Abstract
The three mitochondrial-encoded proteins, COX1, COX2, and COX3, form the core of the cytochrome c oxidase. Upon synthesis, COX2 engages with COX20 in the inner mitochondrial membrane, a scaffold protein that recruits metallochaperones for copper delivery to the CuA-Site of COX2. Here we identified the human protein, TMEM177 as a constituent of the COX20 interaction network. Loss or increase in the amount of TMEM177 affects COX20 abundance leading to reduced or increased COX20 levels respectively. TMEM177 associates with newly synthesized COX2 and SCO2 in a COX20-dependent manner. Our data shows that by unbalancing the amount of TMEM177, newly synthesized COX2 accumulates in a COX20-associated state. We conclude that TMEM177 promotes assembly of COX2 at the level of CuA-site formation.
Keywords: COX2 biogenesis, COX assembly, COX20 chaperone, Mitochondria, Oxidative phosphorylation, Respiratory chain
Highlights
-
•
TMEM177 is a mitochondrial inner membrane protein.
-
•
COX20 associates with TMEM177.
-
•
The TMEM177 protein forms a complex with COX2 and copper chaperones.
1. Introduction
Mitochondria produce the majority of cellular energy in form of ATP by oxidative phosphorylation. Therefore, a proton gradient is generated by the respiratory chain. The required energy to build this proton imbalance is provided by oxidation of NADH and FADH2 in the mitochondrial matrix and electron transport through the respiratory chain. Concomitantly, the F1Fo ATP synthase utilizes the proton gradient to drive ATP synthesis.
The terminal complex of the respiratory chain, cytochrome c oxidase, reduces molecular oxygen to water. This enzyme complex is formed by 14 structural subunits in human [1], [2], [3], [4]. Eleven subunits are encoded in the nucleus and need to be imported into mitochondria. Three highly conserved core subunits (COX1, COX2 and COX3) are encoded by the mitochondrial genome (mtDNA). These core subunits are translated within mitochondria and co-translationally inserted into the inner mitochondrial membrane [5], [6], [7], [8], [9]. COX1 forms the stepping stone of cytochrome c oxidase assembly to which imported and mitochondrial-encoded subunits associate in a sequential manner. The current concept suggests that COX2 and COX3 associate with a specific set of assembly factors first and subsequently associate to the COX1-containing module [2], [10], [11], [12]. Although cytochrome c oxidase maturation is considered as a stepwise process through a series of assembly intermediates, assembly stages of COX2 and COX3 are still ill defined.
Our understanding of the COX2 biogenesis process and the involvement of assembly factors has been mainly obtained from studies in the yeast Saccharomyces cerevisiae (S. cerevisiae). In contrast to human COX2, the yeast homolog is synthesized as a precursor protein [13], [14] and its N-terminal leader sequence processed by the inner membrane peptidase (IMP) complex [15], [16], [17]. Mature Cox2 contains two transmembrane domains and exposes its N- and C-terminus to the intermembrane space. Several Cox2-specific assembly factors have been described in yeast including Pet111, Cox18 and Cox20. While Pet111 acts as Cox2 translational activator [18], [19], [20] and promotes co-translational membrane insertion of the Cox2 N-terminus, Cox18 [21], [22], [23], [24] and Cox20 [14], [25] are involved in C-terminal translocation. In addition, the interaction of Cox20 with Cox2 is required to promote Cox2 association with the metallochaperone Sco1 [26], [27], [28], essential for CuA-side maturation.
In S. cerevisiae Cox20 is present in distinct complexes with different protein compositions [29]. All Cox20-complexes contain Cox2. However, distinct complexes of Cox20 appear to engage with ribosomes and the copper chaperone Sco1. These findings imply that Cox20 fulfills several functions during Cox2 assembly.
SCO1 and SCO2, both human homologs of the yeast Sco1, have been described as COX20-associated proteins and are essential factors for cytochrome c oxidase maturation [30], [31], [32], [33]. Similar to COX20 [34], [35], also SCO1 and SCO2 have been associated with mitochondrial disorders, such as muscle hypotonia, ataxia, encephalocardiomyopathies, hepatopathies and defined cytochrome c oxidase deficiency [36], [37], [38], [39], [40].
COX20 has been found to form a complex with SCO1, SCO2, and newly synthetized COX2 [41]. However, a comprehensive analysis of the interaction network of COX20 has not been carried out. Thus, we used quantitative mass spectrometry to define COX20 interaction partners. Besides COX2 and the expected assembly factors, we identified the uncharacterized human protein TMEM177. Apparently, TMEM177 lacks a clear homolog in yeast. We show that TMEM177 is involved in the COX2 biogenesis pathway. Although TMEM177 is dispensable for cytochrome c oxidase activity, its amount is directly linked to COX20 abundance. Since COX2 is stabilized during depletion and overexpression of TMEM177, we suggest that TMEM177 is involved in COX2 stability and turnover.
2. Materials and methods
2.1. Cell culturing, mutagenesis and generation of cell lines
HEK293T Flp-In™ T-REX™ or HEK293 were cultured in DMEM media, supplemented with 10% (v/v) FBS, 2 mM l-glutamine, 1 mM sodium pyruvate and 50 μg/mL uridine at 37 °C under a 5% CO2 humidified atmosphere [42]. For inhibition of mitochondrial translation, the medium was supplemented with 50 μg/mL thiamphenicol for two days. Stable FLAG-tag TMEM177 and COX20 expressing cell lines were generated as described previously [43] according to sequences at the NCBI-database: NM_001105198.1 (TMEM177), NM_001312871.1 (COX20). The mutant version of COX20FLAG, encoding the amino acid substitution T52P, was generated using the Quickchange™ Lightning Site-Directed Mutagenesis Kit. Transient transfections were performed using GeneJuice® (Novagen) according to manufacturer's instruction. FLAG-constructs were induced with 10 ng/mL doxycycline for 12 h or 72 h prior to harvest. For analysis of cell viability, 0.4% trypan blue (Gibco) was added to the cells collected in PBS. Afterwards, cell numbers were measured directly with a hemocytometer.
2.2. siRNA-mediated knockdown
The following siRNA constructs and concentration were applied: COX20 (5′-GGAGGGUUUAUCUUGGUGA-3′, 33 nM), TMEM177 (5′-GACACUUGUUCCGAAUCAA-3′, 50 nM) (Eurogentec). Transfection reactions were performed on 500,000 cells/25 cm2 with Lipofectamine RNAiMAX (Invitrogen) in OptiMEM medium according to manufacturer's specification. HEK293T cells were analyzed 72 h after transfection [43].
2.3. In vivo labeling of mitochondrial translation products
Labeling of mitochondrial translation products was performed on 500,000 cells/25 cm2 [44]. In pulse-chase experiments, cytosolic translation was inhibited with 100 μg/mL anisomycin and mitochondrial translation products labeled with 0.2 mCi/mL [35S]methionine for 2 h. Afterwards, radioactive media was replaced with standard growth media and cells further incubated under standard growth conditions for 3 h or 12 h. In pulse experiments, anisomycin was substituted with 100 μg/mL emetine and cells were pulsed for 1 h. Cells were harvested in 1 mM EDTA/PBS and further analyzed by SDS-PAGE. Signals were detected by Storage Phosphor Screens and a Storm 820 scanner (GEHealthcare) Signals were quantified with the ImageQuant TL software (GE Healthcare).
2.4. Isolation of mitochondria, fractionation and protein localization analysis
HEK293T cells were collected in 1 mM EDTA/PBS and isolation of mitochondria performed as described [45], [46]. For fractionation analyses, samples of the homogenized cells were taken. Crude mitochondria were separated by centrifugation at 11,000g for 10 min at 4 °C. To separate ER membranes from the cytosol, the post-mitochondrial supernatant was centrifuged at 100,000g for 1 h at 4 °C. Cell equivalents of each fraction were loaded on the gel. To determine the protein localization, isolated mitochondria in SEM buffer (250 mM sucrose, 1 mM EDTA, 10 mM MOPS [morpholinepropanesulfonic acid] pH 7.2) were re-isolated and re-suspended in SEM buffer containing 450 mM KCl and 1% Triton X-100 or their proteins were extracted with 0.1 M Na2CO3 buffer, pH 10.8 or pH 11.5 [47]. Then, samples were centrifuged for 30 min at 100,000g at 4 °C in a TLA-55 rotor (Beckman Coulter). After addition of TCA, the samples were precipitated and analyzed by SDS-PAGE and Western blotting. Submitochondrial localization was determined by protease protection assay [48]. Mitochondria re-suspended in 10 mM MOPS pH 7.2 were either sonicated, osmotically stabilized in SEM buffer or swollen in EM buffer (1 mM EDTA, 10 mM MOPS pH 7.2). Then, samples were treated with proteinase K for 10 min on ice and reactions were stopped by addition of 1 mM PMSF (Phenylmethylsulfonyl fluoride). Samples were analyzed by SDS-PAGE and Western blotting.
2.5. Antibodies
Primary antibodies were purchased (anti-FLAG and anti-COX20, Sigma-Aldrich; anti-COX2, anti-COX18, anti-COX5B Proteintech Group; anti-TMEM177, Abcam) or raised in rabbit and HRP- or fluorophore-coupled secondary antibodies were employed for detection of antibody-protein complexes an enhanced chemiluminescence system detected by X-ray films or laser scanned on an FLA-9000 (Fujifilm).
2.6. Affinity purification
Protein complexes were purified from isolated mitochondria or whole cell extracts bearing FLAG-tagged proteins or not using anti-FLAG-agarose affinity resin as described previously [43]. For this purpose, mitochondria or cells were solubilized (1 mg/mL) in solubilization buffer (50 mM Tris pH 7.4, 150 mM NaCl, 10% v/v Glycerol, 2 mM PMSF, 1 mM EDTA and 1% digitonin) for 30 min on ice. Solubilized material was clarified by centrifugation at maximum speed for 10 min at 4 °C. The supernatant was added to anti-FLAG-agarose affinity resin and incubated for 1 h at 4 °C with shaking. After binding, the beads were washed 10 times with washing buffer (50 mM Tris pH 7.4, 150 mM NaCl, 10% v/v Glycerol, 1 mM PMSF, 1 mM EDTA and 0.3% digitonin) and bound proteins eluted with 0.1 M glycine, pH 2.8, or by competition with FLAG-peptide at 4 °C for 30 min with mild agitation. Subsequently, eluate proteins were analyzed with SDS-PAGE, BN-PAGE or mass spectrometry.
Co-immunoprecipitation of COX20 was carried out following the same procedure. COX20-specific antisera was bound to Protein A-Sepharose using dimethyl pimelimidate as crosslinker as described [49], [50].
2.7. SILAC labeling and mass spectrometry
For SILAC experiments [51], cells were grown for five passages in “SILAC” DMEM media as described previously [43]. Mitochondria were isolated from differentially labeled cells, equally pooled, and solubilized. COX20FLAG complex isolation using anti-FLAG-agarose affinity resin was performed as described above. Four independent replicates including label switch were performed.
Proteins of COX20FLAG complexes were acetone-precipitated, resuspended in urea buffer (8 M urea/50 mM NH4HCO3) and processed for tryptic in-solution digestion including reduction and alkylation of cysteine residues as described previously [52]. Peptide mixtures were analyzed on a Q Exactive mass spectrometer (Thermo Fisher Scientific, Bremen, Germany), which was directly coupled to an UltiMate 3000 RSLCnano HPLC system (Thermo Fisher Scientific, Dreieich, Germany) equipped with PepMap C18 precolumns (5 mm × 300 mm inner diameter; Thermo Scientific) and a C18 reversed-phase nano LC column (Acclaim PepMap RSLC column; 50 cm × 75 mm inner diameter; 2 mm particle size; 100 Å pore size; Thermo Scientific). For peptide elution, a 140-min gradient ranging from 4%–39% (v/v) acetonitrile (ACN) in 0.1% (v/v) formic acid (FA) in 130 min followed by 39%–82% ACN/0.1% FA in 5 min and 5 min at 82% ACN/0.1% FA was applied at a flow rate of 250 nL/min. The Q Exactive was operated in data-dependent mode. Survey scans were acquired in the range of m/z 375–1700 (resolution 70,000 at m/z 200) with an automatic gain control of 3E6 ions and a maximum fill time of 60 ms. A TOP12 method was employed for higher energy collisional dissociation of multiply charged precursor peptides at a normalized collision energy of 28%, an automatic gain control of 1E6 ions, and a maximum fill time of 120 ms. The dynamic exclusion time for previously selected precursor ions was 45 s.
Proteins were identified and quantified using MaxQuant/Andromeda (version 1.4.1.2) [53], [54]. Mass spectrometric raw data were searched against the UniProt Human ProteomeSet database including isoforms (downloaded 01/2014) and a database containing common contaminants applying MaxQuant default settings (unless stated otherwise). Cysteine carbamidomethylation was considered as fixed modification. N-terminal acetylation and methionine oxidation were set as variable modifications and Arg10/Lys8 as heavy labels. Proteins were identified based on ≥ 1 unique peptide and a false discovery rate of 0.01 for peptides and proteins. SILAC-based relative protein quantification was based on unique peptides and ≥ 1 ratio count. COX20FLAG/wild-type ratios were determined, log10-transformed and the mean log10 protein ratios (n ≥ 2) were plotted against the corresponding p-value calculated using a one-sided Student's t-test. Information about proteins identified and quantified in ≥ 2 replicates is provided in Supplementary Table 1.
2.8. Immunofluorescence
Immunofluorescence microscopy was performed on HeLa cells grown on coverslips. Cells were transiently transfected with plasmid expressing TMEM177FLAG and induced for 12 h. Afterwards, the sample was incubated with MitoTracker red for 5 min. Cells were fixed with 4% paraformaldehyde (PFA) for 20 min at 37 °C, washed with PBS and permeabilized with 0.2% Triton X-100 for 20 min at room temperature (RT). Cells were washed once again and the sample was blocked with 1% BSA (bovine serum albumin) for 20 min at RT and incubated with mouse monoclonal anti-FLAG antibody for 1 h at RT. After washing in PBS, the secondary antibody (Alexa Fluor 488 goat anti-mouse) was applied for 30 min at RT. Following a final wash, the sample was fixed in histology mounting medium containing DAPI (Fluoroshield™). Images were analyzed using a Delta Vision Spectris fluorescence microscope at 60 × magnification, equipped with FITC (excitation 475/28, emission 523/36), TRITC (excitation 542/27, emission 594/45) and DAPI (excitation 390/18, emission 435/48) filter set. The image stacks were taken, images deconvoluted and a maximum projection of the stacks generated by merging the individual slices using the softWORx software (Applied Precision).
2.9. Miscellaneous
Standard protocols were used for SDS-PAGE and Western blotting. Protein complexes were separated by BN-PAGE as described [55]. Mitochondria were solubilized in solubilization buffer (1 mg/mL) for 10 min and lysates were clarified by centrifugation at maximum speed for 10 min at 4 °C. 10 × loading was added to the supernatant and samples loaded onto a 4–13% polyacrylamide gel.
Data quantifications are presented as mean ± standard error of the mean.
2.10. Bioinformatics analysis
Prediction of transmembrane spans of TMEM177 was performed using TMpred (http://www.ch.embnet.org/software/TMPRED_form.html) [56]. Mitochondrial targeting sequence was predicted by MitoProt Server [57].
3. Results and discussion
3.1. Mapping the COX20 interaction network
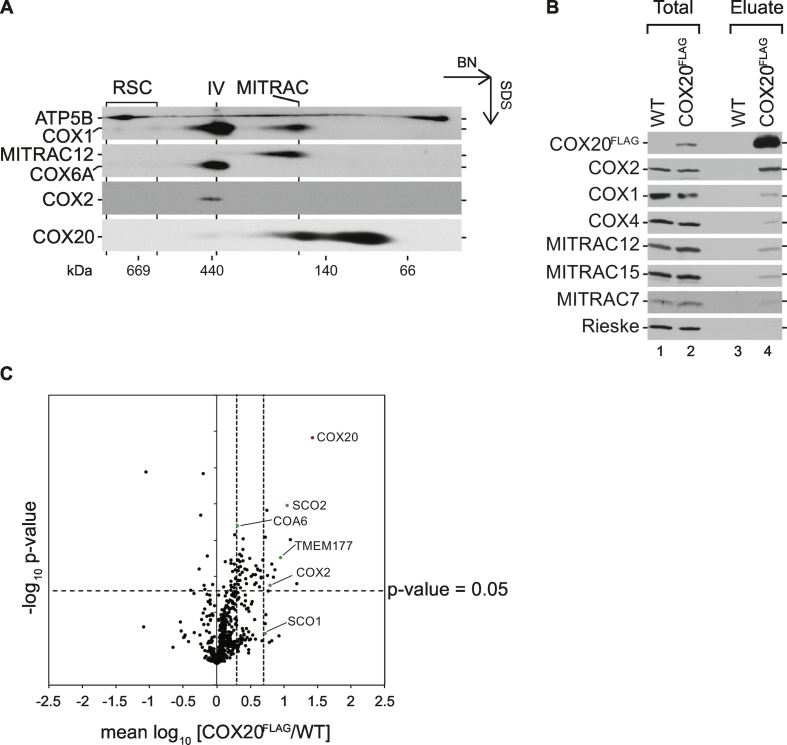
Cytochrome c oxidase assembly in human mitochondria requires a plethora of assembly factors that assist this process at different stages. Many of these factors are functionally not well defined and the number of such proteins is still growing. However, while recent scientific work focused on the assembly pathway and assembly factors involved in the biogenesis of COX1 [58], early steps of COX2 maturation are still ill defined. Among the known assembly factors of the mitochondrial-encoded COX2, the conserved COX20 protein has been described as a COX2-specific chaperone that interacts with copper chaperones required for COX2 copper-center biogenesis [3], [14], [25], [29], [34], [41]. To define COX20-containing complexes and potentially identify new factors involved in COX2 biogenesis, isolated mitochondria from HEK293T cells were solubilized and analyzed by 2D gel analysis (BN-PAGE followed by SDS-PAGE) and Western blotting (Fig. 1A). COX20 formed complexes that migrated in the low molecular range, partially co-migrating with the COX1 assembly intermediate MITRAC complex [42], [43], [59], [60], [61]. To determine components of these complexes, a stable HEK293T cell line, expressing an inducible C-terminally FLAG-tagged COX20 protein (COX20FLAG), was generated. Mitochondria from COX20FLAG and wild-type cells were solubilized and subjected to anti-FLAG affinity purification (Fig. 1B). As expected, COX20FLAG efficiently co-isolated COX2. In contrast, only minute amounts of structural subunits of complex IV, such as COX1 and COX4, as well as COX assembly factors, (MITRAC12, MITRAC15, and MITRAC7) [43], [48], [60], [61], were detected in the eluate.
Fig. 1.
Mass spectrometry analysis identifies an association between COX20 and TMEM177 (A) Endogenous COX20 forms multiple complexes. Isolated mitochondria from HEK293T cells were solubilized in digitonin-containing buffer and complexes were separated by BN-PAGE (4–13%), followed by two-dimensional SDS-PAGE and Western blotting. RSC, respiratory-chain supercomplexes; IV, complex IV; MITRAC (mitochondrial translation regulation assembly intermediate of cytochrome c oxidase), MITRAC complex. (B) COX20 interacts with COX2. Solubilized mitochondria from COX20FLAG-expressing cells were subjected to FLAG-isolation. Eluted proteins were analyzed by SDS-PAGE and Western blotting. COX20FLAG signals were detected using anti-FLAG antibody. Total 1%; Eluate 100%. (C) Quantitative mass spectrometric analysis of COX20FLAG complexes. Mitochondria isolated from differentially SILAC-labeled wild-type and COX20FLAG cells were mixed in equal amounts, solubilized, and subjected to FLAG-immunoprecipitation. Bound proteins were eluted natively using FLAG peptide and analyzed by liquid chromatography-mass spectrometry (n = 4). Red, bait; green, proteins verified by Western blotting. Thresholds: p-value < 0.05, mean log10 ratios > 0.3 or > 0.69 (dashed lines).
In order to define components of COX20-containing complexes in an unbiased manner, we used a quantitative affinity purification-mass spectrometry strategy. Therefore, COX20FLAG and wild-type cells were differentially labeled by stable isotope labeling with amino acids in cell culture (SILAC) [51]. Solubilized mitochondria from both cells cultured in either heavy or light amino acid-containing medium were mixed and subjected to FLAG isolation. Differential labeling of proteins allowed for a quantitative analysis of specifically enriched constituents of COX20 complexes compared to the control (Fig. 1C and Supplementary Table 1). As expected, we observed an enrichment of COX20 and COX2, the copper chaperones COA6 [62], [63], SCO2 and SCO1. The COX2 C-terminal membrane insertase, COX18, was also identified by mass spectrometry (Supplementary Table 1). Due to the lack of COX18 peptides in the control sample, levels of enrichment could not be determined. Moreover, we identified the uncharacterized TMEM177 as a significantly enriched protein (Fig. 1C and Supplementary Table 1). Collectively, these results define the COX20 interaction network in human mitochondria and confirm its known interaction with COX2 and copper chaperones. Moreover, our data revealed TMEM177 as a new COX20 interaction and potential COX2 biogenesis factor.
3.2. TMEM177 is a novel COX20-associated protein
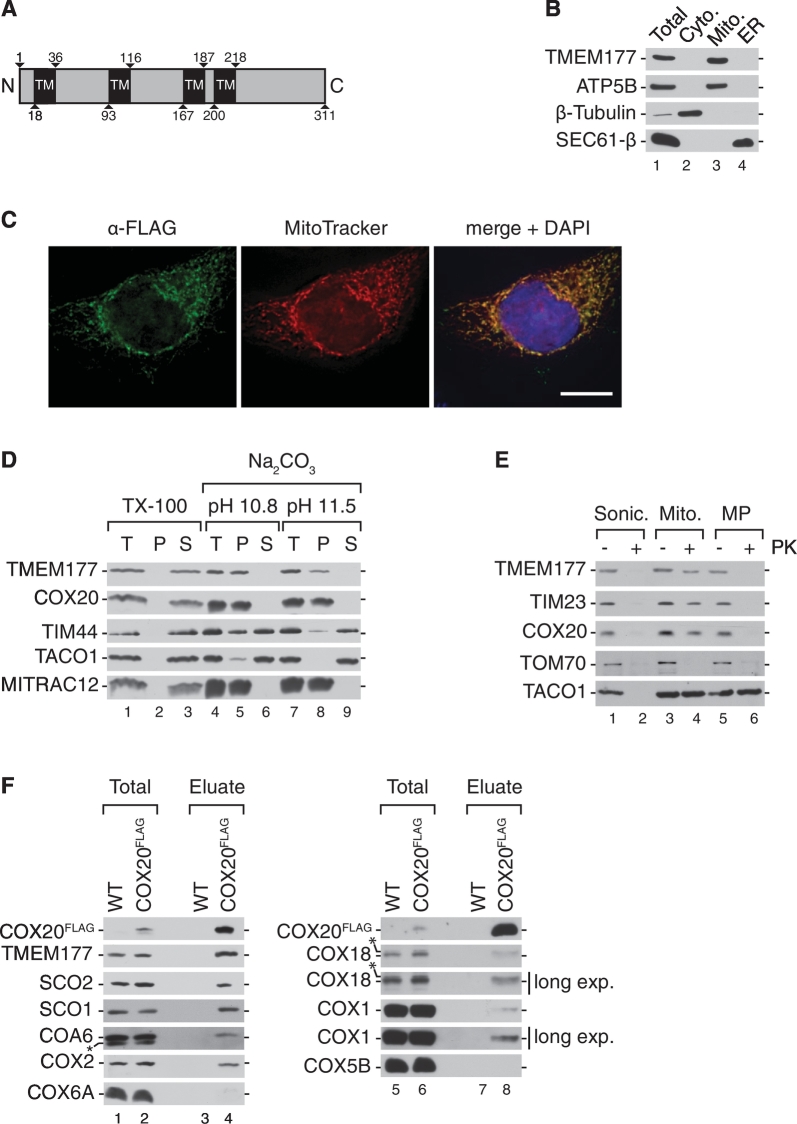
TMEM177 is highly conserved among metazoan. However, a yeast homolog could not be identified in silico (Supplementary Fig. 1). Sequence analyses of TMEM177 predicted four transmembrane spans and an N-terminal mitochondrial presequence necessary for transport to the inner mitochondrial membrane (Fig. 2A and Supplementary Fig. 1). We therefore first confirmed the mitochondrial localization of TMEM177 by cell fractionation (Fig. 2B). The cellular localization of TMEM177 was tested by immunoblotting in different cellular fractions enriched for mitochondria, cytoplasm, and the endoplasmatic reticulum. TMEM177 fractionated exclusively with mitochondria similar to the mitochondrial protein ATP5B (Fig. 2B, lane 2). These data were corroborated by immunofluorescence microscopy. We transiently transfected HeLa cells with a TMEM177FLAG-expressing plasmid and performed immunofluorescence microscopic analyses using an antibody directed against the FLAG-tag. The TMEM177 signal colocalized with mitochondrial networks visualized with MitoTracker red (Fig. 2C). These analyses confirmed that TMEM177 localizes to mitochondria.
Supplementary Fig. 1.
TMEM177 is a conserved protein among metazoa. Alignment of TMEM177 homologs using ClustalW with Blosum 62. Stars indicate 100% similarity; black boxes, 80–100% similarity; gray boxes, 60–80% similarity; light gray boxes, 50–60% similarity. Black arrow indicates the end of the predicted presequence using MitoProt II 1.101.
Fig. 2.
TMEM177 associates with COX20 in the inner mitochondrial membrane (A) A Schematic representation of the primary sequence of TMEM177. The predicted transmembrane spans (TM) are depicted (numbers indicate amino acids residues). (B) TMEM177 localizes to mitochondria. HEK293T cells were fractionated by differential centrifugation. Different fractions were analyzed by SDS-PAGE and Western blotting. Total, total cell extract; Mito., mitochondria; Cyto., cytoplasm; ER, endoplasmatic reticulum. (C) Immunofluorescence microscopy of HeLa cells expressing TMEM177FLAG. Cells were immunolabeled using anti-FLAG antiserum (green) and stained with MitoTracker (red) and DAPI (blue). Scale bar, 10 μm. (D) TMEM177 is an integral membrane protein. Isolated mitochondria from wild-type cells were subjected to carbonate extraction at the indicated pH, or solubilized with Triton X-100. Upon differential centrifugation, different fractions total (T), pellet (P), and supernatant (S) were analyzed by SDS-PAGE and Western blotting. (E) The C-terminus of TMEM177 is exposed to the IMS. Submitochondrial localization analysis using wild-type mitochondria or mitoplast. Sonic., sonication; Mito., mitochondria; MP, mitoplast; PK, proteinase K. (F) COX20 associates with TMEM177. Protein complexes were isolated from mitochondria from wild-type and COX20FLAG-expressing cells by FLAG-immunoprecipitation and analyzed by SDS-PAGE and Western blotting. COX20FLAG signals were detected using anti-FLAG antibody. Total, 1%; Eluate, 100%. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
To define the submitochondrial localization of TMEM177, we performed carbonate extraction analyses of isolated HEK293T mitochondria (Fig. 2D). Peripherally membrane associated proteins such as TIM44 and TACO1 are released from mitochondrial membranes into the supernatant at pH 11.5 (Fig. 2D, lanes 5–6 and 8–9). In contrast, TMEM177 was carbonate extraction resistant and remained in the pellet fraction similar to the integral membrane proteins COX20 and MITRAC12 (Fig. 2D, lanes 5–6 and 8–9). These data indicate that TMEM177 is an integral membrane protein. The membrane topology of TMEM177 was assessed by protease protection analyses. To this end, we tested accessibility of TMEM177 to externally added proteinase K (Fig. 2E). TMEM177 was resistant to protease digestion in mitochondria, whereas the outer membrane protein TOM70 was degraded (Fig. 2E, lanes 3–4). However, upon osmotic disruption of the outer mitochondrial membrane, proteins of the intermembrane space (IMS) become accessible to protease treatment. Under these conditions, the C-terminus of TMEM177 was degraded similar to TIM23 and COX20, suggesting that the C-terminus of this protein, recognized by the antibody, faces the IMS (Fig. 2E, lanes 5–6). As a control, the matrix protein TACO1 became only sensitive to the protease treatment when the inner membrane was disrupted by sonication (Fig. 2E, lane 1–2). Accordingly, TMEM177 is a membrane protein of the mitochondrial inner membrane that exposes its C-terminus into the IMS.
Since TMEM177 was detected by mass spectrometry after COX20 isolation, we confirmed its association with COX20. Therefore, we isolated COX20-associated proteins by FLAG-immunoprecipitation. Known interacting partners, such as SCO2, SCO1, COX18 and COX2 [41], [67], as well as COA6 and TMEM177, were efficiently co-isolated with COX20 (Fig. 2F). In contrast, COX1, COX4, and COX6A could not be detected in the eluate (Fig. 1B and Fig. 2F). Despite the detected enrichment of COX5B by mass spectrometric analyses, the protein could not be identified in the eluate by Western blotting (Fig. 2F). These analyses supported the mass spectrometric data and validated TMEM177 as a novel COX20-associated protein, residing in the inner membrane. The co-isolation of TMEM177 together with COX2 assembly factors suggested a possible role of this protein in the early steps of COX2 biogenesis.
3.3. Addressing the association of TMEM177 with COX20
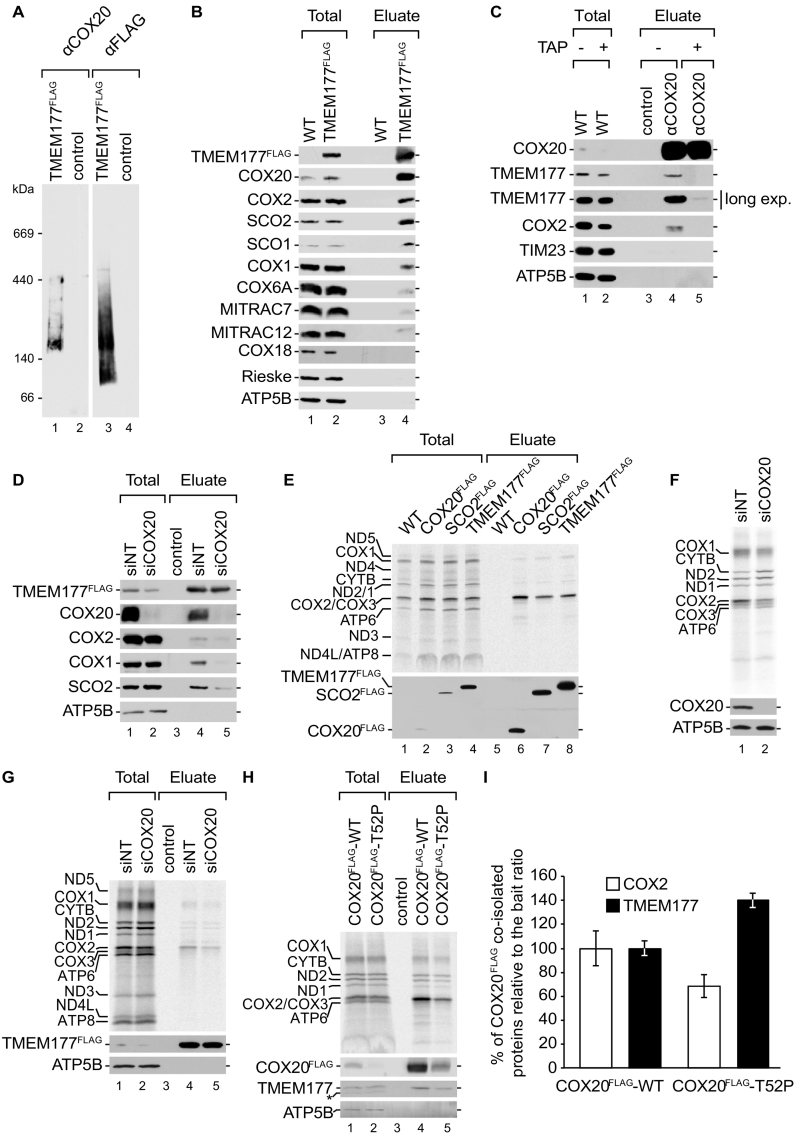
To support the finding that TMEM177 was a constituent of the COX20 interaction network, we isolated TMEM177FLAG-containing complexes and separated the eluate by BN-PAGE. Purified fractions were tested for the presence of TMEM177 and COX20 (Fig. 3A). As expected, COX20 was identified in a trail of complexes that comigrated largely with TMEM177. However, also a free pool of TMEM177 was apparent in the lower molecular weight range of the gel (Fig. 3A). So far our analyses did not define if TMEM177 formed complexes with COX20 alone or in complex with COX2 or the copper chaperones, since we only purified complexes via COX20. Therefore, we performed Western blot analyses of isolated TMEM177FLAG complexes (Fig. 3B). Similar to the COX20 isolation, SCO1, SCO2, COX2, and COX20 co-purified with TMEM177. Only low amounts of COX1 and MITRAC constituents were identified in the eluate. In contrast to the COX20FLAG isolation, COX18 did not co-isolate with TMEM177. These findings support a functional connection of TMEM177 with early steps of COX2 assembly. (Fig. 3B).
Fig. 3.
TMEM177 engages COX2 assembly factors depending on the COX20-COX2 complex (A) TMEM177FLAG-complexes contain COX20. Wild-type and TMEM177FLAG-expressing mitochondria were solubilized in digitonin containing buffer and subjected to FLAG-immunoprecipitation followed by native elution via FLAG peptide. Eluates were separated on 4–13% BN-PAGE and analyzed by Western blotting using specific antibodies against COX20 and TMEM177. (B) TMEM177 interacts with COX2 assembly factors. Mitochondria isolated from wild-type and TMEM177FLAG-expressing cells were solubilized and subjected to FLAG-immunoprecipitation. Samples were analyzed by SDS-PAGE and Western blotting. Total, 1%; Eluate, 100%. (C) TMEM177-COX20 interaction depends on mitochondrial-encoded products. HEK293T cells were treated without (−) or with (+) 50 μg/mL thiamphenicol (TAP) for 2 days. Isolated mitochondria were digitonin-solubilized and subjected to control- or COX20-co-immunoprecipitation. Eluates were analyzed by SDS-PAGE and Western blotting. Long exp., long exposure. Total, 1%; Eluate, 100%. (D) TMEM177 engages with SCO2 in a COX20-dependent manner. TMEM177FLAG-expressing cells were treated with siRNA oligonucleotides against COX20 (siCOX20) or with control (siNT) siRNA for 72 h. Isolated mitochondria were solubilized and subjected to FLAG-immunoprecipitation. Eluates were analyzed by SDS-PAGE and Western blotting. Mitochondria from wild-type cells were used as control. TMEM177FLAG signals were detected using anti-FLAG antibody. Total, 1%; Eluate, 100%. (E) TMEM177 interacts with newly synthesized COX2. Mitochondrial translated products were pulse-labeled by [35S]methionine in TMEM177FLAG, COX20FLAG, SCO2FLAG-expressing cells for 1 h. Cells were lysed in solubilization buffer and anti-FLAG isolation was performed. Eluates were analyzed by SDS-PAGE and Western blotting or autoradiography. Total, 10%; Eluate, 100%. (F) COX20 depleted cells show reduced COX2 stability. Wild-type cells were treated with siNT or siCOX20 for 72 h. Afterwards, mitochondrial-encoded products were labeled for 1 h and cell lysates were analyzed by SDS-PAGE and Western blotting or autoradiography. (G) TMEM177 interacts with newly translated COX2. TMEM177FLAG-expressing cells were transfected with indicated (siNT, siCOX20) siRNA for 72 h. After in vivo labeling of mitochondrial-encoded products, cell lysates were subjected to FLAG-isolation. Eluates were analyzed by SDS-PAGE and Western blotting or digital autoradiography. Wild-type cells were used as control. Anti-FLAG antibody was used to detect TMEM177FLAG signals. Total, 10%; Eluate, 100%. (H) TMEM177 accumulates with mutant COX20. Cells were transiently transfected with plasmid expressing wild-type and mutated (T52P) version of COX20FLAG. 24 h after transfection, mitochondrial-encoded products were pulse labeled and protein complexes were purified from whole cell lysates. Samples were analyzed by SDS-PAGE and Western blotting or digital autoradiography. COX20FLAG signals were detected using anti-FLAG antibody. Asterisk indicates antibody cross-reaction. Total, 10%; Eluate, 100%. (I) Quantification of the amount of newly synthesized COX2 and TMEM177 co-isolated with COX20FLAG-WT and COX20FLAG-T52P, normalized to the isolated ratio (COX20FLAG-T52P/COX20FLAG-WT) amount (mean ± SEM, n = 3).
Next, we asked if the formation of the TMEM177-COX20 complex depended on mitochondrial-encoded proteins (Fig. 3C). To test this, cells were treated with the mitochondrial translation inhibitor thiamphenicol (TAP) prior to COX20 complex isolation. In agreement with the previously observed results (Fig. 2F, lane 4), TMEM177 and COX2 specifically isolated with endogenous COX20 (Fig. 3C, lane 4). Compared to the untreated control, the association of COX20 with TMEM177 was strongly reduced upon thiamphenicol treatment (Fig. 3C). Accordingly, the interaction of TMEM177 with COX20-containing complexes depended on mitochondrial translation products, most probably COX2. Next, we assessed the role of COX20 for the TMEM177-COX2 interaction. Therefore, we conducted an siRNA mediated knockdown of COX20 in control and TMEM177FLAG cells and purified TMEM177-containing complexes via FLAG immunoisolation (Fig. 3D). Upon COX20 knockdown, less COX2 and SCO2 were co-isolated with TMEM177, in agreement with the idea that COX20 recruits TMEM177 to COX2 assembly factors, such as SCO2. Similar to COX2, less COX1 was recovered by TMEM177 in COX20 knockdown cells.
The co-isolation of COX2 assembly factors with TMEM177 and the identified dynamic interaction of TMEM177 with COX20 suggested a role for TMEM177 in the early steps of COX2 biogenesis. Therefore, to address whether TMEM177 associates with newly synthesized COX2, mitochondrial translation products were labeled with radioactive methionine and TMEM177-containing complexes were isolated via FLAG antibody (Fig. 3E). As positive controls for COX2 interaction, COX20 and SCO2 were used. Among the mitochondrial translated products, COX2 was found enriched in the TMEM177FLAG isolation, as well as in the COX20 and SCO2 isolations.
Previous analyses showed that newly synthesized COX2 associates with COX20 and becomes unstable upon loss of COX20 [41] (Fig. 3F). We carried out TMEM177FLAG isolation from control and COX20 knockdown cells after pulse labeling of mitochondrial-encoded proteins to analyze if TMEM177 engaged with newly translated COX2 and whether this interaction required COX20. As expected, newly synthesized COX2 co-isolated with TMEM177. However, less newly synthesized COX2 (approximately 58% less) was found to be associated with TMEM177 in COX20 deficient cells (Fig. 3G). Accordingly, the association of TMEM177 with COX2 depended on the presence of COX20 and the interaction between COX20 and TMEM177 apparently required COX2.
Szklarczyk et al. [34] reported that patient mutation of COX20 resulting in the amino acid exchange T52P led to the accumulation of subassemblies of the cytochrome c oxidase devoid of COX2. To test if the COX20 patient mutation also leads to an accumulation of TMEM177, we transiently transfected HEK293T cells with plasmids expressing wild-type or the mutant version of COX20FLAG. Isolation of COX20-containing complexes was performed after labeling of mitochondrial translation products (Fig. 3H). Obviously, the mutant version of COX20 was much less abundant than the wild type protein. A quantification of the co-isolated COX2 and TMEM177 revealed a decreased interaction of newly synthesized COX2 with the patient version of COX20 but a 40% increase of TMEM177 interaction (Fig. 3I). This differential behaviour could indicate that the T52P exchange in COX20 stalled TMEM177 with COX20 subcomplexes containing lower amounts of COX2.
Taken together, these results defined TMEM177 as a new component of COX20-containing complexes. The formation of thess COX20-TMEM177 complex appears to be dynamic in nature. We conclude, that the association between COX2 assembly factors and TMEM177 appears to require newly translated COX2 and COX20, which seem to link the interaction between them.
3.4. COX20 levels are reduced in the absence of TMEM177
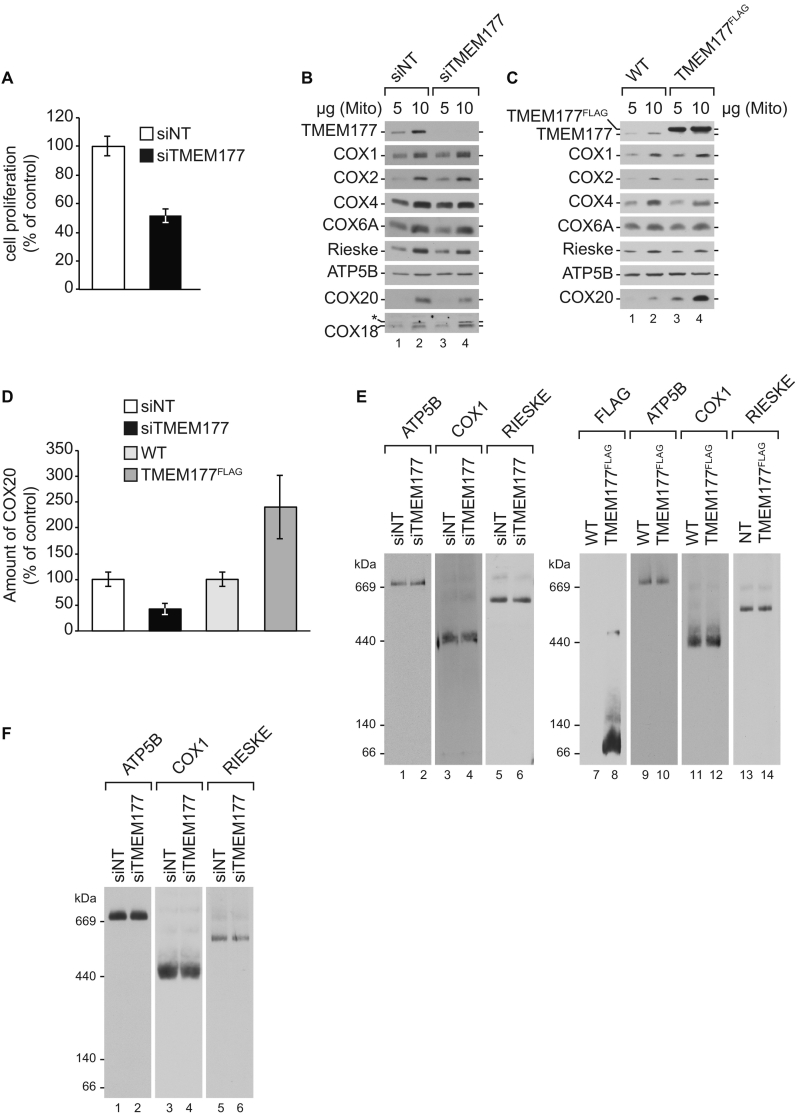
To define the function of TMEM177 we utilized an siRNA-mediated knockdown strategy. Therefore, we generate siRNAs targeting TMEM177 in HEK293T cells. Upon TMEM177 knockdown, the cell number was reduced by approximately 50% after 72 h compared to the non-targeting control (Fig. 4A). A trypan blue exclusion test revealed a decrease in cell viability by approximately 30% in the TMEM177 deficient cells (Fig. 4A). We compared protein steady state levels of mitochondria isolated from non-targeting and TMEM177 knockdown cells. Structural subunits of the oxidative phosphorylation system e.g. complex IV (COX1, COX2, COX4, COX6A), complex III (Rieske), and complex V (ATP5B) appeared to be unaffected under these knockdown conditions (Fig. 4B). However, concomitant to the depletion of TMEM177, we observed a reduction of the levels of COX20, while COX18 levels remained unaffected (Fig. 4B, lanes 2 and 4). Given that COX20 amounts appeared to be dependent on TMEM177 levels, we investigated the phenotype of TMEM177 overexpressing in cells. Mitochondria from wild-type and TMEM177FLAG overexpressing cells were isolated and Western blot analyses performed (Fig. 4C). Surprisingly, upon TMEM177FLAG overexpression, an increase in the amount of COX20 was apparent (Fig. 4C, lanes 2 and 4). As for the TMEM177 knockdown, other structural components of the oxidative phosphorylation system remained unaffected. A quantification of the levels of COX20 reveal a 58% decrease upon TMEM177 knockdown. In contrast, COX20 levels were more than doubled when TMEM177 was overexpressed (Fig. 4C and D).
Fig. 4.
TMEM177 stabilizes COX20 (A) TMEM177 depletion affects cell growth. HEK293T cells were treated with non-targeting (siNT) or TMEM177-specific siRNA (siTMEM177) for 72 h. A trypan blue exclusion test was used to determine cell proliferation. Cell counts of a six-well plate were measured using a hemocytometer. Data are shown as the percentage compared to control (mean ± SEM, n = 3). (B) TMEM177 depletion destabilizes COX20. Protein steady-state levels in TMEM177 depleted cells. Mitochondria were isolated from cells treated with siTMEM177 or siNT for 72 h. Samples were analyzed by SDS-PAGE and Western blotting. (C) Overexpression of TMEM177 stabilizes COX20. Protein steady-state levels in cells overexpressing TMEM177FLAG. Mitochondria were purified from WT and TMEM177FLAG expressing cells treated with doxycycline for 72 h. Samples were analyzed by SDS-PAGE and Western blotting. (D) Quantification of the amount of COX20 in TMEM177 depleted cells from Fig. 4B and cells overexpressing TMEM177FLAG from Fig. 4C. COX20 level was normalized to the amount of ATP5B in each sample. Data are shown as the percentage compared to control, siNT (mean ± SEM, n = 4) or WT (mean ± SEM, n = 3). (E) Solubilized mitochondria from cells treated with siTMEM177 as in Fig. 4B and from cells overexpressing TMEM177FLAG as in Fig. 4C were analyzed by 4–13% BN-PAGE and Western blotting, and detected using the indicated antibodies. (F) Wild-type cells grown in the presence of 0.1% galactose were treated with siTMEM177 and siNT for 72 h. Afterwards, isolated mitochondria from siNT and siTMEM177 were solubilized and analyzed by 4–13% BN-PAGE and Western blotting.
Since COX20-deficient cells display an accumulation of subcomplexes of complex IV [41], we monitored complex IV assembly by BN-PAGE and Western blot analyses in TMEM177 knockdown cells (Fig. 4E, lanes 1–6). No difference in the levels of complex IV, or complex V and III, were apparent, suggesting that residual amounts of COX20 in the TMEM177 knockdown cells are not limiting for complex IV assembly.
Next, mitochondrial protein complexes were analyzed in mitochondria from wild-type and TMEM177FLAG overexpressing cells (Fig. 4E, lanes 7–14). Compared to wild-type, the overexpression of TMEM177 did not affect complex IV, V, and III protein complexes. Hence, loss or overexpression of TMEM177 did not impact the abundance or organization of the respiratory chain complexes. Accordingly, increased levels of COX20 do not affect assembly of the respiratory chain complexes. The TMEM177 growth phenotype was not exacerbated by growing cells on galactose medium. Indeed, respiratory chain complexes remained unaffected in the TMEM177 knockdown (Fig. 4F). In summary, these results suggest that the expression level of TMEM177 is strictly linked to COX20 abundance. However, the levels of the TMEM177 seem to be not critical for assembly of respiratory chain complexes.
3.5. TMEM177 stabilizes newly translated COX2 during assembly
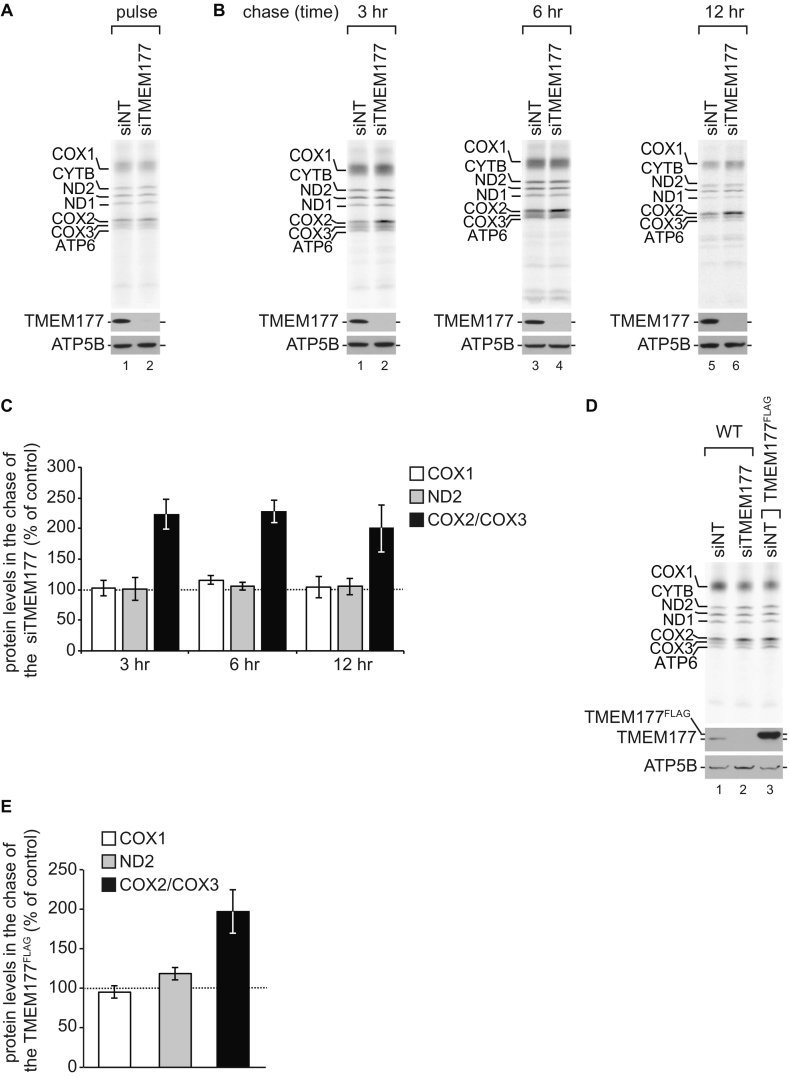
The involvement of TMEM177 in COX20 abundance led us to analyze stability of COX2 in more detail. Therefore, we analyzed COX2 synthesis and degradation in control and TMEM177 knockdown cells by metabolic labeling (Fig. 5A and 5B). After pulse with [35S]methionine, the synthesis of mitochondrial translated proteins remained unaffected in TMEM177 knockdown cells (Fig. 5A). In contrast, the stability of COX2 was clearly increased in TMEM177 depleted cells after a 3, 6, and 12 h chase (Fig. 5B). A quantification of COX2 revealed a two-fold increase of COX2/COX3 levels in knockdown cells as compared to non-targeting cells after 3 h chase, while COX1 and ND2 showed no significant increase. Over the course of the 6 and 12 h chase the levels of COX2/COX3 remained similarly elevated. To investigate if increased amounts of TMEM177 similarly affected COX2 stability, we radioactively labeled mitochondrial translation products and analyzed COX2 levels after a 3 h chase in TMEM177FLAG overexpression cells (Fig. 5D). The COX2 amounts were clearly increased in TMEM177FLAG overexpression cells. A quantification of the mitochondrial-encoded proteins revealed a two-fold increase in stability of COX2/COX3 in TMEM177FLAG overexpression cells, while COX1 and ND2 displayed no significant change (Fig. 5E). It should be noted that the proper amount of TMEM177 in mitochondria appears to be critical and that any unbalancing affects COX20. Therefore, any effort we made to rescue COX2 stability in siRNA knockdown cells with an siRNA-resistant construct failed due to the fact that the exact wild-type-like amount of TMEM177 could not be titrated.
Fig. 5.
TMEM177 stabilizes newly translated COX2 (A) Synthesis of mitochondrial-encoded products remains unaffected in the TMEM177 depleted cells. Mitochondrial translation products were pulse-labeled for 2 h after treatment with TMEM177 siRNA (siTMEM177) or non-targeting siRNA (siNT) for 72 h. Samples were analyzed by SDS-PAGE and Western blotting or digital autoradiography. (B) TMEM177 knockdown shows increase of COX2 stability. Mitochondrial translation products were labeled for 2 h in siNT and siTMEM177 treated cells as in Fig. 5A. Afterwards medium was replaced and cells were cultured in standard medium for additional 3, 6 and 12 h. Cells were lysed and processed as in Fig. 5A. (C) Quantification of COX2/COX3, COX1 and ND2 levels shown in Fig. 5B. Data are shown as the percentage of protein amounts in the chase of the knockdown samples (siTMEM177), normalized to the amounts in the chase of the siNT samples (100%). Each value was internally standardized to ND1 (mean ± SEM, n = 3). (D) Overexpression of TMEM177 stabilizes newly translated COX2. Mitochondrial translation products were radioactively labeled for 2 h and chased for 3 h in WT and TMEM177FLAG expressing cells after treatment with siNT or siTMEM177 in the presence of doxycycline for 72 h. Samples were analyzed by SDS-PAGE and Western blotting or digital autoradiography. (E) Quantification of COX2/COX3, COX1 and ND2 amounts shown in Fig. 5D. Data are shown as the percentage of protein levels in TMEM177FLAG expressing cells, normalized to the WT siNT samples (100%). Each sample was internally standardized to ND1 protein amount (mean ± SEM, n = 3).
We concluded that the levels of TMEM177 are critical for COX2 stability.
In summary, we characterized the novel COX2 assembly factor TMEM177 as a stabilizing factor for COX20 and find that it dynamically interacts with early COX2 subcomplexes in a COX20-dependent manner. We conclude that the COX20-TMEM177 complex stabilizes COX2 during its assembly.
4. Conclusions
Biogenesis of cytochrome c oxidase involves several assembly steps that are characterized by assembly intermediates that contain assembly factors and structural subunits. Progression of the assembly process requires these specific assembly factors that promote complex maturation potentially by stabilizing intermediate folding and interaction states [2]. Accordingly, these assembly factors stabilize assembly intermediates. Defects in assembly factor function lead to cytochrome c oxidase deficiency, which in human is frequently associated with mitochondrial encephalomyopathies [64], [65], [66].
Here, we provide a comprehensive analysis of the COX20 assembly factor interaction network. Our analyses identify TMEM177 as a so far uncharacterized component of COX20 complexes. TMEM177 exhibits a similar interaction network as COX20, it associates with the metallochaperones SCO1 and SCO2 that are required for COX2 maturation [30], [31], [32], [33], [41], [67]. TMEM177 was not found to associate with late assembling subunits of complex IV, such as COX6A, suggesting that it is involved in the early steps of COX2 maturation. While gain or loss of function of TMEM177 did not affect cytochrome c oxidase assembly, we find that TMEM177 affects the abundance of COX20. Moreover, increased or reduced levels of TMEM177 affect COX2 stability. However, the lack of, or increased levels of TMEM177 did not affect function or abundance of complex IV in HEK293T cells. This is reminiscent of MITRAC7, an assembly factor of the COX1 assembly line. MITRAC7, display a stabilizing function on COX1, the core subunit of the complex IV. Similar to TMEM177, MITRAC7 interacts with a newly synthesized core subunit and its assembly factors. However, in contrast to TMEM177, increased or decreased levels of MITRAC7 lead to a reduction of complex IV [48].
Our analyses show that TMEM177 is conserved in higher eukaryotes and forms a complex with newly synthesized COX2, COX20 and the copper-chaperones. TMEM177 may act as a scaffold protein that stabilizes the early COX2-COX20 complex. Moreover, the observed effects of TMEM177 on the levels of COX20 are in agreement with a role of this novel protein in assembly or stability of COX20. However, while a loss of COX20 leads to complex IV deficiency [41], the lack of a clear phenotype, with regard to the amount of complex IV, upon loss of TMEM177, suggests that either loss of TMEM177 can be compensated or that TMEM177 displays a regulatory role in the assembly of COX2 that is not yet apparent.
The following are the supplementary data related to this article.
Results of SILAC-based quantitative mass spectrometry analysis of COX20FLAG complexes. COX20FLAG complexes were affinity-purified from mitochondria isolated from differentially SILAC-labeled wild-type and COX20FLAG-expressing cells mixed in equal amounts and analyzed by liquid chromatography-mass spectrometry (n = 4). Mass spectrometric raw data were processed using MaxQuant/Andromeda (version 1.4.1.2). For detailed information, see “Materials and methods”. Listed are proteins identified and quantified in ≥ 2 replicates.
Transparency document
Transparency document.
Acknowledgments
Acknowledgements
We thank Bettina Knapp for technical assistance. Supported by the Deutsche Forschungsgemeinschaft (BW & PR), SFB1190, GGNB, EXC 294 BIOSS (BW), the European Research Council (ERC) Consolidator Grant 648235 (BW), European Research Council (ERC) Advanced Grant (ERCAdG No. 339580) to PR, MWK FoP 88b (PR), and the Max Planck Society (PR).
Footnotes
The Transparency document associated with this article can be found, in the online version.
Contributor Information
Sven Dennerlein, Email: Sven.Dennerlein@med.uni-goettingen.de.
Peter Rehling, Email: Peter.Rehling@medizin.uni-goettingen.de.
References
- 1.Tsukihara T., Aoyama H., Yamashita E., Tomizaki T., Yamaguchi H., Shinzawa-Itoh K. Structures of metal sites of oxidized bovine heart cytochrome c oxidase at 2.8 A. Science. 1995;269:1069–1074. doi: 10.1126/science.7652554. [DOI] [PubMed] [Google Scholar]
- 2.Soto I.C., Fontanesi F., Liu J., Barrientos A. Biochim. Biophys. Acta. 2012;1817:883–897. doi: 10.1016/j.bbabio.2011.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Herrmann J.M., Funes S. Biogenesis of cytochrome oxidase-sophisticated assembly lines in the mitochondrial inner membrane. Gene. 2005;354:43–52. doi: 10.1016/j.gene.2005.03.017. [DOI] [PubMed] [Google Scholar]
- 4.Fernandez-Vizarra E., Tiranti V., Zeviani M. Assembly of the oxidative phosphorylation system in humans: what we have learned by studying its defects. Biochim. Biophys. Acta. 2009;1793:200–211. doi: 10.1016/j.bbamcr.2008.05.028. [DOI] [PubMed] [Google Scholar]
- 5.Barrientos A., Gouget K., Horn D., Soto I.C., Fontanesi F. Biochim. Biophys. Acta. 2009;1793:97–107. doi: 10.1016/j.bbamcr.2008.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ott M., Amunts A., Brown A. Organization and regulation of mitochondrial protein synthesis. Biochemistry. 2016;85:77–101. doi: 10.1146/annurev-biochem-060815-014334. [DOI] [PubMed] [Google Scholar]
- 7.Hällberg B.M., Larsson N.-G. Making proteins in the powerhouse. Cell Metab. 2014;20:226–240. doi: 10.1016/j.cmet.2014.07.001. [DOI] [PubMed] [Google Scholar]
- 8.Chacinska A., Koehler C.M., Milenkovic D., Lithgow T., Pfanner N. Importing mitochondrial proteins: machineries and mechanisms. Cell. 2009;138:628–644. doi: 10.1016/j.cell.2009.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Richter-Dennerlein R., Dennerlein S., Rehling P. Integrating mitochondrial translation into the cellular context. Nat. Rev. Mol. Cell Biol. 2015;16:586–592. doi: 10.1038/nrm4051. [DOI] [PubMed] [Google Scholar]
- 10.Carr H.S., Winge D.R. Assembly of cytochrome c oxidase within the mitochondrion. Acc. Chem. Res. 2003;36:309–316. doi: 10.1021/ar0200807. [DOI] [PubMed] [Google Scholar]
- 11.Mick D.U., Fox T.D., Rehling P. Inventory control: cytochrome c oxidase assembly regulates mitochondrial translation. Nat. Rev. Mol. Cell Biol. 2011;12:14–20. doi: 10.1038/nrm3029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dennerlein S., Rehling P. Human mitochondrial COX1 assembly into cytochrome c oxidase at a glance. J. Cell Sci. 2015;128:833–837. doi: 10.1242/jcs.161729. [DOI] [PubMed] [Google Scholar]
- 13.Hell K., Herrmann J., Pratje E., Neupert W., Stuart R.A. Oxalp mediates the export of the N- and C-termini of pCoxII from the mitochondrial matrix to the intermembrane space. FEBS Lett. 1997;418:367–370. doi: 10.1016/s0014-5793(97)01412-9. [DOI] [PubMed] [Google Scholar]
- 14.Elliott L.E., Saracco S.A., Fox T.D. Multiple roles of the Cox20 chaperone in assembly of Saccharomyces cerevisiae cytochrome c oxidase. Genetics. 2012;190:559–567. doi: 10.1534/genetics.111.135665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schneider A., Behrens M., Scherer P., Pratje E., Michaelis G., Schatz G. Inner membrane protease I, an enzyme mediating intramitochondrial protein sorting in yeast. EMBO J. 1991;10:247–254. doi: 10.1002/j.1460-2075.1991.tb07944.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nunnari J., Fox T.D., Walter P. A mitochondrial protease with two catalytic subunits of nonoverlapping specificities. Science. 1993;262:1997–2004. doi: 10.1126/science.8266095. [DOI] [PubMed] [Google Scholar]
- 17.Jan P.S., Esser K., Pratje E., Michaelis G. Som1, a third component of the yeast mitochondrial inner membrane peptidase complex that contains Imp1 and Imp2. Mol Gen Genet. 2000;263:483–491. doi: 10.1007/s004380051192. [DOI] [PubMed] [Google Scholar]
- 18.Poutre C.G., Fox T.D. PET111, a Saccharomyces cerevisiae nuclear gene required for translation of the mitochondrial mRNA encoding cytochrome c oxidase subunit II. Genetics. 1987;115:637–647. doi: 10.1093/genetics/115.4.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mulero J.J., Fox T.D. PET111 acts in the 5′-leader of the Saccharomyces cerevisiae mitochondrial COX2 mRNA to promote its translation. Genetics. 1993;133:509–516. doi: 10.1093/genetics/133.3.509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Green-Willms N.S., Butler C.A., Dunstan H.M., Fox T.D. Pet111p, an inner membrane-bound translational activator that limits expression of the Saccharomyces cerevisiae mitochondrial gene COX2. J. Biol. Chem. 2001;276:6392–6397. doi: 10.1074/jbc.M009856200. [DOI] [PubMed] [Google Scholar]
- 21.Saracco S.A., Fox T.D. Cox18p is required for export of the mitochondrially encoded Saccharomyces cerevisiae Cox2p C-tail and interacts with Pnt1p and Mss2p in the inner membrane. Mol. Biol. Cell. 2002;13:1122–1131. doi: 10.1091/mbc.01-12-0580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Souza R.L. Cloning and characterization of COX18, a Saccharomyces cerevisiae PET gene required for the assembly of cytochrome oxidase. J. Biol. Chem. 2000;275:14898–14902. doi: 10.1074/jbc.275.20.14898. [DOI] [PubMed] [Google Scholar]
- 23.Fiumera H.L., Dunham M.J., Saracco S.A., Butler C.A., Kelly J.A., Fox T.D. Translocation and assembly of mitochondrially coded Saccharomyces cerevisiae cytochrome c oxidase subunit Cox2 by Oxa1 and Yme1 in the absence of Cox18. Genetics. 2009;182:519–528. doi: 10.1534/genetics.109.101196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fiumera H.L., Broadley S.A., Fox T.D. Translocation of mitochondrially synthesized Cox2 domains from the matrix to the intermembrane space. Mol. Cell. Biol. 2007;27:4664–4673. doi: 10.1128/MCB.01955-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hell K., Tzagoloff A., Neupert W., Stuart R.A. Identification of Cox20p, a novel protein involved in the maturation and assembly of cytochrome oxidase subunit 2. J. Biol. Chem. 2000;275:4571–4578. doi: 10.1074/jbc.275.7.4571. [DOI] [PubMed] [Google Scholar]
- 26.Nittis T. Yeast Sco1, a protein essential for cytochrome c oxidase function is a Cu(I)-binding protein. J. Biol. Chem. 2001;276:42520–42526. doi: 10.1074/jbc.M107077200. [DOI] [PubMed] [Google Scholar]
- 27.Lode A., Paret C., Rödel G. Molecular characterization of Saccharomyces cerevisiae Sco2p reveals a high degree of redundancy with Sco1p. Yeast. 2002;19:909–922. doi: 10.1002/yea.883. [DOI] [PubMed] [Google Scholar]
- 28.Rigby K., Cobine P.A., Khalimonchuk O., Winge D.R. Mapping the functional interaction of Sco1 and Cox2 in cytochrome oxidase biogenesis. J. Biol. Chem. 2008;283:15015–15022. doi: 10.1074/jbc.M710072200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lorenzi I., Oeljeklaus S., Ronsör C., Bareth B., Warscheid B., Rehling P. The ribosome-associated Mba1 escorts Cox2 from insertion machinery to maturing assembly intermediates. Mol. Cell. Biol. 2016 doi: 10.1128/MCB.00361-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Leary S.C., Kaufman B.A., Pellecchia G., Guercin G.-H., Mattman A., Jaksch M. Human SCO1 and SCO2 have independent, cooperative functions in copper delivery to cytochrome c oxidase. Hum. Mol. Genet. 2004;13:1839–1848. doi: 10.1093/hmg/ddh197. [DOI] [PubMed] [Google Scholar]
- 31.Leary S.C., Sasarman F., Nishimura T., Shoubridge E.A. Human SCO2 is required for the synthesis of CO II and as a thiol-disulphide oxidoreductase for SCO1. Hum. Mol. Genet. 2009;18:2230–2240. doi: 10.1093/hmg/ddp158. [DOI] [PubMed] [Google Scholar]
- 32.Leary S.C., Cobine P.A., Kaufman B.A., Guercin G.-H., Mattman A., Palaty J. The human cytochrome c oxidase assembly factors SCO1 and SCO2 have regulatory roles in the maintenance of cellular copper homeostasis. Cell Metab. 2007;5:9–20. doi: 10.1016/j.cmet.2006.12.001. [DOI] [PubMed] [Google Scholar]
- 33.Horng Y.-C., Leary S.C., Cobine P.A., Young F.B.J., George G.N., Shoubridge E.A. Human Sco1 and Sco2 function as copper-binding proteins. J. Biol. Chem. 2005;280:34113–34122. doi: 10.1074/jbc.M506801200. [DOI] [PubMed] [Google Scholar]
- 34.Szklarczyk R., Wanschers B.F.J., Nijtmans L.G., Rodenburg R.J., Zschocke J., Dikow N. A mutation in the FAM36A gene, the human ortholog of COX20, impairs cytochrome c oxidase assembly and is associated with ataxia and muscle hypotonia. Hum. Mol. Genet. 2013;22:656–667. doi: 10.1093/hmg/dds473. [DOI] [PubMed] [Google Scholar]
- 35.Doss S., Lohmann K., Seibler P., Arns B., Klopstock T., Zühlke C. Recessive dystonia-ataxia syndrome in a Turkish family caused by a COX20 (FAM36A) mutation. J. Neurol. 2014;261:207–212. doi: 10.1007/s00415-013-7177-7. [DOI] [PubMed] [Google Scholar]
- 36.Valnot I., Osmond S., Gigarel N., Mehaye B., Amiel J., Cormier-Daire V. Mutations of the SCO1 gene in mitochondrial cytochrome c oxidase deficiency with neonatal-onset hepatic failure and encephalopathy. Am. J. Hum. Genet. 2000;67:1104–1109. doi: 10.1016/s0002-9297(07)62940-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Leary S.C., Antonicka H., Sasarman F., Weraarpachai W., Cobine P.A., Pan M. Novel mutations in SCO1 as a cause of fatal infantile encephalopathy and lactic acidosis. Hum. Mutat. 2013;34:1366–1370. doi: 10.1002/humu.22385. [DOI] [PubMed] [Google Scholar]
- 38.Papadopoulou L.C., Sue C.M., Davidson M.M., Tanji K., Nishino I., Sadlock J.E. Fatal infantile cardioencephalomyopathy with COX deficiency and mutations in SCO2, a COX assembly gene. Nat. Genet. 1999;23:333–337. doi: 10.1038/15513. [DOI] [PubMed] [Google Scholar]
- 39.Stiburek L., Vesela K., Hansikova H., Hulkova H., Zeman J. Loss of function of Sco1 and its interaction with cytochrome c oxidase. Am. J. Physiol. Cell Physiol. 2009;296:C1218–26. doi: 10.1152/ajpcell.00564.2008. [DOI] [PubMed] [Google Scholar]
- 40.Jaksch M., Ogilvie I., Yao J., Kortenhaus G., Bresser H.G., Gerbitz K.D. Mutations in SCO2 are associated with a distinct form of hypertrophic cardiomyopathy and cytochrome c oxidase deficiency. Hum. Mol. Genet. 2000;9:795–801. doi: 10.1093/hmg/9.5.795. [DOI] [PubMed] [Google Scholar]
- 41.Bourens M., Boulet A., Leary S.C., Barrientos A. Human COX20 cooperates with SCO1 and SCO2 to mature COX2 and promote the assembly of cytochrome c oxidase. Hum. Mol. Genet. 2014;23:2901–2913. doi: 10.1093/hmg/ddu003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Richter-Dennerlein R., Oeljeklaus S., Lorenzi I., Ronsör C., Bareth B., Schendzielorz A.B. Mitochondrial protein synthesis adapts to influx of nuclear-encoded protein. Cell. 2016;167:471–483. doi: 10.1016/j.cell.2016.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mick D.U., Dennerlein S., Wiese H., Reinhold R., Pacheu-Grau D., Lorenzi I. MITRAC links mitochondrial protein translocation to respiratory-chain assembly and translational regulation. Cell. 2012;151:1528–1541. doi: 10.1016/j.cell.2012.11.053. [DOI] [PubMed] [Google Scholar]
- 44.Chomyn A. In vivo labeling and analysis of human mitochondrial translation products. Methods Enzymol. 1996;264:197–211. doi: 10.1016/s0076-6879(96)64020-8. [DOI] [PubMed] [Google Scholar]
- 45.Lazarou M., Smith S.M., Thorburn D.R., Ryan M.T., McKenzie M. Assembly of nuclear DNA-encoded subunits into mitochondrial complex IV, and their preferential integration into supercomplex forms in patient mitochondria. FEBS J. 2009;276:6701–6713. doi: 10.1111/j.1742-4658.2009.07384.x. [DOI] [PubMed] [Google Scholar]
- 46.Callegari S., Oeljeklaus S., Warscheid B., Dennerlein S., Thumm M., Rehling P. Phospho-ubiquitin-PARK2 complex as a marker for mitophagy defects. Autophagy. 2017;13:201–211. doi: 10.1080/15548627.2016.1254852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Callegari S., Richter F., Chojnacka K., Jans D.C., Lorenzi I., Pacheu-Grau D. TIM29 is a subunit of the human carrier translocase required for protein transport. FEBS Lett. 2016;590:4147–4158. doi: 10.1002/1873-3468.12450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dennerlein S., Oeljeklaus S., Jans D., Hellwig C., Bareth B., Jakobs S. MITRAC7 acts as a COX1-specific chaperone and reveals a checkpoint during cytochrome c oxidase assembly. Cell Rep. 2015;12:1644–1655. doi: 10.1016/j.celrep.2015.08.009. [DOI] [PubMed] [Google Scholar]
- 49.Mick D.U., Vukotic M., Piechura H., Meyer H.E., Warscheid B., Deckers M. Coa3 and Cox14 are essential for negative feedback regulation of COX1 translation in mitochondria. J. Cell Biol. 2010;191:141–154. doi: 10.1083/jcb.201007026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bareth B., Nikolov M., Lorenzi I., Hildenbeutel M., Mick D.U., Helbig C. Oms1 associates with cytochrome c oxidase assembly intermediates to stabilize newly synthesized Cox1. Mol. Biol. Cell. 2016;27:1570–1580. doi: 10.1091/mbc.E15-12-0811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ong S.-E., Blagoev B., Kratchmarova I., Kristensen D.B., Steen H., Pandey A. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol. Cell. Proteomics. 2002;1:376–386. doi: 10.1074/mcp.m200025-mcp200. [DOI] [PubMed] [Google Scholar]
- 52.Peikert C.D., Mani J., Morgenstern M., Käser S., Knapp B., Wenger C. Charting organellar importomes by quantitative mass spectrometry. Nat. Commun. 2017;8 doi: 10.1038/ncomms15272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cox J., Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 2008;26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 54.Cox J., Neuhauser N., Michalski A., Scheltema R.A., Olsen J.V., Mann M. Andromeda: a peptide search engine integrated into the MaxQuant environment. J. Proteome Res. 2011;10:1794–1805. doi: 10.1021/pr101065j. [DOI] [PubMed] [Google Scholar]
- 55.Schägger H., von Jagow G. Blue native electrophoresis for isolation of membrane protein complexes in enzymatically active form. Anal. Biochem. 1991;199:223–231. doi: 10.1016/0003-2697(91)90094-a. [DOI] [PubMed] [Google Scholar]
- 56.Hofmann K., Stoffel W. TMbase-A database of membrane spanning protein segments. Biol. Chem. Hoppe Seyler. 1993;374:166. [Google Scholar]
- 57.Claros M.G., Vincens P. Computational method to predict mitochondrially imported proteins and their targeting sequences. Eur. J. Biochem. 1996;241:779–786. doi: 10.1111/j.1432-1033.1996.00779.x. [DOI] [PubMed] [Google Scholar]
- 58.Dennerlein S., Wang C., Rehling P. Plasticity of mitochondrial translation. Trends Cell Biol. 2017 doi: 10.1016/j.tcb.2017.05.004. [DOI] [PubMed] [Google Scholar]
- 59.Szklarczyk R., Wanschers B.F., Cuypers T.D., Esseling J.J., Riemersma M., van den Brand M.A. Iterative orthology prediction uncovers new mitochondrial proteins and identifies C12orf62 as the human ortholog of COX14, a protein involved in the assembly of cytochrome c oxidase. Genome Biol. 2012;13:R12. doi: 10.1186/gb-2012-13-2-r12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Peralta S., Clemente P., Sánchez-Martínez Á., Calleja M., Hernández-Sierra R., Matsushima Y. Coiled coil domain-containing protein 56 (CCDC56) is a novel mitochondrial protein essential for cytochrome c oxidase function. J. Biol. Chem. 2012;287:24174–24185. doi: 10.1074/jbc.M112.343764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Clemente P., Peralta S., Cruz-Bermudez A., Echevarría L., Fontanesi F., Barrientos A. hCOA3 stabilizes cytochrome c oxidase 1 (COX1) and promotes cytochrome c oxidase assembly in human mitochondria. J. Biol. Chem. 2013;288:8321–8331. doi: 10.1074/jbc.M112.422220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Stroud D.A., Maher M.J., Lindau C., Vögtle F.N., Frazier A.E., Surgenor E. COA6 is a mitochondrial complex IV assembly factor critical for biogenesis of mtDNA-encoded COX2. Hum. Mol. Genet. 2015;24:5404–5415. doi: 10.1093/hmg/ddv265. [DOI] [PubMed] [Google Scholar]
- 63.Pacheu-Grau D., Bareth B., Dudek J., Juris L., Vögtle F.N., Wissel M. Cooperation between COA6 and SCO2 in COX2 maturation during cytochrome c oxidase assembly links two mitochondrial cardiomyopathies. Cell Metab. 2015;21:823–833. doi: 10.1016/j.cmet.2015.04.012. [DOI] [PubMed] [Google Scholar]
- 64.Shoubridge E.A. Cytochrome c oxidase deficiency. Am. J. Med. Genet. 2001;106:46–52. doi: 10.1002/ajmg.1378. [DOI] [PubMed] [Google Scholar]
- 65.Smeitink J.A., Zeviani M., Turnbull D.M., Jacobs H.T. Mitochondrial medicine: a metabolic perspective on the pathology of oxidative phosphorylation disorders. Cell Metab. 2006;3:9–13. doi: 10.1016/j.cmet.2005.12.001. [DOI] [PubMed] [Google Scholar]
- 66.Pickrell A.M., Fukui H., Moraes C.T. The role of cytochrome c oxidase deficiency in ROS and amyloid plaque formation. J. Bioenerg. Biomembr. 2009;41:453–456. doi: 10.1007/s10863-009-9245-3. [DOI] [PubMed] [Google Scholar]
- 67.Bourens M., Barrientos A. Human mitochondrial cytochrome c oxidase assembly factor COX18 acts transiently as a membrane insertase within the subunit 2 maturation module. J. Biol. Chem. 2017;292:7774–7783. doi: 10.1074/jbc.M117.778514. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Results of SILAC-based quantitative mass spectrometry analysis of COX20FLAG complexes. COX20FLAG complexes were affinity-purified from mitochondria isolated from differentially SILAC-labeled wild-type and COX20FLAG-expressing cells mixed in equal amounts and analyzed by liquid chromatography-mass spectrometry (n = 4). Mass spectrometric raw data were processed using MaxQuant/Andromeda (version 1.4.1.2). For detailed information, see “Materials and methods”. Listed are proteins identified and quantified in ≥ 2 replicates.
Transparency document.