Abstract
Glycosyltransferases are powerful tools for the synthesis of complex and biologically important carbohydrates. Wild-type glycosyltransferases may not have all the properties and functions that are desired for large-scale production of carbohydrates that exist in nature and those with non-natural modifications. With the increasing availability of crystal structures of glycosyltransferases, especially those in the presence of donor and acceptor analogs, crystal structure-guided rational design has been quite successful in obtaining mutants with desired functionalities. With current limited understanding of the structure-activity relationship of glycosyltransferases, directed evolution continues to be a useful approach for generating additional mutants with functionality that can be screened for in a high-throughput format. Mutating the amino acid residues constituting or close to the substrate binding sites of glycosyltransferases by structure-guided directed evolution further explores the biotechnological potential of glycosyltransferases that can only be realized through enzyme engineering. This mini-review discusses the progress made towards glycosyltransferase engineering and the lessons learned for future engineering efforts and assay development.
Keywords: carbohydrate, directed evolution, enzymatic synthesis, glycosyltransferase, mutagenesis, protein engineering
Introduction
A growing appreciation for the important biological roles and the therapeutic potential of carbohydrates has resulted in an increasing interest in their synthesis. Many advances have been made in chemical glycosylation [1–6]. With an improved understanding of their catalytic mechanisms, enzyme-catalyzed processes using glycosidases, trans-glycosidases, and glycosidase-derived glycosynthases have been developed successfully [7–9]. Glycosyltransferases (GTs), the key enzymes used by nature in producing diverse carbohydrate-containing structures, continue to be important catalysts for accessing complex oligosaccharides and glycoconjugates as well as for glycan modification of glycoconjugates and cell surface glycans [10, 11]. With the elucidation of the genomic sequences of an increasing number of organisms and the continuous effort on functional genomics studies, additional GTs from different species are becoming available. The combination of GTs and sugar nucleotide biosynthetic enzymes in one-pot systems provide facile synthetic routes to naturally existing and non-natural carbohydrates including those with complex structures [6, 11–18].
As with all classes of enzymes, wild-type GTs often do not display the full range of desirable properties for the synthesis of target products. These ideal properties of GTs include: 1) high expression level, preferably in a simple expression system such as Escherichia coli; 2) stability during storage and application; 3) high activity; 4) high stereo- and regio-selectivity; 5) minimal undesirable activities such as donor substrate hydrolysis; and 6) promiscuity towards modified substrates for producing desired analogs. All of these properties can be improved by using enzyme engineering techniques to generate mutants more suitable for the desired application.
The availability of an increasing number of crystal structures of GTs, especially those in the presence of both donor and acceptor analogs, not only provides a better understanding of their catalytic mechanisms [19, 20], but also greatly facilitates rational engineering. For example, some bacterial GTs have been shown to possess side activities that are undesirable for the formation of glycosidic bonds. Crystal structure-based rational design has been used to successfully minimize these side activities [21, 22].
Nevertheless, our understanding of the structure-activity relationship of GTs is still limited. Therefore, directed evolution with effective screening strategies has been shown to be a useful approach for generating mutants that cannot currently be obtained by rational design. Structure-guided directed evolution (SGDE) approaches such as iterative saturation mutagenesis (ISM) [23] combine the predictive elements of rational design with the empirical strategy of directed evolution by specifically randomizing residues most likely to impact the property of interest. SGDE minimizes the cost and necessary throughput of screening and enables more thorough investigation of key residues.
Many potential synthetic applications of GTs, such as glycan remodeling of glycoconjugates, glyco-diversification of small molecules, and efficient combinatorial carbohydrate synthesis will likely require engineered GTs. This mini-review highlights the efforts to engineer GTs for improved or new activities by rational design and directed evolution, as well as additional screening methods that may be applied for high-throughput screening of GTs in future engineering projects.
Structure-based rational design of glycosyltransferases
The Carbohydrate Active enZyme (CAZy) database (http://www.cazy.org) [24, 25] contains over 200,000 GT sequences comprising 94 GT families (excluding GT36, GT46, and GT86 which have been renamed, deleted, or merged) categorized based on protein sequence similarities and close to 3600 non-classified entries (accessed in August 2015). Among these, more than 1,900 GTs have been characterized and structures of 155 from 41 GT families have been reported. Nucleotide sugar-dependent GTs, the major GTs used for enzymatic and chemoenzymatic synthesis of carbohydrates, predominantly adopt one of two structural folds, GT-A [26] or GT-B [27] (Figure 1) [19]. This wealth of sequence and structural information has enabled successful rational design of many mutants with improved or altered functions for GTs that belong to either GT-A or GT-B fold. The following examples are meant to provide insight into sequence and structure-based rational design for GT engineering. A comprehensive list of known examples is beyond the scope of this mini-review.
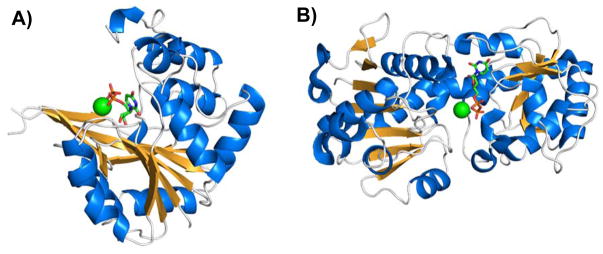
Figure 1.
Structures of (A) GT-A-fold nucleotide-diphosphate-sugar transferase SpsA from Bacillus subtilis (PDB ID 1QGQ) [26] and (B) GT-B-fold bacteriophage T4 β-glucosyltransferase (PDB ID 1JG7) [27]. The magnesium cation (green sphere) and UDP (stick model) in the active site are shown.
The structure of GT-A-fold GTs is usually presented as an open twisted β-sheet surrounded by α-helices on both sides. β-Strands throughout the protein sequence contribute to the continuous central β-sheet, resulting in what appears to be a compact, single-domain Rossmann-like structure [19, 28]. Examples of rationally designed GT-A-fold GTs include β1–4-galactosyltransferase 1 (β4GalT1), human blood group A and B GTs, and β1–3-glucuronyltransferase-I (β3GlcAT-I) (Figure 2) as discussed below.
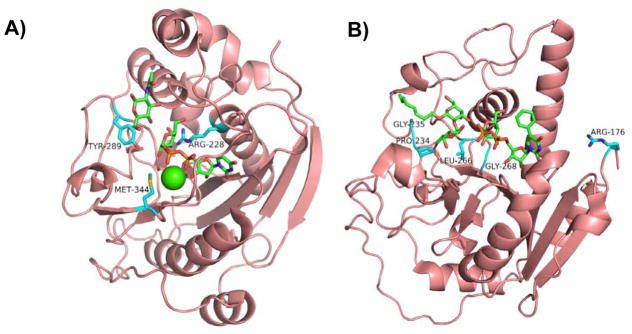
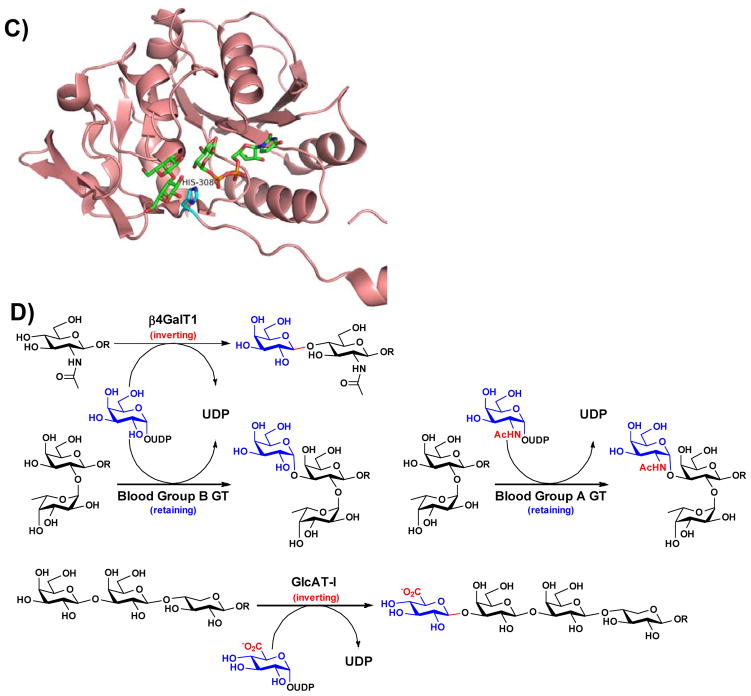
Figure 2.
Structures of representative GT-A-fold enzymes (A–C) highlighting the residues known to influence their substrate specificities as well as reactions (D) catalyzed by these enzymes. (A) Bovine β4GalT1 (PDB IDs: 1NQI and 2FYC) with GlcNAc, UDP-Gal (1), and Ca2+; (B) Human blood group antigen A GT (PDB IDs: 1R81, 3V0L, and 3V0Q) with UDP-Gal (1) analog and H-antigen acceptor; (C) Human β3GlcAT-I (PDB IDs: 1FGG and 1KWS) with Galβ1–3Gal and UDP-GlcA (9). Carbon atoms in the donor and acceptor substrates are shown in green; carbon atoms in amino acid residues are shown in cyan; oxygen atoms are shown in red; nitrogen atoms are shown in blue; and sulfur atom is shown in gold. Images contain elements from multiple PDB structures aligned in PyMOL. (D) β4GalT1 is an inverting GT which catalyzes the transfer of Gal from UDP-Gal (in an α-linkage) with an inversion of stereochemistry at the anomeric carbon of Gal to form β-linked galactosides. Human blood group B GT catalyzes the transfer of Gal from UDP-Gal with a retention of anomeric stereochemistry to form α-linked galactosides. Human blood group A GT is a retaining GT catalyzing the transfer of GalNAc from UDP-GalNAc to blood H antigen acceptor. β3GlcAT-I is an inverting GT catalyzing the transfer of GlcA from UDP-GlcA to Galβ1–3Galβ1–4-Xyl for the formation of the tetrasaccharide core structure of glycosaminoglycans in proteoglycans.
β4GalT1 (Figure 2A) is one of the first mammalian GTs that was cloned and characterized [29, 30]. It is an inverting GT (Figure 2D) belonging to CAZy family GT7. In the absence of α-lactalbumin, a mammary gland-specific protein, it catalyzes the transfer of galactose (Gal) from uridine 5′-diphosphate galactose (UDP-Gal) (1, Figure 3) to N-acetylglucosamine (GlcNAc)-terminated acceptors. It forms a lactose synthase complex with α-lactalbumin which changes its acceptor preference from GlcNAc to glucose (Glc) for the production of lactose [31–33]. α-Lactalbumin also enhances the activity of β4GalT1 in catalyzing the transfer of N-acetylgalactosamine (GalNAc) from UDP-GalNAc (2) to free GlcNAc and chitin oligomers (GlcNAc)n to produce GalNAcβ1–4GlcNAc (LacdiNAc)-terminated structures [34]. β4GalT1 also catalyzes the transfer of Glc from UDP-Glc (3) to GlcNAc-terminated acceptors at 0.3% efficiency [35] which improves over 30-fold to 10% in the presence of α-lactalbumin [36]. Promiscuous donor specificity of bovine β4GalT1 toward UDP-4-deoxyglucose (4), UDP-L-arabinose (5), and UDP-glucosamine (UDP-GlcNH2, 6) at low efficiencies has also been reported [37, 38]. The crystal structures of the β4GalT1 catalytic domain in the presence or the absence of UDP in an open conformation were initially reported in 1999 [39] and the structures of its complex with α-lactalbumin in the presence of Mn2+ and UDP [33], UDP-Glc [36], or UDP-GalNAc [40] in a closed conformation were reported 2–3 years later. The structure of the β4GalT1 catalytic domain in the presence of Mn2+ and UDP-Gal was also reported [41]. Mn2+ was shown to be the preferable metal co-factor for β4GalT1 [42]. Mutating metal-binding Met-344 to His successfully switch the metal cofactor dependence of recombinant bovine β4GalT1 from Mn2+ to Mg2+ [43]. Mutating bovine β4GalT1 Tyr-289, which disrupted UDP-GalNAc binding via steric hindrance, to less bulky Leu, Ile, or Asn residues resulted in relaxed donor specificity and the increase of its GalNAcT activity. The mutants displayed GlcNAcT activity in using UDP-GlcNAc (7) as a donor substrate but no GlcT activity was found [40]. The corresponding Ile to Tyr mutation in Drosophila β1–4-N-acetylgalactosaminyltransferase 1 (β4GalNAcT1) resulted in a strict preference for UDP-Gal [44]. The Y289L mutant of bovine β4GalT1 had enhanced GalNAcT activity and retained its GalT activity [40]. It could also catalyze the transfer of 2-C-acetyl-modified Gal from UDP-Gal2CAc (8) and was found to be useful in labeling GlcNAc-modified glycoproteins [45, 46]. The Y289L/M344H β4GalT1 double mutant combined the metal cofactor switch effect of the M344H mutation with the Gal2CAcT activity of the Y289L mutation to allow tagging and labeling of live cell surface glycans in the presence of more tolerable Mg2+ instead of using potentially cytotoxic levels of Mn2+ needed by the wild-type β4GalT1 [47]. Arg-228 is located near Glu-317, which forms a hydrogen bond between the 4-OH of the Gal in UDP-Gal and is required for the GalT activity [48], and the R228K mutation resulted in 15-fold higher GlcT activity which is further enhanced in the presence of α-lactalbumin to achieve 25% of the wild-type GalT activity [49].
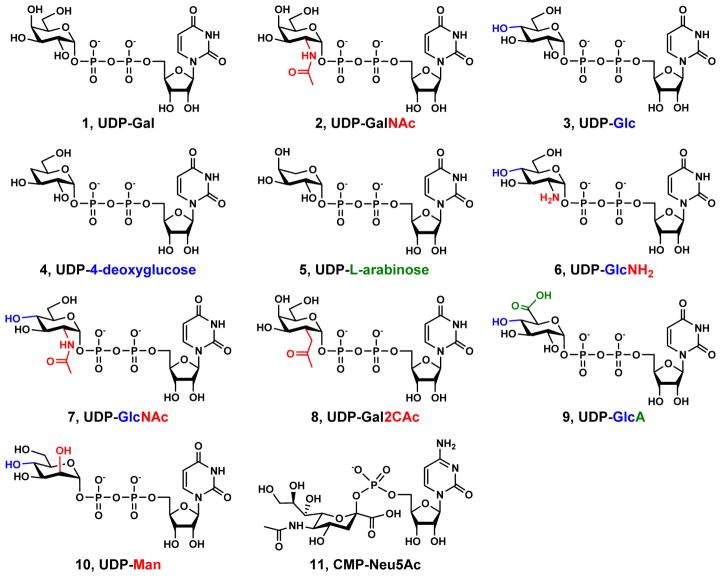
Figure 3.
Structures of UDP-Gal (1), UDP-GalNAc (2), UDP-Glc (3), UDP-4-deoxyglucose (4), UDP-L-arabinose (5), UDP-GlcNH2 (6), UDP-GlcNAc (7), UDP-Gal2CAc (8), UDP-GlcA (9), UDP-Man (10), and CMP-Neu5Ac (11). The structural features that differ other UDP-sugars from UDP-Gal are highlighted in red, blue, and green at C2, C4, and C6 of the Gal in UDP-Gal, respectively.
Human blood group A and B GTs (Figure 2B) are another set of well-studied GTs adopting the GT-A-fold. They are responsible for the formation of terminal GalNAcα1–3Gal and Galα1–3Gal linkages, respectively, on the Fucα1–2GalβOR moieties of blood group O (H) antigens, for the construction of blood group A and B antigens GalNAcα1–3(Fucα1–2)GalβOR and Galα1–3(Fucα1–2)GalβOR, respectively (Figure 2D). They are retaining GTs and are grouped in CAZy family GT6. Wild-type human blood group A and B GTs differ at only four amino acid residues Arg/Gly-176, Gly/Ser-235, Leu/Met-266, and Gly/Ala-268 [50, 51]. Systematic mutagenesis of blood group A GT to introduce blood group B GT residues resulted in the interesting discovery of blood group A GT mutant R176G, which retained blood group A GT donor specificity but gained 11-fold enhanced kcat [52]. Blood group A GT triple mutant R176G/G235S/L266M was a functional hybrid capable of using both UDP-Gal (1) and UDP-GalNAc (2) donors [52]. Interestingly, blood group B GT mutant P234S was found to have blood group A GT donor specificity despite having none of the four distinct blood group A GT mutations [53]. Crystal structures of the catalytic domains of blood group A and B GTs in the presence or the absence of H-antigen disaccharide and UDP were initially reported in 2002 [54]. Two (Leu/Met-266 and Gly/Ala-268) of the four amino acid residues that differ between the two GTs were shown to be close to the binding pocket for the donor substrate and only one (Leu/Met-266) was the major determinant for the donor substrate specificity [54, 55]. Recombinant wild-type blood group GTs also have donor substrate promiscuity. Blood group A and B GTs were able to use UDP-GlcNAc (7) and UDP-Glc (3) as a donor substrates, respectively. A double mutant G235S/L266M of blood group A GT was shown to be able to use both UDP-GlcNAc (7) and UDP-Glc (3) as donor substrate. These properties were used for the synthesis of oligosaccharide analogs of blood group antigens [56].
Human β3GlcAT-I (Figure 2C) is responsible for the synthesis of the tetrasaccharide core structure of glycosaminoglycans (e.g. heparan sulfate, heparin, chondroitin sulfate, and dermatan sulfate) in proteoglycans (Figure 2D) [57]. It is an inverting GT belongs to CAZy family GT43. The crystal structures of its catalytic domain (75–335 aa) in the presence of UDP, Mn2+, and an acceptor analog Galβ1–3Galβ1–4-Xyl [58], and in complex with UDP-GlcA (9) [59] have been reported. Mutation of conserved site His-308 to arginine led to promiscuous activity with UDP-Glc (3), UDP-Man (10), and UDP-GlcNAc (7) as the donor substrates and decreased activity with UDP-GlcA (9) [60].
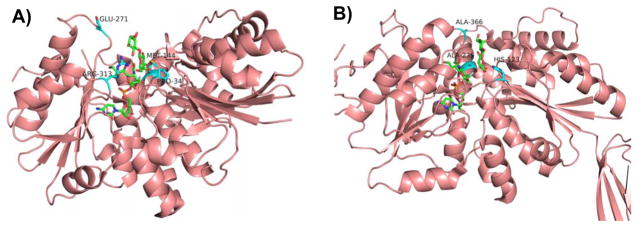
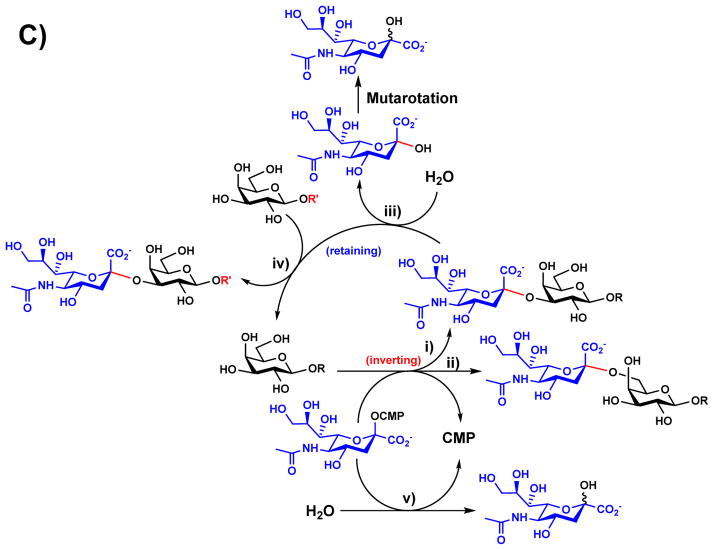
More recent examples of structure-based rational design have been applied to GT-B-fold GTs (Figure 1). The GT-B-fold consists of two Rossmann-like domains including an N-terminal acceptor-binding domain and a C-terminal sugar-nucleotide donor-binding domain [28, 61]. Of particular interest are CAZy family GT80 bacterial sialyltransferases including Pasteurella multocida multifunctional α2–3-sialyltransferase 1 (PmST1) [62], Photobacterium sp. JT-ISH-224 α2–6-sialyltransferase (Psp2,6ST) [63, 64] (Figure 4), Photobacterium damselae α2–6-sialyltransferase (Pd2,6ST) [65, 66], Pasteurella dagmatis α2–3-sialyltransferase (Pd2,3ST) [67], and others. These enzymes are the preferred tools for enzymatic synthesis of α2–3- and α2–6-sialosides due to their high activities, high heterologous over-expression yields in Escherichia coli, and outstanding promiscuity towards substrate modifications [68]. Understanding the complex structure-function relationships of the GT80 sialyltransferases is particularly challenging due to the multiple catalytic functions displayed by each enzyme [22, 62, 67, 69]. This multi-functionality is exemplified by PmST1 (Figure 4C) with i) α2–3-sialyltransferase that catalyzes the transfer of a sialic acid (e.g. the most common sialic acid form N-acetylneuraminic acid, Neu5Ac) from cytidine 5′-monophosphate (CMP)-sialic acid (e.g. CMP-Neu5Ac, 11) to an acceptor to form an α2–3-sialidic linkage; ii) α2–6-sialyltransferase; iii) α2–3-sialidase; iv) α2–3-trans-sialidase; and v) CMP-sialic acid (donor substrate) hydrolysis activities [22, 62]. The α2–6-trans-sialidase activity of a PmST1 analog differing from PmST1 at three sites (N105D, Q135R, and E295G) was also reported [70]. While the α2–3-sialyltransferase and α2–6-sialyltransferase activities involve an inverting mechanism, the α2–3-trans-sialidase and the α2–3-sialidase activities of PmST1 involve a retaining mechanism [21]. Quite interestingly, the mechanism of donor hydrolysis activity of PmST1 seems less stereoselective and appears to involve the combination of both inverting and retaining mechanisms [22]. Since the reports on the crystal structures of PmST1 in the absence or the presence of CMP in 2006 [71] and its binary structures with CMP-3F(axial)Neu5Ac or CMP-3F(equatorial)Neu5Ac and ternary structures with α-lactose and CMP-3F(axial)Neu5Ac or UDP in 2007 [72], a wealth of structural information is now known for CAZy GT80 family enzymes. Synthetically useful PmST1 mutants have been generated to decrease the α2–3-sialidase activity of PmST1, an undesired side function in PmST1-catalyzed sialoside synthesis. Single mutants E271F and R313Y as well as a double mutant E271F/R313Y were designed based on the idea of increasing the hydrophobicity of the substrate/product binding pocket to minimize the access of hydrophilic water molecules [21]. Quite remarkably, the E271F/R313Y double mutant had more than 6000-fold decreased catalytic efficiency for its α2–3-sialidase activity while maintaining wild-type level α2–3-sialyltransferase activity [21]. An E271F/R313Y double mutant of the PmST1 analog with three amino acid difference (N105D, Q135R, and E295G) also displayed decreased α2–3-sialidase and α2–6-trans-sialidase activities and increased α2–3-trans-sialidase activity [73]. PmST1 M144D is another synthetically useful PmST1 mutant developed by crystal structure-based rational design [22]. The mutation of hydrophobic Met-144 to the charged residue aspartate is believed to perturb the conserved catalytic Asp-141, resulting in altered the donor hydrolysis and sialidase activities. M144D was found to decrease the donor hydrolysis activity of the enzyme by 20-fold and the α2–3-sialidase activity by over 5500-fold while retaining the enzyme’s catalytic efficiency toward fucosylated oligosaccharide acceptor substrates such as Lewis x analogs. Unlike most natural α2–3-sialyltransferases which could not use fucosylated acceptor substrates efficiently, the PmST1 M144D enabled the synthesis of sialyl Lewis x structures containing different sialic forms in highly efficient one-pot multienzyme reactions [17, 22]. Although PmST1 adopted a closed conformation in the presence of the donor substrate, intriguingly the M144D mutant was locked in the open conformation [22].
Figure 4.
Amino acid residues known to influence the activities of GT-B-fold GT80 sialyltransferases PmST1 (A) and Psp2,6ST (B) with CMP-3F-Neu5Ac and lactose as well as reactions catalyzed by multifunctional PmST1 (C). Carbon atoms in the donor and acceptor substrates are shown in green; carbon atoms in amino acid residues are shown in cyan; oxygen atoms are shown in red; and nitrogen atoms are shown in blue. Images are constructed in PyMOL from template PDB structures 2IHZ for PmST1 and 2Z4T for Psp2,6ST. The image of the Psp2,6ST structure depicts ligands from 2IHZ within the aligned 2Z4T structure.
Multiple sequence alignments and structural analysis of GT80 sialyltransferases recently led to the identification of the critical site that determines α2–3- or α2–6- regioselectivity. While wild-type Pd2,3ST produced only trace amounts of α2–6-sialoside from lactose at pH 8.0, the P7H mutant produced nearly 95% α2–6-sialoside under the same conditions. The double mutant P7H/M117A was found to be even more selective toward the 6-OH position of the Gal in the acceptors, with a 240- and 180-fold preference toward the 6-OH over the 3-OH of lactose and N-acetyllactosamine (LacNAc), respectively [74]. P7H was found to be the major α2–6-regiospecificity determinant, with M117A (corresponding to Met-144 in PmST1) playing an auxiliary role in discriminating against α2–3-sialylation [74]. In another study, Pd2,6ST A235M (also corresponding to Met-144 in PmST1) was found to have 10-fold improved efficiency with CMP-Neu5Ac as the sugar nucleotide donor and approximately 30% efficiency in using CMP-diacetyllegiominic acid. The Pd2,6ST A235M mutant also resulted in increased donor hydrolysis rates toward both donors [75]. More recently, the Psp2,6ST A366G mutation, predicted to enhance activity with α-GalNAc-containing acceptors, was found to increase the expression level of the enzyme in the soluble active form up to 4-fold and with 1.2–2.2-fold wild-type activity toward various acceptor substrates [76]. Cumulatively, the success of these efforts to engineer GT80 sialyltransferases by rational design are evidence of the important roles played by the amino acid residues close to the substrate binding pockets in regulating the diverse functions of the enzymes. These efforts also indicate the difficulties of predicting the functional consequences of mutation. For example, mutations to PmST1 at Met-144 and the corresponding methionine, alanine, or serine residue of other related GT80 sialyltransferases have resulted in various, unrelated phenotypes including reduced or increased hydrolytic activity toward donor substrate and/or products [22, 75], altered acceptor and donor substrate specificity [22, 75], modified regioselectivity [74], and changes to the enzyme’s preferred conformation [22].
Rational design has not been limited to individual point mutations. Genes of closely related GTs may be shuffled to generate GT chimeras. GT-B-fold GTs appear to be structurally modular with acceptor and donor recognition determined by distinct N-terminal and C-terminal domains respectively. The domain swapping approach can be highly successful when the parent templates share high sequence identity and multiple fusion regions are tested. One should be cautious that the domain-domain interactions responsible for catalysis may be disrupted if the sequences from different GTs are not closely related.
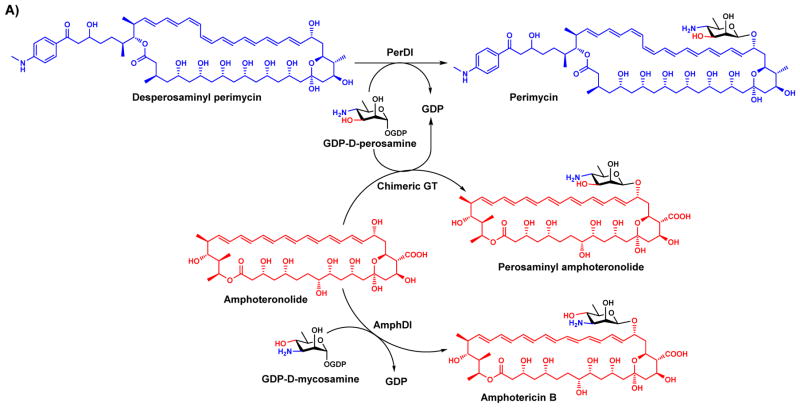
Domain swapping can be applied without crystal structures by using trial and error to identify a suitable crossover point. The strategy has been successfully applied for several GTs that are involved in natural product biosynthesis. Natural product GTs AmphDI and PerDI share 62% sequence identity and are responsible for decorating polyene macrolides with mycosamine and perosamine, respectively, using the corresponding GDP-sugars as the donor substrates (Figure 5A). Although their acceptor and donor substrates are structurally similar, hybrid perosaminyl amphoteronolides could not be produced by either enzyme as AmphDI was unable to recognize GDP-perosamine and PerDI was unable to recognize the amphoteronolide acceptors of AmphDI. A chimera containing the N-terminal domain of AmphDI and the C-terminal domain of PerDI enabled the production of the target perosaminylated amphoteronolide [77].
Figure 5.
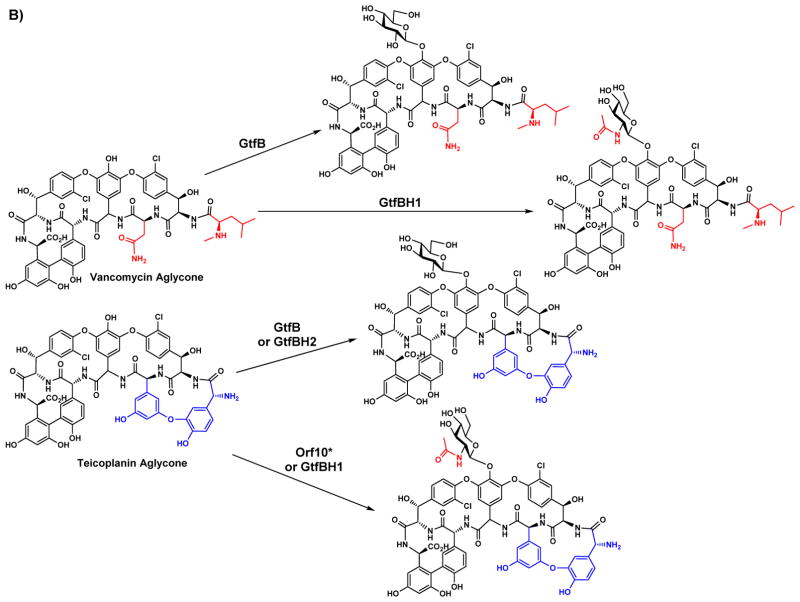
Chimeras of closely related natural product GTs provide access to novel non-natural derivatives of natural products with altered glycosylation patterns. A) A chimera generated from the N-terminal domain of AmphDI and the C-terminal domain of PerDI catalyzes the formation of the hybrid amphoteronolide. B) Chimeras of GtfB and Orf10* display predicted substrate specificities. GtfBH1 consists of the N-terminal domain of GtfB and the C-terminal domain of Orf10*, while GtfBH2 consists of the N-terminal domain of Orf10* and the C-terminal domain of GtfB. Notably, the promiscuous acceptor specificity of GtfB is present in GtfBH1 but not GtBH2.
Another successful example of domain swapping has been shown for GtfB and Orf10* which are glycopeptide natural product GTs that catalyze the transfer of Glc and GlcNAc, respectively, from the corresponding sugar nucleotide donors to suitable acceptors. Orf10* displays strict specificity to the teicoplanin aglycone, but GtfB recognizes both the vancomycin and teicoplanin aglycones (Figure 5B). Chimeras generated from GtfB and Orf10* displayed acceptor substrate specificity determined by the N-terminal domain parent and donor substrate specificity determined by the C-terminal domain parent [78].
Other chimeras have displayed activities that were not predicted from the parent GTs. Seven CAZy GT1 family GTs (UGT71C1, UGT71C2, UGT71E1, UGT85C1, UGT85B1, UGT88B1, and UGT94B1) were used to form twenty different chimeras, and twelve of these chimeras were catalytically active. Their activities toward different acceptor substrates were found to be variable and unpredictable, with both N- and C-terminal domains from different GTs implicated in acceptor specificity [79]. A similar result was found for the role of the C-terminal domain in donor substrate specificity determination for chimeras generated from two pteridine GTs which were able to accept the donor substrates of both parent enzymes [80]. These unexpected results present future opportunities to better understand GT domain-domain interactions.
Directed evolution of glycosyltransferases
Directed evolution is an alternative GT engineering strategy that can succeed without any structural information. Directed evolution relies on iterative rounds of mutagenesis and screening to incrementally improve the enzyme of interest until a mutant with the desired level of change is found [81]. Mutagenesis is most commonly conducted in a random fashion by error-prone polymerase chain reaction (epPCR) or in a semi-rational fashion with saturation mutagenesis, during which one or more codons are randomized by polymerase chain reaction (PCR) using degenerate primers. While saturation mutagenesis typically requires structural information, error-prone PCR libraries can be constructed ab initio. Due to the universality of the genetic code, these mutagenesis techniques can be applied to any gene of interest. However, screens and selections with which mutants are rapidly evaluated for performance must be customized for each class of enzyme. Therefore the availability of a suitable screening method is typically considered to be the bottleneck for conducting directed evolution. GTs have been quite challenging with this respect, as glycosidic bond formation is not easily monitored in a high-throughput fashion [18]. Despite these challenges, several effective fluorescence-based high-throughput screening methods for directed evolution of GTs have been developed in the past decade.
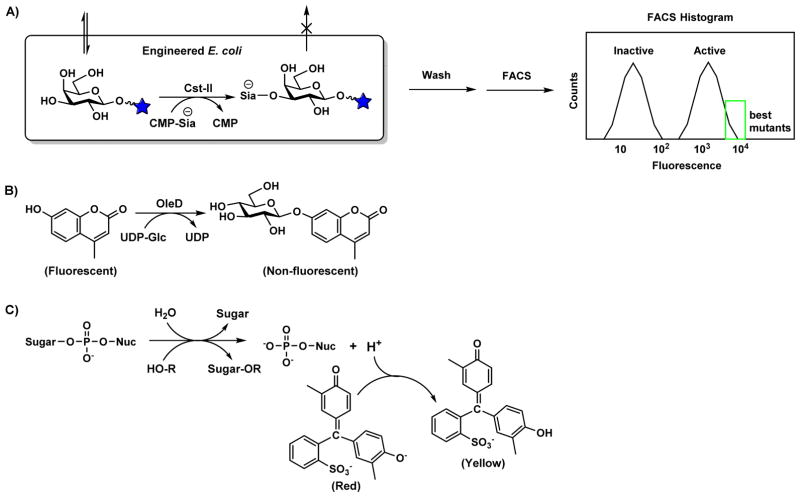
The first successful example of directed evolution of a GT was reported in 2006 using a fluorescence-activated cell sorting (FACS)-based selection for bacterial α2–3/8-sialyltransferase Cst-II catalyzing the transfer of a negatively charged Neu5Ac to a fluorescence-labeled neutral glycan [82]. The negatively charged fluorescence-labeled product was entrapped inside bacterial cells (Figure 6A) to allow ultra-high-throughput screening of mutants with improved sialyltransferase activity by FACS, which could analyze as many as 108 mutants in a day. This FACS-based method remains the highest throughput screen or selection for GT activity to date. However, this throughput came at the cost of precise control of selection parameters and reliance on secondary, lower-throughput screening for the identification of the optimum mutant among the FACS-enriched population of hits. Isolated mutants from FACS-based screening of Cst-II displayed up to 400-fold improved activity toward the fluorescent acceptor galactoside, but this improvement was not seen toward acceptor molecules lacking the fluorophore. Furthermore, product entrapment likely occurs identically for α2–3- and α2–6-sialosides. Additional α2–8-sialyltransferase activity may also take place, but cannot be directly screened for.
Figure 6. Molecular basis for glycosyltransferase mutant screening.
(A) In vivo sialylation of a fluorescently labeled acceptor substrate results in a cell-entrapped fluorescent product. After washing, cells which retain a high fluorescent signal can be isolated by FACS. (B) OleD-catalyzed glucosylation quenches MU fluorescence. (C) Glycosylation releases a proton, causing a pH change that can be detected using Cresol Red, a pH indicator useful across a pH range of 7.2–8.8. Donor hydrolysis, where water acts as the GT acceptor substrate, also results in the release of a proton.
The selective pressure of the method was subsequently more strictly defined to avoid encouraging improvements towards a specific fluorophore by using two fluorescent acceptor substrates [83]. The broad potential scope of this selection was also demonstrated by selecting a bacterial β1–3-galactosyltransferase CgtB catalyzing the transfer of a neutral sugar with improved activity. From over 107 mutants analyzed, a mutant with 300-fold higher activity and a broader substrate specificity than the wild-type was identified. The authors suggested that the transport of the fluorescent acceptor but not the neutrally charged fluorescent product through the cell membrane was a consequence of lactose permease specificity. Heterologous expression of natural or engineered variants of other sugar transport proteins may prove suitable for creating selective entrapment conditions for a wide variety of GT reactions on natural and non-natural acceptors.
Another example of directed evolution of GTs was published in 2007 [84] for a glucosyltransferase (OleD) involved in natural product oleandomycin biosynthesis. OleD was previously reported to be weakly active toward several phenolic acceptor substrates, including the fluorophore 4-methylumbelliferone (MU) [85]. When glycosylated, the fluorescence of the MU acceptor substrate was quenched, allowing an epPCR library of OleD to be screened in microtiter plates (Figure 6B). Several point mutations were found to increase OleD activity, and these mutations were combined to yield a triple mutant P67T/S132F/A242V, which displayed approximately 30-fold higher specific activity toward MU than the wild-type. Fascinatingly, this variant was found to be promiscuous towards a wide range of acceptor and donor substrates. While 15 of 22 donor substrates tested were tolerable by the triple mutant, the wild-type recognized only 3. The mutant was also capable of glucosylating 71 acceptors from a diverse panel of 137 compounds including drug-like scaffolds such as macrolides, flavonoids, isoflavones, aminocoumarins, anthraquinones, indolocarbozoles, polyenes, cardenolides, steroids, beta-lactams, enediynes, and alkaloids. In subsequent work, single-site saturation libraries randomizing sites identified in the initial work were prepared and low-throughput HPLC screening was used to determine activity toward a non-fluorescent acceptor novobiocin [86]. A mutant was found with 300-fold improved activity towards novobiocin and its activity towards 10 of 21 nucleotide sugar donors tested was demonstrated.
When structural information is insufficient for rational design and only low-throughput analysis is capable of determining the desired functional change, an emerging directed evolution strategy known as neutral drift [87] can be used. Instead of direct screening for desired improved or altered activities in traditional directed evolution strategies, in the neutral drift strategy mutants are subjected to a pre-screening process, called a purifying screen, to remove mutants that do not retain functionality. This step can be performed once, or iteratively with additional rounds of mutagenesis. The resulting library of active mutants are then screened for improvements to the activity of interest. This strategy was recently used to engineer Neisseria meningitidis serogroup B polysialyltransferase (polySTNmB) to obtain mutants with narrow size distribution of polysialic acid products [88]. EpPCR was used to create the initial library of polySTNmB mutants, followed by the identification of neutral mutants with a high-throughput screen for polysialic acid production from colonies lysed on nitrocellulose filters [89]. Subsequent rounds of neutral screening were performed on libraries generated via highly mutagenic semi-synthetic gene shuffling and traditional gene shuffling. The 51 variants isolated from neutral drift were highly polymorphic, with 7.1 ± 3.0 amino acid mutations per mutant. Variants containing mutations at Lys-69 displayed a remarkable reduction in product dispersity. Importantly, distinct phenotypic variation was also observed for activity, protein expression, and thermostability, with one mutant displaying a 10 °C increase in Tm. This is an excellent demonstration of the diverse types of mutants that can be obtained by the neutral drift strategy. Neutral drift should be considered when a purifying screen or selection can be performed at significantly higher throughput or lower cost than the screening method needed for the desired improved activity. Additionally, pre-enrichment of mutant libraries by neutral drift may facilitate projects that aim to engineer a single GT along separate evolutionary trajectories.
Structure-guided directed evolution (SGDE) of glycosyltransferases
With available structural information and limited screening throughput, structure-guided directed evolution (SGDE), often described as “semi-rational” engineering [90], is a preferred method for enzyme engineering. The strategy was used to engineer the sialyltransferase activity of PmST1 and Pd2,6ST [91]. Alanine scanning of non-conserved active site residues followed by saturation mutagenesis and screening yielded hits for each sialyltransferase. In subsequent work, three residues at the active site of PmST1 were simultaneously saturated and over 10,000 triple mutants were screened [92]. Only modest kinetic improvements were found in these two directed evolution efforts, perhaps due to a poorly focused selective pressured applied by the high-throughput screen, as was seen for FACS-based directed evolution of Cst-II [82]. This work relied on a previously reported pH-indicator-based assay to detect the proton released during GT reactions [93], but a proton is also released during donor hydrolysis (Figure 6C), a common and undesired side reaction which PmST1 is known to display. Although the poorly focused selective pressure implemented by this screen may limit its use of directed evolution to only those enzymes that do not display donor hydrolysis, this assay may serve as an inexpensive high-throughput screen for GT neutral drift, as both hydrolytic and genuine transferase activities indicate proper folding.
Other assays for glycosyltransferases
In addition to the assays successfully used as screening or selection strategies in GT directed evolution experiments, several other assays appear similarly suitable for the purpose. For example, screening methods have been demonstrated for glycosynthases, which are engineered glycoside hydrolases that catalyze glycosidic bond formation from an activated glycosyl donor such as a glycosyl fluoride. An enzyme-linked immunosorbent assay (ELISA)-based screening technique was used to engineer glycosphingolipid-hydrolyzing enzymes endo-glycoceramidase II (EGC) for glycosynthase activity toward a low-level promiscuous substrate activity [94]. The screen utilizes a microplate-bound acceptor, a cholera toxin B subunit which binds the product, and antibody detection coupled to horseradish peroxidase activity. Activity levels comparable to those for the wild-type substrate were achieved toward an alternative acceptor. Fluorescent-labeled glycan-binding plant lectins can also be used for such purposes [95]. A similar assay was recently developed for polysialyltransferase activity, in which the acceptor substrate was immobilized on plates using click chemistry and the product was detected by a GFP-tagged polysialic acid binding protein [96].
For GTs whose products are not detectable by known carbohydrate binding proteins, the use of bio-orthogonal chemistry can also facilitate product detection. One such example was shown in 2004 for substrate specificity studies of polypeptide α-N-acetylgalactosyltransferases (ppGalNAcTs) using a sugar nucleotide donor with an azido-modified sugar to allow the detection of azido-modified product bound to microtiter plates by Staudinger Ligation in a so called “azido-ELISA approach” [97].
Another interesting screening technique utilized the reversibility of GT reactions to generate an absorption signal [98]. It was determined that the thermodynamic equilibrium for β-glucosyltransferase reactions favored the reverse reaction when 2-chloro-4-nitrophenol was used as the acceptor substrate. By supplying 2-chloro-4-nitrophenol β-glycosides and the appropriate nucleotide diphosphate, the reverse reaction could be detected by the release of the phenolic chromophore. Furthermore, this reaction can be conducted with a second GT in the presence of catalytic amounts of the nucleotide disphosphate so that the chromogenic reverse β-glucosyltransferase reaction is controlled by the rate of consumption of the nucleotide sugar by the second GT. These efforts avoid the necessity of preparing nucleotide sugars. However, as with the pH-indicator based assay, these methods provide a loose selective pressure for directed evolution which may result in enhanced donor hydrolysis rates.
Successful condensation of a glucosyl fluoride donor and glucosyl MU in lysed bacterial colonies expressing glycosynthase mutants could be detected using an endo-cellulase [99]. The endo-cellulase cleaved the disaccharide product, releasing the fluorescent MU. This screen can likely be applied to any oligosaccharide forming enzymes, including GTs, but is restricted to screening for products recognized by available endo-glycosidases. Although the specificity of these endo-glycosidases may limit screening to natural oligosaccharide products, promiscuity may still result from screening for the wild-type reaction. Using this screening system for the directed evolution of Agrobacterium sp. β-glucosidase, the highest activity mutant was discovered to possess relaxed substrate specificity [100].
Several other assays for GT activity measure nucleotide sugar depletion or nucleotide diphosphate formation [3–7]. These types of assays are appealing because they are universal for all GT reactions that use nucleotide diphosphate-sugars. However, GTs are known to be incapable of complete discriminating against water as an acceptor substrate which results in donor hydrolysis [1, 2, 9, 22]. Therefore, a great care must be taken when use any of these assays for screening. When screening enzymes using lysates, negative control plates in the absence of an acceptor substrate should be screened simultaneously. Nevertheless, experimenters should be aware that the negative control may still not be sufficient to remove all false positives from initial screening, as nuclear magnetic resonance (NMR) analysis of human blood group B GT indicated that donor hydrolysis rates were enhanced by the addition of an acceptor substrate analogue [8]. When kinetically characterizing GT mutants, direct quantification of the product formation is essential to avoid mischaracterization.
Conclusions and perspectives
Many wild-type GTs have shown promiscuity towards donor and/or acceptor substrates which can be further enhanced or altered by rational design and directed evolution. These efforts have established that GTs are often highly evolvable. This evolvability suggests that many exciting new phenotypes await discovery as the toolbox for GT engineering develops.
Rational design continues to be a common method for engineering GTs due to the difficulties inherent to high-throughput detection of glycosidic bond formation and the increasing wealth of sequence and structure information for GTs. Nevertheless, numerous screening strategies have been developed for GT activity. Several high-throughput screens can be used in a general fashion for neutral drift enrichment of GT mutant libraries to generate highly polymorphic and active libraries from which improved variants can be found at a higher frequency than in naïve libraries. Although this strategy is in its infancy, the results of neutral drift of polySTNmB indicate that neutral drift may sufficiently enrich libraries to enable hit identification through universal low-throughput screening techniques such as high performance liquid chromatography (HPLC) and liquid chromatography-mass spectrometry (LC-MS). Future methods for high-throughput screening of GTs should avoid detecting donor substrate depletion or byproduct production, which may yield undesirable mutants.
Future advances in GT engineering are likely to come not only from new mutagenesis or screening technologies, but also from a better understanding of GT structure-function relationships. The knowledge necessary to enable rapid identification of key residues from minimal sequence or structural data remains a key objective for GT engineering. Successfully accomplishing this objective will greatly advance the enzyme-catalyzed synthesis and development of carbohydrate-based applications and therapeutics.
Acknowledgments
Funding
The authors are grateful for the financial supports from National Institute of Health grants R01HD065122 and R01GM094523 and from National Science Foundation grants CHE-1012511 and CHE-1300449.
Abbreviations
- CAZy
Carbohydrate Active enZyme
- CMP
cytidine 5′-monophosphate
- EGC
endo-glycoceramidase II
- ELISA
enzyme-linked immunosorbent assay
- epPCR
error-prone polymerase chain reaction
- FACS
fluorescence-activated cell sorting
- Gal
galactose
- Glc
glucose
- GalNAc
N-acetylgalactosamine
- GlcNAc
N-acetylglucosamine
- GT
glycosyltransferase
- HPLC
high performance liquid chromatography
- LC-MS
liquid chromatography-mass spectrometry
- MU
4-methylumbelliferone
- Neu5Ac
N-acetylneuraminic acid, a common sialic acid form
- NMR
nuclear magnetic resonance
- PCR
polymerase chain reaction
- NmBpolyST
Neisseria meningitidis serogroup B polysialyltransferase
- ppGalNAcT
polypeptide α-N-acetylgalactosyltransferase
- SGDE
structure-guided directed evolution
- UDP
uridine 5′-diphosphate
- Cst-II
Campylobacter jejuni α2–3/8-sialyltransferase
- β4GalT1
β1–4-galactosyltransferase 1
- β3GlcAT-I
β1–3-glucuronyltransferase-I
- OleD
a glucosyltransferase involved in natural product oleandomycin biosynthesis
- Pd2
3ST, Pasteurella dagmatis α2–3-sialyltransferase
- Pd2
6ST, Photobacterium damselae α2–6-sialyltransferase
- PmST1
Pasteurella multocida multifunctional α2–3-sialyltransferase 1
- Psp2
6ST, Photobacterium sp. JT-ISH-224 α2–6-sialyltransferase
References
- 1.Solis D, Bovin NV, Davis AP, Jimenez-Barbero J, Romero A, Roy R, Smetana K, Jr, Gabius HJ. A guide into glycosciences: How chemistry, biochemistry and biology cooperate to crack the sugar code. Biochim Biophys Acta. 2015;1850:186–235. doi: 10.1016/j.bbagen.2014.03.016. [DOI] [PubMed] [Google Scholar]
- 2.Bennett CS. Principles of modern solid-phase oligosaccharide synthesis. Org Biomol Chem. 2014;12:1686–1698. doi: 10.1039/c3ob42343c. [DOI] [PubMed] [Google Scholar]
- 3.Kaeothip S, Demchenko AV. Expeditious oligosaccharide synthesis via selective, semi-orthogonal, and orthogonal activation. Carbohydr Res. 2011;346:1371–1388. doi: 10.1016/j.carres.2011.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Seeberger PH. Automated oligosaccharide synthesis. Chem Soc Rev. 2008;37:19–28. doi: 10.1039/b511197h. [DOI] [PubMed] [Google Scholar]
- 5.Wu CY, Wong CH. Programmable one-pot glycosylation. Top Curr Chem. 2011;301:223–252. doi: 10.1007/128_2010_109. [DOI] [PubMed] [Google Scholar]
- 6.Boltje TJ, Buskas T, Boons GJ. Opportunities and challenges in synthetic oligosaccharide and glycoconjugate research. Nat Chem. 2009;1:611–622. doi: 10.1038/nchem.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bissaro B, Monsan P, Faure R, O’Donohue MJ. Glycosynthesis in a waterworld: new insight into the molecular basis of transglycosylation in retaining glycoside hydrolases. Biochem J. 2015;467:17–35. doi: 10.1042/BJ20141412. [DOI] [PubMed] [Google Scholar]
- 8.Wang LX, Huang W. Enzymatic transglycosylation for glycoconjugate synthesis. Curr Opin Chem Biol. 2009;13:592–600. doi: 10.1016/j.cbpa.2009.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shaikh FA, Withers SG. Teaching old enzymes new tricks: engineering and evolution of glycosidases and glycosyl transferases for improved glycoside synthesis. Biochem Cell Biol. 2008;86:169–177. doi: 10.1139/O07-149. [DOI] [PubMed] [Google Scholar]
- 10.Muthana S, Cao H, Chen X. Recent progress in chemical and chemoenzymatic synthesis of carbohydrates. Curr Opin Chem Biol. 2009;13:573–581. doi: 10.1016/j.cbpa.2009.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Koeller KM, Wong CH. Complex carbohydrate synthesis tools for glycobiologists: enzyme-based approach and programmable one-pot strategies. Glycobiology. 2000;10:1157–1169. doi: 10.1093/glycob/10.11.1157. [DOI] [PubMed] [Google Scholar]
- 12.Chang A, Singh S, Phillips GN, Jr, Thorson JS. Glycosyltransferase structural biology and its role in the design of catalysts for glycosylation. Curr Opin Biotechnol. 2011;22:800–808. doi: 10.1016/j.copbio.2011.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yu H, Chen X. Enzymatic synthesis of carbohydrate-containing biomolecules. In: Begley TP, editor. Wiley Encyclopedia of Chemical Biology. John Wiley & Sons, Inc; New Jersey: 2009. pp. 631–652. [Google Scholar]
- 14.Palcic MM. Glycosyltransferases as biocatalysts. Curr Opin Chem Biol. 2011;15:226–233. doi: 10.1016/j.cbpa.2010.11.022. [DOI] [PubMed] [Google Scholar]
- 15.Schmaltz RM, Hanson SR, Wong CH. Enzymes in the synthesis of glycoconjugates. Chem Rev. 2011;111:4259–4307. doi: 10.1021/cr200113w. [DOI] [PubMed] [Google Scholar]
- 16.ACH, XC . Chemical aspects (scope and limiations) A: Enzymatic approaches to O-glycoside introduction: Glycosyltransferases. In: Kamerling JP, Boons G-J, Lee YC, Suzuki A, Taniguchi N, Voragen AGJ, editors. Comprehensive Glycoscience, from Chemistry to Systems Biology. Elsevier; Oxford, UK: 2007. pp. 415–451. [Google Scholar]
- 17.Yu H, Chokhawala HA, Huang S, Chen X. One-pot three-enzyme chemoenzymatic approach to the synthesis of sialosides containing natural and non-natural functionalities. Nat Protoc. 2006;1:2485–2492. doi: 10.1038/nprot.2006.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Williams GJ, Gantt RW, Thorson JS. The impact of enzyme engineering upon natural product glycodiversification. Curr Opin Chem Biol. 2008;12:556–564. doi: 10.1016/j.cbpa.2008.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lairson LL, Henrissat B, Davies GJ, Withers SG. Glycosyltransferases: structures, functions, and mechanisms. Annu Rev Biochem. 2008;77:521–555. doi: 10.1146/annurev.biochem.76.061005.092322. [DOI] [PubMed] [Google Scholar]
- 20.Breton C, Fournel-Gigleux S, Palcic MM. Recent structures, evolution and mechanisms of glycosyltransferases. Curr Opin Struct Biol. 2012;22:540–549. doi: 10.1016/j.sbi.2012.06.007. [DOI] [PubMed] [Google Scholar]
- 21.Sugiarto G, Lau K, Li Y, Khedri Z, Yu H, Le DT, Chen X. Decreasing the sialidase activity of multifunctional Pasteurella multocida alpha2–3-sialyltransferase 1 (PmST1) by site-directed mutagenesis. Mol Biosyst. 2011;7:3021–3027. doi: 10.1039/c1mb05182b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sugiarto G, Lau K, Qu J, Li Y, Lim S, Mu S, Ames JB, Fisher AJ, Chen X. A sialyltransferase mutant with decreased donor hydrolysis and reduced sialidase activities for directly sialylating LewisX. ACS Chem Biol. 2012;7:1232–1240. doi: 10.1021/cb300125k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reetz MT, Carballeira JD. Iterative saturation mutagenesis (ISM) for rapid directed evolution of functional enzymes. Nat Protoc. 2007;2:891–903. doi: 10.1038/nprot.2007.72. [DOI] [PubMed] [Google Scholar]
- 24.Campbell JA, Davies GJ, Bulone V, Henrissat B. A classification of nucleotide-diphospho-sugar glycosyltransferases based on amino acid sequence similarities. Biochem J. 1997;326(Pt 3):929–939. doi: 10.1042/bj3260929u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Coutinho PM, Deleury E, Davies GJ, Henrissat B. An evolving hierarchical family classification for glycosyltransferases. J Mol Biol. 2003;328:307–317. doi: 10.1016/s0022-2836(03)00307-3. [DOI] [PubMed] [Google Scholar]
- 26.Charnock SJ, Davies GJ. Structure of the nucleotide-diphospho-sugar transferase, SpsA from Bacillus subtilis, in native and nucleotide-complexed forms. Biochemistry. 1999;38:6380–6385. doi: 10.1021/bi990270y. [DOI] [PubMed] [Google Scholar]
- 27.Morera S, Lariviere L, Kurzeck J, Aschke-Sonnenborn U, Freemont PS, Janin J, Ruger W. High resolution crystal structures of T4 phage beta-glucosyltransferase: induced fit and effect of substrate and metal binding. J Mol Biol. 2001;311:569–577. doi: 10.1006/jmbi.2001.4905. [DOI] [PubMed] [Google Scholar]
- 28.Breton C, Snajdrova L, Jeanneau C, Koca J, Imberty A. Structures and mechanisms of glycosyltransferases. Glycobiology. 2006;16:29R–37R. doi: 10.1093/glycob/cwj016. [DOI] [PubMed] [Google Scholar]
- 29.Narimatsu H, Sinha S, Brew K, Okayama H, Qasba PK. Cloning and sequencing of cDNA of bovine N-acetylglucosamine (beta 1–4)galactosyltransferase. Proc Natl Acad Sci U S A. 1986;83:4720–4724. doi: 10.1073/pnas.83.13.4720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shaper NL, Shaper JH, Meuth JL, Fox JL, Chang H, Kirsch IR, Hollis GF. Bovine galactosyltransferase: identification of a clone by direct immunological screening of a cDNA expression library. Proc Natl Acad Sci U S A. 1986;83:1573–1577. doi: 10.1073/pnas.83.6.1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brew K, Vanaman TC, Hill RL. The role of alpha-lactalbumin and the A protein in lactose synthetase: a unique mechanism for the control of a biological reaction. Proc Natl Acad Sci U S A. 1968;59:491–497. doi: 10.1073/pnas.59.2.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brodbeck U, Denton WL, Tanahashi N, Ebner KE. The isolation and identification of the B protein of lactose synthetase as alpha-lactalbumin. J Biol Chem. 1967;242:1391–1397. [PubMed] [Google Scholar]
- 33.Ramakrishnan B, Qasba PK. Crystal structure of lactose synthase reveals a large conformational change in its catalytic component, the beta1,4-galactosyltransferase-I. J Mol Biol. 2001;310:205–218. doi: 10.1006/jmbi.2001.4757. [DOI] [PubMed] [Google Scholar]
- 34.Do KY, Do SI, Cummings RD. Alpha-lactalbumin induces bovine milk beta 1,4-galactosyltransferase to utilize UDP-GalNAc. J Biol Chem. 1995;270:18447–18451. doi: 10.1074/jbc.270.31.18447. [DOI] [PubMed] [Google Scholar]
- 35.Andree PJ, Berliner LJ. Glucosyl transferase activity of bovine galactosyl transferase. Biochim Biophys Acta. 1978;544:489–495. doi: 10.1016/0304-4165(78)90323-9. [DOI] [PubMed] [Google Scholar]
- 36.Ramakrishnan B, Shah PS, Qasba PK. alpha-Lactalbumin (LA) stimulates milk beta-1,4-galactosyltransferase I (beta 4Gal-T1) to transfer glucose from UDP-glucose to N-acetylglucosamine. Crystal structure of beta 4Gal-T1 x LA complex with UDP-Glc. J Biol Chem. 2001;276:37665–37671. doi: 10.1074/jbc.M102458200. [DOI] [PubMed] [Google Scholar]
- 37.Berliner LJ, Robinson RD. Structure-function relationships in lactose synthase. Structural requirements of the uridine 5′-diphosphate galactose binding site. Biochemistry. 1982;21:6340–6343. doi: 10.1021/bi00268a003. [DOI] [PubMed] [Google Scholar]
- 38.Palcic MM, Hindsgaul O. Flexibility in the donor substrate specificity of beta 1,4-galactosyltransferase: application in the synthesis of complex carbohydrates. Glycobiology. 1991;1:205–209. doi: 10.1093/glycob/1.2.205. [DOI] [PubMed] [Google Scholar]
- 39.Gastinel LN, Cambillau C, Bourne Y. Crystal structures of the bovine beta4galactosyltransferase catalytic domain and its complex with uridine diphosphogalactose. EMBO J. 1999;18:3546–3557. doi: 10.1093/emboj/18.13.3546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ramakrishnan B, Qasba PK. Structure-based design of beta 1,4-galactosyltransferase I (beta 4Gal-T1) with equally efficient N-acetylgalactosaminyltransferase activity: point mutation broadens beta 4Gal-T1 donor specificity. J Biol Chem. 2002;277:20833–20839. doi: 10.1074/jbc.M111183200. [DOI] [PubMed] [Google Scholar]
- 41.Ramakrishnan B, Balaji PV, Qasba PK. Crystal structure of beta1,4-galactosyltransferase complex with UDP-Gal reveals an oligosaccharide acceptor binding site. J Mol Biol. 2002;318:491–502. doi: 10.1016/S0022-2836(02)00020-7. [DOI] [PubMed] [Google Scholar]
- 42.Powell JT, Brew K. Metal ion activation of galactosyltransferase. J Biol Chem. 1976;251:3645–3652. [PubMed] [Google Scholar]
- 43.Ramakrishnan B, Boeggeman E, Qasba PK. Effect of the Met344His mutation on the conformational dynamics of bovine beta-1,4-galactosyltransferase: crystal structure of the Met344His mutant in complex with chitobiose. Biochemistry. 2004;43:12513–12522. doi: 10.1021/bi049007+. [DOI] [PubMed] [Google Scholar]
- 44.Ramakrishnan B, Qasba PK. Role of a Single Amino Acid in the Evolution of Glycans of Invertebrates and Vertebrates. J Mol Biol. 2007;365:570–576. doi: 10.1016/j.jmb.2006.10.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Khidekel N, Arndt S, Lamarre-Vincent N, Lippert A, Poulin-Kerstien KG, Ramakrishnan B, Qasba PK, Hsieh-Wilson LC. A chemoenzymatic approach toward the rapid and sensitive detection of O-GlcNAc posttranslational modifications. J Am Chem Soc. 2003;125:16162–16163. doi: 10.1021/ja038545r. [DOI] [PubMed] [Google Scholar]
- 46.Boeggeman E, Ramakrishnan B, Kilgore C, Khidekel N, Hsieh-Wilson LC, Simpson JT, Qasba PK. Direct identification of nonreducing GlcNAc residues on N-glycans of glycoproteins using a novel chemoenzymatic method. Bioconjug Chem. 2007;18:806–814. doi: 10.1021/bc060341n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mercer N, Ramakrishnan B, Boeggeman E, Verdi L, Qasba PK. Use of novel mutant galactosyltransferase for the bioconjugation of terminal N-acetylglucosamine (GlcNAc) residues on live cell surface. Bioconjug Chem. 2013;24:144–152. doi: 10.1021/bc300542z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Boeggeman E, Qasba PK. Studies on the metal binding sites in the catalytic domain of beta1,4-galactosyltransferase. Glycobiology. 2002;12:395–407. doi: 10.1093/glycob/cwf045. [DOI] [PubMed] [Google Scholar]
- 49.Ramakrishnan B, Boeggeman E, Qasba PK. Mutation of arginine 228 to lysine enhances the glucosyltransferase activity of bovine beta-1,4-galactosyltransferase I. Biochemistry. 2005;44:3202–3210. doi: 10.1021/bi0479454. [DOI] [PubMed] [Google Scholar]
- 50.Yamamoto F, Clausen H, White T, Marken J, Hakomori S. Molecular genetic basis of the histo-blood group ABO system. Nature. 1990;345:229–233. doi: 10.1038/345229a0. [DOI] [PubMed] [Google Scholar]
- 51.Yamamoto F, Hakomori S. Sugar-nucleotide donor specificity of histo-blood group A and B transferases is based on amino acid substitutions. J Biol Chem. 1990;265:19257–19262. [PubMed] [Google Scholar]
- 52.Seto NO, Palcic MM, Compston CA, Li H, Bundle DR, Narang SA. Sequential interchange of four amino acids from blood group B to blood group A glycosyltransferase boosts catalytic activity and progressively modifies substrate recognition in human recombinant enzymes. J Biol Chem. 1997;272:14133–14138. doi: 10.1074/jbc.272.22.14133. [DOI] [PubMed] [Google Scholar]
- 53.Marcus SL, Polakowski R, Seto NO, Leinala E, Borisova S, Blancher A, Roubinet F, Evans SV, Palcic MM. A single point mutation reverses the donor specificity of human blood group B-synthesizing galactosyltransferase. J Biol Chem. 2003;278:12403–12405. doi: 10.1074/jbc.M212002200. [DOI] [PubMed] [Google Scholar]
- 54.Patenaude SI, Seto NO, Borisova SN, Szpacenko A, Marcus SL, Palcic MM, Evans SV. The structural basis for specificity in human ABO(H) blood group biosynthesis. Nat Struct Biol. 2002;9:685–690. doi: 10.1038/nsb832. [DOI] [PubMed] [Google Scholar]
- 55.Seto NO, Compston CA, Evans SV, Bundle DR, Narang SA, Palcic MM. Donor substrate specificity of recombinant human blood group A, B and hybrid A/B glycosyltransferases expressed in Escherichia coli. Eur J Biochem. 1999;259:770–775. doi: 10.1046/j.1432-1327.1999.00086.x. [DOI] [PubMed] [Google Scholar]
- 56.Seto NO, Compston CA, Szpacenko A, Palcic MM. Enzymatic synthesis of blood group A and B trisaccharide analogues. Carbohydr Res. 2000;324:161–169. doi: 10.1016/s0008-6215(99)00297-9. [DOI] [PubMed] [Google Scholar]
- 57.Kitagawa H, Tone Y, Tamura J, Neumann KW, Ogawa T, Oka S, Kawasaki T, Sugahara K. Molecular cloning and expression of glucuronyltransferase I involved in the biosynthesis of the glycosaminoglycan-protein linkage region of proteoglycans. J Biol Chem. 1998;273:6615–6618. doi: 10.1074/jbc.273.12.6615. [DOI] [PubMed] [Google Scholar]
- 58.Pedersen LC, Tsuchida K, Kitagawa H, Sugahara K, Darden TA, Negishi M. Heparan/chondroitin sulfate biosynthesis. Structure and mechanism of human glucuronyltransferase I. J Biol Chem. 2000;275:34580–34585. doi: 10.1074/jbc.M007399200. [DOI] [PubMed] [Google Scholar]
- 59.Pedersen LC, Darden TA, Negishi M. Crystal structure of beta 1,3-glucuronyltransferase I in complex with active donor substrate UDP-GlcUA. J Biol Chem. 2002;277:21869–21873. doi: 10.1074/jbc.M112343200. [DOI] [PubMed] [Google Scholar]
- 60.Ouzzine M, Gulberti S, Levoin N, Netter P, Magdalou J, Fournel-Gigleux S. The donor substrate specificity of the human beta 1,3-glucuronosyltransferase I toward UDP-glucuronic acid is determined by two crucial histidine and arginine residues. J Biol Chem. 2002;277:25439–25445. doi: 10.1074/jbc.M201912200. [DOI] [PubMed] [Google Scholar]
- 61.Qasba PK, Ramakrishnan B, Boeggeman E. Substrate-induced conformational changes in glycosyltransferases. Trends Biochem Sci. 2005;30:53–62. doi: 10.1016/j.tibs.2004.11.005. [DOI] [PubMed] [Google Scholar]
- 62.Yu H, Chokhawala H, Karpel R, Yu H, Wu B, Zhang J, Zhang Y, Jia Q, Chen X. A multifunctional Pasteurella multocida sialyltransferase: a powerful tool for the synthesis of sialoside libraries. J Am Chem Soc. 2005;127:17618–17619. doi: 10.1021/ja0561690. [DOI] [PubMed] [Google Scholar]
- 63.Tsukamoto H, Takakura Y, Mine T, Yamamoto T. Photobacterium sp. JT-ISH-224 produces two sialyltransferases, alpha-/beta-galactoside alpha2,3-sialyltransferase and beta-galactoside alpha2,6-sialyltransferase. J Biochem. 2008;143:187–197. doi: 10.1093/jb/mvm208. [DOI] [PubMed] [Google Scholar]
- 64.Ding L, Yu H, Lau K, Li Y, Muthana S, Wang J, Chen X. Efficient chemoenzymatic synthesis of sialyl Tn-antigens and derivatives. Chem Commun. 2011;47:8691–8693. doi: 10.1039/c1cc12732b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yamamoto T, Nakashizuka M, Terada I. Cloning and expression of a marine bacterial beta-galactoside alpha2,6-sialyltransferase gene from Photobacterium damsela JT0160. J Biochem. 1998;123:94–100. doi: 10.1093/oxfordjournals.jbchem.a021921. [DOI] [PubMed] [Google Scholar]
- 66.Yu H, Huang S, Chokhawala H, Sun M, Zheng H, Chen X. Highly efficient chemoenzymatic synthesis of naturally occurring and non-natural alpha-2,6-linked sialosides: a P. damsela alpha-2,6-sialyltransferase with extremely flexible donor-substrate specificity. Angew Chem Int Ed Engl. 2006;45:3938–3944. doi: 10.1002/anie.200600572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Schmolzer K, Ribitsch D, Czabany T, Luley-Goedl C, Kokot D, Lyskowski A, Zitzenbacher S, Schwab H, Nidetzky B. Characterization of a multifunctional alpha2,3-sialyltransferase from Pasteurella dagmatis. Glycobiology. 2013;23:1293–1304. doi: 10.1093/glycob/cwt066. [DOI] [PubMed] [Google Scholar]
- 68.Li Y, Chen X. Sialic acid metabolism and sialyltransferases: natural functions and applications. Appl Microbiol Biotechnol. 2012;94:887–905. doi: 10.1007/s00253-012-4040-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cheng J, Huang S, Yu H, Li Y, Lau K, Chen X. Trans-sialidase activity of Photobacterium damsela alpha2,6-sialyltransferase and its application in the synthesis of sialosides. Glycobiology. 2010;20:260–268. doi: 10.1093/glycob/cwp172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Guo Y, Jers C, Meyer AS, Arnous A, Li H, Kirpekar F, Mikkelsen JD. A Pasteurella multocida sialyltransferase displaying dual trans-sialidase activities for production of 3′-sialyl and 6′-sialyl glycans. J Biotechnol. 2014;170:60–67. doi: 10.1016/j.jbiotec.2013.11.013. [DOI] [PubMed] [Google Scholar]
- 71.Ni L, Sun M, Yu H, Chokhawala H, Chen X, Fisher AJ. Cytidine 5′-monophosphate (CMP)-induced structural changes in a multifunctional sialyltransferase from Pasteurella multocida. Biochemistry. 2006;45:2139–2148. doi: 10.1021/bi0524013. [DOI] [PubMed] [Google Scholar]
- 72.Ni L, Chokhawala HA, Cao H, Henning R, Ng L, Huang S, Yu H, Chen X, Fisher AJ. Crystal structures of Pasteurella multocida sialyltransferase complexes with acceptor and donor analogues reveal substrate binding sites and catalytic mechanism. Biochemistry. 2007;46:6288–6298. doi: 10.1021/bi700346w. [DOI] [PubMed] [Google Scholar]
- 73.Guo Y, Jers C, Meyer AS, Li H, Kirpekar F, Mikkelsen JD. Modulating the regioselectivity of a Pasteurella multocida sialyltransferase for biocatalytic production of 3′- and 6′-sialyllactose. Enzyme Microb Technol. 2015;78:54–62. doi: 10.1016/j.enzmictec.2015.06.012. [DOI] [PubMed] [Google Scholar]
- 74.Schmolzer K, Czabany T, Luley-Goedl C, Pavkov-Keller T, Ribitsch D, Schwab H, Gruber K, Weber H, Nidetzky B. Complete switch from alpha-2,3- to alpha-2,6-regioselectivity in Pasteurella dagmatis beta-D-galactoside sialyltransferase by active-site redesign. Chem Commun. 2015;51:3083–3086. doi: 10.1039/c4cc09772f. [DOI] [PubMed] [Google Scholar]
- 75.Watson DC, Wakarchuk WW, Leclerc S, Schur MJ, Schoenhofen IC, Young NM, Gilbert M. Sialyltransferases with enhanced legionaminic acid transferase activity for the preparation of analogs of sialoglycoconjugates. Glycobiology. 2015;25:767–773. doi: 10.1093/glycob/cwv017. [DOI] [PubMed] [Google Scholar]
- 76.Ding L, Zhao C, Qu J, Li Y, Sugiarto G, Yu H, Wang J, Chen X. A Photobacterium sp. alpha2–6-sialyltransferase (Psp2,6ST) mutant with an increased expression level and improved activities in sialylating Tn antigens. Carbohydr Res. 2015;408:127–133. doi: 10.1016/j.carres.2014.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hutchinson E, Murphy B, Dunne T, Breen C, Rawlings B, Caffrey P. Redesign of polyene macrolide glycosylation: engineered biosynthesis of 19-(O)-perosaminyl-amphoteronolide B. Chem Biol. 2010;17:174–182. doi: 10.1016/j.chembiol.2010.01.007. [DOI] [PubMed] [Google Scholar]
- 78.Truman AW, Dias MV, Wu S, Blundell TL, Huang F, Spencer JB. Chimeric glycosyltransferases for the generation of hybrid glycopeptides. Chem Biol. 2009;16:676–685. doi: 10.1016/j.chembiol.2009.04.013. [DOI] [PubMed] [Google Scholar]
- 79.Hansen EH, Osmani SA, Kristensen C, Moller BL, Hansen J. Substrate specificities of family 1 UGTs gained by domain swapping. Phytochemistry. 2009;70:473–482. doi: 10.1016/j.phytochem.2009.01.013. [DOI] [PubMed] [Google Scholar]
- 80.Kim HL, Kim AH, Park MB, Lee SW, Park YS. Altered sugar donor specificity and catalytic activity of pteridine glycosyltransferases by domain swapping or site-directed mutagenesis. BMB Rep. 2013;46:37–40. doi: 10.5483/BMBRep.2013.46.1.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Bershtein S, Tawfik DS. Advances in laboratory evolution of enzymes. Curr Opin Chem Biol. 2008;12:151–158. doi: 10.1016/j.cbpa.2008.01.027. [DOI] [PubMed] [Google Scholar]
- 82.Aharoni A, Thieme K, Chiu CP, Buchini S, Lairson LL, Chen H, Strynadka NC, Wakarchuk WW, Withers SG. High-throughput screening methodology for the directed evolution of glycosyltransferases. Nat Methods. 2006;3:609–614. doi: 10.1038/nmeth899. [DOI] [PubMed] [Google Scholar]
- 83.Yang G, Rich JR, Gilbert M, Wakarchuk WW, Feng Y, Withers SG. Fluorescence Activated Cell Sorting as a General Ultra-High-Throughput Screening Method for Directed Evolution of Glycosyltransferases. J Am Chem Soc. 2010;132:10570–10577. doi: 10.1021/ja104167y. [DOI] [PubMed] [Google Scholar]
- 84.Williams GJ, Zhang C, Thorson JS. Expanding the promiscuity of a natural-product glycosyltransferase by directed evolution. Nat Chem Biol. 2007;3:657–662. doi: 10.1038/nchembio.2007.28. [DOI] [PubMed] [Google Scholar]
- 85.Yang M, Proctor MR, Bolam DN, Errey JC, Field RA, Gilbert HJ, Davis BG. Probing the breadth of macrolide glycosyltransferases: in vitro remodeling of a polyketide antibiotic creates active bacterial uptake and enhances potency. J Am Chem Soc. 2005;127:9336–9337. doi: 10.1021/ja051482n. [DOI] [PubMed] [Google Scholar]
- 86.Williams GJ, Goff RD, Zhang C, Thorson JS. Optimizing glycosyltransferase specificity via “hot spot” saturation mutagenesis presents a catalyst for novobiocin glycorandomization. Chem Biol. 2008;15:393–401. doi: 10.1016/j.chembiol.2008.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Gupta RD, Tawfik DS. Directed enzyme evolution via small and effective neutral drift libraries. Nat Methods. 2008;5:939–942. doi: 10.1038/nmeth.1262. [DOI] [PubMed] [Google Scholar]
- 88.Keys TG, Fuchs HL, Ehrit J, Alves J, Freiberger F, Gerardy-Schahn R. Engineering the product profile of a polysialyltransferase. Nat Chem Biol. 2014;10:437–442. doi: 10.1038/nchembio.1501. [DOI] [PubMed] [Google Scholar]
- 89.Keys TG, Berger M, Gerardy-Schahn R. A high-throughput screen for polysialyltransferase activity. Anal Biochem. 2012;427:60–68. doi: 10.1016/j.ab.2012.04.033. [DOI] [PubMed] [Google Scholar]
- 90.Lutz S. Beyond directed evolution--semi-rational protein engineering and design. Curr Opin Biotechnol. 2010;21:734–743. doi: 10.1016/j.copbio.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Choi YH, Kim JH, Park JH, Lee N, Kim DH, Jang KS, Park IH, Kim BG. Protein engineering of alpha2,3/2,6-sialyltransferase to improve the yield and productivity of in vitro sialyllactose synthesis. Glycobiology. 2014;24:159–169. doi: 10.1093/glycob/cwt092. [DOI] [PubMed] [Google Scholar]
- 92.Kim HC, Kim KS, Kang TJ, Choi JH, Song JJ, Choi YH, Kim BG, Kim DM. Implementing bacterial acid resistance into cell-free protein synthesis for buffer-free expression and screening of enzymes. Biotechnol Bioeng. 2015;112:2630–2635. doi: 10.1002/bit.25671. [DOI] [PubMed] [Google Scholar]
- 93.Park SH, Park HY, Sohng JK, Lee HC, Liou K, Yoon YJ, Kim BG. Expanding substrate specificity of GT-B fold glycosyltransferase via domain swapping and high-throughput screening. Biotechnol Bioeng. 2009;102:988–994. doi: 10.1002/bit.22150. [DOI] [PubMed] [Google Scholar]
- 94.Hancock SM, Rich JR, Caines ME, Strynadka NC, Withers SG. Designer enzymes for glycosphingolipid synthesis by directed evolution. Nat Chem Biol. 2009;5:508–514. doi: 10.1038/nchembio.191. [DOI] [PubMed] [Google Scholar]
- 95.Chokhawala HA, Huang S, Lau K, Yu H, Cheng J, Thon V, Hurtado-Ziola N, Guerrero JA, Varki A, Chen X. Combinatorial chemoenzymatic synthesis and high-throughput screening of sialosides. ACS Chem Biol. 2008;3:567–576. doi: 10.1021/cb800127n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Yu CC, Hill T, Kwan DH, Chen HM, Lin CC, Wakarchuk W, Withers SG. A plate-based high-throughput activity assay for polysialyltransferase from Neisseria meningitidis. Anal Biochem. 2014;444:67–74. doi: 10.1016/j.ab.2013.09.030. [DOI] [PubMed] [Google Scholar]
- 97.Hang HC, Yu C, Pratt MR, Bertozzi CR. Probing Glycosyltransferase Activities with the Staudinger Ligation. J Am Chem Soc. 2004;126:6–7. doi: 10.1021/ja037692m. [DOI] [PubMed] [Google Scholar]
- 98.Gantt RW, Peltier-Pain P, Cournoyer WJ, Thorson JS. Using simple donors to drive the equilibria of glycosyltransferase-catalyzed reactions. Nat Chem Biol. 2011;7:685–691. doi: 10.1038/nchembio.638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Mayer C, Jakeman DL, Mah M, Karjala G, Gal L, Warren RA, Withers SG. Directed evolution of new glycosynthases from Agrobacterium beta-glucosidase: a general screen to detect enzymes for oligosaccharide synthesis. Chem Biol. 2001;8:437–443. doi: 10.1016/s1074-5521(01)00022-9. [DOI] [PubMed] [Google Scholar]
- 100.Kim YW, Lee SS, Warren RA, Withers SG. Directed evolution of a glycosynthase from Agrobacterium sp. increases its catalytic activity dramatically and expands its substrate repertoire. J Biol Chem. 2004;279:42787–42793. doi: 10.1074/jbc.M406890200. [DOI] [PubMed] [Google Scholar]