Abstract
The CXCR4/CXCR7/CXCL12 chemokine axis plays important roles in the migration of tumor cells during cancer development by modulating site‐specific distant metastasis including to regional lymph nodes. We investigated the correlation of these chemokine expressions to prognosis in lymph‐node‐positive non‐small‐cell lung cancer (NSCLC) patients. A total of 140 surgically resected specimens of primary site (PS) and metastatic lymph nodes (MLN) of NSCLC involving hilar and/or mediastinal lymph nodes (N1‐2) were collected. CXCR4, CXCR7 and CXCL12 expressions were evaluated. Cox regression analysis was performed to determine whether these chemokines were independent prognostic factors in N1‐2 NSCLC. High expression of CXCR4 in PS and CXCL12 in MLN was associated with poor overall survival (OS) (P = .025 and .033, respectively). Significant correlations between CXCR4 expression in PS and CXCL12 expression in MLN were observed (P = .040). There was significant difference in OS between 2 groups according to expressions of CXCR4 in PS and CXCL12 in MLN (P = .0033). Expression of CXCL12 in MLN was identified as an independent prognostic factor (HR 1.79, 95% CI 1.08‐3.04, P = .023). CXCL12 in MLN was mainly expressed by tumor cells compared with stromal cells (56% vs 25%, respectively, P < .0001). CXCR4/CXCL12 may play roles in tumor progression in MLN and is associated with poor prognosis of lymph‐node‐positive NSCLC patients.
Keywords: CXCL12, CXCR4, CXCR7, metastatic lymph nodes, non‐small‐cell lung cancer
1. INTRODUCTION
Lung cancer continues to be the leading cause of cancer mortality worldwide.1 Most patients with non‐small‐cell lung cancer (NSCLC) are diagnosed at advanced stage, and even when patients are diagnosed at an early stage and treated surgically, cases usually show metastasis with eventual spread to the lymph nodes, adrenal glands, bone, liver and brain, which might contribute to the high death rate.2
Increasing evidence has demonstrated that tumor‐stromal cell interactions play a critical role in carcinogenesis and proliferation.3 Recently, C‐X‐C chemokine ligand‐12 (CXCL12) and its cognate G‐protein coupled receptor C‐X‐C chemokine receptor‐4 (CXCR4) have been shown to modulate site‐specific distant metastasis of many cancer types.4 C‐X‐C chemokine receptor‐7 (CXCR7) has also been described as a second receptor for CXCL12.5 Previous studies reported that tumor cells expressed a high level of CXCR4 and that tumor metastasis target tissues, such as lung, liver, brain and bone, expressed high levels of CXCL12.6 Furthermore, CXCL12 induced migration of tumor cells to target organs via a CXCL12‐CXCR4 chemotactic gradient. These studies have led to the current CXCL12/CXCR4 “endocrine axis” model, in which CXCR4 upregulation in metastatic cells caused metastasis towards organs that abundantly express CXCL12.7 However, the mechanisms by which these paracrine effects impact cancer progression and the potential role of CXCL12 are still uncertain.
In general, normal lymph node tissue shows high expression of CXCL12.4 Although CXCL12 expression in lymph node stromal cells in pre‐metastatic status is considered to have an important role during the process of tumor metastasis, how CXCL12 expression in lymph nodes is altered in the post‐metastatic status is unknown. Therefore, it is important to analyze metastatic lymph nodes (MLN) and to evaluate the change of CXCL12 expression compared with normal lymph nodes.
High expression of chemokine receptors in the primary site (PS) in cancer suggests the potential effectiveness of diagnostic agents and therapy targeted to chemokine receptor‐overexpressing tumors. However, despite previous reports on chemokine receptor expression in various cancers, only a few clinical studies have been performed to evaluate the clinical significance of chemokine receptor status in NSCLC.8, 9, 10, 11 A previous report showed that high CXCR7 mRNA level might be related to the development of lymph node metastasis in NSCLC,8 and high CXCR4 protein expression was found to be significantly associated with lymph node metastasis, distant metastasis, tumor stage and overall survival (OS) in NSCLC.10 In contrast, in another study, strong CXCR4‐positive nuclear staining was associated with a significantly better outcome in early stage NSCLC compared with CXCR4‐negative nuclear staining.9 The CXCL12/CXCR4 axis was shown to be involved in the dissemination of NSCLC cells into the pleural space.11 Despite these reports, no studies on chemokine receptors and chemokines in MLN in NSCLC have been described.
In this study, we hypothesized that chemokine and chemokine receptor expression in PS as well as MLN affects the prognosis of NSCLC. Here we examined CXCR4 and CXCR7 in PS and CXCL12 expressions in MLN in NSCLC patients and assessed the prognostic impact of the expressions of these factors.
2. MATERIALS AND METHODS
2.1. Patients
From November 2000 to November 2012, a total of 1194 NSCLC patients underwent surgical resection at the Department of Surgery and Science, Kyushu University Hospital, Fukuoka, Japan. Among these patients, we selected 140 patients (11.7%) with resected NSCLC with regional lymph node metastases (pathological N1 or N2). The profiles of these 140 patients are shown in Table 1. These patients consisted of 91 men and 49 women, with a mean age of 67 years (range 29‐83 years) at the time of surgical resection. Based on the World Health Organization (WHO) 2004 classification, histological type was diagnosed by H&E staining. Pathologic staging was undertaken according to the 7th edition of the TNM Classification of Malignant Tumors.12 No patient was treated with chemotherapy or radiotherapy prior to surgery. A total of 76 (54.3%) patients received adjuvant chemotherapy: 23 received oral tegafur and uracil, 28 received platinum‐doublet chemotherapy, 24 participated in a clinical trial for postoperative adjuvant chemotherapy (S‐1 or cisplatin plus S‐1), and 1 received paclitaxel monotherapy. A routine follow‐up involving a physical examination, chest x‐rays, measurements of blood cell counts, serum chemistry, and tumor markers was performed on an outpatient basis 4 times a year for the first 3 years and twice a year thereafter. Computed tomography was performed twice a year for the first 3 years and at least once per year thereafter. Brain magnetic resonance imaging and bone scintigraphy or fluorodeoxyglucose positron‐emission tomography were performed annually. This study was approved by the Kyushu University Institutional Review Board for Clinical Research (No. 28‐186).
Table 1.
Clinicopathological characteristics of all patients
| Factors | All patients (n = 140) |
|---|---|
| Sex (%) | |
| Male | 91 (65) |
| Female | 49 (35) |
| Age (%) | |
| <75 | 118 (84) |
| ≥75 | 22 (16) |
| Smoking (%) | |
| Non‐smoker | 64 (46) |
| Smoker | 76 (54) |
| pT factor (%) | |
| T1 | 43 (31) |
| T2 | 75 (54) |
| T3 | 16 (11) |
| T4 | 6 (4) |
| pN factor (%) | |
| N1 | 66 (47) |
| N2 | 74 (53) |
| pStage (%) | |
| II | 52 (37) |
| III | 88 (63) |
| Surgical procedure (%) | |
| Segmentectomy | 2 (1) |
| Lobectomy | 125 (89) |
| Pneumonectomy | 13 (10) |
| Histology (%) | |
| Adenocarcinoma | 97 (69) |
| SCC | 29 (21) |
| LCC | 8 (6) |
| Sarcomatoid carcinoma | 3 (2) |
| Adenosquamous carcinoma | 2 (1) |
| Neuroendocrine tumors | 1 (1) |
| Histological grade (%) | |
| 1 | 16 (11) |
| 2 | 74 (53) |
| 3 | 35 (25) |
| 4 | 6 (4) |
| Unknown | 9 (7) |
| Intratumoral lymphatic vessel invasion (%) | |
| Negative | 59 (42) |
| Positive | 79 (56) |
| Unknown | 2 (2) |
| Intratumoral vascular invasion (%) | |
| Negative | 41 (29) |
| Positive | 98 (70) |
| Unknown | 1 (1) |
| Metastatic lymph node station (%) | |
| Single | 60 (43) |
| Multiple | 80 (57) |
| CEA, ng/mL (range) | 4.2 (0.4‐128) |
| SUV max (range) | 9.1 (1.1‐23) |
| EGFR status (%) | |
| Wild | 50 (36) |
| Mutation | 20 (14) |
| Unknown | 70 (50) |
EGFR, epidermal growth factor receptor; LCC, large cell carcinoma; SCC, squamous cell carcinoma.
2.2. Tissue preparation and immunohistochemistry analysis
Primary lung carcinoma and lymph node tissues were fixed immediately in 10% (v/v) formalin after resection. After embedding in paraffin, serial 3‐μm sections were prepared from each sample and reserved for H&E staining as well as immunohistochemistry (IHC). We performed IHC for CXCR4, CXCR7 and CXCL12 on the 140 PS and 356 MLN samples. Paraffin‐embedded sections were cut and mounted on poly l‐lysine‐coated slides, dewaxed and rehydrated. Endogenous peroxidase was blocked with 3% hydrogen peroxide in methanol for 30 minute. For antigen retrieval, sections were microwaved at 99°C for 20 minute in 0.01 mL/L citrate buffer (pH 6.0 or pH 9.0). After incubation in a non‐specific stain blocking agent, sections were incubated overnight at 4°C with primary antibody: rabbit monoclonal anti‐human CXCR4 antibody (ab124824; Abcam, Cambridge, UK; 1:100),13 rabbit polyclonal anti‐human CXCR7/RDC‐1 antibody (NBP1‐31309; Novus Biologicals, Littleton, CO, USA; 1:400)14 or mouse monoclonal anti‐human CXCL12 antibody (MAB350; R&D Systems, Minneapolis, MN, USA; 1:20).15 This was followed by an avidin‐biotin procedure using a streptavidin‐biotin‐peroxidase kit (Histofine SABPO kit; Nichirei Bioscience, Tokyo, Japan). Positive controls included a human tonsil specimen stained for CXCR4, CXCR7 and CXCL12. As negative controls, each section was treated with Rabbit IgG, monoclonal [EPR25A]—Isotype Control (ab172730; Abcam; 1:100 or 1:400) and Mouse IgG1 Isotype Control (R&D Systems; 1:20).
2.3. Evaluation of immunohistochemistry
Immunohistochemical staining of CXCR4, CXCR7 and CXCL12 was evaluated by the Allred scoring system.16 Briefly, the total Allred score determined for intensity (absent, 0; weak, 1; moderate, 2; and strong, 3) was added to the score determined for the percentage of both tumor cells and stromal cells for CXCL12 staining (no cells: 0; <1% cells: 1; 1%‐10% each individual. By ROC curve analysis, a total Allred score of 6 was determined for cells: 2; 11%‐33% cells: 3; 34%‐66% cells: 4; and 67%‐100% cells: 5) to yield a total Allred score of 0 or 2‐8. The cut‐off value was determined by performing receiver operating characteristic (ROC) analysis. The area under the curve (AUC) was then calculated as the best diagnosis cut‐off value both in CXCR4 and CXCR7 (PS) (AUC = 0.61, 95% CI 0.044‐1.29 and AUC = 0.61, 95% CI 0.50‐0.87, respectively). Thus, a total Allred score of 0‐5 was regarded as low expression, while a total Allred score of 6‐8 was confirmed as high expression. A total Allred score of 7 was also determined as the best diagnosis cut‐off value in CXCL12 (MLN) (AUC = 0.64, 95% CI 0.89‐1.79) by ROC curve analysis. Thus, a total Allred score of 0‐6 was regarded as low expression, while a score of 7 and 8 was confirmed as high expression (Fig. S1). When multiple lymph node metastases were recognized, the lymph node with the highest expression level was used to determine the total Allred score. IHC results were evaluated independently by 2 of the authors (MK and FS), who were blinded to the clinical data of the patients.
2.4. Indirect immunofluorescence
Lymph node samples were fixed immediately in 10% (v/v) formalin after resection. Samples were incubated with Blocking One Histo (Nakalai Tesque, Kyoto, Japan) to eliminate nonspecific binding, and then incubated overnight at 4°C with primary antibodies: α‐SMA (monoclonal rabbit anti‐human ACTA2/Smooth Muscle Actin; LifeSpan BioSciences, Seattle, WA, USA, 1:200) or CXCL12 (polyclonal goat anti‐human SDF‐1; Gene‐Tex, Irvine, CA, USA; 1:200). The slides were then washed thoroughly with PBS and incubated with the following secondary antibodies for 30 minute: Alexa Fluor 488‐labeled donkey anti‐rabbit IgG H&L (Abcam; 1:2000) or Alexa Fluor 647‐labeled donkey anti‐goat IgG H&L (Abcam; 1:2000). After thoroughly washing with PBS, the slides were mounted in DAPI (Sigma‐Aldrich, St. Louis, MO, USA) and examined using a BZ‐9000 microscope (Keyence, Osaka, Japan).
2.5. RT‐PCR
Of the 140 total cases examined by IHC, 13 patients were examined by RT‐PCR in duplicate with RNA extracted from the paired samples of PS and MLN that had been sampled during surgery and stored in RNAlater solution (Sigma‐Aldrich). All tumor lesions were histologically confirmed to be NSCLC. Total RNA was isolated from each lung tissue sample with the NucleoSpin RNA XS kit (Macherey‐Nagel, Duren, Germany) according to the manufacturer's instructions. RNA was converted into cDNA using the Prime Script RT Master Mix (Takara Bio, Shiga, Japan) according to the manufacturer's protocol. cDNA was synthesized using 500 ng of each mRNA sample with Oligo‐dT‐Primers (Life Technologies, Darmstadt, Germany), Superscript II Reverse Transcriptase (Life Technologies) and dNTP (Fermentas, Waltham, MA, USA) following the manufacturer's instructions. The cDNA was diluted fivefold and used for real‐time PCR analysis with the Thermal Cycler Dice Real Time System II TP‐900 (Takara Bio) using a TaqMan Gene Expression Assay probe and primer mix (Applied Biosystems, Foster City, CA, USA) according to the manufacturer's instructions. PCR reactions were run for 40 cycles with denaturation at 95°C for 5 second and annealing at 60°C for 30 second. Relative gene expression was determined by the ΔΔC t method that is based on the relative expression of the target gene vs a reference gene (β actin) and normalized to the median of the primary lung cancer samples. ΔC t was defined as mRNA (target gene) − mRNA (reference gene). ΔΔC t = ΔC t (target sample: lymph node meta) − ΔC t (control sample: primary lung cancer). RT‐PCR assays were performed in duplicate for each sample, and the mean value was used for calculation of the mRNA expression levels. Primer sets for all genes were purchased from Takara Bio. The following primers were used: CXCR4: Hs00976734m1, ACKR3: Hs00604567m1, CXCL12: Hs03676656 mH and β‐actin: 4310884E.
2.6. Statistical methods
All data are expressed as the means ± SEM. Statistical analysis was performed using JMP Pro statistical software version 12.2.0 (SAS Institute, Cary, NC, USA). The OS rates after surgical treatment were calculated using the Kaplan‐Meier method, and differences were evaluated using the log‐rank test. Clinicopathological analysis of prognostic factors was performed using the χ2‐test, while univariate and multivariate survival analyses were performed using the Cox regression hazards model. A P‐value < .05 was considered statistically significant.
3. RESULTS
3.1. Comparison between total Allred scores and mRNA levels of CXCR4, CXCR7 and CXCL12 in primary sites and metastatic lymph nodes
To confirm our IHC index, we first analyzed the correlation between total Allred score and RT‐PCR quantification. A significant correlation between total Allred score grouped by cut‐off and ΔC t value was found (CXCR4: P < .05; CXCR7: P < .01; and CXCL12: P < .0001) (Figure 1).
Figure 1.
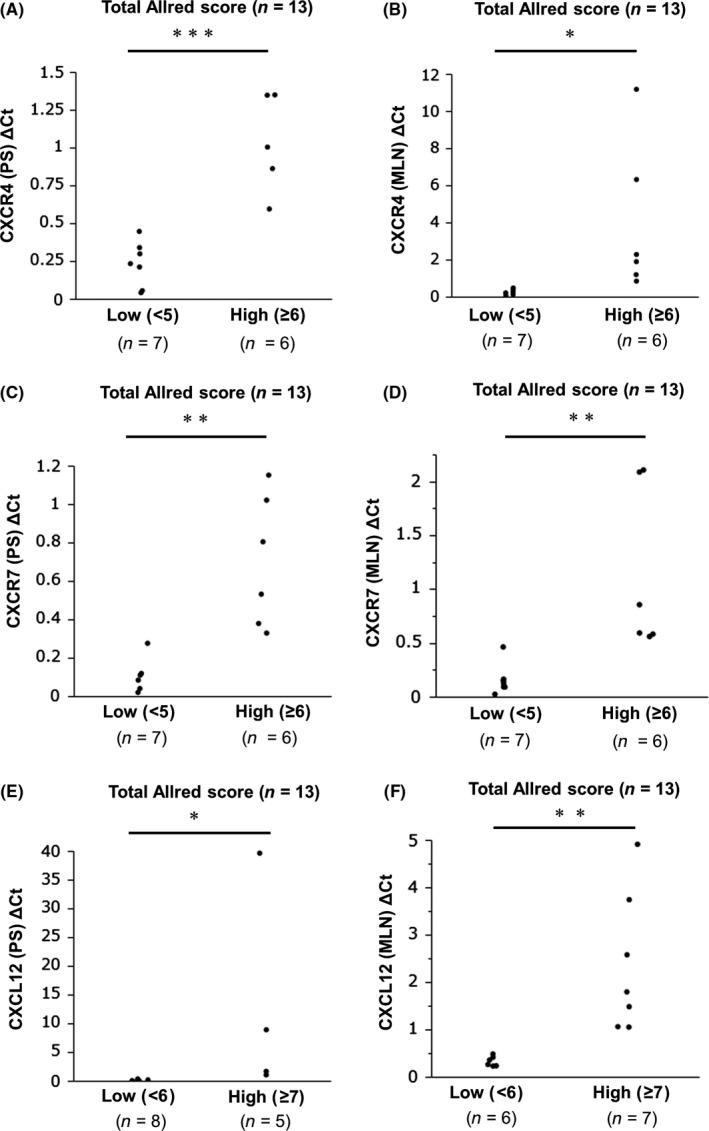
Comparison between total Allred scores and mRNA levels of CXCR4, CXCR7 and CXCL12 in primary sites and metastatic lymph nodes. A,C,E, Comparison of total Allred scores of CXCR4, CXCR7 and CXCL12 in primary site (PS) between low and high groups. B,D,F, Comparison of total Allred scores of CXCR4, CXCR7 and CXCL12 in metastatic lymph nodes (MLN) between low and high groups. C t, threshold cycle. ΔC t, C t of target − C t of reference (β‐actin). The statistical difference was assessed by two‐way analysis of variance. *P < .05, **P < .01, ***P < .0001
3.2. Protein expression of CXCR4 in primary site and CXCR7 and CXCL12 in metastatic lymph nodes
Immunohistochemical analysis revealed CXCR4 expression in the nuclei and/or membrane of tumor cells, whereas CXCR7 expression was found in cytoplasm and/or membrane of tumor cells (Figure 2A,B and Fig. S2A,B). CXCL12 protein was found in the cytoplasm and/or membrane of tumor cells (Figure 2C and Fig. S2C). High CXCR4 protein expression was observed in 60 (42.9%) PS samples. In MLN samples, high CXCR7 and CXCL12 protein expressions were observed in 82 (58.6%) and 79 samples (56.4%) (Figure 2D). The isotype control for CXCL12 in MLN is shown in Fig. S2D. CXCL12 was also expressed on stromal cells in MLN (Fig. S2E).
Figure 2.
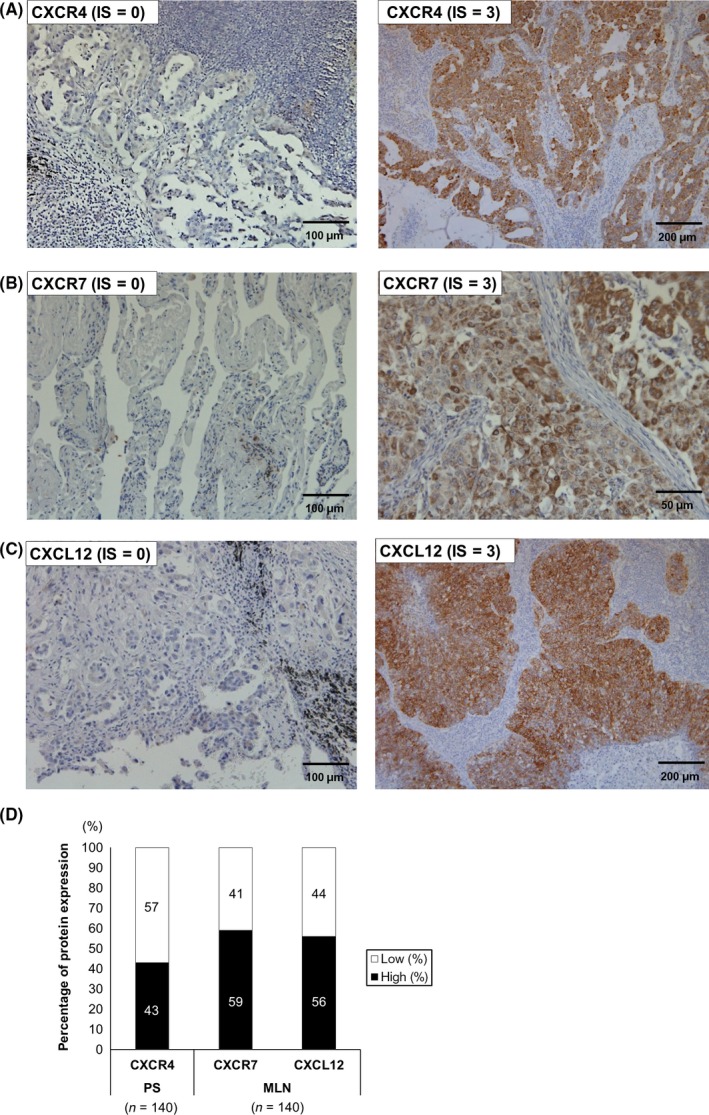
Expression of CXCR4, CXCR7 and CXCL12 in primary sites (PS) and metastatic lymph nodes (MLN). A, CXCR4 immunostaining in lung tumor cells. IS = 0 (left) and IS = 3 (right) (×200 and ×100, respectively). B, CXCR7 immunostaining in lung tumor cells. IS = 0 (left) and IS = 3 (right) (×200 and ×400, respectively). C, CXCL12 immunostaining in lung tumor cells. IS = 0 (left) and IS = 3 (right) (×200 and ×100, respectively). D, The bar graphs show the percentage of expression of each protein in PS or MLN. White bar: low expression group; black bar: high expression group. IS, intensity score; MLN, metastatic lymph node; PS, primary site
3.3. Comparison of protein expression of CXCL12 in stromal cells and tumor cells in metastatic lymph nodes
Figure 3 shows a comparison of CXCL12 protein expression in stromal cells and tumor cells in MLN. Only 25% of stromal cells in MLN expressed CXCL12 protein. Conversely, 56% of tumor cells in MLN expressed CXCL12 protein. There was statistically significant difference between these expression rates (P < .0001). Thus, we evaluated mainly the chemokine expression in tumor cells in MLN.
Figure 3.
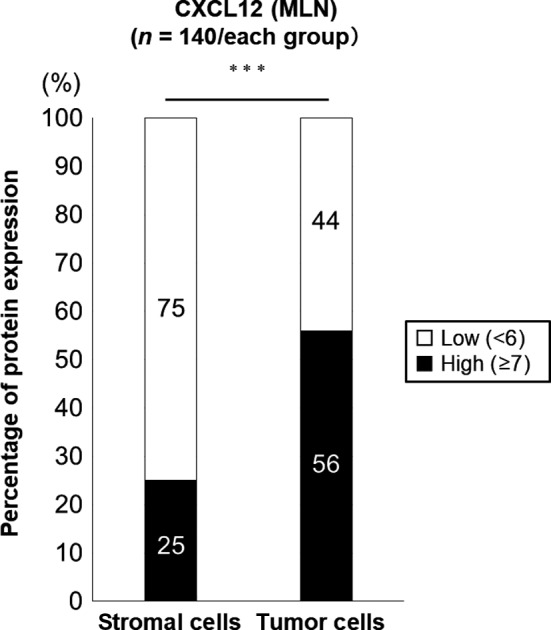
Comparison of CXCL12 total Allred score grouped by cut‐off in stromal cells and tumor cells in metastatic lymph nodes. The bar graphs show the distribution of high and low CXCL12 protein expression grouped by cut‐off in stromal and tumor cells in metastatic lymph nodes (MLN). White bar: low CXCL12 protein expression group; black bar: high CXCL12 protein expression group. ***P < .0001
3.4. Relationships between CXCR4, CXCR7 and CXCL12 protein expression in primary site and metastatic lymph nodes with clinicopathological features
We next assessed the relationships of CXCR4, CXCR7 and CXCL12 protein expression in PS or MLN with clinicopathological features. As shown in Table 2, high CXCR4 protein expression in PS was significantly associated with intratumoral vascular invasion (P = .031). Low CXCR7 protein expression in MLN was associated with histology (P = .011). High CXCL12 protein expression in MLN was not correlated with any clinicopathological features (Table 3). Table S1 shows that high CXCR7 protein expression in PS was significantly associated with histology and histological grade (P = .012 and .027, respectively). High CXCL12 protein expression in PS was significantly associated with intratumoral lymphatic vessel invasion (P = .0031). High CXCR4 protein expression in MLN was associated with sex, pN factor, pStage and histology (P = .0071, .01, .013 and .014, respectively) (Table S2).
Table 2.
Clinicopathological characteristics of patients according to CXCR4 protein expression in the primary site
| Factors | CXCR4 (PS) | P‐value | |
|---|---|---|---|
| Low (n = 80) | High (n = 60) | ||
| Sex (%) | |||
| Male | 50 (63) | 41 (68) | .82 |
| Female | 30 (37) | 19 (32) | |
| Age (%) | |||
| <75 | 68 (85) | 50 (83) | .59 |
| ≥75 | 12 (15) | 10 (17) | |
| Smoking (%) | |||
| Non‐smoker | 48 (60) | 28 (47) | .13 |
| Smoker | 32 (40) | 32 (53) | |
| pT factor (%) | |||
| T1, 2 | 68 (85) | 50 (83) | .82 |
| T3, 4 | 12 (15) | 10 (17) | |
| pN factor (%) | |||
| N1 | 39 (49) | 27 (45) | .73 |
| N2 | 41 (51) | 33 (55) | |
| pStage (%) | |||
| II | 34 (43) | 18 (30) | .16 |
| III | 46 (57) | 42 (70) | |
| Histology (%) | |||
| Adenocarcinoma | 58 (73) | 39 (65) | .11 |
| SCC | 12 (15) | 17 (28) | |
| Others | 10 (12) | 4 (07) | |
| Histological grade (%) | |||
| 1 | 10 (12) | 6 (10) | .29 |
| 2 | 38 (48) | 36 (60) | |
| 3, 4 | 27 (40) | 14 (30) | |
| Intratumoral lymphatic vessel invasion (%) | |||
| Negative | 37 (46) | 22 (37) | .38 |
| Positive | 43 (54) | 36 (63) | |
| Intratumoral vascular invasion (%) | |||
| Negative | 29 (36) | 12 (20) | .031 |
| Positive | 51 (64) | 47 (80) | |
| Metastatic lymph node station (%) | |||
| Single | 35 (44) | 25 (42) | .86 |
| Multiple | 45 (56) | 35 (58) | |
PS, primary site; SCC, squamous cell carcinoma.
Table 3.
Clinicopathological characteristics of patients according to CXCR7 and CXCL12 protein expression in metastatic lymph nodes
| Factors | CXCR7 (MLN) | P‐value | CXCL12 (MLN) | P‐value | ||
|---|---|---|---|---|---|---|
| Low (n = 58) | High (n = 82) | Low (n = 61) | High (n = 79) | |||
| Sex (%) | ||||||
| Male | 33 (57) | 58 (70) | .11 | 41 (67) | 50 (63) | .72 |
| Female | 25 (43) | 24 (30) | 20 (33) | 29 (37) | ||
| Age (%) | ||||||
| <75 | 50 (86) | 68 (83) | .65 | 53 (87) | 65 (82) | .49 |
| ≥75 | 8 (14) | 14 (17) | 8 (13) | 14 (18) | ||
| Smoking (%) | ||||||
| Non‐smoker | 36 (62) | 40 (49) | .13 | 37 (61) | 39 (49) | .23 |
| Smoker | 22 (38) | 42 (51) | 24 (39) | 40 (51) | ||
| pT factor (%) | ||||||
| T1, 2 | 51 (88) | 67 (82) | .36 | 51 (84) | 67 (85) | 1.00 |
| T3, 4 | 7 (12) | 15 (18) | 10 (16) | 12 (15) | ||
| pN factor (%) | ||||||
| N1 | 31 (53) | 35 (43) | .23 | 28 (46) | 38 (48) | .87 |
| N2 | 27 (47) | 47 (57) | 33 (54) | 41 (52) | ||
| pStage (%) | ||||||
| II | 24 (41) | 28 (34) | .48 | 22 (36) | 30 (38) | .86 |
| III | 34 (59) | 54 (66) | 39 (64) | 49 (62) | ||
| Histology (%) | ||||||
| Adenocarcinoma | 37 (64) | 60 (73) | .011 | 45 (74) | 52 (66) | .11 |
| SCC | 10 (17) | 19 (23) | 8 (13) | 21 (27) | ||
| Others | 11 (19) | 3 (04) | 8 (13) | 6 (07) | ||
| Histological grade (%) | ||||||
| 1 | 7 (13) | 9 (12) | .78 | 8 (14) | 8 (11) | .75 |
| 2 | 28 (53) | 46 (59) | 32 (57) | 42 (56) | ||
| 3, 4 | 18 (34) | 23 (29) | 16 (29) | 25 (33) | ||
| Intratumoral lymphatic vessel invasion (%) | ||||||
| Negative | 27 (47) | 32 (40) | .49 | 27 (44) | 32 (42) | .86 |
| Positive | 31 (53) | 48 (60) | 34 (56) | 45 (58) | ||
| Intratumoral vascular invasion (%) | ||||||
| Negative | 18 (31) | 23 (28) | .85 | 16 (26) | 25 (32) | .57 |
| Positive | 40 (69) | 58 (72) | 45 (74) | 53 (68) | ||
| Metastatic lymph node station (%) | ||||||
| Single | 28 (48) | 32 (39) | .3 | 31 (51) | 29 (37) | .12 |
| Multiple | 30 (52) | 50 (61) | 30 (49) | 50 (63) | ||
MLN, metastatic lymph nodes; SCC, squamous cell carcinoma.
3.5. Comparison of total Allred scores and mRNA expression in primary site and metastatic lymph nodes
The total Allred scores of CXCR4, CXCR7 and CXCL12 in both PS and MLN are shown in Fig. S3. The expression of CXCR4 and CXCL12 protein in MLN were significantly higher than those in PS (P < .01 and P < .0001, respectively) (Fig. S3A,C). Conversely, the expression of CXCR7 protein in MLN was significantly lower than that in PS (P < .05) (Fig. S3B). Fig. S3D shows the mRNA levels of CXCR4, CXCR7 and CXCL12 both in PS and MLN. CXCL12 mRNA expression in MLN was significantly higher than that in PS (P = .0077); however, there were no significant differences in CXCR4 or CXCR7 mRNA levels between sites (P = .097 or P = .051, respectively).
3.6. Co‐expression of CXCR4 in primary site, CXCR7 in MLN and CXCL12 on stromal cells in metastatic lymph nodes and in tumor cells in metastatic lymph nodes in each patient
Figure S4 showed, in either high or low CXCR4 in PS, co‐expression of CXCR7 in MLN and CXCL12 in tumor cells in MLN. Thirty‐one patients (52%) showed high expression of all proteins, and it was the most frequent among each group. A significant correlation between CXCR4 protein expression in PS and CXCR7 and CXCL12 protein expression in MLN was observed (P = .0091 and .040, respectively) (Table 4).
Table 4.
Correlations between CXCR4 expression in the primary site and CXCR7 and CXCL12 expression in the metastatic lymph nodes
| CXCR7 (MLN) | P‐value | CXCL12 (MLN) | P‐value | |||
|---|---|---|---|---|---|---|
| Low (n = 58) | High (n = 82) | Low (n = 61) | High (n = 79) | |||
| CXCR4 (PS) (%) | ||||||
| Low | 41 (71) | 39 (48) | .0091 | 41 (67) | 39 (49) | .04 |
| High | 17 (29) | 43 (52) | 20 (33) | 40 (51) | ||
MLN, metastatic lymph nodes; PS, primary site.
3.7. Survival according to CXCR4 protein expression in primary site and CXCL12 protein expression in metastatic lymph nodes
Kaplan‐Meier analysis revealed that patients with high CXCR4 protein expression in PS had a significantly shorter OS than the group with low CXCR4 protein expression in PS (P = .025, log‐rank test) (Figure 4A). Furthermore, the group with high CXCL12 protein expression in MLN had a significantly shorter OS than the group with low CXCL12 protein expression in MLN (P = .033, log‐rank test) (Figure 4B). We observed significant differences between the group with CXCR4 expression in PS and the group with CXCL12 expression in MLN (log‐rank test, P = .0033; Figure 4C). This result indicated that the OS was worse for patients with both high CXCR4 expression in PS and high CXCL12 expression in MLN compared with patients with high expression of CXCR4 or CXCL12 alone.
Figure 4.
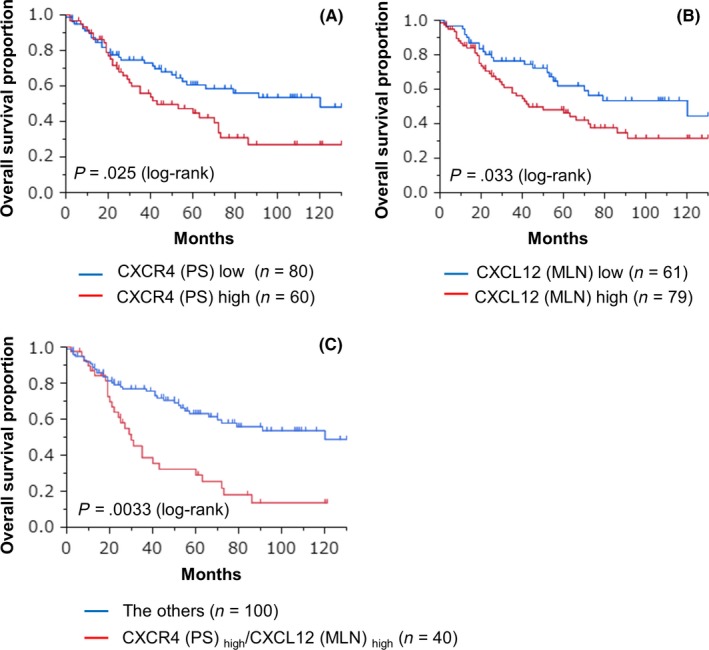
Postoperative overall survival curves of patients with non‐small cell lung cancer (NSCLC) in the primary site and metastatic lymph nodes. A, Kaplan‐Meier curves show overall survival according to CXCR4 in the primary site (PS). Patients with high CXCR4 expression had a poorer prognosis than patients with low CXCR4 expression (P < .05, log‐rank test). B, Kaplan‐Meier curves show overall survival according to CXCL12 expressions in metastatic lymph nodes (MLN). Patients with high CXCL12 expression had a poorer prognosis than patients with low CXCL12 expression (P < .05, log‐rank test). C, Subgroup analysis using both CXCR4 expression in PS and CXCL12 expression in MLN. Patients with both high CXCR4 and high CXCL12 expression had a poorer prognosis than the other patient groups (P < .005)
3.8. Prognostic factors for survival of 140 non‐small‐cell lung cancer patients
Cox regression hazard regression models showed that CXCL12 expression on tumor cells in MLN (HR 1.79; 95% CI 1.08‐3.04; P = .023) as well as advanced age (≥75 years) (HR 1.90; 95% CI 1.01‐3.37), multiple MLN station (HR 1.86; 95% CI 1.12‐3.14; P = .012) and intratumoral vascular invasion (HR 2.30; 95% CI 1.29‐4.34; P = .0041) were independent prognostic factors for OS (Table 5).
Table 5.
Univariate and multivariate analysis of the relationships between overall survival and the clinical factors of the NSCLC patients
| Factors | Univariate analysis | Multivariate analysis | ||
|---|---|---|---|---|
| HR (95% CI) | P‐value | HR (95% CI) | P‐value | |
| Sex | ||||
| Male/female | 1.46 (0.87‐2.55) | .15 | ||
| Age | ||||
| ≥75/<75 | 1.96 (1.04‐3.45) | .038 | 1.90 (1.01‐3.37) | .048 |
| Smoking | ||||
| Smoker/Non‐smoker | 1.52 (0.94‐2.49) | .089 | ||
| Pathological T stage | ||||
| ≥T2/<T2 | 1.05 (0.46‐2.09) | .89 | ||
| Metastatic lymph node station | ||||
| Multiple/single | 1.62 (0.99‐2.70) | .055 | 1.86 (1.12‐3.14) | .012 |
| Intratumoral lymphatic vessel invasion | ||||
| Positive/negative | 1.44 (0.88‐2.43) | .15 | ||
| Intratumoral vascular invasion | ||||
| Positive/negative | 1.85 (1.06‐3.41) | .029 | 2.30 (1.29‐4.34) | .0041 |
| CXCR4 expression (PS) | ||||
| High/low | 1.73 (1.06‐2.82) | .028 | ||
| CXCL12 expression (MLN) | ||||
| High/low | 1.71 (1.04‐2.87) | .033 | 1.79 (1.08‐3.04) | .023 |
MLN, metastatic lymph nodes; NSCLC, non‐small cell lung cancer; PS, primary site.
3.9. CXCL12 expressing cells in the primary site and metastatic lymph nodes
We next performed double immunostaining for CXCL12 and α‐SMA to identify cancer‐associated fibroblasts (CAF) using paraffin‐embedded specimens. In PS, both CXCL12 protein expression and α‐SMA protein expression were predominant (Figure 5A). In MLN, CXCL12 protein expression was found in the membrane of tumor cells and α‐SMA protein expression in the surrounding stroma. Double positive cells were seen at the border area of the tumor cluster. In MLN, CXCL12 protein was expressed at higher levels in tumor cells than in CAF (Figure 5B).
Figure 5.
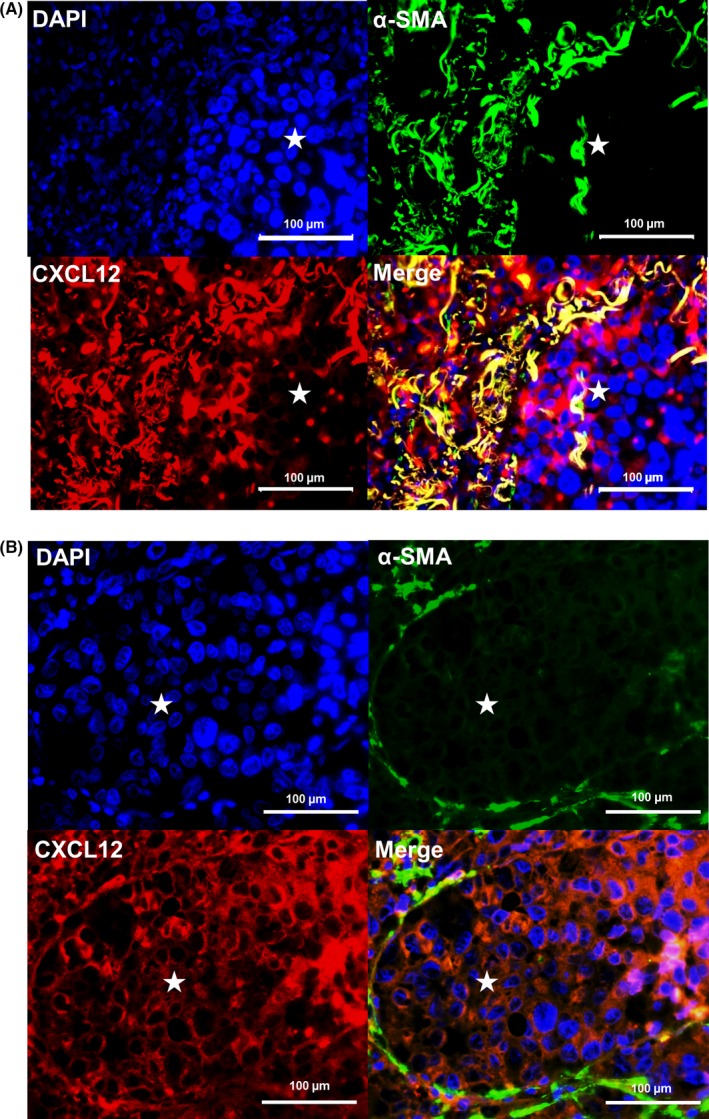
Tumor cells and cancer‐associated fibroblasts produce increased CXCL12 protein in primary sites and metastatic lymph nodes. A, Immunostaining of primary site of invasive human lung cancer for CXCL12 and α‐SMA (×400). B, Immunostaining of metastatic lymph node of invasive human lung cancer for CXCL12 and α‐SMA (×400). DAPI (blue) indicates cell nuclei (upper left), α‐SMA (green, upper right), CXCL12 (red, lower left) and merge (lower right). *Tumor cluster
4. DISCUSSION
In this study, we demonstrated the prognostic impact of CXCR4 expression in PS and CXCL12 expression in MLN in lymph‐node‐positive NSCLC patients. The present study revealed 3 novel findings. First, patients with high CXCR4 protein expression in PS and high CXCL12 protein expression in MLN had a poor prognosis. Second, CXCL12 protein and mRNA expression levels were significantly higher in MLN than in PS. Third, tumor cells in MLN expressed CXCL12 protein similarly to CAF.
The expression of CXCR4 and CXCR7 and their ligand CXCL12 has been shown to be elevated in many types of cancer, such as breast cancer,17 gastric cancer,18 pancreatic cancer,19 ovarian carcinoma,20 cervical carcinoma21 and oral squamous cell carcinoma.22 Kasagi et al.23 previously showed the homing efficiency of CT26 cells to the region highly expressing CXCL12, a chemokine for CXCR4, in a mouse peritoneal dissemination model. CXCL12 is abundantly expressed in the organs that are tumor metastasis target tissues, such as bone, brain, liver, adrenal gland and lymph nodes. Indeed, expression of CXCL12 is found in tumor cells, stromal fibroblasts, multiple immune cells, endothelial cells and stem cells.24 High expression of CXCR4 and CXCL12 was significantly correlated with lymphatic metastases in gastric cancer18 and a low overall survival rate in pancreatic cancer.19 However, whether high CXCL12 expression on tumor cells in MLN might influence the survival of NSCLC patients was unclear.
In general, normal lymph node tissue shows high expression of CXCL12.4 Therefore, it is reasonable to suppose that CXCR4‐positive tumor cells may migrate to regional normal lymph nodes in lung cancer. The exact mechanisms of the upregulation of CXCL12 expression in stromal cells have not yet been revealed. Paget et al.25 proposed that tumor cells might prepare lymph nodes for their later arrival, giving a new interpretation of the seed‐and‐soil hypothesis. However, the chemokine status of lymph nodes with metastasized tumor cells has not been examined. In fact, we found that CXCL12 expression in stromal cells of MLN was low (only 25% of stromal cells in MLN expressed CXCL12). Thus, we considered the following possible explanation for this phenomenon: (i) migration of tumor cells to lymph nodes could be promoted by upregulation of CXCL12 protein expression on stromal cells in pre‐MLN; and (ii) in post‐MLN, tumor cells could regulate CXCL12 expression in MLN by controlling the CXCL12 expression in stromal cells. Although we could not elucidate this mechanism in the present study, we speculate that the CXCL12 expression of stromal cells in lymph nodes might be suppressed by metastatic tumor cells in an autocrine manner. Therefore, we mainly examined the expression of CXCL12 not on stromal cells but on tumor cells in metastatic lymph nodes. The verification of this hypothesis is necessary and we are now planning for further study to address these questions.
In the present study, we showed that CXCL12 expression on tumor cells in MLN was elevated both at protein and mRNA levels. The protein expression results were consistent with mRNA expression findings for only CXCL12. One reason for this may be due to the type of antibody and/or the state of cDNA. Nevertheless, we found that the expression of CXCL12 expression in MLN was significantly higher than CXCL12 expression in PS (Fig. S3). We also showed that CXCL12 in MLN was an independent prognostic factor for survival.
Our results support the observation that abnormal overexpression of CXCR4 is associated with worse OS in NSCLC.26, 27 Su et al.28 report that the migration of NSCLC cells from PS to MLN might be dependent on the level of CXCR4. The findings of this study support the current CXCL12/CXCR4 “endocrine axis” model, in which CXCR4 upregulation by metastatic cells enables these cells to migrate toward organs expressing CXCL12. Furthermore, CXCR4 can also promote tumor vascularization and act as a survival or growth factor.29 We found that CXCR4 exhibited a significant positive correlation with pathological intratumoral vascular invasion (P = .031). Hence, CXCR4 in PS was an independent prognostic factor for survival. Some studies have reported that CXCR4 and CXCR7 are highly expressed in lung cancer tissue and contribute to cancer metastasis.30, 31
Emerging evidence has demonstrated that epithelial‐mesenchymal transition (EMT) plays an important role in cancer metastasis.32 Previous studies have shown that the CXCL12‐CXCR4 signal also plays an important role in EMT.33 CAF can promote EMT by releasing CXCL12, which modulates the stability of EMT transcription factors.34 The mechanisms underlying the upregulation of CXCL12 expression are not completely understood, although previous studies have shown that CAF stimulate tumor progression by CXCL12 secretion.35, 36 Two major mechanisms by which fibroblast‐derived CXCL12 promotes tumor progression have been identified.37 First, CXCL12 facilitates tumor cell growth in a paracrine manner by directly stimulating tumor cell growth via CXCR4. Second, CXCL12 from CAF induces recruitment of endothelial progenitors, which allow for tumor angiogenesis (an endocrine effect of CXCL12).37 Experimental and clinical evidence supports the role of CAF as metastasis promoters in the primary tumor, whereas the role of CAF in lymph nodes during distant metastasis is not clear.38
Finally, one limitation of our study was the retrospective design. Patients received various postoperative adjuvant therapies, and the survival analysis as well as recurrence pattern analysis might have been influenced by these different postoperative treatments. In addition, because the formalin fixation time of tissue samples was not consistent among the previous cases, there is a possibility that this may have impacted the IHC results. Because the research period spans several years, it is difficult to eliminate the possibility of bias in this research. Other limitations were the small number of patients and the fact that this was a single‐center study. The lack of a significant difference in progression‐free survival in patients with lymph node metastases between chemokine receptor and chemokine positive and negative patients might have been due to the small number of patients. Further prospective studies are needed to test our findings in a large population.
In conclusion, the present study demonstrated that high CXCR4 expression in PS and high CXCL12 expression on tumor cells in MLN were critical prognostic factors in lymph‐node‐positive NSCLC patients. We speculate that CXCL12 produced by tumor cells might promote tumor growth as well as CAFs. Thus, CXCR4 in PS and CXCL12 expression on tumor cells in MLN may play an important role in tumor growth of lymph node metastasis in NSCLC.
CONFLICT OF INTEREST
The authors have no conflict of interest to declare.
Supporting information
ACKNOWLEDGMENTS
The authors would like to thank Yoshifumi Wakata for the statistical review. No sources of funding were provided for this study. We thank Gabrielle White Wolf from Edanz Group (www.edanzediting.com/ac) for editing a draft of this manuscript.
Katsura M, Shoji F, Okamoto T, et al. Correlation between CXCR4/CXCR7/CXCL12 chemokine axis expression and prognosis in lymph‐node‐positive lung cancer patients. Cancer Sci. 2018;109:154–165. https://doi.org/10.1111/cas.13422
Funding Information
No sources of funding were provided for this study.
REFERENCES
- 1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66:7‐30. [DOI] [PubMed] [Google Scholar]
- 2. American Cancer Society . Cancer Treatment & Survivorship Facts and Figures 2014‐2015, Vol. 64. Atlanta, GA: American Cancer Society, 2014. [Google Scholar]
- 3. Kamiuska K, Szczylik C, Bielecka ZF, et al. The role of the cell‐cell interactions in cancer progression. J Cell Mol Med. 2015;19:283‐296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Müller A, Homey B, Soto H, et al. Involvement of chemokine receptors in breast cancer metastasis. Nature. 2001;410:50‐56. [DOI] [PubMed] [Google Scholar]
- 5. Balabanian K, Lagane B, Infantino S, et al. The chemokine SDF‐1/CXCL12 binds to and signals through the orphan receptor RDC1 in T lymphocytes. J Biol Chem. 2005;280:35760‐35766. [DOI] [PubMed] [Google Scholar]
- 6. Ben‐Baruch A. Site‐specific metastasis formation. Cell Adh Migr. 2009;3:328‐333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Dai X, Mao Z, Huang J, Xie S, Zhang H. The CXCL12/CXCR4 autocrine loop increases the metastatic potential of non‐small cell lung cancer in vitro. Oncol Lett. 2012;5:277‐282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Takanami I. Overexpression of CCR7 mRNA in nonsmall cell lung cancer: correlation with lymph node metastasis. Int J Cancer. 2003;105:186‐189. [DOI] [PubMed] [Google Scholar]
- 9. Spano JP, Andre F, Morat L, et al. Chemokine receptor CXCR4 and early‐stage non‐small cell lung cancer: pattern of expression and correlation with outcome. Ann Oncol. 2004;15:613‐617. [DOI] [PubMed] [Google Scholar]
- 10. Liang J‐X, Gao W, Liang Y, Zhou X. Chemokine receptor CXCR4 expression and lung cancer prognosis: a meta‐analysis. Int J Clin Exp Med. 2015;8:5163‐5174. [PMC free article] [PubMed] [Google Scholar]
- 11. Oonakahara KI, Matsuyama W, Higashimoto I, Kawabata M, Arimura K, Osame M. Stromal‐derived factor‐1α/CXCL12‐CXCR 4 axis is involved in the dissemination of NSCLC cells into pleural space. Am J Respir Cell Mol Biol. 2004;30:671‐677. [DOI] [PubMed] [Google Scholar]
- 12. Travis WD, Giroux DJ, Chansky K, et al. The IASLC Lung Cancer Staging Project: proposals for the inclusion of broncho‐pulmonary carcinoid tumors in the forthcoming (seventh) edition of the TNM Classification for Lung Cancer. J Thorac Oncol. 2008;3:1213‐1223. [DOI] [PubMed] [Google Scholar]
- 13. Abe P, Mueller W, Schütz D, et al. CXCR7 prevents excessive CXCL12‐mediated downregulation of CXCR4 in migrating cortical interneurons. Development. 2014;141:1857‐1863. [DOI] [PubMed] [Google Scholar]
- 14. Singh RK, Lokeshwar BL. The IL‐8‐regulated chemokine receptor CXCR7 stimulates EGFR signaling to promote prostate cancer growth. Cancer Res. 2011;71:3268‐3277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zhu X, Bai Q, Lu Y, et al. Expression and function of CXCL12/CXCR4/CXCR7 in thyroid cancer. Int J Oncol. 2016;48:2321‐2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Allred DC, Harvey JM, Berardo M, Clark GM. Prognostic and predictive factors in breast cancer by immunohistochemical analysis. Mod Pathol. 1998;11:155‐168. [PubMed] [Google Scholar]
- 17. Liu F, Lang R, Wei J, et al. Increased expression of SDF‐1/CXCR4 is associated with lymph node metastasis of invasive micropapillary carcinoma of the breast. Histopathology. 2009;54:741‐750. [DOI] [PubMed] [Google Scholar]
- 18. Iwasa S, Yanagawa T, Fan J, Katoh R. Expression of CXCR4 and its ligand SDF‐1 in intestinal‐type gastric cancer is associated with lymph node and liver metastasis. Anticancer Res. 2009;29:4751‐4758. [PubMed] [Google Scholar]
- 19. Liang JJ, Zhu S, Bruggeman R, et al. High levels of expression of human stromal cell‐derived factor‐1 are associated with worse prognosis in patients with stage II pancreatic ductal adenocarcinoma. Cancer Epidemiol Biomarkers Prev. 2010;19:2598‐2604. [DOI] [PubMed] [Google Scholar]
- 20. Yu Y, Shi X, Shu Z, et al. Stromal cell‐derived factor‐1 (SDF‐1)/CXCR4 axis enhancess cellular invasion in ovarian carcinoma cells via integrin β1 and β3 expression. Oncol Res. 2014;21:217‐225. [DOI] [PubMed] [Google Scholar]
- 21. Huang Y, Zhang J, Cui ZM, Zhao J, Zheng Y. Expression of the CXCl12/CXCR4 and CXCL16/CXCR6 axes in cervical intraepithelial neoplasia and cervical cancer. Chin J Cancer. 2013;32:289‐296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zhong W, Chen W, Zhang D, et al. CXCL12/CXCR4 axis plays pivotal roles in the organ‐specific metastasis of pancreatic adenocarcinoma: a clinical study. Exp Ther Med. 2012;4:363‐369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kasagi Y, Harada Y, Morodomi Y, et al. Peritoneal dissemination requires an Sp1‐dependent CXCR4/CXCL12 signaling axis and extracellular matrix‐directed spheroid formation. Cancer Res. 2016;76:347‐357. [DOI] [PubMed] [Google Scholar]
- 24. Yu Y, Xiao C‐H, Tan L‐D, Wang Q‐S, Li X‐Q, Feng Y‐M. Cancer‐associated fibroblasts induce epithelial‐mesenchymal transition of breast cancer cells through paracrine TGF‐β signalling. Br J Cancer. 2014;110:724‐732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Paget S. Distribution of secondary growths in cancer of the breast. Lancet. 1889;1:98‐101. [PubMed] [Google Scholar]
- 26. Liu K, Bao C, Yao N, et al. Expression of CXCR4 and non‐small cell lung cancer prognosis: a meta‐analysis. Int J Clin Exp Med. 2015;8:7435‐7445. [PMC free article] [PubMed] [Google Scholar]
- 27. Wagner PL, Hyjek E, Vazquez MF, et al. CXCL12 and CXCR4 in adenocarcinoma of the lung: association with metastasis and survival. J Thorac Cardiovasc Surg. 2009;137:615‐621. [DOI] [PubMed] [Google Scholar]
- 28. Su L, Zhang J, Xu H, et al. Differential expression of CXCR4 is associated with the metastatic potential of human non‐small cell lung cancer cells. Clin Cancer Res. 2005;11:8273‐8280. [DOI] [PubMed] [Google Scholar]
- 29. Ottaiano A, Franco R, Talamanca AA, et al. Overexpression of both CXC chemokine receptor 4 and vascular endothelial growth factor proteins predicts early distant relapse in stage II‐III colorectal cancer patients. Clin Cancer Res. 2006;12:2795‐2803. [DOI] [PubMed] [Google Scholar]
- 30. Miao Z, Luker KE, Summers BC, et al. CXCR7 (RDC1) promotes breast and lung tumor growth in vivo and is expressed on tumor‐associated vasculature. Proc Nat Acad Sci USA. 2007;104:15735‐15740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Iwakiri S, Mino N, Takahashi T, et al. Higher expression of chemokine receptor CXCR7 is linked to early and metastatic recurrence in pathological stage I nonsmall cell lung cancer. Cancer. 2009;115:2580‐2593. [DOI] [PubMed] [Google Scholar]
- 32. Thiery JP, Acloque H, Huang RYJ, Nieto MA. Epithelial‐mesenchymal transitions in development and disease. Cell. 2009;139:871‐890. [DOI] [PubMed] [Google Scholar]
- 33. Liang C‐M. Chemokines and their receptors play important roles in the development of hepatocellular carcinoma. World J Hepatol. 2015;7:1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zhang K, Corsa CA, Ponik SM, et al. The collagen receptor discoidin domain receptor 2 stabilizes SNAIL1 to facilitate breast cancer metastasis. Nat Cell Biol. 2013;15:677‐687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Bremnes RM, Dønnem T, Al‐Saad S, et al. The role of tumor stroma in cancer progression and prognosis: emphasis on carcinoma‐associated fibroblasts and non‐small cell lung cancer. J Thorac Oncol. 2011;6:209‐217. [DOI] [PubMed] [Google Scholar]
- 36. De Veirman K, Rao L, De Bruyne E, et al. Cancer associated fibroblasts and tumor growth: focus on multiple myeloma. Cancers. 2014;6:1363‐1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Burger JA, Stewart DJ, Wald O, Peled A. Potential of CXCR4 antagonists for the treatment of metastatic lung cancer. Expert Rev Anticancer Ther. 2011;11:621‐630. [DOI] [PubMed] [Google Scholar]
- 38. De Wever O, Van Bockstal M, Mareel M, Hendrix A, Bracke M. Carcinoma‐associated fibroblasts provide operational flexibility in metastasis. Semin Cancer Biol. 2014;25:33‐46. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials


