Abstract
Phthoxazolin A, an oxazole-containing polyketide, has a broad spectrum of anti-oomycete activity and herbicidal activity. We recently identified phthoxazolin A as a cryptic metabolite of Streptomyces avermitilis that produces the important anthelmintic agent avermectin. Even though genome data of S. avermitilis is publicly available, no plausible biosynthetic gene cluster for phthoxazolin A is apparent in the sequence data. Here, we identified and characterized the phthoxazolin A (ptx) biosynthetic gene cluster through genome sequencing, comparative genomic analysis, and gene disruption. Sequence analysis uncovered that the putative ptx biosynthetic genes are laid on an extra genomic region that is not found in the public database, and 8 open reading frames in the extra genomic region could be assigned roles in the biosynthesis of the oxazole ring, triene polyketide and carbamoyl moieties. Disruption of the ptxA gene encoding a discrete acyltransferase resulted in a complete loss of phthoxazolin A production, confirming that the trans-AT type I PKS system is responsible for the phthoxazolin A biosynthesis. Based on the predicted functional domains in the ptx assembly line, we propose the biosynthetic pathway of phthoxazolin A.
Introduction
Polyketides and non-ribosomal peptides are structurally diverse classes of natural products representing important therapeutic and agricultural chemicals [1,2]. In bacteria, polyketides are typically produced by polyketide synthase (PKS) assembly lines that are composed of functional modules. Each module is responsible for one step of chain extension of the growing products and consists of a set of domains which dictate the chemical functionality of the incorporated building block [3,4]. The minimal domains of a PKS module are an acyltransferase (AT), ketosynthase (KS) and acyl carrier protein (ACP) domain, where the AT domain loads a coenzyme A-activated dicarboxylic acid extender unit onto the ACP domain and the KS domain of the preceding domain catalyzes condensation of the nascent polyketide with the extender unit. Meanwhile, nonribosomal peptide synthetase (NRPS) also forms multienzyme assembly lines that are similar to those of PKS, and produces peptide-containing compounds [5]. In the NRPS module, an adenylation (A) domain activates an amino acid, and loads onto a peptidyl carrier protein (PCP). Subsequently, a condensation (C) domain generates an amide bond with the growing chain tethered to the upstream PCP domain. Many of the bacteria sequenced to date have been found to possess a “hybrid” PKS/NRPS enzyme [6] and show that both PKS and NRPS function simultaneously in the same assembly line [7], but the products synthesized by most of the hybrid PKS/NRPS enzymes are still unknown.
Oxazole-containing polyketides are typical PKS/NRPS hybrid products, and have various biological activities for medicinal and agricultural purposes; e.g., oxazolomycin is an oxazole-triene antibiotic of Streptomyces albus JA3453 showing antibacterial and antitumor activities [8, 9], rhizoxin is a methyl-oxazole macrolide from the bacterial endosymbiont Burkholderia rhizoxina [10], and conglobatin is an unusual symmetrical macrolide from Streptomyces conglobatus showing antitumor activity [11]. Within these oxazole-containing polyketides, oxazolomycin and rhizoxin are synthesized by the trans-AT type I PKS system together with NRPS machinery [10,12]. One distinct feature of the trans-AT type I PKS system is that the PKS modules lack an AT domain and one or a few free-standing ATs provide extender units for each elongation step [13,14]. This type of PKS modular system has contributed to the expansion of complexity in the polyketide biosynthetic machinery.
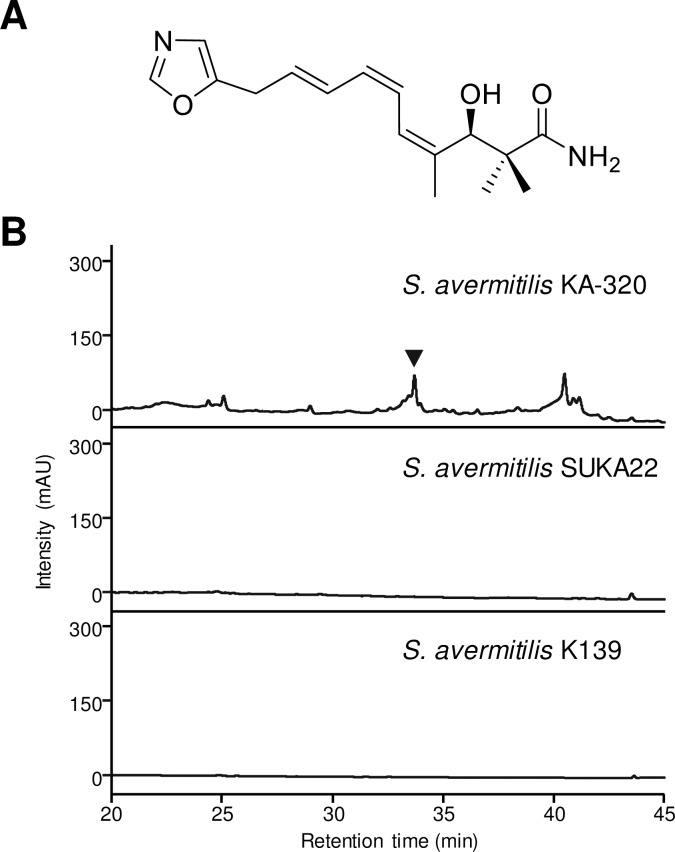
Phthoxazolin A (Fig 1A), one of the oxazole-containing polyketides, is an inhibitor of cellulose biosynthesis and exerts growth-inhibitory activity against plant pathogenic oomycetes [15–17]. The structure of phthoxazolin A includes a unique 5-substituted oxazole ring connected to a triene moiety, which corresponds to the substructure of oxazolomycin. We have previously demonstrated that phthoxazolin A is a cryptic metabolite of Streptomyces avermitilis, the producer of the important anthelmintic agent avermectin [18]. Although the in silico analysis predicted that the genome of S. avermitilis harbors at least 38 gene clusters for secondary metabolites [19], there is no assembly line similar to that for oxazolomycin biosynthesis. Thus, the biosynthetic gene cluster for phthoxazolin A would be hidden somewhere in the S. avermitilis genome.
Fig 1. Phthoxazolin A production in the S. avermitilis progeny.
(A) Chemical structure of phthoxazolin A. (B) HPLC chromatograms of MeOH extracts from S. avermitilis KA-320 (top), S. avermitilis SUKA22 (middle), and S. avermitilis K139 (bottom). mAU, milliabsorbance units at 275 nm. Phthoxazolin A was detected at a retention time of 33.9 min, and is indicated by an inverted triangle.
In this study, we performed genome sequencing, comparative genomic analysis, and mutagenesis to identify a biosynthetic gene cluster for phthoxazolin A, and demonstrated that the gene cluster is laid on the extra genomic region of the original avermectin producer, and phthoxazolin A is synthesized by the trans-AT type I PKS system. The proposed assembly line for phthoxazolin A biosynthesis also suggests a new cleavage system in the PKS/NRPS machinery.
Materials and methods
Bacterial strains, plasmids, and growth conditions
S. avermitilis KA-320 (isogenic to MA-4680, ATCC 31267 and NRRL 8165) [20], K139 (a progeny of KA-320) [21], SUKA22 (K139 as a genetic background) [22] strains were obtained from the culture collection of Kitasato Institute, and S. avermitilis KA-320 ΔavaR3 [20] was used in this study. All these strains were grown on YMS-MC medium for spore formation [20]. Escherichia coli DH5α was used for general DNA manipulation, and E. coli F-dcm Δ(srl-recA) 306::Tn10 carrying pUB307-aph::Tn7 was used for E. coli/Streptomyces conjugation. The plasmids pKU451, pKU470, pKU479, pKU480, and pKU250 were used to construct a vector for gene deletion [23]. The media and general E. coli and Streptomyces manipulations were as described previously [20]. For analysis of phthoxazolin A production, spores (1.0 X 108 CFU) of the S. avermitilis strains were inoculated into 70 mL APM medium in a 500-mL baffled flask, and mycelia were harvested after 48 h of cultivation. The mycelia were washed, re-suspended in fresh APM medium and stored at -80°C until use as a seed culture. All the primers are listed in S1 Table.
Analysis of phthoxazolin A production
The seed culture was inoculated on 2.5 mL YMD solid medium [18], followed by incubation at 28°C for 3 days. The agar culture was diced and extracted with an equal volume of methanol. The methanol extract was analyzed by using a HPLC system as described previously. [18].
Construction of Streptomyces avermitilis large-deletion (SALD) mutant strains
For the SALD-1 mutant, two regions (position 77,219–79,457 nt and 79,479–81,744 nt) were PCR-amplified by the primer pairs (sav68-up-Fw/sav68-up-Re and sav71-dw-Fw/sav71-dw-Re). These fragments were digested by HindIII and SpeI, and inserted to pKU451 resulting in pLT143. This plasmid was digested with SpeI and ligated together with a fragment containing a hygromycin B phosphotransferase gene (hph) with mutant loxP sequence (mut-loxP) (PCR-amplified with pKU480 and the mutloxP-SpeI-Fw/wo-mutloxP-SpeI-Re primer pair) to generate pLT144. A 6.6 kb HindIII fragment, recovered from pLT144, was cloned into pKU250 at the HindIII site to obtain pLT145. pLT145 was introduced by intergeneric conjugation to S. avermitilis KA-320 ΔavaR3 mutant to yield S. avermitilis ΔavaR3/sav71::mut-loxP-hph mutant. Another two regions (582,859–584,876 nt and 587,127–584,906 nt) were also PCR-amplified by the primer pairs (sav432-up-Fw/sav434-up-Re and sav434-dw-Fw/sav434-dw-Re). These fragments were treated with HindIII/SpeI, and ligated to pKU451 to get pLT146. A kanamycin-resistant gene (aphII) with mut-loxP at downstream end was PCR-amplified by the primer pair wo-mutloxP-SpeI-Fw/ mutloxP-SpeI-Re using pKU479 as a template, and introduced into the SpeI site of pLT146 to generate pLT147. A 6.0 kb HindIII fragment, recovered from the resultant plasmid, cloned into at the HindIII site of pKU250 to obtain pLT148. pLT148 was introduced to S. avermitilis ΔavaR3/sav71::mut-loxP-hph mutant, resulting in S. avermitilis ΔavaR3/sav71::mut-loxP-hph mutant/Δsav434::aphII-mut-loxP. The cre expression plasmid pKU470 was introduced into the strain for removal of a 0.51 Mb region covering from sav71 to sav434.
For the SALD-2 mutant, the aphII gene of pLT148 was replaced with a hph gene harboring mut-loxP by SpeI digestion and ligation, resulting in pLT149. pLT149 was introduced into S. avermitilis KA-320 ΔavaR3 mutant to yield S. avermitilis ΔavaR3/sav434::mut-loxP-hph mutant. Two regions (999,468–997,573 nt and 1,004,009–1,002,110 nt) were PCR-amplified by the primer pairs (sav845-up-Fw/sav845-up-Re and sav845-dw-Fw/sav845-dw-Re), and ligated with pKU451 to get pLT150. This plasmid was ligated with the aphII gene prepared previously to generate pLT151. A 5.4 kb HindIII fragment, recovered from pLT151, was cloned into pKU250 at the HindIII site to obtain pLT152. pLT152 was introduced to S. avermitilis ΔavaR3/sav434::mut-loxP-hph mutant, resulting in S. avermitilis ΔavaR3/sav434::mut-loxP-hph mutant/Δsav845::aphII-mut-loxP. pKU470 was introduced into the strain for removal of a 0.42 Mb region covering from sav434 to sav845.
For the SALD-3 mutant, the aphII gene of pLT152 was replaced with a hph gene harboring mut-loxP by SpeI digestion and ligation, resulting in pLT153. pLT153 was introduced into S. avermitilis KA-320 ΔavaR3 mutant to yield S. avermitilis ΔavaR3/sav845::mut-loxP-hph mutant. Two regions (1,271,761–1,273,916 nt and 1,273,956–1,276,187 nt) were PCR-amplified by the primer pairs (sav1007-up-Fw/sav1007-up-Re and sav1007-dw-Fw/sav1007-dw-Re), and ligated with pKU451 to get pLT154. This plasmid was ligated with the aphII gene prepared previously to generate pLT155. A 6.0 kb HindIII fragment, recovered from pLT155, was cloned into pKU250 at the HindIII site to obtain pLT156. pLT156 was introduced to S. avermitilis ΔavaR3/sav845::mut-loxP-hph mutant, resulting in S. avermitilis ΔavaR3/sav845::mut-loxP-hph mutant/Δsav1007::aphII-mut-loxP. pKU470 was introduced into the strain for removal of a 0.28 Mb region covering from sav845 to sav1007.
For the SALD-4 mutant, the aphII gene of pLT156 was replaced with a hph gene harboring mut-loxP by SpeI digestion and ligation, resulting in pLT157. pLT157 was introduced into S. avermitilis KA-320 ΔavaR3 mutant to yield S. avermitilis ΔavaR3/sav1007::mut-loxP-hph mutant. Two regions (1,593,533–1,595,311 nt and 1,595,580–1,597,661 nt) were PCR-amplified by the primer pairs (sav1286-up-Fw/sav1286-up-Re and sav1286-dw-Fw/sav1286-dw-Re), and ligated with pKU451 to get pLT158. This plasmid was ligated with the aphII gene prepared previously to generate pLT159. A 5.5 kb HindIII fragment, recovered from pLT159, was cloned into pKU250 at the HindIII site to obtain pLT160. pLT160 was introduced to S. avermitilis ΔavaR3/sav1007::mut-loxP-hph mutant, resulting in S. avermitilis ΔavaR3/sav1007::mut-loxP-hph mutant/Δsav1286::aphII-mut-loxP. pKU470 was introduced into the strain for removal of a 0.32 Mb region covering from sav1007 to sav1286.
The genotype of candidate strains for the desired large deletion mutation was confirmed by PCR analysis and DNA sequencing. The S. avermitilis KA-320 ΔavaR3 lacking each region (sav71-sav434, sav434-sav845, sav845-sav1007, and sav1007-sav1286) was designated as SALD-1, SALD-2, SALD-3, and SALD-4.
Genome sequencing and bioinformatics analyses
The genomic DNA of S. avermitilis KA-320 was prepared according to the previous procedures [24]. Sequence data of S. avermitilis KA-320 was obtained by assembling both data generated from the old-type DNA sequencer (MegaBACE 1000) and the next-generation DNA sequencer (Illumina). After assembling them, at least 21 contigs were generated. One contig (ca. 164 kb) contained the putative biosynthetic gene cluster for phthoxazolin A (a region around the gene cluster has been deposited in the DDBJ; accession number LC315614). Annotation of open reading frames (ORFs) and gene functions was performed manually by using the FramePlot 4.0beta program (http://nocardia.nih.go.jp/fp4), the 2ndFind program (http://biosyn.nih.go.jp/2ndfind/), the BLAST algorithm and the web-based PKS/NRPS analysis program (http://nrps.igs.umaryland.edu/nrps/).
In-frame deletion of the ptxA gene in the avaR3 mutant
Two 2.0 kb flanking regions of the ptxA gene were PCR-amplified by the primer pairs (ptxA-up-Fw/ptxA-up-Re and ptxA-dw-Re/ptxA-dw-Fw). These fragments were digested by HindIII and SpeI, and inserted into pKU451, resulting in pLT140. This plasmid was digested with SpeI and ligated together with a fragment containing a hph gene (PCR-amplified with pKU480 and the hph-Fw/hph-Re primer pair) to generate pLT141. A 6.2 kb HindIII fragment was recovered from pLT141, and cloned into the HindIII site of pKU250 to obtain pLT142. pLT142 was introduced into the S. avermitilis KA-320 ΔavaR3 mutant by intergeneric conjugation [20], and the DNA region including the ptxA gene was replaced with the disrupted allele by homologous recombination. The genotype of candidate strains for the ptxA mutation was confirmed by PCR analysis and DNA sequencing. The S. avermitilis KA-320 avaR3/ptxA double mutant was abbreviated as ΔavaR3 ΔptxA.
Complementation of the avaR3/ptxA double mutant
A 3.2 kb fragment containing the entire ptxA gene was PCR-amplified by using the primer pair ptxA-comp-Fw/ptxA-comp-Re, and then cloned to the BamHI site of pLT101 [25] using GeneArt Seamless Cloning and Assembly Kit (Life Technologies). The resultant plasmid was introduced into the avaR3/ptxA double mutant by intergeneric conjugation and integration. Integration of the plasmid was confirmed by apramycin resistance and PCR analysis.
Results
Phthoxazolin A production in the S. avermitilis progeny
To identify a biosynthetic gene cluster for phthoxazolin A, in silico screening was performed with the public genome sequence of S. avermitilis using oxazolomycin biosynthetic genes as probes. However, we could not find any assembly line in the genome that was similar to that of the oxazolomycin biosynthesis. Moreover, the genome has no orthologue of OzmO (an NRPS enzyme), which is necessary for the biosynthesis of an oxazole ring moiety. It thus appeared that the public genome sequence contained no clues for identifying the phthoxazolin A biosynthetic gene cluster.
S. avermitilis K139 SUKA22 is genetically constructed by deleting the 1.5 Mb left region of the parental strain [22], and was found to lack the ability to produce phthoxazolin A (Fig 1B), suggesting that the deleted DNA region might contain the phthoxazolin A biosynthetic gene cluster. Thus, we generated several series of large-deletion mutants (SALD mutants) from the S. avermitilis KA320 ΔavaR3 mutant [20], and evaluated phthoxazolin A production in these SALD mutants (S1 Fig). To our surprise, all the large-deletion mutants (SALD-1, SALD-2, SALD-3 and SALD-4 strains) still produced phthoxazolin A at a level comparable to the parental ΔavaR3 mutant strain.
S. avermitilis K139 is one progeny of our wild-type strain (S. avermitilis KA-320, which is isogenic to MA-4680, ATCC 31267 and NRRL 8165), and K139 has been used in the genome sequencing project [18]. The strain K139 also did not produce phthoxazolin A (Fig 1B), suggesting that genetic differences may exist between S. avermitilis KA-320 and K139 to confer ability for the phthoxazolin A biosynthesis.
Identification of the phthoxazolin A biosynthetic gene cluster
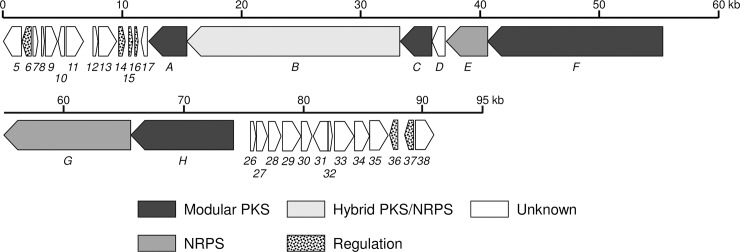
To reveal genetic differences between strains KA-320 and K139, we performed a contour-clamped homogeneous electrical field (CHEF) electrophoresis analysis of these chromosomes, and found that the genome size of strain KA-320 was about 800–1,000 kb larger than that of strain K139 and that the extra region of strain KA-320 was located at the right-hand region of the K139 genome. Based on these results, there was a strong possibility that the extra region of strain KA-320 encodes a biosynthetic gene cluster for phthoxazolin A. Thus, we sequenced the genome of strain KA-320 to find the extra region. By using the same approach as described above, we performed in silico screening with the genome sequencing of S. avermitilis KA-320, and found a possible phthoxazolin A biosynthetic gene cluster that spanned 99.9 kb (Fig 2). Annotation analysis of the sequence and comparison with genes in the public databases revealed 34 ORFs. The genetic organization and proposed functions are shown in Fig 2 and Table 1, respectively.
Fig 2. Genetic organization of the phthoxazolin A biosynthetic gene cluster.
Arrows indicate the direction of transcription and relative gene size. ORFs predicted to participate in phthoxazolin A biosynthesis are shaded. The proposed functions of individual ORFs are indicated here and summarized in Table 1.
Table 1. Deduced functions of ORFs in the phthoxazolin A biosynthetic gene cluster.
| Gene | Sizea | Homologb and origin | Identity/ similarity (%) | Proposed function |
|---|---|---|---|---|
| orf 5 | 504 | GlpK (WP_015654981), Streptomyces davawensis JCM 4913 | 91/95 | Glycerol kinase |
| orf 6 | 255 | ASC56_RS09905 (WP_055490882), Streptomyces sp.TP-A0356 | 92/96 | IclR-family transcriptional regulator |
| orf 7 | 142 | ADL25_RS11400 (WP_059127556), Streptomyces sp. NRRL F-5122 | 67/76 | Histidine kinase |
| orf 8 | 80 | IQ62_RS20385 (WP_037697207), Streptomyces scabiei NCPPB 4086 | 85/91 | Hypothetical protein |
| orf 9 | 333 | Ppk2 (WP_007385561), Streptomyces sviceus ATCC 29083 | 88/93 | Polyphosphate kinase |
| orf 10 | 180 | SAMN05216482_0059 (SEB58800), Streptomyces sp. PAN_FS17 | 78/87 | Hypothetical protein |
| orf 11 | 481 | G412_RS0110405 (WP_02881204), Streptomyces flavidovirens DSM 40150 | 96/98 | Glyceraldehyde 3-phosphate dehydrogenase |
| orf 12 | 137 | NF37_RS0107960 (WP_032755078), Streptomyces alboviridis NRRL B-1579 | 81/91 | Integrase |
| orf 13 | 501 | AWV61_RS50755(WP_060880896), Streptomyces scabiei 95–18 | 83/85 | Transposase |
| orf 14 | 188 | AVL59_RS26005 (WP_067308799), Sreptomyces griseochromogenes ATCC 14511 | 81/86 | Two-component system sensor kinase |
| orf 15 | 113 | CCN44_RS40620 (WP_086704188), Streptomyces tricolor NRRL B-16925 | 86/86 | Two-component system response regulator |
| orf16 | 94 | IG08_RS0113085 (WP_030600335), Streptomyces fulvoviolaceus NRRL B-2870 | 87/88 | LuxR-family transcriptional regulator |
| orf 17 | 169 | BIV24_RS13170 (WP_071366454), Streptomyces sp. MUSC 93 | 74/82 | Polyketide cyclase |
| ptxA | 1065 | OzmM (ABS90474), Streptomyces albus JA3453 | 57/68 | Acetyl transferase |
| ptxB | 5939 | OzmH (ABS90470), S. albus JA3453 | 53/62 | Hybrid NRPS-PKS |
| ptxC | 877 | OzmQ (ABS90478), S. albus JA3453 | 67/76 | Type I PKS |
| ptxD | 362 | OzmP (ABS90477), S. albus JA3453 | 77/88 | Hypothetical protein |
| ptxE | 1154 | OzmO (ABS90476), S. albus JA3453 | 53/62 | NRPS |
| ptxF | 4885 | OzmN (ABS90475), S. albus JA3453 | 51/60 | Type I PKS |
| ptxG | 3542 | NRPS (OMI35273), Streptomyces sparsogenes DSM 40356 | 65/74 | NRPS |
| ptxH | 2860 | PKS 1–1 (ADI03434), Streptomyces bingchenggensis BCW-1 | 63/72 | Type I PKS |
| orf 26 | 155 | SibV (ACN39745), Streptosporangium sibiricum DSM 44039 | 66/77 | Dioxygenase |
| orf 27 | 306 | BZL62_RS04865 (WP_086716558), Streptomyces angustmyceticus NRRL B-2347 | 68/78 | Hypothetical protein |
| orf 28 | 347 | AOK13_RS10670 (WP_055559528), Streptomyces luridiscabiei NRRL B-24455 | 81/87 | IMP dehydrogenase |
| orf 29 | 517 | BZL62_RS04825 (WP_086716551), Streptomyces angustmyceticus NRRL B-2347 | 79/86 | Acetolactate synthase |
| orf 30 | 295 | SAMN05444920_109123 (SEG95653), Nonomuraea solani CGMCC 4.7037 | 55/70 | Hydroxyacid dehydrogenase |
| orf 31 | 410 | BR98_RS37570 (WP_083976095), Kitasatospora azatica KCTC 9699 | 70/82 | Cytochrome P450 |
| orf 32 | 71 | WT80_RS35315 (WP_081087741) Burkholderia stagnalis MSMB774WGS | 43/56 | unknown |
| orf 33 | 526 | KCH_RS21250 (WP_084223811), Kitasatospora cheerisanensis KCTC 2395 | 66/77 | Fatty acid Co A ligase |
| orf 34 | 409 | SAMN05216533_5065 (SEF00159), Streptomyces sp. Ag109_O5-10 | 85/92 | 5-Aminolevulinate synthase |
| orf 35 | 509 | Ann2 (AGY30678), Streptomyces calvus ATCC 13382 | 67/81 | 5-Aminolevulinate synthase |
| orf 36 | 225 | ColR1 (AIL50186), Streptomyces aureus SOK1/5-04 | 50/62 | LuxR-family transcriptional regulator |
| orf 37 | 248 | IF01_RS0119920 (WP_051755722), Streptomyces purpeofuscus NRRL B-1817 | 75/83 | TetR-family transcriptional regulator |
| orf 38 | 516 | OO66_RS31780 (WP_051763676), Streptomyces virginiae NRRL B-8091 | 74/83 | Multidrug MFS (major facilitator superfamily) transporter |
a Numbers refer to amino acid residues.
b Parenthetical codes are National Center for Biotechnology Information accession numbers.
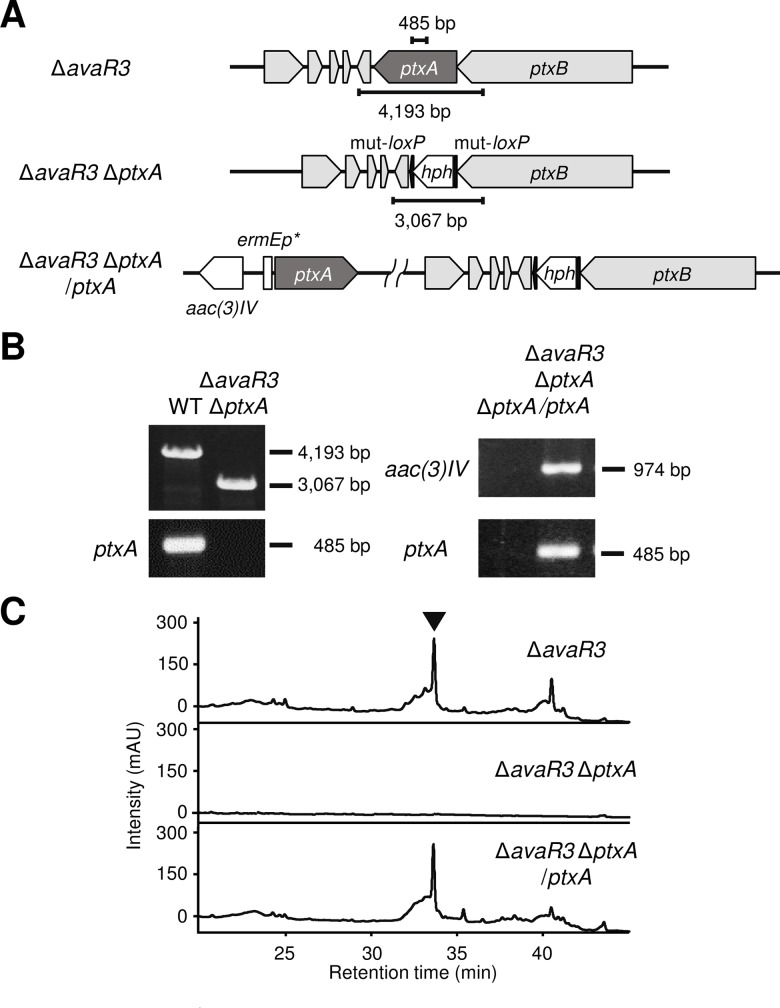
Three PKS genes (ptxC, ptxF, and ptxH), one hybrid NRPS/PKS gene (ptxB), and two NRPS genes (ptxE and ptxG) were identified in the cluster, together with ptxA, which encoded a putative discrete AT enzyme. To investigate the involvement of PtxA in the biosynthesis of phthoxazolin A, we disrupted the ptxA gene by deleting 1,032 amino acids in the ΔavaR3 genetic background. The double mutant (ΔavaR3 ΔptxA) strain was unable to produce phthoxazolin A (Fig 3) and the ptxA-complemented avaR3/ptxA double mutant (ΔavaR3 ΔptxA/ptxA) produced phthoxazolin A to the level in the parental strain (ΔavaR3) (Fig 3), indicating that the discrete AT (PtxA) is essential for the phthoxazolin A biosynthesis. This result also suggested that the PKS genes and the hybrid NRPS/PKS gene, which require enzymatic activity of PtxA, are involved in the biosynthesis of phthoxazolin A (see next section).
Fig 3. Phthoxazolin A production in the avaR3/ptxA double mutant.
(A) Schematic representation of the strategy for the ptxA gene disruption. ΔavaR3, avaR3 mutant; ΔavaR3 ΔptxA, avaR3/ptxA double mutant; ΔavaR3 ΔptxA/ptxA, ptxA-complemented avaR3/ptxA double mutant. (B) PCR analysis to confirm gene-disruption of the ptxA gene and its complementation. With the primer pair ptzA-tFw/ptzA-tRe, a fragment (4,193 bp) containing an intact ptxA gene or a fragment (3,067 bp) containing the mut-loxP-hph-mut-loxP was amplified with PCR. An intact ptxA gene (485 bp) was detected by using the primer pair ptzA-Fw/ptzA-Re. An internal region of aac(3)IV gene (974 bp) was amplified using the primer pair apr-Fw/apr-Re. (C) HPLC chromatograms of MeOH extracts from the avaR3/ptxA double mutant. mAU, milliabsorbance units at 275 nm. Phthoxazolin A is indicated by an inverted triangle.
A gene encoding a discrete AT
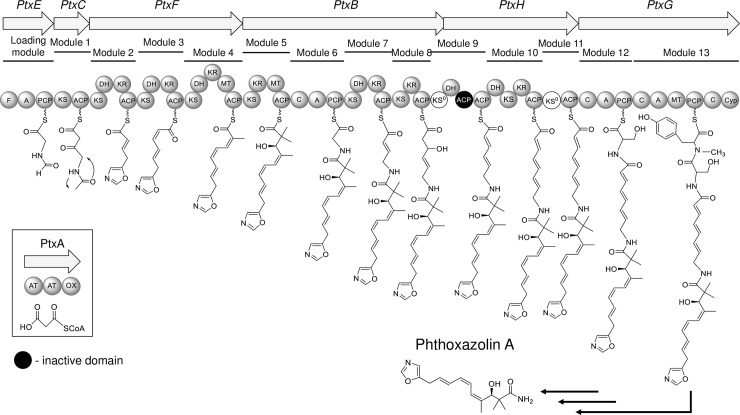
The PtxA protein contains two tandem AT domains together with an oxidoreductase domain, and should function as a discrete AT enzyme in the trans-AT type I PKS gene cluster. In the ptx gene cluster, three PKS genes (ptxC, ptxF, and ptxH) and a hybrid NRPS/PKS gene (ptxB) are embedded to encode 10 PKS modules (modules 1 to 5 and 7 to 11). The PKS modules encoded by ptxB, ptxC, ptxF, and ptxH all lack cognate AT domains, implying that the PtxA protein provides the missing AT activity by acting in trans for these PKS modules. The oxazolomycin pathway has also been established to have the trans-AT type I PKS system with a discrete AT enzyme (OzmM containing the tandem AT domains) [12]. Phylogenetic tree analysis of the AT domains from PtxA (PtxA-AT1 and PtxA-AT2) with other tandem-type discrete AT enzymes revealed that PtxA-AT1 and Ozm-AT1 are positioned in a different clade from PtxA-AT2, Ozm-AT2 and KirC1-AT2 (S2 Fig). Zhao et al. [12] established that OzmM-AT2 is involved in the oxazolomycin biosynthesis, whereas OzmM-AT1 is dispensable. Moreover, KirC1-AT2 loads malonyl-CoA extender units to the ACPs in the kirromycin biosynthesis [26]. With the observation that PtxA-AT1 seems inactive due to the replacement of important amino acid residues [27] (substitution of Glu63 and His91 with His and Ser, respectively), much as in the cases of OzmM-AT1 and KirC1-AT1, it can be concluded that PtxA-AT2 supplies malonyl-CoA units to the Ptx PKS modules.
Genes involved in the polyketide assembly
Ten PKS modules were identified in the ptx gene cluster, encoded by ptxB, ptxC, ptxF, and ptxH. PtxC and PtxF closely resembled OzmQ (67% identity) and OzmN (51% identity), respectively. Both OzmQ and OzmN have PKS modules 2, 3, 4, and 5 in the oxazolomycin assembly line, and render a triene moiety in the structure of oxazolomycin [12]. PKS modules 4 and 5 of PtxB contain two methyltransferase (MT) domains for the addition of methyl groups at C-2 and C-4 of phthoxazolin A.
With respect to the KS domains in the ptx cluster, analysis of the conserved active site (catalytic triad of Cys-His-His) [28] revealed that two KS domains (KS9 and KS11) contain a mutation in the conserved motif (S3 Fig), suggesting that they should be inactive in the polyketide assembly line; these domains were called KS0 domains [14]. Regarding the ACP domains, the Ser residue in the signature motif plays a crucial role as the 4’-phosphopantetheine attachment site [29], but the first ACP domain (ACP9a) of module 9 lacks the Ser residue, while other ACP domains in the Ptx PKS modules retain the conserved Ser residue (S4 Fig), indicating that the PKS module 9 employs two types of ACPs (inactive-type and active-type). Along with the ketoreductase (KR) domains, seven KR domains are predicted in the PKS modules, all of which show a characteristic Rossmann fold for NADP(H)-binding [30] (S5 Fig). In addition, all the KR domains except KR4 and KR5 contain a typical catalytic triad (Ser-Tyr-Asn) [30], while KR4 and KR5 have different sequences (Ser-Tyr-Cys for KR4 and Ser-Tyr-Ser for KR5). Because these minor modifications in the catalytic triad have also been identified in other trans-AT type I PKSs, such as those in OocL and OocR in the oocydin A biosynthesis [31], all the ptx KR domains seem to be active in the PKS assembly line. Regarding the DH domains, all six DH domains possess the conserved His residue in the signature motif HXXXGXXXXP [32] (S6 Fig), indicating that the dehydratase (DH) domains are active, and are likely responsible for the formation of a double bond. Taking these results together, the first 5 PKS modules (modules 1 to 5) appear to be responsible for synthesizing the polyketide moiety of phthoxazolin A, whereas the remaining 5 PKS modules (modules 7, 8, 9, 10, and 11) might be capable of synthesizing an additional triene structure (as discussed later).
Genes involved in the formation of an oxazole ring
The ptxE gene encodes a single module of NRPS comprising a formylation (F) domain, an A domain, and a PCP domain, which probably serve as loading modules to incorporate an initial amino acid into the assembly line. Because the PtxE-A domain is predicted to be a glycine-specific A domain based on bioinformatic analysis using the AntiSMASH database [33], glycine can be considered to be activated by the PtxE-A domain and loaded onto the PtxE-P domain as a glycyl-S-PCP. The presence of an F domain in the loading module has sometimes been identified in other biosynthetic machineries, such as those for the biosynthesis of gramicidin [34], rhizopodin [35], oxazolomycin [12], and conglobatin [11], and the PtxE-F domain can be predicted to catalyze formylation of glycyl-S-PCP to generate formyl-glycyl-S-PCP, from the analogy of the F domain of LgrA1 in the gramicidin biosynthesis.
The downstream region of the ptxE gene includes the ptxD gene, which encodes a protein homologous to OzmP (in oxazolomycin biosynthesis) and CongE (in conglobatin biosynthesis). The gene arrangement (ptxD-ptxE) is identical to those in the biosynthetic gene clusters of oxazolomycin (ozmO-ozmP) and conglobatin (congA-congE). Moreover, these three proteins (PtxD, CongE, and OzmP) have an ATP-pyrophosphatase domain that includes the signature motif SGGKDS in the N-termini for ATP binding [36]. Because CongE has been proposed to activate the amide oxygen by adenyltransfer from ATP with the release of pyrophosphate for formation of the oxazole ring moiety [11], PtxD is likely to be responsible for conversion of the formyl-glycyl intermediate into the oxazole ring by cyclodehydration.
Genes putatively involved in the nonribosomal peptide assembly
The assembly line in the ptx gene cluster possesses four NRPS modules, including a loading module encoded by ptxE. NRPS module 6 is a part of the PKS/NRPS hybrid protein PtxB, and NRPS modules 12 and 13 are encompassed in the PtxG protein. The specificity-conferring codes of the A domains indicated that amino acids activated by PtxB-A6, PtxG-A12 and PtxG-A13 are glycine, serine and tyrosine, respectively. The glycine residue recognized by PtxB-A6 would be incorporated into the structure of phthoxazolin A as a part of a carbamoyl moiety. The terminal module 13 of the PtxG protein includes an N-MT domain and a cytochrome P450 domain. The MT domain probably modifies the Tyr residue incorporated by PtxG-A13. Because the NRPS module 6 is likely to be a final module in the phthoxazolin A assembly line, it is difficult to predict the involvement of the NRPS modules 12 and 13 in phthoxazolin A biosynthesis at present.
Proposed model for phthoxazolin A biosynthesis
The in silico analyses of the Ptx proteins described above allowed us to propose a model for the phthoxazolin A biosynthesis (Fig 4). Phthoxazolin A biosynthesis starts with activation of glycine and the tethering to the PCP domain of PtxE, followed by formylation by the F domain to generate a formyl-glycyl-S-PCP. PKS module 1 of PtxC, representing a minimal module of the trans-AT type I PKS systems, incorporates a malonyl-CoA into the formyl-glycyl intermediate. The cyclodehydration by PtxD on the intermediate would occur after the condensation between the formyl-glycyl-S-PCP and the malonyl-CoA, although the timing of cyclodehydration remains unclear. Subsequently, four malonyl-CoA units and three methyl groups are incorporated by four PKS modules (modules 2 to 5) of PtxF and PtxB. The KR-DH domain pair in the modules 2, 3 and 4 provides three conjugated double bonds (C8-C9, C6-C7 and C4-C5), and generates the trans, cis, and cis configuration of double bonds, respectively. KR5 of PKS module 5 could be classified as a KR domain found in the partially reducing PKS [37], and introduces a hydroxyl group in the R configuration at C-3 of phthoxazolin A. NRPS module 6 activates a glycine residue, and loads it onto the polyketide intermediate to generate a carbamoyl moiety. The remaining five PKS modules could produce a triene moiety, and two NRPS modules could incorporate two additional amino acids (Ser and N-Tyr) into the polyketide assembly line. However, considering the structure of phthoxazolin A, these additional substructures are unnecessary for the biosynthesis of phthoxazolin A. The thioesterase (TE) domain usually catalyzes the release of a completed chain of PKS/NRPS products from the assembly line [38], but no such TE domain was found in the ptx assembly line. Finally, the product generated by the ptx chain assembly should be processed by some enzymatic reactions to complete phthoxazolin A biosynthesis. To date, however, no clear candidate enzyme has been found in the ptx gene cluster or its adjacent regions.
Fig 4. Proposed model for phthoxazolin A biosynthesis.
A, adenylation; ACP, acyl carrier protein; AT, acyltransferase; C, condensation; Cyp, cytochrome P450; DH, dehydratase; F, formylation; KS, ketosynthase; KS0, KS lacking His in the HTGTG motif; KR, ketoreductase; MT, methyltransferase; PCP, peptidyl carrier protein. The presumed inactive ACP domain of module 9 is shaded in black.
Discussion
The oxazole-containing polyketides have the distinct feature of an oxazole-moiety linked with a polyketide structure or a hybrid peptide-polyketide structure, and bestow remarkable biological activities such as antibacterial, antitumor and herbicidal properties. Only a few biosynthetic gene clusters of oxazole-containing polyketides have been identified [10–12,35,39] and their biosynthetic mechanisms have provided us valuable genetic information of the complicated pathways involved in heterocycles formation and sequential extensions by PKSs or PKS/NRPSs. Recently, we identified phthoxazolin A, which is a polyketide compound containing a 5-substituted oxazole ring and a triene moiety, as a cryptic metabolite of the original avermectin producer S. avermitilis KA-320 [18]. In the present study, we have shown that strain KA-320 harbors an extra genomic region, which is absent in the publicly available genome of strain K139, and that a biosynthetic gene cluster for phthoxazolin A is laid on the extra region of strain KA-320. The biosynthetic gene cluster could be classified into a growing number of trans-AT type I PKS systems that utilize a discrete AT enzyme to supply a malonyl-CoA unit into all the PKS modules as opposed to the cis-acting integrated AT domains of canonical PKSs [13,14].
The structure of phthoxazolin A resembles the partial structure of oxazolomycin. Comparison of the functional domains of the biosynthetic gene clusters revealed a virtually complete architectural identity with the corresponding portion of the ptx and ozm PKS-NRPS clusters over the first eight modules, with the exception of a dispensable module (PKS module 7) for the phthoxazolin A biosynthesis. The Ptx proteins are 51%-71% identical to their orthologues in the ozm cluster. Similar to the oxazolomycin biosynthetic pathway, the phthoxazolin A biosynthesis is initiated by formylation of the glycine residue, followed by extension by a malonyl-CoA unit and cyclodehydration to generate an oxazole ring moiety. The biosynthetic maturation of phthoxazolin A should require additional extension by three malonyl-CoA units to yield a triene moiety and termination by glycine incorporation to generate a carbamoyl moiety. The geometries of the conjugated triene moiety of phthoxazolin A are assigned as 4Z,6Z,8E. A PKS module encompassing the pair of a B-type KR domain and a DH domain normally biosynthesize polyketide chain elongation intermediates with a trans (E) double bond [30]. Sequence analysis of KR domains in the ptx cluster demonstrated that KR2, KR3, and KR4 contain the stereochemistry signature “LDD” motif for B-type KR domains [40, 41], although their motifs vary among the individual KR domains. These analyses indicated that the polyketide intermediate of phthoxazolin A is expected to have three trans conjugated double bonds, which are different from those of phthoxazolin A. Whether the change of stereochemistry in the biosynthetic process requires an enzymatic reaction remains obscure. However, this contradictory feature for the formation of conjugated double bonds has also been observed in the biosynthetic pathways of oxazolomycin and chivosazol [12,39].
The ptx gene cluster encodes a series of modules indispensable (from the loading module to NRPS module 6) for the phthoxazolin A biosynthesis, but also includes additional functional modules (PKS module 7 to NRPS module 13) for the biosynthesis of an unidentified compound, which might be a precursor of phthoxazolin A. In particular, PKS modules 8 and 9 include the interesting feature of a non-elongating KS (KS0) domain. These modules are composed of KS-KR-ACP-KS0 domains followed by DH-ACP domains, and are grouped as type A bimodules for dehydration [14,42] In addition, the domain pair of KS0-ACP as a non-elongating module is also embedded in PKS module 11. Both the type A bimodule for dehydration and the domain pair of KS0-ACP are ubiquitously found in the trans-AT type I PKS system [14]. The remaining modules (PKS modules 10 and 11, and NRPS modules 12 and 13) also might be responsible for a polyketide elongation and an incorporation of amino acids, as well as those of the oxazolomycin biosynthesis, suggesting that the ptx gene cluster would produce a larger compound than that of phthoxazolin A; namely, phthoxazolin A is a cleaved compound of the ancestral compound. HPLC analysis of the avaR3/ptxA double mutant demonstrated that, in addition to phthoxazolin A, the strain lost production of several more compounds that are present in the parental strain (Fig 3C): any of the compounds can be the ancestral compounds synthesized by the ptx gene cluster. In the pederin biosynthetic pathway, a putative FAD-dependent monooxygenase (PedG) has been proposed to oxidatively cleave the final product generated by the whole assembly line and produce a pederin precursor [43]. On the other hand, the ptx gene cluster and its flanking regions have no genes encoding an FAD-dependent monooxygenase, suggesting that another cleavage mechanism might be applied to the synthesis of phthoxazolin A.
In conclusion, we have shown that an extra genomic region of the original avermectin producer has a unique trans-AT type I PKS system for the biosynthesis of phthoxazolin A. These findings could provide further insight into the diversity of trans-AT type I PKS systems, which are widely distributed in bacteria [6,14]. Moreover, our finding of additional PKS/NRPS modules in the ptx assembly line suggests that S. avermitilis is capable of producing a larger intermediate than phthoxazolin A, and that the PKS/NRPS machinery incorporates a new cleavage system. Further understanding of the phthoxazolin A biosynthetic pathway should provide interesting perspectives into the engineering of polyketide/non-ribosomal peptide biosynthetic pathways.
Supporting information
(A) Schematic representation for the construction of Streptomyces avermitilis large-deletion (SALD) mutant strains. The details of the construction are described in the Materials and Methods. Gray solid line represents the extra genomic region of S. avermitilis KA-320, and gray dashed lines represent the deleted DNA region in the genome. KA-320, S. avermitilis KA-320; K139, S. avermitilis K139; SUKA22, S. avermitilis SUKA22 (K139 as a genetic background). (B) HPLC chromatograms of MeOH extracts from S. avermitilis SALD mutants. KA-320/ΔavaR3, S. avermitilis ΔavaR3 (KA-320 as genetic background). mAU, milliabsorbance units at 275 nm. Phthoxazolin A is indicated by an inverted triangle.
(PDF)
BaeC (CAG23950), BaeD (CAG23951), and BaeE (CAG23952) from Bacillus amyloliquefaciens FZB42; BatH (ADD82949) and BatJ (ADD82951) from Pseudomonas fluorescens BCCM_ID9359; BryP (ABK51299) from Candidatus endobugula sertula; DifA (CAG23974) from Bacillus amyloliquefaciens FZB42; DszD (AAY32968) from Sorangium cellulosum So ce12; ElsA (WP_012792904) and ElsB (WP_012792903) from Chitinophaga pinensis DSM 2588; KirC1 (CAN89639) from Streptomyces collinus Tu 365; LnmG (AAN85520) from Streptomyces atroolivaceus; LkcD (BAC76473) from Streptomyces rochei; MmpC (AAM12912) from P. fluorescens NCIMB 10586; OzmM (ABS90474) from S. albus JA3453 and OzmM (ADI12766) from Streptomyces bingchenggensis BCW-1; PedC (AAS47559) and PedD (AAS47563) from symbiont bacterium of Paederus fuscipes; PsyH (ADA82589) from an unculturable symbiont of sponge Psammocinia aff. Bulbosa; RhiG (CAL69887) from Burkholderia rhizoxina; SorO (ADN68489) from S. cellulosum So ce12; VirI (BAF50719) from Streptomyces virginiae; FabD (CAA77658) form E. coli; and FabD SAV5788 (BAC73500) from S. avermitilis.
(PDF)
The conserved catalytic triad of C-H-H is marked with an asterisk. The numbers indicate amino acid positions within each domain.
(PDF)
The Ser residue functioning as the phosphopantetheine-binding site is marked with an asterisk. The numbers indicate amino acid positions within each domain.
(PDF)
The conserved catalytic residues are marked with asterisks. The core region for the NADP(H)-binding motif is underlined. The numbers indicate amino acid positions within each domain. The “LDD” motif for B-type KR domains is shown in a box.
(PDF)
The proposed active catalytic His residue is marked with an asterisk. The numbers indicate amino acid positions within each domain.
(PDF)
(PDF)
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This study was supported by the Japan Society for the Promotion of Science (JP15K07358, http://www.jsps.go.jp/index.html, SK); and the Japan Association for Chemical Innovation (New Chemical Technology Research Encouragement Award, http://jaci.or.jp/, SK). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Staunton J, Weissman K J. Polyketide biosynthesis: a millennium review, Nat Prod Rep. 2001; 18: 380–416. [DOI] [PubMed] [Google Scholar]
- 2.Kopp F, Marahiel MA. Where chemistry meets biology: the chemoenzymatic synthesis of nonribosomal peptides and polyketides. Curr Opin Biotechnol. 2007; 18: 513–520. doi: 10.1016/j.copbio.2007.09.009 [DOI] [PubMed] [Google Scholar]
- 3.Hopwood D A. Genetic contributions to understanding polyketide synthases. Chem Rev. 1997; 97: 2465–2497. [DOI] [PubMed] [Google Scholar]
- 4.Katz L. Manipulation of modular polyketide synthases. Chem Rev. 1997; 97: 2557–2575. [DOI] [PubMed] [Google Scholar]
- 5.Finking R, Marahiel MA. Biosynthesis of nonribosomal peptides. Annu Rev Microbiol. 2004; 58: 453–88. doi: 10.1146/annurev.micro.58.030603.123615 [DOI] [PubMed] [Google Scholar]
- 6.Wang H., Fewer DP, Holm L, Rouhiainen L, Sivonen K. Atlas of nonribosomal peptide and polyketide biosynthetic pathways reveals common occurrence of nonmodular enzymes. Proc Natl Acad Sci. USA. 2014; 111: 9259–9254. doi: 10.1073/pnas.1401734111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fisch KM. Biosynthesis of natural products by microbial iterative hybrid PKS–NRPS. RSC Adv. 2013; 3: 18228–18247. [Google Scholar]
- 8.Mori T, Takahashi K, Kashiwabara M, Uemura D. Structure of oxazolomycin, A novel β–lactone antibiotic. Tetrahedron Letters. 1985; 26: 1073–1076. [Google Scholar]
- 9.Grafe U, Kluge H, Thiericke R. Biogenetic studies on oxazolomycin, a metabolite of Streptomyces albus (strain JA3453). Liebigs. Ann. Chem. 1992; 429–432. [Google Scholar]
- 10.Partida-Martinez L, Hertweck C. A gene cluster encoding rhizoxin biosynthesis in “Burkholderia rhizoxinia”, the bacterial endosymbiont of the fungus Rhizopus microsporus. ChemBioChem. 2007; 8: 41–45. doi: 10.1002/cbic.200600393 [DOI] [PubMed] [Google Scholar]
- 11.Zhou Y, Murphy AC, Samborskyy M, Prediger P, Dias LC, Leadlay PF. Iterative mechanism of macrodiolide formation in the anticancer compound conglobatin. Chem Biol. 2015; 22: 745–754. doi: 10.1016/j.chembiol.2015.05.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhao C, Coughlin JM, Ju J, Zhu D, Wendt-Pietkowski E, Zhou X, et al. Oxazolomycin biosynthesis in Streptomyces albus JA3453 featuring an”acyltransferase-less” type I polyketide synthase that incorporated two distinct extender unit. J Biol Chem. 2010; 285: 20097–20108. doi: 10.1074/jbc.M109.090092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cheng YQ, Coughlin JM, Lim SK, Shen B. Type I polyketide synthases that require discrete acyltransferases In Hopwood DA (Editor) Methods in Enzymology vol. 459 Elsevier Inc.; 2009:165–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Helfrich EJN, Piel J. Biosynthesis of polyketides by trans-AT polyketide synthases. Nat Prod Rep. 2016; 33: 231–316. doi: 10.1039/c5np00125k [DOI] [PubMed] [Google Scholar]
- 15.Tanaka Y, Kanaya I, Shiomi K, Tanaka H, Omura S. Phthoxazolin A, a specific inhibitor of cellulose biosynthesis from microbial origin: Isolation, physico-chemical properties, and structure elucidation. J Antibiot (Tokyo). 1993; 46: 1214–1218. [DOI] [PubMed] [Google Scholar]
- 16.Tanaka Y, Sugoh M, Ji W, Iwabuchi J, Yoshida H, Omura S. Screening method for cellulose biosynthesis inhibitor with herbicidal activity. J Antibiot (Tokyo). 1995; 48: 720–724. [DOI] [PubMed] [Google Scholar]
- 17.Omura S, Tanaka Y, Kanaya I, Shinose M, Takahashi Y. Phthoxazolin, a cellulose biosynthesis, produced by a strain of Streptomyces sp. J Antibiot (Tokyo). 1990; 43: 1034–1036. [DOI] [PubMed] [Google Scholar]
- 18.Suroto DA, Kitani S, Miyamoto KT, Sakihama Y, Arai M, Ikeda H, et al. Activation of cryptic phthoxazolin A production in Streptomyces avermitilis by the disruption of autoregulator-receptor homologue AvaR3. J. Biosc Bioeng. 2017; 124: 611–617. https://doi.org/10.1016/j.jbiosc.2017.06.014 [DOI] [PubMed] [Google Scholar]
- 19.Ikeda H, Shin-ya K, Omura S. Genome mining of Streptomyces avermitilis genome and development of genome-minimized host for heterologous expression of biosynthetic gene clusters. J Ind Microbiol Biotechnol. 2014; 41: 233–250. doi: 10.1007/s10295-013-1327-x [DOI] [PubMed] [Google Scholar]
- 20.Miyamoto KT, Kitani S, Komatsu M, Ikeda H, Nihira T. The autoregulator receptor homologue AvaR3 plays a regulatory role in antibiotics production, mycelial aggregation and colony development of Streptomyces avermitilis. Microbiology. 2011; 157: 2266–2275. doi: 10.1099/mic.0.048371-0 [DOI] [PubMed] [Google Scholar]
- 21.Ikeda H, Kotaki H, Omura S. Genetic studies of avermectin biosynthesis in Streptomyces avermitilis. J. Bacteriol. 1987; 169: 5612–5621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Komatsu M, Komatsu K, Koiwai H, Yamada Y, Kozone I, Izumikawa M, et al. Engineered Streptomyces avermitilis host for heterologous expression of biosynthetic gene cluster for secondary metabolites. ACS Synth. Biol. 2013; 2: 384–396. doi: 10.1021/sb3001003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Komatsu M, Uchiyama T, Omura S, Cane DE, Ikeda H. Genome-minimized Streptomyces host for the heterologous expression of secondary metabolism. Proc Nat Acad Sci USA. 2010; 107: 7422–7427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Omura S, Ikeda H, Ishikawa J, Hanamoto A, Takahashi C, Shinose M, et al. Genome sequence of an industrial microorganism Streptomyces avermitilis: Deducing the ability of producing secondary metabolites. Proc Nat Acad Sci USA. 2001; 98: 12215–12220. doi: 10.1073/pnas.211433198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pulsawat N, Kitani S, Fukushima E, Nihira T. Hierarchical control of virginiamycin production in Streptomyces virginiae by three pathway-specific regulators: VmsS, VmsT and VmsR. Microbiology. 2009; 155: 1250–1259. doi: 10.1099/mic.0.022467-0 [DOI] [PubMed] [Google Scholar]
- 26.Musiol EM, Greule A, Härtner T, Kulik A, Wohlleben W, Weber T. The AT2 domain of KirCI loads malonyl extender units to the ACPs of the kirromycin PKS. ChemBioChem. 2013; 14: 1343–1352. doi: 10.1002/cbic.201300211 [DOI] [PubMed] [Google Scholar]
- 27.Yadav G, Gokhale RS, Mohanty D. Computational approach for prediction of domain organization and substrate specificity of modular polyketide synthases. J Mol Biol. 2003; 328: 335–363. [DOI] [PubMed] [Google Scholar]
- 28.He M, Varoglu M, Sherman DH. Structural modeling and site-directed mutagenesis of the actinorhodin β-ketoacyl-acyl carrier protein synthase. J Bacteriol. 2000; 182: 2619–2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lai JR, Koglin A, Walsh CT. Carrier protein structure and recognition in polyketide and nonribosomal peptide. Biochemistry. 2006; 45: 14869–14879. doi: 10.1021/bi061979p [DOI] [PubMed] [Google Scholar]
- 30.Reid R, Piagentini M, Rodriguez E, Ashley G, Visvanathan N, Carney J, et al. A model of structure and catalysis for ketoreductase domains in modular polyketide synthases. Biochemistry. 2003; 42: 72–79. doi: 10.1021/bi0268706 [DOI] [PubMed] [Google Scholar]
- 31.Matilla MA, Stöckmann H, Leeper F J, Salmond GPC. Bacterial biosynthetic gene cluster encoding the anti-cancer haterumalide class of molecules. Biogenesis of the broad spectrum antifungal and anti-oomycete compound, oocydin A. J Biol Chem. 2012; 287: 39125–39138. doi: 10.1074/jbc.M112.401026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu J, Zalesky TJ, Valenzano C, Koshla C, Cane DE. Polyketide double bond biosynthesis. Mechanistic analysis of the dehydratase-containing module 2 of the picromycin /methymycin polyketide synthase. J Am Chem Soc. 2005; 127: 17393–17404. doi: 10.1021/ja055672+ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Weber T, Blin K, Duddela S, Krug D, Kim HU, Bruccoleri R, et al. antiSMASH 3.0—a comprehensive resource for the genome mining of biosynthetic gene clusters, Nucleic Acids Res. 2015; 43: W237–W243. doi: 10.1093/nar/gkv437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schoenafinger G, Schracke N, Linne U, Marahiel MA. Formylation domain: an essential modifying enzyme for the nonribosomal biosynthesis of linear gramicidin. J Am Chem Soc. 2006; 128: 7406–7407. doi: 10.1021/ja0611240 [DOI] [PubMed] [Google Scholar]
- 35.Pistorius D, Müller R. Discovery of the rhizopodin biosynthetic gene cluster in Stigmatella aurantiaca Sg a15 by genome mining. ChemBioChem. 2012; 13: 416–426. doi: 10.1002/cbic.201100575 [DOI] [PubMed] [Google Scholar]
- 36.Bork P, Koonin EV. A P-loop like motif in a widespread ATP pyrophosphatase domain: Implications for the evolution of the sequence motifs and enzyme activity. Protein. 1994; 20: 347–355. [DOI] [PubMed] [Google Scholar]
- 37.Soehano I, Yang L, Ding F, Sun H, Low ZJ, Liu X, et al. Insight into the programmed ketoreductions of partially reducing polyketide synthase: stereo-and substrate-specificity of the ketoreductase domain. Chem Sci. 2012; 00: 1–7. [DOI] [PubMed] [Google Scholar]
- 38.Du L, Lou L. PKS and NRPS release mechanism. Nat Prod Rep. 2010; 27: 255–278. doi: 10.1039/b912037h [DOI] [PubMed] [Google Scholar]
- 39.Perlova O, Gerth K, Kaiser O, Hans A, Müller R. Identification and analysis of chivosazol biosynthetic gene cluster from myxobacterial model strain Sorangium cellulosum Soce56. J Biotechnol. 2006; 121: 174–191. doi: 10.1016/j.jbiotec.2005.10.011 [DOI] [PubMed] [Google Scholar]
- 40.Caffrey P. Conserved amino acid residues correlating with ketoreductase stereo specificity in modular polyketide synthases. ChemBioChem. 2003; 4: 649–662. [DOI] [PubMed] [Google Scholar]
- 41.Keatinge-Clay AT. A tylosin ketoreductase reveals how chirality is determined in polyketides. Chem Biol. 2007; 14: 898–908. doi: 10.1016/j.chembiol.2007.07.009 [DOI] [PubMed] [Google Scholar]
- 42.Wagner DT, Zeng J, Bailey CB, Gay DC, Yuan F, Manion HR, et al. Structural and functional trends in dehydrating bimodules from trans-acyltransferase polyketide synthases. Structure. 2017; 25: 1045–1045. doi: 10.1016/j.str.2017.05.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Piel J. A polyketide synthase-peptide synthetase gene cluster from uncultured bacterial symbiont of Paederus beetles. Proc Natl Acad Sci USA. 2002; 99: 14002–14007. doi: 10.1073/pnas.222481399 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) Schematic representation for the construction of Streptomyces avermitilis large-deletion (SALD) mutant strains. The details of the construction are described in the Materials and Methods. Gray solid line represents the extra genomic region of S. avermitilis KA-320, and gray dashed lines represent the deleted DNA region in the genome. KA-320, S. avermitilis KA-320; K139, S. avermitilis K139; SUKA22, S. avermitilis SUKA22 (K139 as a genetic background). (B) HPLC chromatograms of MeOH extracts from S. avermitilis SALD mutants. KA-320/ΔavaR3, S. avermitilis ΔavaR3 (KA-320 as genetic background). mAU, milliabsorbance units at 275 nm. Phthoxazolin A is indicated by an inverted triangle.
(PDF)
BaeC (CAG23950), BaeD (CAG23951), and BaeE (CAG23952) from Bacillus amyloliquefaciens FZB42; BatH (ADD82949) and BatJ (ADD82951) from Pseudomonas fluorescens BCCM_ID9359; BryP (ABK51299) from Candidatus endobugula sertula; DifA (CAG23974) from Bacillus amyloliquefaciens FZB42; DszD (AAY32968) from Sorangium cellulosum So ce12; ElsA (WP_012792904) and ElsB (WP_012792903) from Chitinophaga pinensis DSM 2588; KirC1 (CAN89639) from Streptomyces collinus Tu 365; LnmG (AAN85520) from Streptomyces atroolivaceus; LkcD (BAC76473) from Streptomyces rochei; MmpC (AAM12912) from P. fluorescens NCIMB 10586; OzmM (ABS90474) from S. albus JA3453 and OzmM (ADI12766) from Streptomyces bingchenggensis BCW-1; PedC (AAS47559) and PedD (AAS47563) from symbiont bacterium of Paederus fuscipes; PsyH (ADA82589) from an unculturable symbiont of sponge Psammocinia aff. Bulbosa; RhiG (CAL69887) from Burkholderia rhizoxina; SorO (ADN68489) from S. cellulosum So ce12; VirI (BAF50719) from Streptomyces virginiae; FabD (CAA77658) form E. coli; and FabD SAV5788 (BAC73500) from S. avermitilis.
(PDF)
The conserved catalytic triad of C-H-H is marked with an asterisk. The numbers indicate amino acid positions within each domain.
(PDF)
The Ser residue functioning as the phosphopantetheine-binding site is marked with an asterisk. The numbers indicate amino acid positions within each domain.
(PDF)
The conserved catalytic residues are marked with asterisks. The core region for the NADP(H)-binding motif is underlined. The numbers indicate amino acid positions within each domain. The “LDD” motif for B-type KR domains is shown in a box.
(PDF)
The proposed active catalytic His residue is marked with an asterisk. The numbers indicate amino acid positions within each domain.
(PDF)
(PDF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.