Abstract
Nine new guaianes (graphostromanes A–I, 1–9) were isolated from the deep-sea-derived fungus Graphostroma sp. MCCC 3A00421, along with four known ones (10–13). The relative configurations were established mainly by detailed analysis of the NMR and HRESIMS data, while the absolute configurations were assigned using the X-ray crystallography and modified Mosher’s method. All isolates were evaluated for their inhibitory effects against lipopolysaccharide (LPS)-induced nitric oxide (NO) production in RAW264.7 macrophages. Graphostromanes F (6) showed remarkable inhibitory effect with an IC50 value of 14.2 μM, which was even stronger than that of aminoguanidine, a positive control with an IC50 value of 23.4 μM.
Introduction
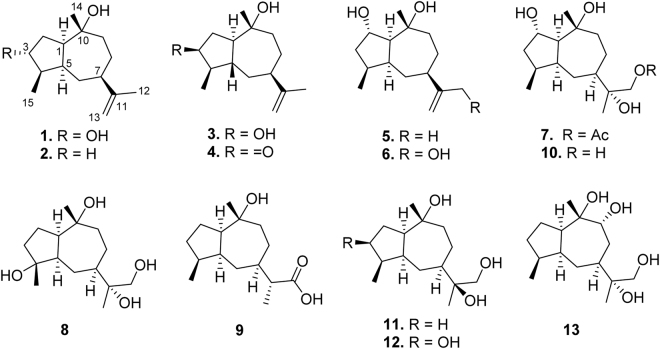
Guaianes are sesquiterpenoids bearing a bicyclo[5.3.0]-decane skeleton with enormously structural diversities, including nor-guaianes1, guaiane alkaloids2,3, guaiane dimers4,5, etc. They possessed a variety of intriguingly biological activities, such as antioxidant2, antimalarial6,7, antinociceptive7, anti-emetic8, antitumor9–11, anti-inflammatory12,13, and antibacteria14,15. Guaianes occur mainly in terrestrial plants16. Very rarely they were found from terrestrial microorganisms and corals17–19. However, they have never been isolated from marine microbes. In our continuing investigation on the deep-sea-derived Graphostroma sp. MCCC 3A0042120, nine new (1–9) and four known (10–13) guaianes were obtained (Fig. 1). Herein, we report the isolation, structure elucidation, and anti-inflammatory activities of these compounds.
Figure 1.
Chemical structures of 1–13.
Results and Discussion
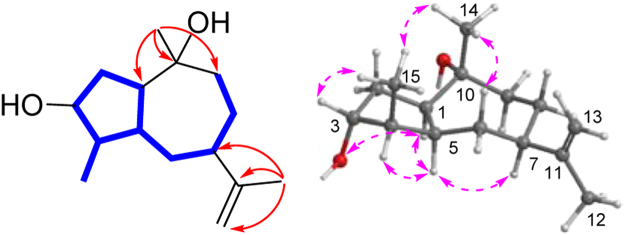
Compound 1 was isolated as a colorless oil. The molecular formula C15H26O2 was established on the basis of the [M + H]+ ionic peak at m/z 239.2044 in its positive HRESIMS, requiring three indices of hydrogen deficiency. The 1H NMR spectroscopic data (Table 1) showed two singlet (δH 1.40 and 1.76) and one doublet (δH 1.08) methyls, one exomethylene (δH 4.83, 4.72), and one oxygenated methine (δH 4.22). These signals were resonance in the 13C NMR data (Table 2) as three methyls (δC 14.4, 19.9, and 29.4), one exocyclic methylene (δC 107.8), one oxygenated methine (δC 77.4). Altogether, the 1H and 13C NMR exhibited 15 carbons attributing to three methyls, five methylenes, five methines, and two nonprotonated carbons (one sp2 at δC 152.7 and one oxygenated sp3 at δC 73.0). Since an olefinic bond accounted for one unsaturation degree, a bicyclic framework was required for 1. In the COSY spectrum, one spin coupling system from H-1 through H2-2 to H-3/H-4/H-5/H2-6/H-7/H2-8/H2-9, from H-5 to H-1, and from H-4 to H3-15 constructed a long fragment. In the HMBC spectrum, correlations from 14-Me (δH 1.40) to C-1 (δC 53.0), C-9 (δC 37.8), and C-10 (δC 73.0) and from 12-Me to C-7, C-11 (δC 152.7), and C-13 (δC 107.8) established 1 as a guaiane sesquiterpene (Fig. 2).
Table 1.
1H NMR spectroscopic data of 1–9 recorded at 400 MHz (δ in ppm, J in Hz within the parenthesis).
| no. | 1 a | 2 b | 3 a | 4 a | 5 b | 6 a | 7 a | 8 a | 9 b |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 2.93, dt (8.3, 9.9) | 2.17, m | 2.61, m | 2.71, m | 2.07, dd (9.9, 7.6) | 2.51, m | 2.55, m | 3.20, m | 2.12, m |
| 2 | 2.22, m | 1.73, m; 1.58, m | 2.67, m; 2.26, m | 3.18, ddd (19.7, 4.2, 1.3); 2.51, dd (19.4, 9.7) | 4.27, q (7.2) | 4.70, q (7.3) | 4.64, q (6.9) | 2.10, m; 1.76, m | 1.72, m; 1.54, m |
| 3 | 4.22, dt (9.5, 3.4) | 1.73, m; 1.28, m | 3.87, dt (6.8, 9.1) | 1.70, m | 1.96, m; 1.91, m | 1.95, m | 2.04, m; 1.92, td (12.8, 3.7) | 1.72, m; 1.25, m | |
| 4 | 2.29, m | 2.03, m | 1.67, m | 2.04, m | 2.21, m | 2.15, m | 2.28, m | 2.02, m | |
| 5 | 2.66, m | 2.04, m | 1.69, m | 2.04, m | 2.38, m | 2.51, m | 2.53, m | 2.54, m | 1.96, m |
| 6 | 1.63, m; 1.47, td (13.3, 2.8) | 1.40, m; 1.27, m | 1.84, br d (12.9); 1.57, br t (11.5) | 1.99, m; 1.48, m | 1.42, m; 1.38, m | 1.64, m | 2.22, m; 1.29, m | 2.63, dd (13.5, 3.2); 1.36, m | 1.44, m; 1.08, m |
| 7 | 2.59, td (10.3, 4.9) | 2.34, m | 2.10, br t (11.0) | 2.29, td (10.9, 2.2) | 2.19, m | 2.39, m | 2.27, m | 2.51, m | 2.05, m |
| 8 | 2.09, m; 1.57, m | 1.87, m; 1.43, m | 1.76, m; 1.64, m | 1.87, m; 1.54, m | 1.75, m; 1.47, m | 1.98, m; 1.68, m | 2.16, m; 1.43, m | 2.28, m; 1.49, m | 1.87, m; 1.35, m |
| 9 | 2.11, m; 1.80, m | 1.91, m; 1.55, m | 2.24, m; 1.98, td (13.5, 3.2) | 2.19, m; 1.91, m | 1.85, m; 1.69, m | 2.15, m; 1.95, m | 2.17, m; 1.91, m | 2.21, m; 1.83, m | 1.90, m; 1.51, m |
| 11 | 2.30, m | ||||||||
| 12 | 1.76, s | 1.70, s | 1.76, s | 1.75, s | 1.70, s | 4.47, t (1.4) | 4.42, s | 3.99, d (10.5); 3.89, d (10.5) | |
| 13 | 4.83, d (1.7); 4.72, m | 4.66, m; 4.58, m | 4.84, d (1.5); 4.75, m | 4.83, m; 4.74, m | 4.65, m; 4.59, m | 5.47, q (1.8); 5.08, m | 1.36, s | 1.41, s | 1.10, d (7.0) |
| 14 | 1.40, s | 1.18, s | 1.50, s | 1.30, s | 1.33, s | 1.61, s | 1.68, s | 1.46, s | 1.15, s |
| 15 | 1.08, d (7.2) | 0.92, d (6.8) | 1.20, d (6.0) | 1.08, d (6.4) | 0.89, d (7.2) | 0.83, d (7.3) | 0.93, d (7.3) | 1.54, s | 0.92, d (7.0) |
| Ac | 1.99, s | ||||||||
| 2-OH | 5.75, br s | ||||||||
| 10-OH | 5.78, br s | 5.63, br s | |||||||
| 11-OH | 5.96, br s |
aMeasured in pyridine-d5. bMeasured in CD3OD.
Table 2.
13C NMR spectroscopic data of compounds 1–9.
| no. | 1 a | 2 b | 3 a | 4 a | 5 b | 6 a | 7 a | 8 a | 9 b |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 53.0, CH | 56.0, CH | 49.6, CH | 47.3, CH | 62.7, CH | 61.2, CH | 62.6, CH | 52.7, CH | 56.1, CH |
| 2 | 36.8, CH2 | 27.1, CH2 | 37.4, CH2 | 40.2, CH2 | 75.2, CH | 74.1, CH | 74.0, CH | 25.9, CH2 | 27.0, CH2 |
| 3 | 77.4, CH | 32.1, CH2 | 77.7, CH | 220.6, C | 43.1, CH2 | 42.5, CH2 | 43.0, CH2 | 39.6, CH2 | 31.8, CH2 |
| 4 | 48.0, CH | 40.1, CH | 49.0, CH | 49.7, CH | 37.0, CH | 35.8, CH | 36.3, CH | 80.9, C | 40.3, CH |
| 5 | 44.9, CH | 47.0, CH | 46.0, CH | 45.5, CH | 46.2, CH | 44.4, CH | 46.0, CH | 54.5, CH | 47.3, CH |
| 6 | 30.4, CH2 | 29.6, CH2 | 37.0, CH2 | 38.3, CH2 | 33.0, CH2 | 34.0, CH2 | 26.2, CH2 | 26.0, CH2 | 26.1, CH2 |
| 7 | 46.7, CH | 47.1, CH | 47.9, CH | 44.9, CH | 48.6, CH | 43.7, CH | 46.4, CH | 45.5, CH | 41.4, CH |
| 8 | 29.7, CH2 | 29.7, CH2 | 32.0, CH2 | 32.5, CH2 | 31.1, CH2 | 31.6, CH2 | 26.2, CH2 | 25.9, CH2 | 28.0, CH2 |
| 9 | 37.8, CH2 | 36.8, CH2 | 46.2, CH2 | 45.0, CH2 | 42.0, CH2 | 43.8, CH2 | 40.7, CH2 | 38.0, CH2 | 35.4, CH2 |
| 10 | 73.0, C | 75.2, C | 73.9, C | 73.3, C | 75.7, C | 74.3, C | 73.9, C | 73.4, C | 75.3, C |
| 11 | 152.7, C | 153.4, C | 152.5, C | 152.1, C | 153.5, C | 157.9, C | 73.4, C | 75.3, C | 47.0, CH |
| 12 | 19.9, CH3 | 20.3, CH3 | 20.1, CH3 | 19.9, CH3 | 20.3, CH3 | 64.1, CH2 | 70.6, CH2 | 68.8, CH2 | 180.2, C |
| 13 | 107.8, CH2 | 108.4, CH2 | 107.8, CH2 | 108.0, CH2 | 108.4, CH2 | 105.8, CH2 | 20.3, CH3 | 19.7, CH3 | 13.5, CH3 |
| 14 | 29.4, CH3 | 30.1, CH3 | 24.1, CH3 | 24.4, CH3 | 28.0, CH3 | 26.9, CH3 | 29.1, CH3 | 29.6, CH3 | 30.5, CH3 |
| 15 | 14.4, CH3 | 16.8, CH3 | 16.4, CH3 | 13.9, CH3 | 16.8, CH3 | 16.7, CH3 | 16.6, CH3 | 25.0, CH3 | 16.6, CH3 |
| Ac | 20.6, CH3 170.7, C |
aMeasured in pyridine-d5 at 100 MHz. bMeasured in CD3OD at 100 MHz.
Figure 2.
Selected COSY ( ), HMBC (
), HMBC ( ), and NOESY (
), and NOESY ( ) correlations of 1.
) correlations of 1.
In the NOESY spectrum, correlations from H-5 (δH 2.66) to H-1/H-4/H-7 (δH 2.59) and from Me-15 to Me-14 and H-3 disclosed that H-1, H-4, H-5, and H-7 were on the same face, whereas H-3, 14-Me, and 15-Me were on the opposite side (Fig. 2). This was unambiguously confirmed by the Cu-Kα X-ray crystallography (Fig. 3). Therefore, 1 was determined to be (1R,3R,4R,5S,7R,10R)-11(13)-en-3,10-dihydroxyguaiene, and named graphostromane A.
Figure 3.
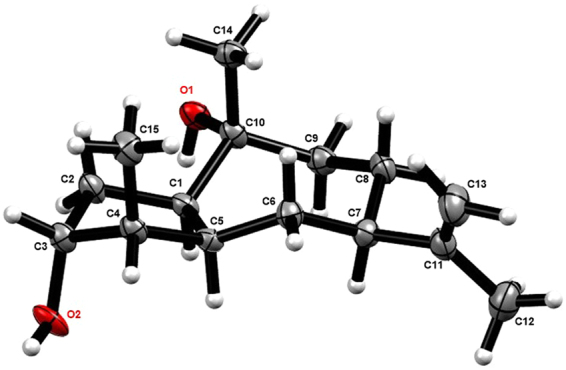
X-ray crystallographic structure of 1.
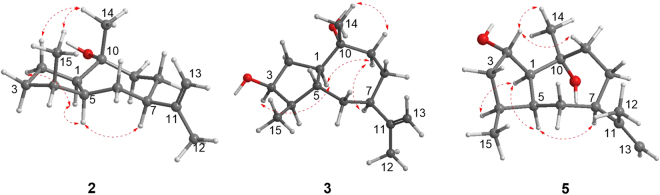
Compound 2 showed the formula molecular of C15H26O as established by the positive HRESIMS at m/z 223.2057 [M + H]+. The 1H and 13C NMR spectra were nearly identical to those of 1 except that a methylene (δC 32.1) instead of an oxygenated methine was located at C-3 position. This observation was evidenced by the HMBC correlation of 15-Me (δH 0.92) to the methylene unit. On the basis of its key NOESY correlations (Fig. 4) and the similar optical rotation value ( −28.5 for 2, while −22.0 for 1), 2 was therefore deduced to be (1R,4S,5S,7R,10R)-11(13)-en-10-hydroxyguaiene, and named graphostromane B.
Figure 4.
Key NOESY correlations of 2, 3, and 5.
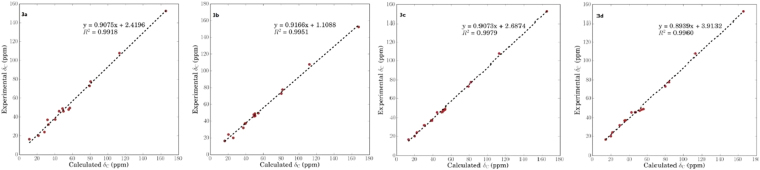
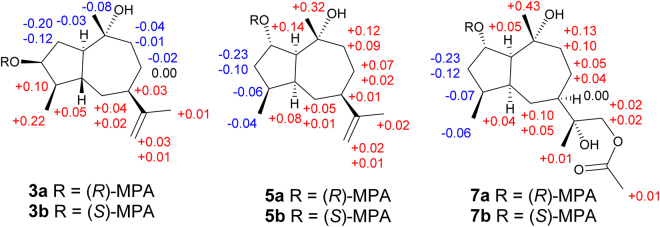
Compound 3 showed the same molecule formula as that of 1 by the positive HRESIMS at m/z 261.1829 (calcd for C15H26O2Na, 261.1830). Interestingly, it also exhibited almost the same 1H and 13C NMR spectra, except for the shielded chemical shift of H-3 (δH 3.87) and its peak pattern, indicating 3 and 1 might be C-3 stereoisomers. The NOESY correlations between H-3/H-1 (δH 2.61), H-1/H-9a (δH 1.98), H-9a/H-7, and H-9b (δH 2.24) to H3-14 (δH 1.50) deduced H-1, H-3, and H-7 were in the α orientations, while H3-14 was β-oriented. However, it is difficult to establish the relative configurations of C-4 and C-5 positions by the NOESY correlations because of the overlap signals of H-4 (δH 1.67) and H-5 (δH 1.69). Therefore, all four possible stereoisomers including a pair of cis-fused C-4 epimers [(1R*,3S*,4R*,5S*,7R*,10R*)-3a and (1R*,3S*,4S*,5S*,7R*,10R*)-3b] and a pair of trans-fused C-4 epimers [(1R*,3S*,4R*,5R*,7R*,10R*)-3c and (1R*,3S*,4S*,5R*,7R*,10R*)-3d] were subjected for the theoretical calculation of the CMR data at mPW1PW91/6-311 + G(2d,p) level using the IEFPCM model in pyridine-d5 by Gaussian 09. As shown in Fig. 5, 3c displayed the smallest deviation, suggesting the relative configuration of 3 to be 1R*,3S*,4R*,5R*,7R*, and 10R*. By the modified Mosher’s method, C-3 was determined to be S configuration (Fig. 6)21. Based on the above evidences, 3 was then established to be (1R,3S,4R,5R,7R,10R)-11(13)-en-3,10-dihydroxyguaiene, and named graphostromane C.
Figure 5.
Calculated 13C NMR spectroscopic data of four possible stereoisomers of 3 (3a, 3b, 3c, and 3d) at mPW1PW91/6-311 + G(2d,p) level in pyridine-d5.
Figure 6.
ΔδH (δHR ‒ δHS) values of (R)- and (S)-MPA esters of 3, 5, and 7 in CDCl3.
Compound 4 was established the molecular formula C15H24O2 on the basis of its HRESIMS. The 1H and 13C NMR spectra were very similar to those of 3 except that a ketone group (δC 220.6) rather than a hydroxy unit was located at C-3. The assumption was corroborated by the HMBC relationships from 15-Me (δH 1.08) to the carbonyl carbon. Therefore, 4 was assigned as (1R,4S,5S,7R,10R)-3-oxo-11(13)-en-10-hydroxyguaiene, and named graphostromane D.
Compound 5 exhibited the same molecular formula as that of 1 according to its positive HRESIMS at m/z 261.1833 [M + Na]+. Close comparison of its 1H and 13C NMR spectra to those of 1 revealed a general similarity except that the hydroxy unit should be attached to C-2 instead of C-3 in 5. This was confirmed by the HMBC correlations of 15-Me to the methylene unit at δC 43.1 of the C-3 position. In the NOESY spectrum, cross-peaks from H-9a (δH 1.85) to Me-14 (δH 1.33) and H-2 revealed H-2 was in β orientation (Fig. 4). By the modified Mosher’s method, C-2 was determined as S-configuration (Fig. 6). Accordingly, 5 was established to be (1S,2S,4S,5S,7R,10R)-11(13)-en-2,10-dihydroxyguaiene, and named graphostromane E.
Compound 6 was assigned the molecular formula C15H26O3 by the HRESIMS at m/z 277.1767 [M + Na]+. The 1H and 13C NMR spectra were nearly identical to those of 5, except for an additional hydroxy unit at C-12 (δC 64.1). The assumption was evidenced by the HMBC correlations from H2-13 (δH 5.47, 5.08) to C-7 (δC 43.7), C-11 (δC 157.9), and C-12. Accordingly, 6 was determined to be (1S,2S,4S,5S,7R,10R)-11(13)-en-2,10,12-trihydroxyguaiene, and named graphostromane F.
Compound 7 gave a molecular formula C17H30O5 from its positive HRESIMS at m/z 337.1979 [M + Na]+. Its 1H and 13C NMR spectra were close to those of (1S,2S,4S,5S,7R,10R)-guaiane-2,10,11,12-tetraol (10)22, except for an additional acetyl group (δH 1.99; δC 20.6, 170.7) at C-12. This was evidenced by the HMBC cross-peaks from H2-12 (δH 4.42) to the carbonyl (δC 170.7) of the acetyl group. The absolute configuration of C-2 was established to be S on the basis of the modified Mosher’s method (Fig. 6), which was further corroborated by the X-ray single-crystal experiment (Fig. 7). Therefore, 7 was unambiguously determined to be (1S,2S,4S,5S,7R,10R,11R)-12-acetyl-2,10,11-trihydroxyguaiane, and named graphostromane G.
Figure 7.
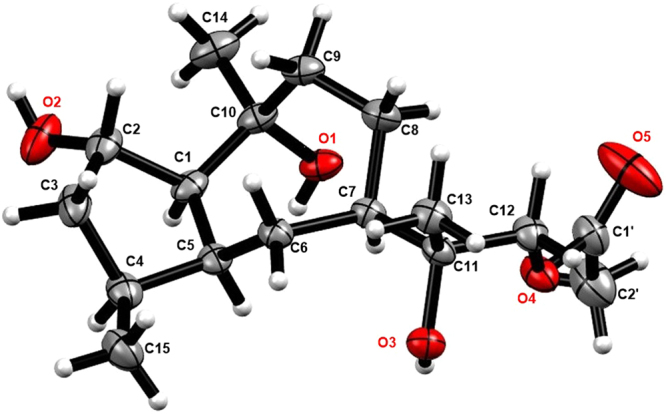
X-ray crystallographic structure of 7.
Compound 8 had the molecular formula of C15H28O4 as established by its HRESIMS. Analysis of the 1D and 2D NMR spectra established the structure of 8 was closely related to 10, except that the hydroxy group was located at C-4 instead of C-2. This was evidenced by the HMBC correlations from 15-Me (δH 1.54) to C-3 (δC 39.6), C-4 (δC 80.9), and C-5 (δC 54.5). The NOE cross-peaks from H-2a (δH 2.10) to 14-Me (δH 1.46) and 15-Me revealed that 4-OH was in α-oriented. Therefore, 8 was identified to be (1R,4R,5R,7R,10R)-4,10,11-trihydroxyguaiane, and named graphostromane H.
Compound 9 was assigned the molecular formula C15H26O3 on the basis of the HRESIMS at m/z 277.1783 [M + Na]+. Its 1H and 13C NMR spectra were very similar to those of (1R,4S,5S,7R,10R,11S)-guaiane-10,11,12-triol (11)15 except that a hydroxy group was absent at C-11, while a carboxyl moiety (δC 180.2) instead of an oxymethylene was attached to C–11. This was corroborated by the HMBC correlations form 13-Me (δH 1.10) to C-7 (δC 41.4), C-11 (δC 47.0), and C–12 (δC 180.2). By the theoretical calculation of CMR spectrum, C-11 was assigned as R-configuration (Fig. 8). Therefore, 9 was established to be (1R,4S,5S,7R,10R,11R)-10-hydroxyguai-12-oic acid, and named graphostromane I.
Figure 8.
Calculated 13C NMR spectroscopic data of a pair of C-11 epimers of (1R,4S,5S,7R,10R,11S)-9a and (1R,4S,5S,7R,10R,11R)-9b at mPW1PW91/6-311 + G(2d,p) level in CD3OD.
By comparison of the NMR, MS, and OR data with those published in the literature, four known guaianes were identified as (1S,2S,4S,5S,7R,10R)-guaiane-2,10,11,12-tetraol (10)22, (1R,4S,5S,7R,10R,11S)-guaiane-10,11,12-triol (11)15, (1R,3S,4R,5S,7R,10R,11S)-guaiane-3,10,11,12-tetraol (12)15, and (1R,4S,5S,7S,9R,10S,11R)-guaiane-9,10,11,12-tetraol (13)15.
All isolates were evaluated for their anti-inflammatory activities against LPS-induced NO production in RAW264.7 macrophages (Table 3). Compound 6 exhibited remarkable anti-inflammatory activity with an IC50 value of 14.2 μM, which was stronger than that of the positive control, aminoguanidine, with an IC50 of 23.4 μM. In addition, 4, 9, and 13 showed weak anti-inflammatory activities with IC50 values of 72.9, 79.1, and 88.2 μM, respectively.
Table 3.
Anti-inflammatory activities of compounds 1–13 against LPS-stimulated NO production by in RAW 264.7 macrophages.
| Compounds | IC50 (μM) | CC50 (μM) |
|---|---|---|
| 1 | 310.1 | >50 |
| 2 | 103.2 | >50 |
| 3 | 112.6 | >50 |
| 4 | 72.9 | >50 |
| 5 | 138.2 | >50 |
| 6 | 14.2 | >50 |
| 7 | 101.0 | >50 |
| 8 | 165.8 | >50 |
| 9 | 79.1 | >50 |
| 10 | 150.4 | >50 |
| 11 | 141.0 | >50 |
| 12 | 122.4 | >50 |
| 13 | 88.2 | >50 |
| Aminoguanidinea | 23.4 | >50 |
aPositive control.
In conclusion, chemical investigation on the deep-sea-derived fungus Graphostroma sp. MCCC 3A00421 led to the isolation of 9 new (graphostromanes A–I, 1–9) and 4 known (10–13) guaianes. They are first examples of guaiane sesquiterpenoids reported from the marine-derived fungi. Additionally, 6 showed potent anti-inflammatory activity against LPS-induced NO production in RAW264.7 macrophages, indicating its potential usage for anti-inflammatory drugs.
Materials and Methods
General Experimental Procedures
An automatic polarimeter Rudolph IV Autopol was used for recording optical rotation data at 25 °C. A Xevo G2 Q-TOF mass spectrometer was used for measuring HRESIMS. A Bruker Avance 400 MHz NMR spectrometer was used for measuring 1H, 13C, HSQC, COSY, HMBC, and NOESY spectra. Chemical shifts (δ) were expressed in ppm referring to the solvent peaks, and coupling constants are in Hz. A Bruker D8 Advance X-ray single-crystal diffractometer was used for measuring X-ray data with Cu Kα radiation. Column chromatography (CC) were performed on Sephadex LH-20 (18–110 μm, Pharmacia, Uppsala, Sweden), silica gel (100–200 or 200–300 mush, Qingdao Marine Chemistry Co. Ltd, Qingdao, China), and ODS (50 μm, Daiso, Japan). TLC precoated silica gel plates (GF254, Qingdao Marine Chemistry Co. Ltd, Qingdao, China) were used for TLC detection. All chemical reagents used were analytical grade.
Fungal Identification and Fermentation
The fungus Graphostroma sp. MCCC 3A00421 was isolated from a hydrothermal sulfide deposit in August 2012 from the Atlantic Ocean (W 13.36°, S 15.17°) at a depth of −2721 m. It was identified to be Graphostroma genus on the basis of comparison of its ITS1-5.8S-ITS2 rRNA gene sequence (KM190888) with those deposited in GenBank of the NCBI using a BLAST searching tool. The voucher strain was deposited at the Marine Culture Collection of China (MCCC) with the accession number MCCC 3A00421.
The working strain was cultured on a PDA plate medium under 25 °C for 3 days. Then the fresh mycelia were inoculated into 30 Erlenmeyer flasks (1 L), each containing 120 mL distilled water and 80 g rice, and then statically fermented for 28 days at 25 °C.
Extraction, Isolation, and Purification
After 28 days, the fermented cultures were extracted with EtOAc for three times. The EtOAc solution was evaporated under reduced pressure to get an organic extract, which was then partitioned between MeOH and petroleum ether (PE). The MeOH fraction was evaporated to get the defatted extract (7.0 g), which was separated by column chromatography (CC) over ODS eluting with gradient MeOH-H2O (5→100%) to yield 24 fractions (F1–F24). Fraction F7 (624 mg) was subjected to CC over Sephadex LH-20 (MeOH), followed by repeated CC on silica gel (CHCl3-MeOH, 15:1; EtOAc-MeOH, 50:1) to yield 10 (48.4 mg) and 12 (10.2 mg). Fraction F9 (420 mg) was purified by CC over Sephadex LH-20 (MeOH) to get three subfractions (SF9-1-9-3). SF9-1 was purified by CC over silica gel using PE-acetone (5:1), followed by Prep. TLC (CHCl3-MeOH, 8:1) to provide 7 (12.6 mg). SF9-3 was subjected to CC over silica gel (CHCl3-MeOH, 15:1) to afford 8 (8.2 mg). Compounds 6 (18.8 mg) and 13 (24.2 mg) were isolated from fraction F10 (603 mg) by CC over Sephadex LH-20 (MeOH) and silica gel (PE-acetone, 3:1; PE-EtOAc, 3:1→1:1). Fraction F14 (389 mg) was subjected to CC over silica gel using gradient PE-EtOAc to give seven subfractions (SF14-1‒SF14-7). SF14-2 was chromatographed on Sephadex LH-20 (MeOH) to yield 4 (26.0 mg). SF14-3 was purified by Prep. TLC (PE-acetone, 2:1) to get 1 (14.7 mg). Compound 3 (10.2 mg) was isolated from SF14-5 by Prep. TLC (EtOAc-acetone, 30:1). SF14-6 was separated by CC over Sephadex LH-20 (MeOH) and silica gel (CHCl3-MeOH, 20:1) to yield 11 (39.2 mg). Fraction F16 (462 mg) was subjected to CC on Sephadex LH-20 (MeOH) and silica gel (CHCl3-MeOH, 100:1→1:1; EtOAc-MeOH, 200:1→10:1; and PE-acetone, 5:1) to yield 2 (23.0 mg), 5 (15.5 mg), and 9 (11.6 mg).
Graphostromane A (1): colorless oil; −22.0 (c 0.58, MeOH); 1H and 13C NMR data, Tables 1 and 2; HRESIMS m/z 239.2044 [M + H]+ (calcd for C15H27O2, 239.2011).
Graphostromane B (2): colorless oil; −28.5 (c 2.16, MeOH); 1H and 13C NMR data, Tables 1 and 2; HRESIMS m/z 223.2057 [M + H]+ (calcd for C15H27O, 223.2062).
Graphostromane C (3): colorless oil; −8.5 (c 0.25, MeOH); 1H and 13C NMR data, Tables 1 and 2; HRESIMS m/z 261.1829 [M + Na]+ (calcd for C15H26O2Na, 261.1830).
Graphostromane D (4): colorless oil; +15.4 (c 0.73, MeOH); 1H and 13C NMR data, Tables 1 and 2; HRESIMS m/z 237.1851 [M + H]+ (calcd for C15H25O2, 237.1855).
Graphostromane E (5): colorless oil; +8.13 (c 0.39, MeOH); 1H and 13C NMR data, Tables 1 and 2; HRESIMS m/z 261.1833 [M + Na]+ (calcd for C15H26O2Na, 261.1830).
Graphostromane F (6): colorless oil; +3.41 (c 0.44, MeOH); 1H and 13C NMR data, Tables 1 and 2; HRESIMS m/z 277.1767 [M + Na]+ (calcd for C15H26O3Na, 277.1780).
Graphostromane G (7): colorless oil; +6.0 (c 0.56, MeOH); 1H and 13C NMR data, Tables 1 and 2; HRESIMS m/z 337.1979 [M + Na]+ (calcd for C17H30O5Na, 337.1991).
Graphostromane H (8): colorless oil; −21.0 (c 0.25, MeOH); 1H and 13C NMR data, Tables 1 and 2; HRESIMS m/z 295.1878 [M + Na]+ (calcd for C15H28O4Na, 295.1885).
Graphostromane I (9): colorless oil; −10.3 (c 0.12, MeOH); 1H and 13C NMR data, Tables 1 and 2; HRESIMS m/z 277.1783 [M + Na]+ (calcd for C15H26O3Na, 277.1780).
Compound 10: 13C NMR (CD3OD, 100 MHz) δC 63.4 (CH-1), 75.1 (CH-2), 43.3 (CH2-3), 37.3 (CH-4), 47.3 (CH-5), 25.8 (CH2-6), 45.7 (CH-7), 26.4 (CH2-8), 39.5 (CH2-9), 75.4 (C-10), 76.6 (C-11), 69.2 (CH2-12), 19.0 (Me-13), 29.6 (Me-14), 16.7 (Me-15).
Preparation of (R)- and (S)-MPA Esters of Compounds 3, 5, and 7
Compound 3 (2.0 mg) was dissolved in CHCl3 (600 µL). Then (R)-MPA (2.5 mg), DCC (2.5 mg), and DMAP (2.5 mg) were added. After stirred 16 h at room temperature, the reactive products were subjected to CC over silica gel (PE-acetone, 3:1) to give the R-MPA ester 3a (1.7 mg). Similarly, the S-MPA ester 3b (1.9 mg) was obtained from (S)-MPA. Analogue treatment of compounds 5 and 7 separately with (R)-MPA and (S)-MPA obtained (R)-MPA esters (5a and 7a) and (S)-MPA esters (5b and 7b), respectively.
(R)-MPA ester of 3 (3a): 1H NMR (CDCl3, 400 MHz) δH 7.32–7.47 (5 H, m, phenyl protons), 4.78 (1 H, s, CH of MPA), 3.43 (3 H, s, OMe of MPA), 2.25 (1 H, m, H-1), 1.44 (1 H, m, H-2a), 2.22 (1 H, m, H-2b), 4.68 (1 H, m, H-3), 1.61 (1 H, m, H-4), 1.66 (1 H, m, H-5), 1.36 (1 H, m, H-6a), 1.69 (1 H, m, H-6b), 1.96 (1 H, dt, J = 2.2, 11.5 Hz, H-7), 1.46 (1 H, m, H-8a), 1.71 (1 H, m, H-8b), 1.63 (1 H, m, H-9a), 1.85 (1 H, ddd, J = 2.6, 4.7, 8.1 Hz, H-9b), 1.70 (3 H, s, Me-12), 4.62 (1 H, br s, H-13a), 4.65 (1 H, br s, H-13b), 1.18 (3 H, s, Me-14), 0.91 (3 H, d, J = 6.2 Hz, Me-15).
(S)-MPA ester of 3 (3b): 1H NMR (CDCl3, 400 MHz) δH 7.33–7.47 (5 H, m, phenyl protons), 4.77 (1 H, s, CH of MPA), 3.44 (3 H, s, OMe of MPA), 2.28 (1 H, m, H-1), 1.64 (1 H, m, H-2a), 2.34 (1 H, m, H-2b), 4.63 (1 H, m, H-3), 1.51 (1 H, m, H-4), 1.61 (1 H, m, H-5), 1.34 (1 H, m, H-6a), 1.65 (1 H, m, H-6b), 1.93 (1 H, t, J = 10.8 Hz, H-7), 1.48 (1 H, m, H-8a), 1.71 (1 H, m, H-8b), 1.64 (1 H, m, H-9a), 1.89 (1 H, m, H-9b), 1.69 (3 H, s, Me-12), 4.61 (1 H, br s, H-13a), 4.64 (1 H, br s, H-13b), 1.26 (3 H, s, Me-14), 0.69 (3 H, d, J = 6.5 Hz, Me-15).
(R)-MPA ester of 5 (5a): 1H NMR (CDCl3, 400 MHz) δH 7.32–7.47 (5 H, m, phenyl protons), 4.78 (1 H, s, CH of MPA), 3.43 (3 H, s, OMe of MPA), 2.35 (1 H, m, H-1), 5.25 (1 H, m, H-2), 1.51 (1 H, m, H-3a), 1.64 (1 H, m, H-3b), 2.16 (1 H, m, H-4), 2.30 (1 H, m, H-5), 1.23 (1 H, m, H-6a), 1.45 (1 H, m, H-6b), 2.11 (1 H, m, H-7), 1.45 (1 H, m, H-8a), 1.75 (1 H, m, H-8b), 1.67 (1 H, m, H-9a), 1.82 (1 H, m, H-9b), 1.69 (3 H, s, Me-12), 4.61 (1 H, br s, H-13a), 4.65 (1 H, br s, H-13b), 1.22 (3 H, s, Me-14), 0.87 (3 H, d, J = 7.0 Hz, Me-15).
(S)-MPA ester of 5 (5b): 1H NMR (CDCl3, 400 MHz) δH 7.31–7.44 (5 H, m, phenyl protons), 4.73 (1 H, s, CH of MPA), 3.41 (3 H, s, OMe of MPA), 2.21 (1 H, m, H-1), 5.15 (1 H, m, H-2), 1.74 (2 H, m, H-3), 2.22 (2 H, m, H-4 and H-5), 1.17 (1 H, m, H-6a), 1.44 (1 H, m, H-6b), 2.10 (1 H, dt, J = 4.1, 11.4 Hz, H-7), 1.38 (1 H, m, H-8a), 1.73 (2 H, m, H-8b and H-9b), 1.55 (1 H, t, J = 10.1 Hz, H-9a), 1.67 (3 H, s, Me-12), 4.60 (1 H, br s, H-13a), 4.63 (1 H, br s, H-13b), 0.90 (3 H, s, Me-14), 0.91 (3 H, d, J = 7.0 Hz, Me-15).
(R)-MPA ester of 7 (7a): 1H NMR (CDCl3, 400 MHz) δH 7.32–7.44 (5 H, m, phenyl protons), 4.74 (1 H, s, CH of MPA), 3.41 (3 H, s, OMe of MPA), 2.31 (1 H, t, J = 6.9 Hz, H-1), 5.17 (1 H, dt, J = 2.9, 7.6 Hz, H-2), 1.45 (1 H, m, H-3a), 1.68 (1 H, m, H-3b), 2.16 (1 H, m, H-4), 2.18 (1 H, m, H-5), 0.91 (1 H, m, H-6a), 1.81 (1 H, m, H-6b), 1.77 (1 H, m, H-7), 1.19 (1 H, m, H-8a), 1.76 (1 H, m, H-8b), 1.59 (1 H, m, H-9a), 1.82 (1 H, m, H-9b), 1.06 (3 H, s, Me-12), 3.97 (1 H, d, J = 11.5 Hz, H-13a), 4.06 (1 H, d, J = 11.5 Hz, H-13b), 1.14 (3 H, s, Me-14), 0.90 (3 H, d, J = 6.5 Hz, Me-15).
(S)-MPA ester of 7 (7b): 1H NMR (CDCl3, 400 MHz) δH 7.32–7.42 (5 H, m, phenyl protons), 4.71 (1 H, s, CH of MPA), 3.40 (3 H, s, OMe of MPA), 2.16 (1 H, m, H-1), 5.05 (1 H, dt, J = 2.8, 7.7 Hz, H-2), 1.68 (1 H, m, H-3a), 1.80 (1 H, m, H-3b), 2.23 (1 H, m, H-4), 2.21 (1 H, m, H-5), 0.86 (1 H, m, H-6a), 1.71 (2 H, m, H-6b and H-8b), 1.77 (1 H, m, H-7), 1.15 (1 H, m, H-8a), 1.46 (1 H, m, H-9a), 1.72 (1 H, m, H-9b), 1.05 (3 H, s, Me-12), 3.95 (1 H, d, J = 11.5 Hz, H-13a), 4.04 (1 H, d, J = 11.5 Hz, H-13b), 0.71 (3 H, s, Me-14), 0.96 (3 H, d, J = 6.8 Hz, Me-15).
X-ray Crystal Data of Compounds 1 and 7
Graphostromane A (1) was obtained as colorless crystals. The monoclinic crystals (0.1 × 0.2 × 0.8 mm3) was recorded on a Bruker D8 Advance X-ray single-crystal diffractometer with Cu Kα radiation. Crystal data of 1: empirical formula C15H26O2, M = 236.36; space group P21, unit cell dimensions a = 6.3808 (4) Å, b = 7.7943 (4) Å, c = 13.9771 (8) Å, α = 90.00°, β = 101.329 (6)°, γ = 90.00°, V = 681.59 (7) Å3, Z = 2, Dcalcd = 1.1516 g/cm3, µ = 0.579 mm−1, F (000) = 260.8; A total of 6871 reflections were collected in the range of 6.44° < 2θ < 124.16°, of which 2128 independent reflections [Rint = 0.0548, Rsigma = 0.0552] were used for the analysis. The structure was solved by the direct methods with the SHELXL-97 program and refined using full-matrix least-squares difference Fourier techniques. The final R indexes [all data] gave R1 = 0.0642, wR2 = 0.1712 and the Flack parameter = −0.1 (3). Crystallographic data of 1 have been deposited in the Cambridge Crystallographic Data Center (deposition number CCDC 1529237).
Colorless crystals of 7 were obtained from MeOH. The monoclinic crystals (0.3 × 0.4 × 0.8 mm3) was measured on a Bruker D8 Advance X-ray single-crystal diffractometer with Cu Kα radiation. Crystal data of 7: empirical formula C17H30O5, M = 314.42; space group C2, unit cell dimensions a = 12.2072 (4) Å, b = 14.3161 (3) Å, c = 13.4457 (4) Å, α = γ = 90.00°, β = 114.561 (4)°, Volume = 2137.16 (12) Å3, Z = 9, Dcalcd = 1.173 g/cm3, µ = 0.774 mm−1, F (000) = 828.0; A total of 11239 reflections were collected in the range of 7.228° < 2θ < 123.85°, of which 3337 independent reflections [Rint = 0.0330, Rsigma = 0.0280] were used for the analysis. The final R indexes [all data] gave R1 = 0.0368, wR2 = 0.1006 and the Flack parameter = 0.04 (7). Crystallographic data of 7 have been deposited in the Cambridge Crystallographic Data Center (deposition number CCDC 1577811).
Theoretical Calculation of CMR Data
The calculated CMR data of 3 and 9 were carried out by Gaussian 0923. Conformational analyses were initially performed using Confab24 with MMFF94 force field for configurations of both compounds. The conformers, which were chosen for CMR calculations with Boltzmann-population over 1%, were firstly optimized at PM6 by semi-empirical theory method to filter some conformers with low Boltzmann-populations. Then, the remaining conformers were optimized at B3LYP/6-31 + G(d,p) in gas phase. The CMR calculation was conducted by the Gauge-Including Atomic Orbitals (GIAO) method at mPW1PW91/6-311 + G(2d,p) level using the IEFPCM model in pyridine for 3, whereas in MeOH for 9, respectively. Finally, the TMS-corrected 13C NMR chemical shift values were averaged according to Boltzmann distribution for each conformer and fitting to the experimental values by linear regression. The calculated CMR chemical shift values of TMS in pyridine and in MeOH were 187.3194 ppm and 187.3772 ppm, respectively.
Anti-Inflammatory Assay
This experiment was conducted according the reported procedure25.
Electronic supplementary material
Acknowledgements
This work was supported by grants from Xiamen Ocean Economic Innovation and Development Demonstration Project (16PZP001SF16), the National Natural Science Foundation of China (41676130 and 21372233), and the China Postdoctoral Science Foundation (2016M602056).
Author Contributions
Xian-Wen Yang designed the project; Siwen Niu isolated compounds and determined their structures; Chun-Lan Xie and Jin-Mei Xia conducted the anti-inflammatory experiments; Zhu-Hua Luo and Zongze Shao isolated and identified the fungus; Siwen Niu and Xian-Wen Yang wrote and revised the paper. All authors reviewed the manuscript.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-017-18841-6.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Chen Z, et al. Rupestonic acids B-G, NO inhibitory sesquiterpenoids from Artemisia rupestris. Bioorg. Med. Chem. Lett. 2014;24:4318–4322. doi: 10.1016/j.bmcl.2014.07.008. [DOI] [PubMed] [Google Scholar]
- 2.Yang YJ, et al. Sesquiterpenoids and tirucallane triterpenoids from the roots of Scorzonera divaricata. Phytochemistry. 2016;124:86–98. doi: 10.1016/j.phytochem.2016.01.015. [DOI] [PubMed] [Google Scholar]
- 3.Yang MC, Choi SU, Choi WS, Kim SY, Lee KR. Guaiane sesquiterpene lactones and amino acid-sesquiterpene lactone conjugates from the aerial parts of Saussurea pulchella. J. Nat. Prod. 2008;71:678–683. doi: 10.1021/np800005r. [DOI] [PubMed] [Google Scholar]
- 4.Ma Cm, Nakamura N, Hattori M, Zhu S, Komatsu K. Guaiane dimers and germacranolide from Artemisia caruifolia. J. Nat. Prod. 2000;63:1626–1629. doi: 10.1021/np000005+. [DOI] [PubMed] [Google Scholar]
- 5.Zhang YL, et al. Xylopiana A, a dimeric guaiane with a case-shaped core from Xylopia vielana: structural elucidation and biomimetic conversion. Org. Lett. 2017;19:3013–3016. doi: 10.1021/acs.orglett.7b01276. [DOI] [PubMed] [Google Scholar]
- 6.Takaya Y, et al. Novel antimalarial guaiane-type sesquiterpenoids from Nardostachys chinensis roots. Tetrahedron Lett. 1998;39:1361–1364. doi: 10.1016/S0040-4039(97)10844-9. [DOI] [Google Scholar]
- 7.Takaya Y, et al. Novel guaiane endoperoxides, nardoguaianone A–D, from Nardostachys chinensis roots and their antinociceptive and antimalarial activities. Tetrahedron. 2000;56:7673–7678. doi: 10.1016/S0040-4020(00)00682-7. [DOI] [Google Scholar]
- 8.Yang Y, et al. Anti-emetic principles of Pogostemon cablin (Blanco) Benth. Phytomedicine. 1999;6:89–93. doi: 10.1016/S0944-7113(99)80041-5. [DOI] [PubMed] [Google Scholar]
- 9.Ratnayake R, Covell D, Ransom TT, Gustafson KR, Beutler JA. Englerin A, a selective inhibitor of renal cancer cell growth, from Phyllanthus engleri. Org. Lett. 2009;11:57–60. doi: 10.1021/ol802339w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang S, et al. Sesquiterpenes from Artemisia argyi: absolute configurations and biological activities. Eur. J. Org. Chem. 2014;2014:973–983. doi: 10.1002/ejoc.201301445. [DOI] [Google Scholar]
- 11.Hyldgaard MG, et al. Guaianolides and a seco-eudesmane from the resinous exudates of cushion bush (Leucophyta brownii) and evaluation of their cytostatic and anti-inflammatory activity. J. Nat. Prod. 2015;78:1877–1885. doi: 10.1021/acs.jnatprod.5b00208. [DOI] [PubMed] [Google Scholar]
- 12.Liu Y, et al. Guaiane-type sesquiterpenes from Curcuma phaeocaulis and their inhibitory effects on nitric oxide production. J. Nat. Prod. 2013;76:1150–1156. doi: 10.1021/np400202f. [DOI] [PubMed] [Google Scholar]
- 13.Li HM, et al. Guaiane-type sesquiterpenoids from Alismatis rhizoma and their anti-inflammatory activity. Chem. Pharm. Bull. 2017;65:403–407. doi: 10.1248/cpb.c16-00798. [DOI] [PubMed] [Google Scholar]
- 14.Chakraborty K, Lipton AP, Paulraj R, Chakraborty RD. Guaiane sesquiterpenes from seaweed Ulva fasciata Delile and their antibacterial properties. Eur. J. Med. Chem. 2010;45:2237–2244. doi: 10.1016/j.ejmech.2010.01.065. [DOI] [PubMed] [Google Scholar]
- 15.Wu SH, et al. Guaiane sesquiterpenes and isopimarane diterpenes from an endophytic fungus Xylaria sp. Phytochemistry. 2014;105:197–204. doi: 10.1016/j.phytochem.2014.04.016. [DOI] [PubMed] [Google Scholar]
- 16.Fraga BM. Natural sesquiterpenoids. Nat. Prod. Rep. 2013;30:1226–1264. doi: 10.1039/c3np70047j. [DOI] [PubMed] [Google Scholar]
- 17.Diep CN, et al. Structures and absolute stereochemistry of guaiane sesquiterpenoids from the gorgonian Menella woodin. Tetrahedron Lett. 2015;56:7001–7004. doi: 10.1016/j.tetlet.2015.10.102. [DOI] [Google Scholar]
- 18.Amand S, et al. Guaiane sesquiterpenes from Biscogniauxia nummularia featuring potent antigerminative activity. J. Nat. Prod. 2012;75:798–801. doi: 10.1021/np2009913. [DOI] [PubMed] [Google Scholar]
- 19.Wei H, et al. Sesquiterpenes and other constituents of Xylaria sp. NC1214, a fungal endophyte of the moss Hypnum sp. Phytochemistry. 2015;118:102–108. doi: 10.1016/j.phytochem.2015.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Niu S, et al. Sesquiterpenes from a deep-sea-derived fungus Graphostroma sp. MCCC 3A00421. Tetrahedron. 2017;73:7267–7273. doi: 10.1016/j.tet.2017.11.013. [DOI] [Google Scholar]
- 21.Freire F, et al. Relative and absolute stereochemistry of secondary/secondary diols: Low-temperature 1H NMR of their bis-MPA esters. J. Org. Chem. 2007;72:2297–2301. doi: 10.1021/jo061939r. [DOI] [PubMed] [Google Scholar]
- 22.Huang R, Xie XS, Fang XW, Ma KX, Wu SH. Five new guaiane sesquiterpenes from the endophytic fungus Xylaria sp. YM 311647 of Azadirachta indica. Chem. Biodivers. 2015;12:1281–1286. doi: 10.1002/cbdv.201400405. [DOI] [PubMed] [Google Scholar]
- 23.Gaussian 09 (Revision D.01) (Gaussian, Inc., Wallingford, CT, 2009).
- 24.O’Boyle NM, Vandermeersch T, Hutchison GR. Confab-generation of diverse low energy conformers. J. Cheminform. 2011;3:3–8. doi: 10.1186/1758-2946-3-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang XW, et al. Anti-inflammatory and anti-tumour effects of Abies georgei extracts. J. Pharm. Pharmacol. 2008;60:937–941. doi: 10.1211/jpp.60.7.0017. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.