Abstract
Non-coding RNA (ncRNA) has been shown to regulate diverse cellular processes and functions through controlling gene expression. Long non-coding RNAs (lncRNAs) act as a competing endogenous RNAs (ceRNAs) where microRNAs (miRNAs) and lncRNAs regulate each other through their biding sites. Interactions of miRNAs and lncRNAs have been reported to trigger decay of the targeted lncRNAs and have important roles in target gene regulation. These interactions form complicated and intertwined networks. Certain lncRNAs encode miRNAs and small nucleolar RNAs (snoRNAs), and may regulate expression of these small RNAs as precursors. SnoRNAs have also been reported to be precursors for PIWI-interacting RNAs (piRNAs) and thus may regulate the piRNAs as a precursor. These miRNAs and piRNAs target messenger RNAs (mRNAs) and regulate gene expression. In this review, we will present and discuss these interactions, cross-talk, and co-regulation of ncRNAs and gene regulation due to these interactions.
Keywords: Long non-coding RNA, Circular RNA, MicroRNA, Competing endogenous RNA, Interaction, Small nucleolar RNA, PIWI-interacting RNAs
Introduction
Recent high-throughput deep sequencing has enabled genomic and transcriptomic sequencing with greater sensitivity and accuracy than the previous technologies. High-throughput RNA sequencing of whole genomes and transcriptomes have revealed that ncRNAs constitute a majority (98%) of the transcriptome with protein-coding RNAs making up the rest (2%). High-throughput sequencing has also enabled us to identify different types of non-coding RNAs (ncRNAs) and quantify their expression levels in different tissues, conditions, or developmental stages. ncRNAs have been demonstrated to have important roles in gene regulation and growing evidence indicates that non-coding RNAs interact with each other and are co-regulated.
Long non-coding RNA–microRNA interaction
LncRNAs are transcribed RNA molecules longer than 200 nucleotides and lncRNA transcripts account for a large proportion of the non-coding transcriptome [1–3]. LncRNAs are differentially expressed in various tissues and have important functions in cellular processes such as cell proliferation, motility, and apoptosis [4–6]. LncRNAs function through diverse molecular mechanisms [7] and a number of lncRNAs associate with chromatin-modifying complexes regulating gene expression [8–12].
MicroRNAs (miRNAs) are highly conserved, single stranded, non-coding RNAs of approximately 21–24 nucleotides that regulate gene expression by post-transcriptional silencing of specific target RNAs. They repress translation or cleave RNA transcripts by binding to the 3′ untranslated region (3′UTR) of target messenger RNAs (mRNAs) through miRNA response elements (MREs) by integrating into the RNA-induced silencing complex (RISC) that contains members of the Argonaute (Ago) family of proteins which silence the integrated RNAs [13–16]. MiRNAs also regulate diverse biological processes such as cell-cycle progression, proliferation, apoptosis, and development [17, 18].
Competitive endogenous RNA (ceRNA)
It has been reported that miRNAs bind to transcribed pseudogenes and lncRNA through MREs, which compete for the binding of these miRNAs to their mRNA-binding sites. These RNAs act as molecular sponges or decoys and suppress the targeting of mRNAs by miRNAs and thus are called competitive endogenous RNAs (ceRNAs). To be able to act as a ceRNA, an MRE in lncRNA requires incomplete complimentary to miRNA binding. Thus, the interactions of lncRNAs with miRNAs do not trigger decay of the interacting RNAs or trigger only slow decays. CeRNA activity regulates diverse cellular processes in development and diseases (Table 1), and lncRNA-miRNA interactions form intertwined and complex regulatory networks. Pioneering works has been published in plants for (lncRNA), human cells (pseudogenes), and viruses (small ncRNAs) as described below.
Table 1.
Non-coding RNA partners in ceRNA regulatory mechanism
| Non-coding RNA | miRNA | mRNA targets of miRNA | Function | Human disease, species, etc. | References |
|---|---|---|---|---|---|
| Pseudogene | |||||
| PTENP1 | miR-20a, miR-19b, miR-21, miR-26a and miR-214 | PTEN | Tumor suppression | Human | [20] |
| KRAS1P | miR-143 and let-7 family | KRAS | Carcinogenesis | Human | [20] |
| PTENP1 | miR-17, miR-19b and miR-20a, | PTEN | Tumor suppression | Hepatocellular carcinoma | [21] |
| Pbcas4 | miR-185 | Bcl2, Il17rd, Pnpla3, Shisa7 and Tapbp | Mouse and human | [22] | |
| HMGA1P7 | miR-15, miR-16, miR-214 and miR-761 | H19 and Igf2 | Carcinogenesis | Mouse | [23] |
| Small ncRNA | |||||
| HSURs 1 and 2 | miR-142-3p, miR-27 and miR-16 | FOXO1 | Viral infection | Human T cells | [24] |
| Circular RNA | |||||
| ciRS-7 (CDR1as) | miR-7 | Brain development | Human and mouse | [40, 41] | |
| Myrip | Diabetes suppression | Human and mouse | [42] | ||
| Sry | miR-138 | Mouse | [40] | ||
| lncRpa and circRar1 | miR-671 | CASP8 and P38 | Apoptosis | Mouse neuronal cell | [110] |
| circHIPK3 | miR-124 | IL6R and DLX2 | Carcinogenesis | Human | [43] |
| circRNA-CER | miR-136 | MMP13 | Osteoarthritis | Human chondrocytes | [111] |
| lncRNA | |||||
| IPS1 | miR-399 | PHO2 | Plant growth | Arabidopsis thaliana | [19] |
| linc-ROR | miR-145 | Oct4, Nanog and Sox2 | Self-renewing | Human embryonic stem cell | [28] |
| miR-205 | ZEB2 | Carcinogenesis | Breast cancer | [26] | |
| HOTAIR | miR-331-3p | HER2 | Carcinogenesis | Gastric cancer | [32] |
| miR-141 | SKA2 | Carcinogenesis | Glioma | [112] | |
| MALAT1 | miR-124 | GRB2 | Carcinogenesis | Cervical cancer | [113] |
| H19 | miR-106a | CDKN1A, DICER1, RB1, ARID4B, ANKRD52 and FAM102A | Myoblast differentiation | Human myoblast | [38] |
| miR-138 and miR-200a | Vimentin, ZEB1, and ZEB2 | Carcinogenesis | Hepatocellular carcinoma | [108] | |
| miR-200b/c and let-7b | Git2 and Cyth3 | Carcinogenesis | Breast cancer | [114] | |
| HULC | miR-372 | PRKACB | Carcinogenesis | Liver cancer | [44] |
| linc-MD1 | miR-133 and miR-135 | MAML1 and MEF2C | Myoblasts differentiation | Mouse and human myoblasts | [45] |
| BACE1-AS | miR-485-5p | BACE1 | Alzheimer’s disease | Human brain | [46] |
| lncRNA-ATB | miR-200 family | ZEB1 and ZEB2 | Carcinogenesis | Hepatocellular carcinoma | [47] |
| CCAT1 | miR-218-5p | Bmi1 | Carcinogenesis | Gallbladder cancer | [115] |
| ZFAS1 | miR-150 | ZEB1, MMP14 and MMP16 | Carcinogenesis | Hepatocellular carcinoma | [116] |
| lncRNA-BGL3 | miR-17, miR-93, miR-20a, miR-20b, miR-106a and miR-106b | PTEN | Tumor suppression | Leukemia | [117] |
CeRNA in plants
The first ceRNA reported was lncRNA IPS1 in plants. In Arabidopsis thaliana, the lncRNA IPS1 has a binding site for the phosphate starvation-induced miR-399 [19]. The mismatched sequence at the potential miRNA-binding site in IPS1 prevents miR-399 from cleaving IPS1. Therefore, IPS1 acts as a ceRNA for PHO2 that encodes an E2 ubiquitin conjugase-related protein, a target of miR-399 [19].
Pseudogene ceRNA
Pseudogenes are genomic DNA sequences similar to normal genes but do not express their gene. The non-coding PTENP1 pseudogene is highly homologous to the tumor suppressor, phosphatase, and tensin homolog (PTEN). Poliseno et al. have reported that the 3′UTR of PTENP1 is targeted by multiple miRNAs (miR-20a, miR-19b, miR-21, miR-26a, and miR-214) that also target the 3′UTR of PTEN. Therefore, PTENP1 functions as a ceRNA (sponge) for PTEN by preventing its repression by these miRNAs. Thus, the 3′UTR of PTENP1 functions as a tumor suppressor [20]. In the normal tissue and prostate tumor samples, the correlation between PTEN and PTENP1 expression was found, suggesting ceRNA regulation in these genes [20]. This study also describes that the 3′UTR of KRAS1P pseudogene has binding sites for miR-143 and the let-7 miRNA family, and acts as a ceRNA for KRAS, thereby functioning as an oncogene [20]. This was the first study to describe that pseudogenes have function and act as ceRNAs. However, the binding sequences in PTENP1 3′UTRs perfectly match the sequences of the miRNA seed regions which are contrasting to ceRNA regulation by IPS1 (1.1.1) and circular RNA ciRS-7 (CDR1as) (1.1.5). These binding may also trigger a decay of PTENP1.
Also PTENP1 decoys oncogenic miRNAs, miR-17, miR-19b, and miR-20a, and reduces downregulation of their target gene, PTEN in hepatocellular carcinoma [21]. PTENP1 also rescues PH domain and leucine-rich repeat protein phosphatase (PHLPP, a negative AKT regulator), autophagy proteins Unc-51 like autophagy activating kinase 1 (ULK1), autophagy-related 7 (ATG7), and sequestosome 1 (p62), from downregulation by the miRNAs [21]. The mouse Pbcas4 pseudogene functions as a ceRNA for human breast carcinoma amplified sequence 4 (BCAS4) by sponging miR-185 and suppressing the repression of miR-185 target genes, Bcl2, Il17rd, Pnpla3, Shisa7, and Tapbp [22]. LncRNA H19 and insulin like growth factor 2 (Igf2) are targeted by high mobility group AT-hook 1 pseudogene 7 (HMGA1P7)-targeting miRNAs (miR-15, miR-16, miR-214, and miR-761), and thus, HMGA1P7 functions as a ceRNA for H19 and Igf2 in mice [23]. In this study, widely used overexpression and knockdown were also applied, which may lead to un-physiological conditions. However, HMGA1P7, H19, and IGF2 expression was found to clearly positively correlate in human breast cancer samples [23], indicating that the ceRNA regulation may potentially occur under physiological condition.
Small ncRNA ceRNA
T cells transformed by Herpesvirus saimiri express small non-coding RNAs called viral U-rich non-coding RNAs (HSURs). HSURs 1 and 2 were found to have potential binding sites for three host-cell miRNAs, miR-142-3p, miR-27, and miR-16. Coimmunoprecipitation shows that HSURs 1 and 2 interact with these miRNAs in T cells transformed by Herpesvirus saimiri. HSUR1 induces degradation of miR-27 in a sequence-specific manner and prevents reduction of expression of forkhead box 1 (FOXO1), a target of miR-27 [24]. This indicates that viral small ncRNAs have potential to manipulate host cells through ceRNA mechanisms. These results indicate that triggering decay of miRNA elicits ceRNA regulation, which is contrasting to other ceRNA mechanisms by which lncRNA are claim to only sequester miRNAs.
lncRNA ceRNA
LincRNA-RoR ceRNA
LincRNA-RoR was identified as a lincRNA whose expression was increased in induced pluripotent stem cells (iPSCs) compared with embryonic stem cells (ESCs) using microarray [25]. It may inhibit cellular stress pathways through the p53 response and promote survival in iPSCs and ESCs [25]. This is the first report which demonstrates that lincRNAs are capable of reprogramming ESCs to iPSCs [25]. LincRNA-RoR has also been reported to function as an oncogene [26, 27]. In stem cell field, an important ceRNA function of linc-RoR was reported. Linc-RoR has binding sites for miR-145 which binds to the 3′UTR of transcription factors, Oct4, Nanog, and Sox2 [28]. Linc-RoR functions as a ceRNA to prevent degradation of these transcription factors by miR-145 targeting, and regulates transcriptional and epigenetic networks in human embryonic stem cell [28].
HOTAIR ceRNA
HOTAIR is an lncRNA which is localized in the Homeobox C (HOXC) gene cluster on chromosome 12. It interacts with the polycomb repressive complex 2 (PRC2) and lysine-specific demethylase 1 (LSD1) complex, enhancing H3K27 trimethylation and H3K4 demethylation, respectively, to suppress expression of multiple genes [29]. HOTAIR has been well studied and has been shown to promote cancer cell invasiveness [29, 30] and to increase cell proliferation, cell-cycle progression, and reduce apoptosis [31]. HOTAIR acts as a ceRNA for human epithelial growth factor receptor 2 (HER2) by binding to miR-331-3p to prevent degradation of HER2, a target of miR-331-3p in gastric cancer [32]. In this study, HOTAIR was also shown to be a target of miR-124 [32]. This is the first HOTAIR ceRNA study and made another milestone in HOTAIR research.
LncRNA H19 ceRNA
LncRNA H19 is one of the most extensively studied lncRNAs. H19 is highly expressed maternally in the developing mouse embryo with the adjacent insulin like growth factor 2 (Igf2) gene being transcribed from the paternal allele [33]. H19 is only abundant in skeletal muscle after birth [33], and H19 is developmentally regulated and is activated very early during muscle cell differentiation [34]. H19 has been reported to be upregulated and promoted various cancers [35]; however, tumor suppressing effects of H19 has also been reported [36, 37].
H19 ceRNA activity in myoblast differentiation has been reported. MiR-17-5p family members including miR-106a were found to bind H19 in HeLa cells and myoblasts using miRNA crosslinking and immunoprecipitation (miR-CLIP) as described in 1.4. During myoblast differentiation, H19 level increases and level of miR-17-5p family members decreases, suggesting that H19 acts as a ceRNA for these miRNAs. Overexpression of miR-106a and H19 RNAi also supported the ceRNA activities [38].
Circular RNA ceRNA
Circular RNA (circRNA) is a non-coding loop RNA in which the 3′ and 5′ ends have been joined together. CircRNAs are resistant to endonuclease enzymatic degradation, since they do not have susceptible 5′ and 3′ ends; therefore, circRNAs are more stable than linear RNAs. The sex determining region Y (SRY) ncRNA was the first-reported mammalian circRNA which was found in mouse testis. Investigation of circRNAs is an early stage, although SRY was discovered about 25 years ago [39].
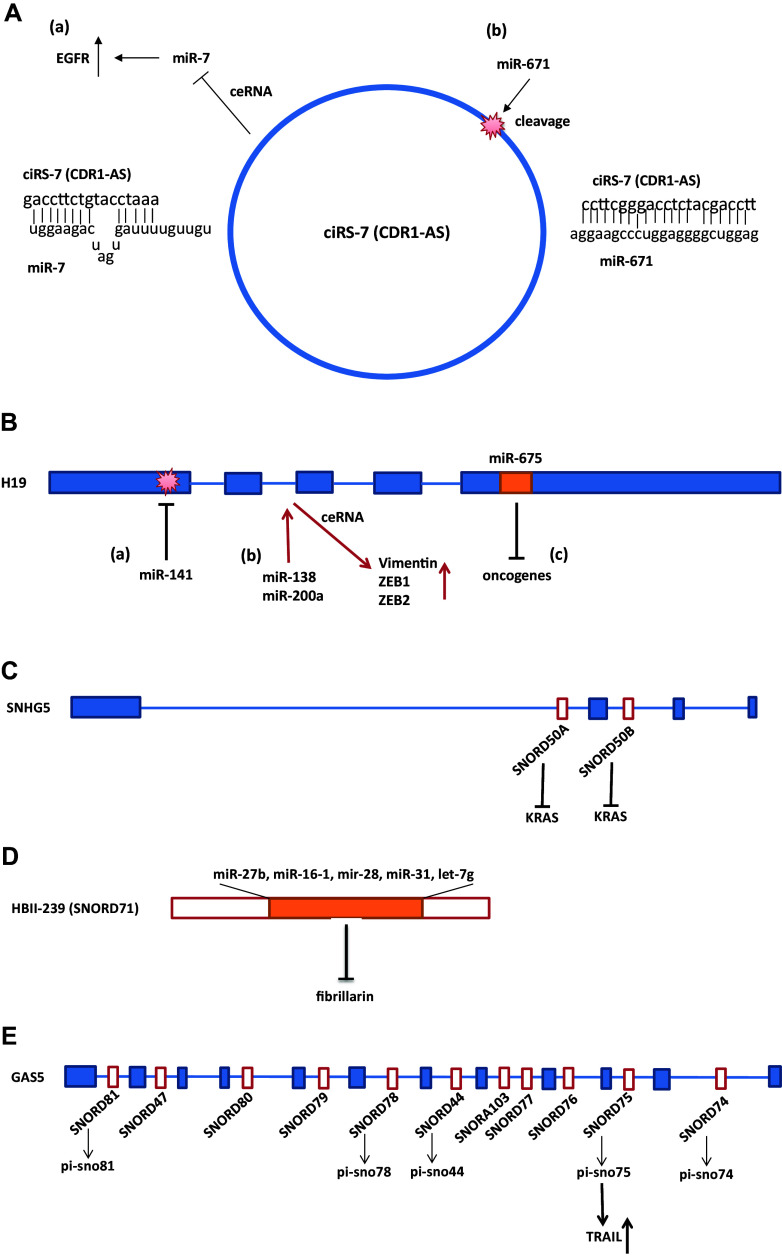
Notable ceRNA studies of circRNA were reported for cerebellar degeneration-related protein 1 (CDR1) antisense transcript, ciRS-7 (CDR1as). CiRS-7 contains more than 60 selectively conserved target sites for miR-7 [40, 41]. A mismatch at the central part of the target sites prevents miRNA-mediated cleavage; thus, ciRs-7 acts as a sponge for miR-7 and increases levels of miR-7 targets (Fig. 1A) [40, 41]. The high number of miR-7 target sites in ciRS-7 may have an advantage for eliciting ceRNA regulation. SRY was also found to function as a miR-138 sponge [40]. The ceRNA functions of CiRS-7 in diabetes have also been examined. MiR-7 targets myosin VIIA and Rab interacting protein (Myrip) which stimulates insulin release from plasma membrane and paired box 6 (Pax6), a transcription factor that activates insulin transcription in pancreatic β cells. CiRS-7 sponges miR-7 to improve β cell function by rescuing Myrip and Pax6 from their downregulation in diabetes [42].
Fig. 1.
Representative interactions and cross-talk between non-coding RNAs. A Interaction and co-regulation between circular RNA, ciRS-7 (CDR1as), and miRNAs. (a) ciRS-7 (CDR1as) contains more than 60 binding sites for miR-7. A mismatch at the central part of the binding region prevents miRNA-mediated cleavage. Thus, ciRs-7 acts as a sponge for miR-7 and increase levels of miR-7 targets [40, 41]. (b) MiR-671 binds to ciRS-7 in a sequence-specific manner and suppresses ciRS-7 expression and function [48]. B Interactions and co-regulation between lncRNA H19 and miRNAs. (a) MiR-141 binds to H19 in a sequence-specific manner and suppresses H19 expression and its oncogenic function [107]. (b) H19 functions as a ceRNA for miR-138 and miR-200a, and reduces suppression of their targets Vimentin, ZEB1, and ZEB2 [108]. (c) MiR-675 which is encoded by H19 targets mRNAs of oncogenes and functions as a tumor suppressor [82, 109]. C LncRNA SNHG5 encodes SNORD50A and SNORD50B which inhibit KRAS function. Expression of these snoRNAs may be dependent on the host lncRNA, SNHG5, and expression [90]. D HBII-239 (SNORD71) encodes miRNAs. Expression of these miRNAs may depend on the expression of the host snoRNA, HBII-239 (SNORD71). The C/D box snoRNA HBII-239 (SNORD71)-derived miRNA precursors bind to fibrillarin protein, a component of a nucleolar small nuclear ribonucleoprotein (snRNP) [94]. E LncRNA GAS5 encodes snoRNAs that generate piwi-interacting RNAs, piRNAs. Pi-snoRNA 75, a piRNA, activates tumor necrosis factor (TNF)-related apoptosis inducing ligand (TRAIL) [97]
Recent study shows that circHIPK3, circRNA derived from the HIPK3 gene Exon2, was found to be significantly abundant in various tissues compared with HIPK3 and upregulated in liver cancer compared with matched normal tissues [43]. The knockdown of circHIPK3 significantly inhibits human cell growth. CircHIPK3 was found to bind to 9 miRNAs with 18 potential binding sites [43]. Because of these high number of binding sites, circHIPK3 may potentially have ceRNA function under natural conditions. In particular, it has been demonstrated that circHIPK3 functions as a ceRNA for miR-124 and inhibits miR-124 targeting of proliferation-promoting genes, IL6R and DLX2 [43].
Other lncRNA ceRNA
Computational analysis has shown that lncRNA HULC contains miR-372-binding sites [44]. HULC may act as a ceRNA, downregulating the activity of miR-372 which downregulates protein kinase C alpha (PRKACB) translation and induces phosphorylation of CREB in liver cancer [44].
Linc-MD1 was found to be expressed during myoblast differentiation, host precursors of muscle-specific miRNAs, miR-206, and miR-133b. Linc-MD1 contains two miR-135 binding sites and one for miR-133. MiR-133 and miR-135 downregulate the expression of their target genes, mastermind like transcriptional coactivator 1 (MAML1), and myocyte enhancer factor 2C (MEF2C), that activate muscle-specific genes in mouse and human myoblasts. Linc-MD1 acts as a ceRNA by binding to miR-133 and miR-135, and promotes muscle differentiation [45]. A pre-miR-133b hairpin structure in linc-MD1 may limit miR-133 accessibility and the smaller number of the binding sites could limit ceRNA activity, which is contrasting to ciRS-7 ceRNA activity.
LncRNA has been show to involve in Alzheimer’s disease. LncRNA BACE1-AS, which is highly upregulated in brain samples from Alzheimer’s patients, acts as a ceRNA for beta-Secretase 1 (BACE1) mRNA by binding to miR-485-5p. A mismatch at the center of the binding site prevents downregulation of BACE1 mRNA, a target of miR-485-5p, which was demonstrated by knockdown of miR-485-5p and BACE1-antisense overexpression experiments [46].
Transforming growth factor beta (TGF-b)-inducing lncRNA-ATB functions as a decoy for the miR-200 family, which upregulates ZEB1 and ZEB2, targets the miR-200 family and induces EMT and invasion in hepatocellular carcinoma [47]. However, lncRNA-ATB binds to the miR-200 family through the perfect-matched seed regions of the miRNAs, which may also decay lncRNA-ATB.
MiRNA targeting of lncRNA
An increasing number of publications demonstrate that miRNAs interact with lncRNAs through MRE, thereby triggering decay of lncRNA or repressing its function (Table 2). The first evidence for an ncRNA as a miRNA target was the finding that MiR‐671 targets and cleaves circRNA, the cerebellar degeneration-related protein 1 (CDR1) antisense transcript, and ciRS-7 (CDR1as) in an Ago2‐dependent manner in human cells [48]. CiR-7 (CDR1as) was found to be formed via non-linear splicing [48]. There is positive correlation between the cerebellar degeneration-related protein 1 (CDR1) mRNA and ciR-7 (CDR1as) expression levels [48]. The decrease of ciR-7 (CDR1as) levels by miR-671 also reduces the CDR1 mRNA levels [48]. As described in 1.1.5, ciRS-7 functions as a ceRNA for miR-7 and also as a target of miR-671 leading to cleavage, which depends on the binding sequences between ciRS-7 and these miRNAs. Cleavage requires perfect or near perfect match of the binding site, while ceRNA mechanisms require mismatches in the binding sequences not to cleave the target (Fig. 1A).
Table 2.
MiRNA targeting of lncRNA by direct interaction
| LncRNA | miRNA | mRNA targets of miRNA | Human disease, species, etc. | References |
|---|---|---|---|---|
| circRNA ciRS-7 (CDR1as) | miR‐671 | Human cells | [48] | |
| Oncogenic lncRNA | ||||
| HOTAIR | miR-130a-3p | Gallbladder cancer | [67] | |
| miR-141 | Kidney cancer | [118] | ||
| miR-152 | HLA-G | Gastric cancer | [119] | |
| miR-34a | Prostate cancer | [120] | ||
| miR-326 | FGF1 | Glioma | [121] | |
| miR-1 | Hepatocellular carcinoma | [122] | ||
| UCA1 | miR-143 | Breast cancer | [123] | |
| miR-507 | FOXM1 | Melanoma | [124] | |
| MALAT1 | miR-217 | Lung cancer | [125] | |
| miR-125b | SIRT7 | Bladder cancer | [126] | |
| miR-9 | Hodgkin lymphoma and glioblastoma | [49] | ||
| miR-9 | Osteosarcoma | [127] | ||
| lincRNA-p21 | let-7 | Cervical carcinoma | [128] | |
| H19 | miR-141 | Gastric cancer | [107] | |
| TreRNA | miR-190a | Hepatoma | [129] | |
| ENST00000515084 | miR-370 | Breast cancer | [130] | |
| HOTTIP | miR-125b | Hepatocellular carcinoma | [131] | |
| miR-192 and miR-204 | Hepatocellular carcinoma | [50] | ||
| lncRNA AC130710 | miR-129-5p | Gastric cancer | [132] | |
| lincRNA NR_024015 | miR-526b | Gastric cancer | [133] | |
| PCGEM1 | miR-145 | Prostate cancer | [134] | |
| XIST | miR-152 | Glioblastoma | [135] | |
| UFC1 | miR-34a | CTNNB1 | Hepatocellular carcinoma | [136] |
| Tumor suppressor lncRNA | ||||
| loc285194 | miR-211 | Colon cancer | [137] | |
| GAS5 | miR-21 | Breast cancer | [138] | |
| TUSC7 | miR-23b | Gastric cancer | [139] | |
| CASC2 | miR-21 | Glioma | [140] | |
| uc003opf.1 | miR-149-3p | Esophageal squamous cell carcinoma | [141] | |
| Muscle differentiation | ||||
| H19 | let-7 miRNA family | Human and mouse | [142] | |
| Neurodegeneration | ||||
| lnc-SCA7 | miR-124 | SCA7 | Mouse | [51] |
| Computer analysis | ||||
| PVT1 (ceRNA) | miR-200 family | Normal breast tissues | [143] | |
| FERIL4 (ceRNA) | miR-106a-5p | PTEN, RB1, RUNX1, VEGFA, CDKN1A, | ||
| E2F1, HIPK3, IL-10, PAK7 and RB1 | Gastric cancer | [144] | ||
| 7sl RNA | miR-125b | Zebra fish | [145] | |
| STXBP5-AS1 (ceRNA) | miR-190b | ERG, STK38L and FNDC3A | Breast cancer | [146] |
LncRNA metastasis associated with lung adenocarcinoma transcript 1 (MALAT1) was originally found to be highly expressed in metastatic non-small-cell lung cancers and is a highly conserved lncRNA [102]. MALAT1 is upregulated in various cancers, and promotes proliferation and metastasis of tumor cells. MiR-9 targets lncRNA MALAT1 through its binding site in an Ago2-dependent manner in the nucleus which was revealed by in situ hybridization and confocal microscopy [49]. This study clearly shows that the miRNA interacts with the lncRNA in the nucleus. MiR-192 and miR-204 were found to target HOTTIP in a sequence-specific and Ago2-dependent manner and inhibited proliferation of hepatocellular carcinoma [50]. Microarrays identified glutaminase (GLS1), whose marked elevation in cancers has been observed as a downstream gene of HOTTIP [50]. In tissue specimens, miR-192 and miR-204 expressions are low and HOTTIP is high in hepatocellular carcinoma compared with normal tissues, showing a negative correlation between these miRNAs and HOTTIP expression [50]. This study indicates that interaction between lncRNA and miRNA is highly relevant to cancer progression.
In neurodegenerative disorders, interaction between miRNA and lncRNA has been reported. A CAG repeat expansion in ATXN7, an essential component of the mammalian transcription coactivator complex, STAGA, causes spinocerebellar ataxia type 7 (SCA7), a neurodegenerative disorder. STAGA induces miR-124 which binds to the 3′UTR of lnc-SCA7 and ATX7, a component of STAGA, and suppresses the expression of these genes in a sequence-specific manner, showing intriguing feedback regulation [51].
Computational analysis of lncRNA–miRNA interaction
There have been a number of reports that demonstrate lncRNA–miRNA interaction identified by computational or data analysis (Table 2). The algorithms for miRNA target search are based on conserved, six-nucleotide interactions between the 5′ end of the miRNA, the seed region, and the 3′UTR of the target mRNA. Since the miRNA target is predicted experimentally and computationally using various algorithms, the results vary in these different algorithms [52]. Bioinformatic prediction of lncRNA-miRNA and ceRNA interactions depends on the algorithms used for miRNA target search; therefore, varied predictions are produced with different algorithms.
Experimental miRNA–lncRNA interactions: methodology and data base
Experimental validation is indispensable for documenting miRNA–lncRNA interactions and ceRNA mechanisms. Crosslinking immunoprecipitation (CLIP) with antibodies against components of the RNA-induced silencing complex (RISC), particularly the Argonaute (AGO) family, has been developed and widely used for RNA interaction studies. CLIP sequencing (CLIP-Seq) techniques, an application of high-throughput sequencing methodologies, have been used extensively to characterize biologically relevant protein–RNA interactions [40].
One of the important methods in miRNA–lncRNA interaction study, miRNA crosslinking, and immunoprecipitation (miR-CLIP), was developed with H19. In this method, miRNA with psoralen and biotin are transfected into cells. After photo-crosslinking, Ago2 immunoprecipitation is performed, followed by streptavidin affinity purification of the miRNA-linked RNAs. The captured miRNA targets are then identified by deep sequencing. MiR-CLIP with pre-miR-106a, a miR-17-5p family member, was performed and H19 was identified as an miR-106 target. MiR-106a targets which TargetScan indicated were significantly enriched by miR-CLIP compared to non-targets, verifying that the method was effective [38].
CLIP-Seq-based approaches have been applied to different types of cells and tissues in RNA research. Advanced CLIP-Seq techniques such as (1) high-throughput sequencing of RNA isolated by crosslinking immunoprecipitation (HITS-CLIP) [53, 54]; (2) photoactivatable-ribonucleoside-enhanced crosslinking and immunoprecipitation (PAR-CLIP) [55]; and (3) crosslinking, ligation, and sequencing of hybrids (CLASH) [56] are able to identify a large number of Argonaute-bound target sequences that contain miRNA-binding sites in targeted RNAs. The databases have been developed by integrating these data and the public data resources for interactions between lncRNA and miRNA and ceRNA as listed in Tables 3 and 4, respectively.
Table 3.
LncRNA–miRNA databases
| Database | URL | Contents | References |
|---|---|---|---|
| DIANA-LncBase v2 | http://www.microrna.gr/LncBase |
Database provides two different miRNA–lncRNA interaction modules. One module is experimentally supported, and the other is in silico predicted interactions Database is supported by low-yield experimental techniques, analysis of more than 150 CLIP-Seq libraries, and publications |
[57, 58] |
| LNCediting | http://bioinfo.life.hust.edu.cn/LNCediting/ | Database provides information about RNA editing in lncRNAs. To predict the functional changes after RNA editing, it shows changes in the secondary structure and miRNA–lncRNA interactions | [59] |
| NPInter v3.0 | http://www.bioinfo.org/NPInter/ |
Database provides information about interactions between ncRNAs (except tRNAs and rRNAs), lncRNAs and others. It offers various types of basic information about the interaction Database is supported by in silico predictions from AGO CLIP-Seq, 991 publications, and high-throughput technologies |
[60–62] |
| lncReg | http://bioinformatics.ustc.edu.cn/lncreg/ |
Database is derived from miRNA–lncRNA interaction information in 259 research articles Database provides basic information about lncRNA, genes, relationships, mechanisms from 1,081 entries, and lncRNA-related regulatory networks |
[63] |
| LNCipedia v4.0 | http://www.lncipedia.org |
Database has comprehensive human lncRNA information which contains RNA sequence, structure, local conservation, and transcript information Database also offers information about possible miRNA–lncRNA interactions. This prediction analysis is supported using the Mir Target2 algorithm |
[63, 65] |
| lncRNASNP | http://bioinfo.life.hust.edu.cn/lncRNASNP/ | Prediction of effects on SNPs in lncRNA secondary structure and lncRNA–miRNA interaction | [66] |
| starBase v2.0 | http://starbase.sysu.edu.cn/ | Database provides comprehensive interaction networks of ncRNAs (lncRNAs, miRNAs, and ceRNAs), mRNA, and RNA-binding proteins in normal tissues and cancer cells based on 108 CLIP-Seq | [68] |
| LncRNAMAP | http://lncRNAMap.mbc.nctu.edu.tw/ |
Database of putative regulatory functions of lncRNAs Database is supported by publicly available deep sequencing data |
[69] |
Table 4.
CeRNA databases
| Database | URL | Content | References |
|---|---|---|---|
| spongeScan | http://spongescan.rc.ufl.edu |
Database predicts miRNA response elements in lncRNAs and presumes lncRNA function as miRNA sponges Sequence complementarity underlies this database |
[70] |
| SomamiR 2.0 | http://compbio.uthsc.edu/SomamiR | Database contains functional analysis of expected miRNA–ceRNA (including mRNAs, circular RNAs and lncRNA) interaction/changes caused by somatic mutations in cancer | [71, 72] |
| ceRNABase | http://starbase.sysu.edu.cn/mrnaCeRNA.php | Part of starBase v2.0 | [68] |
| lnCeDB | http://gyanxet-beta.com/lncedb/ | Database of human lncRNAs that act as ceRNAs. This database assesses lncRNA–mRNA interactions which are potentially controlled by common miRNAs | [73] |
Overlapping of ceRNA regulatory mechanisms and downregulation of lncRNA by direct miRNA–lncRNA interactions
The interactions between lncRNAs and miRNAs which mediate ceRNA mechanisms may trigger silencing of lncRNAs under certain conditions, since lncRNAs may be integrated into the RISC complex and potentially cleaved by Argonaute proteins. The cleavage requires perfect or near perfect match of the binding site. CeRNA mechanisms require mismatches in the binding sequences to avoid immediate cleavage of the target. If the binding is long enough, it may trigger decay, although no information about requirement for binding duration is available. The majority of ceRNA studies show perfect or near perfect matches of biding sequences between lncRNAs and miRNAs. Therefore, these reported ceRNA interactions may also lead to cleavage of lncRNAs in addition to ceRNA regulation. In fact, viral small ncRNA HSUR1 induces degradation of miR-27 in a sequence-specific manner and functions as a ceRNA [24]. Every interaction between miRNA and lncRNA may trigger decay of lncRNA or inhibit lncRNA function and also may function as ceRNA. Therefore, these two mechanisms may overlap each other.
Indirect interaction and co-regulation between lncRNA and miRNA
A number of reports indicate that inverse expression levels between miRNA and lncRNA may occur without direct binding, suggesting cross-talk between these ncRNAs (Table 5).
Table 5.
Indirect interaction between lncRNA and microRNA
| LncRNA | miRNA | mRNA targets of miRNA | Human disease, species, etc. | References |
|---|---|---|---|---|
| Oncogenic lncRNA | ||||
| UCA1 | miR-143 | Bladder cancer | [147] | |
| HULC | miR-9 | PPARA | Hepatoma | [74] |
| HOTAIR | miR-125a-5p | CASP2 | Colon cancer | [148] |
| miR-7 | SETDB1 | Breast cancer stem cells | [149] | |
| miR-568 | NFAT5 | Breast cancer | [150] | |
| H19 | miR-141 | Osteoblasts | [151] | |
| BANCR | miR-9 | NF-kB1 | Gastric cancer | [152] |
| ANRIL | miR-99a and miR-449a | Gastric cancer | [153] | |
| ncNRFR | let-7 | Mouse colonic epithelia | [154] | |
| CCAT2 | miR–17–5p and miR–20a | Colon cancer | [75] | |
| Tumor suppressor lncRNA | ||||
| MEG3 | miR-29a and miR-185 | DNMT1, 3A and 3B | Hepatocellular carcinoma | [155] |
| miR-29a | DNMT1 and 3B | Hepatocellular carcinoma | [156] | |
| miR-148a | DNMT1 | Gastric cancer | [157] | |
| miR-181b | 12/15-LOX | Mouse | [158] | |
Interestingly, lncRNA HULC was found to regulate lipid metabolism. HULC upregulates the transcriptional factor peroxisome proliferator-activated receptor alpha (PPARA) which increases acyl-CoA synthetase long-chain family member 1 (ACSL1), leading to triglycerides and cholesterol production and proliferation of hepatoma cells [74]. Cholesterol upregulates HULC expression through the retinoid receptor RXRA, which also increases HULC expression by activating the HULC promoter, indicating a positive feedback [74]. HULC also increases methylation of CpG islands in the miR-9 promoter and suppressed miR-9 targeting of PPARA. Thus, HULC promotes lipogenesis and malignancy in hepatoma cells [74].
Single-nucleotide polymorphisms (SNP) in lncRNA were reported to affect its function. LncRNA CCAT2 which contains the colorectal cancer-risk-associated rs6983267 SNP transcriptionally upregulates MYC, miR-17-5p, and miR-20a by physically interacting with TCF7L2. This activates the WNT signaling pathway and causes metastatic progression and chromosomal instability in colon cancer [75]. CCAT2 is also a WNT downstream target, suggesting the existence of a feedback loop [75]. The rs6983267 G allele was found to produce more CCAT2 transcript than T allele [75].
Non-coding RNA as precursors for shorter non-coding RNA
High-throughput deep sequencing of transcriptomes shows that certain lncRNAs encode miRNAs and other ncRNAs encode shorter ncRNAs which is discussed below. The expression of these ncRNAs may be correlated.
LncRNA encoding miRNA
Microarray profiling of miRNAs has demonstrated that miRNAs are frequently coexpressed with their host genes [76]. In addition, miRNAs have been found to be located in the exons of ncRNAs and the introns of protein-coding genes [77]. For example, antiPeg11 (antiRtl1) encodes miR-431, miR-433, miR-127, miR-432, and miR-136 [78, 79]. Thus, lncRNAs may regulate their encoding miRNAs as precursors (Table 6).
Table 6.
LncRNA encoding miRNA
| LncRNA | miRNA | mRNA targets of miRNA | Human disease, species, etc. | References |
|---|---|---|---|---|
| antiPeg11 (antiRtl1) | miR-431, miR-433, miR-127, miR-432 and miR-136 | Mouse | [78, 79] | |
| Oncogenic lncRNA | ||||
| H19 | miR-675 | Mouse myoblast | [80] | |
| RUNX1 | Gastric cancer | [159] | ||
| RUNX1 | Gastric cancer | [160] | ||
| Human cancers | [81] | |||
| CALN1 | Gastric cancer | [161] | ||
| c-Cbl and Cbl-b | Breast cancer | [162] | ||
| RB | Colorectal cancer | [163] | ||
| CDH13 | Glioma cell | [164] | ||
| LncRNA Ftx | miR-374b/421 and miR-545/374a | Hepatocellular carcinoma | [84] | |
| NCR143/145 | miR-143 and miR-145 | Human cancer | [85] | |
| LOC554202 | miR-31 | Breast cancer | [165] | |
| MONC | miR-125b-2, miR-99a and let-7c | Acute megakaryoblastic leukemia | [166] | |
| MIR100HG | miR-100, miR-125b-1 and let-7a-2 | Acute megakaryoblastic leukemia | [166] | |
| Tumor suppressor lncRNA | ||||
| H19 | miR-675 | Igf1r | Mouse embryonic and trophoblast stem cells | [82] |
| TGFBI | Prostate cancer | [109] | ||
| LINC00478 | Let-7c, miR99a and miR125b | HER2 | Breast cancer | [83] |
LncRNA H19 encodes miR-675
H19/miR-675 function has been well studied and established. H19 exon1 encodes miRNAs, miR-675-3p, and miR-675-5p, which are induced during skeletal muscle differentiation [80]. Reintroduction of miR-675-3p and miR-675-5p rectifies abnormal skeletal muscle regeneration after injury in H19-deficient mice [80]. miR-675-3p and miR-675-5p promote skeletal muscle differentiation and regeneration by targeting the anti-differentiation Smad transcription factors [80]. This study clearly demonstrated the H19/miR675 role in vivo.
H19/miR-675 has been reported to be an oncogene. TGF-β and hypoxia were shown to induce H19, miR-675, and EMT markers such as Snail and Slug. TGF-β induces Slug, H19, and miR-675 through the PI3K/AKT pathway, while H19 induces Slug and suppresses E-cadherin in cancer cells [81]. H19/miR-675 has also been reported to be a tumor suppressor. The release of miR-675 from H19 is inhibited by the stress-response RNA-binding protein, ELAV Like RNA-Binding Protein 1 (HuR) [82]. MiR-675 potentially targets growth-promoting Igf1r and inhibits proliferation of embryonic and extra-embryonic cell lines [82]. Both tumor suppressor and oncogenic functions have been reported for H19/miR-675, suggesting that its function depends on cell type.
Other lncRNAs which encode miRNAs
MiRNA profiling shows that let-7c and miR125b are encoded in an intron of the lncRNA LINC00478, and are found to be elevated in the estrogen-dependent human breast cancer cell line compared with its estrogen-independent derivative [83]. In breast cancer cell lines, miR125b and let-7c directly target the HER2 3′UTR, downregulating HER2, an oncogene which has been shown to play an important role in the progression of breast cancer [83]. LncRNA Ftx hosts the miR-374b/421 and miR-545/374a clusters in its intron. The miR-545/374a cluster was found to be upregulated in Hepatitis B virus (HBV)-related hepatocellular carcinoma tissues in comparison to matched non-cancerous liver tissue specimens. Overexpression and knockdown of miR-545/374a show that the miR-545/374a cluster may promote tumorigenesis of HBV-related hepatocellular carcinoma [84]. DEAD-box RNA helicase 6, DDX6 (p54/RCK), which is accumulated in processing bodies (P-bodies), promotes the degradation of lncRNA NCR143/145 RNA, a host gene of tumor suppressors miR-143/145. The post-transcriptional downregulation of miR-143/145 promotes malignancy in cancer cells [85]. These studies including H19/miR-675 indicate that the miRNAs encoded in lncRNA have functional roles.
LncRNA encoding small nucleolar RNA
Small nucleolar RNAs (snoRNAs) are non-coding RNAs of approximately 60–200 nucleotides, which chemically modify other RNAs, ribosomal RNAs (rRNAs), transfer RNAs, and small nuclear RNAs, and are primarily required for maturation of rRNAs. Two main classes, box C/D snoRNAs and box H/ACA snoRNAs, have been identified. The box C/D snoRNAs and H/ACA snoRNAs guide by base pairing 2′-O-ribose methylation and pseudouridylation of specific rRNAs, respectively (reviewed in [86]). In addition to these functions, snoRNAs have been reported to have non-canonical functions such as splicing and editing (reviewed in [87]). Dysregulation of snoRNA expression has been demonstrated in various diseases including cancer (reviewed in [88]). LncRNAs have also been found to encode snoRNAs (Table 7).
Table 7.
LncRNA encoding snoRNA and miRNA
| LncRNA | snoRNA and miRNA | Human disease, species etc. | References |
|---|---|---|---|
| SNHG1 | SNORD22, SNORD25, SNORD26, SNORD27, SNORD28, SNORD29, SNORD30, and SNORD31 | ||
| GAS5 (SNHG2) | SNORD44, SNORD47, SNORD74, SNORD75, SNORD76 SNORD77, SNORD78, SNORD79, SNORD80, SNORD81, and SNORA103 | ||
| SNHG3 | SNORA73A and SNORA73B | ||
| SNHG4 | snoRNA U19 (SNORA74A) and SNORA74 | Human | [89] |
| SNHG5 | SNORD50A and SNORD50B | Human cancer | [90] |
| SNHG6 | SNORD87 | ||
| SNHG7 | SNORA17, SNORA17A, SNORA17B and SNORA43 | ||
| SNHG8 | SNORA24 | ||
| SNHG9 | SNORA78 | ||
| SNHG10 | SCARNA13 | ||
| SNHG11 | SNORA60 and SNORA71E | ||
| SNHG12 | SNORA16A, SNORA44, SNORA61, and SNORD99 | ||
| DANCR | SNORA26 | ||
| SNHG14 | SNORD115 cluster, SNORD116 cluster, SNORD109A, and SNORD109B | ||
| SNHG15 | SNORA9 | ||
| SNHG16 | SNORD1A, SNORD1B, and SNORD1C | ||
| SNHG17 | SNORA71, SNORA71A, SNORA71B, SNORA71C, and SNORA71D | ||
| SNHG18 | SNORD123 | ||
| SNHG19 | snoR1 and SNORD60 | ||
| SNHG20 | SCARNA16 and miR-6516 | ||
| SNHG21 | SCARNA15 | ||
| SNHG22 | SCARNA17 and SCARNA18 | ||
| SNHG23 | SNORD113 cluster and SNORD114 cluster | ||
| SNHG24 | SNORD114 cluster | ||
| SNHG25 | SNORD104, SNORA50, SNORA50C, and SNORA76C | ||
| MEG8 | SNORD112, SNORD113-1, SNORD113-2, SNORD113-3, and miR-370 | ||
| ZFAS1 | SNORD12, SNORD12B, and SNORD12C | Breast cancer | [91] |
Based on University of California, Santa Cruz (UCSC) Genome Browser
LncRNA small nucleolar RNA host genes (SNHGs) encode snoRNAs, as indicated in Table 7. SNHG4 hosts small nucleolar RNA (snoRNA) U19 (SNORA74A) in intron 3 [89]. SNHG5 hosts snoRNAs, SNORD50A, and SNORD50B which were found to be deleted in various cancers, also bind KRAS protein, and inhibit RAS-ERK1/ERK2 signaling (Fig. 1C) [90]. In this study, the hosting and function of SNHG5 was not studied; however, significant tumor suppressing effects of SNORD50A and SNORD50B were demonstrated [90]. ZFAS1 which encodes SNORD12, SNORD12B, and SNORD12C functions as a tumor suppressor and is potentially a marker for breast cancer [91]. The expression level between ZFAS1 and the encoded snoRNAs was found to be not correlated [91]. There has been no report regarding functional interaction between lncRNA and its encoding snoRNA as far as we know. However, long ncRNAs with snoRNA ends (sno-lncRNAs) were discovered [92]. Sno-lncRNAs are intron-derived and have snoRNAs on both ends. The genomic region encoding one abundant class of sno-lncRNAs is specifically deleted in Prader–Willi Syndrome (PWS). SnoRNAs are known to localize to Cajal bodies or nucleoli; however, sno-lncRNAs are localized in subnuclear sites near their sites of synthesis in the PWS region. Thus, these sno-lncRNAs interact with Fox2 splicing factor and modify splicing [92]. These results indicate that cooperation of snoRNA and lncRNA enables this function.
SnoRNA encoding miRNA
Deep sequencing has identified short RNAs derived from snoRNAs. H/ACA box snoRNAs generate RNAs of 17–19 nt in length and C/D box snoRNAs generate RNAs longer than 27 nt. SnoRNAs have been found to encode miRNAs and their expression is likely to be dependent on the host snoRNAs. A combination of deep sequencing and bioinformatics has discovered snoRNA-generated miRNAs (sno-miRNAs) (Table 8).
Table 8.
SnoRNA encoding miRNA and piRNA
| snoRNA | miRNA or piRNA | Human disease, species, etc. | References |
|---|---|---|---|
| HBII-239 (SNORD71) | miR-27b, miR-16-1, miR-28, miR-31 and let-7g | HeLa cells | [94] |
| ACA45 | ACA45 sRNA | HEK293 cells | [93] |
| ACA36B (SNORA36B) | miR-664 | Human | [95, 167] |
| ACA34 | miR-1291 | Human | [95, 167] |
| HBI-61 (SNORA81) | miR-1248 | Human | [95, 167] |
| SNORD63 | piR30840 | Human CD4 primary T lymphocytes | [96] |
| SNORD44 | pi-sno44 | Breast cancer | [97] |
| SNORD74 | pi-sno74 | Breast cancer | [97] |
| SNORD75 | pi-sno75 | Breast cancer | [97] |
| SNORD78 | pi-sno78 | Breast cancer | [97] |
| SNORD81 | pi-sno81 | Breast cancer | [97] |
Generation of miRNA AKA45S from the H/ACA box snoRNA ACA45 was found to be DICER-dependent and AKA45S potentially targets the mRNA of Cyclin-Dependent Kinase 19 (CDC2L6) in HEK293 cells [93]. This suggests that AKA45S functions as an miRNA [93]. Computational analyses also identified 84 miRNAs that box C/D snoRNAs or their precursors encode, and have similarity to box C/D snoRNAs. Of these miRNAs, C/D box snoRNA HBII-239 (SNORD71)-derived miRNA precursors, miR-27b, miR-16-1, mir-28, miR-31, and let-7g were found to bind to fibrillarin protein, a component of a nucleolar small nuclear ribonucleoprotein (snRNP), suggesting that these miRNAs evolved from the snoRNA (Fig. 1D) [94]. Deep sequencing revealed that C/D box snoRNAs generated sno-miRNAs [95]. Experimental analysis shows that C/D snoRNAs, ACA36b, HBI-61, and ACA34, encode sno-miRNAs, miR-664, miR-1248, and miR-1291, respectively, which function in gene silencing [95]. These studies indicate that miRNAs may have evolved from snoRNAs and gained their functions.
SnoRNA encoding Piwi-interacting RNA
Piwi-interacting RNAs (piRNAs), 24–31 nucleotides in length, are the largest class of small non-coding RNA molecules expressed in animal cells. PiRNAs form silencing complexes by interacting with piwi proteins, a subfamily of the Argonaute proteins, and the complexes repress transposons and other genetic elements in germ line cells, especially genetic elements in spermatogenesis through transcriptional or post-transcriptional mechanisms. A combination of deep sequencing and bioinformatics also has discovered snoRNA-generated piRNAs (sno-piRNAs) (Table 8).
Two studies have notably demonstrated that snoRNA-derived piRNAs are functional in human cells. Deep sequencing has identified 20 C/D snoRNAs-derived piRNAs (sno-piRNAs) in human CD4 primary T lymphocytes [96]. One sno-piRNA, piR30840, downregulates the expression of IL-4 by binding to the pre-mRNA intron of IL4 in a sequence-specific manner [96]. This downregulation was found to be associated with Piwil4 and Ago4, which further interacts with Trf4–Air2–Mtr4 Polyadenylation (TRAMP) complex [96]. Another study has shown that lncRNA GAS5 hosts several snoRNAs and some of them generate sno-piRNAs. One of these, pi-sno75 upregulates the transcription of tumor necrosis factor (TNF)-related apoptosis inducing ligand (TRAIL), a proapoptotic protein, by binding to its promoter. Pi-sno75 interacts with the PIWIL1/4 proteins and recruits the MLL3/hCOMPASS complex to the promoter region of TRAIL which leads to H3K4 methylation and H3K27 demethylation and induces TRAIL mRNA (Fig. 1E) [97]. Interestingly, pi-sno75 upregulates TRAIL by binding to its promoter; however, it may also have suppression effects on other genes.
The other study has shown that small RNA sequencing of breast cancer and normal breast tissues revealed snoRNAs which were potential biomarkers for overall survival and/or recurrence-free survival. Six snoRNAs were found to host miRNAs and 48 snoRNAs were found to host piRNAs that potentially target oncogenes in breast cancer [98]. These studies have indicated that piRNAs are generated from snoRNA and these snoRNA-derived piRNAs are functional. Study in sno-piRNA is in an early stage and further investigations are awaited.
Conclusion and perspectives
RNA sequencing of whole genomes has revealed that ncRNAs constitute 98% of the human genome. Originally, the majority of DNA and ncRNAs were thought to be non-functional. The discovery of functional roles for these ncRNAs has been remarkable. Networking between ncRNAs has opened a complex regulatory world, adding to their significance in the regulation of diverse biological functions. Integration of networks of protein, non-coding, and coding RNA is of great importance requiring further investigation. Here, we described some of the interactions and cross-talk between ncRNAs.
MiRNA–lncRNA interaction including ceRNA studies has been done using overexpression or knockdown of lncRNA or miRNA. These methods disrupt physiological conditions in cells. Any overexpressed lncRNA may trigger ceRNA regulation or serve as a target of miRNAs because of high copy numbers. In siRNA knockdown, the high concentration of siRNA could bind to off-targets and also perturb the RISC complex. The CRISPR/Cas9-gRNA complex system, a recently developed genome editing system, could disrupt miRNA-binding sites and be used for miRNA–lncRNA interaction studies with a precise targeting, although no experimental method is without its drawbacks.
It has been reported that changes in ceRNAs are too small to influence miRNA-mediated repression, since the number of MREs is too high for the number of targeting miRNAs using miR-122 targets in hepatocytes and livers as a model [99, 100]. It is questionable that a single miR-122 study may be used to generalize to ceRNA regulation. Another study using Ago2 individual-nucleotide resolution UV crosslinking and immunoprecipitation (iCLIP) has demonstrated that highly expressed miRNA families such as miR-294 and let-7 are likely not function in ceRNA mechanisms because of the high miRNA and target concentrations [101]. In contrasted to miR-294 and let-7, high-affinity lncRNA targets of miRNAs of low abundance such as miR-92/25 may function as ceRNAs physiologically [101]. This study is more likely applicable to ceRNA regulation. The ratio between the number of miRNA-binding sites and miRNA molecules is essential for ceRNA regulation and changing the ratio could trigger ceRNA regulation. Since the expression of ncRNAs is specifically regulated with regard to time and location, the changes in cellular status such as disease incidence, development, or stimulation from extracellular sources cause the changes in ncRNA expression and trigger ceRNA regulation.
A number of reports using clinical samples demonstrate that inverse expression levels between lncRNAs and miRNAs which may interact with each other, which suggest that these interactions and targeting take place at physiological levels, although no direct evidence has been presented. Various studies with clinical samples also indicate that ceRNA regulation may occur physiologically, which also has not been directly proven. As discussed in “Overlapping of ceRNA regulatory mechanisms and downregulation of lncRNA by direct miRNA–lncRNA interactions”, the interactions between lncRNAs and miRNAs which mediate ceRNA mechanisms may partially overlap with those of miRNAs and lncRNAs triggering lncRNA decay of or inhibiting lncRNA function. This possible overlap may prevent precise explanation of these experimental data.
MiRNAs are generated in the cytoplasm, bind to the 3′UTR of mRNAs in a sequence-dependent manner, and trigger decay of the mRNAs. In addition to this cytoplasmic activity, miRNAs have been shown to interact with lncRNAs in both the cytoplasm and nucleus. The interaction between miRNA and lncRNA may depend on the concentration of these non-coding RNAs in specific subcellular compartments. These interactions need to be investigated in mouse models for resolving these unsolved issues at physiological levels. Identification of miRNA–target interactions in specific cellular compartments using techniques such as HITS-CLIP and PAR-CLIP and integration of these data with computational analysis would lead to elucidation of ceRNA function and how the intertwined network is regulated.
There have been studies for other ncRNA types. Interestingly, oncogenic lncRNA MALAT1 yields a small tRNA-like cytoplasmic RNA by 3′ end processing [103]. tRNA-derived small RNAs have been reported to significantly silence RNAs by associating with Argonautes 3 and 4 [104]. These studies indicate that tRNA may have gene suppression effects in cancer cells. SnoRNA HBII-52 (SNORD115) has been shown to regulate splicing by binding to the serotonin receptor 2C mRNA [105]. Ultraconserved regions (T-UCRs) are consistently and significantly altered at the genomic level in leukemias and carcinomas, and miRNAs may interact with T-UCRs with significant antisense complementarity [106]. Other small RNAs such as Y RNA, spliced leader RNA (SL RNA), etc. may target other mRNAs and ncRNAs, regulate their expression, and form regulatory networks. Further investigation in the expanding field of ncRNA interaction will reveal its biological function and regulation, and may lead to therapeutic strategies for cancers and other diseases.
Acknowledgements
We thank Dr. Roger Erickson for the support and assistance with the preparation of the manuscript.
Compliance with ethical standards
Conflict of interest
The authors declare no conflict of interest.
Grant support
Mitsuho Imai-Sumida was supported by the Astellas Foundation for Research on Metabolic Disorders fellowship funded by Astellas Pharma, Inc. This work was supported by National Institutes of Health Grant R01CA196848, RO1CA138642, RO1CA160079, RO1CA199694, and RO1CA184966. This study was also supported by Veterans Affairs Program Project BX001604 and Veterans Affairs Merit Review grants.
References
- 1.Bertone P, Stolc V, Royce TE, Rozowsky JS, Urban AE, et al. Global identification of human transcribed sequences with genome tiling arrays. Science. 2004;306:2242–2246. doi: 10.1126/science.1103388. [DOI] [PubMed] [Google Scholar]
- 2.Kapranov P, Cawley SE, Drenkow J, Bekiranov S, Strausberg RL, et al. Large-scale transcriptional activity in chromosomes 21 and 22. Science. 2002;296:916–919. doi: 10.1126/science.1068597. [DOI] [PubMed] [Google Scholar]
- 3.Rinn JL, Euskirchen G, Bertone P, Martone R, Luscombe NM, et al. The transcriptional activity of human Chromosome 22. Genes Dev. 2003;17:529–540. doi: 10.1101/gad.1055203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huarte M, Rinn JL. Large non-coding RNAs: missing links in cancer? Hum Mol Genet. 2010;19:R152–R161. doi: 10.1093/hmg/ddq353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Esteller M. Non-coding RNAs in human disease. Nat Rev Genet. 2011;12:861–874. doi: 10.1038/nrg3074. [DOI] [PubMed] [Google Scholar]
- 7.Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Mol Cell. 2011;43:904–914. doi: 10.1016/j.molcel.2011.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Khalil AM, Guttman M, Huarte M, Garber M, Raj A, et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci USA. 2009;106:11667–11672. doi: 10.1073/pnas.0904715106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang KC, Yang YW, Liu B, Sanyal A, Corces-Zimmerman R, et al. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature. 2011;472:120–124. doi: 10.1038/nature09819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guttman M, Amit I, Garber M, French C, Lin MF, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223–227. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tsai MC, Manor O, Wan Y, Mosammaparast N, Wang JK, et al. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329:689–693. doi: 10.1126/science.1192002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guttman M, Donaghey J, Carey BW, Garber M, Grenier JK, et al. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature. 2011;477:295–300. doi: 10.1038/nature10398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fabian MR, Sonenberg N. The mechanics of miRNA-mediated gene silencing: a look under the hood of miRISC. Nat Struct Mol Biol. 2012;19:586–593. doi: 10.1038/nsmb.2296. [DOI] [PubMed] [Google Scholar]
- 15.Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 17.Croce CM. Causes and consequences of microRNA dysregulation in cancer. Nat Rev Genet. 2009;10:704–714. doi: 10.1038/nrg2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kent OA, Mendell JT. A small piece in the cancer puzzle: microRNAs as tumor suppressors and oncogenes. Oncogene. 2006;25:6188–6196. doi: 10.1038/sj.onc.1209913. [DOI] [PubMed] [Google Scholar]
- 19.Franco-Zorrilla JM, Valli A, Todesco M, Mateos I, Puga MI, et al. Target mimicry provides a new mechanism for regulation of microRNA activity. Nat Genet. 2007;39:1033–1037. doi: 10.1038/ng2079. [DOI] [PubMed] [Google Scholar]
- 20.Poliseno L, Salmena L, Zhang J, Carver B, Haveman WJ, et al. A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature. 2010;465:1033–1038. doi: 10.1038/nature09144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen CL, Tseng YW, Wu JC, Chen GY, Lin KC, et al. Suppression of hepatocellular carcinoma by baculovirus-mediated expression of long non-coding RNA PTENP1 and MicroRNA regulation. Biomaterials. 2015;44:71–81. doi: 10.1016/j.biomaterials.2014.12.023. [DOI] [PubMed] [Google Scholar]
- 22.Marques AC, Tan J, Lee S, Kong L, Heger A, et al. Evidence for conserved post-transcriptional roles of unitary pseudogenes and for frequent bifunctionality of mRNAs. Genome Biol. 2012;13:R102. doi: 10.1186/gb-2012-13-11-r102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.De Martino M, Forzati F, Marfella M, Pellecchia S, Arra C, et al. HMGA1P7-pseudogene regulates H19 and Igf2 expression by a competitive endogenous RNA mechanism. Sci Rep. 2016;6:37622. doi: 10.1038/srep37622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cazalla D, Yario T, Steitz JA. Down-regulation of a host microRNA by a Herpesvirus saimiri noncoding RNA. Science. 2010;328:1563–1566. doi: 10.1126/science.1187197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Loewer S, Cabili MN, Guttman M, Loh YH, Thomas K, et al. Large intergenic non-coding RNA-RoR modulates reprogramming of human induced pluripotent stem cells. Nat Genet. 2010;42:1113–1117. doi: 10.1038/ng.710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hou P, Zhao Y, Li Z, Yao R, Ma M, et al. LincRNA-ROR induces epithelial-to-mesenchymal transition and contributes to breast cancer tumorigenesis and metastasis. Cell Death Dis. 2014;5:e1287. doi: 10.1038/cddis.2014.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang J, Zhang A, Ho TT, Zhang Z, Zhou N, et al. Linc-RoR promotes c-Myc expression through hnRNP I and AUF1. Nucleic Acids Res. 2016;44:3059–3069. doi: 10.1093/nar/gkv1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang Y, Xu Z, Jiang J, Xu C, Kang J, et al. Endogenous miRNA sponge lincRNA-RoR regulates Oct4, Nanog, and Sox2 in human embryonic stem cell self-renewal. Dev Cell. 2013;25:69–80. doi: 10.1016/j.devcel.2013.03.002. [DOI] [PubMed] [Google Scholar]
- 29.Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kogo R, Shimamura T, Mimori K, Kawahara K, Imoto S, et al. Long noncoding RNA HOTAIR regulates polycomb-dependent chromatin modification and is associated with poor prognosis in colorectal cancers. Cancer Res. 2011;71:6320–6326. doi: 10.1158/0008-5472.CAN-11-1021. [DOI] [PubMed] [Google Scholar]
- 31.Kim K, Jutooru I, Chadalapaka G, Johnson G, Frank J, et al. HOTAIR is a negative prognostic factor and exhibits pro-oncogenic activity in pancreatic cancer. Oncogene. 2013;32:1616–1625. doi: 10.1038/onc.2012.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu XH, Sun M, Nie FQ, Ge YB, Zhang EB, et al. Lnc RNA HOTAIR functions as a competing endogenous RNA to regulate HER2 expression by sponging miR-331-3p in gastric cancer. Mol Cancer. 2014;13:92. doi: 10.1186/1476-4598-13-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bartolomei MS, Zemel S, Tilghman SM. Parental imprinting of the mouse H19 gene. Nature. 1991;351:153–155. doi: 10.1038/351153a0. [DOI] [PubMed] [Google Scholar]
- 34.Pachnis V, Brannan CI, Tilghman SM. The structure and expression of a novel gene activated in early mouse embryogenesis. EMBO J. 1988;7:673–681. doi: 10.1002/j.1460-2075.1988.tb02862.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Matouk IJ, DeGroot N, Mezan S, Ayesh S, Abu-lail R, et al. The H19 non-coding RNA is essential for human tumor growth. PLoS One. 2007;2:e845. doi: 10.1371/journal.pone.0000845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hao Y, Crenshaw T, Moulton T, Newcomb E, Tycko B. Tumour-suppressor activity of H19 RNA. Nature. 1993;365:764–767. doi: 10.1038/365764a0. [DOI] [PubMed] [Google Scholar]
- 37.Yoshimizu T, Miroglio A, Ripoche MA, Gabory A, Vernucci M, et al. The H19 locus acts in vivo as a tumor suppressor. Proc Natl Acad Sci USA. 2008;105:12417–12422. doi: 10.1073/pnas.0801540105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Imig J, Brunschweiger A, Brummer A, Guennewig B, Mittal N, et al. miR-CLIP capture of a miRNA targetome uncovers a lincRNA H19–miR-106a interaction. Nat Chem Biol. 2015;11:107–114. doi: 10.1038/nchembio.1713. [DOI] [PubMed] [Google Scholar]
- 39.Capel B, Swain A, Nicolis S, Hacker A, Walter M, et al. Circular transcripts of the testis-determining gene Sry in adult mouse testis. Cell. 1993;73:1019–1030. doi: 10.1016/0092-8674(93)90279-Y. [DOI] [PubMed] [Google Scholar]
- 40.Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- 41.Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–338. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- 42.Xu H, Guo S, Li W, Yu P. The circular RNA Cdr1as, via miR-7 and its targets, regulates insulin transcription and secretion in islet cells. Sci Rep. 2015;5:12453. doi: 10.1038/srep12453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zheng Q, Bao C, Guo W, Li S, Chen J, et al. Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat Commun. 2016;7:11215. doi: 10.1038/ncomms11215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang J, Liu X, Wu H, Ni P, Gu Z, et al. CREB up-regulates long non-coding RNA, HULC expression through interaction with microRNA-372 in liver cancer. Nucleic Acids Res. 2010;38:5366–5383. doi: 10.1093/nar/gkq285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cesana M, Cacchiarelli D, Legnini I, Santini T, Sthandier O, et al. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell. 2011;147:358–369. doi: 10.1016/j.cell.2011.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Faghihi MA, Zhang M, Huang J, Modarresi F, Van der Brug MP, et al. Evidence for natural antisense transcript-mediated inhibition of microRNA function. Genome Biol. 2010;11:R56. doi: 10.1186/gb-2010-11-5-r56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yuan JH, Yang F, Wang F, Ma JZ, Guo YJ, et al. A long noncoding RNA activated by TGF-beta promotes the invasion-metastasis cascade in hepatocellular carcinoma. Cancer Cell. 2014;25:666–681. doi: 10.1016/j.ccr.2014.03.010. [DOI] [PubMed] [Google Scholar]
- 48.Hansen TB, Wiklund ED, Bramsen JB, Villadsen SB, Statham AL, et al. miRNA-dependent gene silencing involving Ago2-mediated cleavage of a circular antisense RNA. EMBO J. 2011;30:4414–4422. doi: 10.1038/emboj.2011.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Leucci E, Patella F, Waage J, Holmstrom K, Lindow M, et al. microRNA-9 targets the long non-coding RNA MALAT1 for degradation in the nucleus. Sci Rep. 2013;3:2535. doi: 10.1038/srep02535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ge Y, Yan X, Jin Y, Yang X, Yu X, et al. MiRNA-192 [corrected] and miRNA-204 directly suppress lncRNA HOTTIP and interrupt GLS1-mediated glutaminolysis in hepatocellular carcinoma. PLoS Genet. 2015;11:e1005726. doi: 10.1371/journal.pgen.1005726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tan JY, Vance KW, Varela MA, Sirey T, Watson LM, et al. Cross-talking noncoding RNAs contribute to cell-specific neurodegeneration in SCA7. Nat Struct Mol Biol. 2014;21:955–961. doi: 10.1038/nsmb.2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Singh NK. miRNAs target databases: developmental methods and target identification techniques with functional annotations. Cell Mol Life Sci. 2017;12:2239–2261. doi: 10.1007/s00018-017-2469-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chi SW, Zang JB, Mele A, Darnell RB. Argonaute HITS-CLIP decodes microRNA–mRNA interaction maps. Nature. 2009;460:479–486. doi: 10.1038/nature08170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Thomson DW, Bracken CP, Goodall GJ. Experimental strategies for microRNA target identification. Nucleic Acids Res. 2011;39:6845–6853. doi: 10.1093/nar/gkr330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hafner M, Landthaler M, Burger L, Khorshid M, Hausser J, et al. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell. 2010;141:129–141. doi: 10.1016/j.cell.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Helwak A, Kudla G, Dudnakova T, Tollervey D. Mapping the human miRNA interactome by CLASH reveals frequent noncanonical binding. Cell. 2013;153:654–665. doi: 10.1016/j.cell.2013.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Paraskevopoulou MD, Georgakilas G, Kostoulas N, Reczko M, Maragkakis M, et al. DIANA-LncBase: experimentally verified and computationally predicted microRNA targets on long non-coding RNAs. Nucleic Acids Res. 2013;41:D239–D245. doi: 10.1093/nar/gks1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Paraskevopoulou MD, Vlachos IS, Karagkouni D, Georgakilas G, Kanellos I, et al. DIANA-LncBase v2: indexing microRNA targets on non-coding transcripts. Nucleic Acids Res. 2016;44:D231–D238. doi: 10.1093/nar/gkv1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gong J, Liu C, Liu W, Xiang Y, Diao L, et al. LNCediting: a database for functional effects of RNA editing in lncRNAs. Nucleic Acids Res. 2017;45:D79–D84. doi: 10.1093/nar/gkw835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hao Y, Wu W, Li H, Yuan J, Luo J, et al. NPInter v3.0: an upgraded database of noncoding RNA-associated interactions. Database (Oxford) 2016;2016:baw057. doi: 10.1093/database/baw057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wu T, Wang J, Liu C, Zhang Y, Shi B, et al. NPInter: the noncoding RNAs and protein related biomacromolecules interaction database. Nucleic Acids Res. 2006;34:D150–D152. doi: 10.1093/nar/gkj025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yuan J, Wu W, Xie C, Zhao G, Zhao Y, et al. NPInter v2.0: an updated database of ncRNA interactions. Nucleic Acids Res. 2014;42:D104–D108. doi: 10.1093/nar/gkt1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Volders PJ, Helsens K, Wang X, Menten B, Martens L, et al. LNCipedia: a database for annotated human lncRNA transcript sequences and structures. Nucleic Acids Res. 2013;41:D246–D251. doi: 10.1093/nar/gks915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhou Z, Shen Y, Khan MR, Li A. LncReg: a reference resource for lncRNA-associated regulatory networks. Database (Oxford) 2015;2015:bav083. doi: 10.1093/database/bav083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Volders PJ, Verheggen K, Menschaert G, Vandepoele K, Martens L, et al. An update on LNCipedia: a database for annotated human lncRNA sequences. Nucleic Acids Res. 2015;43:D174–D180. doi: 10.1093/nar/gku1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gong J, Liu W, Zhang J, Miao X, Guo AY. lncRNASNP: a database of SNPs in lncRNAs and their potential functions in human and mouse. Nucleic Acids Res. 2015;43:D181–D186. doi: 10.1093/nar/gku1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ma MZ, Li CX, Zhang Y, Weng MZ, Zhang MD, et al. Long non-coding RNA HOTAIR, a c-Myc activated driver of malignancy, negatively regulates miRNA-130a in gallbladder cancer. Mol Cancer. 2014;13:156. doi: 10.1186/1476-4598-13-156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yang JH, Li JH, Shao P, Zhou H, Chen YQ, et al. starBase: a database for exploring microRNA–mRNA interaction maps from Argonaute CLIP-Seq and Degradome-Seq data. Nucleic Acids Res. 2011;39:D202–D209. doi: 10.1093/nar/gkq1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chan WL, Huang HD, Chang JG. lncRNAMap: a map of putative regulatory functions in the long non-coding transcriptome. Comput Biol Chem. 2014;50:41–49. doi: 10.1016/j.compbiolchem.2014.01.003. [DOI] [PubMed] [Google Scholar]
- 70.Furio-Tari P, Tarazona S, Gabaldon T, Enright AJ, Conesa A. spongeScan: a web for detecting microRNA binding elements in lncRNA sequences. Nucleic Acids Res. 2016;44:W176–W180. doi: 10.1093/nar/gkw443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bhattacharya A, Cui Y. SomamiR 2.0: a database of cancer somatic mutations altering microRNA–ceRNA interactions. Nucleic Acids Res. 2016;44:D1005–D1010. doi: 10.1093/nar/gkv1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bhattacharya A, Ziebarth JD, Cui Y. SomamiR: a database for somatic mutations impacting microRNA function in cancer. Nucleic Acids Res. 2013;41:D977–D982. doi: 10.1093/nar/gks1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Das S, Ghosal S, Sen R, Chakrabarti J. lnCeDB: database of human long noncoding RNA acting as competing endogenous RNA. PLoS One. 2014;9:e98965. doi: 10.1371/journal.pone.0098965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Cui M, Xiao Z, Wang Y, Zheng M, Song T, et al. Long noncoding RNA HULC modulates abnormal lipid metabolism in hepatoma cells through an miR-9-mediated RXRA signaling pathway. Cancer Res. 2015;75:846–857. doi: 10.1158/0008-5472.CAN-14-1192. [DOI] [PubMed] [Google Scholar]
- 75.Ling H, Spizzo R, Atlasi Y, Nicoloso M, Shimizu M, et al. CCAT2, a novel noncoding RNA mapping to 8q24, underlies metastatic progression and chromosomal instability in colon cancer. Genome Res. 2013;23:1446–1461. doi: 10.1101/gr.152942.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Baskerville S, Bartel DP. Microarray profiling of microRNAs reveals frequent coexpression with neighboring miRNAs and host genes. RNA. 2005;11:241–247. doi: 10.1261/rna.7240905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Rodriguez A, Griffiths-Jones S, Ashurst JL, Bradley A. Identification of mammalian microRNA host genes and transcription units. Genome Res. 2004;14:1902–1910. doi: 10.1101/gr.2722704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Seitz H, Youngson N, Lin SP, Dalbert S, Paulsen M, et al. Imprinted microRNA genes transcribed antisense to a reciprocally imprinted retrotransposon-like gene. Nat Genet. 2003;34:261–262. doi: 10.1038/ng1171. [DOI] [PubMed] [Google Scholar]
- 79.Davis E, Caiment F, Tordoir X, Cavaille J, Ferguson-Smith A, et al. RNAi-mediated allelic trans-interaction at the imprinted Rtl1/Peg11 locus. Curr Biol. 2005;15:743–749. doi: 10.1016/j.cub.2005.02.060. [DOI] [PubMed] [Google Scholar]
- 80.Dey BK, Pfeifer K, Dutta A. The H19 long noncoding RNA gives rise to microRNAs miR-675-3p and miR-675-5p to promote skeletal muscle differentiation and regeneration. Genes Dev. 2014;28:491–501. doi: 10.1101/gad.234419.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Matouk IJ, Raveh E, Abu-lail R, Mezan S, Gilon M, et al. Oncofetal H19 RNA promotes tumor metastasis. Biochim Biophys Acta. 2014;1843:1414–1426. doi: 10.1016/j.bbamcr.2014.03.023. [DOI] [PubMed] [Google Scholar]
- 82.Keniry A, Oxley D, Monnier P, Kyba M, Dandolo L, et al. The H19 lincRNA is a developmental reservoir of miR-675 that suppresses growth and Igf1r. Nat Cell Biol. 2012;14:659–665. doi: 10.1038/ncb2521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Bailey ST, Westerling T, Brown M. Loss of estrogen-regulated microRNA expression increases HER2 signaling and is prognostic of poor outcome in luminal breast cancer. Cancer Res. 2015;75:436–445. doi: 10.1158/0008-5472.CAN-14-1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zhao Q, Li T, Qi J, Liu J, Qin C. The miR-545/374a cluster encoded in the Ftx lncRNA is overexpressed in HBV-related hepatocellular carcinoma and promotes tumorigenesis and tumor progression. PLoS One. 2014;9:e109782. doi: 10.1371/journal.pone.0109782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Iio A, Takagi T, Miki K, Naoe T, Nakayama A, et al. DDX6 post-transcriptionally down-regulates miR-143/145 expression through host gene NCR143/145 in cancer cells. Biochim Biophys Acta. 2013;1829:1102–1110. doi: 10.1016/j.bbagrm.2013.07.010. [DOI] [PubMed] [Google Scholar]
- 86.Kiss T. Small nucleolar RNAs: an abundant group of noncoding RNAs with diverse cellular functions. Cell. 2002;109:145–148. doi: 10.1016/S0092-8674(02)00718-3. [DOI] [PubMed] [Google Scholar]
- 87.Dupuis-Sandoval F, Poirier M, Scott MS. The emerging landscape of small nucleolar RNAs in cell biology. Wiley Interdiscip Rev RNA. 2015;6:381–397. doi: 10.1002/wrna.1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Stepanov GA, Filippova JA, Komissarov AB, Kuligina EV, Richter VA, et al. Regulatory role of small nucleolar RNAs in human diseases. Biomed Res Int. 2015;2015:206849. doi: 10.1155/2015/206849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Bortolin ML, Kiss T. Human U19 intron-encoded snoRNA is processed from a long primary transcript that possesses little potential for protein coding. RNA. 1998;4:445–454. [PMC free article] [PubMed] [Google Scholar]
- 90.Siprashvili Z, Webster DE, Johnston D, Shenoy RM, Ungewickell AJ, et al. The noncoding RNAs SNORD50A and SNORD50B bind K-Ras and are recurrently deleted in human cancer. Nat Genet. 2016;48:53–58. doi: 10.1038/ng.3452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Askarian-Amiri ME, Crawford J, French JD, Smart CE, Smith MA, et al. SNORD-host RNA Zfas1 is a regulator of mammary development and a potential marker for breast cancer. RNA. 2011;17:878–891. doi: 10.1261/rna.2528811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Yin QF, Yang L, Zhang Y, Xiang JF, Wu YW, et al. Long noncoding RNAs with snoRNA ends. Mol Cell. 2012;48:219–230. doi: 10.1016/j.molcel.2012.07.033. [DOI] [PubMed] [Google Scholar]
- 93.Ender C, Krek A, Friedlander MR, Beitzinger M, Weinmann L, et al. A human snoRNA with microRNA-like functions. Mol Cell. 2008;32:519–528. doi: 10.1016/j.molcel.2008.10.017. [DOI] [PubMed] [Google Scholar]
- 94.Ono M, Scott MS, Yamada K, Avolio F, Barton GJ, et al. Identification of human miRNA precursors that resemble box C/D snoRNAs. Nucleic Acids Res. 2011;39:3879–3891. doi: 10.1093/nar/gkq1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Brameier M, Herwig A, Reinhardt R, Walter L, Gruber J. Human box C/D snoRNAs with miRNA like functions: expanding the range of regulatory RNAs. Nucleic Acids Res. 2011;39:675–686. doi: 10.1093/nar/gkq776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Zhong F, Zhou N, Wu K, Guo Y, Tan W, et al. A SnoRNA-derived piRNA interacts with human interleukin-4 pre-mRNA and induces its decay in nuclear exosomes. Nucleic Acids Res. 2015;43:10474–10491. doi: 10.1093/nar/gku1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.He X, Chen X, Zhang X, Duan X, Pan T, et al. An Lnc RNA (GAS5)/SnoRNA-derived piRNA induces activation of TRAIL gene by site-specifically recruiting MLL/COMPASS-like complexes. Nucleic Acids Res. 2015;43:3712–3725. doi: 10.1093/nar/gkv214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Krishnan P, Ghosh S, Wang B, Heyns M, Graham K, et al. Profiling of small nucleolar RNAs by next generation sequencing: potential new players for breast cancer prognosis. PLoS One. 2016;11:e0162622. doi: 10.1371/journal.pone.0162622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Denzler R, Agarwal V, Stefano J, Bartel DP, Stoffel M. Assessing the ceRNA hypothesis with quantitative measurements of miRNA and target abundance. Mol Cell. 2014;54:766–776. doi: 10.1016/j.molcel.2014.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Denzler R, McGeary SE, Title AC, Agarwal V, Bartel DP, et al. Impact of microRNA levels, target-site complementarity, and cooperativity on competing endogenous RNA-regulated gene expression. Mol Cell. 2016;64:565–579. doi: 10.1016/j.molcel.2016.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Bosson AD, Zamudio JR, Sharp PA. Endogenous miRNA and target concentrations determine susceptibility to potential ceRNA competition. Mol Cell. 2014;56:347–359. doi: 10.1016/j.molcel.2014.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Ji P, Diederichs S, Wang W, Boing S, Metzger R, et al. MALAT-1, a novel noncoding RNA, and thymosin beta4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene. 2003;22:8031–8041. doi: 10.1038/sj.onc.1206928. [DOI] [PubMed] [Google Scholar]
- 103.Wilusz JE, Freier SM, Spector DL. 3′ end processing of a long nuclear-retained noncoding RNA yields a tRNA-like cytoplasmic RNA. Cell. 2008;135:919–932. doi: 10.1016/j.cell.2008.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Haussecker D, Huang Y, Lau A, Parameswaran P, Fire AZ, et al. Human tRNA-derived small RNAs in the global regulation of RNA silencing. RNA. 2010;16:673–695. doi: 10.1261/rna.2000810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Kishore S, Stamm S. The snoRNA HBII-52 regulates alternative splicing of the serotonin receptor 2C. Science. 2006;311:230–232. doi: 10.1126/science.1118265. [DOI] [PubMed] [Google Scholar]
- 106.Calin GA, Liu CG, Ferracin M, Hyslop T, Spizzo R, et al. Ultraconserved regions encoding ncRNAs are altered in human leukemias and carcinomas. Cancer Cell. 2007;12:215–229. doi: 10.1016/j.ccr.2007.07.027. [DOI] [PubMed] [Google Scholar]
- 107.Zhou X, Ye F, Yin C, Zhuang Y, Yue G, et al. The interaction between MiR-141 and lncRNA-H19 in regulating cell proliferation and migration in gastric cancer. Cell Physiol Biochem. 2015;36:1440–1452. doi: 10.1159/000430309. [DOI] [PubMed] [Google Scholar]
- 108.Liang WC, Fu WM, Wong CW, Wang Y, Wang WM, et al. The lncRNA H19 promotes epithelial to mesenchymal transition by functioning as miRNA sponges in colorectal cancer. Oncotarget. 2015;6:22513–22525. doi: 10.18632/oncotarget.4154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Zhu M, Chen Q, Liu X, Sun Q, Zhao X, et al. lncRNA H19/miR-675 axis represses prostate cancer metastasis by targeting TGFBI. FEBS J. 2014;281:3766–3775. doi: 10.1111/febs.12902. [DOI] [PubMed] [Google Scholar]
- 110.Nan A, Chen L, Zhang N, Liu Z, Yang T, et al. A novel regulatory network among LncRpa, CircRar1, MiR-671 and apoptotic genes promotes lead-induced neuronal cell apoptosis. Arch Toxicol. 2016;91:1671–1684. doi: 10.1007/s00204-016-1837-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Liu Q, Zhang X, Hu X, Dai L, Fu X, et al. Circular RNA related to the chondrocyte ECM regulates MMP13 expression by functioning as a MiR-136 ‘sponge’ in human cartilage degradation. Sci Rep. 2016;6:22572. doi: 10.1038/srep22572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Bian EB, Ma CC, He XJ, Wang C, Zong G, et al. Epigenetic modification of miR-141 regulates SKA2 by an endogenous ‘sponge’ HOTAIR in glioma. Oncotarget. 2016;7:30610–30625. doi: 10.18632/oncotarget.8895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Liu S, Song L, Zeng S, Zhang L. MALAT1-miR-124-RBG2 axis is involved in growth and invasion of HR-HPV-positive cervical cancer cells. Tumour Biol. 2016;37:633–640. doi: 10.1007/s13277-015-3732-4. [DOI] [PubMed] [Google Scholar]
- 114.Zhou W, Ye XL, Xu J, Cao MG, Fang ZY, et al. The lncRNA H19 mediates breast cancer cell plasticity during EMT and MET plasticity by differentially sponging miR-200b/c and let-7b. Sci Signal. 2017;10:eaak9557. doi: 10.1126/scisignal.aak9557. [DOI] [PubMed] [Google Scholar]
- 115.Ma MZ, Chu BF, Zhang Y, Weng MZ, Qin YY, et al. Long non-coding RNA CCAT1 promotes gallbladder cancer development via negative modulation of miRNA-218-5p. Cell Death Dis. 2015;6:e1583. doi: 10.1038/cddis.2014.541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Li T, Xie J, Shen C, Cheng D, Shi Y, et al. Amplification of long noncoding RNA ZFAS1 promotes metastasis in hepatocellular carcinoma. Cancer Res. 2015;75:3181–3191. doi: 10.1158/0008-5472.CAN-14-3721. [DOI] [PubMed] [Google Scholar]
- 117.Guo G, Kang Q, Zhu X, Chen Q, Wang X, et al. A long noncoding RNA critically regulates Bcr-Abl-mediated cellular transformation by acting as a competitive endogenous RNA. Oncogene. 2015;34:1768–1779. doi: 10.1038/onc.2014.131. [DOI] [PubMed] [Google Scholar]
- 118.Chiyomaru T, Fukuhara S, Saini S, Majid S, Deng G, et al. Long non-coding RNA HOTAIR is targeted and regulated by miR-141 in human cancer cells. J Biol Chem. 2014;289:12550–12565. doi: 10.1074/jbc.M113.488593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Song B, Guan Z, Liu F, Sun D, Wang K, et al. Long non-coding RNA HOTAIR promotes HLA-G expression via inhibiting miR-152 in gastric cancer cells. Biochem Biophys Res Commun. 2015;464:807–813. doi: 10.1016/j.bbrc.2015.07.040. [DOI] [PubMed] [Google Scholar]
- 120.Chiyomaru T, Yamamura S, Fukuhara S, Yoshino H, Kinoshita T, et al. Genistein inhibits prostate cancer cell growth by targeting miR-34a and oncogenic HOTAIR. PLoS One. 2013;8:e70372. doi: 10.1371/journal.pone.0070372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Ke J, Yao YL, Zheng J, Wang P, Liu YH, et al. Knockdown of long non-coding RNA HOTAIR inhibits malignant biological behaviors of human glioma cells via modulation of miR-326. Oncotarget. 2015;6:21934–21949. doi: 10.18632/oncotarget.4290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Su DN, Wu SP, Chen HT, He JH. HOTAIR, a long non-coding RNA driver of malignancy whose expression is activated by FOXC1, negatively regulates miRNA-1 in hepatocellular carcinoma. Oncol Lett. 2016;12:4061–4067. doi: 10.3892/ol.2016.5127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Tuo YL, Li XM, Luo J. Long noncoding RNA UCA1 modulates breast cancer cell growth and apoptosis through decreasing tumor suppressive miR-143. Eur Rev Med Pharmacol Sci. 2015;19:3403–3411. [PubMed] [Google Scholar]
- 124.Wei Y, Sun Q, Zhao L, Wu J, Chen X, et al. LncRNA UCA1-miR-507-FOXM1 axis is involved in cell proliferation, invasion and G0/G1 cell cycle arrest in melanoma. Med Oncol. 2016;33:88. doi: 10.1007/s12032-016-0804-2. [DOI] [PubMed] [Google Scholar]
- 125.Lu L, Luo F, Liu Y, Liu X, Shi L, et al. Posttranscriptional silencing of the lncRNA MALAT1 by miR-217 inhibits the epithelial-mesenchymal transition via enhancer of zeste homolog 2 in the malignant transformation of HBE cells induced by cigarette smoke extract. Toxicol Appl Pharmacol. 2015;289:276–285. doi: 10.1016/j.taap.2015.09.016. [DOI] [PubMed] [Google Scholar]
- 126.Han Y, Liu Y, Zhang H, Wang T, Diao R, et al. Hsa-miR-125b suppresses bladder cancer development by down-regulating oncogene SIRT7 and oncogenic long non-coding RNA MALAT1. FEBS Lett. 2013;587:3875–3882. doi: 10.1016/j.febslet.2013.10.023. [DOI] [PubMed] [Google Scholar]
- 127.Fang D, Yang H, Lin J, Teng Y, Jiang Y, et al. 17beta-estradiol regulates cell proliferation, colony formation, migration, invasion and promotes apoptosis by upregulating miR-9 and thus degrades MALAT-1 in osteosarcoma cell MG-63 in an estrogen receptor-independent manner. Biochem Biophys Res Commun. 2015;457:500–506. doi: 10.1016/j.bbrc.2014.12.114. [DOI] [PubMed] [Google Scholar]
- 128.Yoon JH, Abdelmohsen K, Srikantan S, Yang X, Martindale JL, et al. LincRNA-p21 suppresses target mRNA translation. Mol Cell. 2012;47:648–655. doi: 10.1016/j.molcel.2012.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Wang X, Ren Y, Yang X, Xiong X, Han S, et al. miR-190a inhibits epithelial-mesenchymal transition of hepatoma cells via targeting the long non-coding RNA treRNA. FEBS Lett. 2015;589:4079–4087. doi: 10.1016/j.febslet.2015.11.024. [DOI] [PubMed] [Google Scholar]
- 130.Li N, Zhou P, Zheng J, Deng J, Wu H, et al. A polymorphism rs12325489C>T in the lincRNA-ENST00000515084 exon was found to modulate breast cancer risk via GWAS-based association analyses. PLoS One. 2014;9:e98251. doi: 10.1371/journal.pone.0098251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Tsang FH, Au SL, Wei L, Fan DN, Lee JM, et al. Long non-coding RNA HOTTIP is frequently up-regulated in hepatocellular carcinoma and is targeted by tumour suppressive miR-125b. Liver Int. 2015;35:1597–1606. doi: 10.1111/liv.12746. [DOI] [PubMed] [Google Scholar]
- 132.Xu C, Shao Y, Xia T, Yang Y, Dai J, et al. lncRNA-AC130710 targeting by miR-129-5p is upregulated in gastric cancer and associates with poor prognosis. Tumour Biol. 2014;35:9701–9706. doi: 10.1007/s13277-014-2274-5. [DOI] [PubMed] [Google Scholar]
- 133.Fan QH, Yu R, Huang WX, Cui XX, Luo BH, et al. The has-miR-526b binding-site rs8506G>A polymorphism in the lincRNA-NR_024015 exon identified by GWASs predispose to non-cardia gastric cancer risk. PLoS One. 2014;9:e90008. doi: 10.1371/journal.pone.0090008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.He JH, Zhang JZ, Han ZP, Wang L, Lv YB, et al. Reciprocal regulation of PCGEM1 and miR-145 promote proliferation of LNCaP prostate cancer cells. J Exp Clin Cancer Res. 2014;33:72. doi: 10.1186/s13046-014-0072-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Yao Y, Ma J, Xue Y, Wang P, Li Z, et al. Knockdown of long non-coding RNA XIST exerts tumor-suppressive functions in human glioblastoma stem cells by up-regulating miR-152. Cancer Lett. 2015;359:75–86. doi: 10.1016/j.canlet.2014.12.051. [DOI] [PubMed] [Google Scholar]
- 136.Cao C, Sun J, Zhang D, Guo X, Xie L, et al. The long intergenic noncoding RNA UFC1, a target of microRNA 34a, interacts with the mRNA stabilizing protein HuR to increase levels of beta-catenin in HCC cells. Gastroenterology. 2015;148(415–426):e418. doi: 10.1053/j.gastro.2014.10.012. [DOI] [PubMed] [Google Scholar]
- 137.Liu Q, Huang J, Zhou N, Zhang Z, Zhang A, et al. LncRNA loc285194 is a p53-regulated tumor suppressor. Nucleic Acids Res. 2013;41:4976–4987. doi: 10.1093/nar/gkt182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Zhang Z, Zhu Z, Watabe K, Zhang X, Bai C, et al. Negative regulation of lncRNA GAS5 by miR-21. Cell Death Differ. 2013;20:1558–1568. doi: 10.1038/cdd.2013.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Qi P, Xu MD, Shen XH, Ni SJ, Huang D, et al. Reciprocal repression between TUSC7 and miR-23b in gastric cancer. Int J Cancer. 2015;137:1269–1278. doi: 10.1002/ijc.29516. [DOI] [PubMed] [Google Scholar]
- 140.Wang P, Liu YH, Yao YL, Li Z, Li ZQ, et al. Long non-coding RNA CASC2 suppresses malignancy in human gliomas by miR-21. Cell Signal. 2015;27:275–282. doi: 10.1016/j.cellsig.2014.11.011. [DOI] [PubMed] [Google Scholar]
- 141.Wu H, Zheng J, Deng J, Hu M, You Y, et al. A genetic polymorphism in lincRNA-uc003opf.1 is associated with susceptibility to esophageal squamous cell carcinoma in Chinese populations. Carcinogenesis. 2013;34:2908–2917. doi: 10.1093/carcin/bgt252. [DOI] [PubMed] [Google Scholar]
- 142.Kallen AN, Zhou XB, Xu J, Qiao C, Ma J, et al. The imprinted H19 lncRNA antagonizes let-7 microRNAs. Mol Cell. 2013;52:101–112. doi: 10.1016/j.molcel.2013.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Paci P, Colombo T, Farina L. Computational analysis identifies a sponge interaction network between long non-coding RNAs and messenger RNAs in human breast cancer. BMC Syst Biol. 2014;8:83. doi: 10.1186/1752-0509-8-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Xia T, Liao Q, Jiang X, Shao Y, Xiao B, et al. Long noncoding RNA associated-competing endogenous RNAs in gastric cancer. Sci Rep. 2014;4:6088. doi: 10.1038/srep06088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Jalali S, Bhartiya D, Lalwani MK, Sivasubbu S, Scaria V. Systematic transcriptome wide analysis of lncRNA-miRNA interactions. PLoS One. 2013;8:e53823. doi: 10.1371/journal.pone.0053823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Guo W, Wang Q, Zhan Y, Chen X, Yu Q, et al. Transcriptome sequencing uncovers a three-long noncoding RNA signature in predicting breast cancer survival. Sci Rep. 2016;6:27931. doi: 10.1038/srep27931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Li Z, Li X, Wu S, Xue M, Chen W. Long non-coding RNA UCA1 promotes glycolysis by upregulating hexokinase 2 through the mTOR-STAT3/microRNA143 pathway. Cancer Sci. 2014;105:951–955. doi: 10.1111/cas.12461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Tang L, Shen H, Li X, Li Z, Liu Z, et al. MiR-125a-5p decreases after long non-coding RNA HOTAIR knockdown to promote cancer cell apoptosis by releasing caspase 2. Cell Death Dis. 2016;7:e2137. doi: 10.1038/cddis.2016.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Zhang H, Cai K, Wang J, Wang X, Cheng K, et al. MiR-7, inhibited indirectly by lincRNA HOTAIR, directly inhibits SETDB1 and reverses the EMT of breast cancer stem cells by downregulating the STAT3 pathway. Stem Cells. 2014;32:2858–2868. doi: 10.1002/stem.1795. [DOI] [PubMed] [Google Scholar]
- 150.Li JT, Wang LF, Zhao YL, Yang T, Li W, et al. Nuclear factor of activated T cells 5 maintained by Hotair suppression of miR-568 upregulates S100 calcium binding protein A4 to promote breast cancer metastasis. Breast Cancer Res. 2014;16:454. doi: 10.1186/s13058-014-0454-2. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 151.He P, Zhang Z, Huang G, Wang H, Xu D, et al. miR-141 modulates osteoblastic cell proliferation by regulating the target gene of lncRNA H19 and lncRNA H19-derived miR-675. Am J Transl Res. 2016;8:1780–1788. [PMC free article] [PubMed] [Google Scholar]
- 152.Zhang ZX, Liu ZQ, Jiang B, Lu XY, Ning XF, et al. BRAF activated non-coding RNA (BANCR) promoting gastric cancer cells proliferation via regulation of NF-kappaB1. Biochem Biophys Res Commun. 2015;465:225–231. doi: 10.1016/j.bbrc.2015.07.158. [DOI] [PubMed] [Google Scholar]
- 153.Zhang EB, Kong R, Yin DD, You LH, Sun M, et al. Long noncoding RNA ANRIL indicates a poor prognosis of gastric cancer and promotes tumor growth by epigenetically silencing of miR-99a/miR-449a. Oncotarget. 2014;5:2276–2292. doi: 10.18632/oncotarget.1902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Franklin JL, Rankin CR, Levy S, Snoddy JR, Zhang B, et al. Malignant transformation of colonic epithelial cells by a colon-derived long noncoding RNA. Biochem Biophys Res Commun. 2013;440:99–104. doi: 10.1016/j.bbrc.2013.09.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Zamani M, Sadeghizadeh M, Behmanesh M, Najafi F. Dendrosomal curcumin increases expression of the long non-coding RNA gene MEG3 via up-regulation of epi-miRs in hepatocellular cancer. Phytomedicine. 2015;22:961–967. doi: 10.1016/j.phymed.2015.05.071. [DOI] [PubMed] [Google Scholar]
- 156.Braconi C, Kogure T, Valeri N, Huang N, Nuovo G, et al. microRNA-29 can regulate expression of the long non-coding RNA gene MEG3 in hepatocellular cancer. Oncogene. 2011;30:4750–4756. doi: 10.1038/onc.2011.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Yan J, Guo X, Xia J, Shan T, Gu C, et al. MiR-148a regulates MEG3 in gastric cancer by targeting DNA methyltransferase 1. Med Oncol. 2014;31:879. doi: 10.1007/s12032-014-0879-6. [DOI] [PubMed] [Google Scholar]
- 158.Liu X, Hou L, Huang W, Gao Y, Lv X, et al. The mechanism of long non-coding RNA MEG3 for neurons apoptosis caused by hypoxia: mediated by miR-181b-12/15-LOX signaling pathway. Front Cell Neurosci. 2016;10:201. doi: 10.3389/fncel.2016.00201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Zhuang M, Gao W, Xu J, Wang P, Shu Y. The long non-coding RNA H19-derived miR-675 modulates human gastric cancer cell proliferation by targeting tumor suppressor RUNX1. Biochem Biophys Res Commun. 2014;448:315–322. doi: 10.1016/j.bbrc.2013.12.126. [DOI] [PubMed] [Google Scholar]
- 160.Liu G, Xiang T, Wu QF, Wang WX. Long noncoding RNA H19-derived miR-675 enhances proliferation and invasion via RUNX1 in gastric cancer cells. Oncol Res. 2016;23:99–107. doi: 10.3727/096504015X14496932933575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Li H, Yu B, Li J, Su L, Yan M, et al. Overexpression of lncRNA H19 enhances carcinogenesis and metastasis of gastric cancer. Oncotarget. 2014;5:2318–2329. doi: 10.18632/oncotarget.1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Vennin C, Spruyt N, Dahmani F, Julien S, Bertucci F, et al. H19 non coding RNA-derived miR-675 enhances tumorigenesis and metastasis of breast cancer cells by downregulating c-Cbl and Cbl-b. Oncotarget. 2015;6:29209–29223. doi: 10.18632/oncotarget.4976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Tsang WP, Ng EK, Ng SS, Jin H, Yu J, et al. Oncofetal H19-derived miR-675 regulates tumor suppressor RB in human colorectal cancer. Carcinogenesis. 2010;31:350–358. doi: 10.1093/carcin/bgp181. [DOI] [PubMed] [Google Scholar]
- 164.Shi Y, Wang Y, Luan W, Wang P, Tao T, et al. Long non-coding RNA H19 promotes glioma cell invasion by deriving miR-675. PLoS One. 2014;9:e86295. doi: 10.1371/journal.pone.0086295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Augoff K, McCue B, Plow EF, Sossey-Alaoui K. miR-31 and its host gene lncRNA LOC554202 are regulated by promoter hypermethylation in triple-negative breast cancer. Mol Cancer. 2012;11:5. doi: 10.1186/1476-4598-11-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Emmrich S, Streltsov A, Schmidt F, Thangapandi VR, Reinhardt D, et al. LincRNAs MONC and MIR100HG act as oncogenes in acute megakaryoblastic leukemia. Mol Cancer. 2014;13:171. doi: 10.1186/1476-4598-13-171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Scott MS, Avolio F, Ono M, Lamond AI, Barton GJ. Human miRNA precursors with box H/ACA snoRNA features. PLoS Comput Biol. 2009;5:e1000507. doi: 10.1371/journal.pcbi.1000507. [DOI] [PMC free article] [PubMed] [Google Scholar]