Abstract
The mammalian brain dynamically activates or silences gene programs in response to environmental input and developmental cues. This neuroplasticity is controlled by signaling pathways that modify the activity, localization, and/or expression of transcriptional-regulatory enzymes in combination with alterations in chromatin structure in the nucleus. Consistent with this key neurobiological role, disruptions in the fine-tuning of epigenetic and transcriptional regulation have emerged as a recurrent theme in studies of the genetics of neurodevelopmental and neuropsychiatric disorders. Furthermore, environmental factors have been implicated in the increased risk of heterogeneous, multifactorial, neuropsychiatric disorders via epigenetic mechanisms. Aberrant epigenetic regulation of gene expression thus provides an attractive unifying model for understanding the complex risk architecture of mental illness. Here, we review emerging genetic evidence implicating dysregulation of histone lysine methylation in neuropsychiatric disease and outline advancements in small-molecule probes targeting this chromatin modification. The emerging field of neuroepigenetic research is poised to provide insight into the biochemical basis of genetic risk for diverse neuropsychiatric disorders and to develop the highly selective chemical tools and imaging agents necessary to dissect dynamic transcriptional-regulatory mechanisms in the nervous system. On the basis of these findings, continued advances may lead to the validation of novel, disease-modifying therapeutic targets for a range of disorders with aberrant chromatin-mediated neuroplasticity.
Keywords: epigenetics, chromatin, histone lysine methylation, psychiatric disorder, drug discovery
1. Introduction
A growing repertoire of epigenetic mechanisms have been linked to neural development, stress responses, and synaptic connectivity and neuroplasticity (1,2). The importance of these molecular mechanisms is highlighted by the fact that more than 50 distinct chromatin regulators have been causally implicated in human neurodevelopmental and psychiatric disorders (3–5). Noteworthy epigenetic phenomena in the brain include the dynamic modification of DNA (e.g. 5-hydroxymethylation of cytosine (6,7)), modulation of higher-order chromatin structure (e.g. ATP-dependent remodeling by the BAF (mSWI/SNF) family (3,8)), diverse regulation by non-coding RNAs (e.g. microRNAs (9,10)), and the deposition, detection and removal of over 100 distinct post-translational modifications and variants of histone proteins (11–13). Of these many mechanisms, multiple lines of evidence are converging on the critical importance of histone lysine methylation, in particular methylation of lysine 4 of histone H3 (H3K4), as summarized in Table 1 and recently discussed in several excellent reviews (4,14–17). Importantly, epigenetic regulation of chromatin modifications, including histone methylation, requires active maintenance (18–21), suggesting that the enzymes responsible for chromatin modification may be targets for new and critically-needed therapeutic interventions for mental disorders. This paradigm has been established by an abundance of literature supporting a key role for reversible changes in histone acetylation in the regulation of nervous system function and disease (22). Here, we briefly summarize and provide an update on the rationale for targeting H3K4 methylation in neuropsychiatric disease, with a special focus on the H3K4 demethylase KDM1A/LSD1. We outline a chemical toolkit of approved drugs, pre-clinical compounds, and drugs of abuse known to modulate H3K4 methylation by both direct and indirect mechanisms. Finally, we discuss areas for future development of small-molecule tools towards the detection, prevention, and treatment of psychiatric disease.
Table 1.
Partial list of validated H3K4 methyltransferases and demethylases (4,138). Asterisks indicates mutation associated with neurodevelopment or psychiatric disease (14).
| H3K4 methyltransferases | H3K4 demethylases | ||||
|---|---|---|---|---|---|
|
| |||||
| Systematic name | Alias | Specificity | Systematic name | Alias | Specificity |
| KMT2A* | MLL | me1–3 | KDM1A* | LSD1, AOF2 | me1–2 |
| KMT2B | MLL2/4 | me1–3 | KDM1B | LSD2, AOF1 | me1–2 |
| KMT2C* | MLL3 | me1–3 | KDM5A* | RBP2, JARID1A | me1–3 |
| KMT2D* | MLL2/4 | me1–3 | KDM5B* | PLU1, JARID1B | me1–3 |
| KMT2F* | SETD1A | me1–3 | KDM5C* | SMCX, JARID1C | me1–3 |
| KMT2G | SETD1B | me1–3 | KDM5D | SMCY, JARID1D | me1–3 |
| KMT7 | SETD7 | me1 | |||
2. H3K4 methylation in the brain
Histone lysine methylation is maintained by dynamic opposition of methyltransferase and demethylase enzymes and is detected by methyllysine reader proteins (23,24). Lysine residues can be mono-, di- or tri-methylated, with each modification resulting in different functional outcomes depending on the degree of methylation and its context (25). H3K4 is the most extensively targeted position of the dozens of histone methylation sites (14). This residue is methylated by multi-protein complexes harboring SET-domain-containing histone lysine methyltransferases (26). Demethylation is effected by two KDM families: the flavin-dependent monoamine oxidases, KDM1A/LSD1 and KDM1B/LSD2, and the 2-oxoglutarate-dependent KDM5 hydroxylases (27). Technological advances, including chromatin immunoprecipitation in conjunction with quantification of associated DNA by next-generation sequencing (ChIP-seq), has revealed wide-ranging roles for H3K4 methylation in the neurodevelopment and neuropsychiatric disease. In cultured cells and in the brain, mono-methylation of H3K4 is associated with enhancer regions, whereas di- and tri-methyl H3K4 is associated with the promoters of actively transcribed genes (4,28). Hundreds of transcription start sites, including loci associated with neuropsychiatric susceptibility genes, show differential H3K4 methylation in the prefrontal cortex of humans compared to chimpanzee and macaques, suggesting that coordinated histone methylation plays a role in the gene expression networks that contribute to cognitive function and neurological diseases unique to humans (29).
Dysregulated H3K4 methylation could play an important role in the pathophysiology of psychiatric disease by linking environmental perturbations during development to long-lasting alterations in gene expression (4). External factors known to modulate this mark include fear conditioning (24), maternal care (30), maternal immune activation (31), and exposure to neuroactive drugs (discussed below). Analysis of histones isolated from neurons of humans ranging in age from late in gestation to 80 years of age revealed highly dynamic H3K4 methylation levels during perinatal development and infancy, with continued unidirectional gain or loss of histone methylation with progressively slower kinetics occurring later in life (32). Importantly, the developmental window with the greatest changes in H3K4 methylation overlaps with the time period of greatest risk for mental disorders including schizophrenia and autism. Consistent with this model, hundreds of loci had differential H3K4 methylation and levels of expression in cultured neurons derived from olfactory epithelial biopsies of patients with schizophrenia relative to controls, including genes involved in immune response, oxidative stress, and synaptogenesis (33). Although consistent enrichment or depletion of H3K4 tri-methylation at any particular locus was not detected in prefrontal cortical neurons of patients with autism, subject-specific alterations and, in particular, spreading of methylation away from the transcription start sites and into gene bodies and upstream sequences was observed (34). This finding suggests that epigenetic dysregulation in neuropsychiatric disease may not simply involve relative increases or decreases in H3K4 methylation, but also its changes to spatial profiles of chromatin modifications.
3. Genetics of H3K4 machinery in neuropsychiatric disease
Advances in next-generation sequencing technologies have revived interest in studying the genetic risk factors for neuropsychiatric diseases, leading to three major classes of findings. First, common genetic variants, typically occurring outside of protein coding regions, have been found to confer small increased risk for mental disorders through genome-wide association studies (GWAS). The interpretation of these results is often challenging, and warrants a systematic approach to map the regulatory regions and epigenetic modifications in the human brain (35). Second, rare mutations, typically involving point mutation or truncation of proteins, have been identified by clinical exome sequencing in syndromic cases of intellectual disability, autism and schizophrenia (4,14). Finally, the role of alternative splicing of H3K4-modifying enzymes is beginning to be uncovered. In all cases, variation or mutation in the protein machinery responsible for regulating H3K4 methylation status has been linked to aberrant brain or neuronal development.
Multiple recent discoveries exemplify the large-scale genetic approaches that have uncovered a potential role for H3K4 dysregulation in neuropsychiatric disease. This includes a massive exome sequencing study using data from 4,264 individuals with schizophrenia, 9,343 controls, and 1,077 trios pooled from several independent samples yielded three de novo mutations and seven loss-of-function variants in the H3K4 methyltransferase KMT2F/SETD1A gene providing a large odds ratio and implicating a role for H3K4 demethylation in neurodevelopmental disorders (36). This finding supports early exome sequencing studies in schizophrenia that originally identified multiple de novo loss-of-function variants in KMT2F/SETD1A in a total of 231 trios with at least one affected subject (37). An additional similarly large-scale (3,871 patients and 9,937 ancestry-matched or parental controls) exome sequencing study found enrichment of synaptic, transcriptional, and chromatin modifying variants in autism, including the H3K4 methyltransferase KMT2C/MLL3, the H3K4 demethylase KDM5B/JARID1A/RBP2, and the H3K4 methyllysine binding protein CHD8 (38). Finally, recent analysis by the Psychiatric Genomics Consortium combined GWAS signals observed across schizophrenia, major depression, bipolar disorder, attention-deficit hyperactivity disorder, and autism-spectrum disorders to identify biochemically-meaningful pathways associated with genetic risk in neuropsychiatric disease (39). Regulation of H3K4 methylation emerged as the top pathway to be significantly correlated with adult psychiatric disorders, suggesting that this common etiological mechanism may be an important target for the development of therapeutics and diagnostics (39).
In addition to associations in common variants, rare mutations in H3K4 regulators have also been linked with neurodevelopmental disorders, and are the subject of an excellent review (14). More recently, dominant missense point mutations in KDM1A have been identified and correlated with a new genetic disorder that phenotypically resembles Kabuki syndrome and features skeletal abnormalities and significant cognitive impairment (40). Three cases have been reported, each with a different missense point mutation in the amine oxidase domain responsible for KDM1A demethylase activity. All mutations are heterozygous, indicating that mutant KDM1A results in a dominant phenotype. Biochemical analyses of all three KDM1A mutants found in human disease revealed weaker methylated H3K4 substrate binding and substantially reduced cellular protein stability (41). However, all KDM1A mutants maintained their ability to repress a synthetic reporter gene, indicating that even subtle mutations in KDM1A are associated with cognitive impairment. Intriguingly, up to 70% of cases of Kabuki syndrome studied to date involve mutation or truncation of KMT2D, suggesting that alterations in either H3K4 methyltransferase or demethylase activity results in neurological disease (14,42).
Four mammalian isoforms of KDM1A have been described as resulting from single or double inclusion of two alternatively spliced exons, E2a and E8a. Intriguingly, the isoform retaining the E8a exon (KDM1A+8a) displays a neurospecific pattern of expression, representing one of the few examples of a chromatin-modifying enzyme devoted to neurons (43). The E8a exon is internal to the amine oxidase domain, and encodes the tetrapeptide Asp-Thr-Val-Lys. KDM1A+8a is dynamically regulated during brain development and enhances cortical neuronal maturation when overexpressed (44). Importantly, the E8a exon codes a brain-specific threonine-phosphorylation switch (43). Overexpression of a Thr369bAsp phosphomimetic promotes neurite outgrowth and branching, suggesting that phosphorylation switches KDM1A+8a into a dominant negative isoform to derepress genes required for neuronal maturation. In adult mice, KDM1A+8a is downregulated in response to epileptogenic stimuli, and animals lacking KDM1A+8a are hypoexcitable and have a decreased susceptibility for seizures (43). Furthermore, mice with reduced levels of KDM1A+8a display a low-anxiety phenotype in a panel of behavioral tests (45). Critically, the Thr369bAsp phosphomimetic fails to associate with REST co-repressor 1 (RCOR1), the molecular link between KDM1A and restriction element-1 (RE1)-silencing transcription factor (REST) (43). REST represses neuron-specific genes, such as ion channels, synaptic vesicle proteins, and neurotransmitter receptors, during early development (46), and is induced in the ageing human brain to regulate a network of genes mediating cell death and stress resistance (47). In addition to KDM1A, mutations in several other epigenetic regulators known to associate with REST and its target genes are implicated in neurodevelopmental and psychiatric disorders (48,49), including the BAF chromatin remodeling complex (50), the H3K4 methyltransferase KMT2A/MLL (51), the H3K4 demethylase KDM5C/SMCX (52), and the H3K4 binding protein PHF21/BHC80 (53). With this view of REST target genes as a nexus for dysregulated H3K4 methylation in the brain, alternative splicing of KDM1A to dynamically regulate its association with RCOR1 and REST is likely an important mechanism for fine-tuned control over neuronal gene expression programs in neurodevelopmental disorders.
5. Direct modulation of H3K4 modifying enzymes
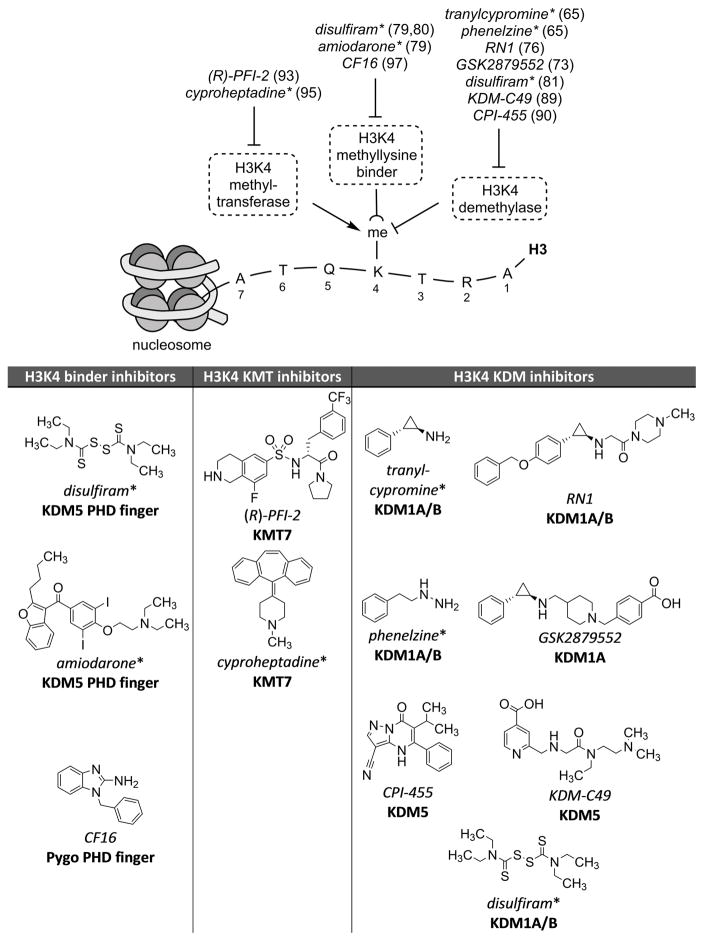
Therapeutic targeting of H3K4 methylation for neuropsychiatric illness remains a distant prospect. However, given the polygenic overlap between neuropsychiatric risk genes and antipsychotic response (54), it is informative to use existing chemical tools to probe disease biology to progress towards new therapeutic agents. Four FDA-approved drugs directly impact the activity of H3K4 modulators in vivo and in the brain, and accelerating advances in probe development have yielded tool compounds with vastly improved potency and selectivity (Figure 1).
Figure 1.
Examples of prototypical small molecules targeting regulators of H3K4 methylation with potential for use in studies of epigenetic regulation in mental disorders. References are indicated next to each compound name, and asterisks indicates FDA-approved status.
Monoamine oxidase (MAO) inhibitors (Phenelzine, Tranylcypromine, and derivatives thereof)
In the early 1950s, arylalkylhydrazines such as the drug iproniazid (Marsilid) were discovered to be anti-tuberculosis agents by phenotypic screening in infected mice (55). It was quickly uncovered that iproniazid inhibits monoamine oxidases A and B (MAO-A/B), enzymes that use a flavin adenine dinucleotide (FAD) cofactor to oxidize their substrates, which include not only the biogenic amines, serotonin and the catecholamines, but other, sympathomimetic amines including tyramine, benzylamine, and phenylethylamine (55). Administration of iproniazid to laboratory animals produced a rapid increase in brain levels of serotonin (56). At the same time, tuberculosis patients treated with iproniazid were reported to have elevated mood, increased sociability, and improved quality of sleep (55). Several years elapsed until these side effects were recognized as representing a new class of drug, an antidepressant, and iproniazid found clinical success (57). The concurrent discovery of the antidepressant qualities of iproniazid and the antipsychotic properties of chlorpromazine and reserpine laid the foundation for much of modern neuropharmacology (58). Medicinal chemistry efforts to further improve the potency of iproniazid lead to the development of three main classes of irreversible monoamine oxidase inhibitors: hydrazines, propargylamines, and cyclopropylamines (55). A posteriori biochemical analysis has revealed that these drugs, including the powerful antidepressant tranylcypromine (TCP; Parnate, a racemic mixture) function through covalent modification of the FAD cofactor (56,59).
Decades later, the notion of irreversible histone lysine methylation was challenged when KIAA0601 (aka KDM1A), a protein with homology to MAO-A/B, was found to associate with several histone deacetylase (HDAC)-containing complexes (60). In 2004, Shi and coworkers provided the first direct evidence that KDM1A functions as an FAD-dependent histone lysine demethylase, with specificity for histone 3 lysine 4 mono- and di-methylated (H3K4me1/2) residues (61). Subsequently, another nuclear amine oxidase with H3K4me1/2 demethylase activity, KDM1B, was discovered by searching for homologous domains in genomic databases (62). Like MAO-A/B, KDM1A and KDM1B couple the reduction of FAD to FADH2 with the oxidation of the C-N methylamine bond to a hydrolytically labile iminium ion, a mechanism only compatible with mono- and di-methylated substrates (61,63,64). Motivated by the similarities in the enzymatic properties of KDM1A and MAO-A/B, McCafferty and coworkers screened a focused group of irreversible MAO inhibitors against KDM1A. The antidepressants TCP and phenelzine were found to weakly inhibit recombinant KDM1A demethylation of nucleosomes, whereas the propargylamines tested were inactive (65). Subsequent studies revealed that the amine oxidase domains of KDM1A/B and MAO-A/B are homologous (37–45% sequence identity) and validated that TCP is a covalent, FAD-directed inhibitor of KDM1A and KDM1B with a mechanism of inactivation similar to that of MAO-A/B (66–69). Importantly, we have shown that clinically-relevant doses of TCP engages KDM1A in the brains of systemically-treated rats (E.R., S.J.H., J.H. manuscript under preparation), raising the intriguing possibility that modulation of H3K4 methylation may, in part, be responsible for the clinical efficacy of this antidepressant. Further testing of this hypothesis will benefit from knowledge of the extent to which, if any, other classes of clinically effective antidepressants, including tricyclic/tetracylic antidepressants (TCAs; e.g. amitriptyline, maprotiline), selective serotonin reuptake Inhibitors (SSRIs; fluoxetine), and dual serotonin-norepinephrine reuptake Inhibitors (SNRIs; e.g. venlafaxine) directly or indirectly modulate histone methylation within the CNS. Here, both in vitro biochemical assays and ex vivo cellular assays will need to be performed to determine if there is a correlation between antidepressant activity and modulation of H3K4 methylation levels in the CNS, through either direct effects on enzymes such as KDM1A or KDM1B, or indirectly through transcriptional or other signaling pathways.
Ultimately, the development of highly-selective, brain penetrant, pharmacological probes of histone methylation epigenetic machinery will allow causal testing of the relationship between changes in H3K4 methylation and changes in neuroplasticity and behavior. Unfortunately, irreversible monoamine oxidase inhibition by TCP and related inhibitors results in accumulation of trace sympathomimetic amines, and strict dietary restrictions to limit tyramine (found in most cheeses, chocolate, and fermented foods) are required to prevent hypertensive crisis. Newer classes of antidepressants, including reversible MAO inhibitors, have greatly improved safety profiles and have relegated TCP to the management of severe and treatment-resistant depression (56). However, inhibition of KDM1A may be therapeutic in many diseases beyond neuropsychiatric disorders (reviewed in (70–72)); accordingly, tremendous drug discovery efforts have been made to identify novel small molecule inhibitors, primarily for application in oncology. The most selective KDM1A inhibitor described to date is a TCP-analog developed by GlaxoSmithKline, GSK2879552, which exerts antitumor effects against some small cell lung carcinoma cell lines without increasing histone methylation levels, although a DNA hypomethylation signature was observed in responsive lines (73). In addition to modifications made to increase selectivity for KDM1A, TCP has also been modified with core scaffolds targeting histone deacetylases or KDM2–7 demethylases to generate dual function inhibitors (74,75), although these compounds tend to be less potent and less selective for KDM1A than typical TCP analogs. Although many FAD-directed inhibitors have been developed, their selectivity against the monoamine oxidases substantially limits their use for studies of KDM1A activity in the brain and other contexts in which neurotransmitters play a physiological role (70). Importantly, a highly brain-penetrant TCP-derivative with greatly improved selectivity for KDM1A, RN1, was found to affect memory in a novel object recognition task, suggesting a role for KDM1A in modulating adult rodent behavior (76).
Disulfiram
Disulfiram is most well-known for its use in the management of alcohol dependence, where it inhibits acetaldehyde dehydrogenase via modification of active site cysteine residues (77). In addition, disulfiram inhibits a number of thiol-reactive proteins with diverse functions, including the O6-methylguanine-DNA methyltransferase MGMT (78) and several histone lysine demethylases. Disulfiram inhibits both KDM4A/JMJD2A and KDM5A/JARID1A not by acting as a ligand but through rather through ejection of structural zinc, thereby blocking the histone-reading function of the PHD finger domain (79,80). In addition to PHD finger disruption, disulfiram inhibits KDM1A/LSD1 by alkylation of a cysteine residue critical for catalytic activity (81). Although disulfiram’s effects on histone lysine methylation are challenging to characterize in a cellular context due to its large number of targets, modulation of H3K4 methylation may becoming increasingly relevant as efforts to repurpose this drug gain traction (82–84).
Emerging tool compounds
Unlike the KDM1 family of amine oxidases, all JmJC lysine demethylases (KDM2–8) utilize 2-oxoglutarate and Fe(II) to generate a reactive Fe(IV)-oxo species which inserts an oxygen atom into the substrate N-methyl C-H bond. The resulting hemi-aminal intermediate fragments to the demethylated lysine and formaldehyde, a catalytic mechanism capable of demethylating mono-, di-, or tri-methylated substrates (85). The KDM5 family of enzymes are often associated with the REST transcriptional complexes (86) and catalyze the removal of all methylation states of H3K4. Although many non-selective inhibitors have been identified, the compact and highly conserved active site of JmJC catalytic domains and lack of structural data has complicated the design of selective demethylase inhibitors (87,88). Recently, the crystal structures of KDM5A and KDM5B were solved and leveraged to develop the selective small molecule inhibitors KDM5-C49 and CPI-455 with excellent selectivity versus other demethylases, with compound treatment leading to genome-wide elevation of H3K4 tri-methylation in cancer cell lines (89,90). Although these compounds remain to be tested in a neurobiological context, these structures provide multiple chemotypes for further rational inhibitor design and future testing in the functional assays of chromatin-mediated neuroplasticity.
Methylation of H3K4 is achieved by proteins containing a conserved Su(var)3–9 and Enhancer of Zeste-Trithorax (SET) domain utilizing S-adenosylmethionine (SAM) as a methyl donor (91). Despite extensive progress towards the development of chemical tools targeting other histone lysine modifiers, including several H3K9 methyltransferases, compounds blocking H3K4 methylation have been more challenging to identify (92). A first-in-class and highly selective inhibitor of the H3K4 mono-methyltransferase KMT7/SETD7, (R)-PFI-2, was discovered by high-throughput screening (93). This compound binds in the active site of the enzyme and displays unusual uncompetitive inhibition with respect to SAM. Importantly, (R)-PFI-2 was shown to engage KMT7 by the cellular thermal shift assay (94), and its enantiomer, (S)-PFI-2, is 500-fold less active and serves as a structurally similar negative control molecule for comparison (93). More recently, cyproheptadine, a clinically approved anti-allergy drug, was discovered to inhibit KMT7 and modulate the transcriptional activity of its non-histone substrate, the estrogen receptor alpha (95). Derivatives of cyproheptadine with improved selectivity for KDM7 versus its anti-allergy targets, the histamine H1 and serotonin 5-HT2A receptors, will have increased utility as tool compounds in neurobiological model systems.
The development of chemical tools to block the histone-reading function of methyllysine binders is relatively underdeveloped compared with inhibitors against methyltransferase and demethylase enzymatic chemistries (96). In addition to disulfiram, the antiarrhythmic agent amiodarone was found to weakly inhibit the histone-reading function of KDM5A’s PHD finger, although unmethylated and trimethylated analogs had similar low potency and cellular activity remains to be determined (79). An NMR fragment screen identified benzimidazoles such as CF16 which compete with H3K4 dimethylated peptide substrate for binding to the PHD finger of Pygo and which may lead to small molecules that displace methyllysine binders (97). Finally, macrocyclic calixarenes were found to act as supramolecular caging compounds, binding directly to methylated H3K4 and inhibiting the interaction between the trimethylated species and the PHD finger of ING2 (98). Although these compounds are not immediately translatable to neurobiologically-revelant contexts, their development points towards greater chemical control over H3K4 methylation by small molecules.
6. Indirect modulators of H3K4 methylation
In addition to compounds that directly target the protein machinery responsible for regulating H3K4 methylation, several therapeutics and drugs of abuse appear to affect these marks indirectly. Although the contribution of H3K4 methylation to the mechanism of action of these compounds is largely speculative, these examples illustrate the plastic nature of H3K4 methylation in the brain in response to neuroactive small molecules.
HDAC inhibitors (Valproate, Butyrate, Trichostatin A, and others)
H3 acetylation and H3K4 di- and tri-methylation are highly correlated epigenetic marks (99) and compounds affecting H3 acetylation, such as histone deacetylases (HDAC) inhibitors, are known to modulate histone methylation and demethylation via several mechanisms. Importantly, HDAC inhibitors are known to affect neuroplasticity, and the FDA-approved drug valproate is a clinically-useful mood stabilizer and antiepileptic (100). KDM1A preferentially demethylates hypoacetylated substrates, which may enable the stepwise and coordinated removal of histone modifications (101,102). Consistent with this observation, purified HDAC-KDM1A complexes have dramatically reduced demethylase activity towards nucleosomal substrates when treated with the HDAC inhibitors sodium butyrate and trichostatin A without alterations in complex stability (103). In contrast, the KMT2 family of methyltransferases are stimulated by acetylated H3, and knock down of KMT2B prevents butyrate-induced H3K4 methylation (104). Thus, HDAC inhibitors potentiate H3K4 methylation by virtue of the fine-tuned substrate specificity of demethylase and methyltransferase enzymes. In addition, HDAC inhibitors increase H3K4 methylation by reducing the expression of multiple demethylases, including KDM1A and KDM5A-C, via the down-regulation of SP1 expression (105). This functional interplay between HDAC inhibitors and H3K4 methylation extends to model systems relevant to psychiatric disease, as treatment of neuronal cultures with valproate increases histone H3K4 trimethylation at the promoters of GABAergic genes, including GAD1 (106). Together, these results raise the possibility that modulation of H3K4 methylation may be partially responsible for the neurobiological effects of HDAC inhibition.
Clozapine
H3K4 methylation progressively increases through puberty at the promoters of GABAergic genes such as GAD1. In patients with schizophrenia, decreased H3K4 trimethylation at the GAD1 promoter and a weakened higher-order chromatin loop between the promoter and its enhancer are associated with reduced expression of GABAergic gene expression in human prefrontal cortex (5,15,106,107). The atypical antipsychotic clozapine, which has a somewhat higher therapeutic efficacy compared with conventional antipsychotics (15,108), upregulates H3K4 trimethylation at the GAD1 promoter in both human patients and in mouse models (106). H3K4 methylation was not affected by the conventional antipsychotic haloperidol, and clozapine-induced methylation was unimpeded in mice lacking the dopamine D2/D3 receptors, indicating that dopamine receptor blockade is not sufficient to explain the epigenetic regulation of GAD1 (106). Notably, mice heterozygous for KMT2A/MLL1 exhibited decreased H3K4 methylation at the promoter of brain GAD1 (106). While the molecular pathways linking clozapine to histone methylation remain unclear, these findings suggest the intriguing possibility that KMT2A, which is highly expressed in GABAergic and other adult cortical neurons, may be targeted for the treatment of psychosis (5,15).
Methamphetamine, cocaine, and ethanol
Epigenetic mechanisms may underpin the long-lived associations between the rewarding effects of a drug of abuse and its contextual stimuli in substance abuse disorders (109). Drug-induced transcriptional regulation in multiple brain regions, including the nucleus accumbens and prefrontal cortex, is critical to the formation of drug-associated memories and is strongly associated with chromatin modification (110,111). Animals treated with methamphetamine in a conditioned place preference paradigm had increased H3K4 di- and tri-methylation in the nucleus accumbens, including at the promoters of the induced genes OXTR (oxytocin receptor) and FOS (112). Importantly, knock down of either KMT2A or KDM5C disrupted methamphetamine-associated memory, highlighting the fine-tuning of histone methylation in drug-related behavioral conditioning (112). Many memory-related genes, including N-methyl-D-aspartate receptor subunits, contain RE1 binding sites, suggesting that disruption of drug-associated memories by H3K4 regulators may be mediated by dysregulated transcription by REST. In addition, exposure of adolescent rats to either cocaine or ethanol decreased and increased H3K4 trimethylation in the prefrontal cortex, respectively (113,114). While considered along with the substantial evidence for H3K9 methylation in the acquisition and persistence of addiction (109–111), these findings are consistent with a role for histone methylation in drug abuse and may be relevant in the comorbidity of substance abuse with other neuropsychiatric diseases.
7. Outlook and areas for future development
Epigenetic control of gene expression is complex and multilayered, and the rational design of small-molecules to modulate chromatin dynamics is far from straightforward (115). A diverse array of small molecule tools will be critical to build an understanding of the therapeutic potential of targeting histone lysine methylation. As the feasibility of large-scale genomic studies increases, it is expected that additional mutations in chromatin regulators will be uncovered, providing additional rationale for developing pharmacological tools for modulating epigenetic function in the brain (3). In addition, development of cell sorting methods to isolate neuronal subpopulations from frozen post-mortem brain samples along with improvements in the performance of ChIP-seq and in data analysis will facilitate a nuanced understanding of the brain-region and cell-type-specific epigenomic differences associated with neuropsychiatric disorders (116–118). For example, significant differences in the distribution of H3K4 methylation is observed in neurons versus non-neurons from the same region of cortical gray matter, indicating that analysis of homogenized tissue may lose critical changes in specific cell types, such as in response to drug treatment (29,119). With systematic efforts by the PsychEncode (35) and related efforts, such as the BrainSeq project (120), to use cutting-edge methodologies for epigenome and genome-wide transcriptome analysis, the multiple facets of epigenetic regulation in the CNS are likely to come into much sharper focus and provide evidence to support the pharmacological targeting of specific epigenetic mechanisms.
Beyond post-mortem studies, with the ability to now generate patient-specific, induced pluripotent stem cell (iPSC) models (121), which can be directed in their differentiation to defined neural and glial subtypes (122), there now exists an opportunity to probe the underlying mechanism of epigenetic dysregulation using genetically accurate human cell models of neuropsychiatric disorders(123,124). Probing the molecular mechanisms of epigenetic regulation over the course of early neurodevelopment has previously not been possible, but early studies with the case of Fragile X syndrome (125) and Rett syndrome (126–128) demonstrate the feasibility of now doing so for multiple neuroepigenetic disorders. Importantly, when combined with genome-editing technologies, such as CRISPR/Cas9 system, to create isogenic, mutation corrected cells, the causal relationship of a genotype-phenotype correlation can be rigorously established (129–131). Additionally, since gain- or loss-of-function mutations (129) or tagged versions of proteins (132) can be introduced without altering the stoichiometry of proteins present in large complexes that can occur with transgenes. Furthermore, since the regulation of endogenous transcripts under epigenetic control can be investigated, including non-coding RNAs that are often poorly conserved between human and lower organisms, such models should help generate more physiologically relevant ex vivo models to investigate the role of epigenetic regulation in mental illness.
In addition to the potential use of CRISPR/Cas9 and other genome-editing techniques for creation of isogenic cell models, as reviewed in Thakore et al. (133) {Thakore, 2016 #140}) and Dominguez et al. (134) {Dominguez, 2016 #139}, the CRISPR/Cas9 system has been ‘repurposed’ to enable site-specific epigenetic and transcriptional regulation involving the targeting of modified forms of Cas9 proteins that include fusions to epigenetic regulatory enzymes, such as KDM1A, that cause local modification of chromatin modifications. Additionally, fusions to transcriptional repressor domains, such as the Kruppel associated box (KRAB) termed CRISPR inhibition (CRISPRi) or to transcriptional activators domains, such as VP64 or the NF-κB complex subunit p65 that can further recruit additional regulatory proteins termed CRISPR activation (CRISPRa) (134) {Dominguez, 2016 #139}. Through the ability to targeting these genetic probes to specific regions of the genome, as well as specific cell types or regions of the brain, conceptually these technologies afford a level of precision not attainable by conventional pharmacological agents. However, one step in this direction of providing more spatially and temporal control of the epigenome comes from the work of Reis et al. who recently reported the development of a new class of ‘optoepigenetic’ probes that enable optical control of epigenome states and cellular gene expression (135). Here, using pharmacological agents incorporating an azobenzene “photoswitch” HDAC activity could be precisely controlled in time and space without the need for expression of any transgene or genome modification. Adaptation of this technique for histone methylation enzymes that is underway could potentially provide additionally highly precise means to probe epigenetic machinery in the nervous system and other tissues.
Finally, there is an emerging direction of ‘neuroepigenetic imaging’ involving the development of in vivo positron emission tomography (PET) imaging tools for probing the expression and function of epigenetic targets as well as advancing the characterization of probes and drug leads. The need for non-invasive, quantitative measurements of epigenetic process (either through expression or function) is paramount when considering how limited our access is to information in the living human brain. To develop a more complete appreciation for gene-environment regulation through mechanisms, including histone modifications and chromatin remodeling over the course of brain development, normal ageing and the development and progression of disease, imaging tool development will be required. Proof-of-concept studies have provided clear evidence that quantification of the epigenetic machinery in the context of density (123,136) and function is possible (137) is possible in primates, and these technologies are now being applied in the context of human brain imaging in healthy volunteers and patients. For example, the class-I HDAC imaging probe, [11C]Martinostat, was progressed to first in human trials (FDA IND #123154) to assess its potential for HDAC quantification and is now being assessed in human neurodegenerative and neuropsychiatric diseases. Here, inherent cross talk between different epigenetic mechanisms and combinatorial functions of histone post-translational modifications may enable indirect read out of changes in epigenetic state, for example changes in KDM activity through assessment of HDAC activity. With future work to link specific imaging agents like [11C]Martinostat to specific chromatin-modifying complexes and specific gene-regulation, non-invasive PET imaging may ultimately be able to measure gene-environment interactions in the human brain as well as to probe biochemical mechanisms implicated as being at the root cause of mental illness and health.
Acknowledgments
We thank members of the Haggarty and Hooker laboratories for their constructive feedback and discussion on the topic. We acknowledge financial support from the US National Institutes of Health (R01DA028301, R01MH095088), and Consortium for Frontotemporal Dementia Research/Bluefield Project.
Footnotes
Disclosure statement
The authors declare no relevant conflicts of interest.
Author contributions
E.L.R., J.M.H., and S.J.H. wrote the text, and all authors agreed on the final version of the manuscript.
References
- 1.McClung CA, Nestler EJ. Neuroplasticity mediated by altered gene expression. Neuropsychopharmacology. 2008;33:3–17. doi: 10.1038/sj.npp.1301544. [DOI] [PubMed] [Google Scholar]
- 2.Qureshi IA, Mehler MF. An evolving view of epigenetic complexity in the brain. Phil Trans R Soc B. 2014;369:20130506. doi: 10.1098/rstb.2013.0506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ronan JL, Wu W, Crabtree GR. From neural development to cognition: unexpected roles for chromatin. Nature Reviews Genetics. 2013;14:347–359. doi: 10.1038/nrg3413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shen E, Shulha H, Weng Z, Akbarian S. Regulation of histone H3K4 methylation in brain development and disease. Philosophical Transactions of the Royal Society of London B: Biological Sciences. 2014;369:20130514. doi: 10.1098/rstb.2013.0514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nestler EJ, Peña CJ, Kundakovic M, Mitchell A, Akbarian S. Epigenetic Basis of Mental Illness. The Neuroscientist. 2015 doi: 10.1177/1073858415608147. 1073858415608147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cheng Y, Bernstein A, Chen D, Jin P. 5-Hydroxymethylcytosine: A new player in brain disorders? Experimental neurology. 2015;268:3–9. doi: 10.1016/j.expneurol.2014.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wen L, Li X, Yan L, Tan Y, Li R, Zhao Y, Wang Y, Xie J, Zhang Y, Song C. Whole-genome analysis of 5-hydroxymethylcytosine and 5-methylcytosine at base resolution in the human brain. Genome Biol. 2014;15:R49. doi: 10.1186/gb-2014-15-3-r49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Son EY, Crabtree GR. American Journal of Medical Genetics Part C: Seminars in Medical Genetics. Wiley Online Library; 2014. The role of BAF (mSWI/SNF) complexes in mammalian neural development. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Issler O, Chen A. Determining the role of microRNAs in psychiatric disorders. Nature Reviews Neuroscience. 2015;16:201–212. doi: 10.1038/nrn3879. [DOI] [PubMed] [Google Scholar]
- 10.Geaghan M, Cairns MJ. MicroRNA and posttranscriptional dysregulation in psychiatry. Biological psychiatry. 2015;78:231–239. doi: 10.1016/j.biopsych.2014.12.009. [DOI] [PubMed] [Google Scholar]
- 11.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 12.Gibney E, Nolan C. Epigenetics and gene expression. Heredity. 2010;105:4–13. doi: 10.1038/hdy.2010.54. [DOI] [PubMed] [Google Scholar]
- 13.Allis CD, Jenuwein T, Reinberg D, Caparros M-L. Epigenetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2007. [Google Scholar]
- 14.Vallianatos CN, Iwase S. Disrupted intricacy of histone H3K4 methylation in neurodevelopmental disorders. Epigenomics. 2015;7:503–519. doi: 10.2217/epi.15.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Peter CJ, Akbarian S. Balancing histone methylation activities in psychiatric disorders. Trends in molecular medicine. 2011;17:372–379. doi: 10.1016/j.molmed.2011.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xu J, Andreassi M. Reversible histone methylation regulates brain gene expression and behavior. Hormones and behavior. 2011;59:383–392. doi: 10.1016/j.yhbeh.2010.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pattaroni C, Jacob C. Histone methylation in the nervous system: functions and dysfunctions. Molecular neurobiology. 2013;47:740–756. doi: 10.1007/s12035-012-8376-4. [DOI] [PubMed] [Google Scholar]
- 18.Black JC, Van Rechem C, Whetstine JR. Histone lysine methylation dynamics: establishment, regulation, and biological impact. Molecular cell. 2012;48:491–507. doi: 10.1016/j.molcel.2012.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zee BM, Levin RS, Xu B, LeRoy G, Wingreen NS, Garcia BA. In vivo residue-specific histone methylation dynamics. Journal of Biological Chemistry. 2010;285:3341–3350. doi: 10.1074/jbc.M109.063784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang Z, Zang C, Cui K, Schones DE, Barski A, Peng W, Zhao K. Genome-wide mapping of HATs and HDACs reveals distinct functions in active and inactive genes. Cell. 2009;138:1019–1031. doi: 10.1016/j.cell.2009.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hathaway NA, Bell O, Hodges C, Miller EL, Neel DS, Crabtree GR. Dynamics and memory of heterochromatin in living cells. Cell. 2012;149:1447–1460. doi: 10.1016/j.cell.2012.03.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gräff J, Tsai LH. Histone acetylation: molecular mnemonics on the chromatin. Nature Reviews Neuroscience. 2013;14:97–111. doi: 10.1038/nrn3427. [DOI] [PubMed] [Google Scholar]
- 23.Akbarian S, Huang HS. Epigenetic regulation in human brain—focus on histone lysine methylation. Biological psychiatry. 2009;65:198–203. doi: 10.1016/j.biopsych.2008.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gupta S, Kim SY, Artis S, Molfese DL, Schumacher A, Sweatt JD, Paylor RE, Lubin FD. Histone methylation regulates memory formation. The Journal of neuroscience. 2010;30:3589–3599. doi: 10.1523/JNEUROSCI.3732-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mosammaparast N, Shi Y. Reversal of histone methylation: biochemical and molecular mechanisms of histone demethylases. Annual review of biochemistry. 2010;79:155–179. doi: 10.1146/annurev.biochem.78.070907.103946. [DOI] [PubMed] [Google Scholar]
- 26.Herz HM, Garruss A, Shilatifard A. SET for life: biochemical activities and biological functions of SET domain-containing proteins. Trends in biochemical sciences. 2013;38:621–639. doi: 10.1016/j.tibs.2013.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kooistra SM, Helin K. Molecular mechanisms and potential functions of histone demethylases. Nature reviews Molecular cell biology. 2012;13:297–311. doi: 10.1038/nrm3327. [DOI] [PubMed] [Google Scholar]
- 28.Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 29.Shulha HP, Crisci JL, Reshetov D, Tushir JS, Cheung I, Bharadwaj R, Chou HJ, Houston IB, Peter CJ, Mitchell AC. Human-specific histone methylation signatures at transcription start sites in prefrontal neurons. PLoS Biol. 2012;10:e1001427. doi: 10.1371/journal.pbio.1001427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bagot RC, Zhang TY, Wen X, Nguyen TTT, Nguyen HB, Diorio J, Wong TP, Meaney MJ. Variations in postnatal maternal care and the epigenetic regulation of metabotropic glutamate receptor 1 expression and hippocampal function in the rat. Proceedings of the National Academy of Sciences. 2012;109:17200–17207. doi: 10.1073/pnas.1204599109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Connor CM, Dincer A, Straubhaar J, Galler JR, Houston IB, Akbarian S. Maternal immune activation alters behavior in adult offspring, with subtle changes in the cortical transcriptome and epigenome. Schizophrenia research. 2012;140:175–184. doi: 10.1016/j.schres.2012.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shulha HP, Cheung I, Guo Y, Akbarian S, Weng Z. Coordinated cell type–specific epigenetic remodeling in prefrontal cortex begins before birth and continues into early adulthood. PLoS Genet. 2013;9:e1003433. doi: 10.1371/journal.pgen.1003433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kano S, Colantuoni C, Han F, Zhou Z, Yuan Q, Wilson A, Takayanagi Y, Lee Y, Rapoport J, Eaton W. Genome-wide profiling of multiple histone methylations in olfactory cells: further implications for cellular susceptibility to oxidative stress in schizophrenia. Molecular psychiatry. 2013;18:740. doi: 10.1038/mp.2012.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shulha HP, Cheung I, Whittle C, Wang J, Virgil D, Lin CL, Guo Y, Lessard A, Akbarian S, Weng Z. Epigenetic signatures of autism: trimethylated H3K4 landscapes in prefrontal neurons. Archives of general psychiatry. 2012;69:314–324. doi: 10.1001/archgenpsychiatry.2011.151. [DOI] [PubMed] [Google Scholar]
- 35.Akbarian S, Liu C, Knowles JA, Vaccarino FM, Farnham PJ, Crawford GE, Jaffe AE, Pinto D, Dracheva S, Geschwind DH. The PsychENCODE project. Nature neuroscience. 2015;18:1707–1712. doi: 10.1038/nn.4156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Singh T, Kurki MI, Curtis D, Purcell SM, Crooks L, McRae J, Suvisaari J, Chheda H, Blackwood D, Breen G. Rare loss-of-function variants in SETD1A are associated with schizophrenia and developmental disorders. Nature neuroscience. 2016 doi: 10.1038/nn.4267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Takata A, Xu B, Ionita-Laza I, Roos JL, Gogos JA, Karayiorgou M. Loss-of-function variants in schizophrenia risk and SETD1A as a candidate susceptibility gene. Neuron. 2014;82:773–780. doi: 10.1016/j.neuron.2014.04.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.De Rubeis S, He X, Goldberg AP, Poultney CS, Samocha K, Cicek AE, Kou Y, Liu L, Fromer M, Walker S. Synaptic, transcriptional and chromatin genes disrupted in autism. Nature. 2014;515:209–215. doi: 10.1038/nature13772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Network, T., and Consortium, P. A. S. o. t. P. G. Psychiatric genome-wide association study analyses implicate neuronal, immune and histone pathways. Nature neuroscience. 2015;18:199–209. doi: 10.1038/nn.3922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tunovic S, Barkovich J, Sherr EH, Slavotinek AM. De novo ANKRD11 and KDM1A gene mutations in a male with features of KBG syndrome and Kabuki syndrome. American Journal of Medical Genetics Part A. 2014;164:1744–1749. doi: 10.1002/ajmg.a.36450. [DOI] [PubMed] [Google Scholar]
- 41.Pilotto S, Speranzini V, Marabelli C, Rusconi F, Toffolo E, Grillo B, Battaglioli E, Mattevi A. LSD1/KDM1A mutations associated to a newly described form of intellectual disability impair demethylase activity and binding to transcription factors. Human Molecular Genetics. 2016:ddw120. doi: 10.1093/hmg/ddw120. [DOI] [PubMed] [Google Scholar]
- 42.Bögershausen N, Wollnik B. Unmasking Kabuki syndrome. Clinical genetics. 2013;83:201–211. doi: 10.1111/cge.12051. [DOI] [PubMed] [Google Scholar]
- 43.Toffolo E, Rusconi F, Paganini L, Tortorici M, Pilotto S, Heise C, Verpelli C, Tedeschi G, Maffioli E, Sala C. Phosphorylation of neuronal Lysine-Specific Demethylase 1LSD1/KDM1A impairs transcriptional repression by regulating interaction with CoREST and histone deacetylases HDAC1/2. Journal of neurochemistry. 2014;128:603–616. doi: 10.1111/jnc.12457. [DOI] [PubMed] [Google Scholar]
- 44.Zibetti C, Adamo A, Binda C, Forneris F, Toffolo E, Verpelli C, Ginelli E, Mattevi A, Sala C, Battaglioli E. Alternative splicing of the histone demethylase LSD1/KDM1 contributes to the modulation of neurite morphogenesis in the mammalian nervous system. The Journal of Neuroscience. 2010;30:2521–2532. doi: 10.1523/JNEUROSCI.5500-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rusconi F, Grillo B, Ponzoni L, Bassani S, Toffolo E, Paganini L, Mallei A, Braida D, Passafaro M, Popoli M. LSD1 modulates stress-evoked transcription of immediate early genes and emotional behavior. Proceedings of the National Academy of Sciences. 2016;113:3651–3656. doi: 10.1073/pnas.1511974113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ballas N, Mandel G. The many faces of REST oversee epigenetic programming of neuronal genes. Current opinion in neurobiology. 2005;15:500–506. doi: 10.1016/j.conb.2005.08.015. [DOI] [PubMed] [Google Scholar]
- 47.Lu T, Aron L, Zullo J, Pan Y, Kim H, Chen Y, Yang TH, Kim HM, Drake D, Liu XS. REST and stress resistance in ageing and Alzheimer/’s disease. Nature. 2014;507:448–454. doi: 10.1038/nature13163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Neale BM, Kou Y, Liu L, Ma’Ayan A, Samocha KE, Sabo A, Lin CF, Stevens C, Wang LS, Makarov V. Patterns and rates of exonic de novo mutations in autism spectrum disorders. Nature. 2012;485:242–245. doi: 10.1038/nature11011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.O’Roak BJ, Vives L, Girirajan S, Karakoc E, Krumm N, Coe BP, Levy R, Ko A, Lee C, Smith JD. Sporadic autism exomes reveal a highly interconnected protein network of de novo mutations. Nature. 2012;485:246–250. doi: 10.1038/nature10989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yoo AS, Staahl BT, Chen L, Crabtree GR. MicroRNA-mediated switching of chromatin-remodelling complexes in neural development. Nature. 2009;460:642–646. doi: 10.1038/nature08139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wynder C, Hakimi MA, Epstein JA, Shilatifard A, Shiekhattar R. Recruitment of MLL by HMG-domain protein iBRAF promotes neural differentiation. Nature cell biology. 2005;7:1113–1117. doi: 10.1038/ncb1312. [DOI] [PubMed] [Google Scholar]
- 52.Tahiliani M, Mei P, Fang R, Leonor T, Rutenberg M, Shimizu F, Li J, Rao A, Shi Y. The histone H3K4 demethylase SMCX links REST target genes to X-linked mental retardation. Nature. 2007;447:601–605. doi: 10.1038/nature05823. [DOI] [PubMed] [Google Scholar]
- 53.Kim HG, Kim HT, Leach NT, Lan F, Ullmann R, Silahtaroglu A, Kurth I, Nowka A, Seong IS, Shen Y. Translocations disrupting PHF21A in the Potocki-Shaffer-syndrome region are associated with intellectual disability and craniofacial anomalies. The American Journal of Human Genetics. 2012;91:56–72. doi: 10.1016/j.ajhg.2012.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ruderfer D, Charney A, Readhead B. Polygenic overlap between schizophrenia risk and antipsychotic response: an analysis of genetic registry data. Lancet Psychiatry. 2015 doi: 10.1016/S2215-0366(15)00553-2. 00553-00552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.López-Muñoz F, Alamo C. Monoaminergic neurotransmission: the history of the discovery of antidepressants from 1950s until today. Current pharmaceutical design. 2009;15:1563–1586. doi: 10.2174/138161209788168001. [DOI] [PubMed] [Google Scholar]
- 56.Frieling H, Bleich S. Tranylcypromine. European archives of psychiatry and clinical neuroscience. 2006;256:268–273. doi: 10.1007/s00406-006-0660-8. [DOI] [PubMed] [Google Scholar]
- 57.Sandler M. Monoamine oxidase inhibitors in depression: history and mythology. Journal of Psychopharmacology. 1990 doi: 10.1177/026988119000400307. [DOI] [PubMed] [Google Scholar]
- 58.Shen WW. A history of antipsychotic drug development. Comprehensive psychiatry. 1999;40:407–414. doi: 10.1016/s0010-440x(99)90082-2. [DOI] [PubMed] [Google Scholar]
- 59.Binda C, Edmondson DE, Mattevi A. Advancing Methods for Biomolecular Crystallography. Springer; 2013. Monoamine Oxidase Inhibitors: Diverse and Surprising Chemistry with Expanding Pharmacological Potential; pp. 309–312. [Google Scholar]
- 60.Shi YG, Tsukada Y-i. The discovery of histone demethylases. Cold Spring Harbor perspectives in biology. 2013;5:a017947. doi: 10.1101/cshperspect.a017947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shi Y, Lan F, Matson C, Mulligan P, Whetstine JR, Cole PA, Casero RA, Shi Y. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell. 2004;119:941–953. doi: 10.1016/j.cell.2004.12.012. [DOI] [PubMed] [Google Scholar]
- 62.Metzger E, Wissmann M, Yin N, Müller JM, Schneider R, Peters AH, Günther T, Buettner R, Schüle R. LSD1 demethylates repressive histone marks to promote androgen-receptor-dependent transcription. Nature. 2005;437:436–439. doi: 10.1038/nature04020. [DOI] [PubMed] [Google Scholar]
- 63.Gaweska H, Henderson Pozzi M, Schmidt DM, McCafferty DG, Fitzpatrick PF. Use of pH and kinetic isotope effects to establish chemistry as rate-limiting in oxidation of a peptide substrate by LSD1. Biochemistry. 2009;48:5440–5445. doi: 10.1021/bi900499w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Karytinos A, Forneris F, Profumo A, Ciossani G, Battaglioli E, Binda C, Mattevi A. A novel mammalian flavin-dependent histone demethylase. Journal of Biological Chemistry. 2009;284:17775–17782. doi: 10.1074/jbc.M109.003087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lee MG, Wynder C, Schmidt DM, McCafferty DG, Shiekhattar R. Histone H3 lysine 4 demethylation is a target of nonselective antidepressive medications. Chemistry & biology. 2006;13:563–567. doi: 10.1016/j.chembiol.2006.05.004. [DOI] [PubMed] [Google Scholar]
- 66.Schmidt DM, McCafferty DG. trans-2-Phenylcyclopropylamine is a mechanism-based inactivator of the histone demethylase LSD1. Biochemistry. 2007;46:4408–4416. doi: 10.1021/bi0618621. [DOI] [PubMed] [Google Scholar]
- 67.Yang M, Culhane JC, Szewczuk LM, Jalili P, Ball HL, Machius M, Cole PA, Yu H. Structural basis for the inhibition of the LSD1 histone demethylase by the antidepressant trans-2-phenylcyclopropylamine. Biochemistry. 2007;46:8058–8065. doi: 10.1021/bi700664y. [DOI] [PubMed] [Google Scholar]
- 68.Binda C, Valente S, Romanenghi M, Pilotto S, Cirilli R, Karytinos A, Ciossani G, Botrugno OA, Forneris F, Tardugno M. Biochemical, structural, and biological evaluation of tranylcypromine derivatives as inhibitors of histone demethylases LSD1 and LSD2. Journal of the American Chemical Society. 2010;132:6827–6833. doi: 10.1021/ja101557k. [DOI] [PubMed] [Google Scholar]
- 69.Fang R, Chen F, Dong Z, Hu D, Barbera AJ, Clark EA, Fang J, Yang Y, Mei P, Rutenberg M. LSD2/KDM1B and its cofactor NPAC/GLYR1 endow a structural and molecular model for regulation of H3K4 demethylation. Molecular cell. 2013;49:558–570. doi: 10.1016/j.molcel.2012.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zheng YC, Ma J, Wang Z, Li J, Jiang B, Zhou W, Shi X, Wang X, Zhao W, Liu HM. A Systematic Review of Histone Lysine-Specific Demethylase 1 and Its Inhibitors. Medicinal research reviews. 2015;35:1032–1071. doi: 10.1002/med.21350. [DOI] [PubMed] [Google Scholar]
- 71.Hayward D, Cole P. LSD1 Histone Demethylase Assays and Inhibition. Methods in Enzymology. 2016 doi: 10.1016/bs.mie.2016.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Stazi G, Zwergel C, Valente S, Mai A. LSD1 inhibitors: a patent review (2010–2015) Expert Opinion on Therapeutic Patents. 2016:1–16. doi: 10.1517/13543776.2016.1165209. [DOI] [PubMed] [Google Scholar]
- 73.Mohammad HP, Smitheman KN, Kamat CD, Soong D, Federowicz KE, Van Aller GS, Schneck JL, Carson JD, Liu Y, Butticello M. A DNA hypomethylation signature predicts antitumor activity of LSD1 inhibitors in SCLC. Cancer cell. 2015;28:57–69. doi: 10.1016/j.ccell.2015.06.002. [DOI] [PubMed] [Google Scholar]
- 74.Kalin J, Wu M, Hayward D, Wang L, Roberts J, Prusevich P, Hancock W, Bradner J, Ryu B, Alani R. CoREST in Peace: Dual Action Inhibitors of Histone Deacetylase and Lysine Specific Demethylase. The FASEB Journal. 2015;29:723–728. [Google Scholar]
- 75.Rotili D, Tomassi S, Conte M, Benedetti R, Tortorici M, Ciossani G, Valente S, Marrocco B, Labella D, Novellino E. Pan-histone demethylase inhibitors simultaneously targeting Jumonji C and lysine-specific demethylases display high anticancer activities. Journal of medicinal chemistry. 2013;57:42–55. doi: 10.1021/jm4012802. [DOI] [PubMed] [Google Scholar]
- 76.Neelamegam R, Ricq EL, Malvaez M, Patnaik D, Norton S, Carlin SM, Hill IT, Wood MA, Haggarty SJ, Hooker JM. Brain-penetrant LSD1 inhibitors can block memory consolidation. ACS chemical neuroscience. 2011;3:120–128. doi: 10.1021/cn200104y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Vallari RC, Pietruszko R. Human aldehyde dehydrogenase: mechanism of inhibition of disulfiram. Science. 1982;216:637–639. doi: 10.1126/science.7071604. [DOI] [PubMed] [Google Scholar]
- 78.Paranjpe A, Zhang R, Ali-Osman F, Bobustuc GC, Srivenugopal KS. Disulfiram is a direct and potent inhibitor of human O6-methylguanine-DNA methyltransferase (MGMT) in brain tumor cells and mouse brain and markedly increases the alkylating DNA damage. Carcinogenesis. 2014;35:692–702. doi: 10.1093/carcin/bgt366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wagner EK, Nath N, Flemming R, Feltenberger JB, Denu JM. Identification and characterization of small molecule inhibitors of a plant homeodomain finger. Biochemistry. 2012;51:8293–8306. doi: 10.1021/bi3009278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Rose NR, Thalhammer A, Seden PT, Mecinović J, Schofield CJ. Inhibition of the histone lysine demethylase JMJD2A by ejection of structural Zn (II) Chemical Communications. 2009:6376–6378. doi: 10.1039/b916357c. [DOI] [PubMed] [Google Scholar]
- 81.Ricq EL, Hooker JM, Haggarty SJ. 2016 Manuscript under review. [Google Scholar]
- 82.Triscott J, Rose Pambid M, Dunn SE. Concise review: bullseye: targeting cancer stem cells to improve the treatment of gliomas by repurposing disulfiram. Stem Cells. 2015;33:1042–1046. doi: 10.1002/stem.1956. [DOI] [PubMed] [Google Scholar]
- 83.Spivak AM, Andrade A, Eisele E, Hoh R, Bacchetti P, Bumpus NN, Emad F, Buckheit R, McCance-Katz EF, Lai J. A pilot study assessing the safety and latency-reversing activity of disulfiram in HIV-1–infected adults on antiretroviral therapy. Clinical infectious diseases. 2013:cit813. doi: 10.1093/cid/cit813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Gough SM, Lee F, Yang F, Walker RL, Zhu YJ, Pineda M, Onozawa M, Chung YJ, Bilke S, Wagner EK. NUP98–PHF23 Is a Chromatin-Modifying Oncoprotein That Causes a Wide Array of Leukemias Sensitive to Inhibition of PHD Histone Reader Function. Cancer discovery. 2014;4:564–577. doi: 10.1158/2159-8290.CD-13-0419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Walport LJ, Hopkinson RJ, Schofield CJ. Mechanisms of human histone and nucleic acid demethylases. Current opinion in chemical biology. 2012;16:525–534. doi: 10.1016/j.cbpa.2012.09.015. [DOI] [PubMed] [Google Scholar]
- 86.Pasini D, Hansen KH, Christensen J, Agger K, Cloos PA, Helin K. Coordinated regulation of transcriptional repression by the RBP2 H3K4 demethylase and Polycomb-Repressive Complex 2. Genes & development. 2008;22:1345–1355. doi: 10.1101/gad.470008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Heinemann B, Nielsen JM, Hudlebusch HR, Lees MJ, Larsen DV, Boesen T, Labelle M, Gerlach LO, Birk P, Helin K. Inhibition of demethylases by GSK-J1/J4. Nature. 2014;514:E1–E2. doi: 10.1038/nature13688. [DOI] [PubMed] [Google Scholar]
- 88.Thinnes CC, England KS, Kawamura A, Chowdhury R, Schofield CJ, Hopkinson RJ. Targeting histone lysine demethylases—Progress, challenges, and the future. Biochimica et Biophysica Acta (BBA)-Gene Regulatory Mechanisms. 2014;1839:1416–1432. doi: 10.1016/j.bbagrm.2014.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Johansson C, Velupillai S, Tumber A, Szykowska A, Hookway ES, Nowak RP, Strain-Damerell C, Gileadi C, Philpott M, Burgess-Brown N. Structural analysis of human KDM5B guides histone demethylase inhibitor development. Nature chemical biology. 2016 doi: 10.1038/nchembio.2087. [DOI] [PubMed] [Google Scholar]
- 90.Vinogradova M, Gehling VS, Gustafson A, Arora S, Tindell CA, Wilson C, Williamson KE, Guler GD, Gangurde P, Manieri W. An inhibitor of KDM5 demethylases reduces survival of drug-tolerant cancer cells. Nature chemical biology. 2016 doi: 10.1038/nchembio.2085. [DOI] [PubMed] [Google Scholar]
- 91.Cheng X, Collins RE, Zhang X. Structural and sequence motifs of protein (histone) methylation enzymes. Annual review of biophysics and biomolecular structure. 2005;34:267. doi: 10.1146/annurev.biophys.34.040204.144452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kaniskan HUm, Konze KD, Jin J. Selective inhibitors of protein methyltransferases. Journal of medicinal chemistry. 2014;58:1596–1629. doi: 10.1021/jm501234a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Barsyte-Lovejoy D, Li F, Oudhoff MJ, Tatlock JH, Dong A, Zeng H, Wu H, Freeman SA, Schapira M, Senisterra GA. (R)-PFI-2 is a potent and selective inhibitor of SETD7 methyltransferase activity in cells. Proceedings of the National Academy of Sciences. 2014;111:12853–12858. doi: 10.1073/pnas.1407358111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Molina DM, Jafari R, Ignatushchenko M, Seki T, Larsson EA, Dan C, Sreekumar L, Cao Y, Nordlund P. Monitoring drug target engagement in cells and tissues using the cellular thermal shift assay. Science. 2013;341:84–87. doi: 10.1126/science.1233606. [DOI] [PubMed] [Google Scholar]
- 95.Takemoto Y, Ito A, Niwa H, Okamura M, Fujiwara T, Hirano T, Handa N, Umehara T, Sonoda T, Ogawa K. Identification of Cyproheptadine as an Inhibitor of SET Domain Containing Lysine Methyltransferase 7/9 (Set7/9) That Regulates Estrogen-Dependent Transcription. Journal of medicinal chemistry. 2016;59:3650–3660. doi: 10.1021/acs.jmedchem.5b01732. [DOI] [PubMed] [Google Scholar]
- 96.Milosevich N, Hof F. Chemical inhibitors of epigenetic methyllysine reader proteins. Biochemistry. 2015 doi: 10.1021/acs.biochem.5b01073. [DOI] [PubMed] [Google Scholar]
- 97.Miller TC, Rutherford TJ, Birchall K, Chugh J, Fiedler M, Bienz M. Competitive binding of a benzimidazole to the histone-binding pocket of the Pygo PHD finger. ACS chemical biology. 2014;9:2864–2874. doi: 10.1021/cb500585s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Ali M, Daze KD, Strongin DE, Rothbart SB, Rincon-Arano H, Allen HF, Li J, Strahl BD, Hof F, Kutateladze TG. Molecular insights into inhibition of the methylated histone-plant homeodomain complexes by calixarenes. Journal of Biological Chemistry. 2015;290:22919–22930. doi: 10.1074/jbc.M115.669333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Nightingale KP, O’Neill LP, Turner BM. Histone modifications: signalling receptors and potential elements of a heritable epigenetic code. Current opinion in genetics & development. 2006;16:125–136. doi: 10.1016/j.gde.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 100.Fass DM, Schroeder FA, Perlis RH, Haggarty SJ. Epigenetic mechanisms in mood disorders: targeting neuroplasticity. Neuroscience. 2014;264:112–130. doi: 10.1016/j.neuroscience.2013.01.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Forneris F, Binda C, Vanoni MA, Battaglioli E, Mattevi A. Human histone demethylase LSD1 reads the histone code. Journal of Biological Chemistry. 2005;280:41360–41365. doi: 10.1074/jbc.M509549200. [DOI] [PubMed] [Google Scholar]
- 102.Shi YJ, Matson C, Lan F, Iwase S, Baba T, Shi Y. Regulation of LSD1 histone demethylase activity by its associated factors. Molecular cell. 2005;19:857–864. doi: 10.1016/j.molcel.2005.08.027. [DOI] [PubMed] [Google Scholar]
- 103.Lee MG, Wynder C, Bochar DA, Hakimi MA, Cooch N, Shiekhattar R. Functional interplay between histone demethylase and deacetylase enzymes. Molecular and cellular biology. 2006;26:6395–6402. doi: 10.1128/MCB.00723-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Nightingale KP, Gendreizig S, White DA, Bradbury C, Hollfelder F, Turner BM. Cross-talk between Histone Modifications in Response to Histone Deacetylase Inhibitors MLL4 LINKS HISTONE H3 ACETYLATION AND HISTONE H3K4 METHYLATION. Journal of Biological Chemistry. 2007;282:4408–4416. doi: 10.1074/jbc.M606773200. [DOI] [PubMed] [Google Scholar]
- 105.Huang PH, Chen CH, Chou CC, Sargeant AM, Kulp SK, Teng CM, Byrd JC, Chen CS. Histone deacetylase inhibitors stimulate histone H3 lysine 4 methylation in part via transcriptional repression of histone H3 lysine 4 demethylases. Molecular pharmacology. 2011;79:197–206. doi: 10.1124/mol.110.067702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Huang HS, Matevossian A, Whittle C, Kim SY, Schumacher A, Baker SP, Akbarian S. Prefrontal dysfunction in schizophrenia involves mixed-lineage leukemia 1-regulated histone methylation at GABAergic gene promoters. The Journal of neuroscience. 2007;27:11254–11262. doi: 10.1523/JNEUROSCI.3272-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Bharadwaj R, Jiang Y, Mao W, Jakovcevski M, Dincer A, Krueger W, Garbett K, Whittle C, Tushir JS, Liu J. Conserved chromosome 2q31 conformations are associated with transcriptional regulation of GAD1 GABA synthesis enzyme and altered in prefrontal cortex of subjects with schizophrenia. The Journal of Neuroscience. 2013;33:11839–11851. doi: 10.1523/JNEUROSCI.1252-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Ibrahim HM, Tamminga CA. Schizophrenia: treatment targets beyond monoamine systems. Annual review of pharmacology and toxicology. 2011;51:189–209. doi: 10.1146/annurev.pharmtox.010909.105851. [DOI] [PubMed] [Google Scholar]
- 109.Nestler EJ. Epigenetic mechanisms of drug addiction. Neuropharmacology. 2014;76:259–268. doi: 10.1016/j.neuropharm.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Renthal W, Nestler EJ. Epigenetic mechanisms in drug addiction. Trends in molecular medicine. 2008;14:341–350. doi: 10.1016/j.molmed.2008.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Robison AJ, Nestler EJ. Transcriptional and epigenetic mechanisms of addiction. Nature reviews neuroscience. 2011;12:623–637. doi: 10.1038/nrn3111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Aguilar-Valles A, Vaissière T, Griggs EM, Mikaelsson MA, Takács IF, Young EJ, Rumbaugh G, Miller CA. Methamphetamine-associated memory is regulated by a writer and an eraser of permissive histone methylation. Biological psychiatry. 2014;76:57–65. doi: 10.1016/j.biopsych.2013.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Black YD, Maclaren FR, Naydenov AV, Carlezon WA, Baxter MG, Konradi C. Altered attention and prefrontal cortex gene expression in rats after binge-like exposure to cocaine during adolescence. The Journal of neuroscience. 2006;26:9656–9665. doi: 10.1523/JNEUROSCI.2391-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Pascual M, Do Couto BR, Alfonso-Loeches S, Aguilar MA, Rodriguez-Arias M, Guerri C. Changes in histone acetylation in the prefrontal cortex of ethanol-exposed adolescent rats are associated with ethanol-induced place conditioning. Neuropharmacology. 2012;62:2309–2319. doi: 10.1016/j.neuropharm.2012.01.011. [DOI] [PubMed] [Google Scholar]
- 115.Szyf M. Prospects for the development of epigenetic drugs for CNS conditions. Nature Reviews Drug Discovery. 2015;14:461–474. doi: 10.1038/nrd4580. [DOI] [PubMed] [Google Scholar]
- 116.Fullard JF, Halene TB, Giambartolomei C, Haroutunian V, Akbarian S, Roussos P. Understanding the genetic liability to schizophrenia through the neuroepigenome. Schizophrenia research. 2016 doi: 10.1016/j.schres.2016.01.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Satterlee JS, Beckel-Mitchener A, Little RA, Procaccini D, Rutter JL, Lossie AC. Neuroepigenomics: resources, obstacles, and opportunities. Neuroepigenetics. 2015;1:2–13. doi: 10.1016/j.nepig.2014.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Jaffe AE. Postmortem human brain genomics in neuropsychiatric disorders—how far can we go? Current opinion in neurobiology. 2016;36:107–111. doi: 10.1016/j.conb.2015.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Cheung I, Shulha HP, Jiang Y, Matevossian A, Wang J, Weng Z, Akbarian S. Developmental regulation and individual differences of neuronal H3K4me3 epigenomes in the prefrontal cortex. Proceedings of the National Academy of Sciences. 2010;107:8824–8829. doi: 10.1073/pnas.1001702107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.BrainSeq AHBGC. BrainSeq: Neurogenomics to Drive Novel Target Discovery for Neuropsychiatric Disorders. Neuron. 2015;88:1078–1083. doi: 10.1016/j.neuron.2015.10.047. [DOI] [PubMed] [Google Scholar]
- 121.Inoue H, Nagata N, Kurokawa H, Yamanaka S. iPS cells: a game changer for future medicine. EMBO J. 2014;33:409–417. doi: 10.1002/embj.201387098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Ho SM, Topol A, Brennand KJ. From “directed differentiation” to “neuronal induction”: modeling neuropsychiatric disease. Biomark Insights. 2015;10:31–41. doi: 10.4137/BMI.S20066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Wang C, Schroeder FA, Wey HY, Borra R, Wagner FF, Reis S, Kim SW, Holson EB, Haggarty SJ, Hooker JM. In vivo imaging of histone deacetylases (HDACs) in the central nervous system and major peripheral organs. J Med Chem. 2014;57:7999–8009. doi: 10.1021/jm500872p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Haggarty SJ, Silva MC, Cross A, Brandon NJ, Perlis RH. Advancing drug discovery for neuropsychiatric disorders using patient-specific stem cell models. Mol Cell Neurosci. 2016;73:104–115. doi: 10.1016/j.mcn.2016.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Sheridan SD, Theriault KM, Reis SA, Zhou F, Madison JM, Daheron L, Loring JF, Haggarty SJ. Epigenetic characterization of the FMR1 gene and aberrant neurodevelopment in human induced pluripotent stem cell models of fragile X syndrome. PLoS One. 2011;6:e26203. doi: 10.1371/journal.pone.0026203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Nageshappa S, Carromeu C, Trujillo CA, Mesci P, Espuny-Camacho I, Pasciuto E, Vanderhaeghen P, Verfaillie CM, Raitano S, Kumar A, Carvalho CM, Bagni C, Ramocki MB, Araujo BH, Torres LB, Lupski JR, Van Esch H, Muotri AR. Altered neuronal network and rescue in a human MECP2 duplication model. Mol Psychiatry. 2016;21:178–188. doi: 10.1038/mp.2015.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Farra N, Zhang WB, Pasceri P, Eubanks JH, Salter MW, Ellis J. Rett syndrome induced pluripotent stem cell-derived neurons reveal novel neurophysiological alterations. Mol Psychiatry. 2012;17:1261–1271. doi: 10.1038/mp.2011.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Kim KY, Hysolli E, Park IH. Neuronal maturation defect in induced pluripotent stem cells from patients with Rett syndrome. Proc Natl Acad Sci U S A. 2011;108:14169–14174. doi: 10.1073/pnas.1018979108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Hsu PD, Lander ES, Zhang F. Development and applications of CRISPR-Cas9 for genome engineering. Cell. 2014;157:1262–1278. doi: 10.1016/j.cell.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Musunuru K. Genome editing of human pluripotent stem cells to generate human cellular disease models. Dis Model Mech. 2013;6:896–904. doi: 10.1242/dmm.012054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Doudna JA, Charpentier E. Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science. 2014;346:1258096. doi: 10.1126/science.1258096. [DOI] [PubMed] [Google Scholar]
- 132.Lackner DH, Carre A, Guzzardo PM, Banning C, Mangena R, Henley T, Oberndorfer S, Gapp BV, Nijman SM, Brummelkamp TR, Burckstummer T. A generic strategy for CRISPR-Cas9-mediated gene tagging. Nat Commun. 2015;6:10237. doi: 10.1038/ncomms10237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Thakore PI, Black JB, Hilton IB, Gersbach CA. Editing the epigenome: technologies for programmable transcription and epigenetic modulation. Nat Methods. 2016;13:127–137. doi: 10.1038/nmeth.3733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Dominguez AA, Lim WA, Qi LS. Beyond editing: repurposing CRISPR-Cas9 for precision genome regulation and interrogation. Nat Rev Mol Cell Biol. 2016;17:5–15. doi: 10.1038/nrm.2015.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Reis SA, Ghosh B, Hendricks JA, Szantai-Kis DM, Tork L, Ross KN, Lamb J, Read-Button W, Zheng B, Wang H, Salthouse C, Haggarty SJ, Mazitschek R. Light-controlled modulation of gene expression by chemical optoepigenetic probes. Nat Chem Biol. 2016;12:317–323. doi: 10.1038/nchembio.2042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Wey HY, Wang C, Schroeder FA, Logan J, Price JC, Hooker JM. Kinetic Analysis and Quantification of [(1)(1)C]Martinostat for in Vivo HDAC Imaging of the Brain. ACS Chem Neurosci. 2015;6:708–715. doi: 10.1021/acschemneuro.5b00066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Yeh HH, Tian M, Hinz R, Young D, Shavrin A, Mukhapadhyay U, Flores LG, Balatoni J, Soghomonyan S, Jeong HJ, Pal A, Uthamanthil R, Jackson JN, Nishii R, Mizuma H, Onoe H, Kagawa S, Higashi T, Fukumitsu N, Alauddin M, Tong W, Herholz K, Gelovani JG. Imaging epigenetic regulation by histone deacetylases in the brain using PET/MRI with (1)(8)F-FAHA. Neuroimage. 2013;64:630–639. doi: 10.1016/j.neuroimage.2012.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Greer EL, Shi Y. Histone methylation: a dynamic mark in health, disease and inheritance. Nature Reviews Genetics. 2012;13:343–357. doi: 10.1038/nrg3173. [DOI] [PMC free article] [PubMed] [Google Scholar]