Abstract
α-Synuclein (α-syn) is the major component of the intraneuronal inclusions called Lewy bodies, which are the pathological hallmark of Parkinson’s disease. α-Syn is capable of self-assembly into many different species, such as soluble oligomers and fibrils. Even though attempts to resolve the structures of the protein have been made, detailed understanding about the structures and their relationship with the different aggregation steps is lacking, which is of interest to provide insights into the pathogenic mechanism of Parkinson’s disease. Here we report the structural flexibility of α-syn monomers and dimers in an aqueous solution environment as probed by single-molecule time-lapse high-speed AFM. In addition, we present the molecular basis for the structural transitions using discrete molecular dynamics (DMD) simulations. α-Syn monomers assume a globular conformation, which is capable of forming tail-like protrusions over dozens of seconds. Importantly, a globular monomer can adopt fully extended conformations. Dimers, on the other hand, are less dynamic and show a dumbbell conformation that experiences morphological changes over time. DMD simulations revealed that the α-syn monomer consists of several tightly packed small helices. The tail-like protrusions are also helical with a small β-sheet, acting as a “hinge”. Monomers within dimers have a large interfacial interaction area and are stabilized by interactions in the non-amyloid central (NAC) regions. Furthermore, the dimer NAC-region of each α-syn monomer forms a β-rich segment. Moreover, NAC-regions are located in the hydrophobic core of the dimer.
I. INTRODUCTION
α-Synuclein (α-syn) is an intrinsically disordered presynaptic protein, the aggregation of which is associated with development of Parkinson’s disease (PD), the second most common neurodegenerative disorder.1 α-Syn may contribute to PD pathogenesis in a number of ways, but it is generally linked to the pathological lesions in the brain, called Lewy bodies, consisting of self-assembled α-syn.2 The aggregation of α-syn is a complex process and the factors and mechanisms of formation of protein nano-assemblies (with different morphologies such as fibrils, spherical oligomers, and nanopores) are poorly understood.3 α-Syn exists as a compact disordered monomer in mammalian cells4 and can, in solution, adopt a natively unfolded monomeric structure.5 Additionally, evidence suggests that it can also exist as an oligomer in the cytosol.6 The protein consists of three domains: The N-terminal domain, spanning residues 1-60, contains five conserved lysine-rich repeats. The non-amyloid central (NAC) region, spanning residues 61-95, contains a sixth lysine-rich repeat and being highly hydrophobic. A key feature of this region is the presence of a segment, spanning residues 71-82, that is required for α-syn aggregation.7 The C-terminal region, spanning residues 96-140, is enriched with proline and acidic residues and thought to regulate α-syn aggregation through auto-inhibitory long-range interactions,8,9 with electrostatic interactions mediated by the acidic residues playing a major role in increasing the fibrillization lag time.10
The ability of α-syn to assemble in aggregates of various types suggests that the protein is dynamic, a feature that was directly confirmed by various single-molecule biophysical techniques. The application of the single-molecule fluorescence resonance energy transfer (FRET) approach demonstrated that α-syn undergoes conformational transitions between a natively unfolded state and multiple α-helical structures.11 Combination of FRET with fast mixing microfluidic methodology made it possible to probe kinetics of conformational transitions of α-syn and characterize the role of lipids in conformational transition of the protein.12 Single-molecule AFM force spectroscopy has also been applied to characterize the assembly of α-syn in dimers.13–17 These studies revealed that specific segments of α-syn enable the assembly of the dimers and the role of mutations in the protein on the pattern of the dimers assembly.17,18 Additionally, the stability of α-syn dimers was measured by AFM force spectroscopy19 and these results coincided with direct measurements performed by the single-molecule fluorescence tethered approach.20
Here we applied the single-molecule time-lapse high-speed AFM (HS-AFM) approach to characterize dynamics of α-syn monomers and self-assembled dimers by imaging the protein in buffer solution. Recently, we demonstrated that this cutting-edge AFM instrument is capable of, at nanoscale, directly visualizing dynamics of SOD1 protein in monomeric and oligomeric forms.21 We find here that although the primary morphology of the α-syn monomer is globular, the protein does not remain in this conformation. Protrusions appear on the globule and demonstrate dynamic behavior with time. Importantly, a globular monomer can adopt fully extended conformations, which is a characteristic feature of intrinsically disordered proteins (IDPs). Dimers, on the other hand, are less dynamics and show a dumbbell conformation that experiences morphological changes over time. Extended protrusions from the dimer globule also appear. To characterize the underlying mechanism of the dynamic behavior, we applied computational modeling, in the form of discrete molecular dynamics (DMD) simulations,22–24 to gain insights into the structural transitions of α-syn.
II. MATERIALS AND METHODS
A. Sample preparation for AFM imaging
Wild-type α-syn protein was provided by Rochet and stock solutions were prepared as described in Ref. 20. Mica disks, 1.5 mm in diameter, were glued to a glass rod of the AFM instrument as described in Refs. 25 and 21 and, after cleavage, were modified with 167 μM aminopropyl silatrane (APS) for 30 min to make APS-mica.26 After thoroughly rinsing the functionalized surface with milli-Q water, followed by phosphate-buffered saline (PBS) buffer (pH 7.4), 2 μl of 30 nM solution of α-syn in the same buffer was deposited on the surface and incubated for 5 min. The surface was then rinsed thoroughly with PBS buffer, which was also used as imaging buffer—the sample was never allowed to dry during preparation or data acquisition. Briefly, 2 μl of buffer was gently placed on the top of the mica disc. The droplet was carefully sucked into kimwipe paper. The steps were repeated several times to remove nonbound protein.
B. HS-AFM procedure
The HS-AFM images were acquired using the HS-AFM instrument (RIBM, Tsukuba, Japan) as described previously.21,25 Electron beam deposition (EBD) modified Olympus BL-AC10DS probes were used for imaging. The spring constants of the probes were between 0.1 and 0.2 N/m, with the resonance frequency 400-700 kHz in aqueous solutions. Data were collected by continuously scanning over an area of 50 nm × 50 nm with scan rate 5 frames/s and resolution of 128 × 128 pixels.
We generated HS-AFM images using home-built software written by Atsushi Miyagi, Y.L.L. laboratory, University of Nebraska Medical Center, Omaha, NE. The contour of the tail was obtained along the tail profile and fitted with parametric splines, see, for example, Fig. S1 of the supplementary material. For two tails’ case, the contour of each tail was calculated separately. Data analysis was performed using Femtoscan Online software (Advanced Technologies Center, Moscow, Russia), with the stiffness of tails formed on the polypeptide chain being calculated according to the following equation, described in Refs. 27–30:
| (1) |
The persistence length (p) represents the stiffness of the polymer, where l is the contour length between the two points on the polymer and r is the end-to-end distance.
For volume calculation, the compact spherical feature was taken as a region of interest (ROI) and the boundaries of ROI were determined by the isoline function in Femtoscan. Afterwards, the volume of this region was calculated, see, for example, Fig. S2 of the supplementary material.
C. Aggregation propensity
The aggregation propensity of the wild-type α-Syn protein was calculated using the Zyggregator software through the website (http://www-mvsoftware.ch.cam.ac.uk/index.php/zyggregator). If the value was above the line at , it was determined as the aggregation-prone region.31
D. Graphic and modeling software
All the figures were plotted using Igor Pro. 6.3.4 (WaveMetrics, Lake Oswego, OR, USA). Simulation snapshots were generated using VMD software.32
E. Discrete molecular dynamics simulations
The structures of the wild-type α-syn monomer and aggregates were obtained using the DMD simulation methodology.22–24 DMD is an event-based simulation engine that at every time step solves a series of two-body collisions, in which colliding atoms’ velocities change instantaneously according to the conservation laws of energy, momentum, and angular momentum. In contrast to conventional MD simulations, DMD uses discrete approximation for continuous inter-atom potentials.33 The Lazaridis-Karplus implicit solvation model34 is adopted to account for the solvation energy, while temperature of the system is controlled with the Andersen thermostat.35
We started our simulations with unfolded conformation of the monomeric α-syn and explore the conformational landscapes of the protein using replica exchange discrete molecular dynamics (REX/DMD) simulations.36 The initial structure was obtained by running a short high temperature DMD simulation of the protein with published nuclear magnetic resonance structure (PDB-ID: 2KKW).37
During REX/DMD simulations, we use the replica exchange approach to efficiently explore the conformational landscape of the protein. In REX/DMD, multiple simulations of the same system at different temperatures (i.e., replicates) are performed in parallel. Different replicates then periodically exchange based on the Metropolis criterion allowing the system to overcome local energetic traps and fully explore free energy landscape.
For the α-syn monomer, we use 24 parallel replicas with temperatures ranging from 0.375 to 0.605 kcal/(mol kB). We run the simulations for 7 × 106 time steps (corresponding to approximately ∼400 ns) per replica. By monitoring the convergence of the heat capacity curves, we determine when the simulations have reached equilibrium.
We assign the first 106 time steps of simulations as system equilibration and omit them from our analyses. In order to determine the relative thermal stability of α-syn structures, we computed their heat capacities using the WHAM algorithm.38 We then isolated the most populated clusters based on monomer conformations from the structures with the lowest energy in our REX/DMD simulations and defined their centroids as representative protein models.
The same strategy was used to explore the conformational landscape of the wild-type dimer of the α-syn protein. We generated the starting structure of the α-syn dimer by aligning two copies of the same NMR structures of monomeric α-syn (PDB-ID: 2KKW37) and then performing DMD.
III. RESULTS
A. Dynamics of -syn monomers
We have recently characterized wild-type α-syn by imaging dried samples of the protein with AFM.21 The protein appeared as a compact globule with the volume corresponding to the expected size of 14 kDa protein. Here, α-syn samples were imaged in aqueous environment in an attempt to visualize potential dynamics of the protein at sub-second scale using HS-AFM.
1. Imaging of -syn monomers with HS-AFM
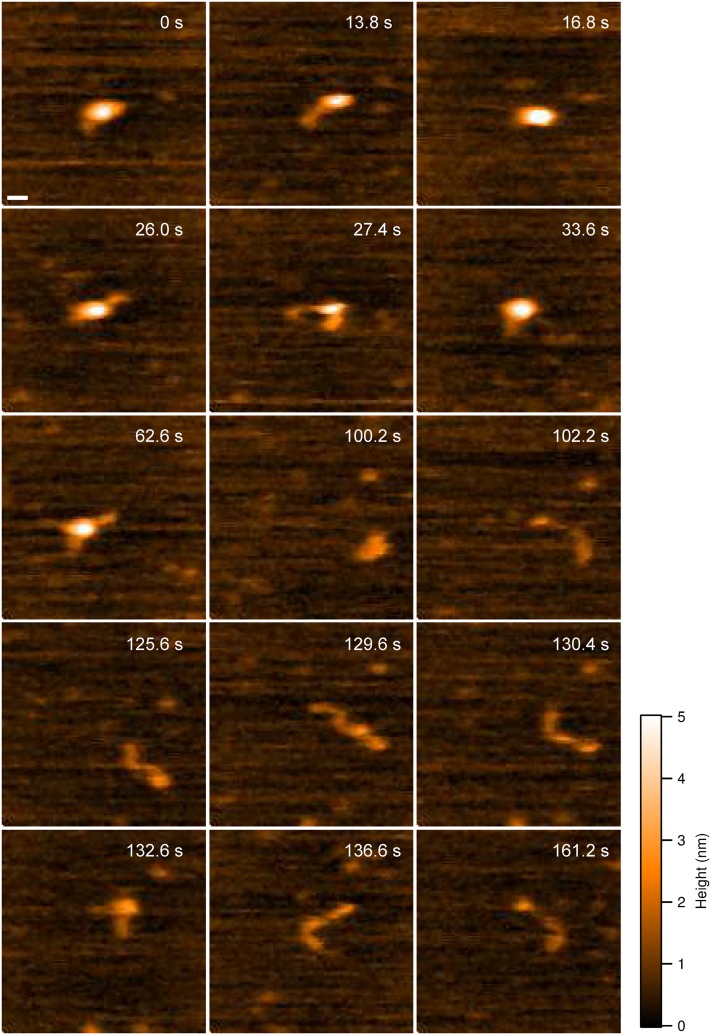
Samples of wild-type α-syn were prepared in PBS buffer (pH 7.4), deposited on APS functionalized mica, and imaged (without being dried) with HS-AFM. Similar to data for dried α-syn samples, we observed that the protein in buffer assumes a compact globular conformation with limited mobility and dynamics. These observations are assembled as movie S1 of the supplementary material, with one frame from the movie shown in Fig. S3A of the supplementary material. In such a conformation, the protein remains spherical over an extended period of time (200 s, n = 59), suggesting that no significant structural changes occur. However, this is not the only conformation of α-syn monomers identified. We observed several other conformations for the α-syn monomer: a one-tail (Fig. S3B of the supplementary material) and two-tail structures (Fig. S3C of the supplementary material) with tails protruding from the main globular segment. The formation of such protrusions from a globular conformation is presented as movie S2 of the supplementary material. Quantitative analysis showed that the length of the tails follows a lognormal distribution, with the main peak at 5.4 ± 0.3 nm (Fig. S3D of the supplementary material). Statistical analysis, using data acquired in multiple experiments, showed that the compact conformation, as shown in Fig. S3A of the supplementary material, is the major species occurring 75% (n = 59, duration time = 200 s) of the time. Since the conformation of the tail-like conformation is quite dynamic (Fig. 1 and movie S3 of the supplementary material) and there are always random transitions between different species, we only take the number of conformations rather than the number of monomers into account for the lifetime estimation in each case. The lifetime of one-tail (n = 68) and two-tail (n = 66) conformations is 3.0 s and 0.6 s, respectively.
FIG. 1.
HS-AFM images of wild-type α-syn monomers. Selected frames from movie S3 of the supplementary material showing the structural transition of α-syn monomer over time. The monomer starts in a globular conformation (0 s) and transitions to one-tailed (13.8 s, 26.0 s, 33.6 s, and 125.6 s), two-tailed (27.4 s, 62.6 s, 129.6 s, and 132.6 s), and extended conformations (130.4 s and 136.6 s). Interspersed with the other conformations, the monomer transitions back to a globular conformation (16.8 s and 100.2 s). The scale bar is 5 nm.
2. Transitions from globular to extended conformations revealed by HS-AFM
The globular conformations described above are able to transition into fully extended conformations. One such transition is shown in movie S3 of the supplementary material as well as frames in Fig. 1. The monomer starts in a globular conformation (Fig. 1, frame 0 s) that transitions to a one-tailed state (Fig. 1, frame 13.8 s), followed by fluctuation between globular and one-tailed conformations (Fig. 1, 16.8 s and 26 s). These events are then followed by the transition to two-tailed structure (Fig. 1, frame 27.4 s); the conversion from the globular directly two-tailed conformations was also observed (Fig. 1, frames 27.4 s, 33.6 s, 62.6 s, and 100.2 s). Interestingly, transition between different conformations with tails and extended structures with compact terminal segments (two heads, Fig. 1, frame 102.2 s) was also observed (Fig. 1, frames 125.6 s, 129.6 s, and 130.4 s). Eventually, the monomer adopts an extended conformation (Fig. 1, frames 136.6 s and 161.2 s).
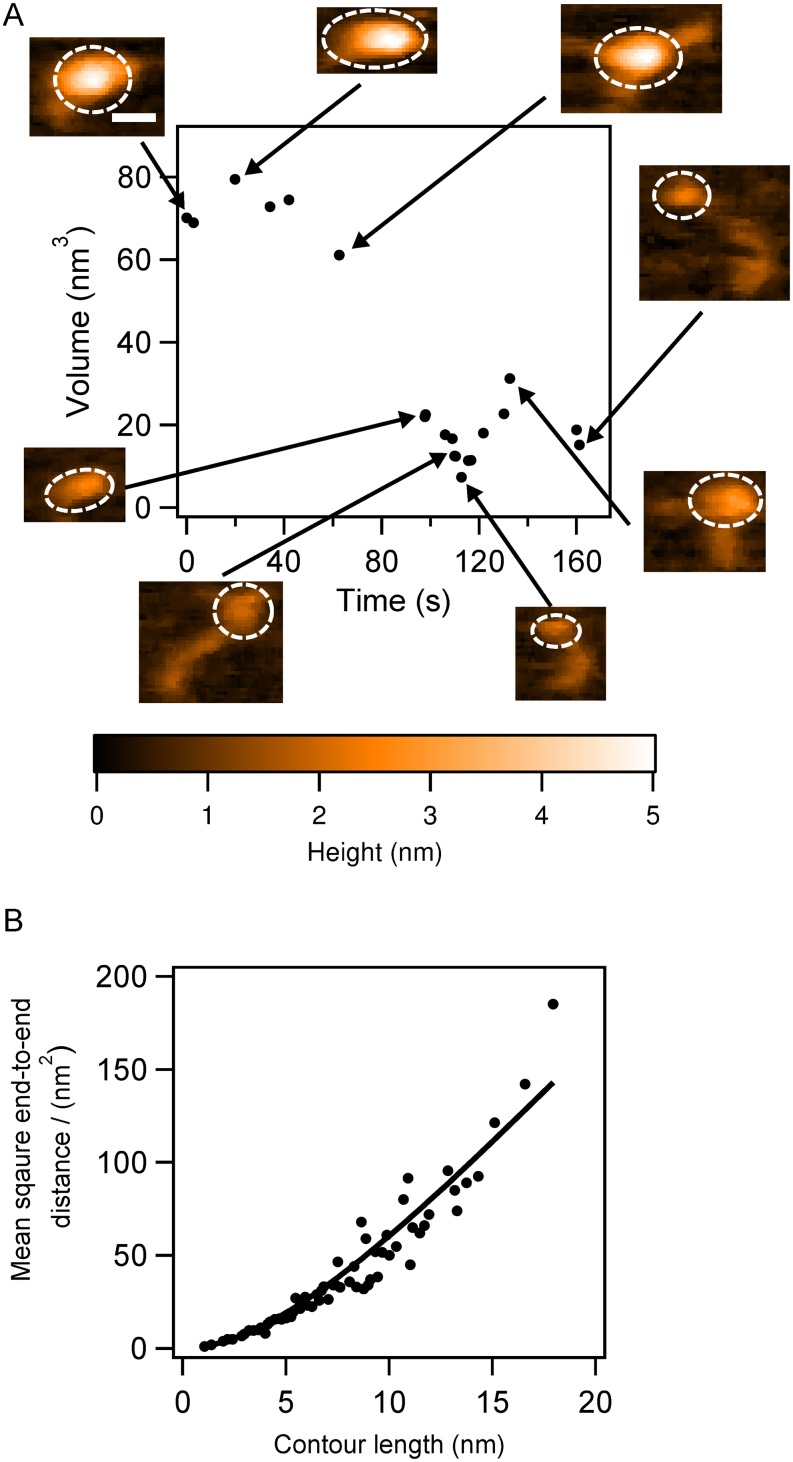
To quantitatively describe the conformational dynamics, we measured the time-dependent change in volume of the globular regions. The relative volume measurements for the monomer shown in Fig. 1 and movie S3 of the supplementary material are graphically displayed in Fig. 2(a). Selected frames are also included in the volume plot to illustrate the different conformations. The volume is initially ca. 70 nm3 [Fig. 2(a), 0 s]; when tails partially protrude from the compact structure, the volume drops to ca. 60 nm3 [Fig. 2(a), 62.6 s]. Further unraveling of the compact structure leads to a drop in volume to ∼20 nm3 [Fig. 2(a), 97.8 s, 106 s, 112.8 s, and 161.2 s]. This trend is reversed when the tails become shorter [Fig. 2(a), 132.6 s].
FIG. 2.
The analysis of the α-syn monomer undergoing structural transition. (a) The monomer volume (black dots) with corresponding frames at specific time points, presented as an inset in the figure, with dashed circles indicating the segments used for volume analysis. (b) The stiffness of the tail-like protrusions in the α-syn monomers. The black dots are raw data and the solid line is the fitted curve; the persistence length is 2.9 nm. The scale bar is 5 nm.
To characterize the properties of the protruding tails, we employed a 2D approach developed to analyze dynamics of extended conformations of intrinsically disordered proteins with AFM time-lapse experiments.28,31 By treating the protruding tails of the α-syn monomers as a polymer on a 2D surface, Eq. (1) allows one to obtain the persistence length of the polymer. We measured the lengths of tails in the different frames, calculated the end-to-end distances, and plotted the dependence of the square of this value against the contour length [Fig. 2(b)]. Fitting the data using Eq. (1) yielded a persistence length, p = 2.9 nm. This value is approximately five-fold smaller than that previously reported for a similar intrinsically disordered chromatin transcription (FACT) protein,28 suggesting that the tails of α-syn monomers are more flexible and that their mechanical properties are different.
3. Extended conformations of -syn conformations revealed by HS-AFM
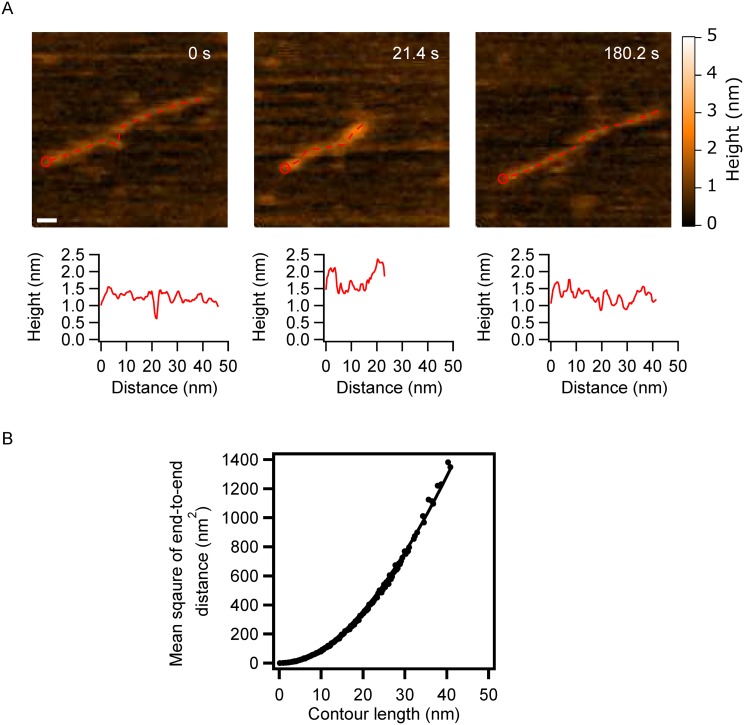
On Fig. 1, in addition to conformations of α-syn with tails, we observe extended conformations in which the protein appears as filaments of different lengths (130.4 s and 136.6 s). Additional experiment revealed that, while not common, these fully extended, filamentous, conformations can occur for an extended time period. Dynamics of such filamentous conformation are presented by selected frames in Fig. 3(a) and movie S4 of the supplementary material. In the initial image (frame 0), α-syn appears as a filament with the length 44 nm, which is slightly shorter than the estimated length of the fully stretched α-syn monomer (ca. 50 nm). Later (21.4 s), the filament compresses approximately two-fold followed by gradual extension to approximately 39 nm, at the end of the observation. Cross sections along the filament were made to compare the morphologies of different segments; they are shown below each frame in Fig. 3(a). The proximal terminal segment of the filament has the highest brightness, which suggests that this part of the protein filament is the most condensed. Due to the high flexibility of the filament and its thermal motion, some segments can transiently dissociate from the surface that leads to their low height values. The filament becomes fully detectable again (21.4 s) in the conformation with compact termini. In the final frame (180.2 s), with the monomer extended, the profile resembles the initial profile (0 s) with the condensed proximal end of the filament.
FIG. 3.
α-Syn monomers with extended conformations. (a) Selected frames showing α-syn monomers in several extended conformations. Cross-sectional length analysis of the monomers is presented below each frame. The starting point of the profile areas indicated by red circles and the profile follows the red dashed line in the images. The scale bar is 5 nm. (b) The stiffness of the extended α-syn monomer. Raw data are plotted as black dots, while the solid line is the fitted curve. The persistence length is 28.6 nm.
The mechanical properties of the extended conformation were investigated using the approach described above. We measured the end-to-end distance of the filaments in each frame and plotted these against the contour length, the data were then fit with Eq. (1) [Fig. 3(b)]. The analysis yielded a persistence length of p = 28.6 nm, which compared to the FACT protein and α-syn monomer tails is two- and 10-fold larger, respectively. The two-fold difference, compared to the FACT protein, is not significant and may be due to the sample preparation procedures and the surface properties. However, compared to the protruding tails, the extended monomer conformation is significantly more rigid. The persistence length in the AFM experiment depends on the interaction of the polymer with the surface. Given that the contact area between the longer polymer and surface is larger than that for the short tail, it can lead to the higher value for the persistence length.
4. Insight into molecular basis of monomer conformations from computational analysis
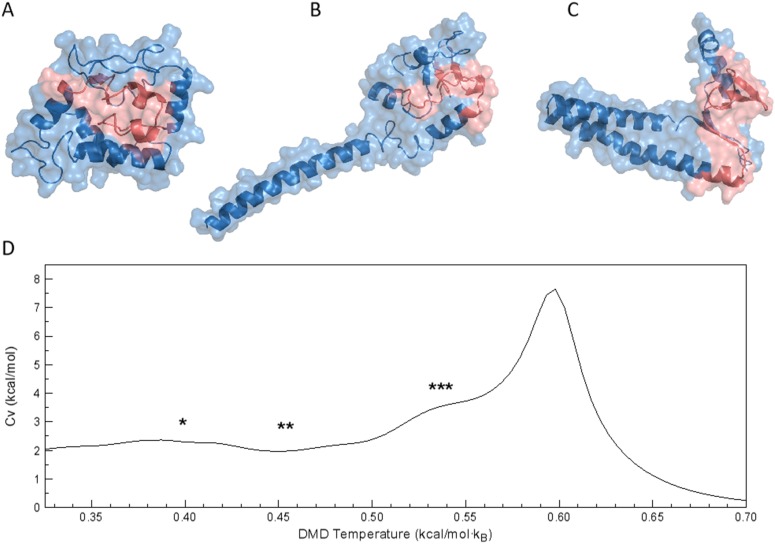
The structural basis of the different conformation of the α-syn monomer was investigated using DMD simulations. Similar to conformations identified in the HS-AFM experiments, DMD simulations yielded both globular and tailed structures. Analyzing the DMD trajectories, we found that the α-syn monomer was present mostly in the compact globular conformation, consisting of several tightly packed small helices [Fig. 4(a)]. This structure represents the most populated lowest energy cluster, accounting for ∼76% of overall population. The second most populated cluster [Fig. 4(b), ∼14% of overall population] has an extended α-helical tail elongating from the core of the protein. The third conformation [Fig. 4(c)] accounts for ∼5% of the overall population and has a small β-sheet, acting as a hinge, between two segments of extended α-helices. The geometry of these two conformations, with extended helices, resembles the one- and two-tailed conformations identified in the HS-AFM experiments. Furthermore, the fractional representation of the different conformations is also closely matched between experiment and simulations. Interestingly, the specific heat plot [Fig. 4(d)], obtained from analysis of REX/DMD simulations, indicates a wide smooth shoulder preceding the main unfolding peak. All predicted models (Fig. 4) correspond to the shoulder, which means that there is no major energy difference between them and that the α-syn monomer is able to freely transition between different conformations. This behavior is usual for IDPs.39
FIG. 4.
DMD models for α-syn monomers. Representative structures of α-syn monomers, determined as the centroids of the most populated lowest energy clusters. Structures from panels (a)–(c) represent the three most populated lowest energy clusters, with population ∼76%, ∼14%, and ∼5%, respectively. NAC-region of the protein, residues 61-95, is colored in red. (d) The heat capacity (Cv) curve for the α-syn monomer obtained using WHAM analysis of REX/DMD trajectories. The biggest peak corresponds to the unfolding of the protein. Asterisks mark temperature regions on the Cv plot corresponding to the temperatures at which representative structures form panel (a)(*), (b)(**), and (c)(***), respectively, were found during the simulation.
B. Dynamics of -syn dimers
1. Dynamics of -syn monomers revealed by HS-AFM
Next, we characterized the dynamics of α-syn dimers, detected in the same sample as monomers. Figure 5 illustrates the dynamics of the dimers, in which two monomers appear as globular structures. This type of dimers, termed type 1, comprises 80% (n = 18) of the observed dimer conformations. The full range of dynamics for type 1 dimers is assembled as movie S5 of the supplementary material. During the entire observation window, the monomers remain in globular conformations; however, the distance between the two monomers fluctuates. Compared to α-syn monomers, type 1 dimers show significantly less structural dynamics. Cross sections, along the primary axis of the dimer, reveal fluctuations in the peak-to-peak distance of the monomers; line profiles are placed below each frame as Fig. 5(b). However, no other dynamic behavior was observed.
FIG. 5.
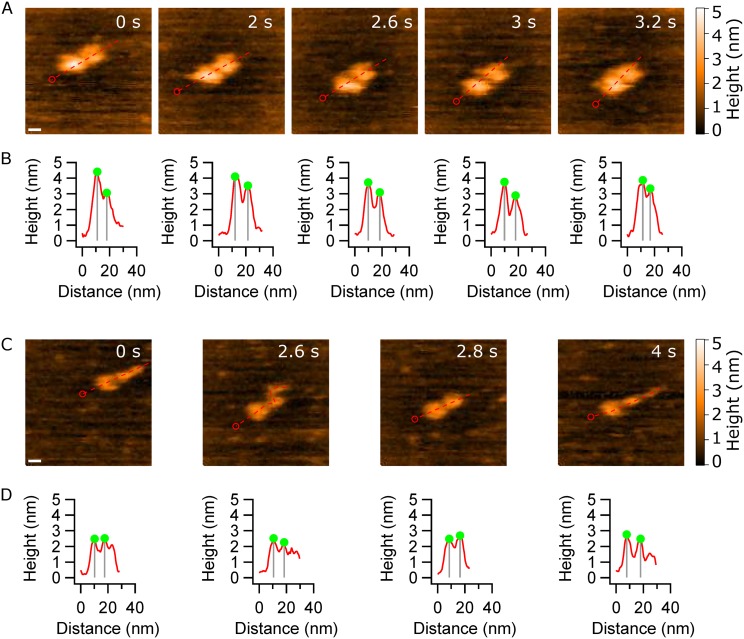
HS-AFM images of α-syn dimers. (a) The α-syn dimer consists of two compact monomers that move apart and come together as the experiment progresses. This type of dimer is termed type 1. The scale bar is 5 nm. (b) Cross-sectional analysis of the dimers clearly shows fluctuations in the peak-to-peak distance between the monomers. (c) The other type of dimer, type 2, consists of a globular monomer and another monomer that is able to form extended protrusion. The scale bar is 5 nm. (d) Cross sections of (c) showing the effect of protrusion in the α-syn monomer. All profiles start at the red circle and follow the dashed line.
The other dimer conformation, accounting for ∼20% (n = 5) of the total population, is termed type 2. In these dimers, one monomer remains in the compact globular conformation, while the other monomer extends forming a flexible tail [Fig. 5(c)]. The captured dynamics of the type 2 dimer is assembled as movie S6 of the supplementary material. At frame 0 s, one monomer is compact, whereas the second monomer forms a protrusion. The next frame (2.6 s) demonstrates that the protrusion is mobile and very dynamic, as seen by the transition back to a compact (2.8 s) and later extended conformation (4 s). This structural flexibility of the monomer within the dimer is similar to observations for isolated monomers (Fig. 1 and movie S4 of the supplementary material). In both cases, the monomer is able to transition from globular conformations and form protrusion and extended conformations. However, even though similar behavior is shown, the dimer is less dynamics than the monomer. Potentially the decreased dynamic behavior may be due to the interactions needed to keep the dimer intact. We also analyzed the cross sections to characterize the type 2 dimers [Fig. 5(d)]. The profiles show that the peak-to-peak distance between the monomers fluctuates from 7.4 nm to 10.1 nm, while a considerable part of one monomer is extends and contracts.
2. Structural elements of the dimer conformations identified by DMD simulations
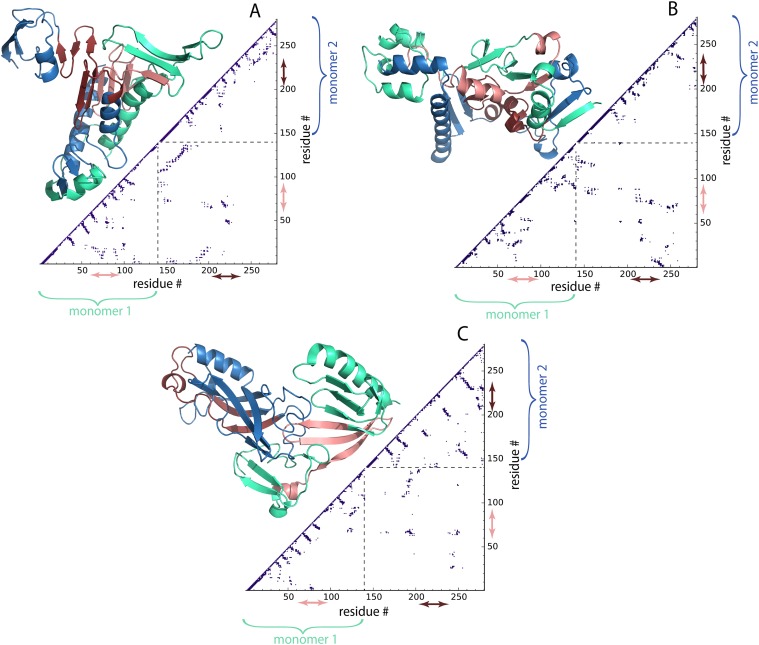
In order to study the structural details of the wild type α-syn, we performed DMD simulations according to the procedure described in Sec. II. DMD simulations of the dimer revealed that the compact dimer is stabilized by interactions in the NAC region and that the monomers have a large interfacial interaction area (Fig. 6). Three representative structures corresponding to the centroids of the most populated lowest energy clusters are shown in Fig. 6. These conformations make-up ∼58%, ∼27%, and ∼11% of the dimer population, respectively. We found that the dimer NAC-region of each α-syn monomer forms a β-rich segment. Moreover, in two of the models, corresponding to the most populated clusters [Figs. 6(a) and 6(b)], NAC-regions are located in the hydrophobic core of the dimer. This is also evident from the Cα contact map of the dimers, where the contact between the dimers is mostly concentrated in the NAC region with contributions from the N- and C-termini as well.
FIG. 6.
Structures of the wild type α-syn dimers predicted by DMD simulations with corresponding contact maps. Structures from panels (a)–(c) represent three most populated lowest energy clusters with population ∼58%, ∼27%, and ∼11%, respectively. Monomer 1 of the dimer is colored green and contains residues from 1 to 140; monomer 2 is colored blue and has residues 141–280. The NAC-regions of the monomer 1, residues 61-95, and monomer 2, residues 201-235, are colored in pink and red, respectfully. Contact maps, lower right of the structures, reveal residue-wise specific contacts within the dimers; Cα atoms within 7.5 Å are considered being in contact. Dashed lines on the plots divide regions corresponding to different monomers forming the dimer. The regions inside the dashed square illustrate contacts between the two monomers in the dimer (141-280 on the horizontal axis and 1-140 on the vertical axis). Red and pink arrows indicate fragments along the sequence corresponding to the NAC-regions of each chain.
Geometrical comparison between DMD simulation results (Fig. 6) and conformations obtained during HS-AFM experiments show good agreement. The compact dimer conformation (type 1 dimer), represented by the dimer in Fig. 6(b), consists of two dense structured regions at each end. From the contact map, it is clear that the individual monomers have extensive intra-peptide interactions; the pattern of these contacts reveals the structural organization within the dimer; the monomers are interacting in an extended mode with the N and C-termini at each end and the NAC regions providing stability. The tailed structures consist of similar secondary structure elements and form two groups with short dense tails [Figs. 6(a) and 6(c)]. The dimers are stabilized by interaction of the NAC regions of the monomers and have an inter-peptide β-sheet structure. However, their monomer-monomer interaction pattern, and thus organization, is different. The type 2 dimer [Fig. 6(a)] is organized in an almost symmetrical conformation, where both monomers contribute equally to the extended segment; this gives the monomers a very large interaction cross section. The type 2 dimer has an extensive helical structure in the extended tail, while the NAC regions form a β-sheet. For type 3 dimers [Fig. 6(c)], the monomers are arranged separately in extended (monomer 1) and compact (monomer 2) conformations. This arrangement may provide the necessary flexibility for one of the monomers to form extended protrusions, which is in line with HS-AFM results where one monomer stays compact at all times while another is dynamic and undergoes extension.
IV. DISCUSSION
Comparison of HS-AFM experimental results with DMD simulations provides insights into the structural dynamics of α-syn monomers and dimers at nanoscale. Previously, structural studies of the α-syn monomer, in the presence of micelles, suggested that residues 3-37 and 45-92 are prone to form helical conformations, while residues 98-140 remained unstructured.40 In comparison, free in solution and under physiological conditions (in absence of binding partners), α-syn monomers are intrinsically unstructured.8,41,42 Our HS-AFM results demonstrate that the WT α-syn monomer adopts compact configurations, which suggests that unlike in fibrils,43–45 the extended β-sheet pattern is not favorable in the monomeric state. The α-Syn primary structure is important for the compaction: N-terminal region, amphipathic α-helices (with repeated KTKEGV motif) from residue 1 to 60; NAC, hydrophobic and highly amyloidogenic non-Aβ component, from residues 61 to 95; C-terminal, highly enriched in acidic residues and prolines, especially from residues 120-140 (8 negative charges).3 The former two regions show high aggregation propensities (Fig. S4 of the supplementary material) and contain the membrane binding domains.46,47 Mutations, A30P, E46K, and A53T, important for the disease development are found in the first region48–50 and alter the structure of α-syn in different ways.51 The C-terminal region participates in protein-protein interactions. Based on the HS-AFM images and DMD modeling, we speculate that the hydrophobic interaction facilitates the compact monomer formation, seen as the blue region in Fig. S4 of the supplementary material. Indeed, deletion of central residues from α-syn can interfere with the fibril formation.7
Conversion between compact and extended morphology was also observed for both monomers and dimers. In particular, as shown in Fig. 1 (monomeric case), over a long observation period (160 s), the monomer fluctuates between compact, tail-like, and an extended conformation. Several studies have proposed to characterize the possibility of intermediate states of α-syn proteins.45,52,53 Specially, tail-like intermediate structures of α-syn have been suggested based on MD simulations, EM, and SAXS experiments.54,55 It is well known that tail-like structures implicate several biological functions in the intrinsically disordered proteins.56 As observed during DMD simulations, in the absence of lipids, residues within N- and C-terminal regions are flexible; we infer that this plays an important role in the frequent structural conversion of the monomers. Interestingly, when two such monomers form a dimer, the compact structure is stabilized, and structural conversions are much less common. Altogether, the variety of monomer and dimer structures further prove the intrinsic heterogeneity of the α-syn protein and can explain conformational transitions during the aggregation process and formation of highly ordered fibrillar structure.
It is noteworthy that the dynamics process of α-syn can be accelerated or retarded in the presence of surfaces.57 Thus, the observed behavior of α-syn may be different from in vivo cases, as illustrated in the previous fibril elongation studies via a “stop-and-go” mechanism.58–60 The interaction of the protein with the surface can lead to protein extension, retarding the α-syn fibril elongation. This is an important issue in terms of the role of membrane and other surfaces on the structure of amyloid proteins, and elucidation of these effects is our future goal.
In the current study, we used a positively charged APS mica surface as a substrate for HS-AFM samples.61 On this surface, we were able to visualize a structural dynamics and transitions of monomers as well as dimers (Figs. 1 and 5). The use of surfaces of different types would allow us to evaluate the role of the surface effect on the α-syn dynamics. AFM is a topographic technique in which images are generated by the scanning tip. Therefore, one should expect the effect of the tip on the sample dynamics in such a way that motion of tails can be altered by the tip. However, this effect is low for HS-AFM as analyzed in our previous work.62 Briefly, the overall energy transferred to the sample due to the tapping is negligible and readily distributed in the surrounding environment, including the water molecules. The second effect, which is the displacement of the sample by the tip, is also very low because of low oscillation amplitude, high oscillation frequency of the tip, and consequently low contact time of the tip with the sample. Therefore, the lateral displacement due to the scanning tip is also negligible. It is illustrated by the images. For instance, as shown in Fig. 1, we can see that the whole protein as well as the tail structure moves in the small area randomly rather than following the direction of tip movement, suggesting that the effect of the tip in these experiments is low, if any.
V. CONCLUSIONS
Our combined experimental and computational studies revealed broad range dynamics of α-syn in a monomeric state. The protein can adopt a fully extended conformation identified earlier in a number of IDP type proteins. Computational modeling suggests that the hydrophobic interaction facilitates the compact monomer formation. Assembly in dimers decreases the range of conformational transitions within monomers enabling them to assemble in a dimer. Computational modeling revealed three types of dimers defined by the monomer-monomer interaction pattern. For example, for type 3 dimers, the monomers are arranged separately in extended and compact conformations. This arrangement may provide the necessary flexibility for one of the monomers to form extended protrusions, which would be in line with HS-AFM results where one monomer stays compact at all times while the other is dynamic and undergoes extension. Altogether, the variety of monomer and dimer structures further prove the intrinsic heterogeneity of the α-syn protein and can explain conformational transitions during the aggregation process and formation of highly ordered fibrillar structure.
SUPPLEMENTARY MATERIAL
See supplementary material for Figs. S1–S4 and Movies S1–S6.
ACKNOWLEDGMENTS
This work was supported by the NIH Grant Nos. GM096039 and R01GM118006 to Y.L.L., the NIH Grant Nos. R01GM11401, R01GM064803, and R01GM123247 to N.V.D., and M.H. was partially supported by the Buckey Memorial Fellowship and the UNMC Graduate Fellowship.
REFERENCES
- 1.Huse D. M., Schulman K., Orsini L., Castelli-Haley J., Kennedy S., and Lenhart G., Mov. Disord. 20(11), 1449–1454 (2005). 10.1002/mds.20609 [DOI] [PubMed] [Google Scholar]
- 2.Spillantini M. G., Schmidt M. L., Lee V. M. Y., Trojanowski J. Q., Jakes R., and Goedert M., Nature 388(6645), 839–840 (1997). 10.1038/42166 [DOI] [PubMed] [Google Scholar]
- 3.Breydo L., Wu J. W., and Uversky V. N., Biochim. Biophys. Acta, Mol. Basis Dis. 1822(2), 261–285 (2012). 10.1016/j.bbadis.2011.10.002 [DOI] [PubMed] [Google Scholar]
- 4.Theillet F.-X., Binolfi A., Bekei B., Martorana A., Rose H. M., Stuiver M., Verzini S., Lorenz D., van Rossum M., Goldfarb D., and Selenko P., Nature 530(7588), 45–50 (2016). 10.1038/nature16531 [DOI] [PubMed] [Google Scholar]
- 5.Weinreb P. H., Zhen W., Poon A. W., Conway K. A., and Lansbury P. T., Biochemistry 35(43), 13709–13715 (1996). 10.1021/bi961799n [DOI] [PubMed] [Google Scholar]
- 6.Bartels T., Choi J. G., and Selkoe D. J., Nature 477(7362), 107–110 (2011). 10.1038/nature10324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Giasson B. I., Murray I. V. J., Trojanowski J. Q., and Lee V. M.-Y., J. Biol. Chem. 276(4), 2380–2386 (2001). 10.1074/jbc.m008919200 [DOI] [PubMed] [Google Scholar]
- 8.Dedmon M. M., Lindorff-Larsen K., Christodoulou J., Vendruscolo M., and Dobson C. M., J. Am. Chem. Soc. 127(2), 476–477 (2005). 10.1021/ja044834j [DOI] [PubMed] [Google Scholar]
- 9.Bertoncini C. W., Jung Y.-S., Fernandez C. O., Hoyer W., Griesinger C., Jovin T. M., and Zweckstetter M., Proc. Natl. Acad. Sci. U. S. A. 102(5), 1430–1435 (2005). 10.1073/pnas.0407146102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Levitan K., Chereau D., Cohen S. I. A., Knowles T. P. J., Dobson C. M., Fink A. L., Anderson J. P., Goldstein J. M., and Millhauser G. L., J. Mol. Biol. 411(2), 329–333 (2011). 10.1016/j.jmb.2011.05.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ferreon A. C. M., Gambin Y., Lemke E. A., and Deniz A. A., Proc. Natl. Acad. Sci. U. S. A. 106(14), 5645–5650 (2009). 10.1073/pnas.0809232106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gambin Y., VanDelinder V., Ferreon A. C. M., Lemke E. A., Groisman A., and Deniz A. A., Nat. Methods 8(3), 239–241 (2011). 10.1038/nmeth.1568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yu J., Malkova S., and Lyubchenko Y. L., J. Mol. Biol. 384(4), 992–1001 (2008). 10.1016/j.jmb.2008.10.006 [DOI] [PubMed] [Google Scholar]
- 14.Yu J. and Lyubchenko Y. L., J. Neuroimmune Pharmacol. 4(1), 10–16 (2009). 10.1007/s11481-008-9115-5 [DOI] [PubMed] [Google Scholar]
- 15.Yu J., Warnke J., and Lyubchenko Y. L., Nanomedicine 7(2), 146–152 (2011). 10.1016/j.nano.2010.08.001 [DOI] [PubMed] [Google Scholar]
- 16.Krasnoslobodtsev A. V., Peng J., Volkov I., Rochet J.-C., and Lyubchenko Y., Biophys. J. 104(2), 388a (2013). 10.1016/j.bpj.2012.11.2163 [DOI] [Google Scholar]
- 17.Krasnoslobodtsev A. V., Volkov I. L., Asiago J. M., Hindupur J., Rochet J. C., and Lyubchenko Y. L., Biochemistry 52(42), 7377–7386 (2013). 10.1021/bi401037z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Krasnoslobodtsev A. V., Peng J., Asiago J. M., Hindupur J., Rochet J. C., and Lyubchenko Y. L., PLoS One 7(5), e38099 (2012). 10.1371/journal.pone.0038099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lyubchenko Y. L., Zhang Y., Krasnoslobodtsev A., and Rochet J. C., in Handbook of Clinical Nanomedicine: From Bench to Bedside, edited by Bawa R., Audette G., and Rubinstein I. (Pan Stanford Publishing, Singapore, 2015), pp. 1500+. [Google Scholar]
- 20.Lv Z., Krasnoslobodtsev A. V., Zhang Y., Ysselstein D., Rochet J. C., Blanchard S. C., and Lyubchenko Y. L., Biophys. J. 108(8), 2038–2047 (2015). 10.1016/j.bpj.2015.03.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Proctor E. A., Fee L., Tao Y., Redler R. L., Fay J. M., Zhang Y., Lv Z., Mercer I. P., Deshmukh M., Lyubchenko Y. L., and Dokholyan N. V., Proc. Natl. Acad. Sci. U. S. A. 113(3), 614–619 (2016). 10.1073/pnas.1516725113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dokholyan N. V., Buldyrev S. V., Stanley H. E., and Shakhnovich E. I., Folding Des. 3(6), 577–587 (1998). 10.1016/s1359-0278(98)00072-8 [DOI] [PubMed] [Google Scholar]
- 23.Ding F., Tsao D., Nie H., and Dokholyan N. V., Structure 16(7), 1010–1018 (2008). 10.1016/j.str.2008.03.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Proctor E. A. and Dokholyan N. V., Curr. Opin. Struct. Biol. 37, 9–13 (2016). 10.1016/j.sbi.2015.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miyagi A., Ando T., and Lyubchenko Y. L., Biochemistry 50(37), 7901–7908 (2011). 10.1021/bi200946z [DOI] [PubMed] [Google Scholar]
- 26.Shlyakhtenko L. S., Gall A. A., Filonov A., Cerovac Z., Lushnikov A., and Lyubchenko Y. L., Ultramicroscopy 97(1), 279–287 (2003). 10.1016/s0304-3991(03)00053-6 [DOI] [PubMed] [Google Scholar]
- 27.Rivetti C., Guthold M., and Bustamante C., J. Mol. Biol. 264(5), 919–932 (1996). 10.1006/jmbi.1996.0687 [DOI] [PubMed] [Google Scholar]
- 28.Miyagi A., Tsunaka Y., Uchihashi T., Mayanagi K., Hirose S., Morikawa K., and Ando T., ChemPhysChem 9(13), 1859–1866 (2008). 10.1002/cphc.200800210 [DOI] [PubMed] [Google Scholar]
- 29.Ando T. and Kodera N., Methods Mol. Biol. 896, 57–69 (2012). 10.1007/978-1-4614-3704-8_4 [DOI] [PubMed] [Google Scholar]
- 30.Lamour G., Yip C. K., Li H., and Gsponer J., ACS Nano 8, 3851 (2014). 10.1021/nn5007013 [DOI] [PubMed] [Google Scholar]
- 31.Pawar A. P., DuBay K. F., Zurdo J., Chiti F., Vendruscolo M., and Dobson C. M., J. Mol. Biol. 350(2), 379–392 (2005). 10.1016/j.jmb.2005.04.016 [DOI] [PubMed] [Google Scholar]
- 32.Humphrey W., Dalke A., and Schulten K., J. Mol. Graphics 14(1), 33–38 (1996). 10.1016/0263-7855(96)00018-5 [DOI] [PubMed] [Google Scholar]
- 33.Yin S., Biedermannova L., Vondrasek J., and Dokholyan N. V., J. Chem. Inf. Model. 48(8), 1656–1662 (2008). 10.1021/ci8001167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lazaridis T. and Karplus M., Curr. Opin. Struct. Biol. 10(2), 139–145 (2000). 10.1016/s0959-440x(00)00063-4 [DOI] [PubMed] [Google Scholar]
- 35.Andersen H. C., J. Chem. Phys. 72(4), 2384–2393 (1980). 10.1063/1.439486 [DOI] [Google Scholar]
- 36.Okamoto Y., J. Mol. Graphics Modell. 22(5), 425–439 (2004). 10.1016/j.jmgm.2003.12.009 [DOI] [PubMed] [Google Scholar]
- 37.Rao J. N., Jao C. C., Hegde B. G., Langen R., and Ulmer T. S., J. Am. Chem. Soc. 132(25), 8657–8668 (2010). 10.1021/ja100646t [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chodera J. D., Swope W. C., Pitera J. W., Seok C., and Dill K. A., J. Chem. Theory Comput. 3(1), 26–41 (2007). 10.1021/ct0502864 [DOI] [PubMed] [Google Scholar]
- 39.Pauwels K., Lebrun P., and Tompa P., Cell. Mol. Life Sci. 74(17), 3185–3204 (2017). 10.1007/s00018-017-2561-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ulmer T. S., Bax A., Cole N. B., and Nussbaum R. L., J. Biol. Chem. 280(10), 9595–9603 (2005). 10.1074/jbc.m411805200 [DOI] [PubMed] [Google Scholar]
- 41.Bernadó P., Bertoncini C. W., Griesinger C., Zweckstetter M., and Blackledge M., J. Am. Chem. Soc. 127(51), 17968–17969 (2005). 10.1021/ja055538p [DOI] [PubMed] [Google Scholar]
- 42.Eliezer D., Kutluay E., R. Bussell, Jr., and Browne G., J. Mol. Biol. 307(4), 1061–1073 (2001). 10.1006/jmbi.2001.4538 [DOI] [PubMed] [Google Scholar]
- 43.Vilar M., Chou H.-T., Lührs T., Maji S. K., Riek-Loher D., Verel R., Manning G., Stahlberg H., and Riek R., Proc. Natl. Acad. Sci. U. S. A. 105(25), 8637–8642 (2008). 10.1073/pnas.0712179105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Heise H., Hoyer W., Becker S., Andronesi O. C., Riedel D., and Baldus M., Proc. Natl. Acad. Sci. U. S. A. 102(44), 15871–15876 (2005). 10.1073/pnas.0506109102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Uversky V. N., Li J., and Fink A. L., J. Biol. Chem. 276(14), 10737–10744 (2001). 10.1074/jbc.m010907200 [DOI] [PubMed] [Google Scholar]
- 46.Fusco G., De Simone A., Gopinath T., Vostrikov V., Vendruscolo M., Dobson C. M., and Veglia G., Nat. Commun. 5, 3827 (2014). 10.1038/ncomms4827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Braun A. R., Lacy M. M., Ducas V. C., Rhoades E., and Sachs J. N., J. Am. Chem. Soc. 136(28), 9962–9972 (2014). 10.1021/ja5016958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zarranz J. J., Alegre J., Gómez-Esteban J. C., Lezcano E., Ros R., Ampuero I., Vidal L., Hoenicka J., Rodriguez O., Atarés B., Llorens V., Tortosa E. G., del Ser T., Muñoz D. G., and de Yebenes J. G., Ann. Neurol. 55(2), 164–173 (2004). 10.1002/ana.10795 [DOI] [PubMed] [Google Scholar]
- 49.Kruger R., Kuhn W., Muller T., Woitalla D., Graeber M., Kosel S., Przuntek H., Epplen J. T., Schols L., and Riess O., Nat. Genet. 18(2), 106–108 (1998). 10.1038/ng0298-106 [DOI] [PubMed] [Google Scholar]
- 50.Polymeropoulos M. H., Lavedan C., Leroy E., Ide S. E., Dehejia A., Dutra A., Pike B., Root H., Rubenstein J., Boyer R., Stenroos E. S., Chandrasekharappa S., Athanassiadou A., Papapetropoulos T., Johnson W. G., Lazzarini A. M., Duvoisin R. C., Di Iorio G., Golbe L. I., and Nussbaum R. L., Science 276(5321), 2045–2047 (1997). 10.1126/science.276.5321.2045 [DOI] [PubMed] [Google Scholar]
- 51.Auluck P. K., Caraveo G., and Lindquist S., Annu. Rev. Cell Dev. Biol. 26, 211–233 (2010). 10.1146/annurev.cellbio.042308.113313 [DOI] [PubMed] [Google Scholar]
- 52.Comellas G., Lemkau L. R., Zhou D. H., George J. M., and Rienstra C. M., J. Am. Chem. Soc. 134(11), 5090–5099 (2012). 10.1021/ja209019s [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhao M., Cascio D., Sawaya M. R., and Eisenberg D., Protein Sci. 20(6), 996–1004 (2011). 10.1002/pro.630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lorenzen N., Nielsen S. B., Buell A. K., Kaspersen J. D., Arosio P., Vad B. S., Paslawski W., Christiansen G., Valnickova-Hansen Z., Andreasen M., Enghild J. J., Pedersen J. S., Dobson C. M., Knowles T. P., and Otzen D. E., J. Am. Chem. Soc. 136, 3859 (2014). 10.1021/ja411577t [DOI] [PubMed] [Google Scholar]
- 55.Tsigelny I. F., Sharikov Y., Wrasidlo W., Gonzalez T., Desplats P. A., Crews L., Spencer B., and Masliah E., FEBS J. 279(6), 1000–1013 (2012). 10.1111/j.1742-4658.2012.08489.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Uversky V. N., FEBS Lett. 587(13), 1891–1901 (2013). 10.1016/j.febslet.2013.04.042 [DOI] [PubMed] [Google Scholar]
- 57.Banerjee S., Hashemi M., Lv Z., Maity S., Rochet J. C., and Lyubchenko Y. L., Sci. Rep. 7, 45592 (2017). 10.1038/srep45592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wördehoff M. M., Bannach O., Shaykhalishahi H., Kulawik A., Schiefer S., Willbold D., Hoyer W., and Birkmann E., J. Mol. Biol. 427(6, Part B), 1428–1435 (2015). 10.1016/j.jmb.2015.01.020 [DOI] [PubMed] [Google Scholar]
- 59.Pinotsi D., Buell A. K., Galvagnion C., Dobson C. M., Kaminski Schierle G. S., and Kaminski C. F., Nano Lett. 14(1), 339–345 (2014). 10.1021/nl4041093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hoyer W., Cherny D., Subramaniam V., and Jovin T. M., J. Mol. Biol. 340(1), 127–139 (2004). 10.1016/j.jmb.2004.04.051 [DOI] [PubMed] [Google Scholar]
- 61.Lyubchenko Y., Gall A., and Shlyakhtenko L., in Electron Microscopy, edited by Kuo J. (Humana Press, 2014), Vol. 1117, pp. 367–384. [Google Scholar]
- 62.Lyubchenko Y. L., Shlyakhtenko L. S., and Ando T., Methods 54(2), 274–283 (2011). 10.1016/j.ymeth.2011.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
See supplementary material for Figs. S1–S4 and Movies S1–S6.