF studies showed that wtS proteins were strongly co-localized with M proteins at the Golgi and were retained intracellularly, while S2A proteins failed to be co-localized and were released to the plasma membrane (McBride et al., 2007). Since the recombinant CT of wtS and S2A could interact equally with M protein in an in vitro binding assay, the data indicated that the ERRS of SARS-CoV S proteins can provide sufficient opportunities to interact with M proteins at the budding site by cycling between the ER and Golgi (McBride et al., 2007). In this study, we further explored the trafficking and subcellular localization of wtS and S2A proteins when M protein was present. To do this, we performed an IF study, a long pulse-chase experiment, and an immunoblotting study at steady state. Combined data suggested that in the presence of M protein, wtS could significantly reduce intracellular trafficking from the ER to the medial-Golgi, and could be retained intracellularly at the pre- and post-medial Golgi compartments and strongly co-localized with M protein at the Golgi, likely by repeated cycling between the ER and Golgi.
In contrast, S2A lacking the ERRS could be partially retained at the pre-medial Golgi compartment but failed to accumulate at the post-medial Golgi compartment. S2A was only partially co-localized with M protein and was largely distributed through the ER at steady state. Moreover, unlike a previous IF study, this study showed that only a few S2A proteins were detected on the cell surface. These data suggested that, under our experimental conditions, S2A protein at or up to the post-medial Golgi compartment appeared to degrade rapidly. Although it remains unclear why S2A proteins that failed to interact with M protein were released to the cell surface in HeLa cells (McBride et al., 2007), they could not in COS-7 and 293T cells and were subjected to rapid degradation. It is possible that newly synthesized S proteins were continuously replenished rather than degraded due to the difference in time during the studies. Alternatively, since we observed that a high concentration of M proteins (e.g. transfection amount of M plasmid from 0.25 to 1.0 µg) seemed to reduce intracellular accumulation of S proteins on COS cells, the ratio of M to S proteins may control whether an S2A protein that failed to interact with M protein was degraded or released to plasma membrane.
In the VLP-incorporation assays, the amount of intracellular accumulation of S proteins at the pre- and post-medial compartments (Endo H-sensitive and -resistant forms, respectively) were correlated with that of their S-incorporation into VLPs. Since S2A lacking ERRS showed reduced incorporation of S protein most significantly at the post-medial Golgi compartment, the intracellular accumulation of S proteins by ERRS would contribute to the incorporation of S proteins, particularly at the post-medial compartments, into VLPs. It is noteworthy that since CoVs bud and assemble at the ERGIC, wtS that first arrived at the ERGIC was able to interact with M protein and become incorporated into VLPs (Endo H-sensitive form), while wtS that passed through the ERGIC became Endo H-resistant in the medial Golgi compartment. However, wtS having the ERRS would return to the ERGIC for subsequent incorporation into VLPs (Endo H-resistant form). In contrast, S2A lacking the ERRS that first arrived at the ERGIC was incorporated into VLPs in similar way of wtS, but S2A that passed through the ERGIC would fail to return to the ERGIC or be subjected to rapid degradation in or prior to the medial Golgi compartment, resulting in low incorporation of the Endo H-resistant form of S2A into VLPs. Thus, the ERRS of SARS-CoV may retrieve S proteins from the medial Golgi to the ERGIC.
Since wtS-VLPs contained both Endo H-sensitive and -resistant forms, while S2A-VLPs showed reduced incorporation of S proteins that contained primarily Endo H-sensitive forms, the receptor binding activity of wtS- and S2A-VLPs was compared. The amount of N protein in VLPs after cell attachment was standardized to that before attachment. S2A-VLPs exhibited a significantly reduced attachment ability compared with wtS-VLPs. However, when standardized to the amount of S protein in the VLPs before attachment, the attachment ability of the S2A-VLPs was comparable, suggesting that the reduced cell attachment of S2A-VLPs was due to the decreased incorporation of S2A protein into VLPs.
In contrast, our previous study showed that mutant PEDV lacking ERRS showed a propensity for growth similar to that of the original virus, and the amount of S incorporation into virions did not differ (Shirato et al., 2011). This difference may have accounted for the inherent retention potency of PEDV S protein, which possessed a tyrosine-dependent localization signal, in addition to an ERRS (Fig. 1b). Although our previous data suggested that the tyrosine signal played a small role in S protein retention (Shirato et al., 2011), another group reported that tyrosine signals of TGEV and IBV could function as a potent retention signal (Schwegmann-Wessels et al., 2004; Winter et al., 2008). Thus, in the maturation of whole virion, the tyrosine signal might compensate for the role of the ERRS. In fact, recombinant IBV lacking ERRS was recovered and showed fewer growth defects, while IBV lacking the tyrosine signal could not be recovered, implying that the tyrosine signal is important for virion formation, rather than the ERRS (Youn et al., 2005).
Although our study showed the importance of SARS-CoV ERRS for S protein intracellular accumulation and incorporation into VLPs, it remains unclear whether ERRS or tyrosine signals of the other CoVs were involved in these functions. Further study is required to better understand the roles of different CoVs.
Methods
Cells.
293T and COS-7 cells were maintained in Dulbecco’s modified Eagle's medium (DMEM) supplemented with 5–10 % FBS.
Antibodies and plasmid.
Rabbit antiserum Sect-2 and S1-2 against soluble ectodomain and the S1 region of SARS-CoV S protein, mouse mAb (SKOT-3) against S protein (Ohnishi et al., 2005), and rabbit antiserum against synthetic peptides of M and N proteins were kindly provided by Drs Morikawa and Tsunetsugu-Yokota (NIID, Japan) and Dr Mizutani (Tokyo University of Agriculture and Technology, Japan), respectively. Anti-calnexin rabbit polyclonal (Abcam), anti-β-actin mAb (Wako), anti-β-actin polyclonal rabbit antibody (Bioss), HRP-conjugated goat anti-rabbit IgG (MP Biomedicals), and Alexa Fluor 594-conjugated goat anti-mouse IgG and FITC-conjugated goat anti-rabbit IgG (both Invitrogen) were purchased. pAcGFP1-Golgi Vector was purchased from Takara.
Site-specific mutagenesis of SARS-CoV S cDNA.
cDNAs encoding SARS-CoV S, M, N and E protein were subcloned into the pCAGGS expression vector (Ujike et al., 2012). The S2A mutant was generated by mutating two alanine residues (amino acids 1251 and 1253) in dibasic ERRS (KxHxx-) into lysine and histidine residues using PCR with the overlap extension technique (Horton & Pease, 1991).
Metabolic labelling and immunoprecipitation.
293T cells in wells of a 24-well plate were transfected with 0.25 µg pCAGGS-SARS-CoV S with or without 0.125 µg pCAGGS-SARS-CoV M using 2.7 µl TransIT 293 (Mirus). At 39 h post-transfection, cells were incubated in methionine- and cysteine-deficient DMEM (DMEM Met/Cys-) for 1 h. Cells were then metabolically labelled with Tran35S-Label (MP Biomedical) (2.5 Ci) in 250 µl MEM Met/Cys- for 20 min at 37 °C and chased with 10 % FBS-DMEM for the times indicated. Cells were lysed with detergent buffer [50 mM NaCl, 50 mM Tris-Cl (pH 8.0), 62.5 mM EDTA, 0.5 % NP-40 and 0.5 % sodium deoxycholate] (Opstelten et al., 1995) on ice for 20 min. After centrifugation, the supernatant was incubated with Sect-2 and protein G-Sepharose beads (GE Healthcare) overnight at 4 °C. Immunocomplexes were washed and released from beads by boiling for 5 min in sample buffer (50 mM Tris, 2 % SDS, 0.1 % bromophenol blue, 10 % glycerol and 1 % 2-mercaptoethanol) and analysed by SDS-PAGE. Dried gels were visualized using a Typhoon FLA 7000 system (GE Healthcare) and quantification of band densities was performed using ImageQuant TL software. For glycosylation profile experiments, washed immunocomplex-bound sepharose beads were digested with peptide-N-glycosidase F (PNGase F) or Endo H (New England Biolabs) according to the manufacturer's instructions.
Detection of cellular S proteins at steady state by immunoprecipitation and immunoblotting.
Cellular S proteins at steady state were detected by immunoprecipitation and immunoblotting to avoid non-specific bands. Transfection, immunoprecipitation, glycosylation digestion and SDS-PAGE were performed as described above, except experiments were performed at 66 h post-transfection and SKOT-3 was used for immunoprecipitation. Proteins were transferred to a PVDF membrane (Merck Millipore) and then incubated with S1-2 and HRP-conjugated goat anti-rabbit antibody. Protein bands were visualized with ECL prime Western Blotting Detection Reagent (GE Healthcare) on an LAS-4000 instrument (Fujifilm) and band densities were quantified with Multi Gauge v3.2 software.
Indirect immunofluorescence microscopy.
COS-7 cells in individual wells of a lumox multiwell 24 (Sarstedt) were co-transfected with 1.0 µg pCAGGS-SARS-CoV S with or without 0.25 µg pCAGGS-SARS-CoV M plasmid using the TransIT-X2 Dynamic Delivery System (Mirus). At 40 h post-transfection, COS-7 cells expressing SARS-CoV S and M protein were fixed with 4 % paraformaldehyde for 30 min at room temperature and permeabilized with 0.1 % Triton X-100 for 20 min at r.t.room temperature Cells were incubated with SKOT-3 and rabbit anti-SARS-CoV M antiserum, and then incubated with Alexa Fluor 594-conjugated goat anti-mouse IgG and FITC-conjugated goat anti-rabbit IgG. To stain S protein on the cell surface, the cells were placed on ice for 20 min and incubated with the primary antibodies at 4 °C for 3 h and then fixed with 4 % paraformaldehyde for 30 min at 4 °C, and finally incubated with secondary antibodies. To confirm the co-localization of S proteins with ER, fixed cells were permeabilized with ice-cold methanol at −20 °C for 10 min and incubated with SKOT-3 and anti-calnexin rabbit polyclonal and then with secondary antibodies. To confirm the co-localization of S proteins with Golgi, cells were co-transfected with 0.25 µg pAcGFP1-Golgi Vector, fixed with 4 % paraformaldehyde and permeabilized, and stained in the same manner except for anti-calnexin rabbit polyclonal antibody. Images were obtained from an Axioscope microscope and LSM 710 laser scanning microscope (Carl Zeiss) using AxioVision SE64 and Zen 2 software (Carl Zeiss), respectively.
VLP formation and detection.
293T cells were placed in six wells of a 24-well plate and co-transfected with 0.75–2.25 µg pCAGGS- SARS-CoV S, with constant 3.5 µg E, 0.75 µg M, 0.75 µg N and 16.2 µl TransIT 293T. At 47 h post-transfection, cells were incubated in DMEM Met/Cys- for 1 h and metabolically labelled with Tran-35S label (2.5 Ci) in 500 µl 5 % FCS-MEM for 18 h at 37 °C. VLPs released into the medium were collected and pelleted through a 20 % sucrose cushion at 45 000 r.p.m. for 2 h (SW55Tirotor; Beckman). The pellet was suspended in sample buffer, boiled and analysed by SDS-PAGE. Detection and quantification of proteins was the same as described above. Cellular S proteins were detected by immunoprecipitation and immunoblotting as described above and N and M proteins were detected with rabbit anti-SARS-CoV N or M protein antiserum, respectively.
VLP cell attachment experiment.
293T cells were co-transfected with 1.5 µg pCAGGS-SARS-CoV S or without S plasmid and other plasmids at the same amount, and VLPs were collected at 66 h post-transfection and pelleted as described above. The pellet was suspended in 100 µl DMEM and added to 293T cells transiently expressing ACE2 (transfected with 0.25 µg pCAGGS-human ACE2 by 0.4 µl TransIT 293) in individual wells of a 96-well plate and incubated on ice for 3 h. Cells were detached from wells and washed with ice-cold 0.2 % BSA-PBS three times. After washing, the cells were suspended in sample buffer, boiled and analysed by SDS-PAGE. Detection and quantification of proteins were the same as described above.
Acknowledgements
This work was supported by a Grant-in-Aid for Young Scientists (B; no. 23790509) (to M. U.) and a Scientific Research grant (C; nos 30107429 and 26460563) (to F. T. and K. S., respectively) from the Japanese Ministry of Education, Culture, Sports, Science and Technology, and supported by Public Health Service grants AI99107 and AI114657 (to S. M.) from the NIH.
References
- Baudoux P., Carrat C., Besnardeau L., Charley B., Laude H.(1998). Coronavirus pseudoparticles formed with recombinant M and E proteins induce alpha interferon synthesis by leukocytes. J Virol 728636–8643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosch B. J., Martina B. E., Van Der Zee R., Lepault J., Haijema B. J., Versluis C., Heck A. J., de Groot R., Osterhaus A. D., Rottier P. J.(2004). Severe acute respiratory syndrome coronavirus (SARS-CoV) infection inhibition using spike protein heptad repeat-derived peptides. Proc Natl Acad Sci U S A 1018455–8460. 10.1073/pnas.0400576101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavanagh D.(1981). Structural polypeptides of coronavirus IBV. J Gen Virol 5393–103. 10.1099/0022-1317-53-1-93 [DOI] [PubMed] [Google Scholar]
- Corse E., Machamer C. E.(2000). Infectious bronchitis virus E protein is targeted to the Golgi complex and directs release of virus-like particles. J Virol 744319–4326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosson P., Letourneur F.(1994). Coatomer interaction with di-lysine endoplasmic reticulum retention motifs. Science 2631629–1631. [DOI] [PubMed] [Google Scholar]
- de Groot R. J., Baker S. C., Baric R., Enjuanes L., Gorbalenya A. E., Holmes K. V., Perlman S., Poon L., Rottier P. J. M., et al. (2012). Virus Taxonomy: Ninth Report of the International Committee on Taxonomy of Viruses: Family Coronaviridae. Edited by King A., Adams M., Cartens E., Lefkowitz E.San Diego, CA: Academic Press. [Google Scholar]
- DeDiego M. L., Alvarez E., Almazán F., Rejas M. T., Lamirande E., Roberts A., Shieh W. J., Zaki S. R., Subbarao K., Enjuanes L.(2007). A severe acute respiratory syndrome coronavirus that lacks the E gene is attenuated in vitro and in vivo. J Virol 811701–1713. 10.1128/JVI.01467-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drosten C., Günther S., Preiser W., van der Werf S., Brodt H. R., Becker S., Rabenau H., Panning M., Kolesnikova L., et al. (2003). Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N Engl J Med 3481967–1976. 10.1056/NEJMoa030747 [DOI] [PubMed] [Google Scholar]
- Fischer F., Stegen C. F., Masters P. S., Samsonoff W. A.(1998). Analysis of constructed E gene mutants of mouse hepatitis virus confirms a pivotal role for E protein in coronavirus assembly. J Virol 727885–7894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fouchier R. A., Kuiken T., Schutten M., van Amerongen G., van Doornum G. J., van den Hoogen B. G., Peiris M., Lim W., Stöhr K., et al. (2003). Aetiology: Koch's postulates fulfilled for SARS virus. Nature 423240. 10.1038/423240a [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heald-Sargent T., Gallagher T.(2012). Ready, set, fuse! The coronavirus spike protein and acquisition of fusion competence. Viruses 4557–580. 10.3390/v4040557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogue B. G., Brian D. A.(1986). Structural proteins of human respiratory coronavirus OC43. Virus Res 5131–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton M. R., Pease L. R.(1991). Recombination and mutagenesis of DNA-sequences using PCR. Directed Mutagenesis: A Practical Approach 217–247. Edited by McPherson M. J.Oxford: Oxford University Press. [Google Scholar]
- Hsieh P. K., Chang S. C., Huang C. C., Lee T. T., Hsiao C. W., Kou Y. H., Chen I. Y., Chang C. K., Huang T. H., et al. (2005). Assembly of severe acute respiratory syndrome coronavirus RNA packaging signal into virus-like particles is nucleocapsid dependent. J Virol 7913848–13855. 10.1128/JVI.79.22.13848-13855.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y., Yang Z. Y., Kong W. P., Nabel G. J.(2004). Generation of synthetic severe acute respiratory syndrome coronavirus pseudoparticles: implications for assembly and vaccine production. J Virol 7812557–12565. 10.1128/JVI.78.22.12557-12565.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King B., Brian D. A.(1982). Bovine coronavirus structural proteins. J Virol 42700–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King B., Potts B. J., Brian D. A.(1985). Bovine coronavirus hemagglutinin protein. Virus Res 253–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klumperman J., Locker J. K., Meijer A., Horzinek M. C., Geuze H. J., Rottier P. J.(1994). Coronavirus M proteins accumulate in the Golgi complex beyond the site of virion budding. J Virol 686523–6534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krijnse-Locker J., Ericsson M., Rottier P. J., Griffiths G.(1994). Characterization of the budding compartment of mouse hepatitis virus: evidence that transport from the RER to the Golgi complex requires only one vesicular transport step. J Cell Biol 12455–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ksiazek T. G., Erdman D., Goldsmith C. S., Zaki S. R., Peret T., Emery S., Tong S., Urbani C., Comer J. A., et al. (2003). A novel coronavirus associated with severe acute respiratory syndrome. N Engl J Med 3481953–1966. 10.1056/NEJMoa030781 [DOI] [PubMed] [Google Scholar]
- Kuo L., Masters P. S.(2002). Genetic evidence for a structural interaction between the carboxy termini of the membrane and nucleocapsid proteins of mouse hepatitis virus. J Virol 764987–4999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M. C., Miller E. A., Goldberg J., Orci L., Schekman R.(2004). Bi-directional protein transport between the ER and Golgi. Annu Rev Cell Dev Biol 2087–123. 10.1146/annurev.cellbio.20.010403.105307 [DOI] [PubMed] [Google Scholar]
- Li F.(2013). Receptor recognition and cross-species infections of SARS coronavirus. Antiviral Res 100246–254. 10.1016/j.antiviral.2013.08.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim K. P., Liu D. X.(2001). The missing link in coronavirus assembly. Retention of the avian coronavirus infectious bronchitis virus envelope protein in the pre-Golgi compartments and physical interaction between the envelope and membrane proteins. J Biol Chem 27617515–17523. 10.1074/jbc.M009731200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lissenberg A., Vrolijk M. M., van Vliet A. L., Langereis M. A., de Groot-Mijnes J. D., Rottier P. J., de Groot R. J.(2005). Luxury at a cost? Recombinant mouse hepatitis viruses expressing the accessory hemagglutinin esterase protein display reduced fitness in vitro. J Virol 7915054–15063. 10.1128/JVI.79.24.15054-15063.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lontok E., Corse E., Machamer C. E.(2004). Intracellular targeting signals contribute to localization of coronavirus spike proteins near the virus assembly site. J Virol 785913–5922. 10.1128/JVI.78.11.5913-5922.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machamer C. E., Mentone S. A., Rose J. K., Farquhar M. G.(1990). The E1 glycoprotein of an avian coronavirus is targeted to the cis Golgi complex. Proc Natl Acad Sci U S A 876944–6948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masters P. S.(2006). The molecular biology of coronaviruses. Adv Virus Res 66193–292. 10.1016/S0065-3527(06)66005-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuyama S., Ujike M., Morikawa S., Tashiro M., Taguchi F.(2005). Protease-mediated enhancement of severe acute respiratory syndrome coronavirus infection. Proc Natl Acad Sci U S A 10212543–12547. 10.1073/pnas.0503203102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride C. E., Li J., Machamer C. E.(2007). The cytoplasmic tail of the severe acute respiratory syndrome coronavirus spike protein contains a novel endoplasmic reticulum retrieval signal that binds COPI and promotes interaction with membrane protein. J Virol 812418–2428. 10.1128/JVI.02146-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayanan K., Maeda A., Maeda J., Makino S.(2000). Characterization of the coronavirus M protein and nucleocapsid interaction in infected cells. J Virol 748127–8134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen V. P., Hogue B. G.(1997). Protein interactions during coronavirus assembly. J Virol 719278–9284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohnishi K., Sakaguchi M., Kaji T., Akagawa K., Taniyama T., Kasai M., Tsunetsugu-Yokota Y., Oshima M., Yamamoto K., et al. (2005). Immunological detection of severe acute respiratory syndrome coronavirus by monoclonal antibodies. Jpn J Infect Dis 5888–94. [PubMed] [Google Scholar]
- Opstelten D. J., Raamsman M. J., Wolfs K., Horzinek M. C., Rottier P. J.(1995). Envelope glycoprotein interactions in coronavirus assembly. J Cell Biol 131339–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rota P. A., Oberste M. S., Monroe S. S., Nix W. A., Campagnoli R., Icenogle J. P., Penaranda S., Bankamp B., Maher K.(2003). Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science 3001394–1399. [DOI] [PubMed] [Google Scholar]
- Schwegmann-Wessels C., Al-Falah M., Escors D., Wang Z., Zimmer G., Deng H., Enjuanes L., Naim H. Y., Herrler G.(2004). A novel sorting signal for intracellular localization is present in the S protein of a porcine coronavirus but absent from severe acute respiratory syndrome-associated coronavirus. J Biol Chem 27943661–43666. 10.1074/jbc.M407233200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirato K., Maejima M., Matsuyama S., Ujike M., Miyazaki A., Takeyama N., Ikeda H., Taguchi F.(2011). Mutation in the cytoplasmic retrieval signal of porcine epidemic diarrhea virus spike (S) protein is responsible for enhanced fusion activity. Virus Res 161188–193. 10.1016/j.virusres.2011.07.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siu Y. L., Teoh K. T., Lo J., Chan C. M., Kien F., Escriou N., Tsao S. W., Nicholls J. M., Altmeyer R., et al. (2008). The M, E, and N structural proteins of the severe acute respiratory syndrome coronavirus are required for efficient assembly, trafficking, and release of virus-like particles. J Virol 8211318–11330. 10.1128/JVI.01052-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tooze J., Tooze S., Warren G.(1984). Replication of coronavirus MHV-A59 in sac- cells: determination of the first site of budding of progeny virions. Eur J Cell Biol 33281–293. [PubMed] [Google Scholar]
- Ujike M., Taguchi F.(2015). Incorporation of spike and membrane glycoproteins into coronavirus virions. Viruses 71700–1725. 10.3390/v7041700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ujike M., Huang C., Shirato K., Matsuyama S., Makino S., Taguchi F.(2012). Two palmitylated cysteine residues of the severe acute respiratory syndrome coronavirus spike (S) protein are critical for S incorporation into virus-like particles, but not for M-S co-localization. J Gen Virol 93823–828. 10.1099/vir.0.038091-0 [DOI] [PubMed] [Google Scholar]
- Ujike M., Nishikawa H., Otaka A., Yamamoto N., Yamamoto N., Matsuoka M., Kodama E., Fujii N., Taguchi F.(2008). Heptad repeat-derived peptides block protease-mediated direct entry from the cell surface of severe acute respiratory syndrome coronavirus but not entry via the endosomal pathway. J Virol 82588–592. 10.1128/JVI.01697-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vennema H., Godeke G. J., Rossen J. W., Voorhout W. F., Horzinek M. C., Opstelten D. J., Rottier P. J.(1996). Nucleocapsid-independent assembly of coronavirus-like particles by co-expression of viral envelope protein genes. EMBO J 152020–2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wege H., Wege H., Nagashima K., ter Meulen V.(1979). Structural polypeptides of the murine coronavirus JHM. J Gen Virol 4237–47. 10.1099/0022-1317-42-1-37 [DOI] [PubMed] [Google Scholar]
- Winter C., Schwegmann-Wessels C., Neumann U., Herrler G.(2008). The spike protein of infectious bronchitis virus is retained intracellularly by a tyrosine motif. J Virol 822765–2771. 10.1128/JVI.02064-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo P. C., Lau S. K., Lam C. S., Lau C. C., Tsang A. K., Lau J. H., Bai R., Teng J. L., Tsang C. C., et al. (2012). Discovery of seven novel mammalian and avian coronaviruses in the genus Deltacoronavirus supports bat coronaviruses as the gene source of Alphacoronavirus and Betacoronavirus and avian coronaviruses as the gene source of Gammacoronavirus and Deltacoronavirus. J Virol 863995–4008. 10.1128/JVI.06540-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youn S., Collisson E. W., Machamer C. E.(2005). Contribution of trafficking signals in the cytoplasmic tail of the infectious bronchitis virus spike protein to virus infection. J Virol 7913209–13217. 10.1128/JVI.79.21.13209-13217.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]