Abstract
Objectives
To provide information about the genetic relationships and mechanism underlying carbapenem resistance in Pseudomonas aeruginosa clinical isolates of a hospital in China.
Materials and methods
One hundred and sixty P. aeruginosa strains were isolated from a hospital in China. Susceptibility to 14 antimicrobial agents was determined by antimicrobial susceptibility testing. Multilocus sequence typing was used to characterize the genetic backgrounds of these clinical isolates. Forty-five strains were randomly selected for further evaluation of their carbapenem resistance mechanism. Their oprD gene was compared with the PAO1 sequence.
Results
Multilocus sequence typing analysis demonstrated that these isolates were highly diverse; 68 sequence types were identified, of which 28 were novel sequence types. Polygenic and eBURST analysis demonstrated genetically similar clones with dissimilar resistance profiles. Among the 45 randomly selected strains associated with carbapenem resistance, 2 were metallo β-lactamase producers; all the 45 strains were not AmpC overproducers. Sequence analysis revealed a high diversity in the oprD sequences among isolates. Strains susceptible to imipenem and meropenem with shortened L7 and L8 loops in oprD were the major strain types observed in this hospital.
Conclusion
This study indicated that oprD provided the main mechanism for carbapenem resistance. The shortened L7 and L8 loops are responsible for carbapenem susceptibility.
Keywords: Pseudomonas aeruginosa, multilocus sequence typing, carbapenem resistance, L7 and L8 loops in OprD
Introduction
The continued high rates of antibiotics usage in hospitals, farms, and agriculture have contributed to antibiotic resistance, forcing a rapid dissemination of multidrug-resistant (MDR) clones worldwide.1 Globally, infectious diseases have become an increasingly important cause of human morbidity and mortality.2 Pseudomonas aeruginosa is one of the most common hospital-acquired pathogens that cause numerous acute and chronic human infections, such as burn wound, urinary tract, and respiratory tract infections; it especially causes infections in intensive care units.3 According to the American Center for Disease Control and Prevention, P. aeruginosa causes about 10% of hospital-acquired infections.4 In recent years, the increasing resistance of P. aeruginosa strains to different antibiotics has led to an increase in the emergence of MDR P. aeruginosa. The treatment of infectious diseases caused by MDR P. aeruginosa is becoming more challenging with each passing year.5
Carbapenems are a class of β-lactam antibiotics that are highly effective for treating infections caused by MDR P. aeruginosa.6 However, resistance of P. aeruginosa to carbapenems is already common in many countries.7,8 Previous studies have shown that the decreased expression of the outer membrane protein OprD, oprD mutations, loss of OprD, and overproduction of metallo β-lactamases (MBLs) and AmpC, and overexpression of the major resistance-nodulation-division efflux pump systems (MexAB-OprM, MexCD-OprJ, MexEF-OprN, and MexXY-OprN) mainly contribute to carbapenem resistance.8–12 OprD is an outer membrane porin protein facilitating the permeation of basic amino acids, small peptides, and carbapenem antibiotics.13,14 A topological model predicts that OprD comprises 16 antiparallel strands connected by eight longer loop regions (L1–L8) on its external surface.15
P. aeruginosa is a ubiquitous microorganism present in many diverse environmental conditions; it can be transmitted by nursing staff, medical equipment, or utensils in hospitals.3,16 Typing of P. aeruginosa isolated from hospital settings is especially important for effective management of hospital-acquired infections and regulation of drug therapy. In this study, 160 P. aeruginosa strains were isolated from a hospital in China. Susceptibility to 14 antimicrobial agents was determined by antimicrobial susceptibility testing. Multilocus sequence typing (MLST) was used to characterize the genetic backgrounds of the clinical isolates. Among the 160 isolates, some carbapenem-resistant strains were selected for further evaluation of their carbapenem resistance mechanism.
The aims of this study were to investigate the drug resistance and genetic background of P. aeruginosa. It is useful to recognize their different mechanisms of resistance to carbapenems and devise a proper infection control strategy.
Materials and methods
Bacterial strains
A total of 160 clinical isolates of P. aeruginosa were collected at Shaanxi Provincial People’s Hospital between July 2016 and January 2017. The clinical isolates were isolated from sputum (n=134), urine (n=8), blood (n=6), endotracheal intubation discharge (n=3), bronchoalveolar lavage fluid (n=5), and skin secretions (n=4). These isolates were identified using the VITEK 2 system (bioMérieux, Marcy l’Etoile, France) and 16S rRNA gene sequencing. P. aeruginosa PAO1 and P. aeruginosa ATCC27853 (kindly donated by The Children’s Hospital of Xi’an City) were used as reference strains.
Antimicrobial susceptibility testing
Antibiotic susceptibility tests of selected strains were performed using the disk diffusion method; the results were interpreted according to the Clinical and Laboratory Standards Institute guidelines.17 The antibiotics used included amikacin, gentamicin, tobramycin, ciprofloxacin, levofloxacin, polymyxin, aztreonam, ceftazidime, cefepime, imipenem (IPM), meropenem (MEM), piperacillin, piperacillin/tazobactam, and cefoperazone/sulbactam.
Multilocus sequence typing
MLST was performed as per previously published protocols.18 Seven housekeeping genes (acsA, aroE, guaA, mutL, nuoD, ppsA, and trpE) were amplified in all isolates using the primers described by Curran et al.18 The polymerase chain reaction products were sequenced and submitted to the P. aeruginosa MLST database (https://pubmlst.org/paeruginosa/) for assignment of allelic numbers. Each isolate was then assigned a sequence type (ST) based on the combination of seven allelic numbers. Any STs or allelic profiles that did not match with those in the existing database were designated as “new”. New allele sequences and the allelic profiles of novel STs were submitted to the P. aeruginosa MLST data-base for assignment of the correct allelic numbers and STs.
Phylogenetic analysis
DnaSP v5 was used to analyze the genetic variability at MLST loci for the 160 isolated strains.19 The mean G+C content, polymorphic sites, difference in number of nucleotides, and the nonsynonymous/synonymous ratio (dN/dS) were calculated.
MLST allelic profiles were analyzed to examine the relationship within the STs of the 160 clinical isolates using eBURST v3, available from http://eburst.mlst.net/. Isolates with five or more identical alleles were considered as part of the same clonal complexes (CCs).
For sequence-based phylogenetic tree analysis, the seven MLST gene fragment sequences of each ST strain were concatenated into a single 2800 bp sequence. The concatenated sequences were aligned and subjected to maximum parsimony analysis using the MEGA 5 software program.20 Rooted neighbor-joining trees based on the concatenated seven-gene sequence data were generated using the Kimura two-parameter model for distance calculations and 1000 bootstrap replicates.21,22
Sequence analysis of the oprD gene and the ampC gene
Primers OprD-F1 (CGCCGACAAGAAGAACTAGC) and OprD-R1 (GTCGATTACAGGATCGACAG) were used for oprD amplification and sequencing, as described previously.23 The sequences were compared with the PAO1 sequence (http://www.pseudomonas.com/) and analyzed with DNA-MAN program, version 5.2.2.
Screening for AmpC and MBL production
MBL production was detected by the double-disk synergy test, performed using 10 μg IPM and 2.5 μM EDTA.24 AmpC production was screened using the cloxacillin double-disk synergy test, as previously described.25
Results
Antimicrobial susceptibility analysis
Using the disk diffusion method as described in the “Materials and methods” section, antimicrobial susceptibility testing was carried out for the collected clinical isolates. Carbapenems are one of the antibiotics considered the last resort against P. aeruginosa infections.6 In the collected 160 isolates, the results indicated that 93 (60%) and 86 (53.7%) strains displayed resistance to IPM and MEM, respectively (Table 1). Rates of resistance to aztreonam, piperacillin, levofloxacin, piperacillin/tazobactam, cefoperazone sodium/sulbactam, cefepime, and ceftazidime were 44.4%, 43.1%, 40.0%, 34.4%, 33.1%, 32.5%, and 30.0%, respectively. Lower rates of resistance were observed against tobramycin (5%), amikacin (8.8%), and gentamicin (11.9%).
Table 1.
The activity of 14 antimicrobial agents against 160 clinically isolated strains
| Antibiotic | Number (%) of 160 strains
|
||
|---|---|---|---|
| Sensitive | Intermediary | Resistant | |
| Aminoglycoside | |||
| Amikacin | 146 (91.2) | 0 (0) | 14 (8.8) |
| Gentamicin | 132 (82.5) | 9 (5.6) | 19 (11.9) |
| Tobramycin | 151 (94.4) | 1 (0.6) | 8 (5) |
| Quinolones | |||
| Ciprofloxacin | 99 (61.9) | 9 (5.6) | 52 (32.5) |
| Levofloxacin | 79 (49.4) | 17 (10.6) | 64 (40.0) |
| Polypeptides | |||
| Ploymixin | 160 (100) | 0 (0) | 0 (0) |
| Monolactams | |||
| Aztreonam | 52 (32.5) | 37 (23.1) | 71 (44.4) |
| Cephalosporins | |||
| Ceftazidime | 103 (64.4) | 9 (5.6) | 48 (30) |
| Cefepime | 84 (52.5) | 24 (15.0) | 52 (32.5) |
| Carbapenems | |||
| Imipenem | 63 (39.4) | 4 (2.5) | 93 (58.1) |
| Meropenem | 67 (41.9) | 7 (4.4) | 86 (53.7) |
| Ureidopenicillins | |||
| Piperacillin | 73 (45.6) | 18 (11.3) | 69 (43.1) |
| β-Lactam/inhibitor complex | |||
| Piperacillin/tazobactam | 85 (53.1) | 20 (12.5) | 55 (34.4) |
| Cefoperazone sodium/sulbactam | 78 (48.8) | 29 (18.1) | 53 (33.1) |
Multilocus sequence typing
MLST was used to distinguish isolated strains.18 According to the P. aeruginosa MLST database, typing of all isolated strains was successful. Three novel alleles were found among the 160 isolates and were submitted to the MLST database. A new allele found for the acsA gene was assigned a new allelic number, 164. Two new alleles were found for the nuoD gene and were assigned the corresponding allelic numbers, 104 and 105. A total of 66 different STs were assigned to the 160 isolates investigated; 132 isolates belonged to 38 known STs and 28 isolates demonstrated 28 new STs (Table 2). P. aeruginosa ATCC27853 and PAO1 were typed as ST155 and ST549, respectively. The ratio of the number of STs to the number of isolates was 0.41. The three predominant STs were ST1639 (22 isolates), ST485 (11 isolates), and ST261 (9 isolates). It was thus demonstrated that the STs of the isolates collected from the hospital were diverse.
Table 2.
The correspondent ST, allelic profile, and frequency of STs among 160 isolated strains, PAO1, and Pseudomonas aeruginosa ATCC27853
| ST | Number | acsA | aroE | guaA | mutL | nuoD | ppsA | trpE |
|---|---|---|---|---|---|---|---|---|
| ST1639 | 22 | 11 | 5 | 6 | 61 | 3 | 7 | 1 |
| ST485 | 11 | 11 | 76 | 5 | 3 | 61 | 14 | 3 |
| ST261 | 9 | 105 | 5 | 30 | 3 | 3 | 4 | 14 |
| ST639 | 7 | 11 | 19 | 19 | 3 | 4 | 4 | 7 |
| ST2375 (new) | 6 | 28 | 5 | 11 | 2 | 4 | 38 | 9 |
| ST882 | 6 | 17 | 5 | 11 | 3 | 4 | 6 | 37 |
| ST209 | 5 | 125 | 5 | 11 | 7 | 1 | 12 | 7 |
| ST357 | 5 | 2 | 4 | 5 | 3 | 1 | 6 | 11 |
| ST277 | 5 | 39 | 5 | 9 | 11 | 27 | 5 | 2 |
| ST291 | 5 | 6 | 5 | 5 | 3 | 52 | 4 | 2 |
| ST16 | 5 | 28 | 5 | 12 | 11 | 27 | 1 | 44 |
| ST2389 (new) | 4 | 164 (new) | 3 | 7 | 5 | 2 | 13 | 3 |
| ST244 | 3 | 17 | 5 | 12 | 3 | 14 | 4 | 7 |
| ST836 | 3 | 6 | 5 | 1 | 5 | 27 | 15 | 7 |
| ST1743 | 3 | 15 | 181 | 1 | 63 | 4 | 15 | 10 |
| ST2374 (new) | 3 | 17 | 5 | 26 | 3 | 4 | 4 | 7 |
| ST274 | 2 | 23 | 5 | 11 | 7 | 1 | 12 | 7 |
| ST1203 | 2 | 33 | 1 | 25 | 6 | 6 | 7 | 5 |
| ST381 | 2 | 11 | 20 | 1 | 65 | 4 | 4 | 10 |
| ST2363 (new) | 2 | 6 | 5 | 11 | 11 | 4 | 4 | 1 |
| ST2380 (new) | 2 | 28 | 5 | 11 | 2 | 4 | 38 | 7 |
| ST2381 (new) | 2 | 17 | 3 | 6 | 3 | 4 | 1 | 18 |
| ST226 | 2 | 11 | 5 | 36 | 3 | 1 | 29 | 7 |
| ST2372(new) | 2 | 125 | 5 | 11 | 7 | 73 | 12 | 7 |
| ST830 | 1 | 5 | 13 | 109 | 5 | 1 | 1 | 47 |
| ST597 | 1 | 17 | 5 | 12 | 3 | 4 | 4 | 7 |
| ST1453 | 1 | 28 | 61 | 3 | 6 | 1 | 33 | 41 |
| ST2100 | 1 | 17 | 8 | 3 | 11 | 1 | 15 | 1 |
| ST270 | 1 | 22 | 3 | 17 | 5 | 2 | 10 | 7 |
| ST1950 | 1 | 40 | 22 | 11 | 14 | 4 | 4 | 7 |
| ST1091 | 1 | 6 | 5 | 11 | 5 | 3 | 6 | 7 |
| ST273 | 1 | 104 | 4 | 36 | 71 | 4 | 4 | 53 |
| ST108 | 1 | 39 | 5 | 20 | 5 | 1 | 6 | 31 |
| ST621 | 1 | 15 | 5 | 20 | 5 | 1 | 4 | 25 |
| ST1226 | 1 | 11 | 66 | 11 | 3 | 29 | 4 | 9 |
| ST527 | 1 | 16 | 52 | 11 | 85 | 59 | 15 | 10 |
| ST408 | 1 | 17 | 5 | 37 | 3 | 1 | 4 | 2 |
| ST664 | 1 | 9 | 5 | 11 | 3 | 4 | 40 | 18 |
| ST676 | 1 | 28 | 5 | 11 | 77 | 3 | 4 | 92 |
| ST612 | 1 | 28 | 5 | 58 | 11 | 4 | 15 | 44 |
| ST845 | 1 | 11 | 5 | 1 | 7 | 4 | 4 | 7 |
| ST554 | 1 | 16 | 5 | 12 | 3 | 2 | 15 | 12 |
| ST792 | 1 | 6 | 5 | 11 | 3 | 2 | 15 | 1 |
| ST207 | 1 | 47 | 4 | 5 | 33 | 1 | 6 | 40 |
| ST1157 | 1 | 1 | 74 | 11 | 5 | 4 | 12 | 7 |
| ST2362 (new) | 1 | 6 | 5 | 1 | 29 | 92 | 4 | 68 |
| ST2364 (new) | 1 | 5 | 5 | 20 | 5 | 1 | 4 | 25 |
| ST2365 (new) | 1 | 11 | 5 | 9 | 11 | 27 | 5 | 2 |
| ST2366 (new) | 1 | 28 | 5 | 6 | 11 | 4 | 4 | 7 |
| ST2367 (new) | 1 | 15 | 5 | 1 | 63 | 4 | 15 | 10 |
| ST2368 (new) | 1 | 11 | 5 | 11 | 3 | 4 | 6 | 37 |
| ST2369 (new) | 1 | 2 | 4 | 5 | 5 | 1 | 6 | 11 |
| ST2370 (new) | 1 | 125 | 5 | 11 | 61 | 1 | 12 | 7 |
| ST2371 (new) | 1 | 17 | 5 | 7 | 3 | 4 | 4 | 7 |
| ST2373 (new) | 1 | 125 | 5 | 11 | 7 | 1 | 5 | 7 |
| ST2376 (new) | 1 | 36 | 27 | 28 | 3 | 52 | 13 | 7 |
| ST2377 (new) | 1 | 4 | 1 | 59 | 6 | 1 | 33 | 42 |
| ST2378 (new) | 1 | 17 | 5 | 11 | 7 | 1 | 12 | 7 |
| ST2379 (new) | 1 | 11 | 5 | 6 | 61 | 3 | 7 | 37 |
| ST2382 (new) | 1 | 28 | 5 | 58 | 61 | 4 | 15 | 44 |
| ST2383 (new) | 1 | 23 | 5 | 11 | 7 | 1 | 4 | 7 |
| ST2388 (new) | 1 | 11 | 5 | 6 | 61 | 104 (new) | 7 | 1 |
| ST2392 (new) | 1 | 164 (new) | 3 | 7 | 3 | 2 | 13 | 3 |
| ST2393 (new) | 1 | 164 (new) | 3 | 7 | 5 | 4 | 13 | 3 |
| ST2405 (new) | 1 | 125 | 5 | 11 | 7 | 105 (new) | 12 | 7 |
| ST2406 (new) | 1 | 29 | 8 | 36 | 67 | 1 | 6 | 3 |
| ST549 (PAO1) | 1 | 7 | 5 | 12 | 3 | 4 | 1 | 7 |
| ST155 (ATCC27853) | 1 | 28 | 5 | 36 | 3 | 3 | 13 | 7 |
Notes: The number assigned to each gene corresponds to the allelic type available in the MLST database. Alleles that did not match with the existing database were designated “new”. Types for strains having seven known alleles that did not match with any known strain type in the database were also designated “new”.
Abbreviations: MLST, multilocus sequence typing; ST, sequence type.
Allelic variation in P. aeruginosa
The characteristics of seven housekeeping genes are summarized in Table 3. Among the 160 strains, the number of distinct alleles for each gene varied from 14 (ppsA) to 24 (acsA), and the polymorphic sites for each gene ranged from 13 (nuoD) to 23 (trpE). The ratio of dN/dS was calculated for all seven genes and found to be equal to 0 for acsA, 0.1621 for nuoD, 0.1507 for guaA, 0.5799 for mutL, 1.6741 for aroE, and 2.3272 for trpE. The acsA gene showed synonymous substitutions, while the ppsA gene showed nonsynonymous substitution. guaA, mutL, nuoD, aroE, and trpE exhibited both synonymous and nonsynonymous substitutions (Table 3).
Table 3.
Characteristics and polymorphism of loci gene of Pseudomonas aeruginosa
| Allele | Length | Haplotype | Polymorphic sites | π | θ | G+C | dN | dS | dN/dS |
|---|---|---|---|---|---|---|---|---|---|
| acsA | 390 | 24 | 18 | 0.01258 | 0.01236 | 0.6871 | 0 | 0.05066 | 0 |
| aroE | 498 | 16 | 21 | 0.01466 | 0.01271 | 0.7044 | 0.02188 | 0.01307 | 1.6741 |
| guaA | 373 | 20 | 14 | 0.00889 | 0.01058 | 0.6568 | 0.0036 | 0.02389 | 0.1507 |
| mutL | 442 | 16 | 18 | 0.00792 | 0.0123 | 0.6715 | 0.00769 | 0.01326 | 0.5799 |
| nuoD | 366 | 15 | 13 | 0.00775 | 0.01092 | 0.6324 | 0.00471 | 0.02904 | 0.1621 |
| ppsA | 370 | 14 | 14 | 0.0087 | 0.0119 | 0.6676 | 0.01221 | 0 | n.a |
| trpE | 443 | 21 | 23 | 0.01403 | 0.01506 | 0.6616 | 0.01771 | 0.00761 | 2.3272 |
| Concatenate | 2882 | 68 | 120 | 0.00748 | 0.00877 | 0.6709 | 0.01318 | 0.00548 | 2.4051 |
Notes: The G+C content was calculated by Geneious 9.1. Haplotype, polymorphic sites, average number of synonymous (dS) and nonsynonymous sites (dN), average nonsynonymous/synonymous ratio (dN/dS), the nucleotide diversity per site (π), and the average number of nucleotide differences per site (θ) were found using DnaSP 5.0.
eBURST analysis
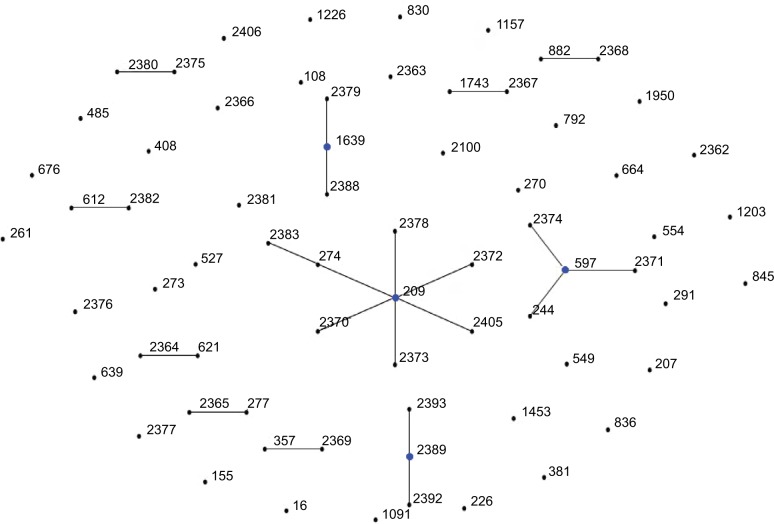
In order to determine the clonal relationship between isolates, 68 STs, including PAO1 and ATCC27853, were clustered by eBURST analysis (http://eburst.mlst.net).26 Figure 1 shows that 68 STs were segregated into 11 CCs because of the sequence identity among five or more alleles. The largest CC consisted of eight STs (ST2378, ST2372, ST274, ST2383, ST2370, ST2373, ST2405, and ST209, with ST209 being the primary founder). The second CC consisted of four STs (ST244, ST2374, ST2371, and ST597, with ST597 being the primary founder). The nine other groups contained two or three STs. Twenty-six STs were classified as singletons (Figure 1).
Figure 1.
Analysis of Pseudomonas aeruginosa STs using eBURST.
Notes: eBURST v3 was used to analyze the 68 unique STs. Each circle corresponds to an ST.
Abbreviation: ST, sequence type.
Phylogenetic analysis of the MLST sequences
To estimate the phylogenetic relationships among the isolates, the concatenated seven-gene sequence of each stain was further analyzed to construct rooted trees using the neighbor-joining method. Sixty-six concatenated sequences from 66 STs and the corresponding sequences from PAO1 and ATCC27853 were used (https://www.ncbi.nlm.nih.gov/). The MLST tree revealed a high genetic diversity in those isolates. The analysis revealed a weak bootstrapping value, especially in major branches. The phylogenetic tree shows that ST2378, ST2372, ST274, ST2383, ST2370, ST2373, ST2405, and ST209 were clustered together (Figure 2). Several close clusters were identified; these were also previously obtained by the eBURST algorithm.
Figure 2.
Rooted tree of the concatenated sequences (2800–2882 bp) of seven housekeeping genes of Pseudomonas aeruginosa using the NJ method based on Kimura 2 correction algorithm.
Abbreviations: NJ, neighbor joining; ST, sequence type.
Sequence analysis of the oprD gene in carbapenem-resistant isolates
To assess the variations in oprD gene in carbapenem-resistant strains isolated from this hospital, oprD gene amplification and sequencing were done in 30 carbapenem-resistant and 15 carbapenem-susceptible isolates. These 45 strains were randomly selected. Of the 30 carbapenem-resistant isolates, there were 8 IPM-resistant and MEM-susceptible strains, 5 IPM-susceptible and MEM-resistant strains, and 17 IPM- and MEM-resistant strains. Fifteen strains with susceptibility to carbapenem were used as controls. The sequences were compared with the wild-type PAO1 sequence. As expected, the results showed a high diversity in the oprD sequences among isolates, especially in IPM- and MEM-resistant strains. The IPM-resistant and MEM-susceptible strains had relatively fewer mutations.
A comparative analysis between IPM-resistant and IPM-susceptible strains strongly suggested that alterations in L7 and L8 loops were responsible for their IPM susceptibility. In 17 of the IPM- and MEM-resistant strains, 14 strains encoded incomplete OprD proteins due to the presence of a premature stop codon at nt 150 (1 isolate), 624 (4 isolates), 714 (1 isolate), 912 (1 isolate), 951 (1 isolate), 1035 (3 isolates), and 1302 (3 isolates), caused by either a frameshift, nonsense mutation or large deletion (Table 4). Five of these 14 strains also carried large deletions at nt 52–61 (1 isolate), 561–624 (3 isolates), and 816–855 (1 isolate), as shown in Table 4. The remaining three strains did not have a stop codon at the end of the oprD sequence. This result demonstrated that the loss or alteration of the original oprD confers resistance to carbapenem in P. aeruginosa.
Table 4.
OprD genotype classification of 45 strains
| Resistance phenotype Number of strains | Number of strains | Stop codon site | Large deletion site (nt) |
|---|---|---|---|
| IPM(R) MEM(R) | |||
| 17 | 1 | 150 | – |
| 3 | 624 | 561–624 | |
| 1 | 624 | – | |
| 1 | 714 | – | |
| 1 | 912 | – | |
| 1 | 951 | 816–855 | |
| 3 | 1035 | – | |
| 3 | 1302 | – | |
| 1 | – | 561–624 | |
| 2 | – | ||
| IPM(R) MEM(S) | |||
| 8 | 4 | 281 | – |
| 1 | 906 | 862–866 | |
| 1 | 1119 | – | |
| 2 | 1302 | – | |
| IPM(S) MEM(R) | |||
| 5 | 2 | 219 | – |
| 1 | 381 | – | |
| 1 | 354 | – | |
| 1 | – | 1116–1118, 1147–1149 | |
| IPM(S) MEM(S) | |||
| 15 | 2 | 162 | – |
| 1 | 219 | – | |
| 1 | 693 | – | |
| 1 | 699 | – | |
| 8 | 1296 | 1116–1118, 1147–1149, 1288–1292 | |
| 1 | 1302 | 348–350, 1147–1149, 1288–1292 | |
| 1 | 1302 | – |
Notes: Forty-five strains were randomly selected: 17 IPM- and MEM-resistant strains, 8 IPM-resistant and MEM-susceptible strains, 5 IPM-susceptible and MEM-resistant strains, and 15 strains with susceptibility to carbapenem used as controls. The sequences were compared with the wild-type PAO1 sequence. “–” Represents “no”.
Abbreviations: IPM, imipenem; MEM, meropenem.
On comparing the PAO1 strain with 15 IPM- and MEM-susceptible strains, 8 strains had 3 bp deletions at nt 1116–1118 and 1147–1149 in L7 and 5 bp deletions at nt 1288–1292 in L8; 1 strain had 3 bp deletions at nt 1147–1149 in L7 and 5 bp deletions at nt 1288–1292 in L8; 1 strain had 3 bp deletions at nt 348–350 in L2, and 1116–1118 and 1147–1149 in L7. The remaining five IPM- and MEM-susceptible isolates had a premature stop codon at nt 162 (2 isolates), 219 (1 isolate), 693 (1 isolate), and 699 (1 isolate) due to various point, insertion, and deletion mutations (Table 4).
Screening for AmpC β-lactamase and MBL production
To characterize the clinical isolates further, all randomly selected P. aeruginosa isolates were tested for MBL and AmpC production. Among the 45 clinical isolates, two MBL-positive strains were identified: one of them was an IPM- and MEM-resistant strain and the other one was an IPM-resistant and MEM-susceptible strain. All 45 isolates were confirmed as AmpC-producing isolates, but not AmpC-overproducing isolates.
Discussion
P. aeruginosa infection in hospitalized patients remains an important issue; it is associated with high rates of mortality and morbidity in immunocompromised patients.27 From a review of the rate of resistance to 14 antibiotics determined in this study, it was concluded that the treatment of P. aeruginosa infection is a challenge. In this study, 58.1% and 53.7% of the clinical isolates were resistant to IPM and MEM, respectively. IPM and MEM resistance rates reported from the surgical intensive care unit of the Peking University First Hospital were 84% and 31%, respectively.28 Data from an earlier study of 65 hospitals from 22 regions in China in 2010 demonstrated that the rates of resistance of isolates to IPM and MEM were 23.1% and 18.1%, respectively.29 There was no obvious association between the antibiotic resistance and geographic origin of the isolates. However, previous studies indicate an increased worldwide prevalence of carbapenem resistance.
MLST is an important tool in the study of the population structure and genetic diversity of P. aeruginosa.18 In this study, 68 STs were distinguished among 160 isolates; 28 of these were new STs. The three predominant STs in these isolates were ST1639, ST485, and ST261. Evidently, these isolates were extraordinarily rich in diversity and contained numerous novel genotypes. We found that the MLST STs and antimicrobial resistance profiles were not correlated in this study. Isolates with the same ST did not show a unique resistance profile pattern. This demonstrated that there is no definitive link between the ST of isolates and their resistance to these 14 antimicrobial agents. Genetic adaptation plays a major role in the successful establishment of a P. aeruginosa infection.30 Analysis of dN/dS ratios showed that the seven housekeeping genes, except aroE and trpE, appeared under purifying selection. aroE and trpE are evolving predominantly by positive selection.
By eBURST and phylogenetic tree analysis of all the clinical isolates in this study, we demonstrated multiple genetically distinct clones in this hospital. Here, we confirm the non-clonal epidemic structure of the population.31 The CCs cluster together in the phylogenetic tree and belong to the same CC in a relaxed eBURST analysis. Mapping of resistance profile data onto the eBURST analysis data and the phylogenetic tree revealed the following: 1) the most similar resistance profiles did not cluster together and 2) isolates with the same STs did not share similar resistance phenotypes. Taken together, this shows that these strains displayed a relatively high degree of genetic variability, demonstrating that antibiotic resistance was most likely determined by individual genetic combinations.
Multiple studies have demonstrated that decreased expression or loss of OprD is the important mechanism of resistance to carbapenem in P. aeruginosa.32 In this study, 14 of 17 IPM- and MEM-resistant strains had a premature stop codon; it demonstrated that the deficient OprD plays an important role in resistance to carbapenem. On the other hand, strains susceptible to IPM and MEM with shortened loops in L7 and L8 were the major strain types in this hospital. This suggested that shortened L7 and L8 loops are responsible for IPM and MEM susceptibility in our clinical isolates. It has been suggested that L5, L7 and L8 may contribute to the constriction of the OprD channel opening; the shortened loop of L7 seems to open the OprD porin channel sufficiently to increase the penetration of MEM in the clinical strains.12,15 In this study, 9 of 15 IPM- and MEM-susceptible strains had modifications in L8 of OprD, but the role of L8 loop on carbapenem susceptibility has not been reported. On the other hand, five carbapenem-susceptible stains carry premature stop codons in the upstream direction (at the following positions: 162, 219, 693, and 699), which indicates that the loss of OprD is not only restricted to carbapenem-resistant clinical isolates but also found in carbapenem-susceptible clinical isolates. It seemed that the issue of carbapenem-susceptible clinical isolates with deficient OprD is not fully explained by the aforementioned carbapenem resistance mechanisms. El Amin et al also found that carbapenem-susceptible clinical isolates had a significant reduction of oprD mRNA and the presence of oprD mutations causing a frameshift or translational stop.33 Ocampo-Sosa et al assessed the impact of the mutations in oprD on carbapenem resistance phenotype in a group of clinical isolates of P. aeruginosa. They believe that the function of OprD has not been completely defined.34 In our clinical isolates, no ampC gene mutations were detected. MBL and AmpC production was also investigated in this study; however, it was demonstrated that only two isolates were MBL overproducers. As a result, MBL and AmpC may not significantly alter carbapenem susceptibility in our clinical strains.
In conclusion, P. aeruginosa isolates from the hospital in this study exhibited multidrug resistance to a variety of drugs and a higher rate of resistance against carbapenem. MLST analysis further revealed the genetic diversity of these clinical isolates of P. aeruginosa. oprD polymorphisms, particularly those resulting in L7 and L8 shortening, are the major cause of carbapenem sensitivity. Moreover, our results also show a high incidence of carbapenem-resistant strains with a loss of OprD, indicating that intact OprD is necessary for resistance to carbapenem. However, the resistance patterns observed in this hospital cannot be fully explained by the currently described carbapenem resistance mechanisms. The role of OprD in carbapenem resistance needs to be intensively studied. These findings will help us to understand the mechanism of carbapenem resistance in P. aeruginosa isolates better.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (31622003, 31670080, and 31500111) and the Natural Science Foundation of Shaanxi Province, China (2017JM3037, 2016JZ007).
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Giske CG, Monnet DL, Cars O, Carmeli Y, ReAct-Action on Antibiotic Resistance Clinical and economic impact of common multidrug-resistant gram-negative bacilli. Antimicrob Agents Chemother. 2008;52(3):813–821. doi: 10.1128/AAC.01169-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Laxminarayan R, Duse A, Wattal C, et al. Antibiotic resistance-the need for global solutions. Lancet Infect Dis. 2013;13(12):1057–1098. doi: 10.1016/S1473-3099(13)70318-9. [DOI] [PubMed] [Google Scholar]
- 3.Yayan J, Ghebremedhin B, Rasche K. Antibiotic resistance of Pseudomonas aeruginosa in Pneumonia at a Single University Hospital Center in Germany over a 10-Year Period. PLoS One. 2015;10(10):e0139836. doi: 10.1371/journal.pone.0139836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tacconelli E, Tumbarello M, Bertagnolio S, et al. Multidrug-resistant Pseudomonas aeruginosa bloodstream infections: analysis of trends in prevalence and epidemiology. Emerg Infect Dis. 2002;8(2):220–221. doi: 10.3201/eid0802.010121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ranjan S, Banashankari G, Babu PS. Comparison of epidemiological and antibiotic susceptibility pattern of metallo-beta-lactamase-positive and metallo-beta-lactamase-negative strains of pseudomonas aeruginosa. J Lab Physicians. 2014;6(2):109–113. doi: 10.4103/0974-2727.141509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pragasam AK, Raghanivedha M, Anandan S, Veeraraghavan B. Characterization of Pseudomonas aeruginosa with discrepant carbapenem susceptibility profile. Ann Clin Microbiol Antimicrob. 2016(15):12. doi: 10.1186/s12941-016-0127-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rodriguez-Martinez JM, Poirel L, Nordmann P. Molecular epidemiology and mechanisms of carbapenem resistance in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 2009;53(11):4783–4788. doi: 10.1128/AAC.00574-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim CH, Kang HY, Kim BR, et al. Mutational inactivation of OprD in carbapenem-resistant Pseudomonas aeruginosa isolates from Korean hospitals. J Microbiol. 2016;54(1):44–49. doi: 10.1007/s12275-016-5562-5. [DOI] [PubMed] [Google Scholar]
- 9.Choudhury D, Talukdar AD, Choudhury MD, et al. Carbapenem non-susceptibility with modified OprD in clinical isolates of Pseudomonas aeruginosa from India. Indian J Med Microbiol. 2017;35(1):137–139. doi: 10.4103/ijmm.IJMM_15_220. [DOI] [PubMed] [Google Scholar]
- 10.Wi YM, Choi JY, Lee JY, et al. Emergence of colistin resistance in Pseudomonas aeruginosa ST235 clone in South Korea. Int J Antimicrob Agents. 2017;49(6):767–769. doi: 10.1016/j.ijantimicag.2017.01.023. [DOI] [PubMed] [Google Scholar]
- 11.Kao CY, Chen SS, Hung KH, et al. Overproduction of active efflux pump and variations of OprD dominate in imipenem-resistant Pseudomonas aeruginosa isolated from patients with bloodstream infections in Taiwan. BMC Microbiol. 2016;16(1):107. doi: 10.1186/s12866-016-0719-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Epp SF, Kohler T, Plesiat P, Michea-Hamzehpour M, Frey J, Pechere JC. C-terminal region of Pseudomonas aeruginosa outer membrane porin OprD modulates susceptibility to meropenem. Antimicrob Agents Chemother. 2001;45(6):1780–1787. doi: 10.1128/AAC.45.6.1780-1787.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lister PD, Wolter DJ, Hanson ND. Antibacterial-resistant Pseudomonas aeruginosa: clinical impact and complex regulation of chromosomally encoded resistance mechanisms. Clin Microbiol Rev. 2009;22(4):582–610. doi: 10.1128/CMR.00040-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hancock RE, Siehnel R, Martin N. Outer membrane proteins of Pseudomonas. Mol Microbiol. 1990;4(7):1069–1075. doi: 10.1111/j.1365-2958.1990.tb00680.x. [DOI] [PubMed] [Google Scholar]
- 15.Huang H, Jeanteur D, Pattus F, Hancock RE. Membrane topology and site-specific mutagenesis of Pseudomonas aeruginosa porin OprD. Mol Microbiol. 1995;16(5):931–941. doi: 10.1111/j.1365-2958.1995.tb02319.x. [DOI] [PubMed] [Google Scholar]
- 16.Feng W, Sun F, Wang Q, et al. Epidemiology and resistance characteristics of Pseudomonas aeruginosa isolates from the respiratory department of a hospital in China. J Glob Antimicrob Resist. 2017;8:142–147. doi: 10.1016/j.jgar.2016.11.012. [DOI] [PubMed] [Google Scholar]
- 17.Clinical and Laboratory Standards Institute . Performance Standards for Antimicrobial Disk Susceptibility Tests. 11th ed. Wayne, PA: CLSI; 2012. [Google Scholar]
- 18.Curran B, Jonas D, Grundmann H, Pitt T, Dowson CG. Development of a multilocus sequence typing scheme for the opportunistic pathogen Pseudomonas aeruginosa. J Clin Microbiol. 2004;42(12):5644–5649. doi: 10.1128/JCM.42.12.5644-5649.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Librado P, Rozas J. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25(11):1451–1452. doi: 10.1093/bioinformatics/btp187. [DOI] [PubMed] [Google Scholar]
- 20.Kumar S, Tamura K, Jakobsen IB, Nei M. MEGA2: molecular evolutionary genetics analysis software. Bioinformatics. 2001;17(12):1244–1245. doi: 10.1093/bioinformatics/17.12.1244. [DOI] [PubMed] [Google Scholar]
- 21.Liu Y, Lai Q, Du J, Shao Z. Genetic diversity and population structure of the Bacillus cereus group bacteria from diverse marine environments. Sci Rep. 2017;7(1):689. doi: 10.1038/s41598-017-00817-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kane RA, Stothard JR, Emery AM, Rollinson D. Molecular characterization of freshwater snails in the genus Bulinus: a role for barcodes? Parasit Vectors. 2008;1(1):15. doi: 10.1186/1756-3305-1-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sun Q, Ba Z, Wu G, Wang W, Lin S, Yang H. Insertion sequence ISRP10 inactivation of the oprD gene in imipenem-resistant Pseudomonas aeruginosa clinical isolates. Int J Antimicrob Agents. 2016;47(5):375–379. doi: 10.1016/j.ijantimicag.2016.02.008. [DOI] [PubMed] [Google Scholar]
- 24.Lee K, Chong Y, Shin HB, Kim YA, Yong D, Yum JH. Modified Hodge and EDTA-disk synergy tests to screen metallo-beta-lactamase-producing strains of Pseudomonas and Acinetobacter species. Clin Microbiol Infect. 2001;7(2):88–91. doi: 10.1046/j.1469-0691.2001.00204.x. [DOI] [PubMed] [Google Scholar]
- 25.Tzouvelekis LS, Markogiannakis A, Psichogiou M, Tassios PT, Daikos GL. Carbapenemases in Klebsiella pneumoniae and other Enterobacteriaceae: an evolving crisis of global dimensions. Clin Microbiol Rev. 2012;25(4):682–707. doi: 10.1128/CMR.05035-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Feil EJ, Li BC, Aanensen DM, Hanage WP, Spratt BG. eBURST: inferring patterns of evolutionary descent among clusters of related bacterial genotypes from multilocus sequence typing data. J Bacteriol. 2004;186(5):1518–1530. doi: 10.1128/JB.186.5.1518-1530.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ozgumus OB, Caylan R, Tosun I, Sandalli C, Aydin K, Koksal I. Molecular epidemiology of clinical Pseudomonas aeruginosa isolates carrying IMP-1 metallo-beta-lactamase gene in a University Hospital in Turkey. Microb Drug Resist. 2007;13(3):191–198. doi: 10.1089/mdr.2007.748. [DOI] [PubMed] [Google Scholar]
- 28.Yi M, Wang P, Liu Y. Molecular typing and resistance mechanisms of carbapenem resistant Pseudomonas aeruginosa isolated from a Chinese surgical intensive care unit. Chin Med J (Engl) 2014;127(6):1071–1076. [PubMed] [Google Scholar]
- 29.Ji J, Wang J, Zhou Z, Wang H, Jiang Y, Yu Y. Multilocus sequence typing reveals genetic diversity of carbapenem- or ceftazidime-nonsusceptible Pseudomonas aeruginosa in China. Antimicrob Agents Chemother. 2013;57(11):5697–5700. doi: 10.1128/AAC.00970-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mena A, Smith EE, Burns JL, et al. Genetic adaptation of Pseudomonas aeruginosa to the airways of cystic fibrosis patients is catalyzed by hypermutation. J Bacteriol. 2008;190(24):7910–7917. doi: 10.1128/JB.01147-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wiehlmann L, Wagner G, Cramer N, et al. Population structure of Pseudomonas aeruginosa. Proc Natl Acad Sci U S A. 2007;104(19):8101–8106. doi: 10.1073/pnas.0609213104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hu YF, Liu CP, Wang NY, Shih SC. In vitro antibacterial activity of rifampicin in combination with imipenem, meropenem and doripenem against multidrug-resistant clinical isolates of Pseudomonas aeruginosa. BMC Infect Dis. 2016;16(1):444. doi: 10.1186/s12879-016-1785-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.El Amin N, Giske CG, Jalal S, Keijser B, Kronvall G, Wretlind B. Carbapenem resistance mechanisms in Pseudomonas aeruginosa: alterations of porin OprD and efflux proteins do not fully explain resistance patterns observed in clinical isolates. APMIS. 2005;113(3):187–196. doi: 10.1111/j.1600-0463.2005.apm1130306.x. [DOI] [PubMed] [Google Scholar]
- 34.Ocampo-Sosa AA, Cabot G, Rodriguez C, et al. Spanish Network for Research in Infectious Diseases (REIPI) Alterations of OprD in carbapenem-intermediate and -susceptible strains of Pseudomonas aeruginosa isolated from patients with bacteremia in a Spanish multicenter study. Antimicrob Agents Chemother. 2012;56(4):1703–1713. doi: 10.1128/AAC.05451-11. [DOI] [PMC free article] [PubMed] [Google Scholar]