SUMMARY
Elimination of lymphoid tissue reservoirs is a key component of HIV eradication strategies. CD8+ T cells play a critical role in control of HIV, but their functional attributes in lymph nodes (LNs) remain unclear. Here, we show that memory, follicular CXCR5+, and HIV-specific CD8+ T cells from LNs do not manifest the properties of cytolytic CD8+ T cells. While the frequency of follicular CXCR5+ CD8+ T cells was strongly inversely associated with peripheral viremia, this association was not dependent on cytolytic CXCR5+ CD8+ T cells. Moreover, the poor cytolytic activity of LN CD8+ T cells was linked to a compartmentalized dissociation between effector programming and the transcription factor T-bet. In line with this, activation of LN CD8+ T cells only partially induced the acquisition of cytolytic functions relative to peripheral blood CD8+ T cells. These results suggest that a state of immune privilege against CD8+ T cell-mediated cytolysis exists in lymphoid tissue, potentially facilitating the persistence of HIV.
In Brief
Reuter et al. show that lymphoid tissue CD8+ T cells from HIV-infected and uninfected individuals do not possess phenotypic, functional, or transcriptional regulatory properties of cytolytic T cells equivalent to those found in circulation. Their findings suggest that the failure to eliminate HIV could be related to compartmentalized CD8+ T cell function favoring noncytolytic responses in lymphoid tissue.
INTRODUCTION
Elimination of viral reservoirs is a major obstacle to the eradication of HIV (Chun et al., 2015). One such reservoir, the lymph node (LN)-resident CD4+ T follicular helper cell (Tfh) compartment, is a major site of ongoing viral replication (Banga et al., 2016; Lindqvist et al., 2012; Perreau et al., 2013; Petrovas et al., 2012). It is established that cytolytic CD8+ T cells are required for effective immune control of HIV and simian immunodeficiency virus (SIV) (Fukazawa et al., 2015; Koup et al., 1994; Schmitz et al., 1999). However, the mechanisms of CD8+ T cell immunosurveillance within lymphoid tissue are not well defined. HIV/SIV-specific CD8+ T cells have been identified in LNs but rarely within the B cell follicles (Chun et al., 2015; Connick et al., 2007, 2014; Folkvord et al., 2005; Oxenius et al., 2001). Recent studies also suggest that LN CD8+ T cells control SIV replication in extra-follicular CD4+ T cells, but not in follicular CD4+ T cells (Fukazawa et al., 2015; Lindqvist et al., 2012; Perreau et al., 2013; Petrovas et al., 2012, 2017). Accordingly, HIV-infected CD4+ Tfh cells are thought to evade immune surveillance largely via segregation from cytolytic CD8+ T cells.
Much of what is known about human CD8+ T cell cytolytic function, phenotype, and transcriptional regulation derives from studies of peripheral blood. In the context of HIV infection, clear associations have been demonstrated between control of HIV and HIV-specific CD8+ T cell cytolytic function, as measured by expression of cytolytic molecules, direct cytolytic killing capacity, and/or expression of the canonical effector function transcription factor T-bet, and control of HIV (Hersperger et al., 2010, 2011b; Migueles et al., 2002, 2008; Sáez-Cirión et al., 2007). However, it is unclear whether CD8+ T cell cytolytic function is manifest in HIV-infected lymphoid tissue. Intuitively, the presence of cytolytic CD8+ T cells in LNs, critical sites of antigen presentation and B/T cell priming, seems counterproductive for the generation and maintenance of immune responses. A number of studies in humans and mice have indeed suggested that CD8+ T cells in lymphoid tissue have limited cytolytic capacity (Andersson et al., 1999; Jöhrens et al., 2006; Quigley et al., 2007; Wolint et al., 2004; Yang et al., 2005). Nonetheless, a systematic evaluation of perforin and granzyme B expression, linked with the regulatory elements T-bet and eomesodermin, has not been reported previously for LN CD8+ T cells.
Here, we examined the expression of cytolytic proteins and their underlying regulatory elements in total, follicular, and HIV-specific CD8+ T cells in LNs. We find that CD8+ T cells in HIV-infected lymphoid tissue, regardless of follicular localization, display low-level, discordant, and dysregulated expression of perforin and granzyme B. These results suggest that the failure of CD8+ T cells to eliminate HIV-infected CD4+ T cells is related not only to physical segregation from infected CD4+ Tfh cells in lymphoid follicles but also to a generalized state of functional immune privilege against cytolytic activity in lymphoid tissue. These findings have broad implications for cure and eradication strategies designed to invoke CD8+ T cell-mediated clearance of HIV-infected CD4+ T cells.
RESULTS
Characteristics of Memory and Cytolytic CD8+ T Cells in HIV-Infected LNs
To define the phenotypic, functional, and cytolytic properties of LN CD8+ T cells, we obtained LNs and peripheral blood mononuclear cells (PBMCs) from HIV−, chronically infected HIV+, and HIV+ individuals on antiretroviral therapy (ART; Table 1). We found significantly fewer memory CD8+ T cells within LNs compared to blood regardless of HIV infection status. However, HIV-infected individuals had expanded memory CD8+ T cells in blood and LNs (Figure S1A). HIV-infected LNs had increased frequencies of effector memory (CD27− CD45RO+) CD8+ T cells compared to HIV− subjects (Figure 1A), but unlike PBMCs, LNs contained few terminally differentiated effector (CD27− CD45RO−) CD8+ T cells (Figure 1A).
Table 1.
Clinical Characteristics of Subjects in Designated Groups
| Status | No. of Subjects |
Time on ART (Months)a |
Viral load RNA (Copies/mL)a | Log Viral Loada | CD4 Count (Cells/µL)a | Sex (M/F) | Age (Years)a | PBMCs/LNMCs (Matched) |
Sample Origin |
|---|---|---|---|---|---|---|---|---|---|
| HIV− | 41 | N/A | N/A | N/A | 1,319 (1,267–1,371), 2 | 15/18 | 38 (17–86), 33 | 23/20 (2) | Ohio (16), Mexico (8), Pennsylvania (15), and Greece (2) |
| Chronic | 27 | N/A | 56,009 (1,439–1,387,603), 27 | 4.75 (3.16–6.14), 27 | 523 (142–1,130), 27 | 25/2 | 31 (20–62), 27 | 22/25 (20) | Mexico (27) |
| Chronic, ART | 17 | 7 (1–108), 15 | 93 (20–7,103), 16 | 1.96 (1.6–3.9), 15 | 301 (199–834), 17 | 15/2 | 32 (22–51), 16 | 16/16 (15) | Mexico (15) and Pennsylvania (2) |
N/A, not applicable.
Data are presented as median (range), number of subjects.
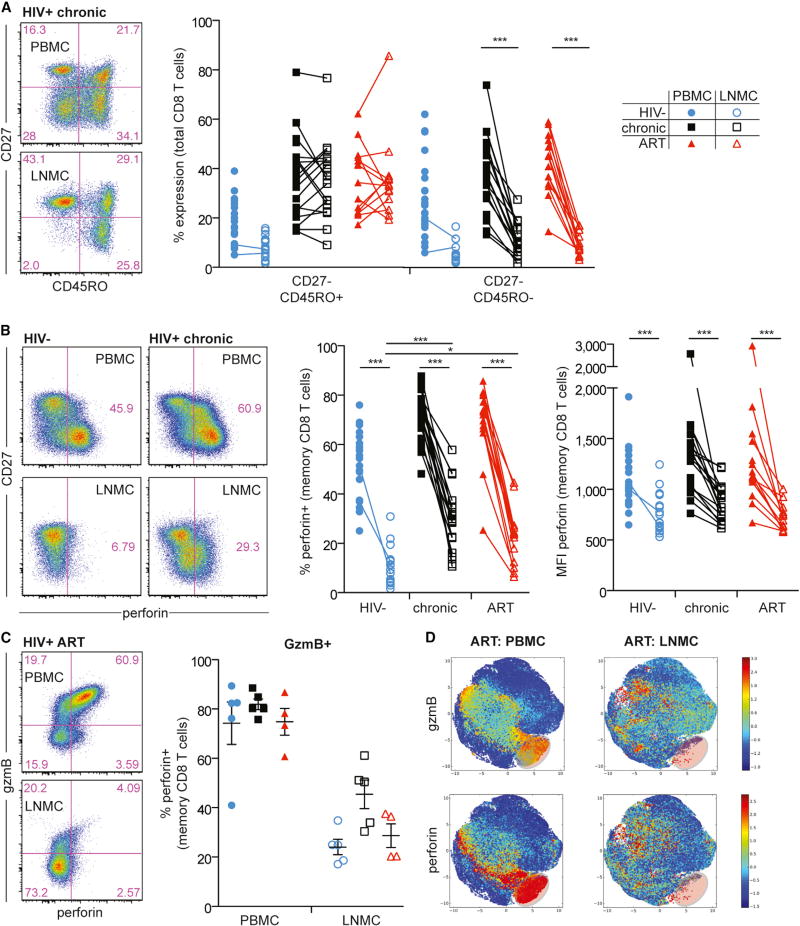
Figure 1. HIV Infection Induces Accumulation of Effector Memory CD8+ T Cells with Low Levels of Perforin Expression in LNs.
(A) Representative flow plots (left) showing CD3+ CD8+ T cells in PBMCs (top) and LNMCs (bottom) from a chronically infected HIV+ individual, and a comparison of memory subsets (right) in PBMCs versus LNMCs.
(B) Representative flow plots (left) showing memory CD8+ T cell expression of perforin in PBMCs (top row) versus LNMCs (bottom row) from an HIV− individual versus a chronically infected HIV+ individual. Perforin frequency and MFI in memory CD8+ T cells in PBMCs versus LNMCs is shown on the right for HIV−, chronic HIV+, and ART-treated HIV+ individuals.
(C) Representative flow plots (left) showing perforin versus gzmB expression in PBMCs (top) versus LNMCs (bottom) from an ART-treated HIV+ individual, and frequency of perforin co-expression in gzmB+ memory CD8+ T cells (right) in PBMCs versus LNMCs.
(D) Cell ACCENSE tSNE plots of perforin and gzmB expression in memory CD8+ T cells comparing PBMCs and LNMCs from ART-treated HIV+ individuals (based on n = 4/group). The highlighted area in lower right of each tSNE plot represents cytolytic effector CD8+ T cells.
The statistical tests used were the Kruskal-Wallis test with Dunn’s post-test (for between-subject groups within a tissue compartment), the Mann-Whitney test (for within a subject group between tissue compartments), and the Wilcoxon matched-pairs two-tailed test (for paired samples). *p < 0.05; **p < 0.01; ***p < 0.001. Blue circle, open blue circle, HIV−; black square, open black square, HIV+ therapy naive; red triangle, open red triangle, HIV+ on ART. Ranges shown on plots represent mean ± SEM. In all figures, closed symbols represent PBMCs and open symbols represent LNMCs. Memory subsets were determined by CD27 and CD45RO expression patterns.
Given the low abundance of effector CD8+ T cells in LNs (Figure 1A), we assessed perforin and granzyme B (gzmB) expression, each linked to control of HIV disease progression (Hersperger et al., 2011b). Compared to peripheral blood, fewer LN CD8+ T cells expressed perforin in HIV− individuals and treated and untreated HIV+ individuals (p < 0.001, Figure 1B). More LN memory CD8+ T cells expressed perforin during chronic and ART-treated HIV infection compared to HIV− individuals (p < 0.001 and p < 0.05, respectively). However, LN memory CD8+ T cells expressed less perforin per cell than PBMCs (p < 0.001; Figure 1B). Similar trends were observed for gzmB (Figure 1C; data not shown). Consistent with previous reports (Chattopadhyay et al., 2009; Hersperger et al., 2011a; Makedonas et al., 2010), perforin and gzmB were co-expressed in blood memory CD8+ T cells (Figure 1C) but notably dissociated in LN memory CD8+ T cells (Figure 1C). This dissociation was somewhat mitigated in chronic HIV infection but remained evident in lymphoid tissue across all subjects. Unbiased computational t-distributed stochastic neighbor embedding (tSNE) analysis (Shekhar et al., 2014) of all collected parameters on memory PBMCs and LN mononuclear cells (LNMCs) in both HIV− and treated and untreated HIV+ individuals further highlighted the dissociated nature of perforin and gzmB expression within LN memory CD8+ T cells and lack of the effector perforin+ population found in blood (Figures 1D and S1E).
Cytolytic Properties of CXCR5+ LN Follicular CD8+ T Cells Are Not Associated with Control of Viremia
Recent studies suggest that follicular CXCR5+ CD8+ T cells are involved in the control of chronic viral infections (He et al., 2016; Im et al., 2016; Leong et al., 2016; Mylvaganam et al., 2017; Petrovas et al., 2017). We therefore assessed whether the CXCR5+ CD8+ T cell population in HIV-infected LNs was present and linked to viremic control. A similar proportion (~20%) of LN memory CD8+ T cells expressed CXCR5 in HIV+ and HIV− individuals (Figure 2A). We found a strong inverse correlation between the frequency of LN CXCR5+ CD8+ T cells and viral load (Figure 2B), suggesting an association with control of HIV. We therefore explored whether CXCR5+ follicular CD8+ T cells expressed perforin and gzmB.
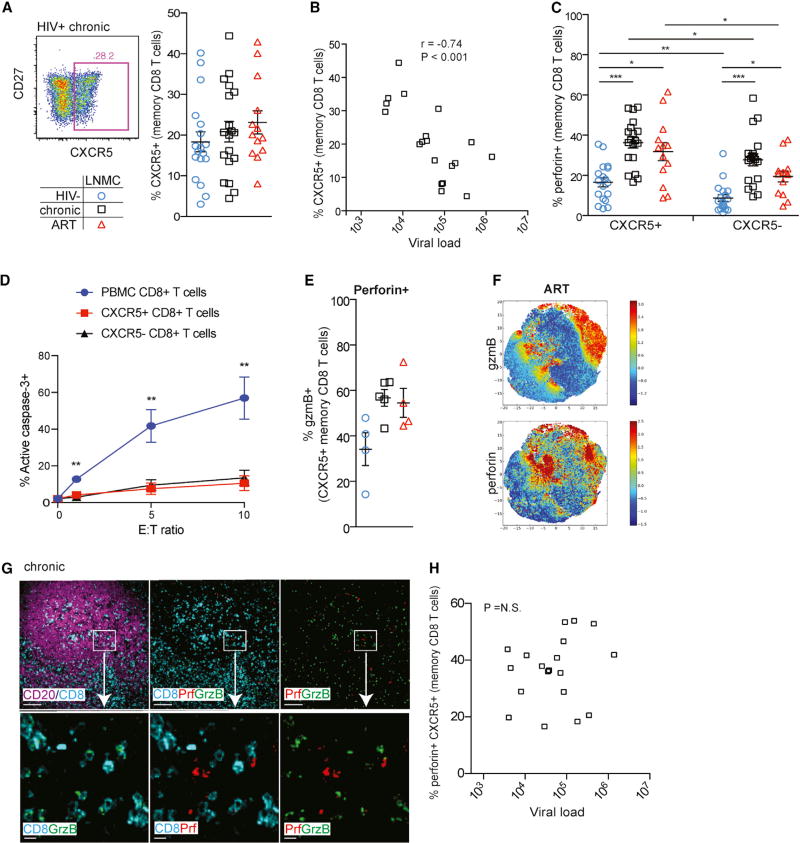
Figure 2. CXCR5+ CD8+ T Cells Are Associated with Control of HIV Independent of Perforin Expression in LNs.
(A) Representative flow plot (left) showing memory CD8+ T cell expression of CXCR5 in LNMCs from a chronically infected HIV+ individual, and frequency of LN CXCR5+ memory CD8+ T cells (right) in HIV−, chronic HIV+, and ART-treated HIV+ individuals.
(B) The frequency of CXCR5+ memory CD8+ T cells in LNMCs is inversely associated with HIV plasma viral load. Statistical significance was determined using Spearman’s test. Data shown were obtained from untreated HIV+ individuals with chronic infection.
(C) Perforin frequency in CXCR5+ and CXCR5− memory CD8+ T cells in LNMCs.
(D) Redirected killing assay measuring the ability of tonsillar CXCR5+ and CXCR5− CD8+ T cells isolated from n = 4 donors to kill anti-CD3 coated P815 target cells using activate caspase-3 as a readout.
(E) Frequency of gzmB co-expression in perforin+ CXCR5+ memory CD8+ T cells in LNMCs.
(F) Cell ACCENSE tSNE plots of perforin and gzmB expression in CXCR5+ memory CD8+ T cells in LNMCs from ART-treated HIV+ individuals (tSNE based on n = 4).
(G) IHC staining of paraffin-embedded tissue sections showing CD8+ T cells in B cell follicles. Top row: low-magnification view of a representative B cell follicle from a chronically infected HIV+ individual (scale bar, 100 µm). Boxes highlight areas for 10× zoom in bottom row (scale bar, 10 µm). Purple, CD19; cyan, CD8; red, perforin; green, gzmB.
(H) The frequency of perforin-expressing CXCR5+ CD8+ T cells in LNMCs is not associated with HIV plasma viral load. Statistical significance was determined using Spearman’s test. Data shown were obtained from untreated HIV+ individuals with chronic infection.
Statistical significance between subject groups was determined using the Kruskal-Wallis test with Dunn’s post-test. Statistical significance within a subject group between tissue compartments was determined using the Mann-Whitney two-tailed test. *p < 0.05; **p < 0.01; ***p < 0.001. Open blue circle, HIV−; open black square, HIV+ therapy naive; open red triangle, HIV+ on ART. Ranges shown on plots represent mean ± SEM.
Similar to CXCR5− CD8+ T cells, the majority of CXCR5+ CD8+ T cells in HIV− LNs did not express perforin (Figure 2C). The frequency of perforin-expressing cells was elevated in both CXCR5+ and CXCR5− memory CD8+ T cells in HIV-infected LNs regardless of treatment status (Figure 2C), with CXCR5+ CD8+ T cells being slightly more likely to express perforin. However, the overall frequency of perforin+ cells was still low compared to peripheral memory CD8+ T cells (Figures 1B and 2C), and the low amount of perforin on a per-cell basis did not differ based on CXCR5 expression or HIV infection status (Figure S1B). Further, to determine whether CXCR5+ CD8+ T cells had a differential ability to kill targets compared to CXCR5− CD8+ T cells, we performed a redirected cell lysis assay (adapted from Kiniry et al., 2017) measuring induction of active caspase-3 using as effector cells CXCR5+ and CXCR5− CD8+ T cells isolated from HIV-infected tonsils (Figure 2D). While CD8+ T cells from control HIV+ PBMCs demonstrated a strong induction of active caspase-3, tonsillar CXCR5+ CD8+ T cells had very little killing ability in comparison. Moreover, contrary to recent reports (Petrovas et al., 2017), we did not find that CXCR5+ CD8+ T cells exhibited stronger cytolytic activity than CXCR5− CD8+ T cells. Similar to memory CD8+ T cells, CXCR5+ CD8+ T cells displayed dissociated perforin and gzmB co-expression patterns (Figure 2E). Unbiased tSNE analysis showed a similar dissociation trend between perforin and gzmB co-expression (Figure 2F). To confirm whether cytolytic molecule expression patterns in CXCR5+ CD8+ T cells matched those physically located in B cell follicles, we performed multispectral confocal imaging analysis. CD8+ T cells within B cell follicles of HIV− individuals rarely expressed perforin and gzmB (Figure S1C). In HIV-infected LNs, irrespective of treatment status, a higher frequency of follicular CD8+ T cells expressed perforin or gzmB (Figures 2G and S1C), but only a small fraction co-expressed both proteins (Figure S1D). In contrast to the inverse association between the frequency of LN CXCR5+ CD8+ T cells and viral load (Figure 2B), we found no association between the frequency of perforin-expressing LN CXCR5+ CD8+ T cells and viral load (Figure 2H). Together, these data suggest that while follicular CXCR5+ CD8+ T cells may be a component of viral control, cytolytic properties are unlikely to be the mechanism by which follicular CD8+ T cells impact peripheral viremia.
Dissociation between Cytolytic CD8+ T Cells and T-Box-Binding Transcription Factors in LNs
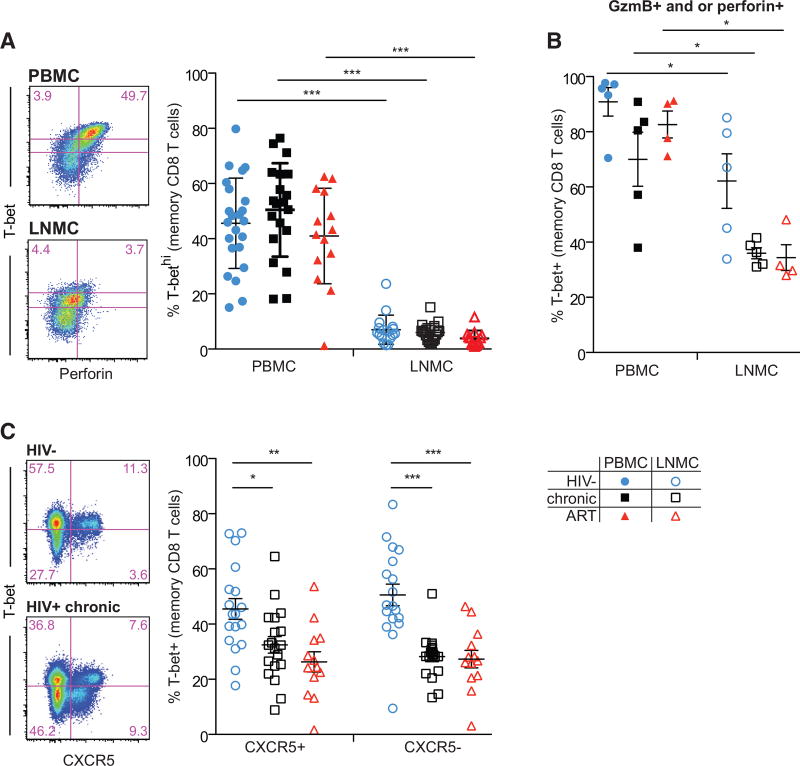
We next addressed the potential mechanisms underlying the differential cytolytic properties of LNs and peripheral blood CD8+ T cells. The transcription factors T-bet and eomesodermin (eomes) regulate effector differentiation and cytolytic function in peripheral blood CD8+ T cells (Cruz-Guilloty et al., 2009; Jenner et al., 2009; Pearce et al., 2003; Pipkin et al., 2010). High levels of T-bet expression have been associated with cytolytic function in human CD8+ T cells (Buggert et al., 2014; Hersperger et al., 2011a). We found that while T-bethigh cells were readily detectable in peripheral blood in all subjects tested regardless of HIV infection status, T-bethigh CD8+ T cells were rare in LNs (p < 0.001; Figures 3A and S2A). When found, ~60% of T-bethigh cells in the LN expressed perforin, substantially lower than in matched peripheral blood (Figure S2G). Because we rarely found T-bethigh CD8+ T cells in LNs, we focused the remainder of our analysis on characterizing all T-bet+ cells rather than separating low versus high expression. As expected, a large proportion of perforin+ CD8+ T cells in blood co-expressed T-bet, but this association was lost in LN CD8+ T cells, especially for HIV+ subjects (Figures 3A, S2A, and S2B), where perforin+ CD8+ T cells were less likely to express T-bet. Eomes was expressed in ~40% of blood and LN memory CD8+ T cells regardless of infection or treatment status (Figure S2A) and was not clearly associated with perforin expression in either blood or LN CD8+ T cells (Figure S2C). Similarly, few T-bet+ CD8+ T cells in LNs expressed gzmB compared to blood (Figures S2D and S2E). We found no clear association between gzmB expression and eomes in LN or peripheral blood CD8+ T cells (Figures S2D–S2F). Finally, most perforin/gzmB+ PBMC CD8+ T cells co-expressed T-bet (Figure 3B), but this association was again absent for LN perforin/gzmB+ CD8+ T cells (Figure 3B). We further assessed T-bet expression in CXCR5+ CD8+ T cells. Memory CXCR5+ and CXCR5− CD8+ T cells had similarly low expression levels of T-bet independent of HIV treatment status (Figure 3C). Together, these data indicate that perforin-expressing memory CD8+ T cells in LNs are transcriptionally and functionally distinct from peripheral blood CD8+ T cells, especially in HIV-infected LNs.
Figure 3. CD8+ T Cells Exhibit Dissociation between Transcriptional Drivers of Cytolytic Function and Expression of Cytolytic Proteins in LNs.
(A) Representative flow plots (left) showing memory CD8+ T cell expression of T-bet and perforin in PBMCs and LNMCs from an ART-treated HIV+ individual, and frequency of T-bethigh memory CD8+ T cells (right) in PBMCs and LNMCs. T-bet gating shown is the sum of the upper gates in the representative flow plots.
(B) T-bet frequency in memory CD8+ T cells expressing perforin and/or gzmB in PBMCs and LNMCs.
(C) Representative flow plots (left) showing memory CD8+ T cell expression of T-bet and CXCR5 in LNMCs from HIV− and HIV+ chronic, and T-bet frequency in CXCR5+ and CXCR5− CD8+ T cells (right) in LNMCs.
Statistical significance between subject groups within a tissue compartment was determined using the Kruskal-Wallis test with Dunn’s post-test. Statistical significance within a subject group between tissue compartments was determined using the Mann-Whitney two-tailed test. *p < 0.05; **p < 0.01; ***p < 0.001. Blue circle, open blue circle, HIV−; black square, open black square, HIV+ therapy-naive; red triangle, open red triangle, HIV+ on ART. Ranges shown on plots represent mean ± SEM.
LN HIV-Specific CD8+ T Cells Express Low Levels of Cytolytic Molecules and T-bet
Most studies to date have characterized bulk or memory CD8+ T cell populations in HIV-infected LNs. However, except for interferon-γ (IFN-γ) ELISpot analysis (Altfeld et al., 2002) and major histocompatibility complex (MHC) class I tetramer quantification (Connick et al., 2007), the functional and phenotypic properties of HIV-specific CD8+ T cells in LNs remain largely undefined. To address this knowledge gap, we screened for immunodominant HLA-A*0201-, HLA-A*2402-, and HLA-B*0702-restricted HIV-specific CD8+ T cell responses in LNs using MHC class I tetramers. Eight paired HIV MHC class I tetramer responses were identified in both HIV-infected LNs and blood from five donors. We found that the frequencies of HIV-specific CD8+ T cells were largely similar between blood and LNs (Figure 4A). Overlapping HIV Gag and Env peptide stimulations confirmed that the frequency of IFN-γ+ and/or tumor necrosis factor (TNF)+ HIV-specific CD8+ T cell responses was comparable between blood and LN irrespective of ART status (Figure S3A). The memory phenotype of LN HIV-specific CD8+ T cells was largely similar to blood, with the exception that effector memory HIV-specific CD8+ T cells were more common in blood compared to LN (Figure 4B). Importantly, LN HIV-specific CD8+ T cells expressed significantly lower levels of gzmB and perforin compared to peripheral blood (Figure 4C). Perforin frequencies (Figure S3B) and median fluorescence intensity (MFI) values (Figure S3C) in HIV-specific cytokine-producing CD8+ T cells were similar between chronically infected and ART-treated HIV+ subjects in both blood and LNs. However, similar to the HIV-tetramer+ CD8+ T cells, blood HIV-specific CD8+ T cells producing IFN-γ and/or TNF demonstrated increased frequency (Figure S3B) and MFI (Figure S3C) for perforin compared to LN HIV-specific CD8+ T cells. Perforin and gzmB expression was almost perfectly correlated in blood HIV-specific CD8+ T cells but completely dissociated in LNs (Figure 4D).
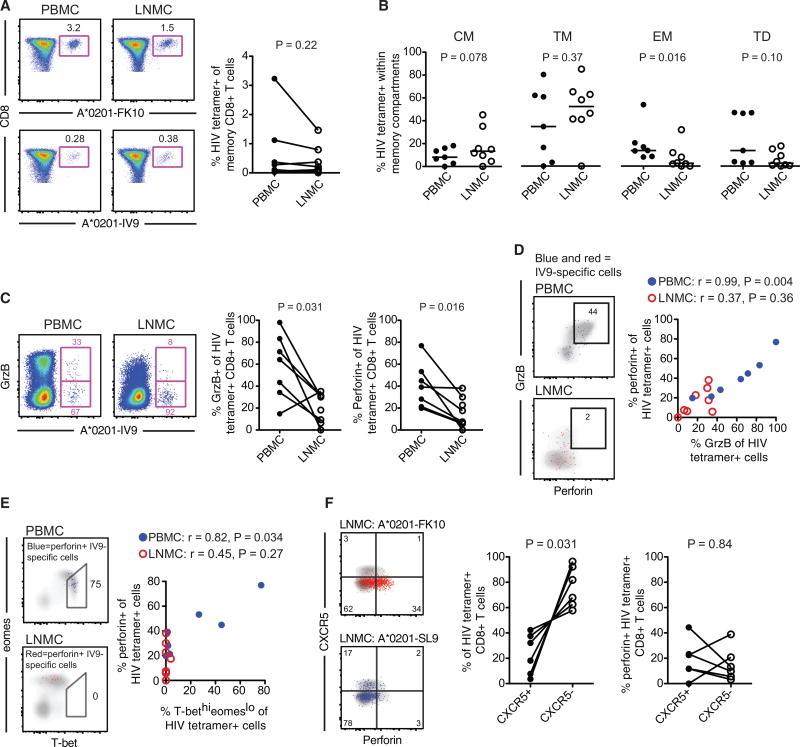
Figure 4. HIV-Specific CD8+ T Cells Are Poorly Cytolytic and Express Low Levels of T-Bet and CXCR5 in LNs.
(A) Flow plots (left) showing FLGKIWPSHK (FK10)/HLA-A*0201 and ILKEPVHGV (IV9)/HLA-A*0201 tetramer stains of PBMCs and LNMCs from the same subject, and frequency of matched HIV-tetramer+ CD8+ T cell responses (right) in PBMCs and LNMCs.
(B) Compartmentalization of HIV-tetramer+ CD8+ T cell responses within different memory subsets: central memory (CM; CCR7+ CD27+ CD45RO+), transitional memory (TM; CCR7− CD27+ CD45RO+), effector memory (EM; CCR7− CD27− CD45RO+), and terminally differentiated (TD; CCR7− CD27− CD45RO−).
(C) Flow cytometry plots (left) showing gzmB expression in HIV-tetramer+ CD8+ T cells in PBMCs and LNMCs from the same subject, and frequency of perforin and gzmB expression for all HIV-tetramer+ CD8+ T cells (right) in PBMCs and LNMCs.
(D) Frequency of perforin and gzmB co-expression in matched HIV-tetramer+ CD8+ T cells (left) in PBMCs (blue) and LNMCs (red), and correlation between perforin+ and gzmB+ HIV-tetramer+ CD8+ T cells (right) in PBMCs (blue) and LNMCs (red).
(E) Frequency of perforin+ HIV-tetramer+ CD8+ T cells within the T-bethigheomesdim population (left) in PBMCs (blue) and LNMCs (red), and correlation between perforin+ and HIV-tetramer+ CD8+ T cells (right) in PBMCs (blue) and LNMCs (red).
(F) Flow plots (left) showing HIV-tetramer+ CD8+ T cell expression of CXCR5 and perforin for two distinct specificities in LNMCs from the same subject, frequency of CXCR5+ versus CXCR5− HIV-tetramer+ CD8+T cells (middle) in LNMCs, and perforin expression in CXCR5+ and CXCR5− HIV-tetramer+ CD8+ T cells (right) in LNMCs. Flow plots are representative of chronically infected HIV+ individuals. Statistical significance within a subject group between tissue compartments was determined using the Mann-Whitney two-tailed test. Statistical significance for correlations was determined using Spearman’s test. Ranges shown on plots represent median and interquartile range (IQR).
We have previously shown that increased effector HIV-specific CD8+ T cell functions in peripheral blood are closely related to high expression levels of T-bet and intermediate levels of eomes (T-bethigheomesdim) (Buggert et al., 2014). In blood, we found that perforin+ HIV-specific CD8+ T cells were primarily T-bethigheomesdim, while perforin+ HIV-specific CD8+ T cells in LN were instead T-betdimeomeshi (Figure 4E). This latter profile has been associated with exhaustion (Buggert et al., 2014). As predicted, the frequency of perforin+ HIV-tetramer+ CD8+ T cells in blood was directly associated with a T-bethigheomesdim phenotype (Figure 4E), but no association was found between perforin expression and the T-bethieomesdim profile in LN HIV-specific CD8+ T cells (Figure 4E).
Finally, we addressed whether CXCR5+ HIV-specific CD8+ T cells express cytolytic molecules. Previous immunohistochemistry studies have demonstrated that LN HIV-specific CD8+ T cells are primarily compartmentalized to the T cell zone (Connick et al., 2007, 2014). In accordance with these reports, we found that most LN HIV-tetramer+ (Figure 4F) and cytokine producing HIV-specific CD8+ T cells were CXCR5− (Figure S3D) regardless of treatment status. We found no difference in the frequency of perforin expression between CXCR5+ and CXCR5− HIV-tetramer+ CD8+ T cells (Figure 4F). Perforin expression was also indistinguishable between chronically infected and ART-treated subjects for CXCR5+ HIV Gag/Env-specific CD8+ T cells (Figure S3E). Together, these data indicate that LN HIV-specific CD8+ T cells, regardless of their follicular localization, do not bear the characteristics of peripheral blood cytolytic CD8+ T cells.
Activation Does Not Fully Restore the Cytolytic Properties of LN HIV-Specific CD8+ T Cells
To determine whether activated LN CD8+ T cells have or can acquire cytolytic properties, we examined whether ex vivo LN CD8+ T cells entering the cell cycle, identified as Ki67+ (Gerdes et al., 1984), express perforin or gzmB. Similar to blood, LN CD8+ T cells displayed higher levels of Ki67 expression in individuals with chronic HIV infection than in HIV− and ART-treated individuals (Figure S4A). Ki67+ LN memory CD8+ T cells in treated and untreated HIV+ individuals co-expressed perforin and/or gzmB more frequently than Ki67+ LN memory CD8+ T cells in HIV− individuals (Figure S4B). However, Ki67− cells in HIV+ individuals also maintained higher levels of perforin and gzmB expression than HIV− subjects. Ki67+ LN memory CD8+ T cells were not more likely to express T-bet compared to Ki67− LN memory CD8+ T cells in any subject group, but eomes trended toward higher expression in both Ki67+ and Ki67− cells in HIV+ individuals compared to HIV− controls (Figures S4D and S4E).
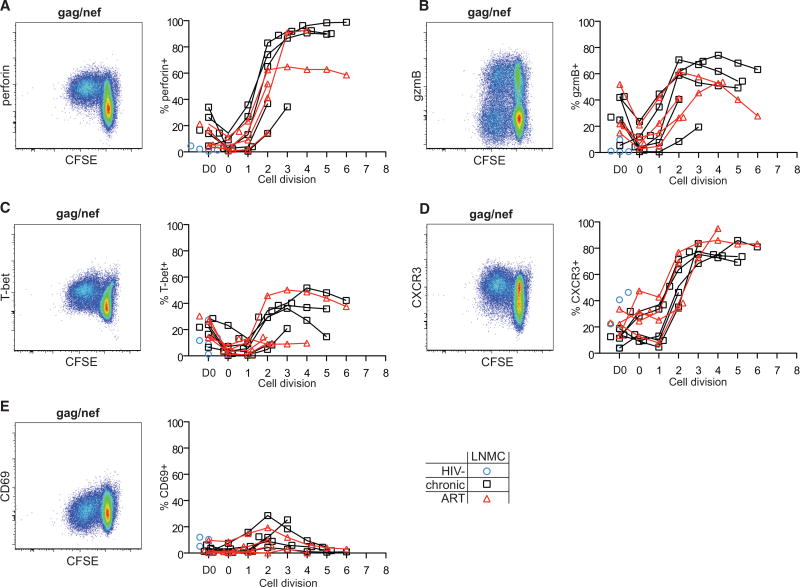
We next performed in vitro carboxyfluorescein succinimidyl ester (CFSE) assays to determine whether proliferating HIV Gag/Nef-specific LN CD8+ T cells acquire cytolytic effector properties. Robust cell division was observed in LN (Figures 5 and S4F) and peripheral blood (Figure S5) CD8+ T cells after stimulation with HIV Gag/Nef in most HIV-infected donors and after nonspecific stimulation with staphylococcal enterotoxin B (SEB) in all donors. While SEB readily induced perforin, gzmB, and T-bet expression in dividing LN and peripheral blood CD8+ T cells after 1 or 2 cell divisions, dividing HIV-specific CD8+ T cells differentially upregulated these proteins. Dividing LN HIV Gag/Nef-specific CD8+ T cells upregulated perforin and gzmB (Figures 5A and 5B), but not as rapidly or consistently as the SEB control (Figure S4G). The amount of perforin on a per cell basis also remained lower for LN CD8+ T cells than peripheral blood CD8+ T cells (Figure S5B). In contrast, SEB-stimulated LN and peripheral blood CD8+ T cells upregulated gzmB similarly (Figures S4G and S5A). Finally, unlike SEB-stimulated LN CD8+ T cells, LN HIV-specific CD8+ T cells did not strongly upregulate T-bet (Figure 5C). Together, these results indicate that LN CD8+ T cells can gain some aspects of cytolytic function after activation but do not under these conditions acquire the cytolytic properties characteristic of circulating effector CD8+ T cells.
Figure 5. Proliferating CD8+ T Cells Do Not Fully Acquire Cytolytic Properties and Express Marker Patterns Associated with Tissue Egress in LNs.
(A–E) Assessment of perforin (A), gzmB (B), T-bet (C), CXCR3 (D), and CD69 (E) expression in CFSE-labeled total CD8+ T cells in LNMCs after 5 days of stimulation with HIV clade B consensus Gag and Nef peptides (n = 5 HIV−, HIV+; n = 4 HIV+ ART). Flow plots (left) show representative data from an ART-treated HIV+ individual, and graphs (right) show protein expression frequency versus cell division (0–8 on the x axis). The gating strategy is shown in Figure S4F. D0 refers to prestimulation protein expression levels. Open blue circle, HIV−; open black square, HIV+ therapy naive; open red triangle, HIV+ on ART. Ranges shown on plots represent mean ± SEM.
Activated LN CD8+ T Cells Acquire LN Egress/Tissue Homing Properties but Do Not Acquire LN Follicular Homing Markers
Induction of effector function in LN CD8+ T cells is associated with upregulation of LN egress and tissue homing proteins (Cyster and Schwab, 2012; Matloubian et al., 2004; Shiow et al., 2006; Wolint et al., 2004). We therefore examined how activated LN CD8+ T cells modulate expression of CXCR3, which regulates tissue trafficking (Bonecchi et al., 1998; Sallusto et al., 1998), and CD69, which prevents tissue egress via interactions with S1PR (Matloubian et al., 2004). Dividing LN CD8+ T cells upregulated CXCR3 within 1 or 2 cell divisions after stimulation with HIV peptides or SEB (Figures 5D and S4H). Concomitantly, CD69 was transiently upregulated after ~2 or 3 cell divisions (Figures 5E and S4H) and then lost. Finally, we examined whether activated LN CD8+ T cells modulate expression of CXCR5. No consistent patterns of CXCR5 upregulation were detected for LN CD8+ T cells dividing in response to either HIV peptides or SEB (Figure S4I). Recent studies have demonstrated that transforming growth factor β (TGF-β) could selectively induce CXCR5 expression on rhesus macaque LN CD8+ T cells (Mylvaganam et al., 2017). We therefore assessed whether human tonsil CD8+ T cells similarly upregulated CXCR5 in response to TGF-β and/or interleukin-12 (IL-12). We did not find that these two cytokines, either alone or in combination, selectively induced CXCR5 expression on dividing cells, but they did each appear to maintain CXCR5 expression on CD8+ T cells compared to cells dividing in the absence of these cytokines (Figure S4J). Together, these results suggest that HIV-specific CD8+ T cells activated in situ would be prompted to leave the LN rather than traffic to LN follicles.
DISCUSSION
The sequestration of CD4+ Tfh cells into B cell follicles during chronic infection represents one mechanism by which CD8+ T cells are unable to eliminate HIV-infected CD4+ T cell reservoirs (Lindqvist et al., 2012; Perreau et al., 2013; Petrovas et al., 2012). Here, we show that CD8+ T cells from human LNs do not possess attributes associated with peripheral cytolytic CD8+ T cell function. In HIV infection, LN HIV-specific CD8+ T cells acquire some hallmarks of cytolytic function but retain characteristics that likely limit in vivo cytolytic activity, including low perforin expression, reduced cytolytic protein co-expression, alternate transcriptional regulation of effector function, and activation-induced tissue trafficking patterns.
Studies of HIV immunopathogenesis have associated CD8+ T cell functionality (Almeida et al., 2007; Betts et al., 2006), specificity (Goulder et al., 1997; Migueles et al., 2000), proliferative capacity (McKinnon et al., 2012; Migueles et al., 2002, 2009), memory phenotype (Addo et al., 2007; Almeida et al., 2007; Jagannathan et al., 2009), and cytolytic potential (Hersperger et al., 2010; Migueles et al., 2002, 2008; Sáez-Cirión et al., 2007) as correlates of protection from disease progression. However, it is unclear whether these peripheral blood CD8+ T cell correlates apply to HIV-infected LN. In line with previous studies (Altfeld et al., 2002; Connick et al., 2007; Oxenius et al., 2001), we did not find substantial differences in the frequency of HIV-specific CD8+ T cells between LNs and peripheral blood. However, profound qualitative differences were apparent between these compartments. While LN HIV-specific CD8+ T cells could individually express perforin, gzmB, and/or degranulate (data not shown) in response to stimulation, only a fraction displayed all three attributes, which are required in unison for cytolytic activity. Limited cytolytic potential is further compounded by restricted follicular access, as only a portion of LN memory and HIV-specific CD8+ T cells express CXCR5. In concordance with this, we found that CD8+ T cells from lymphoid tissue, regardless of whether they express CXCR5, were very limited in their ability to kill target cells. Accordingly, strategies designed to eliminate viral reservoirs in lymphoid tissue by re-invigorating HIV-specific CD8+ T cell cytolytic activity in ART-treated individuals must contend with additional factors beyond the follicular localization of HIV-infected CD4+ T cells.
The transcription factors T-bet (Sullivan et al., 2003; Szabo et al., 2000) and eomes (Cruz-Guilloty et al., 2009; Pearce et al., 2003; Pipkin and Rao, 2009) have been highly associated with cytolytic function in CD8+ T cells. The finding that T-bet was not associated with cytolytic molecule expression in LN CD8+ T cells was unexpected, especially because peripheral blood CD8+ T cells from the same individuals demonstrated coordinate expression of T-bet and perforin. Moreover, T-bet expression was substantially lower in LN CD8+ T cells than in peripheral blood CD8+ T cells and did not associate with effector-like memory CD8+ T cells. Although eomes induces effector CD8+ T cell differentiation in mice (Pipkin et al., 2010), we found no association with cytolytic protein expression in human blood or LNs. However, we did find more eomes-expressing CD8+ T cells in HIV-infected compared to uninfected LNs. These data suggest that transcriptional control of CD8+ T cell cytolytic function is regulated differently in LNs and blood or perhaps that temporal regulation prevents the acquisition of cytolytic function in LNs. In line with this, activated LN CD8+ T cells were able to express perforin, gzmB, and T-bet after cell division, but this gain of function was coupled with a concomitant modulation of trafficking markers to promote LN egress. These findings are consistent with mouse models (Wolint et al., 2004) and predictable based on canonical T cell responses to disease (Masopust and Schenkel, 2013). Together, these findings indicate that we have yet to identify all of the mechanisms that control cytolytic protein expression and effector function in lymphoid tissue.
Recent murine data have demonstrated that follicular CXCR5+ CD8+ T cells are a central component of antiviral immunity (He et al., 2016; Im et al., 2016; Leong et al., 2016). However, it is unclear whether CXCR5+ CD8+ T cells in these studies represent bona fide cytolytic CD8+ T cells. Here, we applied an extensive assessment of cytolytic killing through measurement of the simultaneous expression of perforin and gzmB, killing ability, and the underlying transcriptional programming necessary for cytolytic effector activity directly ex vivo from human lymphoid tissue. In agreement with mouse studies, our results indeed suggest that CXCR5+ CD8+ T cells in lymphoid tissue play a role in the immune response against HIV. However, we also found that the strong inverse correlation between peripheral viremia and the frequency of CXCR5+ CD8+ T cells in lymphoid tissue is not dependent on cytolytic machinery. This finding links conceptually with previous studies implicating non-cytolytic mechanisms as key determinants of immune protection against HIV and SIV (Blackbourn et al., 1996; Klatt et al., 2010; Wong et al., 2010).
In summary, we have shown that CD8+ T cells with cytolytic potential are relatively absent from human LNs. This finding suggests the existence of a biological mechanism that limits CD8+ T cell-mediated immunopathology and enables antigen presentation in a protected environment to facilitate the uninterrupted development of adaptive immune responses. However, the presence of an anatomically defined niche with immune privileges also provides a safe haven for various intracellular pathogens such as HIV. Importantly, these concepts also extend to mucosal tissues, as similar observations of limited cytolytic ability, low perforin expression, and low T-bet expression by mucosal tissue CD8+ T cells are apparent in both uninfected and chronically HIV-infected subjects (Kiniry et al., 2017; Quigley et al., 2006; Shacklett et al., 2004). Novel therapeutic strategies will therefore be required to counteract this natural lack of immune surveillance in the effort to purge and eradicate HIV reservoirs from both lymphoid and nonlymphoid tissues.
EXPERIMENTAL PROCEDURES
Study Groups
Peripheral blood collection and/or LN biopsies were performed in 80 individuals classified as HIV− (n = 40), HIV+ chronic untreated (n = 25), or HIV+ on ART (n = 15). Subject grouping, age, sex, sample collection, and clinical parameters are summarized in Table 1. All enrolled participants gave written informed consent as per protocols approved by the INER-CIENI ethics committee and the Federal Commission for the Protection against Sanitary Risk (COFEPRIS), the institutional review boards of the University of Pennsylvania and Case Western Reserve University, or the University General Hospital of Heraklion, Crete, Greece. Sample sizes were based on the availability of biological samples rather than a prespecified effect size. Exclusion criteria for samples were solely based on cell viability >70%.
Flow Cytometry
PBMC and LNMC stimulations were performed from cryopreserved material as described previously (Hersperger et al., 2010; Makedonas et al., 2010). Cells were incubated at a concentration of 1–2 × 106 cells/mL in complete medium (1 mL per condition) in the presence of costimulatory antibodies (3 µL/mL BD FastImmune α-CD28/CD49d, clones L293 and L25, BD Biosciences) and α-CD107a PE-Cy5 (clone H4A3, eBioscience) to measure degranulation for 6 hr total at 37°C in a 5% CO2 atmosphere with peptide pools representing HIV-1 Gag or Env gp120 (2 µg/mL, clade B, NIH AIDS Reagent Program) or epitopes from cytomegalovirus, Epstein-Barr virus, and influenza virus (1 µg/mL each, NIH AIDS Reagent Program; Kern et al., 1999; Munz, 2005; Wills et al., 1996). As a positive control, cells were stimulated with 1 µg/mL SEB (Sigma). After 1 hr, cultures were supplemented with 1 µg/mL brefeldin A (Sigma) and 0.7 µL/mL BD GolgiStop (BD Biosciences).
Proliferation assays were performed using PBMCs and LNMCs labeled with CellTrace CFSE (Molecular Probes, Invitrogen). Cells were stimulated with peptide pools spanning HIV-1 Gag and Nef (1 µg/mL each, clade B consensus, NIH AIDS Reagent Program, Division of AIDS [DAIDS], National Institute of Allergy and Infectious Diseases [NIAID], NIH) or 1 µg/ml SEB (Sigma) for 5 days in the presence of recombinant human IL-2 (100 U/mL) and plate-bound α-CD28/CD49d (BD Biosciences).
All flow cytometry immunostaining was performed as described previously (Hersperger et al., 2010; Makedonas et al., 2010). The antibodies and MHC tetramers used are described in Supplemental Experimental Procedures. Briefly, cells were washed in PBS and, in some cases, incubated with α-CCR7 at 37°C. Cells were labeled with the amine reactive dye LIVE/DEAD Fixable Aqua (Molecular Probes, Invitrogen) prior to incubation with antibodies against surface proteins. Intracellular proteins and cytokines were labeled after subsequent fixation/permeabilization with BD Cytofix/Cytoperm (BD Biosciences). For tetramer analyses, cells were incubated with tetramers for 10 min before pre-stain. All MHC class I tetramers were produced as described previously (Price et al., 2005). Cells were fixed in paraformaldehyde before analysis.
Cytometric Analyses
For all flow cytometric assays, at least 500,000 events were acquired using a modified LSR-II equipped with FACSDIVA software (BD Biosciences). Data were analyzed with FlowJo software (version 9.8.5, Tree Star). The following gating scheme was used to identify CD8+ T cells of interest: a time gate to ensure sample flow rate was consistent, singlets, lymphocytes, viable (LIVE/DEAD Aqua−), CD14−, CD16−, CD19−, CD3+, CD4−, CD8+. Cell divisions within the proliferation experiments were determined automatically using the FlowJo cell proliferation platform, assuming the default maximal cell division number of 8. Cell division “bins” were then applied to the CFSE dilution of each individual marker of interest in order to define modulation of specific markers with respect to cell division history.
Cell ACCENSE (automatic classification of cellular expression by nonlinear stochastic embedding) analyses were performed to visualize phenotypic relationships within multivariate data obtained from resting cells from 14 individuals (4–5 from each group). This program combines nonlinear dimensionality reduction with density-based partitioning, outputting unbiased protein expression data from all measured parameters as a two-dimensional plot (Shekhar et al., 2014; van der Maaten, 2013). PBMC and LNMC memory CD8+ T cell or LNMC CXCR5+/CXCR5− memory CD8+ T cell data were exported from FlowJo as FCS files. Cell ACCENSE was used to perform a Barnes-Hut-SNE dimensional reduction analysis on a total of 800,000 downsampled events to generate two-dimensional tSNE plots.
Imaging Analysis
Paraffin-embedded sections (6–10 µm) were deparaffinized, and antigen retrieval was performed using a decloaking chamber (125°C, 30 s, Biocare Medical) with Borg Decloaker buffer (Biocare Medical). Tissues were blocked (0.1 M Tris, 0.3% Triton X-100, 1% BSA) for 1 hr at room temperature (RT), stained with titrated amounts of nonconjugated antibodies (α-granzyme B, α-CD8) overnight at 4°C, washed with PBS (3 × 20 minutes), and stained with the appropriate secondary antibodies for 2 hr at RT. After a second blocking step with a 1:1 mixture of normal mouse serum (1 hr at RT), tissues were stained with titrated amounts of directly conjugated antibodies (α-CD20, α-perforin). JoJo staining was performed after a final wash step, and the slides were mounted with Fluoromount G (SouthernBiotech). Images were collected on a Leica SP8 confocal microscope using a 20× 0.75 numerical aperture (NA) or 40× 1.30 NA objective with a 1.5× optical zoom at a density of 1,024 × 1,024 pixels. Additional images were captured using a 63× 1.40 NA objective. Fluorophore spillover was corrected by imaging single-stained tissues and creating a compensation matrix via the Leica LAS-AF Channel Dye Separation module (Leica Microsystems). Compensated images were analyzed with Imaris software (Bitplane Scientific).
Statistical Analyses
Statistical analyses were performed with Prism software (version 5.0a, GraphPad). Non-parametric tests were used to determine significance between groups using the Mann-Whitney two-tailed test (for two groups), the Kruskal-Wallis test with Dunn’s post-test (for three or more groups), or the Wilcoxon matched-pairs two-tailed test (for paired samples) (*p < 0.05; **p < 0.01; ***p < 0.001).
Supplementary Material
Highlights.
CD8+ T cells in quiescent lymphoid tissues do not express markers of cytotoxicity
Lymphoid tissue HIV-specific CD8+ T cells do not possess full cytolytic markers
Noncytolytic CXCR5+ CD8+ T cells in lymphoid tissue associate with viral control
Acknowledgments
This research was supported by NIH R01 grants AI076066, AI118694, and AI108972, the Cleveland VA Geriatric Research Education and Clinical Center, and the Mexican Government (Comisión de Equidad y Género de las legislaturas LX-LXII de la H. Cámara de Diputados de la República Mexicana). D.A.P. is a Wellcome Trust Senior Investigator.
Footnotes
Supplemental Information includes Supplemental Experimental Procedures and five figures and can be found with this article online at https://doi.org/10.1016/j.celrep.2017.11.075.
AUTHOR CONTRIBUTIONS
M.A.R., P.M.D.R.E., M.B., R.A.K., G.R.-T., and M.R.B. designed the study. M.A.R., P.M.D.R.E., M.B., L.K.-C., A.S.J., H.M.G., and S.N. carried out all flow-based assays and analyses. C.P. and S.F.-M. performed all imaging studies and analyses. Y.A.-T. coordinated subject recruitment and obtained all clinical samples from Mexico. D.H.C. coordinated sample collection from Ohio. E.G. and D.A.P. provided custom MHC class I tetramers. P.M.D.R.E., A.R.-A., and D.H.C. processed samples and organized shipments. M.A.R., M.B., D.A.P., and M.R.B. wrote the manuscript. All authors approved the final manuscript.
DECLARATION OF INTERESTS
The authors declare no competing interests.
References
- Addo MM, Draenert R, Rathod A, Verrill CL, Davis BT, Gandhi RT, Robbins GK, Basgoz NO, Stone DR, Cohen DE, et al. Fully differentiated HIV-1 specific CD8+ T effector cells are more frequently detectable in controlled than in progressive HIV-1 infection. PLoS ONE. 2007;2:e321. doi: 10.1371/journal.pone.0000321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almeida JR, Price DA, Papagno L, Arkoub ZA, Sauce D, Bornstein E, Asher TE, Samri A, Schnuriger A, Theodorou I, et al. Superior control of HIV-1 replication by CD8+ T cells is reflected by their avidity, polyfunctionality, and clonal turnover. J. Exp. Med. 2007;204:2473–2485. doi: 10.1084/jem.20070784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altfeld M, van Lunzen J, Frahm N, Yu XG, Schneider C, Eldridge RL, Feeney ME, Meyer-Olson D, Stellbrink HJ, Walker BD. Expansion of pre-existing, lymph node-localized CD8+ T cells during supervised treatment interruptions in chronic HIV-1 infection. J. Clin. Invest. 2002;109:837–843. doi: 10.1172/JCI14789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson J, Behbahani H, Lieberman J, Connick E, Landay A, Patterson B, Sönnerborg A, Loré K, Uccini S, Fehniger TE. Perforin is not co-expressed with granzyme A within cytotoxic granules in CD8 T lymphocytes present in lymphoid tissue during chronic HIV infection. AIDS. 1999;13:1295–1303. doi: 10.1097/00002030-199907300-00005. [DOI] [PubMed] [Google Scholar]
- Banga R, Procopio FA, Noto A, Pollakis G, Cavassini M, Ohmiti K, Corpataux JM, de Leval L, Pantaleo G, Perreau M. PD-1(+) and follicular helper T cells are responsible for persistent HIV-1 transcription in treated aviremic individuals. Nat. Med. 2016;22:754–761. doi: 10.1038/nm.4113. [DOI] [PubMed] [Google Scholar]
- Betts MR, Nason MC, West SM, De Rosa SC, Migueles SA, Abraham J, Lederman MM, Benito JM, Goepfert PA, Connors M, et al. HIV nonprogressors preferentially maintain highly functional HIV-specific CD8+ T cells. Blood. 2006;107:4781–4789. doi: 10.1182/blood-2005-12-4818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackbourn DJ, Mackewicz CE, Barker E, Hunt TK, Herndier B, Haase AT, Levy JA. Suppression of HIV replication by lymphoid tissue CD8+ cells correlates with the clinical state of HIV-infected individuals. Proc. Natl. Acad. Sci. USA. 1996;93:13125–13130. doi: 10.1073/pnas.93.23.13125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonecchi R, Bianchi G, Bordignon PP, D’Ambrosio D, Lang R, Borsatti A, Sozzani S, Allavena P, Gray PA, Mantovani A, Sinigaglia F. Differential expression of chemokine receptors and chemotactic responsiveness of type 1 T helper cells (Th1s) and Th2s. J. Exp. Med. 1998;187:129–134. doi: 10.1084/jem.187.1.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buggert M, Tauriainen J, Yamamoto T, Frederiksen J, Ivarsson MA, Michaëlsson J, Lund O, Hejdeman B, Jansson M, Sönnerborg A, et al. T-bet and Eomes are differentially linked to the exhausted phenotype of CD8+ T cells in HIV infection. PLoS Pathog. 2014;10:e1004251. doi: 10.1371/journal.ppat.1004251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chattopadhyay PK, Betts MR, Price DA, Gostick E, Horton H, Roederer M, De Rosa SC. The cytolytic enzymes granyzme A, granzyme B, and perforin: expression patterns, cell distribution, and their relationship to cell maturity and bright CD57 expression. J. Leukoc. Biol. 2009;85:88–97. doi: 10.1189/jlb.0208107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chun TW, Moir S, Fauci AS. HIV reservoirs as obstacles and opportunities for an HIV cure. Nat. Immunol. 2015;16:584–589. doi: 10.1038/ni.3152. [DOI] [PubMed] [Google Scholar]
- Connick E, Mattila T, Folkvord JM, Schlichtemeier R, Meditz AL, Ray MG, McCarter MD, Mawhinney S, Hage A, White C, Skinner PJ. CTL fail to accumulate at sites of HIV-1 replication in lymphoid tissue. J. Immunol. 2007;178:6975–6983. doi: 10.4049/jimmunol.178.11.6975. [DOI] [PubMed] [Google Scholar]
- Connick E, Folkvord JM, Lind KT, Rakasz EG, Miles B, Wilson NA, Santiago ML, Schmitt K, Stephens EB, Kim HO, et al. Compartmentalization of simian immunodeficiency virus replication within secondary lymphoid tissues of rhesus macaques is linked to disease stage and inversely related to localization of virus-specific CTL. J. Immunol. 2014;193:5613–5625. doi: 10.4049/jimmunol.1401161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz-Guilloty F, Pipkin ME, Djuretic IM, Levanon D, Lotem J, Lichtenheld MG, Groner Y, Rao A. Runx3 and T-box proteins cooperate to establish the transcriptional program of effector CTLs. J. Exp. Med. 2009;206:51–59. doi: 10.1084/jem.20081242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cyster JG, Schwab SR. Sphingosine-1-phosphate and lymphocyte egress from lymphoid organs. Annu. Rev. Immunol. 2012;30:69–94. doi: 10.1146/annurev-immunol-020711-075011. [DOI] [PubMed] [Google Scholar]
- Folkvord JM, Armon C, Connick E. Lymphoid follicles are sites of heightened human immunodeficiency virus type 1 (HIV-1) replication and reduced antiretroviral effector mechanisms. AIDS Res. Hum. Retroviruses. 2005;21:363–370. doi: 10.1089/aid.2005.21.363. [DOI] [PubMed] [Google Scholar]
- Fukazawa Y, Lum R, Okoye AA, Park H, Matsuda K, Bae JY, Hagen SI, Shoemaker R, Deleage C, Lucero C, et al. B cell follicle sanctuary permits persistent productive simian immunodeficiency virus infection in elite controllers. Nat. Med. 2015;21:132–139. doi: 10.1038/nm.3781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerdes J, Lemke H, Baisch H, Wacker HH, Schwab U, Stein H. Cell cycle analysis of a cell proliferation-associated human nuclear antigen defined by the monoclonal antibody Ki-67. J. Immunol. 1984;133:1710–1715. [PubMed] [Google Scholar]
- Goulder PJ, Phillips RE, Colbert RA, McAdam S, Ogg G, Nowak MA, Giangrande P, Luzzi G, Morgan B, Edwards A, et al. Late escape from an immunodominant cytotoxic T-lymphocyte response associated with progression to AIDS. Nat. Med. 1997;3:212–217. doi: 10.1038/nm0297-212. [DOI] [PubMed] [Google Scholar]
- He R, Hou S, Liu C, Zhang A, Bai Q, Han M, Yang Y, Wei G, Shen T, Yang X, et al. Follicular CXCR5− expressing CD8(+) T cells curtail chronic viral infection. Nature. 2016;537:412–428. doi: 10.1038/nature19317. [DOI] [PubMed] [Google Scholar]
- Hersperger AR, Pereyra F, Nason M, Demers K, Sheth P, Shin LY, Kovacs CM, Rodriguez B, Sieg SF, Teixeira-Johnson L, et al. Perforin expression directly ex vivo by HIV-specific CD8 T-cells is a correlate of HIV elite control. PLoS Pathog. 2010;6:e1000917. doi: 10.1371/journal.ppat.1000917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hersperger AR, Martin JN, Shin LY, Sheth PM, Kovacs CM, Cosma GL, Makedonas G, Pereyra F, Walker BD, Kaul R, et al. Increased HIV-specific CD8+ T-cell cytotoxic potential in HIV elite controllers is associated with T-bet expression. Blood. 2011a;117:3799–3808. doi: 10.1182/blood-2010-12-322727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hersperger AR, Migueles SA, Betts MR, Connors M. Qualitative features of the HIV-specific CD8+ T-cell response associated with immunologic control. Curr. Opin. HIV AIDS. 2011b;6:169–173. doi: 10.1097/COH.0b013e3283454c39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Im SJ, Hashimoto M, Gerner MY, Lee J, Kissick HT, Burger MC, Shan Q, Hale JS, Lee J, Nasti TH, et al. Defining CD8+ T cells that provide the proliferative burst after PD-1 therapy. Nature. 2016;537:417–421. doi: 10.1038/nature19330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jagannathan P, Osborne CM, Royce C, Manion MM, Tilton JC, Li L, Fischer S, Hallahan CW, Metcalf JA, McLaughlin M, et al. Comparisons of CD8+ T cells specific for human immunodeficiency virus, hepatitis C virus, and cytomegalovirus reveal differences in frequency, immunodominance, phenotype, and interleukin-2 responsiveness. J. Virol. 2009;83:2728–2742. doi: 10.1128/JVI.02128-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenner RG, Townsend MJ, Jackson I, Sun K, Bouwman RD, Young RA, Glimcher LH, Lord GM. The transcription factors T-bet and GATA-3 control alternative pathways of T-cell differentiation through a shared set of target genes. Proc. Natl. Acad. Sci. USA. 2009;106:17876–17881. doi: 10.1073/pnas.0909357106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jöhrens K, Anagnostopoulos I, Dürkop H, Stein H. Different T-bet expression patterns characterize particular reactive lymphoid tissue lesions. Histopathology. 2006;48:343–352. doi: 10.1111/j.1365-2559.2005.02305.x. [DOI] [PubMed] [Google Scholar]
- Kern F, Surel IP, Faulhaber N, Frömmel C, Schneider-Mergener J, Schönemann C, Reinke P, Volk HD. Target structures of the CD8(+)-T-cell response to human cytomegalovirus: the 72-kilodalton major immediate-early protein revisited. J. Virol. 1999;73:8179–8184. doi: 10.1128/jvi.73.10.8179-8184.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiniry BE, Ganesh A, Critchfield JW, Hunt PW, Hecht FM, Somsouk M, Deeks SG, Shacklett BL. Predominance of weakly cytotoxic, T-bet(Low)Eomes(Neg) CD8(+) T-cells in human gastrointestinal mucosa: implications for HIV infection. Mucosal Immunol. 2017;10:1008–1020. doi: 10.1038/mi.2016.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klatt NR, Shudo E, Ortiz AM, Engram JC, Paiardini M, Lawson B, Miller MD, Else J, Pandrea I, Estes JD, et al. CD8+ lymphocytes control viral replication in SIVmac239-infected rhesus macaques without decreasing the lifespan of productively infected cells. PLoS Pathog. 2010;6:e1000747. doi: 10.1371/journal.ppat.1000747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koup RA, Safrit JT, Cao Y, Andrews CA, McLeod G, Borkowsky W, Farthing C, Ho DD. Temporal association of cellular immune responses with the initial control of viremia in primary human immunodeficiency virus type 1 syndrome. J. Virol. 1994;68:4650–4655. doi: 10.1128/jvi.68.7.4650-4655.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leong YA, Chen Y, Ong HS, Wu D, Man K, Deleage C, Minnich M, Meckiff BJ, Wei Y, Hou Z, et al. CXCR5(+) follicular cytotoxic T cells control viral infection in B cell follicles. Nat. Immunol. 2016;17:1187–1196. doi: 10.1038/ni.3543. [DOI] [PubMed] [Google Scholar]
- Lindqvist M, van Lunzen J, Soghoian DZ, Kuhl BD, Ranasinghe S, Kranias G, Flanders MD, Cutler S, Yudanin N, Muller MI, et al. Expansion of HIV-specific T follicular helper cells in chronic HIV infection. J. Clin. Invest. 2012;122:3271–3280. doi: 10.1172/JCI64314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makedonas G, Hutnick N, Haney D, Amick AC, Gardner J, Cosma G, Hersperger AR, Dolfi D, Wherry EJ, Ferrari G, Betts MR. Perforin and IL-2 upregulation define qualitative differences among highly functional virus-specific human CD8 T cells. PLoS Pathog. 2010;6:e1000798. doi: 10.1371/journal.ppat.1000798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masopust D, Schenkel JM. The integration of T cell migration, differentiation and function. Nat. Rev. Immunol. 2013;13:309–320. doi: 10.1038/nri3442. [DOI] [PubMed] [Google Scholar]
- Matloubian M, Lo CG, Cinamon G, Lesneski MJ, Xu Y, Brinkmann V, Allende ML, Proia RL, Cyster JG. Lymphocyte egress from thymus and peripheral lymphoid organs is dependent on S1P receptor 1. Nature. 2004;427:355–360. doi: 10.1038/nature02284. [DOI] [PubMed] [Google Scholar]
- McKinnon LR, Kaul R, Kimani J, Nagelkerke NJ, Wachihi C, Fowke KR, Ball TB, Plummer FA. HIV-specific CD8(+) T-cell proliferation is prospectively associated with delayed disease progression. Immunol. Cell Biol. 2012;90:346–351. doi: 10.1038/icb.2011.44. [DOI] [PubMed] [Google Scholar]
- Migueles SA, Sabbaghian MS, Shupert WL, Bettinotti MP, Marincola FM, Martino L, Hallahan CW, Selig SM, Schwartz D, Sullivan J, Connors M. HLA B*5701 is highly associated with restriction of virus replication in a subgroup of HIV-infected long term nonprogressors. Proc. Natl. Acad. Sci. USA. 2000;97:2709–2714. doi: 10.1073/pnas.050567397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Migueles SA, Laborico AC, Shupert WL, Sabbaghian MS, Rabin R, Hallahan CW, Van Baarle D, Kostense S, Miedema F, McLaughlin M, et al. HIV-specific CD8+ T cell proliferation is coupled to perforin expression and is maintained in nonprogressors. Nat. Immunol. 2002;3:1061–1068. doi: 10.1038/ni845. [DOI] [PubMed] [Google Scholar]
- Migueles SA, Osborne CM, Royce C, Compton AA, Joshi RP, Weeks KA, Rood JE, Berkley AM, Sacha JB, Cogliano-Shutta NA, et al. Lytic granule loading of CD8+ T cells is required for HIV-infected cell elimination associated with immune control. Immunity. 2008;29:1009–1021. doi: 10.1016/j.immuni.2008.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Migueles SA, Weeks KA, Nou E, Berkley AM, Rood JE, Osborne CM, Hallahan CW, Cogliano-Shutta NA, Metcalf JA, McLaughlin M, et al. Defective human immunodeficiency virus-specific CD8+ T-cell polyfunctionality, proliferation, and cytotoxicity are not restored by antiretroviral therapy. J. Virol. 2009;83:11876–11889. doi: 10.1128/JVI.01153-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munz C. Immune Response and Host Evasion in the Host-EBV Interaction. Caister Academic Press; 2005. [Google Scholar]
- Mylvaganam GH, Rios D, Abdelaal HM, Iyer S, Tharp G, Mavinger M, Hicks S, Chahroudi A, Ahmed R, Bosinger SE, et al. Dynamics of SIV-specific CXCR5+ CD8 T cells during chronic SIV infection. Proc. Natl. Acad. Sci. USA. 2017;114:1976–1981. doi: 10.1073/pnas.1621418114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oxenius A, Yerly S, Ramirez E, Phillips RE, Price DA, Perrin L. Distribution of functional HIV-specific CD8 T lymphocytes between blood and secondary lymphoid organs after 8–18 months of antiretroviral therapy in acutely infected patients. AIDS. 2001;15:1653–1656. doi: 10.1097/00002030-200109070-00007. [DOI] [PubMed] [Google Scholar]
- Pearce EL, Mullen AC, Martins GA, Krawczyk CM, Hutchins AS, Zediak VP, Banica M, DiCioccio CB, Gross DA, Mao CA, et al. Control of effector CD8+ T cell function by the transcription factor Eomesodermin. Science. 2003;302:1041–1043. doi: 10.1126/science.1090148. [DOI] [PubMed] [Google Scholar]
- Perreau M, Savoye AL, De Crignis E, Corpataux JM, Cubas R, Haddad EK, De Leval L, Graziosi C, Pantaleo G. Follicular helper T cells serve as the major CD4 T cell compartment for HIV-1 infection, replication, and production. J. Exp. Med. 2013;210:143–156. doi: 10.1084/jem.20121932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrovas C, Yamamoto T, Gerner MY, Boswell KL, Wloka K, Smith EC, Ambrozak DR, Sandler NG, Timmer KJ, Sun X, et al. CD4 T follicular helper cell dynamics during SIV infection. J. Clin. Invest. 2012;122:3281–3294. doi: 10.1172/JCI63039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrovas C, Ferrando-Martinez S, Gerner MY, Casazza JP, Pegu A, Deleage C, Cooper A, Hataye J, Andrews S, Ambrozak D, et al. Follicular CD8 T cells accumulate in HIV infection and can kill infected cells in vitro via bispecific antibodies. Sci. Transl. Med. 2017;9:eaag2285. doi: 10.1126/scitranslmed.aag2285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pipkin ME, Rao A. SnapShot: Effector and memory T cell differentiation. Cell. 2009;138:606.e1–2. doi: 10.1016/j.cell.2009.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pipkin ME, Sacks JA, Cruz-Guilloty F, Lichtenheld MG, Bevan MJ, Rao A. Interleukin-2 and inflammation induce distinct transcriptional programs that promote the differentiation of effector cytolytic T cells. Immunity. 2010;32:79–90. doi: 10.1016/j.immuni.2009.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price DA, Brenchley JM, Ruff LE, Betts MR, Hill BJ, Roederer M, Koup RA, Migueles SA, Gostick E, Wooldridge L, et al. Avidity for antigen shapes clonal dominance in CD8+ T cell populations specific for persistent DNA viruses. J. Exp. Med. 2005;202:1349–1361. doi: 10.1084/jem.20051357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quigley MF, Abel K, Zuber B, Miller CJ, Sandberg JK, Shacklett BL. Perforin expression in the gastrointestinal mucosa is limited to acute simian immunodeficiency virus infection. J. Virol. 2006;80:3083–3087. doi: 10.1128/JVI.80.6.3083-3087.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quigley MF, Gonzalez VD, Granath A, Andersson J, Sandberg JK. CXCR5+ CCR7− CD8 T cells are early effector memory cells that infiltrate tonsil B cell follicles. Eur. J. Immunol. 2007;37:3352–3362. doi: 10.1002/eji.200636746. [DOI] [PubMed] [Google Scholar]
- Sáez-Cirión A, Lacabaratz C, Lambotte O, Versmisse P, Urrutia A, Boufassa F, Barré-Sinoussi F, Delfraissy JF, Sinet M, Pancino G, Venet A Agence Nationale de Recherches sur le Sida EP36 HIV Controllers Study Group. HIV controllers exhibit potent CD8 T cell capacity to suppress HIV infection ex vivo and peculiar cytotoxic T lymphocyte activation phenotype. Proc. Natl. Acad. Sci. USA. 2007;104:6776–6781. doi: 10.1073/pnas.0611244104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sallusto F, Lenig D, Mackay CR, Lanzavecchia A. Flexible programs of chemokine receptor expression on human polarized T helper 1 and 2 lymphocytes. J. Exp. Med. 1998;187:875–883. doi: 10.1084/jem.187.6.875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz JE, Kuroda MJ, Santra S, Sasseville VG, Simon MA, Lifton MA, Racz P, Tenner-Racz K, Dalesandro M, Scallon BJ, et al. Control of viremia in simian immunodeficiency virus infection by CD8+ lymphocytes. Science. 1999;283:857–860. doi: 10.1126/science.283.5403.857. [DOI] [PubMed] [Google Scholar]
- Shacklett BL, Cox CA, Quigley MF, Kreis C, Stollman NH, Jacobson MA, Andersson J, Sandberg JK, Nixon DF. Abundant expression of granzyme A, but not perforin, in granules of CD8+ T cells in GALT: implications for immune control of HIV-1 infection. J. Immunol. 2004;173:641–648. doi: 10.4049/jimmunol.173.1.641. [DOI] [PubMed] [Google Scholar]
- Shekhar K, Brodin P, Davis MM, Chakraborty AK. Automatic Classification of Cellular Expression by Nonlinear Stochastic Embedding (ACCENSE) Proc. Natl. Acad. Sci. USA. 2014;111:202–207. doi: 10.1073/pnas.1321405111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiow LR, Rosen DB, Brdicková N, Xu Y, An J, Lanier LL, Cyster JG, Matloubian M. CD69 acts downstream of interferon-alpha/beta to inhibit S1P1 and lymphocyte egress from lymphoid organs. Nature. 2006;440:540–544. doi: 10.1038/nature04606. [DOI] [PubMed] [Google Scholar]
- Sullivan BM, Juedes A, Szabo SJ, von Herrath M, Glimcher LH. Antigen-driven effector CD8 T cell function regulated by T-bet. Proc. Natl. Acad. Sci. USA. 2003;100:15818–15823. doi: 10.1073/pnas.2636938100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabo SJ, Kim ST, Costa GL, Zhang X, Fathman CG, Glimcher LH. A novel transcription factor, T-bet, directs Th1 lineage commitment. Cell. 2000;100:655–669. doi: 10.1016/s0092-8674(00)80702-3. [DOI] [PubMed] [Google Scholar]
- van der Maaten L. Barnes-Hut-SNE. arXiv, arXiv:1301.3342. 2013 https://arxiv.org/abs/1301.3342.
- Wills MR, Carmichael AJ, Mynard K, Jin X, Weekes MP, Plachter B, Sissons JGP. The human cytotoxic T-lymphocyte (CTL) response to cytomegalovirus is dominated by structural protein pp65: frequency, specificity, and T-cell receptor usage of pp65-specific CTL. J. Virol. 1996;70:7569–7579. doi: 10.1128/jvi.70.11.7569-7579.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolint P, Betts MR, Koup RA, Oxenius A. Immediate cytotoxicity but not degranulation distinguishes effector and memory subsets of CD8+ T cells. J. Exp. Med. 2004;199:925–936. doi: 10.1084/jem.20031799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong JK, Strain MC, Porrata R, Reay E, Sankaran-Walters S, Ignacio CC, Russell T, Pillai SK, Looney DJ, Dandekar S. In vivo CD8+ T-cell suppression of SIV viremia is not mediated by CTL clearance of productively infected cells. PLoS Pathog. 2010;6:e1000748. doi: 10.1371/journal.ppat.1000748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang OO, Ferbas JJ, Hausner MA, Hultin LE, Hultin PM, McFadden D, Sawicki M, Detels R, Majchrowicz M, Matud JL, et al. Effects of HIV-1 infection on lymphocyte phenotypes in blood versus lymph nodes. J. Acquir. Immune Defic. Syndr. 2005;39:507–518. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.